Abstract
As the human population grows, the anthropogenic impacts from various agricultural and industrial processes produce unwanted contaminants in the environment. The accurate, sensitive and rapid detection of such contaminants is vital for human health and safety. Surface‐enhanced Raman spectroscopy (SERS) is a valuable analytical tool with wide applications in environmental contaminant monitoring. The aim of this review is to summarize recent advancements within SERS research as it applies to environmental detection, with a focus on research published or accessible from January 2021 through December 2021 including early‐access publications. Our goal is to provide a wide breadth of information that can be used to provide background knowledge of the field, as well as inform and encourage further development of SERS techniques in protecting environmental quality and safety. Specifically, we highlight the characteristics of effective SERS nanosubstrates, and explore methods for the SERS detection of inorganic, organic, and biological contaminants including heavy metals, pharmaceuticals, plastic particles, synthetic dyes, pesticides, viruses, bacteria and mycotoxins. We also discuss the current limitations of SERS technologies in environmental detection and propose several avenues for future investigation. We encourage researchers to fill in the identified gaps so that SERS can be implemented in a real‐world environment more effectively and efficiently, ultimately providing reliable and timely data to help and make science‐based strategies and policies to protect environmental safety and public health.
Keywords: analytes of interest, environmental detection, nanosubstrates, surface‐enhanced Raman spectroscopy
Abbreviations
- AFB1
Aflatoxin B1
- AFM
Atomic force microscopy
- AgNDs
silver nanodisks
- AgNPs
Silver nanoparticles
- AOH
Alternariol
- AR18
Acid red 18
- AR26
Acid red 26
- AuNPs
Gold nanoparticles
- CNC
Cellulose nanocrystal
- CRISPR
clustered regularly interspaced short palindromic repeats
- CR
Congo red
- CTAB
cetyltrimethylammonium bromide
- CV
Crystal violet
- dsDNA
double‐stranded DNA
- DTNB
5,5′‐dithiobis (2‐nitrobenzoic acid)
- EF
Enhancement factor
- FB1
Fumonisin B1
- FESEM
Field‐emission scanning electron microscopy
- FZD
Furazolidone
- GaN
gallium nitride
- GNU
Gold nanourchin
- GO
Graphene oxide
- G‐SERS
Graphene‐enhanced Raman scattering
- HBV
Hepatitis B virus
- HIV
Human immunodeficiency virus
- HPV
Human papillomavirus
- IMS
Immunomagnetic separation
- IS
Internal standard
- LMFs
Low‐moisture foods
- LOD
limit of detection
- LSPR
Localized surface plasmon resonance
- MB
Methylene blue
- MG
Malachite green
- MIP
Molecularly imprinted polymers
- ML
Machine learning
- MOFs
Metal‐organic frameworks
- MPLs
Microplastics
- MRA
Molecular recognition agent
- NCs
nanocomposites
- NPGBs
Nanoporous gold bowls
- NPLs
Nanoplastics
- NPs
Nanoparticles
- OII
Acid orange II
- OTA
Ochratoxin A
- PA
Phenylacetylene
- PCA
Principal component analysis
- PCT
paracetamol
- PDA
Polydopamine
- PdNPs
(PSi)‐plated palladium nanoparticles
- PET
Polyethylene terephthalate
- PNA
p‐nitroaniline
- PQ
Paraquat
- PS
Polystyrene
- R6G
Rhodamine 6G
- RMP
Raman molecular probe
- SDME
Single drop microextraction
- SERS
Surface‐enhanced Raman spectroscopy
- SY
Sunset yellow
- TBZ
Thiabendazole
- TC
Tetracycline
- THR
Thiram
- TZ
Tartrazine
- UCL
Upconversion luminescence
- UCNPs
Upconversion nanoparticles
- ZEN
Zearalenone
1. INTRODUCTION
Public health and economic development are often challenged by environmental safety crises. Within 2021 alone, threats to environmental and human health have included the Covid‐19 pandemic, multiple oil spills, vast declines in bee populations due to the widespread use of neonicotinoid pesticides, and the increased occurrence and severity of algal blooms caused by toxic cyanobacteria. Given current widespread concerns over environmental pollutants, there is an increasing need for rapid and sensitive analytical methods to detect and quantify these harmful substances. Traditional methods for contaminant detection include HPLC, GC, MS and UV–Visible absorption. However, these methods have various limitations. For example, HPLC can be expensive and time consuming and may require complex instrumentation. GC is limited by the type of sample that can be analysed, and samples may require extensive purification. MS only gives information about molecular mass and does not differentiate between isomeric compounds. UV‐Vis requires simple instrumentation, but is vulnerable to other contaminants within the sample matrix interfering with the acquired spectra. Also, it is only applicable to analytes of unique light absorbance, which largely limits its application scope. Surface‐enhanced Raman spectroscopy (SERS) is a promising analytical method to address these weaknesses. It is an advanced technique that leverages a unique nano‐induced interfacial phenomenon: enhancement of Raman scattering. Raman scattering was first discovered in 1928 by C. V. Raman, and occurs when light of a specific wavelength inelastically scatters (either decreasing or increasing in vibrational energy). 1 , 2 This scattering is caused by intramolecular vibrations within the analyte, and different substances will have a unique ‘fingerprint’ spectrum with characteristic peaks at certain Raman shift locations. Through the identification of these peaks and their intensities relative to one another, an analyte can be identified with high accuracy, and multiple analytes can be distinguished from each other. 2 Although Raman spectroscopy has many advantages, such as rapid data collection, small sample volume requirement, non‐contact detection and high molecular specificity, there are drawbacks that hinder its ability to be used for environmental monitoring. Raman scattering can be extremely weak, due to the fact that Raman measures only the inelastically scattered photons (approximately one in every one million scattered photons), which largely limits the detection sensitivity. Fluorescence signals are, comparatively, much higher than normal Raman scattering, indicating that if the sample of interest fluoresces, a large background signal could obstruct the Raman scattering signal. 3 These limitations greatly slowed the applications of Raman spectroscopy in environmental detection, and demonstrated a need for techniques to improve its performance.
In 1974, Fleischmann et al. observed an enhancement in the Raman signal through the use of a roughened silver electrode in their experiments. 4 The roughened silver surface helps to enhance the generated Raman scattering and can even quench interfering fluorescence. 3 Further experiments by Jeanmaire and Van Duyne 5 and Albrecht and Creighton 6 in 1977 confirmed the above enhancement phenomena. Meanwhile, Jeanmaire and Van Duyne coined the term surface‐enhanced Raman spectroscopy (SERS). SERS utilizes special substrates, such as roughened electrodes or nanoscale structures (called ‘SERS nanosubstrates’) to enhance the normal Raman signal of target analytes. The enhancement that is observed from SERS nanosubstrates is due to electromagnetic and/or chemical enhancements. The electromagnetic component is the strongest contributor to the enhancement and occurs due to localization of light on the SERS surface when molecules of the analyte are in close physical proximity to the SERS substrate (1‐10 nm). 7 The chemical enhancement is due to the charge transfer that occurs when the analyte molecules are in direct contact with the substrate, commonly via adsorption to the SERS substrate surface. The chemical enhancement depends on the type of molecule adsorbed to the surface when the charge transfer takes place between analyte and substrate surface. 7 , 8 Through electromagnetic and/or chemical enhancements, this technique is able to enhance the original Raman signal by a magnitude up to 1015. 9 , 10 The enhancement is quantified by the enhancement factor (EF), 11 which is defined as the ratio of the Raman signal with SERS enhancement when compared with the normal Raman scattering signal, and is proportional to the intensity of the local electromagnetic field to the fourth power (|E|4). 10 For further in‐depth discussion on these enhancement mechanisms and other theories behind SERS, we direct readers to several excellent articles that have been published. 7 , 12 , 13 , 14
SERS is a rapid, non‐destructive, chemically specific and versatile analytical method that has many advantages over regular Raman spectroscopy. Compared with regular Raman, SERS has higher sensitivity (even single molecules can be detected 15 , 16 , 17 , 18 ) and lower fluorescence interference. 3 The discovery of SERS in the 1970s resulted in a rapid uptick in the development and applications of SERS‐based analytical methods in many fields including environmental monitoring. 19 As shown in Figure 1, the number of articles being published over the past 10 years in relation to SERS and its environmental applications has more than doubled and continues to increase in recent years. In this review, we will look into articles published in 2021 and 2022 (those accessible as of December 2021), with a special focus on newest findings in SERS nanosubstrate development and applications in analysing different chemical and biological pollutants. Additionally, Table 1 provides an overview of these recent publications involving SERS for environmental contaminant detection and Table 2 organizes publications involving pesticide detection. These tables include substrate, analyte, sample or sample matrix, and limit of detection (LOD) if provided.
FIGURE 1.
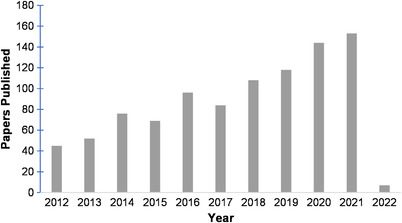
Number of research papers published over the past ten years by using the search parameter of ‘SERS + Environment’ on Web of Science (review papers excluded).
TABLE 1.
Recent publications (2021 or 2022 early access) on the detection of various environmental contaminants using SERS
| Substrate | Analyte | Sample or Sample Matrix | LOD | References |
|---|---|---|---|---|
| SERS aptsensor | Hg2+ | Tap and lake water | 0.11 Fm | [86] |
| IP6@AgNPs probe | Fe3+ | Integrated circuit cleaning solution waste | 0.1 µM | [90] |
| Phenylacetylene‐AgNPs | Hg2+ | Lake water | 87.6 pM | [99] |
| Dual enhancement SERS sensor | Pb2+ | Water | 4.31 pM | [42] |
| Gold nanoparticles | Polystyrene and poly (ethylene terephthalate) | NPL drop cast onto Au‐functionalized glass slides | 10 µg/mL | [115] |
| Silver nanoparticles on regenerated cellulose | Crystal violet and polystyrene | Analyte deposited on SERS substrate | 0.1 mg/mL | [62] |
| Silver @ gold nanostars @ anodized aluminum oxide | Polystyrene | Tap, river and sea water | 0.05 mg/g | [116] |
| Gold nanoparticles | Polystyrene | PS spiked in water | – | [114] |
| Gold nanoparticles @ 4‐MBN | Melamine | Milk | 10 nM | [265] |
| Silver nanoparticles | S. aureus and E. coli | Substrate | – | [235] |
| Gold modified magnetic nanoparticles | Salmonella |
|
4 cfu/mL | [229] |
| Silver/copper oxide nanowires/pyramidal PDMS | Rhodamine 6G, crystal violet and congo red | SERS substrate surface | 10–9, 10–8 and 10–7 M | [51] |
| Graphene oxide nanosheet coated with silver and gold nanoparticles | Beta‐carotene and malachite green | Analyte deposited on SERS substrate | <1 mg/L | [43] |
| Silver nanoparticles ‐ grafted silicon nanocones | Rhodamine 6G, crystal violet, melamine, methyl parathion | AgNPs/SiNC platform, lake water, milk and tap water | 10–14, 10–9, 10–7 and 10–7 M | [49] |
| Sulphur doped MoO2 nanospheres | Rhodamine 6G, rhodamine B, crystal violet | Dropped onto glass slide | 1 × 10–9, 1 × 10–10 and 1 × 10–8 M | [266] |
| Silver‐doped hydroxyapatite nanocomposite | Rhodamine 6G and crystal violet | Dropped on surface of SERS substrate | 10–6 and 10–5 M | [267] |
| Cetyltrimethylammonium bromide (CTAB)/gold nanoparticles | Hydroxyanthraquinones | Mixture of SERS substrate and analyte | – | [40] |
| Silver nanoparticles/glass fibre filter | Rhodamine 6G, malachite green and crystal violet | Simulated sewage solution | 10–10,10–9 and 10–9 M | [71] |
| Silver‐tannin furanic foam | Malachite green | SERS substrate was dipped in 1 mM of analyte solution | – | [120] |
| Silver @ UIO‐66(NH2)/ polydopamine – molecularly imprinted polymer | Orange II | Lake water | 10–10 M | [121] |
| Silver nanoparticles @ cellulose nano crystal | Congo red | Injected into platform | 10–15 M | [122] |
| Silver nanoparticles | Dezocine | Spiked urine, blank urine sample and spike rat serum | 10 µg/mL | [268] |
| Gold multibranched nanoparticles | Ibuprofen | Water | 10–8 M | [102] |
| Molecularly imprinted @ gold– graphene oxide | Biguanides | Health care capsules | 0.10 mg/mL | [105] |
| Nanoporous gold nanoparticles | Pharmaceuticals | Substrate | – | [45] |
| Silver nanodisks decorated filter paper | Tetracycline | Analyte dropped onto substrates surface | 10–9 M | [46] |
| Gold nanoparticles | Sulfadiazine, sulfamerazine and sulfamethazine | Water | 1 and 50 µg/L | [269] |
| Gold nanowires | Furazolidone | Deionized water, running water, river and seawater | 6.83 mg/L | [69] |
| ACE2 sensor | SARS‐CoV‐2 | Purchased spike proteins | 300 nm | [270] |
| Pyramidal nanoholes | Hepatitis A | Water | 13 pg/mL | [219] |
| Silver nanorods | SARS‐CoV‐2 | 23 water samples | – | [218] |
| Raman probe‐functionalized Au nanoparticles (RAuNPs) on the graphene oxide (GO)/triangle gold nanoflower array | Hepatitis B, HPV‐16 and HPV‐18 | Substrate | 1 aM–100 pM | [224] |
| Gold nanoparticles | dsDNA | Duck meat | 100 aM–10 pM | [225] |
| Upconversion nanoparticles | Ochratoxin A | Spiked beer | 3.2 pg/mL | [244] |
| Silica photonic crystal microspheres/gold nanoparticles | Aflatoxin B‐1, ochratoxin A and zearalenone | Rice, corn and wheat | 0.82, 1.43 and 1.00 pg/mL | [271] |
| Gold @ silver nanoparticles and gold nanorods | Ochratoxin A and zearalenone | Spiked corn and wheat | 0.018 and 0.054 ng/mL | [239] |
| Silver nanoparticles | Ochratoxin A | Spiked wine and wheat | – | [272] |
| Iron(II,III)oxide @ gold nanoparticles coupled with gold @ silver nanoparticles | Zearalenone | Spiked beer and wine | 0.001 ng/L | [273] |
| Gold nanorods | Patulin and alternariol | Spiked apple juice | 1 µg/L | [242] |
Articles were obtained by performing the following searches on Web of Science and removing non‐environmental papers: SERS + Environment, SERS + Biological, SERS + Environment + Biological, SERS + Bacteria, SERS + Virus, SERS + Mycotoxin, SERS + Pharmaceutical, SERS + Dyes, SERS + Environment + Dyes, SERS + Microplastics and SERS + Nanoplastics. The table details substrate type, analyte type, the sample or sample matrix used, method LOD and the corresponding reference. Note: a separate table below is provided for pesticides due to the large number of publications (Table 2).
TABLE 2.
Recent publications (2021 or 2022 early access) on the detection of various environmental contaminants using SERS
| Substrates | Analytes | Sample or Sample Matrix | LODs | References |
|---|---|---|---|---|
| Silver @ molecular imprinted polymers | Carbendazim | Water | 1 × 10–9 M | [162] |
| Silver microspheres | Carbendazim | Chinese tea | – | [163] |
| Silver nanoparticles | Carbendazim | Spiked orange juice and kale leaves | <2 mg/L | [164] |
| Gold nanoparticles | Chlorothalonil | Spiked orange peels and standard solution | 0.1 mg/L | [166] |
| Titanium carbide Mxene with bimetallic nanocuboids | Crystal violet, malachite green, methylene blue | Spiked pond water | 10–12 M | [168] |
| Gold nanorods on regenerated cellulose | Crystal violet and thiram | Substrate surface | 1 aM | [161] |
| Quart paper/cellulose nanofibres/silver nanoparticles + gold nanostars | Ferbam | Spike kale | 50 mg/kg | [179] |
| 2‐mercaptoethanol/gold @ silver nanoparticles | Ferbam and thiabendazole | Polluted apple puree | 0.0042 and 0.0064 mg/L | [20] |
| Poly (ethylene terephthalate)/pDA/zinc oxide/silver | Malachite green | Substrate surface | 10–7 M | [169] |
| Parahydrophobic nanoparticles | Malachite green | Water | – | [170] |
| Silver nanowires @ polydimethylsiloxane | Malachite green | Spiked apple and pear juice | 10 nm | [171] |
| Cotton fibre silver nanoparticles | Malachite green | Spiked fish | 0.05 mg/L | [172] |
| Gold‐silver alloy nanochains | Rhodamine 6G, crystal violet and thiram | Analyte on SERS substrate | 0.03 mg/L (thiram) | [127] |
| Methylcellulose decorated silver nanoparticles | Thiram | Spiked tomato and cucumber peels | 2.4 ng/cm2 | [128] |
| Silver/hedgehog like nanosphere array | Thiram | Analyte on SERS substrate | 10–8 M | [129] |
| Gold/zinc oxide nano urchins | Thiram | Analyte on SERS surface | 10 pM | [130] |
| Plasmonic nanocavities | Thiram | Apple pericarp | 10–11 M | [131] |
| Copper oxide @ zinc oxide @ silver biomimetic setaria | Thiram | Apple | Single molecule | [132] |
| Silver nanoparticles/fluorinated ethylene propylene | Thiram | Apple | 0.1 mg/kg | [133] |
| Silver nanoparticles | Thiram | Apple | 0.1 mg/L | [134] |
| Micropyramid array/silver nanoparticles | Thiram | Analyte on SERS substrate | 1 × 10–7 M | [135] |
| Silver/polystyrene nanoparticles | Thiram | Apple peel, mineral water and apple juice | 0.0024 mg/L (apple juice and mineral water) and 600 ng/cm2 (apple peel) | [136] |
| Mesoporous gold @ silver nanowires @ 2,2,6,6‐tetramethylpiperidine‐1‐oxy‐oxidized – cellulose nanofiber | Thiram | Analyte on SERS substrate | 10 fM | [137] |
| Yolk shell gold‐silver nanorods | Thiram | Apple juice | 97 nM | [138] |
| 2D titanium carbide MXene/gold nanorods | Thiram | Mixture of SERS substrate and analyte deposited on Si substrate | 10–8 M | [139] |
| Silver/tungsten oxide nanoflakes | Thiram | Apple peel | 1.3 pM | [140] |
| Silver @ tannic acid @ silicon dioxide | Thiram | Analyte on SERS substrate | 10–8 M | [141] |
| Silver nanoparticles/waterborne polyurethane emulsion | Thiram | Apple | 9.0165 ng/cm2 | [142] |
| Ferrero chocolate like– copper oxide @ silver microspheres | Thiram | Apple peels | 0.018 mg/L | [143] |
| Gold @ silver nanorods | Thiram | Apple | – | [144] |
| Gold nanorods | Thiram | Analyte and SERS substrate | 10–9 M | [145] |
| Silver nanoparticles @ copper (II) hydroxide nanowires/copper mesh | Thiram | Analyte and SERS substrate | 0.1 mM–50 nM | [146] |
| Silver @ tungsten disulfide quantum dots | Thiram | Honey and 4 juices: apple, peach, grape and orange | 50 µg/L | [147] |
| Silver nanoparticles | Thiram | Theoretical | 1.2 × 10–8 M | [148] |
| Silver nanoparticles | Thiram | Analyte on SERS substrate | 1 µM | [149] |
| Silver @ gold nanorods | Thiram, propineb and mancozeb | Romaine lettuce and broccoli | 0.05, 0.1 and 0.2 mg/L | [150] |
| Gold @ silver nanoparticles | Thiabendazole | Apple | 0.001 mg/L | [177] |
| Tape/gold @ silver/poly ethylene terephthalate film | Thiram | Apple, tomato and cucumber peels | 5 ng/cm2 | [61] |
| Polymath methacrylate‐silver / graphite‐like carbon nitride / silver | Thiabendazole and carmine acid | Apple and apple sauce | 2.296 × 10–6 and 6.76 × 10–5 mg/mL | [175] |
| Silver‐nanoparticles @ zinc oxide‐nanowires | Thiram and methyl parathion | Analytes and SERS substrate | 0.79 × 10–9 and 1.51 × 10–8 M | [151] |
| Silver nanoparticles @ free standing porous silicon | Thiram, ammonium nitrate and picric acid | Analytes on SERS substrate | 1, 2 and 1 µM | [152] |
| Gold @ silver nano cuboids | Thiabendazole extracted from pear surface and malachite green in fish pond water | Pear peel and pond water | 0.0105 and 0.87 nM | [173] |
| Gold nanocubes/graphene oxide/silver nanoparticles | Thiram and thiabendazole | Drinking water | 0.37 and 8.3 µg/L | [153] |
| Silver nanoparticles | Thiram and ziram | Apple juice | 10–2 and 10–4 mg/L | [154] |
| Gold @ 4‐mercaptobenzoic acid @ silver nanoparticles | Thiram and thiabendazole | Aqueous and organic phases | 1.58 × 10–9 and 1.26 × 10–7 M | [79] |
| Gold @ silver nanoparticles | Thiabendazole, thiram, endosulfan and malathion | Strawberry extract | 44–88 µg/kg | [155] |
| Silver stars | Rhodamine B, methylene blue, thiram and phosmet | Analytes and SERS substrate | 2.6 × 10–16,1.97 × 10–17, 1.78 × 10–18 and 4.08 × 10–14 M | [156] |
| Silver 4 ‐NTP @ gold nanoparticles | Chlorothalonil, imidacloprid and oxyfluorfen |
|
– | [167] |
| Gold nanoparticles | Thiram, phorate and benthiocarb | Rice, vegetables and fruits | 10–8 g/mL for thiram, 10–6 g/mL for phorate and benthiocarb | [157] |
| 3D‐random crossed woodpile | Thiram, carbaryl, paraquat and fipronil | Analyte on SERS substrate | 5–0.05 µM in 20 µL | [158] |
| Gold nanoparticles | Thiram, imidacloprid and chlorpyrifos | Tap water | – | [159] |
| Mercaptooctane/gold @ silver nanoparticles | Tricyclazole and thiram | Pear extract | 0.005 and 0.003 mg/L | [39] |
| Silver substrate | 2,4,5–trichlorophenoxyacetic acid | Analyte adsorbed on SERS substrate | – | [190] |
| Gold nanorods | Atrazine | Analyte on SERS substrate | 1.8 µM | [182] |
| Silver nanoparticles | Deethylhydroxyatrazine | Humic substances | – | [185] |
| Gold nanostars | Paraquat | Green tea | 0.2 mg/kg | [187] |
| Silver nanoparticles | Paraquat | Lake, tap and drinking water | 1.2 µg/L | [188] |
| Silver nanoparticles | Prometryn and atrazine | Soil |
|
[184] |
| Silver nanoprisms | Atrazine, simazine, irgarol and diuron | Analyte on SERS substrate | 10–3 M | [186] |
| Cyclodextrin‐gold nano satellite | Paraquat, diquat and difenzoquat | Ground apple | 0.05 mg/L | [189] |
| Gold @ silver nanoflowers | 2,4–D and imidacloprid | Milk | 2.73 × 10–4 and 4.25 × 10–4 ng/mL | [191] |
| Silver nanoparticles/poly methyl methacrylate | Parathion and fenitrothion | Tomato and lemon | 10–9 and 10–10 M | [193] |
| Gold nanoparticles | Acetamiprid | Apple and spinach | 1.9 × 10–7 M | [209] |
| Silver nanoparticles | Chlorfenapyr | Chive | 7 mg/L | [210] |
| Silver palladium and gold palladium | Chlorpyrifos | Analyte on SERS substrate | 0.66 mg/kg | [194] |
| Silver @ zinc oxide nanoflowers | Chlorpyrifos | Rice | 0.01 µg/mL | [196] |
| Gold nanoparticles | Chlorpyrifos | Tea | – | [197] |
| Silver nanoparticles | Dichlorvos | Analyte on SERS substrate | 1 mg/L | [211] |
| Gold @ platinum nanoparticles | Dichlorvos | Pear | 20 µg/L | [212] |
| Psi‐Pd NPs | Imidacloprid | Analyte on SERS substrate | 10–9 M | [202] |
| Antigen‐gold nanorods @ silver – 4‐ MBN | Imidacloprid | River water and apple juice | 9.58 nM | [203] |
| Gold nanoparticles | Imidacloprid | Analyte and SERS substrate | – | [204] |
| Gold‐gel | Parathion‐methyl | Analyte on SERS substrate | 10–9 M | [199] |
| Silver nanoparticles | Pymetrozine | Apple and cabbage | 0.01 mg/L | [214] |
| Silver @ DNA/PDA‐cellulose nanofiber | Thiamethoxam | Apple | 0.003 mg/kg | [59] |
| Mesoporous copper (I) oxide @ silver nanoparticles | Pymetrozine and thiram | Tea | 0.1 ng/g | [160] |
| Gold substrate | Acetamiprid and imidacloprid | Plasmon activated water | 5 and 10 µg/L | [205] |
| Ag@D‐TMIP | Carbaryl and thiodicarb | Spiked tap water | – | [208] |
| Silver @ gold nano tetrahedrons | Profenofos, acetamiprid and carbendazim | Real samples | 0.0021, 0.0046 and 0.0061 ng/mL | [165] |
| Silver nanoparticles | Methomyl, acetamiprid and 2,4–dichlorophenoxyacetic acid | Green tea | 5.58 × 10–4, 1.88 × 10–4 and 4.72 × 10–3 µg/mL | [192] |
| Gold nanoparticles | Temephos, acetamiprid, dicofol and fenvalerate | Basil leaf | – | [274] |
| Au/Cys‐Fe3O4/MIL‐101 | Parathion methyl | Juice | 5 mg/L | [200] |
| Gold superlattices particles | Organochlorine (OCP): (4,4′‐DDT, a‐endosulfan, tetradifon and chlordane) | Water | – | [201] |
| Titanium carbide/silicon dioxide/polydimethylsiloxane | Acetamiprid, clothianidin, imidacloprid and thiamethoxam | Apple, wheat, green tea and corn | 7.9, 6.9, 4.8 and 8.2 fM | [206] |
| Aluminum‐titanium dioxide‐metal organic framework‐silver composite sheet | 4‐aminothiophenol | River water | 1 × 10–9 M | [275] |
| Silver/cobalt 2‐methylimida‐zole metal–organic framework/titanium dioxide/copper sheet | 4‐aminothiophenol | Deionized and river water | 5 × 10‐11 M | [276] |
| Gold @ silver‐thioglycolic acid nanoparticles | Thiabendazole and ferbam | Milk | 0.12 and 0.003 mg/L | [178] |
| (S‐MOF@Au) | Tartrazine (TA), chloramphenicol (CP), imidacloprid (IDP) and crystal violet (CV) | Analyte and SERS substrate | 1.0×10–9 M for TA, 8.0 ×10–11 M for CP, 6.4 × 10–11 M for IDP and 5.0 × 10–12 M for CV | [207] |
| Gold @ silver nanorods | Thiabendazole | Apple and peach juice | 0.032 and 0.034 mg/L | [174] |
| Silver nanoparticles | Thiabendazole and 1,2,3,5‐tetrachlorobenzene | Analyte and SERS substrate | 3.7 × 10–7 and 2.3 × 10–5 M | [176] |
| Silver nanoprism/graphene oxide/silicon nanowire arrays | Atrazine | Analyte and SERS substrate | 10–6 M | [183] |
| Gold nanostars / PDMS | Methyl parathion | Apple | 1.946 µg/mL | [198] |
| Gold @ silver nanorods | Nile blue A dye and fenobucarb pesticide | Analyte on SERS substrate | 1.5 × 10−8 M (fenobucarb) | [213] |
Articles were obtained by performing the following search on Web of Science and removing non‐environmental papers: SERS + Pesticide. The table details substrate type, analyte type, the sample or sample matrix used, method LOD and the corresponding reference.
2. SERS NANOSUBSTRATES
The choice of SERS nanosubstrates is of paramount importance in Raman analysis. The SERS substrate used determines the method reliability, accuracy and sensitivity. 20 SERS substrates often include roughened surfaces incorporating gold nanoparticles (AuNPs), silver nanoparticles (AgNPs), CuNPs or their nanocomposites (NCs). 21 As shown in Figure 2A, AgNPs were found to be the most commonly used type in our reviewed articles. In addition to noble metal nanoparticles, non‐metal SERS nanosubstrates have also been developed such as semiconductors, quantum dots and graphene dots. 8 Recently, to enhance the SERS performance and functionality, it has become a trend to integrate nanosubstrates with functional materials, including semiconductors, metal‐organic frameworks (MOFs), molecularly imprinted polymers (MIP), grapheme and cellulose (Figure 2B). In this section, key concepts and methodology related to the recent development of innovative SERS substrates are discussed.
FIGURE 2.
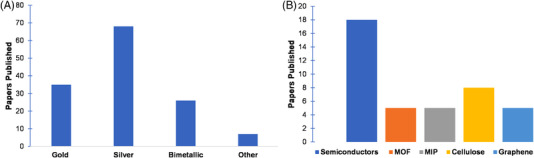
Number of research papers published from 2021 to 2022 on (A) different SERS nanosubstrates and (B) integrated materials to improve SERS performance
2.1. Hot spots
An important consideration in the development of SERS substrates is the creation of ‘hot spots’, or areas of intense localized surface plasmon resonance (LSPR) which comprise the electromagnetic portion of SERS enhancement. Hot spots occur due to the presence of strong electromagnetic fields on the nanometal surface of SERS substrates, which are formed upon excitation from a light source of a particular wavelength. 9 , 22 , 23 Hot spots increase when there are gaps between two or more metallic nanoparticles that are within 10 nm proximity. 24 , 25 A SERS substrate with a higher density of hot spots across the surface is ideal for Raman enhancement. Hot spots can provide enhancement up to 1015 compared with normal Raman scattering, and are practically required for single‐molecule SERS detection. 9 , 10 There are many factors that influence hot spot formation including nanoparticle composition, shape, size and arrangement. It has been suggested that hot spots more readily form at sharp corners and edges due to an increase in surface charge density at these positions. This enhancement is known as the ‘lightning rod’ effect. 10 To produce reproducible substrates with uniformly distributed hot spots, it is necessary to control the shape and size of the metallic nanoparticles. Surfactants have been used successfully to control these factors in AuNPs and AgNPs. 10 , 26 , 27 Control of the nanostructure arrangement helps to configure hot spots on the nanomaterial surface. This can be accomplished through the use of covalent bonds between thiolated ligands and gold which results in the formation of self‐assembled monolayers. 10 , 28 , 29 Many works focus on the development of nanoparticles of a specific shape, size or arrangement that will facilitate the generation of hot spots on the substrate's surface. Due to the frequently heterogeneous nature of SERS substrates, the enhancement from hot spots on the surface can vary greatly, which poses a challenge for quantitative determination of an analyte. 30 , 31 , 32 To combat this issue, internal standard (IS) molecules are often used, and are discussed in greater detail later in this article. A review discussing recently developed quantitative SERS methods was published in 2018. 33
2.2. Composition
Recent work has investigated various new substrate materials or modification of commonly used materials to enhance the signals. It is ideal for substrates to be stable and uniform to provide robust and reproducible SERS detection. In this section, we will discuss recent advancements for SERS substrates that focus on the composition of the materials used including bimetallic compounds, functionalized nanoparticles and graphene‐based SERS substrates.
It has been noted that bi‐metallic nanoparticles often yield better results than single‐element nanoparticles. Gold‐silver core‐shell nanoparticles (Au@AgNPs) are often used, as they take advantage of the high stability of the gold core while obtaining a higher SERS enhancement from the silver shell. 20 , 34 , 35 , 36 In addition, surface modification and functionalization of substrates can enhance the SERS signals, allowing for more sensitive detection through creating more adsorption sites on the substrate for the analyte. 20 , 37 , 38 , 39 One study utilized the beneficial aspects of bi‐metallic nanoparticles while modifying the substrate's surface with a thiol‐containing compound (2‐mercaptoethanol). 20 This compound strongly coordinated with the metals in SERS substrates and acted as a stabilizer, linker and modifier for the surface. The substrate was used to quantify low concentrations of the pesticides ferbam and thiabendazole (TBZ) to 0.0042 and 0.0064 mg/L, respectively. 20 This demonstrated better stability and higher sensitivity than a control substrate made from Au@AgNPs without thiol‐containing ligands. They attributed the higher sensitivity to the creation of more adsorption sites via Au‐S bonds on the surface of core‐shelled NPs, and the higher stability with the thiol‐containing ligands acting as a stabilizer to prevent oxidation of AgNP shells. Similarly, modification of a SERS substrate has also been accomplished through aggregation of AuNPs with cetyltrimethylammonium bromide (CTAB) to obtain more sensitive detection of the synthetic dye, 1,2‐dihydroxyanthraquinone (alizarin). It was suggested that this method could be beneficial for SERS analysis of weakly adsorbing biological molecules. 40
Graphene has been incorporated in SERS substrates and provides an additional enhancement due to G‐SERS or GERS (graphene‐enhanced Raman scattering). Graphene has unique chemical and physical properties that are beneficial for SERS analysis including a tunable band gap, good thermal conductivity, high optical transparency, fluorescence quenching abilities and biocompatibility. 41 , 42 In addition, hot spots are created in gaps between the metal nanoparticles in monolayer graphene or graphene oxide (GO)/AuNP structures. Furthermore, a graphene monolayer consists of sp2‐hybridized carbon atoms and is a π‐conjugated system. This allows analyte molecules to adsorb to the surface and interact electronically with the metal and graphene surface, causing Raman enhancement due to the charge‐transfer resonance between the analyte and the metal or graphene surface. 43 One study utilized G‐SERS to detect DNA bases, malachite green (MG) and beta‐carotene. 43 Through the use of GO nanosheets coated with AgNPs and AuNPs in arrays, small biomolecules were efficiently detected, suggesting that graphene‐based SERS substrates can be used for sensitive and selective detection of particular biological and environmental molecules of interest. Another study used graphene in conjunction with AgNPs to detect p‐nitroaniline (PNA). 44 This work made use of defect‐graphene (DG), which has a higher surface area and porosity, and was created via acid etching of the graphene surface. The DG/Ag substrate was then functionalized with a MIP film that allowed for specific binding to the target molecule and high SERS enhancement (EF = 2.42 × 107). This DG/Ag‐MIP substrate was able to detect PNA in real water samples down to a LOD of 2.5 × 10–15 M and a limit of quantitation of 10–12 M.
2.3. Shape, size and arrangement of nanoparticles
The design of nanoparticles of unique shapes, sizes and arrangements that work well with particular analytes is essential for the development of SERS‐based analytical approaches. For example, Leong et al. accomplished identification and quantification of specific enantiomers through fabrication of nanoporous gold bowls (NPGBs). Figure 3A shows a schematic depicting the ability of the NPGBs to differentiate between enantiomers through utilization of an asymmetric surface structure in conjunction with electrochemical‐SERS. Figure 3B is a high‐magnification SEM image of a NPGB. This enantiospecific adsorption on the substrate resulted in the generation of Raman spectra which were enantiomer‐specific and, thus, allowed for rapid, chemometrics‐driven analysis and identification of enantiomeric compounds. 45 This unique method can provide valuable information regarding enantiomeric ratios of biologically important molecules, such as pharmaceuticals, which may enter in the environment. Another study created a cost‐effective SERS substrate using filter paper and silver nanodisks (AgNDs) to detect the antibiotic tetracycline (TC). 46 The AgNDs were prepared via a seed‐mediated synthesis that used Ag disk‐shaped seeds, N,N‐dimethyldodecylamine N‐oxide (DDAO) as a stabilizing and surfactant agent, and ascorbic acid as a reducing agent. The AgND seeds’ morphology was investigated with atomic force microscopy (AFM), and the disks were determined to have a size between 200‐450 nm with 30‐50 nm thickness. The developed SERS substrate was used effectively for the detection of TC down to 1 nM. The relationship between the TC molar concentration and Raman intensity at 1317 cm–1 was plotted between 1 nM and 1 µM, and the experimental data were fitted to a Freundlich adsorption model 47 (one often used for heterogeneous surfaces). 46 , 48 Silver nanoparticle‐grafted silicon nanocones (AgNPs/SiNC) were developed in a recent work for the detection of trace contaminants in complex liquid environments: crystal violet (CV) in lake water, melamine in milk, methyl parathion in tap water and rhodamine 6G (R6G) in deionized water. 49 To create the nanosubstrate, droplet‐confined electroless deposition was used to deposit AgNPs onto a hydrophobic silicon nanocone array. Due to enrichment of the analytes by ‘super‐hydrophobic delivery’ 50 near hot spots on the substrate surface, analytes were detected at sub‐picomolar concentrations within aqueous solutions. This substrate demonstrates a practical method for monitoring trace contaminants in various liquid environments.
FIGURE 3.
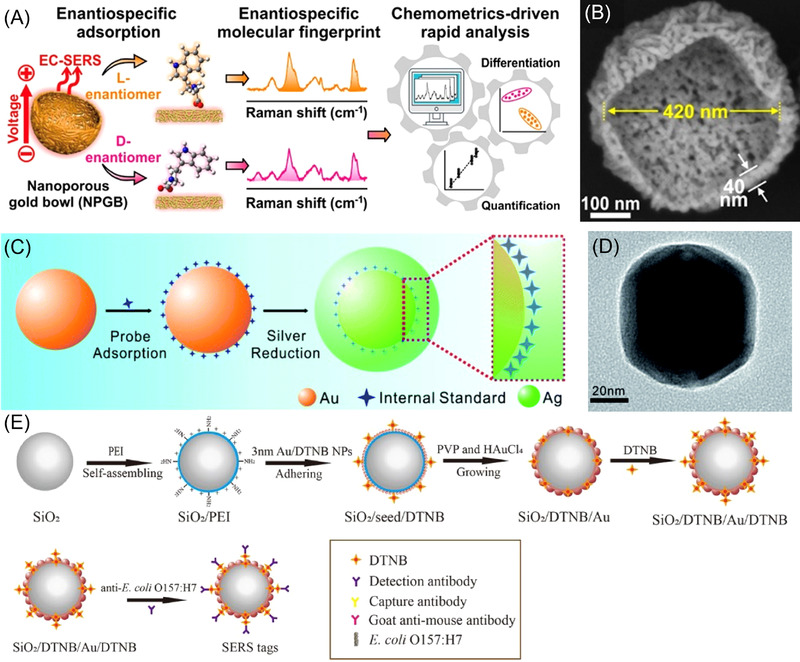
Various methods for creating SERS nanosubstrates are shown here. (A) Schematic showing asymmetric nanoporous gold nanoparticles, which form nanoporous gold bowls, and through electrochemical‐SERS, able to produce enantiospecific molecular fingerprints for a label‐free approach to chiral differentiation (adapted with permission from Ref. [45]. Copyright 2021 American Chemical Society). (B) High‐magnification SEM image of a nanoporous gold bowl (adapted with permission from Ref. [45]). (C) Schematic illustration showing the synthesis of a bimetallic nanoparticle with an embedded internal standard for quantitative detection. The nanoparticle utilizes a gold core, 4‐Mpy as the embedded Raman probe and a silver shell for both protection and enhancement (Au@4‐Mpy@AgNP) (reprinted with permission from Ref. [70]. Copyright 2021 Royal Society of Chemistry). (D) TEM image of a Au@4‐Mpy@AgNP (reprinted with permission from Ref. [70]). (E) Schematic representation for the preparation of the dual DTNB‐modified gold‐shell silica‐core nanoparticles (SiO2/Au NPs) for the quantitative detection of E. coli (adapted with permission from Ref. [232]. Copyright 2020 Shi, Xu, Xiao, Zhou, Wang, Wang and Gu)
Another study created a nanosubstrate of unique shapes for SERS detection: Ag/CuO nanowires (NWs)/pyramidal polydimethylsiloxane (PDMS). 51 A low‐cost fabrication was presented that utilized spin‐coating to apply a PDMS monomer with a cross‐linking agent colloid onto pyramidal Si. Following the removal of the PDMS film, Cu film was deposited via thermal evaporation, followed by immersion in a diluting anti‐formin solution to create CuO NWs. A thin layer of silver was deposited onto the surface via thermal evaporation to gain SERS enhancement. The substrate was tested for uniformity using SERS mapping of R6G signals in a large area. Twenty random points on the substrate were evaluated to determine the RSD of characteristic Raman peaks of R6G (613, 774 and 1365 cm–1), which were 6.76, 7.44 and 8.57%, respectively. 51 In addition to the high uniformity, the method was capable of differentiating R6G, CV and congo red (CR) in a mixture of the three. This substrate shows immense potential for the uniform detection and identification of synthetic dye molecules. 51
Nanoparticle arrays are sometimes used as a technique of arranging nanomaterials into a well‐ordered system that exhibits desirable properties. Their properties can be controlled by engineering the nanoparticle shape, size and distance from other particles. 52 The small gaps between the nanoparticles have enhanced electromagnetic fields, leading to the generation of SERS hot spots across a three‐dimensional (3D) structure. 53 Large areas of 3D nanoparticle arrays have been used successfully for SERS detection. 54 , 55 , 56 , 57 , 58 In a study by Li et al., high area and highly ordered silver ‘urchin‐like’ arrays were created to detect CV and thiram (THR) in real water environments. 53 The nanoparticle array was formed through an easy and cost‐effective double template method involving an ultra‐thin alumina membrane mask and a self‐assembled polystyrene (PS) template. The array demonstrated great SERS enhancement in the gaps between Ag nanocones and at the tips of the nanocones (EF of 1.8 × 106 for R6G detection). This substrate shows great promise to be used with SERS as a sensitive, cost‐effective and quantitative tool for environmental monitoring.
2.4. Flexible SERS nanosubstrates
Due to the natural irregularities of environmental analytes, the fabrication of flexible SERS substrates has become a popular area of interest in recent years. Conventional SERS substrates are often brittle, rigid, expensive or non‐environmental friendly. 59 Cellulose‐ and paper‐based substrates have attracted attention for their high degree of flexibility and 3D structure. 60 , 61 , 62 For further information, we direct readers to some recent reviews published in 2021, one which discusses the development of filter‐based SERS substrates, including a summary of the applications for these substrates in environmental monitoring and food safety, 63 and another which details various advancements in flexible SERS substrates for the detection of hazardous materials.
As a SERS substrate, cellulose has increased sensitivity due to its high porosity and natural wrinkles, allowing for more plasmonic hot spots to form on its surface. 34 Since cellulose is sourced primarily from plant matter, it is a sustainable and renewable resource for SERS substrates. Cellulose that is thin enough (in one or more dimensions) to be measured on the nanometre scale is referred to as nanocellulose. It has been reported previously that nanocellulose is a material which can be self‐assembled into well‐defined structures. 65 , 66 , 67 One study by Jeon et al. used regenerated cellulose (RC) hydrogel films with either gold nanorods (AuNRs) or silver NWs (AgNWs) to detect PS nanoplastics (NPLs), and claimed to have developed one of the first cellulose‐based SERS substrates to detect microplastics (MPLs) and NPLs. 62 Through comparison studies, they found that PS detection was greatly enhanced using the AgNWs/RC film as opposed to the AuNRs/RC film, and suggested that this substrate could be used to detect plastic contamination in water environments. 62 Recently, Xu et al. utilized DNA as a template to create polydopamine (PDA)‐coated, Ag‐modified cellulose nanofibers. These Ag‐modified cellulose nanofibers self‐assembled into a three‐dimensional flexible structure to create the low‐cost substrate, Ag@DNA/PDA‐CNF. 59 This was based on the π‐π conjugation and complementary structure of DNA with PDA, which led to rapid and orderly self‐assembly of PDA nanosheets. 68 There were abundant active sites where AgNPs could attach close to one another, which created large areas of hot spots on the substrate surface. This method could detect R6G with an EF of 1.03 × 108, and the flexibility of the cellulose‐based substrate allowed it to be wiped on fruit peels to detect the pesticide thiamethoxam down to 0.003 mg/kg.
An article‐based substrate with good flexibility was used in a recent work by Sun et al. in which AgNWs were loaded onto filter papers to detect the antibacterial agent furazolidone (FZD) with an EF up to 2.63 × 106. 69 This substrate was modified to be simultaneously hydrophobic and hydrophilic. To do this, a four‐layer matrix was first assembled from adhesive tape, carbon paper, aluminium foil and filter paper (with the filter paper being the top layer). Next, a 2‐mm diameter hole was cut out of the matrix material and coated with hydrophobic fluorosilicone resin. The punched‐out matrix material was then re‐attached in the middle without the hydrophobic coating, and the AgNW solution was dropped onto the central hydrophilic area of the substrate, constituting the completed hydrophilic‐hydrophobic Ag NW‐paper substrate. Methylene blue (MB) was used as a model ligand to test the substrates and it was found that, when MB was dropped into the 2 mm hole in the centre of the developed substrate, there was no ‘coffee ring’ after drying, as there was when MB was dropped onto a 4.5 mm diameter aluminium foil substrate. This substrate allowed an even distribution of analyte molecules across the surface, which is important for accurate SERS analysis. This article‐based hydrophobic‐hydrophilic substrate was able to detect MB and FZD concentrations down to 9.4 × 10–9 mmol/mL and 6.83 mg/L, respectively, with good linearity (R 2 values of 0.991 and 9.995, respectively). The substrate was further tested for the detection of FZD in the presence (and interference) of MB in various water environment and on the surface of fish, and detection recovery was 83.9–111% and 79.5–84.6%, respectively. 69
Cellulose‐ and paper‐derived substrates have the potential to be extremely useful in conjunction with rapid SERS analysis. The substrates are more environmental friendly than conventional SERS substrates, and their flexibility lends to unique applications such as detection of analytes on uneven biological surfaces. These substrates can be cheaply produced and further developed into environmental sensors for specific analytes.
2.5. Internal standards
While SERS is extremely useful and reliable for the identification of analytes, its ability to quantify specific molecules accurately is limited by the reproducibility of the SERS signals on the prepared SERS substrate. A commonly seen issue is that the distribution of hot spots along the substrate's surface is not uniform, resulting in heterogeneous signal strength in various areas. 14 , 33 , 70 Raman mapping can be used to determine the uniformity of a substrate by using two‐dimensional map to show the variation of Raman signal intensity across points within a specified area. If the substrate is relatively uniform, the RSD between the points will be smaller. 42 , 71
Highly uniform SERS nanosubstrates are ideal to ensure SERS reproducibility. However, the synthesis of uniform nanosubstrates can be quite expensive, skill demanding and time consuming. For substrates synthesized with less precision, where uniformity is not guaranteed, it is appropriate to use an IS molecule to increase reproducibility. In utilizing IS molecules, the intensity ratio of the target molecule to the IS molecule is unchanged, even as the overall enhancement across the SERS surface may change due to the heterogeneous nature of substrates with non‐uniform hot spots. 30 , 31 , 32 , 72 Some challenges are that both the analyte and IS need to reach some threshold concentration before the IS provides SERS signals. 73 In some cases, there is a possible competition between the IS molecules and the analyte of interest for the metal SERS substrate surface, potentially decreasing the accuracy of the quantitative measurement. 73 , 74 Many studies used embedded IS molecules, which are bound to the core of the nanosubstrate. A shell is then formed around these to protect the IS molecules. There is no competitive adsorption between the IS molecules and target molecules with embedded IS molecules, so interference from the IS is not a problem for quantitative SERS detection of the target molecule. 70 , 75 , 76 , 77 , 78 A recent study focusing on the detection of the fungicides THR and TBZ utilized 4‐MBA as an IS molecule. 4‐MBA functionalized AuNPs acted as the core in an AgNP‐coated shell, and the core‐molecule‐shell NPs self‐assembled to form a dense nanoparticle array (Au@4‐MBA@Ag NPs). They were able to obtain LODs of 0.38 µg/L for THR and 25 µg/L for TBZ, 79 which produced a significantly more sensitive detection than their previous study, which used an Au@Ag array without an IS (LOD for THR = 1.1 µg/L and for TBZ = 51 µg/L). 80 Similarly, Wang et al. synthesized Au@4‐mercaptopyridine (4‐Mpy)@AgNPs (Figure 3C and D) by first fabricating AuNPs coated with the IS molecule 4‐Mpy, then utilized a silver reduction method to cover these Au@4‐Mpy molecules with an AgNP shell, forming the final core‐shell nanoparticles with an embedded IS. 70 This IS‐embedded nanostructure created a three‐dimensional hot spot matrix SERS platform in water with 2.5% v/v glycerol. Quantitative detection for CV was achieved during evaporation with good sensitivity (down to 5 × 10–9 M) and linearity. The Au@4‐Mpy@AgNPs substrate was also used for quantitative detection of MG and THR through the use of linear fitting plots. It was noted that this substrate was a good candidate to mitigate the effects of Raman signal fluctuation through the use of the embedded IS with a glycerol‐assisted three‐dimensional hot spot platform, and it has great promise for the accurate quantitative detection of various environmental pollutants. 70
Bacteria were recently used in conjunction with Au@Ag core‐shell nanoparticles that were synthesized on the bacterium (Shewanella oneidensis). The bacteria's cell membrane worked as an IS when the Raman signals of both the target molecule and the bacteria were measured, and they maintained a relatively stable ratio for SERS intensity. This method was used to detect several analytes at varying concentrations to create linear curves: R6G (0.01–100 µM), MG (0.001–1 µM) and uric acid (100–500 µM). 32 Competitive adsorption between IS and analyte molecules is avoided with this method. The use of ISs is a promising technique to improve quantitative detection for analytes of interest, especially when using a substrate with a heterogeneous surface with non‐uniform hot spots.
3. ANALYTES OF INTEREST
In this section, we discuss current advancements in the SERS detection of inorganic, organic and biological pollutants. Specifically, we cover how SERS can be used to detect environmental contaminants such as heavy metals, pharmaceuticals, plastic particles, synthetic dyes, pesticides, viruses, bacteria and mycotoxins. In summarizing this topic, our aim is to highlight areas where significant amounts of research are being published, as well as areas that need further investigation. This can also be seen visually in Figure 4, wherein there is a high volume of recent publications for SERS detection of pesticides, but less for other analytes such as plastic particles and viruses.
FIGURE 4.
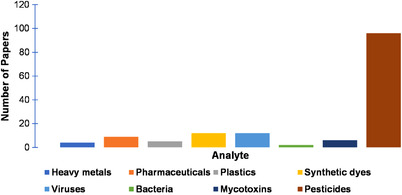
Number of research papers published on different analyte categories discussed in this review from January to December 2021 and early access 2022
3.1. Inorganic pollutants
3.1.1. Detection of heavy metals
Heavy metals are naturally occurring metallic and metalloid elements that have a high atomic weight and high density when compared with water. 81 Some heavy metals are essential in small amounts for physiological functions in living organisms (such as Co, Cu, Fe, Mn, Mo, Ni and Zn, which are considered micronutrients), 82 , 83 while others are extremely toxic and potentially carcinogenic (As, Cd, Cr, Hg and Pb). 84 , 85 There are increasing concerns regarding heavy metal contaminants within the environment, as they pose a threat to both human and ecological health. Certain anthropogenic activities (e.g., mining, battery manufacturing and pesticide use) have greatly increased our exposure in recent years. 84 , 86 For more detailed information on this topic, we direct readers to a recent in‐depth review from 2021 of the detection of heavy metals via SERS. 87 Some recent developments in SERS mainly focus on the detection of lead, iron and mercury in the environment.
Lead is one of the most toxic heavy metal ions and can cause irreversible damage to various organ systems in the body. It is especially dangerous for young children to come into contact with lead contamination, as it can cause life‐long health challenges. 42 , 88 The use of lead in several commercially available products (such as leaded gasoline 89 or lead‐based paints) has significantly decreased in recent years. However, residual lead contamination is still present in soils and waterways. It is of the utmost importance to be able to detect lead contamination selectively and sensitively in our environment. A reusable and cost‐effective SERS sensor was created from monolayer graphene integrated with gold‐silver hybrid nanostructures on a porous gallium nitride (GaN) surface to reliably detect and enhance the Raman signal for Pb2+ down to 4.31 pM. 42 In a two‐dimensional honeycomb arrangement, graphene has shown the ability to provide enhancement for adsorbed analyte molecules when used with SERS due to the GERS effect. 41 , 42 This sensor provided ‘dual enhancement’, as it used graphene with noble metal nanostructures in conjunction with the benefits of enzyme‐linked reactions, to help detect and quantify Pb2+. To accomplish this, DNA was hybridized to Pb2+. Labelled Cy3‐DNAzyme probes with Pb2+‐specific DNA substrate strands were immobilized on the SERS substrate containing metal nanoparticles on porous GaN with monolayer graphene. The Cy3‐labeled thiolated DNAme probe on the substrate produced strong Raman signals. Following this, the complementary DNA strand was attached to the DNAzyme probe to form double‐stranded DNA (dsDNA). This dsDNA yielded weaker Raman signals, because the rigid dsDNA pulled the Cy3 further from the substrate surface. 42 When Pb2+ was present on the substrate, the Pb2+‐specific DNAzyme was activated, which split the DNA strand into two free short chains, allowing the Cy3‐labeled DNAzyme probe to move closer to the SERS surface. This movement is due to π‐π adsorption between the single‐stranded DNA and the graphene surface, and ultimately causes an enhancement in the Raman signal through electromagnetic and chemical enhancements. The Raman signal will increase as Pb2+ concentration increases. 42 It was reported that this sensor was recyclable up to three times with only a slight decrease in background Raman intensity after each use. 42 To reuse the substrate, the SERS sensor was simply washed with PBS to remove dissociated oligonucleotide that had been cleaved by Pb2+. Freshly prepared substrate strand buffer solution was re‐incubated with the sensors for 12 h to prepare them to detect Pb2+ again. The reusability of this sensor makes it more environmental friendly and cost‐effective.
Another recent work focused on the development of a label‐free, environmental‐friendly SERS probe for Fe3+ detection. 90 While small amounts of Fe3+ are essential for living beings, an excess can be harmful and cause ailments such as cancer, 91 liver diseases, 92 and neurodegenerative diseases. 90 , 93 , 94 During various industrial processes, such as those associated with the production or mining of metals, Fe3+ waste is produced and released into the environment. Excess Fe3+ in water systems has toxic effects on aquatic organisms, and may biomagnify and cause harm to humans as well. 95 Thus, it is essential to efficiently monitor Fe3+ in the environment. A recent work focused on a label‐free Raman probe using Tollens’ reagent and phytic acid for the nanoprobe synthesis. 90 Through the Tollens’ reagent method, the AgNPs were synthesized in as little as 30 min, and due to the mild nature of the reagents, they were more homogeneous than those prepared from conventional methods. The phytic acid molecules used in the AgNP synthesis have a strong selectivity for Fe3+, which were detected simply by mixing standards of Fe3+ with the Raman probe, incubating for 30 s, and dropping onto a glass slide for Raman detection. The Fe3+ aggregated the AgNPs and caused a change in the obtained Raman spectra, producing two characteristic peaks. The developed Raman probe was exposed to other metal ions at the same concentration as Fe3+, and it was found that the probe had a strong sensitivity toward Fe3+. While this SERS probe was designed for detection of Fe3+ in integrated circuit cleaning solution waste, it had a low LOD of 0.1 µM, and may be promising for other environmental matrices.
Mercury contamination in the environment has both natural and anthropogenic origins. As exposure to mercury or mercury complexes has several toxic effects on the human body, 96 , 97 , 98 there is a need for trace mercury detection. A SERS‐based Hg2+ sensor was recently developed by using AgNPs functionalized with phenylacetylene (PA), which produced a peak at 1988 cm–1. Detection of Hg2+ occurred when the alkynyl group of the PA coordinated to Hg2+, allowing the alkynyl to separate from the silver surface. The new bond formation lowered the peak at 1988 cm–1 and produced a new peak at 2146 cm–1. This method was successfully used to detect and quantify Hg2+ in lake water samples several orders of magnitude below the USEPA limits for permissible concentrations in drinking water. 99 A different study proposed a novel SERS aptasensor with dual‐recycling amplification for Hg2+ detection, combining SERS with nucleic acid signal amplification to obtain a more sensitive detection. 86 Dual‐recycling amplification involves the immobilization of aptamers onto a substrate and bound with a complementary DNA strand, which will cleave off once the target molecule is introduced and is captured by the aptamer. SERS then detects the released cleavage probes. 100 In this recent work, this method was able to detect Hg2+ from 0.2 to 125 fM, with a LOD of 0.11 fM. The sensor was applied to tap and lake water samples effectively, and shows great potential for the detection of Hg2+ at extremely low concentrations in the environment. Potentially, by altering the aptamer sequences, other analytes may be specifically and sensitively detected by this approach. 86
3.2. Organic pollutants
3.2.1. Detection of pharmaceuticals
Pharmaceutical contamination, particularly in water systems, is a widespread issue which poses a threat not only to human health but also to overall environmental safety. Wastewater treatment facilities are the primary outlet for pharmaceutical contamination, as many pharmaceuticals cannot be successfully removed by current technologies, especially if the pharmaceutical is present as a metabolite or degradate and not as a pure compound. 101 In a recent review conducted by the US Geological Survey, the most common pharmaceutical contaminants listed were diabetes medications (with the most prevalent being the Type II medication metformin) and painkillers such as ibuprofen, tramadol, desvenlafaxine and venlafaxine. 101 Due to the ability of these pharmaceuticals to induce biological changes in non‐target organisms, it is of critical importance to be able to detect these compounds at wastewater treatment plants before they are dispersed into the environment. 101 A recent article by Burtsev et al. established a method for detecting and removing ibuprofen from water samples via microfluidic extraction and then capture by lipophilic functionalized gold multibranched nanoparticles (AuMs). 102 This method did not require sample pre‐treatment, was reliable and reproducible and had a low LOD (1.2 × 10–8 M). Another method detected and degraded pharmaceutical contaminants within one spot, wherein magnetic nanoparticles were hybridized with titanium dioxide and tungsten disulfide to produce Fe3O4@TiO2/WS2 nanoparticles which acted both as a SERS analytical detection agent and a photocatalytic degradation agent. 103 This method was able to achieve removal efficiencies between 68 and 100% for non‐steroidal anti‐inflammatory drugs, antibiotics and cosmetic dyes. Furthermore, the LOD for this method when applied to rhodamine B was as low as 10 nM (with a removal efficiency of 100%), making this a promising method for the detection and photocatalytic degradation of emerging contaminants such as anti‐inflammatories and other pharmaceutical compounds. 103 AgNPs functionalized with graphitic carbon nitride have been used for the detection of acetaminophen and adsorption of organic dyes in drinking water samples, and provided a low‐cost and reusable method for pharmaceutical contaminant detection, though more research is necessary to apply this method to the adsorption and degradation of pharmaceuticals. 104 Two other methods have recently been developed for the detection of pharmaceutical compounds, though neither possessed degradative properties. The first was developed by Sun et al., and utilized a hydrophilic‐hydrophobic silver nanowire paper‐based substrate for the detection of FZD in fish samples. 69 This method provided a lower LOD (10 mg/kg) than comparable methods with SERS, HPLC and ELISA, and had a comparable recovery rate of 79.5–84.6%. 69 The second non‐degradative method used AuNPs immobilized on GO as a substrate, combined with a MIP coated onto the substrate surface to obtain the final MIP@Au‐GO structure. 105 This design was used to detect two pharmaceuticals used in the treatment of hypoglycaemia, metformin hydrochloride (Met HCl) and phenformin hydrochloride (Phen HCl), with a LOD of 0.10 mg/mL. This substrate was reusable, had long‐term stability and provided high recovery rates when applied to real samples of hypoglycaemic dietary supplements (87.5–112.5% for Met HCl and 96.7–115% for Phen HCl). 105 While this method was designed for the detection of Met HCl and Phen HCl used illegally in non‐hypoglycaemic pharmaceuticals, it provides a promising design for pharmaceutical contaminant detection in environmental samples as well. For further discussion of the use of Raman spectroscopy and SERS for the detection of pharmaceuticals, we direct readers to some recent reviews from 2018 and 2017, respectively. 106 , 107
3.2.2. Detection of micro‐ and nanoplastics
The increased use and production of plastic products has resulted in a growing amount of plastic waste. Over time, plastic waste breaks down in the natural environment due to physical, biological and chemical factors. The degradation of macroplastics has led to the generation of MPLs, particles smaller than 5 mm and NPLs, particles smaller than 1 µm, in the environment. There are two forms of MPLs that have been discovered: primary and secondary. Primary MPLs are manufactured plastics, such as those used in cleaning products and personal care products. Secondary MPLs are plastic particles derived from the degradation of macroplastics. Some examples of secondary MPLs include those from road paint, tires and washing synthetic clothing. Primary and secondary MPLs break down into NPLs. MPLs and NPLs have been detected in aquatic environments, seafood, 108 sand, 109 bottled water 110 and table salt. 111 Conventional analytical techniques, such as Raman spectroscopy and FTIR, are incapable of detecting particles less than 1 and 10 µm, respectively, due to their low spatial resolution. 61 A commonly used technique for MPL detection is pyrolysis‐gas chromatography mass spectrometry. However, this technique is destructive and can only analyse a small sample size (e.g., 0.5 mg). 112 Due to the limitations of traditional analytical techniques, recent studies have used SERS as a new approach to detect MPLs and NPLs.
SERS is advantageous for plastic detection as it is highly sensitive and non‐destructive, allows for the detection of particles down to the nanometer size, and has been shown to be capable of detecting particles in complex samples. 113 AgNPs and AuNPs are the most common SERS substrates for MPL and NPL detection. 62 , 109 , 114 , 115 Research on MPLs and NPLs often focuses on detection of PS particles, as these are one of the most commonly found plastics in the natural environment. In a previous study, Zhou et al. analysed 1 µm and 50 nm PS particles in either ultra‐pure water or river water. 114 By mixing AgNPs and PS particles, and dropping the mixture on a silicon water for SERS analysis, this method was effective in detecting both 1 µm and 50 nm PS particles in either water matrix. However, the surface used for analysis was not uniform, leading to poor reproducibility and uneven hot spots. To fix this problem, it is important to prepare more uniform substrates. In addition to the uniformity of the surface, other important factors to consider in MPL/NPL analysis are the shape and size of the nanosubstrates, which have been discussed above in the section of ‘SERS nanosubstrates’. Optimizing factors, such as shape and size, can help provide a greater number of hot spots on a substrate. Some examples that are promising for MPL and NPL detection are AuNPs‐functionalized glass slides, 115 silver NWs on RC 62 and silver‐coated gold nanostars fixed in anodized aluminium oxide nanopores (Ag@AuNSs@AAO). 116 Caldwell et al. prepared AuNPs ‐unctionalised glass slides for detection of nano‐sized PS and polyethylene terephthalate (PET). Using the AuNPs‐functionalized glass slides, it was possible to detect PS particles with sizes of 131 and 62 nm, and PET particles with a size of 62 nm down to 10 µg/mL. 115 This method provides a simple and effective technique for the detection of nano‐sized plastics. Furthermore, this study was able to generate intensity maps from SERS measurements, and such data are imperative for future research as it provides information regarding NPL distribution. As was discussed earlier in this review, using bi‐metallic particles can help increase the efficiency of the SERS active substrate. Lê et al. used silver‐coated gold nanostars placed in anodized aluminium oxide (Ag@AuNSs@AAO) to detect MPLs in the sub‐micrometre size range. 116 A total of 0.4 µm PS particles were tested in varying concentrations in tap, river and sea water. Even in the presence of complex environmental samples, they were able to detect the PS particles using the Ag@AuNSs@AAO and reported a LOD of 0.05 mg/g. 116 In comparison to the study done by Caldwell et al. using gold‐functionalized glass, the bi‐metallic substrate used by Lê et al., was able to detect smaller particles in more complex environmental samples. Furthermore, in a study by Jeon et al., silver NWs or AuNRs were fixed to RC, which was selected for its 3D structure and high flexibility. 62 PS nanoparticles of 84, 444 and 630 nm were synthesized and placed on the prepared AuNRs/RC and AgNWs/RC substrates. The plastic particles were then analysed using a Raman microscope and they found that the AgNWs/RC film was able to detect PS particles at a concentration of 0.1 mg/mL, while the AuNRs/RC film was able to detect these particles down to 0.5 mg/mL. Therefore, they concluded that the AgNWs/RC film is a better SERS substrate than the AuNRs/RC film. 62 While SERS has been shown to be capable of detecting micro‐, sub‐micro‐ and nano‐sized plastic particle standards in proof‐of‐concept experiments, there is little research on its ability to detect a variety of plastic particles of different physical and chemical properties found in the natural environment. Further research is needed to conclusively determine if SERS can be used to detect MPL and NPL particles in complex real‐world environmental samples. A comprehensive review involving recent advancements for detection of MPLs and NPLs via SERS was published earlier this year, and we direct readers searching for more information on this topic to this reference. 117
3.2.3. Detection of synthetic dyes
Synthetic dyes are coloured products chemically derived from petrochemicals. Their widespread use in textiles, paints and printing poses health and environmental risks. 118 In the natural environment, even at low concentrations, synthetic dyes have lasting negative impacts. When present in water, they obstruct the passage of sunlight, inhibiting aquatic plant growth by preventing photosynthesis, increasing oxygen demand. They are reported to be toxic, carcinogenic, mutagenic, bio‐accumulative and decompose at slow rates. 119 In this section, we will discuss recent developments in SERS detection of commonly seen synthetic dyes in environmental settings.
SERS has high potential as a trace analysis technique for the detection of synthetic dyes. Sabathi et al. developed tannin‐furanic foams as SERS substrates. 120 Three different foam substrates were created: standard tannin foam, glyoxal tannin foam and TWEEN pluronic tannin foam. A magnetron sputtering system was used to deposit a 40‐nm thick layer of silver atop the substrates. The SERS performances of these substrates were tested with MG. On the standard tannin‐furanic foam, lower intensity signals were produced. However, intensity was 23.36 times higher and the RSD decreased by 36% on TWEEN pluronic tannin foam relative to standard tannin foam. On glyoxal tannin foam, intensity was 28 times higher, but this was accompanied by a 21% RSD increase. The low RSD and high enhancement for MG on TWEEN pluronic tannin foam were attributed to homogeneity of the hot spot across the substrate surface and the fine pore structure of said substrate. Similar dye tracing work was done by Liu et al., wherein a superhydrophobic AgNPs/glass microfiber filter substrate was developed using magnetron sputtering, annealing and a gas bath. 71 This substrate allowed for the detection of liquid phase analytes with wettability control. The hydrophobic component of the substrate was deemed useful for evenly distributing analyte molecules and making sure they were located in the SERS hot spots, therefore, achieving a more effective enhancement. Excellent Raman enhancement was attained for MG and CV, with EFs of 1.09 × 106 and 1.03 × 106, respectively. Another recent work focused on the detection of the dye acid orange II (OII) in water bodies using a SERS sensor based on PDA altered MOFs coupled with MIP technology. 121 In that work, the MOF UIO‐66(NH2) was synthesized using a solvothermal method, then wrapped in Ag‐loaded PDA to form Ag@UIO‐66(NH2)/PDA (AUP) as the SERS substrate. The AUP‐MIP imprinting material was then synthesized via precipitation polymerization, and allowed for more sensitive detection of OII than the unmodified AUP substrate. This MOFs‐based substrate allowed for a stronger adsorption capacity because of its porous structure and large surface area. At concentrations ranging from 10–8 to 10–10 M, OII exhibited an observable peak at 1596 cm–1. Furthermore, the AUP‐MIP substrate is recyclable through simple rinsing, making it particularly suited to environmental detection.
Chen et al. developed a method to create a multifunctional platform capable of monitoring synthetic dyes using a cellulose nanocrystal (CNC) Ag nanostructure. 122 The CNC‐regulated Ag nanostructure functioned as a platform to detect organic dyes in wastewater. The CNC, acting as both a reducing agent and a stabilizer, reduced silver nitrate to AgNPs and facilitated the formation of a flat nanostructure. The abundance of hydroxyl groups on the surface of CNC allowed AgNPs to form clusters of AgNPs@CNC nanohybrids. These clusters acted as SERS active sites, providing a greater enhancement than single dispersed AgNPs. CR, the analyte of interest, was injected into the sensing platform at varying concentrations. After SERS enhancement with AgNPs and CNC, CR peaks were clearly identifiable in all tested concentrations, with intensity increasing with higher concentration. The detection limit for this analyte was 10–15 M based on the characteristic peak at 1598 cm−1.
Other recent SERS applications in dye detection did not focus on real environmental samples, but rather used them as indicator molecules to evaluate the newly developed methods. Yang et al. reported an approach involving the fabrication of porous octahedral Cu2O onto Cu MOFs to create a novel SERS substrate. 123 SERS sensitivity was notably high for the detection of MB with this newly developed substrate, given that concentrations as low as 5 × 10‐9 M were detected. Vannucci et al. focused on the detection of Acid red 26 (AR26) and Acid red 18 (AR18) with Ag nanostars under varying pH levels. 124 AR26 and AR18 typically undergo tautomerism, meaning that one tautomer is present in greater amounts than the other under different conditions (e.g., pH). Based on the rapid decrease of SERS enhancement as pH increased, it was concluded that only the keto (KH) tautomers of the dyes were detectable with SERS. Raman enhancement was greatest at a pH range of 2–3, allowing for the enhanced detection of AR26 and AR18 at 10–5 M. Additionally, Xu et al. fabricated Ag/CuO NWs/pyramidal PDMS as a plasmon coupling SERS substrate. 51 To make this substrate, the PDMS structure was produced with an elastomer molding method involving spin coating and heating. A Cu film was then deposited onto the PDMS structure and submerged in anti‐formin solution to form NWs. For SERS enhancement, Ag was applied to the CuO NWs/pyramidal PDMS via thermal evaporation. The Ag/CuO NWs/pyramidal PDMS structure was able to detect CV and CR in a range of concentrations: 10–5 to 10–8 M and 10–4 to 10–7 M, respectively. Although these above studies have demonstrated promising laboratory results using newly designed SERS nanosubstrates, how they will perform in actual environmental samples is unknown.
3.2.4. Detection of pesticides
It is vital to monitor pesticides on agricultural products to prevent overuse and to protect human health and the environment. There have been a number of recent developments and improvements in the sensitive and selective SERS detection of environmental pesticides. While it is not possible to discuss all of the published articles using SERS to analyse pesticides from 2021, we have provided a table to organize recent publications (Table 2). An additional review, published in 2021, discusses the detection of organochlorine pesticides via SERS including a tabulation of various SERS strategies for the detection of several analytes. 125 The major recent advancements for SERS‐based detection of fungicides, herbicides and insecticides are discussed below.
Fungicides
Fungicides are a class of pesticides that control fungal growth. These compounds are often used in agricultural settings to protect crops from fungal diseases that may damage them and cause them to be unfit for consumption. While the toxicity of most fungicides is generally low, prolonged exposure to these compounds may lead to adverse health effects such as neurological disruptions. 126 Several recent studies have utilized SERS to detect various fungicides, including tetramethylthiuram disulfide (THR), 39 , 53 , 61 , 70 , 79 , 127 , 128 , 129 , 130 , 131 , 132 , 133 , 134 , 135 , 136 , 137 , 138 , 139 , 140 , 141 , 142 , 143 , 144 , 145 , 146 , 147 , 148 , 149 , 150 , 151 , 152 , 153 , 154 , 155 , 156 , 157 , 158 , 159 , 160 , 161 carbendazim, 162 , 163 , 164 , 165 chlorothalonil, 166 , 167 MG, 168 , 169 , 170 , 171 , 172 , 173 TBZ (a fungicide and parasiticide) 20 , 79 , 153 , 155 , 173 , 174 , 175 , 176 , 177 , 178 and ferbam. 20 , 178 , 179 Among them, THR is the most studied due to its wide agricultural applications and strong binding with SERS nanosubstrates through the thiol group. Here, we will use THR as an example to show the recent advancements in fungicide detection.
THR is a widely used dithiocarbamate fungicide that is applied to various agricultural crops. While THR is able to protect fruits and vegetables from fungal diseases that may destroy the plants, it also causes skin and mucosal disease in humans. 140 , 180 , 181 Numerous recent studies focus on the detection of THR using SERS methods. 39 , 53 , 61 , 70 , 79 , 127 , 128 , 129 , 130 , 131 , 132 , 133 , 134 , 135 , 136 , 137 , 138 , 139 , 140 , 141 , 142 , 143 , 144 , 145 , 146 , 147 , 148 , 149 , 150 , 151 , 152 , 153 , 154 , 155 , 156 , 157 , 158 , 159 , 160 , 161 While we cannot cover each of these references in detail, a few are expanded upon below. Gao et al. have developed a SERS sensor based on vertically aligned AgNPs deposited on tungsten oxide (WO3‐x) nanoflakes that can detect THR down to 1.2 pM. When detecting THR directly on apple peels, spiked THR could be detected down to 2.4 µg/L. 140 The Raman probe, R6G, was used to evaluate the Raman signals from the Ag/WO3‐x nanoflake substrate. SERS mapping revealed a uniform signal distribution across the substrate, and the LOD for R6G was found to be 2.7 × 10–13 M. There is a good linear correlation between Raman intensity at 1365 cm–1 and R6G concentration. The study also showed a strong linear correlation between Raman intensity and THR concentration, with a R2 value of 0.996. 140 Another recent study detected THR through the use of a class of two‐dimensional layered materials, MXenes, which were selected because of their high flexibility and ease of chemical modification. Titanium carbide MXene was used with AuNRs in various ratios to create Raman enhancement at the tips of the nanorods for higher sensitivity. 139 4‐aminothiophenol and 1,2‐bis(4‐pyridyl)ethylene were used to evaluate the ratios of MXene/AuNRs. When the optimal ratio was used, the detection of THR had a LOD of 10–8 M, which is much lower than commercial SERS chips (10–5 M) and Au nanorod arrays (10–6 M). These results suggest that the developed MXene/AuNRs substrate has the potential for sensitive and quantitative detection of pesticides with SERS. 139
Herbicides
Herbicides are a class of pesticides that aim to destroy unwanted vegetation. Several recent studies have used SERS to detect various herbicides present in the environment, including atrazine and its metabolites, 182 , 183 , 184 , 185 , 186 paraquat (PQ), 158 , 187 , 188 , 189 oxyfluorfen, 167 benthiocarb, 157 2,4,5‐trichlorophenoxyacetic acid (2,4,5‐T), 190 prometryn, 184 diquat and difenzoquat, 189 2,4‐Dichlorophenoxyacetic acid (2,4‐D) 191 , 192 and parathion. 193 Daoudi et al. created a novel SERS substrate for the detection of atrazine using a hierarchical silver nanoprism/GO/silicon nanowire array. 183 This substrate combined the benefits of silicon NWs (SiNWs) (cost effectiveness, ultra‐sensitivity, uniformity of the substrate and semi‐conductor properties) with the SERS enhancement from metal nanoparticles to create hot spots for better Raman signals. The SiNWs were produced from silicon wafers using a previously studied chemical etching method. Following this, synthesized GO was spin coated onto the SiNWs. Ag nanoprisms were drop‐cast onto the SiNW substrate and allowed to dry several times to produce a significant Ag nanoprism surface. This surface was characterized using field‐emission scanning electron microscopy (FESEM), AFM and TEM. Common organic pollutants R6G and MB were detected with this substrate to study uniformity and reproducibility. Overall, this substrate was able to provide uniform SERS enhancements. This substrate had a LOD down to micromolar concentrations. 183 Another recent study by Yao et al. used natural lotus leaves as hydrophobic surfaces to detect PQ. 188 AgNPs synthesized through a citrate‐reduction method were mixed with PQ solutions and then applied to the lotus leaves. During evaporation, the AgNPs arranged themselves into arrays on a small area of the lotus leaves, instead of leaving a coffee ring of AgNP aggregates. The LOD for PQ was found to be 1.2 µg/L, with a good linear correlation between PQ concentration and Raman intensity, indicating that this method can be used for quantitative analysis. In evaluating the uniformity and reproducibility of the substrate, the RSD of peak intensity (at 1642 cm–1) for 50 SERS spectra was found to be 11.6%. A ‘traditional SERS sensor’ using AgNPs aggregated with salt was used for comparison, and was found to have an RSD value of 26.9%. The lotus leaf‐based SERS substrates were also found to have Raman signals about 20 times higher than the traditional SERS sensor. The use of lotus leaves as a unique and environmental friendly hydrophobic SERS substrate is a good technique to control the uniformity of AgNPs for more reproducible detection of PQ. The storage of the lotus leaves and the water content within the leaves may have impacted the hydrophobic properties. However, the authors suggested that under appropriate storage and humidity, long‐time storage, up to 30 days, did not impact the hydrophobic properties of the leaves. The hydrophobicity was evaluated on tender, mature and old lotus leaves by measuring the contact angle for droplets on the leaves (150.1 ± 1.0°, 150.0 ± 1.3° and 149.5 ± 0.8°, respectively). It was concluded that all growth stages were ‘superhydrophobic’. 188 This unique SERS substrate has the potential to be used as an environmental friendly material for sensitive and reliable detection of herbicides or other environmental pollutants.
Insecticides
Insecticides are used to kill or repel insects and are often used in agricultural settings to protect crops from destruction. SERS has been used to detect several types of insecticides in the past year such as chlorpyrifos, 159 , 194 , 195 , 196 , 197 methyl parathion, 151 , 198 , 199 , 200 endosulfan, 155 , 201 malathion, 155 phosmet, 156 imidacloprid, 159 , 167 , 191 , 202 , 203 , 204 , 205 , 206 , 207 phorate, 157 carbaryl, 158 , 208 fipronil, 158 fenitrothion, 193 acetamiprid, 165 , 192 , 205 , 206 , 209 chlorfenapyr, 210 dichlorvos, 211 , 212 fenobucarb, 213 thiodicarb, 208 carbendazim, 165 profenofos, 165 methomyl, 192 pymetrozine, 160 , 214 clothianidin and thiamethoxam. 59 , 206 Recently, Sulaiman et al. created a technique for the sensitive detection of the insecticide chlorpyrifos using fabricated bi‐metallic nanoparticle alloys in conjunction with SERS. 194 A pulsed laser‐induced etching was used to create Si nanopillars as the base for the substrate, which has a high density of nanogaps that induce hot spots on the surface. These Si nanopillars were decorated with synthesized bimetallic alloys of Au‐PdNPs and AgPdNPs. Utilization of transition metals, such as Pd, has been previously suggested as a method of enhancing the SERS signal for sensitive detection of analytes. 215 , 216 The SERS sensors were dipped into solutions of chlorpyrifos at different concentrations and dried before SERS detection. The LOD was 0.066 mg/L for Au‐PdNPs and 0.079 mg/L for Ag‐PdNPs, indicating sensitive detection of the target insecticide. The creation of hot spots from the Si nanopillars used with the bi‐metallic alloys served as a promising technique for trace detection of chlorpyrifos. 194 Al‐Syadi et al. also utilized SERS for the detection of trace concentrations of the neonicotinoid insecticide imidacloprid which has had devastating impacts on honeybees. In this work, a cost‐effective SERS substrate was created from porous silicon (PSi)‐plated palladium nanoparticles (PdNPs) through electrochemical anodization and immersion plating. 202 In this recent study, the authors indicated the importance of the porosity of the silicon substrate by comparing low concentrations of imidacloprid (10–6 M) using Si‐Pd NPs (non‐porous Si substrate) with the porous version (PSi‐Pd NPs), and found more intense Raman peaks for the PSi‐Pd NP substrate (with a slight shift in peak position between the two). The PSi‐Pd NP substrate had a LOD of 10–9 M with an EF of 1.2 × 105, which was likely due to the creation of more hot spots on the porous substrate surface. The creation of this substrate for the sensitive detection of imidacloprid may provide a scaffold for further advancements involving porous substrates.
Multiple pesticides
While many studies focus on the detection of only one key pesticide of interest, it is advantageous to develop methods that are able to identify and quantify (sometimes simultaneously) several different pesticides using the same SERS substrate. Several recent studies focus on the detection of multiple pesticides. For example, Asgari et al. integrated the technology of filter‐based microfluidic devices with SERS detection using Au@Ag nanoparticles. 155 Filtration (using a PTFE filter membrane) on the SERS chip allows interfering particulate matter to be removed from the sample to enable a more sensitive detection of target analytes. In this study, two fungicides (TBZ and THR) and two insecticides (endosulfan and malathion) were each detected within a mixture in a strawberry extract (LOD = 55, 44, 88 and 54 µg/kg, respectively). The use of a microfluidic SERS chip requires minimal sample preparation, compared with conventional methods of preparing and purifying samples to remove unwanted particulate matter. The authors presented an optimized filter‐based microfluidic SERS sensor to detect multiple pesticides, establishing the first publication of a filter‐based SERS microchip for detection and quantification of multiple analytes within complex food matrices. 155 Another work reported simultaneous detection of three pesticides, chlorothalonil (fungicide), imidacloprid (insecticide) and oxyfluorfen (herbicide), through the use of a SERS‐based lateral flow assay (SERS‐LFA) test strip and competitive immune binding. 167 In that work, silver nanospheres and silver nanoprisms were used to detect four different pesticides: atrazine (herbicide), simazin (herbicide), irgarol (antimicrobial pesticide) and diuron (herbicide). Three of the pesticides, atrazine, simazin and irgarol, were all able to be detected down to millimolar concentrations. 186 A flexible cellulose nanofiber paper‐based substrate was developed by Li et al. to simultaneously detect four organochlorine pesticides in water samples including 4,4′‐DDT, a‐endosulfan, tetradifon and chlordane. 201 Lu et al. utilized a DNA backbone structure with Ag@Au nanoparticles for the simultaneous detection of profenofos (insecticide), acetamiprid (insecticide) and carbendazim (fungicide). This was achieved using embedded aptamers within the DNA backbone for the three pesticides, and Raman signalling molecules modified onto the Ag@Au NPs. The DNA skeleton deformed when the aptamers recognized any of the three pesticides, and the Ag@Ag NPs moved closer to one another, creating SERS hot spots between them to increase the Raman signals. This technique provided low LODs for the three pesticides, and allowing for the ultra‐sensitive, simultaneous detection of multiple analytes. 165
The simultaneous detection of multiple pesticides is extremely useful as it is common for these compounds to be used in conjunction with one another in agricultural settings. Bioaccumulation of pesticides in ecosystems allows different pesticides to be found in similar locations, so it is important to be able to identify and quantify co‐existing contaminants using one approach. These recent advancements in the simultaneous detection of multiple pesticides are extremely promising for facilitating the development of versatile SERS methods in environmental monitoring.
3.3. Biological pollutants
3.3.1. Detection of viruses
Due to the outbreak and spread of the novel coronavirus, SARS‐CoV‐2, research concerning ultra‐trace SERS detection of viruses is rapidly gaining attention, especially as new methods to detect viruses rapidly, on‐site and with low costs are in high demand. Despite this increase in SERS‐based viral detection methods, many current techniques focus on detection in clinical samples such as blood, faecal matter and live cells. While such detection methods are vital to prevent the spread of disease, they do not provide much information on how viruses may persist and transmit in non‐human biological systems (such as crops or livestock) or non‐biological environments (such as synthetic surfaces). A few recent articles have focused on SARS‐CoV‐2 detection in spiked lab water samples 217 and environmental waters (collected from rivers, hospitals and pipe networks), 218 as well as hepatitis A detection in spiked Milli‐Q water, 219 but to our knowledge there have been no publications in 2020‐2021 which work to detect viruses in food and agricultural samples, or in non‐aqueous environmental samples. Virus detection in food systems is especially important, as food can be easily contaminated at processing facilities, by water and soil pollution, and by contact with infected livestock. 220 The use of magnetic nanoparticles (MagSERS) in conjunction with methods, such as lateral flow immunoassays and digital microfluidics, has been effective in detecting common food‐borne viruses such as influenza A and avian influenza in years prior to 2020, and thus, warrants continued investigation in the future. 220 Magnetic nanoparticles have high water dispersion, uniformity and biocompatibility, making them ideal for the detection and separation of biological molecules. For virus and bacterium detection in particular, immunomagnetic separation (IMS) has gained increasing attention, as the immunological aspect of the assay (usually comprised of an antibody or other molecular recognition agent [MRA] 221 ) confers high specificity, while the magnetic properties of the nanoparticles allow for easy separation of the analyte from the sample matrix, which is especially important for environmental samples. This high specificity between the MRA and the target analyte also allows for a low LOD, often in the picomolar, femtomolar or attomolar ranges for viruses. 220 These properties, combined with the ability to simultaneously detect and remove analytes for subsequent analysis make magSERS a highly promising method for monitoring viruses, such as SARS‐CoV‐2, in the environment.
Another method for the detection of viruses which has been gaining increasing attention is clustered regularly interspaced short palindromic repeats (CRISPR)/Cas‐mediated SERS. CRISPR/Cas technology, initially developed and popularized as a gene editing tool, has become increasingly common in biosensing due to its high sensitivity down to the level of single nucleic acids residues, and programmability to recognize certain nucleic acids. 222 CRISPR/Cas systems can be divided into two classes: Class 1 and Class 2. Class 1 systems are characterized by multi‐protein effector complexes, whereas Class 2 systems utilize a single effector protein, making Class 2 systems simpler and preferable for bio‐sensing applications. Within Class 2, there are three main types of Cas‐guide proteins: Cas9, Cas12a and Cas13a. The guide protein is selected based on the target analyte type. Cas9 can target both DNA and RNA sequences, while Cas12a targets only DNA, and Cas13a targets only RNA. The CRISPR/Cas system is typically coupled with a nucleic acid amplification method, such as recombinase polymerase amplification, loop‐mediated isothermal amplification, or polymerase chain reaction, which greatly increases the detection sensitivity. Furthermore, there are a wide variety of readouts which can be generated, ranging from fluorometric or colorimetric signals detected on a microplate reader to lateral flow strips imaged on a mobile app. 222 In addition, SERS signals from nanoprobes can be monitored by a portable Raman spectrometer, yielding highly sensitive and quantitative detection. This versatility in readout by a portable Raman technology makes CRISPR/Cas systems especially promising for virus detection in the field, where heavy‐duty equipment is not practical. Previously, CRISPR/Cas12a‐assisted SERS platforms have been used to detect human immunodeficiency virus (HIV) in PBS buffer and serum, as well as hepatitis and human papillomavirus (HPV) in cleavage buffer at incredibly low levels (LOD = 0.3 fM for HIV in PBS buffer, 223 LOD = 1 aM for hepatitis and HPV 224 ). CRISPR/Cas12a has also been used in food authenticity screening, and this avenue in particular is especially promising for the detection of pathogens in environmental samples. 225 Future studies may seek to merge these two avenues of CRISPR/Cas research to directly detect viral samples in environmental sources.
Currently, the main drawback of CRISPR/Cas biosensing is that samples require pre‐treatment or nucleic acid extraction, which may be difficult for on‐site analysis. Discovering ways to overcome this pre‐treatment step is an avenue for future work. 222 The amplification step, in particular, often requires equipment that is not practical for field work. Recently, several articles have achieved SERS detection of a variety of samples using amplification‐free CRISPR, demonstrating that CRISPR/Cas is a promising technology for SERS detection in the field. Amplification‐free CRISPR has been used to detect HIV‐1 dsDNA (LOD = 0.3 fM), 223 hepatitis B virus, HPV‐16, and HPV‐18 DNA (LOD = 1 aM), 224 SARS‐CoV‐2 RNA (LOD = 1 fM), 226 SARS‐CoV‐2 N‐gene (LOD = 5 fM) 227 and SARS‐CoV‐2 RNA in clinical samples (LOD = 100 copies/µL). 228 There are multiple ways to bypass the amplification step while still attaining low levels of detection, such as coupling CRISPR with a lateral flow assay, 223 performing detection on a microchamber platform 227 and using multiple different guide RNAs. 227 , 228 Optimizing both substrate and nanoparticle design can confer higher SERS enhancement, as shown by Choi et al., wherein Raman probe‐functionalized AuNPs (RAuNPs) were immobilized by ssDNA on a GO/triangle Au nanoflower (TANF) array. 224 In this design, both the RAuNPs and GO/TANF array individually increased SERS enhancement, so when combined they allowed for sufficient enhancement so that the CRISPR amplification step was not necessary. Future work may, thus, consider how to achieve amplification‐free CRISPR detection by optimizing the nanosubstrate design, designing a microchamber platform, or using multiple guide RNAs, especially for virus detection in the environment.
3.3.2. Detection of bacteria
Pathogenic bacteria, such as Escherichia coli, Staphylococcus aureus and Listeria monocytogenes, pose a significant risk to human and environmental health. Pathogenic bacteria can be found in a wide array of environments, from contaminated drinking water to food produce, as well as agricultural livestock and even medical facilities. As bacterial resistance to antibiotics increases, it is becoming even more important to detect trace amounts of bacteria in environmental samples. SERS is a very promising method for accomplishing this. Here, we will discuss some of the challenges of using SERS to detect bacterial samples in environmental samples as well as recent developments to address these challenges.
One of the main challenges in applying SERS to the environmental detection of bacteria is to identify distinct bacteria in the same sample. 229 Because the cell walls of different bacterial species are made up of the same basic biological components, SERS methods typically cannot distinguish between multiple bacterial species within the same sample unless they use a SERS tag to mark species‐specific bacterial features (also called labelled detection). 229 The most apparent solution to this problem is to use a SERS tag such as antibodies or aptamers. Previously, a study by Yang et al. utilized the established signal amplification method of a DNA walker 230 to detect Salmonella typhimurium in aqueous environments. 229 This labelled method had a LOD lower than 10 CFU/mL, making it a promising method for pathogenic bacteria detection. In another study, L. monocytogenes was detected in milk samples with a LOD of 12 CFU/mL. 231 Having such a low LOD in a milk sample is impressive, as the matrix of milk can interfere with Raman signals much more than water. For this detection method, modified gold‐coated magnetic nanoparticles (Au@Fe3O4) were used as an IMS agent, and modified AuNRs were used as a SERS label. Au@Fe3O4 particles were modified with 11‐mercaptoundecanoic acid (11‐MUA) and avidin (a glycoprotein), allowing for conjugation of the L. monocytogenes antibody and removal of L. monocytogenes bacteria from the solution by IMS. For SERS labelling, AuNRs were modified with the reporter molecule 5,5′‐dithiobis (2‐nitrobenzoic acid) (DTNB or Ellman's reagent) and avidin, and then conjugated with L. monocytogenes antibody. This method was highly effective for the detection of pathogenic bacteria in drink and food samples, and was the first assay to use IMS in conjugation with SERS for the detection of L. monocytogenes. 231 Labelled SERS assays have also been used to develop a rapid and highly sensitive lateral flow immunoassay for the detection of E. coli. 232 As shown in Figure 3E, Shi et al. developed gold‐shell silica‐core nanospheres (SiO2/Au NPs) modified with a DTNB reporter and monoclonal detection antibody. When these SiO2/Au NPs were dropped onto the test line of the nitrocellulose conjugate pad, E. coli molecules became sandwiched between the SERS tags and the capture antibodies located on the test line. The presence of E. coli was then able to be measured quantitatively by obtaining the Raman intensity of the test line. This sandwich complex was not formed on the control line of the conjugate pad, meaning that any E. coli present would not undergo SERS enhancement like those on the test line. This study also found that the use of gold‐coated silica nanospheres rather than colloidal gold improved the stability of the nanoparticles significantly and increased sensitivity 200‐fold, resulting in a LOD as low as 50 cells/mL in PBS solution and 100 cells/mL in tap water, milk, human urine, lettuce extract and beef. 232 Labelled SERS detection is clearly advantageous in liquid samples, but it has also been successfully applied to the detection of pathogenic bacteria in food samples, such as egg powder and black pepper powder, both of which are low‐moisture foods (LMFs). LMFs pose a particular problem for the spread of bacterial pathogens, as such foods are typically considered low risk for bacterial growth, but bacteria which survive in this low moisture state can become more pathogenic upon entrance into the digestive system where moisture is abundant. To detect E. coli in LMFs, Pan et al. used the Dual Immunological Raman‐Enabled Crosschecking Test (DIRECT). 221 DIRECT relies on metallic nanostructures (nanoprobes) functionalized with 4‐ATP as a SERS tag and a MRA, such as an antibody, which allows the metallic particle to bind to a specific feature of the bacterial surface (such as an epitope). When enough of the Raman molecular probes (RMPs) bind to the cell surface, enhancement occurs due to the proximity of nanoparticles to each other, producing a SERS signal of the cell wall or extracellular matrix. Because the signals of the probe and the epitope (or other bacterial surface feature) are being simultaneously enhanced, this method results in dual signals, which are used to confirm the binding of the probe and the bacterium. Furthermore, because enhancement can only occur if the MRA attached to the probe successfully recognizes the target cell feature, no washing or separation step is needed to distinguish between bound and unbound probes. Since this method is highly specific while not requiring a washing step, it allows for the rapid detection of pathogenic bacteria in LMFs with a LOD as low as 102 CFU/g. Furthermore, the use of principal component analysis (PCA) alongside this method allows for the elimination of spectral noise caused by the LMFs, which greatly reduces the complexity of the spectral data and enables the elucidation of bacterial contamination status in environmental samples. 221
The use of labelled SERS methods in pathogen detection is clearly a rapidly growing area of research, but there have also been recent developments in label‐free SERS detection of bacterial pathogens. One method designed by Tadesse et al. detected Gram negative (E. coli and Serratia marcescens) and Gram positive (S. aureus and Staphylococcus epidermidis) bacteria in a liquid environment. 233 AuNR and bacterial concentrations were varied to optimize large‐area enhancement, and this enhancement was proportional to the surface charge density of the bacterium. The surface charge density was unique enough for each bacterial species that the species could be identified simply from the intensity of the SERS signal produced by the nanorods. A second approach to label‐free detection of bacteria was through the use of positively charged nanoparticles combined with multivariate analysis. 234 With this method, silver/gold bi‐metallic nanoparticles (Ag/Au bmNPs) attracted negatively charged bacteria, enhancing their Raman signals. Multivariate analysis methods, such as PCA and canonical discriminant analysis, were used to determine the bacterial species. 234 PCA has also been used to discriminate not only between bacterial species, but also between strains of the same species, making it a powerful tool to analyse different bacteria in complex environmental samples. 235 For a more in‐depth discussion of using SERS for bacteria detection, we direct readers to the following 2021 review by Ahmad et al. 236
3.3.3. Detection of mycotoxins
Mycotoxins are a class of metabolites produced by fungi, such as those in the Aspergillus, Fusarium and Penicillium families. These toxins can grow readily in favourable environmental conditions, are resistant to degradation by high temperatures and may possess additional dangerous properties such as mutagenicity, teratogenicity and carcinogenicity. For these reasons, being able to detect the presence of mycotoxins in food and agricultural products, such as wheat, corn, peanuts and wine, is of critical importance. 237 Sub‐classes of mycotoxins include aflatoxins (AFs), fumonisins and ochratoxins. 238 Previously, challenges to detecting mycotoxins included high costs, training barriers, complex processing of analytes, destruction of the analyte and poor reproducibility resulting from susceptibility to environmental conditions. 239 Furthermore, methods, such as ELISA, TLC, HPLC and LC‐MS/MS, have high accuracy and reproducibility but may not be sustainable due to the amount of reagents needed for detection. 237 Some methods, such as fluorescence or colorimetric detection, may suffer from background interference and false negatives. 237 For these reasons, using SERS to detect mycotoxins has become increasingly common as this method, if used properly, allows for high enhancement and reproducibility in a sustainable and non‐destructive manner. In this review, we will examine recent methods to detect mycotoxins in environmental samples through the use of various substrates designs, such as nanoflowers, nanospheres, nanocubes and nanorods; and through the combination of SERS with aptasensors, which is a rapidly growing research area for detecting biological pollutants.
There are a multitude of SERS substrate designs which have been used in the detection of mycotoxins. Previously, Tegegne et al. used silver nanocubes coated with PDA for the detection of deoxynivalenol, a mycotoxin commonly found in pig feed. 240 This technique, due to substrate uniformity and favourable substrate interactions, was able to reach an EF of 1.82 × 107 and a LOD in the femtomolar range. The PDA‐coated Ag NCs were also more stable than non‐coated Ag NCs. In addition to the higher stability allowed by the Ag NC@PDA structure, the use of filter paper resulted in a higher EF than with a silicon wafer substrate, likely due to the better packing of the Ag NCs observed on the filter paper. 240 Moreover, this method was highly reproducible within the same sample and across different samples, making this method a promising platform for SERS detection of mycotoxins. Another highly reproducible method was developed by Hahm et al. using silver‐embedded silica nanoparticles for the detection of alternariol (AOH). 241 This method had a higher LOD in the nanomolar range, but the reproducibility was higher (lower RSD) than the Ag NC@PDA method. This higher reproducibility was likely due to the substrate structure itself, wherein AgNPs (∼17 nm) were uniformly distributed across the surface of thiolated silicon nanoparticles (∼146 nm). This uniformity within and across particles allowed for reliable and reproducible detection, as well as high enhancement and sensitivity due to the presence of hot spots between AgNPs on the SiO2 particle surface. 241 More recently, the work done by Guo et al. on SERS detection of AOH focused on substrate distribution rather than composition, using AuNRs (80 nm length) to enhance the coffee ring effect which is often observable with liquid analytes. 242 On the coffee ring, the concentration of the analyte was typically higher, leading to higher intensity SERS signals. In this case, both enhancement and sample uniformity were higher within the coffee ring region, making detection results stable and repeatable. 242 The LOD for this method was in the micromolar range, which is significantly higher than the methods discussed previously. To improve the sensitivity, combining the coffee ring effect with optimized substrate design should be explored in the future.
While the above methods are promising, some experiments may require specificity down to the level of a single molecule. Such methods, wherein a SERS probe recognizes and binds to a specific site of the target molecule, are especially helpful for environmental experiments where multiple classes of toxins could be present in the sample. Currently, the primary methods of SERS detection that allow for this level of specificity are ones that use aptamers, or fragments of single‐stranded genetic material which bind with a high affinity to target molecules. 239 This high affinity allows aptamers to act as an alternative to antibodies in many analytical methods, and depending on the experiment may be preferable to antibodies as their smaller size allows for lower steric hindrance and a lower change of a linkage error due to a nucleotide base pair mismatch. 243 Previously, aptamer‐based SERS platforms have been used for the detection of mycotoxins such as ochratoxin A (OTA), zearalenone (ZEN), aflatoxin B1 (AFB1) and fumonisin B1 (FB1). 239 , 244 , 245 , 246 , 247 A study by Chen et al. used gold‐silver core‐shell nanoparticles modified with an aptamer and a Raman reporter molecule (probe 1) and AuNRs modified with cDNA (probe 2) to achieve the simultaneous SERS detection of OTA (LOD = 0.018 ng/mL) and ZEN (LOD = 0.054 ng/mL). 239 Probe 1 was functionalized with aptamers specific to either OTA or ZEN. The aptamer in probe 1 and cDNA in probe 2 could bind with each other to form a bridge, which generated strong Raman enhancement. When the target mycotoxin was present, it outcompeted probe 2 to bind with the corresponding aptamer in probe 1. The competition broke down the bridge and decreased SERS signals significantly. An aptamer‐based method for the detection of OTA in beer utilized upconversion nanoparticles (UCNPs) as a luminescence label and an organic dye as a quencher and Raman reporter on a gold nanourchin (GNU) substrate. UCNPs were modified with cDNA, while GNUs were modified with an aptasensor specific to OTA. When OTA was not present, the cDNA and aptamer were weakly bound together, forming a bridge between the UCNPs and GNUs and producing low upconversion luminescence (UCL) and SERS signals. With OTA present, the aptamers were preferably bound to OTA, releasing the cDNA‐UCNP complex to generate high UCL signals. Conformational alteration of the aptamer after OTA binding also decreased distances between the Raman reporter and GNU, resulting in higher SERS signals. This dual‐mode sensor was able to achieve a LOD of 3.2 pg/mL with UCL and 8.6 pg/mL with SERS. 244 For the detection of AFB1 in peanut oil samples, He et al. used Fe2O4@Au nanoflowers modified with SH‐cDNA as a capture probe, and Au@Ag nanospheres modified with SH‐Apt as a reporter probe (LOD = 0.40 pg/mL). The mechanism for this study is similar to that developed by Chen et al., in which mycotoxin binding caused a linear decrease in SERS intensity. For this method, when AFB1 was not present, the reporter probe bound tightly to the SH‐cDNA‐modified nanoflowers and generated strong SERS signals. When AFB1 was present, it preferably bound to the AFB1 aptamer attached to the reporter probe, releasing it from the capture probe and generating weaker SERS signals. This method was able to achieve a recovery rate of 96.6–115%. 245 AFB1 was detected at the femtomolar level (LOD = 5.07 fg/mL) by Li et al. through the use of an aptamer‐based DNA tweezer, which allowed for highly sensitive control of the distance between AgNPs, and thus, control over the generation of SERS hotspots. 247 The ends of each arm of the DNA tweezer were modified with a single AgNP, and an aptamer specific to AFB1 was bound between the arms in the open state. When AFB1 was not present, the arms were held farther apart, producing low SERS signals. When AFB1 was present, the aptamer preferably bound to it, allowing the arms of the DNA tweezer to move closer together which placed the AgNPs within close proximity to one another and generated higher SERS signals. This method was also successfully applied to the recovery of AFB1 from maize samples, with a recovery rate from 95.6 to 106.4%. Last, in the detection of FB1 on spiked corn samples, He et al. were able to achieve a recovery rate of 92–107% by using platinum‐coated AuNRs modified with cDNA, and aptamers modified with the fluorescent dye and Raman reporter Cy5.5. 246 When FB1 was not present, the aptamer and the cDNA bound, forming a cDNA‐AuNR/aptamer‐Cy5.5 complex which produced strong SERS signals and weak fluorescence signals. The strong SERS signals were generated due to the close proximity of the AuNRs and Cy5.5 reporter molecules, but fluorescence levels were low due to low fluorescence resonance energy transfer between Cy5.5 and the AuNR. When FB1 was present, the aptamer preferably bound to it, taking with it a Cy5.5 molecule and destabilizing the cDNA‐AuNR/aptamer‐Cy5.5 complex. This resulted in lower SERS signals due to the increase in distance between Cy5.5 and the AuNR, but higher fluorescence signals due to greater fluorescence resonance energy transfer energy transfer.
As is clear from the above studies, aptamer‐based methods vary widely, and can implement detection methods in tandem with SERS, such as fluorescence‐ and luminescence‐based methods, allowing for dual signals to differentiate between toxin presence and absence. Aptamer‐based methods specifically have shown great promise in detecting mycotoxins in food samples, with LOD spanning the nanomolar, picomolar and femtomolar range, depending on the toxin detected and the exact method used. 239 Many comparable methods that rely on the use of antibodies have LOD only in the nanomolar range, so aptamer‐based sensors are promising for ultra‐trace detection of mycotoxins in environmental settings. 246
4. SUMMARY AND OUTLOOK
The sensitive and selective detection of pollutants in our environment is important for environmental and ecological welfare as well as human health and safety. In this review, we highlight recent findings on SERS method development and applications in environmental monitoring. We have discussed multiple components of SERS nanosubstrates including hot spots, substrate composition, nanoparticle shape, size, arrangement and ISs. These properties should be optimized for each experiment to achieve the highest possible LOD and EF for a given nanosubstrate and analyte combination. Researchers may seek to optimize hotspot formation by altering nanoparticle size, shape, spatial arrangement or the amount of aggregation between nanoparticles. 10 , 28 , 29 Nanosubstrate composition can vary widely depending on the analyte being detected, but overall bi‐metallic nanoparticles are preferable due to their higher stability, homogeneity and enhancement. 20 , 34 , 35 , 36 Nanoparticle shape can also vary widely, and here we have highlighted a fraction of possible designs, including nanoporous bowls, 45 nanodisks, 46 nanocones 49 and NWs. 51 The spatial arrangement of nanoparticles is also important, and we have detailed how 3D nanoparticle arrays may be used to optimize enhancement 53 and achieve highly sensitive substrates for environmental detection. Aggregation, in particular, can be increased with the use of thiolated ligands, which have also been shown to increase nanoparticle stability by preventing oxidation. 20 , 34 , 35 , 36 The last main aspect of nanoparticle design which we have reviewed is the use of ISs, which are especially beneficial for heterogeneous substrates with non‐uniform hot spot distribution and intensity. 30 , 31 , 32 , 72
In discussing SERS nanosubstrates, we have also covered two types which are being used with increasing frequency: graphene‐based and flexible SERS nanosubstrates. Graphene‐based SERS substrates tend to have higher enhancement due to the formation of hot spots in gaps between nanoparticles and graphene structures. 43 Graphene substrates also confer fluorescence quenching abilities and have good biocompatibility, making them optimal for detecting biomolecules in environmental samples. Flexible SERS substrates are typically cellulose or paper‐based, and provide an environmental friendly option which is still effective due to its high sensitivity. 34 Nanocellulose, in particular, has gained attention for its thin structure and self‐assembling abilities. 65 , 66 , 67 Flexible substrates are promising for environmental detection due to their low cost, minimal preparation, high sensitivity, broad applicability and sustainability, making them an particularly interesting area which warrants further research.
Further, we focused on SERS applications in the detection of inorganic, organic and biological pollutants in various matrices, such as on food, crops and environmental waters. Tables 1 and 2 summarize recent publications covering SERS‐based detection of different analytes discussed in this review. In applying SERS for the detection of inorganic pollutants, we have focused on SERS detection of heavy metals, such as lead, which pose a significant environmental threat. We have highlighted a labelled method which utilizes graphene‐based SERS and enzyme‐linked reactions to both detect and quantify Pb2+, 42 as well as an aptamer‐based labelled method for Hg2+ detection. 86 For label‐free methods of detection, we have discussed an AgNP substrate 90 for the detection of Fe3+ and PA‐functionalized AgNPs 99 for the detection of Hg2+. Some of these methods also implemented environmental friendly design principles, such as reusability 42 and the use of mild reagents, 90 making them especially promising for environmental applications.
For organic pollutants, we have summarized SERS methods applied to the detection of pharmaceuticals, MPLs and NPLs, synthetic dyes and various classes of pesticides. Pharmaceutical contamination, particularly in waterways, is a growing environmental issue which requires fast and accurate detection methods to solve. We have summarized methods with the following features: no pre‐treatment requirement, 102 simultaneous detection and degradation of pollutants, 103 reusability 104 and pharmaceutical detection in real samples (such as environmental water, 104 fish 69 and supplemental medications 105 ). While each of these methods has their own limitations, they provide a promising starting point for future research, which may seek to integrate these beneficial properties to create more advanced approaches for pharmaceutical contaminant detection. For the detection of plastic contamination, we have discussed the capabilities of SERS for detecting particles smaller than 1 µm 62 , 114 , 115 , 116 and plastic particles in various types of water, such as tap, river and sea water. 114 , 116 While previous methods were able to detect PS nanoparticles, few articles analysed a second type of plastic. PET is of particular concern, as it is commonly used in water bottle and food packaging. However, a majority of the research on SERS and plastic detection focuses on PS. In the future, it is imperative to develop SERS methods to detect MPLs and NPLs of a variety of compositions, such as PET, in environmental samples. Regarding the detection of synthetic dyes, SERS has been proven to be a useful trace analysis technique. Synthetic dyes commonly contaminate water bodies like wastewater, groundwater and rivers as a result of industrial processes. Recent work has allowed for the sensitive detection of commonly used dyes in these locations. These advancements have been made possible with the development of new SERS substrates that promote signal enhancement. Dyes that have received recent attention because of enhanced SERS signals and low limits of detection include MG, 71 , 120 CV, 51 , 71 OII, 121 CR, 51 , 122 MB, 123 AR26 and AR18. 124 Some of the newly developed SERS substrates involve the use of tannin‐furanic foams, 120 superhydrophobic NP/glass microfiber filters, 71 MOFs, 121 , 123 CNC‐regulated nanostructures 122 and NWs/pyramidal PDMS structures. 51 Last, in discussing SERS detection of pesticides, we have covered fungicides, herbicides and insecticides, and discussed methods that allow for the detection of multiple pesticides on the same substrate. Within fungicide detection, we evaluated promising methods for THR detection, such as the use of Ag/WO3‐x substrates 140 and MXene/AuNR substrates, 139 both of which allow for highly sensitive detection. For herbicide detection, we highlighted a novel method for atrazine quantification using a silver nanoprism/GO/SiNW array, 183 as well as an AgNP array‐based substrate which avoided the coffee ring effect by using hydrophobic lotus leaves. 188 In applying SERS to insecticide detection, multiple methods have been able to achieve trace detection, such as the Au‐PdNPs, Ag‐PdNPs, 194 and PSi‐PD NPs 202 substrates discussed previously. Last, for SERS detection of multiple pesticides, we highlighted the innovative use of a filter‐based microfluidic SERS chip for detection in complex matrices, 155 a SERS‐LFA test strip, 167 and an aptamer‐based method using a DNA backbone decorated with Ag@AuNPs for ultra‐trace detection, 165 among others. From these methods alone, it is clear that there is a wide variety of platforms possible for pesticide detection, and we encourage future research to both expand upon these platforms and develop novel ones, especially as pesticide contamination becomes an increasingly important and complex environmental issue.
Finally, for SERS detection of biological pollutants, we have discussed advances made in detecting viruses, bacteria and mycotoxins. There are a wide variety of SERS methods that may be used for virus detection, but currently few have been applied to detect environmental samples. Promising avenues for future research include magnetic nanoparticles (MagSERS) in conjunction with immunoassays and digital microfluidics, 220 , 221 as well as amplification‐free Class 2 CRISPR/Cas systems. 223 , 224 , 226 , 227 , 228 For the detection of bacteria with SERS, there are a number of available methods, and many have already been applied to environmental safety research. For labelled methods of bacterial detection, previous studies have utilized aptamers 229 as well as antibody‐based 221 , 222 , 223 , 224 , 225 , 226 , 227 , 228 , 229 , 230 , 231 , 232 platforms to detect bacteria in environmental waters, milk, food samples and LMFs. Label‐free methods also show great promise for environmental detection of bacterial samples, as they require less pre‐processing and may, therefore, be easier to adapt to field studies. Previously, label‐free methods have not been able to effectively detect analyte molecules in real samples which contain multiple species of bacteria, but recent studies have been able to overcome this barrier by elucidating bacterial species based on surface charge density, 233 and by combining spectroscopic data with multivariate analysis methods to distinguish between species 234 or even strains of bacteria. 235 The last class of biological pollutant we have discussed here is mycotoxins, which pose a significant threat to human and environmental health due to their toxic properties and ability to persist in a wide variety of environments. Previous studies have used substrates, such as thin‐film PDA‐coated silver nanocubes, 240 silver‐embedded silica nanoparticles, 241 and AuNRs, 242 to detect mycotoxins with high sensitivity and reproducibility. More promising, though, is the use of aptamer‐based platforms, which have successfully been applied to detect multiple classes of mycotoxins. 239 , 244 , 245 , 246 , 247 Aptamer‐based methods, due to their extremely high level of specificity, 243 have been particularly successful in analysing mycotoxins in real samples such as beer, 244 peanut oil, 245 corn, 246 and maize 247 and warrant further investigation in future research.
Despite these significant advancements in SERS technology and its applications to environmental detection, there are several analytical gaps that should be addressed in the future. We discuss these gaps in current research below, and aim to provide concise topics which future research may address.
4.1. Improvement of reproducibility
Reproducibility can limit the ability to use SERS to detect contaminants consistently in environmental samples. Low reproducibility is often associated with the uniformity of the SERS nanosubstrate. Substrates with low uniformity have heterogeneous hotspots, making it difficult to obtain similar results along different areas of the substrate. Furthermore, if the substrate is not uniform, it is difficult to obtain quantitative results due to the large variations in signal intensity. To evaluate uniformity of fabricated substrates, SERS scans in large areas have been successfully used to map the spatial distribution of SERS signals. Recently, more uniform substrates have been produced through the control of the size, position and orientation of nanostructures on surfaces, leading to predictable and reproducible hot spots. 45 , 46 , 49 , 51 , 53 However, further work is needed in this area, as the uniformity of substrates aids in the reproducibility of analyte detection. To combat uniformity issues, IS can be used, such as the use of plasmonic electronic Raman scattering peak, 248 or 4‐MBA 79 and 4‐Mpy, 70 as internal reporter molecules encased between the core‐shell layers of SERS nanosubtrates. The use of IS is extremely promising for future SERS research, as it eliminates the need for perfectly homogeneous substrates, and allows for quantitative detection of analytes with high accuracy.
4.2. Minimizing interference from environmental matrices
Recent advances in SERS technologies have made it possible to detect and quantify various contaminants in our environment. However, given that real‐world samples often contain interfering components, there is a need to develop SERS methods which can identify target analytes within complex matrices such as soil samples or biological tissues. Several publications claim that their methods have real‐world environmental applications, but it has been shown that samples within complex matrices that contain interference often do not yield Raman enhancement as high as in simple laboratory samples. For example, the SERS substrate developed by Yao et al. was able to detect THR down to 10–9 M in a simple water matrix, but on apple peels, it was only able to detect THR at concentration of 1.2 × 10–7 M. 143 Future studies should aim to improve SERS detection in a variety of complex matrices in both natural and man‐made environments. One strategy of minimizing matrix interference is using more advanced sample purification methods. Previously, single drop microextraction (SDME) has been used to pre‐concentrate analytes in simple sample matrices before SERS detection, allowing for more concentrated samples and, thus, significantly lower limits of detection. 249 , 250 However, the amount of research combining SDME with SERS is very limited, and to our knowledge there have been no studies which investigated SDME's abilities to improve SERS detection in complex environmental matrices, which is a promising area for future research. Another strategy to reduce matrix interference is integrating materials with purification and enrichment properties into SERS nanosubstrates. SERS‐active‐charged microgels have been used for the detection of biological samples without any pre‐treatment steps, and help to protect the analyte from contamination due to protein adsorption. 251 Smart SERS sensors formed from charged poly(vinyl alcohol) microgels have previously been developed for the detection of charged pesticides in seawater, and show great promise for detection in other complex matrices. 252 This smart sensor could selectively concentrate analytes of a particular charge without pre‐treatment of the sea water samples. Clearly, applying methods, such as those described above to a variety of complex matrices, is of particular interest for environmental pollutant analysis. We propose that future research integrate advanced sample purification strategies into SERS‐based analytical methods to minimize interference from complex environmental matrices and improve detection performance.
4.3. Reusable and other multi‐functional SERS substrates
To make SERS‐based analytical methods more accessible in low‐resource settings, reusable, sustainable, low‐cost and versatile SERS substrates are necessary. Reusable SERS substrates are a relatively new but rapidly growing area which warrants further research. The main advantages of reusable substrates are that they are less costly and less wasteful since they can be used multiple times. They can also utilize environmental friendly components in their design such as green synthesized nanosubstrates. 253 Recently, reusable SERS substrates were utilized to detect different analytes such as lead (II), 42 melamine, 254 azo dyes, 255 antibiotic residues, 256 R6G, 257 dipicolinic acid, 2,4‐dinitrotoluene, and picric acid. 258 Reusable platforms have also been used for the simultaneous detection of paracetamol (PCT), also known as acetaminophen, and FZD, 259 antibiotics 256 and synthetic dyes. 257 The materials used to fabricate reusable substrates can vary widely, but recent publications have utilized platforms such as graphene monolayers, 42 a zinc oxide/GO/silver (ZnO/GO/Ag) hybrid substrate, 254 a carbon cloth supporting material combined with a leaf‐like MOF and electrodeposited AgNPs (CC‐L‐MOF@F‐Ag), 255 silver‐coated AuNRs (AgNPs/GNRs), 256 AgNPs deposited on flower‐like zinc oxide microcrystals (ZnO@Ag), 257 one‐dimensional silver/gold/silver‐chloride NWs (Ag/Au/AgCl NWs), 259 Au@Cu2O–AgNC 260 and AuNPs distributed on a laser‐textured Au sheet. 258 A schematic depicting the determination of the reusability of the laser‐textured Au nanostructures decorated with AuNPs is shown in Figure 5A. This substrate was used for the ultra‐sensitive detection of several hazardous materials including dipicolinic acid, 2,4‐dinitrotoluene, and picric acid down to LODs of 0.83, 3.6 and 2.3 pg/L, respectively. The substrate was tested for its reusability using washing cycles by immersing them in ethanol and sonicating for 15 min, then testing the remaining signal using Raman spectroscopy to ensure picric acid was completely removed. After a fourth washing cycle, the Raman spectrum did not show any characteristic peaks for picric acid, and the substrate could subsequently be used again to detect different analytes. 258 In a study by Barveen et al., Ag NWs were utilized as a reusable SERS substrate for the detection of antibiotics and analgesics (Figure 5B‐K). 259 These NWs were used as a template for photoreduction with an Au precursor to produce AgCl and AuNPs. To assess the reusability of this material, SERS spectra were taken before and after the photodegradation process. In Figure 5C and E, the retention rates are shown for each analyte based on the normalized SERS intensity (a.u.) for PCT (at 1342 cm–1) FZD (at 1359 cm–1), respectively, after several photodegradation cycles. Figure 5C shows retention rates for PCT in the second, third and fourth cycles at 91.00, 82.00 and 69.99%, respectively, while Figure 5E shows retention rates for FZD in the second, third, fourth and fifth cycles at 94.4, 91.7, 85.6 and 81.5%, respectively, indicating the self‐reviving nature of this material. Another study from Barveen et al., developed hybrid flower‐shaped ZnO@Ag nanostructures as reusable SERS probes for the ultra‐sensitive detection of synthetic dyes R6G, sunset yellow (SY) and tartrazine (TZ) (Figure 5L‐W). 257 After four photocatalytic degradation cycles, 90.9% of the original R6G SERS peak at 1367 cm–1 is retained, 76% of the original SY peak at 1393 cm–1 is retained and 80.1% of the original TZ peak at 1353 cm–1 is retained, demonstrating the substrate's reusability.
FIGURE 5.
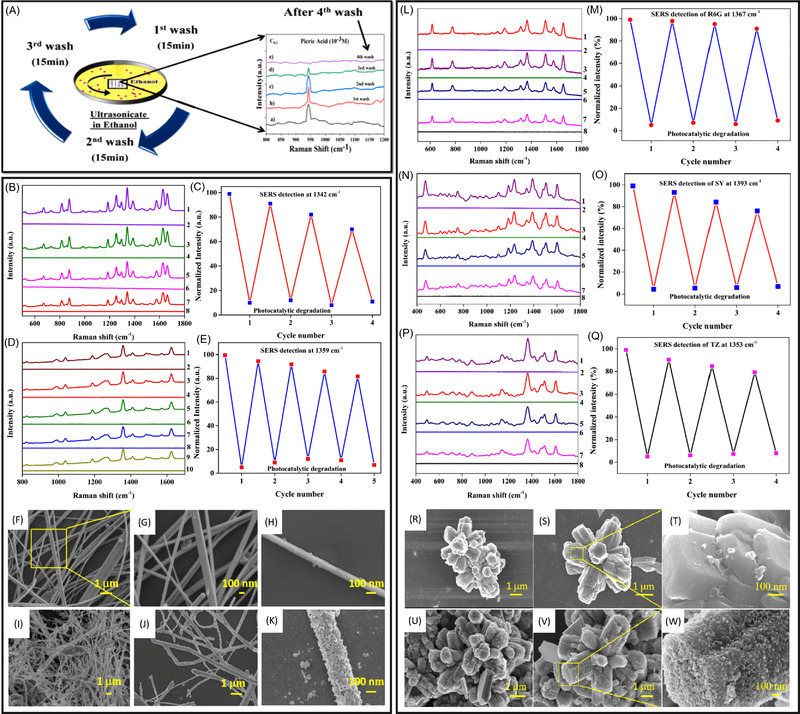
Different reusable SERS substrates cited within this review. (A) Diagram showing the determination of reusability of the developed SERs substrate: laser‐textured Au nanostructures decorated with AuNPs for the detection of multiple hazardous materials (reproduced with permission from Ref. [258]). (F, G, H) FESEM images of silver nanowires developed by Barveen et al., which were used as a template for photoreduction with Au precursor solution, producing AgCl and AuNPs. (I, J, K) FESEM images showing Ag/Au/AgCl heterostructure after a 30‐min photoreduction. (B) and (D) Show the SERS spectra of paracetamol (PCT) and furazolidone (FZD), respectively, adsorbed on the Ag/Au/AgCl structure before and after photodegradation. (C) Using the Raman peak of PCT at 1342 cm‐1 and (E) using the FZD peak of 1359 cm‐1 as the normalized SERS intensity, the retention rates of the Raman peak intensity are shown after several photodegradation cycles for the Raman probe molecules. (C) Shows retention rates for PCT in the second, third and fourth cycles at 91.00, 82.00 and 69.99%, respectively, and (E) shows retention rates for FZD in the second, third, fourth and fifth cycles at 94.4, 91.7, 85.6 and 81.5%, respectively (reproduced with permission from Ref. [259]). From another study by Barveen et al., (R, S, T) show FESEM images of flowerlike ZnO microcrystals and (U, V, W) show the flower‐like ZnO@Ag nanostructures used to detect synthetic dyes rhodamine 6G (R6G), sunset yellow (SY) and tartrazine (TZ). Ag particles were deposited on ZnO after a 30‐ min photoreduction. (L,N,P) Show the SERS spectra of R6G, SY and TZ, respectively, through four cycles of the ‘self‐reviving’ process. (M,O, Q) Show the photocatalytic degradation of the three dyes at characteristic Raman peaks over four cycles, demonstrating the substrate's reusability (reproduced with permission from Ref. [257])
The mechanism by which re‐usable substrates can be regenerated is also important and warrants further investigation. Currently, the most common methods for regeneration of the substrate are UV‐assisted degradation/UV light irradiation, 254 , 257 , 259 sonication with ethanol 258 and cyclical electrodeposition/stripping. 256 The Au@Cu2O–Ag NC substrate developed by Wu et al. allowed for visible light irradiation rather than UV‐light irradiation, which is especially helpful for detection in environmental samples where UV light may damage the analyte (particularly biological analytes). 260 Most notably, only one recent article has, to our knowledge, been able to regenerate the substrate with water. 255 Inspired by superhydrophobic fish scales, Wang et al. developed a CC‐L‐MOF@F‐Ag framework which was chemically modified to confer superhydrophobicity, allowing the substrate to be easily regenerated with de‐ionized water due to its anti‐wetting abilities. While research regarding re‐usable SERS materials has made significant progress and is promising for environmental detection, we have identified several limitations in current research. First is the number of cycles for which a substrate is reusable. Though the AgNPs/GNRs developed by Peng et al. 256 had excellent renewability for ten cycles, the majority of recently reported reusable substrates can be used for fewer cycles, typically ranging from three to six. 42 , 254 , 255 , 258 , 259 , 260 Thus, a primary goal for future research should be increasing the cycle count of reusable substrates, especially ones which may be implemented in the field. A second goal is to design substrates which can be regenerated by easily accessible means such as water. More research is needed to not only increase the cycle count of the substrate, but also allow the substrate to be regenerated by simple means, so that re‐usable substrates can be used in the field more easily and efficiently.
Re‐usable SERS substrates are a specific category of multi‐functional substrates known as regeneration‐enhancement‐in‐one substrates. However, there are several other categories of multi‐functional substrates, including flexible substrates, separation‐enhancement‐in‐one substrates, calibration‐enhancement‐in‐one substrates and regeneration‐enhancement‐in‐one substrates. 261 Flexible substrates have been covered previously in the section of ‘SERS nanosubstrates’ in this review, so here we will focus on the other three categories. Separation‐enhancement‐in‐one substrates allow for direct detection in complex matrices, as they selectively enhance the Raman signals of the analyte by separating the analyte from the matrix based on its properties such as particle size or polarity. Calibration‐enhancement‐in‐one substrates utilize IS to detect analytes within complex matrices without pre‐treatment. Last, regeneration‐enhancement‐in‐one substrates are substrates which can be utilized multiple times, and include substrates which can be regenerated by simple washing with solvents, photocatalytic regeneration (and degradation), superhydrophobic self‐cleaning and reversible intermolecular interactions. For more details on each of these categories, as well as detailed information on regeneration mechanisms, please see the in‐depth review by Li et al. from 2021. 261 There are a plethora of ways for reusable substrates to be multi‐functional outside of their reusability, but one area of particular interest is reusable substrates with contaminant removal abilities. The previously mentioned Au@Cu2O–Ag NC substrate developed by Wu et al. was re‐usable and also multi‐functional as it allowed for the simultaneous detection and degradation of MG. 260 Nanosubstrates may also possess additional properties, such as anti‐bacterial properties, which allow for bacteria detection and removal at the same time. Work by Soleymani et al. developed green‐synthesized reduced GO nanosheets functionalized with AgNPs from celery seeds (RGO/Ag). While the reusable potential of this RGO/Ag substrate was not assessed, it did allow for both the simultaneous detection and removal of pathogenic bacteria due to the Raman signal enhancement and anti‐bacterial effect of the green‐synthesized nanoparticles. Since the target bacteria could be removed by the nanosubstrate, there is a high potential that this type of substrates is reusable, which constitutes an innovative direction for future research, particularly concerning the use of green synthesis in developing multi‐functional materials. 253 Researchers may build upon these findings to increase the number of environmental friendly and versatile SERS platforms available which can detect multiple analytes simultaneously, or perform multiple functions (such as flexibility, separation, calibration or degradation), within the same substrate.
4.4. Advanced data analysis
Some data analysis methods have been mentioned in previous sections of this review, such as PCA and canonical discriminant analysis, which are different forms of multi‐variate analysis. PCA, in particular, has been useful in detecting specific species or even strains of bacteria within complex environmental samples. 234 , 235 Continuing to develop SERS methods which utilize PCA and other multi‐variate analysis techniques is critically important, particularly for environmental detection, as these techniques can help identify multiple similar analytes simultaneously, and in doing this can eliminate pre‐processing steps which may otherwise be necessary for environmental detection. However, there are other advanced methods of data analysis which should also be investigated in future research, and here we aim to briefly discuss how machine learning (ML) can be applied to future SERS studies. ML has already been used in SERS research in a multitude of ways, and have been particularly useful in optimizing substrate design by predicting new Raman metasurfaces, as well as changes within analytes (such as viral mutations). 262 ML also allows for faster data processing with lower error rates compared to human‐analysed data. Advanced algorithms such as the support vector machines‐stochastic gradient descent (SVM‐SGD) algorithm and k‐nearest neighbours algorithm, can even be used to evaluate spectral data quality and classify obtained spectra in settings where trained technicians or researchers are not available (e.g., clinical settings), improving the applicability and accessibility of SERS platforms. 263 , 264 While such advancements in ML have significantly benefited SERS research in clinical settings, there has currently been very little research applying advanced algorithms (like SVM‐SGD and k‐nearest neighbours) to environmental detection. With the benefits that even relatively simple analysis techniques, such as PCA, can confer to SERS detection, it is clear that using more advanced methods, such as ML, could improve environmental SERS detection methods significantly. Thus, we propose two avenues which future research may aim to address: (1) applying current ML algorithms to environmental safety research and (2) developing new algorithms which are uniquely suited for addressing complex environmental matrices.
CONFLICT OF INTEREST
The authors have declared no conflict of interest.
AUTHOR CONTRIBUTIONS
CRediT statement: Lynn R Terry: lead writing and editing (equal). Sage Sanders: lead writing and editing (equal). Rebecca H Potoff: writing, editing, figures. Jacob W Kruel: writing and editing. Manan Jain: writing and editing. Huiyuan Guo: conceptualization, guidance, and editing.
ACKNOWLEDGEMENTS
The authors acknowledge financial support from The Transdisciplinary Areas of Excellence (TAE) Seed Grant Program and Interdisciplinary Collaborations Grants (ICG) Program at SUNY Binghamton.
Terry LR, Sanders S, Potoff RH, Kruel JW, Jain M, Guo H. Applications of surface‐enhanced Raman spectroscopy in environmental detection. Anal Sci Adv. 2022;3:113–145. 10.1002/ansa.202200003
DATA AVAILABILITY STATEMENT
Data sharing not applicable—no new data generated.
REFERENCES
- 1. Raman CV. A change of wave‐length in light scattering. Nature. 1928; 121(3051): 619‐619. [Google Scholar]
- 2. Raman CV, Krishnan KS. A new type of secondary radiation. Nature. 1928; 121(3048): 501‐502. [Google Scholar]
- 3. Kagan MR, McCreery RL. Reduction of fluorescence interference in Raman spectroscopy via analyte adsorption on graphitic carbon. Anal Chem. 1994; 66(23): 4159‐4165. [Google Scholar]
- 4. Fleischmann M, Hendra PJ, McQuillan AJ. Raman spectra of pyridine adsorbed at a silver electrode. Chem Phys Lett. 1974;26(2):163‐166. [Google Scholar]
- 5. Jeanmaire DL, Van Duyne RP. Surface Raman spectroelectrochemistry. J Electroanal Chem Interfacial Electrochem. 1977; 84(1): 1‐20. [Google Scholar]
- 6. Albrecht MG, Creighton JA. Anomalously intense Raman spectra of pyridine at a silver electrode. J Am Chem Soc. 1977; 99(15): 5215‐5217. [Google Scholar]
- 7. Pilot R, Signorini R, Durante C, Orian L, Bhamidipati M, Fabris L. A review on surface‐enhanced Raman scattering. Biosensors. 2019; 9(2): 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Neng J, Zhang Q, Sun P. Application of surface‐enhanced Raman spectroscopy in fast detection of toxic and harmful substances in food. Biosens Bioelectron. 2020;167:112480. [DOI] [PubMed] [Google Scholar]
- 9. Anderson DJ, Moskovits M. A SERS‐active system based on silver nanoparticles tethered to a deposited silver film. J Phys Chem B. 2006; 110(28): 13722‐13727. [DOI] [PubMed] [Google Scholar]
- 10. Maher RC. SERS hot spots. In: Kumar CSSR, ed. Raman Spectroscopy for Nanomaterials Characterization. Berlin, Heidelberg: Springer, 2012: 215‐260. 10.1007/978-3-642-20620-7_10 [DOI] [Google Scholar]
- 11. Zheng XS, Jahn JI, Weber K, Cialla‐May D, Popp J. Label‐free SERS in biological and biomedical applications: recent progress, current challenges and opportunities. Spectrochim Acta A Mol Biomol Spectrosc. 2018;197:56‐77. [DOI] [PubMed] [Google Scholar]
- 12. Pilot R, Signorini R, Fabris L. Surface‐enhanced Raman Spectroscopy: principles, substrates, and applications. In: Deepak FL, ed. Metal Nanoparticles and Clusters: Advances in Synthesis, Properties and Applications. Berlin, Germany: Springer International Publishing; 2018: 89‐164. [Google Scholar]
- 13. Muehlethaler C, Leona M, Lombardi JR. Review of surface enhanced Raman scattering applications in forensic science. Anal Chem. 2016; 88(1): 152‐169. [DOI] [PubMed] [Google Scholar]
- 14. Pérez‐Jiménez AI, Lyu D, Lu Z, Liu G, Ren B. Surface‐enhanced Raman spectroscopy: benefits, trade‐offs and future developments. Chem Sci. 2020; 11(18): 4563‐4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kneipp K, Kneipp H, Itzkan I, Dasari RR, Feld MS. Single molecule detection using near infrared surface‐enhanced Raman scattering. Single Molecule Spectroscopy. 2001;67: 144‐160. Published online. [Google Scholar]
- 16. Nie S, Emory SR. Probing single molecules and single nanoparticles by surface‐enhanced Raman scattering. Science. 1997; 275(5303): 1102‐1106. [DOI] [PubMed] [Google Scholar]
- 17. Pettinger B. Single‐molecule surface‐and tip‐enhanced Raman spectroscopy. Mol Phys. https://www.tandfonline.com/doi/abs/10.1080/00268976.2010.506891?casa_token=GjVB73MwVVYAAAAA:3TvvUHMRoP9CcoK‐r5ELxaSfashM1hRNKLKoHbji_gHtokzR2oZX4QmwuRpiZafw8CG8hQ7_lq0hAXM. Published online 2010. [Google Scholar]
- 18. Etchegoin PG, Le Ru EC. A perspective on single molecule SERS: current status and future challenges. Phys Chem Chem Phys. 2008; 10(40): 6079‐6089. [DOI] [PubMed] [Google Scholar]
- 19. Cialla D, März A, Böhme R, et al. Surface‐enhanced Raman spectroscopy (SERS): progress and trends. Anal Bioanal Chem. 2012; 403(1): 27‐54. [DOI] [PubMed] [Google Scholar]
- 20. Hussain N, Pu H, Sun DW. Synthesis of bimetallic core‐shelled nanoparticles modified by 2‐mercaptoethanol as SERS substrates for detecting ferbam and thiabendazole in apple puree. Food Addit Contam Part A Chem Anal Control Expo Risk Assess. 2021; 38(8): 1386‐1399. [DOI] [PubMed] [Google Scholar]
- 21. Mosier‐Boss PA. Review of SERS substrates for chemical sensing. Nanomaterials (Basel). 2017; 7(6):142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Le Ru EC, Etchegoin PG. Sub‐wavelength localization of hot‐spots in SERS. Chem Phys Lett. 2004; 396(4): 393‐397. [Google Scholar]
- 23. Pu H, Xiao W, Sun DW. SERS‐microfluidic systems: a potential platform for rapid analysis of food contaminants. Trends Food Sci Technol. 2017; 70: 114‐126. [Google Scholar]
- 24. Wustholz KL, Henry AI, McMahon JM, et al. Structure‐activity relationships in gold nanoparticle dimers and trimers for surface‐enhanced Raman spectroscopy. J Am Chem Soc. 2010; 132(31): 10903‐10910. [DOI] [PubMed] [Google Scholar]
- 25. Asiala SM, Schultz ZD. Characterization of hotspots in a highly enhancing SERS substrate. Analyst. 2011; 136(21): 4472‐4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wiley B, Sun Y, Xia Y. Synthesis of silver nanostructures with controlled shapes and properties. Acc Chem Res. 2007; 40(10): 1067‐1076. [DOI] [PubMed] [Google Scholar]
- 27. Sardar R, Funston AM, Mulvaney P, Murray RW. Gold nanoparticles: past, present, and future. Langmuir. 2009; 25(24): 13840‐13851. [DOI] [PubMed] [Google Scholar]
- 28. Freeman RG, Grabar KC, Allison KJ, et al. Self‐assembled metal colloid monolayers: an approach to SERS substrates. Science. 1995; 267(5204): 1629‐1632. [DOI] [PubMed] [Google Scholar]
- 29. Daniel MC, Astruc D. Gold nanoparticles: assembly, supramolecular chemistry, quantum‐size‐related properties, and applications toward biology, catalysis, and nanotechnology. Chem Rev. 2004; 104(1): 293‐346. [DOI] [PubMed] [Google Scholar]
- 30. Cai K, Xiao X, Zhang H, et al. Universal chitosan‐assisted synthesis of Ag‐including heterostructured nanocrystals for label‐free in situ SERS monitoring. Nanoscale. 2015; 7(45): 18878‐18882. [DOI] [PubMed] [Google Scholar]
- 31. Bell SEJ, Sirimuthu NMS. Quantitative surface‐enhanced Raman spectroscopy. Chem Soc Rev. 2008; 37(5): 1012‐1024. [DOI] [PubMed] [Google Scholar]
- 32. Liu J, Hong Z, Yang W, et al. Bacteria inspired internal standard SERS substrate for quantitative detection. ACS Appl Bio Mater. 2021; 4(3): 2009‐2019. [DOI] [PubMed] [Google Scholar]
- 33. Goodacre R, Graham D, Faulds K. Recent developments in quantitative SERS: moving towards absolute quantification. Trends Analyt Chem. 2018; 102: 359‐368. [Google Scholar]
- 34. Asgari S, Sun L, Lin J, et al. Nanofibrillar cellulose/Au@Ag nanoparticle nanocomposite as a SERS substrate for detection of paraquat and thiram in lettuce. Mikrochim Acta. 2020; 187(7): 390. [DOI] [PubMed] [Google Scholar]
- 35. Jana NR. Silver coated gold nanoparticles as new surface enhanced Raman substrate at low analyte concentration. Analyst. 2003; 128(7): 954‐956. [Google Scholar]
- 36. Yang Y, Liu J, Fu ZW, Qin D. Galvanic replacement‐free deposition of Au on Ag for core‐shell nanocubes with enhanced chemical stability and SERS activity. J Am Chem Soc. 2014; 136(23): 8153‐8156. [DOI] [PubMed] [Google Scholar]
- 37. Gao J, Huang X, Liu H, Zan F, Ren J. Colloidal stability of gold nanoparticles modified with thiol compounds: bioconjugation and application in cancer cell imaging. Langmuir. 2012; 28(9): 4464‐4471. [DOI] [PubMed] [Google Scholar]
- 38. Mazloomi‐Rezvani M, Salami‐Kalajahi M, Roghani‐Mamaqani H, Pirayesh A. Effect of surface modification with various thiol compounds on colloidal stability of gold nanoparticles. Appl Organomet Chem. 2018; 32(2): e4079. [Google Scholar]
- 39. Hussain N, Pu H, Sun DW. Core size optimized silver coated gold nanoparticles for rapid screening of tricyclazole and thiram residues in pear extracts using SERS. Food Chem. 2021; 350: 129025. [DOI] [PubMed] [Google Scholar]
- 40. Yeo J, Lee D, Pang Y. Surface adsorption of hydroxyanthraquinones on CTAB‐modified gold nanosurfaces. Spectrochim Acta A Mol Biomol Spectrosc. 2021; 251: 119408. [DOI] [PubMed] [Google Scholar]
- 41. Xu W, Xiao J, Chen Y, Chen Y, Ling X, Zhang J. Graphene‐veiled gold substrate for surface‐enhanced Raman spectroscopy. Advanced Materials. 2013; 25(6): 928‐933. [DOI] [PubMed] [Google Scholar]
- 42. He Q, Han Y, Huang Y, et al. Reusable dual‐enhancement SERS sensor based on graphene and hybrid nanostructures for ultrasensitive lead (Ⅱ) detection. Sens Actuators B Chem. 2021; 341: 130031. [Google Scholar]
- 43. Gupta S, Banaszak A. Detection of DNA bases and environmentally relevant biomolecules and monitoring ssDNA hybridization by noble metal nanoparticles decorated graphene nanosheets as ultrasensitive G‐SERS platforms. J Raman Spectroscop. 2021; 52(5): 930‐948. [Google Scholar]
- 44. Chen S, Bu M, You X, Dai Z, Shi J. High‐performance detection of p‐nitroaniline on defect‐graphene SERS substrate utilizing molecular imprinting technique. Microchem J. 2021; 168: 106536. [Google Scholar]
- 45. Leong SX, Koh CSL, Sim HYF, et al. Enantiospecific molecular fingerprinting using potential‐modulated surface‐enhanced Raman scattering to achieve label‐free chiral differentiation. ACS Nano. 2021; 15(1): 1817‐1825. [DOI] [PubMed] [Google Scholar]
- 46. Pagano R, Ottolini M, Valli L, Bettini S, Giancane G. Ag nanodisks decorated filter paper as a SERS platform for nanomolar tetracycline detection. Colloids Surf A Physicochem Eng Asp. 2021; 624: 126787. [Google Scholar]
- 47. Saadi R, Saadi Z, Fazaeli R, Fard NE. Monolayer and multilayer adsorption isotherm models for sorption from aqueous media. Korean J Chem Eng. 2015; 32(5): 787‐799. [Google Scholar]
- 48. Ng KC, Burhan M, Shahzad MW, Ismail AB. A universal isotherm model to capture adsorption uptake and energy distribution of porous heterogeneous surface. Sci Rep. 2017; 7(1): 10634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang Z, Zhu Q, Wang Y, Dou S, Chen Q, Lu N. Silver‐nanoparticle‐grafted silicon nanocones for reproducible Raman detection of trace contaminants in complex liquid environments. Spectrochim Acta A Mol Biomol Spectrosc. 2021; 251: 119447. [DOI] [PubMed] [Google Scholar]
- 50. De Angelis F, Gentile F, Mecarini F, et al. Breaking the diffusion limit with super‐hydrophobic delivery of molecules to plasmonic nanofocusing SERS structures. Nat Photonics. 2011; 5(11): 682‐687. [Google Scholar]
- 51. Xu J, Si Y, Li Z, et al. Multiscale structure enabled effective plasmon coupling and molecular enriching for SERS detection. Appl Surf Sci. 2021; 544: 148908. [Google Scholar]
- 52. Mangold MA, Holleitner AW, Agustsson JS, Calame M. Nanoparticle arrays. In: Aliofkhazraei M, ed. Handbook of Nanoparticles.Berlin, Germany: Springer International Publishing; 2016: 565‐601. [Google Scholar]
- 53. Li K, Tang X, Liu G, et al. An efficient double template strategy to construct large‐area and highly ordered silver “urchin‐like” arrays for sensitive SERS analysis. Appl Surf Sci. 2021; 570: 151069. [Google Scholar]
- 54. Zhang X, Zhang X, Luo C, et al. Volume‐enhanced Raman scattering detection of viruses. Small. 2019; 15(11): e1805516. [DOI] [PubMed] [Google Scholar]
- 55. Li K, Liu G, Zhang S, et al. A porous Au‐Ag hybrid nanoparticle array with broadband absorption and high‐density hotspots for stable SERS analysis. Nanoscale. 2019; 11(19): 9587‐9592. [DOI] [PubMed] [Google Scholar]
- 56. Zhang X, Xiao X, Dai Z, et al. Ultrasensitive SERS performance in 3D “sunflower‐like” nanoarrays decorated with Ag nanoparticles. Nanoscale. 2017; 9(9): 3114‐3120. [DOI] [PubMed] [Google Scholar]
- 57. Mao P, Liu C, Favraud G, et al. Broadband single molecule SERS detection designed by warped optical spaces. Nat Commun. 2018; 9(1): 5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Guan Y, Wang Z, Gu P, Wang Y, Zhang W, Zhang G. An in situ SERS study of plasmonic nanochemistry based on bifunctional “hedgehog‐like” arrays. Nanoscale. 2019; 11(19): 9422‐9428. [DOI] [PubMed] [Google Scholar]
- 59. Xu X, Hu X, Fu F, Liu L, Liu X. DNA‐induced assembly of silver nanoparticle decorated cellulose nanofiber: a flexible surface‐enhanced Raman spectroscopy substrate for the selective charge molecular detection and wipe test of pesticide residues in fruits. ACS Sustainable Chem Eng. 2021; 9(14): 5217‐5229. [Google Scholar]
- 60. Liang Y, Lin C, Shen A, Guo R, Li Y. Porous POM/PLLA membranes decorated with gold nanoparticles as flexible and efficient plasmonic substrates for surface‐enhanced Raman scattering. Appl Surf Sci. 2019; 498: 143856. [Google Scholar]
- 61. Wang K, Sun DW, Pu H, Wei Q. Polymer multilayers enabled stable and flexible Au@Ag nanoparticle array for nondestructive SERS detection of pesticide residues. Talanta. 2021; 223(Pt 2): 121782. [DOI] [PubMed] [Google Scholar]
- 62. Jeon Y, Kim D, Kwon G, et al. Detection of nanoplastics based on surface‐enhanced Raman scattering with silver nanowire arrays on regenerated cellulose films. Carbohydr Polym. 2021; 272: 118470. [DOI] [PubMed] [Google Scholar]
- 63. Xu F, Xuan M, Ben Z, Shang W, Ma G. Surface enhanced Raman scattering analysis with filter‐based enhancement substrates: a mini review. Rev Anal Chem. 2021; 40(1): 75‐92. [Google Scholar]
- 64. Bharati MSS, Soma VR. Flexible SERS substrates for hazardous materials detection: recent advances. Opto‐Electron Adv. 2021; 4(11): 210048‐210048. [Google Scholar]
- 65. Ye D, Lei X, Li T, et al. Ultrahigh tough, super clear, and highly anisotropic nanofiber‐structured regenerated cellulose films. ACS Nano. 2019; 13(4): 4843‐4853. [DOI] [PubMed] [Google Scholar]
- 66. Zhang X, Elsayed I, Navarathna C, Schueneman GT, Hassan EB. Biohybrid hydrogel and aerogel from self‐assembled nanocellulose and nanochitin as a high‐efficiency adsorbent for water purification. ACS Appl Mater Interfaces. 2019; 11(50): 46714‐46725. [DOI] [PubMed] [Google Scholar]
- 67. Chen J, Huang M, Kong L, Lin M. Jellylike flexible nanocellulose SERS substrate for rapid in‐situ non‐invasive pesticide detection in fruits/vegetables. Carbohydr Polym. 2019; 205: 596‐600. [DOI] [PubMed] [Google Scholar]
- 68. Tokura Y, Harvey S, Chen C, Wu Y, Ng DYW, Weil T. Fabrication of defined polydopamine nanostructures by DNA origami‐templated polymerization. Angew Chem Int Ed. 2018; 57(6): 1587‐1591. https://onlinelibrary.wiley.com/doi/pdf/10.1002/anie.201711560%4010.1002/%28ISSN%291521‐3773.MaxPlanckDay. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sun H, Li X, Hu Z, et al. Hydrophilic‐hydrophobic silver nanowire‐paper based SERS substrate for in‐situ detection of furazolidone under various environments. Appl Surf Sci. 2021; 556: 149748. [Google Scholar]
- 70. Wang XA, Shen W, Zhou B, et al. The rationality of using core–shell nanoparticles with embedded internal standards for SERS quantitative analysis based glycerol‐assisted 3D hotspots platform. RSC Adv. 2021; 11(33): 20326‐20334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Liu C, Lei F, Wei Y, et al. Preparation of a superhydrophobic AgNP/GF substrate and its SERS application in a complex detection environment. Opt Express. 2021; 29(21): 34085‐34096. [DOI] [PubMed] [Google Scholar]
- 72. Shen W, Lin X, Jiang C, et al. Reliable quantitative SERS analysis facilitated by core‐shell nanoparticles with embedded internal standards. Angew Chem Int Ed Engl. 2015; 54(25): 7308‐7312. [DOI] [PubMed] [Google Scholar]
- 73. Guo P, Zeng W, Tian S, Chen H, Liu W, Chen C. Quantitative detection of nanomolar drug using surface‐enhanced Raman scattering combined with internal standard method and two‐step centrifugation method. Microchem J. 2020; 158: 105202. [Google Scholar]
- 74. Cowcher DP, Xu Y, Goodacre R. Portable, quantitative detection of Bacillus bacterial spores using surface‐enhanced Raman scattering. Anal Chem. 2013; 85(6): 3297‐3302. [DOI] [PubMed] [Google Scholar]
- 75. Li M, Wang JY, Chen QQ, et al. Background‐free quantitative surface enhanced Raman spectroscopy analysis using core–shell nanoparticles with an inherent internal standard. Anal Chem. 2019; 91(23): 15025‐15031. [DOI] [PubMed] [Google Scholar]
- 76. Lin X, Wang Y, Wang L, et al. Interference‐free and high precision biosensor based on surface enhanced Raman spectroscopy integrated with surface molecularly imprinted polymer technology for tumor biomarker detection in human blood. Biosensor Bioelectronic. 2019; 143: 111599. https://www.sciencedirect.com/science/article/pii/S0956566319306785?casa_token=WfJldzDAfD4AAAAA:iEwuGwml0bDEDU8GhZhK84sRlz0aBZ5sez0NXJC3EqxgHeAYoHFDCIInd7_T9ir1NdENmQKDNaBB. [DOI] [PubMed] [Google Scholar]
- 77. Yuan K, Mei Q, Guo X, et al. Antimicrobial peptide based magnetic recognition elements and Au@ Ag‐GO SERS tags with stable internal standards: a three in one biosensor for isolation, discrimination and killing of multiple bacteria in whole blood. Chem Sci. 2018; 9(47): 8781‐8795. https://pubs.rsc.org/doi/c8sc04637a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Xu Y, Liu H, Jiang T. Reliable quantitative SERS analysis mediated by Ag nano coix seeds with internal standard molecule. J Nanopart Res. 2019; 21(5). [Google Scholar]
- 79. Wang K, Li J. Reliable SERS detection of pesticides with a large‐scale self‐assembled Au@4‐MBA@Ag nanoparticle array. Spectrochim Acta A Mol Biomol Spectrosc. 2021; 263: 120218. [DOI] [PubMed] [Google Scholar]
- 80. Wang K, Sun DW, Pu H, Wei Q. Two‐dimensional Au@Ag nanodot array for sensing dual‐fungicides in fruit juices with surface‐enhanced Raman spectroscopy technique. Food Chem. 2020; 310: 125923. [DOI] [PubMed] [Google Scholar]
- 81. Fergusson JE. The Heavy Elements: Chemistry, Environmental Impact and Health Effects. Amsterdam, Netherlands: Elsevier Science Limited; 1990. https://play.google.com/store/books/details?id=qRlSAAAAMAAJ. [Google Scholar]
- 82. Rengel Z. In: Prasad MNV, ed. Heavy Metal Stress in Plants: From Biomolecules to Ecosystems. Berlin, Heidelberg: Springer; 2004: 271‐294. Heavy Metals as Essential Nutrients. [Google Scholar]
- 83. Walzel E. Trace elements in human nutrition and health. XVIII und 343 Seiten, zahlr. Abbildungen und Tabellen, 3 Farbtafeln. World Health Organization, Geneva 1996. Preis: 85,‐ Sfr/76.50 US $. Food/Nahrung. 1997; 41(3): 183‐184. [Google Scholar]
- 84. Tchounwou PB, Yedjou CG, Patlolla AK, Sutton DJ. Heavy metal toxicity and the environment. Experientia Suppl. 2012; 101: 133‐164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Waalkes MP, Misra RR, Chang LW. Toxicology of Metals. New York, NY: CRC; 1996. [Google Scholar]
- 86. Tian C, Zhao L, Zhu J, Zhang S. Ultrasensitive detection of trace Hg2+ by SERS aptasensor based on dual recycling amplification in water environment. J Hazard Mater. 2021; 416: 126251. [DOI] [PubMed] [Google Scholar]
- 87. Xu G, Song P, Xia L. Examples in the detection of heavy metal ions based on surface‐enhanced Raman scattering spectroscopy. Nanophotonics. 2021; 10(18): 4419‐4445. [Google Scholar]
- 88. Reuben A, Caspi A, Belsky DW, et al. Association of childhood blood lead levels with cognitive function and socioeconomic status at age 38 years and with IQ change and socioeconomic mobility between childhood and adulthood. JAMA. 2017; 317(12): 1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Gamillo E, Leaded gasoline use in vehicles has now officially ended worldwide. Smithsonian Magazine. Published August 31, 2021. Accessed December 8, 2021. https://www.smithsonianmag.com/smart‐news/worldwide‐use‐leaded‐gasoline‐vehicles‐now‐completely‐phased‐out‐180978549/
- 90. Zou Q, Ai X, Xue T. Synthesis of environment‐friendly and label‐free SERS probe for Iron(III) detection in integrated circuit cleaning solution waste. Microchem J. 2021; 169: 106549. [Google Scholar]
- 91. Toyokuni S. Role of iron in carcinogenesis: cancer as a ferrotoxic disease. Cancer Sci. 2009; 100(1): 9‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Eisenbach C, Gehrke SG, Stremmel W. Iron, the HFE gene, and hepatitis C. Clin Liver Dis. 2004; 8(4): 775‐785. vii‐viii. [DOI] [PubMed] [Google Scholar]
- 93. Li K, Reichmann H. Role of iron in neurodegenerative diseases. J Neural Transm. 2016; 123(4): 389‐399. [DOI] [PubMed] [Google Scholar]
- 94. Schneider SA, Bhatia KP. Excess iron harms the brain: the syndromes of neurodegeneration with brain iron accumulation (NBIA). J Neural Transm. 2013; 120(4): 695‐703. [DOI] [PubMed] [Google Scholar]
- 95. Ding X, Song L, Han Y, et al. Effects of Fe3+ on acute toxicity and regeneration of planarian (Dugesia japonica) at different temperatures. Biomed Res Int. 2019; 2019: 8591631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Driscoll CT, Mason RP, Chan HM, Jacob DJ, Pirrone N. Mercury as a global pollutant: sources, pathways, and effects. Environ Sci Technol. 2013; 47(10): 4967‐4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Virtanen JK, Rissanen TH, Voutilainen S, Tuomainen TP. Mercury as a risk factor for cardiovascular diseases. J Nutr Biochem. 2007; 18(2): 75‐85. [DOI] [PubMed] [Google Scholar]
- 98. Gu Z, Zhao M, Sheng Y, Bentolila LA, Tang Y. Detection of mercury ion by infrared fluorescent protein and its hydrogel‐based paper assay. Anal Chem. 2011; 83(6): 2324‐2329. [DOI] [PubMed] [Google Scholar]
- 99. Xu G, Zhang Q, Gao C, Ma L, Song P, Xia L. A label‐free SERS sensor for the detection of Hg2+ based on phenylacetylene functionalized Ag nanoparticles. Microchem J. 2021; 168: 106504. [Google Scholar]
- 100. Huang S, Gan N, Li T, Zhou Y, Cao Y, Dong Y. Electrochemical aptasensor for multi‐antibiotics detection based on endonuclease and exonuclease assisted dual recycling amplification strategy. Talanta. 2018; 179: 28‐36. [DOI] [PubMed] [Google Scholar]
- 101. Bradley PM, Journey CA, Button DT, et al. Multi‐region assessment of pharmaceutical exposures and predicted effects in USA wadeable urban‐gradient streams. PLoS One. 2020; 15(1): e0228214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Burtsev V, Erzina M, Guselnikova O, et al. Detection of trace amounts of insoluble pharmaceuticals in water by extraction and SERS measurements in a microfluidic flow regime. Analyst. 2021; 146(11): 3686‐3696. [DOI] [PubMed] [Google Scholar]
- 103. Low bandgap microsphere‐like magnetic nanocomposite: an enhanced photocatalyst for degradation of organic contaminants and fabrication of SERS‐active surfaces. Colloids Surf A Physicochem Eng Asp. 2020;589:124436. [Google Scholar]
- 104. Mohanraj J, Durgalakshmi D, Saravanan R. Water‐soluble graphitic carbon nitride for clean environmental applications. Environ Pollut. 2021; 269: 116172. [DOI] [PubMed] [Google Scholar]
- 105. Lu Q, Qi Z, Wang S, Tian X, Xu X. Rapid detection of illegal biguanides in hypoglycemic health products using molecular imprinting combined with SERS technology. Microchem J. 2021; 169: 106523. [Google Scholar]
- 106. Alula MT, Mengesha ZT, Mwenesongole E. Advances in surface‐enhanced Raman spectroscopy for analysis of pharmaceuticals: a review. Vib Spectrosc. 2018; 98: 50‐63. [Google Scholar]
- 107. Cailletaud J, De Bleye C, Dumont E, et al. Critical review of surface‐enhanced Raman spectroscopy applications in the pharmaceutical field. J Pharm Biomed Anal. 2018; 147: 458‐472. [DOI] [PubMed] [Google Scholar]
- 108. Smith M, Love DC, Rochman CM, Neff RA. Microplastics in seafood and the implications for human health. Curr Environ Health Rep. 2018; 5(3): 375‐386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Yaranal NA, Subbiah S, Mohanty K. Distribution and characterization of microplastics in beach sediments from Karnataka (India) coastal environments. Mar Pollut Bull. 2021; 169: 112550. [DOI] [PubMed] [Google Scholar]
- 110. Oßmann BE, Sarau G, Holtmannspötter H, Pischetsrieder M, Christiansen SH, Dicke W. Small‐sized microplastics and pigmented particles in bottled mineral water. Water Res. 2018; 141: 307‐316. [DOI] [PubMed] [Google Scholar]
- 111. Goncharuk VV. Drinking water: factors affecting the quality of drinking water. Drinking Water: 105‐245. 10.1007/978-3-319-04334-0_4. Published online 2014. [DOI] [Google Scholar]
- 112. Dümichen E, Eisentraut P, Bannick CG, Barthel AK, Senz R, Braun U. Fast identification of microplastics in complex environmental samples by a thermal degradation method. Chemosphere. 2017; 174: 572‐584. [DOI] [PubMed] [Google Scholar]
- 113. Lv L, He L, Jiang S, et al. In situ surface‐enhanced Raman spectroscopy for detecting microplastics and nanoplastics in aquatic environments. Sci Total Environ. 2020; 728: 138449. [DOI] [PubMed] [Google Scholar]
- 114. Zhou XX, Liu R, Hao LT, Liu JF. Identification of polystyrene nanoplastics using surface enhanced Raman spectroscopy. Talanta. 2021; 221: 121552. [DOI] [PubMed] [Google Scholar]
- 115. Caldwell J, Taladriz‐Blanco P, Rothen‐Rutishauser B, Petri‐Fink A. Detection of sub‐micro‐ and nanoplastic particles on gold nanoparticle‐based substrates through surface‐enhanced Raman scattering (SERS) spectroscopy. Nanomaterials. 2021; 11(5): 1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Lê QT, Ly NH, Kim MK, et al. Nanostructured Raman substrates for the sensitive detection of submicrometer‐sized plastic pollutants in water. J Hazard Mater. 2021; 402: 123499. [DOI] [PubMed] [Google Scholar]
- 117. Vélez‐Escamilla LY, Contreras‐Torres FF. Latest advances and developments to detection of micro‐ and nanoplastics using surface‐enhanced Raman spectroscopy. Part Part Syst Charact. 2100217. 10.1002/ppsc.202100217. Published online January 31, 2022. [DOI] [Google Scholar]
- 118. Gulzar T, Farooq T, Kiran S, Ahmad I, Hameed A. 1 ‐ Green chemistry in the wet processing of textiles. In: Shahid‐ul‐Islam, Butola BS, eds. The Impact and Prospects of Green Chemistry for Textile Technology. Woodhead Publishing, 2019: 1‐20. 10.1016/B978-0-08-102491-1.00001-0. [DOI] [Google Scholar]
- 119. Ardila‐Leal LD, Poutou‐Piñales RA, Pedroza‐Rodríguez AM, Quevedo‐Hidalgo BE. A brief history of colour, the environmental impact of synthetic dyes and removal by using laccases. Molecules. 2021; 26(13):3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Sabathi G, Reyer A, Cefarin N, et al. Tannin‐furanic foams used as biomaterial substrates for SERS sensing in possible wastewater filter applications. Mater Res Express. 2021; 8(11): 115404. [Google Scholar]
- 121. Xue Y, Shao J, Sui G, Ma Y, Li H. Rapid detection of orange II dyes in water with SERS imprinted sensor based on PDA‐modified MOFs@Ag. J Environment Chem Eng. 2021; 9(6): 106317. [Google Scholar]
- 122. Chen Z, Wang Y, Yang Y, Yang X, Zhang X. Multifunctional sensing platform based on green‐synthesized silver nanostructure and microcrack architecture. Chem Eng J. 2021; 403: 126388. [Google Scholar]
- 123. Yang Y, Gao X, Yang S, Shen Y, Xie A. Synthesis and superior SERS performance of porous octahedron Cu2O with oxygen vacancy derived from MOFs. J Mater Sci. 2021; 56(16): 9702‐9711. [Google Scholar]
- 124. Vannucci G, Cañamares MV, Prati S, Sanchez‐Cortes S. Analysis of the tautomeric equilibrium of two red monoazo dyes by UV–Visible, Raman and SERS spectroscopies. Spectrochimica Acta Part A: Mol Biomol Spectroscop. 2021; 261: 120007. [DOI] [PubMed] [Google Scholar]
- 125. Moldovan R, Iacob BC, Farcău C, Bodoki E, Oprean R. Strategies for SERS detection of organochlorine pesticides. Nanomaterials (Basel). 2021; 11(2):304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Pearson BL, Simon JM, McCoy ES, Salazar G, Fragola G, Zylka MJ. Identification of chemicals that mimic transcriptional changes associated with autism, brain aging and neurodegeneration. Nat Commun. 2016; 7: 11173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Dong J, Zhao K, Wang Q, et al. Plasmonic alloy nanochains assembled via dielectrophoresis for ultrasensitive SERS. Opt Express. 2021; 29(22): 36857‐36870. [DOI] [PubMed] [Google Scholar]
- 128. Zhang Q, Xu G, Guo N, Wang T, Song P, Xia L. In‐situ synthesis of methyl cellulose film decorated with silver nanoparticles as a flexible surface‐enhanced Raman substrate for the rapid detection of pesticide residues in fruits and vegetables. Materials. 2021; 14(19):5750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Liu Y, Li X, Cheng J, et al. SERS devices with “hedgehog‐like” nanosphere arrays for detection of trace pesticides. J Innov Opt Health Sci. 2021; 14(04): 2141005. [Google Scholar]
- 130. Barbillon G, Graniel O, Bechelany M. Assembled Au/ZnO nano‐urchins for SERS sensing of the pesticide thiram. Nanomaterials (Basel). 2021; 11(9):2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Zhang Y, Zhu A, Wang Y, Zhang X. Plasmonic structure with nanocavity cavities for SERS detection of pesticide thiram. Nanotechnology. 2021; 32(13): 135301. [DOI] [PubMed] [Google Scholar]
- 132. Han C, Wei Y, Lei F, et al. Heterostructured CuO@ZnO@Ag biomimetic setaria as wettability‐switchable difunctional SERS substrate for trace pesticide and DNA detections. Nanophotonics. 2021; 10(10): 2671‐2682. [Google Scholar]
- 133. Xu L, Liu H, Zhou H, Hong M. One‐step fabrication of metal nanoparticles on polymer film by femtosecond LIPAA method for SERS detection. Talanta. 2021; 228: 122204. [DOI] [PubMed] [Google Scholar]
- 134. Liu S, Guo J, Hinestroza JP, Kong X, Yu Q. Fabrication of plasmonic absorbent cotton as a SERS substrate for adsorption and detection of harmful ingredients in food. Microchem J. 2021; 170: 106662. [Google Scholar]
- 135. Zhang C, Chen S, Jiang Z, Shi Z, Wang J, Du L. Highly sensitive and reproducible SERS substrates based on ordered micropyramid array and silver nanoparticles. ACS Appl Mater Interfaces. 2021; 13(24): 29222‐29229. [DOI] [PubMed] [Google Scholar]
- 136. Martins NCT, Fateixa S, Fernandes T, Nogueira HIS, Trindade T. Inkjet Printing of Ag and polystyrene nanoparticle emulsions for the one‐step fabrication of hydrophobic paper‐based surface‐enhanced Raman scattering substrates. ACS Appl Nano Mater. 2021; 4(5): 4484‐4495. [Google Scholar]
- 137. Kim D, Kim J, Henzie J, et al. Mesoporous Au films assembled on flexible cellulose nanopaper as high‐performance SERS substrates. Chem Eng J. 2021; 419: 129445. [Google Scholar]
- 138. Zhu J, Zhang S, Weng GJ, Li JJ, Zhao JW. Spiky yolk‐shell AuAg bimetallic nanorods with uniform interior gap for the SERS detection of thiram residues in fruit juice. Spectrochim Acta A Mol Biomol Spectrosc. 2021; 262: 120108. [DOI] [PubMed] [Google Scholar]
- 139. Wang T, Dong P, Zhu C, et al. Fabrication of 2D titanium carbide MXene/Au nanorods as a nanosensor platform for sensitive SERS detection. Ceram Int. 2021; 47(21): 30082‐30090. [Google Scholar]
- 140. Gao F, Kong W, He G, et al. SERS‐active vertically aligned silver/tungsten oxide nanoflakes for ultrasensitive and reliable detection of thiram. Microchem J. 2021; 165: 106046. [Google Scholar]
- 141. Wan M, Zhao H, Wang Z, Zou X, Zhao Y, Sun L. Fabrication of Ag modified SiO2 electrospun nanofibrous membranes as ultrasensitive and high stable SERS substrates for multiple analytes detection. Colloid Interfac Sci Communicat. 2021; 42: 100428. [Google Scholar]
- 142. Bai F, Dong J, Qu J, Zhang Z. Construction of flexible, transparent and mechanically robust SERS‐active substrate with an efficient spin coating method for rapidin‐situtarget molecules detection. Nanotechnology. 2021; 32(38). [DOI] [PubMed] [Google Scholar]
- 143. Yao C, Gao X, Liu X, Shen Y, Xie A. In‐situ preparation of Ferrero® chocolate‐like Cu2O@Ag microsphere as SERS substrate for detection of thiram. J Mater Res Technol. 2021; 11: 857‐865. [Google Scholar]
- 144. Pu H, Huang Z, Xu F, Sun DW. Two‐dimensional self‐assembled Au‐Ag core‐shell nanorods nanoarray for sensitive detection of thiram in apple using surface‐enhanced Raman spectroscopy. Food Chem. 2021; 343: 128548. [DOI] [PubMed] [Google Scholar]
- 145. Qian‐Qian D, Fu‐Hua W, Hong‐Hua Z, Yun‐Peng D, Ting W, Ying‐Yi W. Detection of Thirm based on highly uniform au nanorods as surface‐enhanced Raman spectroscopy substrate. Chinese J Inorganic Chemist. 2021; 37(7): 1315‐1321. [Google Scholar]
- 146. Zhou N, Meng G, Huang Z, Zhang X, Zhu C, Ke Y. Ag‐coated 3D Cu(OH)2 nanowires on the woven copper mesh as a cost‐effective surface‐enhanced Raman scattering substrate. Surf Coat Technol. 2021; 415: 127132. [Google Scholar]
- 147. Song Y, Huang HC, Lu W, et al. Ag@WS2 quantum dots for Surface Enhanced Raman Spectroscopy: enhanced charge transfer induced highly sensitive detection of thiram from honey and beverages. Food Chem. 2021; 344: 128570. [DOI] [PubMed] [Google Scholar]
- 148. Oliveira MJS, Martin CS, Rubira RJG, Batagin‐Neto A, Constantino CJL, Aroca RF. Surface‐enhanced Raman scattering of thiram: quantitative and theoretical analyses. J Raman Spectrosc. 2021; 52(12): 2557‐2571. [Google Scholar]
- 149. Dao DQ, Truong DH, Nguyen TLA, Ngo TC, An NTT . Insight into SERS chemical enhancement mechanism of fungicide Thiram adsorbed on silver nanoparticles. J Cluster Sci. 10.1007/s10876-021-02197-z. Published online November 18, 2021. [DOI] [Google Scholar]
- 150. Tsen CM, Yu CW, Chen SY, Lin CL, Chuang CY. Application of surface‐enhanced Raman scattering in rapid detection of dithiocarbamate pesticide residues in foods. Appl Surf Sci. 2021; 558: 149740. [Google Scholar]
- 151. Huo D, Chen B, Li M, Meng G, Lei Y, Zhu C. Template‐assisted fabrication of Ag‐nanoparticles@ZnO‐nanorods array as recyclable 3D surface enhanced Raman scattering substrate for rapid detection of trace pesticides. Nanotechnology. 2021; 32(14): 145302. [DOI] [PubMed] [Google Scholar]
- 152. Vendamani VS, Beeram R, Nageswara Rao SVS, Pathak AP, Soma VR. Trace level detection of explosives and pesticides using robust, low‐cost, free‐standing silver nanoparticles decorated porous silicon. Opt Express. 2021; 29(19): 30045‐30061. [DOI] [PubMed] [Google Scholar]
- 153. Zhu C, Zhao Q, Wang X, Li Z, Hu X. Ag‐nanocubes/graphene‐oxide/Au‐nanoparticles composite film with highly dense plasmonic hotspots for surface‐enhanced Raman scattering detection of pesticide. Microchem J. 2021; 165: 106090. [Google Scholar]
- 154. Wei Q, Zhang L, Song C, Yuan H, Li X. Quantitative detection of dithiocarbamate pesticides by surface‐enhanced Raman spectroscopy combined with an exhaustive peak‐seeking method. Anal Methods. 2021; 13(12): 1479‐1488. [DOI] [PubMed] [Google Scholar]
- 155. Asgari S, Wu G, Aghvami SA, Zhang Y, Lin M. Optimisation using the finite element method of a filter‐based microfluidic SERS sensor for detection of multiple pesticides in strawberry. Food Addit Contam Part A Chem Anal Control Expo Risk Assess. 2021; 38(4): 646‐658. [DOI] [PubMed] [Google Scholar]
- 156. Verma AK, Soni RK. Multi‐spiked silver stars for ultrasensitive and multiplexed SERS detection of analytes. J Phys D Appl Phys. 2021; 54(47): 475107. [Google Scholar]
- 157. Yu H, Lyu Q, Chen X, et al. Nylon membranes modified by gold nanoparticles as surface‐enhanced Raman spectroscopy substrates for several pesticides detection. RSC Adv. 2021; 11(39): 24183‐24189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Huang CT, Jan FJ, Chang CC. A 3D plasmonic crossed‐wire nanostructure for Surface‐Enhanced Raman scattering and plasmon‐enhanced fluorescence detection. Molecules. 2021; 26(2):281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Ibáñez D, González‐García MB, Hernández‐Santos D, Fanjul‐Bolado P. Detection of dithiocarbamate, chloronicotinyl and organophosphate pesticides by electrochemical activation of SERS features of screen‐printed electrodes. Spectrochim Acta A Mol Biomol Spectrosc. 2021; 248: 119174. [DOI] [PubMed] [Google Scholar]
- 160. Xu Y, Hassan MM, Ali S, Li H, Ouyang Q, Chen Q. Self‐cleaning‐mediated sers chip coupled chemometric algorithms for detection and photocatalytic degradation of pesticides in food. J Agric Food Chem. 2021; 69(5): 1667‐1674. [DOI] [PubMed] [Google Scholar]
- 161. Kim D, Lee K, Jeon Y, et al. Plasmonic nanoparticle‐analyte nanoarchitectronics combined with efficient analyte deposition method on regenerated cellulose‐based SERS platform. Cellulose. 2021; 28(18): 11493‐11502. [Google Scholar]
- 162. Ren X, Feng X, Li X, Li X. Preparation of silver with an ultrathin molecular imprinted layer for detection of carbendazim by SERS. Chem Pap. 2021; 75(12): 6477‐6485. [Google Scholar]
- 163. He J, Li H, Zhang L, et al. Silver microspheres aggregation‐induced Raman enhanced scattering used for rapid detection of carbendazim in Chinese tea. Food Chem. 2021; 339: 128085. [DOI] [PubMed] [Google Scholar]
- 164. Shen Z, Fan Q, Yu Q, Wang R, Wang H, Kong X. Facile detection of carbendazim in food using TLC‐SERS on diatomite thin layer chromatography. Spectrochim Acta A Mol Biomol Spectrosc. 2021; 247: 119037. [DOI] [PubMed] [Google Scholar]
- 165. Lu Y, Tan Y, Xiao Y, Li Z, Sheng E, Dai Z. A silver@gold nanoparticle tetrahedron biosensor for multiple pesticides detection based on surface‐enhanced Raman scattering. Talanta. 2021; 234: 122585. [DOI] [PubMed] [Google Scholar]
- 166. Yu H, Xu L, Yang F, et al. Rapid surface‐enhanced raman spectroscopy detection of chlorothalonil in standard solution and orange peels with pretreatment of ultraviolet irradiation. Bull Environ Contam Toxicol. 2021; 107(2): 221‐227. [DOI] [PubMed] [Google Scholar]
- 167. Sheng E, Lu Y, Xiao Y, Li Z, Wang H, Dai Z. Simultaneous and ultrasensitive detection of three pesticides using a surface‐enhanced Raman scattering‐based lateral flow assay test strip. Biosens Bioelectron. 2021; 181: 113149. [DOI] [PubMed] [Google Scholar]
- 168. Miao Y, Yang K, Zong S, Wang Z, Cui Y. Au/Ag bimetallic nanocuboid superlattices coated with Ti3C2 nanosheets for surface‐enhanced Raman spectroscopy detection of fish drug residues in pond water. ACS Appl Nano Mater. 2021; 4(7): 6844‐6851. [Google Scholar]
- 169. Cheng D, Zhang Y, Yan C, et al. Polydopamine‐assisted in situ growth of three‐dimensional ZnO/Ag nanocomposites on PET films for SERS and catalytic properties. J Mol Liq. 2021; 338: 116639. [Google Scholar]
- 170. Bo G, Yu‐dong Y, Qian Z, et al. A SERS substrate for on‐site detection of trace pesticide molecules based on parahydrophobic nanostructures. Spectroscop Spectrl Anal. 2021; 41(8): 2499‐2504. [Google Scholar]
- 171. Luo J, Wang Z, Li Y, et al. Durable and flexible Ag‐nanowire‐embedded PDMS films for the recyclable swabbing detection of malachite green residue in fruits and fingerprints. Sens Actuators B Chem. 2021; 347: 130602. [Google Scholar]
- 172. Tian X, Fan Q, Guo J, Yu Q, Xu L, Kong X. Surface‐enhanced Raman scattering of flexible cotton fiber‐Ag for rapid adsorption and detection of malachite green in fish. Spectrochim Acta A Mol Biomol Spectrosc. 2021; 263: 120174. [DOI] [PubMed] [Google Scholar]
- 173. Li J, Wang Q, Wang J, et al. Quantitative SERS sensor based on self‐assembled Au@Ag heterogeneous nanocuboids monolayer with high enhancement factor for practical quantitative detection. Anal Bioanal Chem. 2021; 413(16): 4207‐4215. [DOI] [PubMed] [Google Scholar]
- 174. Chen Z, Sun Y, Shi J, et al. Facile synthesis of Au@Ag core‐shell nanorod with bimetallic synergistic effect for SERS detection of thiabendazole in fruit juice. Food Chem. 2022; 370: 131276. [DOI] [PubMed] [Google Scholar]
- 175. Li X, Liu H, Chen Y, et al. Construction of reusable PMMA–Ag/g‐C3N4/Ag hybrid substrates with plasmonic‐enhanced intrinsic Raman signals for quantitative SERS detection and green degradation. ACS Sustainable Chem Eng. 2021; 9(38): 12885‐12898. [Google Scholar]
- 176. Park E, Jin S, Park Y, Guo S, Chang H, Jung YM. Trapping analytes into dynamic hot spots using Tyramine‐medicated crosslinking chemistry for designing versatile sensor. J Colloid Interface Sci. 2022; 607(Pt 1): 782‐790. [DOI] [PubMed] [Google Scholar]
- 177. Li H, Mehedi Hassan M, Wang J, et al. Investigation of nonlinear relationship of surface enhanced Raman scattering signal for robust prediction of thiabendazole in apple. Food Chem. 2021; 339: 127843. [DOI] [PubMed] [Google Scholar]
- 178. Hussain A, Pu H, Hu B, Sun DW. Au@Ag‐TGANPs based SERS for facile screening of thiabendazole and ferbam in liquid milk. Spectrochim Acta A Mol Biomol Spectrosc. 2021; 245: 118908. [DOI] [PubMed] [Google Scholar]
- 179. Sun L, Yu Z, Alsammarraie FK, et al. Development of cellulose nanofiber‐based substrates for rapid detection of ferbam in kale by surface‐enhanced Raman spectroscopy. Food Chem. 2021; 347: 129023. [DOI] [PubMed] [Google Scholar]
- 180. Duan J, Qiu Z, Li L, Feng L, Huang L, Xiao G. Inkjet printed silver nanoparticles on hydrophobic papers for efficient detection of thiram. Spectrochim Acta A Mol Biomol Spectrosc. 2020; 243: 118811. [DOI] [PubMed] [Google Scholar]
- 181. Walia S, Sharma RK, Parmar BS. Isolation and simultaneous LC analysis of thiram and its less toxic transformation product in DS formulation. Bull Environ Contam Toxicol. 2009; 83(3): 363‐368. [DOI] [PubMed] [Google Scholar]
- 182. Albarghouthi N, MacMillan P, Brosseau CL. Optimization of gold nanorod arrays for surface enhanced Raman spectroscopy (SERS) detection of atrazine. Analyst. 2021; 146(6): 2037‐2047. [DOI] [PubMed] [Google Scholar]
- 183. Daoudi K, Gaidi M, Columbus S, Shameer M, Alawadhi H. Hierarchically assembled silver nanoprism‐graphene oxide‐silicon nanowire arrays for ultrasensitive surface enhanced Raman spectroscopy sensing of atrazine. Mater Sci Semicond Process. 2022; 138: 106288. [Google Scholar]
- 184. Rubira RJG, Camacho SA, Constantino CJL, Sanchez‐Cortes S. Increasing the sensitivity of surface‐enhanced Raman scattering detection for s‐triazine pesticides by taking advantage of interactions with soil humic substances. J Raman Spectrosc. 2022;53:40–48. Published online October 4, 2021. [Google Scholar]
- 185. Zanasi G, Rubira RJG, Francioso O, Cañamares MV, Constantino CJL, Sanchez‐Cortes S. Sensing atrazine herbicide degradation products through their interactions with humic substances by surface‐enhanced Raman scattering. Chemosensors. 2021; 9(6): 148. [Google Scholar]
- 186. Mikac L, Kovačević E, Ukić Š, et al. Detection of multi‐class pesticide residues with surface‐enhanced Raman spectroscopy. Spectrochim Acta A Mol Biomol Spectrosc. 2021; 252: 119478. [DOI] [PubMed] [Google Scholar]
- 187. Lin MH, Sun L, Kong F, Lin M. Rapid detection of paraquat residues in green tea using surface‐enhanced Raman spectroscopy (SERS) coupled with gold nanostars. Food Control. 2021; 130: 108280. [Google Scholar]
- 188. Yao L, Dai P, Ouyang L, Zhu L. A sensitive and reproducible SERS sensor based on natural lotus leaf for paraquat detection. Microchem J. 2021; 160: 105728. [Google Scholar]
- 189. Koh EH, Moon JY, Kim SY, et al. A cyclodextrin‐decorated plasmonic gold nanosatellite substrate for selective detection of bipyridylium pesticides. Analyst. 2021; 146(1): 305‐314. [DOI] [PubMed] [Google Scholar]
- 190. Dao DQ, Ngo TC, Le TTH, et al. SERS chemical enhancement of 2,4,5‐trichlorophenoxyacetic acid adsorbed on silver substrate. J Phys Chem A. 2021; 125(39): 8529‐8541. [DOI] [PubMed] [Google Scholar]
- 191. Zhu A, Xu Y, Ali S, Ouyang Q, Chen Q. Au@Ag nanoflowers based SERS coupled chemometric algorithms for determination of organochlorine pesticides in milk. LWT. 2021; 150: 111978. [Google Scholar]
- 192. Hassan MM, Zareef M, Jiao T, et al. Signal optimized rough silver nanoparticle for rapid SERS sensing of pesticide residues in tea. Food Chem. 2021; 338: 127796. [DOI] [PubMed] [Google Scholar]
- 193. Wang TJ, Barveen NR, Liu ZY, Chen CH, Chou MH. Transparent, flexible plasmonic Ag NP/PMMA substrates using chemically patterned ferroelectric crystals for detecting pesticides on curved surfaces. ACS Appl Mater Interfaces. 2021; 13(29): 34910‐34922. [DOI] [PubMed] [Google Scholar]
- 194. Sulaiman D, Alwan AM, Hamoudi WK. Enhanced pesticides’ limit of detection using bimetallic alloys nanoparticles. J Mater Sci: Mater Electron. 2021; 32(14): 18689‐18698. [Google Scholar]
- 195. Sanaeifar A, Li X, He Y, Huang Z, Zhan Z. A data fusion approach on confocal Raman microspectroscopy and electronic nose for quantitative evaluation of pesticide residue in tea. Biosystems Eng. 2021; 210: 206‐222. [Google Scholar]
- 196. Jiang L, Mehedi Hassan M, Jiao T, Li H, Chen Q. Rapid detection of chlorpyrifos residue in rice using surface‐enhanced Raman scattering coupled with chemometric algorithm. Spectrochim Acta A Mol Biomol Spectrosc. 2021; 261: 119996. [DOI] [PubMed] [Google Scholar]
- 197. Zhu X, Li W, Wu R, et al. Rapid detection of chlorpyrifos pesticide residue in tea using surface‐enhanced Raman spectroscopy combined with chemometrics. Spectrochim Acta A Mol Biomol Spectrosc. 2021; 250: 119366. [DOI] [PubMed] [Google Scholar]
- 198. Ma X, Xie J, Wang Z, Zhang Y. Transparent and flexible AuNSs/PDMS‐based SERS substrates for in‐situ detection of pesticide residues. Spectrochim Acta A Mol Biomol Spectrosc. 2022; 267(Pt 2): 120542. [DOI] [PubMed] [Google Scholar]
- 199. Zhou L, Peng Y, Zhang N, et al. Size‐tunable gold aerogels: a durable and misfocus‐tolerant 3D substrate for multiplex SERS detection. Adv Opt Mater. 2021; 9(17): 2100352. [Google Scholar]
- 200. Zhu A, Xuan T, Zhai Y, et al. Preparation of magnetic metal organic framework: a magnetically induced improvement effect for detection of parathion‐methyl. Sens Actuators B Chem. 2021; 339: 129909. [Google Scholar]
- 201. Li R, Chen M, Yang H, et al. Simultaneous in situ extraction and self‐assembly of plasmonic colloidal gold superparticles for SERS detection of organochlorine pesticides in water. Anal Chem. 2021; 93(10): 4657‐4665. [DOI] [PubMed] [Google Scholar]
- 202. Al‐Syadi AM, Faisal M, Harraz FA, Jalalah M, Alsaiari M. Immersion‐plated palladium nanoparticles onto meso‐porous silicon layer as novel SERS substrate for sensitive detection of imidacloprid pesticide. Sci Rep. 2021; 11(1): 9174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Sun Y, Zhang N, Han C, et al. Competitive immunosensor for sensitive and optical anti‐interference detection of imidacloprid by surface‐enhanced Raman scattering. Food Chem. 2021; 358: 129898. [DOI] [PubMed] [Google Scholar]
- 204. Hermsen A, Lamers D, Schoettl J, Mayer C, Jaeger M. In‐field detection method for imidacloprid by surface enhanced Raman spectroscopy. Toxicol Environ Chem: 1‐19. 10.1080/02772248.2021.1991929. Published online October 10, 2021. [DOI] [Google Scholar]
- 205. Kao WY, Yu SH, Mai FD, Tsai HY, Liu YC. Innovative strategy on improved surface‐enhanced Raman scattering sensing by using plasmon‐activated water dissolving analyte. J Electroanal Chem. 2021; 891: 115287. [Google Scholar]
- 206. Gao ZF, Li YX, Dong LM, et al. Photothermal‐induced partial Leidenfrost superhydrophobic surface as ultrasensitive surface‐enhanced Raman scattering platform for the detection of neonicotinoid insecticides. Sens Actuators B Chem. 2021; 348: 130728. [Google Scholar]
- 207. Wang Q, Zhao Y, Bu T, et al. Semi‐sacrificial template growth‐assisted self‐supporting MOF chip: a versatile and high‐performance SERS sensor for food contaminants monitoring. Sens Actuators B Chem. 2022; 352: 131025. [Google Scholar]
- 208. Cheshari EC, Ren X, Li X. Core–shell Ag‐dual template molecularly imprinted composite for detection of carbamate pesticide residues. Chem Pap. 2021; 75(7): 3679‐3693. [Google Scholar]
- 209. Wang P, Sun Y, Wang L, Li X, Liu M, Li G. Facile detection and quantification of acetamiprid using a portable Raman spectrometer combined with self‐assembled gold nanoparticle array. Chemosensors. 2021; 9(11): 327. [Google Scholar]
- 210. Liu J, Siavash Moakhar R, Mahshid S, Vasefi F, Wachsmann‐Hogiu S. Multimodal electrochemical and SERS platform for chlorfenapyr detection. Appl Surf Sci. 2021; 566: 150617. [Google Scholar]
- 211. Augustine S, Sooraj KP, Pachchigar V, Murali Krishna C, Ranjan M. SERS based detection of Dichlorvos pesticide using silver nanoparticles arrays: influence of array wavelength/amplitude. Appl Surf Sci. 2021; 544: 148878. [Google Scholar]
- 212. Yu H, Wang M, Cao J, et al. Determination of Dichlorvos in pears by Surface‐Enhanced Raman Scattering (SERS) with catalysis by platinum coated gold nanoparticles. Anal Lett. 1‐11. 10.1080/00032719.2021.1938104. Published online July 5, 2021. [DOI] [Google Scholar]
- 213. Thi Dang L, Le Nguyen H, Van Pham H, Nguyen MTT. Shell thickness‐controlled synthesis of Au@Ag core‐shell nanorods structure for contaminants sensing by SERS. Nanotechnology. 2021; 33(7): 075704. [DOI] [PubMed] [Google Scholar]
- 214. Pan TT, Guo MT, Guo W, Lu P, Hu DY. A sensitive SERS method for determination of pymetrozine in apple and cabbage based on an easily prepared substrate. Foods. 2021; 10(8):1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Jabbar AA, Alwan AM. Efficient detecting of TNT molecules using palladium nanoparticles/cross shape pores like structure porous silicon. Vibrational Spectroscop. 2019; 103: 102933. [Google Scholar]
- 216. Wu DY, Li JF, Ren B, Tian ZQ. Electrochemical surface‐enhanced Raman spectroscopy of nanostructures. Chem Soc Rev. 2008; 37(5): 1025‐1041. [DOI] [PubMed] [Google Scholar]
- 217. Yang Y, Peng Y, Lin C, et al. Human ACE2‐functionalized gold “virus‐trap” nanostructures for accurate capture of SARS‐CoV‐2 and single‐virus SERS detection. Nanomicro Lett. 2021; 13: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Zhang D, Zhang X, Ma R, et al. Ultra‐fast and onsite interrogation of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS‐CoV‐2) in waters via surface enhanced Raman scattering (SERS). Water Res. 2021; 200: 117243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Palermo G, Rippa M, Conti Y, et al. Plasmonic metasurfaces based on pyramidal nanoholes for high‐efficiency SERS biosensing. ACS Appl Mater Interfaces. 2021; 13(36): 43715‐43725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Zhang C, Huang L, Pu H, Sun DW. Magnetic surface‐enhanced Raman scattering (MagSERS) biosensors for microbial food safety: fundamentals and applications. Trends Food Sci Technol. 2021; 113: 366‐381. [Google Scholar]
- 221. Pan C, Zhu B, Yu C. A dual immunological Raman‐enabled crosschecking test (DIRECT) for detection of bacteria in low moisture food. Biosensors. 2020; 10(12):200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Yin L, Man S, Ye S, Liu G, Ma L. CRISPR‐Cas based virus detection: recent advances and perspectives. Biosens Bioelectron. 2021; 193: 113541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Pang Y, Li Q, Wang C, Zhen S, Sun Z, Xiao R. CRISPR‐cas12a mediated SERS lateral flow assay for amplification‐free detection of double‐stranded DNA and single‐base mutation. Chem Eng J. 2022;429:132109. [Google Scholar]
- 224. Choi JH, Shin M, Yang L, et al. Clustered regularly interspaced short palindromic repeats‐mediated amplification‐free detection of viral DNAs using surface‐enhanced raman spectroscopy‐active nanoarray. ACS Nano. 10.1021/acsnano.1c03975. Published online August 9, 2021. [DOI] [PubMed] [Google Scholar]
- 225. Liu J, Chen J, Wu D, et al. CRISPR‐/Cas12a‐mediated liposome‐amplified strategy for the Surface‐Enhanced Raman Scattering and naked‐eye detection of nucleic acid and application to food authenticity screening. Anal Chem. 2021; 93(29): 10167‐10174. [DOI] [PubMed] [Google Scholar]
- 226. Liang J, Teng P, Xiao W, et al. Application of the amplification‐free SERS‐based CRISPR/Cas12a platform in the identification of SARS‐CoV‐2 from clinical samples. J Nanobiotechnology. 2021; 19(1): 273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227. Shinoda H, Taguchi Y, Nakagawa R, et al. Amplification‐free RNA detection with CRISPR‐Cas13. Commun Biol. 2021; 4(1): 476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Fozouni P, Son S, Díaz de León Derby M, et al. Amplification‐free detection of SARS‐CoV‐2 with CRISPR‐Cas13a and mobile phone microscopy. Cell. 2021; 184(2): 323‐333.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. Yang E, Li D, Yin P, et al. A novel surface‐enhanced Raman scattering (SERS) strategy for ultrasensitive detection of bacteria based on three‐dimensional (3D) DNA walker. Biosensor Bioelectron. 2021; 172: 112758. [DOI] [PubMed] [Google Scholar]
- 230. Yang X, Tang Y, Mason SD, Chen J, Li F. Enzyme‐powered three‐dimensional DNA nanomachine for DNA walking, payload release, and biosensing. ACS Nano. 2016; 10(2): 2324‐2330. [DOI] [PubMed] [Google Scholar]
- 231. Yeğenoğlu Akçinar H, Aslim B, Torul H, et al. Immunomagnetic separation and Listeriamonocytogenes detection with surface‐enhanced Raman scattering. Turk J Med Sci. 2020; 50(4): 1157‐1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232. Shi L, Xu L, Xiao R, et al. Rapid, Quantitative, high‐sensitive detection of O157:h7 by gold‐shell silica‐core nanospheres‐based Surface‐Enhanced Raman Scattering lateral flow immunoassay. Front Microbiol. 2020; 11: 596005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. Tadesse LF, Ho CS, Chen DH, et al. Plasmonic and electrostatic interactions enable uniformly enhanced liquid bacterial Surface‐Enhanced Raman Scattering (SERS). Nano Lett. 2020; 20(10): 7655‐7661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234. Prakash O, Sil S, Verma T, Umapathy S. Direct detection of bacteria using positively charged Ag/Au bimetallic nanoparticles: a label‐free surface‐enhanced Raman scattering study coupled with multivariate analysis. J Phys Chem C Nanomater Interfaces. 2020; 124(1): 861‐869. [Google Scholar]
- 235. Andrei CC, Moraillon A, Larquet E, et al. SERS characterization of aggregated and isolated bacteria deposited on silver‐based substrates. Anal Bioanal Chem. 2021; 413(5): 1417‐1428. [DOI] [PubMed] [Google Scholar]
- 236. Ahmad W, Wang J, Li H, Jiao T, Chen Q. Trends in the bacterial recognition patterns used in surface enhanced Raman spectroscopy. Trends Analyt Chem. 2021; 142: 116310. [Google Scholar]
- 237. Wu Z, Pu H, Sun DW. Fingerprinting and tagging detection of mycotoxins in agri‐food products by surface‐enhanced Raman spectroscopy: principles and recent applications. Trends Food Sci Technol. 2021; 110: 393‐404. [Google Scholar]
- 238. Zhang W, Tang S, Jin Y, et al. Multiplex SERS‐based lateral flow immunosensor for the detection of major mycotoxins in maize utilizing dual Raman labels and triple test lines. J Hazard Mater. 2020; 393: 122348. [DOI] [PubMed] [Google Scholar]
- 239. Chen R, Li S, Sun Y, et al. Surface‐enhanced Raman spectroscopy aptasensor for simultaneous determination of ochratoxin A and zearalenone using Au@Ag core‐shell nanoparticles and gold nanorods. Mikrochim Acta. 2021; 188(8): 281. [DOI] [PubMed] [Google Scholar]
- 240. Tegegne WA, Mekonnen ML, Beyene AB, Su WN, Hwang BJ. Sensitive and reliable detection of deoxynivalenol mycotoxin in pig feed by surface enhanced Raman spectroscopy on silver nanocubes@polydopamine substrate. Spectrochim Acta A Mol Biomol Spectrosc. 2020; 229: 117940. [DOI] [PubMed] [Google Scholar]
- 241. Hahm E, Kim YH, Pham XH, Jun BH. Highly reproducible Surface‐Enhanced Raman Scattering Detection of alternariol using silver‐embedded silica nanoparticles. Sensors. 2020; 20(12):3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242. Guo Z, Chen P, Wang M, et al. Rapid enrichment detection of patulin and alternariol in apple using surface enhanced Raman spectroscopy with coffee‐ring effect. Lebenson Wiss Technol. 2021; 152(112333): 112333. [Google Scholar]
- 243. Yan Q, Cai M, Zhou L, et al. Using an RNA aptamer probe for super‐resolution imaging of native EGFR. Nanoscale Adv. 2019; 1(1): 291‐298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244. Lin X, Li C, He C, et al. Upconversion nanoparticles assembled with gold nanourchins as luminescence and Surface‐Enhanced Raman Scattering dual‐mode aptasensors for detection of ochratoxin A. ACS Applied Nano Materials. 2021; 4(8): 8231‐8240. [Google Scholar]
- 245. He H, Sun DW, Pu H, Huang L. Bridging Fe3O4@Au nanoflowers and Au@Ag nanospheres with aptamer for ultrasensitive SERS detection of aflatoxin B1. Food Chemistry. 2020; 324: 126832. [DOI] [PubMed] [Google Scholar]
- 246. He D, Wu Z, Cui B, Xu E. Aptamer and gold nanorod‐based fumonisin B1 assay using both fluorometry and SERS. Mikrochim Acta. 2020; 187(4): 215. [DOI] [PubMed] [Google Scholar]
- 247. Li J, Wang W, Zhang H, et al. Programmable DNA tweezer‐actuated SERS probe for the sensitive detection of AFB1. Analytical Chemistry. 2020; 92(7): 4900‐4907. [DOI] [PubMed] [Google Scholar]
- 248. Nam W, Zhao Y, Song J, et al. Plasmonic electronic Raman scattering as internal standard for spatial and temporal calibration in quantitative Surface‐Enhanced Raman Spectroscopy. J Phys Chem Lett. 2020; 11(22): 9543‐9551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249. Tang S, Qi T, Ansah PD, et al. Single‐drop microextraction. Trends Analyt Chem. 2018; 108: 306‐313. [Google Scholar]
- 250. Santos EB, Valsecchi C, Gonçalves JLS, Ávila LF, Menezes JW. Coupling single‐drop microextraction with SERS: a demonstration using p‐MBA on gold nanohole array substrate. Sensors. 2019; 19(20):4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251. Kim DJ, Park SG, Kim DH, Kim SH. SERS‐active‐charged microgels for size‐ and charge‐selective molecular analysis of complex biological samples. Small. 2018; 14(40): e1802520. [DOI] [PubMed] [Google Scholar]
- 252. Zhang Q, Li D, Cao X, Gu H, Deng W. Self‐assembled microgels arrays for electrostatic concentration and Surface‐Enhanced Raman Spectroscopy detection of charged pesticides in seawater. Anal Chem. 2019; 91(17): 11192‐11199. [DOI] [PubMed] [Google Scholar]
- 253. Soleymani AR, Rafigh SM, Hekmati M. Green synthesis of RGO/Ag: as evidence for the production of uniform mono‐dispersed nanospheres using microfluidization. Appl Surf Sci. 2020; 518(146264): 146264. [Google Scholar]
- 254. Chen H, Liu H, Chen Y, Li X, Gu C, Jiang T. Surface‐enhanced Raman scattering detection of additive by a reusable ZnO/GO/Ag hybrid substrate. Mater Chem Phys. 2021; 273(125083): 125083. [Google Scholar]
- 255. Wang Q, Xu Z, Zhao Y, et al. Bio‐inspired self‐cleaning carbon cloth based on flower‐like Ag nanoparticles and leaf‐like MOF: a high‐performance and reusable substrate for SERS detection of azo dyes in soft drinks. Sens Actuators B Chem. 2021; 329(129080): 129080. [Google Scholar]
- 256. Peng X, Li D, Li Y, Xing H, Deng W. Plasmonic tunable Ag‐coated gold nanorod arrays as reusable SERS substrates for multiplexed antibiotics detection. J Mater Chem B Mater Biol Med. 2021; 9(4): 1123‐1130. [DOI] [PubMed] [Google Scholar]
- 257. Barveen NR, Wang TJ, Chang YH, Yuan‐Liu Z. Ultrasensitive and reusable SERS probe for the detection of synthetic dyes in food industry through hybrid flower‐shaped ZnO@Ag nanostructures. J Alloys Compd. 2021; 861(157952): 157952. [Google Scholar]
- 258. Naqvi TK, Bajpai A, Bharati MSS, et al. Ultra‐sensitive reusable SERS sensor for multiple hazardous materials detection on single platform. J Hazard Mater. 2021; 407: 124353. [DOI] [PubMed] [Google Scholar]
- 259. Barveen RN, Wang TJ, Chang YH. Photochemical synthesis of Ag/Au/AgCl heterostructure from Ag nanowires as a reusable SERS substrate for ultrasensitive detection of analgesics and antibiotics. Chem Eng J. 2021; 423(130191): 130191. [Google Scholar]
- 260. Wu T, Zheng H, Kou Y, et al. Self‐sustainable and recyclable ternary Au@CuO‐Ag nanocomposites: application in ultrasensitive SERS detection and highly efficient photocatalysis of organic dyes under visible light. Microsyst Nanoeng. 2021; 7: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261. Li C, Huang Y, Li X, et al. Towards practical and sustainable SERS: a review of recent developments in the construction of multifunctional enhancing substrates. J Mater Chem. 2021; 9(35): 11517‐11552. [Google Scholar]
- 262. Tadesse LF, Safir F, Ho CS, et al. Toward rapid infectious disease diagnosis with advances in surface‐enhanced Raman spectroscopy. J Chem Phys. 2020; 152(24): 240902. [DOI] [PubMed] [Google Scholar]
- 263. Rojalin T, Antonio D, Kulkarni A, Carney RP. Machine learning‐assisted sampling of SERS substrates improves data collection efficiency. Appl Spectrosc. 37028211034543. 10.1177/00037028211034543. Published online August 3, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264. Uysal Ciloglu F, Saridag AM, Kilic IH, Tokmakci M, Kahraman M, Aydin O. Identification of methicillin‐resistant Staphylococcus aureus bacteria using surface‐enhanced Raman spectroscopy and machine learning techniques. Analyst. 2020; 145(23): 7559‐7570. [DOI] [PubMed] [Google Scholar]
- 265. Sun D, Cao F, Wang H, et al. SERS hydrogel pellets for highly repeatable and reliable detections of significant small biomolecules in complex samples without pretreatment. Sens Actuators B Chem. 2021; 327: 128943. [Google Scholar]
- 266. Zhou X, Zhao X, Gu S, Xie F, Wang X, Tang Z. Sulfur doped MoO 2 hollow nanospheres as a highly sensitive SERS substrate for multiple detections of organic pollutants. Anal Methods. 2021; 13(24): 2679‐2687. [DOI] [PubMed] [Google Scholar]
- 267. Lin Y, Zheng M, Zhao X, et al. Quantitative detection of crystal violet using a surface‐enhanced Raman scattering based on a flower‐like HAp/Ag nanocomposite. Anal Methods. 2021; 13(36): 4143‐4149. [DOI] [PubMed] [Google Scholar]
- 268. Han S, Zhang C, Chen Z, Sha X, Hasi W. Rapid detection of Dezocine in biological fluids based on SERS technology. Anal Sci. 2021; 37(2): 315‐320. [DOI] [PubMed] [Google Scholar]
- 269. Zhou ZM, Zheng H, Liu T, et al. Improving SERS sensitivity toward trace sulfonamides: the key role of trade‐off interfacial interactions among the target molecules, anions, and cations on the SERS active surface. Anal Chem. 2021; 93(24): 8603‐8612. [DOI] [PubMed] [Google Scholar]
- 270. Payne TD, Klawa SJ, Jian T, et al. Catching COVID: engineering peptide‐modified Surface‐Enhanced Raman Spectroscopy Sensors for SARS‐CoV‐2. ACS Sens. 2021; 6(9): 3436‐3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271. Sun J, Li W, Zhu X, et al. A novel multiplex mycotoxin Surface‐Enhanced Raman Spectroscopy immunoassay using functional gold nanotags on a silica photonic crystal microsphere biochip. J Agric Food Chem. 2021; 69(38): 11494‐11501. [DOI] [PubMed] [Google Scholar]
- 272. Rojas LM, Qu Y, He L. A facile solvent extraction method facilitating surface‐enhanced Raman spectroscopic detection of ochratoxin A in wine and wheat. Talanta. 2021; 224: 121792. [DOI] [PubMed] [Google Scholar]
- 273. Chen R, Sun Y, Huo B, et al. Development of Fe3O4@Au nanoparticles coupled to Au@Ag core‐shell nanoparticles for the sensitive detection of zearalenone. Analytica Chimica Acta. 2021; 1180: 338888. [DOI] [PubMed] [Google Scholar]
- 274. Tang J, Zhang Q, Zhou J, Fang H, Yang H, Wang F. Investigation of pesticide residue removal effect of gelatinized starch using surface‐enhanced Raman scattering mapping. Food Chem. 2021; 365: 130448. [DOI] [PubMed] [Google Scholar]
- 275. Chen Q, Qin L, Shi C, Kang SZ, Li X. A stable and plug‐and‐play aluminium/titanium dioxide/metal‐organic framework/silver composite sheet for sensitive Raman detection and photocatalytic removal of 4‐aminothiophenol. Chemosphere. 2021; 282: 131000. [DOI] [PubMed] [Google Scholar]
- 276. Shi C, Qin L, Wu S, Kang SZ, Li X. Highly sensitive SERS detection and photocatalytic degradation of 4‐aminothiophenol by a cost‐effective cobalt metal–organic framework‐based sandwich‐like sheet. Chem Eng J. 2021; 422: 129970. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing not applicable—no new data generated.


