Abstract
Transcription of the vaccinia virus genome is mediated by a virus-encoded multisubunit DNA-dependent RNA polymerase in conjunction with early-, intermediate-, and late-stage-specific factors. Previous studies indicated that two virus-encoded proteins (capping enzyme and VITF-1) and one unidentified cellular protein (VITF-2) are required for specific transcription of an intermediate promoter template in vitro. We have now extensively purified an additional virus-induced intermediate transcription factor with a native mass of approximately 100 kDa.
Vaccinia virus (VV) transcription is temporally regulated within the cytoplasm of infected cells (11). The three classes of genes—early, intermediate, and late—have stage-specific promoters and cognate transcription factors that act in conjunction with the virus-encoded multisubunit DNA-dependent RNA polymerase. The enzymes and factors for early transcription are packaged within the virus particle and activated immediately after infection; intermediate factors are all synthesized before viral DNA replication, whereas some late factors are made afterwards. The current status of our knowledge regarding VV transcription factors can be summarized as follows. A heterodimeric early transcription factor and an RNA polymerase-associated protein are required for early transcription (2, 4); capping enzyme, a virus-encoded early protein, and an unidentified cellular protein are needed for intermediate transcription (13, 14, 18, 19); and four virus-encoded proteins and an unidentified cellular protein are known to be involved in late transcription (9, 10, 21). Additional viral proteins participate in transcription elongation and termination as well as capping, methylation, and polyadenylylation (5–7, 15, 16).
The present communication is concerned with the factors required for intermediate transcription. Initial studies indicated that all of the components necessary to transcribe a template regulated by a VV intermediate promoter are present in extracts from HeLa cells infected with VV in the presence of an inhibitor of DNA replication (20). Vos and coworkers (18) resolved the transcription components into a partially purified RNA polymerase fraction and two factors called VITF-A and VITF-B. VITF-A was subsequently identified as capping enzyme (19) and shown to have a transcriptional role independent of RNA guanylylation (8). VITF-B was not extensively purified but was shown to complement virion extracts, which contain RNA polymerase and capping enzyme (18). Rosales and coworkers (13, 14) purified two intermediate transcription factors, called VITF-1 and VITF-2, in addition to capping enzyme and RNA polymerase. VITF-1 was purified to homogeneity as a 30-kDa monomeric protein corresponding to RPO30, a subunit of the VV RNA polymerase with homology to the eucaryotic transcription factor TFIIS (1, 3). The finding that VITF-1 and RPO30 are products of the same gene but have different functional roles and physical states is similar to the situation for the VV RNA 2′-O-methyltransferase, which exists as a monomer and as a processivity factor subunit of the poly(A) polymerase (7, 15). The second factor, VITF-2, was partially purified from both infected and uninfected cell extracts and has a molecular mass of approximately 68 kDa, as estimated by glycerol gradient sedimentation (14). Whether VITF-B (18) corresponds to VITF-1, VITF-2, a different factor, or a combination of factors is unclear. Using a modified protocol, we now report the purification of an additional intermediate transcription factor that can be distinguished from the previously identified VITF-1, VITF-2, and capping enzyme.
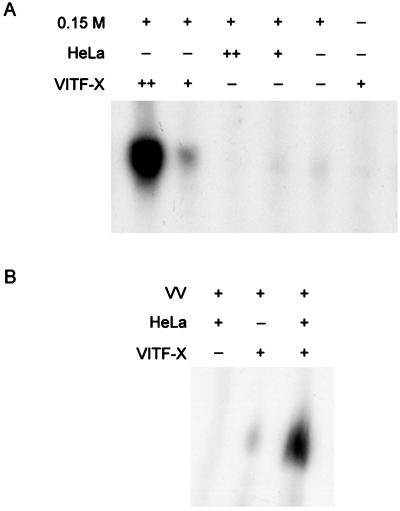
Transcription factors were purified from approximately 2.6 × 1011 (400 liters) HeLa S3 cells (National Cell Culture Center, Minneapolis, Minn.) that were infected with VV (10 PFU/cell) in the presence of the DNA replication inhibitor AraC (40 μg/ml). After 20 h at 37°C, the cells were lysed by Dounce homogenization and the cytoplasmic fraction was centrifuged at 100,000 × g for 45 min. The proteins in the supernatant fraction were precipitated with 0.35 g of ammonium sulfate/ml, resuspended in buffer A (40 mM Tris [pH 8.0], 0.2 mM EDTA, 2 mM dithiothreitol, 15% glycerol, 0.5 mM phenylmethylsulfonyl fluoride) containing 0.05 M NaCl, dialyzed against the same buffer, and applied to a diethylaminoethyl (DEAE)-cellulose column. The elution procedure, as described below, differed in several aspects from that previously reported (13). The flowthrough fraction and the fractions eluting after steps of 0.15 and 1 M NaCl in buffer A were collected. In vitro assays indicated that the 0.15 M fraction still contained all the factors needed for transcription of a template with an intermediate promoter. The 0.15 M fraction was reapplied to a second DEAE-cellulose column that had been equilibrated with 0.15 M NaCl in buffer A; the flowthrough was collected, and the bound proteins were eluted with buffer A containing 1 M NaCl and combined with the 1 M fraction from the first DEAE-cellulose column. The 0.15 M flowthrough fraction from the second DEAE-cellulose column did not support transcription, even though it contained RNA polymerase and capping enzyme, but could complement the active component(s), which was provisionally called VITF-X, of the combined 1 M NaCl fractions (Fig. 1A).
FIG. 1.
Evidence for a new intermediate transcription factor. In vitro transcription was carried out as previously described (13) for 30 min at 37°C in a total volume of 20 μl with 0.1 μg of uncleaved plasmid (containing the G8R intermediate promoter followed by a template lacking G residues), ribonucleoside triphosphates including [α-32P]UTP, and additional protein components as indicated in the figure. The RNA was analyzed on a 4% polyacrylamide gel, which was then dried and autoradiographed. Symbols: −, no addition; +, 1× addition; ++, 10× addition. (A) Protein components of the reaction mixture were the 0.15 M NaCl fraction from the second DEAE-cellulose column (0.15 M), a total uninfected HeLa cell extract (HeLa), and the pooled 1 M NaCl fractions from the first and second DEAE-cellulose columns (VITF-X). (B) Protein components were an extract of purified vaccinia virions (VV), a total extract of uninfected HeLa cells (HeLa), and VITF-X purified by DEAE-cellulose, phosphocellulose, SP Sepharose, and single-stranded DNA agarose chromatography.
VITF-X could not be replaced either by an extract (14) of uninfected HeLa cells (Fig. 1A) or by a 1 M NaCl DEAE-cellulose fraction from uninfected cells (data not shown), suggesting that the 0.15 M NaCl fraction from virus-infected cells contained sufficient VITF-2 to mediate transcription. This interpretation was supported by the use of extracts of purified virions, containing RNA polymerase and capping enzyme, that had been depleted of DNA by passage through a DEAE-cellulose column (12) in place of the 0.15 M fraction. In this case, intermediate transcription was dependent on VITF-X and was greatly stimulated by VITF-2 supplied in the uninfected HeLa cell extract (Fig. 1B). Thus, VITF-X does not contain significant VITF-2 activity and must be a different factor. The presence of VITF-1 activity in the 0.15 M NaCl fraction and the virion extract was more difficult to evaluate, since the product of the E4L gene exists as both a subunit of RNA polymerase and monomeric VITF-1. Free E4L gene product could be detected in both samples, however, by Western blotting after RNA polymerase was separated by column chromatography (data not shown). Nevertheless, we could not determine whether the E4L protein was transcriptionally active or present in an amount sufficient to rule out the possibility that VITF-X is or includes VITF-1.
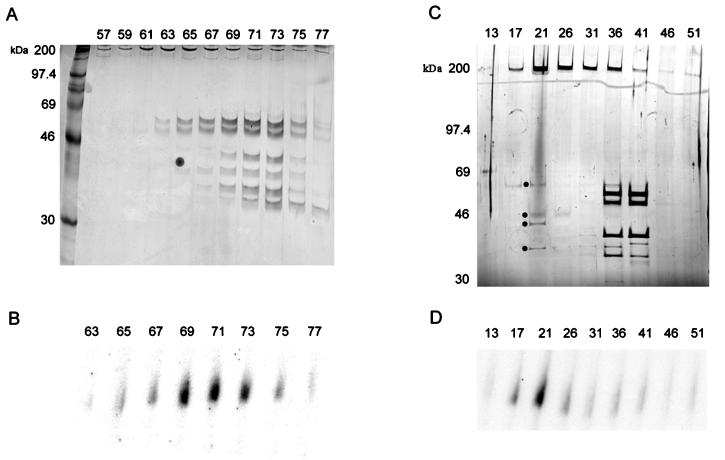
Further purification of VITF-X, from the 1 M DEAE-cellulose fraction, was achieved by chromatography on phosphocellulose (P11; Whatman), SP Sepharose (Pharmacia), single-stranded DNA (Sigma), HQ (Poros), heparin (Poros), HS (Poros), and CM (Poros) columns. VITF-X activity was assayed by complementation with the 0.15 M NaCl fraction from the second DEAE-cellulose column, and the extent of purification was monitored by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). All traces of RNA polymerase, VITF-1, and H5R late transcription factor were removed during purification, as determined by Western blotting, and no capping enzyme was detectable by labeling with [α-32P]GTP followed by SDS-PAGE (17). At least 10 polypeptides eluted in the active fractions of the penultimate column (Fig. 2A), and 5 of these, with masses of 200, 65, 50, 45, and 35 kDa, were detected in the active fractions of the final column (Fig. 2B). At this stage, the activity of VITF-X was limiting and the low background of the 0.15 M complementing fraction became significant, precluding further purification. Although insufficient protein was available for microsequencing, the purification served to distinguish VITF-X from previously recognized intermediate transcription factors.
FIG. 2.
Elution of VITF-X from the final two chromatography columns. Samples (20 μl) from HS (A) or CM (C) column fractions were analyzed by SDS-PAGE on 10% or 4 to 20% polyacrylamide gels, respectively, and silver stained. Samples—1 μl from the HS column (B) and 5 μl from the CM column (D)—were tested for VITF-X activity as described in the legend to Fig. 1. Fraction numbers are indicated at the top of each panel, and the positions and masses (in kilodaltons) of markers are indicated on the left. In panel C, dots are placed next to the 65-, 50-, 45-, and 35-kDa bands in the most active fraction. In panel A, the dot in lane 65 is a silver staining artifact.
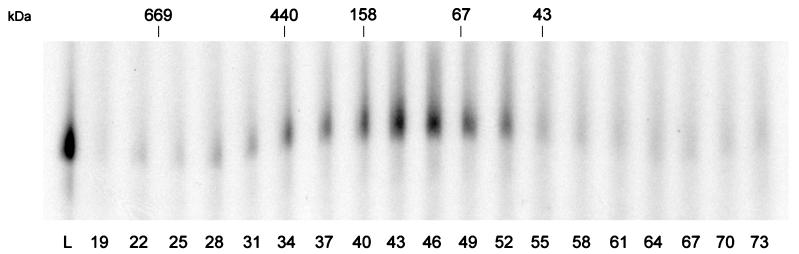
To determine the mass of the native protein, partially purified VITF-X was applied to a calibrated S300 Sephacryl column. VITF-X activity eluted mostly between fractions 43 and 46, corresponding to a globular protein of about 100 kDa (Fig. 3). This result makes it very unlikely that the 200-kDa polypeptide detected by SDS-PAGE (Fig. 2C) is VITF-X and raises the possibility that the factor is an oligomer of one or more of the smaller polypeptides.
FIG. 3.
Gel filtration of VITF-X. Partially purified VITF-X was applied to a 1.6- by 60-cm S300 Sephacryl column equilibrated with 0.15 M NaCl, 40 mM Tris-HCl (pH 8.0), 5% glycerol, 5 mM imidazole, and 0.5 mM phenylmethylsulfonyl fluoride. Samples—0.1 μl of loading material (L) or 2 μl of column fractions (numbered)—were assayed for VITF-X activity as described in the legend to Fig. 1. The masses (in kilodaltons) and elution positions of standard proteins (thyroglobulin, 669 kDa; ferritin, 440 kDa; aldolase, 158 kDa; serum albumin, 67 kDa; and ovalbumin, 43 kDa) (Pharmacia) used to calibrate the column are indicated at the top.
Determination of the number and identities of the stage-specific transcription factors is crucial to fully understanding the regulation of poxvirus gene expression. Of the three transcriptional stages, the intermediate stage has proven to be the most recalcitrant to investigation. Whereas the purification of the early transcription factor was facilitated by its presence within virus particles (4) and three late transcription factors were identified by a reverse genetic screen (9), neither of these approaches is applicable to intermediate transcription factors. Furthermore, no temperature-sensitive mutant with a specific defect in intermediate transcription has been described. Because the amounts of the intermediate transcription factors are low, large numbers of infected cells are required for purification. Moreover, since there are multiple factors, it is difficult to perform complementation assays. Thus, VITF-X was presumably an unrecognized minor component of other partially purified RNA polymerase or factor preparations. For example, VITF-B (18) or VITF-1 and VITF-2 (13) were able to complement virion extracts without additions, suggesting that they may have included VITF-X.
In conclusion, the present data indicating the existence of an additional intermediate transcription factor, VITF-X, should accelerate further studies on the regulation of VV gene expression. The new 100-kDa intermediate transcription factor is present in cells infected with VV in the presence of AraC but not in uninfected cells, suggesting that it is virus encoded or is a cellular protein that is virus induced or virus activated.
Acknowledgments
We thank N. Cooper for VV stocks, C. Cassetti for the virion extract, N. Harris and G. R. Kovacs for helpful discussions, and T. Kristie for comments on the manuscript.
REFERENCES
- 1.Ahn B-Y, Gershon P D, Jones E V, Moss B. Identification of rpo30, a vaccinia virus RNA polymerase gene with structural similarity to a eukaryotic transcription factor. Mol Cell Biol. 1990;10:5433–5441. doi: 10.1128/mcb.10.10.5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahn B-Y, Moss B. RNA polymerase-associated transcription specificity factor encoded by vaccinia virus. Proc Natl Acad Sci USA. 1992;89:3536–3540. doi: 10.1073/pnas.89.8.3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Broyles S S, Pennington M J. Vaccinia virus gene encoding a 30-kilodalton subunit of the viral DNA-dependent RNA polymerase. J Virol. 1990;64:5376–5382. doi: 10.1128/jvi.64.11.5376-5382.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Broyles S S, Yuen L, Shuman S, Moss B. Purification of a factor required for transcription of vaccinia virus early genes. J Biol Chem. 1988;263:10754–10760. [PubMed] [Google Scholar]
- 5.Condit R C, Xiang Y, Lewis J I. Mutation of vaccinia virus gene G2R causes suppression of gene A18R ts mutants: implication for control of transcription. Virology. 1996;220:10–19. doi: 10.1006/viro.1996.0280. [DOI] [PubMed] [Google Scholar]
- 6.Deng L, Shuman S. An ATPase component of the transcription elongation complex is required for factor-dependent transcription termination by vaccinia RNA polymerase. J Biol Chem. 1996;271:29386–29392. doi: 10.1074/jbc.271.46.29386. [DOI] [PubMed] [Google Scholar]
- 7.Gershon P D, Ahn B Y, Garfield M, Moss B. Poly(A) polymerase and a dissociable polyadenylation stimulatory factor encoded by vaccinia virus. Cell. 1991;66:1269–1278. doi: 10.1016/0092-8674(91)90048-4. [DOI] [PubMed] [Google Scholar]
- 8.Harris N, Rosales R, Moss B. Transcription initiation factor activity of vaccinia virus capping enzyme is independent of mRNA guanylylation. Proc Natl Acad Sci USA. 1993;90:2860–2864. doi: 10.1073/pnas.90.7.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keck J G, Baldick C J, Moss B. Role of DNA replication in vaccinia virus gene expression: a naked template is required for transcription of three late transactivator genes. Cell. 1990;61:801–809. doi: 10.1016/0092-8674(90)90190-p. [DOI] [PubMed] [Google Scholar]
- 10.Kovacs G R, Moss B. The vaccinia virus H5R gene encodes late gene transcription factor 4: purification, cloning, and overexpression. J Virol. 1996;70:6796–6802. doi: 10.1128/jvi.70.10.6796-6802.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moss B. Poxviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2637–2671. [Google Scholar]
- 12.Rohrmann G, Moss B. Transcription of vaccinia virus early genes by a template-dependent soluble extract of purified virions. J Virol. 1985;56:349–355. doi: 10.1128/jvi.56.2.349-355.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosales R, Harris N, Ahn B-Y, Moss B. Purification and identification of a vaccinia virus-encoded intermediate stage promoter-specific transcription factor that has homology to eukaryotic transcription factor SII (TFIIS) and an additional role as a viral RNA polymerase subunit. J Biol Chem. 1994;269:14260–14267. [PubMed] [Google Scholar]
- 14.Rosales R, Sutter G, Moss B. A cellular factor is required for transcription of vaccinia viral intermediate stage genes. Proc Natl Acad Sci USA. 1994;91:3794–3798. doi: 10.1073/pnas.91.9.3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schnierle B S, Gershon P D, Moss B. Cap-specific mRNA (nucleoside-O2′-)-methyltransferase and poly(A) polymerase stimulatory activities of vaccinia virus are mediated by a single protein. Proc Natl Acad Sci USA. 1992;89:2897–2901. doi: 10.1073/pnas.89.7.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shuman S, Broyles S S, Moss B. Purification and characterization of a transcription termination factor from vaccinia virions. J Biol Chem. 1987;262:12372–12380. [PubMed] [Google Scholar]
- 17.Shuman S, Hurwitz J. Mechanism of mRNA capping by vaccinia virus guanylyltransferase: characterization of an enzyme-guanylate intermediate. Proc Natl Acad Sci USA. 1981;78:187–191. doi: 10.1073/pnas.78.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vos J C, Sasker M, Stunnenberg H G. Promoter melting by a stage-specific vaccinia virus transcription factor is independent of the presence of RNA polymerase. Cell. 1991;65:105–114. doi: 10.1016/0092-8674(91)90412-r. [DOI] [PubMed] [Google Scholar]
- 19.Vos J C, Sasker M, Stunnenberg H G. Vaccinia virus capping enzyme is a transcription initiation factor. EMBO J. 1991;10:2553–2558. doi: 10.1002/j.1460-2075.1991.tb07795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vos J C, Stunnenberg H G. Derepression of a novel class of vaccinia virus genes upon DNA replication. EMBO J. 1988;7:3487–3492. doi: 10.1002/j.1460-2075.1988.tb03224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wright C F, Hubbs A E, Gunasinghe S K, Oswald B W. A vaccinia virus late transcription factor copurifies with a factor that binds to a viral late promoter and is complemented by extracts from uninfected Hela cells. J Virol. 1998;72:1446–1451. doi: 10.1128/jvi.72.2.1446-1451.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]