Abstract
Congenital heart disease (CHD) is a prevalent birth defect and a significant contributor to childhood mortality. The major characteristics of CHD include cardiovascular malformations and hemodynamical disorders. However, the impact of CHD extends beyond the circulatory system. Evidence has identified dysbiosis of the gut microbiome in patients with CHD. Chronic hypoxia and inflammation associated with CHD affect the gut microbiome, leading to alterations in its number, abundance, and composition. The gut microbiome, aside from providing essential nutrients, engages in direct interactions with the host immune system and indirect interactions via metabolites. The abnormal gut microbiome or its products can translocate into the bloodstream through an impaired gut barrier, leading to an inflammatory state. Metabolites of the gut microbiome, such as short‐chain fatty acids and trimethylamine N‐oxide, also play important roles in the development, treatment, and prognosis of CHD. This review discusses the role of the gut microbiome in immunity, gut barrier, neurodevelopment, and perioperative period in CHD. By fostering a better understanding of the cross‐talk between CHD and the gut microbiome, this review aims to contribute to improve clinical management and outcomes for CHD patients.
Keywords: cardiovascular surgery, congenital heart disease, immunoinflammatory homeostasis imbalance, intestinal microbiology, neurodevelopmental injury
Children with congenital heart disease (CHD) experience gut microbiome dysbiosis in their early life. The gut microbiome affects CHD via immune stimulation and microbiome metabolites, influencing immunity, gut barrier, and inflammation. The bidirectional interaction between the gut microbiome and CHD plays an important role in CHD's development, treatment, and prognosis.
Highlights
Children with CHD experience gut microbiome dysbiosis in their early life.
The gut microbiome affects CHD via immune stimulation and microbiome metabolites, influencing immunity, gut barrier, and inflammation.
The bidirectional interaction between the gut microbiome and CHD plays an important role in CHD's development, treatment, and prognosis.
INTRODUCTION
Congenital heart disease (CHD) is a common birth defect, afflicting approximately 2% of live newborns globally, and remains a significant contributor to infant morbidity and mortality [1, 2]. Despite advancements in medical technologies that have led to a decline in CHD‐related deaths, patients with CHD face an elevated risk of neurodevelopmental disabilities, immune dysfunctions, infections, and cancer [3, 4, 5, 6]. These potential complications have severe implications for the long‐term well‐being of CHD‐affected individuals and underscore the ongoing challenges in managing this complex congenital disease.
Research has revealed the vastness of the microbial population inhabiting the gastrointestinal tract, with estimates indicating that their numbers exceed 1014 and the genome is approximately 100 times more abundant than the human genome [7]. The complex interplay between the gut microbiome and its host extends far beyond the provision of essential nutrients. This cross‐talk is crucial to maintaining homeostasis, with studies suggesting its important role in the development of various physiological systems, including but not limited to the nervous, immune, and endocrine systems [8, 9, 10]. Moreover, the gut microbiome is involved in the progression of a spectrum of pathological conditions, such as intestinal epithelial barrier dysfunction (EBD) and systemic inflammation [11, 12].
In recent years, researchers have identified the substantial influence of gut microbiome composition and diversity on an individual's susceptibility to various diseases [13, 14, 15]. The gut microbiome has been demonstrated to have associations with various cardiovascular diseases [16, 17, 18, 19, 20, 21]. In our previous study, we provided the initial evidence that an aberrant gut microbiome, coupled with metabolic disruptions, was associated with immune imbalances and unfavorable clinical outcomes in neonates afflicted with critical CHD [22]. This finding underscores the significance of restoring an optimal gut microbiome to uphold host metabolic and immunological homeostasis. Additionally, some pathophysiological features of CHD, such as chronic hypoxia, are associated with changes in the composition and homeostasis of the gut microbiome [21, 23]. Cardiac operations, especially those requiring cardiopulmonary bypass (CPB) usage, are related to microbiome disorder and EBD, ultimately leading to the systemic inflammatory response postoperatively [16, 24, 25]. Furthermore, notable shifts in microbiome composition have been observed in patients receiving cardiac intensive care, which is relevant to clinical outcomes [26].
Studies focusing on the gut microbiome and CHD have identified the significance of the gut microbiome in CHD. We believe that the meaningful role of the gut microbiome in CHD needs further emphasis and exploration. This review focuses on the role of the gut microbiome in pediatric immunity, gut barrier, neurodevelopment, cardiac surgery, and postoperative pediatric intensive care unit (PICU). We bring together the relationship of the gut microbiome in multiple aspects of CHD, aiming to understand the underlying mechanisms and implications of such associations with a focus on identifying potential therapeutic interventions and directions for future research (Figure 1).
Figure 1.
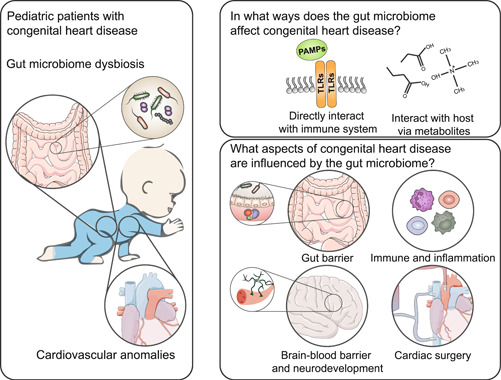
Relationship between CHD and the gut microbiome. The gut microbiome exerts its influence on the body through the production of metabolites and immune stimulations. In the development of CHD, the gut microbiome plays crucial roles in altering gut barrier function, modulating the immune system, and influencing the extent of injury and inflammation resulting from cardiac surgery. CHD, congenital heart disease; PAMP, pathogen‐associated molecular pattern; TLR, Toll‐like receptor.
FORMATION AND DEVELOPMENT OF THE EARLY LIFE GUT MICROBIOME
The formation of the gut microbiome undergoes dynamic changes from fetal to adulthood. The Firmicutes to Bacteroidetes ratio (F/B ratio) varies at different stages of a lifetime: 0.4 in infants, 10.9 in adults, and 0.6 in older people [27]. Studies have demonstrated the presence of the microbiome in the meconium, which originated from uteri and was associated with gut colonization [28]. Bacilli and other Firmicutes are the dominant bacteria groups in meconium [29]. Similarly, the most abundant phyla in infants are Firmicutes and Bacteroidetes, followed by Actinobacteria and Proteobacteria [30]. The species diversity in infant fecal microbiomes is highest within the first 24 h after birth and decreases over the first‐week postpartum [31]. This change may be attributed to some microbiomes from maternal or other sources that do not colonize the newborn's digestive tract. In the early years of life, Bifidobacteriaceae typically increases and becomes the dominant microbiome in infant feces [32]. However, the proportion of phylotypes belonging to Bifidobacterium longum declines with age [33]. In healthy adult individuals, Bacteroidetes and Firmicutes exhibit the highest abundance [34].
The intrapersonal variation of the gut microbiome is more significant in children compared with adults [33]. During early life, various factors can influence the composition of the gut microbiome. Maternal obesity is linked to the richness of neonates' Firmicutes and elevated risk of becoming overweight [35]. A meta‐analysis pointed out that intrapartum antibiotic use, maternal overweight/obesity, and gestational weight gain were associated with reduced diversity in the infant gut microbiome [36]. Premature infants, in contrast to full‐term infants, exhibit higher levels of facultative anaerobic microbes and lower levels of strict anaerobes, such as Bifidobacterium, Bacteroides, and Atopobium [37]. The comparison of the fecal samples of infants and their mothers indicates that 72% of the species of early life microbiomes in vaginally delivered infants matches those microbiomes of their mothers, whereas this proportion is 41% in C‐section newborns [30]. The mother‐to‐child microbiome transmission via human milk is an important way for infants to gain microbiomes, such as Bifidobacterium and Staphylococci [38]. The human milk oligosaccharides (HMO) may partially contribute to the microbiological variation and the gut barrier function for infants fed with breast milk [39]. Certain microorganisms, including B. longum subsp. infantis, Bacteroides fragilis, and Bacteroides vulgatus strains, can easily metabolize HMO [40, 41]. Additionally, formula feeding leads to alterations in the microbiome associated with obesity, potentially attributed to the higher relative abundance of Lachnospiraceae at 3–4 months after birth [42]. It is worth noting that the gut microbiome of breastfed infants originates not only from breast milk itself but also from the skin of the areola. It was reported that breastfed infants acquired 27.7% of their bacteria from breast milk and 10.3% from the areolar skin [32]. The induction of solid food is associated with an elevated abundance of bacteria producing butyrate and a reduced amount of Enterobacteriaceae and Staphylococcus [43]. Another factor affecting the infant microbiome is antibiotic use, which has been linked to the reduction of microbiome richness and diversity [44]. The administration of intrapartum antibiotic use influences microbial communities and antimicrobial genes in the gut microbiome of infants [45]. A randomized trial examining broad‐spectrum antibiotics against sepsis indicated a decreased abundance of Bifidobacterium and an increased abundance of Klebsiella and Enterococcus [46].
MICROBIOME METABOLITES AND CARDIOVASCULAR DISEASES
Microbiome metabolites play important roles in various physiological processes and have been found to have significant effects on cardiovascular diseases (Figure 2) [47]. Some of the metabolites include short‐chain fatty acids (SCFAs), trimethylamine (TMA), and trimethylamine N‐oxide (TMAO). SCFAs are fatty acids with less than six carbon atoms, produced by saccharolytic fermentation of undigested carbohydrates, mainly consisting of acetate, propionate, and butyrate [48, 49]. SCFAs can target signal receptors on various cells within the cardiovascular system as well as other systems, like, the immune system and adipose tissue [50]. SCFAs regulate blood pressure by acting as the ligand to the G‐protein coupled receptors (GPCRs), including GPR41, GPR43, GPR109A, OR51E2, and OR51E1 [51]. The connection between SCFAs and GPCRs affects vascular endothelial cells and immune cells, leading to increased peripheral vascular resistance, vascular remodeling, and renal sodium absorption [51]. SCFAs have a protective effect on atherosclerotic cardiovascular disease (ACVD). Feeding apolipoprotein E knockout‐deficient mice with propionate inhibits angiotensin II‐induced atherosclerosis [52].
Figure 2.
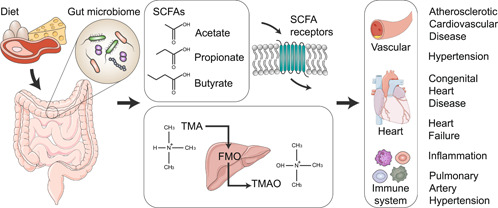
Associations between microbiome metabolites and cardiovascular diseases. Short‐chain fatty acids (SCFAs) are generated through the fermentation of indigestible polysaccharides by the gut microbiome and act on specific SCFAs receptors. Trimethylamine (TMA), a metabolite of the gut microbiome, undergoes an oxidation process in the liver via flavin‐containing monooxygenases (FMOs) and is subsequently converted into trimethylamine N‐oxide (TMAO). These metabolites exert both direct and indirect effects on the cardiovascular system, and they have been associated with certain cardiovascular diseases.
The gut microbiome breaks down compounds containing TMA, such as red meat, resulting in the production of TMA. This TMA can then undergo further oxidation in the host liver to generate TMAO [53, 54]. Elevated plasma TMAO level has been associated with an increased incidence of all‐cause cardiovascular mortality and major adverse cardiac and cerebrovascular events [55]. TMAO has a proatherogenic property, typically by enhancing vascular endothelial cell inflammation, inhibiting reverse cholesterol efflux, and promoting platelet aggregation and thrombosis [54]. TMAO activates NLRP3 inflammasome and then triggers vascular inflammation [56]. Moreover, a high circulating TMAO level is related to poor prognosis in various types of pulmonary arterial hypertension (PAH), including PAH associated with CHD [57].
The gut microbiome affects the metabolism of bile acids, especially secondary bile acids. The primary bile acids are synthesized in the liver and then conjugated with glycine or taurine to form conjugated bile acids. The conjugated bile acids are secreted into bile and then into the gut, where they are deconjugated by the gut microbiome and transformed into secondary bile acids. The primary bile acid‐to‐secondary bile acid ratio may be an important influencing factor in cardiovascular diseases [58]. Patients with chronic heart failure have a decreased primary bile acids level and an increased secondary bile acids level [59]. Bile acids are proven to be associated with lipid metabolism, glucose metabolism, and inflammation, which may be the reason why bile acids metabolism is important in cardiovascular disease [60].
Numerous studies have shown that the disruption of the host gut microbiota can significantly impact the occurrence and development of various cardiovascular diseases. Alterations in the maternal gut microbiome, characterized by a reduction in alpha diversity and distinctive microbiome composition, along with changes in plasma metabolites, were observed to correlate with an increased risk of offspring CHD [61]. The decreased composition of certain probiotics may affect fetal development through impaired gut barrier, chronic inflammation, and insulin resistance [61]. The disorder of gut microbiome and metabolism was observed in patients with CHD and heart failure, which was characterized by decreasing microbiome diversity, richness, and downregulation of retinol metabolism [62]. The serum level of bile acids in patients with repaired Tetralogy of Fallot (TOF) has a positive correlation with indexed right ventricular end‐diastolic volume, which may suggest impaired liver function due to right ventricular dysfunction and systemic congestion [63]. However, it is worth noting that the impact of gut microbiota dysbiosis on the phenotypes of these diseases related to CHD has not yet been systematically and comprehensively summarized.
CHD, GUT MICROBIOME, AND IMMUNITY
The immune system starts to develop in the fetus alongside the formation of the heart and intestine. Functional thymus and the production of circulating T cells commence at 10–11 weeks of gestation, and B cells can be detected in the spleen between the 12th and 23rd weeks of gestation [64]. The birthing process exposes the infant to a wide range of microorganisms, initiating the colonization of beneficial bacteria on the skin and in the gastrointestinal tract. Studies have highlighted the role of the immune system in CHD, which is primarily characterized by systemic inflammation and immune alterations [65].
CHD patients have shown the presence of circulating markers, primarily cytokines. Fan et al. reported the observation of immune activation in patients with CHD, which was evident through an elevation in inflammatory cytokines and a reduction in anti‐inflammatory cytokines [66]. These cytokine levels returned to normal following transcatheter CHD treatment [66]. Various studies have reported elevated levels of tumor necrosis factor‐alpha (TNF‐α) and interleukin‐6 (IL‐6) in CHD patients [67, 68, 69]. It has been established that there is a positive correlation between the levels of growth differentiation factor‐15, β2‐microglobulin, and the severity of chronic heart failure [70]. Additionally, the elevated circulating inflammatory cytokines may be associated with diaphragm dysfunction and restrictive ventilation disorder in CHD patients [71]. Other circulating markers, such as IL‐1, IL‐8, plasma endotoxin, and ghrelin, also indicate systemic inflammation of CHD [65].
Patients with CHD, especially cyanotic CHD, experience persistent exposure to hypoxia‐caused myocardial stress, which triggers the activation of the innate immune system at the cellular level, leading to the activation of proinflammatory cells. [72]. The elevated Neutrophil‐Lymphocyte Ratio (NLR) has been identified in patients with CHD [73, 74]. Other components of the innate immune system, including monocytes, natural killer cells, and mast cells are also activated [65]. T cells are key components of the adaptive immune response as they possess the ability to recognize and selectively target pathogens. Upon maturation, T cells circulate throughout the body, patrolling the blood and lymphatic system in search of foreign invaders. As T cells undergo maturation within the thymus, a byproduct known as T cell receptor excision circles (TRECs) is produced, serving as biomarkers of lymphopoiesis [75]. Compared with the normal population, a reduced level of TRECs in children with CHD has been observed, and a combination of reduced and premature TRECs copies was associated with infection‐caused hospitalization [76]. However, no correlation was found between the TRECs level and the severity of CHD [76].
Surgical repair of cardiac anomalies usually occurs within the first several years of a lifetime. Thymus removal is a procedure often performed during CHD operations to enhance the visibility of the heart and facilitate the surgical intervention, while this process may also have implications for the ongoing immune development of children with CHD [65, 77]. Early thymectomy in CHD patients is associated with changes in the T cell compartment, including decreased total T cells, CD4+ T cells, CD8+ T cells, and a reduced diversity of the T cell receptor [78]. The peripheral naive T cell subset in thymectomy patients exhibits similarities to the immune profile seen in elderly individuals with thymic involution, characterized by decreased counts of CD4+CD45RA+CD62L+ T cells [79]. Besides, elevated levels of the antinuclear antibody and the antineutrophil cytoplasmic antibody are correlated with a higher percentage of CD4+ memory T cells after early thymectomy [80].
The pathogen‐associated molecular patterns (PAMPs), which consist of surface layer proteins, flagella, pili, and capsular polysaccharides of the microbiome, are specifically identified by pattern recognition receptors (PRRs) [81]. Typically, continuous communication does not result in inflammation; on the contrary, it enhances host immune function [82]. However, when commensal microorganisms are misidentified, a consistently activated immune response occurs, which can be detrimental to the host [83]. The gut microbiome can substantially affect host immune homeostasis in early life, play an essential role in the development of postnatal intestinal endotoxin tolerance, and have an impact on immune cells [84, 85]. A higher relative abundance of Bifidobacteria has been reported to be associated with improved CD4+ T‐cell responses and vaccine memory [86]. SCFAs have a systemic anti‐inflammatory effect by upregulating anti‐inflammatory while downregulating proinflammatory cytokines [87]. SCFAs regulate the mitogen‐activated protein kinase and nuclear factor kappa‐B (NF‐κB) signaling pathways through the activation of free fatty acid receptors type 2/3, GPR109A, as well as the inhibition of histone deacetylases [88]. SCFAs produced by commensal microbiomes facilitate the extrathymic generation of regulatory T cells (Tregs) and ameliorate the development of intestinal inflammation [89, 90]. Another metabolite, 12,13‐diHOME, a monohydroxy fatty acid, influences the development of Tregs in early life [91].
HYPOXIA AND HYPOPERFUSION AFFECT THE GUT BARRIER
A symbolic pathophysiological characteristic of CHD is chronic hypoxia. Patients with CHD may experience hypoxemia or tissue hypoxia due to structural abnormalities in their hearts or large vessels. The extent of hypoxia mainly depends on the type and severity of the cardiac defect, including shunting (e.g., patent ductus arteriosus), blood flow obstruction (e.g., coarctation of the aorta), inadequate pumping (e.g., dilated cardiomyopathy), and cyanotic CHDs (e.g., TOF).
It is well known that hypoxia is associated with inflammation. The myocardial response to chronic hypoxia parallels that of bacterial infection, leading to the generation of molecules, such as reactive oxygen species (ROS) [72]. Hypoxia induces ROS production at mitochondrial Complex III, activating inflammatory pathways and stimulating the production of proinflammatory cytokines [92]. Despite the lower oxygen concentration in the digestive tract, especially the large intestine, pathophysiologic hypoxia still causes intestinal inflammation and gut barrier injury [23, 93]. Under inflammatory conditions, oxidative stress generated by neutrophils results in the disruption of interendothelial junctions and facilitates the migration of inflammatory cells across the endothelial barrier, contributing to the clearance of pathogens and leading to tissue injury [94]. Children with CHD face an increased risk of intestinal barrier injury. The overgrowth of Enterococcus in CHD neonates has been associated with gut barrier dysfunction and stimulated inflammatory responses, evidenced by increased levels of intestinal fatty acid binding protein (FABP) and d‐lactate [22]. Steck et al. indicated Enterococcus compromises the gut barrier via gelatinase, a metalloprotease secreted by Enterococcus faecalis [95].
The hypoxia‐inducible factor (HIF) is a protein dimer that orchestrates the expression of several genes in adaptive responses to oxygen deprivation (Figure 3A). As the key molecule in adapting to chronic hypoxia, HIF‐1α upregulated glucose utilization and downregulated fatty acid utilization to maintain adenosine triphosphate supply and cardiac function [96]. Mutations in EPAS1 (gene encoding HIF‐2α) have been identified in patients with CHD residing in high‐altitude regions [97]. These EPAS1 mutations are associated with angiogenesis, demonstrating adaption to chronic hypoxia.
Figure 3.
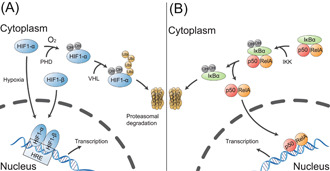
Regulation of HIF and NF‐κB. (A) Regulation of HIF in hypoxic and normoxic conditions. In hypoxic conditions, HIF‐α stabilizes and forms a complex with HIF‐β. This HIF complex translocates to the nucleus, binds to hypoxia response elements (HREs), and activates genes that help the cell adapt to low oxygen. HIF‐α is hydroxylated by prolyl hydroxylases (PHDs) under normoxic conditions. The hydroxylated HIF‐α is then catalyzed by the von Hippel–Lindau (VHL) protein, leading to its subsequent degradation through the proteasome pathway. (B) The canonical activation process of NF‐κB. NF‐κB is located in the cytoplasm and binds with the inhibitory protein IκBα. When IκB kinase (IKK) is activated, the IκBα is phosphorylated and degraded via the ubiquitination pathway. With the degradation of IκBα, the p50, and RelA dimer translocates to the nucleus, where it activates the transcription of multiple target genes. HIF, hypoxia‐inducible factor; NF‐κB, nuclear factor kappa‐B.
One of the significant roles of HIF lies in maintaining gut barrier function via regulating molecules related to the tight junction (claudin‐1) and epithelial surface repair (Trefoil factor‐3) [98]. The microbiome maintains the gut barrier by generating metabolites and interacting with PRR in the gut mucosa, thus preserving gut homeostasis [99]. Healthy diets are metabolized by microbiomes to yield beneficial metabolites that interact with particular receptors located on the membrane or nucleus, regulating HIF and enhancing barrier function [100]. For example, microbiome‐derived butyrate stabilizes HIF by inhibiting HIF prolyl hydroxylases [101].
Chronic hypoxia increases the level of ROS and is also associated with the activation of NF‐κB [102]. NF‐κB exists as a heterodimer of p50 and RelA subunits. When it is activated, the NF‐κB dimer is translocated into the nucleus and regulates the transcription of certain inflammatory genes (Figure 3B) [103]. The gut microbiome can affect NF‐κB‐mediated inflammatory pathways. Streptococcus salivarius inhibits the activation of the NF‐κB and its downstream inflammatory cytokines in intestinal epithelial cells [104, 105].
Similar to hypoxemia, reduced gut perfusion is also a risk factor for intestinal EBD, resulting in increased uptake of lipopolysaccharide (LPS) or other PAMPs [106, 107]. Impaired mucosal immunity and weakened intestinal barrier can lead to the translocation of bacteria and/or their products from the intestinal lumen into adjacent tissues, triggering an amplified inflammatory response and subsequent injury to the mucosal epithelium (Figure 4). CHD represents a risk factor for necrotizing enterocolitis (NEC) due to gut hypoperfusion. Reduced gut perfusion initiates inflammation, which, in turn, triggers secondary vasoconstriction that worsens gut hypoperfusion, thereby establishing a cycle of hypoperfusion and inflammation [108]. In patients with right heart dysfunction, systemic veinous congestion leads to intestinal edema and facilitates the endotoxin translocation from the intestinal tract into the bloodstream [109, 110].
Figure 4.
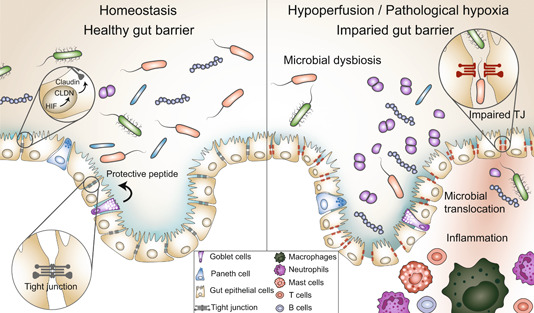
Pathological hypoxia/hypoperfusion induces microbiome dysbiosis and gut barrier dysfunction. Pathological hypoxia/hypoperfusion can result in an alteration of gut barrier integrity. The protective peptide in the mucus barrier and the tight junction between epithelial cells are important components for maintaining gut homeostasis. A weakened gut barrier may facilitate the translocation of bacteria and/or their byproducts from the gut lumen into the surrounding tissues, triggering an amplified inflammatory response. CLDN, claudin; HIF, hypoxia‐inducible factor; TJ, tight junction.
GUT MICROBIOME AND NEURODEVELOPMENT OF CHILDREN WITH CHD
A concern in CHD patients is the occurrence of neurodevelopmental disability (NDD). NDD is observed in nearly half of the children who have undergone cardiac intervention, leading to various challenges, such as cognitive dysfunction, attention and hyperactivity difficulties, motor functioning deficits, impaired language and communication skills [111]. NDD begins at gestation, as the complex circulation of CHD impacts the blood delivery to the fetal brain. Take transposition of the great arteries (TGA) as an example, the fetal brain of patients with TGA receives venous blood with decreased oxygen and metabolic substrates. TGA fetuses had lower brain‐to‐body ratios and higher cerebellar‐to‐total brain ratios compared with normal fetuses with similar gestational age and body growth, indicating immature brain development [112]. Risk factors such as prematurity, chronic hypoxia, and surgical interventions likely have a cumulative effect on the progression of NDD from the fetal period through childhood, with potential persistence into school age and even adulthood [113, 114]. Hemodynamically impaired CHD children have deficiencies in skills related to gross motor, fine motor, and language [115]. Moreover, children who underwent CHD operation with CPB have worse performance for cognition and fine motor skills [116].
The mechanisms underlying brain injury in CHD patients have not been fully studied. Tissue hypoxia and systemic inflammation can lead to the dysfunction of the blood–brain barrier (BBB) and subsequent injury to the nervous system, as indicated by the presence of the phosphorylated form of high molecular weight neurofilament protein [117]. Researchers have attempted to identify morphological differences between the brains of CHD patients and those of healthy individuals. It has been found a delayed brain maturation and a smaller gestational age‐ and weight‐adjusted total brain volumes in fetuses with CHD [118]. Additionally, children with CHD have smaller hippocampal volumes and surface area, and these morphologic alterations are correlated to executive function [119].
Delayed brain development increases susceptibility to acquired white matter injury (WMI). A cohort study involving fetuses/neonates with TGA who underwent preoperative magnetic resonance imaging (MRI) scans revealed that the brain volume of the fetus was associated with acquired WMI after birth [120]. The immaturity of the brain increases the write matter vulnerability to hypoxic‐ischemic brain injury [121, 122]. Early in the 1990s, acquired neuropathology, such as hypoxic‐ischemic lesions and intracranial hemorrhage, was found to be related to CPB procedures involving hypothermic total circulatory arrest lasting longer than 40 min [123]. The CPB‐induced oligodendrocyte dysmaturation and microglial expansion have been observed in a porcine model, suggesting brain damage [124]. Preterm infants exposed to general anesthesia are at an increased risk of brain abnormalities, as evidenced by findings from postoperative MRI scans [125].
The gut microbiome is involved in neurodevelopment in early life and the integrity of BBB, which contributes to NDD in patients with CHD. The gut microbiome can influence BBB integrity through microbe‐stimulated cytokines or metabolites derived from gut microbiomes [126]. Germ‐free mice exhibit higher BBB permeability, which begins with intrauterine life and maintains adulthood. However, exposure to a pathogen‐free gut microbiome has been shown to restore the permeability of BBB and enhance the expression of tight junction proteins, such as occludin and claudin‐5 [127]. The neuroprotective effect of the gut microbiome can be mediated by metabolites. Microbiomes that produce abundant SCFAs, such as Clostridium tyrobutyricum, have been found to improve BBB integrity in mice [127]. Moreover, TMAO has been shown to enhance the integrity of BBB, protect BBB from inflammatory injury, and improve cognitive function through the tight junction regulator Annexin A1 [128]. Another protective metabolite is p‐cresol glucuronide, the gut metabolite of amino acids tyrosine and phenylalanine. This metabolite promotes BBB integrity by antagonizing the LPS receptor TLR4 [129].
The gut microbiome has a significant impact on neuroimmune during brain development. Signals generated from the maternal microbiome may shape the development of fetal microglia [130, 131]. The microbiome metabolites have been shown to influence microglia maturity and functioning [132]. Acetate has been identified as an essential SCFA for microglia maturation and for maintaining their homeostatic metabolic state [133]. Moreover, maternal perinatal probiotic intake leads to an alteration of offspring's astrocyte metabolism, marked by the increased expression of prefrontal cortex PFKFB3 [134].
GUT MICROBIOME AND PEDIATRIC CARDIOVASCULAR SURGERY
Cardiovascular surgery leads to significant alterations in the gut microbiome. Aardema H et al. analyzed fecal samples from patients who underwent coronary artery bypass graft and/or valve surgery [26]. The 16s RNA gene sequencing identified a significant increase of pathobionts (e.g., Eggerthella, Enterococcus, Rothia, and Peptococcus) and a decrease in strictly anaerobic SCFA‐producing gut bacteria (e.g., Anaerostipes, Faecalibacterium, Blautia, and Roseburia). The imbalance of the gut microbiome during the perioperative period may be associated with patients' clinical features and postoperative complications. The postoperative dysbiosis of the gut microbiome and metabolic abnormalities can impact patients' susceptibility to cardiac surgery‐associated acute kidney injury (AKI) [135, 136]. Analysis has identified Escherichia coli, Rothia mucilaginosa, and Clostridium innocuum as AKI‐related microbiomes [136]. Similarly, by analyzing fecal samples from cardiac surgery patients with postoperative pseudopsia, the increased count of Staphylococcus and Pseudomonas were identified [137].
CPB is a crucial and challenging technique employed in cardiac surgery to temporarily assume cardiac and pulmonary functions, ensuring continuous blood flow and oxygen supply. Most CHD surgery involves CPB, while CPB introduces extra risks to the surgery, such as ischemia‐perfusion injury (IRI) [138]. SCFAs have demonstrated the ability to improve cardiac function after IRI [48]. Propionate can act as a protector against myocardial IRI by alleviating the increased Angiotensin II levels through GPR41 [139]. Treatment with butyrate has been shown to significantly improve myocardial IRI via a gut–brain neural circuit, which can be inhibited by subdiaphragmatic vagotomy [140]. Other cardioprotective metabolites including Urolithin B, protect against IRI via the p62/Keap1/Nrf2 signaling pathway [141].
CPB is known to induce inflammation, both at the intestinal and systemic levels [142]. In the early stages following CHD operation, intermediate monocytes exhibit increased levels of CD64, TLR2, and TLR4, which may be associated with impaired postoperative recovery and organ dysfunction [143]. The classical perioperative factor NLR serves as a reflection of systemic inflammation and has been correlated with the composition of the gut microbiome [73, 74]. Elevated NLR is often a risk factor for unfavorable outcomes of CHD operation, including prolonged ventilation duration, extended ICU and hospital stay after TOF repair, reduced cardiac output, and worse single ventricular physiology after the bidirectional Glenn procedure [74, 144, 145, 146]. Compared with pediatric patients without cardiac surgery (without CPB), children who received CHD operation (with CPB) exhibit a greater relative abundance of Actinobacteria and Proteobacteria, as well as significant intestinal barrier dysfunction [16]. An animal experiment further revealed CPB‐mediated dysbiosis of the gut microbiome, SCFAs levels reduction, inflammatory cytokines expression, and EBD [147]. EBD is a important contributor of CPB‐induced systemic inflammation. The dysregulation of claudin‐2 and claudin‐3 after CPB may suggest impaired tight junction and elevated intestinal permeability [147]. However, the loss of gut barrier function seemed to begin preoperatively, marked by the elevated concentration of intestinal FABP2 [148]. Pediatric patients with CHD experienced a disrupted microbiome at baseline, which includes large proportions of proinflammatory, LPS‐expressing bacteria and reduction of gut health‐promoting bacteria [16]. These disruptions may be attributed to chronic hypoxia associated with CHD, and their severity may be exacerbated by subsequent CPB procedures.
Risk factors for impaired gut barrier not only include abnormal circulation, hypoxemia, and poor intestinal perfusion, but also include surgery‐related impairment. Studies have pointed out that children who underwent CHD surgery exhibit abnormal gut permeability [149]. The impaired permeability leads to postoperative endotoxemia, systemic inflammation, and organ dysfunction. Several pathways are activated after the CHD operation, including pathogen‐sensing pathways, antigen‐processing pathways, and immune‐suppressing pathways [150]. The Fontan procedure, a palliative intervention for complex CHD, creates a connection between the right atrium and the pulmonary arteries and redirects the oxygen‐depleted blood directly to the lungs without passing through the impaired right ventricle. Fontan circulation significantly improves the survival of certain CHD patients; however, it has several serious long‐term complications, such as protein‐losing enteropathy (PLE). Fontan‐PLE may be related to elevated resistance of mesenteric vasculature [151]. Patients with Fontan‐PLE exhibit abnormal intestinal permeability and elevated systemic proinflammatory cytokines, marked by intestinal FABP, TNF‐α, and IL‐6 [152].
Postoperative antibiotic use reduces both the richness and diversity of the gut microbiome and increases the relative abundance of Enterococcus and Lactobacillus [153]. Compared with preoperative antibiotic use, postoperative antibiotic use continuously impacts the gut microbiome, which may need an antibiotic withdrawal of more than 2 weeks to diminish.
DISCUSSION
While numerous studies have elucidated the critical role of the gut microbiome in various aspects of life, including growth, aging, and various diseases, there is a paucity of research focused on CHD. It is evident that there exists a bidirectional cross‐talk between CHD and the gut microbiome. Nonetheless, the detailed interplay has primarily been explored through research conducted in other related fields, particularly studies on pediatric and adult cardiovascular surgery.
Another important CHD‐relevant field is prematurity. Although CHD and prematurity are distinct conditions, neonates with CHD are more likely to be born prematurely, and preterm neonates have increased possibilities of comorbidities with CHD [154]. CHD infants and premature infants share certain similarities, such as respiratory challenges, extended hospital stays, proinflammatory environment, immature immune system, and intestinal dysbiosis [155].
Preterm infants encounter sustained inflammatory challenges since birth. In terms of the immune defense, preterm infants primarily rely on nonspecific innate immunity, the functionality of T cell responses, including those mediated by T helper cells responsible for regulating inflammation, may be less effective. [156]. The immature hearts of preterm infants, when exposed to systemic inflammation, may experience perturbations in epigenetic modifications of genes involved in cardiac development [157]. In premature infants, the intestinal environment fosters the expansion of facultative anaerobes due to several factors, such as patchy mucous layer, increased permeability of tight junctions, and reduced antimicrobial peptides [158]. Dysbiosis of the gut microbiome may be associated with various pathological conditions. Take NEC as an example. Infants with NEC tend to exhibit a significantly lower relative abundance of B. longum and a higher relative abundance of Enterobacter cloacae in their gut microbiome profiles [159]. Premature infants have a high susceptibility to NEC due to the challenges their immature gut faces in managing enteral feeding and establishing a healthy bacterial environment [160]. In patients with critical CHD, there is also a noted decrease in the proportion of probiotics and an increase in the proportion of opportunistic pathogens [22]. Several studies have reported that the administration of probiotics can effectively reduce inflammation of preterm infants, resembling their gut environment to that of full‐term infants [161, 162, 163]. The above evidence underscores the importance of future research aimed at investigating microecological therapies targeting probiotic metabolites to improve many clinical phenotypes in children with CHD.
Despite the microbiome homeostasis in the gut, the microbiome in other systems is also worth investigating. For instance, oral microbiome homeostasis of CHD patients. Patients with CHD and caries have elevated abundance of Fusobacterium, Prevotella, Capnocytophaga, and Oribacterium, compared to healthy individuals [164]. It was reported that the immune dysregulation, production of proinflammatory cytokines, and progressive inflammation due to periodontal pathogens were responsible for ACVD [165]. Preventing oral microbiome dysbiosis may serve as an effective approach to reduce the risk of systemic inflammation and infection. Maternal microbiome, as they can affect offspring microbiome, should also be valued in research related to CHD. It has been identified that the maternal microbiome is associated with neurodevelopment, immunity, and inflammation [166, 167]. During pregnancy, microbes derived within the maternal gut are captured by dendritic cells and translocated to the fetus, then participate in the development of the offspring [168]. Furthermore, the maternal microbiome also plays a significant role in pregnancy‐related diseases, such as hypertensive disorder of pregnancy and insulin resistance, increasing the risk for fetal development and potential teratogenic effect [61, 169].
Several research gaps persist in our understanding of the relationship between the gut microbiome and CHD. While various studies have identified the direct involvement of the gut microbiome in cardiovascular diseases, limited research provides direct evidence of its role in CHD. The oral microbiome and maternal microbiome should also be valued as they play key roles in CHD occurrence and progression. The direct impact of metabolites should be carefully investigated in various aspects of CHD, such as growth, neurodevelopment, and immune development. Cells and molecules associated with inflammation, such as immune cells, cytokines, HIF, and NF‐κB within this cross‐talk are also essential areas of exploration. The relationship between the gut microbiome and noncoding RNAs, such as micro‐RNAs also warrants comprehensive examination. Existing studies have identified micro‐RNAs as regulators in both cardiovascular diseases and the gut microbiome. Several biological processes associated with CHD, such as cardiac development, myocardial protection during hypoxia, and adaptive right ventricular hypertrophy in response to PAH, have been linked to certain micro‐RNAs [170].
The therapeutic potential of the gut microbiome in the treatment of CHD is also a promising area for future research. Various laboratory and clinical evidence have underscored the impact of the gut microbiome on the treatment of cardiovascular disease. For example, the gut microbiome can affect the bioavailability and effectiveness of drugs by metabolizing drug compounds, regulating host genes related to drug transport, and influencing the intestinal reuptake process [171]. Individualized analysis of the microbiome based on metabolomic profiling, holds the potential for guiding dietary choices and optimizing perioperative management [172]. Dietary and nutritional strategies can modify the gut microbiome and accelerate patients' recovery, potentially serving as a complementary approach to enhanced recovery after surgery [173]. Cohort studies and clinical trials are vital for evaluating the therapeutic potential of the microbiome in CHD treatment. This may encompass interventions like probiotic therapy in PICU and strategies to optimize the composition of patients' gut microbiome during the perioperative period. Additionally, innovative approaches like fecal microbiome transplantation have the potential to reshape the gut microbial ecology to produce beneficial metabolites. By maintaining gut microbiome homeostasis, it may be feasible to regulate systemic inflammation resulting from surgical injury or CPB. Reconstituting balanced host‐microbiome interactions in patients with CHD may ameliorate the metabolic and immunity disorder, thus preventing certain clinical outcomes and improving prognosis.
AUTHOR CONTRIBUTIONS
Shoujun Li provided direction and guidance throughout the preparation of this manuscript. Yuze Liu and Yuan Huang collected and interpreted studies and were major contributors to the writing and editing of the manuscript. Qiyu He, Zheng Dou, Min Zeng, and Xu Wang reviewed and made significant revisions to the manuscript. All authors have read and approved the final manuscript.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
ACKNOWLEDGMENTS
This work was supported by grants from the National High‐Level Hospital Clinical Research Funding (Grant Nos. 2022‐GSP‐GG‐19 and 2023‐GSP‐QN‐23) and the National Natural Science Foundation of China (Grant No. 82300345).
Liu, Yuze , Huang Yuan, He Qiyu, Dou Zheng, Zeng Min, Wang Xu, and Li Shoujun. 2023. “From Heart to Gut: Exploring the Gut Microbiome in Congenital Heart Disease.” iMeta 2, e144. 10.1002/imt2.144
Yuze Liu and Yuan Huang contributed equally to this work.
DATA AVAILABILITY STATEMENT
This manuscript does not generate any code or data. Supplementary materials (graphical abstracts, slides, videos, Chinese translated version, and update materials) may be found in the online DOI or iMeta Science http://www.imeta.science/.
REFERENCES
- 1. Roth, Gregory A. , Mensah George A., Johnson Catherine O., Addolorato Giovanni, Ammirati Enrico, Baddour Larry M., Barengo Noël C., et al. 2020. “Global Burden of Cardiovascular Diseases and Risk Factors, 1990–2019: Update From the GBD 2019 Study.” Journal of the American College of Cardiology 76: 2982–3021. 10.1016/j.jacc.2020.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Su, Zhanhao , Zou Zhiyong, Hay Simon I., Liu Yiwei, Li Shoujun, Chen Huiwen, Naghavi Mohsen, et al. 2022. “Global, Regional, and National Time Trends in Mortality for Congenital Heart Disease, 1990–2019: An Age‐Period‐Cohort Analysis for the Global Burden of Disease 2019 Study.” eClinicalMedicine 43: 101249. 10.1016/j.eclinm.2021.10124910/gqmnc5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bagge, Carina N. , Smit Jesper, Madsen Nicolas L., and Olsen Morten. 2020. “Congenital Heart Disease and Risk of Central Nervous System Infections: A Nationwide Cohort Study.” Pediatric Cardiology 41: 869–876. 10.1007/s00246-020-02324-z [DOI] [PubMed] [Google Scholar]
- 4. Diller, Gerhard‐Paul , Kempny Aleksander, Alonso‐Gonzalez Rafael, Swan Lorna, Uebing Anselm, Li Wei, Babu‐Narayan Sonya, et al. 2015. “Survival Prospects and Circumstances of Death in Contemporary Adult Congenital Heart Disease Patients Under Follow‐Up at a Large Tertiary Centre.” Circulation 132: 2118–2125. 10.1161/CIRCULATIONAHA.115.017202 [DOI] [PubMed] [Google Scholar]
- 5. Marino, Bradley S. , Lipkin Paul H., Newburger Jane W., Peacock Georgina, Gerdes Marsha, Gaynor J. William, Mussatto Kathleen A., et al. 2012. “Neurodevelopmental Outcomes in Children With Congenital Heart Disease: Evaluation and Management: A Scientific Statement from the American Heart Association.” Circulation 126: 1143–1172. 10.1161/CIR.0b013e318265ee8a [DOI] [PubMed] [Google Scholar]
- 6. Hill, Matthew C. , Kadow Zachary A., Long Hali, Morikawa Yuka, Martin Thomas J., Birks Emma J., Campbell Kenneth S., et al. 2022. “Integrated Multi‐Omic Characterization of Congenital Heart Disease.” Nature 608: 181–191. 10.1038/s41586-022-04989-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Thursby, Elizabeth , and Juge Nathalie. 2017. “Introduction to the Human Gut Microbiota.” The Biochemical Journal 474: 1823–1836. 10.1042/BCJ20160510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tognini, Paola . 2017. “Gut Microbiota: A Potential Regulator of Neurodevelopment.” Frontiers in Cellular Neuroscience 11: 25. 10.3389/fncel.2017.00025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rowland, Ian , Gibson Glenn, Heinken Almut, Scott Karen, Swann Jonathan, Thiele Ines, and Tuohy Kieran. 2018. “Gut Microbiota Functions: Metabolism of Nutrients and Other Food Components.” European Journal of Nutrition 57: 1–24. 10.1007/s00394-017-1445-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang, Shugui , Harvey Louise, Martin Rocio, van der Beek Eline M., Knol Jan, Cryan John F., and Renes Ingrid B.. 2018. “Targeting the Gut Microbiota to Influence Brain Development and Function in Early Life.” Neuroscience & Biobehavioral Reviews 95: 191–201. 10.1016/j.neubiorev.2018.09.002 [DOI] [PubMed] [Google Scholar]
- 11. Kim, Seungbum , Goel Ruby, Kumar Ashok, Qi Yanfei, Lobaton Gil, Hosaka Koji, Mohammed Mohammed, et al. 2018. “Imbalance of Gut Microbiome and Intestinal Epithelial Barrier Dysfunction in Patients With High Blood Pressure.” Clinical Science (London, England: 1979) 132: 701–718. 10.1042/CS20180087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shi, Na , Li Na, Duan Xinwang, and Niu Haitao. 2017. “Interaction Between the Gut Microbiome and Mucosal Immune System.” Military Medical Research 4: 14. 10.1186/s40779-017-0122-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Miyauchi, Eiji , Shimokawa Chikako, Steimle Alex, Desai Mahesh S., and Ohno Hiroshi. 2023. “The Impact of the Gut Microbiome on Extra‐Intestinal Autoimmune Diseases.” Nature Reviews. Immunology 23: 9–23. 10.1038/s41577-022-00727-y [DOI] [PubMed] [Google Scholar]
- 14. Gupta, Arpana , Osadchiy Vadim, and Mayer Emeran A.. 2020. “Brain‐Gut‐Microbiome Interactions in Obesity and Food Addiction.” Nature Reviews. Gastroenterology & Hepatology 17: 655–672. 10.1038/s41575-020-0341-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yang, Ge , Wei Jinlong, Liu Pinyi, Zhang Qihe, Tian Yuan, Hou Guowen, Meng Lingbin, Xin Ying, and Jiang Xin. 2021. “Role of the Gut Microbiota in Type 2 Diabetes and Related Diseases.” Metabolism: Clinical and Experimental 117: 154712. 10.1016/j.metabol.2021.154712 [DOI] [PubMed] [Google Scholar]
- 16. Salomon, Jeffrey , Ericsson Aaron, Price Amber, Manithody Chandrashekhara, Murry Daryl J., Chhonker Yashpal S., Buchanan Paula, and Jain Ajay K.. 2021. “Dysbiosis and Intestinal Barrier Dysfunction in Pediatric Congenital Heart Disease Is Exacerbated Following Cardiopulmonary Bypass.” JACC: Basic to Translational Science 6: 311–327. 10.1016/j.jacbts.2020.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li, Jing , Zhao Fangqing, Wang Yidan, Chen Junru, Tao Jie, Tian Gang, Wu Shouling, et al. 2017. “Gut Microbiota Dysbiosis Contributes to the Development of Hypertension.” Microbiome 5: 14. 10.1186/s40168-016-0222-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Barrington, William T. , and Lusis Aldons J.. 2017. “Association Between the Gut Microbiome and Atherosclerosis.” Nature Reviews Cardiology 14: 699–700. 10.1038/nrcardio.2017.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tang, W. H. Wilson , Li Daniel Y., and Hazen Stanley L.. 2019. “Dietary Metabolism, the Gut Microbiome, and Heart Failure.” Nature Reviews Cardiology 16: 137–154. 10.1038/s41569-018-0108-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shi, Bozhong , Zhang Xiaoyang, Song Zhiying, Dai Zihao, Luo Kai, Chen Bo, Zhou Zijie, et al. 2023. “Targeting Gut Microbiota‐Derived Kynurenine to Predict and Protect the Remodeling of the Pressure‐Overloaded Young Heart.” Science Advances 9: eadg7417. 10.1126/sciadv.adg7417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xing, Junyue , Ying Yongquan, Mao Chenxi, Liu Yiwei, Wang Tingting, Zhao Qian, Zhang Xiaoling, Yan Fuxia, and Zhang Hao. 2018. “Hypoxia Induces Senescence of Bone Marrow Mesenchymal Stem Cells Via Altered Gut Microbiota.” Nature Communications 9: 2020. 10.1038/s41467-018-04453-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Huang, Yuan , Lu Wenlong, Zeng Min, Hu Xiaoyue, Su Zhanhao, Liu Yiwei, Liu Zeye, et al. 2022. “Mapping the Early Life Gut Microbiome in Neonates With Critical Congenital Heart Disease: Multiomics Insights and Implications for Host Metabolic and Immunological Health.” Microbiome 10: 245. 10.1186/s40168-022-01437-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pral, Laís P. , Fachi José L., Corrêa Renan O., Colonna Marco, and Vinolo Marco A. R.. 2021. “Hypoxia and HIF‐1 as Key Regulators of Gut Microbiota and Host Interactions.” Trends in Immunology 42: 604–621. 10.1016/j.it.2021.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xia, Xiong , Ni Jiangjin, Yin Shengnan, Yang Zhipeng, Jiang Haini, Wang Chao, Peng Jian, Wei Hongkui, and Wang Xingyu. 2021. “Elevated Systemic and Intestinal Inflammatory Response are Associated With Gut Microbiome Disorder After Cardiovascular Surgery.” Frontiers in Microbiology 12: 686648. 10.3389/fmicb.2021.686648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dela Cruz, M. , Littmann E., Nayak R., Lehmann C., Keskey R., Baker T., Lin H., et al. 2021. “The Gut Microbiome in Heart Transplantation: A Prospective Pilot Study.” The Journal of Heart and Lung Transplantation 40: S25. 10.1016/j.healun.2021.01.1797 [DOI] [Google Scholar]
- 26. Aardema, Heleen , Lisotto Paola, Kurilshikov Alexander, Diepeveen Janneke R. J., Friedrich Alex W., Sinha Bhanu, de Smet Anne Marie G. A., and Harmsen Hermie J. M.. 2020. “Marked Changes in Gut Microbiota in Cardio‐Surgical Intensive Care Patients: A Longitudinal Cohort Study.” Frontiers in Cellular and Infection Microbiology 9: 467. 10.3389/fcimb.2019.00467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mariat, D. , Firmesse O., Levenez F., Guimarăes Vd, Sokol H., Doré J., Corthier G., and Furet J.‐P.. 2009. “The Firmicutes/Bacteroidetes Ratio of the Human Microbiota Changes With Age.” BMC microbiology 9: 123. 10.1186/1471-2180-9-123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gosalbes, M. J. , Llop S., Vallès Y., Moya A., Ballester F., and Francino M. P.. 2013. “Meconium Microbiota Types Dominated by Lactic Acid or Enteric Bacteria are Differentially Associated With Maternal Eczema and Respiratory Problems in Infants.” Clinical & Experimental Allergy 43: 198–211. 10.1111/cea.12063 [DOI] [PubMed] [Google Scholar]
- 29. Moles, Laura , Gómez Marta, Heilig Hans, Bustos Gerardo, Fuentes Susana, de Vos Willem, Fernández Leónides, Rodríguez Juan M., and Jiménez Esther. 2013. “Bacterial Diversity in Meconium of Preterm Neonates and Evolution of Their Fecal Microbiota During the First Month of Life.” PLoS ONE 8: e66986. 10.1371/journal.pone.0066986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bäckhed, Fredrik , Roswall Josefine, Peng Yangqing, Feng Qiang, Jia Huijue, Kovatcheva‐Datchary Petia, Li Yin, et al. 2015. “Dynamics and Stabilization of the Human Gut Microbiome During the First Year of Life.” Cell Host & Microbe 17: 690–703. 10.1016/j.chom.2015.04.004 [DOI] [PubMed] [Google Scholar]
- 31. Ferretti, Pamela , Pasolli Edoardo, Tett Adrian, Asnicar Francesco, Gorfer Valentina, Fedi Sabina, Armanini Federica, et al. 2018. “Mother‐to‐Infant Microbial Transmission from Different Body Sites Shapes the Developing Infant Gut Microbiome.” Cell Host & Microbe 24: 133–145.e5. 10.1016/j.chom.2018.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pannaraj, Pia S. , Li Fan, Cerini Chiara, Bender Jeffrey M., Yang Shangxin, Rollie Adrienne, Adisetiyo Helty, et al. 2017. “Association Between Breast Milk Bacterial Communities and Establishment and Development of the Infant Gut Microbiome.” JAMA Pediatrics 171: 647–654. 10.1001/jamapediatrics.2017.0378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yatsunenko, Tanya , Rey Federico E., Manary Mark J., Trehan Indi, Dominguez‐Bello Maria Gloria, Contreras Monica, Magris Magda, et al. 2012. “Human Gut Microbiome Viewed Across Age and Geography.” Nature 486: 222–227. 10.1038/nature11053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Qin, Junjie , Li Ruiqiang, Raes Jeroen, Arumugam Manimozhiyan, Burgdorf Kristoffer Solvsten, Manichanh Chaysavanh, Nielsen Trine, et al. 2010. “A Human Gut Microbial Gene Catalogue Established by Metagenomic Sequencing.” Nature 464: 59–65. 10.1038/nature08821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tun, Hein M. , Bridgman Sarah L., Chari Radha, Field Catherine J., Guttman David S., Becker Allan B., Mandhane Piush J., et al. 2018. “Roles of Birth Mode and Infant Gut Microbiota in Intergenerational Transmission of Overweight and Obesity from Mother to Offspring.” JAMA Pediatrics 172: 368–377. 10.1001/jamapediatrics.2017.5535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Grech, Allison , Collins Clare E., Holmes Andrew, Lal Ravin, Duncanson Kerith, Taylor Rachael, and Gordon Adrienne. 2021. “Maternal Exposures and the Infant Gut Microbiome: A Systematic Review With Meta‐Analysis.” Gut Microbes 13: 1–30. 10.1080/19490976.2021.1897210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Arboleya, Silvia , Binetti Ana, Salazar Nuria, Fernández Nuria, Solís Gonzalo, Hernández‐Barranco Ana, Margolles Abelardo, Los Reyes‐Gavilán Clara G., and Gueimonde Miguel. 2012. “Establishment and Development of Intestinal Microbiota in Preterm Neonates.” FEMS Microbiology Ecology 79: 763–772. 10.1111/j.1574-6941.2011.01261.x [DOI] [PubMed] [Google Scholar]
- 38. Kapourchali, Fatemeh Ramezani , and Cresci Gail A. M.. 2020. “Early‐Life Gut Microbiome‐The Importance of Maternal and Infant Factors in Its Establishment.” Nutrition in Clinical Practice 35: 386–405. 10.1002/ncp.10490 [DOI] [PubMed] [Google Scholar]
- 39. Ihekweazu, Faith D. , and Versalovic James. 2018. “Development of the Pediatric Gut Microbiome: Impact on Health and Disease.” The American Journal of the Medical Sciences 356: 413–423. 10.1016/j.amjms.2018.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Granger, Claire L. , Embleton Nicholas D., Palmer Jeremy M., Lamb Christopher A., Berrington Janet E., and Stewart Christopher J.. 2021. “Maternal Breastmilk, Infant Gut Microbiome and the Impact on Preterm Infant Health.” Acta Paediatrica 110: 450–457. 10.1111/apa.15534 [DOI] [PubMed] [Google Scholar]
- 41. Marcobal, Angela , Barboza Mariana, Froehlich John W., Block David E., German J. Bruce, Lebrilla Carlito B., and Mills David A.. 2010. “Consumption of Human Milk Oligosaccharides by Gut‐Related Microbes.” Journal of Agricultural and Food Chemistry 58: 5334–5340. 10.1021/jf9044205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Forbes, Jessica D. , Azad Meghan B., Vehling Lorena, Tun Hein M., Konya Theodore B., Guttman David S., Field Catherine J., et al. 2018. “Association of Exposure to Formula in the Hospital and Subsequent Infant Feeding Practices With Gut Microbiota and Risk of Overweight in the First Year of Life.” JAMA Pediatrics 172: e181161. 10.1001/jamapediatrics.2018.1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Martin, Rocio , Makino Hiroshi, Cetinyurek Yavuz Aysun, Ben‐Amor Kaouther, Roelofs Mieke, Ishikawa Eiji, Kubota Hiroyuki, et al. 2016. “Early‐Life Events, Including Mode of Delivery and Type of Feeding, Siblings and Gender, Shape the Developing Gut Microbiota.” PLoS ONE 11: e0158498. 10.1371/journal.pone.0158498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. McDonnell, Lucy , Gilkes Alexander, Ashworth Mark, Rowland Victoria, Harries Timothy Hugh, Armstrong David, and White Patrick. 2021. “Association Between Antibiotics and Gut Microbiome Dysbiosis in Children: Systematic Review and Meta‐Analysis.” Gut Microbes 13: 1–18. 10.1080/19490976.2020.1870402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Garcia, Vanessa R . 2021. “Impact of Intrapartum Antibiotic Prophylaxis for Group B Streptococcus on the Term Infant Gut Microbiome: A State of the Science Review.” Journal of Midwifery & Women's Health 66: 351–359. 10.1111/jmwh.13245 [DOI] [PubMed] [Google Scholar]
- 46. Reyman, Marta , van Houten Marlies A., Watson Rebecca L., Chu Mei Ling J. N., Arp Kayleigh, de Waal Wouter J., Schiering Irene, et al. 2022. “Effects of Early‐Life Antibiotics on the Developing Infant Gut Microbiome and Resistome: A Randomized Trial.” Nature Communications 13: 893. 10.1038/s41467-022-28525-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Xu, Jing , Yang Yicheng, Li Xin, Ding Shusi, Zheng Lemin, Xiong Changming, and Yang Yuejin. 2023. “Pleiotropic Activities of Succinate: The Interplay Between Gut Microbiota and Cardiovascular Diseases.” iMeta 2: e124. 10.1002/imt2.124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hu, Tongtong , Wu Qingqing, Yao Qi, Jiang Kebing, Yu Jiabin, and Tang Qizhu. 2022. “Short‐Chain Fatty Acid Metabolism and Multiple Effects on Cardiovascular Diseases.” Ageing Research Reviews 81: 101706. 10.1016/j.arr.2022.101706 [DOI] [PubMed] [Google Scholar]
- 49. Morrison, Douglas J. , and Preston Tom. 2016. “Formation of Short Chain Fatty Acids by the Gut Microbiota and Their Impact on Human Metabolism.” Gut Microbes 7: 189–200. 10.1080/19490976.2015.1134082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chen, Xiao‐Feng , Chen Xiangqi, and Tang Xiaoqiang. 2020. “Short‐Chain Fatty Acid, Acylation and Cardiovascular Diseases.” Clinical Science (London, England: 1979) 134: 657–676. 10.1042/CS20200128 [DOI] [PubMed] [Google Scholar]
- 51. Xu, Jiaojiao , Moore Brittni N., and Pluznick Jennifer L.. 2022. “Short‐Chain Fatty Acid Receptors and Blood Pressure Regulation: Council on Hypertension Mid‐Career Award for Research Excellence 2021.” Hypertension 79: 2127–2137. 10.1161/HYPERTENSIONAHA.122.18558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bartolomaeus, Hendrik , Balogh András, Yakoub Mina, Homann Susanne, Markó Lajos, Höges Sascha, Tsvetkov Dmitry, et al. 2019. “Short‐Chain Fatty Acid Propionate Protects from Hypertensive Cardiovascular Damage.” Circulation 139: 1407–1421. 10.1161/CIRCULATIONAHA.118.036652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ufnal, Marcin , Zadlo Anna, and Ostaszewski Ryszard. 2015. “TMAO: A Small Molecule of Great Expectations.” Nutrition 31: 1317–1323. 10.1016/j.nut.2015.05.006 [DOI] [PubMed] [Google Scholar]
- 54. Wang, Zeneng , and Zhao Yongzhong. 2018. “Gut Microbiota Derived Metabolites in Cardiovascular Health and Disease.” Protein & Cell 9: 416–431. 10.1007/s13238-018-0549-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Schiattarella, Gabriele Giacomo , Sannino Anna, Toscano Evelina, Giugliano Giuseppe, Gargiulo Giuseppe, Franzone Anna, Trimarco Bruno, Esposito Giovanni, and Perrino Cinzia. 2017. “Gut Microbe‐Generated Metabolite Trimethylamine‐N‐Oxide as Cardiovascular Risk Biomarker: A Systematic Review and Dose‐Response Meta‐Analysis.” European Heart Journal 38: 2948–2956. 10.1093/eurheartj/ehx342 [DOI] [PubMed] [Google Scholar]
- 56. Zhang, Xiao‐Nan , Yu Zong‐Liang, Chen Ji‐Ye, Li Xiao‐Ya, Wang Ze‐Ping, Wu Min, and Liu Long‐Tao. 2022. “The Crosstalk Between NLRP3 Inflammasome and Gut Microbiome in Atherosclerosis.” Pharmacological Research 181: 106289. 10.1016/j.phrs.2022.106289 [DOI] [PubMed] [Google Scholar]
- 57. Yang, Yicheng , Yang Beilan, Li Xin, Xue Lin, Liu Bingyang, Liang Yanru, Zhao Zhihui, et al. 2022. “Higher Circulating Trimethylamine N‐Oxide Levels are Associated With Worse Severity and Prognosis in Pulmonary Hypertension: a Cohort Study.” Respiratory Research 23: 344. 10.1186/s12931-022-02282-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kazemian, Negin , Mahmoudi Morteza, Halperin Frank, Wu Joseph C., and Pakpour Sepideh. 2020. “Gut Microbiota and Cardiovascular Disease: Opportunities and Challenges.” Microbiome 8: 36. 10.1186/s40168-020-00821-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mayerhofer, Cristiane C. K. , Ueland Thor, Broch Kaspar, Vincent Royce P., Cross Gemma F., Dahl Christen P., Aukrust Pål, Gullestad Lars, Hov Johannes R., and Trøseid Marius. 2017. “Increased Secondary/Primary Bile Acid Ratio in Chronic Heart Failure.” Journal of Cardiac Failure 23: 666–671. 10.1016/j.cardfail.2017.06.007 [DOI] [PubMed] [Google Scholar]
- 60. Tang, W. H. Wilson , Kitai Takeshi, and Hazen Stanley L.. 2017. “Gut Microbiota in Cardiovascular Health and Disease.” Circulation Research 120: 1183–1196. 10.1161/CIRCRESAHA.117.309715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wang, Tingting , Chen Lizhang, Huang Peng, Yang Tubao, Zhang Senmao, Zhao Lijuan, Chen Letao, Ye Ziwei, Luo Liu, and Qin Jiabi. 2021. “Association of Maternal Gut Microbiota and Plasma Metabolism With Congenital Heart Disease in Offspring: A Multi‐Omic Analysis.” Scientific Reports 11: 5339. 10.1038/s41598-021-84901-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zhang, Qi‐Liang , Chen Xiu‐Hua, Zhou Si‐Jia, Lei Yu‐Qing, Huang Jiang‐Shan, Chen Qiang, and Cao Hua. 2023. “Relationship Between Disorders of the Intestinal Microbiota and Heart Failure in Infants With Congenital Heart Disease.” Frontiers in Cellular and Infection Microbiology 13: 1152349. 10.3389/fcimb.2023.1152349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Grangl, Gernot , Zöhrer Evelyn, Köstenberger Martin, Jud Alexandra, Fauler Günter, Scharnagl Hubert, Stojakovic Tatjana, Marterer Robert, Gamillscheg Andreas, and Jahnel Jörg. 2015. “Serum Bile Acids in Repaired Tetralogy of Fallot: A Marker for Liver and Heart?” PLoS ONE 10: e0144745. 10.1371/journal.pone.0144745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hossain, Zakir , Reza A. H. M. Mohsinul, Qasem Wafaa A., Friel James K., and Omri Abdelwahab. 2022. “Development of the Immune System in the Human Embryo.” Pediatric Research 92: 951–955. 10.1038/s41390-022-01940-0 [DOI] [PubMed] [Google Scholar]
- 65. Wienecke, Laura M. , Cohen Sarah, Bauersachs Johann, Mebazaa Alexandre, and Chousterman Benjamin G.. 2022. “Immunity and Inflammation: The Neglected Key Players in Congenital Heart Disease?” Heart Failure Reviews 27: 1957–1971. 10.1007/s10741-021-10187-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Fan, Shunyang , Li Kefang, Zhang Deyin, and Liu Fuyun. 2018. “JNK and NF‐κB Signaling Pathways are Involved in Cytokine Changes in Patients With Congenital Heart Disease Prior to and After Transcatheter Closure.” Experimental and Therapeutic Medicine 15: 1525–1531. 10.3892/etm.2017.5595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zhang, Sai , Guo Gong‐Liang, Yang Li‐Li, and Sun Li‐Qun. 2017. “Elevated Serum Levels of Ghrelin and TNF‐α in Patients With Cyanotic and Acyanotic Congenital Heart Disease.” World Journal of Pediatrics 13: 122–128. 10.1007/s12519-016-0068-0 [DOI] [PubMed] [Google Scholar]
- 68. Afify, M. F. , Mohamed G. B., El‐Maboud M. A., and Abdel‐Latif E. A.. 2009. “Serum Levels of Ghrelin, Tumor Necrosis Factor‐ and Interleukin‐6 in Infants and Children With Congenital Heart Disease.” Journal of Tropical Pediatrics 55: 388–392. 10.1093/tropej/fmp036 [DOI] [PubMed] [Google Scholar]
- 69. Wang, Dandan , Fang Jian, Wang Ruigeng, Sun Dongming, Xia Kun, Yin Wei, Zhang Sai, and Sun Liqun. 2016. “Elevated Serum Ghrelin, Tumor Necrosis Factor‐α and interleukin‐6 in Congenital Heart Disease.” Pediatrics International 58: 259–264. 10.1111/ped.12773 [DOI] [PubMed] [Google Scholar]
- 70. Zhou, X.‐J. , Zhang X., Zhang J., Zhou L., Zhou T.‐T., and Zhang J.‐W.. 2020. “Diagnostic Value of Growth Differentiation Factor‐15 and β2‐Microglobulin in Children With Congenital Heart Disease Combined With Chronic Heart Failure and Its Relationship With Cardiac Function.” European Review for Medical and Pharmacological Sciences 24: 8096–8103. 10.26355/eurrev_202008_22494 [DOI] [PubMed] [Google Scholar]
- 71. Spiesshoefer, Jens , Orwat Stefan, Henke Carolin, Kabitz Hans‐Joachim, Katsianos Stratis, Borrelli Chiara, Baumgartner Helmut, et al. 2020. “Inspiratory Muscle Dysfunction and Restrictive Lung Function Impairment in Congenital Heart Disease: Association With Immune Inflammatory Response and Exercise Intolerance.” International Journal of Cardiology 318: 45–51. 10.1016/j.ijcard.2020.06.055 [DOI] [PubMed] [Google Scholar]
- 72. Manuel, Valdano , Miana Leonardo A., and Jatene Marcelo B.. 2022. “Neutrophil‐Lymphocyte Ratio in Congenital Heart Surgery: What is Known and What is New?” World Journal for Pediatric & Congenital Heart Surgery 13: 208–216. 10.1177/21501351211064143 [DOI] [PubMed] [Google Scholar]
- 73. Kain, Vasundhara , Van Der Pol William, Mariappan Nithya, Ahmad Aftab, Eipers Peter, Gibson Deanna L., Gladine Cecile, et al. 2019. “Obesogenic Diet in Aging Mice Disrupts Gut Microbe Composition and Alters Neutrophi:Lymphocyte Ratio, Leading to Inflamed Milieu in Acute Heart Failure.” The FASEB Journal 33: 6456–6469. 10.1096/fj.201802477R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Iliopoulos, Ilias , Alder Matthew N., Cooper David S., Villarreal Enrique G., Loomba Rohit, Sahay Rashmi D., Fei Lin, Steele Paul E., and Flores Saul. 2020. “Pre‐Operative Neutrophil–Lymphocyte Ratio Predicts Low Cardiac Output in Children After Cardiac Surgery.” Cardiology in the Young 30: 521–525. 10.1017/S1047951120000487 [DOI] [PubMed] [Google Scholar]
- 75. Yuki, Koichi , and Koutsogiannaki Sophia. 2021. “Neutrophil and T Cell Functions in Patients With Congenital Heart Diseases: A Review.” Pediatric Cardiology 42: 1478–1482. 10.1007/s00246-021-02681-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Davey, Brooke T. , Elder Robert W., Cloutier Michelle M., Bennett Nicholas, Lee Ji Hyun, Wang Zhu, Manning Adrienne, et al. 2019. “T‐Cell Receptor Excision Circles in Newborns With Congenital Heart Disease.” The Journal of Pediatrics 213: 96–102.e2. 10.1016/j.jpeds.2019.05.061 [DOI] [PubMed] [Google Scholar]
- 77. Roosen, Jorg , Oosterlinck Wouter, and Meyns Bart. 2015. “Routine Thymectomy in Congenital Cardiac Surgery Changes Adaptive Immunity Without Clinical Relevance.” Interactive Cardiovascular and Thoracic Surgery 20: 101–106. 10.1093/icvts/ivu343 [DOI] [PubMed] [Google Scholar]
- 78. Cavalcanti, Nara Vasconcelos , Palmeira Patrícia, Jatene Marcelo Biscegli, de Barros Dorna Mayra, and Carneiro‐Sampaio Magda. 2021. “Early Thymectomy Is Associated With Long‐Term Impairment of the Immune System: A Systematic Review.” Frontiers in Immunology 12: 774780. 10.3389/fimmu.2021.774780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Prelog, Martina , Keller Michael, Geiger Ralf, Brandstätter Anita, Würzner Reinhard, Schweigmann Ulrich, Zlamy Manuela, Zimmerhackl Lothar Bernd, and Grubeck‐Loebenstein Beatrix. 2009. “Thymectomy in Early Childhood: Significant Alterations of the CD4+CD45RA+CD62L+ T Cell Compartment in Later Life.” Clinical Immunology 130: 123–132. 10.1016/j.clim.2008.08.023 [DOI] [PubMed] [Google Scholar]
- 80. van den Broek, Theo , Madi Asaf, Delemarre Eveline M., Schadenberg Alvin W. L., Tesselaar Kiki, Borghans José A. M., Nierkens Stefan, et al. 2017. “Human Neonatal Thymectomy Induces Altered B‐Cell Responses and Autoreactivity.” European Journal of Immunology 47: 1970–1981. 10.1002/eji.201746971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Liu, Qing , Yu Zhiming, Tian Fengwei, Zhao Jianxin, Zhang Hao, Zhai Qixiao, and Chen Wei. 2020. “Surface Components and Metabolites of Probiotics for Regulation Of Intestinal Epithelial Barrier.” Microbial Cell Factories 19: 23. 10.1186/s12934-020-1289-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Round, June L. , and Mazmanian Sarkis K.. 2009. “The Gut Microbiota Shapes Intestinal Immune Responses During Health and Disease.” Nature Reviews. Immunology 9: 313–323. 10.1038/nri2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Fawkner‐Corbett, David , Simmons Alison, and Parikh Kaushal. 2017. “Microbiome, Pattern Recognition Receptor Function in Health and Inflammation.” Best Practice & Research Clinical Gastroenterology 31: 683–691. 10.1016/j.bpg.2017.11.001 [DOI] [PubMed] [Google Scholar]
- 84. Zhang, Chenguang , Liu Huifeng, Sun Lei, Wang Yue, Chen Xiaodong, Du Juan, Sjöling Åsa, Yao Junhu, and Wu Shengru. 2023. “An Overview of Host‐Derived Molecules That Interact With Gut Microbiota.” iMeta 2: e88. 10.1002/imt2.88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Cuna, Alain , Morowitz Michael J., Ahmed Ishfaq, Umar Shahid, and Sampath Venkatesh. 2021. “Dynamics of the Preterm Gut Microbiome in Health and Disease.” American Journal of Physiology—Gastrointestinal and Liver Physiology 320: G411–G419. 10.1152/ajpgi.00399.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Davis, Erin C. , Castagna Vanessa P., Sela David A., Hillard Margaret A., Lindberg Samantha, Mantis Nicholas J., Seppo Antti E., and Järvinen Kirsi M.. 2022. “Gut Microbiome and Breast‐Feeding: Implications for Early Immune Development.” Journal of Allergy and Clinical Immunology 150: 523–534. 10.1016/j.jaci.2022.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Martin‐Gallausiaux, Camille , Marinelli Ludovica, Blottière Hervé M., Larraufie Pierre, and Lapaque Nicolas. 2021. “SCFA: Mechanisms and Functional Importance in the Gut.” Proceedings of the Nutrition Society 80: 37–49. 10.1017/S0029665120006916 [DOI] [PubMed] [Google Scholar]
- 88. Li, Meng , van Esch Betty C. A. M., Wagenaar Gerry T. M., Garssen Johan, Folkerts Gert, and Henricks Paul A. J.. 2018. “Pro‐ and Anti‐Inflammatory Effects of Short Chain Fatty Acids on Immune and Endothelial Cells.” European Journal of Pharmacology 831: 52–59. 10.1016/j.ejphar.2018.05.003 [DOI] [PubMed] [Google Scholar]
- 89. Furusawa, Yukihiro , Obata Yuuki, Fukuda Shinji, Endo Takaho A., Nakato Gaku, Takahashi Daisuke, Nakanishi Yumiko, et al. 2013. “Commensal Microbe‐Derived Butyrate Induces the Differentiation of Colonic Regulatory T Cells.” Nature 504: 446–450. 10.1038/nature12721 [DOI] [PubMed] [Google Scholar]
- 90. Arpaia, Nicholas , Campbell Clarissa, Fan Xiying, Dikiy Stanislav, van der Veeken Joris, de Roos Paul, Liu Hui, et al. 2013. “Metabolites Produced by Commensal Bacteria Promote Peripheral Regulatory T‐Cell Generation.” Nature 504: 451–455. 10.1038/nature12726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Levan, Sophia R. , Stamnes Kelsey A., Lin Din L., Panzer Ariane R., Fukui Elle, McCauley Kathryn, Fujimura Kei E., et al. 2019. “Elevated Faecal 12,13‐diHOME Concentration in Neonates at High Risk for Asthma Is Produced by Gut Bacteria and Impedes Immune Tolerance.” Nature Microbiology 4: 1851–1861. 10.1038/s41564-019-0498-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Chandel, Navdeep S. , McClintock David S., Feliciano Carlos E., Wood Teresa M., Melendez J. Andres, Rodriguez Ana M., and Schumacker Paul T.. 2000. “Reactive Oxygen Species Generated at Mitochondrial Complex III Stabilize Hypoxia‐Inducible Factor‐1α During Hypoxia.” Journal of Biological Chemistry 275: 25130–25138. 10.1074/jbc.M001914200 [DOI] [PubMed] [Google Scholar]
- 93. Fagundes, Raphael R. , and Taylor Cormac T.. 2017. “Determinants of Hypoxia‐Inducible Factor Activity in the Intestinal Mucosa.” Journal of Applied Physiology 123: 1328–1334. 10.1152/japplphysiol.00203.2017 [DOI] [PubMed] [Google Scholar]
- 94. Mittal, Manish , Siddiqui Mohammad Rizwan, Tran Khiem, Reddy Sekhar P., and Malik Asrar B.. 2014. “Reactive Oxygen Species in Inflammation and Tissue Injury.” Antioxidants & Redox Signaling 20: 1126–1167. 10.1089/ars.2012.5149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Steck, Natalie , Hoffmann Micha, Sava Irina G., Kim Sandra C., Hahne Hannes, Tonkonogy Susan L., Mair Katrin, et al. 2011. “ Enterococcus faecalis Metalloprotease Compromises Epithelial Barrier and Contributes to Intestinal Inflammation.” Gastroenterology 141: 959–971. 10.1053/j.gastro.2011.05.035 [DOI] [PubMed] [Google Scholar]
- 96. Liu, Yiwei , Luo Qipeng, Su Zhanhao, Xing Junyue, Wu Jinlin, Xiang Li, Huang Yuan, et al. 2021. “Suppression of Myocardial Hypoxia‐Inducible Factor‐1α Compromises Metabolic Adaptation and Impairs Cardiac Function in Patients With Cyanotic Congenital Heart Disease During Puberty.” Circulation 143: 2254–2272. 10.1161/CIRCULATIONAHA.120.051937 [DOI] [PubMed] [Google Scholar]
- 97. Pan, Hong , Chen Qiuhong, Qi Shenggui, Li Tengyan, Liu Beihong, Liu Shiming, Ma Xu, and Wang Binbin. 2018. “Mutations in EPAS1 in Congenital Heart Disease in Tibetans.” Bioscience Reports 38: BSR20181389. 10.1042/BSR20181389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Singhal, Rashi , and Shah Yatrik M.. 2020. “Oxygen Battle in the Gut: Hypoxia and Hypoxia‐Inducible Factors in Metabolic and inflammatory Responses in the Intestine.” Journal of Biological Chemistry 295: 10493–10505. 10.1074/jbc.REV120.011188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Wells, Jerry M. , Brummer Robert J., Derrien Muriel, MacDonald Thomas T., Troost Freddy, Cani Patrice D., Theodorou Vassilia, et al. 2017. “Homeostasis of the Gut Barrier and Potential Biomarkers.” American Journal of Physiology—Gastrointestinal and Liver Physiology 312: G171–G193. 10.1152/ajpgi.00048.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Ghosh, Sweta , Whitley Caleb Samuel, Haribabu Bodduluri, and Jala Venkatakrishna Rao. 2021. “Regulation of Intestinal Barrier Function by Microbial Metabolites.” Cellular and Molecular Gastroenterology and Hepatology 11: 1463–1482. 10.1016/j.jcmgh.2021.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Wang, Ruth X. , Henen Morkos A., Lee J. Scott, Vögeli Beat, and Colgan Sean P.. 2021. “Microbiota‐Derived Butyrate Is an Endogenous HIF Prolyl Hydroxylase Inhibitor.” Gut Microbes 13: 1938380. 10.1080/19490976.2021.1938380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Korbecki, Jan , Simińska Donata, Gąssowska‐Dobrowolska Magdalena, Listos Joanna, Gutowska Izabela, Chlubek Dariusz, and Baranowska‐Bosiacka Irena. 2021. “Chronic and Cycling Hypoxia: Drivers of Cancer Chronic Inflammation Through HIF‐1 and NF‐κB Activation: A Review of the Molecular Mechanisms.” International Journal of Molecular Sciences 22: 10701. 10.3390/ijms221910701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Eltzschig, Holger K. , and Carmeliet Peter. 2011. “Hypoxia and Inflammation.” New England Journal of Medicine 364: 656–665. 10.1056/NEJMra0910283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Kaci, Ghalia , Goudercourt Denise, Dennin Véronique, Pot Bruno, Doré Joël, Ehrlich S. Dusko, Renault Pierre, Blottière Hervé M., Daniel Catherine, and Delorme Christine. 2014. “Anti‐Inflammatory Properties of Streptococcus salivarius, a Commensal Bacterium of the Oral Cavity and Digestive Tract.” Applied and Environmental Microbiology 80: 928–934. 10.1128/AEM.03133-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Kaci, Ghalia , Lakhdari Omar, Doré Joël, Ehrlich S. Dusko, Renault Pierre, Blottière Hervé M., and Delorme Christine. 2011. “Inhibition of the NF‐κB Pathway in Human Intestinal Epithelial Cells by Commensal Streptococcus salivarius .” Applied and Environmental Microbiology 77: 4681–4684. 10.1128/AEM.03021-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Opal, Steven M . 2011. “Endotoxemia Before and After Surgical Repair for Congenital Heart Disease.” American Journal of Respiratory and Critical Care Medicine 184: 1223–1224. 10.1164/rccm.201108-1432ED [DOI] [PubMed] [Google Scholar]
- 107. Feng, Dan , Christensen Jason T., Yetman Anji T., Lindsey Merry L., Singh Amar B., and Salomon Jeffrey D.. 2021. “The Microbiome's Relationship With Congenital Heart Disease: More Than a Gut Feeling.” Journal of Congenital Cardiology 5: 5. 10.1186/s40949-021-00060-4 [DOI] [Google Scholar]
- 108. Bowker, Rakhee M. , Yan Xiaocai, and De Plaen Isabelle G.. 2018. “Intestinal Microcirculation and Necrotizing Enterocolitis: The Vascular Endothelial Growth Factor System.” Seminars in Fetal & Neonatal Medicine 23: 411–415. 10.1016/j.siny.2018.08.008 [DOI] [PubMed] [Google Scholar]
- 109. Sharma, Rakesh , Bolger Aidan P., Li Wei, Davlouros Periklis A., Volk Hans‐Dieter, Poole‐Wilson Philip A., Coats Andrew J. S., Gatzoulis Michael A., and Anker Stefan D.. 2003. “Elevated Circulating Levels of Inflammatory Cytokines and Bacterial Endotoxin in Adults With Congenital Heart Disease.” The American Journal of Cardiology 92: 188–193. 10.1016/s0002-9149(03)00536-8 [DOI] [PubMed] [Google Scholar]
- 110. Valentova, Miroslava , von Haehling Stephan, Bauditz Juergen, Doehner Wolfram, Ebner Nicole, Bekfani Tarek, Elsner Sebastian, et al. 2016. “Intestinal Congestion and Right Ventricular Dysfunction: A Link With Appetite Loss, Inflammation, and Cachexia in Chronic Heart Failure.” European Heart Journal 37: 1684–1691. 10.1093/eurheartj/ehw008 [DOI] [PubMed] [Google Scholar]
- 111. Verrall, Charlotte E. , Blue Gillian M., Loughran‐Fowlds Alison, Kasparian Nadine, Gecz Jozef, Walker Karen, Dunwoodie Sally L., et al. 2019. “Big Issues' in Neurodevelopment for Children and Adults With Congenital Heart Disease.” Open Heart 6: e000998. 10.1136/openhrt-2018-000998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Jørgensen, Ditte E. S. , Tabor Ann, Rode Line, Dyre Liv, Ekelund Charlotte K., Hellmuth Signe G., Macgowan Christopher K., et al. 2018. “Longitudinal Brain and Body Growth in Fetuses With and Without Transposition of the Great Arteries: Quantitative Volumetric Magnetic Resonance Imaging Study.” Circulation 138: 1368–1370. 10.1161/CIRCULATIONAHA.118.034467 [DOI] [PubMed] [Google Scholar]
- 113. Wehrle, Flavia M. , Bartal Timm, Adams Mark, Bassler Dirk, Hagmann Cornelia F., Kretschmar Oliver, Natalucci Giancarlo, and Latal Beatrice. 2022. “Similarities and Differences in the Neurodevelopmental Outcome of Children With Congenital Heart Disease and Children Born Very Preterm at School Entry.” The Journal of Pediatrics 250: 29–37.e1. 10.1016/j.jpeds.2022.05.047 [DOI] [PubMed] [Google Scholar]
- 114. Howell, Heather B. , Zaccario Michele, Kazmi Sadaf H., Desai Purnahamsi, Sklamberg Felice E., and Mally Pradeep. 2019. “Neurodevelopmental Outcomes of Children With Congenital Heart Disease: A Review.” Current Problems in Pediatric and Adolescent Health Care 49: 100685. 10.1016/j.cppeds.2019.100685 [DOI] [PubMed] [Google Scholar]
- 115. Polat, Selda , Okuyaz Cetin, Hallıoğlu Olgu, Mert Ertan, and Makharoblidze Khatuna. 2011. “Evaluation of Growth and Neurodevelopment in Children With Congenital Heart Disease: Congenital Heart Disease.” Pediatrics International: Official Journal of the Japan Pediatric Society 53: 345–349. 10.1111/j.1442-200X.2010.03230.x [DOI] [PubMed] [Google Scholar]
- 116. Schultz, Amy H. , Ittenbach Richard F., Gerdes Marsha, Jarvik Gail P., Wernovsky Gil, Bernbaum Judy, Solot Cynthia, et al. 2017. “Effect of Congenital Heart Disease on 4‐year Neurodevelopment Within Multiple‐Gestation Births.” The Journal of Thoracic and Cardiovascular Surgery 154: 273–281.e2. 10.1016/j.jtcvs.2017.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Pironkova, Rossitza P. , Giamelli Joseph, Seiden Howard, Parnell Vincent A., Gruber Dorota, Sison Cristina P., Kowal Czeslawa, and Ojamaa Kaie. 2017. “Brain Injury With Systemic Inflammation in Newborns With Congenital Heart Disease Undergoing Heart Surgery.” Experimental and Therapeutic Medicine 14: 228–238. 10.3892/etm.2017.4493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Limperopoulos, Catherine , Tworetzky Wayne, McElhinney Doff B., Newburger Jane W., Brown David W., Robertson Richard L., Guizard Nicolas, et al. 2010. “Brain Volume and Metabolism in Fetuses With Congenital Heart Disease: Evaluation With Quantitative Magnetic Resonance Imaging and Spectroscopy.” Circulation 121: 26–33. 10.1161/CIRCULATIONAHA.109.865568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Fontes, Kimberly , Rohlicek Charles V., Saint‐Martin Christine, Gilbert Guillaume, Easson Kaitlyn, Majnemer Annette, Majnemer Annette, et al. 2019. “Hippocampal Alterations and Functional Correlates in Adolescents and Young Adults With Congenital Heart Disease.” Human Brain Mapping 40: 3548–3560. 10.1002/hbm.24615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Peyvandi, Shabnam , Lim Jessie Mei, Marini Davide, Xu Duan, Reddy V. Mohan, Barkovich A. James, Miller Steven, McQuillen Patrick, and Seed Mike. 2021. “Fetal Brain Growth and Risk of Postnatal White Matter Injury in Critical Congenital Heart Disease.” The Journal of Thoracic and Cardiovascular Surgery 162: 1007–1014.e1. 10.1016/j.jtcvs.2020.09.096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Beca, John , Gunn Julia K., Coleman Lee, Hope Ayton, Reed Peter W., Hunt Rodney W., Finucane Kirsten, et al. 2013. “New White Matter Brain Injury After Infant Heart Surgery Is Associated With Diagnostic Group and the Use of Circulatory Arrest.” Circulation 127: 971–979. 10.1161/CIRCULATIONAHA.112.001089 [DOI] [PubMed] [Google Scholar]
- 122. Morton, Paul D. , Ishibashi Nobuyuki, and Jonas Richard A.. 2017. “Neurodevelopmental Abnormalities and Congenital Heart Disease: Insights Into Altered Brain Maturation.” Circulation Research 120: 960–977. 10.1161/CIRCRESAHA.116.309048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Glauser, Tracy A. , Rorke Lucy B., Weinberg Paul M., and Clancy Robert R.. 1990. “Acquired Neuropathologic Lesions Associated With the Hypoplastic Left Heart Syndrome.” Pediatrics 85: 991–1000. 10.1542/peds.85.6.991 [DOI] [PubMed] [Google Scholar]
- 124. Stinnett, Gary R. , Lin Stephen, Korotcov Alexandru V., Korotcova Ludmila, Morton Paul D., Ramachandra Shruti D., Pham Angeline, et al. 2017. “Microstructural Alterations and Oligodendrocyte Dysmaturation in White Matter After Cardiopulmonary Bypass in a Juvenile Porcine Model.” Journal of the American Heart Association 6: e005997. 10.1161/JAHA.117.005997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Kojima, Katsuaki , Liu Chunyan, Ehrlich Shelley, Kline‐Fath Beth M., Jain Shipra, and Parikh Nehal A.. 2023. “Early Surgery in Very Preterm Infants Is Associated With Brain Abnormalities on Term MRI: A Propensity Score Analysis.” Journal of Perinatology 43: 877–883. 10.1038/s41372-023-01645-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Logsdon, Aric F. , Erickson Michelle A., Rhea Elizabeth M., Salameh Therese S., and Banks William A.. 2018. “Gut Reactions: How the Blood–Brain Barrier Connects the Microbiome and the Brain.” Experimental Biology and Medicine (Maywood) 243: 159–165. 10.1177/1535370217743766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Braniste, Viorica , Al‐Asmakh Maha, Kowal Czeslawa, Anuar Farhana, Abbaspour Afrouz, Tóth Miklós, Korecka Agata, et al. 2014. “The Gut Microbiota Influences Blood–Brain Barrier Permeability in Mice.” Science Translational Medicine 6: 263ra158. 10.1126/scitranslmed.3009759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Hoyles, Lesley , Pontifex Matthew G., Rodriguez‐Ramiro Ildefonso, Anis‐Alavi M. Areeb, Jelane Khadija S., Snelling Tom, Solito Egle, et al. 2021. “Regulation of Blood–Brain Barrier Integrity by Microbiome‐Associated Methylamines and Cognition by Trimethylamine N‐Oxide.” Microbiome 9: 235. 10.1186/s40168-021-01181-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Stachulski, Andrew V. , Knausenberger Tobias B.‐A., Shah Sita N., Hoyles Lesley, and McArthur Simon. 2023. “A Host‐Gut Microbial Amino Acid Co‐Metabolite, p‐Cresol Glucuronide, Promotes Blood–Brain Barrier Integrity In Vivo.” Tissue Barriers 11: 2073175. 10.1080/21688370.2022.2073175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Abdel‐Haq, Reem , Schlachetzki Johannes C. M., Glass Christopher K., and Mazmanian Sarkis K.. 2019. “Microbiome‐Microglia Connections via the Gut–Brain Axis.” The Journal of Experimental Medicine 216: 41–59. 10.1084/jem.20180794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Thion, Morgane Sonia , Low Donovan, Silvin Aymeric, Chen Jinmiao, Grisel Pauline, Schulte‐Schrepping Jonas, Blecher Ronnie, et al. 2018. “Microbiome Influences Prenatal and Adult Microglia in a Sex‐Specific Manner.” Cell 172: 500–516.e16. 10.1016/j.cell.2017.11.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Osadchiy, Vadim , Martin Clair R., and Mayer Emeran A.. 2019. “The Gut–Brain Axis and the Microbiome: Mechanisms and Clinical Implications.” Clinical Gastroenterology and Hepatology 17: 322–332. 10.1016/j.cgh.2018.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Erny, Daniel , Dokalis Nikolaos, Mezö Charlotte, Castoldi Angela, Mossad Omar, Staszewski Ori, Frosch Maximilian, et al. 2021. “Microbiota‐Derived Acetate Enables the Metabolic Fitness of the Brain Innate Immune System During Health and Disease.” Cell Metabolism 33: 2260–2276.e7. 10.1016/j.cmet.2021.10.010 [DOI] [PubMed] [Google Scholar]
- 134. Radford‐Smith, Daniel E. , Probert Fay, Burnet Philip W. J., and Anthony Daniel C.. 2022. “Modifying the Maternal Microbiota Alters the Gut–Brain Metabolome and Prevents Emotional Dysfunction in the Adult Offspring of Obese Dams.” Proceedings of the National Academy of Sciences 119: e2108581119. 10.1073/pnas.2108581119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Bai, Yunpeng , Huang Wendong, Jiang Xinyi, Xu Wang, Li Ying, Wang Yirong, Huang Sumei, et al. 2023. “Metabolomic Interplay Between Gut Microbiome and Plasma Metabolome in Cardiac Surgery‐Associated Acute Kidney Injury.” Rapid Communications in Mass Spectrometry 37: e9504. 10.1002/rcm.9504 [DOI] [PubMed] [Google Scholar]
- 136. Li, Ying , Jiang Xinyi, Chen Jingchun, Hu Yali, Bai Yunpeng, Xu Wang, He Linling, Wang Yirong, Chen Chunbo, and Chen Jimei. 2023. “Evaluation of the Contribution of Gut Microbiome Dysbiosis to Cardiac Surgery‐Associated Acute Kidney Injury by Comparative Metagenome Analysis.” Frontiers in Microbiology 14: 1119959. 10.3389/fmicb.2023.1119959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Maekawa, Masaki , Yoshitani Kenji, Yahagi Musashi, Asahara Takashi, Shishido Yoshiyuki, Fukushima Satsuki, Tadokoro Naoki, Fujita Tomoyuki, and Ohnishi Yoshihiko. 2020. “Association Between Postoperative Changes in the Gut Microbiota and Pseudopsia After Cardiac Surgery: Prospective Observational Study.” BMC Surgery 20: 247. 10.1186/s12893-020-00907-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Squiccimarro, Enrico , Labriola Cataldo, Malvindi Pietro Giorgio, Margari Vito, Guida Pietro, Visicchio Giuseppe, Kounakis Georgios, et al. 2019. “Prevalence and Clinical Impact of Systemic Inflammatory Reaction After Cardiac Surgery.” Journal of Cardiothoracic and Vascular Anesthesia 33: 1682–1690. 10.1053/j.jvca.2019.01.043 [DOI] [PubMed] [Google Scholar]
- 139. Deng, Fan , Zhang Liang‐Qing, Wu Han, Chen Yu, Yu Wen‐Qian, Han Rong‐Hui, Han Yuan, et al. 2022. “Propionate Alleviates Myocardial Ischemia‐Reperfusion Injury Aggravated by Angiotensin II Dependent on Caveolin‐1/ACE2 Axis Through GPR41.” International Journal of Biological Sciences 18: 858–872. 10.7150/ijbs.67724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Yu, Zhiyao , Han Jiapeng, Chen Huaqiang, Wang Yueyi, Zhou Liping, Wang Meng, Zhang Rong, et al. 2021. “Oral Supplementation With Butyrate Improves Myocardial Ischemia/Reperfusion Injury via a Gut–Brain Neural Circuit.” Frontiers in Cardiovascular Medicine 8: 718674. 10.3389/fcvm.2021.718674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Zheng, Dechong , Liu Zumei, Zhou You, Hou Ning, Yan Wei, Qin Yuan, Ye Qianfang, et al. 2020. “Urolithin B, a Gut Microbiota Metabolite, Protects Against Myocardial Ischemia/Reperfusion Injury via p62/Keap1/Nrf2 Signaling Pathway.” Pharmacological Research 153: 104655. 10.1016/j.phrs.2020.104655 [DOI] [PubMed] [Google Scholar]
- 142. Halter, Jeffrey , Steinberg Jay, Fink Gregory, Lutz Charles, Picone Anthony, Maybury Rubie, Fedors Nathan, et al. 2005. “Evidence of Systemic Cytokine Release in Patients Undergoing Cardiopulmonary Bypass.” The Journal of Extra‐Corporeal Technology 37: 272–277. 10.1051/ject/200537272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Merbecks, Moritz B. , Ziesenitz Victoria C., Rubner Tobias, Meier Noëmi, Klein Berthold, Rauch Helmut, Saur Patrick, et al. 2020. “Intermediate Monocytes Exhibit Higher Levels of TLR2, TLR4 and CD64 Early After Congenital Heart Surgery.” Cytokine 133: 155153. 10.1016/j.cyto.2020.155153 [DOI] [PubMed] [Google Scholar]
- 144. Manuel, Valdano , Miana Leonardo A., Guerreiro Gustavo Pampolha, Turquetto Aida, Santos Rômullo Medeiros, Fernandes Natália, Tenório Davi Freitas, Caneo Luiz Fernando, Jatene Fabio B., and Jatene Marcelo Biscegli. 2021. “Preoperative Neutrophil‐Lymphocyte Ratio Can Predict Outcomes for Patients Undergoing Tetralogy of Fallot Repair.” Brazilian Journal of Cardiovascular Surgery 36: 607–613. 10.21470/1678-9741-2020-0408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Gao, Peng , Liu Jinping, Wang Xu, Zhang Peiyao, Jin Yu, Bai Liting, and Li Yixuan. 2021. “The Association Between Neutrophillymphocyte Ratio and Poor Outcomes Following Infant Cardiac Surgery.” BMC Cardiovascular Disorders 21: 529. 10.1186/s12872-021-02345-3 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 146. Manuel, Valdano , Miana Leonardo A., Guerreiro Gustavo P., Tenório Davi F., Turquetto Aida, Penha Juliano G., Massoti Maria R., et al. 2020. “Prognostic Value of the Preoperative Neutrophil‐Lymphocyte Ratio in Patients Undergoing the Bidirectional Glenn Procedure.” Journal of Cardiac Surgery 35: 328–334. 10.1111/jocs.14381 [DOI] [PubMed] [Google Scholar]
- 147. Salomon, Jeffrey D. , Qiu Haowen, Feng Dan, Owens Jacob, Khailova Ludmila, Lujan Suzanne Osorio, Iguidbashian John, et al. 2022. “Piglet Cardiopulmonary Bypass Induces Intestinal Dysbiosis and Barrier Dysfunction Associated With Systemic Inflammation.” Disease Models & Mechanisms 16: 049742. 10.1242/dmm.049742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Typpo, Katri V. , Larmonier Claire B., Deschenes Jendar, Redford Daniel, Kiela Pawel R., and Ghishan Fayez K.. 2015. “Clinical Characteristics Associated With Postoperative Intestinal Epithelial Barrier Dysfunction in Children With Congenital Heart Disease.” Pediatric Critical Care Medicine 16: 37–44. 10.1097/PCC.0000000000000256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Malagon, I. , Onkenhout W., Klok G., van der Poel P. F. H., Bovill J. G., and Hazekamp M. G.. 2005. “Gut Permeability in Paediatric Cardiac Surgery.” British Journal of Anaesthesia 94: 181–185. 10.1093/bja/aei014 [DOI] [PubMed] [Google Scholar]
- 150. Pathan, Nazima , Burmester Margarita, Adamovic Tanja, Berk Maurice, Ng Keng Wooi, Betts Helen, Macrae Duncan, et al. 2011. “Intestinal Injury and Endotoxemia in Children Undergoing Surgery for Congenital Heart Disease.” American Journal of Respiratory and Critical Care Medicine 184: 1261–1269. 10.1164/rccm.201104-0715OC [DOI] [PubMed] [Google Scholar]
- 151. Patel, Jyoti K. , Loomes Kathleen M., Goldberg David J., Mercer‐Rosa Laura, Dodds Kathryn, and Rychik Jack. 2016. “Early Impact of Fontan Operation on Enteric Protein Loss.” The Annals of Thoracic Surgery 101: 1025–1030. 10.1016/j.athoracsur.2015.09.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Rodríguez de Santiago, Enrique , Téllez Luis, Garrido‐Lestache rodríguez‐Monte Elvira, Garrido‐Gómez Elena, Aguilera‐Castro Lara, Álvarez‐Fuente María, Del Cerro María Jesús, and Albillos Agustín. 2020. “Fontan Protein‐Losing Enteropathy is Associated With Advanced Liver Disease and a Proinflammatory Intestinal and Systemic State.” Liver International 40: 638–645. 10.1111/liv.14375 [DOI] [PubMed] [Google Scholar]
- 153. Xue, Ling , Ding Yinglong, Qin Qiong, Liu Linsheng, Ding Xiaoliang, Zhou Yi, Liu Kun, et al. 2023. “Assessment of the Impact of Intravenous Antibiotics Treatment on Gut Microbiota in Patients: Clinical Data from Pre‐and Post‐Cardiac Surgery.” Frontiers in Cellular and Infection Microbiology 12: 1043971. 10.3389/fcimb.2022.1043971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Reddy, Reshma K. , McVadon Deani H., Zyblewski Sinai C., Rajab Taufiek K., Diego Ellen, Southgate W. Michael, Fogg Kristi L., and Costello John M.. 2022. “Prematurity and Congenital Heart Disease: A Contemporary Review.” NeoReviews 23: e472–e485. 10.1542/neo.23-7-e472 [DOI] [PubMed] [Google Scholar]
- 155. Tirone, Chiara , Pezza Lucilla, Paladini Angela, Tana Milena, Aurilia Claudia, Lio Alessandra, D'Ippolito Silvia, et al. 2019. “Gut and Lung Microbiota in Preterm Infants: Immunological Modulation and Implication in Neonatal Outcomes.” Frontiers in Immunology 10: 2910. 10.3389/fimmu.2019.02910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Humberg, Alexander , Fortmann Ingmar, Siller Bastian, Kopp Matthias Volkmar, Herting Egbert, Göpel Wolfgang, and Härtel Christoph. 2020. “Preterm Birth and Sustained Inflammation: Consequences for the Neonate.” Seminars in Immunopathology 42: 451–468. 10.1007/s00281-020-00803-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Roseboom, T. J. , van der Meulen J. H., Osmond C., Barker D. J., Ravelli A. C., Schroeder‐Tanka J. M., van Montfrans G. A., Michels R. P., and Bleker O. P.. 2000. “Coronary Heart Disease After Prenatal Exposure to the Dutch Famine, 1944–45.” Heart (British Cardiac Society) 84: 595–598. 10.1136/heart.84.6.595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Healy, David B. , Ryan C. Anthony, Ross R. Paul, Stanton Catherine, and Dempsey Eugene M.. 2022. “Clinical Implications of Preterm Infant Gut Microbiome Development.” Nature Microbiology 7: 22–33. 10.1038/s41564-021-01025-4 [DOI] [PubMed] [Google Scholar]
- 159. Masi, Andrea C. , Embleton Nicholas D., Lamb Christopher A., Young Gregory, Granger Claire L., Najera Julia, Smith Daniel P., et al. 2021. “Human Milk Oligosaccharide DSLNT and Gut Microbiome in Preterm Infants Predicts Necrotising Enterocolitis.” Gut 70: 2273–2282. 10.1136/gutjnl-2020-322771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Bering, Stine Brandt . 2018. “Human Milk Oligosaccharides to Prevent Gut Dysfunction and Necrotizing Enterocolitis in Preterm Neonates.” Nutrients 10: 1461. 10.3390/nu10101461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Alcon‐Giner, Cristina , Dalby Matthew J., Caim Shabhonam, Ketskemety Jennifer, Shaw Alex, Sim Kathleen, Lawson Melissa A. E., et al. 2020. “Microbiota Supplementation With Bifidobacterium and Lactobacillus Modifies the Preterm Infant Gut Microbiota and Metabolome: An Observational Study.” Cell Reports Medicine 1: 100077. 10.1016/j.xcrm.2020.100077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Asbury, Michelle R. , Shama Sara, Sa Jong Yup, Bando Nicole, Butcher James, Comelli Elena M., Copeland Julia K., et al. 2022. “Human Milk Nutrient Fortifiers Alter the Developing Gastrointestinal Microbiota of Very‐Low‐Birth‐Weight Infants.” Cell Host & Microbe 30: 1328–1339.e5. 10.1016/j.chom.2022.07.011 [DOI] [PubMed] [Google Scholar]
- 163. Samara, Jumana , Moossavi Shirin, Alshaikh Belal, Ortega Van A., Pettersen Veronika Kuchařová, Ferdous Tahsin, Hoops Suzie L., et al. 2022. “Supplementation With a Probiotic Mixture Accelerates Gut Microbiome Maturation and Reduces Intestinal Inflammation in Extremely Preterm Infants.” Cell Host & Microbe 30: 696–711.e65. 10.1016/j.chom.2022.04.005 [DOI] [PubMed] [Google Scholar]
- 164. Schulz‐Weidner, Nelly , Weigel Markus, Turujlija Filip, Komma Kassandra, Mengel Jan Philipp, Schlenz Maximiliane Amelie, Bulski Julia Camilla, Krämer Norbert, and Hain Torsten. 2021. “Microbiome Analysis of Carious Lesions in Pre‐School Children With Early Childhood Caries and Congenital Heart Disease.” Microorganisms 9: 1904. 10.3390/microorganisms9091904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Slocum, C. , Kramer C., and Genco C. A.. 2016. “Immune Dysregulation Mediated by the Oral Microbiome: Potential Link to Chronic Inflammation and Atherosclerosis.” Journal of Internal Medicine 280: 114–128. 10.1111/joim.12476 [DOI] [PubMed] [Google Scholar]
- 166. Lim, Ai Ing , McFadden Taryn, Link Verena M., Han Seong‐Ji, Karlsson Rose‐Marie, Stacy Apollo, Farley Taylor K., et al. 2021. “Prenatal Maternal Infection Promotes Tissue‐Specific Immunity and Inflammation in Offspring.” Science 373: eabf3002. 10.1126/science.abf3002 [DOI] [PubMed] [Google Scholar]
- 167. Vuong, Helen E. , Pronovost Geoffrey N., Williams Drake W., Coley Elena J. L., Siegler Emily L., Qiu Austin, Kazantsev Maria, et al. 2020. “The Maternal Microbiome Modulates Fetal Neurodevelopment in Mice.” Nature 586: 281–286. 10.1038/s41586-020-2745-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Tian, Min , Li Qihui, Zheng Tenghui, Yang Siwang, Chen Fang, Guan Wutai, and Zhang Shihai. 2023. “Maternal Microbe‐Specific Modulation of the Offspring Microbiome and Development During Pregnancy and Lactation.” Gut Microbes 15: 2206505. 10.1080/19490976.2023.2206505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Beckers, Kalie F. , and Sones Jenny L.. 2020. “Maternal Microbiome and the Hypertensive Disorder of Pregnancy, Preeclampsia.” American Journal of Physiology—Heart and Circulatory Physiology 318: H1–H10. 10.1152/ajpheart.00469.2019 [DOI] [PubMed] [Google Scholar]
- 170. Hoelscher, Sarah C. , Doppler Stefanie A., Dreßen Martina, Lahm Harald, Lange Rüdiger, and Krane Markus. 2017. “micro‐RNAs: Pleiotropic Players in Congenital Heart Disease and Regeneration.” Journal of Thoracic Disease 9: S64–S81. 10.21037/jtd.2017.03.149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Chakaroun, Rima Mohsen , Olsson Lisa M., and Bäckhed Fredrik. 2023. “The Potential of Tailoring the Gut Microbiome to Prevent and Treat Cardiometabolic Disease.” Nature Reviews Cardiology 20: 217–235. 10.1038/s41569-022-00771-0 [DOI] [PubMed] [Google Scholar]
- 172. Ahmad, Adilah F. , Dwivedi Girish, O'Gara Fergal, Caparros‐Martin Jose, and Ward Natalie C.. 2019. “The Gut Microbiome and Cardiovascular Disease: Current Knowledge and Clinical Potential.” American Journal of Physiology—Heart and Circulatory Physiology 317: H923–H938. 10.1152/ajpheart.00376.2019 [DOI] [PubMed] [Google Scholar]
- 173. Liu, Zaoqu , Li Na, Dang Qin, Liu Long, Wang Libo, Li Huanyun, and Han Xinwei. 2023. “Exploring the Roles of Intestinal Flora in Enhanced Recovery After Surgery.” iScience 26: 105959. 10.1016/j.isci.2023.105959 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This manuscript does not generate any code or data. Supplementary materials (graphical abstracts, slides, videos, Chinese translated version, and update materials) may be found in the online DOI or iMeta Science http://www.imeta.science/.


