Abstract
Genetic studies of nematodes have been dominated by Caenorhabditis elegans as a model species. A lack of genomic resources has limited the expansion of genetic research to other groups of nematodes. Here, we report a draft genome assembly of a mermithid nematode, Mermis nigrescens. Mermithidae are insect parasitic nematodes with hosts including a wide range of terrestrial arthropods. We sequenced, assembled, and annotated the whole genome of M. nigrescens using nanopore long reads and 10X Chromium link reads. The assembly is 524 Mb in size consisting of 867 scaffolds. The N50 value is 2.42 Mb, and half of the assembly is in the 30 longest scaffolds. The assembly BUSCO score from the eukaryotic database (eukaryota_odb10) indicates that the genome is 86.7% complete and 5.1% partial. The genome has a high level of heterozygosity (6.6%) with a repeat content of 83.98%. mRNA-seq reads from different sized nematodes (≤2 cm, 3.5–7 cm, and >7 cm body length) representing different developmental stages were also generated and used for the genome annotation. Using ab initio and evidence-based gene model predictions, 12,313 protein-coding genes and 24,186 mRNAs were annotated. These genomic resources will help researchers investigate the various aspects of the biology and host–parasite interactions of mermithid nematodes.
Keywords: Mermis nigrescens, nematode, hybrid genome assembly, repeatome, genome annotation
Introduction
Nematodes are a highly diverse group with over 1 million estimated species (Scheffers et al. 2012; Smythe et al. 2019). They are ubiquitous, occurring in habitats ranging from deep oceans to mountain peaks (Schratzberger et al. 2019). Most animals and plants species are host to at least one species of parasitic nematode (Blaxter and Koutsovoulos 2015). Despite representing an important component of all natural ecosystems (Ferris 2010; Cardoso et al. 2016), most nematode species remain undescribed (Dobson et al. 2008).
Nematodes fall into two broad classes based on both molecular and morphological systematics: Chromadorea and Enoplea (Blaxter et al. 1998; Liu et al. 2013). They have a conserved body plan with similar structural organization and cellular morphologies (Basyoni and Rizk 2016). However, comparative analyses have highlighted the molecular and physiological diversity within the phylum (Mitreva et al. 2005; International Helminth Genomes Consortium 1975). Caenorhabditis elegans is a Chromadorean that has been at the forefront of genetic research as a model species. In contrast, there exists very little genomic information from the members of class Enoplea. Most studies on Enoplea focus on Trichinella spiralis (Enoplea: Trichocephalida) because of its importance as a mammalian parasite (Mitreva et al. 2011). Mermithida represents an order within Enoplea where most members are endoparasites of arthropods. However, among Mermithida, only the genome of Romanomermis culicivorax (Dorylaimia: Mermithidae) is publicly available (Schiffer et al. 2013).
Mermithids are obligate endoparasitic nematodes. They represent a large portion of the understudied nematode class Enoplea. There are over 100 described mermithid species; however, their biology and genetics are scarcely studied (International Helminth Genomes Consortium 1975; Presswell et al. 2015). Mermithids are parasitic in different kinds of invertebrates like insects, spiders, leeches, and crustaceans. However, insects are by far the most common hosts, ∼15 different orders of insects are host to different mermithids (Nickle 1972). These nematodes lack a functional gut and absorb nutrients through their outer cuticle from the insect's hemolymph and store them in their trophosome. The free-living form will rely on those stored reserves, so they depend heavily on their host for their nutritional requirements (Petersen 1985). They are relatively large nematodes with a wide distribution spanning the Americas, Eurasia, Australia, and New Zealand. Mermis subnigrescens was reported from New Zealand as early as 1989 by Thomas (cited by Presswell et al. 2015). Later M. subnigrescens was synonymized with M. nigrescens (Nickle 1972).
Mermithids are reported to be facultatively parthenogenetic (Christie 1937). Environmental signaling is thought to play a primary role in their sex determination (Christie 1929). The first individual establishing in a host usually develops as female, with the sex of individuals arriving later determined in response to others in the pool. However, the genetic mechanisms of sex determination for mermithid individuals in a population are unknown.
Several mermithids manipulate their host's behavior for their own benefit (Herbison et al. 2019). Adult mermithids are free-living; after completion of their development within an arthropod host, they must emerge from the host to mate and lay eggs. As they are prone to desiccation, they must emerge in water. Since many mermithids parasitize terrestrial arthropod hosts, the need to emerge from the host in water has selected in several mermithids the ability of inducing water-seeking behavior in their host. For example, the mermithid nematode Strelkovimermis spiculatus modifies the behavior of its adult female mosquito host to make it seek water instead of a blood meal, providing a dispersal means for the nematodes and ensuring their return to a suitable habitat for reproduction (Allahverdipour et al. 2019). Similarly, Mermis nigrescens induces water-seeking behavior in its terrestrial arthropod host, enabling the nematodes to emerge into a favorable environment to continue their life cycle (Presswell et al. 2015; Herbison et al. 2019).
Here, we present a high-quality genome assembly of M. nigrescens. The life cycle, morphological, and molecular characteristics of M. nigrescens have been well studied (Baker and Capinera 1997; Presswell et al. 2015). However, the lack of whole-genome information greatly limits the genetic investigations of its biology, behavior, and adaptations. This genome will also fill the knowledge and resource gap for Enoplean genomes. It will further facilitate the application of a range of molecular tools and approaches to study the genetic underpinnings of successful host exploitation by mermithids.
Materials and methods
Sampling and nucleotide extractions
Nematodes (M. nigrescens) were dissected out of European earwigs (Forficula auricularia) collected from the Dunedin Botanic Garden (latitude: −45° 51′ 27.59″ S; longitude: 170° 31′ 15.56″ E) and reared in a temperature-controlled room (temperature: cycling from 15 to 12°C, day/night; photoperiod of L:D 16:8) in the Department of Zoology, University of Otago, Dunedin. Nematodes thus obtained were snap-frozen in liquid nitrogen and stored at −80°C until further use. Individual nematodes were used for each nucleotide extraction. Individuals were not sexed prior to sequencing due to the difficulty in achieving this in a manner timely enough to maintain high-quality RNA and DNA samples. DNA extracted using DNeasy Blood & Tissue Kit (Qiagen, USA) was used for nanopore sequencing. DNA extracted using Nanobind Tissue Big DNA kit (Circulomics, USA) was used for 10x Chromium linked-read library preparation. RNase treatment to remove RNA was performed using 4μl of RNase A (10 mg/ml) per 200μl of template following DNA extraction. DNA samples were quantified, and quality assessed using a Qubit 2.0 Fluorometer (Life Technologies, USA) and Nanodrop. High-quality DNA samples were stored at −20°C and were used within a week of extraction.
Total RNA from different sized nematodes (≤2 cm, 3.5–7 cm, and >7 cm body length) representing different developmental stages was extracted using Direct-zol RNA MicroPrep kit (Zymo Research) following the manufacturer's instruction and stored at −80°C until further use. RNA from 5 individuals from each size category (small, medium, and large) were individually extracted and later pooled at equimolar concentration to make a single sequencing library for each category. Samples were quantified and quality checked on a Qubit 2.0 Fluorometer (Life Technologies, USA) and Nanodrop. RNA integrity for the pooled samples was assessed using a Fragment Analyzer (Advanced Analytical Technologies Inc., USA) at the Otago Genomics Facility (OGF), University of Otago, Dunedin, New Zealand. The RNA quality number values ranged from 8.3 to 9.5, indicating high-quality samples.
Library preparation and sequencing
A long-read sequencing library for Oxford minion was prepared with 416.5 ng of genomic DNA using a ligation sequencing kit (SQK-LSK109) (Oxford Nanopore Technologies, Oxford, UK) following the manufacturer's instructions. Lambda phage (DNA CS) was used as a positive control. The prepared library was sequenced with R9 chemistry MinION flow cell (FLO-MIN106) (Oxford Nanopore Technologies, UK).
A linked-read library was prepared at the Genetic Analysis Services (GAS), University of Otago (Dunedin, New Zealand). DNA was size selected for fragments over 40 kbp using a Blue Pippin (Sage Science, USA). A 10x Chromium linked-read (10x Genomics, USA) library was prepared following the manufacturer's instructions. The library was sequenced on the Illumina Nova-seq platform to generate paired-end reads (2 × 151 bp) at the Garvan Institute, Australia.
TruSeq stranded mRNA libraries were prepared and sequenced at OGF as 2 × 100 bp paired-end reads across 2 Rapid V2 flowcell lanes of HiSeq 2500.
Genome size estimation
Genome size was estimated with flow cytometry analysis at Flowjoanna (Palmerston North, New Zealand). Two whole worm samples were prepared following a protocol described in our earlier study (Bhattarai et al. 2022). Rooster red blood cells from the domestic chicken (Gallus gallus) were used a reference. Samples were then processed on a FACSCalibur (BD Biosciences, USA) and the data were analyzed using Flowjo (BD Biosciences, USA).
Bioinformatic pipeline
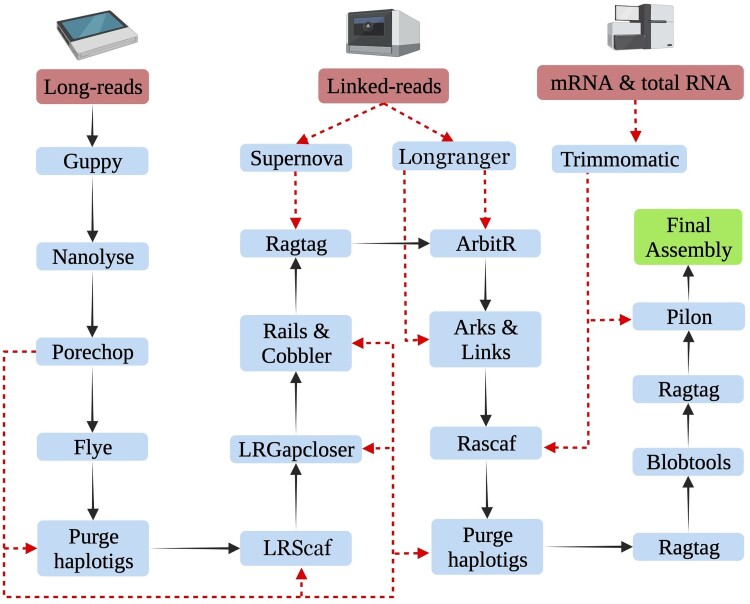
We used various software and packages to assemble and refine the genome (Fig. 1). The pipeline and the scripts used in this project are available on GitHub (https://github.com/upendrabhattarai/Nematode_Genome_Project). The bioinformatics software and packages were run in New Zealand eScience Infrastructure (NeSI).
Fig. 1.
Schematic representation of the assembly pipeline for the Mermis nigrescens genome. The black arrows represent the workflow, and the red dotted lines represent the input data in the pipeline (created with BioRender.com).
Genome and transcriptome assembly
All the Nanopore reads were basecalled using Guppy (v.5.0.7) post-sequencing. Quality control of the long-read data involved Nanolyse (v1.2.0) (De Coster et al. 2018) and Porechop (v.0.2.4) (Wick et al. 2017). Reads were assembled using Flye (v.2.9) assembler (Kolmogorov et al. 2019) with default parameters. The paired-end reads from the chromium library were assembled using supernova (Weisenfeld et al. 2017). BUSCO (v.4.1.4) (Simão et al. 2015) with eukaryota_odb10 and nematoda_odb10 databases and Quast (v.5.0.2) (Gurevich et al. 2013) were used to evaluate the assembly statistics. The assembly from long reads outperformed the linked reads for the BUSCO completeness and assembly contiguity. Therefore, long-read assembly was used as a primary assembly, and the linked-read assembly was used for scaffolding later in the pipeline (Fig. 1). Multiple scaffolding and gap-closing steps were performed using Lrscaf (v.1.1.11) (Qin et al. 2019), Rails (v.1.5.1) and Cobbler (v.0.6.1) (Warren 2016), Ragtag (v.2.1.0) (Alonge et al. 2019), ArbitR (v.0.2) (Hiltunen et al. 2021), Arks (v.1.0.4) (Coombe et al. 2018), Links (v.1.8.7) (Warren et al. 2015), and Lrgapcloser (Xu et al. 2019). Linked reads were mapped to the assembly with Longranger (v.2.2.2) (Marks et al. 2019) prior to scaffolding. mRNA-seq reads obtained for genome annotation purposes, and total RNA-seq reads sequenced for another project (manuscript under preparation) were also used for scaffolding the assembly with Rascaf (Song et al. 2016). Purgehaplotigs (Roach et al. 2018) removed haplotigs from the assembly. Removed haplotig sequences were used to further scaffold the assembly with Ragtag (v.2.1.0). Blobtools2 (v.2.6.4) (Laetsch and Blaxter 2017) was used to remove and discard bacterial reads and remove low coverage (<5× coverage) and short (<1,000 bp) scaffolds. Low coverage and short scaffolds were then used to scaffold the filtered assembly with Ragtag (v.2.1.0) (Alonge et al. 2019). Finally, Pilon (v.1.24) was used to polish the assembly using mRNA-seq data.
The adapters and low-quality reads from mRNA-seq data were filtered using Trimmomatic (v.0.38) (Bolger et al. 2014) with options HEADCROP:15 LEADING:3 TRAILING:3 SLIDINGWINDOW:4:20 MINLEN:35. Clean data were de novo assembled with Trinity (v.2.13.2) (Grabherr et al. 2011) using all the default parameters.
Repetitive content masking
A custom repeat library was generated using de novo and homology-based identifiers, including LTRharvest (Ellinghaus et al. 2008), LTRdigest (Steinbiss et al. 2009), RepeatModeler (Flynn et al. 2020), TransposonPSI (Brian Haas 2010), SINEBase (Vassetzky and Kramerov 2013), and MITE-Tracker (Crescente et al. 2018). Individual libraries from each software were concatenated, and sequences with more than 80% similarity were merged to remove redundancy using usearch (v.11.0.667) (Edgar 2010). The library was then classified using RepeatClassifier. Sequences with unknown categories in the library were mapped against the reviewed nematode database from UniProtKB/Swiss-Prot database (e < 1e−01); if not annotated as repeat sequences, they were removed from the library. The final repeat library was used in RepeatMasker (v.4.1.0) (Chen 2004) to mask the repeats and generate a report. The repeat library was used as an input to the MAKER2 pipeline (Holt and Yandell 2011). Tandem repeats were also identified using the TRF (v. 4.09.1) pipeline (Benson 1999).
Genome annotation
The assembly was annotated using evidence-based and ab initio gene model predictions through MAKER2 (v.2.31.9) (Holt and Yandell 2011) and Braker (v.2.16) (Hoff et al. 2019) pipelines. The first round of MAKER2 was run with 180,630 mRNA transcripts de novo assembled with the Trinity (v.2.13.2) pipeline (Grabherr et al. 2011) along with 511,117 mRNA and 1,148,233 protein sequences from all the Dorylaimia species available in NCBI and WormBase (https://wormbase.org) databases. mRNA-seq reads and output from GenMark-ES were used to train Augustus with the Braker pipeline. SNAP was trained after each round of MAKER2. Trained SNAP and Augustus were used for 2 more iterations of the MAKER2 pipeline.
We used InterProScan (v.5.51–85.0) (Jones et al. 2014) for the functional annotation on the predicted protein sequences from MAKER and retrieved InterPro ID, PFAM domains, and Gene Ontology (GO) terms. Furthermore, the Uniprot database with BLAST was used to assign gene descriptors to each transcript based on the best hit.
Results and discussion
Genome size estimates
The flow cytometry estimate of the nematode genome size is 814.185 ± 24.2 Mb (mean ± SD). However, the nematode samples yielded very low nuclei counts, i.e. as low as <200 total nuclei for 1 of the samples. Given that this is a whole animal digest, and the very low nuclei counts in the samples, the calculated pg/nucleus may not be accurate. To get better genome size estimates by flow cytometry in this nematode, we recommend increasing the sample size and replicated measurements from the nuclei suspension in future experiments.
Genome and transcriptome assembly
A total of 6.9 Gb of sequencing data was generated from a Nanopore minion flowcell run consisting of 530,888 reads with an N50 length of 17,849 bp (Supplementary Table 1). Flye (v.2.7.1) produced the primary long-read assembly with 16,414 contigs with N50 of 94,974 bp and a total length of ∼714 Mb. Quast reported 88.45% complete BUSCO from the Eukaryote database for this assembly (Table 1).
Table 1.
Statistics for the genome assembly of Mermis nigrescens.
| Assembly length | No. of scaffolds | N50 | L50 | Ns per 100 kbp | BUSCO% Quast (eukaryota_odb10) | ||
|---|---|---|---|---|---|---|---|
| Complete | Partial | ||||||
| Flye | 714,265,163 | 16,414 | 94,974 | 2,079 | 0.45 | 88.45 | 4.29 |
| Supernova | 582,450,576 | 147,205 | 9,664 | 12,318 | 732.59 | 20.13 | 21.45 |
| Final assembly | 524,220,005 | 867 | 2,429,002 | 30 | 240.96 | 86.70 | 5.10 |
The 10x Chromium library yielded 450.3 million paired-end reads. The Supernova assembler produced an assembly of 582.5 Mb with 147,205 scaffolds and N50 of 9,664 bp. Quast reported only 20.13% of complete BUSCO genes (Table 1). Comparing the contiguity and completeness of the long-read vs linked-read assemblies, we used long-read assembly as the primary assembly and the linked-read assembly was used later for scaffolding (Fig. 1).
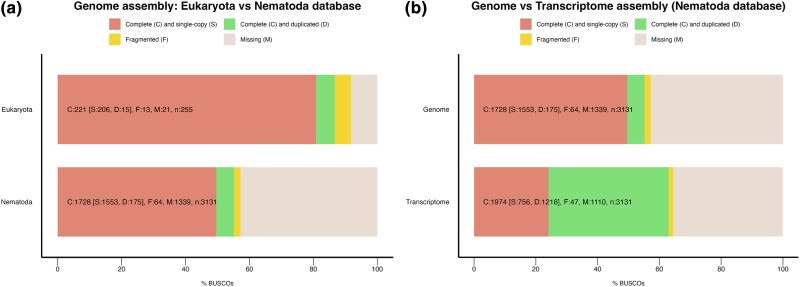
The assembly statistics after each round of processing are presented in Supplementary Table 2. The final assembly has a size of 524.2 Mb with 867 scaffolds. The assembly has N50 of 2,429,002 bp, and 50% of the assembly is in the 30 longest scaffolds. Gaps in the assembly account for 240.97 bp per 100 kbp length. The final assembly has a complete BUSCO score of 86.7% and a partial BUSCO score of 5.1% to the eukaryotic database (Fig. 2a).
Fig. 2.
The BUSCO (v.4.1.4) analysis of the genome and transcriptome assembly of Mermis nigrescens. a) Comparison between the Eukaryote (eukaryota_odb10) and the Nematode (nematoda_odb10) database for BUSCO scores for the assembled genome. b) Comparison between genome and transcriptome assemblies for BUSCO scores with the nematoda_odb10 database.
The mRNA-seq libraries generated 288.9 million paired-end reads. They were de novo assembled using Trinity (v.2.13.2) (Grabherr et al. 2011) producing 180,630 transcripts with N50 of 1,362 bp including 103,144 trinity genes. The BUSCO score from the nematoda_odb10 database was 63% for the transcriptome assembly and 55.2% complete for the genome assembly (Fig. 2b). This suggests that the nematode BUSCO database (nematoda_odb10) does not provide a good representation of the M. nigrescens genome, further highlighting the high genetic diversity among different nematode clades.
R. culicivorax is the only other mermithid nematode with a publicly available whole-genome assembly (Schiffer et al. 2013) to date. Its genome assembly has a 322 Mb size with N50 of 17,632 bp. It has 62,537 scaffolds and 37.3% complete BUSCO genes (nematoda_odb10, n = 3131). Between these 2 assemblies, the M. nigrescens’ genome assembly is more contiguous and complete and can be used as a better representative of mermithid nematodes.
Genes and repeats annotation
A total of 12,313 protein-coding genes and 24,186 mRNAs including their isoforms were identified in the genome. The mean gene length is 12,224 bp, and the total gene length is 150.5 Mb. The longest gene annotated is 206,129 bp (Table 2). Functional annotation resulted in 7,496 genes annotated to the InterPro and Pfam databases and 4,623 annotated to GO terms (Supplementary Table 3). Quality of annotation was measured using Annotation Edit Distance (AED) score, and 96% of the annotated genes have an AED score of 0.5 or less, ensuring highly confident gene prediction (Supplementary Fig. 1).
Table 2.
Genome annotation summary for Mermis nigrescens.
| Total sequence length (bp) | 524,220,005 |
| Number of genes | 12,313 |
| Number of mRNAs (isoforms counted) | 24,185 |
| Number of exons | 152,563 |
| Number of introns | 128,378 |
| Number of CDS | 24,185 |
| Total gene length (bp) | 150,457,507 |
| Total mRNA length (bp) | 298,270,450 |
| Total exon length (bp) | 30,890,614 |
| Total intron length (bp) | 267,636,592 |
| Total CDS length (bp) | 23,759,814 |
| Longest gene (bp) | 206,129 |
| Longest mRNA (bp) | 206,129 |
| Longest exon (bp) | 6,519 |
| Longest intron (bp) | 129,245 |
| Longest CDS (bp) | 19,164 |
| Mean gene length (bp) | 12,219 |
| Mean mRNA length (bp) | 12,333 |
| Mean exon length (bp) | 202 |
| Mean intron length (bp) | 2,085 |
| Mean CDS length (bp) | 982 |
The repeat contents in the genome amount to 440.21 Mb comprising 83.98% of the whole assembly. It includes 17.86% of retroelements, 21.97% of DNA transposons, 1.09% rolling circles, 37.18% unclassified repeatomes, and 5.23% of tandem repeats (Supplementary Table 4). Compared to other nematode species, this is a very high proportion of repeats in the genome. For example, the repeat content in R. culicivorax genome is 47% (Schiffer et al. 2013), in T. spiralis genome it is 19.8% (Mitreva et al. 2011) and in C. elegans genome it is 17% (C. elegans Sequencing Consortium 1998). At present, we do not know the significance of such a highly repetitive genome in M. nigrescens. However, further investigations might shed light on its biological and evolutionary significance.
Conclusions
This study presented a high-quality genome assembly, annotation, and repeat analysis of M. nigrescens. The genome was assembled using the long and linked-read sequencing data. Transcriptomic data from different developmental stages of the nematode were also generated. The M. nigrescens genome showed very high level of repeat content compared to other nematode genomes. These new resources and information will contribute to the better understanding of the genomic architecture and biology of mermithid nematodes and their adaptation to a broad range of insect host.
Supplementary Material
Acknowledgments
We would like to than the New Zealand eScience Infrastructure (NeSI) support team for their help with running various software on their HPC platforms.
Contributor Information
Upendra R Bhattarai, Department of Anatomy, University of Otago, Dunedin 9016, New Zealand; Department of Organismic & Evolutionary Biology, Harvard University, Cambridge, MA 02138, USA.
Robert Poulin, Department of Zoology, University of Otago, Dunedin 9016, New Zealand.
Neil J Gemmell, Department of Anatomy, University of Otago, Dunedin 9016, New Zealand.
Eddy Dowle, Department of Anatomy, University of Otago, Dunedin 9016, New Zealand.
Data availability
The genome sequencing reads, RNA-seq reads, and the genome assembly for M. nigrescens were submitted to NCBI under the BioProject accession number: PRJNA802644. The genome assembly and the annotation can also be accessed from FigShare: https://doi.org/10.6084/m9.figshare.21761501.v1. The pipeline and the scripts used in this project are available on GitHub: https://github.com/upendrabhattarai/Nematode_Genome_Project.
Supplemental material available at G3 online.
Funding
This study was funded by the Royal Society Te Apārangi Marsden Fund grant (16-UOO-152).
Literature cited
- Allahverdipour HH, Talaei-Hassanloui R, Karimi J, Wang Y, Rochlin I, Gaugler R. 2019. Behavior manipulation of mosquitoes by a mermithid nematode. J Invertebr Pathol. 168:107273. doi: 10.1016/j.jip.2019.107273. [DOI] [PubMed] [Google Scholar]
- Alonge M, Soyk S, Ramakrishnan S, Wang X, Goodwin S, Sedlazeck FJ, Lippman ZB, Schatz MC. 2019. RaGOO: fast and accurate reference-guided scaffolding of draft genomes. Genome Biol. 20(1):224. doi: 10.1186/s13059-019-1829-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker GL, Capinera JL. 1997. Nematodes and nematomorphs as control agents of grasshoppers and locusts. Mem Ent Soc Can. 129(S171):157–211. doi: 10.4039/entm129171157-1. [DOI] [Google Scholar]
- Basyoni MMA, Rizk EMA. 2016. Nematodes ultrastructure: complex systems and processes. J Parasit Dis. 40(4):1130–1140. doi: 10.1007/s12639-015-0707-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27(2):573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattarai UR, Katuwal M, Poulin R, Gemmell NJ, Dowle E. 2022. Genome assembly and annotation of the European earwig Forficula auricularia (subspecies B). G3 (Bethesda). 12(10):jkac199. doi: 10.1093/g3journal/jkac199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaxter ML, De Ley P, Garey JR, Liu LX, Scheldeman P, Vierstraete A, Vanfleteren JR, Mackey LY, Dorris M, Frisse LM, et al. 1998. A molecular evolutionary framework for the phylum Nematoda. Nature. 392(6671):71–75. doi: 10.1038/32160. [DOI] [PubMed] [Google Scholar]
- Blaxter M, Koutsovoulos G. 2015. The evolution of parasitism in Nematoda. Parasitology. 142 Suppl 1(Suppl 1):S26–S39. doi: 10.1017/S0031182014000791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brian Haas . 2010. TransposonPSI: An Application of PSI-Blast to Mine (Retro-) Transposon ORF Homologies. https://transposonpsi.sourceforge.net/.
- Cardoso MSO, Pedrosa EMR, Ferris H, Rolim MM, Oliveira LSC. 2016. Nematode fauna of tropical rainforest in Brazil: a descriptive and seasonal approach. J Nematol. 48(2):116–125. doi: 10.21307/jofnem-2017-017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- C. elegans Sequencing Consortium. 1998. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 282(5396):2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- Chen N. 2004. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr Protoc Bioinformatics. Chapter 4:Unit 4.10. doi: 10.1002/0471250953.bi0410s05. [DOI] [PubMed] [Google Scholar]
- Christie JR. 1929. Some observations on sex in the Mermithidae. J Exp Zool. 53(1):59–76. doi: 10.1002/jez.1400530106. [DOI] [Google Scholar]
- Christie JR. 1937. Mermis subnigrescens, a nematode parasite of grasshoppers. J Agric Res. 55:353–364. https://books.google.com/books?hl=en&lr=&id=bePUAAAAMAAJ&oi=fnd&pg=PA353&dq=Christie+JR.+1937.+Mermis+subnigrescens,+a+nematode+parasite+of+grasshoppers.+Journal+of+Agricultural+Research+55:+353-364.&ots=t0ldSKeULd&sig=6wDtYCOEP56fx5GpOojml9EfRK8#v=onepage&q&f=false. [Google Scholar]
- Coombe L, Zhang J, Vandervalk BP, Chu J, Jackman SD, Birol I, Warren RL. 2018. ARKS: chromosome-scale scaffolding of human genome drafts with linked read kmers. BMC Bioinformatics. 19(1):234. doi: 10.1186/s12859-018-2243-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crescente JM, Zavallo D, Helguera M, Vanzetti LS. 2018. MITE Tracker: an accurate approach to identify miniature inverted-repeat transposable elements in large genomes. BMC Bioinformatics. 19(1):348. doi: 10.1186/s12859-018-2376-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Coster W, D’Hert S, Schultz DT, Cruts M, Van Broeckhoven C. 2018. NanoPack: visualizing and processing long-read sequencing data. Bioinformatics. 34(15):2666–2669. doi: 10.1093/bioinformatics/bty149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson A, Lafferty KD, Kuris AM, Hechinger RF, Jetz W. 2008. Colloquium paper: homage to Linnaeus: how many parasites? How many hosts? Proc Natl Acad Sci U S A. 105 Suppl 1(Suppl 1):11482–11489. doi: 10.1073/pnas.0803232105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 26(19):2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Ellinghaus D, Kurtz S, Willhoeft U. 2008. LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinformatics. 9:18. doi: 10.1186/1471-2105-9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris H. 2010. Contribution of nematodes to the structure and function of the soil food web. J Nematol. 42(1):63–67. https://pubmed.ncbi.nlm.nih.gov/22736838/. [PMC free article] [PubMed] [Google Scholar]
- Flynn JM, Hubley R, Goubert C, Rosen J, Clark AG, Feschotte C, Smit AF. 2020. RepeatModeler2 for automated genomic discovery of transposable element families. Proc Natl Acad Sci U S A. 117(17):9451–9457. doi: 10.1073/pnas.1921046117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 29(7):644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich A, Saveliev V, Vyahhi N, Tesler G. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 29(8):1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbison REH, Evans S, Doherty J-F, Poulin R. 2019. Let's go swimming: mermithid-infected earwigs exhibit positive hydrotaxis. Parasitology. 146(13):1631–1635. doi: 10.1017/S0031182019001045. [DOI] [PubMed] [Google Scholar]
- Hiltunen M, Ryberg M, Johannesson H. 2021. ARBitR: an overlap-aware genome assembly scaffolder for linked reads. Bioinformatics. 37(15):2203–2205. doi: 10.1093/bioinformatics/btaa975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoff KJ, Lomsadze A, Borodovsky M, Stanke M. 2019. Whole-genome annotation with BRAKER. Methods Mol Biol. 1962:65–95. doi: 10.1007/978-1-4939-9173-0_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt C, Yandell M. 2011. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics. 12:491. doi: 10.1186/1471-2105-12-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Helminth Genomes Consortium . 2019. Comparative genomics of the major parasitic worms. Nat Genet. 51(1):163–174. doi: 10.1038/s41588-018-0262-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P, Binns D, Chang H-Y, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, et al. 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics. 30(9):1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolmogorov M, Yuan J, Lin Y, Pevzner PA. 2019. Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol. 37(5):540–546. doi: 10.1038/s41587-019-0072-8. [DOI] [PubMed] [Google Scholar]
- Laetsch DR, Blaxter ML. 2017. BlobTools: interrogation of genome assemblies. F1000Res. 6:1287. doi: 10.12688/f1000research.12232.1. [DOI] [Google Scholar]
- Liu G-H, Shao R, Li J-Y, Zhou D-H, Li H, Zhu X-Q. 2013. The complete mitochondrial genomes of three parasitic nematodes of birds: a unique gene order and insights into nematode phylogeny. BMC Genomics. 14:414. doi: 10.1186/1471-2164-14-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks P, Garcia S, Barrio AM, Belhocine K, Bernate J, Bharadwaj R, Bjornson K, Catalanotti C, Delaney J, Fehr A, et al. 2019. Resolving the full spectrum of human genome variation using Linked-Reads. Genome Res. 29(4):635–645. doi: 10.1101/gr.234443.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitreva M, Blaxter ML, Bird DM, McCarter JP. 2005. Comparative genomics of nematodes. Trends Genet. 21(10):573–581. doi: 10.1016/j.tig.2005.08.003. [DOI] [PubMed] [Google Scholar]
- Mitreva M, Jasmer DP, Zarlenga DS, Wang Z, Abubucker S, Martin J, Taylor CM, Yin Y, Fulton L, Minx P, et al. 2011. The draft genome of the parasitic nematode Trichinella spiralis. Nat Genet. 43(3):228–235. doi: 10.1038/ng.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickle WR. 1972. A contribution to our knowledge of the Mermithidae (Nematoda). J Nematol. 4(2):113–146. https://pubmed.ncbi.nlm.nih.gov/19319257. [PMC free article] [PubMed] [Google Scholar]
- Petersen JJ. 1985. Nematodes as biological control agents: part I. Mermithidae. Adv Parasitol. 24:307–346. doi: 10.1016/S0065-308X(08)60565-5. [DOI] [PubMed] [Google Scholar]
- Poinar GO. 1975. Entomogenous Nematodes: A Manual and Host List of Insect-Nematode Associations. Leiden, Netherlands: Brill Archive. [Google Scholar]
- Presswell B, Evans S, Poulin R, Jorge F. 2015. Morphological and molecular characterization of Mermis nigrescens Dujardin, (Nematoda: Mermithidae) parasitizing the introduced European earwig (Dermaptera: Forficulidae) in New Zealand. J Helminthol. 89(3):267–276. doi: 10.1017/S0022149X14000017. [DOI] [PubMed] [Google Scholar]
- Qin M, Wu S, Li A, Zhao F, Feng H, Ding L, Ruan J. 2019. LRScaf: improving draft genomes using long noisy reads. BMC Genomics. 20(1):955. doi: 10.1186/s12864-019-6337-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach MJ, Schmidt SA, Borneman AR. 2018. Purge Haplotigs: allelic contig reassignment for third-gen diploid genome assemblies. BMC Bioinformatics. 19(1):460. doi: 10.1186/s12859-018-2485-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffers BR, Joppa LN, Pimm SL, Laurance WF. 2012. What we know and don’t know about Earth's missing biodiversity. Trends Ecol Evol. 27(2):501–510. doi: 10.1016/j.tree.2012.05.008. [DOI] [PubMed] [Google Scholar]
- Schiffer PH, Kroiher M, Kraus C, Koutsovoulos GD, Kumar S, Camps JI, Nsah NA, Stappert D, Morris K, Heger P, et al. 2013. The genome of Romanomermis culicivorax: revealing fundamental changes in the core developmental genetic toolkit in Nematoda. BMC Genomics. 14(1):923. doi: 10.1186/1471-2164-14-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schratzberger M, Holterman M, van Oevelen D, Helder J. 2019. A worm's world: ecological flexibility pays off for free-living nematodes in sediments and soils. Bioscience. 69(11):867–876. doi: 10.1093/biosci/biz086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 31(19):3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- Smythe AB, Holovachov O, Kocot KM. 2019. Improved phylogenomic sampling of free-living nematodes enhances resolution of higher-level nematode phylogeny. BMC Evol Biol. 19(1):121. doi: 10.1186/s12862-019-1444-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Shankar DS, Florea L. 2016. Rascaf: improving genome assembly with RNA sequencing data. Plant Genome. 9(3):0027. doi: 10.3835/plantgenome2016.03.0027. [DOI] [PubMed] [Google Scholar]
- Steinbiss S, Willhoeft U, Gremme G, Kurtz S. 2009. Fine-grained annotation and classification of de novo predicted LTR retrotransposons. Nucleic Acids Res. 37(21):7002–7013. doi: 10.1093/nar/gkp759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassetzky NS, Kramerov DA. 2013. SINEBase: a database and tool for SINE analysis. Nucleic Acids Res. 41(D1):D83–D89. doi: 10.1093/nar/gks1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren RL. 2016. RAILS and Cobbler: scaffolding and automated finishing of draft genomes using long DNA sequences. J Open Source Softw. 1(7):116. doi: 10.21105/joss.00116. [DOI] [Google Scholar]
- Warren RL, Yang C, Vandervalk BP, Behsaz B, Lagman A, Jones SJM, Birol I. 2015. LINKS: scalable, alignment-free scaffolding of draft genomes with long reads. Gigascience. 4(1):35. doi: 10.1186/s13742-015-0076-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisenfeld NI, Kumar V, Shah P, Church DM, Jaffe DB. 2017. Direct determination of diploid genome sequences. Genome Res. 27(5):757–767. doi: 10.1101/gr.214874.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Completing bacterial genome assemblies with multiplex MinION sequencing. Microb Genom. 3(10):e000132. doi: 10.1099/mgen.0.000132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G-C, Xu T-J, Zhu R, Zhang Y, Li S-Q, Wang H-W, Li J-T. 2019. LR_Gapcloser: a tiling path-based gap closer that uses long reads to complete genome assembly. Gigascience. 8(10):giy157. doi: 10.1093/gigascience/giy157. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genome sequencing reads, RNA-seq reads, and the genome assembly for M. nigrescens were submitted to NCBI under the BioProject accession number: PRJNA802644. The genome assembly and the annotation can also be accessed from FigShare: https://doi.org/10.6084/m9.figshare.21761501.v1. The pipeline and the scripts used in this project are available on GitHub: https://github.com/upendrabhattarai/Nematode_Genome_Project.
Supplemental material available at G3 online.