Abstract
RNA viruses (realm: Riboviria), including RNA phages and eukaryote‐infecting RNA viruses, are essential components of marine ecosystems. A large number of marine RNA viruses have been discovered in the last two decades because of the rapid development of next‐generation sequencing (NGS) technology. Indeed, the combination of NGS and state‐of‐the‐art meta‐omics methods (viromics, the study of all viruses in a specific environment) has led to a fundamental understanding of the taxonomy and genetic diversity of RNA viruses in the sea, suggesting the complex ecological roles played by RNA viruses in this complex ecosystem. Furthermore, comparisons of viromes in the context of highly variable marine niches reveal the biogeographic patterns and ecological impact of marine RNA viruses, whose role in global ecology is becoming increasingly clearer. In this review, we summarize the characteristics of the global marine RNA virosphere and outline the taxonomic hierarchy of RNA viruses with a specific focus on their ancient evolutionary history. We also review the development of methodology and the major progress resulting from its applications in RNA viromics. The aim of this review is not only to provide an in‐depth understanding of multifaceted aspects of marine RNA viruses, but to offer future perspectives on developing a better methodology for discovery, and exploring the evolutionary origin and major ecological significance of marine RNA virosphere.
Keywords: ecology, marine RNA viruses, metagenomics, metatranscriptomics, taxonomy, virus evolution
Advances in meta‐omics provide a surge of marine RNA virus information. Deep marine virus discovery contributes greatly to the megataxonomy of RNA viruses. Understanding diversity and biogeography helps reveal the significance of marine RNA viruses, including evolutionary trajectories, hierarchical taxonomy, and ecological importance.
Highlights
Advances in meta‐omics provide a surge of marine RNA virus information.
Deep marine virus discovery contributes greatly to the megataxonomy of RNA viruses.
Understanding diversity and biogeography helps reveal the significance of marine RNA viruses, including evolutionary trajectories, hierarchical taxonomy, and ecological importance.
INTRODUCTION
The first documented marine virus was discovered in a harbor crab (Liocarcinus depurator) in 1966 [1]; since then, numerous efforts have been made to explore viral pathogens in the ocean [2, 3, 4, 5]. Viruses were found to exist in high abundance in aquatic environments [6, 7], up to 2.5 × 108 virions per milliliter, and to serve as an important component of the marine ecosystem through cell lysis and geochemical cycling [8, 9]. Despite extensive information on DNA viruses in natural environments, the features of RNA viruses in marine ecosystems, including their abundance, prevalence, distribution, ecology, and evolutionary patterns and trajectories, are poorly understood and summarized. Recently, studies to explore the global RNA virosphere [10] have shed light on their significance and function in natural niches [11, 12, 13, 14].
Covering over 70% of Earth's surface, marine habitats, with unique characteristics (e.g., hypoxia, limited light, hydrostatic pressure, and high inorganic chemical concentrations), provide a complex and changeable environment for aquatic organisms [15]. On the basis of depth, the open ocean is subdivided into five layers (i.e., epipelagic, mesopelagic, bathypelagic, abyssopelagic, and hadopelagic zone from the sea surface to the bottom), with significant differences in the environment due to unequal sunlight exposure [16]. Notably, similarity and heterogeneity of marine RNA virosphere between and within biogeochemically distinct layers have been revealed, and attempts have been made to predict how these relationships change along with environmental factors [17]. Notably, since the early 1990s, viruses have been found to impact the marine system, beyond as pathogens of marine cellular organisms, by affecting the host community and functioning as contributors to biogeochemical cycles. For example, the biological pump, transporting organic matter from the epipelagic layer to the deep zone, functions in global carbon cycling [18] in which viruses might play a vital role as they are estimated to kill numerous organisms per day [19]. Specifically, a recent study found a potential connection between marine carbon cycling and RNA viruses, since they probably infect marine hosts as a critical component of the biological pump and encode auxiliary metabolic genes (AMGs) involved in nutrient transport and photosynthesis. In addition, the abundance of some specific marine RNA viruses may strongly predict ocean carbon flux [20].
Recently, advanced high‐throughput sequencing technology has made it possible to explore the marine virome via a series of methods independent of culturing [21, 22]. In 2015, 5476 populations of marine viruses, most of which were phages, were reported [23] during a global scientific expedition, and this number was expanded to 15,222 in 2016 and 195,728 in 2019 [24, 25]. Several studies focusing on marine RNA viruses in recent years have been productive, and the amount of RNA virus information in public databases has increased sharply [26, 27, 28], bringing about challenges involving detecting divergent viruses and determining their taxonomy [29]. However, this dilemma is to some extent alleviated by deep phylogenetic analysis relying on RNA‐dependent RNA polymerase (RdRp), which has been used for sequence‐to‐sequence comparison and taxonomic assignment. And for viruses in the Kingdom of Orthornaviriae, multifaceted evidence like structure comparison and RdRp‐based hidden Markov model (HMM) can be used parallelly to address the problem of extreme sequence divergence of RdRp due to the ancient origin and high evolutionary change rates [10, 26, 30, 31, 32, 33].
Although the complete picture of marine RNA viruses is still emerging, minimal efforts have been made to outline the ecology and significance of marine RNA viruses in their specific niches due to biases in sampling and culturing techniques. Accordingly, it is becoming increasingly clear that our knowledge of marine RNA viruses needs to be considered beyond their clinical and economical effects and in the global context. This review examines our current knowledge of marine RdRp‐encoding RNA viruses (kingdom: Orthornavirae), particularly since the advent of next‐generation sequencing (NGS) methods, and highlights their basic features and a novel taxonomic system suitable for the era of meta‐omics (Figure 1). Finally, we further discuss the theoretical and computational approaches for revealing the marine RNA virus world as well as future challenges and prospects from various perspectives.
Figure 1.
Various perspectives about marine RNA viruses. Many experimental efforts have already been made to characterize biological and epidemiological features and the host range of marine RNA viruses. Nowadays, a large number of marine RNA viruses have been discovered owing to the advent and advance of meta‐omics, including both metagenomics and metatranscriptomics, contributing to the hierarchical taxonomy and deep evolutionary process of RNA virosphere. Furthermore, with global sampling and comparative study, the ecological significance of marine RNA viruses is becoming clear. The taxonomy is generated from the International Committee on Taxonomy of Viruses (ICTV) website (https://ictv.global/taxonomy).
CHARACTERISTICS OF MARINE RNA VIRUSES
Biological characteristics
Marine RNA viruses have complex genomic features and morphological characteristics. The genome carries the biological information of an organism, and the morphological structure of viral particles is important for the study of the infection mechanism of viruses.
RNA viruses use RNA as genetic material. Their genomes include diverse, complex structures, such as mixes of segmented, unsegmented, and circular genomes. Traditionally, RNA viruses can be divided into single‐stranded RNA (ssRNA) viruses and double‐stranded RNA (dsRNA) viruses. One of the major differences between RNA viruses is their gene expression pattern: positive‐sense ssRNA viruses are translated directly, and negative‐sense ssRNA viruses are translated after conversion to positive‐sense RNA, and the dsRNA may transcribe positive‐sense RNA, which can be used as messenger RNA and replicated to create a new dsRNA viral genome [34]. The ancient gene, RdRp, is the only sequence domain conserved in all RNA viruses [35] and is considered the ancestor of several polymerases and one of the first genes in the peptide‐RNA world, widely used to study the origin of viruses [36, 37]. RNA viral genomes are typically 4–12 kb in size, and the largest known RNA viral genome is that of coronaviruses (41 kb). The main factor limiting RNA viral genome size is low replication fidelity, with the RdRp introducing approximately 10−4 errors per nucleotide [38, 39]. This property of RdRp allows RNA viruses to produce different descendants with different genotypes in a short generation time [40]. The high mutation rate leads to overlapping genes in the genomes of RNA viruses, thus leading to genome compression [41]. The biochemical basis for the low replication fidelity of RNA viruses is the lack of proofreading, repair, and postreplication error correction mechanisms during replication, as well as the lack of a replicase system, which makes RNA viruses unstable and prone to mutation [42, 43]. They often show frequent virus–host recombination, horizontal gene transfer, gene gain or loss, and complex rearrangements, which greatly complicate their genetic diversity. This instability makes RNA viruses more susceptible to mutation and, at the same time, makes RNA viruses more unfavorable to study, either by isolation and culture or by metagenomic approaches [44].
RNA viruses in different families vary in their morphology, such as capsid diameter and tail length. Methodologically, variance in virus capsid diameter and tail length can be observed by quantitative transmission electron microscopy [45]. The high degree of order (up to the Å scale) in the secondary/tertiary/quaternary structure of viral capsid proteins constitutes the different morphologies of the viral capsid—spherical/icosahedral and cylindrical/helical [46]. The spontaneous assembly process of the external capsid of RNA viruses is driven by electrostatic interactions between the positive charge on the capsid protein and the negative charge on the genome [47]. Encapsulation of RNA by artificially designed viral capsid proteins could be a new means of protecting and delivering RNA information [48]. The average capsid size of nontailed viruses in the ocean is 54 nm, and according to the nontailed virus cortovirus PM2 currently found in the ocean, they have capsid‐associated lipids [49]. As previously reported, nontailed viruses are dominant (79% of all viruses on average) in surface water, and RNA viruses may account for 16%–100% of the nontailed viruses observed [45, 50]. Because of the difficulty in isolating and culturing RNA viruses from the marine environment, more research is needed for RNA virus morphology.
Hosts of marine RNA viruses
Viruses cannot exist without a host, and they may be present in every cellular life form [51]. Marine RNA viruses have been found in both eukaryotes and prokaryotes. Upon infection of the host, the virus either directly lyses cells and multiplies or forms RNA phages (only in prokaryotes), and the multiplication of phages occurs mainly through lysis or lysogenic infection [52, 53]. Lytic infection of RNA virus means that the virus replicates after entering the host cell, leading to lytic infection of the host cell. Lysogeny means that the virus gene is integrated with the host gene and does not produce progeny virus particles, but the virus gene can replicate with the host gene and pass along with the division of the host [54]. There are several ways to identify the host of RNA viruses: One is through experimental observation, and the other is through reasonable prediction from the data set. The following methods are commonly used for metagenomic host prediction: (1) detecting whether the host and virus taxa contain consistent clustered regularly interspaced short palindromic repeat (CRISPR) spacer sequences; (2) determining the host based on the host of sequences encoding tRNAs and AMGs in the viral genome; (3) determining the host based on the similarity between viral genomic fragments and potential host gene fragments; (4) predicting hosts based on the abundance and expression levels of viruses and potential hosts in the samples; (5) predicting hosts based on similar tetranucleotide frequencies between viruses and potential hosts [24, 55, 56]. RNA viruses interact with their hosts. RNA viruses bind to antiviral competent restriction factors in the host during their life cycle, inhibiting viral replication and expression [57]. Conversely, RNA viruses induce the production of proteins that degrade restriction factors to inhibit host gene expression during the life cycle of the infected host [58]. RNA viruses also induce host expression of proviral replication RNAs, such as lncRNA‐ACOD1 [59]. RNA virus–host interactions play a vital role in the understanding of virus evolution and ecological changes. The presence of shared RNA viruses in hosts such as protozoa, animals, and plants explains horizontal viral transfer between different hosts as a central aspect of RNA virus evolution [44]. It has been suggested that the cosegregation of RNA viruses with their hosts provides the basis for cross‐species transmission and explains the macroevolution of RNA viruses [60]. Virus–host interactions can affect ecosystems, disease transmission, and other areas relevant to human life. For instance, the interactions between viruses and their hosts can control prokaryotic mortality, release inert organic matter, and compensate for suppressed host metabolic pathways in deep‐sea ecosystems [2, 61, 62, 63]. And at deep‐sea hydrothermal vents, RNA viruses can lyse host cells to control the size of host populations and affect host community structure by killing specific microorganisms. However, viruses also affect the host by manipulating bacterial metabolism to compensate for suppressed host metabolic pathways, thus helping the host to adapt to extreme environments [64].
Abundance of RNA viruses in the marine ecosystem
Viruses are an integral part of ecosystems and are found in environments, such as soil, humans, and oceans. Just as RNA viruses infect human health, marine RNA viruses affect ocean health and the planet's well‐being [65]. Unlike other environments, the ocean is regarded as the origin of life, and RNA viruses are capable of rapid gene transfer in flowing seawater. Deciphering marine viruses' role, behavior, and function of marine viruses is “one of the great mysteries of this century” [66]. Temperature, ocean currents, host distribution, and biogeography can all impact host abundance and virus abundance [67, 68]. In the marine environment, RNA viruses from low‐ to high‐productivity systems ranged from 108 virus particles per liter to over 1011 virus particles per liter [69], and the distribution of RNA viruses is influenced by sea latitude. For example, Taraviricota has a high abundance in temperate and tropical zones, and the ‐ssRNA phylum “Arctiviricota” has the highest abundance in Atlantic Arctic waters [26]. Abundant RNA viruses in the ocean have reshaped our understanding of the early evolution of viruses. Moreover, researchers have identified AMGs in marine RNA viruses that have important effects on marine carbon export [20]. Marine biofouling causes damage to biodiversity and marine ecosystems, through which RNA viruses can be transferred to the global ocean. Spreads of abundant RNA viruses introduce diseases, affect human marine activities, and compromise sustainable human development [70]. With the expansion of RNA virus studies, many problems emerged, such as host sampling for many RNA viruses is insufficient [26], and there are limitations to environmental RNA virus abundance measurements. Solving these issues will require more virus and sample data.
Adaptation of viruses in the marine environment
Extreme marine environments, such as deep‐sea, polar, and hot regions, hydrothermal vents, and areas of high pressure or salinity, are close to the limits of life [71]. Virus communities adapt to extreme environments by skewing the coding frequencies of specific amino acid residues [72, 73]. Extreme environments do not increase the mutation rate of viruses, but selection pressure from the local environment can increase the abundance of certain types of viruses [74]. The good environmental adaptability of RNA viruses in extreme environments is associated with their high mutation rate. The high adaptive capacity of RNA viruses facilitates the generation of populations consisting of mutant profiles with broad phenotypic and genetic heterogeneity [75, 76]. In these populations, mutant variants that are dominant under selection pressure can be retained as minority variants and rapidly selected when the virus is again exposed to the same selection pressure. Such properties make RNA viruses more adaptable to fluctuating selection pressures [76, 77]. The unique ecological stresses of extreme marine environments may allow viruses and their microbial host communities to synthesize new compounds with different biological activities, and these communities play an important role in biogeochemical cycles [71].
DEEP MARINE VIRUS DISCOVERY CONTRIBUTES TO THE MEGATAXONOMY OF RNA VIRUSES
Substantial efforts have been made to understand pathogenic viruses that infect humans, economically or socially important animals, and plants [78, 79, 80], and the development of NGS at a spectacularly rapid pace has led to a new age of virus discovery and taxonomy [81]. It is widely recognized that viral information is extremely abundant in the biosphere and public databases worldwide, far beyond what virologists have captured in laboratory studies [82]. Accordingly, viral taxonomy, including the practice and science of virus hierarchical classification, has changed dynamically over the last two decades, during which RNA viruses have received frequent and distinct taxonomic updates (Table 1).
Table 1.
History and updates of the megataxonomy of RNA viruses
| In five‐rank hierarchy | In 15‐rank hierarchy | |||
|---|---|---|---|---|
| Five phyla | Proposed 10 phyla | |||
| Criteria for | Virus biology | |||
| classification | Epidemiology | Phenotypes according to genome and metagenome data | ||
| Host factors | Genetic relationship inferred from sequence similarity | |||
| Sequence relationship from genome analysis | Biological data are not essential | |||
| Top rank of RNA viruses | Order | Phylum | Phylum | |
|---|---|---|---|---|
| Number of species in Orthornavirae | 1825 (2016)a | 4060 (2021)b | More than 5000 new species discovered | |
| Duplornaviricota | ||||
| Assigned or | None | Duplornaviricota | Kitrinoviricota | |
| proposed | Kitrinoviricota | Lenarviricota | ||
| phylum | Lenarviricota | Negarnaviricota | ||
| Negarnaviricota | Pisuviricota | |||
| Pisuviricota | Arctiviricota | |||
| Paraxenoviricota | ||||
| Pomiviricota | ||||
| Taraviricota | ||||
| Wamoviricota | ||||
Derived from International Committee on Taxonomy of Viruses (ICTV) Master Species List 2016 v1.3, https://talk.ictvonline.org/files/master-species-lists/m/msl/6776.
Derived from ICTV Master Species List 2021.v1, https://talk.ictvonline.org/files/master-species-lists/m/msl/13425.
Early five‐rank hierarchy of viruses
Global viruses replicate and express their genomes by several different strategies, which were used by Baltimore in 1971 to classify all known viruses into six groups and later used to introduce a seventh group [83]. The Baltimore framework, based on forms, polarity, and expression of the viruses' nucleic acids, classifies RNA viruses as dsRNA viruses, positive‐sense RNA [(+)RNA] viruses, negative‐sense RNA [(−)RNA] viruses, and RNA reverse‐transcribing viruses. Since this system provides insights into both the replication and transcription of viruses, it is still widely used [84]. However, recent evidence illustrates that viruses in the same Baltimore class do not always share a common ancestor [85], indicating that the system does not necessarily reflect the actual evolutionary relationships between and among different groups.
The earliest research on viruses emphasized molecular virology after laboratory isolation and culture, and the ratification of a new species by the International Committee on Taxonomy of Viruses (ICTV) during the 1970s–1990s was mainly dependent on biological characteristics, such as their properties in vitro, virion structure, and antigenic relationships [86]. Meanwhile, many host factors (e.g., virulence, pathogenicity, host range, and epidemiology) were also considered. On that basis, the ICTV then classified viruses into two different ranks and expanded the virus taxonomic classification system to include a five‐rank hierarchical structure of species, genus, subfamily, family, and order [84]. This hierarchy was in place until 2017, at which time nine orders, 122 families (37 subfamilies), 735 genera, and 4404 species were listed (ICTV Master Species List 2016 v1.3, https://talk.ictvonline.org/files/master-species-lists/m/msl/6776).
Fifteen hierarchical ranks and a five‐branch hierarchy of riboviria
With advances in Sanger sequencing techniques and the understanding of virus genetics, viruses can be sequenced for their nucleic acid, even before their biological attributes are characterized [87, 88]. Accordingly, inferences from virus sequences via divergence estimation and phylogeny construction were combined with experimental and epidemiological factors to assign viruses to taxonomic groups [89, 90, 91]. Heterosigma akashiwo RNA virus (HaRNAV), for example, was the first marine RNA virus isolated in coastal British Columbia and was characterized to infect the toxic red‐tide‐forming photosynthetic alga raphidophyte H. akashiwo. HaRNAV was the first classified member of the newly named family Marnaviridae in the order Picornavirales according to its host, morphology, virion size, genome size, distinct domain organization, and phylogenetic relationship with viruses in related families [92, 93].
Given that viruses lack common hallmark genes that can be used to construct a unified phylogenetic tree encompassing all viruses, the taxonomy of viruses differs to some degree from that of cellular organisms, particularly for viruses at different higher ranks and in different Baltimore classes. For RNA viruses, the presence of homologous RdRp enables inference of sequence distances and evolutionary relationships between families, which may provide support for novel ranks [84].
In the era of metagenomics, potential viruses in environments or living organisms that show limited similarity with known viruses or cannot be classified outnumber those recognized by the ICTV. A study profiling the transcriptome of marine and inland invertebrate species identified 1445 RNA viruses, some of which were significantly divergent from known families [35]. Despite the fact that contigs derived from mixed virus populations are at risk of artificial chimeras and cannot recover bipartite or multipartite genomes (often seen in RNA viruses), advanced computational and experimental methods (e.g., generating longer reads) increase the potential to resolve these problems [94, 95, 96]. This is why a workshop of the ICTV Executive Committee was held, with experts invited to discuss whether and how viruses discovered by metagenomics can be absorbed into the ICTV taxonomy. As a consequence, a consensus was reached that the proposed taxonomy can be applied to virus genome and metagenome sequence data. Alternatively, phenotypes predicted from genome analysis and distances inferred from sequence comparisons function as fundamental elements for virus taxonomy, while biological data are no longer essential [86].
A 15‐rank hierarchy, placing the global virosphere into eight primary ranks and seven secondary ranks, was discussed and approved by the ICTV Working Group in 2016 as the best equivalent of the complete Linnaean taxonomic system and a novel system for virus taxonomy [84]. The primary ranks include four already existing (order, family, genus, and species) and four newly incorporated ranks (realm, kingdom, phylum, and class), while the secondary ranks include the previously used subfamily rank and six new ranks derived from the primary ranks (subrealm, subphylum, subkingdom, subclass, suborder, and subgenus). This updated taxonomic system serves as a dynamic framework for approving virus taxonomic assignment as virus discovery continues and is inclusive of numerous viruses that remained outside of the former virus taxonomic system and thus reflected biases toward some sampling environments and organisms as well as different virus lineages.
The only realm established later, Riboviria, included RdRp‐encoding RNA viruses from all three Baltimore groups (https://talk.ictvonline.org/taxonomy/p/taxonomy-history?taxnode_id=202107095). A comparative genomic study carried out to reinvestigate the evolutionary relationships within RNA viruses through deep phylogenetic analysis and sequence similarity networks of virus hallmark genes supported the RNA virus RdRp phylogeny containing five major branches [30], which were proposed as phyla Lenarviricota (branch 1), Pisuviricota (branch 2), Kitrinoviricota (branch 3), Duplornaviricota (branch 4), and Negarnaviricota (already established, branch 5), respectively, in the proposed kingdom Orthornavirae (RdRp‐encoding RNA viruses contrasting with RT‐encoding RNA viruses). Though important issues of extreme divergence of RdRp sequences and subjectivity in the process of manual curation have received intense discussion, several complementary methods like using RdRp primary sequence, HMM models, and three‐dimensional (3D) structure supported the practicability of RdRp‐based sequence analysis with some refinement and supplements [10, 26, 31]. The creation of the Riboviria realm and subsequent 5‐branch taxonomic partitioning of viruses was considered the first step in applying the newly proposed rank structure of virus taxonomy and further helped to clarify an obvious relationship between RNA viruses, which facilitated the discovery, classification, and nomenclature of emerging RNA viruses [85].
Ten proposed phyla of global RNA viruses
More large‐scale endeavors have been made to further uncover the marine virosphere. In an up‐to‐date attempt to expand RNA virus diversity in global oceans, Ahmed A. Zayed and colleagues recovered over 44,000 RdRp‐coding contigs and 6686 complete or near‐complete RdRps via an optimized iterative search approach from 771 Tara Ocean metatranscriptomic resources globally collected from the world's five oceans and 143 new metatranscriptomic data points from the Arctic Ocean [26]. More importantly, this groundbreaking and highly representative work hinted that six newly established “megaclusters” may correspond to five new phyla (named “Arctiviricota,” “Paraxenoviricota,” “Pomiviricota,” “Taraviricota,” and “Wamoviricota”), doubling the number of known phyla in the kingdom Orthornavirae.
Additionally, revision of the origin and early relationships of reported orthornaviran phyla was considered based on analysis at the phylogenetic, structural, and genomic levels, thus complementing the primary sequence‐inferred phylogeny and representing a relatively robust roadmap. These analyses suggested polyphyly of viruses in the phylum Duplornaviricota (dsRNA viruses), while a previous study indicated that these dsRNA viruses evolved from (+)RNA viruses, with the former possibility supported by more evidence and revealing three different phyla. Meanwhile, the newly proposed phylum “Taraviricota,” placed in a phylogenetic position linking retroelements and orthornavirans, was considered a potential early evolutionary origin of other orthornaviran phyla.
Although challenges and biases toward sample types remain, continuous methodological and intellectual advances, especially in deep metagenomics and evolutionary virology, shed light on the discovery and taxonomy of global viruses and reveal the secrets of the marine RNA virosphere.
META‐OMICS OF VIRUS DISCOVERY AND ANNOTATION
Before the modern metagenomic era
The earliest report on a marine RNA virus was initially based on the isolation of RNA viruses infecting marine animals economically important for aquaculture [97]. Since then, several protistan RNA viruses in the sea have been discovered and found to have a wide host range [98, 99]. Although instructive and informative, these virological studies predominantly focused on molecular characteristics [100, 101, 102], indicating that an understanding of the ecology of marine RNA viruses remains to be reached, including their diversity, biogeography, interactions with hosts, evolutionary status, and environmental contributions.
Environmental RNA virus discovery by isolation‐independent approaches subsequently arose and revealed the diversity and crypticity of the RNA viral community in the sea [103, 104, 105]. For viral genomes, unlike the genomes of cellular organisms, no universal and conserved genes are shared by all seven Baltimore classes of viruses. However, polymerase chain reaction (PCR) targeting broadly conserved hallmark genes has been applied to amplify and discover diverse groups of viruses. As early as 2003, degenerate primers targeting RdRps combined with reverse transcription‐PCR (RT‐PCR) were used to survey the presence and diversity of picorna‐like viruses in seawater [106]. Four monophyletic clades of picorna‐like viruses probably representing at least two new viral families were identified using sequence analysis, showing the potential of a single‐gene survey for RNA viral discovery in marine environments [107]. In addition to viral discovery, more recent studies based on RdRp‐targeted RT‐PCR also revealed the temporal and spatial variation of picorna‐like ssRNA viruses in the coastal areas of British Columbia by combining location and time‐series sampling [108], the predominant role of picornavirads in the pool of RNA viruses in tropical coastal seawater samples through additional metagenomic methods [109], and the impact of RNA viruses on the community composition of the potential host with extra amplicon sequencing of marker genes [108, 110]. Notably, these data gave an initial glimpse into the diversity, richness, and ecology of a particular RNA virus group in the ocean, but this method provided little information about genetic diversity (i.e., other viral genes) and is inapplicable to the global and comprehensive assessment of RNA viruses, especially at the genome and function levels, because of underrepresentation of all viral groups and the lack of complete genetic materials.
Marine RNA viromics based on viral particle enrichment
The limitations of the culture‐dependent approach and single‐gene probing motivated the advent of modern metagenomics, a concept first defined as the study of collective genetic material cloned from natural environmental samples [111] and subsequently expanded to the direct study of microbial communities without laboratory isolation and cultivation of individual species [112]. Viromic studies of inchoate marine viruses characterized by underestimated sampling sites and with only double‐stranded DNA viruses revealed [113, 114] and necessitated large‐scale sampling and sequencing efforts such as the Sargasso Sea project [103] to provide raw data and insights into the community dynamics of marine viruses [99, 112].
The first RNA viral community‐focused study investigated the community structure of uncultivated RNA viruses in four aquatic environments by constructing random reversed‐transcribed whole‐genome shotgun libraries and shed light on the persistent and widespread existence of RNA virus assemblages (including viruses in the orders Picornavirales, Mulpavirales, and Tolivirales and other unclassified and unknown viruses) with relatively short genomes in the sea [104]. This study verified the feasibility of RNA viral metagenomics as a practicable approach for unveiling the hidden marine RNA virosphere. Compared with traditional methods, marine viromics has shown superiority in recovering viral contigs bypassing complex isolation and culturing procedures, thus the number of documented uncultivated viruses has exceeded isolated viruses as early as 2016 [28]. Meanwhile, marine virus discovery through meta‐omics is competent to avoid sampling biases and provide global landscape and ecological perspectives [20, 26, 115]. In the years that followed, an increasing number of projects were carried out to explore viruses in different environments, such as tropical seawaters [109, 116], Atlantic coastal seas [117, 118], the southern Indian Ocean [119], the Canadian Arctic [120], the Baltic Sea [121], and the deepest trenches [68], and from marine organisms, such as diatoms [122], oomycetes, marine arthropods, and eukaryotic phytoplankton [27]. Likewise, RNA viral metagenomics contributed greatly to expanding our understanding of RNA ecological characteristics, including the abundance and diversity [7], temporal dynamics [118], biogeographic distribution [120], and host interactions [120] of marine RNA viruses.
RNA viromics contributed significantly to viral discoveries for many research priorities, consisting of capturing viral genetic materials directly without the need for isolation and cultivation, removing background DNA, and providing a metastable approach with no wet laboratory necessary. Operationally, a representative particle‐based RNA viromic workflow entails the following steps: sampling and cellular fraction filtration, viral fraction concentration, nucleic acid extraction and amplification, library preparation and high‐throughput sequencing, and finally, downstream personalized data analysis [24, 25, 123, 124]. Among these main procedures, several important biases exist, indicating that careful consideration must be made to avoid misinterpretation. A viral concentration and purification process are generally performed to reduce the sample volume and facilitate subsequent steps, with tangential flow filtration (TFF), iron chloride flocculation, ultracentrifugation, and CsCl gradient centrifugation frequently used [125]. Through different cutoffs [126, 127], these methods maintain the integrity of viruses but introduce bias in the virus types, as they present diverse viability and efficiency levels for different types of viruses [128, 129]. Exogenous nucleic acid contamination, probably derived from samples, reagents [85], and sequencing platforms [130], should be safely controlled and minimized to guarantee the accuracy of the result via DNA/RNA nuclease treatment, experimental control design, and cautious consideration of viruses with low abundance (Figure 2).
Figure 2.
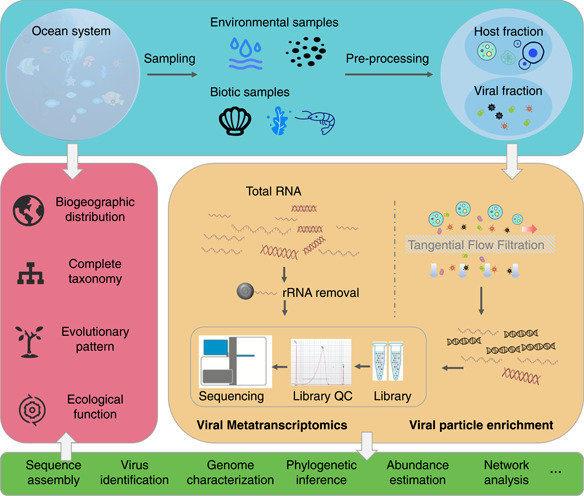
Summary of a marine viral meta‐omic pipeline. Step (1): experimental design and protocol. This step is often placed little importance in the meta‐omics study but could provide substantial information if carried out carefully. Step (2): different approaches to capturing viral nucleic acid provide discrepant information about marine RNA viruses and their hosts. Step (3): various and personalized analyses after assembling virus genomes. Step (4): detailed explanation in the context of viruses, hosts, and environment and inference from a global view.
Marine viral metatranscriptomics
Various factors, including accumulating RNA sequences in databases, better experimental protocols, and more sensitive in silico tools, brought about the application of metatranscriptomics—the total transcriptome in a given niche—to RNA virus discoveries over the past decade, which is superior in offering information about RNA viruses (with RNA‐based genomes) as well as the transcriptome of organisms with a DNA‐based genome (i.e., cellular lifeforms and DNA viruses) [60]. A typical RNA viromics protocol includes sampling (from either environments or organismal tissues), cell lysis, DNA removal, and RNA extraction and concentration (via mRNA isolation or rRNA removal), followed by reverse transcription, sequencing, and data analysis (Figure 2) [44, 131, 132].
By virtue of being unbiased, widely available, and cost‐efficient, metatranscriptomics has helped uncover cryptic and diverse RNA viruses among a wide range of sample habitats, including environmental samples [133, 134], microorganisms [135], plants [136], invertebrates [17, 78], and vertebrates [137]. These studies made predominant contributions to the knowledge of the hidden diversity [17], evolutionary scenarios [44, 131], genetic and phylogenetic diversity, and biogeographic distribution of marine RNA viruses. Furthermore, as a multifaceted tool providing information from both viruses and their hosts, this technique has been used to detect active viral infections and infer virus–host relationships [138] and interactions [1] in situ.
Along with the accumulation of viral metadata in public databases, several advanced bioinformatic tools based on different principles [139] can detect and classify RNA viruses after identifying proteins from recovered metagenomic or metatranscriptomic contigs, including similarity‐based Basic Local Alignment Search Tool searches [140], the k‐mer approach in VirFinder [141] and Kraken2 [142], and Vibrant [143] and VirSorter2 [144], which use domain abundance and/or key homologous genes. These approaches have been benchmarked for their performance in discovering viruses in terms of precision, recall, and F1 scores across simulation conditions [139]. Both contigs length and taxonomic complexity can influence the average performance of these tools and researchers can select the appropriate tool according to their own needs. Notably, correction of possible errors is usually achieved by laboratory experiments (RT‐PCR based on designed primers [11, 35, 115]) and computational confirmations (considering authenticity in the context reference RdRps from known RNA viruses [115, 131], or mapping reads into viral contigs and estimating abundance [11, 14]). Simultaneously, deep phylogenetic analysis centered on RdRp proteins appears to be informative and practicable in inferring the evolution and taxonomy of marine RNA viruses [10], although it does face challenges in cases of extreme sequence divergence and is robust only in the case of distinct primary sequence similarity [31]. Notably, it is suggested that conclusions drawn from RdRp‐based phylogenies be carefully considered in the context of other evidence, such as RdRp 3D structure and clustering, existence and permutation of other domains, and whole‐genome features. Additionally, the ecological roles and biogeographic patterns of marine RNA viruses can be explored when sufficient niche background information is available.
Future challenges and perspectives
NGS and viromics have revolutionized virus discovery and led to an explosion of known and unknown viral sequences, which, to a certain degree, complicates an accurate understanding of the actual marine RNA virosphere. Unlike defining prokaryotic DNA virus–host relationships, it is usually not adaptable to detecting eukaryotic RNA virus hosts via approaches based on CRISPR spacer matches or sequence similarity [145]. Even when sampling tissues directly, the inferred species cannot be verified as the host since the viruses may actually infect internal microbes or a dietary component. Studies aiming at exploring promising mechanisms (like infection‐induced sequence features and coexistence relationships) or new technical innovations are encouraged to define virus–host associations and to improve accuracy and applicability. Second, improvement of viral genome completeness is urgent, necessitating greater sequencing depths, advances in long‐read sequencing, and better assembler tools. Another challenge is that few bioinformatic software programs can be utilized to precisely classify RNA viruses showing little or no similarity with known viruses [146]. Last, large technical gaps remain in the isolation of RNA viral particles from environmental and poorly studied biological samples, as well as in viral culturomics and molecular verification. Progresses in this field will contribute to a deeper understanding of RNA virus biological features and broader application of RNA viruses by human beings at the community and/or ecosystem levels.
Virus discovery through meta‐omic sequencing may represent a field without an obvious trajectory and help illuminate underexplored RNA viral “dark matter,” representing unknown aspects regarding marine RNA viruses, like, taxonomic, metabolic, and functional diversity. The evolutionary processes of different types of viruses and the broad taxonomy of the virus world are expected to be refined through large‐scale and unbiased viromics. In addition, as previous ecological function studies focused mainly on DNA phages, there has been less evidence for the roles of RNA viruses. Considering their abundance and host range, marine RNA viruses have been reported to be associated with the modulation of host diversity [121], algal bloom [138], host metabolic reprogramming, and ocean carbon export [20]. So, it is of utmost importance to clarify what impacts marine virosphere and what marine viruses impact. Particularly, the roles of marine RNA viral groups involved in carbon, nitrogen, sulfur, and other earth element cycles can be determined at the genomic and metabolic levels, by integrating the metabolite spectrum and geographical‐climatic characteristics, thus providing a theoretical basis for the development of marine viral resources and the use of RNA viruses to determine and regulate land‐ocean‐atmosphere carbon fluxes. More studies are still needed to support a more complete functional characterization of these RNA viruses.
CONCLUSION
In this review, we highlight that the global search for RNA virosphere components via meta‐omics has helped to unravel cryptic and diverse RNA viral populations in marine environments and shed light on their biogeographic distribution and evolutionary patterns. However, these findings also hint at the situation where gaps still exist, particularly in the current virus hierarchical taxonomic system in the era of information explosion and functional annotation in this specific ecosystem. Additionally, more advanced sequencing (generation of longer reads) and computational methods (assembly of viral genomes as completely as possible) are needed to further improve our understanding of marine RNA viruses both already known and yet to be discovered. With a large number of RNA viruses and their relationships with hosts and environmental elements revealed by using multifaceted approaches, the potential roles that marine RNA viruses play, like shaping carbon cycling in soil, might emerge and thus redefine their ecological status in nature.
AUTHOR CONTRIBUTIONS
Meng‐en Liao and Jie Cui conceived the idea. Meng‐en Liao, Mang Shi, and Jie Cui discussed the content. Meng‐en Liao, Yunyi Xie, and Jie Cui wrote the first draft of the manuscript. Jie Cui contributed to critical revision.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
ACKNOWLEDGMENTS
This study was supported by the National Natural Science Foundation of China (31970176), Collaborative Research Grant KLMVI‐OP‐202002 of CAS Key Laboratory of Molecular Virology & Immunology, Institut Pasteur of Shanghai, Chinese Academy of Sciences.
Liao, Meng‐en , Xie Yunyi, Shi Mang, and Cui Jie. 2022. “Over two decades of research on the marine RNA virosphere.” iMeta 1, e59. 10.1002/imt2.59
DATA AVAILABILITY STATEMENT
This manuscript does not generate any script and data. All authors read and approved the final manuscript. Supplementary materials (figures, tables, scripts, graphical abstract, slides, videos, Chinese translated version, and updated materials) may be found in the online DOI or iMeta Science http://www.imeta.science/.
REFERENCES
- 1. Van Eynde, Benigna , Christiaens Olivier, Delbare Daan, Shi Chenyan, Vanhulle Emiel, Yinda Claude Kwe, Matthijnssens Jelle, and Smagghe Guy. 2020. “Exploration of the Virome of the European Brown Shrimp (Crangon crangon).” The Journal of General Virology 101: 651–66. 10.1099/jgv.0.001412 [DOI] [PubMed] [Google Scholar]
- 2. Wigington, Charles H. , Sonderegger Derek, Brussaard Corina P. D., Buchan Alison, Finke Jan F., Fuhrman Jed A., Lennon Jay T., et al. 2016. “Re‐examination of the Relationship Between Marine Virus and Microbial Cell Abundances.” Nature Microbiology 1: 15024. 10.1038/nmicrobiol.2015.24 [DOI] [PubMed] [Google Scholar]
- 3. Lara, Elena , Vaqué Dolors, Sà Elisabet Laia, Boras Julia A., Gomes Ana, Borrull Encarna, Díez‐Vives Cristina, et al. 2017. “Unveiling the Role and Life Strategies of Viruses from the Surface to the Dark Ocean.” Science Advances 3: e1602565. 10.1126/sciadv.1602565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wei, Wei , Chen Xiaowei, Weinbauer Markus G., Jiao Nianzhi, and Zhang Rui. 2022. “Reduced Bacterial Mortality and Enhanced Viral Productivity During Sinking in the Ocean.” The ISME Journal 16: 1668–75. 10.1038/s41396-022-01224-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. De Corte, Daniele , Sintes Eva, Yokokawa Taichi, Lekunberri Itziar, and Herndl Gerhard J.. 2016. “Large‐Scale Distribution Of Microbial and Viral Populations in the South Atlantic Ocean.” Environmental Microbiology Reports 8: 305–15. 10.1111/1758-2229.12381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bergh, Øivind , Børsheim Knut Yngve, Bratbak Gunnar, and Heldal Mikal. 1989. “High Abundance of Viruses Found in Aquatic Environments.” Nature 340: 467–8. 10.1038/340467a0; https://pubmed.ncbi.nlm.nih.gov/2755508 [DOI] [PubMed] [Google Scholar]
- 7. Steward, Grieg F. , Culley Alexander I., Mueller Jaclyn A., Wood‐Charlson Elisha M., Belcaid Mahdi, and Poisson Guylaine. 2013. “Are We Missing Half of the Viruses in the Ocean? The ISME Journal 7: 672–9. 10.1038/ismej.2012.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Knowles, Ben , Bailey Barbara, Boling Lance, Breitbart Mya, Cobián‐Güemes Ana, del Campo Javier, Edwards Rob, et al. 2017. “Variability and Host Density Independence in Inductions‐Based Estimates of Environmental Lysogeny.” Nature Microbiology 2: 17064. 10.1038/nmicrobiol.2017.64 [DOI] [PubMed] [Google Scholar]
- 9. Middelboe, Mathias , and Brussaard Corina. 2017. “Marine Viruses: Key Players in Marine Ecosystems.” Viruses 9: 302. 10.3390/v9100302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wolf, Yuri I. , Kazlauskas Darius, Iranzo Jaime, Lucía‐Sanz Adriana, Kuhn Jens H., Krupovic Mart, Dolja Valerian V., and Koonin Eugene V.. 2019. “Reply to Holmes and Duchêne, “Can Sequence Phylogenies Safely Infer the Origin of the Global Virome?”: Deep Phylogenetic Analysis of RNA Viruses Is Highly Challenging but Not Meaningless.” MBio 10: e00542‐19. 10.1128/mBio.00542-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shi, Mang , Lin Xian‐Dan, Chen Xiao, Tian Jun‐Hua, Chen Liang‐Jun, Li Kun, Wang Wen, et al. 2018. “The Evolutionary History of Vertebrate RNA Viruses.” Nature 556: 197–202. 10.1038/s41586-018-0012-7 [DOI] [PubMed] [Google Scholar]
- 12. Tokarz, Rafal , Sameroff Stephen, Tagliafierro Teresa, Jain Komal, Williams Simon H., Cucura D. Moses, Rochlin Ilia, et al. 2018. “Identification of Novel Viruses in Amblyomma Americanum, Dermacentor Variabilis, and Ixodes Scapularis Ticks.” MSphere 3: e00614‐17. 10.1128/mSphere.00614-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bejerman, Nicolás , Debat Humberto, and Dietzgen Ralf G.. 2020. “The Plant Negative‐Sense RNA Virosphere: Virus Discovery Through New Eyes.” Frontiers in Microbiology 11: 588427. 10.3389/fmicb.2020.588427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mahar, Jackie E. , Shi Mang, Hall Robyn N., Strive Tanja, and Holmes Edward C.. 2020. “Comparative Analysis of RNA Virome Composition in Rabbits and Associated Ectoparasites.” Journal of Virology 94: e02119‐19. 10.1128/JVI.02119-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shu, Wen‐Sheng , and Huang Li‐Nan. 2022. “Microbial Diversity in Extreme Environments.” Nature Reviews Microbiology 20: 219–35. 10.1038/s41579-021-00648-y [DOI] [PubMed] [Google Scholar]
- 16. Robinson, Carol , Steinberg Deborah K., Anderson Thomas R., Arístegui Javier, Carlson Craig A., Frost Jessica R., Ghiglione Jean‐François, et al. 2010. “Mesopelagic Zone Ecology and Biogeochemistry—A Synthesis.” Deep Sea Research Part II: Topical Studies in Oceanography 57: 1504–18. 10.1016/j.dsr2.2010.02.018 [DOI] [Google Scholar]
- 17. Zhang, Yu‐Yi , Liao Meng‐En, Wu Fengxia, Chen Yicong, Song Yuhe, Sun Qinglei, Jiang Shuai, Huang Honghui, and Cui Jie. 2021. “Comparative Study of the Malacostraca Viromes Between Deep Sea and Shallow Water.” Science Bulletin 66: 2458–61. 10.1016/j.scib.2021.09.008 [DOI] [PubMed] [Google Scholar]
- 18. Sigman, Daniel M. , and Boyle Edward A.. 2000. “Glacial/Interglacial Variations in Atmospheric Carbon Dioxide.” Nature 407: 859–69. 10.1038/35038000; https://pubmed.ncbi.nlm.nih.gov/11057657 [DOI] [PubMed] [Google Scholar]
- 19. Tran, Patricia Q. , and Anantharaman Karthik. 2021. “Biogeochemistry Goes Viral: Towards a Multifaceted Approach to Study Viruses and Biogeochemical Cycling.” MSystems 6: e0113821. 10.1128/mSystems.01138-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dominguez‐Huerta, Guillermo , Zayed Ahmed A., Wainaina James M., Guo Jiarong, Tian Funing, Pratama Akbar Adjie, Bolduc Benjamin, et al. 2022. “Diversity and Ecological Footprint of Global Ocean RNA Viruses.” Science 376: 1202–8. 10.1126/science.abn6358 [DOI] [PubMed] [Google Scholar]
- 21. Carroll, Dennis , Daszak Peter, Wolfe Nathan D., Gao George F., Morel Carlos M., Morzaria Subhash, Pablos‐Méndez Ariel, Tomori Oyewale, and Mazet Jonna A. K.. 2018. “The Global Virome Project.” Science 359: 872–4. 10.1126/science.aap7463 [DOI] [PubMed] [Google Scholar]
- 22. Carroll, Dennis , Watson Brooke, Togami Eri, Daszak Peter, Mazet Jonna A.k., Chrisman Cara J., Rubin Edward M., et al. 2018. “Building a Global Atlas of Zoonotic Viruses.” Bulletin of the World Health Organization 96: 292–4. 10.2471/BLT.17.205005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Brum, Jennifer R. , Ignacio‐Espinoza J. Cesar, Roux Simon, Doulcier Guilhem, Acinas Silvia G., Alberti Adriana, Chaffron Samuel, et al. 2015. “Patterns and Ecological Drivers of Ocean Viral Communities.” Science 348: 1261498. 10.1126/science.1261498 [DOI] [PubMed] [Google Scholar]
- 24. Roux, Simon , Brum Jennifer R., Dutilh Bas E., Sunagawa Shinichi, Duhaime Melissa B., Loy Alexander, Poulos Bonnie T., et al. 2016. “Ecogenomics and Potential Biogeochemical Impacts of Globally Abundant Ocean Viruses.” Nature 537: 689–93. 10.1038/nature19366 [DOI] [PubMed] [Google Scholar]
- 25. Gregory, Ann C. , Ahmed A. Zayed, Conceição‐Neto Nádia, Temperton Ben, Bolduc Ben, Alberti Adriana, Ardyna Mathieu, et al. 2019. “Marine DNA Viral Macro‐ and Microdiversity from Pole to Pole.” Cell 177: 1109‐1123. 10.1016/j.cell.2019.03.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zayed, Ahmed A. , Wainaina James M., Dominguez‐Huerta Guillermo, Pelletier Eric, Guo Jiarong, Mohssen Mohamed, Tian Funing, et al. 2022. “Cryptic and Abundant Marine Viruses at the Evolutionary Origins of Earths RNA Virome.” Science 376: 156–62. 10.1126/science.abm5847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wolf, Yuri I. , Silas Sukrit, Wang Yongjie, Wu Shuang, Bocek Michael, Kazlauskas Darius, Krupovic Mart, et al. 2020. “Doubling of the Known Set of RNA Viruses by Metagenomic Analysis of an Aquatic Virome.” Nature Microbiology 5: 1262–70. 10.1038/s41564-020-0755-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Roux, Simon , Adriaenssens Evelien M., Dutilh Bas E., Koonin Eugene V., Kropinski Andrew M., Krupovic Mart, Kuhn Jens H., et al. 2019. “Minimum Information About an Uncultivated Virus Genome (MIUViG).” Nature Biotechnology 37: 29–37. 10.1038/nbt.4306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cobbin, Joanna C. A. , Charon Justine, Harvey Erin, Holmes Edward C., and Mahar Jackie E.. 2021. “Current Challenges to Virus Discovery by Meta‐Transcriptomics.” Current Opinion in Virology 51: 48–55. 10.1016/j.coviro.2021.09.007 [DOI] [PubMed] [Google Scholar]
- 30. Wolf, Yuri I. , Kazlauskas Darius, Iranzo Jaime, Lucía‐Sanz Adriana, Kuhn Jens H., Krupovic Mart, Dolja Valerian V., and Koonin Eugene V.. 2018. “Origins and Evolution of the Global RNA Virome.” MBio 9: e02329‐18. 10.1128/mBio.02329-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Holmes, Edward C. , and Duchêne Sebastián. 2019. “Can Sequence Phylogenies Safely Infer the Origin of the Global Virome?” MBio 10: e00289‐19. 10.1128/mBio.00289-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Koonin, Eugene V. , Dolja Valerian V., and Krupovic Mart. 2022. “The Logic of Virus Evolution.” Cell Host & Microbe 30: 917–29. 10.1016/j.chom.2022.06.008 [DOI] [PubMed] [Google Scholar]
- 33. Sanjuán, Rafael , Nebot Miguel R., Chirico Nicola, Mansky Louis M., and Belshaw Robert. 2010. “Viral Mutation Rates.” Journal of Virology 84: 9733–48. 10.1128/JVI.00694-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wickner, Reed B. 1993. “Double‐Stranded RNA Virus Replication and Packaging.” Journal of Biological Chemistry 268: 3797–800. 10.1016/S0021-9258(18)53539-0; https://pubmed.ncbi.nlm.nih.gov/8440674 [DOI] [PubMed] [Google Scholar]
- 35. Shi, Mang , Lin Xian‐Dan, Tian Jun‐Hua, Chen Liang‐Jun, Chen Xiao, Li Ci‐Xiu, Qin Xin‐Cheng, et al. 2016. “Redefining the Invertebrate RNA Virosphere.” Nature 540: 539–43. 10.1038/nature20167 [DOI] [PubMed] [Google Scholar]
- 36. Carter, Charles W. 2015. “What RNA World? Why a Peptide/RNA Partnership Merits Renewed Experimental Attention.” Life (Basel, Switzerland) 5: 294–320. 10.3390/life5010294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pereira dos Santos, Ariosvaldo, Junior , José Marco V., and Torres de Farias Sávio. 2021. “From RNA to DNA: Insights About the Transition of Informational Molecule in the Biological Systems Based on the Structural Proximity Between the Polymerases.” BioSystems 206: 104442. 10.1016/j.biosystems.2021.104442 [DOI] [PubMed] [Google Scholar]
- 38. Agranovsky, Alexey A. 2021. “Structure and Expression of Large (+)RNA Genomes of Viruses of Higher Eukaryotes.” Biochemistry (Moscow) 86: 248–56. 10.1134/S0006297921030020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Saberi, Amir , Gulyaeva Anastasia A., Brubacher John L., Newmark Phillip A., and Gorbalenya Alexander E.. 2018. “A Planarian Nidovirus Expands the Limits of RNA Genome Size.” PLoS Pathogens 14: e1007314. 10.1371/journal.ppat.1007314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mandary, Madiiha Bibi , Masomian Malihe, and Poh Chit Laa. 2019. “Impact of RNA Virus Evolution on Quasispecies Formation and Virulence.” International Journal of Molecular Sciences 20: 4657. 10.3390/ijms20184657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Belshaw, Robert , Pybus Oliver G., and Rambaut Andrew. 2007. “The Evolution of Genome Compression and Genomic Novelty in RNA Viruses.” Genome Research 17: 1496–504. 10.1101/gr.6305707; https://pubmed.ncbi.nlm.nih.gov/17785537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schekman, Randy , Weiner Alan, and Kornberg Arthur. 1974. “Multienzyme Systems of DNA Replication.” Science 186: 987–93. 10.1126/science.186.4168.987; https://pubmed.ncbi.nlm.nih.gov/4620044 [DOI] [PubMed] [Google Scholar]
- 43. Domingo, Esteban , Escarmís Cristina, Sevilla Noemi, Moya Andres, Elena Santiago F., Quer Josep, Novella Isabel S., and Holland John J.. 1996. “Basic Concepts in RNA Virus Evolution.” The FASEB Journal 10: 859–64. 10.1096/fasebj.10.8.8666162; https://pubmed.ncbi.nlm.nih.gov/8666162 [DOI] [PubMed] [Google Scholar]
- 44. Dolja, Valerian V. , and Koonin Eugene V.. 2018. “Metagenomics Reshapes the Concepts of RNA Virus Evolution by Revealing Extensive Horizontal Virus Transfer.” Virus Research 244: 36–52. 10.1016/j.virusres.2017.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Brum, Jennifer R. , Schenck Ryan O., and Sullivan Matthew B.. 2013. “Global Morphological Analysis of Marine Viruses Shows Minimal Regional Variation and Dominance of Non‐Tailed Viruses.” The ISME Journal 7: 1738–51. 10.1038/ismej.2013.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Knobler, Charles M. , and Gelbart William M.. 2022. “How and Why RNA Genomes Are (Partially) Ordered in Viral Capsids.” Current Opinion In Virology 52: 203–10. 10.1016/j.coviro.2021.11.014 [DOI] [PubMed] [Google Scholar]
- 47. Panahandeh, Sanaz , Li Siyu, Dragnea Bogdan, and Zandi Roya. 2022. “Virus Assembly Pathways Inside a Host Cell.” ACS Nano 16: 317–27. 10.1021/acsnano.1c06335 [DOI] [PubMed] [Google Scholar]
- 48. Tetter, Stephan , Terasaka Naohiro, Steinauer Angela, Bingham Richard J., Clark Sam, Scott Andrew J. P., Patel Nikesh, et al. 2021. “Evolution of a Virus‐Like Architecture and Packaging Mechanism in a Repurposed Bacterial Protein.” Science 372: 1220–4. 10.1126/science.abg2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ignacio‐Espinoza, Julio Cesar , and Fuhrman Jed A.. 2018. “A Non‐Tailed Twist in the Viral Tale.” Nature 554: 38–9. 10.1038/d41586-018-00923-8 [DOI] [PubMed] [Google Scholar]
- 50. Karsenti, Eric , Acinas Silvia G., Bork Peer, Bowler Chris, De Vargas Colomban, Raes Jeroen, Sullivan Matthew, et al. 2011. “A Holistic Approach to Marine Eco‐Systems Biology.” PLoS Biology 9: e1001177. 10.1371/journal.pbio.1001177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Koonin, Eugene V. , Senkevich Tatiana G., and Dolja Valerian V.. 2006. “The Ancient Virus World and Evolution of Cells.” Biology Direct 1: 29. 10.1186/1745-6150-1-29; https://pubmed.ncbi.nlm.nih.gov/16984643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Evans, Claire , and Brussaard Corina P. D.. 2012. “Regional Variation in Lytic and Lysogenic Viral Infection in the Southern Ocean and its Contribution to Biogeochemical Cycling.” Applied and Environmental Microbiology 78: 6741–8. 10.1128/AEM.01388-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Weinbauer, Markus G. 2004. “Ecology of Prokaryotic Viruses.” FEMS Microbiology Reviews 28: 127–81. 10.1016/j.femsre.2003.08.001; https://pubmed.ncbi.nlm.nih.gov/15109783 [DOI] [PubMed] [Google Scholar]
- 54. Sime‐Ngando, Télesphore , and Colombet Jonathan. 2009. “Virus and Prophages in Aquatic Ecosystems.” Canadian Journal of Microbiology 55: 95–109. 10.1139/w08-099 [DOI] [PubMed] [Google Scholar]
- 55. Gu, Xiaoqiong , Tay Qi Xiang Martin, Te Shu Harn, Saeidi Nazanin, Goh Shin Giek, Kushmaro Ariel, Thompson Janelle R., and Gin Karina Yew‐Hoong. 2018. “Geospatial distribution of viromes in tropical freshwater ecosystems.” Water Research 137: 220–32. 10.1016/j.watres.2018.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Paez‐Espino, David , Eloe‐Fadrosh Emiley A., Pavlopoulos Georgios A., Thomas Alex D., Huntemann Marcel, Mikhailova Natalia, Rubin Edward, Ivanova Natalia N., and Kyrpides Nikos C.. 2016. “Uncovering Earths Virome.” Nature 536: 425–30. 10.1038/nature19094; https://pubmed.ncbi.nlm.nih.gov/27533034 [DOI] [PubMed] [Google Scholar]
- 57. Iselin, Louisa , Palmalux Natasha, Kamel Wael, Simmonds Peter, Mohammed Shabaz, and Castello Alfredo. 2022. “Uncovering Viral RNA‐host Cell Interactions on a Proteome‐Wide Scale.” Trends In Biochemical Sciences 47: 23–38. 10.1016/j.tibs.2021.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lyles, Douglas S. 2000. “Cytopathogenesis and Inhibition of Host Gene Expression by RNA Viruses.” Microbiology and Molecular Biology Reviews 64: 709–24. 10.1128/MMBR.64.4.709-724.2000; https://pubmed.ncbi.nlm.nih.gov/11104816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wang, Pin , Xu Junfang, Wang Yujia, and Cao Xuetao. 2017. “An Interferon‐Independent lncRNA Promotes Viral Replication by Modulating Cellular Metabolism.” Science 358: 1051–5. 10.1126/science.aao0409 [DOI] [PubMed] [Google Scholar]
- 60. Zhang, Yong‐Zhen , Shi Mang, and Holmes Edward C.. 2018. “Using Metagenomics to Characterize an Expanding Virosphere.” Cell 172: 1168–72. 10.1016/j.cell.2018.02.043 [DOI] [PubMed] [Google Scholar]
- 61. Danovaro, Roberto , Dell'Anno Antonio, Corinaldesi Cinzia, Magagnini Mirko, Noble Rachel, Tamburini Christian, and Weinbauer Markus. 2008. “Major Viral Impact on the Functioning of Benthic Deep‐Sea Ecosystems.” Nature 454: 1084–7. 10.1038/nature07268 [DOI] [PubMed] [Google Scholar]
- 62. Hurwitz, Bonnie L. , Hallam Steven J., and Sullivan Matthew B.. 2013. “Metabolic by Viruses in the Sunlit and Dark Ocean.” Genome Biology 14: R123. 10.1186/gb-2013-14-11-r123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hurwitz, Bonnie L. , Brum Jennifer R., and Sullivan Matthew B.. 2015. “Depth‐Stratified Functional and Taxonomic Niche Specialization in the ‘Core’ and ‘Flexible’ Pacific Ocean Virome.” The ISME Journal 9: 472–84. 10.1038/ismej.2014.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. He, Tianliang , Li Hongyun, and Zhang Xiaobo. 2017. “Deep‐Sea Hydrothermal Vent Viruses Compensate for Microbial Metabolism in Virus–Host Interactions.” MBio 8: e00893‐17. 10.1128/mBio.00893-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Marx, Vivien . 2022. “Why the Ocean Virome Matters.” Nature Methods 19: 924–7. 10.1038/s41592-022-01567-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Marx, Vivien . 2022. “Trawling the Ocean Virome.” Nature Methods 19: 928–31. 10.1038/s41592-022-01568-2 [DOI] [PubMed] [Google Scholar]
- 67. Jover, Luis F. , Effler T. Chad, Buchan Alison, Wilhelm Steven W., and Weitz Joshua S.. 2014. “The Elemental Composition of Virus Particles: Implications for Marine Biogeochemical Cycles.” Nature Reviews Microbiology 12: 519–28. 10.1038/nrmicro3289 [DOI] [PubMed] [Google Scholar]
- 68. Jian, Huahua , Yi Yi, Wang Jiahua, Hao Yali, Zhang Mujie, Wang Siyuan, Meng Canxing, et al. 2021. “Diversity and Distribution of Viruses Inhabiting the Deepest Ocean on Earth.” The ISME Journal 15: 3094–110. 10.1038/s41396-021-00994-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Clasen, Jessica L. , Brigden Sean M., Payet Jérôme P., and Suttle Curtis A.. 2008. “Evidence That Viral Abundance Across Oceans and Lakes Is Driven by Different Biological Factors.” Freshwater Biology 53: 1090–100. 10.1111/j.1365-2427.2008.01992.x [DOI] [Google Scholar]
- 70. Liu, Xiaobo , Yang Jin‐Long, Rittschof Daniel, Maki James S., and Gu Ji‐Dong. 2022. “Redirecting Marine Antibiofouling Innovations from Sustainable Horizons.” Trends In Ecology & Evolution 37: 469–72. 10.1016/j.tree.2022.02.009 [DOI] [PubMed] [Google Scholar]
- 71. Giordano, Daniela . 2020. “Bioactive Molecules from Extreme Environments.” Marine Drugs 18: 640. 10.3390/md18120640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Alarcón‐Schumacher, Tomás , Guajardo‐Leiva Sergio, Martinez‐Garcia Manuel, and Díez Beatriz. 2021. “Ecogenomics and Adaptation Strategies of Southern Ocean Viral Communities.” MSystems 6: e0039621. 10.1128/mSystems.00396-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Fortunato, Caroline S. , Larson Benjamin, Butterfield David A., and Huber Julie A.. 2018. “Spatially Distinct, Temporally Stable Microbial Populations Mediate Biogeochemical Cycling at and Below the Seafloor in Hydrothermal Vent Fluids.” Environmental Microbiology 20: 769–84. 10.1111/1462-2920.14011 [DOI] [PubMed] [Google Scholar]
- 74. de Wit, Rutger , and Bouvier Thierry. 2006. “Everything Is Everywhere, But, the Environment Selects; What Did Baas Becking and Beijerinck Really Say? Environmental Microbiology 8: 755–8. 10.1111/j.1462-2920.2006.01017.x; https://pubmed.ncbi.nlm.nih.gov/16584487 [DOI] [PubMed] [Google Scholar]
- 75. Domingo, Esteban , Ruiz‐Jarabo Carmen M., Sierra Saleta, Arias Armando, Pariente Nonia, Baranowski Eric, and Escarmís Cristina. 2001. “Emergence and Selection of RNA Virus Variants: Memory and Extinction.” Virus Research 82: 39–44. 10.1016/S0168-1702(01)00385-9; https://pubmed.ncbi.nlm.nih.gov/11885948 [DOI] [PubMed] [Google Scholar]
- 76. Briones, Carlos , and Domingo Esteban. 2008. “Minority Report: Hidden Memory Genomes in HIV‐1 Quasispecies and Possible Clinical Implications.” AIDS Reviews 10: 93–109. https://pubmed.ncbi.nlm.nih.gov/18615120 [PubMed] [Google Scholar]
- 77. Ruiz‐Jarabo, Carmen M. , Arias Armando, Baranowski Eric, Escarmís Cristina, and Domingo Esteban. 2000. “Memory in Viral Quasispecies.” Journal of Virology 74: 3543–7. 10.1128/JVI.74.8.3543-3547.2000; https://pubmed.ncbi.nlm.nih.gov/10729128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Liu, Qiyan , Zhang Song, Mei Shiqiang, Zhou Yan, Wang Jianhua, Han Guan‐Zhu, Chen Lei, Zhou Changyong, and Cao Mengji. 2021. “Viromics Unveils Extraordinary Genetic Diversity of the Family Closteroviridae in Wild Citrus.” PLoS Pathogens 17: e1009751. 10.1371/journal.ppat.1009751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Feng, Yun , Gou Qin‐Yu, Yang Wei‐Hong, Wu Wei‐Chen, Wang Juan, Holmes Edward C., Liang Guodong, and Shi Mang. 2022. “A Time‐Series Meta‐Transcriptomic Analysis Reveals the Seasonal, Host, and Gender Structure of Mosquito Viromes.” Virus Evolution 8: veac006. 10.1093/ve/veac006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. He, Wan‐Ting , Hou Xin, Zhao Jin, Sun Jiumeng, He Haijian, Si Wei, Wang Jing, et al. 2022. “Virome Characterization of Game Animals in China Reveals a Spectrum of Emerging Pathogens.” Cell 185: 1117‐1129. 10.1016/j.cell.2022.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Kuhn, Jens H. , Wolf Yuri I., Krupovic Mart, Zhang Yong‐Zhen, Maes Piet, Dolja Valerian V., and Koonin Eugene V.. 2019. “Classify Viruses—The Gain Is Worth the Pain.” Nature 566: 318–20. 10.1038/d41586-019-00599-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Roux, Simon , Páez‐Espino David, Chen I. Min A., Palaniappan Krishna, Ratner Anna, Chu Ken, Reddy T. B. K., et al. 2021. “IMG/VR v3: An Integrated Ecological and Evolutionary Framework for Interrogating Genomes of Uncultivated Viruses.” Nucleic Acids Research 49: D764–75. 10.1093/nar/gkaa946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Baltimore, David. 1971. “Expression Animal Virus Genomes.” Bacteriological Reviews 35: 235–41. 10.1128/br.35.3.235-241.1971; https://pubmed.ncbi.nlm.nih.gov/4329869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. International Committee on Taxonomy of Viruses Executive Committee . 2020. “The New Scope of Virus Taxonomy: Partitioning the Virosphere into 15 Hierarchical Ranks.” Nature Microbiology 5: 668–74. 10.1038/s41564-020-0709-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Koonin, Eugene V. , Dolja Valerian V., Krupovic Mart, Varsani Arvind, Wolf Yuri I., Yutin Natalya, Zerbini F. Murilo, and Kuhn Jens H.. 2020. “Global Organization and Proposed Megataxonomy of the Virus World.” Microbiology and Molecular Biology Reviews: MMBR 84: e00061‐19. 10.1128/MMBR.00061-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Simmonds, Peter , Adams Mike J., Benkő Mária, Breitbart Mya, Brister J. Rodney, Carstens Eric B., Davison Andrew J., et al. 2017. “Virus Taxonomy in the Age of Metagenomics.” Nature Reviews Microbiology 15: 161–8. 10.1038/nrmicro.2016.177 [DOI] [PubMed] [Google Scholar]
- 87. Li, Shihao , Lv Xinjia, Yu Yang, Zhang Xiaojun, and Li Fuhua. 2020. “Molecular and Functional Diversity of Crustin‐Like Genes in the Shrimp.” Marine Drugs 18: 361. 10.3390/md18070361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Oidtmann, Birgit , and Stentiford Grant D.. 2011. “White Spot Syndrome Virus (WSSV) Concentrations in Crustacean Tissues—A Review of Data Relevant to Assess the Risk Associated with Commodity Trade: WSSV Concentrations in Crustacean Tissues.” Transboundary and Emerging Diseases 58: 469–82. 10.1111/j.1865-1682.2011.01231.x [DOI] [PubMed] [Google Scholar]
- 89. Brum, Jennifer R. , and Sullivan Matthew B.. 2015. “Rising to the Challenge: Accelerated Pace of Discovery Transforms Marine Virology.” Nature Reviews Microbiology 13: 147–59. 10.1038/nrmicro3404 [DOI] [PubMed] [Google Scholar]
- 90. Yan, Yang , Cui Huachun, Jiang Songshan, Huang Youhua, Huang Xiaohong, Wei Shina, Xu Weiyi, and Qin Qiwei. 2011. “Identification of a Novel Marine Fish Virus, Singapore Grouper Iridovirus‐Encoded microRNAs Expressed in Grouper Cells by Solexa Sequencing.” PLoS ONE 6: e19148. 10.1371/journal.pone.0019148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Aswad, Amr , and Katzourakis Aris. 2017. “A Novel Viral Lineage Distantly Related to Herpesviruses Discovered within Fish Genome Sequence Data.” Virus Evolution 3: vex016. 10.1093/ve/vex016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Lang, Andrew S. , Culley Alexander I., and Suttle Curtis A.. 2004. “Genome Sequence and Characterization of a Virus (HaRNAV) Related to Picorna‐Like Viruses That Infects the Marine Toxic Bloom‐Forming Alga Heterosigma akashiwo .” Virology 320: 206–17. 10.1016/j.virol.2003.10.015; https://pubmed.ncbi.nlm.nih.gov/15016544 [DOI] [PubMed] [Google Scholar]
- 93. Vlok, Marli , Suttle Curtis A., and Lang Andrew S.. 2021. “Algal Marnaviruses (Marnaviridae).” Encyclopedia of Virology 4: 671–6. 10.1016/b978-0-12-809633-8.21323-x [DOI] [Google Scholar]
- 94. Nielsen, H. Bjørn , Almeida Mathieu, Juncker Agnieszka Sierakowska, Rasmussen Simon, Li Junhua, Sunagawa Shinichi, Plichta Damian R., et al. 2014. “Identification and Assembly of Genomes and Genetic Elements in Complex Metagenomic Samples without Using Reference Genomes.” Nature Biotechnology 32: 822–8. 10.1038/nbt.2939 [DOI] [PubMed] [Google Scholar]
- 95. Meyer, Fernando , Fritz Adrian, Deng Zhi‐Luo, Koslicki David, Lesker Till Robin, Gurevich Alexey, Robertson Gary, et al. 2022. “Critical Assessment of Metagenome Interpretation: The Second Round of Challenges.” Nature Methods 19: 429–40. 10.1038/s41592-022-01431-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Johansen, Joachim , Plichta Damian R., Nissen Jakob Nybo, Jespersen Marie Louise, Shah Shiraz A., Deng Ling, Stokholm Jakob, et al. 2022. “Genome Binning of Viral Entities from Bulk Metagenomics Data.” Nature Communications 13: 965. 10.1038/s41467-022-28581-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Culley, Alexander . 2018. “New Insight into the RNA Aquatic Virosphere via Viromics.” Virus Research 244: 84–9. 10.1016/j.virusres.2017.11.008 [DOI] [PubMed] [Google Scholar]
- 98. Lang, Andrew S. , Rise Matthew L., Culley Alexander I., and Steward Grieg F.. 2009. “RNA Viruses in the Sea.” FEMS Microbiology Reviews 33: 295–323. 10.1111/j.1574-6976.2008.00132.x [DOI] [PubMed] [Google Scholar]
- 99. Edwards, Robert A. , and Rohwer Forest. 2005. “Viral Metagenomics.” Nature Reviews Microbiology 3: 504–10. 10.1038/nrmicro1163; https://pubmed.ncbi.nlm.nih.gov/15886693 [DOI] [PubMed] [Google Scholar]
- 100. Jackson, William T. , Giddings Thomas H., Taylor Matthew P., Mulinyawe Sara, Rabinovitch Marlene, Kopito Ron R., and Kirkegaard Karla. 2005. “Subversion of Cellular Autophagosomal Machinery by RNA Viruses.” PLoS Biology 3: e156. 10.1371/journal.pbio.0030156; https://pubmed.ncbi.nlm.nih.gov/15884975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Lawrence, Janice E. , Brussaard Corina P. D., and Suttle Curtis A.. 2006. “Virus‐Specific Responses of Heterosigma akashiwo to Infection.” Applied and Environmental Microbiology 72: 7829–34. 10.1128/AEM.01207-06; https://pubmed.ncbi.nlm.nih.gov/17041155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Miller, Sven , and Krijnse‐Locker Jacomine. 2008. “Modification of Intracellular Membrane Structures for Virus Replication.” Nature Reviews Microbiology 6: 363–74. 10.1038/nrmicro1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Venter, J. Craig , Remington Karin, Heidelberg John F., Halpern Aaron L., Rusch Doug, Eisen Jonathan A., Wu Dongying, et al. 2004. “Environmental Genome Shotgun Sequencing of the Sargasso Sea.” Science 304: 66–74. 10.1126/science.1093857; https://pubmed.ncbi.nlm.nih.gov/15001713 [DOI] [PubMed] [Google Scholar]
- 104. Culley, Alexander I. , Lang Andrew S., and Suttle Curtis A.. 2006. “Metagenomic Analysis of Coastal RNA Virus Communities.” Science 312: 1795–8. 10.1126/science.1127404; https://pubmed.ncbi.nlm.nih.gov/16794078 [DOI] [PubMed] [Google Scholar]
- 105. Moran, Mary Ann , Satinsky Brandon, Gifford Scott M., Luo Haiwei, Rivers Adam, Chan Leong‐Keat, Meng Jun, et al. 2013. “Sizing up Metatranscriptomics.” The ISME Journal 7: 237–43. 10.1038/ismej.2012.94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Culley, Alexander I. , Lang Andrew S., and Suttle Curtis A.. 2003. “High Diversity of Unknown Picorna‐Like Viruses in the Sea.” Nature 424: 1054–7. 10.1038/nature01886; https://pubmed.ncbi.nlm.nih.gov/12944967 [DOI] [PubMed] [Google Scholar]
- 107. Culley, Alexander I. , and Steward Grieg F.. 2007. “New Genera of RNA Viruses in Subtropical Seawater, Inferred from Polymerase Gene Sequences.” Applied and Environmental Microbiology 73: 5937–44. 10.1128/AEM.01065-07; https://pubmed.ncbi.nlm.nih.gov/17644642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Gustavsen, Julia A. , Winget Danielle M., Tian Xi, and Suttle Curtis A.. 2014. “High Temporal and Spatial Diversity in Marine RNA Viruses Implies That They Have an Important Role in Mortality and Structuring Plankton Communities.” Frontiers In Microbiology 5: 703. 10.3389/fmicb.2014.00703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Culley, Alexander I. , Mueller Jaclyn A., Belcaid Madhi, Wood‐Charlson Elisha M., Poisson Guylaine, and Steward Grieg F.. 2014. “The Characterization of RNA Viruses in Tropical Seawater Using Targeted PCR and Metagenomics.” MBio 5: e01210–4. 10.1128/mBio.01210-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Gustavsen, Julia A. , and Suttle Curtis A.. 2021. “Role of Phylogenetic Structure in the Dynamics of Coastal Viral Assemblages.” Applied and Environmental Microbiology 87: e02704‐20. 10.1128/AEM.02704-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Handelsman, Jo , Rondon Michelle R., Brady Sean F., Clardy Jon, and Goodman Robert M.. 1998. “Molecular Biological Access to the Chemistry of Unknown Soil Microbes: A New Frontier for Natural Products.” Chemistry & Biology 5: R245–9. https://pubmed.ncbi.nlm.nih.gov/9818143 [DOI] [PubMed] [Google Scholar]
- 112. Chen, Kevin , and Pachter Lior. 2005. “Bioinformatics for Whole‐Genome Shotgun Sequencing of Microbial Communities.” PLoS Computational Biology 1: e24. 10.1371/journal.pcbi.0010024; https://pubmed.ncbi.nlm.nih.gov/16110337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Breitbart, Mya , Salamon Peter, Andresen Bjarne, Mahaffy Joseph M., Segall Anca M., Mead David, Azam Farooq, and Rohwer Forest. 2002. “Genomic Analysis of Uncultured Marine Viral Communities.” Proceedings of the National Academy of Sciences 99: 14250–5. 10.1073/pnas.202488399; https://pubmed.ncbi.nlm.nih.gov/12384570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Breitbart, Mya , Felts Ben, Kelley Scott, Mahaffy Joseph M., Nulton James, Salamon Peter, and Rohwer Forest. 2004. “Diversity and Population Structure of a Near‐Shore Marine‐Sediment Viral Community.” Proceedings of the Royal Society of London. Series B: Biological Sciences 271: 565–74. 10.1098/rspb.2003.2628; https://pubmed.ncbi.nlm.nih.gov/15156913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Chen, Yan‐Mei , Sadiq Sabrina, Tian Jun‐Hua, Chen Xiao, Lin Xian‐Dan, Shen Jin‐Jin, Chen Hao, et al. 2022. “RNA Viromes from Terrestrial Sites Across China Expand Environmental Viral Diversity.” Nature Microbiology 7: 1312–23. 10.1038/s41564-022-01180-2 [DOI] [PubMed] [Google Scholar]
- 116. Brum, Jennifer R. , Culley Alexander I., and Steward Grieg F.. 2013. “Assembly of a Marine Viral Metagenome After Physical Fractionation.” PLoS ONE 8: e60604. 10.1371/journal.pone.0060604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Andrews‐Pfannkoch, Cynthia , Fadrosh Douglas W., Thorpe Joyce, and Williamson Shannon J.. 2010. “Hydroxyapatite‐Mediated Separation of Double‐Stranded DNA, Single‐Stranded DNA, and RNA Genomes from Natural Viral Assemblages.” Applied and Environmental Microbiology 76: 5039–45. 10.1128/AEM.00204-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Miranda, Jaclyn A. , Culley Alexander I., Schvarcz Christopher R., and Steward Grieg F.. 2016. “RNA Viruses as Major Contributors to Antarctic Virioplankton.” Environmental Microbiology 18: 3714–27. 10.1111/1462-2920.13291 [DOI] [PubMed] [Google Scholar]
- 119. Williamson, Shannon J. , Allen Lisa Zeigler, Lorenzi Hernan A., Fadrosh Douglas W., Brami Daniel, Thiagarajan Mathangi, McCrow John P., et al. 2012. “Metagenomic Exploration of Viruses Throughout the Indian Ocean.” PLoS ONE 7: e42047. 10.1371/journal.pone.0042047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Vlok, Marli , Lang Andrew S., and Suttle Curtis A.. 2019. “Marine RNA Virus Quasispecies Are Distributed Throughout the Oceans.” MSphere 4: e00157‐19. 10.1128/mSphereDirect.00157-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Zeigler Allen, Lisa , McCrow John P., Ininbergs Karolina, Dupont Christopher L., Badger Jonathan H., Hoffman Jeffery M., Ekman Martin, et al. 2017. “The Baltic Sea Virome: Diversity and Transcriptional Activity of DNA and RNA Viruses.” MSystems 2: e00125‐16. 10.1128/mSystems.00125-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Sadeghi, Mohammadreza , Tomaru Yuji, and Ahola Tero. 2021. “RNA Viruses in Aquatic Unicellular Eukaryotes.” Viruses 13: 362. 10.3390/v13030362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Rastrojo, Alberto , and Alcamí Antonio. 2017. “Aquatic Viral Metagenomics: Lights and Shadows.” Virus Research 239: 87–96. 10.1016/j.virusres.2016.11.021 [DOI] [PubMed] [Google Scholar]
- 124. Hall, Richard J. , Wang Jing, Todd Angela K., Bissielo Ange B., Yen Seiha, Strydom Hugo, Moore Nicole E., et al. 2014. “Evaluation of Rapid and Simple Techniques for the Enrichment of Viruses Prior to Metagenomic Virus Discovery.” Journal of Virological Methods 195: 194–204. 10.1016/j.jviromet.2013.08.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Thurber, Rebecca V. , Haynes Matthew, Breitbart Mya, Wegley Linda, and Rohwer Forest. 2009. “Laboratory Procedures to Generate Viral Metagenomes.” Nature Protocols 4: 470–83. 10.1038/nprot.2009.10 [DOI] [PubMed] [Google Scholar]
- 126. Furtak, Vyacheslav , Roivainen Merja, Mirochnichenko Olga, Zagorodnyaya Tatiana, Laassri Majid, Zaidi Sohail Z., Rehman Lubna, et al. 2016. “Environmental Surveillance of Viruses by Tangential Flow Filtration and Metagenomic Reconstruction.” Euro Surveillance: Bulletin Europeen Sur Les Maladies Transmissibles = European Communicable Disease Bulletin 21: pii=30193. 10.2807/1560-7917.ES.2016.21.15.30193 [DOI] [PubMed] [Google Scholar]
- 127. Sun, Guowei , Xiao Jinzhou, Wang Hongming, Gong Chaowen, Pan Yingjie, Yan Shuling, and Wang Yongjie. 2014. “Efficient Purification and Concentration of Viruses from a Large Body of High Turbidity Seawater.” MethodsX 1: 197–206. 10.1016/j.mex.2014.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Hurwitz, Bonnie L. , Li, Deng , Deng Li, Poulos Bonnie T., and Sullivan Matthew B.. 2013. “Evaluation of Methods to Concentrate and Purify Ocean Virus Communities Through Comparative, Replicated Metagenomics.” Environmental Microbiology 15: 1428–40. 10.1111/j.1462-2920.2012.02836.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Székely, Anna J. , and Breitbart Mya. 2016. “Single‐Stranded DNA Phages: From Early Molecular Biology Tools to Recent Revolutions in Environmental Microbiology.” FEMS Microbiology Letters 363: fnw027. 10.1093/femsle/fnw027 [DOI] [PubMed] [Google Scholar]
- 130. van der Valk, Tom , Vezzi Francesco, Ormestad Mattias, Dalén Love, and Guschanski Katerina. 2020. “Index Hopping on the Illumina HiseqX Platform and Its Consequences for Ancient DNA Studies.” Molecular Ecology Resources 20: 1171–81. 10.1111/1755-0998.13009 [DOI] [PubMed] [Google Scholar]
- 131. Zhang, Yu‐Yi , Chen Yicong, Wei Xiaoman, and Cui Jie. 2022. “Viromes in Marine Ecosystems Reveal Remarkable Invertebrate RNA Virus Diversity.” Science China. Life Sciences 65: 426–37. 10.1007/s11427-020-1936-2 [DOI] [PubMed] [Google Scholar]
- 132. Pettersson, John H. O. , Shi Mang, Eden John‐Sebastian, Holmes Edward C., and Hesson Jenny C.. 2019. “Meta‐Transcriptomic Comparison of the RNA Viromes of the Mosquito Vectors Culex pipiens and Culex Torrentium in Northern Europe.” Viruses 11: 1033. 10.3390/v11111033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Zhou, Ying‐Li , Mara Paraskevi, Cui Guo‐Jie, Edgcomb Virginia P., and Wang Yong. 2022. “Microbiomes in the Challenger Deep Slope and Bottom‐Axis Sediments.” Nature Communications 13: 1515. 10.1038/s41467-022-29144-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Ter Horst, Anneliek M. , Santos‐Medellín Christian, Sorensen Jackson W., Zinke Laura A., Wilson Rachel M., Johnston Eric R., Trubl Gareth, et al. 2021. “Minnesota Peat Viromes Reveal Terrestrial and Aquatic Niche Partitioning for Local and Global Viral Populations.” Microbiome 9: 233. 10.1186/s40168-021-01156-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Neri, Uri , Wolf Yuri I., Roux Simon, Camargo Antonio Pedro, Lee Benjamin, Kazlauskas Darius, Chen I. Min, et al. 2022. “A Five‐Fold Expansion of the Global RNA Virome Reveals Multiple New Clades of RNA Bacteriophages.” Cell S0092‐8674: 01118–7. 10.1016/j.cell.2022.08.023 [DOI] [PubMed] [Google Scholar]
- 136. Dolja, Valerian V. , Krupovic Mart, and Koonin Eugene V.. 2020. “Deep Roots and Splendid Boughs of the Global Plant Virome.” Annual Review of Phytopathology 58: 23–53. 10.1146/annurev-phyto-030320-041346 [DOI] [PubMed] [Google Scholar]
- 137. Geoghegan, Jemma L. , Di Giallonardo Francesca, Wille Michelle, Ortiz‐Baez Ayda Susana, Costa Vincenzo A., Ghaly Timothy, Mifsud Jonathon C. O., et al. 2021. “Virome Composition in Marine Fish Revealed by Meta‐Transcriptomics.” Virus Evolution 7: veab005. 10.1093/ve/veab005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Moniruzzaman, Mohammad , Wurch Louie L., Alexander Harriet, Dyhrman Sonya T., Gobler Christopher J., and Wilhelm Steven W.. 2017. “Virus–Host Relationshipsof Marine Single‐Celled Eukaryotes Resolved from Metatranscriptomics.” Nature Communications 8: 16054. 10.1038/ncomms16054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Glickman, Cody , Hendrix Jo, and Strong Michael. 2021. “Simulation Study and Comparative Evaluation of Viral Contiguous Sequence Identification Tools.” BMC Bioinformatics 22: 329. 10.1186/s12859-021-04242-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Altschul, Stephen F. , Gish Warren, Miller Webb, Myers Eugene W., and Lipman David J.. 1990. “Basic Local Alignment Search Tool.” Journal of Molecular Biology 215: 403–10. 10.1016/S0022-2836(05)80360-2; https://pubmed.ncbi.nlm.nih.gov/2231712 [DOI] [PubMed] [Google Scholar]
- 141. Ren, Jie , Ahlgren Nathan A., Lu Yang Young, Fuhrman Jed A., and Sun Fengzhu. 2017. “VirFinder: A Novel k‐Mer Based Tool for Identifying Viral Sequences from Assembled Metagenomic Data.” Microbiome 5: 69. 10.1186/s40168-017-0283-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Garcia, Benjamin J. , Simha Ramanuja, Garvin Michael, Furches Anna, Jones Piet, Gazolla Joao G. F. M., Hyatt P. Doug, et al. 2021. “A k‐Mer Based Approach for Classifying Viruses without Taxonomy Identifies Viral Associations In Human Autism and Plant Microbiomes.” Computational and Structural Biotechnology Journal 19: 5911–9. 10.1016/j.csbj.2021.10.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Kieft, Kristopher , Zhou Zhichao, and Anantharaman Karthik. 2020. “VIBRANT: Automated Recovery, Annotation and Curation of Microbial Viruses, and Evaluation of Viral Community Function from Genomic Sequences.” Microbiome 8: 90. 10.1186/s40168-020-00867-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Guo, Jiarong , Bolduc Ben, Zayed Ahmed A., Varsani Arvind, Dominguez‐Huerta Guillermo, Delmont Tom O., Pratama Akbar Adjie, et al. 2021. “VirSorter2: A Multi‐Classifier, Expert‐Guided Approach to Detect Diverse DNA and RNA Viruses.” Microbiome 9: 37. 10.1186/s40168-020-00990-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Lu, Congyu , Zhang Zheng, Cai Zena, Zhu Zhaozhong, Qiu Ye, Wu Aiping, Jiang Taijiao, Zheng Heping, and Peng Yousong. 2021. “Prokaryotic Virus Host Predictor: A Gaussian Model for Host Prediction of Prokaryotic Viruses in Metagenomics.” BMC Biology 19: 5. 10.1186/s12915-020-00938-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Dutilh, Bas E. , Varsani Arvind, Tong Yigang, Simmonds Peter, Sabanadzovic Sead, Rubino Luisa, Roux Simon, et al. 2021. “Perspective on Taxonomic Classification of Uncultivated Viruses.” Current Opinion In Virology 51: 207–15. 10.1016/j.coviro.2021.10.011 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This manuscript does not generate any script and data. All authors read and approved the final manuscript. Supplementary materials (figures, tables, scripts, graphical abstract, slides, videos, Chinese translated version, and updated materials) may be found in the online DOI or iMeta Science http://www.imeta.science/.


