Abstract
The prevalence of cadmium (Cd)‐polluted agricultural soils is increasing globally, and arbuscular mycorrhizal fungi (AMF) can reduce the absorption of heavy metals by plants and improve mineral nutrition. However, the immobilization of the rhizosphere on cadmium is often overlooked. In this study, Glomus mosseae and Medicago sativa were established as symbiotes, and Cd migration and environmental properties in the rhizosphere were analyzed. AMF reduced Cd migration, and Cd2+ changed to an organic‐bound state. AMF symbiosis treatment and Cd exposure resulted in microbial community variation, exhibiting a distinct deterministic process (|βNTI| > 2), which ultimately resulted in a core microbiome function of heavy metal resistance and nutrient cycling. AMF increased available N and P, extracellular enzyme activity (LaC, LiP, and CAT), organic matter content (TOC, EOC, and GRSP), and Eh of the rhizosphere soil, significantly correlating with decreased Cd migration (p < 0.05). Furthermore, AMF significantly affected root metabolism by upregulating 739 metabolites, with flavonoids being the main factor causing microbiome variation. The structural equation model and variance partial analysis revealed that the superposition of the root metabolites, microbial, and soil exhibited the maximum explanation rate for Cd migration reduction (42.4%), and the microbial model had the highest single explanation rate (15.5%). Thus, the AMF in the rhizosphere microenvironment can regulate metabolite–soil–microbial interactions, reducing Cd migration. In summary, the study provides a new scientific explanation for how AMF improves plant Cd tolerance and offers a sustainable solution that could benefit both the environment and human health.
Keywords: arbuscular mycorrhizal fungi, cadmium, microbiome reassembly, rhizosphere, soil microbial interaction
Arbuscular mycorrhizal fungi (AMF) reduced Cd migration in the rhizosphere through microbial community variation, exhibiting a distinct deterministic process, which ultimately resulted in a core microbiome function of heavy metal resistance and nutrient cycling. The above process is related to upregulating root metabolites by AMF, with flavonoids, coumarin, fatty acids and so on being the main factor causing microbiome variation. Additionally, the superposition of the root metabolites, microbial, and soil make contributed to Cd migration reduction, and the microbial model had the highest single explanation rate. Thus, the AMF in the rhizosphere microenvironment can regulate metabolite‐soil‐microbial interactions, reducing Cd migration.
Highlights
Arbuscular mycorrhizal fungi (AMF) resulted in microbial community assembling to migration capacity.
AMF promotes recruitment of metal‐resistant microbial community.
AMF changes root metabolites to affect rhizosphere bacterial community assembly.
AMF passivates rhizosphere Cd based on regulating ecological characteristics.
INTRODUCTION
Cadmium (Cd) is easily absorbed by crops and threatens human health as it remains in the soil and cannot be degraded owing to its long half‐life [1]. According to the Agency for Toxic Substances and Disease Registry, the global average level of Cd pollution in agricultural soil is 0.01–2 mg/kg; however, the pollution rate in some areas exceeds this level. Therefore, Cd is the first harmful chemical substance of global significance [2]. Owing to the limited amount of cultivated land globally, the absorption of pollutants should be reduced while simultaneously improving crop resistance to stress [3]. The artificial inoculation of functional microorganisms effectively solves these problems [4]. Arbuscular mycorrhizal fungi (AMF) are rhizosphere microorganisms that exist widely in nature and can form symbiotes with more than 90% of plants. Importantly, AMF can maintain symbiotic relationships with plants in severely polluted soils and regulate the rhizosphere environment [5], improving plant mineral nutrition and reducing plant absorption and heavy metal (HM) accumulation [6]. The resistance of Medicago sativa for Cd considerably improves after inoculation with AMF and reduces Cd transfer from the soil to aboveground organs, which is of great significance for herbage harvested from aboveground parts [3]. AMF can cause the “Bio‐dilution” of HMs in vivo by increasing plant biomass to improve plant tolerance, but the concentration of HMs absorbed tends to increase owing to the improvement of root morphology and function [7]. The alfalfa root biomass was increased after AMF inoculation (compared with without inoculation). Importantly, the root surface area (potential absorption area) increased by 24% (p < 0.05) without any significant increase in total Cd content uptake (p < 0.05) [3]. Therefore, we speculate that the AMF regulation of rhizosphere microecological characteristics prevents the absorption of Cd by roots, which is the “first line of defense” against Cd absorption.
Cd absorbed by plants mainly occurs in the rhizosphere, which is considered one of the most complex ecological regions and hotspots in the fields of biology and ecology [8]. The rhizosphere microenvironment is related to the chemical characteristics of pollutants, as well as the occurrence region of plant–soil feedback [9], among which the living microbial community is a key hub for achieving the above interaction [10]. Rhizosphere microbiome is the main driver of soil material cycling and conversion and an important factor affecting Cd bioavailability [11]. Changes in the metabolic function of microbes can decrease Cd bioavailability by modulating soil properties (aggregates, water content, organic matter, Eh, and pH) and possible microbial core functions toward HM resistance [12]. This process is closely related to the species and function of the keystone (core microbial or dominant species) [13]. Previous studies have reported microbial community rebuilding by AMF in the plant rhizosphere [5, 14, 15]; however, its regulatory pathways and hub signals have not been clarified, although the stimulator is considered to be phytogenic rhizosphere secretion [16]. Root exudates maintain the interaction between plants and microorganisms as signals [17], and AMF inoculation can cause changes (composition and content) in host plant root exudates [18], such as the acceleration of the synthesis of plant organic oxides, vitamins, polyphenols, plant hormones, flavonoids, and other metabolites, eventually leading to variations in the microbial community [19]. Determining the hub components of root exudates that affect rhizosphere microbial construction is difficult [16]. Although defective mutants of key genes for specific secretion synthesis can be constructed, the secretion components affecting rhizosphere microbial construction are indistinguishable, and most plants cannot obtain the corresponding mutants [20]. Therefore, the relationship of metabolites associated with the microbial community assembly is difficult to identify. The joint application of the topological overlap matrix (TOM) algorithm of weighted correlation network analysis (WGCNA) and the important prediction of variables in the random forest model, identifying the “key” metabolites induce rhizosphere microbiome assembly and reflect the microbial community characteristics (keystone, dominant species, and Bray–Curtis distance) in the “rebuild” state [21]. In addition, root exudates directly impact the soil, further influencing the interactions with microorganisms. Thus, understanding the pathways involved in this complex rhizosphere system can contribute to revealing the mechanisms through which Cd is passivated.
High microbial biomass and activity in the rhizosphere make the root interface an important hot spot for carbon and nutrient cycling [22]. However, in previous standardized experimental procedures, soil microbial diversity or enzyme activity was measured after soil homogenization, disregarding the difference in microbial activity between hot and nonhot spot areas and also causing disturbance to the in situ rhizosphere microbial environment, resulting in a large experimental deviation. The root box method can obtain rhizosphere soil in situ, achieving undisturbed sampling and displaying migration characteristics [23]. This study established Glomus mosseae and alfalfa as a symbiotic system and the root box‐Cd2+ membrane blotting combined assay. The Cd properties in the rhizosphere environment were analyzed in situ, and changes in root metabolomics, soil physicochemical properties, and microbial characteristics in situ under Cd stress induced by AMF inoculation were compared. This study aimed to address the following questions: (1) whether AMF reduces the Cd migration by regulating the rhizosphere environment; (2) the role of rhizosphere metabolites in the phylogeny of microbial, and the “signaling substances (metabolites)” that trigger microbial community rebuilding; and (3) regulation of root metabolite–soil–microbial interactions by AMF in the rhizosphere to reduce Cd uptake by plants. Therefore, in the present study, we elucidated causal interactions between the rhizosphere microbiota, host metabolism variation, and soil factor influencing root absorbing Cd under Cd pollution.
RESULTS
AMF drives the reduction of migration of Cd in the rhizosphere
Soil Cd distribution showed a uniform distribution, gradual subsidence, and reduced distribution during 0–20 days of alfalfa transplantation into the root box (Figure 1A). Until Day 27, Cd was distributed along the plant roots, and hot spots of Cd accumulation appeared around the roots on Day 33 (red area, unit pixel Cd >4.05 mg/kg). However, the hotspot percentage in the AMCd group was significantly lower (p < 0.05) than that in the Cd‐exposed group (Figure 1B). Cd2+ fluorescence localization scanning also showed that the median frequency distribution of pixel sites and hotspot proportion of the AMCd group (larger than 4 mg/kg) were lower than those of the Cd exposure group (Figure 1C) (p < 0.05). Moreover, colocalisation analysis (image of root and Cd2+ fluorescence distribution) based on the Mantel test also revealed that Cd2+ aggregation in the rhizosphere after Cd exposure (r = 0.21, p = 0.032) was significantly higher than that after AMCd treatment (r = 0.17, p = 0.041) (p < 0.05). The Cd concentration in the Cd exposure (4.23 mg/kg) was higher than that in the AMCd treatment (3.92 mg/kg) in the rhizosphere (Figure 1D). In addition, the content of available Cd in the AMCd treatment was significantly lower by approximately 1/3 than that in Cd exposure (p < 0.05). However, the organic‐bound Cd increased significantly (p < 0.05), and the change of the organic‐bound Cd and available Cd had an evident correlation with Cd migration ability (Figure 1E,F).
Figure 1.
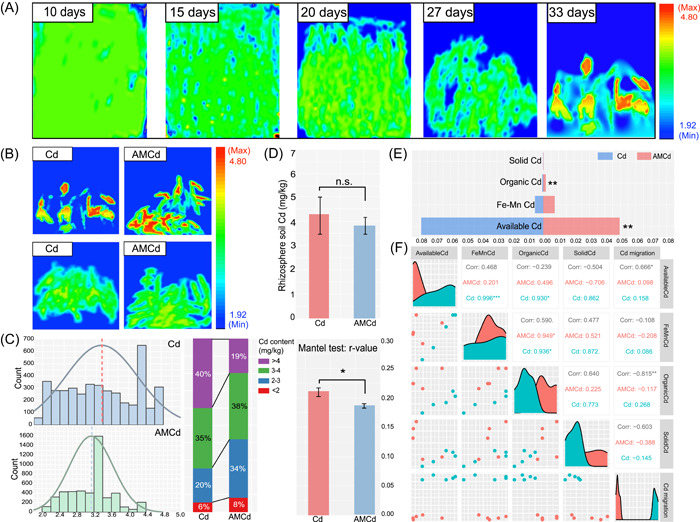
Migration and chemical states of Cd in the rhizosphere treated by Cd exposure and AMCd treatment. (A) Changes in Cd fluorescence imprinting in root boxes of groups treated for 0–33 days in Cd exposure. (B) After 33 days of Cd exposure, Cd fluorescence imprinting was performed on the rhizosphere of the Cd exposure and AMCd treatment group. (C) Frequency distribution of scanning values of fluorescent pixel sites in Cd and AMCd processing groups and colocation analysis based on Mantel test. (D) Cd content in rhizosphere soil of Cd exposure and AMCd treatment. (E) Chemical states of Cd in rhizosphere soil by Cd exposure and AMCd treatment. (F) Spearman correlation analysis between Cd chemical states and Cd migration characteristics (percentage of hot spots). Five biological replicates were performed for each index, and the error bars are standard errors. *Significant differences (p < 0.05) among each treatment. **Significant differences (p < 0.01) among each treatment. ***Significant differences (p < 0.001) among each treatment.
Changes in rhizosphere soil bacteria community
First, 5187 OTUs were obtained by 16S sequencing (Supporting Information: Figure S2). Classification analysis showed that the rhizosphere bacteria mainly comprised six phyla (Figure 2A), of which Proteobacteria was the largest (67.65%), followed by Firmicutes (9.08%), Patescibacteria (8.06%), Bacteroidetes (7.36%), Actinobacteriota (2.08%), and Acidobacteriota (1.5%). Proteobacteria (61.67%) and Actinobacteriota (2.2%) decreased significantly after Cd exposure compared to CK (p < 0.05), whereas Firmicutes (11.72%), Patescibacteria (11.54%), Bacteroidetes (7.16%), Actinobacteriota (1.76%) were upregulated (p < 0.05). Proteobacteria (64.41%), Firmicutes (9.51%), and Bacteroidetes (8.67%) were more abundant in the AMCd‐treated group than in the Cd‐exposed group (p < 0.05). The top 45 genera which have the most relative abundance are shown in Figure 2B, the dominant being Dyella (30.59%), LWQ8 (27.69%), Burkholderia (23.83%), Massilia (23.08%), Rhodanobacter (16.65%), Sphingomonas (16.44%), Novosphingobium (13.39%), Enterococcus (7.5%), Bacillus (5.7%), and Micropepsis (4.8%). Compared with CK, the abundance of Dyella and Micropepsis in the Cd exposure group decreased by 35% and 6.5%, respectively (p < 0.05). Burkholderia, Massilia, Rhodanobacter, and Novosphingobium increased (p < 0.05), and Massilia increased nearly threefold. In addition, Dyella and Sphingomonas in the AMCd treatment were significantly increased by 45% and 39%, respectively, compared to Cd (p < 0.05); however, Sphingomonas was higher than in CK (p < 0.05), and Dyella was not significantly different from CK. The rarefaction curve shows (Figure S3) that the Shannon, OTUs, Simpson, and Chao1 indices of the tested samples tended to be flat with increased sequencing quantity, indicating the high reliability of the results. In Cd exposure, Shannon, OTUs, Simpson, and Chao1 indices decreased significantly compared to those in CK (p < 0.05). Chao1 and OTUs in the AMCd treatment significantly improved with Cd exposure (p < 0.05), and the Shannon and Simpson indices increased slightly but not significantly (p > 0.05) (Figure 2C). Based on the paired comparison of Bray–Curtis distance and the complete data set of ANOSIM analysis (Figure 2D), CK versus Cd (r = 0.388, p = 0.047), Cd versus AMCd (r = 0.224, p = 0.039), CK versus AM (r = 0.280, p = 0.048), and CK versus AMCd (r = 0.580, p = 0.005). These results indicate that AM treatment and Cd exposure resulted in variations in the bacterial community. Microbial community assembly process analysis (Figure 2E) revealed that CK and AMCd treatments were random processes (|βNTI| < 2), and Cd and AM treatments were deterministic processes (|βNTI| > 2). Further, exploring the community change process (paired comparison βNTI) found that CK versus Cd, CK versus AM were |βNTI| > 2, and CK versus AMCd were |βNTI| < 2, Cd and AM were both the primary factors in the bacteria community assembly tending to the deterministic process; however, the former was for homogenous selection, the latter for variable selection (Figure 2F).
Figure 2.
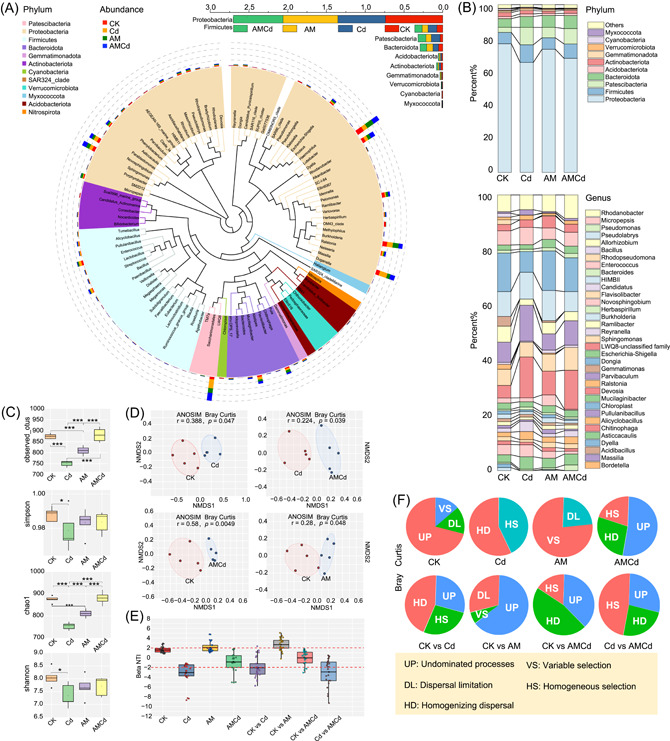
Composition and diversity (α and β) of alfalfa rhizosphere soil bacterial community under CK, Cd, AM, and AMCd treatments. (A) Maximum‐likelihood phylogenetic tree of the bacterial genome bins detected and the relative abundance of dominant phyla (top 10) in all soil samples under four treatments. (B) Bacteria in the soil at the phylum and genus levels under four treatments. (C) Indices of Shannon, Simpson, Chao1, and OTUs of bacteria in the rhizosphere soil. (D) Nonmetric multidimensional scaling (NMDS) ordinations based on Bray–Curtis distances and Bonferroni based on ANOSIM in rhizosphere soil bacterial communities. (E) Contributions of deterministic and stochastic processes in community assembly within collected rhizosphere soil samples. βNTI calculation of phylogenetic turnover among each treatment. (F) The relative influence of each community assembly process among four treatments was defined by the percentage of site pairs governed by each process. Five biological replicates were performed for each index, and the error bars are standard errors. *Significant differences (p < 0.05) among each treatment. **Significant differences (p < 0.01) among each treatment. ***Significant differences (p < 0.001) among each treatment.
Bacterial biomarker and keystone
The soil bacterial microbiome in the rhizosphere varied among the four treatments, as shown by LDA effect size (LEFSe) analysis (Figure 3A). The paired t‐test comparison showed that the significant differences in CK versus Cd were Ramlibacter and Rhodopseudomonas (Figure 3B) and that in Cd versus AMCd were Asticcacaulis and Bacteroides. Paired similarity percentage (SIMPER) analysis showed that the top five contributors of CK versus Cd and Cd versus AMCd were LWQ8, Massilia, Dyella, Burkholderia, and Enterococcus (Figure 3C). The topological features of the co‐occurrence network are shown in Figure 3D. The average network degrees of the CK and AM were 10.643 and 10.857, respectively, and the average weight degrees were 0.421 and 0.405, respectively. The average degree of Cd and AMCd were 13.57 and 13.032. The eigenvector centrality of the bacteria in the Cd exposure group was higher than CK, and the modularity coefficients were also increased. Compared with Cd, AMCd showed a higher modularity coefficient and eigenvector centrality, indicating a closer relationship between AMCd treatments. The highest scores of Page ranks or eigencentrality were identified as keystones, and Proteobacteria, Bacteroidetes, and Firmicutes with the highest ranking were identified as keystones (Supporting Information: Table S1). The keystone of CK was Asticcacaulis, Methylobacterium, and Sphingomonas, including nine genera (Page ranks = 0.007, Eigencentrality = 1), and that of Cd exposure was Rhodanobacter, Dyella, and Burkholderia in Cd exposure (Page ranks = 0.0058, Eigencentrality = 1) (Supporting Information: Table S1). Parvibaculum, Massilia, and Alistipes, among others, including eight keystone genera, were treated with AMCd (Page ranks = 0.0067, Eigencentrality = 1), and some keystones overlapped with CK (LWQ8, Ramlibacter) and Cd (Massilia). Thus, AMF and Cd can affect bacterial communities by recruiting preferred keystones.
Figure 3.
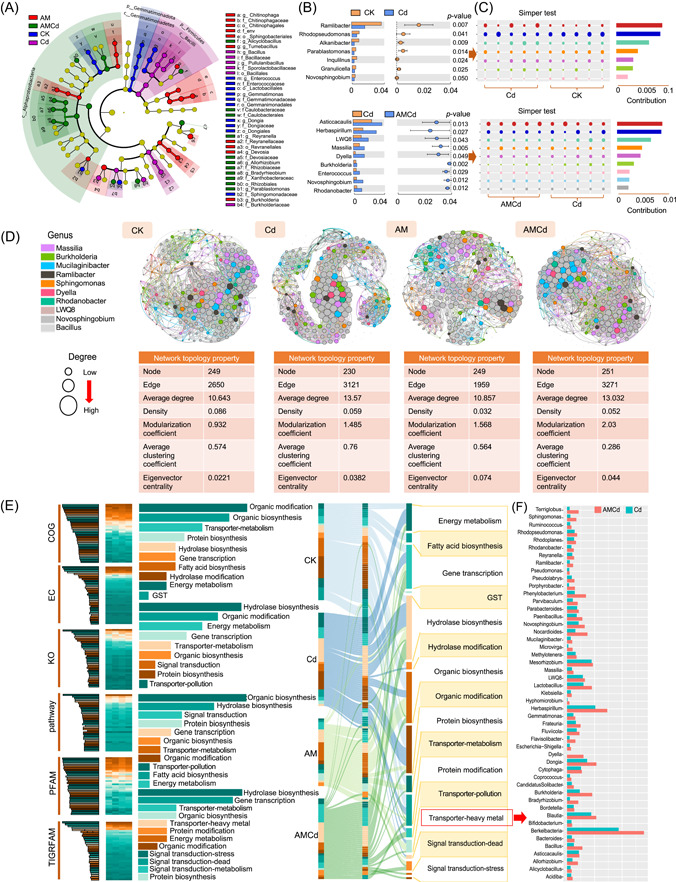
Prediction of keystone and function of alfalfa rhizosphere soil bacterial community under CK, Cd, AM, and AMCd treatments. (A) Bacteria biomarkers in the different treatments based on the LEfSe analysis. Different colors represent different treatments, and the circles from inside to outside correspond with phylum and genus. The color‐coded one within the cladogram denotes the taxa with a significantly higher relative abundance in the treatment as analyzed by the Kruskal–Wallis test with p < 0.05 and a logarithmic LDA score >3.5. Genera with a relative abundance of less than 0.1% were not included. (B) The main bacteria (genus level) differs significantly between CK versus Cd and Cd versus AMCd groups based on Welch's t‐test. (C) A similarity percentage (SIMPER) analysis based on the decomposition of the Bray–Curtis difference index between CK versus Cd and Cd versus AMCd groups. (D) Visual network and topology statistics of microbial co‐occurrence in the four treatments. (E) Cluster analysis of relative abundance based on PICRUSt2 functional annotation to soil bacteria under the four treatments. (F) Heavy metal (pollutant resistance) related 49 bacteria were selected in Cd versus AMCd treatment groups based on PICRUSt2 functional annotation, and their relative abundance was ranked.
AMF recruits bacteria capable of passivating HM
Functional prediction of rhizosphere bacteria under the four treatments based on databases revealed that the four most abundant groups (accounting for 75%) were lipid metabolism, signal transduction, synthesis, and organic matter modification, and substance transport (Figure 3E). The functional preferences of rhizosphere bacteria were (from high to low) organic synthesis and modification, metabolite transport, hydrolytic enzyme synthesis, gene transcription, energy metabolism, and N absorption and assimilation in the CK. Hydrolases, organic synthesis and modification, energy metabolism, gene transcription, signal transduction, protein modification, and pollutant transport were observed upon Cd exposure. Meanwhile, the functions of stress signal transduction, programmed apoptosis signals, HM transport, and metabolite transport were increased in the AMCd treatment compared to Cd exposure. The microbial increase in the AMCd treatment with the HM transport (pollutant removal) function was selected. Approximately 139 bacteria could be negatively correlated with changes in Cd hotspots (p < 0.05), belonging to 101 genera (Supporting Information: Table S2), including Dyella, LWQ8, Ruminococcus, Reyranella, Allorhizobium, Pseudolabrys, Novosphingobium, and Flavisolibacter. Forty‐nine genera showed significant increases in abundance (Cd vs. AMCd; Figure 3F) and were compared using (Web of Science, CNKI, ELSEVIER, and KEGG). The above bacteria could phagocytose, adsorb, or embed HM; therefore, they were named HM‐removers.
Soil factors associated with reduced Cd hot spots
Cd exposure reduced the available nutrient (N, P, and K) content in the rhizosphere soil compared with CK (p < 0.05) (Supporting Information: Figure S4); NH4‐N decreased by approximately 40% (p < 0.05), but EOC and GRSP were significantly increased (p < 0.05). However, AMCd treatment increased available N content (including NH4‐N and NO3‐N), available K, TOC, EOC, GRSP, and MBC compared to Cd exposure (p < 0.05). AMF improved rhizospheric soil nutrient concentration. In addition, the extracellular enzyme activity in the rhizosphere soil changed significantly. Compared with CK, Cd exposure significantly reduced enzyme activities related to N and P cycles in the rhizosphere soil (including acid phosphatase, laccase, urease, phenol oxidase, and nitrogenase, p < 0.05), whereas the C cycle‐related enzymes (β‐glucanase, β‐glucosidase, xylanase, leucine aminopeptidase, luciferase diacetate, and lignin peroxidase) were significantly increased (p < 0.05) (Supporting Information: Figure S5). Compared with Cd, the activities of N and P cycle‐related enzymes (including acid phosphatase, laccase, urease, phenol oxidase, and nitrogenase) in the AMCd treatment were increased, p < 0.05), whereas β‐glucanase, luciferase diacetate, and lignin peroxidase in the above C cycle related enzymes were decreased (p < 0.05). However, they all were higher than CK (p > 0.05). These results indicate that AMF improved the rhizospheric soil nutrient concentration by increasing extracellular enzyme activity (Supporting Information: Figure S6). Mantel test and the Spearman correlation model were used to screen the soil factors and bacterial groups related to Cd passivation based on “Cd‐soil attributes‐microorganisms” as the main axis (threshold: r > 0.6, p < 0.05). Acid phosphatase, urease, NO3‐N, NH4‐N, available P, β‐glucanase, and phenol oxidase may be related to the Cd passivation process (Figure 4A), and the above factors were significantly correlated with bacteria (Figure 4B). Acidobacteriota, Actinobacteriota, Bacteroidota, Firmicutes, Gemmatimonadota, Patescibacteria, Proteobacteria, and Verrucomicrobiota were significantly correlated with the above six soil factors (Figure 4C), and AM inoculation significantly increased the abundance of the above bacteria (Cd vs. AMCd). Approximately 63% of these bacteria were HM‐removers and influence soil and Cd migration indirectly or directly (Figure 4D). Therefore, changes in soil properties can reduce Cd migration.
Figure 4.
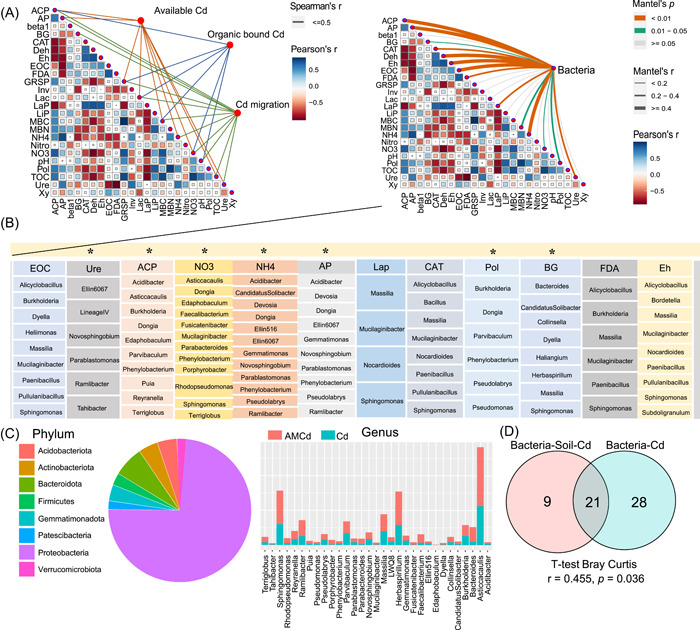
The interaction between soil and bacteria in the rhizosphere was investigated with “Cd–soil–bacteria” as the main axis. (A) Two Mantel tests of Cd property–soil property and bacteria–soil property, respectively (p < 0.05). (B) A Spearman correlation model between bacteria and soil factors, which both have Cd passivation ability. *Significant correlation (p < 0.05). (C) The selected bacteria were counted at phylum and genus levels, and their relative abundance under AM‐ and non‐AM treatment groups were compared. (D) A community similarity test based on Bray–Curtis distance for bacteria groups that can influence Cd properties and simultaneously affect Cd and soil.
Changes in root metabolites and their effects on rhizosphere microecology
Root metabolomics showed significant differences under different treatments (p < 0.05; Supporting Information: Figure S7A), with the Cd versus AMCd treatment group showing the largest difference (containing 301 different metabolites), followed by the AM versus AMCd treatment group (containing 235 different metabolites). Based on the ANOSIM analysis of the Bray–Curtis distance in the four treatments, AMF principally caused differences in metabolic substances (top Bray–Curtis ranks, Supporting Information: Figure S7B). In addition, it may upregulate the metabolic substance content through flavonoid synthesis, amino acid metabolism, phenyl propyl synthesis, and secondary metabolism (based on the comparison between the AM and non‐AMCd groups, Supporting Information: Figure S7C). Moreover, as mentioned above, AMF can affect the bacterial community to decrease Cd migration; therefore, weighted metabolite–microbial coexpression network analysis was used to explore the main factors affecting microbial community variation. The metabolites were divided into modules by hierarchical clustering (module corresponding color), and the model was adapted to microbial keystone and overall microbial community expression (model parameters: correlation >0.2, p < 0.05) (Figure 5A,B). The keystone and community matrix (Bray–Curtis distance) of microorganisms under different treatments were closely correlated with flavonoid metabolites (weight and module connectivity were highest) (Figure 5C). Therefore, flavonoid metabolites were considered the key hubs affecting community change, and AMF changed the bacterial community by increasing the flavonoid content (Figure 5C). In addition, using weighted metabolite–microbial coexpression network analysis, Spearman correlation model, and Mantel test, metabolites that can affect bacterial and soil properties (screened for Cd passivation ability) and reduce Cd migration were identified (Figure 5D–F). Eleven metabolites had ternary overlap, and 14 substances had binary overlap, indicating that metabolites, microbial, and soil properties could directly passivate Cd and had complex interactions among them (Figure 5G).
Figure 5.
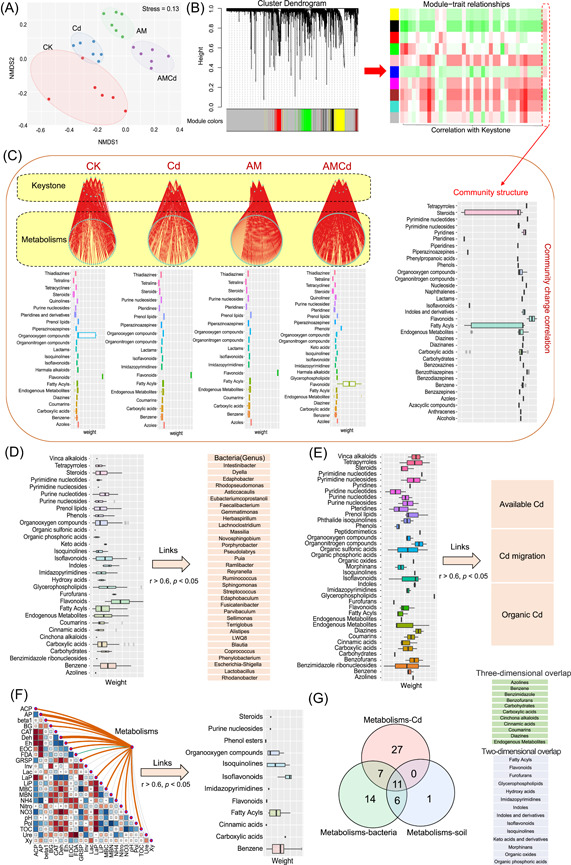
Root metabolome and rhizosphere microbiome interactions. (A) Nonmetric multidimensional scaling (NMDS) ordinations based on Bray–Curtis distances and Bonferroni based on ANOSIM in alfalfa root metabolisms. (B) The module division of root metabolites, the hierarchical clustering tree with module identification, the allocated modules, and the metabolite information contained in the modules. The colors red to green indicate the correlation between metabolite modules and microbial information from strong to moderate, among which only red represents a significant influence (p < 0.05). (C) A Spearman correlation model of the bacterial keystone and the Bray–Curtis distance information representing the community characteristic structure under four treatments and weight ranking of high effects metabolites were obtained by prediction. Spearman's correlation was calculated by relating the eigenmetabolite of each module to the relative abundance of each taxon using the WGCNA trait function. (D) A Spearman correlation model (r > 0.6, p < 0.05) between HM‐resistant bacteria and metabolite, which both increased by AM inoculation (AM vs. non‐AM, t‐test, p < 0.05) and a metabolite weight ranking. (E) A Spearman correlation model (r > 0.6, p < 0.05) between Cd property and metabolite, which increased by AM inoculation (AM vs. non‐AM, t‐test, p < 0.05) and a metabolite weight ranking. (F) A Mantel test (r > 0.6, p < 0.05) between soil property and metabolite, which increased by AM inoculation (AM vs. non‐AM, t‐test, p < 0.05) and a metabolite weight ranking. (G) A Venn diagram based on the statistics of the frequent occurrence of the same metabolites in the three groups of metabolic versus soil, metabolic versus Cd, and metabolic versus bacteria. WGCNA, weighted correlation network analysis.
AMF passivates rhizosphere Cd based on the regulation of metabolic–microbial–soil
A composite SEM clarified the interactions among root exudates, soil factors, microbes, and Cd (Figure 6A). After AMF inoculation, Cd migration capacity was reduced in three ways, “metabolite–microbe–soil–Cd,” and “metabolite–soil–Cd,” two indirect actions, and “metabolite–Cd“ direct action mode. Complementary VPA found that the combined effect of the three could reduce the maximum explanation rate of Cd migration (42.4%), suggesting the importance of combining the three for Cd toxicity mitigation (Figure 6B). Regarding the single interpretation rate, the microbial population was the largest (15.5%), whereas the total explanation rate of microbes (including single microbes and their superimposed effects) reached 72%, verifying that rhizosphere microbial remodeling played an important role in reducing Cd migration. The rhizosphere soil is affected by root metabolites and soil microbial secretions; therefore, soil extracellular enzymes (LaC, LiP, and CAT), organic matter (TOC, EOC, and GRSP), and the REDOX potential are the key cadmium passivation factors (Figure 4A–C). Based on our results, we can fully understand the interactions of metabolites and soil microbes in the rhizosphere as well as minimize the risks of collinearity and pseudo‐correlation, which will help resolve how AMF regulates Cd in rhizosphere microspheres.
Figure 6.
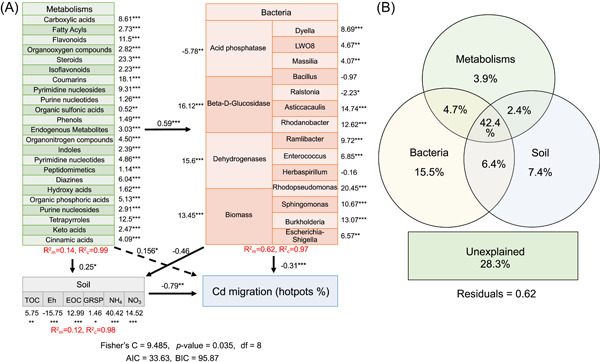
Process of cadmium passivation by AMF‐alfalfa symbiont. (A) Structural equation model of the interaction between multidimensional data sets of metabolites, microorganisms, soils, and Cd. (B) Variance decomposition analysis of the effect of metabolites, microbial, and soil on Cd migration. *Significant differences (p < 0.05) among each treatment. **Significant differences (p < 0.01) among each treatment. ***Significant differences (p < 0.001) among each treatment. AMF, arbuscular mycorrhizal fungi.
DISCUSSION
Recruitment of “functional community” by rhizosphere bacteria rebuilding
Using ANOSIM analysis, it was found that AM and Cd‐induced rebuilding of the rhizosphere microbial community (Figure 2D). Furthermore, the AMCd treatment group was formed by 49 genera of bacteria composed of HM‐remover (Figure 3E). This is one of the reasons for the Cd passivation of microorganisms. Therefore, how are these resistant microbial communities formed? We first analysed whether the AM experimental group (no Cd exposure) could also recruit function microbial. A total of 32 common rhizosphere growth‐promoting bacteria (AM vs. non‐AM) were recruited by AMF, of which 19 were nitrogen fixing, 10 were phosphorus solubilizing, and three had both capacities (Supporting Information: Figure S8). This explains why AM inoculation can effectively alleviate the N and P limitations caused by Cd pollution in rhizosphere soil (Supporting Information: Figure S4–S6). The Spearman correlation model (r > 0.6, p < 0.01), random forest model prediction (Importance > ShadowMax), and HM‐remover in the inoculation treatment group were obtained (Figure 3F), of which 10 were plant growth‐promoting rhizobacteria (PGPR) (Supporting Information: Table S3). Their mechanisms of action on HM are divided into three categories based on the following studies. The first category includes the phagocytosis of HM (Supporting Information: Table S4), which has 19 groups, including Bacteroides, Burkholderia, and Candidatus solibacter [24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41], can directly ingest (phagocytose) HM ions. The second has surface adsorption ability (Supporting Information: Table S4), including 15 groups such as Asticcacaulis, Bacillus, and Bacteroides. Abundant functional groups, such as –OH, C═O, and –NH existed on the biofilm surface of the above bacteria, which could be directly complexed with Pb, Cu, Hg, Cd, and other bivalent HM ions [42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55]. Therefore, this indicates that organic‐bound Cd increased (Figure 1E). The third achieves Cd degradation through secretion production, including 10 bacteria of Acidibacillus, Alicyclobacillus, and Allorhizobium (Supporting Information: Table S4). Acidibacillus, Alicyclobacillus, and Bordetella secretions reduce metal ions to mineral‐bound and residual states [56, 57, 58]. While Parvibaculum, Phenylobacterium, and Porphyrobacter produce extracellular polymers, metallothionein, and porphyrins, respectively, which are all strong metal complexes [28, 59, 60, 61, 62, 63, 64]. These microbes indirectly decrease Cd migration by altering soil properties (extracellular enzymes, GRSP, MBC, and Eh). Thus, the recruited “functional community” reduced Cd migration in several ways.
Nonaccidental behavior by AMF recruitment of resistance microbial community
AMF can recruit PGPR [65]; however, the randomness of the recruitment of HM‐removing bacteria requires verification. Therefore, a meta‐analysis was conducted by integrating literature on the relationship between AMF and rhizosphere bacteria into 34 parallel experiments (The methods and results are detailed in the supplementary materials, and references are available at Supporting Information: Table S5). The screening of Cd‐resistant bacteria in the above literature and the HM‐remover strains in this study mostly coincided (Supporting Information: Table S6). Among them, 33 taxa, including Rhodoplanes, Burkholderia, and Sphingomonas, frequently appeared (ranking in the top 25%) (Supporting Information: Figure S9). Indigenous microbial species (OTUs) were the main cause of the frequency differences (p < 0.05) (Supporting Information: Figure S10), and the random forest model results were also consistent with the above (Supporting Information: Figure S11). Specifically, there was a significant positive correlation between the two (r = 0.53, p = 0.0011) (linear fitting model, Supporting Information: Figure S12), and the regulation was consistent with grassland, garden, farmland, marsh, and wasteland ecosystems (p < 0.05) (Supporting Information: Figure S13). This effectively proved that AMF could recruit HM‐resistant bacteria, but the number and type of recruitment depended on the species of indigenous microorganisms (OTUs), and this explains the difference in coincidence rates (p < 0.05) (Supporting Information: Figure S10).
Conditions for metal‐resistant bacteria play a key function
Although the above analysis proved that resistant bacteria recruited by AMF were universal, this did not mean an evident HM‐resistant functional microbiome. In contrast, some studies suggested that AMF could promote bacteria involved in nitrification and denitrification processes, nitrogen fixation, and nodulation to become keystones. This is because most of the experiments were based on AMF inoculation when the soil was poor in N and P, ultimately making the core function of the rhizosphere microbiome incline to the nutrient acquisition strategy [14, 15, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79]. Therefore, they play an important role only when environmental stress causes the recruited functional microbes to become keystones. After AMF randomly recruiting bacteria, most bacteria have functional redundancy, and when encountering various stresses, functional microbial relief plays a leading role, making them keystones [80]. Simultaneously, if the stress of adversity is too high, the recruitment process will be accompanied by the death of bacteria, and only resistant microbes with strong vitality will be retained [81]. Similar to our study, AMF and Cd could recruit their preferred keystones (Figure 3D). In the AMCd treatment, keystone bacteria were increased, such as Parvibaculum, Alistipes, Lactobacillus, Duganella, Ramlibacter, Bacteroides, and LWQ8, based on AM inoculation (compared to the AM treatment group) (Supporting Information: Table S1). These seven bacteria belong to HM‐remover (Supporting Information: Table S2), which may be one of the reasons for the HM‐remover feature group in AMCd. Meanwhile, through ggClusterNet, modules related to Cd migration and chemical state change were clearly distinguished, and the above keystone was also an important hub of modules (Supporting Information: Figure S14). In conclusion, the combination of AMF's extensive recruitment of microorganisms and Cd's screening effect on them formed a functional module of HM resistance in the AMCd treatment.
Root metabolites affect rhizosphere bacterial community assembly
Plants can release 20%–40% of photo‐assimilated carbon through root secretions [17], and the composition and content are related to macromolecular metabolites and passively diffused low‐molecular‐weight compounds produced by plant metabolism [16]. Specific metabolites of plant roots (coumarins, flavones, and benzene) as secretions affect the rhizosphere microbiome and the specific species enrichment [20]. In this study, weighted metabolite–microbial coexpression network analysis was used for prediction, and sugar, flavone, coumarin, phenol, fatty acids, and amino acids were significantly correlated with the microbial community composition (Bray–Curtis distance) and keystones (Supporting Information: Figure S7A–C). Based on the random forest model, flavonoids had the greatest influence on the dominant microbial species and keystones (the largest weight). Moreover, supplementary experiments have confirmed that it can be secreted into the extraneous environment in large quantities (Supporting Information: Figure S15). Therefore, flavonoids are signaling regulatory factors that affect bacterial community reassembly. In particular, phytogenic flavonoids regulate rhizosphere microbial communities [82]. The interactions between plants and microorganisms occur in two stages. First, plants secrete chemical signals into the rhizosphere that can encourage, limit, or inhibit microbial activity and proliferation. Flavonoids are often accepted as signaling molecules [82] and can selectively promote the growth and propagation of PGPR (such as Azotobacter) [83]. The keystone and dominant species in the AMCd treatment were mostly PGPR (including HM‐remover), possibly owing to the high contribution of flavonoids (Figure 5C). The second stage is a quorum sensing (QS) process, in which microbes detect low‐molecular‐weight compounds released by plant roots (or other microorganisms) and regulate various microbial population activities through a signal cascade [20]. Phytogenic flavonoids may be the regulatory molecules in the AHL family signaling pathways (AHL‐mediated QS system is the first “language family”) [17]. Moreover, phylogenetic lipids, phenols, alcohols, ketones, aldehydes, keystones, and terpenes are used as QS signaling substances to comprehensively regulate microbial communities [84], consistent with our prediction of metabolite species (Figure 5C–F). Polypeptides, coumarins, fatty acids, benzene, phenols, secondary metabolites, and other substances contributed significantly to microbiome assembly (p < 0.01). The above can be effectively secreted into the extraneous environment (Supporting Information: Figure S15). In summary, AMF regulates rhizosphere microbial rebuilding through the superimposed effect of flavonoid metabolites as the dominant metabolites accompanied by other small molecules. Although the model was used to predict the relationship between metabolites and microorganisms (high threshold background), accurate single/combination addition of metabolites is required to verify the relationship between the two.
CONCLUSION
The study indicates that controlled metabolic activities in alfalfa roots are critical for beneficial interactions with the rhizosphere microbiota. Under Cd toxicity, AMF specifically assembles bacteria of the taxon HM‐resistant microbiome in the rhizosphere, and this in turn decreases Cd migration. A full understanding of the causal relationships between the rhizosphere microbiota, host metabolism regulation, and soil factors change could offer a sustainable solution that could benefit both the environment and human health. Based on our results, the flavones increased by AMF in the alfalfa rhizosphere might recruit HM‐remover bacteria. Hence, it provides a new scientific explanation for how AMF improves plant Cd tolerance from the perspective of plant and microbial ecological interactions.
METHODS
Alfalfa growth conditions
The topsoil (0–20 cm) was collected from soil (Cd background content of 0.017 mg/kg) of a corn field (126°48′E, 45°16′N), passed through a 2 mm sieve, and mixed with sterilized flower soil and vermiculite at 3:3:1 to form a composite medium. Furthermore, 50 g/kg activated and inactivated AMF bacterium (G. mosseae L.) (provided by Laboratory of Restoration Ecology, Heilongjiang University) were added to form two treatments: a control (CK) and an AMF inoculation, approximately 35 spores/g soil. They were placed into self‐made root boxes, and young alfalfa (M. sativa L.) seedlings (courtesy of the Chinese Academy of Agricultural Sciences) were planted in each box. The root box comprised a (polypropylene) PP plate with 20 × 20 × 0.3 cm dimensions. The door was movable, and the bottom was made of 600‐mesh nylon mesh. The soil in the root box absorbed water from the bottom up such that the relative position of soil Cd could not sediment with the flow. The root box was tilted at 45–50° to enable the roots to fit the door at the back of the box owing to ground growth, as shown in Supporting Information: Figure S1A–C. The root infection rate was determined every 10 days during seedling growth. Infection was considered successful when the infection rate reached 90% (Supporting Information: Figure S1D,F). Then, the two groups were treated with 2 mg/kg CdCl2 (the concentration obtained according to supplement materials) and sterile distilled water, and a total of four groups were formed: CK, Cd, AM, AMCd, and cultured for 25 days.
Rhizosphere Cd fluorescence imprinting
The experimental process of root box in situ imprinting is shown in Supporting Information: Figure S1E (applied for a patent in the State Intellectual Property Office: CN114518347A) [85]. Root boxes treated with Cd and AMCd were randomly selected every 5 days for the Cd2+ fluorescence imprinting test. A standard 600‐mesh nylon film was cut to an appropriate size and soaked in LeadmiumTM fluorescent dye (Molecular ProbesTM; Thermo Fisher Scientific) for approximately 1 h. The root box was opened, and the saturated membrane was tightly fitted onto the surface, covered, and returned. Approximately 0.5 h later, the root box was opened, the imprinted membrane was removed, and sand and mud particles on the surface were carefully removed using tweezers. It was then placed on an ultraviolet imager (GelDoc XR Biorad; Bio‐Rad Laboratories), and the SYBR Green fluorescence imaging mode was selected for imaging and taking photos. Digital images were analyzed using the ImageJ software 1.53 (MathWorks). An RGB image was converted into black and white, and each pixel was assigned a grayscale value corresponding to 0 for black and 256 for white. The average gray value of each circular calibration film was calculated. The Cd concentration in the calibration film is a function of the gray values. This function was applied to digital images of the fluorescence spectrum. To illustrate the results explicitly, we used color to describe the value of the gray image, where red corresponded to the highest gray value (value 256), and blue corresponded to the lowest gray value (value 0). The color area 25% higher than the average gray value of the pixel points was set as red, indicating the hotspot of Cd aggregation [23]. The Cd speciations (exchangeable Cd, carbonate‐bound Cd, Fe‐Mn oxides Cd, organic matter‐bound Cd, and residual Cd) in rhizosphere soil were analysed using Tessier's sequential extraction procedure [86]. Cd content (including different speciations) was determined using a Perkin Elmer 900 T atomic absorption UV spectrophotometer (Perkin Elmer).
Measurement of soil properties
Rhizosphere soil sampling is shown in Supporting Information: Figure S1G. Soil pH values were measured using a pH meter with a soil‐water ratio of 1:5 (PHS‐3C; LEICI). Organic matter content was determined by an element analyser (Vario EL‐III; Elementar), and total carbon and total nitrogen (N) contents were determined by a dry combustion carbon and nitrogen analyser (JC‐NY4B; Jingcheng Instrument). Soil nitrate (NO3 −‐N) and ammonia (NH4 +‐N) were analyzed using Lachat 8000 flow injection analyser (Lachat QC5000; LACHAT). The soil oxidation–reduction potential (REDOX) was determined using the platinum electrode‐reference electrode method. The above methods refer to Xu et al. [86]. The glomalin‐related soil protein (GRSP) content was determined using the Battini et al. [87] method. Acid phosphatase, β‐1,4‐glucanase, β‐glucosidase, xylanase, catalase, dehydrogenase, invertase, laccase, urease, polyphenol oxidase, leucine aminopeptidase, lignin peroxidase, and FDA hydrolase were determined by microplate method. The above methods refer to Wang et al. [88]. Soil chemical properties were recorded using Excel (v.2019), and significance was tested using SPSS (v.19) analysis of variance.
Extraction and polymerase chain reaction amplification of genomic DNA from rhizosphere soil bacteria
Total microbial community DNA was extracted from the samples using a Fast DNA Spin Kit for Soil (MP, Biomedicals). The bacterial 16S rRNA gene fragments were amplified with the universal primers 799F (5′‐GTGCC AGCMGCCGCGGTAA‐3′) and 1193R (5′‐CCCCGYCAATTCMTTTRAGT‐3′) fused with unique barcode [89]. Gel‐purified polymerase chain reaction products were mixed with equal molar following Illumina sequencing using the platform of Novaseq. 6000. All sequence data have been submitted to the National Center for Biotechnology Information (NCBI) Sequence Read Archive under Bioproject PRJNA975279.
Microbial informatics analysis
QIIME2 software 2017.6 was used to calculate amplicon sequence variant (ASV), Shannon, Simpson, and Chao1 indices, and a Rarefaction Curve was drawn. QIIME2 software was used to calculate the Unifrac distance and plot the nonmetric multidimensional scaling (NMDS) dimension reduction graphs using packages ade4 and ggplot2 in R software. Subsequently, the ANOSIM function in the QIIME2 software was used to analyse significant differences between the groups' community structures. Finally, species with significant differences between the groups were analyzed using LEfSe or R software. LEfSe analysis was performed using LEfSe software and LDA threshold value should be greater than 3.5. By default, the linear discriminant analysis score threshold was four. R software was used to test the differences in the MetaStat analysis between the two comparison groups at six classification levels: phylum, class, order, family, genus, and species, and p‐values were obtained. Species with a p < 0.05 were identified as significantly different species. The t‐test also used R software to analyze significant differences in species at various taxonomic levels. ASV for microbes was conducted using PICRUSt2. The gene function spectrum was inferred from the gene information of the tree and ASV, and the gene function spectra of other unknown species in the Greengene database were used to construct the gene function prediction spectrum for the entire lineage of the bacterial domain. Finally, the bacterial community composition obtained by sequencing was “mapped” to a database to predict bacterial metabolic functions [90]. The bacterial network topology parameters were calculated using Cytoscape v 3.8.0 and Gpehi v 0.9.2.
Correlation analysis of microbial‐environmental factors
Spearman correlation analysis was conducted between species and environmental factors, their significance was evaluated using Corr. test function in the R Psych package, and visualization was performed using the Pheatmap function. The vegan package in R was used for the Mantel test. Based on the species matrix and the provided environmental factor data matrix, the Vegdist function was first used to convert the distance matrix of the two types of data, and then the Mantel function was used to conduct Spearman correlation analysis on the two types of matrices to obtain r and p‐values [21].
Determination of root metabolites
An appropriate amount of sample was accurately weighed into a 2 mL centrifuge tube, 600 μL MeOH (stored at −20°C) was added containing 2‐amino‐3‐(2‐chloro‐phenyl)‐propionic acid (4 ppm) and vortexed for 30 s. Glass beads (100 mg) were added, and samples were placed in a tissue grinder for 90 s at 60 Hz. After centrifugation for 10 min at 12,000 rpm and 4°C, the supernatant was filtered by a 0.22 μm membrane and transferred into a detection bottle for LC‐MS detection. The LC analysis was performed using a UHPLC System (Vanquish; Thermo Fisher Scientific). Mass spectrometric detection of metabolites was performed on Q Exactive (Q Exactive; Thermo Fisher Scientific) with an ESI ion source [91].
Host root metabolome correlates with changes in the microbiome
We used the R package based on WGCNA to replace gene expression in metabolite ion scanning lineages with data sources to identify correlations between quantitative data sets: (1) Metabolite coexpression modules between alfalfa roots under four treatments; (2) Quantitative value of modular characteristic metabolites–microbial community association and in‐module hub metabolites. WGCNA is an unsupervised assay that clusters metabolites based on the expression profiles of 180 substance‐specific samples. To construct coexpression networks robustly, we reserved chromatographic peak areas of ≥5 mapped reads in corresponding regions of at least one substance. The substance expression was normalized to log2 (FPKM + 1). Soft threshold power β calculated adjacency. To minimize the effects of noise and false associations, we converted adjacency into a TOM and calculated the corresponding dissimilarity (dissTOM) as 1‐TOM. We used dissTOM as a distance measure for hierarchical clustering and set the minimum module size (number of genes) to 28 to detect the modules. To quantify the coexpression similarity of all modules, the characteristic genes were calculated and clustered according to their correlations, which were subsequently used to study the crosstalk between metabolite expression differences and microbial traits. We chose 0.8 value relatives for the cluster module. Modular characteristic metabolites were used to represent the patterns of substance expression within a module. The relative abundance of refined bacterial operational taxonomic unit (OTU) tables with taxonomic information was further imported into the WGCNA data framework, and each taxonomic taxon was regarded as a “feature interval.” The module characteristic metabolites were most significantly correlated with these microbial traits. Then, a paired comparison of the expression values of all modular characteristic genes of Pearson's correlation and their associated p‐values was generated. Bonferroni adjustment was used to correct multiple comparisons. We further studied those modules highly correlated with microbial taxa to identify the types of highly connected module ‘hub’ metabolites associated with microbial taxa. The above method can be referred to Yu et al. [16]. Rhizosphere secretion extraction experiments were conducted to verify that the key compounds identified above can be released into the extracellular environment in large quantities. See supplementary material for the methods and results.
Relationship between metabolites, microbes, soil factors, and the Cd hot spots
Spearman and Mantel tests were separately used to determine the correspondence between the percentage of metabolite‐Cd aggregation hotspots and metabolite‐soil factors, and the threshold was set as r > 0.6 and p < 0.01 to improve accuracy. A random forest model was used to compare the results, and duplicate items were retained. The random forest algorithm adopts the “Boruta” algorithm to calculate in RStudio. To improve the accuracy, we set the importance threshold of the variables to >ShadowMax. Mantel test uses dplyr and linkET program packages and sets the threshold as r > 0.6 and p < 0.01. After excluding nonsignificant factors in different datasets with the above algorithm, the composite structural equation model was used to fit the relationship between variables in the four data sets, which was performed using the nlme, lme4, piecewiseSEM, and QuantPsyc packages. The above method can be referred to Tan et al. and Liu et al. [92, 93]. To determine the relative contribution rates of metabolites, microorganisms, and soil factors to the percentage of Cd aggregation hotspots, variance partial analysis was used to quantify the explanatory amount of single data factors and their interactions on the percentage of Cd aggregation hotspots.
AUTHOR CONTRIBUTIONS
Hong‐Rui Wang: Conceptualisation; methodology; software; formal analysis; investigation; writing—original draft. Xin‐Ran Du: Software; formal analysis; investigation. Zhuo‐Yun Zhang: Methodology; software; formal analysis. Fu‐Juan Feng: Writing—review and editing; funding acquisition. Jia‐Ming Zhang: Software; formal analysis.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
Supporting information
Supporting information.
Supporting information.
ACKNOWLEDGMENTS
This study was financially supported by the National Natural Science Foundation of China (413223003) and Fundamental Research Funds for the Central Universities (2572021AW13).
Wang, Hong‐Rui , Du Xin‐Ran, Zhang Zhuo‐Yun, Feng Fu‐Juan, and Zhang Jia‐Ming. 2023. “Rhizosphere Interface Microbiome Reassembly by Arbuscular Mycorrhizal Fungi Weakens Cadmium Migration Dynamics.” iMeta 2, e133. 10.1002/imt2.133
DATA AVAILABILITY STATEMENT
All raw sequence data and code in this study are available from the corresponding author on request. Nucleic acid sequence data have been submitted to the National Centre for Biotechnology Information (NCBI) Sequence Read Archive under Bioproject PRJNA975279 https://www.ncbi.nlm.nih.gov/sra/?term=PRJNA975279/. Supplementary materials (figures, tables, scripts, graphical abstract, slides, videos, Chinese translated version and update materials) may be found in the online DOI or iMeta Science http://www.imeta.science/.
REFERENCES
- 1. Xuan, Wen , Chen Qian, Li Zhen Sun, and Wang Zhao Jun. 2022. “Spatial Distribution Characteristics and Pollution Evaluation of Heavy Metals in Arable Land Soil of China.” Environmental Science 41: 2822–2833. 10.13227/j.hjkx.201910075 [DOI] [PubMed] [Google Scholar]
- 2. Tucker, Pamela G . 2008. “Case Studies in Environmental Medicine—Cadmium Toxicity.” Agency for Toxic Substances and Diseases Registry WB1096: 1‐63. https://www.atsdr.cdc.gov/csem/cadmium/docs/cadmium.pdf
- 3. Wang, Hong‐Rui , Zhao Xin‐Yu, Zhang Jia‐Ming, Lu Chang, and Feng Fu‐Juan. 2022. “Arbuscular Mycorrhizal Fungus Regulates Cadmium Accumulation, Migration, Transport, and Tolerance in Medicago sativa .” Journal of Hazardous Materials 435: 129077. 10.1016/j.jhazmat.2022.129077 [DOI] [PubMed] [Google Scholar]
- 4. Vallejos‐Torres, Geomar , Torres Samy C., Gaona‐Jimenez Nery, Saavedra Jorge, Tuesta Juan C., Tuesta Oscar A., Becerra Alejandra G., Marín César, and del Mar Alguacil María. 2022. “The Combined Effect of Arbuscular Mycorrhizal Fungi and Compost Improves Growth and Soil Parameters and Decreases Cadmium Absorption in Cacao (Theobroma cacao L.) Plants.” Journal of Soil Science and Plant Nutrition 22: 5174–5182. 10.1007/s42729-022-00992-9 [DOI] [Google Scholar]
- 5. Fan, Xiaoxu , Chang Wei, Sui Xin, Liu Yufei, Song Ge, Song Fuqiang, and Feng Fujuan. 2020. “Changes in Rhizobacterial Community Mediating Atrazine Dissipation by Arbuscular Mycorrhiza.” Chemosphere 256: 127046. 10.1016/j.chemosphere.2020.127046 [DOI] [PubMed] [Google Scholar]
- 6. Wang, Hong‐Rui , Che Yan‐Hui, Wang Zi‐Han, Zhang Bei‐Ning, Huang Dan, Feng Fu‐Juan, and Ao Hong. 2021. “The Multiple Effects of Hydrogen Sulfide on Cadmium Toxicity in Tobacco May be Interacted With CaM Signal Transduction.” Journal of Hazardous Materials 403: 123651. 10.1016/j.jhazmat.2020.123651 [DOI] [PubMed] [Google Scholar]
- 7. Nogueira, Marco A. , Magalhães Giuliana C., and Cardoso Elke J. B. N.. 2004. “Manganese Toxicity in Mycorrhizal and Phosphorus‐Fertilized Soybean Plants.” Journal of Plant Nutrition 27: 141–156. 10.1081/PLN-120027552 [DOI] [Google Scholar]
- 8. Nie, Ming , Xian Naixing, Fu Xiaohua, Chen Xiaofeng, and Li Bo. 2010. “The Interactive Effects of Petroleum‐Hydrocarbon Spillage and Plant Rhizosphere on Concentrations and Distribution of Heavy Metals in Sediments in the Yellow River Delta, China.” Journal of Hazardous Materials 174: 156–161. 10.1016/j.jhazmat.2009.09.030 [DOI] [PubMed] [Google Scholar]
- 9. Wu, Hongmiao , Fang Changxun, Malacrinò Antonino, Winkelmann Traud, and Xiong Wu. 2022. “Editorial: Rhizosphere Conversation Among the Plant‐Plant Microbiome‐Soil Under Consecutive Monoculture Regimes.” Frontiers in Microbiology 13: 1061427. 10.3389/fmicb.2022.1061427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. DeWolf, Ella , Brock Marcus T., Calder William John, Kliebenstein Daniel J., Katz Ella, Li Baohua, Morrison Hilary G., Maïgnien Lois, and Weinig Cynthia. 2023. “The Rhizosphere Microbiome and Host Plant Glucosinolates Exhibit Feedback Cycles in Brassica rapa .” Molecular Ecology 32: 741–751. 10.1111/mec.16782 [DOI] [PubMed] [Google Scholar]
- 11. Teixeira, Catarina , Almeida C. Marisa R., Nunes da Silva Marta, Bordalo Adriano A., and Mucha Ana P.. 2014. “Development of Autochthonous Microbial Consortia for Enhanced Phytoremediation of Salt‐Marsh Sediments Contaminated With Cadmium.” Science of The Total Environment 493: 757–765. 10.1016/j.scitotenv.2014.06.040 [DOI] [PubMed] [Google Scholar]
- 12. Shentu, Jia‐Li , He Zhen‐Li, Yang Xiao‐E, and Li Ting‐Qiang. 2008. “Microbial Activity and Community Diversity in a Variable Charge Soil as Affected by Cadmium Exposure Levels and Time.” Journal of Zhejiang University Science B 9: 250–260. 10.1631/jzus.B0710630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fan, Qiao , Chen Yeqiang, Xu Rui, and Guo Zhaohui. 2022. “Characterization of Keystone Taxa and Microbial Metabolic Potentials in Copper Tailing Soils.” Environment and Science Pollution Research 30: 1231. 10.1007/s11356-022-22294-4 [DOI] [PubMed] [Google Scholar]
- 14. Zhang, Lin , Shi Ning, Fan Jiequn, Wang Fei, George Timothy S., and Feng Gu. 2018. “Arbuscular Mycorrhizal Fungi Stimulate Organic Phosphate Mobilization Associated With Changing Bacterial Community Structure Under Field Conditions.” Environmental Microbiology 20: 2639–2651. 10.1111/1462-2920.14289 [DOI] [PubMed] [Google Scholar]
- 15. Cao, Jia , Wang Chong, Dou Zhengxia, Liu Mengli, and Ji Dingge. 2018. “Hyphospheric Impacts of Earthworms and Arbuscular Mycorrhizal Fungus on Soil Bacterial Community to Promote Oxytetracycline Degradation.” Journal of Hazardous Materials 341: 346–354. 10.1016/j.jhazmat.2017.07.038 [DOI] [PubMed] [Google Scholar]
- 16. Yu, Peng , He Xiaoming, Baer Marcel, Beirinckx Stien, Tian Tian, Moya Yudelsy A. T., Zhang Xuechen, et al. 2021. “Plant Flavones Enrich Rhizosphere Oxalobacteraceae to Improve Maize Performance Under Nitrogen Deprivation.” Nature Plants 7: 481–499. 10.1038/s41477-021-00897-y [DOI] [PubMed] [Google Scholar]
- 17. Hassan, Samira , and Mathesius Ulrike. 2012. “The Role of Flavonoids in Root‐Rhizosphere Signalling: Opportunities and Challenges for Improving Plant‐Microbe Interactions.” Journal of Experimental Botany 63: 3429–3444. 10.1093/jxb/err430 [DOI] [PubMed] [Google Scholar]
- 18. Sun, Chenyu , Zeng Yanhong, Ma Junqing, Liu Lu, Wang Wenqi, and Huang Jinghua. 2020. “Effects of Arbuscular Mycorrhizal Fungi on Artemisia annua L.Growth and Chemical Composition of Root Exudates.” Chinese Journal Tropical Crops 41: 1831–1837. 10.3969/j.issn.1000-2561.2020.09.016 [DOI] [Google Scholar]
- 19. Chen, Bao‐Dong , Sun Yu‐Qing, Zhang Xin, and Wu Song‐Lin. 2015. “Underlying Mechanisms of the Heavy Metal Tolerance of Mycorrhizal Fungi.” Huanjing Kexue 36: 1123–1132. 10.13227/j.hjkx.2015.03.048 [DOI] [PubMed] [Google Scholar]
- 20. Macabuhay, Allene , Arsova Borjana, Walker Robert, Johnson Alexander, Watt Michelle, and Roessner Ute. 2022. “Modulators or Facilitators? Roles of Lipids in Plant Root–Microbe Interactions.” Trends in Plant Science 27: 180–190. 10.1016/j.tplants.2021.08.004 [DOI] [PubMed] [Google Scholar]
- 21. Zhang, Zeyu , Xiao Yang Sean, Zhan Yabin, Zhang Zengqiang, Liu Youzhou, Wei Yuquan, Xu Ting, and Li Ji. 2022. “Tomato Microbiome Under Long‐Term Organic and Conventional Farming.” iMeta 1: 1–13. 10.1002/imt2.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Araujo, Ademir Sergio Ferreira , Miranda Ana Roberta Lima, de Araujo Pereira Arthur Prudêncio, de Melo Wanderley José, Melo Vania Maria Maciel, Ventura Sabrina Hermelindo, Eudemio Sousa Brito Junior, Jr. , et al. 2023. “Microbial Communities In the Rhizosphere of Maize and Cowpea Respond Differently to Chromium Contamination.” Chemosphere 313: 137417. 10.1016/j.chemosphere.2022.137417 [DOI] [PubMed] [Google Scholar]
- 23. Spohn, Marie , Carminati Andrea, and Kuzyakov Yakov. 2013. “Soil Zymography—A Novel in Situ Method for Mapping Distribution of Enzyme Activity in Soil.” Soil Biology and Biochemistry 58: 275–280. 10.1016/j.soilbio.2012.12.004 [DOI] [Google Scholar]
- 24. Caldwell, Daniel R. , and Arcand Caryn. 1974. “Inorganic and Metal‐Organic Growth Requirements of the Genus Bacteroides .” Journal of Bacteriology 120: 322–333. 10.1128/JB.120.1.322-333.1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yang, Zhihui , Shi Wei, Yang Weichun, Liang Lifen, Yao Wenbin, Chai Liyuan, Gao Shikang, and Liao Qi. 2018. “Combination of Bioleaching by Gross Bacterial Biosurfactants and Flocculation: A Potential Remediation for the Heavy Metal Contaminated Soils.” Chemosphere 206: 83–91. 10.1016/j.chemosphere.2018.04.166 [DOI] [PubMed] [Google Scholar]
- 26. Meng, Xiao‐Lin , Li Shuai, Qin Chao‐Bin, Zhu Zhen‐Xiang, Hu Wen‐Pan, Yang Li‐Ping, Lu Rong‐Hua, Li Wen‐Jun, and Nie Guo‐Xing. 2018. “Intestinal Microbiota and Lipid Metabolism Responses In the Common Carp (Cyprinus carpio L.) Following Copper Exposure.” Ecotoxicology and Environmental Safety 160: 257–264. 10.1016/j.ecoenv.2018.05.050 [DOI] [PubMed] [Google Scholar]
- 27. Puthusseri, Rinu Madhu , Nair Harisree Paramel, Johny Tina Kollannoor, and Bhat Sarita Ganapathy. 2021. “Insights Into the Response of Mangrove Sediment Microbiomes to Heavy Metal Pollution: Ecological Risk Assessment and Metagenomics Perspectives.” Journal of Environmental Management 298: 113492. 10.1016/j.jenvman.2021.113492 [DOI] [PubMed] [Google Scholar]
- 28. Kumar Rai, Prabhat . 2010. “Heavy Metal Pollution in Lentic Ecosystem of Sub‐Tropical Industrial Region and Its Phytoremediation.” International Journal of Phytoremediation 12: 226–242. 10.1080/15226510903563843 [DOI] [PubMed] [Google Scholar]
- 29. Lin, Hai , Liu Chenjing, Li Bing, and Dong Yingbo. 2021. “ Trifolium repens L. Regulated Phytoremediation of Heavy Metal Contaminated Soil by Promoting Soil Enzyme Activities and Beneficial Rhizosphere Associated Microorganisms.” Journal of Hazardous Materials 402: 123829. 10.1016/j.jhazmat.2020.123829 [DOI] [PubMed] [Google Scholar]
- 30. Aguilar‐Romero, Inés , van Dillewijn Pieter, Nesme Joseph, Sørensen Søren J., Nogales Rogelio, Delgado‐Moreno Laura, and Romero Esperanza. 2022. “A Novel and Affordable Bioaugmentation Strategy With Microbial Extracts to Accelerate the Biodegradation of Emerging Contaminants in Different Media.” Science of the Total Environment 834: 155234. 10.1016/j.scitotenv.2022.155234 [DOI] [PubMed] [Google Scholar]
- 31. Johnson, D. Barrie , Rolfe Stewart, Hallberg Kevin B., and Iversen Eigil. 2001. “Isolation and Phylogenetic Characterization of Acidophilic Microorganisms Indigenous to Acidic Drainage Waters at an Abandoned Norwegian Copper Mine.” Environmental Microbiology 3: 630–637. 10.1046/j.1462-2920.2001.00234.x [DOI] [PubMed] [Google Scholar]
- 32. Chen, Fan , Lv Miao, Ye Yin, Miao Shiyu, Tang Xin, Liu Yang, Liang Bin, et al. 2022. “Insights on Uranium Removal by Ion Exchange Columns: The Deactivation Mechanisms, and an Overlooked Biological Pathway.” Chemical Engineering Journal 434: 134708. 10.1016/j.cej.2022.134708 [DOI] [Google Scholar]
- 33. Roosa, Stéphanie , Wauven Corinne Vander, Billon Gabriel, Matthijs Sandra, Wattiez Ruddy, and Gillan David C.. 2014. “The Pseudomonas Community in Metal‐Contaminated Sediments as Revealed by Quantitative PCR: A Link With Metal Bioavailability.” Research in Microbiology 165: 647–656. 10.1016/j.resmic.2014.07.011 [DOI] [PubMed] [Google Scholar]
- 34. Skouri‐Panet, Fériel , Benzerara Karim, Cosmidis Julie, Férard Céline, Caumes Géraldine, De Luca Gilles, Heulin Thierry, and Duprat Elodie. 2018. “In Vitro and in Silico Evidence of Phosphatase Diversity in the Biomineralizing Bacterium Ramlibacter tataouinensis .” Frontiers in Microbiology 8: 2592. 10.3389/fmicb.2017.02592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hong, Chen , Si Yanxiao, Xing Yi, and Li Yang. 2015. “Illumina MiSeq Sequencing Investigation on the Contrasting Soil Bacterial Community Structures in Different Iron Mining Areas.” Environmental Science and Pollution Research 22: 10788–10799. 10.1007/s11356-015-4186-3 [DOI] [PubMed] [Google Scholar]
- 36. Tao, Xuanyu , Zhou Aifen, Kempher Megan L., Liu Jiantao, Peng Mu, Li Yuan, Michael Jonathan P., et al. 2022. “Development of a Markerless Deletion Mutagenesis System in Nitrate‐Reducing Bacterium Rhodanobacter denitrificans .” Applied and Environmental Microbiology 88: e14. 10.1128/aem.00401-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kasemodel, Mariana Consiglio , Kimiko Sakamoto Isabel, Varesche Maria Bernadete Amancio, and Rodrigues Valeria Guimares Silvestre. 2019. “Potentially Toxic Metal Contamination and Microbial Community Analysis in an Abandoned Pb and Zn Mining Waste Deposit.” Science of The Total Environment 675: 367–379. 10.1016/j.scitotenv.2019.04.223 [DOI] [PubMed] [Google Scholar]
- 38. Zhou, Maohong , Zhang Xuejun, and Zhao Xiaowei. 2003. “A Few Biological Effects of Some Heavy Metal Ions on Photosynthetic Bacterium Rhodopseudomonas Palustrk.” Microbiology 30: 64–68. 10.13344/j.microbiol.china.2003.03.016 [DOI] [Google Scholar]
- 39. Zhu, Yueming , Men Yan, Bai Wei, Li Xiaobo, Zhang Lili, Sun Yuanxia, and Ma Yanhe. 2012. “Overexpression of D‐psicose 3‐epimerase From Ruminococcus sp. in Escherichia coli and Its Potential Application in D‐Psicose Production.” Biotechnology Letters 34: 1901–1906. 10.1007/s10529-012-0986-4 [DOI] [PubMed] [Google Scholar]
- 40. Xu, Guo‐Xiang , Li Xu‐Qiang, and Chen Wen‐Bin. 2013. “Equilibrium Sorption Isotherms of Cr(III) on Sphingomonas sp. XJ2.” Asian Journal of Chemistry 25: 3687–3691. 10.14233/ajchem.2013.13718 [DOI] [Google Scholar]
- 41. Klümper, Uli , Dechesne Arnaud, Riber Leise, Brandt Kristian K., Gülay Arda, Sørensen Søren J, and Smets Barth F.. 2017. “Metal Stressors Consistently Modulate Bacterial Conjugal Plasmid Uptake Potential in a Phylogenetically Conserved Manner.” The ISME Journal 11: 152–165. 10.1038/ismej.2016.98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Banach, Artur M. , Kuźniar Agnieszka, Grządziel Jarosław, and Wolińska Agnieszka. 2020. “ Azolla filiculoides L. as a Source of Metal‐Tolerant Microorganisms.” Plos One 15: e0232699. 10.1371/journal.pone.0232699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Noriega, Luis , Margolles Abelardo, and de los Reyes‐Gavilan Clara G.. 2007. “Susceptibility of Bifidobacterium Strains To Some Metal Salts.” Milchwissenschaft 62: 146–148. [Google Scholar]
- 44. Yausheva, Еlena , Miroshnikov Sergey, and Sizova Еlena. 2018. “Intestinal Microbiome of Broiler Chickens After Use of Nanoparticles and Metal Salts.” Environmental Science and Pollution Research 25: 18109–18120. 10.1007/s11356-018-1991-5 [DOI] [PubMed] [Google Scholar]
- 45. Zhang, Xiaorong , Gong Zongqiang, Allinson Graeme, Li Xiaojun, and Jia Chunyun. 2022. “Joint Effects of Bacterium and Biochar in Remediation of Antibiotic‐Heavy Metal Contaminated Soil and Responses of Resistance Gene and Microbial Community.” Chemosphere 299: 134333. 10.1016/j.chemosphere.2022.134333 [DOI] [PubMed] [Google Scholar]
- 46. Stein, Lisa Y. , La Duc Myron T., Grundl Timothy J., and Nealson Kenneth H.. 2001. “Bacterial and Archaeal Populations Associated With Freshwater Ferromanganous Micronodules and Sediments.” Environmental Microbiology 3: 10–18. 10.1046/j.1462-2920.2001.00154.x [DOI] [PubMed] [Google Scholar]
- 47. Huynh, Uyen , Qiao Muxin, King John, Trinh Brittany, Valdez Juventino, Haq Marium, and Zastrow Melissa L.. 2022. “Differential Effects of Transition Metals on Growth and Metal Uptake for Two Distinct Lactobacillus Species.” Microbiology Spectrum 10: e01006. 10.1128/spectrum.01006-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tapase, Savita R. , and Kodam Kisan M.. 2018. “Assessment of Arsenic Oxidation Potential of Microvirga indica S‐MI1b sp nov in Heavy Metal Polluted Environment.” Chemosphere 195: 1–10. 10.1016/j.chemosphere.2017.12.022 [DOI] [PubMed] [Google Scholar]
- 49. Tang, Jingwei , Huang Jing, Qiao Zixu, Wang Rui, and Wang Gejiao. 2016. “ Mucilaginibacter pedocola sp nov., isolated from a Heavy‐Metal‐Contaminated Paddy Field.” International Journal of Systematic Evolutionary Microbiology 66: 4033–4038. 10.1099/ijsem.0.001306 [DOI] [PubMed] [Google Scholar]
- 50. Çolak, Ferdağ , Olgun Asim, Atar Necip, and Yazıcıoğlu Demet. 2013. “Heavy Metal Resistances and Biosorptive Behaviors of Paenibacillus polymyxa: Batch and Column Studies.” Journal of Industrial and Engineering Chemistry 19: 863–869. 10.1016/j.jiec.2012.11.001 [DOI] [Google Scholar]
- 51. Yu, Jiafeng , Liu Jian, Senthil Kumar P., Wei Yunwei, Zhou Meng, Vo Dai‐Viet N., and Xiao Leilei. 2022. “Promotion of Methane Production by Magnetite Via Increasing Acetogenesis Revealed by Metagenome‐Assembled Genomes.” Bioresource Technology 345: 126521. 10.1016/j.biortech.2021.126521 [DOI] [PubMed] [Google Scholar]
- 52. Kou, Shumeng , Vincent Gilles, Gonzalez Emmanuel, Pitre Frederic E., Labrecque Michel, and Brereton Nicholas J. B.. 2018. “The Response of a 16S Ribosomal RNA Gene Fragment Amplified Community to Lead, Zinc, and Copper Pollution in a Shanghai Field Trial.” Frontiers in Microbiology 9: 366. 10.3389/fmicb.2018.00366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hayat, Rifat , Ali Safdar, Amara Ummay, Khalid Rabia, and Ahmed Iftikhar. 2010. “Soil Beneficial Bacteria and Their Role In Plant Growth Promotion: A Review.” Annals of Microbiology 60: 579–598. 10.1007/s13213-010-0117-1 [DOI] [Google Scholar]
- 54. Parades‐Aguilar, Jonathan , Reyes‐Martínez Viviana, Bustamante Guadalupe, Almendáriz‐Tapia Francisco J., Martínez‐Meza Guadalupe, Vílchez‐Vargas Ramiro, Link Alexander, Certucha‐Barragán María T., and Calderón Kadiya. 2021. “Removal of Nickel (II) From Wastewater Using a Zeolite‐Packed Anaerobic Bioreactor: Bacterial Diversity and Community Structure Shifts.” Journal of Environmental Management 279: 111558. 10.1016/j.jenvman.2020.111558 [DOI] [PubMed] [Google Scholar]
- 55. Hao, Xiuli , Lin Yanbing, Johnstone Laurel, Baltrus David A., Miller Susan J., Wei Gehong, and Rensing Christopher. 2012. “Draft Genome Sequence of Plant Growth‐Promoting Rhizobium Mesorhizobium Amorphae, Isolated From Zinc‐Lead Mine Tailings.” Journal of Bacteriology 194: 736–737. 10.1128/JB.06475-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Holanda, Roseanne , Hedrich Sabrina, Ňancucheo Ivan, Oliveira Guilherme, Grail Barry M., and Johnson D. Barrie. 2016. “Isolation and Characterisation of Mineral‐Oxidising ‘Acidibacillus’ Spp. From Mine Sites and Geothermal Environments in Different Global Locations.” Research in Microbiology 167: 613–623. 10.1016/j.resmic.2016.04.008 [DOI] [PubMed] [Google Scholar]
- 57. Gobbi, Emanuela , Falasconi Matteo, Concina Isabella, Mantero Giovanni, Bianchi Federica, Mattarozzi Monica, Musci Marilena, and Sberveglieri Giorgio. 2010. “Electronic Nose and Alicyclobacillus Spp. Spoilage of Fruit Juices: An Emerging Diagnostic Tool.” Food Control 21: 1374–1382. 10.1016/j.foodcont.2010.04.011 [DOI] [Google Scholar]
- 58. Kazuoka, Takayuki , Takigawa Shouhei, Arakawa Noriaki, Hizukuri Yoshiyuki, Muraoka Ikuo, Oikawa Tadao, and Soda Kenji. 2003. “Novel Psychrophilic and Thermolabile L‐Threonine Dehydrogenase From Psychrophilic Cytophaga Sp Strain KUC‐1.” Journal of Bacteriology 185: 4483–4489. 10.1128/JB.185.15.4483-4489.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yang, Wenling , Du Zhimin, Sun Zhaohua, Zhou Fuzhong, An Mingli, Lei Gao, Wei Chuanjun, and Gong Tao. 2021. “Research Progress of Bacillus in Remediation of Heavy Metal Contaminated Soil.” Environmental Pollution & Control 43: 759–763. 10.15985/j.cnki.1001-3865.2021.06.017 [DOI] [Google Scholar]
- 60. McCall, Crystal A. , Jordan Katerina S., Habash Marc B., and Dunfield Kari E.. 2015. “Monitoring Bacteroides Spp. Markers, Nutrients, Metals and Escherichia coli in Soil and Leachate After Land Application of Three Types of Municipal Biosolids.” Water Research 70: 255–265. 10.1016/j.watres.2014.12.004 [DOI] [PubMed] [Google Scholar]
- 61. Sharma, Gaurav , and Merz Kenneth M.. 2021. “Formation of the Metal‐Binding Core of the ZRT/IRT‐like Protein (ZIP) Family Zinc Transporter.” Biochemistry 60: 2727–2738. 10.1021/acs.biochem.1c00415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bianucci, Eliana , Fabra Adriana, and Castro Stella. 2011. “Cadmium Accumulation and Tolerance in Bradyrhizobium spp. (Peanut Microsymbionts).” Current Microbiology 62: 96–100. 10.1007/s00284-010-9675-5 [DOI] [PubMed] [Google Scholar]
- 63. Natagarn, Tongphanpharn , Chou Chihhuang, Guan Chuangyu, and Yu Changping. 2021. “Plant Microbial Fuel Cells With Oryza rufipogon and Typha Orientalis for Remediation of Cadmium Contaminated Soil.” Environmental Technology & Innovation 24: 102030. 10.1016/j.eti.2021.102030 [DOI] [Google Scholar]
- 64. Fan, Miaochun , Lin Yanbing, Huo Haibo, Liu Yang, Zhao Liang, Wang Entao, Chen Weimin, and Wei Gehong. 2016. “Microbial Communities in Riparian Soils of a Settling Pond for Mine Drainage Treatment.” Water Research 96: 198–207. 10.1016/j.watres.2016.03.061 [DOI] [PubMed] [Google Scholar]
- 65. Dwivedi, Deepti , Johri Bhavdish N., Ineichen Kurt, Wray Victor, and Wiemken Andres. 2009. “Impact of Antifungals Producing Rhizobacteria on the Performance of Vigna radiata in the Presence of Arbuscular Mycorrhizal Fungi.” Mycorrhiza 19: 559–570. 10.1007/s00572-009-0253-2 [DOI] [PubMed] [Google Scholar]
- 66. Picard, Christine , Baruffa Elisa, and Bosco Marco. 2008. “Enrichment and Diversity of Plant‐Probiotic Microorganisms in the Rhizosphere of Hybrid Maize During Four Growth Cycles.” Soil Biology & Biochemistry 40: 106–115. 10.1016/j.soilbio.2007.07.011 [DOI] [Google Scholar]
- 67. Aggangan, Nelly Siababa , Cortes Angelbert D., Opulencia Rina B., Jomao‐As Joshua G., and Yecyec Romualdo P.. 2019. “Effects of Mycorrhizal Fungi and Bamboo Biochar on the Rhizosphere Bacterial Population and Nutrient Uptake of Cacao (Theobroma cacao L.) Seedlings.” Philippine Journal of Crop Science 44: 1–9. https://www.researchgate.net/publication/332230710 [Google Scholar]
- 68. Marro, Nicolás , Grilli Gabriel, Soteras Florencia, Caccia Milena, Longo Silvana, Cofré Noelia, et al. 2022. “The Effects of Arbuscular Mycorrhizal Fungal Species and Taxonomic Groups on Stressed and Unstressed Plants: A Global Meta‐Analysis.” New Phytologist 235: 320–332. 10.1111/nph.18102 [DOI] [PubMed] [Google Scholar]
- 69. Cristiana, Sbrana , Agnolucci Monica, Avio Luciano, Giovannini Luca, Palla Michela, Turrini Alessandra, and Giovannetti Manuela. 2022. “Mycorrhizal Symbionts and Associated Bacteria: Potent Allies to Improve Plant Phosphorus Availability and Food Security.” Frontiers Microbiology 12: 1–6. 10.3389/fmicb.2021.797381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Gui, Heng , Purahong Witoon, Hyde Kevin D., Xu Jianchu, and Mortimer Peter E.. 2017. “The Arbuscular Mycorrhizal Fungus Funneliformis Mosseae Alters Bacterial Communities In Subtropical Forest Soils During Litter Decomposition.” Frontiers in Microbiology 8: 1–11. 10.3389/fmicb.2017.01120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ordoñez, Yuli Marcela , Fernandez Belen Rocio, Lara Lidia Susana, Rodriguez Alia, Uribe‐Vélez Daniel, and Sanders Ian R.. 2016. “Bacteria With Phosphate Solubilizing Capacity Alter Mycorrhizal Fungal Growth Both Inside and Outside the Root and in the Presence of Native Microbial Communities.” PLoS One 11: e0154438. 10.1371/journal.pone.0154438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Li, Yunliang , Gan Yantai, Lupwayi Newton, and Hamel Chantal. 2019. “Influence of Introduced Arbuscular Mycorrhizal Fungi and Phosphorus Sources on Plant Traits, Soil Properties, and Rhizosphere Microbial Communities in Organic Legume‐Flax Rotation.” Plant and Soil 443: 87–106. 10.1007/s11104-019-04213-8 [DOI] [Google Scholar]
- 73. Zhang, Haoqiang , Tang Ming, Chen Hui, Tian Zhiqiang, Xue Yaoqin, and Feng Ye. 2010. “Communities of Arbuscular Mycorrhizal Fungi and Bacteria in the Rhizosphere of Caragana Korshinkii and Hippophae rhamnoides in Zhifanggou Watershed.” Plant and Soil 326: 415–424. 10.1007/s11104-009-0022-1 [DOI] [Google Scholar]
- 74. Battini, Fabio , Grønlund Mette, Agnolucci Monica, Giovannetti Manuela, and Jakobsen Iver. 2017. “Facilitation of Phosphorus Uptake in Maize Plants by Mycorrhizosphere Bacteria Article.” Science Reports 7: 1–11. 10.1038/s41598-017-04959-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Xu, Jing , Liu Shijun, Song Shurui, Guo Hanling, Tang Jianjun, Yong Jean Wanhong, Ma Yuandan, and Chen Xin. 2018. “Arbuscular Mycorrhizal Fungi Influence Decomposition and the Associated Soil Microbial Community Under Different Soil Phosphorus Availability.” Soil Biology & Biochemistry 120: 181–190. 10.1016/j.soilbio.2018.02.010 [DOI] [Google Scholar]
- 76. Nuccio, Erin E. , Hodge Angela, Pett‐Ridge Jennifer, Herman Donald J., Weber Peter K., and Firestone Mary K.. 2013. “An Arbuscular Mycorrhizal Fungus Significantly Modifies the Soil Bacterial Community and Nitrogen Cycling During Litter Decomposition.” Environmental Microbiology 15: 1870–1881. 10.1111/1462-2920.12081 [DOI] [PubMed] [Google Scholar]
- 77. Ujvári, Gergely , Turrini Alessandra, Avio Luciano, and Agnolucci Monica. 2021. “Possible Role Of Arbuscular Mycorrhizal Fungi and Associated Bacteria in the Recruitment of Endophytic Bacterial Communities by Plant Roots.” Mycorrhiza 31: 527–544. 10.1007/s00572-021-01040-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Liu, Ning , Shao Cai, Sun Hai, Liu Zhengbo, Guan Yiming, Wu Lianju, Zhang Linlin, et al. 2020. “Arbuscular Mycorrhizal Fungi Biofertilizer Improves American Ginseng (Panax quinquefolius L.) Growth Under the Continuous Cropping Regime.” Geoderma 363: 114155. 10.1016/j.geoderma.2019.114155 [DOI] [Google Scholar]
- 79. Tang, Haoqi , Zhang Na, Sun Bo, and Liang Yuting. 2020. “Effect of Interaction Between Arbuscular Mycorrhizal Fungi and Rhizosphere Bacteria in Farmland Soils on Nutrients Utilization.” Acta Microbiology Sinica 60: 1117–1129. 10.13343/j.cnki.wsxb.20200204 [DOI] [Google Scholar]
- 80. Herren, Cristina M. , and McMahon Katherine D.. 2018. “Keystone Taxa Predict Compositional Change in Microbial Communities.” Environmental Microbiology 20: 2207–2217. 10.1111/1462-2920.14257 [DOI] [PubMed] [Google Scholar]
- 81. Wang, Binhao , Zheng Xiafei, Zhang Hangjun, Xiao Fanshu, He Zhili, and Yan Qingyun. 2020. “Keystone Taxa of Water Microbiome Respond to Environmental Quality and Predict Water Contamination.” Environmental Research 187: 109666. 10.1016/j.envres.2020.109666 [DOI] [PubMed] [Google Scholar]
- 82. Mierziak, Justyna , Kostyn Kamil, and Kulma Anna. 2014. “Flavonoids as Important Molecules of Plant Interactions With the Environment.” Molecules 19: 16240–16265. 10.3390/molecules191016240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Beauregard, Pascale B . 2015. “Not Just Sweet Talkers: How Roots Stimulate Their Colonization by Beneficial Bacteria.” Plant Microbe interactions 75: 1–20. 10.1016/bs.abr.2015.07.001 [DOI] [Google Scholar]
- 84. Pan, Jiachuan , and Ren Dacheng. 2009. “Quorum Sensing Inhibitors: A Patent Overview.” Expert Opinion on Therapeutic Patents 19: 1581–1601. 10.1517/13543770903222293 [DOI] [PubMed] [Google Scholar]
- 85. Wang, Hong‐Rui , Zhang Zhuo‐Yun, and Feng Fu‐Juan. 2022. “A Device for the Determination of Plant Rhizosphere Pollutants and a Method for the Determination of Plant Rhizosphere Pollutants.” Chinese patent: CN114518347A. https://kns.cnki.net/kcms2/article/abstract?v=kxaUMs6x7-4I2jr5WTdXti3zQ9F92xu0nlgSAA876Br4k7Yiof5ge4xcIcW09x-BERALFQChyqqKoipfDaJPJVc0yyEXAvKA%26uniplatform=NZKPT
- 86. Xu, Zhimin , Lu Ziyan, Zhang Liangshi, Fan Hanyun, Wang Yifan, Li Junwei, Lin Yanlin, et al. 2021. “Red Mud Based Passivator Reduced Cd Accumulation in Edible Amaranth by Influencing Root Organic Matter Metabolism and Soil Aggregate Distribution.” Environmental Pollution 275: 116543. 10.1016/j.envpol.2021.116543 [DOI] [PubMed] [Google Scholar]
- 87. Battini, Fabio , Grønlund Mette, Agnolucci Monica, Giovannetti Manuela, and Jakobsen Iver. 2017. “Facilitation of Phosphorus Uptake in Maize Plants by Mycorrhizosphere Bacteria.” Scientific Reports 7: 4686. 10.1038/s41598-017-04959-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Wang, Yanhong , Li Ping, Jiang Zhou, Sinkkonen Aki, Wang Shi, Tu Jin, Wei Dazhun, Dong Hailiang, and Wang Yanxin. 2016. “Microbial Community of High Arsenic Groundwater in Agricultural Irrigation Area of Hetao Plain, Inner Mongolia.” Frontiers in Microbiology 7: 1917. 10.3389/fmicb.2016.01917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Caporaso, J. Gregory , Lauber Christian L., Walters William A., Berg‐Lyons Donna, Lozupone Catherine A., Turnbaugh Peter J., Fierer Noah, and Knight Rob. 2011. “Global Patterns of 16S rRNA Diversity at a Depth Of Millions of Sequences per Sample.” Proceedings of the National Academy of Sciences 108: 4516–4522. 10.1073/pnas.1000080107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Bokulich, Nicholas A. , Kaehler Benjamin D., Rideout Jai Ram, Dillon Matthew, Bolyen Evan, Knight Rob, Huttley Gavin A., and Gregory Caporaso J.. 2018. “Optimizing Taxonomic Classification of Marker‐Gene Amplicon Sequences With QIIME 2's q2‐Feature‐Classifier Plugin.” Microbiome 6: 90. 10.1186/s40168-018-0470-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Xia, Jianguo , and Wishart David S.. 2011. “Web‐Based Inference of Biological Patterns, Functions and Pathways From Metabolomic Data Using MetaboAnalyst.” Nature Protocols 6: 743–760. 10.1038/nprot.2011.319 [DOI] [PubMed] [Google Scholar]
- 92. Liu, Shengen , Garcia‐Palacios Pablo, Tedersoo Leho, Guirado Emilio, van der Heijden Marcel G. A., van der Heijden Cameron, Chen Dima, et al. 2022. “Phylotype Diversity Within Soil Fungal Functional Groups Drives Ecosystem Stability.” Nature Ecology & Evolution 6: 900. 10.1038/s41559-022-01756-5 [DOI] [PubMed] [Google Scholar]
- 93. Tan, Lin , Gu Songsong, Li Shi, Ren Zuohua, Deng Ye, Liu Zhonghua, Gong Zhihua, Xiao Wenjun, and Hu Qiulong. 2019. “Responses of Microbial Communities and Interaction Networks to Different Management Practices in Tea Plantation Soils.” Sustainability 11: 4428. 10.3390/su11164428 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information.
Supporting information.
Data Availability Statement
All raw sequence data and code in this study are available from the corresponding author on request. Nucleic acid sequence data have been submitted to the National Centre for Biotechnology Information (NCBI) Sequence Read Archive under Bioproject PRJNA975279 https://www.ncbi.nlm.nih.gov/sra/?term=PRJNA975279/. Supplementary materials (figures, tables, scripts, graphical abstract, slides, videos, Chinese translated version and update materials) may be found in the online DOI or iMeta Science http://www.imeta.science/.


