Abstract
Reference genome assemblies have been created from multiple lineages within the Canidae family; however, despite its phylogenetic relevance as a basal genus within the clade, there is currently no reference genome for the gray fox (Urocyon cinereoargenteus). Here, we present a chromosome-level assembly for the gray fox (U. cinereoargenteus), which represents the most contiguous, non-domestic canid reference genome available to date, with 90% of the genome contained in just 34 scaffolds and a contig N50 and scaffold N50 of 59.4 and 72.9 Megabases, respectively. Repeat analyses identified an increased number of simple repeats relative to other canids. Based on mitochondrial DNA, our Vermont sample clusters with other gray fox samples from the northeastern United States and contains slightly lower levels of heterozygosity than gray foxes on the west coast of California. This new assembly lays the groundwork for future studies to describe past and present population dynamics, including the delineation of evolutionarily significant units of management relevance. Importantly, the phylogenetic position of Urocyon allows us to verify the loss of PRDM9 functionality in the basal canid lineage, confirming that pseudogenization occurred at least 10 million years ago.
Keywords: gray fox, Canidae, PRDM9, heterozygosity
Reference genome assemblies have been created from multiple lineages within the Canidae family, however, despite its phylogenetic relevance as a basal genus within the clade, there is currently no reference genome for the gray fox (Urocyon cinereoargenteus). Here, Armstrong et al. present a chromosome-level assembly for the gray fox (U. cinereoargenteus), which represents the most contiguous, non-domestic canid reference genome available to date, with 90% of the genome contained in just 34 scaffolds and a contig N50 and scaffold N50 of 59.4 and 72.9 Megabases (Mb), respectively. Repeat analyses identified an increased number of simple repeats relative to other canids. Based on mitochondrial DNA, our Vermont sample clusters with other gray fox samples from the northeastern US and contains slightly lower levels of heterozygosity than gray foxes on the west coast of California. This new assembly lays the groundwork for future studies to describe past and present population dynamics, including the delineation of evolutionarily significant units of management relevance. Importantly, the phylogenetic position of Urocyon allows us to verify the loss of prdm9 functionality in the basal canid lineage, confirming that pseudogenization occurred at least 10 million years ago.
Introduction
Reference genomes have become valuable tools in conservation science and decision-making (Supple and Shapiro 2018; Formenti et al. 2022; Paez et al. 2022). While mammals tend to be the most well-represented amongst large-scale consortia endeavors (e.g. Zoonomia; Zoonomia Consortium 2020), progress on generating data for the mammalian family Canidae has lagged, with only 6 of the 13 extant genera having publicly available, representative reference genome assemblies. Canidae currently contains 39 extant species that vary in size, ecology, and distribution and diverged from other carnivoran families ∼40–60 million years ago (MYA) (Wayne 1993; Nyakatura and Bininda-Emonds 2012).
Within Canidae, the genus Urocyon has historically been difficult to place, but it is thought to represent the sister lineage to all other living canids (Tedford et al. 1995; Lindblad-Toh et al. 2005; Nyakatura and Bininda-Emonds 2012). The genus contains only two species: the gray fox (Urocyon cinereoargenteus, Schreber 1775), which is found from southern Canada through northern South America, and the island fox (Urocyon littoralis), which is restricted to the California Channel Islands. Gray foxes are grizzled in appearance and have a number of scansorial (climbing) adaptations that facilitate their use of deciduous forests (Fritzell and Haroldson 1982). However, there is significant variation in their responses to habitat alterations across their North American range, with some studies suggesting they are tolerant of habitat disturbance and extremely abundant, while others suggest they are highly sensitive and occur at lower population densities (McAlpine et al. 2008; Bauder et al. 2020; Allen et al. 2021).
The lack of precise ecological knowledge concerning the gray fox across its range is compounded by uncertainty in the species' historical distribution and genetic structure, and the perceived expansion of gray foxes into urban ecosystems and new geographic areas is accompanied by the potential for human–wildlife conflict and disease spillover. For example, a gray fox in New England was diagnosed with concurrent infections of antibiotic-resistant Listeria monocytogenes, skunk adenovirus-1, and canine distemper virus (Needle et al. 2020). Both mitogenomes and genotyping by sequencing approaches suggest that western and eastern North American gray fox populations diverged in the early–mid-Pleistocene (∼0.8 MYA) and now form a secondary contact zone of eastern and western lineages at the Great Plains Suture (Reding et al. 2021; Kierepka et al. 2023), displaying a distinct pattern from many other North American carnivores, and disagreeing with previously held morphological subspecies designations. Eastern populations of the gray fox are phylogenetically distinct from all other gray fox populations, and the island fox (U. littoralis) is nested within the western gray fox clade (Preckler-Quisquater et al. 2023). This cryptic divergence pattern, coupled with recent heterogenous range expansions and population declines (e.g. McAlpine et al. 2008), underscores the need for additional genomic resources for this species.
The gene PRDM9 directs the majority of meiotic recombination events in humans and most mammals by directing the location of double-stranded breaks in the genome (Baudat et al. 2010; Myers et al. 2010; Parvanov et al. 2010). Nevertheless, PRDM9 is pseudogenized across wild (e.g. Ethiopian wolf, red fox, coyote, and dhole) and domestic canids (Munoz-Fuentes et al. 2011; Axelsson et al. 2012; Mooney et al. 2023) as well as other vertebrate species. Though the mechanisms that direct recombination in canids are unknown, most recombination events tend to be directed toward promoters and GC-rich regions (Auton et al. 2013). More recent work has posited that the loss occurred in the branch leading to Canidae, ∼14–46 MYA (Cavassim et al. 2022). However, the authors did not have access to sequence data from the most basal canid lineage (U. cinereoargenteus) to verify the loss. Given the high quality of our new reference assembly, we sought to determine whether the pseudogenization of PRDM9 occurred before or after the differentiation of Canidae. We built a chromosome-level reference assembly for gray fox from a male individual from Vermont, which can be used as a foundation to answer pressing questions about the phylogenetic relevance of Urocyon as a basal genus within Canidae, its importance in a One Health context, the uncertainty regarding the antiquity of the island fox (Hofman et al. 2016; Sacks et al. 2022), and whether the Eastern and Western clades of North American gray fox may represent distinct species (Goddard et al. 2015; Hofman et al. 2015; Reding et al. 2021). We assessed the quality of our assembly relative to other available canid genomes, contextualized our mitogenome and heterozygosity within available data for Urocyon, and assessed the functionality of the PRDM9 gene. This reference genome expands the possibilities for future studies of gray fox chromosomal architecture, population-level diversity, and disease ecology.
Methods and materials
Sample collection, sequencing, and assembly
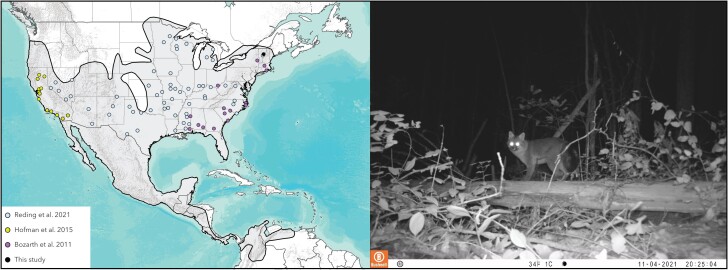
In October 2021, the Vermont Department of Fish and Wildlife obtained a liver sample from a deceased adult male gray fox (U. cinereoargenteus) donated by licensed fur trappers as part of ongoing wildlife health surveillance studies (Fig. 1). The sample was shipped on wet ice to Cantata Bio (Santa Cruz, CA, USA). Liver tissue (100 mg) was combined with 10 ml of G2 buffer + RNase producing 60 µg of DNA after incubation. A Maxi column was used to spin the DNA down, followed by a wash with ethanol. The resulting pellet of DNA was dissolved in TE.
Fig. 1.
Left: Map showing the location of gray fox samples used in some genetic studies and the location of our new reference genome from Vermont, overlaid on the current gray fox range (IUCN). Map sources: Esri, USGS, and NOAA. Right: A gray fox explores a commonly used hiking trail on the Middlebury College campus (photo by Andrew Ng, Middlebury College).
DNA samples were quantified using a Qubit 2.0 Fluorometer (Life Technologies, Carlsbad, CA, USA). The PacBio SMRTbell library (∼20 kb) for PacBio Sequel was constructed using the SMRTbell Express Template Prep Kit 2.0 (Pacific Biosciences, Menlo Park, CA, USA) following the manufacturer-recommended protocol. The library was bound to polymerase using the Sequel II Binding Kit 2.0 (PacBio) and loaded onto the PacBio Sequel II. Sequencing was performed on PacBio Sequel II 8M SMRT cells.
PacBio CCS (124 Gb in total) reads were used as an input to Hifiasm v0.15.4-r347 (Cheng et al. 2021) with default parameters. BLAST (ncbi-blast+/2.11.0; Camacho et al. 2009) results of the Hifiasm output assembly (hifiasm.p_ctg.fa) against the nucleotide database were used as input for blobtools2 v1.1.1 (Laetsch and Blaxter 2017) and contigs identified as possible contamination were removed from the assembly (filtered.asm.cns.fa). Finally, purge_dups v1.2.5 (Guan et al. 2020) was used to remove haplotigs and contig overlaps (purged.fa).
Chromatin from liver samples was fixed in place with formaldehyde in the nucleus and then extracted to construct each Dovetail Omni-C library. Fixed chromatin was digested with DNAse I, and chromatin ends were repaired and ligated to a biotinylated bridge adapter followed by proximity ligation of adapter-containing ends. After proximity ligation, crosslinks were reversed and the DNA was purified. Purified DNA was treated to remove biotin that was not internal to ligated fragments. Sequencing libraries were generated using NEBNext Ultra enzymes and Illumina-compatible adapters. Biotin-containing fragments were isolated using streptavidin beads before PCR enrichment of each library. The library was sequenced on an Illumina HiSeqX platform to produce ∼30× sequence coverage.
The de novo assembly produced by Hifiasm and Dovetail OmniC library reads were used as input data for HiRise (accessed September 2022), a software pipeline designed specifically for using proximity ligation data to scaffold genome assemblies (Putnam et al. 2016). Dovetail OmniC library sequences were aligned to the draft input assembly using BWA-MEM v0.7.17-r1188 (Li and Durbin 2009) The separations of Dovetail OmniC read pairs mapped (Map Quality > 50) within draft scaffolds were analyzed by HiRise to produce a likelihood model for genomic distance between read pairs, and the model was used to identify and break putative misjoins, score prospective joins, and make joins above a threshold.
Assembly quality and continuity
To assess the size distribution of contigs and scaffolds, as well as the quality and continuity of our genome assembly, we used scripts from Assemblathon2 (Bradnam et al. 2013). We compared our assembly to assemblies of other representative canids with available genome assemblies (Table 1).
Table 1.
Assembly statistics for representative published Canidae assemblies and the gray fox genome from this study.
| Species | Assembly accession | 2N | Assembly length | Number of contigs | Contig N50 | Number of scaffolds | Scaffold N50 | Scaffold L90 | Ns |
|---|---|---|---|---|---|---|---|---|---|
|
Domestic dog
Canis lupus familiaris |
GCF_011100685.1 (Wang et al. 2021) | 78 | 2.48 Gb | 2,782 | 14,840,767 | 2,197 | 64,299,765 | 35 | 58.5k |
|
Gray wolf
Canis lupus |
GCA_905319855.2 (Darwin Tree of Life) | 78 | 2.45 Gb | 248 | 34,375,412 | 82 | 65,778,685 | 33 | 45.8k |
|
Dingo
Canis lupus dingo |
GCF_003254725.2 | 78 | 2.35 Gb | 228 | 40,716,615 | 159 | 64,250,934 | 33 | 33.7k |
|
African wild dog
Lycaon pictus |
DNAZoo | 78 | 2.35 Gb | 25,566 | 193,716 | 1,919 | 62,683,413 | 33 | 58.7M |
|
Maned wolf
Chrysocyon brachyurus |
GCA_028533335.1 | 76 | 2.34 Gb | 157,193 | 91,425 | 120,371 | 60,785,871 | 35 | 9.91M |
|
Bush dog
Speothos venaticus |
GCA_023170115.1 | 74 | 2.32 Gb | 139,953 | 44,841 | 52,603 | 111,912 | 21,243 | 22.7M |
|
Raccoon dog
Nyctereutes procyonoides |
GCF_905146905.1 (Lan et al. 2022) | 54 + B | 2.39 Gb | 879 | 35,077,230 | 811 | 53,959,811 | 46 | 500k |
|
Bat-eared fox
Otocyon megalotis |
DNAZoo | 64 | 2.38 Gb | 13,740 | 617,182 | 6,963 | 68,620,662 | 33 | 3.35M |
|
Corsac fox
Vulpes corsac |
GCA_030463245.1 (Zhang et al. 2023) | 36 | 2.35 Gb | 309 | 41,624,634 | 202 | 132,204,642 | 16 | 10.7k |
|
Tibetan sand fox
Vulpes ferrilata |
GCA_024500485.1 (Lyu et al. 2022) | 36 | 2.38 Gb | 379 | 52,909,674 | 234 | 133,960,477 | 16 | 34.1k |
|
Arctic fox
Vulpes lagopus |
GCF_018345385.1 (Peng et al. 2021) | 50 | 2.35 Gb | 1,456 | 33,460,300 | 929 | 131,537,142 | 20 | 52.7k |
|
Red fox
Vulpes vulpes |
DNAZoo | 34 + (0–8)B | 2.42 Gb | 175,019 | 63,228 | 82,277 | 139,030,359 | 16 | 71.8M |
|
Island fox
Urocyon littoralis |
DNAZoo | 66 | 2.54 Gb | 710,357 | 125,395 | 685,071 | 65,046,675 | 21,594 | 9.17 M |
|
Gray fox
Urocyon cinereoargenteus |
This study | 66 | 2.66 Gb | 924 | 59,386,518 | 898 | 72,908,786 | 34 | 2,600 |
We assessed the completeness of genes using the compleasm v0.2.2 program (Huang and Li 2023), which is a faster application of BUSCO (Simão et al. 2015; Waterhouse et al. 2018). Compleasm leverages the BUSCO framework to search genomes for highly conserved sets of orthologs and evaluates whether they are present in the genome in a single copy or are duplicated, fragmented, or missing. We utilized the provided carnivora_odb10 gene set to query the canid genomes.
Finally, we investigated chromosomal rearrangements and chromosomal contiguity. We aligned the genomic sequence of the Gray fox to the Arctic fox (GCA_018345385; Peng et al. 2021) and the domestic dog (GCF_011100685; Wang et al. 2021) using minimap2 v2.24 (Li 2018), followed by visualization with CIRCA v1.2.3 software (https://omgenomics.com/circa/), which allows for easy visualization of syntenic regions between the genomes. Prior to plotting, we used the pafr program v0.0.2 (https://github.com/dwinter/pafr) to read in and filter PAF files and subsequently convert them to CSV in R v4.2.1 (R Core Team 2021). We discarded alignments with a length <25,000 base pairs (bp) and a map quality <60. Alignments were also restricted to autosomes and the X chromosome, as some assemblies were female and others did not have identified Y chromosomes.
Mitochondrial phylogenetics
We used minimap2 to align the PacBio raw reads from U. cinereoargenteus against a reference mitogenome (MW600067.1; Reding et al. 2021). The resulting SAM file was then converted to a BAM file using SAMtools v1.16.1 (Li et al. 2009; Danecek et al. 2021), and a consensus sequence called using ANGSD v0.940 (Korneliussen et al. 2014) with the flags “-doFasta 2” and “-doCounts 1” to produce a consensus mitochondrial genome for the fox. Then, existing complete mitochondrial genome data from U. cinereoargenteus was downloaded from NCBI genbank (N = 102, NCBI Popset 2239967418 and 765367839; Hofman et al. 2015; Reding et al. 2021). The FASTA files were concatenated alongside the consensus mitogenome of the gray fox sampled in this study. The resulting FASTA file underwent multiple sequence alignment using the MAFFT v7.515 program (Katoh and Standley 2013). The output was an alignment file (.aln) which was used to generate a Bayesian phylogenetic tree with the outgroup as the root of the tree using IQ-TREE v1.6.12 (Minh et al. 2020) with flags “-b 100” and “-m TEST”, which run 100 bootstrap replicates and find the best nucleotide substitution model, respectively. The tree was subsequently visualized using FigTree v1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/), rooted in the Western clade, and colored according to the region of origin.
Heterozygosity
All additional U. cinereoargenteus and U. littoralis whole-genome sequences available at the time of this study (January 2023) were obtained from NCBI (Robinson et al. 2016, 2018) bioprojects PRJNA312115 and PRJNA478450. Short-read data were mapped to the gray fox genome assembly using BWA-MEM and converted to BAM format, sorted, and indexed using SAMtools. We estimated the site frequency spectrum (SFS) using ANGSD by inputting the BAM files, along with flags “-anc”, “-ref”, “-fold 1”, “-dosaf 1”, “-GL 2”, “-C 50”, “-minq 20”, and “-minmapq 30”. The reference gray fox genome was provided for the “-anc” and “-ref” flags. The “-fold” flag was used to assign the reference sequence as the ancestral allele and fold the SFS. Flags “-C 50”, “-minq 20”, and “-minmapq 30” were used to remove reads and bases with low mapping and genotype call qualities. Subsequently, we ran realSFS (within ANGSD) and estimated the number of heterozygotes using the folded spectra in compliance with ANGSD guidelines (see http://www.popgen.dk/angsd/index.php/Heterozygosity).
Repetitive elements
We estimated repeat content in our de novo gray fox assembly as well as other representative canid genomes (Table 1) using TETools v1.7 (Lerat et al. 2017). For each genome, we first built a database using the BuildDatabase command. Subsequently, we used the RepeatModeler command, which uses the RepeatModeler v2.0.2 tool (Flynn et al. 2020) to locate and annotate repeats de novo in sequences and genome assemblies. Last, we used the command RepeatMasker v4.1.4 (Flynn et al. 2020) within TETools, which takes the output from RepeatModeler, performs additional screening for repeats within the genome based on homology using the dfam database v3.6 (Storer et al. 2021), and outputs a summary of the repeats in the input sequence.
Functionality of PRDM9
We used Seqtk v1.3-r117 (https://github.com/lh3/seqtk) to extract the human PRDM9 gene sequence from the composite sequence dataset used in Mooney et al. (2023). Then, we used this human PRDM9 sequence data to locate the PRDM9 ortholog within the gray fox genome using BLAST (ncbi-blast+ v2.7.1). We identified the likely start and end of the PRDM9 gene within the gray fox genome and used Seqtk to extract this sequence, adding ∼10 kb of buffer on either side to ensure that the totality of the gene sequence was extracted. Then, we concatenated the gray fox PRDM9-like sequence back into a dataset from Mooney et al. (2023), which contained PRDM9 sequences from the human, Ethiopian wolf (Canis simensis), two populations of gray wolf (Arctic wolf, Isle Royale wolf; Canis lupus), and a number of domestic dog breeds including pug, labrador retriever, Tibetan mastiff, and border collie. We used MAFFT to align all sequences from the new concatenated file, searching for any loss-of-function mutations in our alignment to reveal information about the functionality of PRDM9 in gray foxes. Finally, GeneWise (Birney et al. 2004, https://www.ebi.ac.uk/Tools/psa/genewise/) was used to align the DNA sequence from gray fox to human PRDM9 protein sequence, which allowed us to visually identify any insertions or frameshift errors (see Supplementary Fig. 1). A file with all nucleotide sequences can be found on the GitHub repository.
Results and discussion
Assembly quality and continuity
We assembled a genome for a male gray fox using PacBio HiFi and Dovetail OmniC data. The final assembly totaled 2,658,766,243 bp in length across 898 scaffolds. The contig and scaffold L90 were 51 and 34, respectively, and the contig and scaffold N50 were 59.4 and 72.9 Megabases, respectively (Table 1). Given the statistics of the final assembly, the gray fox genome was of notably better or of equal quality compared to the other canid assemblies (Table 1), despite karyotypic differences. The scaffold L90 was close to the total number of chromosomes expected in the gray fox (32 autosomes and an X and Y chromosome; Graphodatsky et al. 2008), indicating that it is likely that most autosomes and the X chromosome are contained in approximately one scaffold.
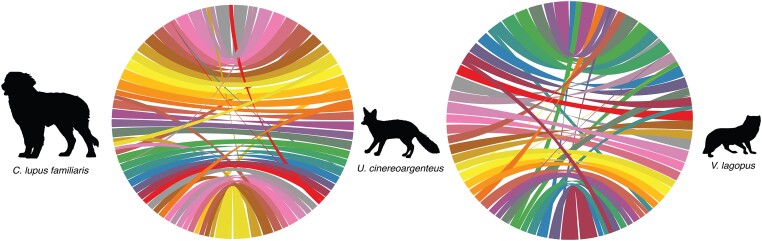
We identified linkage groups using whole-genome alignment in conjunction with information obtained from previous physical mapping studies (Graphodatsky et al. 2008). Whole-genome alignments confirmed that the expected linkage groups for all autosomes and the X chromosome were present (Fig. 2). As the Y chromosome is difficult to assemble due to its repetitive nature (Burgoyne 1982), we did not identify any Y chromosome scaffolds or contigs as part of this work; there are only limited and incomplete representations of canid Y chromosomes that are publicly available. However, we were able to observe multiple autosomal fissions and fusions relative to the domestic dog karyotype which have been previously identified in karyotypic studies (Fig. 2; Nie et al. 2012; Perelman et al. 2012).
Fig. 2.
Whole-genome alignments for a) the domestic dog (GCF_011100685.1) and gray fox and b) the Arctic fox (GCF_018345385.1) and the gray fox. Colored ribbons originate on the gray fox chromosomes (center), so that each unique color is assigned to one gray fox chromosome. In each alignment, the X chromosome is positioned at the bottom of the alignment.
We also assessed the quality of the genome assembly using compleasm (Huang and Li 2023). Broadly, canid assemblies scored relatively high with most assemblies having over 90% of the genes queried in complete and single copies, with the exception of the African wild dog and the bush dog (Table 2). The gray fox genome assembled here had the highest single-copy score of any canid genome assembly assessed, which confirms that most gene regions were assembled well.
Table 2.
Compleasm results for representative published Canidae assemblies and the gray fox genome from this study.
| Species | Single | Duplicate | Fragmented | Incomplete | Missing |
|---|---|---|---|---|---|
|
Domestic dog
Canis lupus familiaris |
97.39% | 1.73% | 0.41% | 0.00% | 0.47% |
|
Gray wolf
Canis lupus |
97.59% | 1.54% | 0.41% | 0.00% | 0.46% |
|
Dingo
Canis lupus dingo |
97.63% | 1.5% | 0.41% | 0.00% | 0.45% |
|
African wild dog
Lycaon pictus |
87.57% | 1.41% | 0.77% | 0.00% | 10.26% |
|
Maned wolf
Chrysocyon brachyurus |
96.61% | 0.97% | 0.97% | 0.00% | 1.45% |
|
Bush dog
Speothos venaticus |
81.51% | 1.27% | 6.89% | 0.01% | 10.67% |
|
Raccoon dog
Nyctereutes procyonoides |
96.60% | 2.38% | 0.42% | 0.00% | 0.60% |
|
Bat-eared fox
Otocyon megalotis |
96.21% | 1.3% | 0.87% | 0.00% | 1.63% |
|
Corsac fox
Vulpes corsac |
97.52% | 1.57% | 0.41% | 0.00% | 0.50% |
|
Tibetan sand fox
Vulpes ferrilata |
97.48% | 1.60% | 0.41% | 0.00% | 0.51% |
|
Arctic fox
Vulpes lagopus |
96.96% | 1.78% | 0.47% | 0.00% | 0.79% |
|
Red fox
Vulpes vulpes |
97.01% | 1.36% | 0.78% | 0.00% | 0.86% |
|
Island fox
Urocyon littoralis |
94.12% | 1.70% | 1.28% | 0.00% | 2.90% |
|
Gray fox
Urocyon cinereoargenteus |
97.70% | 1.28% | 0.41% | 0.00% | 0.62% |
Repeat elements
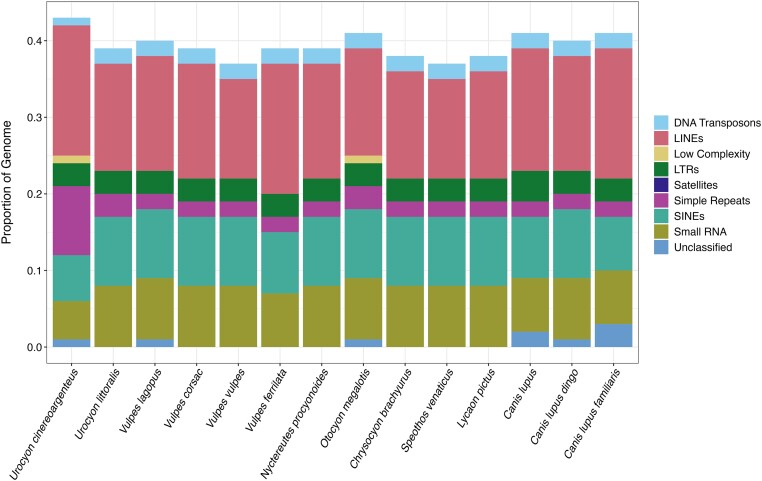
We found that the quantity and classification of repeat elements were relatively similar across the Canidae family. However, gray foxes—but not island foxes—have an increased number of simple sequence repeats (SSRs) compared to Arctic foxes, dogs, gray wolves, and red foxes (Fig. 3). SSRs, also known as microsatellites, are short repeating motifs of 6 bp or less. SSRs are generally thought to evolve through the process of slippage (Schlötterer and Tautz 1992; Ellegren 2004) and have been broadly leveraged to query the diversity and identity of individuals across the tree of life through microsatellite typing (Schlötterer and Pemberton 1994; Wright and Bentzen 1995). They have been implicated in many processes, such as transcription regulation (Hancock and Simon 2005; Kashi and King 2006) and disease (Hancock and Simon 2005), and expansions of these elements may also contribute broadly to genomic instability (Khristich and Mirkin 2020).
Fig. 3.
Repeat element proportions for members of the Canidae family with reference genome assemblies. Species are ordered phylogenetically. Proportions correspond to the proportion of bp in each repeat category as identified by TETools.
It is unclear whether the increased number of SSRs in the gray fox is the ancestral state and SSRs were subsequently lost in the remainder of the canids, or whether the expansion was more recent, at least after the establishment of the western gray fox or island fox lineage. Additionally, the island fox genome assembly, despite being in chromosomes, has a very low contig N50 since it was assembled using short-read data (Table 1). It is thus unclear if the absence of SSRs in the island fox genome assembly (or the difference relative to what we found in the gray fox) is due to an assembly artifact. However, previous studies comparing genomes which have been assembled using short-read data (similar to the island fox genome assembly) have in general suggested that even highly fragmented assemblies will yield accurate repeat content statistics (Armstrong et al. 2020). Additional investigation into the timing of the SSR expansion, and whether it is indeed absent in the island species, may help to elucidate the timing of diversification of the lineages and provide information on the canid ancestral genome.
Fossil record, range, and phylogeographic patterns
The genus Urocyon extends back into the Pliocene (Hemiphillian North American Land Mammal Age) (Kurten and Anderson 1980). Shifts in glacial/interglacial cycles (e.g. Sangamonian, Wisconsin) could have resulted in range expansions and contractions, events which could be used to test contemporary genomic structure and dispersal patterns. For example, our observations of heterozygosity (see below) are consistent with the hypothesis that there was a recent expansion of gray foxes from a southern refugium into the northeast post-Pleistocene (Bozarth et al 2011); however, additional samples from populations in the eastern and northeastern United States have not been explored, even in recent papers which generated genome-level population data for the species (Kierepka et al. 2023; Preckler-Quisquater et al. 2023).
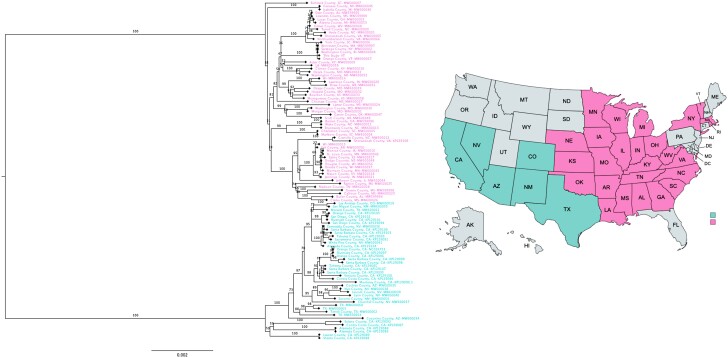
Our mitochondrial phylogeny recovered a division between the eastern and western haplotypes of gray fox, consistent with previous results (Goddard et al. 2015; Reding et al. 2021; Kierepka et al. 2023; Fig. 4). The consensus mitochondrial genome for the Vermont gray fox sample fell within the eastern clade and clustered with the single other Vermont mitochondrial sample available from the literature (Fig. 4). We observed no intermixing between samples originating from the eastern and western United States, supporting previous evidence that admixture between groups is likely limited (Bozarth et al. 2011; Reding et al. 2021). In several recent studies, novel genomic data revealed a contact zone between the eastern and western clades in the plains region in Texas and Oklahoma (Kierepka et al. 2023, Preckler-Quisquater et al. 2023). These studies did not contain samples from most of the northeastern range of the foxes outside of some samples in Tennessee and South Carolina, which will be important in understanding the range of wide diversity and structure of the gray fox. Future analyses using this reference genome, combined with a reference genome recently released for the Santa Catalina island fox (U. littoralis catalinae) (Hendricks et al. 2022), will be important for understanding the taxonomic relationship between these two lineages, with consequences for species delimitation and management.
Fig. 4.
Phylogenetic tree generated from an analysis of 104 unique U. cinereoargenteus mitogenome haplotypes. Coloring of the sample codes reflect the location origin of the sample. Turquoise = Western United States (N = 42), Pink = Eastern United States (N = 61).
The natural history literature contains a number of contrasting statements regarding the baseline range of gray foxes in the northeastern United States, particularly New England. Some have hypothesized that gray fox populations declined extensively in the 19th century and has since been followed by expansions in the past 50 years linked to a warming climate (Banfield 1974; Finley and Godin 1978). Recent genomic analyses did not investigate the most northeastern parts of the gray fox range, thus overlooking archaeological and paleontological records from this region. For example, in 1635 Ce, pilgrims in Massachusetts described “two or three kinds of fox, one a great yellow Fox, another Grey, who will climb up into trees” (Keay 1901) and is known from the precontact zooarchaeological record of Martha's Vineyard (∼400–1,100 years before present) (Huntington 1959). By 1931 CE, the “Mammals of Hampshire County, Massachusetts” noted that the species is common but was perceived as less common than the red fox because of its dense forest association and less desirable fur (Crane 1931).
However, not all areas of New England have a long record of gray fox presence. Moving northward, the “Notes on New Hampshire Mammals” does not include the gray fox in the list of native mammals occurring from 1915 to 1920 Ce (Jackson 1922). The earliest gray fox pelt in a museum collection from Vermont is from 1910 Ce (MCZ:Mamm:64310). Osgood (1938) noted in “The Mammals of Vermont” that the subspecies U. c. borealis “reaches its northern limit in Vermont”, with the farthest north occurrence considered in Whiting (Champlain Valley) in western Vermont and Woodstock in eastern Vermont and, by 1938, reported confirmed skulls from Rutland and Springfield. In contrast with Massachusetts, the gray fox is not present in the zooarchaeological record of Vermont, despite the Holocene presence of other carnivores such as red foxes, fishers, and martens (Mychajliw et al. 2023). The hypothesis that gray foxes are a relatively new addition to the canid community of Vermont, starting in the early 20th century, has yet to be tested with genomic data.
Heterozygosity
The Vermont gray fox had slightly lower levels of heterozygosity compared to those known from California (Table 3). Congruent with previous results (Robinson et al. 2016, 2018), we found that gray foxes exhibited higher levels of heterozygosity than island foxes restricted to the California Channel Islands (U. littoralis) (Table 3). These results are consistent with a recent study showing that gray fox populations east of the contact zone had significantly lower heterozygosity than foxes from areas to the west of the contact zone (Preckler-Quisquater et al. 2023) and that diversity decreased with increasing latitude (Kierepka et al. 2023). Our preliminary results here suggest that diversity may only be slightly lower at these expansion fronts. Though our sample sizes limit our ability to make inferences about the patterns observed, they reinforce the need for sampling gray foxes across their northern and eastern range extents to fully reconstruct possible ancient refugia and recent expansion patterns.
Table 3.
Mean heterozygosity of gray (U. cinereoargenteus) and island fox (U. littoralis) as estimated in ANGSD.
| Species | Location | Sample size | Mean heterozygosity per bp | Interquartile range |
|---|---|---|---|---|
| U. cinereoargenteus | Vermont | 1 | 0.002126 | N/A |
| U. cinereoargenteus | California | 2 | 0.002277 | 8.315e-07 |
| U. littoralis | San Miguel Island | 2 | 0.000417 | 5.55805e-05 |
| U. littoralis | Santa Rosa Island | 3 | 0.001840 | 0.001684175 |
| U. littoralis | Santa Cruz Island | 2 | 0.000835 | 3.108e-06 |
| U. littoralis | San Nicolas Island | 3 | 0.000635 | 0.000342932 |
| U. littoralis | Santa Catalina Island | 2 | 0.001333 | 0.000163839 |
| U. littoralis | San Clemente Island | 2 | 0.000655 | 0.0001764175 |
Functionality of PRDM9
Using our new reference genome, we identified four frameshift mutations and an additional stop codon in the zinc-finger region of PRDM9 in the gray fox using GeneWise (Supplementary Fig. 1). Recent work on examining PRMD9 functionality demonstrated that PRDM9 function was lost across all wolf-like canids, including both the Ethiopian wolf and Dhole (Mooney et al. 2023), but the loss had yet to be confirmed in other canids, including the gray fox. Prior to this work, Axelson and colleagues had postulated that PRDM9 was pseudogenized across all of Canidae, but their study lacked sequence data from the most divergent lineage, Urocyon (Axelsson et al. 2012). Our data contribute to the existing strong evidence that PRDM9 pseudogenization occurred at least 9–10 MYA prior to the divergence of Urocyon and the remainder of the modern canid family (Lindblad-Toh et al. 2005; Eizirik et al. 2010; Matzke and Wright 2016).
Relevance to conservation
In the United States, the gray fox (U. cineoargenteus) is managed as a furbearing mammal and is regularly harvested in many states. Within Vermont, gray foxes are listed as an S5 common species with both open hunting and trapping seasons in the fall-winter, though they are not as commonly trapped as other canids. The gray fox is also involved in human–wildlife conflict: over 1,000 gray foxes across the United States were killed/euthanized by the USDA APHIS Wildlife Services program in 2022 alone (Program Data Report G, 2022; https://www.aphis.usda.gov/aphis/ourfocus/wildlifedamage/pdr/?file=PDR-G_Report&p=2022:INDEX:). Given their expansion into suburban and urban areas and co-occurrence with multiple canids including domestic dogs, gray foxes may serve as models to understand the ecology and genomic basis of virus transmission and disease susceptibility in mesocarnivores (Henn et al. 2007). For example, a distinct clade of canine distemper virus in New England is now shared across multiple carnivore species, including the gray fox (Needle et al. 2019).
The gray fox's wide geographic range and apparent population stability is contrasted by reports of regional declines in the Midwest (Bluett 2006; Willingham 2008; Alessi et al. 2012) and inconsistencies in fur harvest records (Bauder et al. 2020). A recent petition called for the listing of the prairie gray fox subspecies (though, we note prior that these subspecies designations are not supported by genetic data), U. c. ocythous, under the Endangered Species Act because of declines across its range in Iowa, Arkansas, Missouri, and Minnesota (https://www.fws.gov/species-publication-action/90-day-finding-petition-list-prairie-gray-fox-plains-spotted-skunk-and) (Department of the Interior, 2012; https://www.federalregister.gov/documents/2012/12/04/2012-29188/endangered-and-threatened-wildlife-and-plants-90-day-finding-on-a-petition-to-list-the-prairie-gray). A dearth of data across their range, both at comparable scales and types, hinders our ability to contextualize regional patterns to see a larger picture for this species (Allen et al. 2021). Such regional declines may be driven by interference competition and intraguild predation with expanding coyotes (Egan et al. 2021). Gray foxes may have different tolerances to human activities and landscape alterations, with the advantage of a generalist diet outweighed by its more specific habitat needs (Morin et al. 2022), and may be extirpated where it cannot shift its spatial resource use (Levi and Wilmers 2012). The presence of domestic dogs may also exacerbate this competitive tension, as gray foxes shift their diel activity patterns and decrease in abundance where dogs are present (Royle and Nichols 2003). Tree cover is likely important for facilitating coexistence with increasing coyote populations, particularly, in suburban areas. Parsons et al. (2022) suggest a management benchmark of 50% forest cover in a 1 km radius to allow for the coexistence of gray foxes and coyotes (Parsons et al. 2022).
The gray fox is currently listed by the International Union for the Conservation of Nature (IUCN) as Least Concern (Roemer and Cypher 2016), but such a listing could change if taxonomic revisions are made based on additional genomic research. Already, the 0.8 MY divergence timing of eastern and western gray fox clades is deeper than most intraspecific splits of other North American carnivores, such as black bears (Puckett et al. 2015). This divergence time exceeds that of interspecific splits within multiple genera in Carnivora, including the gray fox and Channel Island fox (Hofman et al. 2016; Sacks et al. 2022), as well as within two genera of South American canids, the genus Dusicyon (including the Falkland Islands wolf) (Austin et al. 2013) and the genus Lycalopex (including the culpeo and Darwin's fox) (Favarini et al. 2022). The gray fox has 16 subspecies described based on morphological features (Fritzell and Haroldson 1982), but mitochondrial haplotype data have consistently disagreed with these subspecies designations, instead suggesting more cryptic divergence patterns (Reding et al. 2021) and the potential for management unit revaluation under legislation such as the Endangered Species Act in the United States. Within Canada (where gray foxes are thought to have recently expanded), the species is currently listed as threatened under the federal Species at Risk Act. In fact, Pelee Island, Ontario, contains Canada's only breeding population, with fewer than 300 individuals (McAlpine et al. 2008). Our reference genome provides an important tool in facilitating whole-genome, nuclear studies to resolve discrepancies between morphological and mitochondrial DNA to the benefit of conserving and managing this species across its range.
Supplementary Material
Acknowledgments
We would like to thank David Guertin for assistance with the high-performance computing cluster (“Ada”) at Middlebury College; this material is based upon the work supported by the National Science Foundation under Grant No. 1827373. We thank M. Daly and T. Swale of Cantata Bio for their contributions and facilitation of this work. We thank Dave Allen and the Middlebury Department of Biology for facilitating the Praxis course for January term students. Last, we are indebted to Alex Feltus, David Clark, and the Praxis AI team for support during the course. Figure 2 depiction of gray fox created by Gabriela Palomo-Munoz (CC BY-NC 3.0), courtesy of phylopic.org.
Contributor Information
Ellie E Armstrong, School of Biological Sciences, Washington State University, Pullman, WA 99164, USA.
Ky L Bissell, Department of Biology, Middlebury College, Middlebury, VT 05753, USA.
H Sophia Fatima, Department of Biology, Middlebury College, Middlebury, VT 05753, USA.
Maya A Heikkinen, Department of Biology, Middlebury College, Middlebury, VT 05753, USA.
Anika Jessup, Department of Biology, Middlebury College, Middlebury, VT 05753, USA.
Maryam O Junaid, Department of Biology, Middlebury College, Middlebury, VT 05753, USA.
Dong H Lee, Department of Biology, Middlebury College, Middlebury, VT 05753, USA.
Emily C Lieb, Department of Biology, Middlebury College, Middlebury, VT 05753, USA.
Josef T Liem, Department of Biology, Middlebury College, Middlebury, VT 05753, USA.
Estelle M Martin, Department of Biology, Middlebury College, Middlebury, VT 05753, USA.
Mauricio Moreno, Department of Biology, Middlebury College, Middlebury, VT 05753, USA.
Khuslen Otgonbayar, Department of Biology, Middlebury College, Middlebury, VT 05753, USA.
Betsy W Romans, Department of Biology, Middlebury College, Middlebury, VT 05753, USA.
Kim Royar, Vermont Department of Fish and Wildlife, Montpelier, VT 05620, USA.
Mary Beth Adler, Vermont Department of Fish and Wildlife, Montpelier, VT 05620, USA.
David B Needle, Department of Molecular, Cellular, and Biomedical Sciences, University of New Hampshire, Durham, NH 03824, USA.
Alex Harkess, HudsonAlpha Institute for Biotechnology, Huntsville, AL 35806, USA.
Joanna L Kelley, School of Biological Sciences, Washington State University, Pullman, WA 99164, USA; Department of Ecology and Evolutionary Biology, University of California, Santa Cruz, Santa Cruz, CA 95064, USA.
Jazlyn A Mooney, Department of Quantitative and Computational Biology, University of Southern California, Los Angeles, CA 90007, USA.
Alexis M Mychajliw, Department of Biology, Middlebury College, Middlebury, VT 05753, USA; Program in Environmental Studies, Middlebury College, Middlebury, VT 05753, USA.
Data availability
All code used to process the genome can be found at https://github.com/ellieearmstrong/GrayFox_Middlebury/. Raw reads and assembly files are available under NCBI Bioproject PRJNA1005958. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables.
Supplemental material available at G3 online.
Funding
The support for this project was provided by Revive and Restore, Cantata Bio, and Dovetail Genomics as part of an “AG4” program award to EEA, JLK, and AMM. EEA was supported by a Washington Research Foundation Postdoctoral Fellowship.
Author contributions
EMM, JTL, ECL, HSF, KLB, MM, AJ, WHL, BWR, MAH, KO, and MAJ performed research and analyzed data with the guidance of EEA and AH. JAM provided data and advice for PRDM9 analyses. EEA, JAM, and AMM designed research, performed research, analyzed data, and wrote the paper. KR, MBA, and DN contributed samples and provided guidance on the project and JLK provided guidance on the project.
Literature cited
- Alessi MG, Campbell LK, Miller CA. 2012. 2011–2012 Illinois hunter harvest report. Job completion report, federal AID in wildlife restoration W-112-R-21. Champaign (IL): Illinois Natural History Survey. Human Dimensions Research Program Report HR-12-01/INHS Technical Report (23). https://publish.illinois.edu/human-dimensions/files/2021/01/2011-12-Illinois-Hunter-Harvest-Results.pdf
- Allen ML, Avrin AC, Farmer MJ, Whipple LS, Alexander EP, Cervantes AM, Bauder JM. 2021. Limitations of current knowledge about the ecology of grey foxes hamper conservation efforts. J Threat Taxa. 13(8):19079–19092. doi: 10.11609/jott.7102.13.8.19079-19092. [DOI] [Google Scholar]
- Armstrong EE, Taylor RW, Miller DE, Kaelin CB, Barsh GS, Hadly EA, Petrov D. 2020. Long live the king: chromosome-level assembly of the lion (Panthera leo) using linked-read, Hi-C, and long-read data. BMC Biol. 18(1):1–14. doi: 10.1186/s12915-019-0734-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin JJ, Soubrier J, Prevosti FJ, Prates L, Trejo V, Mena F, Cooper A. 2013. The origins of the enigmatic Falkland Islands wolf. Nat Commun. 4(1):1552. doi: 10.1038/ncomms2570. [DOI] [PubMed] [Google Scholar]
- Auton A, Rui Li Y, Kidd J, Oliveira K, Nadel J, Holloway JK, Hayward JJ, Cohen PE, Greally JM, Wang J, et al. 2013. Genetic recombination is targeted towards gene promoter regions in dogs. PLoS Genet. 9(12):e1003984. doi: 10.1371/journal.pgen.1003984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelsson E, Webster MT, Ratnakumar A, Consortium LUPA, Ponting CP, Lindblad-Toh K. 2012. Death of PRDM9 coincides with stabilization of the recombination landscape in the dog genome. Genome Res. 22(1):51–63. doi: 10.1101/gr.124123.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banfield AWF. 1974. The Mammals of Canada. Toronto: University of Toronto Press. [Google Scholar]
- Baudat F, Buard J, Grey C, Fledel-Alon A, Ober C, Przeworski M, Coop G, De Massy B. 2010. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science. 327(5967):836–840. doi: 10.1126/science.1183439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauder JM, Allen ML, Ahlers AA, Benson TJ, Miller CA, Stodola KW. 2020. Identifying and controlling for variation in Canid harvest data. J Wildl Manage. 84(7):1234–1245. doi: 10.1002/jwmg.21919. [DOI] [Google Scholar]
- Birney E, Clamp M, Durbin R. 2004. GeneWise and Genomewise. Genome Res. 14(5):988–995. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bluett B. Archery deer hunter survey. Illinois Department of Natural Resources Wildlife Diversity Program Note. 2006. 06-4.
- Bozarth CA, Lance SL, Civitello DJ, Glenn JL, Maldonado JE. 2011. Phylogeography of the gray fox (Urocyon cinereoargenteus) in the eastern United States. J Mammal. 92(2):283–294. doi: 10.1644/10-MAMM-A-141.1. [DOI] [Google Scholar]
- Bradnam KR, Fass JN, Alexandrov A, Baranay P, Bechner M, Birol I, Boisvert S, Chapman JA, Chapuis G, Chikhi R, et al. 2013. Assemblathon 2: evaluating de novo methods of genome assembly in three vertebrate species. Gigascience. 2(1):10. doi: 10.1186/2047-217X-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgoyne PS. 1982. Genetic homology and crossing over in the X and Y chromosomes of mammals. Hum Genet. 61(2):85–90. doi: 10.1007/BF00274192. [DOI] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinform. 10(1):1–9. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavassim MIA, Baker Z, Hoge C, Schierup MH, Schumer M, Przeworski M. 2022. PRDM9 losses in vertebrates are coupled to those of paralogs ZCWPW1 and ZCWPW2. Proc Natl Acad Sci U S A. 119(9):e2114401119. doi: 10.1073/pnas.2114401119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Concepcion GT, Feng X, Zhang H, Li H. 2021. Haplotype-resolved de novo assembly using phased assembly graphs with Hifiasm. Nat Methods. 18(2):170–175. doi: 10.1038/s41592-020-01056-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane J. 1931. Mammals of Hampshire County, Massachusetts. J Mammal. 12(3):267–273. doi: 10.2307/1373876. [DOI] [Google Scholar]
- Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, Whitwham A, Keane T, McCarthy SA, Davies RM, et al. 2021. Twelve years of SAMtools and BCFtools. Gigascience. 10(2):giab008. doi: 10.1093/gigascience/giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan ME, Day CC, Katzner TE, Zollner PA. 2021. Relative abundance of coyotes (Canis latrans) influences gray fox (Urocyon cinereoargenteus) occupancy across the eastern United States. Can J Zool. 99(2):63–72. doi: 10.1139/cjz-2019-0246. [DOI] [Google Scholar]
- Eizirik E, Murphy WJ, Koepfli K-P, Johnson WE, Dragoo JW, Wayne RK, O’Brien SJ. 2010. Pattern and timing of diversification of the mammalian order Carnivora inferred from multiple nuclear gene sequences. Mol Phylogenet Evol. 56(1):49–63. doi: 10.1016/j.ympev.2010.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H. 2004. Microsatellites: simple sequences with complex evolution. Nat Rev Genet. 5(6):435–445. doi: 10.1038/nrg1348. [DOI] [PubMed] [Google Scholar]
- Favarini MO, Simão TLL, Macedo GS, Garcez FS, Oliveira LR, Cárdenas-Alayza S, Cardeña Mormontoy M, Angulo F, Kasper CB, Johnson WE, et al. 2022. Complex evolutionary history of the South American fox genus Lycalopex (Mammalia, Carnivora, Canidae) inferred from multiple mitochondrial and nuclear markers. Diversity (Basel). 14(8):642. doi: 10.3390/d14080642. [DOI] [Google Scholar]
- Finley RB, Godin AJ. 1978. Wild mammals of New England. J Range Manage. 31(4):319. doi: 10.2307/3897615. [DOI] [Google Scholar]
- Flynn JM, Hubley R, Goubert C, Rosen J, Clark AG, Feschotte C, Smit AF. 2020. RepeatModeler2 for automated genomic discovery of transposable element families. Proc Natl Acad Sci U S A. 117(17):9451–9457. doi: 10.1073/pnas.1921046117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formenti G, Theissinger K, Fernandes C, Bista I, Bombarely A, Bleidorn C, Ciofi C, Crottini A, Godoy JA, Höglund J, et al. 2022. The era of reference genomes in conservation genomics. Trends Ecol Evol. 37(3):197–202. doi: 10.1016/j.tree.2021.11.008. [DOI] [PubMed] [Google Scholar]
- Fritzell EK, Haroldson KJ. 1982. Urocyon cinereoargenteus. Mamm Species. 189(189):1–8. doi: 10.2307/3503957. [DOI] [Google Scholar]
- Goddard NS, Statham MJ, Sacks BN. 2015. Mitochondrial analysis of the most basal canid reveals deep divergence between eastern and western North American gray foxes (Urocyon spp.) and ancient roots in Pleistocene California. PLoS One. 10(8):e0136329. doi: 10.1371/journal.pone.0136329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graphodatsky AS, Perelman PL, Sokolovskaya NV, Beklemisheva VR, Serdukova NA, Dobigny G, O’Brien SJ, Ferguson-Smith MA, Yang F, 2008. Phylogenomics of the dog and fox family (Canidae, Carnivora) revealed by chromosome painting. Chromosome Res. 16(1):129–143. doi: 10.1007/s10577-007-1203-5. [DOI] [PubMed] [Google Scholar]
- Guan D, McCarthy SA, Wood J, Howe K, Wang Y, Durbin R. 2020. Identifying and removing haplotypic duplication in primary genome assemblies. Bioinformatics. 36(9):2896–2898. doi: 10.1093/bioinformatics/btaa025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendricks SA, King JL, Duncan CL, Vickers W, Hohenlohe PA, Davis BW. 2022. Genomic assessment of cancer susceptibility in the threatened catalina island fox (Urocyon littoralis catalinae). Genes. 13(8):1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock JM, Simon M. 2005. Simple sequence repeats in proteins and their significance for network evolution. Gene. 345(1):113–118. doi: 10.1016/j.gene.2004.11.023. [DOI] [PubMed] [Google Scholar]
- Henn JB, Gabriel MW, Kasten RW, Brown RN, Theis JH, Foley JE, Chomel BB. 2007. Gray foxes (Urocyon cinereoargenteus) as a potential reservoir of a Bartonella clarridgeiae-like bacterium and domestic dogs as part of a sentinel system for surveillance of zoonotic arthropod-borne pathogens in northern California. J Clin Microbiol. 45(8):2411–2418. doi: 10.1128/JCM.02539-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofman CA, Rick TC, Hawkins MTR, Funk WC, Ralls K, Boser CL, Collins PW, Coonan T, King JL, Morrison SA, et al. 2015. Mitochondrial genomes suggest rapid evolution of dwarf California Channel Islands foxes (Urocyon littoralis). PLoS One. 10(2):e0118240. doi: 10.1371/journal.pone.0118240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofman CA, Rick TC, Maldonado JE, Collins PW, Erlandson JM, Fleischer RC, Smith C, Sillett TS, Ralls K, Teeter W, et al. 2016. Tracking the origins and diet of an endemic island canid (Urocyon littoralis) across 7300 years of human cultural and environmental change. Quat Sci Rev. 146:147–160. doi: 10.1016/j.quascirev.2016.06.010. [DOI] [Google Scholar]
- Huang N, Li H. 2023. Compleasm: a faster and more accurate reimplementation of BUSCO. Bioinformatics. 39:btad595. doi: 10.1093/bioinformatics/btad595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntington EG. 1959. An archaeological study from Martha’s vineyard. Dukes Cty Intell. 1(2):1–21. [Google Scholar]
- Jackson CF. 1922. Notes on New Hampshire mammals. J Mammal. 3(1):13–15. doi: 10.2307/1373445. [DOI] [Google Scholar]
- Kashi Y, King DG. 2006. Simple sequence repeats as advantageous mutators in evolution. Trends Genet. 22(5):253–259. doi: 10.1016/j.tig.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keay FE. 1901. The animals which our fathers found in New England. New England Magazine. 24:535–545. [Google Scholar]
- Khristich AN, Mirkin SM. 2020. On the wrong DNA track: molecular mechanisms of repeat-mediated genome instability. J Biol Chem. 295(13):4134–4170. doi: 10.1074/jbc.REV119.007678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierepka EM, Preckler-Quisquater S, Reding DM, Piaggio AJ, Riley SPD, Sacks BN. 2023. Genomic analyses of gray fox lineages suggest ancient divergence and secondary contact in the southern Great Plains. J Hered. 114(2):110–119. doi: 10.1093/jhered/esac060. [DOI] [PubMed] [Google Scholar]
- Korneliussen TS, Albrechtsen A, Nielsen R. 2014. ANGSD: analysis of next generation sequencing data. BMC Bioinform. 15(1):356. doi: 10.1186/s12859-014-0356-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurten B, Anderson E. 1980. Pleistocene Mammals of North America. New York: Columbia University Press. [Google Scholar]
- Laetsch DR, Blaxter ML. 2017. BlobTools: interrogation of genome assemblies. F1000Res. 6:1287. doi: 10.12688/f1000research.12232.1. [DOI] [Google Scholar]
- Lan T, Li H, Yang S, Shi M, Han L, Sahu SK, Lu Y, Wang J, Zhou M, Liu H, et al. 2022. The chromosome-scale genome of the raccoon dog: insights into its evolutionary characteristics. Iscience. 25(10):105117. doi: 10.1016/j.isci.2022.105117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerat E, Fablet M, Modolo L, Lopez-Maestre H, Vieira C. 2017. TEtools facilitates big data expression analysis of transposable elements and reveals an antagonism between their activity and that of piRNA genes. Nucleic Acids Res. 45(4):e17. doi: 10.1093/nar/gkw953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levi T, Wilmers CC. 2012. Wolves-coyotes-foxes: a cascade among carnivores. Ecology. 93(4):921–929. doi: 10.1890/11-0165.1. [DOI] [PubMed] [Google Scholar]
- Li H. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 34(18):3094–3100. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblad-Toh K, Wade CM, Mikkelsen TS, Karlsson EK, Jaffe DB, Kamal M, Clamp M, Chang JL, Kulbokas EJ, Zody MC. 2005. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 438(7069):803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- Lyu TS, Wei QG, Wang LD, Zhou SY, Shi LP, Dong YH, Dou H-S, Sha W-L, Ga T, Zhang HH. 2022. High-quality chromosome-level genome assembly of Tibetan fox (Vulpes ferrilata). Zool Res. 43(3):362–366. doi: 10.24272/j.issn.2095-8137.2021.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke NJ, Wright A. 2016. Inferring node dates from tip dates in fossil Canidae: the importance of tree priors. Biol Lett. 12(8):20160328. doi: 10.1098/rsbl.2016.0328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAlpine D, Martin JD, Libby C. 2008. First occurrence of the grey fox, Urocyon cinereoargenteus, in New Brunswick: a climate-change mediated range expansion? Can Field Nat. 122(2):169–171. doi: 10.22621/cfn.v122i2.578. [DOI] [Google Scholar]
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37(5):1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney JA, Marsden CD, Yohannes A, Wayne RK, Lohmueller KE. 2023. Long-term small population size, deleterious variation, and altitude adaptation in the Ethiopian wolf, a severely endangered canid. Mol Biol Evol. 40(1):msac277. doi: 10.1093/molbev/msac277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin DJ, Lesmeister DB, Nielsen CK, Schauber EM. 2022. Asymmetrical intraguild interactions with coyotes, red foxes, and domestic dogs may contribute to competitive exclusion of declining gray foxes. Ecol Evol. 12(7):e9074. doi: 10.1002/ece3.9074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Fuentes V, Di Rienzo A, Vila C. 2011. Prdm9, a major determinant of meiotic recombination hotspots, is not functional in dogs and their wild relatives, wolves and coyotes. PLoS One. 6(11):e25498. doi: 10.1371/journal.pone.0025498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mychajliw AM, Hsi AY, An-Pham D, Olson OL, Carder N, Crock JG, Robinson F“J”W. 2023. Zooarchaeological assemblages contextualize the historical ecology and harvest of fur-bearing mammals in Vermont. Front Ecol Evol. 11:1065567. doi: 10.3389/fevo.2023.1065567. [DOI] [Google Scholar]
- Myers S, Bowden R, Tumian A, Bontrop RE, Freeman C, MacFie TS, McVean G, Donnelly P. 2010. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science. 327(5967):876–879. doi: 10.1126/science.1182363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needle DB, Burnell VC, Forzán MJ, Dubovi EJ, Schuler KL, Bernier C, Hollingshead NA, Ellis JC, Stevens BA, Tate P, et al. 2019. Infection of eight mesocarnivores in New Hampshire and Vermont with a distinct clade of canine distemper virus in 2016–2017. J Vet Diagn Invest. 31(4):562–567. doi: 10.1177/1040638719847510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needle DB, Marr JL, Park CJ, Andam CP, Wise AG, Maes RK, Wilkes RP, Anis EA, Sidor IF, Agnew D, et al. 2020. Concurrent infection of skunk Adenovirus-1, Listeria monocytogenes, and a regionally specific clade of canine distemper virus in one gray fox (Urocyon cinereoargenteus) and concurrent listeriosis and canine distemper in a second gray fox. Pathogens. 9(7):591. doi: 10.3390/pathogens9070591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie W, Wang J, Su W, Wang D, Tanomtong A, Perelman PL, Graphodatsky AS, Yang F. 2012. Chromosomal rearrangements and karyotype evolution in carnivores revealed by chromosome painting. Heredity (Edinb). 108(1):17–27. doi: 10.1038/hdy.2011.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyakatura K, Bininda-Emonds ORP. 2012. Updating the evolutionary history of Carnivora (Mammalia): a new species-level supertree complete with divergence time estimates. BMC Biol. 10(1):12. doi: 10.1186/1741-7007-10-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osgood FL. 1938. The mammals of Vermont. J Mammal. 19(4):435–441. doi: 10.2307/1374228. [DOI] [Google Scholar]
- Paez S, Kraus RHS, Shapiro B, Gilbert MTP, Jarvis ED, Al-Ajli FO, Ceballos G, Crawford AJ, Fedrigo O, Johnson RN, et al. 2022. Reference genomes for conservation. Science. 377(6604):364–366. doi: 10.1126/science.abm8127. [DOI] [PubMed] [Google Scholar]
- Parsons AW, Kellner KF, Rota CT, Schuttler SG, Millspaugh JJ, Kays RW. 2022. The effect of urbanization on spatiotemporal interactions between gray foxes and coyotes. Ecosphere. 13(3):e3993. doi: 10.1002/ecs2.3993. [DOI] [Google Scholar]
- Parvanov ED, Petkov PM, Paigen K. 2010. Prdm9 controls activation of mammalian recombination hotspots. Science. 327(5967):835–835. doi: 10.1126/science.1181495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Li H, Liu Z, Zhang C, Li K, Gong Y, Geng L, Su J, Guan X, Liu L, et al. 2021. Chromosome-level genome assembly of the Arctic fox (Vulpes lagopus) using PacBio sequencing and Hi-C technology. Mol Ecol Resour. 21(6):2093–2108. doi: 10.1111/1755-0998.13397. [DOI] [PubMed] [Google Scholar]
- Perelman PL, Beklemisheva VR, Yudkin DV, Petrina TN, Rozhnov VV, Nie W, Graphodatsky AS. 2012. Comparative chromosome painting in Carnivora and Pholidota. Cytogenet Genome Res. 137(2–4):174–193. doi: 10.1159/000341389. [DOI] [PubMed] [Google Scholar]
- Preckler-Quisquater S, Kierepka EM, Reding DM, Piaggio AJ, Sacks BN. 2023. Can demographic histories explain long-term isolation and recent pulses of asymmetric gene flow between highly divergent grey fox lineages? Mol Ecol. 32(19):5323–5337. doi: 10.1111/mec.17105. [DOI] [PubMed] [Google Scholar]
- Puckett EE, Etter PD, Johnson EA, Eggert LS. 2015. Phylogeographic analyses of American black bears (Ursus americanus) suggest four glacial refugia and complex patterns of postglacial admixture. Mol Biol Evol. 32(9):2338–2350. doi: 10.1093/molbev/msv114. [DOI] [PubMed] [Google Scholar]
- Putnam NH, O’Connell BL, Stites JC, Rice BJ, Blanchette M, Calef R, Troll CJ, Fields A, Hartley PD, Sugnet CW, et al. 2016. Chromosome-scale shotgun assembly using an in vitro method for long-range linkage. Genome Res. 26(3):342–350. doi: 10.1101/gr.193474.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . 2021. R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. https://www.R-project.org/. [Google Scholar]
- Reding DM, Castañeda-Rico S, Shirazi S, Hofman CA, Cancellare IA, Lance SL, Beringer J, Clark WR, Maldonado JE. 2021. Mitochondrial genomes of the United States distribution of gray fox (Urocyon cinereoargenteus) reveal a major phylogeographic break at the Great Plains suture zone. Front Ecol Evol. 9:666800. doi: 10.3389/fevo.2021.666800. [DOI] [Google Scholar]
- Robinson JA, Brown C, Kim BY, Lohmueller KE, Wayne RK. 2018. Purging of strongly deleterious mutations explains long-term persistence and absence of inbreeding depression in island foxes. Curr Biol. 28(21):3487–3494.e4. doi: 10.1016/j.cub.2018.08.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JA, Ortega-Del Vecchyo D, Fan Z, Kim BY, von Holdt BM, Marsden CD, Lohmueller KE, Wayne RK. 2016. Genomic flatlining in the endangered island fox. Curr Biol. 26(9):1183–1189. doi: 10.1016/j.cub.2016.02.062. [DOI] [PubMed] [Google Scholar]
- Roemer G, Cypher B. 2016. Urocyon cinereoargenteus. The IUCN red list of threatened species 2016: e.T22780A46178068.
- Royle JA, Nichols JD. 2003. Estimating abundance from repeated presence–absence data or point counts. Ecology. 84(3):777–790. doi: 10.1890/0012-9658(2003)084[0777:EAFRPA]2.0.CO;2. [DOI] [Google Scholar]
- Sacks BN, Statham MJ, Serieys LEK, Riley SPD. 2022. Population genetics of California gray foxes clarify origins of the island fox. Genes (Basel). 13(10):1859. doi: 10.3390/genes13101859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlötterer C, Pemberton J. 1994. The use of microsatellites for genetic analysis of natural populations. In: Schierwater B, Streit B, Wagner GP, DeSalle R, editors. Molecular Ecology and Evolution: Approaches and Applications. Basel: Birkhäuser. p. 203–214. [Google Scholar]
- Schlötterer C, Tautz D. 1992. Slippage synthesis of simple sequence DNA. Nucleic Acids Res. 20(2):211–215. doi: 10.1093/nar/20.2.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 31(19):3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- Storer J, Hubley R, Rosen J, Wheeler TJ, Smit AF. 2021. The Dfam community resource of transposable element families, sequence models, and genome annotations. Mob DNA. 12(1):2. doi: 10.1186/s13100-020-00230-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supple MA, Shapiro B. 2018. Conservation of biodiversity in the genomics era. Genome Biol. 19(1):131. doi: 10.1186/s13059-018-1520-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedford RH, Taylor BE, Wang X. 1995. Phylogeny of the Caninae (Carnivora, Canidae): the living taxa. American Museum Novitates. no. 3146. [Google Scholar]
- Wang C, Wallerman O, Arendt M-L, Sundström E, Karlsson Å, Nordin J, Mäkeläinen S, Pielberg GR, Hanson J, Ohlsson Å, et al. 2021. A novel canine reference genome resolves genomic architecture and uncovers transcript complexity. Commun Biol. 4(1):185. doi: 10.1038/s42003-021-01698-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse RM, Seppey M, Simão FA, Manni M, Ioannidis P, Klioutchnikov G, Kriventseva EV, Zdobnov EM. 2018. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol Biol Evol. 53(3):543–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne RK. 1993. Molecular evolution of the dog family. Trends Genet. 9(6):218–224. doi: 10.1016/0168-9525(93)90122-X. [DOI] [PubMed] [Google Scholar]
- Willingham AN. Emerging factors associated with the decline of a gray fox population and multi-scale land cover associations of mesopredators in the Chicago metropolitan area. [Doctoral dissertation, The Ohio State University]. 2008.
- Wright JM, Bentzen P. 1995. Microsatellites: genetic markers for the future. In: Carvalho GR, Pitcher TJ, editors. Molecular Genetics in Fisheries. Dordrecht: Springer Netherlands. p. 117–121. [Google Scholar]
- Zhang Z, Xia T, Zhou S, Yang X, Lyu T, Wang L, Fang J, Wang Q, Dou H, Zhang H. 2023. High-quality chromosome-level genome assembly of the Corsac fox (Vulpes corsac) reveals adaptation to semiarid and harsh environments. Int J Mol Sci. 24(11):9599. doi: 10.3390/ijms24119599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoonomia Consortium . 2020. A comparative genomics multitool for scientific discovery and conservation. Nature. 587(7833):240–245. doi: 10.1038/s41586-020-2876-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All code used to process the genome can be found at https://github.com/ellieearmstrong/GrayFox_Middlebury/. Raw reads and assembly files are available under NCBI Bioproject PRJNA1005958. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables.
Supplemental material available at G3 online.