Abstract
Programmable gene editing tools have transformed the life sciences and have shown potential for the treatment of genetic disease. Among the CRISPR–Cas technologies that can currently make targeted DNA changes in mammalian cells, prime editors offer an unusual combination of versatility, specificity and precision. Prime editors do not require double-strand DNA breaks and can make virtually any substitution, small insertion and small deletion within the DNA of living cells. Prime editing minimally requires a programmable nickase fused to a polymerase enzyme, and an extended guide RNA that both specifies the target site and templates the desired genome edit. In this Review, we summarize prime editing strategies to generate programmed genomic changes, highlight their limitations and recent developments that circumvent these bottlenecks, and discuss applications and future directions.
Graphical Abstract
In this Review, Chen and Liu discuss the latest developments in prime editing systems, including improvements to their editing efficiency and capabilities, as well as diverse emerging applications in research and preclinical therapeutic studies.
Introduction
Technologies that can make desired sequence changes at specified sites in the DNA of living cells have revolutionized the biomedical sciences, driven innovation in biotechnology, and shown clinical promise for the therapeutic correction of genetic disorders. An ideal gene editing technology can convert any targeted DNA to any other chosen sequence with high yield, minimal undesired byproducts at the target locus, and minimal unintended changes to off-target genomic loci. Developing gene editing tools with high efficiency, versatility, product purity and sequence specificity has therefore been a longstanding goal of the life sciences. The landmark discovery of RNA-programmable CRISPR systems1–4 has led to the creation of three classes of technologies that can edit the genomes of mammalian cells at a broad range of target sites and in a wide variety of cell types and organisms: CRISPR-associated (Cas) nucleases, base editors, and prime editors. These technologies differ extensively in their capabilities and limitations, which determine their optimal usage for precision genome manipulation.
Cas nucleases such as Cas9 or Cas12 stimulate target DNA modification by making a double-strand DNA break (DSB) at a target sequence specified by a guide RNA (gRNA)5,6 (Fig. 1a). As nucleases do not inherently possess the ability to directly alter the sequence of DNA, cellular pathways, such as non-homologous end joining (NHEJ) or microhomology-mediated end joining (MMEJ), repair this DSB intermediate to yield a mixture of insertion and deletion (indel) outcomes7,8. As the majority of these indel outcomes cause frameshifts in coding sequence targets8, Cas nucleases are well-suited for gene disruption. However, although these indels can occasionally coincide with desired editing outcomes9–12, the sequence of indels cannot be specified by the researcher or precisely controlled.
Figure 1 |. Precision genome editing in mammalian cells.
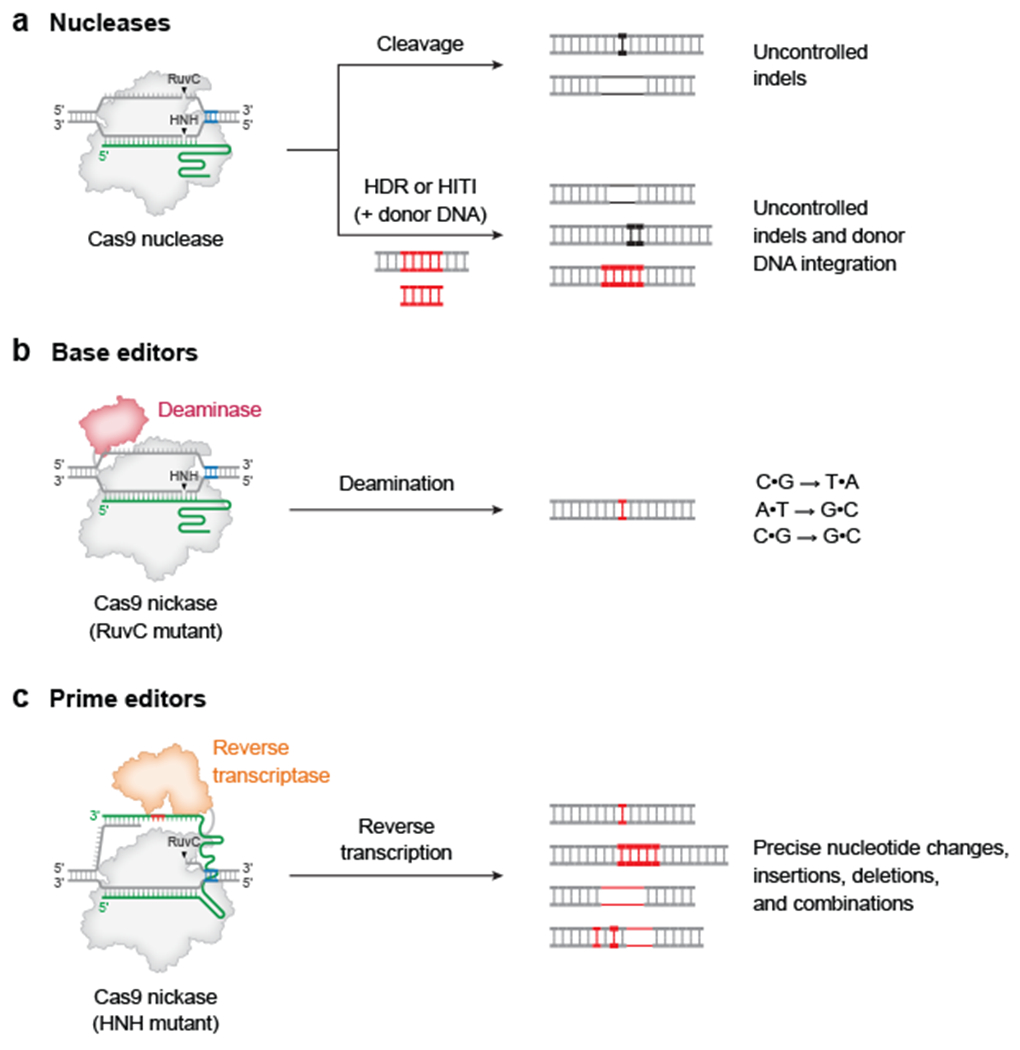
a | Cas nucleases can induce target DNA disrupt via formation of small insertions or deletions (indels), or DNA integration typically accompanied by substantial frequencies of undesired indel byproducts. b | Base editing can mediate C•G-to-T•A, C•G-to-G•C, and A•T-to-G•C conversions with few indel outcomes. Base editors canonically use a Cas9 nickase that only cuts the complementary strand. c | Prime editing can program any type of precise nucleotide substitutions, as well as insertions or deletions of up to hundreds of bases. Prime editors canonically use a Cas9 nickase that only cuts the non-complementary strand. Red DNA represents precisely edited sequence, and black DNA represents undesired outcomes. Blue DNA bases show the position of the protospacer-adjacent motif (PAM) required for Cas9 targeting. RuvC and HNH represent nuclease domains of Cas9. HDR, homology-directed repair; HITI, homology-independent targeted integration
To make specified DNA changes, nucleases can be co-delivered with an exogenous donor DNA template that contains the desired edit flanked by sequence homologous to the genomic target site13,14 (Fig. 1a). Following DSB generation, cellular homology-directed repair (HDR) can then recombine the DNA template into the DSB site. Although this approach can in principle make almost any type of edit (e.g. point mutations, insertions or deletions), HDR is only active in mitotic cells and is typically outcompeted by NHEJ for processing of the DSB15–17. These factors result in frequent indel byproducts that compromise the purity of the precisely corrected editing outcome. Alternatively, in an approach called homology-independent targeted integration (HITI), DNA donors that lack homologous sequence can be inserted at a DSB site through NHEJ, but the orientation and number of insertions cannot be controlled, and undesired indel outcomes typically outnumber the precise intended edit18 (Fig. 1a). Moreover, DSBs generated by nucleases can cause large deletions, chromosomal translocations, chromothripsis [G] , retrotransposon insertion and activation of p53 that can enrich oncogenic cells19–26. Taken together, the prodigious challenges and undesired consequences of using nucleases for precision editing have inspired the development of other technologies that can mediate precise gene correction without the creation of DSBs.
Base editing is one such technology. Base editors can make C•G-to-T•A, A•T-to-G•C, and, in some cases, C•G-to-G•C point mutations without directly generating DSBs or requiring DNA donors (Fig. 1b). Base editors consist of a programmable DNA-binding protein, such as a catalytically impaired Cas nuclease27–29 or a transcription activator-like effector (TALE) repeat array30–32, fused to a deaminase enzyme that converts one base to a different base. For CRISPR base editors, a guide RNA targets the base editor to bind a matching sequence within genomic DNA. The Cas protein domain of the base editor displaces single-stranded DNA (ssDNA) at the target site, triggering deamination by the tethered ssDNA-specific deaminase enzyme. Cytosine base editors (CBEs) and adenine base editors (ABEs) contain deaminases that catalyze C•G-to-T•A and A•T-to-G•C changes, respectively27–31. C•G-to-G•C base editors (CGBEs) are similar to CBEs, but stimulate replacement of the deaminated cytosine base with guanine, albeit with lower typical efficiencies and product purities compared to CBEs and ABEs33–36. Compared to Cas nucleases, base editors exhibit substantially greater efficiency with few indel byproducts, and show far fewer undesired consequences of DSBs than nucleases in side-by-side comparisons19,21,23–26,37,38.
Because base editors deaminate within a small window of 4–5 nucleotides (nt) canonically, C or A nucleotides very close to the target C or A can also undergo conversion, resulting in ‘bystander editing’. The application of base editors to make changes in the coding sequence of genes usually results in only synonymous mutations within a typical base editing activity window39 due to the frequency of transition mutations being silent in the genetic code. Nevertheless, undesired base editing of bystander nucleotides can be challenging to avoid in some cases. Base editing activity is also restricted by the targeting scope of the Cas domain, which requires the presence of a protospacer adjacent motif (PAM) sequence at a specific distance range (typically 15±2 nt) from the target nucleotide. Moreover, some base editors can induce off-target mutations in DNA and RNA40–42. Although engineering efforts have mitigated many of these drawbacks43–51, current base editors can only make six of the 12 possible types of point mutations, leaving many other classes of possible DNA edits, such as insertions, deletions and most transversions, beyond the reach of base editing.
Motivated by the need for a precise and highly versatile gene editing technology, prime editing enables all types of DNA substitutions, small insertions, and small deletions to be installed at targeted sites in the genomes of living cells without directly forming DSBs52 (Fig. 1c). Prime editing minimally requires a prime editor (PE) protein, which is typically a fusion of a nickase Cas9 [G] (nCas9) and a reverse transcriptase (RT), along with a prime editing guide RNA (pegRNA), which specifies the target site for the edit and contains a programmable RNA template for the desired DNA sequence change. To mediate editing, PEs nick the target site on the genome and extend new DNA sequence from this nick using the pegRNA as a template (Fig. 2). This edited DNA strand is then incorporated into the genome through endogenous cellular processes that can be promoted by also nicking the non-edited strand52.
Figure 2 |. Original prime editing systems.
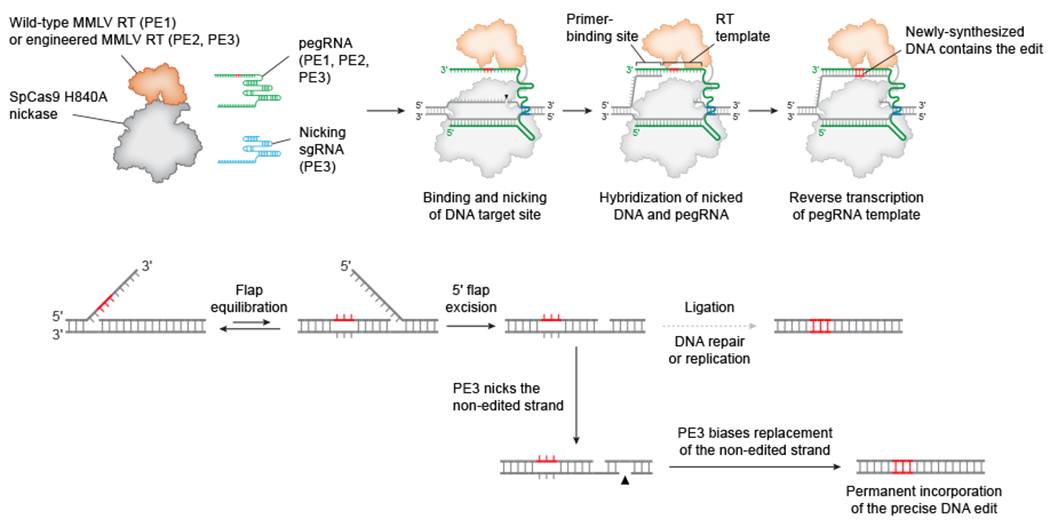
Prime editor 1 (PE1) editing systems use a fusion of Streptococcus pyogenes Cas9 (SpCas9) nickase and Moloney murine leukaemia virus reverse transcriptase (MMLV RT) in complex with a prime editing guide RNA (pegRNA) to nick the non-complementary genomic strand and template the synthesis of an edited DNA flap52. The PE2 prime editor uses an engineered MMLV RT with improved efficiency and stability52. The edited 3ʹ flap is processed by endogenous cellular pathways to permanently copy the edited sequence to the non-edited strand. PE3 editing systems use an additional single-guide RNA (sgRNA) to direct the PE2 enzyme to nick the non-edited strand and stimulate replacement of the non-edited strand, which enhances permanent incorporation of the edited sequence52.
Through this mechanism of directly rewriting target DNA, prime editing offers extraordinarily high versatility, editing purity and DNA target specificity in comparison to base editing and HDR with nucleases. However, the original prime editing systems are primarily limited by inconsistent editing efficiency that can vary widely across different desired edits, target sites and cell types. Recent insights into the cellular determinants of prime editing efficiency have resulted in many advances to prime editing that address this limitation53,54. In addition, prime editing strategies that utilize multiple pegRNAs have expanded the size of possible targeted sequence insertions and deletions55–57. Moreover, combining the use of prime editors with site-specific recombinases have enabled the first gene-sized (>5 kb) RNA-programmed gene insertion or inversion in mammalian cells56,58. In this Review, we describe the capabilities and limitations of prime editing, highlight improvements to its performance and scope, and discuss applications and opportunities for further enhancement.
Development and capabilities of prime editing
Origin of prime editing.
Liu and co-workers created the original prime editor (PE1) by fusing a Streptococcus pyogenes Cas9 (SpCas9) H840A nickase mutant to the wild-type RT from Moloney murine leukaemia virus (MMLV)52 (Fig. 2, Table 1). Prime editing also uses a pegRNA, which is an engineered guide RNA that contains a spacer sequence for directing PE1 to the genomic target site, as well as a critical 3ʹ extension that templates the desired edit sequence. Within a cell, the prime editor–pegRNA complex binds the genomic target site complementary to the pegRNA spacer, forming an R loop [G] in which the displaced ssDNA is nicked by the prime editor (Fig. 2). This nick releases a 3ʹ DNA end that can hybridize to the pegRNA extension and prime reverse transcription of the template region of the pegRNA. DNA polymerization via primer extension of the target DNA strand then generates a 3ʹ DNA flap that contains the edit and homology to the downstream genomic sequence. Next, the 3ʹ flap displaces an adjacent strand of genomic DNA through flap interconversion53,59. Excision of the displaced 5ʹ flap followed by ligation of the remaining nick results in a heteroduplex [G] in which one genomic strand contains the edit. Finally, DNA repair or replication copies the edit to the complementary strand and makes the prime edit permanent.
Table 1 |.
Prime editor and pegRNA architectures
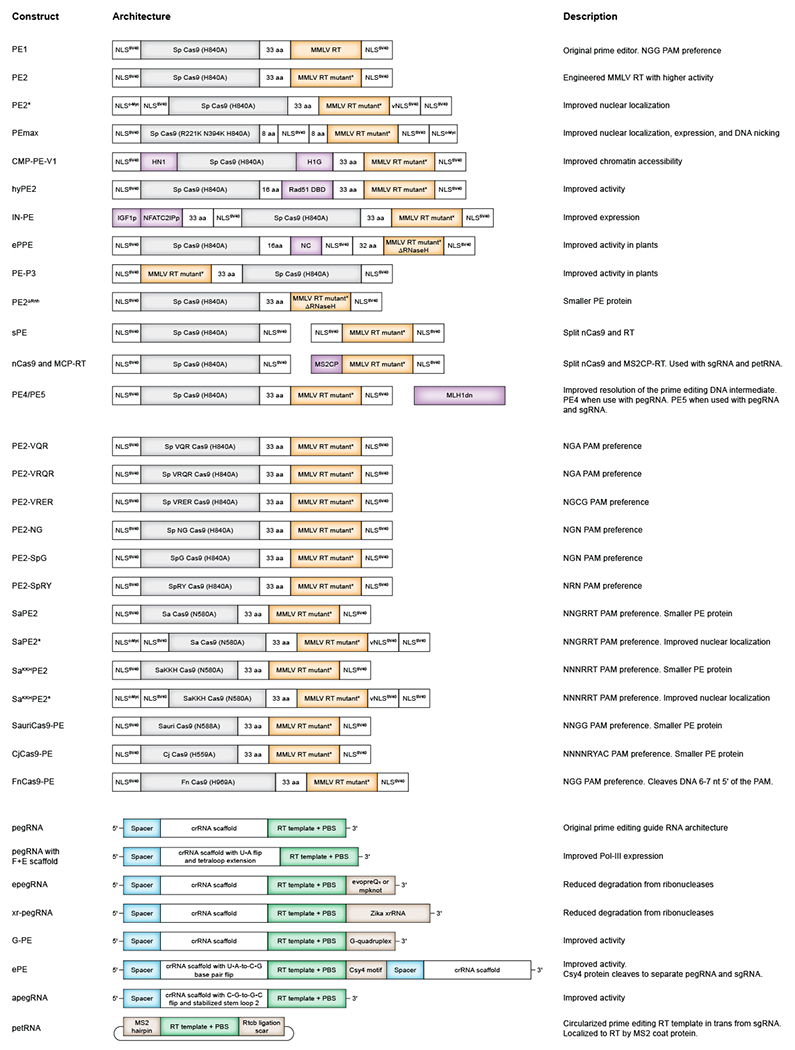
|
PE1 can mediate single-base substitutions, small insertion and small deletion edits in human cells, but the efficiency of this first-generation system is modest; typically <5% of targeted alleles are converted to the desired sequence52. To improve editing efficiency, Liu and co-workers introduced five mutations to MMLV RT known to enhance its thermostability, processivity, and binding to template–primer complexes, yielding a second-generation PE2 editor52 (Fig. 2, Table 1). These additions resulted in a 1.6- to 5.1-fold increase in editing efficiency compared to PE1 in human cells.
Despite its improvement, PE2 still relies on endogenous repair processes to copy the edit from the newly synthesized DNA strand to the complementary strand. To bias cellular replacement of the non-edited strand, Liu and co-workers engineered third-generation prime editing systems (PE3), which use an additional single-guide RNA [G] (sgRNA) to direct the prime editor enzyme to nick the non-edited strand52 (Fig. 2). Nicking the complementary strand 40–100 base pairs (bp) downstream of the pegRNA-directed nick site further improved editing efficiencies by 1.5- to 4.2-fold over PE2 in HEK293T cells. However, PE3 also induced modest indel formation, presumably from the occasional presence of simultaneous nicks on both DNA strands that can lead to DSBs. Although the frequency of the desired edit from PE3 is still substantially higher than the frequency of indels in most cases, these indel byproducts could be virtually eliminated without compromising editing efficiency using the PE3b system, in which the complementary-strand nick is specific for the edited sequence to minimize coincident nicks on both genomic strands52. Together, PE3 and PE3b support highly versatile targeted substitutions, insertions and deletions in multiple human cell types, including in primary, non-dividing mouse cortical neurons52.
Capabilities and advantages of prime editing.
Compared to Cas nucleases and base editors, prime editing offers a unique set of advantages for genome editing. Because the desired edit is programmed within the pegRNA template (Fig. 2), a large diversity of edit types can be installed with high precision52. These include all twelve types of single-base substitutions, multiple base substitutions, small insertions (up to dozens of nucleotides for the original PE3 system), small deletions (up to hundreds of nucleotides for the original PE3 system), and combinations thereof. In addition, prime editing can change bases far (at least 33 bp) from the site of the initial PE-mediated nick52. As a result, prime editing is less restricted than base editing or Cas nuclease-mediated HDR by the requirement of a PAM sequence near the site of the desired edit.
For applications that require high editing precision, prime editing with the PE2 system, which does not nick both DNA strands, can mediate efficient editing and very rarely generates indel byproducts (typically <0.5% of editing outcomes) (Fig. 2). As noted above, PE3 typically achieves higher levels of gene editing by nicking the non-edited strand, which may create DNA intermediates with nicks on both strands. Despite the possibility of forming DSBs, the frequency of PE3 indel byproducts is generally much lower compared to those generated with Cas nucleases. Optimizing the position of the complementary-strand nick or the use of the PE3b strategy can also reduce prime editing indel byproducts52.
Lastly, prime editing rarely induces DNA changes at off-target genomic loci. Unlike Cas nucleases and base editors, prime editing requires three checkpoints of complementary base pairing for productive editing. First, the prime editor binds and nicks the on-target locus that is complementary to the pegRNA spacer (Fig. 2). Second, the primer binding site (PBS) of the pegRNA must hybridize to the nicked 3ʹ end of the target DNA to prime reverse transcription. Finally, the reverse-transcribed 3ʹ DNA flap is required to hybridize with the downstream genomic sequence to enable incorporation into the genome. In addition, in the PE3 system, the nicking sgRNA guides the prime editor to nick the non-edited strand at the sequence complementary to the spacer. The requirement for these multiple base-pairing events results in unusually high prime editing specificity compared to other gene editing technologies. Indeed, prime editing has been found to induce virtually no detected off-target genomic changes in mammalian cells60, mammalian organoids61,62, mouse embryos63–66 and plants67.
Prime editing limitations and improvements
Enhanced prime editor effector proteins.
Successful prime editing occurs through many concerted steps. To engage the target DNA, the prime editor must first be expressed, transported into the nucleus, and complex with a pegRNA (Fig. 3a). Next, the prime editor must efficiently find the target site, nick the exposed DNA strand, and prime reverse transcription of the pegRNA. The resulting edited DNA strand then must be copied onto the non-edited strand through cellular DNA repair processes, making the edit permanent. Efforts to improve prime editing performance have therefore focused on enhancing the efficiency of these steps.
Figure 3 |. Advancements in prime editing systems.
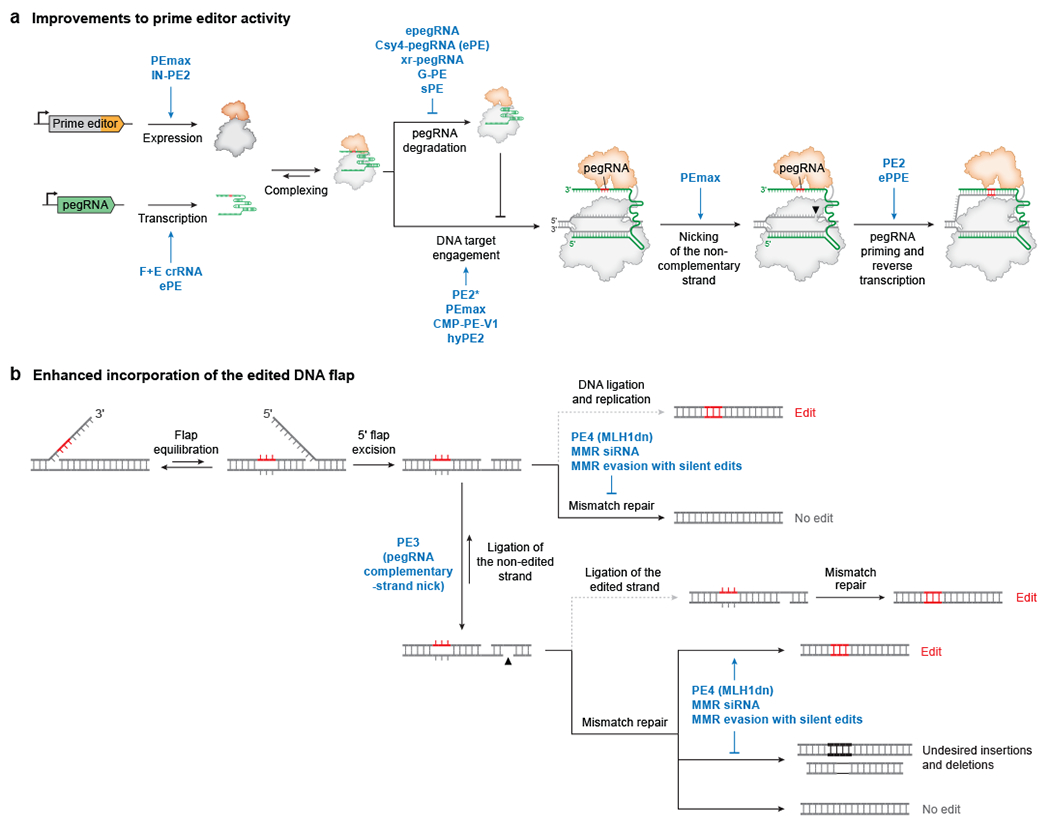
a | Variants of prime editors or prime editing guide RNAs (pegRNAs) that ultimately enhance the creation of the edited 3ʹ DNA flap. PEmax53, IN-PE270, pegRNAs carrying the F+E crRNA scaffold54,78, and ePE pegRNAs77 improve prime editor and pegRNA expression. epegRNAs54, ePE pegRNAs77, xr-pegRNAs74, G-PE pegRNAs75, and sPE pegRNAs76 reduce pegRNA degradation. PE2*68, PEmax53, CMP-PE-V163, and hyPE271 improve localization and/or DNA targeting of the prime editor complex. PEmax53 improves nicking of the genomic DNA strand. ePPE assists pegRNA-primer annealing72. The engineered MMLV RT in PE252 strongly enhances DNA flap synthesis. b | Strategies that promote permanent incorporation of the edited 3ʹ flap into genomic DNA. DNA ligation of the 3ʹ nicked heteroduplex intermediate followed by DNA replication successfully incorporates the desired prime edit, but mismatch repair of this intermediate excises and replaces the edited strand, resulting in no prime editing. Nicking the non-edited DNA strand with PE3 promotes copying of the edit to both genomic strands52. Cellular mismatch repair excises the nicked strand of DNA heteroduplexes, commonly leading to removal of the edit. Transient inhibition of mismatch repair with MLH1dn (PE4 and PE5)53 or with small interfering RNAs (siRNAs)53,59, or mismatch repair evasion through judicious design of the 3ʹ flap53, enhances conversion to the desired prime editing product and reduces undesired formation of small insertions or deletions (indels).
The originally reported PE2 prime editor protein contains N- and C-terminal bipartite SV40 nuclear localization signals (NLS) and a flexible, 32-amino acid peptide linking the SpCas9 H840A and engineered MMLV RT domains52. Xue, Liu, and co-workers independently identified poor nuclear localization as a limiting factor for prime editing efficacy and engineered enhanced PE2 architectures, PE2* and PEmax, by adding additional NLS tags53,68 (Fig. 3a, Table 1). PEmax also contains an engineered MMLV RT domain that has been codon optimized for human expression, as well as SpCas9 mutations previously shown to enhance Cas9 nuclease activity69. In cultured mammalian cells, PEmax improves editing efficiency over PE2 by up to 3-fold on average. Sherwood and co-workers also increased cellular PE2 expression upon tethering peptides to PE270. Using a high-throughput approach called PepSEq to identify candidate peptide fusions, they designed an IN-PE2 editor construct that shows improvement in mouse and human cell types (Fig. 3a, Table 1).
Many groups have optimized the prime editor architecture by inserting DNA- or RNA-interacting proteins between the Cas9 and RT domains. K. Kim, H. Kim, and their respective co-workers engineered PEs with an additional high-mobility group peptide (CMP-PE-V1)63 or hRad51 DNA binding domain (hyPE2)71 within the linker, which shows modest improvements over PE2 in mammalian cell culture (Fig. 3a, Table 1). Gao and co-workers similarly constructed an engineered plant prime editor (ePPE) by removing the RNaseH domain from MMLV RT and inserting a viral nucleocapsid protein between Cas9 and RT domains, which together increases editing by an average 5.8-fold in rice protoplasts72 (Fig. 3a, Table 1). Interestingly, Yang and co-workers found that a flipped MMLV RT–SpCas9 H840A architecture (PE-P3) can outperform PE2 in rice but not in human cells73 (Fig. 3a, Table 1). Future efforts to characterize the structure of the PE–pegRNA–DNA target complex may inform the design of more active prime editor architectures or suggest tailored prime editor variants that are optimized for cell types or target sites of interest.
Improved pegRNAs for prime editing.
The 3ʹ extension of a pegRNA is critical for priming reverse transcription and templating the desired prime edit, but may be exposed to ribonucleases within the PE–pegRNA complex. Degradation of the 3ʹ extension can result in defective PE–pegRNA ribonucleoproteins (RNPs) that compete with competent PE–pegRNAs for binding to target DNA and thus weaken prime editing efficiency (Fig. 3a). Liu and co-workers addressed this issue by adding RNA structural motifs, evopreQ1 or mpknot, to the 3ʹ end of a pegRNA that protect the 3ʹ extension from exonucleases54. These engineered pegRNAs (epegRNAs) enhance editing efficiency by 1.5- to 4-fold compared to canonical pegRNAs, particularly for non-synthetic pegRNAs that do not carry nuclease-resistant chemical modifications (Fig. 3a, Table 1). In similar independent studies, other groups have added a Zikavirus exoribonuclease-resistant RNA motif (xr-pegRNA), a G-quadruplex (G-PE), or a stem-loop aptamer (sPE) to the pegRNA 3ʹ extension and have shown comparable improvements to prime editing in mammalian cell culture74–76 (Fig. 3a, Table 1). Furthermore, Wang and co-workers designed extended pegRNAs containing a 3ʹ Csy4 recognition motif hairpin, which are likely to enhance prime editing through the same mechanism77. In their enhanced prime editing system (ePE), these extended pegRNAs are also fused to a nicking sgRNA and co-delivered with Csy4 nuclease, which can cleave the Csy4 recognition site and separate the two guide RNAs within the target cell (Table 1). Collectively, these studies stress the importance of protecting the pegRNA 3ʹ extension for prime editing. Moreover, because self-complementarity between the pegRNA spacer and extension can reduce editing efficiency77, it is tempting to speculate whether these 3ʹ structured RNA elements may reduce pegRNA self-annealing.
Improving pegRNA transcription or stability can also enhance prime editing efficiency. Multiple groups have reported modestly higher editing with pegRNA CRISPR RNA (crRNA) scaffold sequences that interrupt a putative RNA polymerase III terminator (4 consecutive Us) with a U•A-to-C•G base pair flip or with a U•A-to-A•U flip and extension of the crRNA tetraloop54,77,78 (Fig. 3a, Table 1). Alternatively, expressing pegRNAs with RNA polymerase II also removes the need to avoid poly(U) termination signals79. In addition, Chen and co-workers showed higher prime editing efficiency with apegRNAs that stabilize the secondary structure of the second stem-loop within the pegRNA scaffold80 (Table 1). However, other pegRNA scaffold variants with even lower folding energies did not improve prime editing80, suggesting that the sequence determinants of pegRNA efficiency are complex. Additional exploration of pegRNA scaffold variants may therefore provide an opportunity for further prime editing improvement.
Although linking the spacer and template sequences within a single pegRNA ensures their proximity during prime editing, separating these components can also support prime editing. Sontheimer and co-workers designed a split pegRNA system composed of a normal sgRNA that targets the PE and a linear or circular prime editing template RNA (petRNA) that contains the PBS and RT template81 (Table 1). Importantly, the petRNA also harbours an MS2 hairpin that facilitates its recruitment to the RT by binding a fused MS2 coat protein. Ma and co-workers independently established a similar strategy, called Split pegRNA prime editors (SnPEs)76. Both studies showed that prime editing with a separate sgRNA and template RNA is possible, but usually results in lower efficiency than prime editing with a canonical pegRNA76,81.
Prime editing byproducts and improved DNA repair.
After PEs synthesize an edited 3ʹ DNA flap at a target site, endogenous cellular pathways process this intermediate to either reject the edit, incorporate the edit into the genome, or generate unwanted indel outcomes. PE2 and PE3b editing systems, which do not concurrently nick both DNA strands at the target site, rarely produce indel byproducts52. By contrast, PE3 generates a modest frequency of undesired indels, which commonly include deletions or tandem duplications of the sequence between the pegRNA- and sgRNA-programmed nicks53,82. Notably, pegRNA and sgRNA pairs with inward-facing PAMs (PAM-in orientation) tend to result in deletion byproducts, whereas pairs with outward-facing PAMs (PAM-out orientation) have a propensity to form tandem duplication byproducts53,83.
Using CRISPR interference screens that reveal the impact of many DNA repair gene knockdowns on prime editing outcomes, Liu and co-workers discovered that the DNA mismatch repair (MMR) pathway strongly antagonizes prime editing and stimulates indel byproducts53. Loizou and co-workers also reported that knocking out MMR increases prime editing efficiency and fidelity59. Together, these findings support a model in which MMR reverts the nicked heteroduplex formed when the edited 3ʹ DNA flap anneals to the genome (Fig. 3b). Consistent with this model, MMR only impedes prime edits that form mismatches known to be repaired by MMR, including single-base substitutions and insertions or deletions of less than 13 nt53. G•C-to-C•G edits or substitutions of multiple contiguous bases evade MMR activity and are therefore installed with higher efficiency with PEs.
These insights inspired two approaches to improve prime editing outcomes. First, a dominant negative variant of an MMR protein, MLH1dn, was engineered to transiently inhibit MMR and enhance prime editing efficiency53 (Fig. 3b, Table 1). Co-expression of MLH1dn with prime editing agents yielded fourth- and fifth-generation systems (PE4 = PE2 + MLH1dn, PE5 = PE3 + MLH1dn), which respectively enhance editing by 7-fold over PE2 and 2-fold over PE3. PE5 also reduces indel byproducts by 2-fold compared to PE3, particularly for large indel byproducts. Similarly, MLH1 knockdown with small interfering RNAs (siRNAs) or chemically inducible degradation of MLH1 within cell lines carrying a degron-tagged MLH1 can also transiently inhibit MMR and improve prime editing53,59 (Fig. 3b). Although evidence suggests that transient inhibition of MMR may not strongly impact genome stability53,79, the off-target consequences of DNA repair modulation need to be more thoroughly characterized for clinical applications.
Second, programming additional silent mutations near the intended edit can create a heteroduplex intermediate with more mismatches that evades MMR recognition, increasing prime editing efficiency without globally perturbing MMR activity53 (Fig. 3b). Yang, Chen, and their respective co-workers similarly report that introducing multiple substitutions enhances prime editing efficiency73,80. However, identifying the ideal set of silent mutations to evade MMR requires intuition and experimental optimization, and is not currently predictable. Independent of their effect on DNA repair, installing additional silent mutations may also improve the folding or stability of the pegRNA.
Recent studies have also found that transient inhibition of p53 can improve prime editing efficiency in human embryonic stem cells (hESCs), although the mechanism of this improvement has not been fully characterized79. Notably, p53 inhibition has also been reported to enhance the efficiency of Cas nuclease in human pluripotent stem cells (hPSCs)24. Investigating the basis of this improvement and evaluating the safety of perturbing the p53 DNA damage response are required to assess the potential applicability of this approach.
Prime editing can also generate an additional type of byproduct in which pegRNA scaffold sequence is incorporated into the genome52. Although infrequent, these outcomes arise from reverse transcription of the pegRNA RT template into the crRNA scaffold to generate an extended 3ʹ DNA flap. Liu and co-workers found that recoding the 3ʹ end of the pegRNA scaffold to reduce sequence homology with the genomic target can reduce the incidence of this byproduct53. Alternatively, PE2 or PE4 systems, which do not nick the non-edited strand, generate very few editing outcomes with scaffold sequence incorporation compared to PE3 or PE553.
Chromatin accessibility.
Successful prime editing requires that the genomic target site is sufficiently accessible for engagement by a PE–pegRNA complex. Kim and co-workers showed that a PE variant with chromatin modulating peptides (CMP-PE-V1) can open the local chromatin at the target locus and improve prime editing in mammalian cells63 (Fig. 3a, Table 1). They also found that targeting PE to bind nearby the edit site via truncated, dead sgRNAs also increased chromatin accessibility and editing efficiency. Consistent with these results, chemical inhibition of HDAC7 to promote an open chromatin state was observed to enhance the frequency of prime editing insertions and deletions84. Collectively, these findings suggest that chromatin remodelling can influence prime editing.
Prime editor variants.
Although prime editing does not require a precisely positioned PAM at the target site, editing nucleotides far from the PE target site (>40 nt away) can still pose a challenge. Prime editing a distant base requires the creation of a long pegRNA, synthesis of a long 3ʹ DNA flap containing the edit, and incorporation of this flap into the genome. As a result, it may be difficult to edit DNA far from the NGG PAM sequence preference of canonical PE enzymes.
To expand the range of sequences that can be efficiently prime edited, researchers have constructed PEs that utilize other Cas9 variants with non-NGG PAMs or alternative nuclease cut sites. The discovery and development of these Cas9 variants and orthologues has been reviewed extensively elsewhere6. Kim and co-workers described a set of PEs with engineered PAM variants of SpCas9, including PE2-VQR, PE2-VRQR, PE2-VRER, PE2-NG, PE2-SpG, and PE2-SpRY85 (Table 1). Multiple groups have also developed other PEs that use smaller Cas9 variants, such as SaCas9, SaCas9KKH, SauriCas9, and CjCas9, which each have unique PAM preferences53,68,86 (Table 1). In addition, prime editors show activity with FnCas9, which recognizes an NGG PAM but nicks the target DNA strand farther upstream of the PAM compared to SpCas9, resulting in a shifted window of editable bases87 (Table 1). Circular permutant SpCas9 variants have also been tested in prime editors, but these variants edited less efficiently than the canonical PE2 effector86. Although this collection of PE variants widens the scope of target DNA that can be edited by PEs, future work is needed to improve their activity. PEs that use other Cas9 variants typically exhibit reduced editing efficiency, suggesting that prime editing is highly sensitive to Cas9–pegRNA complexing, DNA target engagement or DNA nicking kinetics. Further underscoring the importance of improving Cas domain compatibility with PEs, using smaller Cas proteins may also facilitate viral packaging or delivery of PEs using methods that prefer or require smaller cargos.
In addition to exchanging the Cas9 domain, PE variants that utilize alternative RTs have also been established. Multiple groups have observed that the RNaseH domain of the engineered MMLV RT in PE2 is dispensable and can be removed without affecting editing levels72,86,88,89. Researchers have also constructed PEs using the CaMV RT from cauliflower mosaic virus, a retron-derived RT from E. coli BL21, the Eubacterium rectale maturase RT (MarathonRT) and GsI-IIC RT (TGIRT III)81,90. Although these RT variants support measurable levels of prime editing, they exhibit substantially weaker activity than the engineered MMLV RT used in PE2. Nevertheless, identifying or engineering novel RTs and RT variants with improved processivity, efficiency and substrate engagement has the potential to enhance prime editing potency. Smaller RT variants may also yield smaller PEs that are easier to deliver.
Predicting pegRNA design and efficiency.
Optimization of pegRNAs is critical for maximizing prime editing, because editing efficiency varies widely across pegRNAs with different PBS and RT template lengths52. Depending on the desired edit, researchers commonly screen pegRNAs with PBS and RT templates of 8–15 nt and 10–74 nt in length, respectively91. Given the expense and time required for these optimizations, multiple groups have developed programs for designing pegRNAs92–98. However, only some of these tools can accurately predict well-performing pegRNA PBS and RT template lengths for a given target site and edit. Gao and co-workers observed that the optimal melting temperature between PBS and genomic DNA strand is 30°C in plants99, but this guideline may not apply to mammalian cells cultured at 37°C. To this end, Kim and co-workers performed pooled pegRNA–target site library screens in human cells to train a deep learning model, DeepPE, that predicts optimal pegRNA PBS and RT template lengths100. Although DeepPE only optimizes pegRNAs for one type of edit (a G•C-to-C•G change of a PAM nucleotide), they identified sequence determinants of prime editing efficiency, such as Cas9 nuclease efficiency and GC content of the PBS. Cheng and co-workers also used the dataset from Kim and co-workers to train their own deep learning model, Easy-Prime95. These computational tools, as well as models for other types of prime edits that may be developed in the future, have the potential to simplify the successful use of prime editing and broaden its applicability.
Newer prime editing variants
Prime editing with paired pegRNAs.
Compared to traditional prime editing systems that use a pegRNA and an optional sgRNA (Fig. 2), using two pegRNAs to simultaneously edit both DNA strands expands the size and type of genomic manipulations possible with prime editing. Many prime editing approaches that use two pegRNAs have been reported, including dual-pegRNA, HOPE, PRIME-Del, PEDAR, twinPE, GRAND, Bi-PE, bi-WT-PE and PETI55–57,99,101–105 (Fig. 4a). These approaches use pegRNA pairs in a PAM-in orientation to template 3ʹ flaps on opposite genomic strands, but differ in the design of the synthesized flaps and in how they cut the target DNA.
Figure 4 |. Prime editing variants.
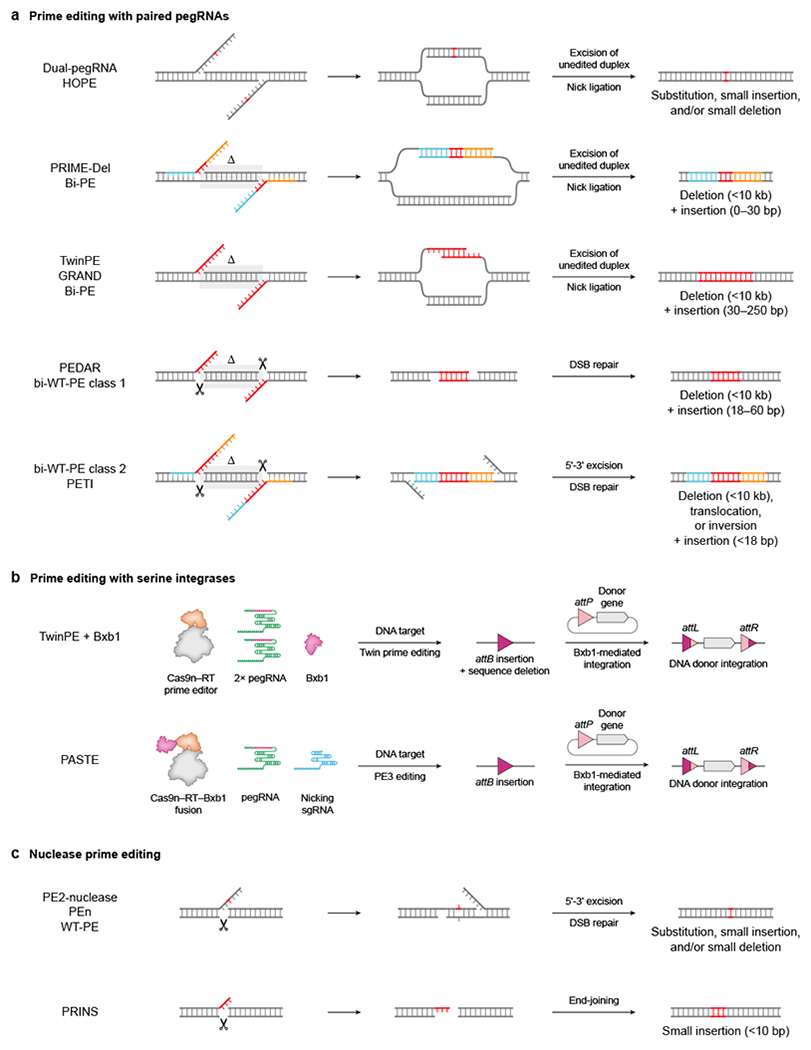
a | Prime editing with paired prime editing guide RNAs (pegRNAs) that template two 3ʹ DNA flaps containing the edit55–57,99,101–105. DNA intermediates formed after prime editor (PE)-mediated nicking and flap extension at the pegRNA target sites are shown on the left. Annealing of the complementary flaps, excision of the original genomic duplex, and ligation of the resulting nicks can efficiently mediate small edits or large insertions and deletions. b | Prime editing with serine integrases enables the targeted insertion of gene-sized (>1 kilobase) DNA segments. Twin prime editing or PASTE first installs an attB site into the genome using prime editing, then uses Bxb1 recombinase to mediate integration of donor DNA into the site56,58. c | Prime editors containing Cas9 nuclease create double-strand breaks (DSBs) with an edited 3ʹ overhang that can be incorporated into the genome but also result in frequent formation of small insertion or deletion (indel) byproducts104,106,107. Red DNA bases represent edited DNA with heterologous sequence. Blue and orange DNA bases represent homologous sequence. ∆ specifies DNA bases deleted by the edit. Scissors denote sites of Cas9-induced DSBs.
The dual-pegRNA and HOPE systems can install substitutions, small insertions and small deletions at modestly higher efficiency and precision than traditional prime editing systems99,101 (Fig. 4a). These approaches use two pegRNAs targeted in close proximity (<50 bp from one another) to generate 3ʹ flaps that each contain the intended edit and some homology with downstream genomic sequence. The resulting 3ʹ flaps can anneal to one another to form a duplex containing the edit. Subsequent 5ʹ excision of the unedited genomic strands and ligation of the nicks then results in permanent incorporation of the edit into both strands of DNA (Fig. 4a). Each edited 3ʹ flap can also independently anneal to the genome and lead to successful editing through a traditional PE2 mechanism.
Large sequence deletions and insertions.
Targeting pegRNA pairs at a much greater distance from one another unlocks the potential to program much larger sequence changes with prime editing. Developed by Shendure and co-workers, PRIME-Del can mediate large deletions (<10 kb) with efficiencies up to 25% at endogenous genomic sites55 (Fig. 4a). PRIME-Del uses pegRNAs targeted far apart that are each homologous to the genomic sequence past the opposing nick. As a result, PRIME-Del deletes the entire sequence between the pegRNA-directed nick sites, which limits its flexibility. The pair of pegRNAs can also optionally encode a short insertion (<30 bp) that is simultaneously installed at the deletion junction. Like dual-pegRNA and HOPE systems, each 3ʹ flap for PRIME-Del can independently facilitate the programmed edit, suggesting that the 3ʹ flaps may not need to be concurrently resolved for editing.
In a similar approach, Liu and co-workers developed twin prime editing (twinPE), which can replace a long DNA sequence between pegRNA-directed nicks with a desired insertion up to 113 bp in length56 (Fig. 4a). Unlike PRIME-Del, twinPE uses pegRNAs that template complementary 3ʹ flaps, each with the desired sequence insertion but without designed homology to the genome. Importantly, because the newly synthesized 3ʹ flaps are typically dissimilar to the target site, they hybridize to each other with minimal competition from genomic sequence. These flaps also do not need to fully anneal to each other and only require at least 20 nt of overlap on their 3ʹ ends. Annealing of these 3ʹ flaps, excision of the unedited homoduplex, and subsequent ligation can then integrate the edited DNA strands into the genome. TwinPE avoids multiple DNA repair steps that can reject the edit for traditional prime editing, including 3ʹ flap annealing to the genome, 5ʹ flap displacement, and heteroduplex resolution. By circumventing these steps, twinPE can direct sequence replacement with efficiency as high as 90% and with few indel byproducts56. In an independent study, Yin and co-workers developed GRAND editing, which can simultaneously insert and delete large DNA sequences in an identical fashion as twinPE102 (Fig. 4a). GRAND editing was demonstrated to efficiently install 250 bp DNA sequences with up to 27% conversion at endogenous loci and in non-dividing cells. In other similar work, Yao and co-workers established Bi-PE approaches which are equivalent to PRIME-Del and twinPE103 (Fig. 4a).
In addition to replacement of DNA sequences, twinPE can also mediate precise deletions of less than 1 kb with greater flexibility than PRIME-Del. Specifically, single-anchor twinPE (SA-twinPE) fixes one deletion junction at a pegRNA nick site, and hybrid-anchor twinPE (HA-twinPE) allows flexibility for both deletion junctions56. The synthesized 3ʹ flaps for SA-twinPE and HA-twinPE are fully complementary to one another, and are also homologous to the genomic sequence around the deletion junctions. However, despite the greater flexibility for programming large deletions, Sa-twinPE and HA-twinPE are typically less efficient than PRIME-Del56.
In an approach similar to twinPE, PEDAR was developed by Xue and co-workers to install large sequence deletions (<10 kb) with a short insertion (<60 bp) at the junction57 (Fig. 4a). Although pegRNA design is identical between the two approaches, PEDAR uses a Cas9 nuclease instead of nCas9in the prime editor effector. Consequently, PEDAR pegRNAs direct Cas9–RT to make two DSBs with complementary 3ʹ overhangs containing only the desired short insertion. MMEJ or single-strand annealing pathways then process this intermediate, leading to the intended sequence replacement outcome. However, although PEDAR can make these desired editing outcomes with up to 27% efficiency, they are typically outnumbered by indel byproducts. In independent studies, Yao, Kim, and their respective co-workers developed bi-directional WT-PE (bi-WT-PE) and PETI systems that use a Cas9 nuclease prime editor similar to PEDAR104,105 (Fig. 4a). They showed that templating flaps with some homology at the opposing nick site (such as in PRIME-Del) can support the targeted deletion of hundreds of bases and targeted inter-chromosomal translocations. PETI can also program targeted inversions using pegRNAs designed with flaps homologous to the inverted sequence. Despite the complexity of targeted genomic rearrangements possible with PEDAR, bi-WT-PE, and PETI, these approaches suffer from low editing purity and possess off-target consequences from generating multiple DSBs.
Collectively, PRIME-Del, twinPE, GRAND, bi-PE, PEDAR, bi-WT-PE, and PETI can precisely delete large stretches of target DNA (<10 kb), or replace them with DNA inserts of 250 bp at high efficiency. In their current form, most of these approaches use pegRNA-directed nick sites to define the junctions for deletion, which may restrict their flexibility for precision edits. Future work is needed to assess these approaches with paired pegRNAs that program shifted deletion windows or with Cas9 PAM variants that increase pegRNA targeting scope.
Gene-sized manipulations with prime editing and integrases.
Despite their versatility, traditional prime editing and paired pegRNA prime editing systems are unable to efficiently install DNA cargo larger than a few hundred base pairs. To enable the insertion of longer sequences, Liu and co-workers combined twinPE with site-specific serine integrases, which catalyze recombination between attB and attP attachment sites in a directional manner56 (Fig. 4b). Researchers have previously used serine integrases, such as Bxb1, to insert plasmids containing an att site into the complementary att site that is pre-installed within the genome. TwinPE can insert 38-bp attB or 50-bp attP sequences at specified target DNA sites in mammalian cells with high efficiency, followed by Bxb1-mediated integration of a 5.6-kb plasmid donor at this landing pad site. These prime editing and integration steps can be performed sequentially or as a single transfection to achieve up to 6% knock-in of DNA cargo56. In addition, installing both attB and attP sites with twinPE enables the deletion or inversion of very long intervening genomic sequence with Bxb1. Liu and co-workers used this approach to precisely invert 40 kb of DNA between the IDS and IDS2 genes with 10% efficiency56.
In an independent study, Gootenberg, Abudayyeh, and co-workers developed PASTE, which similarly uses prime editing to insert landing pad sites for Bxb1-catalyzed knock-in of donor DNAs up to 10 kb in size58 (Fig. 4b). However, the prime editing and integration steps in PASTE are performed by a single protein fusion of Cas9 nickase, RT, and Bxb1 recombinase. PASTE uses traditional PE3 editing (with a pegRNA and nicking sgRNA) to install attB. As a result, although PASTE is less efficient than twinPE at inserting the attB sequence56,58, PASTE does not delete any genomic sequence during the prime editing step and therefore only leaves attL and attR sequence scars from Bxb1 recombination during donor knock-in (Fig. 4b). Taken together, twinPE and PASTE can make targeted insertions, deletions, and inversions of gene-sized (>5 kb) DNA sequences, which substantially expands the capabilities of prime editing and targeted gene integration in mammalian cells.
Prime editors with nucleases that make DSBs.
Traditional prime editing systems such as PE2–PE5 rely on several DNA repair processes to incorporate the edit, including 3ʹ flap annealing, 5ʹ flap displacement, 5ʹ flap excision and heteroduplex resolution. These steps present multiple opportunities to reject the edit. Creating a DSB instead of a DNA nick at the junction of flap synthesis may circumvent these steps and engage other DNA repair pathways to resolve the intermediate towards the intended edit, albeit with the undesired consequence of inducing more indel byproducts from end-joining processes. As discussed above, PEDAR, bi-WT-PE, and PETI use this approach to mediate large sequence deletions, inversions and translocations57,104,105.
To test traditional prime editing with a pegRNA-directed DSB, multiple groups engineered a PE2-nuclease editor by reverting the SpCas9 H840A mutation in PE2106 (Fig. 4c). To prevent the formation of multiple DSBs from pegRNAs and sgRNAs, these PE2-nuclease and WT-PE systems only used pegRNAs. Compared to PE3 with the canonical prime editor nickase, PE2-nuclease and WT-PE installed precise edits at a similar frequency. However, as expected these outcomes were outnumbered by indels that predominantly contain partial tandem duplications from insertion of the reverse-transcribed RT template, consistent with NHEJ of the synthesized 3ʹ overhang at the DSB junction.
Maresca and co-workers also constructed a PE2-nuclease editor, termed PEn, and similarly observed many partial tandem duplication outcomes from editing107 (Fig. 4c). Disrupting NHEJ with a DNA protein kinase (DNA-PK) inhibitor eliminated these partial tandem duplications, and knocking out POLQ, a mediator of alternative end-joining, eliminated all other indel byproducts. To leverage NHEJ for targeting sequence insertions, Maresca and co-workers also developed the PRINS system, which uses PEn and a pegRNA that templates a 3ʹ DNA flap containing only the desired sequence insertion without any homology to the genome (Fig. 4c). In cell culture, PRINS can insert small (<10 bp) sequences with 10–50% efficiency with modest indel byproducts, but imprecise insertions account for roughly half of editing outcomes. In addition, PRINS exhibits the higher off-target propensity of Cas9 nuclease and can only insert sequences at the precise DSB junction programmed by the pegRNA. Despite these limitations, PRINS may offer an editing approach that bypasses DNA repair pathways, such as MMR, that impede traditional prime editing.
Delivery of prime editing components
DNA transfection and viral delivery.
Efficient delivery or expression of PE protein and guide RNAs in the target cell is required for productive prime editing. A common strategy is to deliver DNA encoding prime editing components into target cells and exploit the cells’ endogenous transcription and translation machinery to produce PE RNPs. In many cultured mammalian cell lines, transient lipid-mediated transfection or electroporation of prime editor plasmids can mediate high editing efficiencies, and selection for transfected cells can further improve editing performance52,53,91. Hydrodynamic tail-vein injection of plasmid DNA can also deliver PEs into mouse hepatocytes in vivo57,68,81,88,108. These approaches induce transient expression of prime editing agents in mammalian cells and thereby limit undesired editing at off-target loci. Nevertheless, DNA delivery carries the risk of exogenous DNA integration into the genome.
Some prime editing applications, such as pooled screening, require stable cellular expression of PE from a genomically integrated cassette. PiggyBac transposase and lentiviruses can stochastically insert DNA encoding PEs and pegRNAs into the genome and mediate high editing efficiencies in human cell lines, induced pluripotent stem cells (iPSCs) and mouse cortical neurons52,55,109–111. Similarly, human iPSC lines have been generated with inducible PE2 at the AAVS1 safe harbour locus by HDR-mediated integration82,112,113. Efficient editing (>20%) can be induced in these iPSC lines when transiently transfected with pegRNAs and sgRNAs. Lastly, single integration of pegRNAs with lentiviral transduction has also been demonstrated to enable screening with pooled pegRNA libraries58,71,100,108. Taken together, stable expression of prime editor components may facilitate high-throughput genetic perturbation and library experiments with prime editing.
Viruses offer many advantages for delivery, including efficient transduction and tissue selectivity, making them an attractive modality for delivering PEs in vivo. As discussed above, lentiviruses can genomically integrate PEs, but can induce oncogenesis from insertional mutagenesis and elevate off-target editing from persistent expression114. Transient expression reduces these risks and is therefore favoured for therapeutic applications. Among DNA viruses with a lower risk of integration, adeno-associated virus (AAV), adenovirus and herpes simplex virus (HSV) have been previously used for transient transduction with success114.
Because of their low immunogenicity and broad tropism, AAVs are a promising approach for prime editor delivery. However, AAV can only package DNA cargo up to ~4.7 kb in length, which is substantially smaller than the size of a prime editor and pegRNA (7 kb). As one approach to circumvent this limitation, multiple groups have divided the prime editor into two protein halves that are each fused to half of a trans-splicing intein68,86,88,89,115. After co-infection with AAVs expressing each PE–split-intein half, the full-length prime editor is reconstituted by in trans protein splicing. In a separate approach to bypass the AAV packaging limit, Kim and co-workers encoded each prime editor half on a trans RNA-splicing AAV (tsAAV)108. Homologous recombination between the inverted terminal repeats (ITR) sequences of the tsAAVs then generates the full-length transcript, leading to PE expression. Lastly, Sontheimer and co-workers demonstrated that delivering untethered nCas9 and RT (sPE) in two different AAVs can mediate efficient prime editing81. Encouragingly, these strategies can induce modest editing in the mouse liver and retina, but attaining therapeutic levels of in vivo prime editing with AAV transduction (≥20%) has posed a challenge thus far.
Compared to AAV, adenovirus and HSV stimulate a much stronger inflammatory response in vivo, raising safety concerns with their use in the clinic114. However, adenovirus and HSV have larger packaging capacities (8.5 kb and 40 kb, respectively) and can therefore fit the prime editor and pegRNA into a single vector for transduction. In addition, adenovirus can efficiently transduce most tissue types whereas HSV possesses strong tropism for neurons. Given these advantages over AAV, Schwank and co-workers found that adenoviral delivery of a single PE construct outperforms dual AAV delivery and can mediate up to 60% editing in the mouse liver86. Gonçalves and co-workers also demonstrated high levels of prime editing in cell culture with high-capacity adenoviral vectors, which are devoid of viral coding elements and have a 30 kb packaging limit116. Lastly, Berger and co-workers have also delivered PEs, pegRNAs, and nicking sgRNAs in a single baculovirus vector to perform prime editing with many pegRNAs in mammalian cell culture117. Innate immune responses to baculovirus have thus far limited the efficiency and potency of this approach in vivo. Although several strategies show promise, optimizing viral vectors for transducing PEs in vivo will be critical for enabling therapeutic prime editing in human patients.
mRNA delivery.
Compared to DNA transfection and viral transduction, delivery of PE-encoding messenger RNA (mRNA) eliminates the possibility of DNA recombination into the genome and can reduce off-target editing by narrowing the duration of PE expression. Co-electroporation of PE mRNA and chemically modified or in vitro-transcribed guide RNAs has been shown to mediate efficient prime editing in cultured cell lines, primary human T cells and human stem cells, in most cases with greater efficacy than with plasmid transfection53,54,81,91,113,118. In addition, enhanced PE4 and PE5 editing systems are particularly suited for delivery by mRNA, which can limit the potential mutagenic consequences of the MMR inhibitor, MLH1dn53 (Fig. 3b). Multiple groups also demonstrated that microinjection of PE mRNA can stimulate prime editing in mouse embryos63–65, but may generate a high level of indel byproducts when using PE3 in the absence of MMR inhibition119. RNA delivery of PE is therefore an effective strategy for editing cell culture, embryos, and hematopoietic cells ex vivo. Lipid nanoparticles (LNPs) are also commonly used to encapsulate and deliver mRNAs encoding genome editing agents into mice and humans in vivo120, but their use to deliver prime editors has not yet been reported.
RNP delivery.
Delivery of a PE–pegRNA RNP can shorten the duration of exposure of target cells to PE agents, minimizing off-target editing. Moreover, endogenous transcription and translation machinery in the target cell is not required to produce PEs when they are delivered by RNP. In the first demonstration of prime editing with RNPs, Yeh and co-workers purified PE2 protein complexed with pegRNAs and microinjected these RNPs into zebrafish embryos, which yielded modest editing78. These PE RNPs were also electroporated into cultured cell lines and primary human T cells, but the resulting prime editing efficiencies were substantially lower than what has been previously observed from plasmid or mRNA delivery53. Others have also tested PE RNPs in human iPSCs and observed similarly low editing efficiencies81,113. Thus, despite the theoretical advantages offered by RNP delivery, current PE RNP delivery methods lag in efficiency compared to DNA and mRNA delivery.
Applications of prime editing
Study and treatment of disease.
The biomedical research community has shown strong interest in using prime editing to model and potentially correct genetic mutations that cause human illnesses. Approximately 90% of human pathogenic genetic variants are single-base mutations or insertions and deletions fewer than a dozen base pairs121, which are types of DNA changes that are well within the capabilities of prime editing systems. In cultured cell lines, PEs have been shown to directly correct the 4-base insertion in HEXA that causes Tay–Sachs disease, the HBB E6V mutation responsible for sickle cell disease, and a single-base deletion that leads to CDKL5 deficiency disorder52,53. PEs have also shown the ability to fix mutations associated with DGAT1 deficiency, Wilson’s disease, and cystic fibrosis in human organoid models61,62.
As a direct consequence of its versatility, prime editing can also make precise genetic manipulations that indirectly ameliorate or confer protection from disease. For example, Olson and co-workers used PEs to insert 2 bp to restore the DMD reading frame in a human iPSC model for Duchenne muscular dystrophy that contains an exon 51 deletion122. Liu and co-workers have analogously used twinPE to completely excise this exon, which can partially rescue DMD function56. Prime editing has also been shown to rescue full-length SMN expression in a human iPSC model of spinal muscular atrophy by precisely deleting the intronic splicing silencer within SMN123. Multiple groups have also applied prime editing to install genetic alleles protective for prion disease, HIV infection, and cardiovascular disease52,53,88,89. These examples highlight that the therapeutic scope of prime editing extends beyond the direct correction of pathogenic mutations.
Growing evidence has emerged that many human illnesses, such as coronary artery disease, are polygenic diseases [G]124. Whereas most genome editing tools lack the flexibility to simultaneously program multiple precise DNA changes, prime editing can make several desired edits using tandem arrays of pegRNAs111,117,125. In addition, because PE3 and twinPE systems can edit a stretch of DNA at least 34 bp and 64 bp long52,56, respectively, a single pegRNA or pegRNA pair could be used to correct multiple different variants within a mutational hotspot of a gene. The same PE treatment therefore has the potential to cure heterogeneous patient populations with different mutations.
Early demonstrations of in vivo prime editing have shown promise for correcting pathogenic mutations in animals. Multiple groups have rescued disease phenotypes in post-natal mice by targeting the liver and eye with PEs. Kim, Sontheimer, and their respective co-workers corrected a Fah transversion mutation within a mouse model of tyrosinemia type I using AAV delivery and hydrodynamic tail-vein injection of PEs81,108. These treatments rescued weight loss in mice, despite a modest 11.5% editing in the liver from AAV and 1.3% editing from hydrodynamic injection. Also targeting the liver, Schwank and co-workers used PE adenoviruses to repair the causal Pah F263S mutation in a mouse model of phenylketonuria86. Correction of the mutation with 11% efficiency was sufficient to strongly reduce toxic buildup of phenylalanine in the blood. Finally, Kim and co-workers injected PE AAVs into mouse retina and retinal pigment epithelium to correct Leber congenital amaurosis, a genetic eye disease108. These prime editor injections reversed the causal C•G-to-T•A mutation in RPE65 with 6.4% efficiency and improved visual function in these mice. Although these demonstrations illustrate the potential of prime editing for correcting genetic disorders in human patients, delivery into other organs such as the heart, muscle, lungs and central nervous system, remains a major challenge for therapeutic prime editing applications. Innovating delivery technologies and incorporating recent improvements to PE protein and pegRNA components described above will be critical for maximizing the potency of prime editing in animals.
Generating animal models.
Engineering the DNA of model organisms is useful for elucidating the function of genes and their encoded biomolecules in complex systems. To generate animal models with a desired genotype, genome editing is typically performed in early embryos or germ cells to minimize the chance of mosaicism [G]. Prime editing exhibits substantially higher editing purity than HDR with CRISPR–Cas nucleases and greater edit flexibility than base editing, making it well-suited for creating transgenic organisms with precise genomic changes.
Multiple groups have demonstrated efficient prime editing in mouse embryos by microinjecting zygotes with PE2-encoding mRNA, pegRNAs and sgRNAs63–66. Although these studies have observed editing frequencies as high as 47%, Feng and co-workers reported that PE3 editing in mouse embryos resulted in substantial indel byproducts, whereas PE2 (without a nicking sgRNA) preserves high editing purity119. Other groups have also edited rabbit embryos by PE microinjection and dog embryos through nuclear transfer of PE-corrected fibroblasts126,127.
Prime editing is also capable of creating non-mammalian animal models. Yeh and co-workers made zebrafish models carrying pathogenic kras and tyr mutations with efficiencies up to 6.5% by injection of PE2 RNPs into zebrafish embryos78. Interestingly, both PE2 and PE3 editing in these zebrafish experiments yielded similar frequencies of indel byproducts, and PE3 did not improve editing efficiencies over PE2, which suggests differences in the DNA repair of these embryos compared to cultured cells. In addition, Perrimon and co-workers generated prime edited Drosophila melanogaster by crossing transgenic flies that express guide RNAs and PE2 protein in germ cells128. They also injected fly embryos with plasmids encoding PE components to produce transgenic flies.
Together, these studies demonstrate that prime editing can facilitate the creation of transgenic animal models with high precision, although with modest editing efficiency in some cases. Efficient embryo prime editing would facilitate multiplex editing for modelling polygenic traits in animals, and could allow researchers to directly phenotype edited animals without the need for outcrossing. Using improved prime editing systems discussed above and elucidating the cellular determinants of embryo editing have the potential to enhance prime editing performance and may enable some of these applications.
Functional screens of genetic variants.
Introducing genetic perturbations and assessing their consequences by functional characterization or enrichment is a ubiquitous approach for illuminating biological pathways and mechanisms. To this end, Cas nucleases have been widely used for gene knockout in pooled and arrayed screens129,130. However, for making precise genomic changes, the inefficiency and low product purity of HDR with Cas nucleases in many cell types restricts its applicability for screening. Base editors have shown utility for tiling mutations across genes of interest131–133, but are limited primarily to C•G-to-T•A and A•T-to-G•C transition edits. Due to its precision and versatility, prime editing is therefore well suited for making genetic mutations for functional screens.
In a proof of concept, Cohn and co-workers used prime editing to assess the function of genetic variants of unknown significance within NPC1, which underlies lysosomal storage disorder Niemann–Pick disease type C1134. They installed these variants with PEs, sorted the resulting pool of edited cells by lysosomal expansion, and sequenced the targeted NPC1 loci following enrichment to quantify the phenotypic effect of these variants. In a similar approach, Wei and co-workers mutagenized OsACC1 in rice plants with prime editing and identified herbicide resistant variants after selection135. They used pegRNAs that install random codons (NNN) to induce site-specific mutagenesis, analogous to the ‘Random-PE’ strategy established by Yao and co-workers136. However, in these approaches, the DNA changes made by prime editors were identified by sequencing the edited locus, which limits the region of mutagenesis to a single gene. Instead, sequencing the pegRNA can also identify the induced prime edits, but without imposing restrictions on the location of the edits. Continued advances in prime editing systems will facilitate genetic screens of gene variants or even combinations of gene variants across multi-component pathways.
Tagging endogenous genes.
The ability to tag endogenous genes is broadly useful for selectively detecting or manipulating proteins in cells. The original PE3 prime editing systems can program the insertion of 18-bp 6×His tags or 24-bp FLAG tags (DYKDDDDK) with frequencies as high as 70% in cell culture52,54, which suggests that other tags of comparable sizes, such as HiBiT luciferase137, GFP11138 or cellular localization signals, could be installed. In addition, next-generation prime editing systems have been shown to mediate the installation of larger tags. Using a protein linker sequence compatible with the attR scar from Bxb1 recombination, PASTE was demonstrated to make in-frame GFP fusions to ACTB, SRRM, NOLC1 and LMNB158. While these results establish the ability of prime editing to install protein tags or peptide handles onto endogenous genes of interest, improving the efficiency of longer insertions by PEs will unlock additional gene tagging applications.
Molecular recording and lineage tracing.
Recording cellular activity as permanent changes in the sequence of genomic DNA enables non-destructive tracking of biological events and represents a powerful approach for interrogating complex systems. Typically, cellular events or lineages are coupled to gene editing at a target site that serves as a record that can be decoded by sequencing. In contrast to Cas9 nuclease-mediated end joining and base editing, prime editing can efficiently insert DNA information in a unidirectional manner, making it uniquely capable of encoding the type, duration and order of cellular signals over time.
In the first demonstration of a prime editing molecular recorder, Shendure and co-workers developed a DNA Typewriter system that sequentially writes information into a tandem array of truncated PE target sites139. The target sites are designed such that only one site can be edited at a time. To transcribe activity in a directional manner, short PE-mediated insertions encode the cellular signal being recorded and also complete the protospacer sequence of the adjacent target site, which converts the adjacent site into a viable prime editing target for the next recording event. Shendure and co-workers used this system to encode text messages and reconstruct cellular lineages, displaying the potential of prime editing molecular recorders for biotechnology and basic research.
Prime editing in agriculture.
Gene editing is poised to have a major impact in agriculture. As drought, salinity, nutrient deficiency and bacteria pose a challenge for crop production, engineering plants tolerant to disease, herbicide and growth conditions can dramatically improve crop yields. Towards these aims, precision genome editing in plants can enable the generation of crops with desired properties at much greater speed and ease than traditional plant-breeding techniques140,141. As such, CRISPR–Cas nucleases and base editors have been widely used to study and engineer plants. However, the unique capabilities of prime editing summarized above have motivated the application of prime editing in plants.
In the first demonstrations of prime editing in monocot plants, multiple groups showed that PEs can precisely edit the genomes of rice protoplasts and wheat with modest efficiencies typically less than 10%90,142,143. Later work substantially improved upon these prime editing efficiencies in rice by using a dual pegRNA approach or enhanced PE and pegRNA architectures72,73,99,144,145. Furthermore, multiplex prime editing is possible in rice using tandem arrays of pegRNAs and nicking sgRNAs125. Wei and co-workers also used PEs to make rice mutagenesis libraries and identify plants with herbicide-resistant OsACC1 alleles135, establishing the potential of prime editing for crop engineering. Importantly, Gao and co-workers validated that prime editing in plants is highly specific, with few DNA or RNA off-targets67.
Prime editing has also shown activity in dicotyledon plants, which are more difficult to transform than monocots. Zhu and co-workers found that a PE optimized for plant expression can mediate up to 3% gene conversion in diploid tomatoes146. Nogué and co-workers also demonstrated successful prime editing in the tetraploid potatoes, albeit also with low efficiency147. Prime editing has also been demonstrated in legumes, such as peanuts, chickpeas and cowpeas148. While even modest prime editing efficiencies in plants can be sufficient to generate cultivars with the desired traits following small-scale screening, achieving higher than 10% desired prime editing in plants has so far been a challenge, highlighting a need for future improvement. Inefficient transformation of constructs into plant cells has long represented a major bottleneck for agricultural gene editing and probably limits the potency of prime editing in plants. Additionally, in contrast to editing in mammalian cells, using a complementary-strand nick (PE3) has not been reported to reliably enhance prime editing in plants. A deeper understanding of the cellular factors that influence prime editing outcomes in plants may yield new insights to further expand the scope of prime editing in agriculture.
Conclusions and future perspectives
The ability to manipulate genomes in a precise and programmable manner has had a transformative impact on the life sciences. Compared to other gene editing technologies, prime editing offers much greater versatility, product purity and/or target sequence specificity. The potential of prime editing has driven enormous innovation to elevate its efficiency, expand its capabilities and demonstrate its application for basic research and therapy.
Several remaining challenges for prime editing present opportunities for further development. Current prime editors typically struggle to insert or replace more than ~100 nt of DNA efficiently without the assistance of a recombinase. Although more than 90% of known pathogenic mutations are smaller than 30 bp121, extending the length of possible sequence insertions or recoding with prime editing may enable gene-sized integrations without the requirements of donor DNA or recombinase enzymes. Investigating the cellular determinants of additional types of prime edits, such as insertions, deletions and dual pegRNA approaches, could reveal pathways that can be manipulated for further enhancement. Improving the catalytic activity or DNA target engagement of serine recombinases could also increase the efficiency of large payload integration with twinPE or similar systems. Lastly, the delivery of PEs into target cell types and tissues remains a major constraint for in vivo prime editing. Improving delivery will be undoubtedly critical for realizing the full potential of prime editing for disease modelling and therapeutic correction.
Advancing the capabilities of prime editing may also enable diverse applications that have not yet been fully explored. Highly robust prime editing systems may one day support pooled screens in which genetic perturbations are identified by sequencing singly integrated pegRNAs. Such a screening platform could in theory allow the high-throughput characterization of genetic variants across pathways or the genome. Finally, extending the prime editing system to edit the genomes of mitochondria and chloroplasts, or even to directly edit RNA sequences using analogous strategies, would initiate many new classes of applications.
Acknowledgements
D.R.L. acknowledges support from US National Institutes of Health awards U01 AI142756, RM1 HG009490, R35 GM118062, the Bill and Melinda Gates Foundation, and the Howard Hughes Medical Institute. P.J.C. acknowledges support from a US National Science Foundation (NSF) graduate research fellowship. The authors thank A. Anzalone for helpful comments.
Glossary terms
- Chromothripsis
A process in which tens to thousands of chromosomal rearrangements occur in a single event
- Nickase Cas9
Cas9 that has either its HNH or RuvC nuclease domain catalytically inactivated, resulting in a Cas9 enzyme that can only cut one strand of targeted double-stranded DNA
- R loop
A three-stranded nucleic acid structure that contains a DNA:RNA hybrid and a displaced strand of DNA
- Heteroduplex
Double-stranded DNA in which the sequences of the strands are not perfectly complementary
- Single-guide RNA (sgRNA)
A single guide RNA molecule, composed of a CRISPR RNA (crRNA) fused to its corresponding trans-activating CRISPR RNA (tracrRNA) scaffold sequence, that directs the binding and nuclease activity of Cas9 enzymes
- Polygenic diseases
Diseases that are mediated by numerous genetic variants that each individually contribute small effects
- Mosaicism
A condition in which an animal contains multiple cell lineages with different genotypes
Footnotes
Competing interests
The authors have filed patent applications on gene editing technologies through the Broad Institute of MIT and Harvard. P.J.C. is currently an employee of Prime Medicine. D.R.L. is a consultant and equity owner of Beam Therapeutics, Pairwise Plants, Prime Medicine, Chroma Medicine, and Nvelop Therapeutics, companies that use or deliver genome editing or genome engineering technologies.
References
- 1.Barrangou R. et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712, doi: 10.1126/science.1138140 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Marraffini LA & Sontheimer EJ CRISPR interference limits horizontal gene transfer in Staphylococci by targeting DNA. Science 322, 1843–1845, doi: 10.1126/science.1165771 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deltcheva E. et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471, 602–607, doi: 10.1038/nature09886 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jinek M. et al. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816, doi: 10.1126/science.1225829 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pickar-Oliver A & Gersbach CA The next generation of CRISPR–Cas technologies and applications. Nature Reviews Molecular Cell Biology 20, 490–507, doi: 10.1038/s41580-019-0131-5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anzalone AV, Koblan LW & Liu DR Genome editing with CRISPR–Cas nucleases, base editors, transposases and prime editors. Nature Biotechnology 38, 824–844, doi: 10.1038/s41587-020-0561-9 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Lieber MR The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annual Review of Biochemistry 79, 181–211, doi: 10.1146/annurev.biochem.052308.093131 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Overbeek M. et al. DNA repair profiling reveals nonrandom outcomes at Cas9-mediated breaks. Molecular Cell 63, 633–646, doi: 10.1016/j.molcel.2016.06.037 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Shen MW et al. Predictable and precise template-free CRISPR editing of pathogenic variants. Nature 563, 646–651, doi: 10.1038/s41586-018-0686-x (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allen F. et al. Predicting the mutations generated by repair of Cas9-induced double-strand breaks. Nature Biotechnology 37, 64–72, doi: 10.1038/nbt.4317 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen W. et al. Massively parallel profiling and predictive modeling of the outcomes of CRISPR/Cas9-mediated double-strand break repair. Nucleic Acids Research 47, 7989–8003, doi: 10.1093/nar/gkz487 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iyer S. et al. Precise therapeutic gene correction by a simple nuclease-induced double-stranded break. Nature 568, 561–565, doi: 10.1038/s41586-019-1076-8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rouet P, Smih F & Jasin M Expression of a site-specific endonuclease stimulates homologous recombination in mammalian cells. Proceedings of the National Academy of Sciences 91, 6064–6068, doi: 10.1073/pnas.91.13.6064 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rouet P, Smih F & Jasin M Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Molecular and Cellular Biology 14, 8096–8106, doi: 10.1128/mcb.14.12.8096-8106.1994 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heyer W-D, Ehmsen KT & Liu J Regulation of homologous recombination in eukaryotes. Annual Review of Genetics 44, 113–139, doi: 10.1146/annurev-genet-051710-150955 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Komor AC, Badran AH & Liu DR CRISPR-Based Technologies for the Manipulation of Eukaryotic Genomes. Cell 168, 20–36, doi: 10.1016/j.cell.2016.10.044 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paquet D et al. Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature 533, 125–129, doi: 10.1038/nature17664 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Suzuki K et al. In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature 540, 144–149, doi: 10.1038/nature20565 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kosicki M, Tomberg K & Bradley A Repair of double-strand breaks induced by CRISPR–Cas9 leads to large deletions and complex rearrangements. Nature Biotechnology 36, 765–771, doi: 10.1038/nbt.4192 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cullot G et al. CRISPR-Cas9 genome editing induces megabase-scale chromosomal truncations. Nat Commun 10, 1136, doi: 10.1038/s41467-019-09006-2 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alanis-Lobato G et al. Frequent loss of heterozygosity in CRISPR-Cas9 edited early human embryos. Proceedings of the National Academy of Sciences 118, e2004832117, doi:doi: 10.1073/pnas.2004832117 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leibowitz ML et al. Chromothripsis as an on-target consequence of CRISPR–Cas9 genome editing. Nature Genetics 53, 895–905, doi: 10.1038/s41588-021-00838-7 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tao J, Wang Q, Mendez-Dorantes C, Burns KH & Chiarle R Frequency and mechanisms of LINE-1 retrotransposon insertions at CRISPR/Cas9 sites. Nature Communications 13, doi: 10.1038/s41467-022-31322-3 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ihry RJ et al. p53 inhibits CRISPR–Cas9 engineering in human pluripotent stem cells. Nature Medicine 24, 939–946, doi: 10.1038/s41591-018-0050-6 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Haapaniemi E, Botla S, Persson J, Schmierer B & Taipale J CRISPR–Cas9 genome editing induces a p53-mediated DNA damage response. Nature Medicine 24, 927–930, doi: 10.1038/s41591-018-0049-z (2018). [DOI] [PubMed] [Google Scholar]
- 26.Enache OM et al. Cas9 activates the p53 pathway and selects for p53-inactivating mutations. Nature Genetics 52, 662–668, doi: 10.1038/s41588-020-0623-4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Komor AC, Kim YB, Packer MS, Zuris JA & Liu DR Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424, doi: 10.1038/nature17946 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nishida K et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science 353, aaf8729, doi:doi: 10.1126/science.aaf8729 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Gaudelli NM et al. Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature 551, 464–471, doi: 10.1038/nature24644 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mok BY et al. A bacterial cytidine deaminase toxin enables CRISPR-free mitochondrial base editing. Nature 583, 631–637, doi: 10.1038/s41586-020-2477-4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cho S-I et al. Targeted A-to-G base editing in human mitochondrial DNA with programmable deaminases. Cell 185, 1764–1776.e1712, doi: 10.1016/j.cell.2022.03.039 (2022). [DOI] [PubMed] [Google Scholar]
- 32.Yang L et al. Engineering and optimising deaminase fusions for genome editing. Nature Communications 7, 13330, doi: 10.1038/ncomms13330 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kurt IC et al. CRISPR C-to-G base editors for inducing targeted DNA transversions in human cells. Nature Biotechnology 39, 41–46, doi: 10.1038/s41587-020-0609-x (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao D et al. Glycosylase base editors enable C-to-A and C-to-G base changes. Nature Biotechnology 39, 35–40, doi: 10.1038/s41587-020-0592-2 (2021). [DOI] [PubMed] [Google Scholar]
- 35.Chen L et al. Programmable C:G to G:C genome editing with CRISPR-Cas9-directed base excision repair proteins. Nature Communications 12, 1384, doi: 10.1038/s41467-021-21559-9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koblan LW et al. Efficient C•G-to-G•C base editors developed using CRISPRi screens, target-library analysis, and machine learning. Nature Biotechnology 39, 1414–1425, doi: 10.1038/s41587-021-00938-z (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ferrari S et al. Efficient gene editing of human long-term hematopoietic stem cells validated by clonal tracking. Nature Biotechnology 38, 1298–1308, doi: 10.1038/s41587-020-0551-y (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song Y et al. Large-fragment deletions induced by Cas9 cleavage while not in the BEs System. Molecular Therapy - Nucleic Acids 21, 523–526, doi: 10.1016/j.omtn.2020.06.019 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rees HA & Liu DR Base editing: precision chemistry on the genome and transcriptome of living cells. Nature Reviews Genetics 19, 770–788, doi: 10.1038/s41576-018-0059-1 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jin S et al. Cytosine, but not adenine, base editors induce genome-wide off-target mutations in rice. Science 364, 292–295, doi:doi: 10.1126/science.aaw7166 (2019). [DOI] [PubMed] [Google Scholar]
- 41.Zuo E et al. Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science 364, 289–292, doi:doi: 10.1126/science.aav9973 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grünewald J et al. Transcriptome-wide off-target RNA editing induced by CRISPR-guided DNA base editors. Nature 569, 433–437, doi: 10.1038/s41586-019-1161-z (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gehrke JM et al. An APOBEC3A-Cas9 base editor with minimized bystander and off-target activities. Nature Biotechnology 36, 977–982, doi: 10.1038/nbt.4199 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim YB et al. Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions. Nat Biotechnol 35, 371–376, doi: 10.1038/nbt.3803 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nishimasu H et al. Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science 361, 1259–1262, doi:doi: 10.1126/science.aas9129 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang TP et al. Circularly permuted and PAM-modified Cas9 variants broaden the targeting scope of base editors. Nature Biotechnology 37, 626–631, doi: 10.1038/s41587-019-0134-y (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walton RT, Christie KA, Whittaker MN & Kleinstiver BP Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science 368, 290–296, doi: 10.1126/science.aba8853 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miller SM et al. Continuous evolution of SpCas9 variants compatible with non-G PAMs. Nature Biotechnology 38, 471–481, doi: 10.1038/s41587-020-0412-8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Doman JL, Raguram A, Newby GA & Liu DR Evaluation and minimization of Cas9-independent off-target DNA editing by cytosine base editors. Nature Biotechnology 38, 620–628, doi: 10.1038/s41587-020-0414-6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rees HA, Wilson C, Doman JL & Liu DR Analysis and minimization of cellular RNA editing by DNA adenine base editors. Science Advances 5, eaax5717, doi:doi: 10.1126/sciadv.aax5717 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grünewald J et al. CRISPR DNA base editors with reduced RNA off-target and self-editing activities. Nature Biotechnology 37, 1041–1048, doi: 10.1038/s41587-019-0236-6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anzalone AV et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157, doi: 10.1038/s41586-019-1711-4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen PJ et al. Enhanced prime editing systems by manipulating cellular determinants of editing outcomes. Cell 184, 5635–5652.e5629, doi: 10.1016/j.cell.2021.09.018 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nelson JW et al. Engineered pegRNAs improve prime editing efficiency. Nature Biotechnology 40, 402–410, doi: 10.1038/s41587-021-01039-7 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Choi J et al. Precise genomic deletions using paired prime editing. Nature Biotechnology 40, 218–226, doi: 10.1038/s41587-021-01025-z (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Anzalone AV et al. Programmable deletion, replacement, integration and inversion of large DNA sequences with twin prime editing. Nature Biotechnology 40, 731–740, doi: 10.1038/s41587-021-01133-w (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jiang T, Zhang X-O, Weng Z & Xue W Deletion and replacement of long genomic sequences using prime editing. Nature Biotechnology 40, 227–234 doi: 10.1038/s41587-021-01026-y (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ioannidi EI et al. Drag-and-drop genome insertion without DNA cleavage with CRISPR-directed integrases. bioRxiv, 2021.2011.2001.466786, doi: 10.1101/2021.11.01.466786 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ferreira Da Silva J et al. Prime editing efficiency and fidelity are enhanced in the absence of mismatch repair. Nature Communications 13, doi: 10.1038/s41467-022-28442-1 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gao R et al. Genomic and Transcriptomic Analyses of Prime Editing Guide RNA–Independent Off-Target Effects by Prime Editors. The CRISPR Journal 5, 276–293, doi: 10.1089/crispr.2021.0080 (2022). [DOI] [PubMed] [Google Scholar]
- 61.Schene IF et al. Prime editing for functional repair in patient-derived disease models. Nature Communications 11, doi: 10.1038/s41467-020-19136-7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Geurts MH et al. Evaluating CRISPR-based prime editing for cancer modeling and CFTR repair in organoids. Life Science Alliance 4, e202000940, doi: 10.26508/lsa.202000940 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Park S-J et al. Targeted mutagenesis in mouse cells and embryos using an enhanced prime editor. Genome biology 22, doi: 10.1186/s13059-021-02389-w (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu Y et al. Efficient generation of mouse models with the prime editing system. Cell Discovery 6, doi: 10.1038/s41421-020-0165-z (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gao P et al. Prime editing in mice reveals the essentiality of a single base in driving tissue-specific gene expression. Genome biology 22, doi: 10.1186/s13059-021-02304-3 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lin J et al. Modeling a cataract disorder in mice with prime editing. Molecular Therapy - Nucleic Acids 25, 494–501, doi: 10.1016/j.omtn.2021.06.020 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jin S et al. Genome-wide specificity of prime editors in plants. Nature Biotechnology 39, 1292–1299, doi: 10.1038/s41587-021-00891-x (2021). [DOI] [PubMed] [Google Scholar]
- 68.Liu P et al. Improved prime editors enable pathogenic allele correction and cancer modelling in adult mice. Nature Communications 12, doi: 10.1038/s41467-021-22295-w (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Spencer JM & Zhang X Deep mutational scanning of S. pyogenes Cas9 reveals important functional domains. Scientific Reports 7, doi: 10.1038/s41598-017-17081-y (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Velimirovic M et al. Peptide fusion improves prime editing efficiency. Nature Communications 13, doi: 10.1038/s41467-022-31270-y (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Song M et al. Generation of a more efficient prime editor 2 by addition of the Rad51 DNA-binding domain. Nature Communications 12, doi: 10.1038/s41467-021-25928-2 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zong Y et al. An engineered prime editor with enhanced editing efficiency in plants. Nature Biotechnology 40, 1394–1402, doi: 10.1038/s41587-022-01254-w (2022). [DOI] [PubMed] [Google Scholar]
- 73.Xu W et al. A design optimized prime editor with expanded scope and capability in plants. Nature Plants 8, 45–52, doi: 10.1038/s41477-021-01043-4 (2022). [DOI] [PubMed] [Google Scholar]
- 74.Zhang G et al. Enhancement of prime editing via xrRNA motif-joined pegRNA. Nature Communications 13, doi: 10.1038/s41467-022-29507-x (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li X et al. Enhancing prime editing efficiency by modified pegRNA with RNA G-quadruplexes. Journal of Molecular Cell Biology 14, doi: 10.1093/jmcb/mjac022 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Feng Y et al. Enhancing prime editing efficiency and flexibility with tethered and split pegRNAs. Protein & Cell, pwac014, doi: 10.1093/procel/pwac014 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu Y et al. Enhancing prime editing by Csy4-mediated processing of pegRNA. Cell Research 31, 1134–1136, doi: 10.1038/s41422-021-00520-x (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Petri K et al. CRISPR prime editing with ribonucleoprotein complexes in zebrafish and primary human cells. Nature Biotechnology 40, 189–193, doi: 10.1038/s41587-021-00901-y (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Huang S et al. Broadening prime editing toolkits using RNA-Pol-II-driven engineered pegRNA. Molecular Therapy 30, 2923–2932, doi: 10.1016/j.ymthe.2022.07.002 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li X et al. Highly efficient prime editing by introducing same-sense mutations in pegRNA or stabilizing its structure. Nature Communications 13, doi: 10.1038/s41467-022-29339-9 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu B et al. A split prime editor with untethered reverse transcriptase and circular RNA template. Nature Biotechnology 40, 1388–1393, doi: 10.1038/s41587-022-01255-9 (2022). [DOI] [PubMed] [Google Scholar]
- 82.Habib O, Habib G, Hwang G-H & Bae S Comprehensive analysis of prime editing outcomes in human embryonic stem cells. Nucleic Acids Research 50, 1187–1197, doi: 10.1093/nar/gkab1295 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bothmer A et al. Characterization of the interplay between DNA repair and CRISPR/Cas9-induced DNA lesions at an endogenous locus. Nature Communications 8, 13905, doi: 10.1038/ncomms13905 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu N et al. HDAC inhibitors improve CRISPR/Cas9 mediated prime editing and base editing. Molecular Therapy - Nucleic Acids 29, 36–46, doi: 10.1016/j.omtn.2022.05.036 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kweon J et al. Engineered prime editors with PAM flexibility. Molecular Therapy 29, 2001–2007, doi: 10.1016/j.ymthe.2021.02.022 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Böck D et al. In vivo prime editing of a metabolic liver disease in mice. Science Translational Medicine 14, eabl9238, doi:doi: 10.1126/scitranslmed.abl9238 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Oh Y et al. Expansion of the prime editing modality with Cas9 from Francisella novicida. Genome biology 23, doi: 10.1186/s13059-022-02644-8 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zheng C et al. A flexible split prime editor using truncated reverse transcriptase improves dual-AAV delivery in mouse liver. Molecular Therapy 30, 1343–1351, doi: 10.1016/j.ymthe.2022.01.005 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gao Z et al. A truncated reverse transcriptase enhances prime editing by split AAV vectors. Molecular Therapy 30, 2942–2951, doi: 10.1016/j.ymthe.2022.07.001 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lin Q et al. Prime genome editing in rice and wheat. Nature Biotechnology 38, 582–585, doi: 10.1038/s41587-020-0455-x (2020). [DOI] [PubMed] [Google Scholar]
- 91.Doman JL, Sousa AA, Randolph PB, Chen PJ & Liu DR Designing and executing prime editing experiments in mammalian cells. Nature Protocols, doi: 10.1038/s41596-022-00724-4 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bhagwat AM et al. multicrispr: gRNA design for prime editing and parallel targeting of thousands of targets. Life Science Alliance 3, e202000757, doi: 10.26508/lsa.202000757 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chow RD, Chen JS, Shen J & Chen S A web tool for the design of prime-editing guide RNAs. Nature Biomedical Engineering 5, 190–194, doi: 10.1038/s41551-020-00622-8 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hwang G-H et al. PE-Designer and PE-Analyzer: web-based design and analysis tools for CRISPR prime editing. Nucleic Acids Research 49, W499–W504, doi: 10.1093/nar/gkab319 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Li Y, Chen J, Tsai SQ & Cheng Y Easy-Prime: a machine learning–based prime editor design tool. Genome biology 22, doi: 10.1186/s13059-021-02458-0 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Standage-Beier K, Tekel SJ, Brafman DA & Wang X Prime editing guide RNA design automation using PINE-CONE. ACS Synthetic Biology 10, 422–427, doi: 10.1021/acssynbio.0c00445 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Siegner SM, Karasu ME, Schröder MS, Kontarakis Z & Corn JE PnB Designer: a web application to design prime and base editor guide RNAs for animals and plants. BMC Bioinformatics 22, doi: 10.1186/s12859-021-04034-6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hsu JY et al. PrimeDesign software for rapid and simplified design of prime editing guide RNAs. Nature Communications 12, 1034, doi: 10.1038/s41467-021-21337-7 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lin Q et al. High-efficiency prime editing with optimized, paired pegRNAs in plants. Nature Biotechnology 39, 923–927, doi: 10.1038/s41587-021-00868-w (2021). [DOI] [PubMed] [Google Scholar]
- 100.Kim HK et al. Predicting the efficiency of prime editing guide RNAs in human cells. Nature Biotechnology 39, 198–206, doi: 10.1038/s41587-020-0677-y (2021). [DOI] [PubMed] [Google Scholar]
- 101.Zhuang Y et al. Increasing the efficiency and precision of prime editing with guide RNA pairs. Nature Chemical Biology 18, 29–37, doi: 10.1038/s41589-021-00889-1 (2021). [DOI] [PubMed] [Google Scholar]
- 102.Wang J. et al. Efficient targeted insertion of large DNA fragments without DNA donors. Nature Methods 19, 331–340, doi: 10.1038/s41592-022-01399-1 (2022). [DOI] [PubMed] [Google Scholar]
- 103.Tao R et al. Bi-PE: bi-directional priming improves CRISPR/Cas9 prime editing in mammalian cells. Nucleic Acids Research 50, 6423–6434, doi: 10.1093/nar/gkac506 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tao R et al. WT-PE: Prime editing with nuclease wild-type Cas9 enables versatile large-scale genome editing. Signal Transduction and Targeted Therapy 7, doi: 10.1038/s41392-022-00936-w (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kweon J et al. Targeted genomic translocations and inversions generated using a paired prime editing strategy. Molecular Therapy, doi: 10.1016/j.ymthe.2022.09.008 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Adikusuma F et al. Optimized nickase- and nuclease-based prime editing in human and mouse cells. Nucleic Acids Research 49, 10785–10795, doi: 10.1093/nar/gkab792 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Peterka M et al. Harnessing DSB repair to promote efficient homology-dependent and -independent prime editing. Nature Communications 13, doi: 10.1038/s41467-022-28771-1 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jang H et al. Application of prime editing to the correction of mutations and phenotypes in adult mice with liver and eye diseases. Nature Biomedical Engineering 6, 181–194, doi: 10.1038/s41551-021-00788-9 (2022). [DOI] [PubMed] [Google Scholar]
- 109.Eggenschwiler R et al. A selectable all-in-one CRISPR prime editing piggyBac transposon allows for highly efficient gene editing in human cell lines. Scientific Reports 11, doi: 10.1038/s41598-021-01689-2 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wolff JH, Haldrup J, Thomsen EA, Andersen S & Mikkelsen JG piggyPrime: High-efficacy prime editing in human cells using piggyBac-based DNA transposition. Frontiers in Genome Editing 3, doi: 10.3389/fgeed.2021.786893 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yuan Q & Gao X Multiplex base- and prime-editing with drive-and-process CRISPR arrays. Nature Communications 13, doi: 10.1038/s41467-022-30514-1 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bharucha N, Ataam JA, Gavidia AA & Karakikes I Generation of AAVS1 integrated doxycycline-inducible CRISPR-Prime Editor human induced pluripotent stem cell line. Stem Cell Research 57, 102610, doi: 10.1016/j.scr.2021.102610 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Li H et al. Highly efficient generation of isogenic pluripotent stem cell models using prime editing. eLife 11, doi: 10.7554/elife.79208 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Thomas CE, Ehrhardt A & Kay MA Progress and problems with the use of viral vectors for gene therapy. Nature Reviews Genetics 4, 346–358, doi: 10.1038/nrg1066 (2003). [DOI] [PubMed] [Google Scholar]
- 115.Zhi S et al. Dual-AAV delivering split prime editor system for in vivo genome editing. Molecular Therapy 30, 283–294, doi: 10.1016/j.ymthe.2021.07.011 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wang Q et al. Broadening the reach and investigating the potential of prime editors through fully viral gene-deleted adenoviral vector delivery. Nucleic Acids Research 49, 11986–12001, doi: 10.1093/nar/gkab938 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Aulicino F et al. Highly efficient CRISPR-mediated large DNA docking and multiplexed prime editing using a single baculovirus. Nucleic Acids Research 50, 7783–7799, doi: 10.1093/nar/gkac587 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sürün D et al. Efficient generation and correction of mutations in human iPS cells utilizing mRNAs of CRISPR base editors and prime editors. Genes 11, 511, doi: 10.3390/genes11050511 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Aida T et al. Prime editing primarily induces undesired outcomes in mice. bioRxiv, 2020.2008.2006.239723, doi: 10.1101/2020.08.06.239723 (2020). [DOI] [Google Scholar]
- 120.Hou X, Zaks T, Langer R & Dong Y Lipid nanoparticles for mRNA delivery. Nature Reviews Materials 6, 1078–1094, doi: 10.1038/s41578-021-00358-0 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Landrum MJ et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Research 44, D862–D868, doi: 10.1093/nar/gkv1222 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chemello F et al. Precise correction of Duchenne muscular dystrophy exon deletion mutations by base and prime editing. Science Advances 7, eabg4910, doi: 10.1126/sciadv.abg4910 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zhou M et al. Targeted-deletion of a tiny sequence via prime editing to restore SMN expression. International Journal of Molecular Sciences 23, 7941, doi: 10.3390/ijms23147941 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Torkamani A, Wineinger NE & Topol EJ The personal and clinical utility of polygenic risk scores. Nature Reviews Genetics 19, 581–590, doi: 10.1038/s41576-018-0018-x (2018). [DOI] [PubMed] [Google Scholar]
- 125.Li H et al. Multiplex precision gene editing by a surrogate prime editor in rice. Molecular Plant 15, 1077–1080, doi: 10.1016/j.molp.2022.05.009 (2022). [DOI] [PubMed] [Google Scholar]
- 126.Qian Y et al. Efficient and precise generation of Tay–Sachs disease model in rabbit by prime editing system. Cell Discovery 7, doi: 10.1038/s41421-021-00276-z (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kim DE et al. Prime editor-mediated correction of a pathogenic mutation in purebred dogs. Scientific Reports 12, doi: 10.1038/s41598-022-17200-4 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Bosch JA, Birchak G & Perrimon N Precise genome engineering in Drosophila using prime editing. Proceedings of the National Academy of Sciences 118, e2021996118, doi: 10.1073/pnas.2021996118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Doench JG Am I ready for CRISPR? A user’s guide to genetic screens. Nature Reviews Genetics 19, 67–80, doi: 10.1038/nrg.2017.97 (2018). [DOI] [PubMed] [Google Scholar]
- 130.Bock C et al. High-content CRISPR screening. Nature Reviews Methods Primers 2, doi: 10.1038/s43586-021-00093-4 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hanna RE et al. Massively parallel assessment of human variants with base editor screens. Cell 184, 1064–1080.e1020, doi: 10.1016/j.cell.2021.01.012 (2021). [DOI] [PubMed] [Google Scholar]
- 132.Kim Y et al. High-throughput functional evaluation of human cancer-associated mutations using base editors. Nature Biotechnology 40, 874–884, doi: 10.1038/s41587-022-01276-4 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Cuella-Martin R et al. Functional interrogation of DNA damage response variants with base editing screens. Cell 184, 1081–1097.e1019, doi: 10.1016/j.cell.2021.01.041 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Erwood S et al. Saturation variant interpretation using CRISPR prime editing. Nature Biotechnology 40, 885–895, doi: 10.1038/s41587-021-01201-1 (2022). [DOI] [PubMed] [Google Scholar]
- 135.Xu R, Liu X, Li J, Qin R & Wei P Identification of herbicide resistance OsACC1 mutations via in planta prime-editing-library screening in rice. Nature Plants 7, 888–892, doi: 10.1038/s41477-021-00942-w (2021). [DOI] [PubMed] [Google Scholar]
- 136.Jiao Y et al. Random-PE: an efficient integration of random sequences into mammalian genome by prime editing. Molecular Biomedicine 2, doi: 10.1186/s43556-021-00057-w (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Oh-hashi K, Furuta E, Fujimura K & Hirata Y Application of a novel HiBiT peptide tag for monitoring ATF4 protein expression in Neuro2a cells. Biochemistry and Biophysics Reports 12, 40–45, doi: 10.1016/j.bbrep.2017.08.002 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Leonetti MD, Sekine S, Kamiyama D, Weissman JS & Huang B A scalable strategy for high-throughput GFP tagging of endogenous human proteins. Proceedings of the National Academy of Sciences 113, E3501–E3508, doi:doi: 10.1073/pnas.1606731113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Choi J et al. A time-resolved, multi-symbol molecular recorder via sequential genome editing. Nature 608, 98–107, doi: 10.1038/s41586-022-04922-8 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Bortesi L & Fischer R The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnology Advances 33, 41–52, doi: 10.1016/j.biotechadv.2014.12.006 (2015). [DOI] [PubMed] [Google Scholar]
- 141.Yin K, Gao C & Qiu J-L Progress and prospects in plant genome editing. Nature Plants 3, 17107, doi: 10.1038/nplants.2017.107 (2017). [DOI] [PubMed] [Google Scholar]
- 142.Tang X et al. Plant prime editors enable precise gene editing in rice cells. Molecular Plant 13, 667–670, doi: 10.1016/j.molp.2020.03.010 (2020). [DOI] [PubMed] [Google Scholar]
- 143.Xu R et al. Development of plant prime-editing systems for precise genome editing. Plant Communications 1, 100043, doi: 10.1016/j.xplc.2020.100043 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Li J et al. Development of a highly efficient prime editor 2 system in plants. Genome biology 23, doi: 10.1186/s13059-022-02730-x (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Jiang Y et al. Optimized prime editing efficiently generates glyphosate-resistant rice plants carrying homozygous TAP-IVS mutation in EPSPS. Molecular Plant, doi: 10.1016/j.molp.2022.09.006 (2022). [DOI] [PubMed] [Google Scholar]
- 146.Lu Y et al. Precise genome modification in tomato using an improved prime editing system. Plant Biotechnology Journal 19, 415–417, doi: 10.1111/pbi.13497 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Perroud P-F et al. Prime editing in the model plant Physcomitrium patens and its potential in the tetraploid potato. Plant Science 316, 111162, doi: 10.1016/j.plantsci.2021.111162 (2022). [DOI] [PubMed] [Google Scholar]
- 148.Biswas S, Bridgeland A, Irum S, Thomson MJ & Septiningsih EM Optimization of Prime Editing in Rice, Peanut, Chickpea, and Cowpea Protoplasts by Restoration of GFP Activity. International Journal of Molecular Sciences 23, 9809, doi: 10.3390/ijms23179809 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]


