Abstract
Avian infectious laryngotracheitis virus (ILTV) possesses an alphaherpesvirus type D DNA genome of ca. 155 kbp. Completion of our previous sequence analyses (W. Fuchs and T. C. Mettenleiter, J. Gen. Virol. 77:2221–2229, 1996) of the right end of the unique long (UL) genome region revealed the presence of two adjacent, presumably ILTV-specific genes, which were named UL0 and UL[−1] because of their location upstream of the conserved UL1 (glycoprotein L) gene. Transcriptional analyses showed that both genes are abundantly expressed during the late phase of the viral replication cycle and that both mRNAs are spliced by the removal of short introns close to their 5′ ends. Furthermore, the deduced gene products exhibit a moderate but significant homology of 28% to each other. The newly identified ILTV genes encode proteins of 63 kDa (UL0) and 73 kDa (UL[−1]), which both are predominantly localized in the nuclei of virus infected chicken cells. In summary, our results indicate that duplication of a spliced ILTV-specific gene encoding a nuclear protein has occurred during evolution of ILTV.
Infectious laryngotracheitis is an economically important respiratory disease of chickens and is caused by infectious laryngotracheitis virus (ILTV), also designated gallid herpesvirus 1 (2, 53). Based on biological properties such as its rapid lytic replication in respiratory epithelial tissues and its ability to establish latent infections in sensory neurons (2, 63), ILTV was classified as a member of the Alphaherpesvirinae subfamily of the Herpesviridae (53). However, in contrast to most other alphaherpesviruses, ILTV exhibits both in vivo and in vitro, a very narrow host range which is restricted almost exclusively to chicken and chicken-derived cells (2).
Early molecular analyses demonstrated that ILTV possesses a herpesvirus type D genome of ca. 155 kbp with a G+C content of 45% (28, 43). During the last years, the DNA sequence of ca. 50% of the ILTV genome has been determined. Most of the identified ILTV genes were shown to be conserved and found in collinear arrangement compared to the completely sequenced alphaherpesvirus genomes of herpes simplex virus type 1 (HSV-1) (44), varicella-zoster virus (VZV) (17), equine herpesvirus 1 (EHV-1) (59), and bovine herpesvirus 1 (BHV-1) (56). The characterized parts of the ILTV genome include the entire unique short (US) region (62), most of the adjoining inverted internal repeat (IRS) and terminal repeat (TRS) sequences encoding the major immediate-early protein ICP4 (31), and several segments of the unique long (UL) region. Identified viral gene products comprise glycoproteins B, C, and G (gB, gC, and gG) and gp60 (36, 37, 38, 50). Adjacent to the recently characterized left genome end, the ILTV homologs of the UL54, UL53 (gK), and UL52 genes of HSV-1 were localized (29, 32); close to the right end of the UL region, the UL1 (gL) to UL5 genes of ILTV were found (22). These findings indicate that in the type D genomes of ILTV, VZV and EHV-1, the UL region is fixed in opposite orientation to the prototypic isomer of the HSV-1 type E genome (17, 54, 59). Another sequenced genome part encompasses the ILTV homologs of the UL50 to UL45 genes located close to the UL22 (gH) to UL27 (gB) genes (24, 25, 64). In a different part of the UL region, the UL21 gene is located immediately downstream of the UL44 (gC) gene, which indicates that the ILTV genome contains a large internal inversion compared to most other alphaherpesvirus genomes (36, 64). Remarkably, a related gene rearrangement was also found in the pseudorabies virus (PrV) genome (4). Additional specific characteristics of the ILTV genome are the translocation of the conserved UL47 gene from the UL to the US region and the presence of several nonconserved, presumably ILTV-specific genes in both the UL and US regions (22, 62, 64). These observations, as well as phylogenetic analyses of conserved protein coding sequences (24, 30, 47), indicated that ILTV is only distantly related to the better-characterized mammalian alphaherpesviruses but is also clearly distinct from avian Marek’s disease virus (MDV) and herpesvirus of turkeys (HVT).
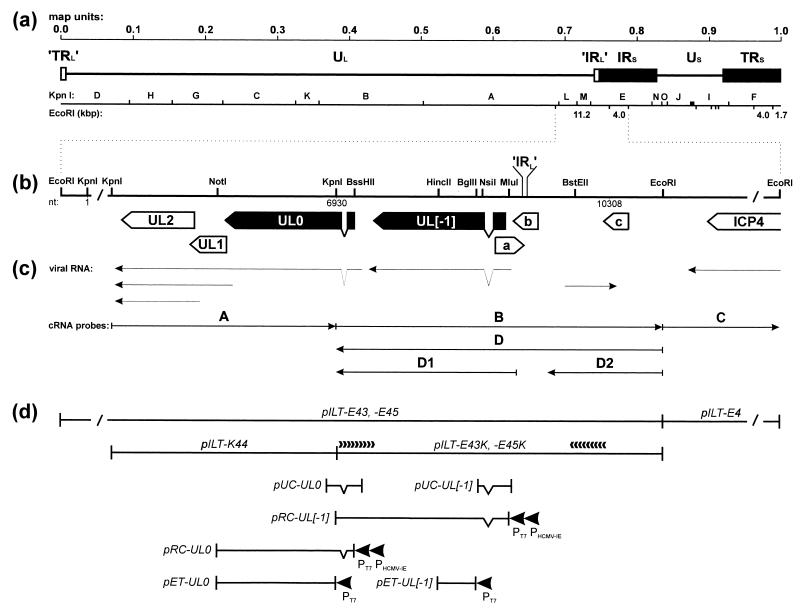
From our previous sequence analyses of the right part of the UL genome region, we obtained evidence for the presence of a unique ILTV gene, which was localized upstream of the conserved UL5 to UL1 genes and was therefore designated UL0 (22). However, since the known DNA sequence contained only the 3′-terminal part of this open reading frame (ORF), the presence of conserved domains at the N terminus of the predicted protein could not be excluded. Therefore, after cloning of the 11.2-kbp EcoRI fragment B of ILTV DNA, we completed sequence analysis of the very right end of the ILTV UL genome region including the junction to the IRS sequences, which contain the ICP4 gene (31) (Fig. 1a, b, and d).
FIG. 1.
Genome structure of ILTV. (a) Diagram of the type D genome of ILTV with KpnI restriction fragment map. Locations and sizes of relevant EcoRI restriction fragments are indicated. The US region is flanked by extended inverted repeat sequences (IRS and TRS). Very short (17-bp) inverted repetitions (’IRL’ and ’TRL’) were also found at both ends of the UL region. (b) Enlarged map of the investigated region with relevant restriction sites. Close to either end, the DNA fragments are not plotted to scale (/). Location of the ’IRL’ sequence separating the UL and IRS regions is indicated. Nucleotide numbers refer to the DNA sequence deposited in GenBank (accession no. X97256). Locations and orientations of ORFs are depicted by pointed rectangles, and splice sites are indicated by triangles. Conserved genes are named according to their homologs in HSV-1, and the analyzed ILTV-specific genes are highlighted in black. (c) Arrows indicate identified viral transcripts and the in vitro-transcribed cRNA probes which were used for hybridization as shown in Fig. 2. (d) Plasmid maps. pILT-E43, -E45, -E4, and -K44 contain EcoRI or KpnI fragments of ILTV DNA. Plasmids pILT-E43K and -E45K were used for nested deletion subcloning and directed DNA sequencing as indicated by arrowheads. pUC-UL0 and pUC-UL[−1] are cDNA clones encompassing the 5′ termini of either mRNA. They were used for reconstitution of the processed UL0 and UL[−1] ORFs under control of the bacteriophage T7 (PT7) and HCMV immediate-early (PHCMV-IE) promoters in pRC-UL0 and pRC-UL[−1]. For immunization, bacterial fusion proteins were expressed from pET-UL0 and pET-UL[−1] under the control of the T7 promoter (PT7).
To obtain the investigated plasmids (Fig. 1d), viral DNA of a pathogenic ILTV strain (obtained from D. Lütticken, Boxmeer, The Netherlands) was prepared essentially as described previously (22), and EcoRI or KpnI restriction fragments were inserted into vector pBS(−) (Stratagene, Heidelberg, Germany). The inserts of two independently obtained clones of the ILTV EcoRI fragment B (pILT-E43 and pILT-E45) were shortened to 4.0 kbp by KpnI digestion and religation. To permit bidirectional sequencing, the resulting plasmids pILT-E43K and pILT-E45K (Fig. 1d) were digested with HindIII and BamHI and with EcoRI and BstEII, respectively, and sets of unidirectionally truncated subclones were generated and analyzed with vector-specific M13 or M13 reverse primers as described previously (22). Finally, the DNA sequences of the fragment ends were determined from pILT-E43 and pILT-E45 with deduced ILTV-specific primers (purchased from GibcoBRL), and sequences were assembled and analyzed by using the Genetics Computer Group (GCG) package (18) in UNIX version 9.1.
Sequence analyses demonstrated that the 11.2-kbp EcoRI fragment B of ILTV DNA includes the previously characterized KpnI fragments L and M (22) and at the left end extends our published DNA sequence by 338 bp, which presumably contain the 5′-terminal part of the ILTV UL6 gene (data not shown). At the right end, a 702-bp overlap with known DNA sequences from the ICP4 gene region of ILTV (31) was identified. Since the newly determined DNA sequence of 3,378 bp begins within the functional UL0 gene (see below), it was added to our previously published sequence (GenBank accession no. X97256). The combined ILTV DNA sequence is now 10,308 bp in size and overlaps the complementary strand of the ICP4 gene sequence (31) by 43 nucleotides (nt), including one C-to-T transition at position 10269. The sequences of the two analyzed ILTV DNA clones were completely identical, with the exception of a presumably noncoding oligo(dG) stretch (positions 9838 to 9847), which comprised 10 nt in pILT-E45 and 9 nt in pILT-E43.
Southern blot hybridization of EcoRI-digested ILTV virion DNA with the labeled plasmid pILT-E45 detected the viral EcoRI fragment B and an additional 1.7-kbp viral DNA fragment (results not shown). The latter fragment represents the right end of the ILTV genome, suggesting that the sequences presented in this study include the boundary between the UL region and the IRS sequences (Fig. 1a). To confirm this, we compared our DNA sequence with that of the left genome end of ILTV (32) and with that of a joined fragment containing both genome ends, which was cloned from replicative, concatemeric ILTV DNA (23). From position 9239 to the end the sequence was indeed found to be identical to the joined fragment. Interestingly, between positions 9239 and 9255 it also corresponds to the complementary nt 3 to 19 from the left genome end. This finding demonstrates that the UL region of the ILTV genome is actually flanked by very short inverted repeat sequences comprising 17 bp (’TRL’ and ’IRL’ [Fig. 1a]). Comparably short repeats are also present at both ends of the UL regions of the alphaherpesvirus type D genomes of VZV and EHV-1 (17, 59). These repetitive sequence elements might be related to the much longer TRL and IRL sequences in the type E genomes of HSV-1 and -2, MDV, and HVT (12, 46). According to our data, the IRS of the ILTV genome starts at nt 9256 of the presented DNA sequence.
In addition to the 5′ end of the described UL0 gene (22), one large ORF, which was named UL[−1] (Fig. 1b), was detected. Upstream from this gene were found several smaller ORFs, three of them encompassing more than 100 codons (ORFs a [nt 8898 to 9254], b [reverse of nt 9124 to 9429], and c [reverse of nt 10240 to 10542] [Fig. 1a]). However, database searches using the GCG programs FASTA and BLAST (1, 27) revealed no significant homologies between the deduced products of these five ORFs and any known herpesvirus proteins.
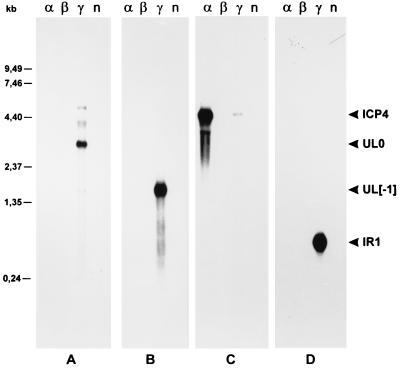
To investigate whether the identified ILTV-specific ORFs might represent functional genes, transcription of the sequenced genome part was monitored under different conditions of virus infection. Monolayers of primary chicken kidney cells were infected with ILTV at a multiplicity of infection (MOI) of 5. After 1 h at 37°C, the inoculum was replaced by fresh medium and incubation was continued. For accumulation of viral immediate-early (α) RNA, protein synthesis was blocked by addition of cycloheximide (100 μg/ml) to both virus inoculum and medium, and cells were harvested 6 h after infection. To obtain viral early (β) transcripts, phosphonoacetic acid (250 μg/ml) was added to the inoculum and medium to inhibit DNA synthesis, and the infected cells were incubated for 16 h at 37°C. Viral late (γ) RNA was prepared 16 h after infection in the absence of any inhibitors. Total RNA was isolated from the cells by guanidinium thiocyanate-phenol-chloroform extraction (15), separated on 1% formaldehyde–0.8% agarose gels, transferred to nylon membranes, and hybridized with 32P-labeled, strand-specific RNA probes, which were transcribed in vitro with T7 or T3 RNA polymerases from cloned ILTV DNA fragments as described previously (22). The hybridization probes used for the Northern blots shown in Fig. 2 are indicated in Fig. 1c (cRNA probes). Probe C was used as a control to detect the immediate-early ICP4 mRNA (31). As expected, the 4.5-kb ICP4 transcript accumulated in the presence of cycloheximide, whereas in the absence of inhibitors only a weak signal was detectable (Fig. 2C). In contrast, the 3.1-kb transcript of the UL0 ORF (22) was not detected under immediate-early conditions but was abundantly present during the late phase of virus replication (Fig. 2A). In agreement with earlier results (22), probe A showed additional faint reactions with the viral UL1 and UL2 mRNAs of 2.0 and 1.3 kb, which are transcribed 3′ coterminally with UL0 (Fig. 1c), and with several larger transcripts of unknown origin (Fig. 2A). Hybridization probes B and D, which represent both strands of the newly characterized part of the ILTV genome, were synthesized from plasmid pILT-E45K (Fig. 1c and d). Probe B recognized an abundant viral late RNA of 1.7 kbp, which corresponds in size and orientation to the UL[−1] ORF (Fig. 2B; Table 1). After overexposure of the blot, an additional RNA of 3.1 kb was detectable (not shown), indicating that transcription of the UL0 gene starts upstream of the KpnI site at the end of the previously described DNA sequence (Fig. 1b). With probe D, a 0.7-kb viral RNA which is transcribed in an orientation opposite to that of the UL0, UL[−1], and ICP4 genes was detected (Fig. 2D). This RNA was named IR1, since hybridization with probes D1 and D2 (Fig. 1c) demonstrated that it originates from the IRS sequences and therefore cannot represent the mRNA of any of the identified ORFs (Fig. 1b). Consequently, the small ORFs a, b, and c do not appear to be expressed at detectable levels. Correlating with the localization of copies of the consensus polyadenylation signal AATAAA (61) downstream of the UL[−1] ORF, as well as downstream of the coterminal UL0, UL1, and UL2 transcription unit (Table 1), the 3.1- and 1.7-kb transcripts are polyadenylated (data not shown). In contrast, the IR1 transcript of ILTV is not polyadenylated, since it was absent from oligo(dT)-cellulose-purified RNA preparations. A common feature of many alphaherpesviruses is the expression of a restricted set of latency-associated transcripts (LATs) originating from the right end of the UL region or the IRL and IRS sequences (11, 14, 41, 60). The majority of these LATs are nonpolyadenylated nuclear RNAs, which are transcribed in an opposite orientation to that of and partly overlap with viral immediate-early genes. Besides their presence in latently infected tissues, LATs are also detectable in lytically infected cells. The 0.7-kb IR1 transcript meets most of these criteria. Analysis of latently infected chicken will demonstrate whether this RNA is indeed a new member of the LAT family.
FIG. 2.
Northern analyses of RNA from ILTV-infected and noninfected (n) chicken kidney cells. Cells were infected at an MOI of 5 and incubated for 6 h in the presence of cycloheximide (100 μg/ml) (α), for 16 h in the presence of phosphonoacetic acid (250 μg/ml) (β), or for 16 h without drugs (γ). Total RNA (5 μg per lane) was separated in 1% formaldehyde–0.8% agarose gels, transferred to nylon membranes, and hybridized with the probes depicted in Fig. 1c (A to D). Molecular sizes of RNA markers are indicated on the left. The sizes of the detected ILTV-specific transcripts (ICP4, UL0, UL[−1], IR1) are given in the text.
TABLE 1.
Properties of UL0 and UL[−1] ORFa
| Gene | mRNA
|
Position in ORF
|
Protein
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Positiona
|
Size (kb)
|
Exon 1 | Exon 2 | No. of amino acids | pI | Molecular mass (kDa)
|
|||||||
| TATA box | Start site | Intron | Poly(A)+ signal | Expected | Detected | Expected | In vitro | In vivo | |||||
| UL0 | 7247–7242 | 7216 | 7067–6990 | 4208–4203 | 2.95 + poly(A) | 3.1 | 7152–7068 | 6989–5554 | 506 | 6.59 | 57.0 | 61 | 63 |
| UL[−1] | ND | ≤9127 | 8880–8760 | 7381–7376 | ≥1.6 + poly(A) | 1.7 | 9022–8881 | 8759–7396 | 501 | 6.54 | 57.6 | 73 | 73 |
Nucleotide numbers refer to the DNA sequence deposited in GenBank; all listed sequence patterns are located on the reverse strand. The expected transcript sizes were calculated from the distances between initiation sites and polyadenylation signals, minus the sizes of the introns. Expected molecular masses and isoelectric points of the proteins were calculated from the deduced amino acid sequences. Molecular masses of in vitro translation products and viral proteins (in vivo) were estimated from SDS–10% polyacrylamide gels. ND, not detected.
The 5′ ends of the UL0 and UL[−1] mRNAs were determined by primer extension experiments. The results of these studies implied transcriptional initiation sites located about 340 (UL0) or 260 (UL[−1]) nt upstream of the first in-frame start codons of the respective ORFs (data not shown). Since the presence of extensive 5′ nontranslated sequences is uncommon in alphaherpesvirus RNAs, cDNAs of the 5′-terminal parts of the UL0 and UL[−1] mRNAs were generated with a 5′-RACE (rapid amplification of 5′ cDNA ends) system and custom primers (GibcoBRL, Eggenstein, Germany). The gene-specific primers UL0-EP1 (GenBank accession no. X97256; nt 6696 to 6730) and UL[−1]-EP1 (nt 8621 to 8648) were used for reverse transcription of total late RNA from ILTV-infected cells. After oligo(dC) tailing, the first-strand DNAs were amplified by PCR with an oligo(dG) primer and the gene-specific primer UL0-EP2 (nt 6730 to 6764) or UL[−1]-EP2 (nt 8638 to 8665). For the UL[−1] cDNA, an additional nested PCR with primer UL[−1]-EP3 (nt 8715 to 8734) was required. The ca. 450-bp (UL0) and 320-bp (UL[−1]) PCR products were treated with Klenow polymerase and polynucleotide kinase, isolated from agarose gels (Qiaquick gel extraction kit; Qiagen, Hilden, Germany), and inserted into SmaI-digested vector pUC18 (Pharmacia, Freiburg, Germany). Several independently obtained clones of each of the plasmids pUC-UL0 and pUC-UL[−1] (Fig. 1d) were sequenced and found to be distinct from the genomic ILTV DNA sequence. All UL0 cDNA clones contained a 78-bp deletion from nt 6990 to 7067 of the viral DNA sequence, whereas all UL[−1] cDNA plasmids lacked 121 bp between nt 8760 and 8880 (Table 1). The 5′ (GT) and 3′ (AG) ends of both deletions match the consensus sequences for eukaryotic splice donor and acceptor sites (6), indicating that the viral transcripts are processed at cellular spliceosomes. By intron removal a stop codon within the UL0 gene is eliminated, and a frameshift is introduced into the UL[−1] gene. Both events lead to 5′-terminally extended coding sequences compared to the ORFs as predicted from the ILTV genomic sequence. Sequencing of different cDNA plasmids revealed that the UL0 mRNA initiates with an A residue which is the reverse of nt 7216 of the DNA sequence. This finding is consistent with the identification of the upstream sequence motif TAATAA (reverse of nt 7242 to 7247), which probably serves as a TATA-box element (6). In contrast, the 5′ end of the UL[−1] mRNA could not be determined exactly, since all investigated cDNA plasmids start at different positions between nt 9094 and 9127. One possible reason for this inaccuracy might be the absence of an upstream TATA-box-like sequence element, which is required for site-specific initiation of transcription in most eukaryotic promoters (6). Taking into account the determined 5′ ends, the removed introns, and the location of polyadenylation signals resulting in addition of poly(A) tails of ∼100 nt each, the expected sizes of the UL0 and UL[−1] mRNAs (Table 1) are in good agreement with those found in Northern blot analyses (Fig. 2).
Splicing of alphaherpesvirus mRNAs is rare, and only four HSV-1 genes were shown to be expressed from spliced transcripts. Remarkably, besides the UL15, US1, and US12 genes, they also include the ICP0 gene (55), which is a positional homolog of the ILTV-specific UL0 and/or UL[−1] genes. Although this could be indicative of a phylogenetic relationship, it should be mentioned that besides HSV-1 ICP0, all other known alphaherpesvirus ICP0 homologs (19) are expressed from intronless genes.
As a result of mRNA splicing, the UL0 and UL[−1] ORFs of ILTV contain 506 and 501 codons, respectively (Table 1). The nucleotides surrounding the proposed start codons (underlined) of UL0 (AGAATGA) and UL[−1] (GAAATGG) agree with consensus sequences for efficient initiation of translation (40). The deduced UL0 and UL[−1] proteins possess similar neutral isoelectric points but are characterized by extended hydrophilic stretches correlating with their relatively high contents of 27.5% (UL0) and 35% (UL[−1]) of charged amino acids. By computer analyses with the GCG program motifs, no conserved domains indicating a possible function of these proteins were found. Moreover, database searches with the GCG programs FASTA and BLAST did not reveal any related proteins. In particular, neither the UL0 nor the UL[−1] protein showed amino acid sequence homology to members of the ICP0 transactivator family (19). However, repeated searches performed after addition of both predicted ILTV gene products to the protein databases showed UL0 as the closest cognate of UL[−1], and vice versa. This homology could be confirmed by pairwise comparison of the proteins, which revealed a similarity of 38%, including 28% identical residues. Remarkably, the homology is restricted to sequences of both proteins encoded by the second exons and is most pronounced within their N-terminal parts (data not shown).
To demonstrate the presence of these unique ILTV proteins, parts of the UL0 and UL[−1] ORFs were expressed as fusion proteins in Escherichia coli (Fig. 1d) and used for the generation of monospecific rabbit antisera. A 1,478-bp EcoRI-NotI fragment, which encompasses codons 49 to 506 of UL0 preceded by a short stretch (12 bp) of coding vector sequences, was isolated from pILT-K44 and inserted into expression vector pET-23a(+) (Novagen, Madison, Wis.). For expression of codons 79 to 235 of UL[−1], a 472-bp BglII-HincII fragment of pILT-E45K was cloned into plasmid pET-23c(+) (Novagen), which had been digested with BamHI and HincII. After induction of fusion protein expression, bacterial cell lysates were separated on discontinuous sodium dodecyl sulfate (SDS)–10% polyacrylamide gels (42), stained for 20 min in 0.2% Serva blue R (Serva, Heidelberg, Germany)–0.5% acetic acid–20% methanol, and destained in 30% methanol. The 53-kDa (UL0) and 22-kDa (UL[−1]) fusion proteins were excised, equilibrated in SDS electrophoresis buffer (42), electroeluted into Centricon ultrafiltration units (Amicon, Witten, Germany) for 16 h at 100 V, concentrated, and finally resuspended in phosphate-buffered saline (PBS). Two rabbits were immunized four times at 3-week intervals by intramuscular injection of mineral oil emulsions containing 100 μg of the UL0 or UL[−1] fusion protein.
The sera collected after immunization reacted specifically with the corresponding fusion proteins (data not shown). As additional controls, reactivity with the in vitro translation products of the cloned UL0 and UL[−1] genes was analyzed (Fig. 3). Since both ILTV genes are spliced, the authentic ORFs had first to be reconstituted from cDNA and genomic DNA clones (Fig. 1d). After removal of noncoding sequences from pUC-UL0 by double digestion with SphI and BssHII, Klenow treatment, and religation, the 3′ part of the UL0 gene was added by insertion of the genomic KpnI fragment M derived from pILT-K44 in the correct orientation. From the resulting plasmid the complete UL0 gene was recloned as a 1,626-bp HindIII-NotI fragment into the expression vector pRc-CMV (Invitrogen, Leek, The Netherlands). The UL[−1] gene was assembled by insertion of a 1,812-bp NsiI-KpnI fragment from pILT-E45K into pUC-UL[−1], which had previously been shortened by SphI-MluI double digestion, Klenow treatment, and religation. Finally, the UL[−1] gene was also recloned as a 2,023-bp HindIII-KpnI fragment into pRc-CMV, which permits expression of the cloned genes in cell-free systems from the T7 promoter, as well as in eukaryotic cells from the human cytomegalovirus (HCMV) immediate-early promoter (Fig. 1d). The obtained plasmids pRC-UL0 and pRC-UL[−1] were transcribed and translated in vitro with a coupled reticulocyte lysate system (Promega, Mannheim, Germany) in the presence of 35S-labeled l-methionine (ICN, Eschwege, Germany). Immunoprecipitations were performed essentially as described previously (34, 35). Proteins were separated in discontinuous SDS–10% polyacrylamide gels, which were then incubated in En3Hance (NEN DuPont, Boston, Mass.), dried, and exposed to X-ray films (Hyperfilm MP; Amersham, Braunschweig, Germany).
FIG. 3.
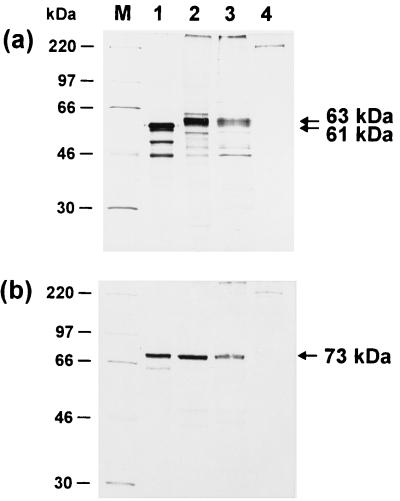
Identification of the UL0 and UL[−1] proteins. Radiolabeled proteins were incubated with monospecific anti-UL0 (a) and anti-UL[−1] sera (b) and immunoprecipitated. The in vitro translation products of pRC-UL0 and pRC-UL[−1] (lanes 1) were compared to proteins from primary chicken kidney cells transfected with the respective plasmid (lanes 2). In addition, lysates of chicken kidney cells were analyzed 24 h after infection with ILTV at an MOI of 5 (lanes 3). Results from precipitation of noninfected cell lysates are shown in lanes 4. In the depicted fluorograms of discontinuous SDS–10% polyacrylamide gels, the molecular masses of marker proteins (M) are indicated on the left, and locations of UL0 (61 and 63 kDa) and UL[−1] (73 kDa) gene products are marked by arrows.
After immunoprecipitation with the anti-UL0 serum, a major 61-kDa translation product of pRC-UL0 was detected in addition to several smaller proteins (Fig. 3a, lane 1). These probably represent products from internal translation initiation at downstream in-frame start codons. Precipitation with the anti-UL[−1] serum yielded a 73-kDa translation product of pRC-UL[−1] (Fig. 3b, lane 1). Remarkably, the apparent molecular masses of both proteins after gel electrophoresis are higher than the calculated masses of the deduced viral gene products, a difference most pronounced for the UL[−1] protein (Table 1).
For detection of the authentic viral UL0 and UL[−1] proteins, primary chicken kidney cells were labeled 4 h after ILTV infection (MOI = 5) with 100 μCi of Tran[35S]-label (ICN) per ml. Cells were also labeled 24 h after transfection with pRC-UL0 or pRC-UL[−1] (20 μg of DNA per 106 cells; mammalian transfection kit; Stratagene). The cells were lysed after a 24-h labeling period and analyzed by immunoprecipitation as described above.
With the anti-UL0 serum, we detected a protein of 63 kDa in infected cell lysates (Fig. 3a, lane 3) which was not present in lysates of noninfected cells (Fig. 3a, lane 4). The 63-kDa protein was not precipitated by the preimmune serum from the same rabbit, and specific precipitation of the viral protein was inhibited by pretreatment of the antiserum with the bacterial UL0-fusion protein (data not shown). With the anti-UL[−1] serum, a 73-kDa protein was precipitated from ILTV-infected cells but not from noninfected cells (Fig. 3b, lanes 3 and 4), and the specificity of this reaction could also be confirmed by competition studies (data not shown). Viral proteins of similar sizes were also identified in Western blot analyses (data not shown). As for the in vitro translation products, the estimated molecular masses of both ILTV proteins were again significantly higher than calculated (Table 1). The apparent molecular mass of the authentic UL0 protein (63 kDa; Fig. 3a, lane 3) was slightly higher than that of the in vitro translation product (61 kDa; Fig. 3a, lane 1). However, the protein expressed from the HCMV immediate-early promoter after transfection of chicken kidney cells with the same plasmid (pRC-UL0) appeared identical to the mature viral gene product (Fig. 3a, lanes 2 and 3). This could indicate minor posttranslational modifications of the UL0 protein. In contrast, the apparent molecular mass of the UL[−1] protein after either in vitro translation or transient expression was indistinguishable from the mature viral gene product (Fig. 3b, lanes 1 to 3). Although the amino acid sequences of the UL0 and UL[−1] gene products contain several putative N-glycosylation sites (39), our results do not indicate glycosylation of either protein.
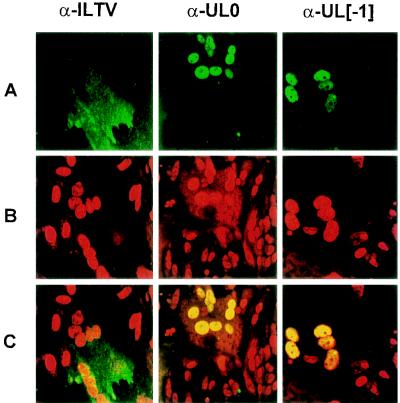
To localize the UL0 and UL[−1] gene products in ILTV-infected cells, indirect immunofluorescence experiments were evaluated by confocal laser scan microscopy. Monolayers of primary chicken kidney cells were grown on coverslips, fixed 18 h after ILTV infection (MOI = 0.01) with methanol-acetone (1:1) for 15 min, and washed three times with PBS. Cells were then incubated for 30 min with the rabbit UL0 and UL[−1] antisera or with a hyperimmune serum from an ILTV-infected chicken (obtained from D. Lütticken), all diluted in PBS. After three washes in PBS, the cells were incubated for 30 min with fluorescein-conjugated anti-rabbit or anti-chicken immunoglobulins, respectively (Dianova, Hamburg, Germany), and washed again as described above. Finally, the cells were overlaid with 10% PBS–90% glycerol containing 1,4-diazabicyclo[2.2.2.]octane (25 mg/ml) and propidium iodide (1 μg/ml). Fluorescence was analyzed in a confocal laser scan microscope (LSM 510; Zeiss, Jena, Germany). In ILTV-infected cells, the formation of virus-induced syncytia was detectable, and propidium iodide staining indicated local accumulations of nuclei, which appear swollen and seem to contain increased amounts of nucleic acids (Fig. 4B). The anti-ILTV hyperimmune serum mainly detected cytoplasmic antigens, as shown by the reaction of fluorescein-conjugated secondary antibodies with large syncytial areas (Fig. 4A, α-ILTV). In contrast, both monospecific antisera recognized viral proteins predominantly in the nuclei of infected cells (Fig. 4A, α-UL0 and α-UL[−1]). Vertical scanning of the samples revealed an almost even intranuclear distribution of the UL0 and UL[−1] proteins, but excluding the nucleoli (data not shown). Combined fluorescence demonstrated a perfect colocalization of either of the ILTV proteins with intranuclear DNA and/or RNA (Fig. 4C, α-UL0 and α-UL[−1]). These results were controlled by monitoring reactivity of the tested antisera with noninfected cells and reaction of the respective preimmune sera with infected cells (data not shown).
FIG. 4.
Subcellular localization of the UL0 and UL[−1] proteins. ILTV-infected (MOI = 0.01) chicken kidney cells were fixed after 18 h and incubated with a chicken hyperimmune serum (α-ILTV) or monospecific rabbit antisera (α-UL0 or α-UL[−1]). Confocal laser scan microscopy was performed after subsequent incubation with fluorescein-conjugated secondary antibodies (row A) and staining of nuclear DNA with propidium iodide (row B). The combined green and red fluorescence (row C) demonstrates colocalization of the UL0 and UL[−1] proteins with nuclear DNA.
Prediction of nuclear localization signals is difficult, but usually they consist of short stretches of basic amino acids (51, 57). The UL0 and UL[−1] polypeptides of ILTV are rich in arginine residues (UL0, 10%; UL[−1], 12%) which are clustered in the C-terminal parts and might be responsible for targeting of the proteins into the nucleus. Since ILTV, like other herpesviruses, replicates in the nucleus of the host cell (26, 52), many viral gene products are expected to be present in this cell compartment. Kinetics of UL0 and UL[−1] expression exclude roles in early regulation of viral gene expression, unless these proteins are incorporated into virions and thereby introduced with viral particles into host cells, as has been shown for the HSV-1 tegument protein VP16 (10). Enzymes which are directly involved in viral DNA synthesis also have to be expressed during the early phase of replication (55). Therefore, it is more likely that the UL0 and UL[−1] gene products of ILTV are involved in modulation of host cell gene expression or in cleavage and encapsidation of viral DNA, or that they represent structural components of nucleocapsids assembled in the host cell nucleus. However, so far it is unclear whether the UL0 and UL[−1] proteins are present in ILTV virions.
The UL0 and UL[−1] genes are located upstream from the conserved UL1 gene (22) in parallel orientation. At collinear positions in the genomes of VZV, EHV-1, BHV-1, and PrV, genes encoding homologs of the transactivator protein ICP0 are located (14, 20, 21, 48). In the HSV-1 type E genome, ICP0 is encoded by two gene copies within the IRL and TRL sequences (49). Although overall amino acid sequence homology between ICP0-related proteins is only moderate, there are two characteristics which indicate a function different from that of the ILTV UL0 and UL[−1] proteins. First, ICP0 homologs possess a characteristic zinc binding RING finger domain close to the N terminus (19), which is absent from the deduced UL0 and UL[−1] polypeptides. Furthermore, ICP0 genes are expressed during the immediate-early or, as in PrV (14), the early phase of virus replication. In contrast, UL0 and UL[−1] mRNAs accumulate only during the late phase of virus replication, and newly synthesized proteins were detectable in immunoprecipitation experiments from 8 h postinfection (data not shown). Since the presented sequence closes the gap between the UL5 to UL1 genes (22) at one side and the ICP4 gene (31) at the other side, our results demonstrate that ILTV possesses no positionally conserved ICP0 gene. Whether this gene is translocated to a yet uncharacterized part of the genome or whether it is completely missing has yet to be elucidated. The absence of an ICP0 gene from the ILTV genome appears possible since HSV-1 ICP0 was shown to be dispensable for virus replication (8, 58), and no ICP0 homologs have been identified in avian MDV and HVT (9).
The adjacent UL0 and UL[−1] genes of ILTV are closely related with respect to expression kinetics, mRNA structures, and subcellular localization of the proteins. This correlates with a significant amino acid sequence homology of 28%, suggesting the duplication of one ancestral gene. Similar duplication events were also proposed for the evolution of the alphaherpesvirus genes coding for glycoproteins gG, gD, gJ, gI, and gE, which are clustered within the US genome regions (45). However, the homology between UL0 and UL[−1] is hardly detectable at the level of coding DNA sequences, indicating that the gene duplication is an ancient event. Thus, acquisition and duplication of the identified genes presumably occurred after separation of the ILTV lineage from that of beta- and gamma-herpesviruses as well as from mammalian alphaherpesviruses, since no characterized member of these subfamilies encodes homologs of these proteins. Although available sequence data from avian MDV and HVT are limited, no UL0 or UL[−1] homologous genes have been found, at least in a corresponding genome region. This again highlights the isolated phylogenetic position of ILTV among alphaherpesviruses (24, 30, 47).
UL0 and UL[−1] are not the first ILTV-specific genes to be described. Besides two novel genes in the US region (62), a set of five unique ORFs is located between two rearranged clusters of conserved genes within the UL region (64). Finally, at the left end of the ILTV genome, a nonconserved region of ca. 10 kbp was recently analyzed by DNA sequencing (32). Differences in gene content were also identified between other alphaherpesviruses. The ORF1 gene, for example, is found only in PrV and EHV-1 (3), and the UL3.5 gene, which is conserved in ILTV and most mammalian viruses (22), is absent from HSV-1 and -2. The gene content of the US genome regions in particular exhibits some variability between avian and mammalian alphaherpesviruses (7, 62) and also between the more closely related human alphaherpesviruses (16). The tumorigenic MDV possesses a set of specific genes within the IRL and TRL sequences, including several putative oncogenes (5, 13, 33). Remarkably, the long repeat sequences of the MDV type E herpesvirus genome (12) positionally correspond to the ends of the UL region in the type D genomes of other alphaherpesviruses, including ILTV. Therefore, these regions might be considered preferred sites of recombination and acquisition of species-specific virus genes. Possibly the UL0 and UL[−1] genes of ILTV, like the unique MDV genes, are involved in determination of host tropism and specific pathogenicity mechanisms of the respective viruses. To elucidate the functions of UL0 and UL[−1] of ILTV, a major topic of future studies will be the generation and characterization of virus mutants lacking these genes.
Nucleotide sequence accession number.
The sequence obtained in this study has been assigned GenBank accession no. X97256.
Acknowledgments
This study was supported by a grant from Intervet Intl. B.V.
We thank R. Riebe for providing primary chicken cells, E. Mundt for help with preparation of antisera, and N. Osterrieder for confocal microscopy.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Bagust T J, Guy J S. Laryngotracheitis. In: Calnek B W, Barnes H J, Beard C W, McDougald L R, Saif Y M, editors. Diseases of poultry. 10th ed. Ames, Iowa: Iowa State University Press; 1997. pp. 527–539. [Google Scholar]
- 3.Baumeister J, Klupp B G, Mettenleiter T C. Pseudorabies virus and equine herpesvirus 1 share a nonessential gene which is absent in other herpesviruses and located adjacent to a highly conserved gene cluster. J Virol. 1995;69:5560–5567. doi: 10.1128/jvi.69.9.5560-5567.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ben-Porat T, Veach R A, Ihara S. Localization of the regions of homology between the genomes of herpes simplex virus type 1 and pseudorabies virus. Virology. 1983;127:194–204. doi: 10.1016/0042-6822(83)90383-5. [DOI] [PubMed] [Google Scholar]
- 5.Bradley G, Hayashi M, Lancz G, Tanaka A, Nonoyama M. Structure of the Marek’s disease virus BamHI-H gene family: genes of putative importance for tumor induction. J Virol. 1989;63:2534–2542. doi: 10.1128/jvi.63.6.2534-2542.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Breathnach R, Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- 7.Brunovskis P, Velicer L F. The Marek’s disease virus (MDV) unique short region: alphaherpesvirus-homologous, fowlpox virus-homologous, and MDV-specific genes. Virology. 1995;206:324–338. doi: 10.1016/s0042-6822(95)80048-4. [DOI] [PubMed] [Google Scholar]
- 8.Cai W, Astor T L, Liptak L M, Cho C, Coen D M, Schaffer P A. The herpes simplex virus type 1 regulatory protein ICP0 enhances virus replication during acute infection and reactivation from latency. J Virol. 1993;67:7501–7512. doi: 10.1128/jvi.67.12.7501-7512.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calnek B W, Witter R L. Marek’s disease. In: Calnek B W, Barnes H J, Beard C W, McDougald L R, Saif Y M, editors. Diseases of poultry. 10th ed. Ames, Iowa: Iowa State University Press; 1997. pp. 369–413. [Google Scholar]
- 10.Campbell M E M, Palfreyman J W, Preston C M. Identification of herpes simplex virus DNA sequences which encode a trans-acting polypeptide responsible for stimulation of immediate early transcription. J Mol Biol. 1984;180:1–19. doi: 10.1016/0022-2836(84)90427-3. [DOI] [PubMed] [Google Scholar]
- 11.Cantello J L, Anderson A S, Morgan R W. Identification of latency-associated transcripts that map antisense to the ICP4 homolog gene of Marek’s disease virus. J Virol. 1994;68:6280–6290. doi: 10.1128/jvi.68.10.6280-6290.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cebrian J, Kaschka-Dierich C, Berthelot N, Sheldrick P. Inverted repeat nucleotide sequences in the genomes of Marek’s disease virus and the herpesvirus of turkeys. Proc Natl Acad Sci USA. 1982;79:555–558. doi: 10.1073/pnas.79.2.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen X, Sondermeijer P J A, Velicer L F. Identification of a unique Marek’s disease virus gene which encodes a 38-kilodalton phosphoprotein and is expressed in both lytically infected cells and latently infected lymphoblastoid tumor cells. J Virol. 1992;66:85–94. doi: 10.1128/jvi.66.1.85-94.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheung A K. Cloning of the latency gene and the early protein 0 gene of pseudorabies virus. J Virol. 1991;65:5260–5271. doi: 10.1128/jvi.65.10.5260-5271.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 16.Davison A J, McGeoch D J. Evolutionary comparisons of the S segments in the genomes of herpes simplex virus type 1 and varicella-zoster virus. J Gen Virol. 1986;67:597–611. doi: 10.1099/0022-1317-67-4-597. [DOI] [PubMed] [Google Scholar]
- 17.Davison A J, Scott J E. The complete DNA sequence of varicella-zoster virus. J Gen Virol. 1986;67:1759–1816. doi: 10.1099/0022-1317-67-9-1759. [DOI] [PubMed] [Google Scholar]
- 18.Devereux J P, Haeberli P, Smithies O. A comprehensive set of sequence analysis for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Everett R, Barlow P, Milner A, Luisi B, Orr A, Hope R, Lyon D. A novel arrangement of zinc binding residues and secondary structure in the C3HC4 motif of an alpha herpes virus protein family. J Mol Biol. 1993;234:1038–1047. doi: 10.1006/jmbi.1993.1657. [DOI] [PubMed] [Google Scholar]
- 20.Everett R, Orr A, Elliott M. The equine herpesvirus 1 gene 63 RING finger protein partially complements Vmw110, its herpes simplex virus type 1 counterpart. J Gen Virol. 1995;76:2369–2374. doi: 10.1099/0022-1317-76-9-2369. [DOI] [PubMed] [Google Scholar]
- 21.Fraefel C, Zeng J, Choffat Y, Engels M, Schwyzer M, Ackermann M. Identification and zinc dependence of the bovine herpesvirus 1 transactivator protein BICP0. J Virol. 1994;68:3154–3162. doi: 10.1128/jvi.68.5.3154-3162.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fuchs W, Mettenleiter T C. DNA sequence and transcriptional analysis of the UL1 to UL5 gene cluster of infectious laryngotracheitis virus. J Gen Virol. 1996;77:2221–2229. doi: 10.1099/0022-1317-77-9-2221. [DOI] [PubMed] [Google Scholar]
- 23.Fuchs, W. Unpublished data.
- 24.Griffin A M, Boursnell M E G. Analysis of the nucleotide sequence of DNA from the region of the thymidine kinase gene of infectious laryngotracheitis virus; potential evolutionary relationships between the herpes virus subfamilies. J Gen Virol. 1990;71:841–850. doi: 10.1099/0022-1317-71-4-841. [DOI] [PubMed] [Google Scholar]
- 25.Griffin A M. The nucleotide sequence of the glycoprotein gB gene of infectious laryngotracheitis virus: analysis and evolutionary relationships to the homologous gene from other herpesviruses. J Gen Virol. 1991;72:393–398. doi: 10.1099/0022-1317-72-2-393. [DOI] [PubMed] [Google Scholar]
- 26.Guo P, Scholz E, Turek J, Nodgreen R, Maloney B. Assembly pathway of avian infectious laryngotracheitis virus. Am J Vet Res. 1993;54:2031–2039. [PubMed] [Google Scholar]
- 27.Henikoff S, Henikoff J G. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson M A, Prideaux C T, Kongsuwan K, Sheppard M, Fahey K J. Gallid herpesvirus 1 (infectious laryngotracheitis virus): cloning and physical maps of the SA-2 strain. Arch Virol. 1991;119:181–198. doi: 10.1007/BF01310669. [DOI] [PubMed] [Google Scholar]
- 29.Johnson M A, Prideaux C T, Kongsuwan K, Tyack S G, Sheppard M. ICP27 immediate early gene, glycoprotein K (gK) and DNA helicase homologues of infectious laryngotracheitis virus (gallid herpesvirus 1) SA-2 strain. Arch Virol. 1995;140:623–634. doi: 10.1007/BF01309954. [DOI] [PubMed] [Google Scholar]
- 30.Johnson M A, Tyack S G. Molecular evolution of infectious laryngotracheitis virus (ILTV; gallid herpesvirus 1): an ancient example of the alphaherpesviridae? Vet Microbiol. 1995;46:221–231. doi: 10.1016/0378-1135(95)00086-p. [DOI] [PubMed] [Google Scholar]
- 31.Johnson M A, Tyack S G, Prideaux C, Kongsuwan K, Sheppard M. Nucleotide sequence of infectious laryngotracheitis virus (gallid herpesvirus 1) ICP4 gene. Virus Res. 1995;35:193–204. doi: 10.1016/0168-1702(94)00096-u. [DOI] [PubMed] [Google Scholar]
- 32.Johnson M A, Tyack S G, Prideaux C T, Kongsuwan K, Sheppard M. Nucleotide sequence of the left-terminus of infectious laryngotracheitis virus (gallid herpesvirus 1) SA-2 strain. Arch Virol. 1997;142:1903–1910. doi: 10.1007/s007050050209. [DOI] [PubMed] [Google Scholar]
- 33.Jones D, Lee L, Liu J L, Kung H J, Tillotson J K. Marek’s disease virus encodes a basic-leucine zipper gene resembling the fos/jun oncogenes that is highly expressed in lymphoblastoid tumors. Proc Natl Acad Sci USA. 1992;89:4042–4046. doi: 10.1073/pnas.89.9.4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keil G M, Fibi M R, Koszinowski U H. Characterization of the major immediate-early polypeptides encoded by murine cytomegalovirus. J Virol. 1985;54:422–428. doi: 10.1128/jvi.54.2.422-428.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kessler S W. Use of protein A-bearing staphylococci for the immune precipitation and isolation of antigens from cells. Methods Enzymol. 1981;73:442–459. doi: 10.1016/0076-6879(81)73084-2. [DOI] [PubMed] [Google Scholar]
- 36.Kingsley D H, Hazel J W, Keeler C L., Jr Identification and characterization of the infectious laryngotracheitis virus glycoprotein C gene. Virology. 1994;203:336–343. doi: 10.1006/viro.1994.1492. [DOI] [PubMed] [Google Scholar]
- 37.Kongsuwan K, Johnson M A, Prideaux C T, Sheppard M. Identification of an infectious laryngotracheitis virus gene encoding an immunogenic protein with a predicted Mr of 32 kilodaltons. Virus Res. 1993;29:125–140. doi: 10.1016/0168-1702(93)90054-q. [DOI] [PubMed] [Google Scholar]
- 38.Kongsuwan K, Johnson M A, Prideaux C T, Sheppard M. Use of λgt11 and monoclonal antibodies to map the gene for the 60,000 dalton glycoprotein of infectious laryngotracheitis virus. Virus Genes. 1993;7:297–303. doi: 10.1007/BF01702590. [DOI] [PubMed] [Google Scholar]
- 39.Kornfeld R, Kornfeld S. Assembly of asparagin linked oligosaccharides. Annu Rev Biochem. 1985;54:631–664. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- 40.Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eucaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 41.Kutish G, Mainprize T, Rock D. Characterization of the latency-related transcriptionally active region of the bovine herpesvirus 1 genome. J Virol. 1990;64:5730–5737. doi: 10.1128/jvi.64.12.5730-5737.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 43.Leib D A, Bradbury J M, Hart C A, McCarthy K. Genome isomerism in two alphaherpesviruses: herpesvirus saimiri-1 (herpesvirus tamarinus) and avian infectious laryngotracheitis virus. Arch Virol. 1987;93:287–294. doi: 10.1007/BF01310982. [DOI] [PubMed] [Google Scholar]
- 44.McGeoch D J, Dalrymple M A, Davison A J, Dolan A, Frame M C, McNab D, Perry L J, Scott J E, Taylor P. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J Gen Virol. 1988;69:1531–1574. doi: 10.1099/0022-1317-69-7-1531. [DOI] [PubMed] [Google Scholar]
- 45.McGeoch D J. Evolutionary relationships of virion glycoprotein genes in the S regions of alphaherpesvirus genomes. J Gen Virol. 1990;71:2361–2367. doi: 10.1099/0022-1317-71-10-2361. [DOI] [PubMed] [Google Scholar]
- 46.McGeoch D J, Cunningham C, McIntyre G, Dolan A. Comparative sequence analysis of the long repeat regions and adjoining parts of the unique long regions in the genome of herpes simplex viruses types 1 and 2. J Gen Virol. 1991;72:3057–3075. doi: 10.1099/0022-1317-72-12-3057. [DOI] [PubMed] [Google Scholar]
- 47.McGeoch D J, Cook S. Molecular phylogeny of the alphaherpesvirinae subfamily and a proposed evolutionary timescale. J Mol Biol. 1994;238:9–22. doi: 10.1006/jmbi.1994.1264. [DOI] [PubMed] [Google Scholar]
- 48.Moriuchi H, Moriuchi M, Smith H A, Straus S E, Cohen J I. Varicella-zoster virus open reading frame 61 protein is functionally homologous to herpes simplex virus type 1 ICP0. J Virol. 1992;66:7303–7308. doi: 10.1128/jvi.66.12.7303-7308.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perry L J, Rixon F J, Everett R D, Frame M C, McGeoch D J. Characterization of the IE110 gene of herpes simplex virus type 1. J Gen Virol. 1986;67:2365–2380. doi: 10.1099/0022-1317-67-11-2365. [DOI] [PubMed] [Google Scholar]
- 50.Poulsen D J, Keeler C L., Jr Characterization of the assembly and processing of infectious laryngotracheitis virus glycoprotein B. J Gen Virol. 1997;78:2945–2951. doi: 10.1099/0022-1317-78-11-2945. [DOI] [PubMed] [Google Scholar]
- 51.Powers M A, Forbes D J. Cytosolic factors in nuclear transport: what’s importin? Cell. 1994;79:931–934. doi: 10.1016/0092-8674(94)90024-8. [DOI] [PubMed] [Google Scholar]
- 52.Prideaux C T, Kongsuwan K, Johnson M A, Sheppard M, Fahey K J. Infectious laryngotracheitis virus growth, DNA replication, and protein synthesis. Arch Virol. 1992;123:181–192. doi: 10.1007/BF01317148. [DOI] [PubMed] [Google Scholar]
- 53.Roizman B, Derosiers R C, Fleckenstein B, Lopez C, Minson A C, Studdert M J. The family Herpesviridae: an update. Arch Virol. 1992;123:425–449. doi: 10.1007/BF01317276. [DOI] [PubMed] [Google Scholar]
- 54.Roizman B. Herpesviridae. In: Fields B N, Knipe D M, Howley P M, editors. Virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2221–2230. [Google Scholar]
- 55.Roizman B, Sears A E. Herpes simplex viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2231–2295. [Google Scholar]
- 56.Schwyzer M, Ackermann M. Molecular virology of ruminant herpesviruses. Vet Microbiol. 1996;53:17–29. doi: 10.1016/s0378-1135(96)01231-x. [DOI] [PubMed] [Google Scholar]
- 57.Silver P A. How proteins enter the nucleus. Cell. 1991;64:489–497. doi: 10.1016/0092-8674(91)90233-o. [DOI] [PubMed] [Google Scholar]
- 58.Stow N D, Stow E C. Isolation and characterization of a herpes simplex virus type 1 mutant containing a deletion within the gene encoding the immediate early protein Vmw 110. J Gen Virol. 1986;67:2571–2585. doi: 10.1099/0022-1317-67-12-2571. [DOI] [PubMed] [Google Scholar]
- 59.Telford E A, Watson M S, McBride K, Davison A J. The DNA sequence of equine herpesvirus-1. Virology. 1992;189:304–316. doi: 10.1016/0042-6822(92)90706-u. [DOI] [PubMed] [Google Scholar]
- 60.Wechsler S L, Nesburn A B, Watson R J, Slanina S, Ghiasi H. Fine mapping of the latency-related gene of herpes simplex virus type 1: alternative splicing produces distinct latency-related RNAs containing open reading frames. J Virol. 1988;62:4051–4058. doi: 10.1128/jvi.62.11.4051-4058.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wickens M. How the messenger got its tail: addition of poly(A) in the nucleus. Trends Biochem Sci. 1990;15:277–281. doi: 10.1016/0968-0004(90)90054-f. [DOI] [PubMed] [Google Scholar]
- 62.Wild M A, Cook S, Cochran M. A genomic map of infectious laryngotracheitis virus and the sequence and organization of genes present in the unique short and flanking regions. Virus Genes. 1996;12:107–116. doi: 10.1007/BF00572949. [DOI] [PubMed] [Google Scholar]
- 63.Williams R A, Bennet M, Bradbury J M, Gaskell R M, Jones R C, Jordan F T W. Demonstration of sites of latency of infectious laryngotracheitis virus using the polymerase chain reaction. J Gen Virol. 1992;73:2415–2420. doi: 10.1099/0022-1317-73-9-2415. [DOI] [PubMed] [Google Scholar]
- 64.Ziemann K, Mettenleiter T C, Fuchs W. Gene arrangement within the unique long genome region of infectious laryngotracheitis virus is distinct from that of other alphaherpesviruses. J Virol. 1998;72:847–852. doi: 10.1128/jvi.72.1.847-852.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]