Figure 4.
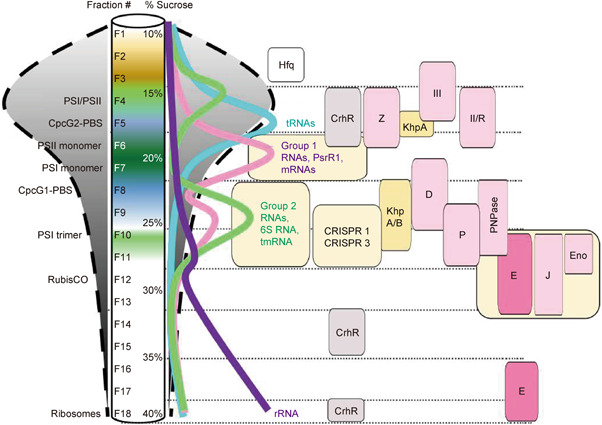
Grad‐seq analysis aids the analysis of ribonucleases, auxiliary proteins, and ribonucleoprotein complexes. A typical sedimentation profile obtained in the analysis of the cyanobacterium Synechocystis PCC 6803 is shown 190 . The positions of several major macromolecular complexes as determined by mass spectrometry are given to the left, the respective fraction numbers and sucrose percentages are indicated along the gradient. The different colors result from the native pigmentation of protein–pigment complexes involved in photosynthesis. The distribution of distinct groups of RNAs is sketched by the colored lines to the right of the gradient. The positions of abundant RNA–protein complexes, such as two of the three CRISPR complexes 193 and noncoding RNA–ribonucleoprotein complexes containing 6S RNA or transfer‐messenger RNA (tmRNA) are shown. Note that characterized regulatory small RNAs such as PsrR1 59 peaked in fraction 7 (F7) together with the bulk of mRNAs, but there were secondary peaks in mRNA abundance in other fractions (for details, see Riediger et al. 190 ). Several proteins and RNA‐protein complexes involved in RNA metabolism, such as RNase D (D), RNase J (J), RNase E (E), RNase P (P), PNPase, enolase (Eno), and CrhR occur in the higher molecular fractions, indicating their association with larger complexes. Most RNAs were detected in overlapping fractions as well, indicating their likely direct association with such complexes. The striking overlap in the in‐gradient distribution of PNPase, enolase, RNase E and J, consistent with their possible colocalization into degradosomes is boxed.
Two different gene products were detected for RNase II/R (II/R) and RNase III (III) in the lighter fractions, while Mini‐III was not detected at all. The strong correlation between RNase Z (Z) and the bulk of tRNAs is consistent with the role of this enzyme in tRNA maturation. Note that Hfq was found only in very light fractions, consistent with its non‐RNA binding character in cyanobacteria. Candidates for alternative RNA chaperones are the KhpA/B homologs Slr0287 and Slr1472. See Table 1 for the gene IDs of all other mentioned proteins. The entire data set is available at https://sunshine.biologie.uni-freiburg.de/GradSeqExplorer/. Reprinted in modified form with permission from Riediger et al. 190 .
