Abstract
The dynamin-related guanosine triphosphatase, Drp1 (encoded by Dnm1l), plays a central role in mitochondrial fission and is requisite for numerous cellular processes; however, its role in muscle metabolism remains unclear. Here, we show that, among human tissues, the highest number of gene correlations with DNM1L is in skeletal muscle. Knockdown of Drp1 (Drp1-KD) promoted mitochondrial hyperfusion in the muscle of male mice. Reduced fatty acid oxidation and impaired insulin action along with increased muscle succinate was observed in Drp1-KD muscle. Muscle Drp1-KD reduced complex II assembly and activity as a consequence of diminished mitochondrial translocation of succinate dehydrogenase assembly factor 2 (Sdhaf2). Restoration of Sdhaf2 normalized complex II activity, lipid oxidation, and insulin action in Drp1-KD myocytes. Drp1 is critical in maintaining mitochondrial complex II assembly, lipid oxidation, and insulin sensitivity, suggesting a mechanistic link between mitochondrial morphology and skeletal muscle metabolism, which is clinically relevant in combatting metabolic-related diseases.
Drp1-driven translocation of Sdhaf2 to mitochondria controls complex II assembly and skeletal muscle metabolism.
INTRODUCTION
Skeletal muscle is critical for locomotion and is a primary tissue controlling glucose homeostasis and lipid metabolism in humans and rodents (1, 2). A failure to maintain muscle function is often observed in aging and metabolic-related diseases (3–5). As essential organelles of energy metabolism in skeletal muscle, mitochondria regulate numerous cellular processes, including apoptosis, Ca2+ signaling, and redox homeostasis (6, 7). Mitochondrial dysfunction in skeletal muscle is linked with a wide range of metabolic diseases including obesity and type 2 diabetes (8, 9).
To maintain a healthy network and respond to changes in metabolic demand, mitochondria rapidly undergo fusion and fission remodeling events, primarily mediated by a family of guanosine triphosphatases (GTPases), including mitofusin 1 (Mfn1), mitofusin 2 (Mfn2), optic atrophy 1 (Opa1), and dynamin-related protein 1 (Drp1) (7, 10–12). Specifically, mitochondrial division is controlled by Drp1 binding to outer mitochondrial membrane (OMM)–docking proteins while engaging actin and myosin IIA to drive oligomerization and organelle scission (13–16). Phosphorylation of Drp1 at serine-616 promotes mitochondrial translocation of Drp1 from the cytosol to the OMM to drive fission, while phosphorylation at serine-637 is inhibitory of fission consequent to cytosolic retention of Drp1 (17). We show that Drp1 serine-616 phosphorylation is increased with acute physical activity, and this is linked with exercise-induced fatty acid oxidation (18). Conversely, in a model of impaired fatty acid oxidation and insulin resistance, our laboratory showed that Drp1 translocation incompetence was linked with reduced serine-616 but increased serine-637 Drp1 phosphorylation in skeletal muscle (19). Similarly, reduced Drp1 signaling and mitochondrial fission were shown to inhibit long-chain fatty acid oxidation by impairment of carnitine O-palmitoyltransferase 1 activation (20–22). Experimental inhibition of mitochondrial fission in several cell types, including brown adipose tissue and liver, studied by Shirihai and colleagues, produced metabolic dysfunction and accumulation of tissue triglyceride (21, 23).
Although global as well as cardiac-specific deletion of Drp1 is embryonically lethal (24), acute loss and gain of Drp1 expression by viral approaches are shown to govern muscle growth (25) and mass (26). Collectively, the findings appear to suggest that the impact of Drp1 expression on metabolism is influenced by cell specificity, age, and environmental factors including diet and activity.
We have previously shown that muscle-specific knockdown of Drp1 reduces maximal running speed and muscle endurance capacity (18). Here, we investigated the role of Drp1 in regulating skeletal muscle fatty acid metabolism and insulin action using genetic and pharmacological models of Drp1 inactivation. We demonstrate that skeletal muscle Drp1 is essential for maintaining fatty acid oxidation and insulin action by facilitating the mitochondrial translocation and import of succinate dehydrogenase assembly factor 2 (Sdhaf2). We find that Drp1 is requisite for mitochondrial complex II assembly and activity. Overexpression of Sdhaf2 in the context of Drp1 knockdown in myocytes (Drp1KD) restored mitochondrial respiration, fatty acid oxidation, and muscle insulin action. These findings provide insight into the role of Drp1 on metabolism and provide a mechanistic link between mitochondrial morphology and the regulation of skeletal muscle metabolism.
RESULTS
Association of Drp1 with obesity and type 2 diabetes in humans and rodents
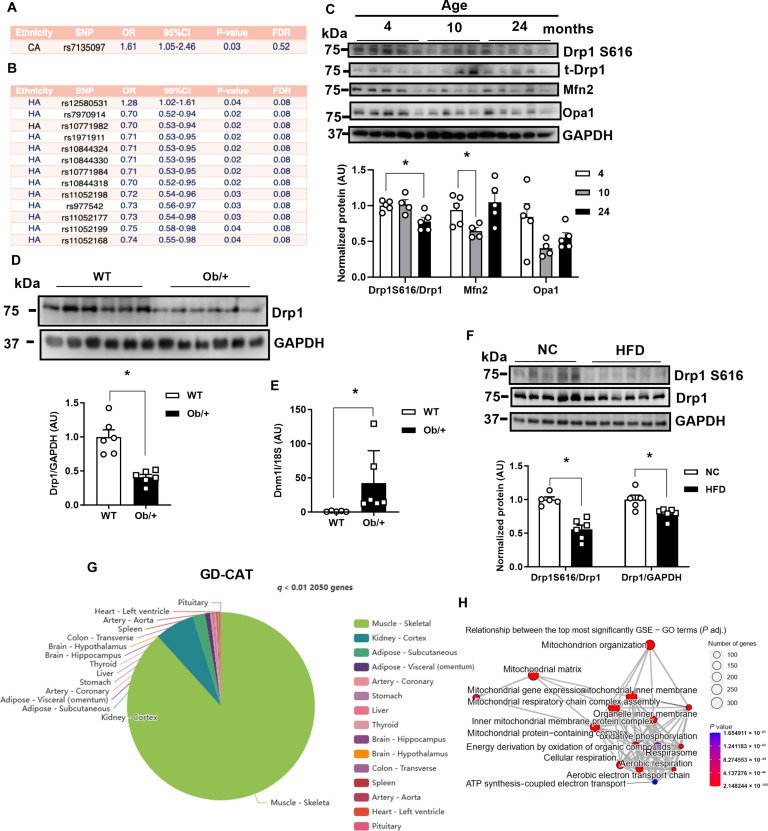
Genome-wide association studies from the Women’s Health Initiative identified a DNM1L single-nucleotide polymorphism (SNP) associated with obesity as well as 13 SNPs associated with type 2 diabetes (Fig. 1, A and B). Thirteen of the 14 SNPs affected women of Hispanic American descent. These findings aligned with our previous observations associating Dnm1l SNPs with obesity/type 2 diabetes (19). Moreover, we found that active Drp1 phosphorylation at serin-616 was significantly decreased in skeletal muscle from aged rodents (Fig. 1C; up to 24 months). We observed discordance of Dnm1l transcript and Drp1 protein in genetic obesity, as Drp1 protein levels were markedly reduced; however, transcript levels were significantly increased in skeletal muscle of mice heterozygous for the leptin mutation (Ob/+) compared to age-matched wild-type (WT) mice (Fig. 1, D and E, and fig. S1A). Overnutrition by high-fat diet (HFD) feeding reduced total Drp1 protein level and Drp1 activation at serine-616, without changing mitochondrial fission and fusion gene expression in skeletal muscle of male WT C57Bl/6J mice (Fig. 1F and fig. S1B). Our prior work has shown that both pharmacological inhibition (Mdivi-1) and genetic deletion of Drp1 in C2C12 myotubes (Drp1KD) reduce insulin-stimulated phosphorylation of protein kinase B (Akt) (19). Similar to these findings, incubation of C2C12 myotubes with the mitochondrial fusion promoter M1 (27) (induces mitochondria elongation) impairs insulin action (fig. S1C). We mined Genotype-Tissue Expression (GTEx) data to identify transcripts correlated with DNM1L expression in skeletal muscle (28). Using the gene-derived correlations across tissue (GD-CAT) (https://pipeline.biochem.uci.edu/gtex/) computational tool, we found that, of the 2050 genes significantly associated with DNM1L, skeletal muscle had a high abundance of gene-gene correlations compared to all other metabolic tissues, and these gene correlations were also minimally overlapping with other metabolic tissues (P < 0.001; Fig. 1G). Gene ontology terms associated with human skeletal muscle DNM1L expression include mitochondrion organization, mitochondrial matrix, cellular respiration, organelle inner membrane, and mitochondrial respiratory chain complex assembly (Fig. 1H). Specific gene correlations that reached statistical significance include voltage-dependent anion channel 2 (VDAC2), mitochondrial import inner membrane translocase subunit Tim17-A (TIMM17A), mitochondrial adenosine 5′-triphosphate synthase F1 subunit beta (ATP5B), NADH:ubiquinone oxidoreductase subunit A5 (NDUFA5), prohibitin 1 (PHB), and inner membrane mitochondrial protein (IMMT/MIC60) (P < 0.01; table S1). Collectively, these findings show that DNM1L expression and specific gene variants are associated with metabolic health and insulin action. As we hypothesized, correlation matrices show that DNM1L is highly correlated with mitochondrial inner and outer membrane remodeling as well as oxidative metabolism.
Fig. 1. Skeletal muscle Dnm1l/Drp1 is associated with transcripts regulating mitochondrial inner membrane assembly and oxidative metabolism.
(A and B) DNM1L genetic variant association with (A) obesity and (B) type 2 diabetes from the Women’s Health Initiative. HA, Hispanic American. (C) Phospho-Drp1 S616, Drp1, Mfn2, and Opa1 protein levels in skeletal muscle from male wild-type (WT) mice aged 4, 10, and 24 months (n = 4 to 5 mice per age). (D) Drp1 protein level and (E) gene expression in the skeletal muscle of male WT and Ob/+ mice (n = 6, ages 18 to 20 weeks). (F) Immunoblots and densitometry of Dnm1L/Drp1 in muscle from high-fat diet (HFD)–fed male mice (n = 6, ages 18 to 20 weeks). AU, arbitrary units; GAPDH, glyceraldehyde-3-phosphate dehydrogenase. (G) Pie chart reflecting GTEx data from male participants (n = 210, ages 20 to 79 years) showing the number of DNM1L-associated transcripts in skeletal muscle and the distribution of these across various metabolic tissues (P < 0.01; generated in GDCAT.org). (H) Gene ontology (GO) analysis showing top pathways associated with DNM1L expression in skeletal muscle (human vastus lateralis; P < 0.01). All values are presented as means ± SEM; *P < 0.05. Unpaired Student’s t test, two-tailed.
Muscle-specific Drp1 heterozygous null mice develop glucose and insulin intolerance
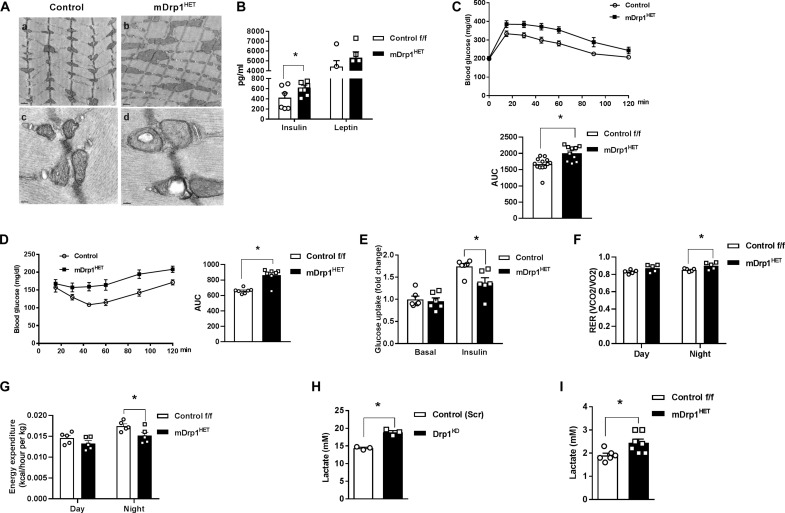
To investigate the impact of Dnm1l expression on metabolic homeostasis and insulin action in muscle, we generated a conventional muscle-specific heterozygous knockdown of Drp1 (mDrp1HET) mouse model. Drp1 protein was reduced by a ~30% selectively in skeletal muscle, but Drp1 protein abundance remained identical in cardiac muscle between the genotypes (fig. S2A) (18). Drp1 deletion is known to induce mitochondrial fission incompetency. Enlarged and elongated mitochondria were observed in skeletal muscle of mDrp1HET mice and Drp1KD myocytes (Fig. 2A and fig. S2, B and C). Despite marked alteration of mitochondrial morphology, histology of skeletal muscle from mDrp1HET appeared similar to that of control f/f mice (fig. S2D).
Fig. 2. Skeletal muscle Drp1 maintains glucose homeostasis and metabolic flexibility in male mice.
(A) Enlarged and elongated mitochondria in soleus muscle of mDrp1HET versus control f/f mice shown by transmission electron microscopy. (B) Plasma insulin and leptin in male mDrp1HET versus control f/f mice fed a normal chow (NC) diet (n = 5 to 8 mice per genotype). (C) Intraperitoneal glucose tolerance test and area under the curve (AUC) for NC-fed control f/f versus mDrp1HET mice (n = 10 to 13 mice per genotype). (D) Intraperitoneal insulin tolerance test and AUC in NC-fed control f/f versus mDrp1HET mice (n = 7 per genotype). (E) Ex vivo soleus muscle glucose uptake (fold change from basal; n = 6 mice per genotype). (F) Respiratory exchange ratio (RER) and (G) energy expenditure (EE) of NC-fed control f/f versus mDrp1HET mice (n = 6 per genotype). (H) Lactate levels in the culture medium of differentiated control (Scr) and Drp1KD myotubes and (I) in the plasma of control f/f and mDrp1HET mice (n = 6 mice per genotype). All values are presented as means ± SEM; *P < 0.05 determined by unpaired, two-tailed Student’s t test.
Elevated plasma insulin levels and impairment of glucose homeostasis and insulin action were observed in normal chow (NC)–fed mDrp1HET versus control f/f mice (Fig. 2, B to D). Reduction in ex vivo insulin–stimulated soleus muscle 2-deoxyglucose uptake for mDrp1HET versus control mice indicates substantial insulin resistance induced by Dnm1l gene reduction (Fig. 2E). Although no difference in adiposity was observed between the genotypes during NC feeding, in the context of HFD feeding, we observed an increased adiposity (ratio of epididymal white adipose tissue to body weight) in male mDrp1HET compared to control (fig. S2E).
To evaluate the impact of Drp1 expression on energy homeostasis, we individually housed NC-fed mice in metabolic chambers. The respiratory exchange ratio (RER) was significantly elevated, but energy expenditure (EE), oxygen consumption rate (VO2), and carbon dioxide production rate (VCO2) were lower in mDrp1HET mice with no change in activity or food and water intake compared with control f/f mice (Fig. 2, F and G, and fig. S2, I to P). Consistent with increased circulating lactate concentration in a human patient harboring a heterozygous dominant-negative mutation (A395D) of DNM1L, lactate levels were markedly elevated in both the culture medium of Drp1KD cells and plasma of mDrp1HET versus control f/f mice (Fig. 2, H and I) (29). Together, these findings indicate that muscle Drp1 plays an important role in regulating metabolic homeostasis and insulin action in male mice.
Drp1 deletion impairs lipid oxidation in skeletal muscle
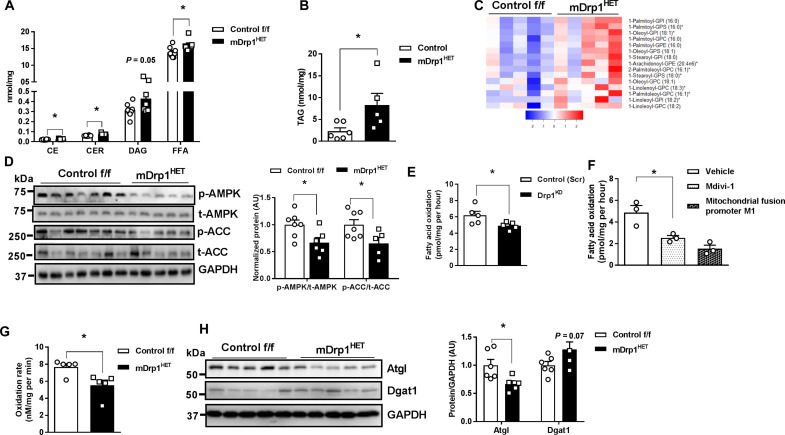
Intramyocellular lipid content inversely correlates with peripheral insulin sensitivity in healthy sedentary individuals (30). In skeletal muscle of mDrp1HET mice, cholesterol esters, ceramides, intramuscular diglyceride, free fatty acids, and triacylglycerol (TAG) levels were significantly increased compared with control f/f animals (Fig. 3, A and B, and fig. S3, A and B). In addition, neutral lipids were elevated in Drp1KD versus control-scrambled (Scr) myocytes as shown by Oil Red O staining (fig. S3C). Untargeted metabolomic analysis showed a reduction of fatty acid metabolism and elevation in a subset of lipids in skeletal muscle of mDrp1HET mice including phospholysolipids, long-chain fatty acids, polyunsaturated fatty acids, and monoacylglycerols compared with control f/f mice (Fig. 3C, fig. S3D, and table S4).
Fig. 3. Skeletal muscle Drp1 deletion impairs fatty acid oxidation promoting lipid accumulation.
(A) Intramuscular cholesterolesters (CE), ceramides (CER), diacylglycerol (DAG), free fatty acid (FFA), and (B) triacylglycerol levels of male control f/f and mDrp1HET mice (n = 7 to 8 mice per genotype). (C) Elevated phospholysolipids in skeletal muscle of mDrp1HET versus control f/f mice (n = 5 mice per genotype). GPI, glycerophosphatidylinositol; GPS, glycerophosphatidylserine; GPC, glycerophosphatidylcholine; GPE, glycerophosphatidylethanolamine. (D) Immunoblot analysis of p-AMPK Thr172/total AMPK, and p-ACC Ser79/total ACC in muscle of male control f/f and mDrp1HET mice (n = 6 to 7 mice per genotype). Densitometric analysis of phosphorylated proteins normalized to total protein. (E) Fatty acid oxidation measured by [14C]-labeled palmitate in control (Scr) and Drp1KD myotubes, (F) differentiated WT C2C12 myotubes incubated with vehicle, Drp1 inhibitor (Mdivi-1, 50 μM, 6 hours), or mitochondrial elongation promoter M1 (M1, 5 μM, 24 hours). (G) Fatty acid oxidation in isolated mitochondria from muscles of control f/f and mDrp1HET mice (n = 6 to 7 mice per genotype). (H) Immunoblot analysis of Atgl and Dgat1 in control f/f and mDrp1HET mice (n = 5 to 6 mice per genotype). Densitometric analysis of Atgl and Dgat1 normalized to GAPDH. All values are presented as means ± SEM; *P < 0.05 determined by unpaired Student’s t test, two-tailed.
To explore the mechanisms underlying lipid accumulation, we assessed lipid metabolism signaling and rates of fatty acid oxidation. Adenosine 5′-monophosphate (AMP)–activated protein kinase (AMPK) is an energy sensor that regulates skeletal muscle lipid metabolism by suppressing its downstream target acetyl–coenzyme A (CoA) carboxylase (ACC) (31). AMPK phosphorylation at Thr172 and downstream ACC-Ser79 phosphorylation were reduced in the muscle of mDrp1HET versus control f/f mice (Fig. 3D). Next, fatty acid oxidation was measured using [14C]-labeled palmitate ex vivo and in vitro. Fatty oxidation was reduced in Drp1KD versus control (Scr) myotubes, C2C12 myotubes incubated with either Drp1 inhibitor (Mdivi-1) or mitochondrial fusion promoter M1 versus vehicle and in the isolated mitochondria from skeletal muscle of mDrp1HET mice versus control f/f mice (Fig. 3, E to G).
To support these findings, we examined the expression of transcripts associated with lipid metabolism in control (Scr) and Drp1KD myocytes. Notably, there was reduced expression of fatty acid oxidation-related (Acadl, Acadm, Acox1, Abca1, and Acsl1) and increased expression of lipid transport-related transcripts (Fabp4 and Lpl) in Drp1KD versus control (Scr) myotubes (fig. S3E). Furthermore, reduced adipose triglyceride lipase (Atgl) protein levels and elevated diglyceride acyltransferase (Dgat1) protein levels in both Drp1KD cells and mDrp1HET muscle versus control cells and mice were likely contributors to Drp1KD-associated lipid accumulation (Fig. 3H and fig. S3F). Thus, impaired insulin action was paralleled by diminished fatty acid oxidation and elevated lipid accumulation in Drp1-deficient muscle and myocytes.
Muscle Drp1 deletion impairs complex II assembly and activity
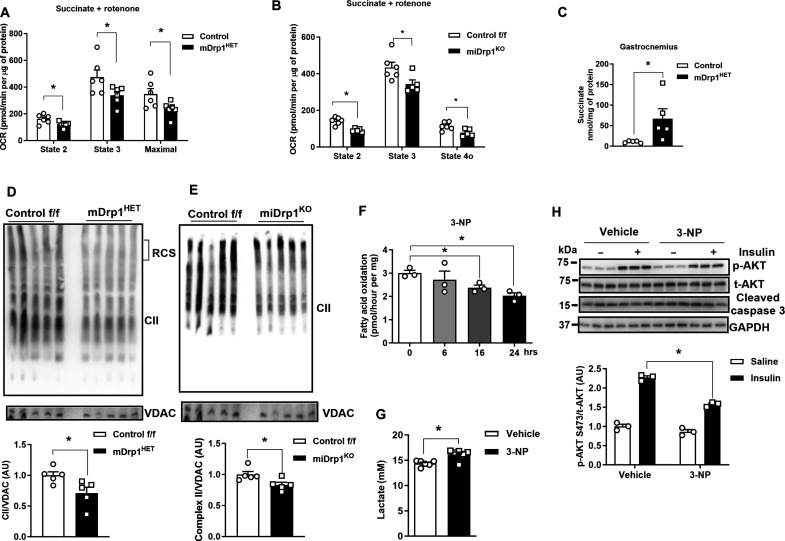
To identify sites of metabolic impairment, we performed an integrated computational assessment of metabolomics and RNA sequencing of muscle from mDrp1HET versus control f/f mice. The citric acid cycle (TCA cycle) and mitochondrial dysfunction emerged as top pathways from gene ontology analyses of differentially expressed transcripts between the genotypes (fig. S4, A to C, and tables S4 and S5). Therefore, we investigated mitochondrial OCR in C2C12 myocytes and found a substantial decrease in OCR of Drp1KD versus control (Scr) myocytes (fig. S4D). Next, we analyzed the activity of mitochondria isolated from skeletal muscle of control f/f and mDrp1HET mice that were incubated with either pyruvate and malate or succinate and rotenone. We observed a marked reduction in OCR (state 2, state3, and maximal) in the mitochondria from mDrp1HET versus control f/f mouse muscles incubated with succinate and rotenone but detected no difference in OCR between the genotypes when mitochondria were incubated with pyruvate and malate (Fig. 4B and fig. S4E). Consistently, a notable reduction of OCR (state 2, state 3, and state 4) was only observed in isolated mitochondria from skeletal muscle of miDrp1KO (acute skeletal muscle–specific Drp1 knockout) versus control f/f mouse muscles incubated with succinate and rotenone (3 weeks after Dnm1l deletion; Fig. 4B and fig. S4F). These findings from two different Drp1-KD mouse models indicate that deletion of Drp1 in skeletal muscle impairs complex II [succinate dehydrogenase (SDH)] activity specifically. Not only complex II participates in the electron transport chain (ETC), but also SDH oxidizes succinate to fumarate in the TCA cycle (32). Notably, elevated succinate was observed in the quadriceps muscles of mDrp1HET versus control mice as well as Drp1-KD Drosophila, which substantiates impairment of complex II activity as a consequence of Drp1 deletion (Fig. 4C).
Fig. 4. Drp1 deletion reduces mitochondrial Complex II assembly and activity in muscle from male mice.
(A) Oxygen consumption rate (OCR) of mitochondria isolated from gastrocnemius muscles of NC-fed control f/f and mDrp1HET mice, and (B) mitochondria from control f/f and miDrp1KO mice treated with the succinate and rotenone (n = 5 per genotype). (C) Succinate levels in skeletal muscle of NC-fed control f/f and mDrp1HET mice (n = 5 per genotype, 5 months of age). (D) Representative Blue Native polyacrylamide gel electrophoresis (BN-PAGE) gels showing complex II assembly (blot for SDHA) of the gastrocnemius muscles of control f/f and mDrp1HET mice, and (E) muscle from control f/f and miDrp1KO mice (n = 5 per genotype). (F) Fatty acid oxidation of C2C12 myotubes with 3-NP administration (n = 3 biological replicates). (G) Lactate levels in the culture medium of differentiated control (Scr) and Drp1KD myotubes treated with 3-NP (100 μM, 24 hours). (H) Immunoblot of p-Akt serine-473, total Akt, and cleaved caspase 3 in C2C12 myotubes incubated with vehicle or 3-NP (complex II inhibitor, 100 μM, 24 hours) with and without insulin (10 nM, 15 min) (n = 3 biological replicates). All values are presented as means ± SEM; *P < 0.05 determined by unpaired Student’s t test, two-tailed.
Complex II is composed of four nuclear-encoded SDH complex subunits (SDHA/B/C/D). The reduction in complex II observed in skeletal muscle of mDrp1HET and Drp1KD cells was not paralleled by a reduction in transcript or protein abundance or by impairment in mitochondrial translocation of SDHA and SDHB subunits (fig. S4, G to J). Although no difference in SDHA and SDHB total protein in muscle lysates was observed between the genotypes, Drp1 knockdown reduced the protein abundance and stability of SDHC and SDHD in mitochondrial fractions from myocytes (fig. S4K).
These findings led us to examine the assembly of complex II in Drp1-deficient cells and muscle. Using Blue Native (BN) polyacrylamide gel electrophoresis (BN-PAGE) gel analysis, we observed decreased assembly of complex II, but not complex I, III, or IV in Drp1KD myocytes, as well as mDrp1HET and conditional miDrp1KO mouse muscles compared with control (Fig. 4, D and E, and fig. S4, L to T). Collectively, these findings suggest that muscle-specific Drp1 deletion impairs complex II activity by disrupting its assembly.
Disruption of complex II impairs insulin action and fatty acid oxidation
The assembly of complex II serves as a pivotal regulator of cellular metabolism (33). To determine whether complex II dysfunction phenocopies an impairment in fatty acid oxidation and insulin action observed in Drp1-deficient myocytes, we used 3-nitropropionic acid (3-NP) to irreversibly and competitively inhibit SDH in C2C12 myotubes (34, 35). As expected, incubation of C2C12 myotubes with 3-NP diminished fatty acid oxidation, elevated lactate levels in the culture medium, and impaired insulin action (Fig. 4, F to H). Consistently, insulin-stimulated Akt phosphorylation at serine-473 was decreased in SDHAKD myocytes (fig. S4U). Thus, our findings support that Drp1 controls fatty acid oxidation and insulin sensitivity in myocytes and skeletal muscle by regulating complex II action.
Drp1 interacts with Sdhaf2, promoting its translocation to mitochondria
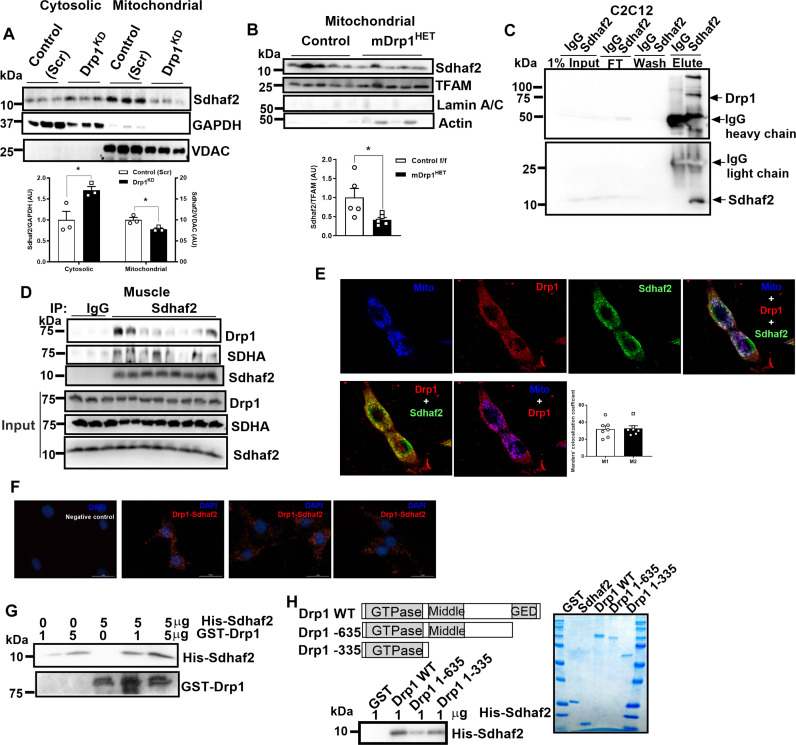
We observed no difference of SDHA and SDHB protein abundance in either muscle lysates or mitochondrial fractions from mDrp1HET versus control f/f mice. To investigate the mechanisms of Drp1-controlled complex II assembly in muscle, we analyzed the mRNA expression and protein levels of SDH complex assembly factors 1, 2, and 4 (Sdhaf1, Sdhaf2, and Sdhaf4, respectively) in the muscle of mDrp1HET versus control f/f mice. While protein and mRNA levels of these assembly factors in muscle homogenates were identical between the genotypes (fig. S5, A and B), a notable reduction in Sdhaf2 protein abundance was observed in mitochondrial fractions from both Drp1KD myotubes and mDrp1HET muscles (Fig. 5, A and B, and fig. S5C). These findings indicate reduced mitochondrial translocation of Sdhaf2 in Drp1-deficient muscle and myocytes in culture.
Fig. 5. Drp1 interacts with Sdhaf2 and enhances its mitochondrial translocation.
(A) Immunoblot of Sdhaf2 in cytosolic and mitochondrial fractions of control (Scr) and Drp1KD myotubes (n = 3 biological replicates). Densitometric quantification analysis of Sdhaf2 protein normalized to GAPDH (cytosolic fraction) or VDAC (mitochondrial fraction). (B) Immunoblot of Sdhaf2 in the mitochondrial fraction of gastrocnemius muscles from control f/f and mDrp1HET mice (n = 5 per genotype). Densitometric quantification of Sdhaf2 protein normalized to the mitochondrial transcription factor A (TFAM). (C) Sdhaf2 coimmunoprecipitated with Drp1 in WT C2C12 myotubes and (D) in skeletal muscle [n = 3 mice in immunoglobulin G (IgG) group, n = 8 mice in Sdhaf2 group]. (E) Confocal microscopy analysis of the colocalization of Sdhaf2, Drp1, and mitochondria in C2C12 myocytes. Manders’ Colocalization Coefficient, M1 (the extent of the fluorescence of colocalizing objects relative to the total Drp1 fluorescence) and M2 (the extent of the fluorescence of colocalizing objects relative to the total Sdhaf2 fluorescence). (F) Proximity labeling assay of Drp1 with Sdhaf2 in WT C2C12 myocytes. The Drp1-Sdhaf2 interaction was shown in red. (G) In vitro binding assay using recombinant full length, and (H) truncated human Drp1 and Sdhaf2. All values are presented as means ± SEM; *P < 0.05 determined by unpaired Student’s t test, two-tailed.
Given the substantial reduction in Drp1 protein levels (~60%) observed in the muscle of Ob/+ versus WT mice, we hypothesized that similar defects in Sdhaf2 mitochondrial translocation and complex II assembly in mitochondria may be observed in skeletal muscle of genetically obese animals. We found that, although Sdhaf2 protein abundance was markedly increased in the total lysate, its abundance was reduced in mitochondrial fractions from muscle of Ob/+ versus WT mice (fig. S5, D and E). Furthermore, two-dimensional BN/SDS gel electrophoresis on muscle mitochondria revealed an additional spot in Ob/+ versus WT mice (arrow), indicative of an irregular complex II architecture (fig. S5, F and G). These observations confirmed that Drp1, Sdhaf2, and complex II assembly are inextricably linked.
Although both mouse and human Sdhaf2 proteins have a mitochondrial localization signal, we hypothesized that Drp1 interacts with Sdhaf2 and enhances its mitochondrial translocation (fig. S5H). As posited, coimmunoprecipitation studies showed that Sdhaf2 associates with Drp1 in myocytes and mouse muscle (Fig. 5, C and D). To provide additional evidence of this protein interaction, we visualized the colocalization of Drp1 and Sdhaf2 in WT C2C12 myoblast cells using laser scanning microscopy with Airyscan detector (Fig. 5E). Imaris analysis detected interaction rates of Drp1-Sdhaf2 colocalization at mitochondria approximating 74% (fig. S5I). The interaction between Drp1 and Sdhaf2 was also confirmed in C2C12 myocytes by Duolink proximity labeling (Fig. 5F). Moreover, in vitro binding assays using recombinant human Drp1 and Sdhaf2 proteins show Drp1 protein physically interacts with Sdhaf2 (Fig. 5G). Using truncated proteins, we determined that the GTPase and C-terminal domains of Drp1 as well as amino acids 41 to 164 of Sdhaf2 are essential for the protein-protein interaction (Fig. 5H and fig. S5J). Together, we demonstrated that Drp1 acts as a protein escort for Sdhaf2 in the regulation of complex II assembly and activity.
Fission incompetency by Drp1 K38A mutation fails to disrupt mitochondrial translocation of Sdhaf2
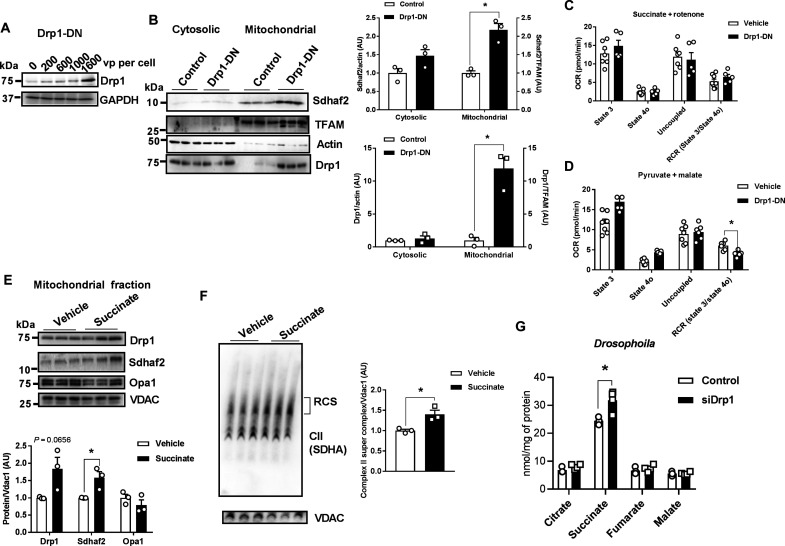
The K38A mutation in Drp1 inhibits GTP binding and prevents mitochondrial fission (12). Counter to our original hypothesis, overexpression of Drp1 K38A [Drp1-dominant negative (Drp-DN)] in C2C12 myotubes increased mitochondrial translocation of Drp1 and Sdhaf2. This led to a reduction of the respiratory control ratio (RCR) of complex I, but no alteration of complex II activity compared with control (Scr) myotubes (Fig. 6, A to D).
Fig. 6. Metabolic impact of Drp1 K38A in C2C12 murine myocytes and Drosophila.
(A) Dose-response of AAV-Drp1-DN on Drp1 protein level in C2C12 myotubes (n = 3 biological replicates). (B) Immunoblots of Sdhaf2 and Drp1 in the cytosolic and mitochondrial fractions of C2C12 myotubes treated with AAV-Drp1-DN [1600 viral particle (vp) per cell]. Densitometric quantification was performed by normalizing Sdhaf2 and Drp1 proteins to TFAM or actin (n = 3 biological replicates). (C) OCR of C2C12 myotubes treated with vehicle or AAV-Drp1-DN includes the following substrates pyruvate and malate or (D) succinate and rotenone (n = 8 biological replicates). (E) Succinate treatment elevates both Drp1 and Sdhaf2 protein in the mitochondrial fraction of C2C12 myotubes (n = 3 biological replicates). (F) Succinate elevates complex II containing respiratory supercomplexes (n = 3). (G) TCA cycle intermediates and related metabolites in control and siDrp1 Drosophila (n = 4 to 5). All values are presented as means ± SEM; *P < 0.05 determined by unpaired Student’s t test two-tailed.
Considering the excess succinate production in Drp1-deficient muscle, we explored the impact of succinate treatment on Sdhaf2 protein and complex II assembly. Succinate treatment increased Drp1 and Sdhaf2 protein abundance in mitochondrial fractions and promoted mitochondrial respiratory chain supercomplex formation (Fig. 6, E and F).
Next, we investigated the impact of Drp1-DN and Drp1 silencing (siDrp1) in Drosophila. Consistent with findings in Drp1KD cells and mouse muscle, we found that siDrp1 significantly increased succinate levels in flies, while succinate in Drp1-DN overexpressed flies was identical to control. Of interest, Drp1-DN drove a unique metabolomic signature including reduction of specific amino acids (glycine, leucine, isoleucine, phenylalanine, and lysine) and increased citrate, fumarate, and malate concentrations (Fig. 6E and fig. S6, A to C). Overexpression of Mfn in Drosophila reduced succinate levels suggesting that impaired mitochondrial fission or enhanced mitochondrial fusion regulates succinate turnover (fig. S6, D and E). These findings indicate that Drp1K38 is dispensable for Sdhaf2-mediated complex II assembly and activity.
Sdhaf2 overexpression elevates fatty acid oxidation and insulin sensitivity
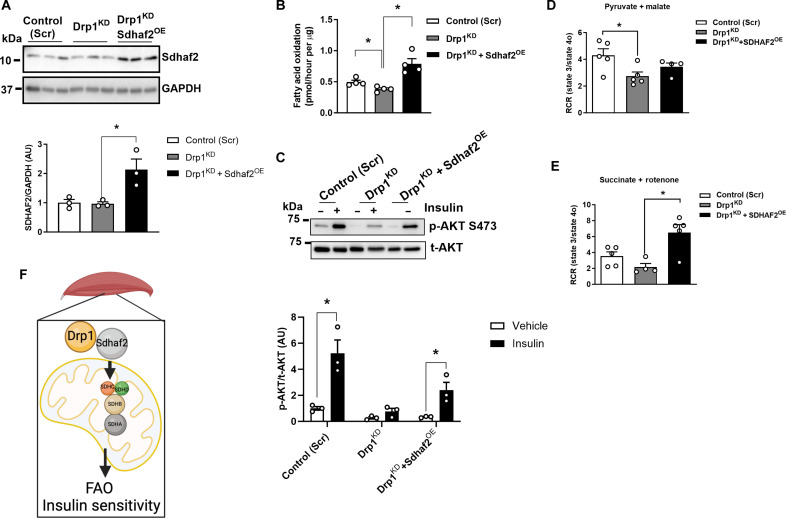
Because reduced mitochondrial translocation of Sdhaf2 impaired complex II assembly and activity in Drp1-deficient muscle, we determined whether overexpression of Sdhaf2 in Drp1KD myocytes could reverse impairment of insulin action and fatty acid oxidation. Using a CRISPR activation plasmid, we successfully overexpressed Sdhaf2 in Drp1KD myocytes (Drp1KD and Sdhaf2OE) (Fig. 7A). Sdhaf2 overexpression robustly elevated the rate of fatty acid oxidation and insulin-stimulated Akt phosphorylation at serine-473 in Drp1KD cells while concurrently reducing lactate levels in the culture medium compared with control (Scr) cells (Fig. 7, B and C, and fig. S7, A to C).
Fig. 7. Sdhaf2 overexpression restores fatty acid oxidation and insulin action in DrpKD myocytes.
(A) Immunoblot of Sdhaf2 in control (Scr), Drp1KD, and Drp1KD and Sdhaf2OE myocytes. Densitometric quantification of Sdhaf2 protein normalized to GAPDH (n = 3 biological replications). (B) Fatty acid oxidation analysis of control (Scr), Drp1KD, and Drp1KD and Sdhaf2OE myocytes (n = 4 biological replicates). (C) Immunoblot of phospho-Akt serine-473 and total-Akt in control (Scr), Drp1KD, and Drp1KD and Sdhaf2OE myotubes with and without insulin (n = 3 biological replications). Respiratory control ratio (RCR) of control (Scr) versus Drp1KD versus Drp1KD and Sdhaf2OE myocytes with (D) pyruvate and malate or (E) succinate and rotenone (n = 4 to 5 per genotype). (F) Schematic overview of Drp1-Sdhaf2 interaction promoting mitochondrial translocation to enhance fatty acid oxidation (FAO) and insulin sensitivity in skeletal muscle. All values are presented as means ± SEM; *P < 0.05 determined by unpaired Student’s t test, two-tailed.
Moreover, we observed that Sdhaf2 overexpression significantly increased the RCR in Drp1KD myocytes, but only in the presence of succinate, not pyruvate (Fig. 7, D and E). This finding indicates an enhanced capacity for succinate oxidation and ATP turnover in Sdhaf2-overexpressing cells. In addition, overexpression of Sdhaf2 markedly decreased mitochondrial aspect ratio, increased mitochondrial circularity, and transformed mitochondrion into donut-shaped organelles in Drp1KD myocytes (fig. S7D). Collectively, we determined that overexpression of Sdhaf2 alters mitochondrial morphology and enhances mitochondrial function and lipid oxidation in Drp1-deficient muscle cells. Together, Drp1 protein is critical for the control of mitochondrial remodeling, which is linked with the assembly of complex II for the complete oxidation of fatty acids and the maintenance of insulin action (Fig. 7F).
Discussion
Over the past decade, the field of mitochondrial biology has experienced a remarkable uptick in research, establishing links between dysregulation of mitochondrial dynamics and human disease pathobiology. Mitochondrial division is a key process required for organelle quality control, mitochondrial DNA replication, calcium and iron homeostasis, and shifts in oxidative metabolism. Drp1 is the central GTPase governing mitochondrial fission, and many variants in Dnm1l have been linked to a spectrum of clinical phenotypes including lethality (15, 36). To date, a variety of mechanisms are shown to impair mitochondrial and peroxisomal dynamics, including Drp1 protein instability, and impaired GTPase activity and oligomerization (16, 37–40). However, the metabolic consequences of specific genetic variants of Drp1 remain inadequately explored.
Our interest in Drp1 and its control of metabolism first arose when we observed a substantial reduction in Drp1 action in two independent mouse models of muscle insulin resistance, heat shock protein 72 knockout and muscle-specific estrogen receptor alpha knockouts (19, 41). In both genetically engineered mouse models, we observed hyperfused mitochondrial architectures, lipid accumulation, and insulin resistance. These phenotypes were recapitulated in the muscle-specific Drp1 deletion model described herein. We identified one unreported DNM1L gene SNP associated with obesity and multiple previously unreported SNPs associated with type 2 diabetes. Our studies have shown that a variety of factors, including aging, physical activity, as well as genetic and diet-induced obesity, affect muscle Drp1 phosphorylation status, Drp1 protein level, and mitochondrial morphology.
To determine the molecular and metabolic consequences of Drp1 deletion that recapitulate human DNM1L variants encoding loss of Drp1 function, we generated a conventional muscle-specific heterozygous Drp1-knockdown mouse model. The critical role of Drp1 in skeletal muscle was reinforced by the failed generation of muscle homozygous null Dnm1l animals. Here, we reported that heterozygous knockdown of Drp1 is sufficient to promote marked disruption of mitochondrial architecture and reduction of EE. These alterations in muscle metabolism promoted adipose tissue weight gain and skeletal muscle insulin resistance. Consistent with the findings in individuals with DNM1L mutations, we detected elevated circulating lactate levels in mDrp1HET mice, indicating a reliance on glycolysis to meet energy demand (29, 42–44). Considering that mitochondrial dynamics govern metabolic programming across cell types and tissues (45–47), elevation of RER in mDrp1HET mice reflects a shift in reliance on carbohydrates as an energy source. Both in vitro and in vivo studies show decreased mitochondrial OCRs and impaired fatty acid oxidation in the context of Drp1 knockdown (48). Reduced AMPK/ACC phosphorylation, diminished Atgl protein abundance, and elevated Dgat1 protein levels in skeletal muscle corroborated impairment in lipid oxidation. These data are internally consistent with lipid accumulation in Drp1-deficient myocytes and muscle. We posit that these defects contribute to impaired insulin action in skeletal muscle (49, 50).
It has been proposed that Drp1-depleted HeLa cells have defects in complex II, but not complex I, of the respiratory chain (51). Consistently, our in vitro and in vivo studies show that Drp1 plays a unique role in regulating mitochondrial complex II function. complex II uniquely links the TCA cycle with oxidative phosphorylation (52). Complex II is the only ETC complex member to lack subunits encoded by the mitochondrial genome and the only respiratory complex that does not pump protons across the inner mitochondrial membrane during its catalytic cycle. Moreover, complex II is also involved in redox control as it is responsible for shuttling electrons from succinate to ubiquinone; thus, impairment in complex II will hamper electron flow to complex III and the quinone pool, thereby promoting oxidative stress. Previous studies have established a negative correlation between complex II activity in visceral adipose tissue and hemoglobin A1c levels in humans (20). Similar findings were observed in mouse myotubes, where complex II inhibition by 3-NP impaired insulin action and fatty acid oxidation, without promoting apoptosis (34, 35). Furthermore, we identified an interaction between Drp1 and Sdhaf2, a protein crucial for the flavination of SDHA and complex II assembly (33, 52, 53). Although Sdhaf2 has a mitochondrial localization signal, its association with Drp1 facilitated its mitochondrial localization for complex II assembly. Loss of Drp1 GTPase activity by K38A point mutation does not diminish but instead increases the mitochondrial translocation of both Drp1 and Sdhaf2. This positive correlation of Drp1 and Sdhaf2 in mitochondrial fractions further supports the notion that Drp1 acts as a chaperone of Sdhaf2. We showed that, when Sdhaf2 expression is experimentally increased in Drp1-deficient myocytes, fatty acid oxidation, succinate oxidation, insulin action, and ATP turnover are restored. Thus, our findings reveal an essential role for Drp1-Sdhaf2 in complex II assembly and activity, which governs lipid metabolism and glucose homeostasis in skeletal muscle.
Complex II catalyzes the oxidation of succinate to fumarate. Succinate levels were increased in Drp1-deficient skeletal muscle as well as Drp1-silenced Drosophila. These findings indicate a link between Drp1 expression and succinate production in the context of altered complex II activity. Excessive succinate in cells, tissues, and plasma is linked with inflammation, insulin resistance, and cardiovascular disease risk (54, 55). Succinylation is a posttranslational modification (PTM) where succinyl moieties (-CO-CH2-CH2-CO2H) are added to lysine residues of proteins or histones. Succinylation reverses protein charge from positive to negative and contributes a sizable mass to the modified protein compared to other PTMs, which presumably contribute, in part, to the alteration in target action. Succinylation of histone 3 at lysine residue 122 (H3K122succ) reduces nucleosome stability, indicating an important role for histone succinylation in modulating chromatin dynamics. Together, our findings provide a framework for how Drp1 action alters mitochondrial dynamics and controls metabolic flux, as well as transcriptional efficacy in the maintenance of cellular health.
Sdhaf2 overexpression in Drp1KD myocytes substantially modified mitochondrial architecture, leading to donut-like shape organelles. Studies in healthy skeletal muscle show that mitochondrial donut holes in glycolytic muscle can be filled with sarcoplasmic reticulum, while holes residing in oxidative fibers were filled with lipid droplets. These observations suggests distinct roles for mitochondrial architectures in different muscle types (56). It is hypothesized that donuts have an advantage over linear structures in tolerating matrix swelling and are protective of membrane curvature (57). The formation of “donuts” from an elongated mitochondrial architecture supports cristae curvature to control the ratio of ETC complexes to ATP synthase and ATP production. This could be resultant of the strong correlation between Drp1 and expression of inner membrane regulators, including IMMT/MIC60 (56). Both complete and transient fusion events driving donut architecture are thought to serve as a protective mechanism to preserve organelle function under conditions of metabolic stress. In addition, it has been posited that circular mitochondria readily detach from microtubule tracks and associate with lipid droplets, especially in oxidative fibers (56). This type of altered morphology and localization likely underpins unique functionalities characteristic of distinct mitochondrial pools within different metabolic cell types.
In conclusion, our research shows that Drp1 interacts with Sdhaf2 and enhances its mitochondrial translocation in skeletal muscle to govern oxidative metabolism. Overexpression of Sdhaf2 in the context of Drp1 deletion normalizes complex II assembly and activity and restores skeletal muscle lipid oxidation and insulin action. Our findings collectively demonstrate the multifunctional actions of Drp1 in governing skeletal muscle metabolism and implicates its functional importance in the context of metabolic-related disease.
MATERIALS AND METHODS
GTEx analysis using GD-CAT
We used the most comprehensive pan-tissue dataset in humans (GTEx), which was filtered for individuals where most metabolic tissues were sequenced (58). Collectively, this dataset contains 310 individuals, consisting of 210 male and 100 female (self-reported) participants between the ages of 20 to 79 years. Gene correlation structure showed strong overlap with known physiologic roles of given endocrine proteins. We have previously shown using the hybrid mouse diversity panel that adopting a gene-centric approach to surveying genetic correlation structure can inform mechanism of coordination between metabolic tissues. All analyses, datasets, and scripts used to generate the associated web tool (GD-CAT) can be accessed via https://github.com/mingqizh/GD-CAT or within the associated docker image. In addition, access to the GD-CAT web tool is also available through the web portal gdcat.org (28). This portal was created to provide a user-friendly interface for accessing and using the GD-CAT tool without the need to download or install any software or packages. Users can simply visit the website, process data, and start using the tool. Corresponding tutorial and the other resources were made available to facilitate the utilization of the web tool on GitHub. The interface and server of the web were built and linked on the basis of the shiny package using R (v. 4.2.0). Shiny package provides a powerful tool for building interactive web applications using R, allowing for fast and flexible development of custom applications with minimal coding required.
Animal
All animal care and experimentation procedures were approved by the University of California, Los Angeles, Institutional Animal Care and Use Committee. Mice were maintained on a 12-hour light/dark cycle from 6 a.m. to 6 p.m. at ambient temperature (~22°C) with controlled humidity (~45%) in pathogen-free conditions. Food consumption, mouse activity, and health were monitored daily by the Division of Laboratory Animal Medicine at UCLA. Before organ harvest, mice were euthanized by isoflurane overdose followed by cervical dislocation. This is an approved method according to the recommendations of the panel on Euthanasia of the American Veterinary Medical Association. The floxed Drp1 mice (a gift from H. Sesaki, Johns Hopkins University) (36) were first backcrossed into the C57BL/6J mice for at least 10 generations. Then, the floxed Drp1 mice were crossed with a muscle-specific Cre transgenic mouse line (MCK-Cre, the Jackson Laboratory, no. 006475) to generate muscle-specific heterozygous Drp1-knockdown mice mDrp1HET, with a tamoxifen-inducible skeletal muscle–specific Cre mouse line (MerCreMer, the Jackson Laboratory, no. 006475) to generate inducible skeletal muscle Drp1 knockout mice (miDrp1KO). miDrp1KO mice develop muscle atrophy starting 6 weeks after gene deletion. Therefore, we studied miDrp1KO mice at 3 weeks after tamoxifen-induced gene deletion before detectable muscle damage. Lean WT and obese mice (heterozygous, Ob/+) were obtained from Jackson Laboratories at 8 weeks of age and studied at 12 weeks of age. Control (flox/+ or flox/flox), mDrp1HET, and miDrp1KO male mice were maintained on a NC diet (LabDiet 5053) or 45% HFD (Research Diets, D12451) and studied after 6 hours fasting with or without insulin stimulation. Additional cohorts of control and genetically engineered mice were used for ex vivo assessment of insulin sensitivity (soleus muscle 2-deoxyglucose uptake assays). Body mass was examined by nuclear magnetic resonance scanning. All mice studied were in C57BL/6J background about 4 to 6 months of age. Blood from 6-hour-fasted male mice was analyzed for circulating factors: Glucose (HemoCue), inflammatory cytokines, and chemokines (Meso Scale Discovery, CA, catalog no. K15048D) were performed on mice after 6 hours of fasting. Intraperitoneal insulin tolerance tests (0.7 U/kg) and intraperitoneal glucose tolerance tests (dextrose, 1 g/kg) were performed on mice after 1 week of recovery between tests.
Indirect calorimetry
Oxygen consumption, carbon dioxide production, food and water consumption, and ambulatory movement were determined in NC-fed male mice using the metabolic chambers (Oxymax metabolic chambers, Columbus Instruments).
Cell culture and treatments
C2C12 cells were maintained in high-glucose Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum and penicillin/streptomycin. To obtain C2C12 myotubes, cells were allowed to reach confluence, and the medium was switched for high-glucose DMEM supplemented with 2% horse serum and penicillin/streptomycin for 5 to 7 days. Drp1KD and Sdhaf2OE myocytes were generated by transfecting Sdhaf2 CRISPR activation plasmid (SC-425801-ACT, Santa Cruz Biotechnology) into Drp1KD cells using Lipofectamine 2000. Transfected myocytes were selected by hygromycin B (300 μg/ml) and blasticidin (3 μg/ml) as described by the manufacturer (Invitrogen). SDHAKD myocytes were generated by transfecting mouse SDHA CRISPR Double Nickase Plasmid (SC-426283, Santa Cruz Biotechnology) into C2C12 cells using Lipofectamine 3000. For insulin treatment, differentiated myotubes were treated with 10 nM insulin (Sigma-Aldrich, I0516) for 15 min before homogenization.
Lentiviral-induced Drp1 KD in C2C12 myocytes
To achieve knockdown of Drp1 (Drp1KD) in myocytes, lentiviral particles (Sigma-Aldrich) carrying short hairpin RNA targeted to Dnm1l/Drp1 were used to transduce C2C12 myoblasts. After selecting positive transformants with puromycin (5 mg/ml), the selected clones were expanded and analyzed for KD efficiency as measured by quantitative reverse transcription polymerase chain reaction (PCR) and immunoblotting. The resulting cultures were then used for subsequent assays in the undifferentiated and differentiated states.
Immunoblot analysis
Immediately snap-frozen mouse tissue samples were homogenized in radioimmunoprecipitation assay lysis buffer containing freshly added protease (cOmplete EDTA-free, Roche) and phosphatase inhibitors (Sigma-Aldrich). All lysates were clarified, centrifuged, and resolved by SDS-PAGE. After transfer, polyvinylidene difluoride membranes were cut in half and subsequently probed with the listed antibodies (table S2) and then imaged separately using a Bio-Rad ChemiDoc XRS imaging system. The exposure time was adjusted, and the densitometric analysis was performed using the BioRad Quantity One or ImageLab software (Bio-Rad). Phospho-protein abundance is normalized to its total protein level as indicated in the graph. The cropped images were organized to form a figure in GraphPad Prism10 software.
Quantitative reverse transcription PCR
Tissues were first homogenized using TRIzol reagent (Invitrogen), and RNA was isolated and further cleaned using RNeasy columns (Qiagen) with DNase digestion. The RNeasy Plus Kit was used for RNA isolation from cells as per the manufacturer’s instructions. Complementary DNA synthesis was performed using 1 mg of RNA with SuperScript II Reverse Transcriptase (Invitrogen). PCRs were prepared using PowerUP SYBR Green Master Mix (Thermo Fisher Scientific). All PCRs were performed using QuantStudio 5 (Invitrogen). Quantification of a given gene, expressed as relative mRNA level compared with control, was calculated after normalization to a standard housekeeping gene (18S or Ppia). We performed separate control experiments to ensure that the efficiencies of target and reference amplification were equal, as described in the User Bulletin 2 from Applied Biosystems. Primer pairs (table S3) were designed using Primer Express 2.0 software (Applied Biosystems) or previously published sequences. Primer sets were selected spanning at least one exon-exon junction when possible and were checked for specificity using BLAST (Basic Local Alignment Search Tool; National Center for Biotechnology Information). The specificity of the PCR amplification was confirmed by melting curve analysis ensuring that a single product with its characteristic melting temperature was obtained.
Ex vivo skeletal muscle glucose uptake
Whole-muscle ex vivo glucose uptake was assessed using 2-deoxyglucose as previously described (19). Briefly, soleus muscles were carefully excised from anesthetized animals and immediately incubated for 30 min in complete Krebs-Henseleit buffer with or without insulin (60 μU/ml) at 35°C. Muscles were then transferred to the same buffer containing [3H]2-deoxyglucose (3 mCi/ml) and [14C] mannitol (0.053 mCi/ml) and incubated for exactly 20 min before being blotted and snap-frozen. Muscles were homogenized in lysis buffer and counted for radioactivity or analyzed by immunoblotting. Glucose uptake was standardized to the nonspecific uptake of mannitol and estimated as micromoles of glucose uptake per gram of tissue.
Coimmunoprecipitation
Coimmunoprecipitation was performed as before (59). Briefly, C2C12 myocytes (~5 × 107 cells) or skeletal muscle lysates (5 mg) were lysed in five packed cell pellet volumes of lysis buffer [10 mM Hepes-KOH (pH 7.9), 0.5% NP-40, 140 mM NaCl, 10 mM KCl, 1.5 mM MgCl2, and protease inhibitors] using a Dounce homogenizer on ice. The lysate (same total protein amount between groups) was centrifuged at 16,000g for 15 min at 4°C, and the supernatant was used for immunoprecipitation. After preincubation of each supernatant with 30 μl of Protein A/G magnetic beads (Thermo Fisher Scientific, 88802) at 4°C for 1 hour, the precleared supernatant was incubated with 3 μg of anti-Sdhaf2 antibody or normal immunoglobulin G at 4°C for overnight with end-to-end rotation. The beads were then washed four times with lysis buffer, and the bound proteins were eluted from the beads by boiling in 2× Laemmli sample buffer with 2-mercaptoethanol (BME). The eluted proteins were analyzed by Western blotting.
In vitro binding assay
Recombinant glutathione S-transferase (GST) tag, GST-Drp1, GST-Drp1 1-635, GST-Drp1 1-335, and His-Sdhaf2 were purified from Escherichia coli and purchased from GenScript Biotech. Briefly, the indicated amounts of recombinant proteins were incubated with 40 μl of 50% slurry of glutathione magnetic agarose beads (Thermo Fisher Scientific, 78602) in the binding buffer [140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, (pH 7.3), 0.1% Triton X-100, and protease inhibitor] at 4°C for overnight. The associated proteins were eluted by the elution buffer [50 mM tris-HCl (pH 8.0) and 10 mM reduced glutathione] after washing with the binding buffer four times. The eluted proteins were separated by SDS-PAGE gel for immunoblotting analysis.
Duolink proximity ligation assay
Duolink proximity ligation assay (PLA) was examined by Duolink In Situ Detection Reagents Red (DUO90228, Sigma-Aldrich). Briefly, cells were seeded into the Nunc Lab-Tek II Chamber Slide System (Thermo Fisher Scientific) and cultured at 37°C overnight in the cell culture incubator. The next day, cells were fixed by 4% paraformaldehyde (PFA) at room temperature for 10 min, permeabilized by digitonin (5 μg/ml), and blocked by Duolink blocking solution in a heated humidity chamber for 60 min at 37°C. Then, each well was incubated with or without primary antibodies followed by Duolink PLA probes in a heated humidity chamber for 60 min at 37°C. Cells were incubated with freshly made ligase and application polymerase for 60 min at 37°C. The signals were captured by BioTek Lionheart LX automated microscope (Agilent Technologies) after washing.
Fatty acid oxidation analysis
Fatty acid oxidation rates were determined in myotubes and isolated mitochondria from skeletal muscle, as adapted from the method previously described (19). Briefly, cells were incubated in low-glucose DMEM containing 1% fatty acid–free bovine serum albumin (BSA) and 0.5 mM palmitate for 1 hour. The medium was then replaced with the addition of [14C]-palmitate (1 mCi/ml) and allowed to incubate for an additional 2 hours. A similar procedure was performed on isolated mitochondria from fresh skeletal muscle (60). Briefly, isolated mitochondria were incubated with oxidation reaction mixture [100 mM sucrose, 10 mM tris-HCl (pH 7.4), 5 mM KH2PO4, 0.2 mM EDTA (pH 8), 80 mM KCl, 1 mM MgCl2, 2 mM l-carnitine, 0.1 mM malate, 0.05 mM CoA, 2 mM ATP, 1 mM dithiothreitol, 0.7% BSA/500 μM palmitate/0.4 μCi 14C-palmitate] at 37°C for 30 min. Complete oxidation of palmitate was measured by benzethonium hydroxide captured 14CO2, liberated from acid-treated culture medium. Calculations were normalized to the specific activity of [14C]-palmitate in the medium.
Isolated mitochondria respirometry
All procedures were performed with prechilled buffers, equipment, and consumables. Isolated mitochondria were resuspended in respiration buffer [100 mM KCl, 10 mM KH2POa, 2 mM MgCl2, 5 mM Hepes, 1 mM EGTA, 0.1% BSA, and 1 mM guanosine diphosphate (pH 7.2)] and kept on ice as described previously (61). Four micrograms of mitochondria per well were loaded into Seahorse XF96 microplate in 20-ml volume containing substrates. The loaded plate was centrifuged at 2000g for 5 min at 4°C, and an additional 115 ml of buffer and substrate was added to each well. Substrate concentrations were as follows: 5 mM pyruvate and 5 mM malate, and 5 mM succinate and 2 mM rotenone. Adenosine 5′-diphosphate was injected at port A (3.5 mM final concentration), oligomycin at port B (3.5 mM), carbonyl cyanide p-trifluoromethoxyphenylhydrazone at port C (4 mM), and antimycin A at port D (4 mM). Mix and measure times were 0.5 and 4 min, respectively. A 2-min wait time was included for oligomycin-resistant respiration measurements.
BN gel electrophoresis
Mitochondrial proteins were solubilized by adding 8 mg of digitonin/mg of mitochondrial protein and incubation on ice for 1 hour. Because commercial digitonin is only 50% pure, we used 16 mg of crude powder to achieve 8 mg (no recrystallization). Digitonin was dissolved in phosphate-buffered saline (PBS) by boiling and stored at 4°C until use. Solubilized samples were centrifuged at maximal speed in a microcentrifuge (Thermo Fisher Scientific) for 30 min at 4°C. Pellets were discarded, and supernatant was combined with 1 ml of 2.5% Coomassie G-250. Samples were loaded into Native PAGE 3 to 12% bis-tris gel and electrophoresed at 4°C in xCell SureLock (Novex) in constant voltage at 150 V for 120 min or until the dye front exited the gel.
Immunofluorescence
C2C12 myocytes were cultured on coverslips and stained with 100 nM MitoTracker Deep Red FM (Invitrogen, CA) for 30 min before fixation with 4% v/v PFA for 15 min at room temperature. After washing in PBS, cells were incubated in permeabilization buffer [0.1% digitonin (Sigma-Aldrich) in PBS (pH 7.4)] for 15 min at room temperature. Cells were then blocked with 3% BSA for 1 hour at room temperature and incubated with 1:100 primary antibody at 4°C overnight. The next day, cells were washed in PBS and incubated with 1:1000 anti-mouse Alexa Fluor 488 or anti-Rabbit Alexa Fluor 546 secondary antibodies for 1 hour at room temperature. After washing in PBS, coverslips were mounted in mounting medium (H-1000, Vector) on a glass slide, air-dried, and stored at 4°C.
Fluorescence microscopy
All imaging was performed using a Zeiss LSM880 confocal microscope. Super-resolution imaging was performed with a 63× apochromatic oil-immersion lens and an AiryScan super-resolution detector. All image analysis was performed using Fiji (ImageJ, National Institutes of Health).
Electron microscopy
Muscles were harvested and immediately immersed in 2% glutaraldehyde in PBS for 2 hours at room temperature and then at 4°C overnight (19). Fixed tissues were washed and postfixed in a solution of 1% OsO4 for 2 hours. After further washes in buffer, tissues were dehydrated through serial immersions in graded ethanol solutions (50 to 100%), passed through propylene oxide, and infiltrated in mixtures of Epon 812 and propylene oxide 1:1, and then 2:1 for 2 hours each and then in pure Epon 812 overnight. Embedding was then performed in pure Epon 812, and curing was done at 60°C for 48 hours. Longitudinal sections of muscle, 60-nm thickness, were cut using an ultramicrotome (RMC MTX). Sections were double-stained in aqueous solutions of 8% uranyl acetate (25 min at 60°C) and lead citrate (3 min at room temperature). Thin sections were subsequently examined with a 100CX JEOL electron microscope.
Metabolomic analysis
Quadriceps femoris muscle samples collected from control f/f and mDrp1HET mice were used for metabolomic analysis (Metabolon Inc., Morrisville, NC).
TAG assay
Triglyceride levels in soleus muscles were examined by a triglyceride quantification assay kit (Abcam, ab65336). Briefly, muscles were washed in cold PBS and homogenized in 5% NP-40 solution using a Dounce homogenizer. The samples were heated to 95°C in a water bath for 5 min twice before centrifuge. Supernatants were diluted 10-fold and incubated with lipase and triglyceride reaction mix for 1 hour. Optical density was measured at 570 nm.
RNA sequencing
Frozen muscle samples were homogenized using a tissue homogenizer in TRIzol (62). RNA was isolated using the Qiagen RNeasy Mini QIAcube Kit following the manufacturer’s instructions. The concentration and purity of isolated RNA were determined using a NanoDrop (Invitrogen) and the Agilent TapeStation separately. Only samples with RNA integrity number (RIN) > 7.0 were used. Libraries were prepared using the kits of TruSeq Stranded Total RNA Library Prep and RiboZero Gold following the manufacturer’s instructions. The resulting libraries were combined into two pools and sequenced on an Illumina Sequencing by Synthesis HiSeq 4000 analyzer within the UCLA Neuroscience Genomics Core facility. Quality of raw reads was examined using FastQC, aligned to the Mus musculus GRCm38, and counted using M. musculus GRCm38 version 97. Alignment and counting occurred using Rsubread v 2.4.2. Raw counts were then analyzed for differential gene expression using the DESeq2 v1.30.0.
Integrated metabolomics and transcriptomics analysis
Integrated metabolomics and transcriptomics analysis was performed using the log2 fold change of mDrp1HET versus control as input into MetaboAnalystR 2.0 R Package as described previously (63).
Statistics
Values presented are expressed as means ± SEM unless otherwise indicated. Statistical analyses were performed using Student’s t test when comparing two groups of samples or two-way analysis of variance (ANOVA) for identification of significance within and between groups using GraphPad Prism 10 (GraphPad Software). Significance was set a priori at P < 0.05.
Acknowledgments
We would like to take this opportunity to thank H. Sesaki for the gift of the floxed Drp1 mice. We would like to thank the University of California, San Diego, Cellular and Molecular Medicine Electron Microscopy Core (UCSD-CMM-EM Core, RRID: SCR_022039) for equipment access and technical assistance.
Funding: This work was supported by the National Institutes of Health, grants P30DK063491 and DK109724 (A.L.H.), DK125354 (Z.Z.), and AG037514 and AG049157 (D.W.W.). The UCSD-CMM-EM Core is supported, in part, by the National Institutes of Health, award number S10OD023527.
Author contributions: Conceptualization: Z.Z., T.M.M., O.S., R.C.-W., A.R.S., A.M.v.d.B., D.W.W., and A.L.H. Methodology: Z.Z., D.M.W., N.Y., P.T., M.S., A.R.S., W.R., K.F., J.W., M.L., L.S., S.M., M.H., T.M.M., D.W.W., and A.L.H. Investigation: Z.Z., A.M., D.M.W., N.Y., P.T., M.S., A.R.S., W.R., J.W., R.A.-P., L.S., S.M., T.M.M., M.O.G., and M.H. Visualization: T.M.M., M.S., Z.Z., A.R.S., M.H., D.M.W., K.F., and A.L.H. Supervision: A.L.H., M.L., C.M.M., A.R.S., D.W.W., and Z.Z. Writing—original draft: Z.Z., T.M.M., A.R.S., J.W., and A.L.H. Writing—review and editing: Z.Z., T.M.M., A.R.S., W.R., M.S., M.O.G., M.H., and A.L.H.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The raw data for tables S4 and S5 are available online with the digital object identifier (DOI): doi:10.5061/dryad.wstqjq2sz (https://datadryad.org/stash/share/ahVIeqoLJSBTJZw9BvGgw96m5iM1NJ9RwLAR-bmLumQ). The RNA sequencing data used are also publicly available at Gene Expression Omnibus through GEO series accession number GSE248081. Floxed Drp1 mice are available by material transfer agreement (MTA) with the expressed consent of H. Sesaki.
Supplementary Materials
This PDF file includes:
Figs. S1 to S7
Tables S1 to S3
Legends for tables S4 and S5
Other Supplementary Material for this manuscript includes the following:
Tables S4 and S5
REFERENCES AND NOTES
- 1.DeFronzo R. A., Tripathy D., Skeletal muscle insulin resistance is the primary defect in type 2 diabetes. Diabetes Care 32, S157–S163 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Herman M. A., Kahn B. B., Glucose transport and sensing in the maintenance of glucose homeostasis and metabolic harmony. J. Clin. Invest. 116, 1767–1775 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.D'Souza D. M., Al-Sajee D., Hawke T. J., Diabetic myopathy: Impact of diabetes mellitus on skeletal muscle progenitor cells. Front. Physiol. 4, 379 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hernandez-Ochoa E. O., Llanos P., Lanner J. T., The underlying mechanisms of diabetic myopathy. J. Diabetes Res. 2017, 7485738 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park S. W., Goodpaster B. H., Lee J. S., Kuller L. H., Boudreau R., de Rekeneire N., Harris T. B., Kritchevsky S., Tylavsky F. A., Nevitt M., Cho Y. W., Newman A. B.; Health, Aging, and Body Composition Study , Excessive loss of skeletal muscle mass in older adults with type 2 diabetes. Diabetes Care 32, 1993–1997 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duchen M. R., Szabadkai G., Roles of mitochondria in human disease. Essays Biochem. 47, 115–137 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Youle R. J., van der Bliek A. M., Mitochondrial fission, fusion, and stress. Science 337, 1062–1065 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sivitz W. I., Yorek M. A., Mitochondrial dysfunction in diabetes: From molecular mechanisms to functional significance and therapeutic opportunities. Antioxid. Redox Signal. 12, 537–577 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Montgomery M. K., Turner N., Mitochondrial dysfunction and insulin resistance: An update. Endocr. Connect. 4, R1–R15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mishra P., Chan D. C., Metabolic regulation of mitochondrial dynamics. J. Cell Biol. 212, 379–387 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wai T., Langer T., Mitochondrial dynamics and metabolic regulation. Trends Endocrinol. Metab. 27, 105–117 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Smirnova E., Griparic L., Shurland D. L., van der Bliek A. M., Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol. Biol. Cell 12, 2245–2256 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tilokani L., Nagashima S., Paupe V., Prudent J., Mitochondrial dynamics: Overview of molecular mechanisms. Essays Biochem. 62, 341–360 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klecker T., Bockler S., Westermann B., Making connections: Interorganelle contacts orchestrate mitochondrial behavior. Trends Cell Biol. 24, 537–545 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Archer S. L., Mitochondrial dynamics—Mitochondrial fission and fusion in human diseases. N. Engl. J. Med. 369, 2236–2251 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Lewis S. C., Uchiyama L. F., Nunnari J., ER-mitochondria contacts couple mtDNA synthesis with mitochondrial division in human cells. Science 353, aaf5549 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Otera H., Ishihara N., Mihara K., New insights into the function and regulation of mitochondrial fission. Biochim. Biophys. Acta 1833, 1256–1268 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Moore T. M., Zhou Z., Cohn W., Norheim F., Lin A. J., Kalajian N., Strumwasser A. R., Cory K., Whitney K., Ho T., Ho T., Lee J. L., Rucker D. H., Shirihai O., van der Bliek A. M., Whitelegge J. P., Seldin M. M., Lusis A. J., Lee S., Drevon C. A., Mahata S. K., Turcotte L. P., Hevener A. L., The impact of exercise on mitochondrial dynamics and the role of Drp1 in exercise performance and training adaptations in skeletal muscle. Mol. Metab. 21, 51–67 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ribas V., Drew B. G., Zhou Z., Phun J., Kalajian N. Y., Soleymani T., Daraei P., Widjaja K., Wanagat J., de Aguiar Vallim T. Q., Fluitt A. H., Bensinger S., Le T., Radu C., Whitelegge J. P., Beaven S. W., Tontonoz P., Lusis A. J., Parks B. W., Vergnes L., Reue K., Singh H., Bopassa J. C., Toro L., Stefani E., Watt M. J., Schenk S., Akerstrom T., Kelly M., Pedersen B. K., Hewitt S. C., Korach K. S., Hevener A. L., Skeletal muscle action of estrogen receptor alpha is critical for the maintenance of mitochondrial function and metabolic homeostasis in females. Sci. Transl. Med. 8, 334ra354 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ngo D. T. M., Sverdlov A. L., Karki S., Macartney-Coxson D., Stubbs R. S., Farb M. G., Carmine B., Hess D. T., Colucci W. S., Gokce N., Oxidative modifications of mitochondrial complex II are associated with insulin resistance of visceral fat in obesity. Am. J. Physiol. Endocrinol. Metab. 316, E168–E177 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liesa M., Shirihai O. S., Mitochondrial dynamics in the regulation of nutrient utilization and energy expenditure. Cell Metab. 17, 491–506 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ngo J., Choi D. W., Stanley I. A., Stiles L., Molina A. J. A., Chen P. H., Lako A., Sung I. C. H., Goswami R., Kim M. Y., Miller N., Baghdasarian S., Kim-Vasquez D., Jones A. E., Roach B., Gutierrez V., Erion K., Divakaruni A. S., Liesa M., Danial N. N., Shirihai O. S., Mitochondrial morphology controls fatty acid utilization by changing CPT1 sensitivity to malonyl-CoA. EMBO J. 42, e111901 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Steffen J., Ngo J., Wang S. P., Williams K., Kramer H. F., Ho G., Rodriguez C., Yekkala K., Amuzie C., Bialecki R., Norquay L., Nawrocki A. R., Erion M., Pocai A., Shirihai O. S., Liesa M., The mitochondrial fission protein Drp1 in liver is required to mitigate NASH and prevents the activation of the mitochondrial ISR. Mol. Metab. 64, 101566 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song M., Franco A., Fleischer J. A., Zhang L., Dorn G. W., Abrogating mitochondrial dynamics in mouse hearts accelerates mitochondrial senescence. Cell Metab. 26, 872–883.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Touvier T., de Palma C., Rigamonti E., Scagliola A., Incerti E., Mazelin L., Thomas J. L., D'Antonio M., Politi L., Schaeffer L., Clementi E., Brunelli S., Muscle-specific Drp1 overexpression impairs skeletal muscle growth via translational attenuation. Cell Death Dis. 6, e1663 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Favaro G., Romanello V., Varanita T., Andrea Desbats M., Morbidoni V., Tezze C., Albiero M., Canato M., Gherardi G., de Stefani D., Mammucari C., Blaauw B., Boncompagni S., Protasi F., Reggiani C., Scorrano L., Salviati L., Sandri M., DRP1-mediated mitochondrial shape controls calcium homeostasis and muscle mass. Nat. Commun. 10, 2576 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang D., Wang J., Bonamy G. M. C., Meeusen S., Brusch R. G., Turk C., Yang P., Schultz P. G., A small molecule promotes mitochondrial fusion in mammalian cells. Angew. Chem. Int. Ed. Engl. 51, 9302–9305 (2012). [DOI] [PubMed] [Google Scholar]
- 28.M. Zhou, I. Tamburini, C. Van, J. Molendijk, C. M. Nguyen, I. Y.-Y. Chang, C. Johnson, L. M. Velez, Y. Cheon, R. Yeo, H. Bae, J. Le, N. Larson, R. Pulido, C. H. V. Nascimento-Filho, C. Jang, I. Marazzi, J. Justice, N. Pannunzio, A. L. Hevener, L. Sparks, E. Kershaw, D. Nicholas, B. L. Parker, S. Masri, M. M. Seldin, Leveraging genetic correlation structure to target discrete signaling mechanisms across metabolic tissues. 12, RP88863 (2023).
- 29.Waterham H. R., Koster J., van Roermund C. W. T., Mooyer P. A. W., Wanders R. J. A., Leonard J. V., A lethal defect of mitochondrial and peroxisomal fission. N. Engl. J. Med. 356, 1736–1741 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Samuel V. T., Petersen K. F., Shulman G. I., Lipid-induced insulin resistance: Unravelling the mechanism. Lancet 375, 2267–2277 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jorgensen S. B., Richter E. A., Wojtaszewski J. F., Role of AMPK in skeletal muscle metabolic regulation and adaptation in relation to exercise. J. Physiol. 574, 17–31 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hederstedt L., Structural biology. Complex II is complex too. Science 299, 671–672 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Bezawork-Geleta A., Wen H., Dong L. F., Yan B., Vider J., Boukalova S., Krobova L., Vanova K., Zobalova R., Sobol M., Hozak P., Novais S. M., Caisova V., Abaffy P., Naraine R., Pang Y., Zaw T., Zhang P., Sindelka R., Kubista M., Zuryn S., Molloy M. P., Berridge M. V., Pacak K., Rohlena J., Park S., Neuzil J., Alternative assembly of respiratory complex II connects energy stress to metabolic checkpoints. Nat. Commun. 9, 2221 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liot G., Bossy B., Lubitz S., Kushnareva Y., Sejbuk N., Bossy-Wetzel E., Complex II inhibition by 3-NP causes mitochondrial fragmentation and neuronal cell death via an NMDA- and ROS-dependent pathway. Cell Death Differ. 16, 899–909 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang L. S., Sun G., Cobessi D., Wang A. C., Shen J. T., Tung E. Y., Anderson V. E., Berry E. A., 3-nitropropionic acid is a suicide inhibitor of mitochondrial respiration that, upon oxidation by complex II, forms a covalent adduct with a catalytic base arginine in the active site of the enzyme. J. Biol. Chem. 281, 5965–5972 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wakabayashi J., Zhang Z., Wakabayashi N., Tamura Y., Fukaya M., Kensler T. W., Iijima M., Sesaki H., The dynamin-related GTPase Drp1 is required for embryonic and brain development in mice. J. Cell Biol. 186, 805–816 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ji W. K., Hatch A. L., Merrill R. A., Strack S., Higgs H. N., Actin filaments target the oligomeric maturation of the dynamin GTPase Drp1 to mitochondrial fission sites. eLife 4, e11553 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Din S., Mason M., Völkers M., Johnson B., Cottage C. T., Wang Z., Joyo A. Y., Quijada P., Erhardt P., Magnuson N. S., Konstandin M. H., Sussman M. A., Pim-1 preserves mitochondrial morphology by inhibiting dynamin-related protein 1 translocation. Proc. Natl. Acad. Sci. U.S.A. 110, 5969–5974 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rehklau K., Hoffmann L., Gurniak C. B., Ott M., Witke W., Scorrano L., Culmsee C., Rust M. B., Cofilin1-dependent actin dynamics control DRP1-mediated mitochondrial fission. Cell Death Dis. 8, e3063 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kalia R., Wang R. Y. R., Yusuf A., Thomas P. V., Agard D. A., Shaw J. M., Frost A., Structural basis of mitochondrial receptor binding and constriction by DRP1. Nature 558, 401–405 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Drew B. G., Ribas V., le J. A., Henstridge D. C., Phun J., Zhou Z., Soleymani T., Daraei P., Sitz D., Vergnes L., Wanagat J., Reue K., Febbraio M. A., Hevener A. L., HSP72 is a mitochondrial stress sensor critical for Parkin action, oxidative metabolism, and insulin sensitivity in skeletal muscle. Diabetes 63, 1488–1505 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu X., Zhang Z., Li D., Lei M., Li Q., Liu X., Zhang P., DNM1L-related mitochondrial fission defects presenting as encephalopathy: A case report and literature review. Front. Pediatr. 9, 626657 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ladds E., Whitney A., Dombi E., Hofer M., Anand G., Harrison V., Fratter C., Carver J., Barbosa I. A., Simpson M., Jayawant S., Poulton J., De novo DNM1L mutation associated with mitochondrial epilepsy syndrome with fever sensitivity. Neurol. Genet. 4, e258 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Whitley B. N., Lam C., Cui H., Haude K., Bai R., Escobar L., Hamilton A., Brady L., Tarnopolsky M. A., Dengle L., Picker J., Lincoln S., Lackner L. L., Glass I. A., Hoppins S., Aberrant Drp1-mediated mitochondrial division presents in humans with variable outcomes. Hum. Mol. Genet. 27, 3710–3719 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kulkarni S. S., Joffraud M., Boutant M., Ratajczak J., Gao A. W., Maclachlan C., Hernandez-Alvarez M. I., Raymond F., Metairon S., Descombes P., Houtkooper R. H., Zorzano A., Cantó C., Mfn1 deficiency in the liver protects against diet-induced insulin resistance and enhances the hypoglycemic effect of metformin. Diabetes 65, 3552–3560 (2016). [DOI] [PubMed] [Google Scholar]
- 46.Buck M. D., O’Sullivan D., Klein Geltink R. I., Curtis J. D., Chang C. H., Sanin D. E., Qiu J., Kretz O., Braas D., van der Windt G. J. W., Chen Q., Huang S. C. C., O’Neill C. M., Edelson B. T., Pearce E. J., Sesaki H., Huber T. B., Rambold A. S., Pearce E. L., Mitochondrial dynamics controls T cell fate through metabolic programming. Cell 166, 63–76 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mahdaviani K., Benador I. Y., Su S., Gharakhanian R. A., Stiles L., Trudeau K. M., Cardamone M., Enríquez-Zarralanga V., Ritou E., Aprahamian T., Oliveira M. F., Corkey B. E., Perissi V., Liesa M., Shirihai O. S., Mfn2 deletion in brown adipose tissue protects from insulin resistance and impairs thermogenesis. EMBO Rep. 18, 1123–1138 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.King W. T., Axelrod C. L., Zunica E. R. M., Noland R. C., Davuluri G., Fujioka H., Tandler B., Pergola K., Hermann G. E., Rogers R. C., López-Domènech S., Dantas W. S., Stadler K., Hoppel C. L., Kirwan J. P., Dynamin-related protein 1 regulates substrate oxidation in skeletal muscle by stabilizing cellular and mitochondrial calcium dynamics. J. Biol. Chem. 297, 101196 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Savage D. B., Petersen K. F., Shulman G. I., Disordered lipid metabolism and the pathogenesis of insulin resistance. Physiol. Rev. 87, 507–520 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kelley D. E., He J., Menshikova E. V., Ritov V. B., Dysfunction of mitochondria in human skeletal muscle in type 2 diabetes. Diabetes 51, 2944–2950 (2002). [DOI] [PubMed] [Google Scholar]
- 51.Estaquier J., Arnoult D., Inhibiting Drp1-mediated mitochondrial fission selectively prevents the release of cytochrome c during apoptosis. Cell Death Differ. 14, 1086–1094 (2007). [DOI] [PubMed] [Google Scholar]
- 52.Bezawork-Geleta A., Rohlena J., Dong L., Pacak K., Neuzil J., Mitochondrial complex II: At the crossroads. Trends Biochem. Sci. 42, 312–325 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rasheed M., Tarjan G., Succinate dehydrogenase complex: An updated review. Arch. Pathol. Lab. Med. 142, 1564–1570 (2018). [DOI] [PubMed] [Google Scholar]
- 54.Osuna-Prieto F. J., Martinez-Tellez B., Ortiz-Alvarez L., di X., Jurado-Fasoli L., Xu H., Ceperuelo-Mallafré V., Núñez-Roa C., Kohler I., Segura-Carretero A., García-Lario J. V., Gil A., Aguilera C. M., Llamas-Elvira J. M., Rensen P. C. N., Vendrell J., Ruiz J. R., Fernández-Veledo S., Elevated plasma succinate levels are linked to higher cardiovascular disease risk factors in young adults. Cardiovasc. Diabetol. 20, 151 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang Y., Tapias V., Acosta D., Xu H., Chen H., Bhawal R., Anderson E. T., Ivanova E., Lin H., Sagdullaev B. T., Chen J., Klein W. L., Viola K. L., Gandy S., Haroutunian V., Beal M. F., Eliezer D., Zhang S., Gibson G. E., Altered succinylation of mitochondrial proteins, APP and tau in Alzheimer’s disease. Nat. Commun. 13, 159 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Glancy B., Kim Y., Katti P., Willingham T. B., The functional impact of mitochondrial structure across subcellular scales. Front. Physiol. 11, 541040 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu X., Hajnoczky G., Altered fusion dynamics underlie unique morphological changes in mitochondria during hypoxia-reoxygenation stress. Cell Death Differ. 18, 1561–1572 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.GTEx consortium , Genetic effects on gene expression across human tissues. Nature 550, 204–213 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou Z., Zhou J., Du Y., Estrogen receptor alpha interacts with mitochondrial protein HADHB and affects beta-oxidation activity. Mol. Cell. Proteomics 11, M111 011056 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huynh F. K., Green M. F., Koves T. R., Hirschey M. D., Measurement of fatty acid oxidation rates in animal tissues and cell lines. Methods Enzymol. 542, 391–405 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Benador I. Y., Veliova M., Mahdaviani K., Petcherski A., Wikstrom J. D., Assali E. A., Acín-Pérez R., Shum M., Oliveira M. F., Cinti S., Sztalryd C., Barshop W. D., Wohlschlegel J. A., Corkey B. E., Liesa M., Shirihai O. S., Mitochondria bound to lipid droplets have unique bioenergetics, composition, and dynamics that support lipid droplet expansion. Cell Metab. 27, 869–885.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moore T. M., Cheng L., Wolf D. M., Ngo J., Segawa M., Zhu X., Strumwasser A. R., Cao Y., Clifford B. L., Ma A., Scumpia P., Shirihai O. S., Vallim T. Q. A., Laakso M., Lusis A. J., Hevener A. L., Zhou Z., Parkin regulates adiposity by coordinating mitophagy with mitochondrial biogenesis in white adipocytes. Nat. Commun. 13, 6661 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chong J., Xia J., MetaboAnalystR: An R package for flexible and reproducible analysis of metabolomics data. Bioinformatics 34, 4313–4314 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S7
Tables S1 to S3
Legends for tables S4 and S5
Tables S4 and S5