Abstract
The E1 protein from bovine papillomavirus has site-specific DNA binding activity, DNA helicase activity, and DNA-dependent ATPase activity consistent with the properties of an initiator protein. Here we have identified and characterized a novel oligomeric form of E1 that is associated with the ATPase and DNA helicase activities and whose formation is strongly stimulated by single-stranded DNA. This oligomeric form corresponds to a hexamer of E1.
DNA helicases constitute an interesting class of proteins with important functions in a large number of biological processes. These proteins, by virtue of their ability to separate the two complementary strands of the double helix, are essential for all processes that require exposure of the base sequence, including transcription, DNA replication, and recombination (for reviews, see references 12 and 16). The initiator proteins of the small DNA tumor viruses simian virus 40 (SV40) and polyomavirus form an interesting subgroup of hexameric DNA helicases. These proteins, in addition to helicase activity, also have other DNA replication-related activities, such as sequence-specific DNA binding activity for ori recognition (for reviews, see references 3 and 8). While most DNA helicases require a region of single-stranded DNA for entry, these proteins can initiate unwinding from completely double-stranded DNA, presumably by causing helix melting and entry of the DNA helicase onto a single-stranded region. A consequence of these three activities is the fact that the initiator proteins have the unique capability of initiating unwinding on a circular double-stranded template in a sequence-specific manner.
Bovine papillomavirus (BPV) has been studied intensively as a model system for papillomavirus DNA replication (for a review, see reference 25). In vivo DNA replication studies have demonstrated that two viral proteins are required for viral DNA replication (27). These proteins, encoded from the E1 and E2 open reading frames, bind to the viral origin of replication, which contains specific binding sites for both proteins (28, 31, 32). The BPV E1 protein belongs to the group of viral initiator proteins whose best-studied member is SV40 large T antigen (1, 6, 14, 17, 22, 26, 33). Thus, BPV E1 has ori-specific DNA-binding activity, DNA-dependent ATPase activity, and DNA helicase activity (11, 13, 22, 26, 28, 31, 33).
Here, we describe the characterization of a novel oligomeric E1 complex. This complex forms spontaneously in the presence of single-stranded DNA and appears to contain six molecules of E1 together with one molecule of single-stranded DNA. The dependence on DNA for the formation of this complex may reflect a dependence on DNA for the assembly of the individual E1 subunits into a specific configuration required for enzymatic activity.
DNA-dependent ATPase resides in an oligomeric form of E1.
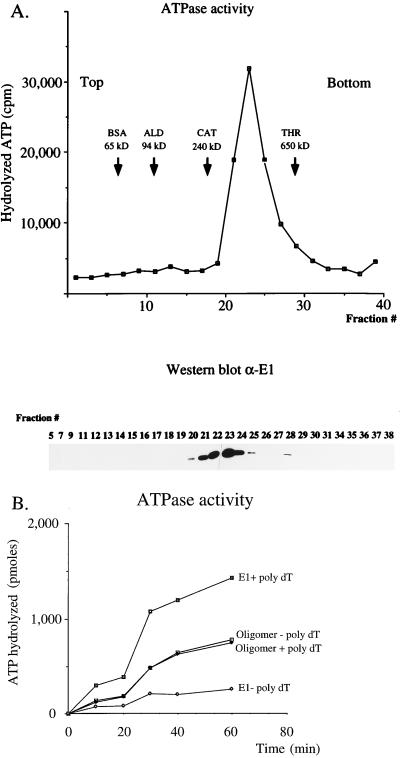
A hallmark of DNA helicases is the fact that the helicase activity is invariably associated with DNA-dependent nucleoside triphosphatase activity. Consistent with this finding, the BPV E1 protein has been demonstrated to have DNA-dependent ATPase activity as well as DNA helicase activity (22, 33). To determine in which form the ATPase and helicase activities resided, we assembled reactions under the conditions used for ATPase assays. Complexes were formed in 200 μl of buffer A (25 mM HEPES-Na [pH 7.6], 100 mM NaCl, 7 mM MgCl2, 100 μM ATP, 1 mM dithiothreitol) containing 4 mM ATP and 0.1 mg of bovine serum albumin (BSA)/ml, with 12 μg of E1 protein (21) and 2 μg of a single-stranded 28-mer oligonucleotide. The mixture was incubated for 10 min at room temperature, mixed with marker proteins, loaded onto a 15 to 30% glycerol gradient, and centrifuged in an SW55 Ti rotor (Beckman Instruments) at 50,000 rpm for 10 h at 4°C. Forty fractions were collected and assayed for the presence of E1 by Western blotting with a monoclonal antibody directed against E1 as well as for ATPase activity by a standard ATPase assay as described previously (30). The purified E1 protein in the absence of ATP, magnesium, and oligonucleotide sedimented close to the BSA marker protein, indicating that the protein was monomeric (data not shown). Strikingly, incubation in the presence of the oligonucleotide and ATP resulted in a quantitative conversion of E1 from the monomeric form to a much larger form that sedimented as a sharp peak between the marker proteins catalase (240 kDa) and thyroglobulin (650 kDa), as judged by Western blotting (Fig. 1A). These same fractions also contained all the ATPase activity that we could detect in the gradient. Under these conditions, no E1 could be detected elsewhere in the gradient.
FIG. 1.
Incubation of E1 in the presence of single-stranded DNA and ATP results in the quantitative conversion of E1 into an oligomeric complex. (A) E1 was incubated under conditions used for DNA-dependent ATPase assays and was subsequently loaded onto a 15 to 30% glycerol gradient and sedimented as described in Materials and Methods. The gradient was fractionated into 40 fractions, and every other fraction was analyzed for ATPase activity. The glycerol gradient fractions were analyzed for the presence of E1 by Western blotting with a monoclonal antibody directed against E1. ALD, aldolase; CAT, catalase; THR, thyroglobulin. (B) The ATPase activity of the oligomeric E1 complex is not stimulated by DNA. Material from the oligomeric peak in the glycerol gradient in panel A was analyzed for ATPase activity in the absence or presence of poly(dT) over a 60-min time course. In parallel, 0.2 μg of purified, unfractionated E1 protein was analyzed under the same conditions.
ATPase activity associated with the oligomeric E1 complex is not stimulated by DNA.
We were interested in whether the ATPase activity recovered from the glycerol gradient had properties similar to those of the unfractionated ATPase activity. Specifically, we wanted to determine if the ATPase activity of the oligomeric form of E1 was stimulated by the addition of DNA, like that of the unfractionated, monomeric E1 protein. We therefore performed ATPase assays in the absence and presence of added DNA with monomeric E1 as well as with the oligomeric E1 from the peak fraction in the gradient, as shown in Fig. 1B. As expected, purified E1 showed a low level of ATPase activity that was significantly stimulated by the addition of poly(dT). However, the ATPase activity of the oligomeric E1 isolated from the gradient was not stimulated upon addition of DNA. Several explanations for this difference are possible. Sufficient quantities of oligonucleotide may be present nonspecifically throughout the gradient to mask the DNA dependence of the ATPase, or the DNA may be required for the formation of the oligomeric complex and not directly for activity.
Single-stranded DNA is strongly stimulatory for E1 oligomerization and is stably associated with the E1 oligomer.
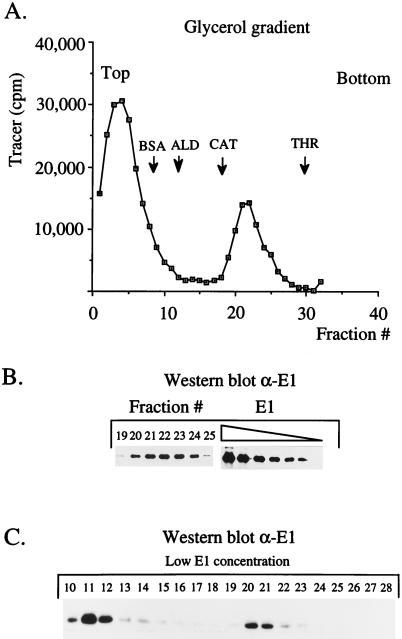
To address these questions, we monitored both E1 and the oligonucleotide in the gradient. We assembled reaction mixtures as described above but included small quantities of 32P-labeled oligonucleotide. After sedimentation, as shown in Fig. 2A, the 32P-oligonucleotide was present in two discrete peaks, one at the top of the gradient corresponding to free oligonucleotide and one peak that coincided with the oligomeric E1 peak that we had observed previously. Western analysis of the gradient fractions demonstrated that these fractions indeed corresponded to the oligomeric E1 peak (Fig. 2B). These results demonstrated that the oligonucleotide was specifically and stably associated with the E1 oligomer and not distributed throughout the gradient. The association with oligonucleotide did not exhibit any obvious sequence specificity: several different oligonucleotides of similar sizes but with different sequences appeared to function equally well for oligomer formation, although a shorter, 17-mer oligonucleotide was less active for E1 oligomerization (data not shown).
FIG. 2.
The oligonucleotide is stably associated with the oligomeric E1 complex. (A) E1 was incubated in the presence of a 27-mer oligonucleotide and sedimented on a gradient as described in the legend to Fig. 1, except that a small fraction of 32P-labeled 27-mer oligonucleotide was added. The gradient fractions were analyzed for the presence of 32P counts. ALD, aldolase; CAT, catalase; THR, thyroglobulin. (B) Western blot analysis of the oligomeric E1 peak. The E1 protein in the oligomer peak was quantitated by comparison to a titration of known quantities (50, 25, 12, 8, 6, and 4 ng) of purified E1 protein. (C) E1 was incubated in the presence of oligonucleotide as described in the legend to Fig. 1, except that 10-fold lower quantities of E1 protein (1.2 μg) and oligonucleotide (0.2 μg) were used, followed by glycerol gradient sedimentation. The fractions were analyzed by Western blotting.
We used the results from this experiment to estimate the stoichiometry of binding of E1 and oligonucleotide. We determined the approximate quantity of E1 in the oligomeric peak by comparing the intensity of the signal in the Western blot to those of standard quantities of the purified E1 that was loaded onto the gradient (Fig. 2B). The chemiluminescent signal was detected by exposure to X-ray film, and light exposures were used for quantitation with a digital imaging system. The oligomeric peak accounts for at least 90% of the input E1 protein. We also determined the fraction of input radioactive oligonucleotide that was present in the oligomer peak, from which we could calculate the molar quantity of oligonucleotide present in the oligomer peak. Using these quantitations, we could calculate a molar ratio of E1 to oligonucleotide. In this experiment this ratio was found to be 5.4 mol of E1 per mol of oligonucleotide.
When identical reaction mixtures were assembled in the absence of oligonucleotide, we failed to detect an oligomeric peak of E1 by Western analysis, demonstrating that at least under these specific conditions, the presence of oligonucleotide was required for the formation of the E1 oligomer (data not shown). Instead, in the presence of ATP and magnesium, but in the absence of oligonucleotide, the majority of the E1 protein could be recovered from the pellet, consistent with the formation of very large E1 complexes (data not shown). At high E1 concentrations the oligonucleotide may therefore stimulate oligomer formation partly by preventing extensive aggregation. At lower E1 concentrations, the oligonucleotide appears to stimulate oligomer formation by nucleation. At low E1 concentrations, in the presence of limiting concentrations of oligonucleotide, two E1 peaks corresponding to monomeric E1 and the oligomer can be observed simultaneously in the gradient (Fig. 2C). These were the only conditions under which we could observe both these forms of E1 simultaneously.
The oligomeric E1 complex has DNA helicase activity.
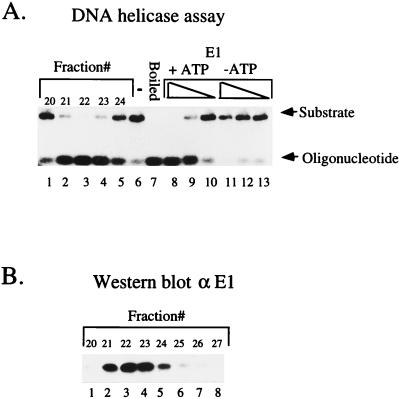
To determine if the oligomeric form of E1, isolated from glycerol gradients, was active in a DNA helicase assay, we analyzed the material from the gradient peak in an oligonucleotide displacement assay, as described by Seo and Hurwitz (23). A 50-mer oligonucleotide with partial complementarity to the plasmid pET 11C was synthesized, generating a substrate with a 28-nucleotide-long double-stranded region and a 22-nucleotide-long single-stranded 3′ tail. The oligonucleotide was 5′ labeled and annealed to the single-stranded plasmid at 55°C for 30 min in a buffer containing 20 mM Tris-HCl (pH 7.5), 1 mM EDTA, and 0.5 M NaCl. The substrate was separated from free oligonucleotide by gel filtration on a Sepharose 6B CL column. Gradient fractions, or purified E1, were incubated with substrate in a buffer containing 50 mM Tris-HCl (pH 7.9), 3 mM MgCl2, 2 mM dithiothreitol, 1 mM ATP, and 0.2 mg of BSA/ml at 37°C for 15 min. After the incubation, sodium dodecyl sulfate was added to 0.1% and the sample was loaded onto a 1.5% agarose gel in TAE buffer (0.04 M Tris-acetone, 1 mM EDTA).
In the absence of added E1, only a very small fraction of free oligonucleotide could be detected in the sample (Fig. 3A, lane 6). When the sample was heated to 100°C, all the labeled oligonucleotide was released and migrated at the bottom of the gel (Fig. 3A, lane 7). When purified E1 protein was incubated in the presence of ATP, the oligonucleotide was also displaced (Fig. 3A, lanes 8 to 10), while under the same conditions in the absence of ATP, no displacement was observed (Fig. 3A, lanes 11 to 13), indicating that, as expected, the displacement of the oligonucleotide by E1 was ATP dependent. The gradient fractions 20 to 25 had detectable helicase activity. These fractions coincided exactly with the E1 oligomer peak, as measured by Western blotting with a monoclonal E1 antibody (compare Fig. 3A, lanes 1 to 5 to Fig. 3B, lanes 1 to 5). Whether the helicase activity resides in the oligomer or possibly results from the rearrangement of the oligomer upon incubation is unclear; however, the stable nature of the E1 oligomer, which allows sedimentation, may indicate that the preformed oligomer is indeed the active helicase.
FIG. 3.
Fractions containing the oligomeric E1 complex have DNA helicase activity. (A) Glycerol gradient fractions containing the peak of tracer oligonucleotide were analyzed for DNA helicase activity by an oligonucleotide displacement assay (lanes 1 to 5). Purified E1 (300, 30, or 3 ng) was used in the same assay as a positive control with the same substrate in the presence (lanes 8 to 10) and absence (lanes 11 to 13) of ATP. (B) Western blot of the same gradient fractions that were used in the helicase assay probed with a monoclonal antibody directed against E1.
The multimeric E1 complex has a molecular mass consistent with that of an E1 hexamer.
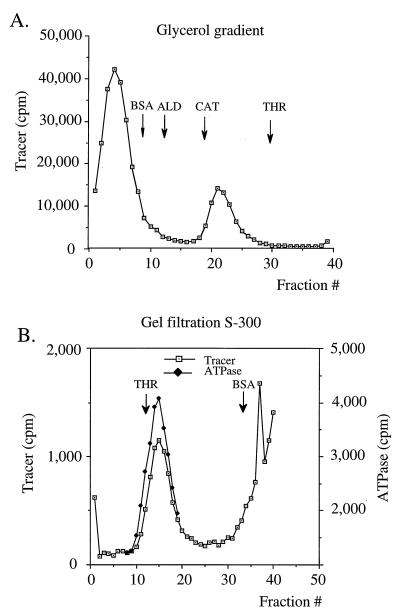
To estimate the relative molecular mass of the oligomeric form of E1, we performed glycerol gradient centrifugation experiments to determine the sedimentation rate of the oligomeric E1 complex relative to those of marker proteins under the conditions described above. The sedimentation values relative to those of the marker proteins were determined for four separate gradient runs and averaged. We also determined the Stokes’ radius of the complex relative to those of marker proteins by gel filtration. Gel filtration chromatography was performed on a low-pressure 1-cm2 by 50-cm Sephacryl S-300 column equilibrated with buffer A. The sample was generated in 200 μl of buffer A containing 4 mM ATP and 0.1 mg of BSA/ml with 12 μg of E1 protein and 2 μg of 28-mer oligonucleotide. The elution of the marker proteins was determined by Bradford protein assays. The thyroglobulin peak fraction corresponded to the void volume (18 ml). The mixture was incubated for 10 min at room temperature before it was loaded onto the column. The Stokes’ radius of the complex relative to those of marker proteins was determined from six different gel filtration experiments and averaged. An example of these analyses is shown in Fig. 4. The results from these experiments demonstrated that the oligomeric E1 complex sedimented at 12.7S ± 0.5S with a Stokes’ radius of 82 ± 5 Å. Using these values, we could calculate the relative molecular mass of the oligomeric E1 complex to be 420 ± 40 kDa. This would correspond well to a stoichiometry of six E1 molecules (68 kDa each) in the oligomeric E1 complex. This would be a far from surprising result; several helicases characterized to date form hexameric complexes. These results also indicate that the stoichiometry of binding between E1 and the oligonucleotide, which we have determined to be approximately 6 to 1, most likely means that the E1 oligomer consists of a hexamer of E1 bound to one molecule of oligonucleotide.
FIG. 4.
The relative molecular mass of the oligomeric E1 complex is consistent with that of a hexamer of E1. The relative molecular mass of the oligomeric E1 complex was estimated by using a combination of glycerol gradient centrifugation (A) and gel filtration analysis (B) to determine the S value and Stokes’ radius for the oligomeric complex relative to those of marker proteins. The Stokes’ radius for the oligomeric E1 complex was determined to be 82 ± 5 Å, and the sedimentation rate was 12.7S ± 0.5S, resulting in an estimated molecular mass of 420 ± 40 kDa. The marker proteins were BSA (35 Å; 4.2S), aldolase (ALD) (46 Å; 8.3S), catalase (CAT) (52 Å; 11.3S), and thyroglobulin (THR) (85 Å; 18.5S).
Under conditions where ATPase activity of E1 can be detected, purified monomeric E1 protein is quantitatively converted into a large oligomeric form with a molecular mass consistent with that of a hexamer. Fractions containing this complex show both ATPase and DNA helicase activities, suggesting that both of these activities reside in the hexameric form of E1. This is consistent with results obtained for SV40 large T antigen, where similar experiments have indicated that a hexamer has helicase activity (15, 29). It is difficult, however, to completely rule out the possibility that a rearrangement of the isolated hexamers can occur after isolation and that this rearrangement results in the active enzyme. A stimulatory effect of oligonucleotide on the formation of hexameric helicases is not unprecedented; it has been demonstrated, for the T7 gene 4 protein as well as for DnaB, that single-stranded oligonucleotides are strongly stimulatory for hexamer formation and that the stoichiometry of binding is very close to one DNA molecule per hexamer (4, 5, 10, 19). A likely possibility is that the helicase assembles on its cognate substrate. Like SV40 large T antigen, the E1 protein is capable of inducing KMnO4 sensitivity in the sequences flanking its binding site (2, 9, 18, 20). A simple model for the loading of the DNA helicase is a situation where a region of single-stranded DNA in a structurally distorted ori could serve as a specific inducer and site of formation for the hexamer. If the E1 hexamer forms a ring-like structure, as has been suggested for hexameric helicases, one implication of this model would be that E1 encircles one DNA strand (7, 24).
Acknowledgments
This work was supported by National Institutes of Health grant CA 13106 to A.S.
REFERENCES
- 1.Bonne-Andrea C, Santucci S, Clertant P, Tillier F. Bovine papillomavirus E1 protein binds specifically DNA polymerase α but not replication protein A. J Virol. 1995;69:2341–2350. doi: 10.1128/jvi.69.4.2341-2350.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borowiec J A, Hurwitz J. Localized melting and structural changes in the SV40 origin of DNA replication induced by T-antigen. EMBO J. 1988;7:3149–3158. doi: 10.1002/j.1460-2075.1988.tb03182.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borowiec J A, Dean F B, Bullock P, Hurwitz J. Binding and unwinding—how T-antigen engages the SV40 origin of replication. Cell. 1990;60:181–184. doi: 10.1016/0092-8674(90)90730-3. [DOI] [PubMed] [Google Scholar]
- 4.Bujalowski W, Klonowska M M, Jezewska M J. Oligomeric structure of Escherichia coli primary replicative helicase DnaB protein. J Biol Chem. 1994;269:31350–31358. [PubMed] [Google Scholar]
- 5.Bujalowski W, Jezewska M J. Interactions of Escherichia coli primary replicative helicase DnaB protein with single stranded DNA. The nucleic acid does not wrap around the protein hexamer. Biochemistry. 1995;34:8513–8519. doi: 10.1021/bi00027a001. [DOI] [PubMed] [Google Scholar]
- 6.Clertant P, Seif I. A common function for polyoma virus large-T and papillomavirus E1 proteins? Nature (London) 1984;311:276–279. doi: 10.1038/311276a0. [DOI] [PubMed] [Google Scholar]
- 7.Egelman E H, Yu X, Wild R, Hingorani M M, Patel S. Bacteriophage T7 helicase/primase proteins form rings around single-stranded DNA that suggests a general structure for hexameric helicases. Proc Natl Acad Sci USA. 1995;92:3869–3873. doi: 10.1073/pnas.92.9.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fanning E, Knippers R. Structure and function of simian virus 40 large tumor antigen. Annu Rev Biochem. 1992;61:55–85. doi: 10.1146/annurev.bi.61.070192.000415. [DOI] [PubMed] [Google Scholar]
- 9.Gillette T G, Lusky M, Borowiec J A. Induction of structural changes in the bovine papillomavirus type 1 origin of replication by the viral E1 and E2 proteins. Proc Natl Acad Sci USA. 1994;91:8846–8850. doi: 10.1073/pnas.91.19.8846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hingorani M M, Patel S S. Interactions of bacteriophage T7 primase/helicase protein with single-stranded and double-stranded DNAs. Biochemistry. 1993;32:12478–12487. doi: 10.1021/bi00097a028. [DOI] [PubMed] [Google Scholar]
- 11.Holt S E, Wilson V G. Mutational analysis of the 18-base-pair inverted repeat element at the bovine papillomavirus origin of replication: identification of critical sequences for E1 binding and in vivo replication. J Virol. 1995;69:6525–6532. doi: 10.1128/jvi.69.10.6525-6532.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lohman T M, Bjornson K P. Mechanisms of helicase-catalyzed unwinding. Annu Rev Biochem. 1996;65:169–214. doi: 10.1146/annurev.bi.65.070196.001125. [DOI] [PubMed] [Google Scholar]
- 13.MacPherson P, Thorner L, Botchan M. The bovine papillomavirus E1 protein has ATPase activity essential to viral DNA replication and efficient transformation in cells. Virology. 1994;204:403–408. doi: 10.1006/viro.1994.1544. [DOI] [PubMed] [Google Scholar]
- 14.Mansky K C, Batiza A, Lambert P F. Bovine papillomavirus type 1 E1 and simian virus 40 large T antigen share regions of sequence similarity required for multiple functions. J Virol. 1997;71:7600–7608. doi: 10.1128/jvi.71.10.7600-7608.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mastrangelo I A, Hough P V C, Wall J S, Dodson M, Dean F B, Hurwitz J. ATP-dependent assembly of soluble hexamers of SV40 T-antigen at the viral origin of DNA replication. Nature (London) 1989;338:658–662. doi: 10.1038/338658a0. [DOI] [PubMed] [Google Scholar]
- 16.Matson S, Kaiser-Rogers K A. DNA helicases. Annu Rev Biochem. 1990;59:289–329. doi: 10.1146/annurev.bi.59.070190.001445. [DOI] [PubMed] [Google Scholar]
- 17.Park P, Copeland W, Yang L, Wang T, Botchan M R, Mohr I. The cellular DNA polymerase α-primase is required for papillomavirus DNA replication and associates with the viral E1 helicase. Proc Natl Acad Sci USA. 1994;91:8700–8704. doi: 10.1073/pnas.91.18.8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parsons R, Anderson M E, Tegtmeyer P. Three domains in the simian virus 40 core origin orchestrate the binding, melting, and DNA helicase activities of T antigen. J Virol. 1990;64:509–518. doi: 10.1128/jvi.64.2.509-518.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patel S S, Hingorani M M. Oligomeric structure of bacteriophage T7 DNA primase/helicase proteins. J Biol Chem. 1993;268:10668–10675. [PubMed] [Google Scholar]
- 20.Sanders, C. M., and A. Stenlund. ATP-dependent assembly of the BPV-1 replication initiation complex catalyzed by viral transcription factor E2. Submitted for publication.
- 21.Sedman T, Sedman J, Stenlund A. Binding of the E1 and E2 proteins to the origin of replication of bovine papillomavirus. J Virol. 1997;71:2887–2896. doi: 10.1128/jvi.71.4.2887-2896.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seo Y-S, Muller F, Lusky M, Hurwitz J. Bovine papillomavirus (BPV)-encoded E1 protein contains multiple activities required for BPV DNA replication. Proc Natl Acad Sci USA. 1993;90:702–706. doi: 10.1073/pnas.90.2.702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seo Y-S, Hurwitz J. Isolation of helicase α, a DNA helicase from HeLa cells stimulated by a fork structure and single-stranded DNA-binding proteins. J Biol Chem. 1993;268:10282–10295. [PubMed] [Google Scholar]
- 24.Stasiak A, Tsaneva I R, West S C, Benson C J B, Yu X, Egelman E H. The Escherichia coli RuvB branch migration protein forms double hexameric rings around DNA. Proc Natl Acad Sci USA. 1994;91:7618–7622. doi: 10.1073/pnas.91.16.7618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stenlund A. Papillomavirus DNA replication. In: DePamphilis M L, editor. DNA replication in eukaryotic cells. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 679–699. [Google Scholar]
- 26.Thorner L, Lim D, Botchan M. DNA binding domain of bovine papillomavirus type 1 E1 helicase: structural and functional aspects. J Virol. 1993;67:6000–6014. doi: 10.1128/jvi.67.10.6000-6014.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ustav M, Stenlund A. Transient replication of BPV-1 requires two viral polypeptides encoded by the E1 and E2 open reading frames. EMBO J. 1991;10:449–457. doi: 10.1002/j.1460-2075.1991.tb07967.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ustav M, Ustav E, Szymanski P, Stenlund A. Identification of the origin of replication of bovine papillomavirus and characterization of the viral origin recognition factor E1. EMBO J. 1991;10:4321–4329. doi: 10.1002/j.1460-2075.1991.tb05010.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wessel R, Schweizer J, Stahl H. Simian virus 40 T-antigen DNA helicase is a hexamer which forms a binary complex during bidirectional unwinding from the viral origin of DNA replication. J Virol. 1992;66:804–815. doi: 10.1128/jvi.66.2.804-815.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wickner S, Hurwitz J. Interaction of Escherichia coli dna B and dna C (D) gene products in vitro. Proc Natl Acad Sci USA. 1975;72:921–925. doi: 10.1073/pnas.72.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilson V, Ludes-Meyers M J. A bovine papillomavirus E1-related protein binds specifically to bovine papillomavirus DNA. J Virol. 1991;65:5314–5322. doi: 10.1128/jvi.65.10.5314-5322.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang L, Li R, Mohr I, Clark R, Botchan M R. Activation of BPV-1 replication in vitro by the transcription factor E2. Nature (London) 1991;353:628–633. doi: 10.1038/353628a0. [DOI] [PubMed] [Google Scholar]
- 33.Yang L, Mohr I, Fouts E, Lim D A, Nohaile M, Botchan M. The E1 protein of the papillomavirus BPV-1 is an ATP dependent DNA helicase. Proc Natl Acad Sci USA. 1993;90:5086–5090. doi: 10.1073/pnas.90.11.5086. [DOI] [PMC free article] [PubMed] [Google Scholar]