Abstract
There are multiple habitats in the oral cavity with bacteria, fungi, viruses, and protozoa residing in, which together constitute the oral micro‐ecosystem. These microflorae in the oral cavity primarily include saliva, supragingival dental plaque, subgingival dental plaque, submucosal plaque around implants, plaque in root canals, and plaque on the mucosal surface. The interest and knowledge of the microbiome have dynamically increased with the advancement of technology. Therefore, a reliable, feasible, and practical sampling strategy for the oral microbiome is required for the investigation. This paper introduced the sampling strategy of oral microorganisms, consisting of sample collection, transport, processing, and storage. The materials and devices involved in this study are all commonly used in clinical practice or laboratory. The feasibility and reliability of the sampling methods described in this paper have been verified by multiple studies.
Keywords: oral cavity, microbiome, saliva, plaque, sampling strategy
Highlights
Oral microbiome attracts substantial attention, and a reliable, feasible, and practical sampling strategy for the oral microbiome is required.
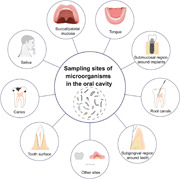
The habitats of microorganisms in the oral cavity primarily include saliva, supragingival area, subgingival/submucosal area, infected root canals, and mucosal surface.
The sampling strategy of oral microbiota is described in this study, which consists of sample collection, transport, processing, and storage.
The sampling sites in the oral cavity primarily include saliva, supragingival area, subgingival/submucosal area, infected root canals, and mucosal surface.
INTRODUCTION
The oral cavity is home to a wide variety of microorganisms, making it the second‐largest microbial community in the human body, next to the gut [1]. Oral microbiota is comprised of bacteria, fungi, viruses, and protozoa, of which bacteria are the dominant component. The microorganisms differ among individuals and alter dynamically under different clinical conditions, microenvironments, and other influencing factors. It is deeply involved in oral health, even systemic health. Multiple studies have confirmed microorganisms to be the etiological factor of a variety of oral diseases, such as periodontitis [2], caries [2], and mucositis [3]. It is also considered to have a potential association with a number of systemic diseases. Oral microorganisms may play a role in the development of diabetes [4], cardiovascular diseases [5], cancer [6], Alzheimer's disease [7], and other disorders. In turn, oral microorganisms may be influenced by systemic factors to some extent [8].
Despite the fact that oral microbiota has been studied for decades, it still attracts considerable critical attention. Technology has dramatically increased our understanding of the morphological traits, composition, function, and interacting mechanism of oral microorganisms, allowing us to better explore the characteristics of the oral microbial community [9]. High‐throughput sequencing, for example, could provide a global view of the composition of microbial community [10]; fluorescence in situ hybridization (FISH) could present the spatial structure of plaque [11]; proteome and metabolism could help to reveal the metabolites of microorganisms; advanced laboratory techniques are available to investigate their function and pathogenicity. With the advancement of technology, the pathogenic theories of periodontitis, peri‐implantitis, caries, and mucosal disorders have also altered. The developments in oral microbiology have heightened the requirement for the sampling strategy of the oral microbiome.
Every microenvironment accommodates a specific microbial community; therefore, different samples type could provide distinct information. Saliva comes from a variety of sources and immerses practically all the surfaces of hard and soft tissue. It could be used to investigate multiple oral diseases, as well as some systemic diseases. On the surfaces of oral hard and soft tissue, microorganisms exist as biofilm, which forms a sticky deposit on the surface of teeth called dental plaque. Supragingival dental plaque could be used to investigate the microbiota associated with the external tooth surface. Subgingival/submucosal dental plaque could aid in exploring the pathogenesis of periodontal/peri‐implant diseases. Dental plaque in root canals could provide information on canal infection or periapical infection. The samples on the oral mucosa may facilitate the investigation of mucosal diseases.
THE SAMPLING METHOD FOR SALIVA
Saliva is an affluent body fluid, containing water, minerals, electrolytes, enzymes, cytokines, immunoglobulins, as well as shed cells, food residues, and microbes [12]. Healthy adults produce 600–700 ml of saliva per day [13], and it covers all the surfaces of the oral cavity, including teeth, gingiva, tongue, palate, and mucosa. Saliva acts as a “reservoir” of oral microbiota, collecting microorganisms from multiple habitats and accommodating them. Saliva, in turn, transfers and releases microorganisms into these environments. Saliva is a noninvasive and easily accessible biofluid that permits the investigation of the oral microbiome.
The saliva flow rate fluctuates during the day, and diet and oral hygiene measures have an impact on the salivary microbiome. Saliva flow decreases when sleeping and progressively increases after waking up in the morning [14]. Therefore, collecting samples before cleaning teeth in the morning or between meals is recommended. Salivary flow is classified into two types: unstimulated saliva and stimulated saliva. Unstimulated saliva is obtained under physiological and natural conditions, whereas stimulated saliva is obtained following masticatory stimulation or gustatory stimulation. Stimulated saliva was initially used to evaluate the function of salivary glands. The differences in microbial composition between stimulated and unstimulated saliva have been reported [15, 16]. Considering that additional stimulating substances may have impacts on salivary microorganisms, unstimulated saliva is generally recommended for the investigation of oral microorganisms.
Materials and devices
Cup container; syringe; stimulant (gum base, cotton puff, rubber bands, sour candy, citric acid, or video), ice box or drikold bucket; sterile centrifuge tube; centrifuge; pipettor; ultra‐low temperature freezer.
Collection of saliva
There are four methods for collecting unstimulated saliva:
Method 1: Ask the volunteer to hold the sterile containers; sit still, head down, mouth slightly open, eyes open, head slightly forward. Avoid swallowing and allow saliva to flow naturally into the container [17].
Method 2: Ask the volunteer to sit still with his/her head up, and avoid swallowing. Concentrate the saliva on the floor of the mouth before spitting them into a container [17].
Method 3: Ask patients to sit on the dental chair and open their mouths. Suction off the saliva that accumulates in the mouth with a syringe.
Method 4: The whole saliva can also be sampled by placing sterile absorbent cotton or other absorbent materials in the mouth. Then, squeeze the saliva into the container.
Collecting unstimulated 1‐ml saliva typically takes 5–10 min. Collecting saliva with absorbent material should avoid wiping away the plaque on the mucosal surfaces.
There are three methods for collecting stimulated saliva:
Method 1: Ask volunteers to chew stimulants, such as paraffin wax [15, 18], unflavored chewing gum base, cotton puff, or rubber bands for 30 s. Then ask them to spit the accumulated saliva into the container (mastication stimulation) [17].
Method 2: Drop the sour candy or 4% citric acid on the tongue dorsum and wait for 1 min. Ask the volunteer to spit the accumulated saliva into the container (gustatory stimulation) [19].
Method 3: Watching a video of acidic foods or imagining some acidic foods can also help while collecting saliva. Ask the volunteer to spit the accumulated saliva into the container (imaginary stimulation).
Collecting stimulated 1‐milliliter saliva typically takes 1–5 min.
Sample transport and processing
The samples could be put into an ice box or drikold bucket to transfer, preferably within 3 h. The collected saliva was separated into a 1.5 ml sterile centrifuge tube and centrifuged (10,000–16,000 g, 4℃, 15 min). The supernatant was removed and sediment was kept for further detection [20].
Sample storage
The samples should be sent for subsequent detection and analysis as soon as possible. If an immediate analysis is not available, the samples could be stored at −80°C ultra‐low temperature freezer or temporally stored at −20°C for 1 month. To harvest DNA from bacteria, centrifuge and form a bacterial pellet before freezing [20]. A cryoprotective agent (such as 80% glycerol [21] or 10%–20% skim milk [22]) should be added if the sample is kept for subsequent bacteria culture (Figure 1).
Figure 1.
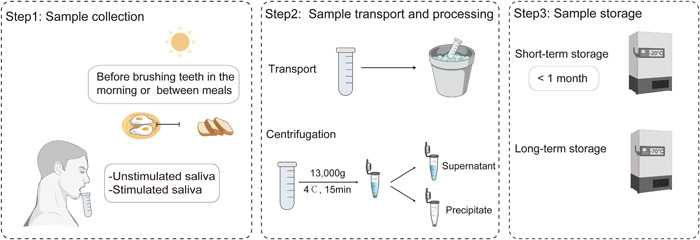
Sampling process of saliva.
THE SAMPLING METHOD OF SUPRAGINGIVAL DENTAL PLAQUE
The bacteria colonize on the tooth surface with the medium acquired pellicle, which is predominantly generated by salivary proteins and glycoproteins [23]. The supragingival dental plaque is an ecosystem existing as a biofilm on the tooth, which is influenced by various factors, consisting of food, mastication, muscular activity, saliva rinsing, and oral hygiene measures [24]. The buildup of biofilm and the accumulation of acidogenic microorganisms are widely acknowledged as the etiology of caries [25]. Dental plaque produces acids from carbohydrates, which leads to caries. Mutans Streptococci have been identified as the primary pathogens of dental caries, and they are thought to induce caries by generating acid and water‐insoluble glucan [26]. The proportions and numbers of acid‐base‐producing bacteria are considered the core of dental caries activity [27]. The analysis of supragingival microorganisms could facilitate the investigation of oral health.
Materials and devices
Oral cavity check‐up trays with forceps, probe, and mouth mirror; sterile cotton rolls or rubber dam; sample collector on the tooth surface (sterile probe, swab, or scraper); sample collector in the tooth cavity (sterile probe, curette, excavator, floss, or fine wire); sterile centrifuge tube; buffer solution (RNA protect reagent, TE buffer, Tris–HCl, EDTA, normal saline, PBS, or double‐distilled water); ice box or drikold bucket; centrifuge; pipettor; ultra‐low temperature freezer.
Sample collection
The sampling procedure is carried out on the dental chair with sterile equipment. Before collecting the sample, rinse the mouth with purified water to removeany food residue left in the mouth.
Dental plaque on the tooth surface: ask the volunteer to open his/her mouth, and isolate the sampling site from saliva with sterile cotton rolls or a rubber dam before sampling. Scrape the plaque samples from the target tooth surface using a sterile probe, swab, or scraper. After that, transfer the samples to a 0.6 ml sterile centrifugal tube. Dental plaque can also be collected using a sterile small brush or swab, and then eluted the plaque by scrubbing them in the prepared buffer for about 1 min.
Dental plaque in caries: The tool for collecting dental plaque in caries depends on the location and profile of the tooth cavity: sterile probe and curette are generally used to collect dental plaque on the occlusal surface and axial surface; sterile probe, floss, or fine wire can be used on proximal surfaces; sterile curettes can be used for collecting the samples in root caries; sterile excavators could be used in soft and moist cavity. After sampling, elute plaque in prepared buffer (scrub for about 1 min).
The buffers for plaque collection could be RNA protect reagent, TE buffer, 10 mM Tris–HCl, 1 mM EDTA, phosphate‐buffered saline (PBS), normal saline, and double‐distilled water, depending on the research goal. All of these solutions support subsequent preservation and detection of the microbiome. RNA protect reagent, TE buffer, Tris–HCl, and EDTA could protect RNA and DNA from cleavage. Dealt with PBS buffer (pH = 7.4) could facilitate subsequent analysis of protein, cytokines in the supernatant; Dealt with double distilled water allow the subsequent detection of metabolites of microorganisms. In addition, pooled samples of multiple index teeth can be used to investigate patient‐level microbiome. Pooled samples of multiple sites with the same clinical condition can be used to investigate the characteristics of the microbiome correspondingly [28].
Sample transport and processing
The collected samples could be transported to the laboratory within an ice box or drikold bucket. The collected sample should be centrifuged (10,000–16,000 g, 4℃, 15 min) to obtain the sediment for further detection (Figure 2).
Figure 2.
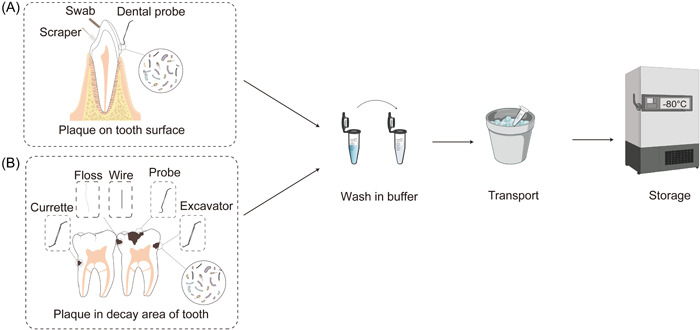
Sampling process of supragingival dental plaque. (A) Supragingival dental plaque on the tooth surface. (B) Dental plaque in the decayed area of the tooth.
Sample storage
The samples should be sent for subsequent detection and analysis as soon as possible. If an immediate analysis is not available, the samples could be stored at −80°C ultra‐low temperature freezer or temporally stored at −20°C within 1 month. To harvest DNA from bacteria, centrifuge and form a bacterial pellet before freezing [20]. A cryoprotective agent (such as 80% glycerol [21] or 10%–20% skim milk [22]) should be added if the sample is kept for subsequent bacteria culture.
THE SAMPLING METHOD OF SUBGINGIVAL PLAQUE AROUND TEETH AND SUBMUCOSAL PLAQUE AROUND IMPLANTS
Subgingival dental plaque has been widely accepted as the initial factor of periodontitis for long [29]. The accumulation of plaque also has been identified as the etiological factor of peri‐implant diseases [30]. Based on similar anatomical structure and immunological characteristics, the pathogenic mechanism of periodontal and peri‐implant diseases is comparable [31]. Meanwhile, the inflammatory internal wall of periodontal/peri‐implant pockets opens a window for systemic infection. Therefore, the investigation of the subgingival microbiome and submucosal microbiome could help to evaluate the key pathogenic factor of periodontal diseases and peri‐implant diseases. It could also expedite our understanding of the relationship between oral infection and systemic conditions.
Materials and devices
Oral cavity check‐up trays with forceps, probe, and mouth mirror; sterile cotton rolls; sterile curette; paper strips/points; 0.6 and 1.5 ml sterile centrifuge tube; buffer solution (RNA protect reagent, TE buffer, Tris‐HCl, EDTA, normal saline, PBS, or double distilled water); ice box or drikold bucket; alcohol lamp; oscillator; centrifuge; pipettor; ultra‐low temperature freezer.
Sample collection
According to the research design, the sample could be collected at the site level, tooth/implant level, and patient level. Site‐specific samples could provide information on local lesions. Tooth‐/implant‐level investigation generally needs to sample 1–6 sites. The sampling sites are often selected from the mesial and distal buccal sides of the target teeth, which can avoid saliva contamination as much as possible. For a tooth with shallow gingival sulcus or implant with shallow mucosal sulcus, collecting samples from multiple sites could help to obtain a sufficient quantity of microorganisms. A patient‐level investigation needs to pool the samples of several teeth or implants. Ramfjord index teeth are considered to be representative to assess overall periodontal condition [32]. Ramfjord index teeth consist of 16, 21, 24, 36, 41, 44 [32]. If the index teeth are lost, adjacent teeth in the same region will be sampled.
Before sample collection, ask volunteers to rinse their mouths to remove food residue in the mouth. After scraping the calculus, food residue, plaque, and soft debris on the supragingival tooth surface with a probe or scaler or small cotton ball, the saliva was isolated with sterile cotton around sampled sites.
Sampling collection of subgingival plaque or submucosal plaque is carried out on the dental treatment chair with sterile sampling equipment. The commonly used methods for subgingival plaque and submucosal plaque around implants include the curetting method (Figure 3a) and the adsorption method (paper strips, Figure 3b). Both of them are reliable and widely used. However, the curetting method could mainly collect attached plaque on the surface of teeth or implants, while the adsorption method has more advantages in collecting unattached bacteria in periodontal or peri‐implant pockets.
Figure 3.
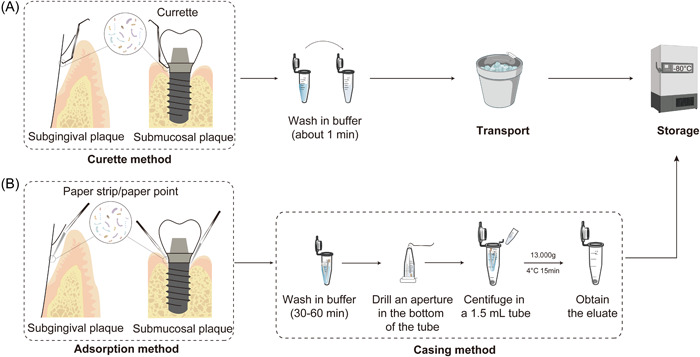
The sampling process of subgingival plaque and submucosal plaque. (A) The sampling of subgingival plaque around the tooth or submucosal plaque around the implant with the curette method. (B) The sampling method of subgingival plaque around the tooth or submucosal plaque around the implant with the adsorption method.
Curette method (plaque method): Subgingival plaque in the periodontal pocket or submucosal plaque around the implant was collected with a sterile periodontal curette and placed in a 0.6 ml sterile centrifugal tube for transport in an ice box or drikold bucket [28, 33].
Adsorption method (gingival crevicular fluid (GCF) method): For subgingival/submucosal sampling, 2 × 10 mm filter paper strips (Whatman, United Kingdom) and 0.02 35# absorbent paper points (GAPADENT, China) with 0.5–1 cm tip cut off is recommended. The strips should be processed using a sterile tweezer to the proper size, and accept high‐pressure steam sterilization and drying before sampling. Hold absorbent paper points or filter paper strips, insert them into the periodontal pockets along the tooth surface, hold on for 30 s when resistance was encountered, and then put it into a 0.6 ml sterile centrifugal tube for transport in an ice box or drikold bucket. Paper strips or paper points should be placed into the bottom of the sulcus or pockets to obtain the plaque at the frontier of disease progress. For healthy peri‐implant tissue with a tight cuff, the curette could be very difficult to insert in without damaging the mucosa. In that case, the adsorption method with finer strips would be a better choice. The paper strips should be carefully inserted into the sulcus or pockets along the margin of the gingiva or mucosa [34, 36].
Sample processing
The collected samples could be transported to the laboratory within an ice box or drikold bucket.
Curette method: The collected sample should be centrifuged (10,000–16,000 g, 4℃, 15 min) to obtain the sediment for further detection.
Adsorption method: The samples collected by paper strips or paper points need to be processed by the casing (pipe‐in‐pipe) method. The detailed procedure is as follows: Add 100 μL buffer solution into the 0.6 ml centrifuge tube with sterile filter paper strips or absorbent paper points sample, and vibrate at 4℃ for 30–60 min. Invert the 0.6 ml centrifuge tube, quickly pierce and pull it out with a red‐hot needle tip in the center of the bottom to make a small aperture (<the diameter of filter paper strips or absorbent paper points). Then, put the tube into a 1.5 ml sterile tube and centrifuged symmetrically (10,000–16,000 g, 4℃, 15 min). The supernatant should be gently absorbed and placed into another centrifugal tube for retention, and the precipitation could be obtained as the plaque sample. The sample could also be cryopreserved before sample processing (Figure 3).
Sample storage
The samples should be sent for subsequent detection and analysis as soon as possible. If an immediate analysis is not available, the samples could be stored at −80°C ultra‐low temperature freezer or temporally stored at −20°C for 1 month. To harvest DNA from bacteria, centrifuge and form a bacterial pellet before freezing [20]. A cryoprotective agent (such as 80% glycerol [21] or 10%–20% skim milk [22]) should be added if the sample is kept for subsequent bacteria culture.
THE SAMPLING METHOD OF DENTAL PLAQUE IN ROOT CANALS
The colonization of bacteria in the root canal could result in pulpitis [37], periapical periodontitis [38], and even osteomyelitis. The pathogenesis of microorganisms in the root canal system determines the degree of inflammation, symptoms, and clinical treatment effect. Therefore, eliminating bacteria from the infected root canal and preventing reinfection of the tooth is the key point of root canal treatment to preserve the teeth. For prolonged and refractory infections or cases of risk of systemic spread of infection, root canal sampling for microbiological diagnostics is recommended.
Materials and devices
Oral cavity check‐up trays with forceps, probe, and mouth mirror; sterile cotton rolls or rubber dam; sterile curette; paper points; 0.6 and 1.5 ml sterile centrifuge tube; buffer solution (RNA protect reagent, TE buffer, Tris–HCl, EDTA, normal saline, PBS, or double distilled water); ice box or drikold bucket; alcohol lamp; centrifuge; pipettor; ultra‐low temperature freezer.
Sample collection
The adsorption sampling method with sterile paper points is usually used for plaque samples in the infected root canal. If there is much exudation, springe with a fine point could also be used to collect the sample. The following operations are carried out on the dental treatment chair with sterile equipment. Before sampling, remove food residue, calculus, plaque, and soft debris from the crown of teeth to avoid potential contamination; cut the canal wall with a root canal preparation instrument to release biofilm. In addition, sample collection requires a clear approach to the medullary cavity and root canal (avoid plaque damage caused by excessive flushing).
Use sterile cotton to isolate sampling sites. Insert sterile hygroscopic paper points (paper points should be chosen according to the taper and thickness of the root canal) into the root canal and retain them for 30 s. Then remove and put them into a 0.6 ml sterile centrifugal tube for transport in an ice box or drikold bucket. The wet cusp of paper strips may indicate that the collection of samples is successful. If the root canal is desiccative making the dental plaque difficult to be absorbed with paper points, we can inject some sterile solution (like saline or double‐distilled water), then extract the fluid in the root canal using syringe or paper points.
Sample processing
The casing (pipe‐in‐pipe) method is described above. Add 100 μl buffer solution into a 0.6 ml centrifuge tube with paper strips or paper points, and vibrate for 30–60 min at 4℃. Invert the 0.6 ml centrifuge tube, quickly pierce and pull it out with a red‐hot needle tip in the center of the bottom to make a small aperture (<the diameter of absorbent paper points or filter paper strips). Then, place the above 0.6 ml centrifuge tube in a 1.5 ml sterile tube and centrifuge symmetrically (10,000–16,000 g, 4℃, 15 min). The supernatant should be gently removed and placed into another centrifugal tube for retention, and the precipitation should be obtained as plaque (Figure 4).
Figure 4.
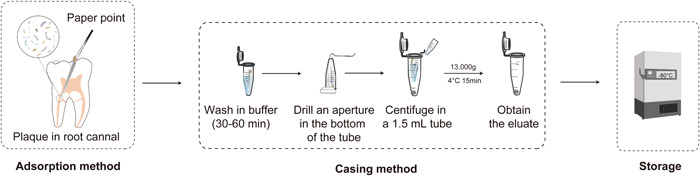
Sampling process of plaque in the root canal.
Sample storage
The samples should be sent for subsequent detection and analysis as soon as possible. If an immediate analysis is not available, the samples could be stored at −80°C ultra‐low temperature freezer or temporally stored at −20°C for 1 month. To harvest DNA from bacteria, centrifuge and form a bacterial pellet before freezing [20]. A cryoprotective agent (such as 80% glycerol [21] or 10%–20% skim milk [22]) should be added if the sample is kept for subsequent bacteria culture.
THE SAMPLING METHOD OF PLAQUE ON THE SURFACE OF THE ORAL MUCOSA
Oral mucosa is laminated squamous epithelium lining the inside of the mouth, consisting of lining mucosa, masticatory mucosa, and special mucosa [39]. Healthy epithelia with cell–cell junction only have straticulate biofilm on them due to the muscle movement and saliva scour, while ulcerated mucosa generally has many microorganisms accumulating on it and infiltrating into tissue [39]. The tongue dorsum harbors abundant nipples on the surface of the epithelium, which provides a favorable microenvironment for microorganisms to colonize and reproduce [40]. In addition to the tongue dorsum, coating mucosa in the oral cavity serves as soft tissue surface, lateral and abdominal sides of the tongue, buccal mucosa, hard palate, soft palate, gingiva, and tonsils. The microorganism on the mucosa is closely related to mucosal health and diseases. Halitosis [41], herpes gingivostomatitis [42], hand‐foot‐and‐mouth disease [43], and oral candidiasis [44] are caused by pathogenic microorganisms, consisting of bacteria, viruses, and fungus. The analysis of the microbiome on the mucosa can also assist in the diagnosis and evaluation of some systemic infections with oral manifestations, such as oral tuberculosis [45] and syphilis [46].
Materials and devices
Oral cavity check‐up trays with forceps, probe, and mouth mirror; sterile cotton swab, brush, or batten; 0.6 and 1.5 ml sterile centrifuge tube; buffer solution (RNA protect reagent, TE buffer, Tris‐HCl, EDTA, normal saline, PBS, or double‐distilled water); ice box or drikold bucket; centrifuge; pipettor; ultra‐low temperature freezer.
Sample collection and transport
Sampling sites on oral mucosa mainly include tongue dorsum, buccal mucosa, and palate. Volunteers should rinse their mouths with purified water to remove any food residue before sampling.
Tongue dorsum: Ask volunteers to open their mouth and extend their tongue slightly, brush with a sterile brush from one side to the other side in an imbricate shape on the back of the tongue, cut off the head of the brush with a plaque on it, and put it into a sterile tube. A sterile scraper and cotton swab could also be used to collect the plaque on the tongue dorsum [47].
Other oral coating mucosae (lining mucosa and masticatory mucosa): Scrape the plaque on the mucosal surface with a sterile batten or cotton swab [28], put it into the sterile tube, and wash it repeatedly in the tube with buffer solution. The sample should be transported within an ice box or drikold bucket.
Sample processing
Add freshly prepared buffer solution. The brush was removed from the liquid, centrifuged at low temperature (10,000–16,000 g, 4℃, 15 min), and the supernatant was gently absorbed and discarded to obtain the precipitate as a tongue dorsal plaque sample (Figure 5).
Figure 5.
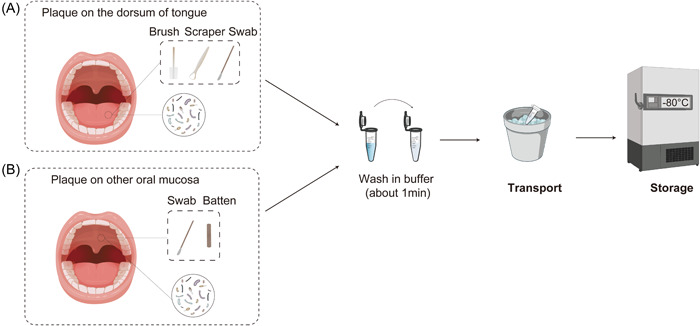
The sampling process of plaque on the oral mucosa. (A) Sampling of plaque on tongue dorsum with a sterile brush, scrape, or cotton swab. (B) The sampling method from other mucosal surfaces with sterile batten or cotton swab.
Sample storage
The samples should be sent for subsequent detection and analysis as soon as possible. If an immediate analysis is not available, the samples could be stored at −80°C ultra‐low temperature freezer or temporally stored at −20°C within 1 month. To harvest DNA from bacteria, centrifuge and form a bacterial pellet before freezing [20]. A cryoprotective agent (such as 80% glycerol [21] or 10%–20% skim milk [22]) should be added if the sample is kept for subsequent bacteria culture.
THE SAMPLING METHOD OF OTHER SITES
There are retention, hydrops, effusion, and abscess in the oral cavity, which may be a result of infection. The sample in these sites could be obtained with a sterile syringe or sterile paper strips depending on the quantity and position. Sometimes surgery may be required to create an approach for sample collection.
SUMMARY
Sampling procedures could be conducted by dentists, dental hygienists, or trained researchers. The most important thing for sample collection is to avoid injuring volunteers. Gingiva is extremely vulnerable and could get hurt during the collection of subgingival/submucosal dental plaque. Another important thing is to avoid potential contamination of the nearby microbial ecosystem. Collecting subgingival/submucosal dental plaque is very easy to be polluted by supragingival dental plaque because of the confined access. Saliva is also very easy to be polluted by plaque from the mucosal surface.
With some microorganisms firmly colonizing the oral cavity serving as the fingerprint, the oral microbiome could reflect individual features to some extent. On the other hand, the oral cavity is a complex and incessantly changing environment, which is also affected by a variety of external factors and systemic factors. Therefore, only the sample in real‐time could provide precise information about the dynamical oral microbiome. The characteristic of the microbiome is influenced by several factors, such as age, gender, circadian rhythm, diet, smoking, physiological status, oral hygiene measures, and intake of medicine. Therefore, investigators should keep an eye on the influencing factors and choose appropriate sampling procedures according to the purpose of the research.
AUTHOR CONTRIBUTIONS
Hongye Lu wrote the manuscript and revised the figures. Peihui Zou designed and constructed the diagrams. Yifei Zhang and Qian Zhang revised the manuscript. Zhibin Chen and Feng Chen supervised this project. All authors have read the final manuscript and approved it for publication.
CONFLICTS OF INTEREST
This study was authorized by Bio‐protocol.
ACKNOWLEDGMENT
This study was supported by the Special Foundation of China Postdoctoral Science Foundation (Grant No. 2021TQ0277).
Lu, Hongye , Zou Peihui, Zhang Yifei, Zhang Qian, Chen Zhibin, and Chen Feng. 2022. “The Sampling Strategy of Oral Microbiome.” iMeta 1, e23. 10.1002/imt2.23
Contributor Information
Zhibin Chen, Email: kqyehui21@bjmu.edu.cn.
Feng Chen, Email: chenfeng2011@hsc.pku.edu.cn.
DATA AVAILABILITY STATEMENT
Supplementary materials (figures, tables, scripts, graphical abstract, slides, videos, Chinese translated version, and updated materials) may be found in the online DOI or iMeta Science http://www.imeta.science/.
REFERENCES
- 1. Verma, Digvijay , Garg Pankaj K., and Dubey Ashok K.. 2018. “Insights into the Human Oral Microbiome.” Archives of Microbiology 200: 525–40. 10.1007/s00203-018-1505-3 [DOI] [PubMed] [Google Scholar]
- 2. Wade, William G. 2013. “The Oral Microbiome in Health and Disease.” Pharmacological Research 69: 137–43. 10.1016/j.phrs.2012.11.006 [DOI] [PubMed] [Google Scholar]
- 3. Hong, Bo‐Young , Sobue Takanori, Choquette Linda, Dupuy Amanda K., Thompson Angela, Burleson Joseph A., Salner Andrew L., et al. 2019. “Chemotherapy‐Induced Oral Mucositis is Associated with Detrimental Bacterial Dysbiosis.” Microbiome 7: 66. 10.1186/s40168-019-0679-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Xiao, E , Mattos Marcelo, Vieira Gustavo H. A., Chen Shanshan, Corrêa Jôice D., Wu Yingying, Albiero Mayra L., Bittinger Kyle, Graves Dana T.. 2017. “Diabetes Enhances IL‐17 Expression and Alters the Oral Microbiome to Increase its Pathogenicity.” Cell Host Microbe 22: 120–8.e124. 10.1016/j.chom.2017.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schenkein, Harvey A. , Papapanou Panos N., Genco Robert, and Sanz Mariano. 2020. “Mechanisms Underlying the Association between Periodontitis and Atherosclerotic Disease.” Periodontology 2000 83: 90–106. 10.1111/PRD.12304 [DOI] [PubMed] [Google Scholar]
- 6. Fan, Xiaozhou , Alekseyenko Alexander V., Wu Jing, Peters Brandilyn A., Jacobs Eric J., Gapstur Susan M., Purdue Mark P., et al. 2018. “Human Oral Microbiome and Prospective Risk for Pancreatic Cancer: A Population‐based Nested Case‐Control Study.” Gut 67: 120–7. 10.1136/gutjnl-2016-312580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ryder, Mark I. 2020. “ Porphyromonas gingivalis and Alzheimer Disease: Recent Findings and Potential Therapies.” Journal of Periodontology 91(Suppl 1): S45–S49. 10.1002/jper.20-0104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Graves, Dana T. , D. Corrêa Jôice, and A. Silva Teógenes. 2019. “The Oral Microbiota Is Modified by Systemic Diseases.” Journal of Dental Research 98: 148–56. 10.1177/0022034518805739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gao, Chuqi , Li Xuantao, Zhao Xiaole, Yang Peiyue, Wang Xiao, Chen Xiaoli, Chen Ning, and Chen Feng. 2022. “Standardized Studies of the Oral Microbiome: From Technology‐Driven to Hypothesis‐Driven.” iMeta 1: e19. 10.1002/imt2.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Emanuel, Ezekiel J. 2013. “Reconsidering the Declaration of Helsinki.” The Lancet 381: 1532–3. 10.1016/S0140-6736(13)60970-8 [DOI] [PubMed] [Google Scholar]
- 11. Geier, Benedikt , Sogin Emilia M., Michellod Dolma, Janda Moritz, Kompauer Mario, Spengler Bernhard, Dubilier Nicole, and Liebeke Manuel. 2020. “Spatial Metabolomics of In Situ Host‐Microbe Interactions at the Micrometre Scale.” Nature Microbiology 5: 498–510. 10.1038/S41564-019-0664-6 [DOI] [PubMed] [Google Scholar]
- 12. Marsh, Philip D. , Do Thuy, Beighton David, and Devine Deirdre A.. 2016. “Influence of Saliva on the Oral Microbiota.” Periodontol 2000 70: 80–92. 10.1111/prd.12098 [DOI] [PubMed] [Google Scholar]
- 13. Watanabe, Seiya , and Dawes Colin. 1988. “The Effects of Different Foods and Concentrations of Citric Acid on the Flow Rate of Whole Saliva in Man.” Archives of Oral Biology 33: 1–5. 10.1016/0003-9969(88)90089-1 [DOI] [PubMed] [Google Scholar]
- 14. Ferguson, David B. , and Botchway C. A.. 1980. “A Comparison of Circadian Variation in the Flow Rate and Composition of Stimulated Human Parotid, Submandibular and Whole Salivas from the Same Individuals.” Archives of Oral Biology 25: 559–68. 10.1016/0003-9969(80)90068-0 [DOI] [PubMed] [Google Scholar]
- 15. Jo, Ryutaro , Nishimoto Yuichiro, Umezawa Kouta, Yama Kazuma, Aita Yuto, Ichiba Yuko, and Murakami Shinnosuke, et al. 2019. “Comparison of Oral Microbiome Profiles in Stimulated and Unstimulated Saliva, Tongue, and Mouth‐Rinsed Water.” Scientific Reports 9: 16124. 10.1038/s41598-019-52445-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gomar‐Vercher, Sonia , Simón‐Soro Aurea, Montiel‐Company José M., Almerich‐Silla José M., and Mira Alex. 2018. “Stimulated and Unstimulated Saliva Samples have Significantly Different Bacterial Profiles.” PLoS One 13: e0198021. 10.1371/journal.pone.0198021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Justino, Allisson Benatti , Teixeira Renata R., Peixoto Leonardo G., Jaramillo Olga L. Bocanegra, and Espindola Foued S.. 2017. “Effect of Saliva Collection Methods and Oral Hygiene on Salivary Biomarkers.” Scandinavian Journal of Clinical and Laboratory Investigation 77: 415–22. 10.1080/00365513.2017.1334261 [DOI] [PubMed] [Google Scholar]
- 18. Almhöjd, Ulrica , Cevik‐Aras Hulya, Karlsson Niclas, Chuncheng Jin, and Almståhl Annica. 2021. “Stimulated Saliva Composition in Patients with Cancer of the Head and Neck Region.” BMC Oral Health 21: 509. 10.1186/s12903-021-01872-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Büttler, Rahel M. , Bagci Esra, Brand Henk S., den Heijer Martin, Blankenstein Marinus A., Heijboer Annemieke C.. 2018. “Testosterone, Androstenedione, Cortisol and Cortisone Levels in Human Unstimulated, Stimulated and Parotid Saliva.” Steroids 138: 26–34. 10.1016/j.steroids.2018.05.013 [DOI] [PubMed] [Google Scholar]
- 20. Lu, Hongye , He Lu, Jin Dongsiqi, Zhu Yunxuan, and Meng Huanxin. 2021. “Effect of Adjunctive Systemic Antibiotics on Microbial Populations Compared with Scaling and Root Planing alone for the Treatment of Periodontitis: A Pilot Randomized Clinical Trial.” Journal of Periodontology 93: 570–83. 10.1002/jper.20-0764 [DOI] [PubMed] [Google Scholar]
- 21. Koh, Cheryl M. 2013. “Storage of bacteria and yeast.” Methods Enzymol 533: 15–21. 10.1016/b978-0-12-420067-8.00002-7 [DOI] [PubMed] [Google Scholar]
- 22. Cody, William L. , Wilson James W., Hendrixson David R., McIver Kevin S., Hagman Kayla E., Ott C. M., Nickerson Cheryl A., and Schurr Michael J.. 2008. “Skim Milk Enhances the Preservation of Thawed −80 Degrees C Bacterial Stocks.” Journal of Microbiological Methods 75: 135–8. 10.1016/j.mimet.2008.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hannig, Christian , and Matthias Hannig Thomas A.. 2005. “Enzymes in the Acquired Enamel Pellicle.” European Journal of Oral Sciences 113: 2–13. 10.1111/j.1600-0722.2004.00180.x [DOI] [PubMed] [Google Scholar]
- 24. Haffajee, Anne D. , Teles Ricardo P., Patel Michele R., Song X., Veiga Nancy, and Socransky Sigmund S.. 2009. “Factors Affecting Human Supragingival Biofilm Composition. I. Plaque Mass.” Journal of Periodontal Research 44: 511–9. 10.1111/j.1600-0765.2008.01154.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Takahashi, Nobuhiro , and Nyvad Bente. 2011. “The Role of Bacteria in the Caries Process: Ecological Perspectives.” Journal of Dental Research 90: 294–303. 10.1177/0022034510379602 [DOI] [PubMed] [Google Scholar]
- 26. Tanzer, Janson M. , Livingston Jill, and Thompson Angela M.. 2001. “The Microbiology of Primary Dental Caries in Humans.” Journal of Dental Education 65: 1028–37. 10.1002/j.0022-0337.2001.65.10.tb03446.x [DOI] [PubMed] [Google Scholar]
- 27. Kleinberg, Israel . 2002. “A Mixed‐Bacteria Ecological Approach to Understanding the Role of the Oral Bacteria in Dental Caries Causation: An Alternative to Streptococcus Mutans and The Specific‐plaque Hypothesis.” Critical Reviews in Oral Biology & Medicine 13: 108–25. 10.1177/154411130201300202 [DOI] [PubMed] [Google Scholar]
- 28. Hussein, Heba . 2021. “Oral Sampling Techniques.” Methods in Molecular Biology 2327: 17–29. 10.1007/978-1-0716-1518-8_2 [DOI] [PubMed] [Google Scholar]
- 29. Kinane, Denis F. , Panagiota G. Stathopoulou, Papapanou Panos N.. 2017. “Periodontal Diseases.” Nature Reviews Disease Primers 3: 17038. 10.1038/nrdp.2017.38 [DOI] [PubMed] [Google Scholar]
- 30. Schwarz, Frank , Derks Jan, Monje Alberto, and Wang Hom‐Lay. 2018. “Peri‐Implantitis.” Journal of Periodontology 89(Suppl 1): S267–S290. 10.1002/jper.16-0350 [DOI] [PubMed] [Google Scholar]
- 31. Ivanovski, Saso , Lee Ryan. 2018. “Comparison of peri‐implant and periodontal marginal soft tissues in health and disease.” Periodontol 2000 76: 116–30. 10.1111/prd.12150 [DOI] [PubMed] [Google Scholar]
- 32. Ramfjord, Sigurd P. 1959. “Indices for Prevalence and Incidence of Periodontal Disease.” Journal of Periodontology 30: 51–9. 10.1902/JOP.1959.30.1.51 [DOI] [Google Scholar]
- 33. Lu, Hongye , He Lu, Xu Jingling, Song Wenli, Feng Xianghui, Zhao Yibing, and Meng Huanxin. 2020. “Well‐Maintained Patients with a History of Periodontitis Still Harbor a More Disbiotic Microbiome than Health.” Journal of Periodontology 91: 1584–94. 10.1002/JPER.19-0498 [DOI] [PubMed] [Google Scholar]
- 34. Lu, Hongye , Zhao Yibing, Feng Xianghui, He Lu, and Meng Huanxin. 2019. “Microbiome in Maintained Periodontitis and its shift Over a Single Maintenance Interval of 3 Months.” Journal of Clinical Periodontology 46: 1094–104. 10.1111/jcpe.13177 [DOI] [PubMed] [Google Scholar]
- 35. Lu, Hongye , Yan Xia, Zhu Bin, Zhang Li, Feng Xianghui, Piao Muzi, and Huang Baoxin, et al. 2021. “The Occurrence of Peri‐Implant Mucositis Associated with the Shift of Submucosal Microbiome in Patients with a History of Periodontitis During the First Two Years.” Journal of Clinical Periodontology 48: 441–54. 10.1111/jcpe.13410 [DOI] [PubMed] [Google Scholar]
- 36. Wang, Qi , Lu Hongye, Zhang Li, Yan Xia, Zhu Bin, and Meng Huanxin. 2020. “Peri‐Implant Mucositis Sites with Suppuration have Higher Microbial Risk than Sites Without Suppuration.” Journal of Periodontology 91: 1284–94. [DOI] [PubMed] [Google Scholar]
- 37. Zargar, Nazanin , Ashraf Hengameh, Marashi S. Amin, Sabeti Mohammad, Aziz Alireza. 2020. “Identification of Microorganisms in Irreversible Pulpitis and Primary Endodontic Infections with Respect to Clinical and Radiographic Findings.” Clinical Oral Investigations 24: 2099–108. 10.1007/s00784-019-03075-9 [DOI] [PubMed] [Google Scholar]
- 38. Siqueira, José F. , and N. Rôças Isabela. 2013. “Microbiology and Treatment of Acute Apical Abscesses.” Clinical Microbiology Reviews 26: 255–73. 10.1128/cmr.00082-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Groeger, Sabine , Meyle Joerg. 2019. “Oral Mucosal Epithelial Cells.” Frontiers in Immunology 10: 208. 10.3389/fimmu.2019.00208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wilbert, Steven A. , Mark Welch Jessica L., and Borisy Gary G.. 2020. “Spatial Ecology of the Human Tongue Dorsum Microbiome.” Cell Reports 30: 4003–15. e4003. 10.1016/j.celrep.2020.02.097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bernardi, Sara , Karygianni Lamprini, Filippi Andreas, Anderson Annette C., Zürcher Andrea, Hellwig Elmar, Vach Kirstin, Macchiarelli Guido, and Al‐Ahmad Ali. 2020. “Combining Culture and Culture‐Independent Methods Reveals New Microbial Composition of Halitosis Patients' Tongue Biofilm.” Microbiologyopen 9: e958. 10.1002/mbo3.958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Petti, Stefano , Lodi Giovanni. 2019. “The Controversial Natural History of Oral Herpes Simplex Virus Type 1 Infection.” Oral Diseases 25: 1850–65. 10.1111/odi.13234 [DOI] [PubMed] [Google Scholar]
- 43. Saguil, Aaron , Kane Shawn F., Lauters Rebecca, and Mercado Michael G.. 2019. “Hand‐Foot‐and‐Mouth Disease: Rapid Evidence Review.” American Family Physician 100: 408–14. [PubMed] [Google Scholar]
- 44. Millsop, Jillian W. , and Fazel Nasim. 2016. “Oral Candidiasis.” Clinics in Dermatology 34: 487–94. 10.1016/j.clindermatol.2016.02.022 [DOI] [PubMed] [Google Scholar]
- 45. Wu, Yu‐Hsueh , Chang Julia Yu‐Fong, Sun Andy, and Chiang Chun‐Pin. 2017. “Oral Tuberculosis.” Journal of the Formosan Medical Association 116: 64–5. 10.1016/j.jfma.2016.10.016 [DOI] [PubMed] [Google Scholar]
- 46. de Arruda, José Alcides Almeida , Valle Isabella B. do, Mesquita Ricardo A., and Silva Tarcília A.. 2021. “Oral Syphilis.” Journal of the American Academy of Dermatology 84: e101–e102. 10.1016/j.jaad.2020.09.067 [DOI] [PubMed] [Google Scholar]
- 47. Stephen, Abish S. , Dhadwal Narinder, Nagala Vamshidhar, Gonzales‐Marin Cecilia, Gillam David G., Bradshaw David J., Burnett Gary R., and Allaker Robert P.. 2021. “Interdental and Subgingival Microbiota may Affect the Tongue Microbial Ecology and Oral Malodour in Health, Gingivitis and Periodontitis.” Journal of Periodontal Research 56: 1174–84. 10.1111/jre.12931 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Supplementary materials (figures, tables, scripts, graphical abstract, slides, videos, Chinese translated version, and updated materials) may be found in the online DOI or iMeta Science http://www.imeta.science/.


