Impact statement
Methane oxidizing microbes play a key role in reducing the emission of this potent greenhouse gas to the atmosphere. The known versatility of the recently discovered anaerobic Methylomirabilota methanotrophs is limited. Here, we report a novel uncultured Methylomirabilis species, Candidatus Methylomirabilis iodofontis, with the genetic potential of iodate respiration from biofilm in iodine‐rich cavern spring water. Star‐like cells resembling Methylomirabilis oxyfera were directly observed from the biofilm and a high‐quality metagenome‐assembled genome (MAG) of Ca. M. iodofontis was assembled. In addition to oxygenic denitrification and aerobic methane oxidation pathways, the M. iodofontis MAG also indicated its iodate‐reducing potential, a capability that would enable the bacterium to use iodate other than nitrite as an electron acceptor, a hitherto unrecognized metabolic potential of Methylomirabilota methanotrophs. The results advance the current understanding of the ecophysiology of anaerobic Methylomirabilota methanotrophs and may suggest an additional methane sink, especially in iodate‐rich ecosystems.
Methane oxidizing microbes are essential in controlling methane emissions from various environments. In addition to aerobic methanotrophs within the Proteobacteria and Verrucomicrobiota, anaerobic methantrophic archaea (the ANMEs) and bacteria within the Methylomirabilota (previously NC10 phylum), capable of anaerobic oxidation of methane (AOM), have been discovered during the last two decades. ANME archaea are suggested to oxidize methane via reverse methanogenesis 1 , using different electron acceptors, such as sulfate 2 , iron oxides 3 , nitrate and nitrite 4 , 5 , with or without a syntrophic partner. In contrast, bacteria within the methanotrophic Methylomirabilota oxidize methane via a canonical methane monooxygenase‐dependent aerobic pathway, exclusively using nitrite as electron acceptor 6 , 7 . Methylomirabilota methanotrophs are proposed to generate their own intracellular oxygen supply via nitric oxide (NO) dismutation into O2 and N2, catalyzed by a putative NO dismutase 8 . NO dismutase (nod) genes are widely distributed among diverse microbial lineages 9 . In addition to this peculiar metabolism, Methylomirabilis oxyfera was reported to display a characteristic polygonal cell shape in electron micrographs 10 . However, it remains to be shown whether other Methylomirabilota methanotrophs also show similar morphologies.
The diversity of Methylomirabilota methanotrophs as inferred from functional marker genes, such as particulate methane monooxygenase (pmoA) 11 or nod genes 12 seems limited, especially in comparison to the diversity of Methylomirabilota derived from 16S rRNA sequences 12 , 13 . Hitherto, the dominant bacteria in denitrifying AOM cultures, for example 14 , 15 , 16 , as well as environmental microbes with supposed denitrifying methane‐oxidizing capability 17 , 18 , were all closely related to M. oxyfera. Other environmental metagenome‐assembled genomes (MAGs) affiliated with the Methylomirabilota phylum did not indicate a denitrifying potential linked to methanotrophy 19 , 20 . Recently, denitrifying AOM enrichment cultures containing Methylomirabilota bacteria were reported to reduce selenate 21 or chlorate 22 under methane oxidation. However, there was no direct evidence for the involvement of Methylomirabilota in these processes. Hence, our current understanding of the diversity and metabolic versatility of Methylomirabilota methanotrophs remains very limited.
Here, we report the MAG of a novel Methylomirabilota bacterium, Candidatus Methylomirabilis iodofontis, from methane‐oxidizing biofilms sampled under iodine‐rich mineral water in a subsurface spring cavern in Sulzbrunn, Germany. Iodine‐rich (>20 mg l−1) formation water from the subalpine Lower Marine Molasse enters the spring together with thermogenic methane, which accumulated up to 3000 ppm in the undisturbed microoxic cavern atmosphere 23 . Within the submersed biofilm at the cavern wall, transmission electron microscopy revealed peculiar star‐shaped microbial morphologies, resembling that of M. oxyfera (Figure 1A). In addition, 16S rRNA gene sequences related to that of Methylomirabilis spp. were retrieved via targeted PCR and cloning (Figure S1), consistent with previous results of 16S rRNA gene amplicon sequencing of the respective submersed biofilms, where reads of the Methylomirabilota (NC10) accounted for up to 10% 23 . These lines of evidence all indicate the presence of Methylomirabilis methanotrophs in the cave biofilm.
Figure 1.
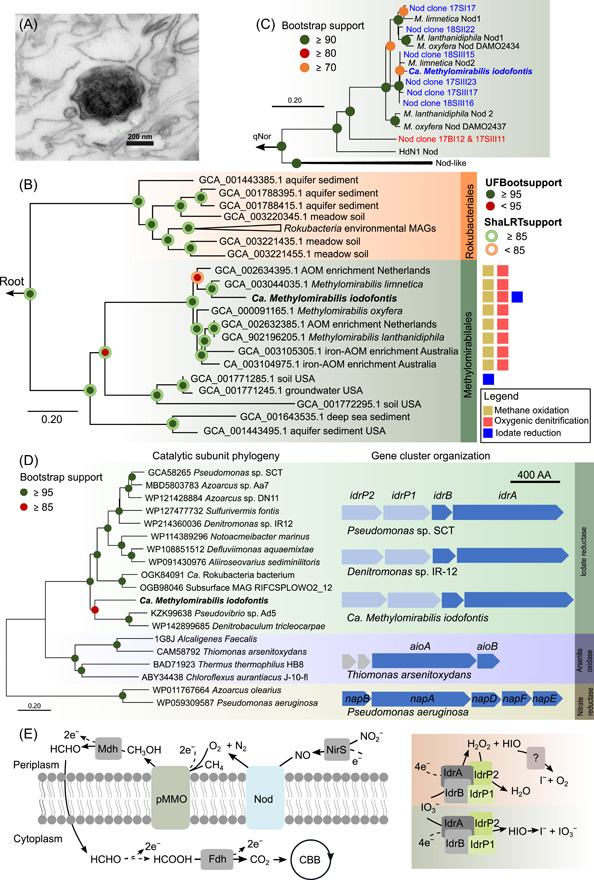
Cell morphology, phylogenetic analysis, gene cluster organization, and key respiratory pathways. (A) TEM image of Methylomirabilis oxyfera‐shaped cell from the submersed biofilm. (B) Phylogenomic analysis of Methylomirabilota phylum bacteria and MAGs, including Methylomirabilis iodofontis and other Methylomirabilis species and Rocubacteriales. (C) Nod phylogenetic tree including cloned Nod sequences from submersed biofilm and assembled Nod in Candidatus Methylomirabilis iodofontis genome. (D) Phylogenetic tree of the catalytic subunit of iodate reductase (IdrA), arsenite oxidase (AioA), and periplasmic nitrate reductase (NapA). IdrA encoded in the M. iodofontis is in bold, and the gene cluster organization of iodate reductase in Pseudomonas sp. SCT, Denitromonas sp. IR‐12 and Ca. M. iodofontis, and arsenite oxidase, nitrate reductase in other microbes are shown. (E) Key respiratory pathways in M. iodofontis according to genetic analysis. Both proposed iodate reduction routes taking place in periplasmic space are illustrated.
Therefore, we sequenced the metagenome of the submersed biofilm and assembled a putative Methylomirabilota genomic bin (bin48), which was over 70% complete and with very low contamination (1.52%) (Table S1). In total, 4780 Methylomirabilota 16S rRNA gene reads were retrieved, accounting for 14.3% of all 16S rRNA reads detected in the metagenomic library, representing one of the most abundant (sub)phylum‐level populations (Table S2). All Methylomirabilota 16S rRNA reads were assembled into one consensus full‐length 16S rRNA gene, which showed >99% similarity to that of Methylomirabilis limnetica (Figure S1). Yet, the pairwise average amino acid identity (AAI) and the average nucleotide identity (ANI) between M. limnetica genome and bin48 were only 85.8% and 91.3%, respectively, suggesting the newly binned MAG to represent a novel Methylomirabilis species, which was tentatively named Ca. M. iodofontis. Phylogenomic analysis based on 121 concatenated protein markers further supported that M. iodofontis was closely related to other Methylomirabilis species, forming a monophyletic clade within the order Methylomirabilales of the Methylomirabilota phylum (Figure 1B).
In the MAG of M. iodofontis, a pyrroloquinoline quinone (PQQ)‐dependent methanol‐dehydrogenase and a formate‐dehydrogenase highly similar to those in M. oxyfera and M. limnetica were also present. However, a particulate methane monooxygenase (pMMO) operon was missing (Table 1), possibly due to the incompleteness of the MAG. The presence of a complete methane‐oxidizing pathway in the MAG was statistically assessed using MetaPOAP 24 , and the false‐positive and false‐negative probabilities were 7.524e−10 and 0.069, respectively, suggesting that the pMMO genes are likely present in the source genome. Moreover, M. iodofontis harbored a complete Calvin−Benson−Bassham (CBB) cycle, except for the Rubisco small unit gene (Table 1), indicating an autotrophic lifestyle like M. oxyfera 25 . The Rubisco large subunit of M. iodofontis clustered closely to that of other Methylomirabilis spp., all falling in the type IC/D group (Figure S2). The high similarity between M. iodofontis and other Methylomirabilis methanotrophs on the whole‐genome level as well as for key methane‐oxidizing enzyme genes (Table 1) also strongly argues for a methane‐oxidizing capability in M. iodofontis. Like other Methylomirabilis species, a complete oxygenic denitrification pathway was present, although a second nod (DAMO2434‐like) gene 12 was not identified in the MAG (Table 1). Yet, nod‐targeted PCR and cloning recovered two Nod clusters as known for other Methylomirabilis spp., and a distantly related Nod (Figure 1C), indicating that the M. iodofontis genome likely also harbors two distinct nod gene homologs. The M. iodofontis Nod possessed all characteristic substitutions known for other Nod sequences (Figure S3). In comparison, reconstructed genomes of other members of the Methylomirabilales 19 , 20 , distantly related to Methylomirabilis spp., neither indicated methane oxidation nor oxygenic denitrification capacities (Figure 1B). Likely, the denitrifying methanotrophic lifestyle is restricted to the genus Methylomirabilis within the Methylomirabilota.
Table 1.
Key metabolic pathways and CBB cycle‐associated genes in Ca. Methylomirabilis iodofontis.
| Pathway | Locus tag | Gene | Enzyme | Similaritya to gene of M. oxyfera (%) | Top hit (similarity)a |
|---|---|---|---|---|---|
| Oxygenic denitrification | bin‐48‐10‐cds15 | napB | Nitrate reductase cytochrome c‐type subunit NapB | 77.0 | M. limnetica (83.5%) |
| bin‐48‐10‐cds16 | napA | Periplasmic nitrate reductase NapA | 89.1 | M. limnetica (92.5%) | |
| bin‐48‐132‐cds1 | nirS | Nitrite reductase (NO‐forming) | 88.5 | M. limnetica (97.8%) | |
| bin‐48‐55‐cds1b | nod | Putative nitric oxide dismutase | 83.8 | M. limnetica (93.4%) | |
| bin‐48‐326‐cds1b | nod | Putative nitric oxide dismutase | 91.0 | M. limnetica (99.3%) | |
| Methane oxidation | Missing | pmoCAB | Particulate methane monooxygenase | ||
| bin‐48‐119‐cds7 | mxaF | Methanol dehydrogenase | 96.4 | M. limnetica (97%) | |
| Missing | fea | Formaldehyde activating enzyme | |||
| bin‐48‐146‐cds2 | fhcD | Formylmethanofuran tetrahydromethanopterin formyltransferase | 93.4 | M. oxyfera (93.4%) | |
| bin‐48‐153‐cds5 | folD | Methylene H4F dehydrogenase | 89.6 | M. limnetica (94.7%) | |
| bin‐48‐154‐cds2 | fdhA | Formate dehydrogenase major subunit | 87.9 | M. limnetica (91.1%) | |
| bin‐48‐7‐cds14 | fdhD | Formate dehydrogenase accessory protein | 89.2 | M. limnetica (93.3%) | |
| CBB cycle | bin‐48‐50‐cds5 | rbcL | Ribulose bisphosphate carboxylase, large chain, N‐terminal | 87.5 | M. limnetica (92.4%) |
| bin‐48‐99‐cds1 | rbcL | Ribulose‐bisphosphate carboxylase, large chain | 96.9 | M. limnetica (97.6%) | |
| Missing | rbcS | Ribulose‐bisphosphate carboxylase, small chain | |||
| bin‐48‐166‐cds1 | pgk | Phosphoglycerate kinase | 94.1 | M. oxyfera (94.1%) | |
| bin‐48‐242‐cds3 | pgk | Phosphoglycerate kinase | 87.2 | M. limnetica (95.2%) | |
| bin‐48‐108‐cds1 | gap | Glyceraldehyde 3‐phosphate dehydrogenase | 87.2 | M. limnetica (96.6%) | |
| bin‐48‐166‐cds2 | gap | Glyceraldehyde‐3‐phosphate dehydrogenase (NAD(P)) | 88.0 | M. limnetica (98.8%) | |
| bin‐48‐242‐cds2 | tpi | Triosephosphate isomerase | 81.9 | M. limnetica (93.4%) | |
| bin‐48‐242‐cds3 | tpi | Triosephosphate isomerase | 87.2 | M. limnetica (95.2%) | |
| bin‐48‐99‐cds4 | fbb | Fructose‐bisphosphate aldolase | ND | M. limnetica (95.6%) | |
| bin‐48‐99‐cds3 | fbp | Fructose‐1,6‐bisphosphatase I | 87.3 | M. limnetica (94.4%) | |
| bin‐48‐108‐cds3 | glpX | Fructose‐1,6‐bisphosphatase II | 91.1 | AOM enrichment (92.0%) | |
| bin‐48‐96‐cds4 | tkt | Transketolase | ND | AOM enrichment (78.6) | |
| bin‐48‐108‐cds2 | tkt | Transketolase | 89.3 | M. limnetica (95.5) | |
| bin‐48‐17‐cds6 | xfp | Xylulose‐5‐phosphate/fructose‐6‐phosphate phosphoketolase | ND | AOM enrichment (85.0) | |
| bin‐48‐99‐cds2 | rpiA | Ribose 5‐phosphate isomerase A | 89.0 | M. limnetica (91.8) | |
| bin‐48‐34‐cds1 | prk | Phosphoribulokinase | 94.8 | M. limnetica (97.4) | |
| Iodate reduction | bin‐48‐25‐cds2 | idrP2 | Cytochrome c peroxidase | 34.1 | Environmental MAG (58.5%) |
| bin‐48‐25‐cds3 | idrP1 | Cytochrome c peroxidase | 34.1 | Chloroflexi bac. (61.1%) | |
| bin‐48‐25‐cds4 | idrB | Arsenite oxidase small subunit | ND | Rhodocyclaceae bac. (55.7%) | |
| bin‐48‐25‐cds5 | idrA | Arsenite oxidase large subunit | 25.2 | Plancetomycetaceae bac. (65.4%) |
Based on amino acid sequence.
The two Nod sequences have 26 residual overlap and can be assembled, resulting in one complete M. iodofontis Nod. ND, no significant similarity found.
Interestingly, the cave spring water only contained low nitrate concentrations (<0.2 mg l−1) and nitrite was undetectable 23 . Thus, a potential for respiring other electron acceptors by M. iodofontis was assessed within the MAG. Remarkably, the corresponding MAG also harbored a gene cluster encoding cytochrome c peroxidases (IdrP1 and IdrP2) and an iodate reductase (IdrBA), the activity of which was recently demonstrated for Pseudomonas sp. SCT 26 and Denitromonas sp. IR‐12 27 . The GC content and sequencing depth of the contig (bin48_25), where the iodate reductase gene cluster was located, was comparable to that of other contigs in the MAG, supporting its origin from M. iodofontis (Figure S4). Phylogenetic analysis demonstrated that the catalytic subunit of the iodate reductase (IdrA) of M. iodofontis was clearly placed within a cluster of iodate reductases (Figure 1D). The organization of this iodate reductase gene cluster (idrP2,P1,B,A) in Ca. M. iodofontis was also the same as in Pseudomonas sp. SCT and Denitromons sp. IR‐12 (Figure 1D). This organization seems characteristic among iodate reductases, distinct from more distantly related arsenite oxidases and periplasmic nitrate reductase encoding gene clusters 27 . These results strongly suggest that M. iodofontis carries a functional iodate reductase. Notably, M. iodofontis iodate reductase genes had no significant hits in genomes of other Methylomirabilis species (Table 1). An incomplete operon (idrP1,B,A) was detected on a contig of another subsurface Methylomirabilota MAG (GCA_001771285.1) (Figure 1B), which also belonged to the order Methylomirabilales but was not placed within the Methylomirabilis clade and lacked oxygenic denitrification and methane oxidation pathways (Figure 1B). This may indicate that M. iodofontis could have acquired iodate reductase genes via lateral gene transfer, as also proposed for other iodate‐reducing bacteria 27 .
SignalP analysis 28 revealed that both IdrP1 and IdrP2 possess the Sec and IdrB possesses a twin‐arginine translocation (TAT) signal peptide, suggesting a periplasmic location of the M. iodofontis iodate reductase. This was also shown for Pseudomonas sp. SCT and Denitromonas sp. IR‐12 26 , 27 . It has been proposed that in Denitromonas sp. IR‐12, IdrAB first reduces iodate to hypoiodous acid (HIO), which is chemically unstable and undergoes abiotic disproportionation to I− and IO3 −. The latter is subsequently cycled back to the enzymatic reduction 27 . In Pseudomonas sp. SCT, iodate reduction by IdrAB to hydrogen peroxide (H2O2) and HIO was proposed. The resulting H2O2 is detoxified by cytochrome c peroxidase (IdrP1 and IdrP2) to water and HIO is presumably disproportionated into O2 and iodide by a chlorite dismutase like (Cld‐like) enzyme 26 . Both Denitromonas sp. IR‐12 and Pseudomonas sp. SCT oxidize acetate to fuel iodate reduction; however, the potential electron donor for this reaction in M. iodofontis is still unclear. Notably, iodate reduction via the second proposed pathway would also allow for an oxygen‐dependent methane oxidation in M. iodofontis (Figure 1E), via the following redox reaction:
However, this metagenome‐derived physiology of M. iodofontis clearly awaits validation via labeling experiments in biofilm samples and enrichment cultures under laboratory conditions.
In summary, we report the MAG of a novel, yet uncultured Methylomirabilota methanotroph, Ca. M. iodofontis. Consistent with the specific biogeochemical setting of the iodine‐ and methane‐rich mineral spring cave, genetic and phylogenomic analyses suggest a capacity for methane oxidation, oxygenic denitrification, as well as iodate reduction in M. iodofontis (Figure 1E). This expands our perspective of the metabolic versatility of Methylomirabilota methanotrophs. Due to the ubiquity of iodate in ocean waters 29 , such ecophysiologies might be widely distributed and represent an overlooked methane sink in marine ecosystems.
AUTHOR CONTRIBUTIONS
Clemens Karwautz, Baoli Zhu, and Tillmann Lueders obtained samples and did sequencing; Baoli Zhu, Stefan Andrei, and Clemens Karwautz analyzed the data; Andreas Klingl did EM; Baoli Zhu wrote the manuscript with inputs from Jakob Pernthaler and Tillmann Lueders. All authors read and approved the final manuscript.
ETHICS STATEMENT
This study did not involve any human participant or animal subject.
CONFLICT OF INTERESTS
The authors declare no conflict of interests.
Supporting information
Supporting information.
ACKNOWLEDGMENTS
Baoli Zhu was supported by the National Natural Science Foundation of China (42177104). This research was also supported by the European Research Council (ERC) under the European Union's Seventh Framework Program (FP7/2007‐2013), grant agreement 616644 (POLLOX) to Tillmann Lueders. Metagenomic sequencing was done via a Census of Deep Life (CoDL) metagenome project awarded by the Deep Carbon Observatory (DCO) under the project name DCO_LUE.
Zhu B, Karwautz C, Andrei S, Klingl A, Pernthaler J, Lueders T. A novel Methylomirabilota methanotroph potentially couples methane oxidation to iodate reduction. mLife. 2022;1:323–328. 10.1002/mlf2.12033
Edited by Fengping Wang, Shanghai Jiao Tong University, China
Contributor Information
Baoli Zhu, Email: baoli.zhu@isa.ac.cn.
Tillmann Lueders, Email: tillmann.lueders@uni-bayreuth.de.
DATA AVAILABILITY
The metagenome sequences of this project were deposited at NCBI with accession number PRJNA825327.
REFERENCES
- 1. Evans PN, Boyd JA, Leu AO, Woodcroft BJ, Parks DH, Hugenholtz P, et al. An evolving view of methane metabolism in the Archaea. Nat Rev Microbiol. 2019;17:219–32. [DOI] [PubMed] [Google Scholar]
- 2. Boetius A, Ravenschlag K, Schubert CJ, Rickert D, Widdel F, Gieseke A, et al. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature. 2000;407:623–6. [DOI] [PubMed] [Google Scholar]
- 3. Ettwig KF, Zhu B, Speth D, Keltjens JT, Jetten MSM, Kartal B. Archaea catalyze iron‐dependent anaerobic oxidation of methane. Proc Natl Acad Sci USA. 2016;113:12792–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Haroon MF, Hu S, Shi Y, Imelfort M, Keller J, Hugenholtz P, et al. Anaerobic oxidation of methane coupled to nitrate reduction in a novel archaeal lineage. Nature. 2013;500:567–70. [DOI] [PubMed] [Google Scholar]
- 5. Raghoebarsing AA, Pol A, van de Pas‐Schoonen KT, Smolders AJ, Ettwig KF, Rijpstra WI, et al. A microbial consortium couples anaerobic methane oxidation to denitrification. Nature. 2006;440:918–21. [DOI] [PubMed] [Google Scholar]
- 6. Ettwig KF, Butler MK, Le Paslier D, Pelletier E, Mangenot S, Kuypers MM, et al. Nitrite‐driven anaerobic methane oxidation by oxygenic bacteria. Nature. 2010;464:543–8. [DOI] [PubMed] [Google Scholar]
- 7. Reimann J, Jetten MSM, Keltjens JT. Metal enzymes in “impossible” microorganisms catalyzing the anaerobic oxidation of ammonium and methane. In: Kroneck PMH, Sosa Torres ME, editors. Sustaining life on planet earth: metalloenzymes mastering dioxygen and other chewy gases. Vol. 15. Cham, Switzerland: Springer International Publishing; 2015. p. 257–313.
- 8. Ettwig KF, Speth DR, Reimann J, Wu ML, Jetten MS, Keltjens JT. Bacterial oxygen production in the dark. Front Microbiol. 2012;3:273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhu B, Bradford L, Huang S, Szalay A, Leix C, Weissbach M, et al. Unexpected diversity and high abundance of putative nitric oxide dismutase (Nod) genes in contaminated aquifers and wastewater treatment systems. Appl Environ Microbiol. 2017;83:e02750‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wu ML, van Teeseling MC, Willems MJ, van Donselaar EG, Klingl A, Rachel R, et al. Ultrastructure of the denitrifying methanotroph “Candidatus Methylomirabilis oxyfera,” a novel polygon‐shaped bacterium. J Bacteriol. 2012;194:284–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Luesken FA, Zhu B, van Alen TA, Butler MK, Diaz MR, Song B, et al. pmoA primers for detection of anaerobic methanotrophs. Appl Environ Microbiol. 2011;77:3877–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhu B, Wang J, Bradford LM, Ettwig K, Hu B, Lueders T. Nitric oxide dismutase (nod) genes as a functional marker for the diversity and phylogeny of methane‐driven oxygenic denitrifiers. Front Microbiol. 2019;10:1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ettwig KF, van Alen T, van de Pas‐Schoonen KT, Jetten MS, Strous M. Enrichment and molecular detection of denitrifying methanotrophic bacteria of the NC10 phylum. Appl Environ Microbiol. 2009;75:3656–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. He Z, Geng S, Cai C, Liu S, Liu Y, Pan Y, et al. Anaerobic oxidation of methane coupled to nitrite reduction by halophilic marine NC10 bacteria. Appl Environ Microbiol. 2015;81:5538–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhu B, van Dijk G, Fritz C, Smolders AJ, Pol A, Jetten MS, Ettwig KF. Anaerobic oxidization of methane in a minerotrophic peatland: enrichment of nitrite‐dependent methane‐oxidizing bacteria. Appl Environ Microbiol. 2012;78:8657–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Versantvoort W, Guerrero‐Cruz S, Speth DR, Frank J, Gambelli L, Cremers G, et al. Comparative genomics of Candidatus Methylomirabilis species and description of Ca. Methylomirabilis lanthanidiphila. Front Microbiol. 2018;9:1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Graf JS, Mayr MJ, Marchant HK, Tienken D, Hach PF, Brand A, et al. Bloom of a denitrifying methanotroph, ‘Candidatus Methylomirabilis limnetica’, in a deep stratified lake. Environ Microbiol. 2018;20:2598–614. [DOI] [PubMed] [Google Scholar]
- 18. Deutzmann JS, Stief P, Brandes J, Schink B. Anaerobic methane oxidation coupled to denitrification is the dominant methane sink in a deep lake. Proc Natl Acad Sci USA. 2014;111:18273–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hug LA, Thomas BC, Sharon I, Brown CT, Sharma R, Hettich RL, et al. Critical biogeochemical functions in the subsurface are associated with bacteria from new phyla and little studied lineages. Environ Microbiol. 2016;18:159–73. [DOI] [PubMed] [Google Scholar]
- 20. Anantharaman K, Brown CT, Hug LA, Sharon I, Castelle CJ, Probst AJ, et al. Thousands of microbial genomes shed light on interconnected biogeochemical processes in an aquifer system. Nat Commun. 2016;7:13219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Luo JH, Chen H, Hu SH, Cai C, Yuan ZG, Guo JH. Microbial selenate reduction driven by a denitrifying anaerobic methane oxidation biofilm. Environ Sci Technol. 2018;52:4006–12. [DOI] [PubMed] [Google Scholar]
- 22. Li ZY, Li X, Tan B, Lv PL, Zhao HP. NC10 bacteria promoted methane oxidation coupled to chlorate reduction. Biodegradation. 2020;31:319–29. [DOI] [PubMed] [Google Scholar]
- 23. Karwautz C, Kus G, Stockl M, Neu TR, Lueders T. Microbial megacities fueled by methane oxidation in a mineral spring cave. ISME J. 2018;12:87–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ward LM, Shih PM, Fischer WW. MetaPOAP: presence or absence of metabolic pathways in metagenome‐assembled genomes. Bioinformatics. 2018;34:4284–6. [DOI] [PubMed] [Google Scholar]
- 25. Rasigraf O, Kool DM, Jetten MSM, Damste JSS, Ettwig KF. Autotrophic carbon dioxide fixation via the Calvin−Benson−Bassham cycle by the denitrifying methanotroph “Candidatus Methylomirabilis oxyfera”. Appl Environ Microbiol. 2014;80:2451–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yamazaki C, Kashiwa S, Horiuchi A, Kasahara Y, Yamamura S, Amachi S. A novel dimethylsulfoxide reductase family of molybdenum enzyme, Idr, is involved in iodate respiration by Pseudomonas sp. SCT. Environ Microbiol. 2020;22:2196–212. [DOI] [PubMed] [Google Scholar]
- 27. Reyes‐Umana V, Henning Z, Lee K, Barnum TP, Coates JD. Genetic and phylogenetic analysis of dissimilatory iodate‐reducing bacteria identifies potential niches across the world's oceans. ISME J. 2022;16:38–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Almagro Armenteros JJ, Tsirigos KD, Sønderby CK, Petersen TN, Winther O, Brunak S, et al. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol. 2019;37:420–3. [DOI] [PubMed] [Google Scholar]
- 29. He P, Hou XL, Aldahan A, Possnert G, Yi P. Iodine isotopes species fingerprinting environmental conditions in surface water along the northeastern Atlantic Ocean. Sci Rep. 2013;3:2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information.
Data Availability Statement
The metagenome sequences of this project were deposited at NCBI with accession number PRJNA825327.


