Abstract
All animals must maintain genome and proteome integrity, especially when experiencing endogenous or exogenous stress. To cope, organisms have evolved sophisticated and conserved response systems: unfolded protein responses (UPRs) ensure proteostasis, while DNA damage responses (DDRs) maintain genome integrity. Emerging evidence suggests that UPRs and DDRs crosstalk, but this remains poorly understood. Here, we demonstrate that depletion of the DNA primases pri-1 or pri-2, which synthesize RNA primers at replication forks and whose inactivation causes DNA damage, activates the UPR of the endoplasmic reticulum (UPR-ER) in Caenorhabditis elegans, with especially strong activation in the germline. We observed activation of both the inositol-requiring-enzyme 1 (ire-1) and the protein kinase RNA-like endoplasmic reticulum kinase (pek-1) branches of the (UPR-ER). Interestingly, activation of the (UPR-ER) output gene heat shock protein 4 (hsp-4) was partially independent of its canonical activators, ire-1 and X-box binding protein (xbp-1), and instead required the third branch of the (UPR-ER), activating transcription factor 6 (atf-6), suggesting functional redundancy. We further found that primase depletion specifically induces the (UPR-ER), but not the distinct cytosolic or mitochondrial UPRs, suggesting that primase inactivation causes compartment-specific rather than global stress. Functionally, loss of ire-1 or pek-1 sensitizes animals to replication stress caused by hydroxyurea. Finally, transcriptome analysis of pri-1 embryos revealed several deregulated processes that could cause (UPR-ER) activation, including protein glycosylation, calcium signaling, and fatty acid desaturation. Together, our data show that the (UPR-ER), but not other UPRs, responds to replication fork stress and that the (UPR-ER) is required to alleviate this stress.
Keywords: UPR, BiP, DNA replication, DNA damage, primase
Introduction
The endoplasmic reticulum (ER) of eukaryotes is a dynamic membrane network required for many cellular processes, and ER homeostasis is critical to cellular and organismal health. ER homeostasis is disturbed by both impaired proteostasis and by ER membrane lipid disequilibrium (Gardner et al. 2013; Senft and Ronai 2015; Xu and Taubert 2021; Celik et al. 2023). To ensure ER function and cell viability, a conserved adaptive mechanism has evolved that restores ER homeostasis during stress: the ER unfolded protein response (UPR-ER) (Walter and Ron 2011; Gardner et al. 2013; Senft and Ronai 2015; Hetz et al. 2020; Xu and Taubert 2021). In higher eukaryotes, the UPR-ER consists of 3 parallel ER stress sensing and transducing branches: the inositol-requiring-enzyme 1 (IRE1/IRE-1, also known as ER to nucleus signaling 1 or ERN1 in mammals) branch (Adams et al. 2019), the protein kinase RNA-like ER kinase (PERK/PEK-1, also known as eukaryotic translation initiation factor 2α kinase 3 or EIF2AK3) branch (McQuiston and Diehl 2017), and the activating transcription factor 6 (ATF6/ATF-6) branch (Hillary and FitzGerald 2018). Together, they alleviate ER stress by reprograming transcription and translation to promote protein folding, degradation, and transport, as well as lipid synthesis and remodeling. If ER stress cannot be resolved, the UPR-ER switches from promoting survival and adaptation to triggering apoptosis, ensuring tissue and organism integrity (Walter and Ron 2011; Hetz et al. 2020).
Like ER homeostasis, genome integrity is paramount for cellular and organismal health. Cells have to safeguard against DNA damage caused by endogenous and exogenous agents that induce different types of DNA lesions. To repair and mitigate DNA damage, cells activate DNA repair pathways and modulate cell cycle progression and apoptosis, a response collectively known as the DNA damage response (DDR) (Jackson and Bartek 2009; Gartner and Engebrecht 2021; McClure et al. 2022). Repairing this damage is critical to ensuring faithful DNA replication and thus cell division and organism development, growth, maintenance, and aging.
Interestingly, there is crosstalk between the UPR-ER and the DDR (González-Quiroz et al. 2020; Bolland et al. 2021). In yeast, IRE1 promotes DNA repair via several different pathways, and deletion of IRE1 sensitizes yeast to genotoxic stress and causes chromosome loss even in unstressed conditions (Henry et al. 2010). Moreover, activation of the checkpoint pathway by DNA damage upregulates the key UPR-ER output transcription factor Hac1p (Tao et al. 2011). Similarly, in cultured human cell lines, IRE-1 promotes genome integrity through the downstream effector X-box binding protein 1 (XBP1; the human ortholog of Hac1p), which directly regulates several DDR pathways, and through noncanonical regulated IRE-1-dependent decay (RIDD) of mRNA (Dufey et al. 2020). Consequently, loss of XBP1 correlates with increased DNA damage (Acosta-Alvear et al. 2007; Argemí et al. 2017; Lyu et al. 2019; González-Quiroz et al. 2020). Moreover, DNA damaging agents such as camptothecin and ionizing radiation trigger UPR-ER activation in cancer cell lines through the conserved DDR sensor (Ataxia-telangiectasia mutated; Hotokezaka et al. 2020), and the genome integrity regulator p53 mediates ER structure remodeling in chemically induced genotoxicity (Zheng et al. 2018). The UPR-ER, especially the IRE-1 branch, is therefore both activated by DNA damage and functionally required to repair such damage.
Whether integration of the DDR and the UPR-ER also occurs in animals, and how different tissues respond in this context, is less well understood. In the nematode worm Caenorhabditis elegans, DNA damage in the adult germline promotes stress resistance in the postmitotic soma via MAP kinase signaling, innate immune responses, and the ubiquitin proteasome system (Ermolaeva et al. 2013), which, like the UPR-ER, maintains proteostasis (Papaevgeniou and Chondrogianni 2014; Zhang et al. 2022). Moreover, C. elegans xbp-1 is required to express DNA repair genes (Shen et al. 2005). Recently, increased stress resistance to the ER stressors dithiothreitol (DTT) and tunicamycin was observed in C. elegans exposed to UV, which increased the activity of the IRE-1–XBP-1 branch by elevating the levels of unsaturated phosphatidylcholine (Deng et al. 2021), a key ER membrane lipid (van Meer et al. 2008). However, this study predominantly analyzed DDR and UPR-ER signaling in the glp-1 mutant, which lacks a germline, is long-lived and stress resistant, and shows dysregulation of DDR genes (Arantes-Oliveira et al. 2002; Boyd et al. 2010; TeKippe and Aballay 2010; Ratnappan et al. 2014; Goh et al. 2018). The interaction of the UPR-ER and the DDR in wild-type animals with an intact germline, which is the primary tissue of active DNA repair, therefore remains incompletely understood.
In a screen for genes whose inactivation causes UPR-ER activation in wild-type C. elegans, we identified two DNA primase genes (Ho et al. 2020). Eukaryotic primase complexes synthesize short RNA primers required for initiating lagging strand DNA replication and also contribute to DNA repair and possibly transcription (Guilliam et al. 2015; Yoon et al. 2018). Abnormal primase function causes stalled replication forks, leading to DNA damage and genome instability. This, therefore, provided us with the opportunity to study the relationship between primase function, replication stress, and the UPR-ER in animals with an intact germline. Here, we show that primase inactivation and UV–C irradiation activate both the IRE-1 and the PEK-1 branches of the UPR-ER, with stronger induction in the germline than in the soma. Interestingly, activation of the heat shock protein 4 (hsp-4) gene, which canonically requires the IRE-1–XBP-1 axis, required atf-6 in the germline, suggesting differential regulatory mechanisms. We further found that primase inactivation selectively activated the UPR-ER, but not the cytosolic or mitochondrial UPRs, arguing for a specific role of the UPR-ER in maintaining genome integrity. We also showed that loss of ire-1 or pek-1 sensitizes C. elegans to replication stress, showing that the UPR-ER is functionally protective. RNA-sequencing (RNA-seq) analysis revealed several pathways comprising both proteostasis and lipidostasis that could underlie UPR-ER activation following replication stress. Collectively, our data show that the UPR-ER plays important roles in ensuring genome integrity in C. elegans.
Materials and methods
Worm strains
The following worm strains were used: N2 wild-type, SJ4005 zcIs4 [hsp-4p::gfp] V (Calfon et al. 2002), SJ17 xbp-1(zc12) III; zcIs4 [hsp-4p::gfp] V (Calfon et al. 2002), SJ30 ire-1(zc14) II; zcIs4 [hsp-4p::gfp] V (Calfon et al. 2002), SJ4100 zcIs13 [hsp-6p::gfp] V (Yoneda et al. 2004), TJ375 gpIs1 [hsp-16.2p::gfp] (Henderson and Johnson 2001), xbp-1(tm2482) III (Richardson et al. 2011), RB545 pek-1(ok275) X (Consortium 2012), RB925 ire-1(ok799) II (Consortium 2012), RB772 atf-6(ok551) X (Consortium 2012), PHX2824 hsp-4::gfp(syb2824) II (generated by SunyBiotech Co., Ltd., Fujian, China), and STE142 hsp-4::gfp(syb2824) II;atf-6(ok551) X (this study). All strains were backcrossed 6 times to the laboratory N2 wild-type background before use.
Worm growth conditions
We cultured C. elegans strains at 20°C on nematode growth medium (NGM)-lite agar plates with E. coli OP50 as food source, except for RNA interference (RNAi), for which HT115 strain was used (Ho et al. 2020). To developmentally synchronize worm populations, gravid adult worms were treated with alkaline sodium hypochlorite solution to extract embryos, which were washed twice with M9, and then plated onto an unseeded NGM-lite plate to allow hatching overnight. When imaging worms, adult worms were bleached >10 minutes until autofluorescent mother bodies disappeared. The resulting synchronized L1 larvae were transferred onto OP50 NGM-lite plates or RNAi plates (NGM-lite plates containing 25 μg/mL carbenicillin (BioBasic CDJ469), 2 mM Isopropyl ß-D-1-thiogalactopyranoside (Santa Cruz sc-202185B), and 12.5 μg/mL tetracycline (BioBasic TB0504)). RNAi plates were seeded twice with the appropriate HT115 RNAi bacteria (Ahringer library, Source BioScience); RNAi clones were sequenced prior to use to ensure construct identity. Synchronized L1 worms were placed on RNAi plates and grown until they reached the desired developmental stage.
Differential interference contrast and fluorescence microscopy
Worms were mounted onto 2% (w/v) agarose pads containing a drop of 20 mM sodium azide (NaN3) for microscopy. Eggs were picked from plates onto 2% (w/v) agarose pads containing a drop of M9 for microscopy. Worms were imaged using differential interference contrast (DIC) and fluorescence optics through a CoolSnap HQ camera (Photometrics, Tucson, AZ, USA) on a Zeiss Axioplan 2 compound microscope (Carl Zeiss Microscopy, Thornwood, NY, USA). All GFP images were taken at the same exposure time (300 ms). Using the ImageJ software, the images in the GFP channel were adjusted to the same brightness (maximum display value = 4095, minimum = 201; these parameters were applied to all GFP images used for quantification in this study) and contrast levels for subsequent display and quantification purposes. Analysis of overall fluorescence intensity of individual worms was performed by tracing the outline of the worms on the corresponding DIC images, and then normalizing for area and background fluorescence, as described (Shomer et al. 2019).
Protein extraction and immunoblots
Whole-worm protein extracts were generated by sonication in radioimmunoprecipitation assay (RIPA) lysis buffer with cOmpleteTM Protease Inhibitor Cocktail (Roche #4693116001) and β-glycerophosphate (Sigma-Aldrich G6251). Protein concentrations were determined using the reducing agent and detergent compatible (RCDC) protein assay kit (Bio-Rad #500-0121), and sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE) analysis and immunoblotting were performed as described (Hou et al. 2014), using anti-Ser51-Phospho-eIF2α rabbit antibody (Cell Signaling Technologies #9721), anti-α-tubulin mouse antibody (Sigma #T9026), and anti-rabbit HRP-conjugated (New England Biolabs [NEB] #7074) and anti-mouse HRP-conjugated (Cell Signaling Technologies #7076) secondary antibodies. Detection was done using ECL (Pierce #32109).
Exposure to genotoxic agents
For UV–C exposure, synchronized populations of day-1 adult C. elegans were placed on NGM-lite plates seeded with a thin layer of OP50. Uncovered NGM-lite plates were then placed in a Stratalinker 2400 UV Crosslinker (Stratagene) and irradiated with wavelength 254 nm light at 400 J/m2. After 24 h of recovery at 20°C, worms and embryos were mounted and imaged.
For hydroxyurea (HU) exposure L1 recovery experiments, age-synchronized L1 populations were grown for 72 h on NGM-lite or on NGM-lite containing either 5 or 10 mM HU (Sigma-Aldrich H8627). Then, worms were transferred to OP50-seeded NGM-lite plates for recovery and egg laying. After 4 h, the number of eggs was counted for each genotype and condition, as indicated.
For HU exposure L4 recovery experiments, synchronized L1 worms were grown for 48 h. Then, age-synchronized L4 populations were transferred to and maintained on either NGM-lite plates or NGM-lite plates containing 20 mM HU for 24 h. Then, adult worms were transferred to OP50 plates for recovery and egg laying. After 4 h, the number of eggs for each genotype and condition was counted.
To measure developmental rate, synchronized L1 populations were grown for 48 h on NGM-lite plates containing DMSO vehicle or 15 mM HU. Then, the number of L4 or older worms and the total number of worms were counted for each genotype and condition.
For body size quantification, synchronized L1 stage worms were grown for 72 h on NGM-lite plates containing DMSO or 15 mM HU, before >10 worms for each genotype and condition were imaged.
RNA-Sequencing and data analysis
Synchronized L1 N2 worm populations fed empty vector (EV) or pri-1 RNAi were grown for 96 h at 20°C and allowed to lay eggs. Plates were washed with M9 twice to remove adults and hatched worms before eggs were harvested with a cell scraper. The collected eggs were washed twice with M9 to remove bacteria, and then flash-frozen in an ethanol-dry ice bath. For total RNA extraction, eggs were thawed in Trizol and sonicated. Total RNA was extracted using Trizol and 1-bromo-3-chloropropane, as described (Doering et al. 2022). RNA integrity and quantity were assessed on an Agilent Technologies 2100 Bioanalyzer System.
Library preparation and sequencing was performed by The Center for Applied Genomics, SickKids, Toronto, ON (http://www.tcag.ca). Briefly, RNA was prepared for sequencing using the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina (NEB #E7760). Sequencing was performed on an Ilumina NovaSeq 6000 instrument equipped with an S4 flow cell generating 150 bp paired-end reads. Low quality reads and adapter sequences were trimmed using Trimmomatic 0.36 (Bolger et al. 2014) with parameters LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:36. The trimmed reads were quantified to the C. elegans Ensembl transcriptome build WBcel235 using Salmon ver1.4.0 (Patro et al. 2017) with the parameters -l A -p 8 –gcBias –validateMappings. Then, transcript-level read counts were imported into R and summed into gene-level read counts using tximport (Soneson et al. 2016) (genes listed in Supplementary Table 1). Genes not expressed at a level greater than 10 reads in at least 3 of the samples were excluded from further analysis. Differential expression analysis was performed with quasi-likelihood F-test with the generalized linear model (GLM) approach in edgeR (Robinson et al. 2010). Genes with P-value <0.05 and False Discovery Rate < 0.05 were considered differentially expressed. RNA-seq data were deposited in the Gene Expression Omnibus under the accession number GSE225569. Gene set enrichment analysis (GSEA) using either the biological process (BP (Chagoyen and Pazos 2010)) or the Kyoto encyclopedia of genes and genomes (KEGG (Kanehisa 1997; Kanehisa et al. 2020)) as underlying databases was performed with eVITTA (Cheng et al. 2021), using Pval < 0.05 and Padj < 0.25 as cutoffs.
Statistical analysis
P values were calculated using 2-tailed Student's t-tests, Welch's t-tests, 1-way ANOVA tests, or 2-way ANOVA tests using GraphPad Prism 9 or 10, as reported in the figure legends. Scatter plots were generated in GraphPad Prism 9 or 10. Error bars denote standard deviation; the number of independent experiments performed and number of animals studied are indicated in the figure legends.
Results
Knockdown of the C. elegans primase genes pri-1 or pri-2 activates the ire-1 branch of the UPR-ER in embryos
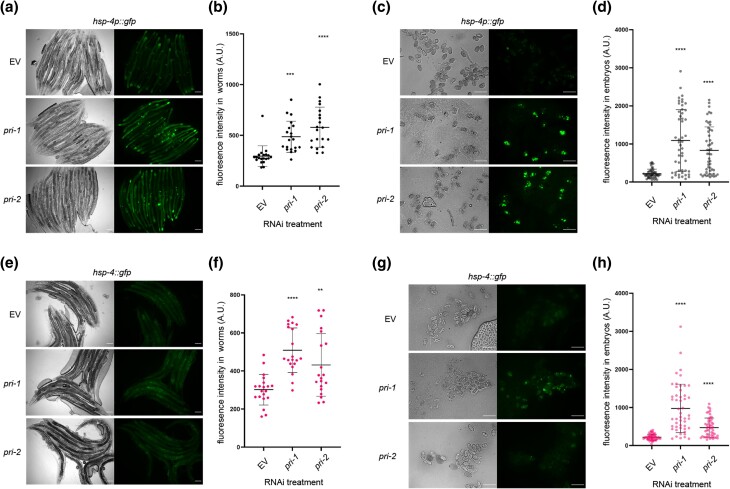
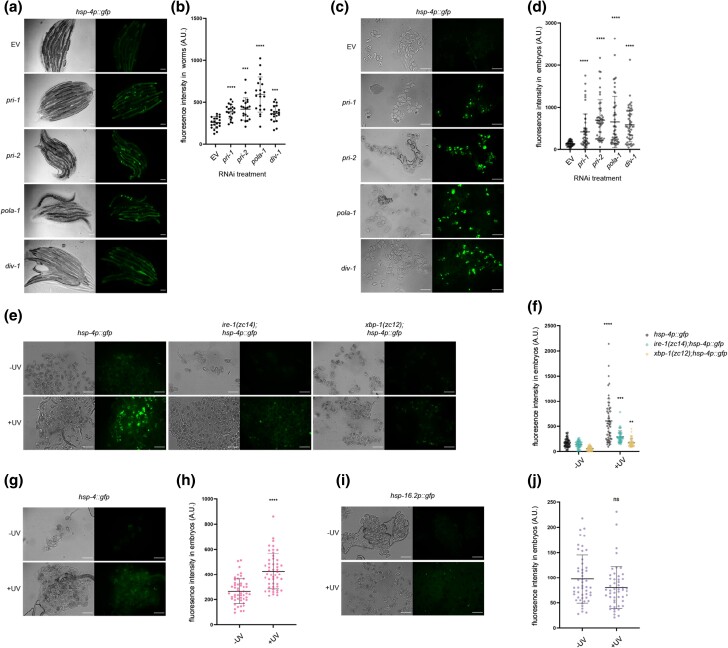
We previously showed that RNAi against the 2 DNA primase subunit genes of C. elegans, pri-1 or pri-2, caused activation of the UPR-ER (Ho et al. 2020). To validate this finding, we quantified the induction of hsp-4p::gfp, a widely used transcriptional reporter for the ER stress-inducible, ire-1- and xbp-1-activated hsp-4 gene promoter (Calfon et al. 2002; Hou et al. 2014; Ho et al. 2020). We found that pri-1 or pri-2 RNAi induced sp-4p::gfp fluorescence in the worm soma ∼1.5–2-fold (Fig. 1, a and b). Interestingly, we observed a larger increase in hsp-4p::gfp activity (∼4-fold) in F1 embryos from RNAi-fed P0 adults (Fig. 1, c and d). This phenotype manifests despite the fact that F1 eggs from pri-1 or pri-2 RNAi-treated P0 adults never hatch, but rather arrest at the early embryogenesis/pre-morphogenetic stage due to persisting replication fork stalling, which causes double-stranded DNA breaks (DSBs) (Zeman and Cimprich 2014). UPR-ER activation in the F1 generation by pri-1 or pri-2 RNAi in P0 suggested a link between the UPR-ER and genotoxic stress in embryos.
Fig. 1.
pri-1 or pri-2 knockdown induces IRE-1 branch activity in the soma and embryos of C. elegans. a and b) The figure shows representative micrographs (a) and whole-worm GFP quantification (b) of hsp-4p::gfp adult worms fed EV, pri-1, or pri-2 RNAi (n = 3 experiments totaling >20 animals per RNAi treatment). c and d) The figure shows representative micrographs (c) and GFP quantification (d) of F1 embryos laid by hsp-4p::gfp adult worms fed EV, pri-1, or pri-2 RNAi (n = 3 experiments totaling >50 embryos per RNAi treatment). e and f) The figure shows representative micrographs (e) and whole-worm GFP quantification (f) of hsp-4::gfp adult worms fed EV, pri-1, or pri-2 RNAi (n = 2 experiments totaling >20 animals per RNAi treatment). g and h) The figure shows representative micrographs (g) and GFP quantification (h) of embryos laid by hsp-4::gfp adult worms fed EV, pri-1, or pri-2 RNAi (n = 2 experiments totaling >50 embryos per treatment). In all micrographs, the scale bar represents 100 μm. In dot plots, each dot represents the signal detected in one individual worm or embryo; the error bars represent standard deviation. Statistical analysis for b, d, f, h: **P < 0.01, ***P < 0.001, ****P < 0.0001 vs. EV RNAi-treated control (Brown–Forsythe and Welch ANOVA test corrected for multiple comparisons using the Dunnett T3 method).
To confirm activation of the endogenous UPR-ER by pri-1 or pri-2 RNAi, we studied a genome-edited strain wherein the 3′ end of the hsp-4 coding sequence is tagged with gfp, resulting in a C-terminal HSP-4::GFP fusion protein (hereafter referred to as hsp-4::gfp). To validate this strain, we examined GFP intensity following proteotoxic stress by tunicamycin and lipotoxic stress by mdt-15 RNAi, both established UPR-ER inducers (Calfon et al. 2002; Hou et al. 2014). As expected, we observed elevated GFP intensity in hsp-4::gfp worms challenged with tunicamycin or mdt-15 RNAi (Supplementary Fig. 1), suggesting that this strain faithfully reports on the regulation of endogenous hsp-4 by different stresses. Next, we treated the hsp-4::gfp reporter strain with pri-1 or pri-2 RNAi and studied GFP fluorescence in the soma of P0 adult worms and in F1 embryos. We observed strong induction of endogenous HSP-4::GFP in F1 embryos, and weaker induction in P0 somatic cells (Fig. 1, e–h). These data suggest that loss of primase function and subsequent replication defects trigger UPR-ER activation in embryos and in somatic cells of C. elegans, with stronger induction in embryos.
Knockdown of pri-1 or pri-2 induces embryonic hsp-4 partially independently of ire-1 and xbp-1
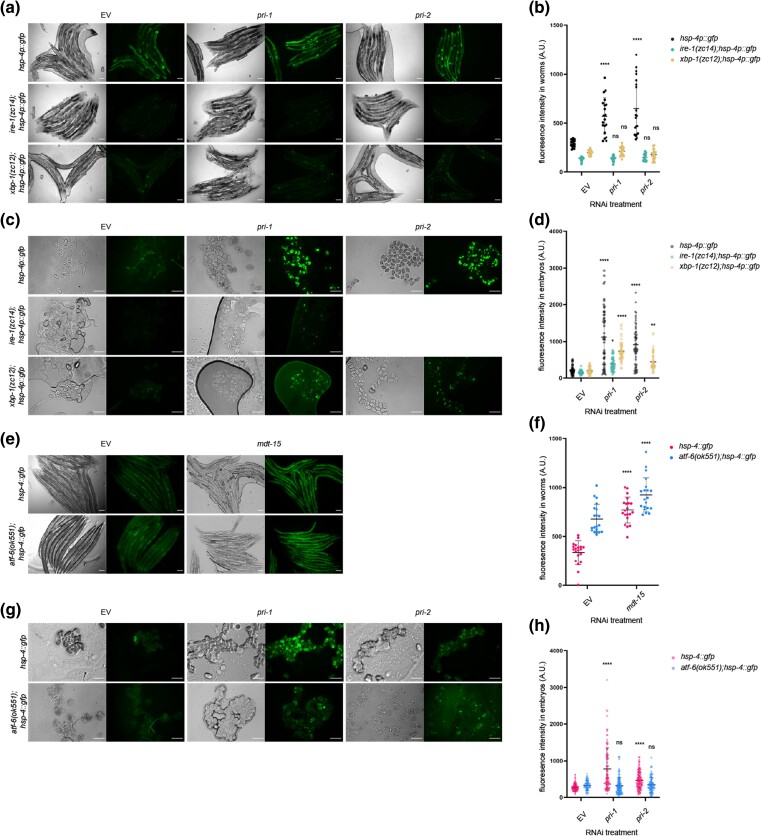
Canonical hsp-4 induction requires the transmembrane ER stress sensor ire-1 and the downstream transcription factor xbp-1 (Shen et al. 2001; Richardson et al. 2010). Thus, we studied hsp-4 induction in ire-1; hsp-4p::gfp, and xbp-1; hsp-4p::gfp worms treated with pri-1 or pri-2 RNAi. Consistent with canonical UPR-ER induction in somatic cells, increased fluorescence in pri-1- or pri-2-treated worms depended completely on ire-1 and xbp-1 (Fig. 2, a and b). By contrast, significant induction of hsp-4p::gfp remained in embryos when ire-1 or xbp-1 was deleted (Fig. 2, c and d). This suggests that additional genes are required to induce hsp-4 in embryos experiencing replication stress.
Fig. 2.
Activation of hsp-4 by pri-1 or pri-2 RNAi requires ire-1, xbp-1, and atf-6. a and b) The figure shows representative micrographs (a) and whole-worm GFP quantification (b) of hsp-4p::gfp, ire-1(zc14);hsp-4p::gfp, and xbp-1(zc12);hsp-4p::gfp adult worms fed EV, pri-1, or pri-2 RNAi (n = 3 experiments totaling >20 individual animals per RNAi treatment). c and d) The figure shows representative micrographs (c) and GFP quantification (d) of embryos laid by hsp-4p::gfp, ire-1(zc14);hsp-4p::gfp, and xbp-1(zc12);hsp-4p::gfp adult worms fed EV, pri-1, or pri-2 RNAi (n = 3 experiments totaling >50 individual embryos per RNAi treatment; note that pri-2 caused lethality in this experiment, preventing experimental assessment). e and f) The figure shows representative micrographs (e) and whole-worm GFP quantification (f) of hsp-4::gfp and atf-6 (ok551);hsp-4::gfp adult worms fed EV or mdt-15 RNAi (n = 3 experiments totaling >20 individual animals per RNAi treatment). g and h) The figure shows representative micrographs (g) and GFP quantification (h) of embryos laid by hsp-4::gfp and atf-6(ok551);hsp-4::gfp adult worms fed EV, pri-1, or pri-2 RNAi (n = 3 experiments totaling >75 individual embryos per treatment). In all micrographs, the scale bar represents 100 μm. In dot plots, each dot represents the signal detected in one individual worm or embryo; the error bars represent standard deviation. Statistical analysis: b, d: ns P > 0.05, *P < 0.05, **P < 0.01, ****P < 0.0001, vs. EV RNAi-treated control of the same genotype (2-way ANOVA test corrected for multiple comparisons using Sidak's method).
atf-6 is required for hsp-4 induction in pri-1 or pri-2 RNAi-treated embryos
In mammals, ATF6 is required for the transcription of XBP1 mRNA (Yoshida et al. 2001; Lee et al. 2002). Thus, we tested if C. elegans atf-6 is required for replication-stress-induced hsp-4 induction. We crossed the hsp-4::gfp translational reporter into a strain bearing the atf-6(ok551) null allele, and treated atf-6; hsp-4::gfp worms with pri-1 or pri-2 RNAi. As a control, we first studied mdt-15 RNAi, which caused hsp-4::gfp induction in the soma despite the atf-6 mutation (Fig. 2, e and f), suggesting that hsp-4 induction in somatic tissues does not require atf-6; unexpectedly, atf-6 loss alone induced hsp-4 expression in the soma (Fig. 2, e and f). Notably, atf-6 deletion reduced the increased fluorescence in embryos treated with pri-1 or pri-2 RNAi (Fig. 2, g and h). This indicates that hsp-4 induction in pri-1 or pri-2 RNAi-treated embryos depends on atf-6.
pri-1 or pri-2 knockdown activates the pek-1 branch of the UPR-ER
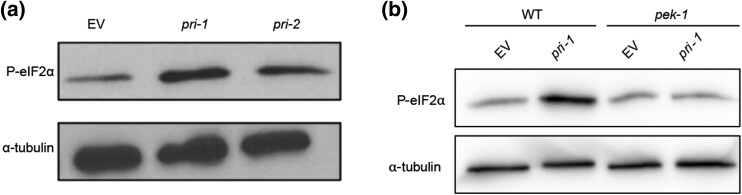
The C. elegans UPR-ER also features a branch controlled by the kinase PEK-1, which causes phosphorylation of the eukaryotic translation initiation factor eIF2α and subsequent activation of the transcription factor ATF-4 (McQuiston and Diehl 2017). As a readout of PEK-1 activity, we performed immunoblots on embryos to detect phospho-Ser51 on eIF2α, a marker for activated PEK-1 (Nukazuka et al. 2008). We observed increased levels of phospho-Ser51 in pri-1 or pri-2 RNAi-treated embryos (Fig. 3a, Supplementary Fig. 2). Critically, this induction was dependent on pek-1 (Fig. 3b, Supplementary Fig. 2), implicating canonical signaling via this branch of the UPR-ER. Thus, pri-1 or pri-2 RNAi activates both the ire-1 and pek-1 branches of the UPR-ER in the soma and in the embryos.
Fig. 3.
pri-1 or pri-2 RNAi activate the PEK-1 branch in the embryos of C. elegans. a and b) The immunoblot depicts the levels of phospho-Ser51 eIF2α (P-eIF2α) and α-tubulin in EV, pri-1, or pri-2 RNAi-treated wild-type or pek-1 mutant worm embryos (n = 2–3, for additional repeats, please see Supplementary Fig. 2).
The cytosolic and the mitochondrial UPRs are not substantially induced by pri-1 or pri-2 knockdown
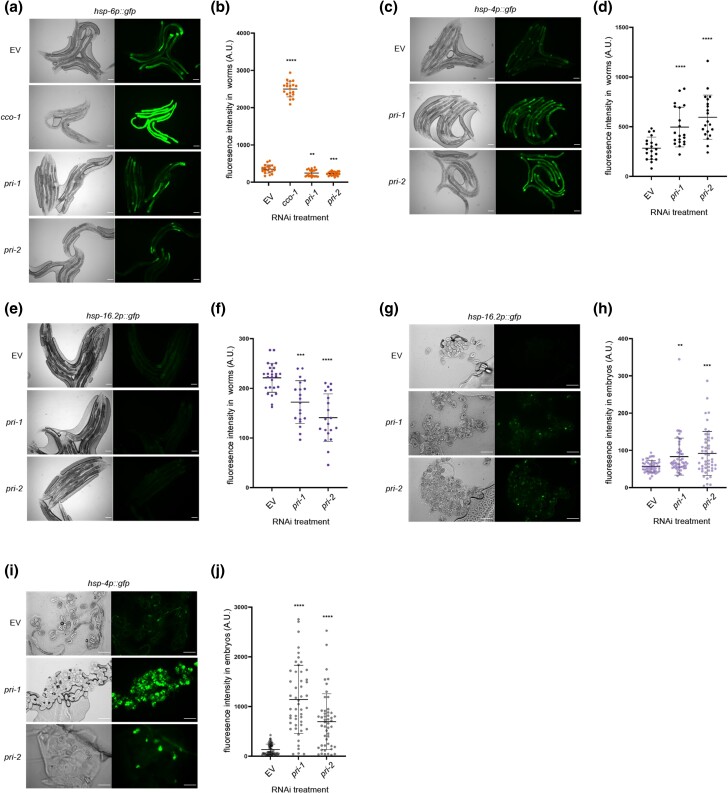
Induction of the UPR-ER due to pri-1 or pri-2 RNAi might reflect general protein misfolding in terminally arrested embryos. Thus, we monitored the activity of the cytosolic and mitochondrial UPRs with their well-established reporters, hsp-16.2p::gfp and hsp-6p::gfp, respectively (Rea et al. 2005; Bennett et al. 2014). In the worm soma, heat shock strongly induced hsp-16.2p::gfp (Supplementary Fig. 3a, b), and positive control cco-1 RNAi strongly induced hsp-6p::gfp (Fig. 4, a and b), as expected (Bennett et al. 2014). By contrast, although pri-1 or pri-2 RNAi effectively induced hsp-4p::gfp (Fig. 4, c and d), it did not induce hsp-6p::gfp or hsp-16.2p::gfp in somatic tissues; the activity of both reporters was in fact reduced (Fig. 4, a, b, e, and f). Similarly, whereas heat stress strongly (∼5-fold) induced hsp-16.2p::gfp throughout the embryo (Supplementary Fig. 3, c and d), pri-1 or pri-2 RNAi only weakly (less than 2-fold), albeit still significantly, induced hsp-16.2p::gfp fluorescence in the embryos (Fig. 4, g and h), while strongly inducing hsp-4p::gfp (Fig. 4, i and j). Thus, pri-1 or pri-2 RNAi-induced replication stress appears to predominantly trigger the UPR-ER.
Fig. 4.
pri-1 or pri-2 RNAi preferentially induces the UPR-ER. a and b) The figure shows representative micrographs (a) and whole-worm GFP quantification (b) of hsp-6p::gfp adult worms fed EV, cco-1, pri-1, or pri-2 RNAi (n = 3 experiments totaling >20 individual animals per RNAi treatment). c and d) The figure shows representative micrographs (a) and whole-worm GFP quantification (b) of hsp-4p::gfp adult worms fed EV, pri-1, or pri-2 RNAi (n = 3 experiments totaling >20 individual animals per RNAi treatment). e and f) The figure shows representative micrographs (e) and whole-worm GFP quantification (f) of hsp-16.2p::gfp adult worms fed EV, pri-1, or pri-2 RNAi (n = 2 experiments totaling >20 individual animals per RNAi treatment). g and h) The figure shows representative micrographs (g) and GFP quantification (h) of embryos laid by hsp-16.2p::gfp adult worms fed EV, pri-1, or pri-2 RNAi (n = 3 experiments totaling >50 individual embryos per treatment). i and j) The figure shows representative micrographs (i) and GFP quantification (j) of embryos laid by hsp-4p::gfp adult worms fed EV, pri-1, or pri-2 RNAi (n = 3 experiments totaling >50 individual embryos per treatment). In all micrographs, the scale bar represents 100 μm. In dot plots, each dot represents the signal detected in one individual worm or embryo; the error bars represent standard deviation. Statistical analysis for b, d, f, h, j: **P < 0.01, ***P < 0.001, ****P < 0.0001 vs. EV RNAi-treated control (Brown–Forsythe and Welch ANOVA test corrected for multiple comparisons using the Dunnett T3 method).
Inactivation of other polymerase α primase complex genes phenocopies pri-1 or pri-2 RNAi
Four genes encode C. elegans primase complex subunits: the DNA polymerase α catalytic subunit gene pola-1, the DNA polymerase α accessory subunit gene div-1, and the primase subunit genes pri-1 and pri-2 (Guilliam et al. 2015; Yoon et al. 2018). We asked if RNAi knockdown of pola-1 or div-1 phenocopied pri-1 or pri-2 RNAi. Indeed, pola-1 and div-1 RNAi activated hsp-4p::gfp in both the soma of he P0 worms and in F1 embryos, with stronger activation in embryos than in the soma (Fig. 5, a–d). This suggests that UPR-ER induction likely results from replication stress caused by defective polymerase α primase complex function.
Fig. 5.
Knockdown of polymerase α primase complex subunits and UV–C irradiation cause UPR-ER activation. a and b) The figure shows representative micrographs (a) and whole-worm GFP quantification (b) of hsp-4p::gfp adult worms fed EV, pri-1, pri-2, pola-1, or div-1 RNAi (n = 3 experiments totaling >20 individual animals per RNAi treatment). c, d) The figure shows representative micrographs (c) and GFP quantification (d) of embryos laid by hsp-4p::gfp adult worms fed EV, pri-1, pri-2, pola-1, or div-1 RNAi (n = 3 experiments totaling >50 individual embryos per RNAi treatment). e and f) The figure shows representative micrographs (e) and GFP quantification (f) of embryos laid by hsp-4p::gfp, ire-1(zc14);hsp-4p::gfp, and xbp-1(zc12);hsp-4p::gfp adult worms irradiated with 400 J/m2 UV–C (n = 3 experiments repeats, totaling >50 individual embryos for each sample). g and h) The figure shows representative micrographs (g) and GFP quantification (h) of embryos laid by hsp-4::gfp adult worms irradiated with 400 J/m2 UV–C (n = 3 experiments totaling >50 individual embryos for UV-irradiated and nonirradiated samples). i and j) The figure shows representative micrographs (i) and GFP quantification (j) of embryos laid by hsp-16.2p::gfp adult worms irradiated with 400 J/m2 UV–C (n = 3 experiments totaling >50 individual embryos for UV-irradiated and nonirradiated samples). In all micrographs, the scale bar represents 100 μm. In dot plots, each dot represents the signal detected in one individual worm or embryo; the error bars represent standard deviation. Statistical analysis: b, d: ***P < 0.001, ****P < 0.0001 vs. EV RNAi control (Brown–Forsythe and Welch ANOVA test corrected for multiple comparisons using the Dunnett T3 method); f: **P < 0.01, ***P < 0.001, ****P < 0.0001 vs. nonirradiated embryos of the same genotype (ordinary 2-way ANOVA test corrected for multiple comparisons using Sidak's method); h, j: ns P > 0.05, ***P < 0.001, ****P < 0.0001 vs. nonirradiated embryos (Welch's t-test).
UV–C treatment phenocopies pri-1 or pri-2 RNAi
Like replication block, ultraviolet C (UV–C) light induces DSBs if the resulting bipyrimidine photoproducts are not resolved by the nucleotide excision repair (NER) pathway (Stergiou et al. 2011). Thus, we tested if UV–C exposure in early embryos phenocopies pri-1 or pri-2 RNAi treatment. We irradiated day 1 P0 adult worms with 400J/m2 UV–C and studied F1 embryos 24 h thereafter. Like pri-1 or pri-2 RNAi, UV–C treatment strongly activated hsp-4p::gfp (Fig. 5, e and f), in line with previously published observations (Deng et al. 2021). As observed for pri-1 or pri-2 RNAi, UV–C-induced hsp-4p::gfp upregulation was partially independent of ire-1 and xbp-1 in embryos (Fig. 5, e and f). The hsp-4::gfp translational reporter was also induced by UV–C (Fig. 5, g and h). By contrast, the cytosolic UPR reporter hsp-16.2p::gfp was not activated (Fig. 5, i and j), suggesting that UV–C specifically activates the UPR-ER in embryos. In the soma, UV–C caused ∼2-fold activation of hsp-16.2p::gfp, whereas hsp-4p::gfp, hsp-4::gfp, and hsp-6p::gfp were not activated (Supplementary Fig. 4). Collectively, these observations suggest that UV–C irradiation, like pri-1 or pri-2 RNAi, primarily activates the UPR-ER, especially in the embryo.
Inactivating components of the DNA repair machinery does not activate the UPR-ER in somatic cells
The abovementioned data raised the possibility that genotoxic stress in general activates the UPR-ER. To test this hypothesis, we used RNAi to inactivate several DNA repair genes, which should cause increased DNA damage, specifically: msh-2 (mismatch repair (Degtyareva et al. 2002)), xpf-1 (NER (Saito et al. 2009)), him-1 (a cohesin, whose loss results in chromosomal segregation defects in mitosis and meiosis (Chan et al. 2003)), mus-81 (replicative repair (O’Neil et al. 2013)), dog-1 and him-6 (whose loss causes formation of R-loops or G4 structures, causing deletions in poly-G tracts and genome instability (Cheung et al. 2002; Youds et al. 2006)), and cid-1 (DNA damage checkpoint, whose loss reverts HU-induced developmental arrest and activates hsp-4p::gfp (Olsen et al. 2006)). Unlike pri-1 or pri-2 RNAi, the knockdown of none of these genes activated any UPR-ER reporter in somatic cells (Supplementary Fig. 5). Notably, in our hands, cid-1 RNAi failed to induce hsp-4 (Supplementary Fig. 5), possibly because we initiate RNAi in synchronized L1 stage larvae and not in embryos. In embryos, inactivation of him-1 activated the UPR-ER (Supplementary Fig. 6), but RNAi of the other tested DNA repair genes did not. We conclude that inactivation of DDR and repair machinery genes does not consistently activate the UPR-ER.
The UPR-ER is not required to protect the germline against HU-induced replication stress
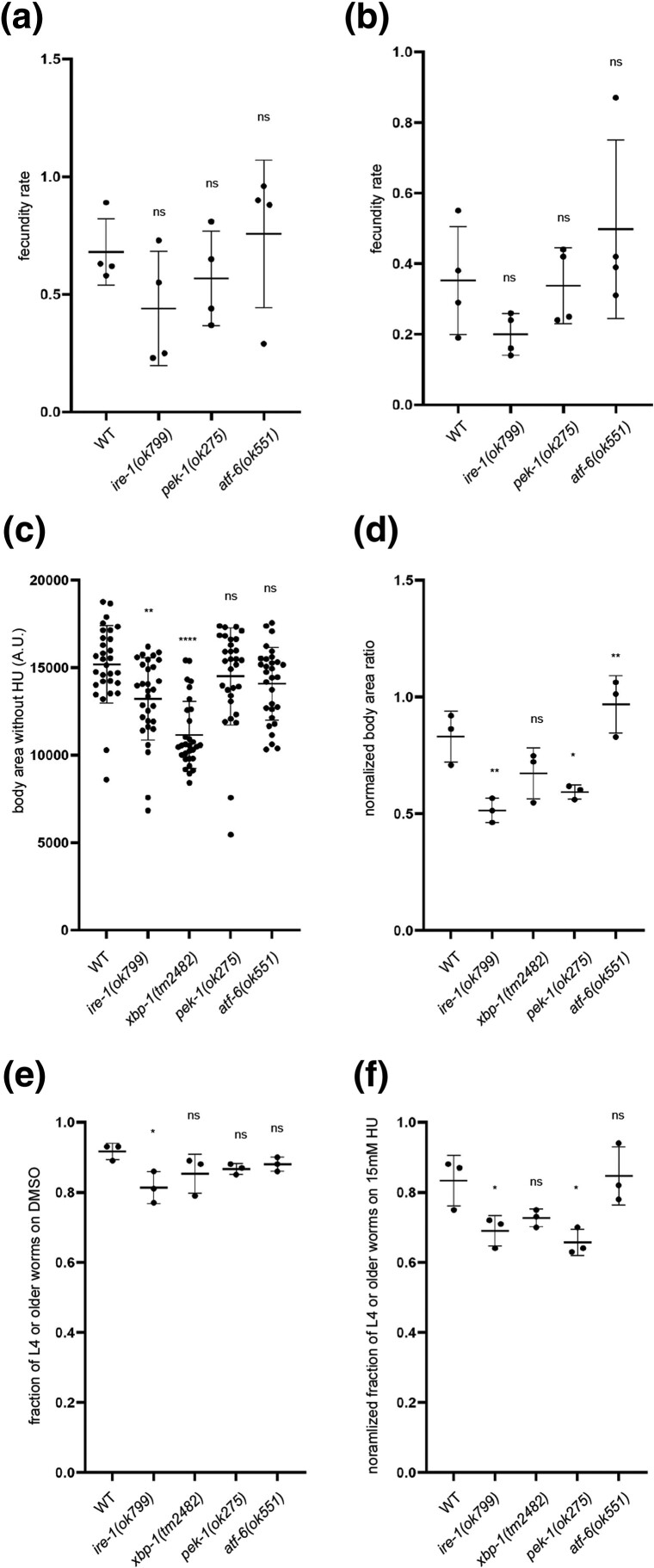
Because the UPR-ER is activated in somatic cells and embryos after pri-1 or pri-2 RNAi, we hypothesized that the UPR-ER protects worms from replication stress and the resulting DNA damage. To test this hypothesis, we used HU, a widely used chemical that inhibits ribonucleotide reductase, which reduces ribonucleosides into deoxyribonucleosides for DNA synthesis (Craig et al. 2012). In C. elegans, HU exposure leads to S-phase arrest, causing oversized nuclei in the mitotic germline, an extension of the duration of the first cell cycle in early embryos, and germline apoptosis (MacQueen and Villeneuve 2001; Garcia-Muse and Boulton 2005; Stevens et al. 2016). To quantify functional requirements of UPR-ER genes in response to replication inhibition, we compared the number of eggs laid per HU-exposed worm during a 4-h recovery period to the number of eggs laid by an unstressed worm of the same genotype, and also after prolonged replication stress by chronic HU exposure from the L1 stage onward. Acute and prolonged HU exposure both caused fecundity defects, but neither was exacerbated in the tested UPR-ER gene mutants (Fig. 6, a and b). Therefore, the UPR-ER is apparently dispensable to protect the C. elegans germline from replication stress.
Fig. 6.
ire-1 and pek-1 are required to protect C. elegans against HU-induced replication stress. a and b) The graphs show the relative fecundity of wild-type, ire-1(ok799), pek-1(ok275), or atf-6(ok551) worms subjected to 20 mM HU for 24 h from late L4 stage (a) or 10 mM HU for 72 h from L1 stage (b). Relative fecundity is calculated as follows: average number of eggs laid by an HU-treated worm during a 4-h postexposure period/average number of eggs laid by an unstressed worm of the same genotype during a 4-h period. c) Body size quantification of wild-type, ire-1(ok799), xbp-1(tm2482), pek-1(ok275), or atf-6(ok551) adult worms grown under unstressed conditions (n = 3 experiments totaling >30 animals per genotype). d) The graph shows the normalized average body area ratio of wild-type, ire-1(ok799), xbp-1(tm2482), pek-1(ok275), or atf-6(ok551) adult worms, calculated as follows: average body area of worms grown on 15 mM HU (n = 3 experiments totaling >30 individual HU-treated animals per genotype)/average body area of worms of the same genotype on DMSO. e) The graph shows the fractions of wild-type, ire-1(ok799), xbp-1(tm2482), pek-1(ok275), or atf-6(ok551) worms grown past L4 stage on DMSO at 48 h posthatching (n = 3 experiments totaling >90 individual animals per genotype). f) The graph shows normalized fraction of wild-type, ire-1(ok799), xbp-1(tm2482), pek-1(ok275), or atf-6(ok551) worms grown past L4 stage on 15 mM HU at 48 h posthatching, calculated as follows: fraction of worms past L4 on 15 mM HU/fraction of worms past L4 on DMSO (n = 3 experiments totaling >120 individual animals per genotype). In all graphs, the error bar represents standard deviation. Statistical analysis: ns P > 0.05, *P < 0.05, **P < 0.01, ****P < 0.0001 (ordinary 1-way ANOVA test corrected for multiple comparisons using Dunnett's method).
ire-1 and pek-1 are required to protect the soma from HU-induced replication stress
To test whether the UPR-ER protects somatic growth and development from damage caused by prolonged HU exposure, we measured the body area of worms grown on 15 mM HU from the L1 stage for 72 h. Because ire-1 and xbp-1 mutant worms have a smaller body size than wild-type worms (Fig. 6c), we normalized body size within each genotype (stressed/unstressed condition). We found that ire-1 and pek-1 mutant worms showed a reduced average body size ratio when exposed to HU (Fig. 6d). This suggests that the ire-1 and pek-1 branches of the UPR-ER are required for worms to tolerate or resolve prolonged replication stress to achieve normal somatic growth, while the atf-6 branch is dispensable.
In addition to body size, we also quantified developmental success of worm mutants in the absence of stress and following HU exposure. Loss of ire-1 caused developmental delay in unstressed conditions, whereas loss of other UPR-ER components did not (Fig. 6e). When exposed to 15 mM HU from the L1 stage on, ire-1 or pek-1 mutant worms showed a reduced ability to progress past the L4 stage within 48 h, whereas atf-6 or xbp-1 mutation had no effect (Fig. 6f). Collectively, these data show that the ire-1 and pek-1 branches, but not the atf-6 branch, are required to maintain somatic resistance to replication stress.
Transcriptome analysis of pri-1 RNAi-treated embryos suggests deregulated glycosylation, calcium signaling, and fatty acid desaturation as potential sources of ER stress
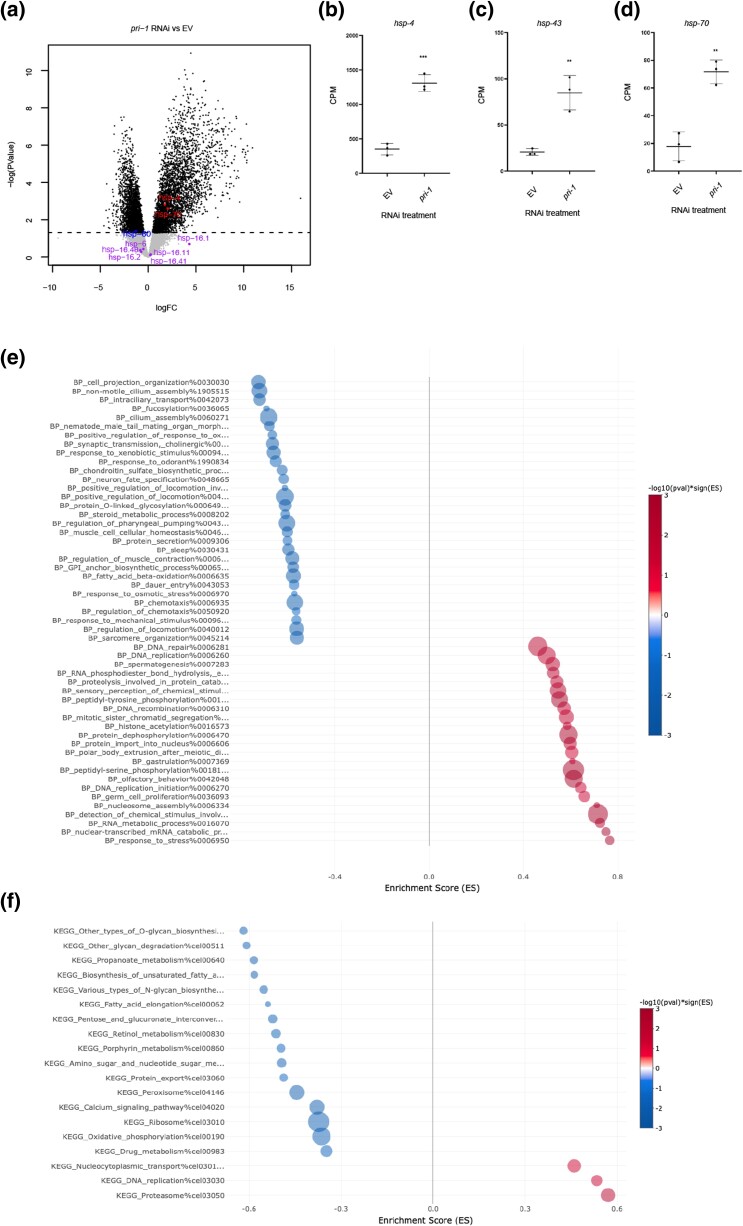
To identify genes and processes altered by replication fork stalling, we studied the transcriptomes of wild-type embryos treated with EV or pri-1 RNAi using RNA-seq. We identified 2785 genes that were up- and 1738 genes that were downregulated following pri-1 depletion (Fig. 7a; Supplementary Tables 1–3 and Fig. 7). In line with the abovementioned data, hsp-4 was significantly induced following pri-1 depletion, as were two other chaperones, hsp-43 and hsp-70 (Fig. 7, b–d; Supplementary Tables 1 and 2); others have reported that hsp-70 is induced by tunicamycin in an xbp-1-dependent fashion (Urano et al. 2002; Lim et al. 2014), suggesting that it is an effector chaperone of the UPR-ER. By contrast, neither the mitochondrial UPR chaperones hsp-6 and hsp-60, nor any of the cytoplasmic UPR chaperones of the hsp-16 family (hsp-16.1, hsp-16.11, hsp-16.2, hsp-16.41, and hsp-16.48) were induced (Supplementary Tables 1–3 and Fig. 8; Fig. 7a). Hence, unbiased transcriptome profiling confirms that the UPR-ER is specifically activated in embryos experiencing replication fork stress, whereas other UPRs are not.
Fig. 7.
Replication stress in embryos alters protein glycosylation, calcium signaling, and fatty acid desaturation. a) The volcano plot shows the expression of all detected genes in EV and pri-1 RNAi-treated embryos. X-axis, logFC; Y-axis, -log10(P-value). Black, P-value <0.05; gray, P-value ≥0.05; blue, highlighted and significantly downregulated; red, highlighted and significantly downregulated; purple, highlighted but not significant. b–d) The graph shows average transcript levels in counts per million (CPM) of hsp-4, hsp-43, and hsp-70 mRNA in EV or pri-1 RNAi-treated embryos; the error bars represent standard deviation (n = 3 experiments). Statistics: **P < 0.01, ***P < 0.001 (unpaired Student's t-test). e and f) The bubble plots show processes enriched negatively (blue, enrichment score <0) and positively (red, enrichment score >0) in pri-1 RNAi-treated worms, based on the BP (e) and KEGG (f) databases. Bubbles represent the top 30 or fewer gene sets determined to be statistically significant (cutoffs P < 0.05, Padj < 0.25), as determined by analysis with the easyGSEA function of the eVITTA webserver (Cheng et al. 2021). The size of the bubble corresponds to the number of genes represented in each gene set. X-axis: enrichment score (ES).
To delineate how replication fork stress could induce the UPR-ER, we performed GSEA using the BP and KEGG databases. As expected, terms relating to the UPR-ER stress response were enriched, e.g. the terms “BP_response_to_stress%0006950” (which includes hsp-4 and hsp-70) and “BP_PERK-mediated_unfolded_protein_response%0036499” (Fig. 7, e–f, Supplementary Tables 4 and 5). Furthermore, we observed an enrichment of terms related to DNA replication and DNA repair (Fig. 7, e and f, Supplementary Tables 4 and 5), as expected in worms experiencing replication stress. For example, “KEGG_DNA_replication%cel03030” was one of only three upregulated terms when using the KEGG database for analysis, while upregulated terms identified with the BP database included terms such as “BP_DNA_replication_initiation%000627”, “BP_DNA_repair%0006281”, “BP_DNA_recombination%0006310”, and “BP_double-strand_break_repair_via_homologous_recombination%0000724”. Finally, our analysis identified several processes whose downregulation could indicate the source of UPR-ER activation in pri-1 RNAi-treated worms. Specifically, when using analysis with the KEGG database, of the only 13 downregulated terms, three relate to protein N- and O-glycosylation (“KEGG_Various_types_of_N-glycan_biosynthesis%cel00513”, “KEGG_Other_glycan_degradation%cel00511”, “KEGG_Other_types_of_O-glycan_biosynthesis%cel00514”), 1 relates to calcium signaling (“KEGG_Calcium_signaling_pathway%cel04020”), and 2 relate to the biosynthesis of unsaturated fatty acids (“KEGG_Fatty_acid_elongation%cel00062”, “KEGG_Biosynthesis_of_unsaturated_fatty_acids%cel01040”). We conclude that the dysregulation of multiple cellular processes by replication fork stalling after pri-1 RNAi likely activates the UPR-ER in the embryos.
Discussion
Animals such as C. elegans consistently experience and must handle diverse stresses in their environment. Optimal adaptation to such insults requires the deployment of multiple response pathways, and recent studies have identified functional crosstalk between stress responses such as the UPR-ER and the DDR. Here, we show that replication fork stalling strongly induces two branches of the UPR-ER in C. elegans embryos. In turn, the UPR-ER is required to protect worms from the deleterious effects of stalled replication forks. Surprisingly, analysis of transcriptional reporters and transcriptome data suggest that it is primarily the UPR-ER that is induced by this stress, whereas other UPRs are only mildly activated. Our data suggest that replication fork stalling specifically causes ER dysfunction, possibly by disturbing cellular processes that are unique, or especially important, to ER function.
Replication fork stalling selectively activates the UPR-ER
The UPR-ER, the cytosolic UPR, and the mitochondrial UPR are interconnected adaptive pathways that ensure homeostasis in the face of stress. Conditions that impair general protein folding such as oxidative stress and protein degradation defects induce all three UPRs in C. elegans (Rodrigues et al. 2011; Hou et al. 2014; Bartoszewska and Collawn 2020; Taylor et al. 2021). Here, we observed robust induction of 2 UPR-ER reporters by pri-1 or pri-2 RNAi or UV–C irradiation, but did not observe the induction of cytosolic or mitochondrial UPR reporters. This lack of induction was confirmed by our unbiased transcriptome profiling. Together, these data indicate that ER proteostasis or membrane lipid equilibrium, but not cytosolic or mitochondrial proteostasis, is disturbed by replication stress. This selectivity appears to rule out a mechanism whereby DNA replication fork stalling causes protein misfolding, aggregation, and proteotoxicity in all organelles. A clue regarding the specific mechanisms underlying UPR-ER activation was revealed by transcriptome profiling following pri-1 depletion, which revealed dysregulated molecular processes and pathways with important links to the ER. These include protein glycosylation, which is important for the modification of secreted, ER-synthesized proteins; calcium metabolism, which is vital for protein folding in the ER and whose interference via thapsigargin is widely used to study ER protein processing; and fatty acid desaturation, which is essential for maintaining normal ER membrane lipid composition and is monitored by the UPR-ER (Hou et al. 2014; Denzel and Antebi 2015; Burkewitz et al. 2020; Ho et al. 2020; Deng et al. 2021; Xu and Taubert 2021). None of these processes are known to impact the activity of the cytoplasmic or mitochondrial UPRs, which may explain why replication fork stalling selectively activates the UPR-ER. However, it remains unclear why genes in these pathways are dysregulated by replication fork stalling.
The outer nuclear membrane is continuous with the ER and linked to the lumen of the ER, suggesting that ER stress responses may be directly linked to disturbances in nuclear processes such as DNA replication and transcription. For example, UPR-ER activation indirectly helps removing stalled replication protein complexes, and thus restarts replication by helping replication fork turnover; although such a mechanism may be less relevant in UV–C-treated animals with DNA damage, it could be especially relevant in animals with reduced primase complex activity.
Replication stress induces noncanonical, atf-6-dependent hsp-4 expression in embryos
The hsp-4p::gfp reporter is widely used in C. elegans to monitor the activity of the UPR-ER and to infer the presence of ER stress (Calfon et al. 2002; Urano et al. 2002; Ho et al. 2020). Canonical activation of this reporter depends strictly on the IRE-1-XBP-1 pathway. Here, we observed only partially ire-1- and xbp-1-dependent activation of hsp-4 after pri-1 or pri-2 depletion in embryos. This is surprising because IRE-1 is the only known UPR-ER sensor that processes the unspliced xbp-1u mRNA into the mature xbp-1s product, which is then translated into XBP-1, the transcription factor that upregulates hsp-4 expression (Walter and Ron 2011; Gardner et al. 2013; Senft and Ronai 2015; Kopp et al. 2018; Hetz et al. 2020; Xu and Taubert 2021). Because ATF6 is required to express XBP1 mRNA in humans, we studied atf-6, which represents the third branch the UPR-ER in C. elegans but is thought to be largely dispensable in this organism for stress-induced UPR-ER activity, as many ER stress-activated genes do not require atf-6 for induction (Shen et al. 2005; Lee et al. 2007). Interestingly, atf-6 loss of function significantly diminished pri-1 or pri-2 RNAi-induced hsp-4p::gfp activation in embryos. In somatic cells, transient hsp-4 induction that is independent of ire-1 and xbp-1 occurs during the differentiation of stem-like seam cells into alae-secreting cells (Zha et al. 2019). Although the transcriptional factor B-lymphocyte–induced maturation protein 1 (blmp-1) is required to suppress hsp-4 in this context (Zha et al. 2019), how it is activated is unknown. Our data suggest that atf-6 may be involved in this process, in line with the view that atf-6 plays important roles in C. elegans development. In sum, our data identify an important new nuance of the mechanisms that fine-tune UPR-ER activation in C. elegans.
ire-1 and pek-1 are required for resistance to replication fork stress
A bidirectional crosstalk between the DDR and the UPR-ER has begun to emerge (González-Quiroz et al. 2020; Bolland et al. 2021). Yeast IRE1 is required for survival on HU (Zha et al. 2019). We found that the IRE-1 branch is required to protect C. elegans during prolonged HU exposure initiated at an early developmental stage. By contrast, short-term acute HU exposure at a later developmental stage was tolerated, similarly to what has been reported about treating ire-1 worms at L4 stage with rad-51 RNAi to induce DNA damage (Levi-Ferber et al. 2014).
Little evidence exists for the roles of the other two branches in response to genotoxicity. We report here that the PEK-1 branch is activated by replication fork stalling and that pek-1 is required for somatic resistance to HU. By contrast, the atf-6 branch was not required to protect the soma or germline from HU. This was surprising because, as noted above, atf-6 is required to induce hsp-4 in embryos. Nevertheless, our data implicate the UPR-ER as a whole in response to replication fork stalling. Future work will be required to define ire-1- and pek-1-dependent processes that promote survival and growth in genotoxic conditions.
Contributor Information
Jiaming Xu, Graduate Program in Cell & Developmental Biology, The University of British Columbia, 950 W 28th Ave, Vancouver, BC V5Z 4H4, Canada; Centre for Molecular Medicine and Therapeutics, The University of British Columbia, 950 W 28th Ave, Vancouver, BC V5Z 4H4, Canada; British Columbia Children’s Hospital Research Institute, 950 W 28th Ave, Vancouver, BC V5Z 4H4, Canada.
Brendil Sabatino, Centre for Molecular Medicine and Therapeutics, The University of British Columbia, 950 W 28th Ave, Vancouver, BC V5Z 4H4, Canada; British Columbia Children’s Hospital Research Institute, 950 W 28th Ave, Vancouver, BC V5Z 4H4, Canada; Department of Medical Genetics, The University of British Columbia, 950 W 28th Ave, Vancouver, BC V5Z 4H4, Canada.
Junran Yan, Centre for Molecular Medicine and Therapeutics, The University of British Columbia, 950 W 28th Ave, Vancouver, BC V5Z 4H4, Canada; British Columbia Children’s Hospital Research Institute, 950 W 28th Ave, Vancouver, BC V5Z 4H4, Canada; Edwin S.H. Leong Centre for Healthy Aging, The University of British Columbia, 117-2194 Health Sciences Mall, Vancouver, BC V6T 1Z3, Canada; Department of Medical Genetics, The University of British Columbia, 950 W 28th Ave, Vancouver, BC V5Z 4H4, Canada.
Glafira Ermakova, Centre for Molecular Medicine and Therapeutics, The University of British Columbia, 950 W 28th Ave, Vancouver, BC V5Z 4H4, Canada; British Columbia Children’s Hospital Research Institute, 950 W 28th Ave, Vancouver, BC V5Z 4H4, Canada; Edwin S.H. Leong Centre for Healthy Aging, The University of British Columbia, 117-2194 Health Sciences Mall, Vancouver, BC V6T 1Z3, Canada; Department of Medical Genetics, The University of British Columbia, 950 W 28th Ave, Vancouver, BC V5Z 4H4, Canada.
Kelsie R S Doering, Centre for Molecular Medicine and Therapeutics, The University of British Columbia, 950 W 28th Ave, Vancouver, BC V5Z 4H4, Canada; British Columbia Children’s Hospital Research Institute, 950 W 28th Ave, Vancouver, BC V5Z 4H4, Canada; Edwin S.H. Leong Centre for Healthy Aging, The University of British Columbia, 117-2194 Health Sciences Mall, Vancouver, BC V6T 1Z3, Canada; Department of Medical Genetics, The University of British Columbia, 950 W 28th Ave, Vancouver, BC V5Z 4H4, Canada.
Stefan Taubert, Graduate Program in Cell & Developmental Biology, The University of British Columbia, 950 W 28th Ave, Vancouver, BC V5Z 4H4, Canada; Centre for Molecular Medicine and Therapeutics, The University of British Columbia, 950 W 28th Ave, Vancouver, BC V5Z 4H4, Canada; British Columbia Children’s Hospital Research Institute, 950 W 28th Ave, Vancouver, BC V5Z 4H4, Canada; Edwin S.H. Leong Centre for Healthy Aging, The University of British Columbia, 117-2194 Health Sciences Mall, Vancouver, BC V6T 1Z3, Canada; Department of Medical Genetics, The University of British Columbia, 950 W 28th Ave, Vancouver, BC V5Z 4H4, Canada.
Data availability
The data described in this study are available in the main manuscript, the Supplementary material, or in a public repository. Supplementary Figs. 1, 3, 4, 5, and 6 describe additional experiments using GFP reporters, Supplementary Fig. 2 contains additional repeats of immunoblots, and Supplementary Figs. 7 and 8 provide additional description of the RNA-seq analysis. Supplementary Tables 1–3 contain lists describing gene expression data identified by RNA-seq; Supplementary Tables 4–5 contain lists describing processes identified by RNA-seq analysis. Supplementary material is available Xu J, Sabatino B, Yan J, Ermakova G, Doering KRS, Taubert S; 2023; Supplemental Material for Xu et al., 2023; Figshare: https://doi.org/10.25387/g3.24941133. Raw and processed RNA-seq files have been deposited: Sabatino B, Xu J, Taubert S; 2023; Effect of pri-1 (DNA primase) RNAi on gene expression in embryos of Caenorhaditis elegans; Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/); GSE225569. See the methods for information on reagents and strains. C. elegans strains described for the first time in this study can be requested from the authors.
Funding
This work was funded by grant support from The Canadian Institutes of Health Research (CIHR; PJT-153199, PJT-186144 to S.T.) and the Natural Sciences and Engineering Research Council of Canada (NSERC; RGPIN-2018-05133 to S.T.). J.X. and J.Y. were supported by scholarships from BC Children’s Hospital Research Institute, J.X. by a scholarships from the Pei-Huang Tung and Tan-Wen Tung Graduate Fellowships of The University of British Columbia (UBC) and from the UBC Cell & Developmental Biology Graduate Program, B.S. and G.E. by scholarships from the UBC Medical Genetics Graduate Program, and J.Y. by a UBC 4-year fellowship (4YF). Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440).
Literature cited
- Acosta-Alvear D, Zhou Y, Blais A, Tsikitis M, Lents NH, Arias C, Lennon CJ, Kluger Y, Dynlacht BD. 2007. XBP1 controls diverse cell type- and condition-specific transcriptional regulatory networks. Mol Cell. 27(1):53–66. doi: 10.1016/j.molcel.2007.06.011. [DOI] [PubMed] [Google Scholar]
- Adams CJ, Kopp MC, Larburu N, Nowak PR, Ali MMU. 2019. Structure and molecular mechanism of ER stress signaling by the unfolded protein response signal activator IRE1. Frontiers Mol Biosci. 6:11. doi: 10.3389/fmolb.2019.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arantes-Oliveira N, Apfeld J, Dillin A, Kenyon C. 2002. Regulation of life-span by germ-line stem cells in Caenorhabditis elegans. Science. 295(5554):502–505. doi: 10.1126/science.1065768. [DOI] [PubMed] [Google Scholar]
- Argemí J, Kress TR, Chang HCY, Ferrero R, Bértolo C, Moreno H, González-Aparicio M, Uriarte I, Guembe L, Segura V, et al. 2017. X-box binding protein 1 regulates unfolded protein, acute-phase, and DNA damage responses during regeneration of mouse liver. Gastroenterology. 152(5):1203–1216.e15. doi: 10.1053/j.gastro.2016.12.040. [DOI] [PubMed] [Google Scholar]
- Bartoszewska S, Collawn JF. 2020. Unfolded protein response (UPR) integrated signaling networks determine cell fate during hypoxia. Cell Mol Biol Lett. 25(1):18. doi: 10.1186/s11658-020-00212-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett CF, Wende HV, Simko M, Klum S, Barfield S, Choi H, Pineda VV, Kaeberlein M. 2014. Activation of the mitochondrial unfolded protein response does not predict longevity in Caenorhabditis elegans. Nat Commun. 5(1):3483. doi: 10.1038/ncomms4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics. 30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolland H, Ma TS, Ramlee S, Ramadan K, Hammond EM. 2021. Links between the unfolded protein response and the DNA damage response in hypoxia: a systematic review. Biochem Soc T. 49(3):1251–1263. doi: 10.1042/bst20200861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd WA, Crocker TL, Rodriguez AM, Leung MCK, Wade Lehmann D, Freedman JH, Van Houten B, Meyer JN. 2010. Nucleotide excision repair genes are expressed at low levels and are not detectably inducible in Caenorhabditis elegans somatic tissues, but their function is required for normal adult life after UVC exposure. Mutat Res Fundam Mol Mech Mutagen. 683(1–2):57–67. doi: 10.1016/j.mrfmmm.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkewitz K, Feng G, Dutta S, Kelley CA, Steinbaugh M, Cram EJ, Mair WB. 2020. Atf-6 regulates lifespan through ER-mitochondrial calcium homeostasis. Cell Rep. 32(10):108125. doi: 10.1016/j.celrep.2020.108125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- C. elegans Deletion Mutant Consortium . 2012. Large-Scale screening for targeted knockouts in the Caenorhabditis elegans genome. G3 (Bethesda). 2(11):1415–1425. doi: 10.1534/g3.112.003830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Clark SG, Ron D. 2002. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 415(6867):92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- Celik C, Lee SYT, Yap WS, Thibault G. 2023. Endoplasmic reticulum stress and lipids in health and diseases. Prog Lipid Res. 89:101198. doi: 10.1016/j.plipres.2022.101198. [DOI] [PubMed] [Google Scholar]
- Chagoyen M, Pazos F. 2010. Quantifying the biological significance of gene ontology biological processes—implications for the analysis of systems-wide data. Bioinformatics. 26(3):378–384. doi: 10.1093/bioinformatics/btp663. [DOI] [PubMed] [Google Scholar]
- Chan RC, Chan A, Jeon M, Wu TF, Pasqualone D, Rougvie AE, Meyer BJ. 2003. Chromosome cohesion is regulated by a clock gene paralogue TIM-1. Nature. 423(6943):1002–1009. doi: 10.1038/nature01697. [DOI] [PubMed] [Google Scholar]
- Cheng X, Yan J, Liu Y, Wang J, Taubert S. 2021. eVITTA: a web-based visualization and inference toolbox for transcriptome analysis. Nucleic Acids Res. 49(W1):W207–W215. doi: 10.1093/nar/gkab366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung I, Schertzer M, Rose A, Lansdorp PM. 2002. Disruption of dog-1 in Caenorhabditis elegans triggers deletions upstream of guanine-rich DNA. Nat Genet. 31(4):405–409. doi: 10.1038/ng928. [DOI] [PubMed] [Google Scholar]
- Craig AL, Moser SC, Bailly AP, Gartner A. 2012. Methods for studying the DNA damage response in the Caenorhabdatis elegans germ line. Methods Cell Biol. 107:321–352. doi: 10.1016/b978-0-12-394620-1.00011-4. [DOI] [PubMed] [Google Scholar]
- Degtyareva NP, Greenwell P, Hofmann ER, Hengartner MO, Zhang L, Culotti JG, Petes TD. 2002. Caenorhabditis elegans DNA mismatch repair gene msh-2 is required for microsatellite stability and maintenance of genome integrity. Proc Natl Acad Sci. 99(4):2158–2163. doi: 10.1073/pnas.032671599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J, Bai X, Tang H, Pang S. 2021. DNA damage promotes ER stress resistance through elevation of unsaturated phosphatidylcholine in Caenorhabditis elegans. J Biol Chem. 296:100095. doi: 10.1074/jbc.ra120.016083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denzel MS, Antebi A. 2015. Hexosamine pathway and (ER) protein quality control. Curr Opin Cell Biol. 33:14–18. doi: 10.1016/j.ceb.2014.10.001. [DOI] [PubMed] [Google Scholar]
- Doering KR, Cheng X, Milburn L, Ratnappan R, Ghazi A, Miller DL, Taubert S. 2022. Nuclear hormone receptor NHR-49 acts in parallel with HIF-1 to promote hypoxia adaptation in Caenorhabditis elegans. Elife. 11:e67911. doi: 10.7554/elife.67911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufey E, Pedro JMB-S, Eggers C, González-Quiroz M, Urra H, Sagredo AI, Sepulveda D, Pihán P, Carreras-Sureda A, Hazari Y, et al. 2020. Genotoxic stress triggers the activation of IRE1α-dependent RNA decay to modulate the DNA damage response. Nat Commun. 11(1):2401–2413. doi: 10.1038/s41467-020-15694-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermolaeva MA, Segref A, Dakhovnik A, Ou H-L, Schneider JI, Utermöhlen O, Hoppe T, Schumacher B. 2013. DNA damage in germ cells induces an innate immune response that triggers systemic stress resistance. Nature. 501(7467):416–420. doi: 10.1038/nature12452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Muse T, Boulton SJ. 2005. Distinct modes of ATR activation after replication stress and DNA double-strand breaks in Caenorhabditis elegans. EMBO J. 24(24):4345–4355. doi: 10.1038/sj.emboj.7600896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner BM, Pincus D, Gotthardt K, Gallagher CM, Walter P. 2013. Endoplasmic reticulum stress sensing in the unfolded protein response. Cold Spring Harb Perspect Biol. 5(3):a013169. doi: 10.1101/cshperspect.a013169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartner A, Engebrecht J. 2021. DNA repair, recombination, and damage signaling. Genetics. 220(2):iyab178. doi: 10.1093/genetics/iyab178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh GYS, Winter JJ, Bhanshali F, Doering KRS, Lai R, Lee K, Veal EA, Taubert S. 2018. NHR-49/HNF4 integrates regulation of fatty acid metabolism with a protective transcriptional response to oxidative stress and fasting. Aging Cell. 17(3):e12743. doi: 10.1111/acel.12743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Quiroz M, Blondel A, Sagredo A, Hetz C, Chevet E, Pedeux R. 2020. When endoplasmic reticulum proteostasis meets the DNA damage response. Trends Cell Biol. 30(11):881–891. doi: 10.1016/j.tcb.2020.09.002. [DOI] [PubMed] [Google Scholar]
- Guilliam TA, Keen BA, Brissett NC, Doherty AJ. 2015. Primase-polymerases are a functionally diverse superfamily of replication and repair enzymes. Nucleic Acids Res. 43(14):6651–6664. doi: 10.1093/nar/gkv625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson ST, Johnson TE. 2001. daf-16 integrates developmental and environmental inputs to mediate aging in the nematode Caenorhabditis elegans. Curr Biol. 11(24):1975–1980. doi: 10.1016/S0960-9822(01)00594-2. [DOI] [PubMed] [Google Scholar]
- Henry KA, Blank HM, Hoose SA, Polymenis M. 2010. The unfolded protein response is not necessary for the G1/S transition, but it is required for chromosome maintenance in Saccharomyces cerevisiae. PLoS One. 5(9):e12732. doi: 10.1371/journal.pone.0012732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetz C, Zhang K, Kaufman RJ. 2020. Mechanisms, regulation and functions of the unfolded protein response. Nat Rev Mol Cell Bio. 21(8):421–438. doi: 10.1038/s41580-020-0250-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillary RF, FitzGerald U. 2018. A lifetime of stress: ATF6 in development and homeostasis. J Biomed Sci. 25(1):48. doi: 10.1186/s12929-018-0453-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho N, Yap WS, Xu J, Wu H, Koh JH, Goh WWB, George B, Chong SC, Taubert S, Thibault G. 2020. Stress sensor Ire1 deploys a divergent transcriptional program in response to lipid bilayer stress. J Cell Biol. 219(7):e201909165. doi: 10.1083/jcb.201909165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotokezaka Y, Katayama I, Nakamura T. 2020. ATM-associated signalling triggers the unfolded protein response and cell death in response to stress. Commun Biol. 3(1):378. doi: 10.1038/s42003-020-1102-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou NS, Gutschmidt A, Choi DY, Pather K, Shi X, Watts JL, Hoppe T, Taubert S. 2014. Activation of the endoplasmic reticulum unfolded protein response by lipid disequilibrium without disturbed proteostasis in vivo. Proc Natl Acad Sci U S A. 111(22):E2271–E2280. doi: 10.1073/pnas.1318262111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SP, Bartek J. 2009. The DNA-damage response in human biology and disease. Nature. 461(7267):1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M. 1997. A database for post-genome analysis. Trends Genet. 13(9):375–376. doi: 10.1016/s0168-9525(97)01223-7. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Furumichi M, Sato Y, Ishiguro-Watanabe M, Tanabe M. 2020. KEGG: integrating viruses and cellular organisms. Nucleic Acids Res. 49(D1):D545–D551. doi: 10.1093/nar/gkaa970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp MC, Nowak PR, Larburu N, Adams CJ, Ali MM. 2018. In vitro FRET analysis of IRE1 and BiP association and dissociation upon endoplasmic reticulum stress. Elife. 7:e30257. doi: 10.7554/elife.30257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, Singaravelu G, Park B-J, Ahnn J. 2007. Differential requirement of unfolded protein response pathway for calreticulin expression in Caenorhabditis elegans. J Mol Biol. 372(2):331–340. doi: 10.1016/j.jmb.2007.06.071. [DOI] [PubMed] [Google Scholar]
- Lee K, Tirasophon W, Shen X, Michalak M, Prywes R, Okada T, Yoshida H, Mori K, Kaufman RJ. 2002. IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 16(4):452–466. doi: 10.1101/gad.964702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levi-Ferber M, Salzberg Y, Safra M, Haviv-Chesner A, Bülow HE, Henis-Korenblit S. 2014. It's all in your mind: determining germ cell fate by neuronal IRE-1 in C. elegans. PLoS Genet. 10(10):e1004747. doi: 10.1371/journal.pgen.1004747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim Y, Lee D, Kalichamy K, Hong S-E, Michalak M, Ahnn J, Kim DH, Lee SK. 2014. Sumoylation regulates ER stress response by modulating calreticulin gene expression in XBP-1-dependent mode in Caenorhabditis elegans. Int J Biochem Cell Biol. 53:399–408. doi: 10.1016/j.biocel.2014.06.005. [DOI] [PubMed] [Google Scholar]
- Lyu X, Zhang M, Li G, Cai Y, Li G, Qiao Q. 2019. Interleukin-6 production mediated by the IRE1-XBP1 pathway confers radioresistance in human papillomavirus-negative oropharyngeal carcinoma. Cancer Sci. 110(8):2471–2484. doi: 10.1111/cas.14094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQueen AJ, Villeneuve AM. 2001. Nuclear reorganization and homologous chromosome pairing during meiotic prophase require C. elegans chk-2. Gene Dev. 15(13):1674–1687. doi: 10.1101/gad.902601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure AW, Canal B, Diffley JFX. 2022. A DNA replication fork-centric view of the budding yeast DNA damage response. DNA Repair (Amst). 119:103393. doi: 10.1016/j.dnarep.2022.103393. [DOI] [PubMed] [Google Scholar]
- McQuiston A, Diehl JA. 2017. Recent insights into PERK-dependent signaling from the stressed endoplasmic reticulum. F1000Res. 6:1897. doi: 10.12688/f1000research.12138.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nukazuka A, Fujisawa H, Inada T, Oda Y, Takagi S. 2008. Semaphorin controls epidermal morphogenesis by stimulating mRNA translation via eIF2α in Caenorhabditis elegans. Gene Dev. 22(8):1025–1036. doi: 10.1101/gad.1644008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neil NJ, Martin JS, Youds JL, Ward JD, Petalcorin MIR, Rose AM, Boulton SJ. 2013. Joint molecule resolution requires the redundant activities of MUS-81 and XPF-1 during Caenorhabditis elegans meiosis. PLoS Genet. 9(7):e1003582. doi: 10.1371/journal.pgen.1003582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen A, Vantipalli MC, Lithgow GJ. 2006. Checkpoint proteins control survival of the postmitotic cells in Caenorhabditis elegans. Science. 312(5778):1381–1385. doi: 10.1126/science.1124981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaevgeniou N, Chondrogianni N. 2014. The ubiquitin proteasome system in Caenorhabditis elegans and its regulation. Redox Biol. 2:333–347. doi: 10.1016/j.redox.2014.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. 2017. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 14(4):417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnappan R, Amrit FRG, Chen S-W, Gill H, Holden K, Ward J, Yamamoto KR, Olsen CP, Ghazi A. 2014. Germline signals deploy NHR-49 to modulate fatty-acid β-oxidation and desaturation in somatic tissues of C. elegans. PLoS Genet. 10(12):e1004829. doi: 10.1371/journal.pgen.1004829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea SL, Wu D, Cypser JR, Vaupel JW, Johnson TE. 2005. A stress-sensitive reporter predicts longevity in isogenic populations of Caenorhabditis elegans. Nat Genet. 37(8):894–898. doi: 10.1038/ng1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson CE, Kinkel S, Kim DH. 2011. Physiological IRE-1-XBP-1 and PEK-1 signaling in Caenorhabditis elegans larval development and immunity. PLoS Genet. 7(11):e1002391. doi: 10.1371/journal.pgen.1002391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson CE, Kooistra T, Kim DH. 2010. An essential role for XBP-1 in host protection against immune activation in C. elegans. Nature. 463(7284):1092–1095. doi: 10.1038/nature08762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. Edger: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues AJ, Neves-Carvalho A, Teixeira-Castro A, Rokka A, Corthals G, Logarinho E, Maciel P. 2011. Absence of ataxin-3 leads to enhanced stress response in C. elegans. PLoS One. 6(4):e18512. doi: 10.1371/journal.pone.0018512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito TT, Youds JL, Boulton SJ, Colaiácovo MP. 2009. Caenorhabditis elegans HIM-18/SLX-4 interacts with SLX-1 and XPF-1 and maintains genomic integrity in the germline by processing recombination intermediates. PLoS Genet. 5(11):e1000735. doi: 10.1371/journal.pgen.1000735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senft D, Ronai ZA. 2015. UPR, autophagy, and mitochondria crosstalk underlies the ER stress response. Trends Biochem Sci. 40(3):141–148. doi: 10.1016/j.tibs.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Ellis RE, Lee K, Liu CY, Yang K, Solomon A, Yoshida H, Morimoto R, Kurnit DM, Mori K, et al. 2001. Complementary signaling pathways regulate the unfolded protein response and are required for C. elegans development. Cell. 107(7):893–903. doi: 10.1016/S0092-8674(01)00612-2. [DOI] [PubMed] [Google Scholar]
- Shen X, Ellis RE, Sakaki K, Kaufman RJ. 2005. Genetic interactions due to constitutive and inducible gene regulation mediated by the unfolded protein response in C. elegans. PLoS Genet. 1(3):e37. doi: 10.1371/journal.pgen.0010037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shomer N, Kadhim AZ, Grants JM, Cheng X, Alhusari D, Bhanshali F, Poon AFY, Lee MYY, Muhuri A, Park JI, et al. 2019. Mediator subunit MDT-15/MED15 and nuclear receptor HIZR-1/HNF4 cooperate to regulate toxic metal stress responses in Caenorhabditis elegans. PLoS Genet. 15(12):e1008508. doi: 10.1371/journal.pgen.1008508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soneson C, Love MI, Robinson MD. 2016. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res. 4:1521. doi: 10.12688/f1000research.7563.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stergiou L, Eberhard R, Doukoumetzidis K, Hengartner MO. 2011. NER and HR pathways act sequentially to promote UV-C-induced germ cell apoptosis in Caenorhabditis elegans. Cell Death Differ. 18(5):897–906. doi: 10.1038/cdd.2010.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens H, Williams AB, Michael WM. 2016. Cell-Type specific responses to DNA replication stress in early C. elegans embryos. PLoS One. 11(10):e0164601. doi: 10.1371/journal.pone.0164601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao R, Chen H, Gao C, Xue P, Yang F, Han JDJ, Zhou B, Chen YG. 2011. Xbp1-mediated histone H4 deacetylation contributes to DNA double-strand break repair in yeast. Cell Res. 21(11):1619–1633. doi: 10.1038/cr.2011.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SKB, Minhas MH, Tong J, Selvaganapathy PR, Mishra RK, Gupta BP. 2021. C. elegans electrotaxis behavior is modulated by heat shock response and unfolded protein response signaling pathways. Sci Rep. 11(1):3115. doi: 10.1038/s41598-021-82466-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TeKippe M, Aballay A. 2010. C. elegans germline-deficient mutants respond to pathogen infection using shared and distinct mechanisms. PLoS One. 5(7):e11777. doi: 10.1371/journal.pone.0011777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano F, Calfon M, Yoneda T, Yun C, Kiraly M, Clark SG, Ron D. 2002. A survival pathway for Caenorhabditis elegans with a blocked unfolded protein response. J Cell Biol. 158(4):639–646. doi: 10.1083/jcb.200203086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Meer G, Voelker DR, Feigenson GW. 2008. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol. 9(2):112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, Ron D. 2011. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 334(6059):1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- Xu J, Taubert S. 2021. Beyond proteostasis: lipid metabolism as a new player in ER homeostasis. Metabolites. 11(1):52. doi: 10.3390/metabo11010052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneda T, Benedetti C, Urano F, Clark SG, Harding HP, Ron D. 2004. Compartment-specific perturbation of protein handling activates genes encoding mitochondrial chaperones. J Cell Sci. 117(18):4055–4066. doi: 10.1242/jcs.01275. [DOI] [PubMed] [Google Scholar]
- Yoon DS, Cha DS, Alfhili MA, Keiper BD, Lee M. 2018. Subunits of the DNA polymerase alpha-primase complex promote notch-mediated proliferation with discrete and shared functions in C. elegans germline. FEBS J. 285(14):2590–2604. doi: 10.1111/febs.14512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. 2001. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 107(7):881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- Youds JL, O’Neil NJ, Rose AM. 2006. Homologous recombination is required for genome stability in the absence of DOG-1 in Caenorhabditis elegans. Genetics. 173(2):697–708. doi: 10.1534/genetics.106.056879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeman MK, Cimprich KA. 2014. Causes and consequences of replication stress. Nat Cell Biol. 16(1):2–9. doi: 10.1038/ncb2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zha J, Ying M, Alexander-Floyd J, Gidalevitz T. 2019. HSP-4/BiP expression in secretory cells is regulated by a developmental program and not by the unfolded protein response. PLoS Biol. 17(3):e3000196. doi: 10.1371/journal.pbio.3000196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang WH, Koyuncu S, Vilchez D. 2022. Insights into the links between proteostasis and aging from C. elegans. Frontiers Aging. 3:854157. doi: 10.3389/fragi.2022.854157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng P, Chen Q, Tian X, Qian N, Chai P, Liu Bing, Hu J, Blackstone C, Zhu D, Teng J, et al. 2018. DNA damage triggers tubular endoplasmic reticulum extension to promote apoptosis by facilitating ER-mitochondria signaling. Cell Res. 28(8):833–854. doi: 10.1038/s41422-018-0065-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data described in this study are available in the main manuscript, the Supplementary material, or in a public repository. Supplementary Figs. 1, 3, 4, 5, and 6 describe additional experiments using GFP reporters, Supplementary Fig. 2 contains additional repeats of immunoblots, and Supplementary Figs. 7 and 8 provide additional description of the RNA-seq analysis. Supplementary Tables 1–3 contain lists describing gene expression data identified by RNA-seq; Supplementary Tables 4–5 contain lists describing processes identified by RNA-seq analysis. Supplementary material is available Xu J, Sabatino B, Yan J, Ermakova G, Doering KRS, Taubert S; 2023; Supplemental Material for Xu et al., 2023; Figshare: https://doi.org/10.25387/g3.24941133. Raw and processed RNA-seq files have been deposited: Sabatino B, Xu J, Taubert S; 2023; Effect of pri-1 (DNA primase) RNAi on gene expression in embryos of Caenorhaditis elegans; Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/); GSE225569. See the methods for information on reagents and strains. C. elegans strains described for the first time in this study can be requested from the authors.