Abstract
The discovery of Asgard archaea and the exploration of their diversity over the last 6 years have deeply impacted the scientific community working on eukaryogenesis, rejuvenating an intense debate on the topology of the universal tree of life (uTol). Here, we discuss how this debate is impacted by two recent publications that expand the number of Asgard lineages and eukaryotic signature proteins (ESPs). We discuss some of the main difficulties that can impair the phylogenetic reconstructions of the uTol and suggest that the debate about its topology is not settled. We notably hypothesize the existence of horizontal gene transfers between ancestral Asgards and proto‐eukaryotes that could result in the observed abnormal behaviors of some Asgard ESPs and universal marker proteins. This hypothesis is relevant regardless of the scenario considered regarding eukaryogenesis. It implies that the Asgards were already diversified before the last eukaryotic common ancestor and shared the same biotopes with proto‐eukaryotes. We suggest that some Asgards might be still living in symbiosis today with modern Eukarya.
Keywords: Asgard, horizontal gene transfer, molecular phylogeny, tree of life
INTRODUCTION
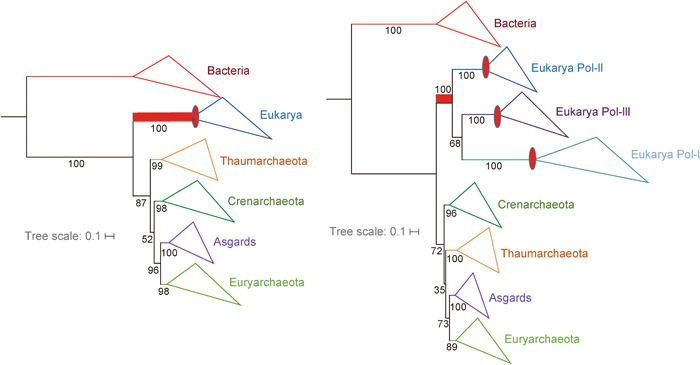
The discovery of Asgard archaea has revitalized an intense debate about the universal tree of life (uTol) topology 1 , 2 . Proponents of the two primary domain (2D) uTol propose a scenario in which Eukarya emerged within Archaea, with Eukarya being a subgroup of the Asgard superphylum 3 , 4 , whereas others still support a uTol in which all three domains (3D) are monophyletic, with Asgards nested within Archaea 5 , 6 , 7 (Figure 1).
Figure 1.
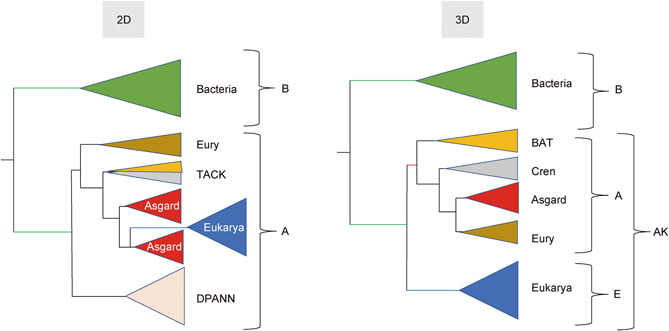
Two vs. three domains models for the universal tree of life. In the two domains (2D) model, Eukarya (E) branch within Archaea (A) and should be formally considered to be Archaea themselves (otherwise Archaea being paraphyletic are not a valid taxon). This schematic tree is based on one possible result of Liu et al. 13 in 2021, in which eukaryotes belong to the Asgard superphylum. In the three domains, model Archaea (A) and Eukarya (E) form a clade dubbed Arkarya (AK) in Forterre 8 . The bacterial branch (green) is much longer than the eukaryotic and archaeal branches (blue and red, respectively). The archaeal branch (red) is very short and only present in the three domains (3D) model. This schematic tree is based on Da Cunha et al. 5 . Eury(eury): Euryarchaeota; Cren: Crenarchaeota. TACK is the acronym for the clade grouping Thaumarchaea, Aigarchaea, Crenarchaea, and Korarchaea; DPANN is the acronym for the clade grouping Diapherotrites, Parvarchaea, Aenigmarchaea, Nanohaloarchaea, and Nanoarchaea; BAT is the acronym for the clade grouping Bathyarchaea, Aigarchaea, and Thaumarchaea 9 . The trees are rooted in the bacterial branch, based on comparative analysis of ribosomal protein distribution 8 .
The number of proposed Asgard phyla has continuously increased in recent years, from 1 in 2015 (Lokiarchaeota) to 18 in late 2021 (Figure 2) 1 , 2 , 10 , 11 , 12 , 13 , 14 , 15 , 16 . In a recent study, Liu et al. 13 described six new Asgard phyla from an analysis of 75 metagenome‐assembled genomes (MAGs), while three additional phyla were described by Xie et al. 14 from an analysis of 128 new MAGs. The characterization of these new Asgard lineages led to the identification of several new eukaryotic signature proteins (ESPs) but also emphasizes their patchy distribution among the various Asgard lineages.
Figure 2.
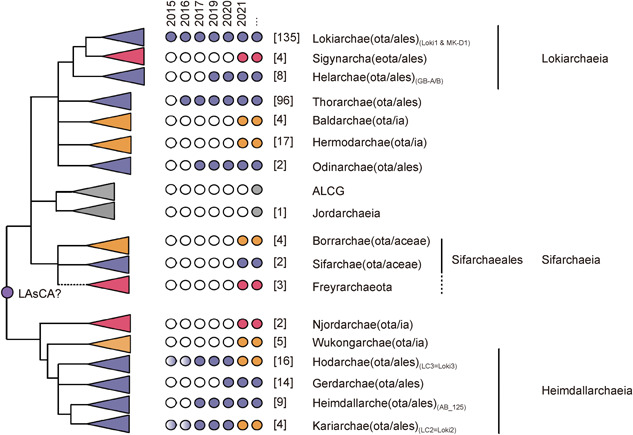
Schematic representation of the known diversity of the Asgard archaea including the recently published lineages. In this schema, we indicate in orange and red the newly discovered lineages introduced in the two publications of Liu et al. 13 and Xie et al. 14 , respectively, and in grey new lineages not present in these publications corresponding to the Sifarchaea 12 , the Jordarchaeia 16 and Asgard Lake Cootharaba 16 . The schema was designed combining the trees of all these publications. The suffix within brackets indicates that, depending on the authors, these lineages are considered to be Phylum (ota), Families (aceae), or Orders (ales). We locate the position of LAsCA (last Asgard archaeal common ancestor) based on the root observed in most phylogenetic trees, and used by Liu et al. to discuss the ESPs distribution. For each Asgard lineage, we also indicate the year of publication of the genome of its first representative, and changes in taxonomy are indicated by color changes of the dots. The light purple dots indicate that Asgards of the Karia and Hodar and lineages were described before 2017 as Loki 2 and Loki 3, respectively 1 . In addition, the number of genomes available for each lineage is indicated within brackets.
Liu and colleagues obtained a 2D uTol, with Eukarya most likely branching within Asgards as a sister group to an extended clade grouping the Wukong lineages with lineages previously considered to be Heimdallarchaeota 13 . Their phylogenetic analysis was based on a concatenation of 29 universal proteins that only partially overlap with the lists of universal markers previously used 1 , 2 , 4 , 17 . Xie and colleagues also obtained a 2D uTol but with Eukarya as sister group to the Njord lineage (a close relative of Wukong and Heimdall); their analysis was based on 21 universal proteins previously used by Williams et al. 4 , who also obtained a 2D tree.
Although both teams concluded in favor of a 2D tree, Liu et al. 13 cautiously mentioned that “further phylogenomic study with an even broader representation of diverse archaeal lineages, extended sets of phylogenetic markers and—possibly—more sophisticated evolutionary models are required to clarify the relationships between Archaea and eukaryotes”.
We discuss here these assumptions in light of our previous results and new data that have suggested various biases that could favor 2D topologies. We also propose that some Asgard ESPs and universal proteins could have been recruited from proto‐eukaryotes, possibly explaining the patchy distribution of ESPs and the atypical placement of some Asgard lineages in single‐gene uTols.
Rinke and colleagues recently proposed to consider the whole Asgard clade as a single phylum and the various Asgard lineages either as families or orders, with some of them being grouped in classes 16 . Since it is still an ongoing debate at the time of writing this review 18 , we will here use the more neutral term “lineage” for the various subgroups of Asgards (e.g., Heimdall lineage for Heimdallarchaeota/ales).
BROADER REPRESENTATION OF ARCHAEAL LINEAGES CAN FAVOR 2D MODELS
Compared to previous studies, Liu, Xie, and their respective colleagues used an expanded species data set, especially enriched in Archaea and Asgards. This can be problematic since simulation experiments have shown that overrepresentation of Archaea in uTols can favor 2D models 19 . This could be due to the very short internal branch defining the archaeal monophyly in the 3D uTol (in red in Figure 1) since internal branches become progressively shorter as taxonomic sampling increases 20 . Overbalanced representation of diverse archaeal lineages could thus introduce a bias if the representation of Bacteria and Eukarya is not increased accordingly 19 . In a recent reconstruction of phylogenetic trees, including Archaea and Bacteria, the authors noted that removal of some genomes can improve the phylogenetic inference, and that larger sets do not necessarily improve phylogenetic accuracy 21 . They noticed that oversampling of some groups relative to others can influence the placement of other groups in the tree.
It is also important to keep in mind that phylogenetic trees are the final output of successive analytical steps. Among them, multiple sequence alignments are critical, and despite undeniable improvements in their methods, the risk of misalignment increases along with the size of the data set. Misalignments of universal proteins due to domain inversion/substitution or mismatch have been detected by Nasir et al. 7 in 42% of the universal markers used in the first Asgard paper. Since some of these markers are also present in the data sets of Liu et al. and of Xie et al. (reference 22 and personal observation), it will be important to check the new alignments for possible mistakes. Notably, the risk of misalignment is increased in the case of taxon oversampling and with the inclusion of fast‐evolving species, two criteria frequently observed in uTol studies 7 , 23 .
THE CONUNDRUM OF FAST‐EVOLVING SPECIES
A major feature of the uTol is the difference between the lengths of its different stem branches (Figure 1). The topology of the tree is therefore very sensitive to long branch attraction (LBA) artifacts. Simulation experiments have shown that even the best models cannot cope with LBA when the branch of the outgroup is especially long 24 , which is the case for the bacterial branch in uTols (green in Figure 1) 5 , 25 , 26 . The presence of many fast‐evolving Archaea, such as the Diapherotrites, Parvarchaea, Aenigmarchaea, Nanohaloarchaea, Nanoarchaea (DPANN) and Methanopyrus kandleri in the data set of Liu et al., can thus be problematic. We have previously shown that removing fast‐evolving Archaea from the species data set of the first Asgard paper dramatically increased the number of single‐protein uTols with 3D topology 5 . [Correction added on June 6, 2022, after first online publication: A citation for reference 17 has been deleted from this sentence.] The presence of fast‐evolving Archaea could probably favor a 2D topology through their attraction by the long branch of Bacteria (Figure 3). Considering the possible close evolutionary relationships between some DPANN and Euryarchaeota 27 , this could result in the branching of Eukarya within Archaea, transforming a 3D uTol into a 2D uTol (Figure 3).
Figure 3.
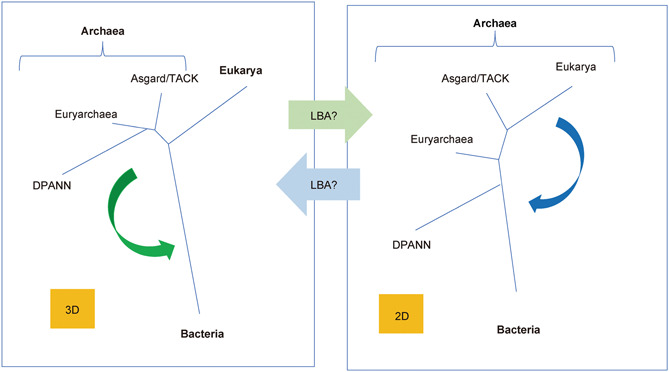
Two possible long branch attraction effects that could transform a 3D uTol into a 2D uTol and vice versa. On the left, the green arrow suggests that the attraction of DPANN and Euryarchaeota by Bacteria could transform the correct 3D tree into the 2D tree on the right. On the right, the blue arrow suggests that the attraction of eukaryotes by Bacteria could transform the correct 2D tree into the 3D tree on the left.
It is likely that solving the uTol topology is a difficult exercise because of a complex interplay of branch attraction effects between Bacteria, Euryarchaeota, Asgards, and Eukarya. This highlights the importance of taxon sampling since the incorporation of poorly sampled groups, as well as the oversampling of some groups relative to others, may indeed lead to LBA 21 .
THE ANOMALOUS BEHAVIOR OF ASGARD PROTEINS
Liu et al. 13 reported that the position of Eukarya was variable in single‐protein trees, branching either within Asgards, as sister group to all Asgards, or as sister group to other Archaea. We previously observed the same phenomenon with the first three published Asgards (Loki 1, 2, and 3) in 36 single‐protein trees 1 , 5 . The Asgards were also often paraphyletic in these trees, whereas the monophyly of other major archaeal clades was usually recovered (table 1 in Da Cunha et al. 5 ). This suggested that the scattered positions of the Asgards were unlikely due to a lack of resolution. Anomalous behavior of Asgard proteins was also reported by Garg et al. 33 for ribosomal proteins (r‐proteins). These authors suggested that the dispersal of Asgard lineages in r‐protein uTols could reflect mistakes in MAG reconstruction. However, this probably cannot explain every situation. Notably, we observed the same type of anomalous behavior in single‐protein trees with universal proteins encoded by fast‐evolving Archaea of the DPANN superphylum (see fig. S1 in Da Cunha et al. 5 ). This raises the possibility that the variable positions of Asgards in single‐protein trees could be due to a fast‐evolving phenotype. Metabolic reconstructions have suggested that Asgards are dependent on symbiotic interactions for both anabolism and catabolism, possibly explaining why they are so difficult to cultivate 34 . It is thus possible that the adaptation of the Asgards to their partners increased the evolutionary rate of some of their proteins.
THE IMPORTANCE OF THE PROTEIN SAMPLING
Another problem that can bias the topology of the uTol is the composition of the marker data set. Liu et al. 13 noticed that removing a eukaryotic protein of bacterial origin, the YchF ATPase, from their initial data set of 30 proteins, transformed a 3D tree into a 2D tree. Similarly, we have previously observed that removing the elongation factor EF2 from a data set of 36 proteins transformed a 2D into a 3D tree (fig. 5 in Da Cunha et al. 5 ). The fact that a single protein could determine the topology of the uTol based on several dozens of markers highlights the importance of carefully analyzing single‐protein trees before performing the concatenation of their alignments.
Figure 5.
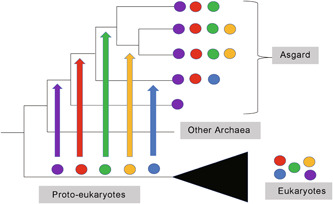
Explaining the patchy distribution of eukaryotic signature proteins (ESPs) in Asgards. Each color circle represents an ESP. Arrows with a similar color indicate their possible transfer from proto‐eukaryotes to Asgard at different periods of Asgard evolution. This is a schematic tree that emphasizes these specific transfers. The present patchy distribution of ESPs probably originated from a more complex pattern involving losses of ESPs in some archaeal lineages and horizontal gene transfer (HGT) between Asgard lineages. HGT of ESPs most likely also occurred between Asgard and other Archaea, as well as from Asgard to proto‐eukaryotes.
The choice of the markers is also critical. We have shown that large proteins tend to support 3D trees, whereas short ones tend to support 2D trees 5 . It is possible that short proteins do not harbor enough informative positions to detect the signal corresponding to the monophyly of Archaea. The data sets used by Liu, Xie, and their colleagues are both enriched in short r‐proteins (80% and 40%, respectively), potentially favoring the 2D topology, and lack several large proteins that gave 3D trees in our previous analysis 6 (Table 1). In particular, they both lack the large RNA polymerase B subunit. In a recent study of markers conserved between Archaea and Bacteria, Martinez‐Gutierrez and Aylward 21 have shown that RNA polymerase large subunits have the highest phylogenetic signal (i.e., they more accurately predict a true line of vertical ancestry) and outperformed their full set of 41 conserved archaeal and bacterial markers, despite having a shorter overall alignment length. In contrast, r‐proteins individually tend to carry a low phylogenetic signal. The phylogenetic signal obtained with the concatenation of r‐proteins was much higher than those obtained with single r‐proteins, but remained lower than the signal obtained with the concatenation of the two RNA polymerase large subunits 21 .
Table 1.
Presence/absence of universal markers giving either 2D or 3D trees.
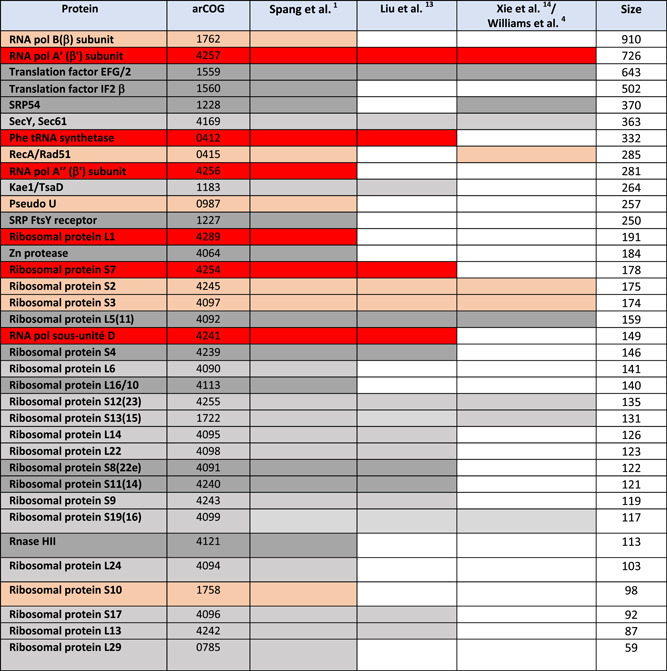
|
Lists of protein markers used by Spang and colleagues 1 to construct uTol and reanalyzed by Da Cunha et al. 5 that were present (colored boxes) or absent (white boxes) in the datasets of, Liu and colleagues 13 or Xie and colleagues 14 taken from Williams et al. 4 . In red boxes, proteins giving 3D tree supported by AU test in Da Cunha et al. 5 ; in light orange boxes, proteins giving 3D tree not supported by AU test in Da Cunha et al. 5 ; in grey boxes, proteins giving 2D tree supported by AU test in Da Cunha et al. 5 ; in light grey boxes, proteins giving 2D tree not supported by AU test in Da Cunha et al. 5 . The sizes correspond to the number of amino acids in the trimmed alignment of each marker.
We have previously obtained a robust 3D uTol with the concatenation of the two large RNA polymerase subunits with non‐homogeneous models in the Bayesian framework 5 . Notably, Williams et al. 4 also obtained a 3D RNA polymerase uTol with our data set and a different non‐homogeneous model (fig. S1 in Williams et al. 4 ). They only obtained a 2D uTol after recoding the amino acids of the multiple sequence alignments. Since amino acid recoding strongly reduces the signal in phylogenetic analysis 35 , it is possible that the switch of the RNA polymerase uTol from a 3D to a 2D tree after recoding is due to a lower signal supporting the monophyly of Archaea.
It has been suggested that 3D trees could be due to the attraction of Eukarya by Bacteria (Figure 3) 4 . However, the inclusion of the three eukaryotic RNA polymerases in the RNA polymerase uTol, to reduce the length of the eukaryotic branch, did not produce a 2D uTol 28 , suggesting that the 3D RNA polymerase uTol is not biased by an attraction between Eukarya and Bacteria (Figure 4).
Figure 4.
RNA polymerase tree remains 3D after shortening the eukaryal branch. Maximum‐likelihood (ML) phylogenetic trees were constructed based on the concatenated two largest DNA‐dependent RNA polymerase subunits. ML phylogenetic trees are of the Bacteria, eukaryotes, and Archaea, including Asgard sequences from Lokiarchaeota (1 genome), Heimdallarchaeota (2 genomes), Odinarchaeota (1 genome), and Thorarchaeota (3 genomes). The tree on the left is from data set 5 , while the tree on the right corresponds to the same data set and parameters but after the inclusion of sequences of the eukaryotic RNA polymerases I and III from data set 28 . For consistency, the same method has been applied to both and is described in Da Cunha et al. 5 . Briefly, the sequences were aligned with MAFFT v7 (default settings 29 ) and trimmed with BMGE (BLOSUM30 matrix 30 ). PhyML v3 31 was used for tree reconstruction with the BEST option for the tree search topology operations after the model was chosen according to the Akaike information criterion from ProtTest v3 32 . The scale bars represent the average number of substitutions per site. Supports at branch correspond to nonparametric bootstrap (out of 100).
More globally, an important step in phylogenetic reconstructions is the filtering of ambiguous sites through various tools to reduce the noise that can be generated when aligning universal markers, especially with large data sets. This step can also deeply affect the final phylogenetic tree 23 , 36 and considering the difficulty to find an appropriate balance between signal and noise filtering, it is possible that excessive trimming algorithms would tend to lead toward 2D uTols by decreasing an already relatively low archaeal‐specific signal carried within short protein sequences.
HGT BETWEEN ASGARDS AND PROTO‐EUKARYOTES COULD EXPLAIN THE PATCHY DISTRIBUTION OF SOME ESPS
The characterization of new Asgard lineages led to the identification of several new ESPs 13 , 14 . Notably, both teams noticed that many ESPs are lineage‐specific and that many were missing in the Asgard lineages that are sister groups to Eukarya in their analyses. This patchy distribution has already been observed 37 , 38 , but is even more visible with the ongoing expansion of the new Asgard lineages. Intriguingly, despite this major expansion, several ESPs are still only present in a few Asgard lineage 13 , 14 . This is for instance the case of tubulin, which is only present in the Odin lineage 2 , 14 .
In both 2D and 3D models, the extremely patchy phyletic distribution of ESPs requires to involve many losses and/or transfers between archaeal lineages. In the 2D scenario, one should further assume that Eukarya emerged from an extinct Asgard lineage that acquired the whole set of ESPs now distributed among the various Asgard lineages 37 . In both models, it is especially difficult to explain the existence of ESPs presently restricted to a single or few Asgard lineages.
An alternative hypothesis that could more easily explain the patchy distribution of Asgard ESPs is that some ESPs were recruited by Asgards from proto‐eukaryotes (i.e., members of the eukaryotic lineages that predated the last eukaryotic common ancestor [LECA]). Such horizontal gene transfer (HGTs) might have occurred either early in Asgard evolution, with the corresponding ESPs being present in most or all Asgards, or later, during the diversification of Asgard lineages, explaining the restricted distribution of the corresponding ESPs (Figure 5). Notably, such proto‐eukaryotic HGT hypothesis has been previously proposed to explain the presence of ESPs in a few Bacteria, such as actin or tubulin, very similar to the eukaryotic ones but branching as a sister group to Eukarya 39 , 40 , 41 , 42 .
Importantly, this proto‐eukaryotic HGT hypothesis would explain why some Asgard ESPs are much more similar to their eukaryotic homologs than to archaeal ones. This is the case of Odin tubulin, which is much more closely related to eukaryotic tubulins than to the tubulin found in Thaumarchaeota (Extended Data fig. 6 in Zaremba‐Niedzwiedzka 2 ). Similarly, most Asgard actins are much more similar to eukaryotic actins and actin‐related proteins (ARPs) than to archaeal crenactins 43 , 44 . In the case of actin, the proto‐eukaryotic HGT hypothesis is strongly supported by phylogenetic analyses showing that the various forms of actins present in Asgards branch between the various clades of eukaryotic actin paralogs (cytoplasmic actin and ARPs) that were already present in LECA 43 , 44 . Also, corroborating this hypothesis, Nasir et al. 7 concluded from a comparative analysis of conserved folds that ESPs are of relatively recent origin and could correspond to late HGTs between Archaea and Eukarya.
Notably, the proto‐eukaryotic HGT hypothesis is relevant in both the 2D and 3D scenarios. This implies that, independently of the correct scenario, the study of ESPs could provide important information about eukaryogenesis by identifying intermediate steps in the evolution of these proteins. Moreover, this hypothesis implies that ancient Asgards were already diversified before LECA and shared their biotopes with proto‐eukaryotes. Consequently, studying the environmental distribution of modern Asgards could provide critical insights into the nature of the biotopes in which proto‐eukaryotes were thriving.
In the modern biosphere, HGTs between Eukarya and Bacteria are especially prevalent between species living in close association. For instance, some intracellular Bacteria have acquired many eukaryotic proteins from their hosts 45 . Symbiotic associations between Archaea and Eukarya have been also described, such as methanogens thriving in protists from different eukaryotic lineages 46 or Cenarchaeum symbiosum inhabiting in marine sponges 47 . It is thus possible that some Asgard ancestors were symbionts of proto‐eukaryotes, exchanging genes with their hosts. Interestingly, the first cultivated Asgard, Candidatus Prometheoarchaeum syntrophicum, is a symbiotic organism living in association with a methanogen and a bacterium 34 . It could be worth exploring if some Asgards are living in symbiosis with modern Eukarya by looking for Asgard signatures in various types of eukaryotic cells.
HGT BETWEEN ASGARDS AND PROTO‐EUKARYOTES COULD ALSO EXPLAIN THE ODD POSITION OF SOME ASGARD LINEAGES IN SINGLE PROTEIN TREES
If HGTs took place between proto‐eukaryotes and Asgards, one can wonder if they also involve universal markers. Examining the single‐protein trees obtained by Liu and colleagues, we found a possible illustration of such a scenario in their uTol of the Kae1/TsaD protein (Figure 6A). Kae1/TsaD catalyzes the synthesis of threonylcarbamoyladenosine (t6A), a universal modification of transfer RNAs 48 . We previously obtained a 3D phylogeny for this protein before the discovery of Asgards 49 , whereas Liu et al. 13 obtained a 2D tree. In this tree, Asgards are paraphyletic with the Heimdall and Karia lineages branching as sister groups to Eukarya, whereas other Asgards form three distinct clades branching within Archaea (Figure 6A). This suggests that the latter correspond to the ancestral archaeal version of Kae1/TsaD that was replaced by a proto‐eukaryotic‐like version in a common ancestor to the Heimdall and Karia lineages (blue arrow). We also noticed that Archaea were rooted within DPANN and that M. kandleri did not branch with other Euryarchaeota (in green), testifying again for the difficulty to correctly position fast‐evolving species. When we removed these species from the data set, the 2D tree then obtained was essentially a 3D tree with Heimdall and Karia, still branching with Eukarya, and the rest of Asgards being nested within monophyletic Archaea (Figure 6B). Predictably, removing the Heimdall and Karia lineages from the uTol produced a 3D tree (Figure 6C) instead of a 2D tree.
Figure 6.
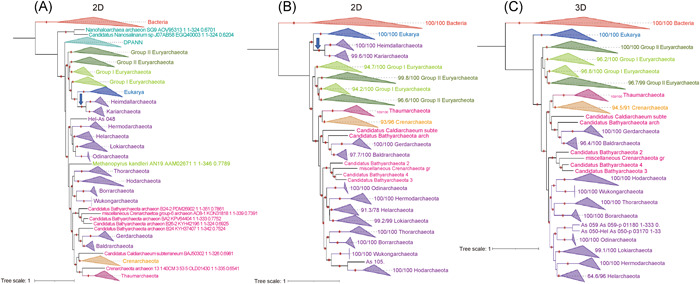
Phylogenetic trees of the universal protein Kae1/TsaD highlight the existence of different versions of Asgard proteins. From left to right. Initial 2D tree with DPANN at the base of Archaea and the fast‐evolving euryarchaeon Methanopyrus kandleri branching within paraphyletic Asgard (A), the 2D tree after removing fast‐evolving species (DPANN, M. kandleri) with Asgard divided into two monophyletic groups, one (two groups) sister group to Eukarya and the other (10 groups) branching between Euryarchaeota and the TACK clade (B) and the 3D tree obtained after further removing the two Asgard clades sister group to Eukarya (C). We observed in red several MAGs annotated as putative Bathyarchaota or Aigarchaeota that could be results of HGT or new groups of Asgard (C). The trimmed alignment provided of the Kae1/TsaD protein was selected from https://doi.org/10.5281/zenodo.4624280 13 . The maximum likelihood trees were constructed using IQ‐TREE version 1.6.1239 under the model of evolution according to the MFP option for model selection. Values at nodes represent branch supports calculated with the Shimodaira–Hasegawa‐like approximate likelihood ratio test (aLRT) (10,000 replicates) and UFBoot (10,000 replicates), and a dot is indicated if the aLRT >80 or UFBoot >95. The scale bar represents the average number of substitutions per site. The trees will be available on figshare (https://figshare.com/s/9e93072a77c3e014efd0).
Surprisingly, MAGs of Bathyarchaea (in red) did not form a robust clade with Thaumarchaeota in the Kae1/TsaD trees, as in most archaeal phylogenies 13 , but were interspersed with Asgards (Figure 6). This could reflect mistakes in MAG annotation or HGTs between Asgards and Bathyarchaea, which co‐occur in various environments 11 .
We suspect that the HGT hypothesis could also explain our previous observation of the strong impact of EF2 from one Asgard species of the Hodar lineage (formerly Loki 3) on the uTol 5 . This would explain why Hodar EF2 contained specific eukaryotic‐like insertions that were missing in all other Asgards 5,50 . Remarkably, these EF2 branched as sister group to various eukaryal EF2 paralogs in phylogenetic analyses, whereas all other Asgards EF2 branched as a monophyletic group within Archaea (fig. 2 in Narrowe et al. 50 [Correction added on June 6, 2022, after first online publication: The citation, ‘Hecker et al.49’ has been amended to ‘Narrowe et al.50’ in this sentence.] see also fig. 1 in Da Cunhua et al. 5 ), reminiscent of the situation previously described for Kae1/TsaD. This suggests that the archaeal version of EF2 was replaced in a common ancestor of the Hodar lineage by a proto‐eukaryotic version.
The identification in specific Asgard lineages of universal proteins that are much more similar to their eukaryotic homologs than to their homologs in all other Asgard lineages indicates that HGTs between proto‐eukaryotes and Asgard could be another confounding factor favoring 2D over 3D uTols.
CONCLUSION
Many recent observations about Asgards have been systematically interpreted in the 2D scenario framework. For instance, Avci et al. 51 suggested that condensed nucleoid observed in some Asgard cells could correspond to transition steps in the formation of the nucleus of eukaryotic cells, whereas Rambo et al. 52 concluded that Asgard Caudoviricetes have both archaeovirus and eukaryovirus characteristics because they encode informational proteins with homologs in eukaryotic viruses of the phylum Nucleocytoviricota. However, some Bacteria also have condensed nucleoid 53 and homologs of proteins encoded by Nucleocytoviricota are also present in bacterial Caudoviricetes 54 , 55 , 56 . Moreover, the recent characterization of Asgard viruses revealed three groups of viruses typical of other Archaea, including two groups (Caudoviricetes and Tectiliviricetes) that are present in Bacteria but not in Eukarya 57 , 58 . Asgard cells are rather small, with typical archaeal lipids, hence do not exhibit obvious intracellular complexity and probably lack phagocytosis ability 59 . The discovery of Asgards thus did not reduce the formidable divide existing between the archaeal and eukaryotic phenotypes. The transformation of an archaeon into a eukaryote still involves many transitions that have never been observed in nature 60 , 61 , 62 . As pointed out by Nasir et al. 7 , all known organisms forming intimate endosymbiotic or ectosymbiotic associations have maintained their domain identity. They noticed that “even mitochondria are still recognizable as highly‐reduced Bacteria despite billions of years of coexistence inside the eukaryotic cell.”
Despite the apparent congruence between 2D phylogenies displaying Eukarya branching within the Asgards and the abundance of ESPs in these latter, we argue that the debate between the 2D and 3D uToLs is not closed. We have here described several factors that could artifactually favor the 2D over the 3D topology in uTol analyses and hypothesized the existence of HGTs between Archaea and proto‐eukaryotes that could easily explain the presence of some ESPs in the Asgards. Taking all these observations and hypotheses into consideration, Box 1 summarizes our advices to improve future phylogenetic analyses and finally fulfill Darwin's dream to reconstruct the tree of life.
Box 1. .
Use a similar number of species for each domain and for each phylum within the domain based on more recent domain‐specific phylogenies. Avoid overrepresentation of some groups.
Do not include fast‐evolving species, such as M. kandleri, Korarchaeota, or DPANN in Archaea or candidate phylum radiation in Bacteria. The position of these species in their respective domain should be tested independently of the uTol.
Search for new universal proteins from the literature. Some of them are removed from automatic program searches because of paralogy or because they are split into two or more proteins in some groups. This probably explains why some data sets do not include the B subunit of RNA polymerase.
Examine single‐protein trees to detect paralogy and HGT between and within domains based on previous knowledge of domain phylogeny. Remove species affected by HGT and fast‐evolving paralogous groups.
Search markers with the best phylogenetic signal and congruence using tools such as the Tree Certainty metric 21 . Best markers should recover the monophyly of well‐established clades within domains.
For concatenation, test different combinations of subsets of proteins to detect markers that could bias the signal.
Test different trimming with different sets of species/markers and verify the absence of domain inversion/substitution mismatches. Apply different models for tree reconstruction.
Supporting information
Supporting information.
ACKNOWLEDGMENTS
V. D. C., M. G., and P. F. were supported by a European Research Council (ERC) grant from the European Union's Seventh Framework Program (FP/2007‐2013)/Project EVOMOBIL‐ERC Grant Agreement No. 340440.
Edited by Wen‐Jun Li, Sun Yat‐Sen University, China
REFERENCES
- 1. Spang A, Saw JH, Jørgensen SL, Zaremba‐Niedzwiedzka K, Martijn J, Lind AE, et al. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature. 2015;521:173–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zaremba‐Niedzwiedzka K, Caceres EF, Saw JH, Bäckström D, Juzokaite L, Vancaester E, et al. Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature. 2017;541:353–8. [DOI] [PubMed] [Google Scholar]
- 3. Spang A, Eme L, Saw JH, Caceres EF, Zaremba‐Niedzwiedzka K, Lombard J, et al. Asgard archaea are the closest prokaryotic relatives of eukaryotes. PLoS Genet. 2018;14:e1007080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Williams TA, Cox CJ, Foster PG, Szöllősi GJ, Embley TM. Phylogenomics provides robust support for a two‐domains tree of life. Nat Ecol Evol. 2020;4:138–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Da Cunha V, Gaia M, Gadelle D, Nasir A, Forterre P. Lokiarchaea are close relatives of Euryarchaeota, not bridging the gap between prokaryotes and eukaryotes. PLoS Genet. 2017;13:e1006810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Da Cunha V, Gaia M, Nasir A, Forterre P. Asgard archaea do not close the debate about the universal tree of life topology. PLoS Genet. 2018;14:e1007215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nasir A, Mughal F, Caetano‐Anollés G. The tree of life describes a tripartite cellular world. BioEssays. 2021;43:e2000343. [DOI] [PubMed] [Google Scholar]
- 8. Forterre P. The universal tree of life: an update. Front Microbiol. 2015;6:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gaïa M, Da Cunha V, Forterre P. The tree of life. In: Rampelotto PH, editor. Molecular Mechanisms of Microbial Evolution, Grand Challenges in Biology and Biotechnology. Springer; 2018. [Google Scholar]
- 10. Seitz KW, Lazar CS, Hinrichs K‐U, Teske AP, Baker BJ. Genomic reconstruction of a novel, deeply branched sediment archaeal phylum with pathways for acetogenesis and sulfur reduction. ISME J. 2016;10:1696–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cai M, Richter‐Heitmann T, Yin X, Huang WC, Yang Y, Zhang C, et al. Ecological features and global distribution of Asgard archaea. Sci Total Environ. 2021;758:143581. [DOI] [PubMed] [Google Scholar]
- 12. Farag IF, Zhao R, Biddle JF. “Sifarchaeota,” a novel Asgard Phylum from Costa Rican sediment capable of polysaccharide degradation and anaerobic methylotrophy. Appl Environ Microbiol. 2021;87:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu Y, Makarova KS, Huang W‐C, Wolf YI, Nikolskaya AN, Zhang X, et al. Expanded diversity of Asgard archaea and their relationships with eukaryotes. Nature. 2021;593:553–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xie R, Wang Y, Huang D, Hou J, Li L, Hu H, et al. Expanding Asgard members in the domain of Archaea sheds new light on the origin of eukaryotes. Sci China Life Sci. 2021. 10.1007/s11427-021-1969-6 [DOI] [PubMed] [Google Scholar]
- 15. Seitz KW, Dombrowski N, Eme L, Spang A, Lombard J, Sieber JR, et al. Asgard archaea capable of anaerobic hydrocarbon cycling. Nat Commun. 2019;10:1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sun J, Evans PN, Gagen EJ, Woodcroft BJ, Hedlund BP, Woyke T, et al. Recoding of stop codons expands the metabolic potential of two novel Asgardarchaeota lineages. bioRxiv. 2021. 10.1101/2021.02.19.431964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jay ZJ, Beam JP, Dlakić M, Rusch DB, Kozubal MA, Inskeep WP. Marsarchaeota are an aerobic archaeal lineage abundant in geothermal iron oxide microbial mats. Nat Microbiol. 2018;3:732–40. [DOI] [PubMed] [Google Scholar]
- 18. Tahon G, Ettema TJG. Expanding Archaeal diversity and phylogeny: past and future. Annu Rev Microbiol. 2021;75:359–81. [DOI] [PubMed] [Google Scholar]
- 19. Nasir A, Kim KM, Da Cunha V, Caetano‐Anollés G. Arguments reinforcing the three‐domain view of diversified cellular life. Archaea. 2016;2016:1851865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rangel LT, Fournier GP. Fast‐evolving alignment sites are highly informative for reconstructions of deep Tree of Life phylogenies. bioRxiv. 2019. 10.1101/835504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Martinez‐Gutierrez CA, Aylward FO. Phylogenetic signal, congruence, and uncertainty across Bacteria and Archaea. Mol Biol Evol. 2021;38:5514–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Caetano‐Anollés G, Mughal F. The Tree of Life describes a tripartite cellular world: neglected support from genome structure and codon usage and the fallacy of alignment‐dependent phylogenetic interpretations. BioEssays. 2021;43:e2100130. [DOI] [PubMed] [Google Scholar]
- 23. Ranwez V, Chantret NN, Ranwez V, Strengths NC, Sequence M, Ranwez V, et al. Strengths and limits of multiple sequence alignment and filtering methods. In: Scornavacca C, Delsuc F, Galtier N, editors. Phylogenetics in the Genomic Era Chapter 2.2. 2020. p. 2.2:1–2:36. [Google Scholar]
- 24. Gouy R, Baurain D, Philippe H. Rooting the tree of life: the phylogenetic jury is still out. Philos Trans R Soc Lond B. 2015;370:20140329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Berkemer SJ, McGlynn SE. A new analysis of Archaea–Bacteria domain separation: variable phylogenetic distance and the tempo of early evolution. Mol Biol Evol. 2020;27:2332–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Moody ERR, Mahendrarajah TA, Dombrowski N, Clark JW, Petitjean C, Offre P, et al. An estimate of the deepest branches of the tree of life from ancient vertically‐evolving genes. eLife. 2022;11:e66695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Brochier‐Armanet C, Forterre P, Gribaldo S. Phylogeny and evolution of the Archaea:2 one hundred genomes later. Curr Opin Microbiol. 2011;14:274–81. [DOI] [PubMed] [Google Scholar]
- 28. Guglielmini J, Woo AC, Krupovic M, Forterre P, Gaia M, Gaïa M. Diversification of giant and large eukaryotic dsDNA viruses predated the origin of modern eukaryotes. Proc Natl Acad Sci USA. 2019;116:19535–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Criscuolo A, Gribaldo S. BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol Biol. 2010;10:210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum‐likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–21. [DOI] [PubMed] [Google Scholar]
- 32. Darriba D, Taboada GL, Doallo R, Posada D. ProtTest‐HPC: fast selection of best‐fit models of protein evolution. Bioinformatics. 2011;27:1164–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Garg SG, Kapust N, Lin W, Knopp M, Tria FDK, Nelson‐Sathi S, et al. Anomalous phylogenetic behavior of ribosomal proteins in metagenome‐assembled Asgard Archaea. Genome Biol Evol. 2021;13:evaa238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Imachi H, Nobu MK, Nakahara N, Morono Y, Ogawara M, Takaki Y, et al. Isolation of an archaeon at the prokaryote–eukaryote interface. Nature. 2020;577:519–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hernandez AM, Ryan JF. Six‐state amino acid recoding is not an effective strategy to offset compositional heterogeneity and saturation in phylogenetic analyses. Syst Biol. 2021;70:1200–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tan G, Muffato M, Ledergerber C, Herrero J, Goldman N, Gil M, et al. Current methods for automated filtering of multiple sequence alignments frequently worsen single‐gene phylogenetic inference. Syst Biol. 2015;64:778–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Eme L, Spang A, Lombard J, Stairs CW, Ettema TJG. Archaea and the origin of eukaryotes. Nat Rev Microbiol. 2017;15:711–23. [DOI] [PubMed] [Google Scholar]
- 38. Knopp M, Stockhorst S, van der Giezen M, Garg SG, Gould SB. The Asgard Archaeal—unique contribution to protein families of the eukaryotic common ancestor was 0.3. Genome Biol Evol. 2021;13:evab085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schlieper D, Oliva MA, Andreu JM, Löwe J. Structure of bacterial tubulin BtubA/B: evidence for horizontal gene transfer. Proc Natl Acad Sci USA. 2005;102:9170–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shiratori T, Suzuki S, Kakizawa Y, Ishida K. Phagocytosis‐like cell engulfment by a planctomycete bacterium. Nat Commun. 2019;10:5529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Martin‐Galiano AJ, Oliva MA, Sanz L, Bhattacharyya A, Serna M, Yebenes H, et al. Bacterial tubulin distinct loop sequences and primitive assembly properties support its origin from a eukaryotic tubulin ancestor. J Biol Chem. 2011;286:19789–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Guljamow A, Jenke‐kodama H, Saumweber H, Quillardet P, Frangeul L, Castets AM, et al. Horizontal gene transfer of two cytoskeletal elements from a eukaryote to a cyanobacterium. Curr Biol. 2007;17:757–9. [DOI] [PubMed] [Google Scholar]
- 43. Stairs CW, Ettema TJG. The Archaeal roots of the eukaryotic dynamic actin cytoskeleton. Curr Biol. 2020;30:R521–6. [DOI] [PubMed] [Google Scholar]
- 44. Da Cunha V, Gaia M, Ogata H, Jaillon O, Delmont TO, Forterre P Giant viruses encode actin related proteins. Mol Biol Evol . 2022;39:msac022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Best A, Kwaik YA. Evolution of the arsenal of Legionella pneumophila effectors to modulate protist hosts. mBio. 2018;9:e01313–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Husnik F, Tashyreva D, Boscaro V, George EE, Lukeš J, Keeling PJ. Bacterial and archaeal symbioses with protists. Curr Biol. 2021;31:R862–77. [DOI] [PubMed] [Google Scholar]
- 47. Preston CM, Wu KY, Molinski TF, DeLong EF. A psychrophilic crenarchaeon inhabits a marine sponge: Cenarchaeum symbiosum gen. nov., sp. nov. Proc Natl Acad Sci USA. 1996;93:6241–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Thiaville PC, Iwata‐Reuyl D, DeCrécy‐Lagard V. Diversity of the biosynthesis pathway for threonylcarbamoyladenosine (t6A), a universal modification of tRNA. RNA Biol. 2014;11:1529–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hecker A, Leulliot N, Gadelle D, Graille M, Justome A, Dorlet P, et al. An archaeal orthologue of the universal protein Kae1 is an iron metalloprotein which exhibits atypical DNA‐binding properties and apurinic‐endonuclease activity in vitro . Nucleic Acids Res. 2007;35:6042–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Narrowe AB, Spang A, Stairs CW, Caceres EF, Baker BJ, Miller CS, et al. Complex evolutionary history of translation elongation 2 and diphthamide biosynthesis in Archaea and Parabasalids. Genome Biol Evol. 2018;10:2380–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Avcı B, Brandt J, Nachmias D, Elia N, Albertsen M, Ettema TJG, et al. Spatial separation of ribosomes and DNA in Asgard archaeal cells. ISME J. 2022;16:606–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rambo IM, Anda VDe, Langwig MV, Baker BJ. Unique viruses that infect Archaea related to eukaryotes. bioRxiv. 2021. 10.1101/2021.07.29.454249 [DOI] [Google Scholar]
- 53. Floc'h K, Lacroix F, Servant P, Wong YS, Kleman JP, Bourgeois D, et al. Cell morphology and nucleoid dynamics in dividing Deinococcus radiodurans . Nat Commun. 2019;10:3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Forterre P, Gadelle D. Phylogenomics of DNA topoisomerases: their origin and putative roles in the emergence of modern organisms. Nucleic Acids Res. 2009;37:679–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kazlauskas D, Krupovic M, Guglielmini J, Forterre P, Venclovas CS. Diversity and evolution of B‐family DNA polymerases. Nucleic Acids Res. 2020;48:10142–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pan J, Lian K, Sarre A, Leiros HKS, Williamson A. Bacteriophage origin of some minimal ATP‐dependent DNA ligases: a new structure from Burkholderia pseudomallei with striking similarity to Chlorella virus ligase. Sci Rep. 2021;11:18693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Medvedeva S, Sun J, Yutin N, Koonin EV, Nunoura T, Rinke C, et al. Viruses of Asgard archaea. bioRxiv. 2021. 10.1101/2021.07.29.453957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tamarit D, Caceres EF, Krupovic M, Nijland R, Eme L, Robinson P, et al. A closed Candidatus Odinarchaeum genome exposes Asgard archaeal viruses. bioRxiv. 2021. 10.1101/2021.09.01.458545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Burns JA, Pittis AA, Kim E. Gene‐based predictive models of trophic modes suggest Asgard archaea are not phagocytotic. Nat Ecol Evol. 2018;2:697–704. [DOI] [PubMed] [Google Scholar]
- 60. Forterre P. The common ancestor of archaea and eukarya was not an archaeon. Archaea. 2013;2013:372396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Forterre P, Gaia M, Da Cunha V. Engineered bacterium fuels evolution debate. Nature. 2019;571:326. [DOI] [PubMed] [Google Scholar]
- 62. Novoa EM, Pavon‐Eternod M, Pan T, Ribas De Pouplana L. A role for tRNA modifications in genome structure and codon usage. Cell. 2012;149:202–13. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information.


