Abstract
This review delves into the strategies for early detection and characterization of Naegleria fowleri infections leading to primary amoebic meningoencephalitis (PAM). The study provides an in-depth analysis of current diagnostic approaches, including cerebrospinal fluid analysis, brain tissue examination, immunostaining techniques, and culture methods, elucidating their strengths and limitations. It explores the geographical distribution of N. fowleri, with a focus on regions near the equator, and environmental factors contributing to its prevalence. The review emphasizes the crucial role of early detection in PAM management, discussing the benefits of timely identification in treatment, personalized care, and prevention strategies. Genomic profiling techniques, such as conventional PCR, nested PCR, multiplex PCR, and real-time PCR, are thoroughly examined as essential tools for accurate and prompt diagnosis. Additionally, the study explores advanced microscopic imaging techniques to characterize N. fowleri’s morphology and behavior at different infection stages, enhancing our understanding of its life cycle and pathogenic mechanisms. In conclusion, this review underscores the potential of these strategies to improve our ability to detect, understand, and combat N. fowleri infections, ultimately leading to better patient outcomes and enhanced public health protection.
Keywords: diagnostic biomarkers, early detection, genomic profiling, microscopic imaging, Naegleria fowleri, primary amoebic meningoencephalitis (PAM)
Introduction
Highlights
Early detection is critical for the effective management of primary amoebic meningoencephalitis (PAM).
Various diagnostic methods, including cerebrospinal fluid analysis, tissue examination, immunostaining, and culture techniques, play a crucial role in confirming Naegleria fowleri infections.
N. fowleri is globally distributed, with higher prevalence in equatorial regions due to warmer climates.
Genomic profiling techniques, particularly PCR-based methods, offer high sensitivity and specificity for accurate diagnosis.
Advanced microscopic imaging methods provide insights into N. fowleri’s morphology and behavior during different infection stages.
These strategies hold promise for improving the detection, understanding, and management of N. fowleri infections leading to PAM.
Naegleria fowleri, a pathogenic and opportunistic microorganism, is an aerobic, mitochondrial, eukaryotic protist found worldwide, presenting a potential infection risk to humans and animals1. Belonging to the family Vahlkampfiidae in the Heterolobosea class, N. fowleri is a free-living, thermophilic, and pathogenic flagellate amoeba commonly found in warm freshwater lakes and rivers1,2. During the warmer seasons, N. fowleri flourishes by withstanding temperatures as high as 45°C and predominantly nourishes itself by consuming bacteria found in freshwater ecosystems2. Its adaptable nature allows it to switch between a free-living existence in nature and a parasitic mode within host tissues, which has led to its classification as an amphizoic amoeba3.
Among the 47 recognized Naegleria species, N. fowleri is the sole causative agent of a fatal brain infection known as primary amoebic meningoencephalitis (PAM)4. While PAM is infrequent, it is a highly lethal human ailment, with a mortality rate ranging from 95 to 97%5. The fatality usually occurs within about a week and is characterized by a rapid and destructive form of meningoencephalitis with hemorrhaging5. PAM is observed in individuals with robust immune systems and healthy children and young adults recently exposed to recreational freshwater activities. PAM is primarily transmitted through water; therefore, most cases are linked to recreational activities involving exposure to contaminated water sources5. These activities include swimming, diving in pools with lower chlorine levels, and engaging in water sports like water skiing in polluted canals, spas, and environmental water sources. Another potential infection route is using neti pots for nasal cleansing and ablution6.
Historical perspective of PAM
In 1965, a pivotal moment occurred when Fowler and Carter documented the emergence of PAM. This deadly disease claimed the lives of four individuals at the Adelaide Children’s Hospital in Australia. The culprit behind their demise was identified as an amoeba that infiltrated their meninges, wreaking havoc by causing extensive brain damage and triggering severe inflammation7–9. Since that ominous discovery, PAM has cast its ominous shadow across the globe, with reported cases spanning various countries. While it is said that ~400 individuals have fallen victim to this affliction worldwide, the actual tally remains uncertain. This ambiguity arises from potential misdiagnoses and unreported instances, hinting that the true magnitude of PAM’s reach may surpass current knowledge.
Strategies for improved PAM management
The rapid onset and progression of symptoms in PAM underscore the critical importance of early detection and understanding. Early detection of PAM allows for the timely administration of appropriate medical treatments, enhancing recovery chances and preventing treatment delays due to misdiagnosis. Additionally, early identification and comprehension of PAM support personalized care, potentially leading to better outcomes. Furthermore, it aids researchers in developing more effective treatment and prevention strategies. Early detection prompts public health measures in affected areas to safeguard the community’s well-being. In summary, early detection and improved understanding of PAM can enhance patient outcomes, advance research, and protect a larger population from this disease.
The research is dedicated to deepening our understanding of early detection and characterization of N. fowleri infections, particularly those culminating in PAM. This study encompasses three primary objectives: first, the identification of noninvasive diagnostic biomarkers detectable in samples like cerebrospinal fluid (CSF) or nasal swabs to enable swift diagnosis and timely treatment of N. fowleri infections; second, the comprehensive genomic profiling of diverse N. fowleri strains to pinpoint genetic variations influencing virulence, pathogenicity, and drug resistance, shedding light on the pathogen’s diversity and evolution; and lastly, the application of advanced microscopic imaging techniques to characterize N. fowleri’s morphology and behavior at different infection stages, enhancing our understanding of its life cycle and pathogenic mechanisms. This research holds significant promise for improving our capability to detect, understand, and combat N. fowleri infections leading to PAM.
Epidemiology
Geographical distribution of N. fowleri
N. fowleri’s presence spans every continent except for Antarctica, with a notable prevalence in countries near the equator, where warmer climates prevail10. In temperate nations, like those in Northern Europe, N. fowleri tends to inhabit waters that have been artificially heated, such as those in power station cooling towers or naturally warmed, as seen in geothermal areas. Instances of PAM primarily occur in regions with higher average annual temperatures, typically within the range of 15–18°C. Numerous reports suggest that PAM exhibits a pronounced seasonality, particularly in areas distant from the equator11. Interestingly, the distribution of PAM closely mirrors the distribution of human populations inhabiting warmer regions.
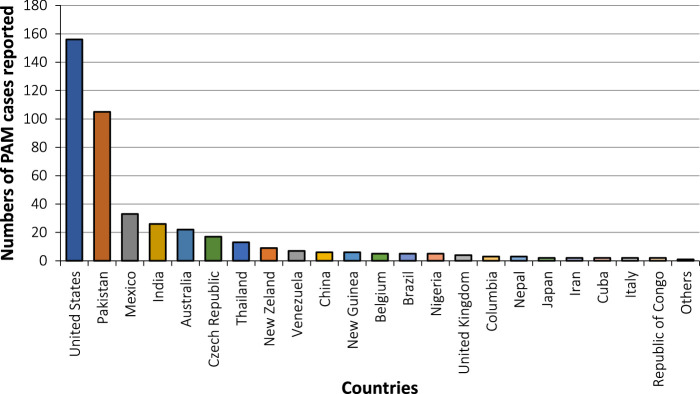
Based on the latest information, N. fowleri infections have been documented in 39 nations (Figs 1 and 2)12. Nevertheless, a select group of countries, including the United States of America (USA), Pakistan, Mexico, Australia, the Czech Republic, and India, have experienced a higher incidence of these infections. One possible explanation for this heightened susceptibility in these regions could be their consistently warm climates throughout the year and accessible contaminated water sources12. Understanding what factors dictate the distribution and prevalence of N. fowleri in the environment. These factors encompass variables such as temperature, salinity, suitable prey organisms, and amoeba pathogens, which may include certain bacteria and viruses. A further, less-explored consideration is competition with other amoebae, especially other Naegleria species13.
Figure 1.
Worldwide documented cases of Naegleria fowleri infections until 2018.
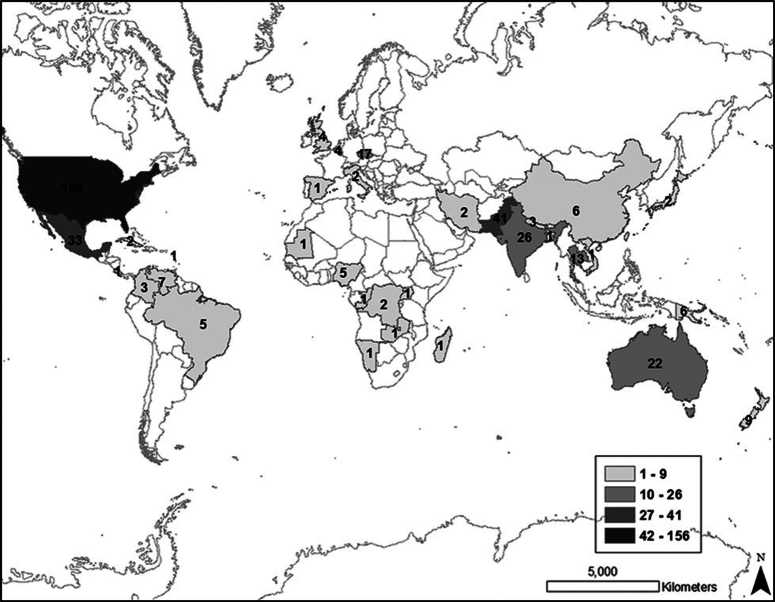
Figure 2.
Reported cases of primary amoebic meningoencephalitis (n=381) by country of exposure.
Prevalence and underreporting in tropical regions
Given the organism’s propensity to thrive and reproduce in warm water, most cases would be concentrated in tropical regions. However, it is noteworthy that while PAM infections may be more prevalent in tropical areas, most documented cases originate from subtropical or even temperate zones. The apparent underreporting of infections in tropical regions likely stems from the comparatively more robust public health services available in temperate zones. In tropical areas, PAM infections might quickly go unnoticed amidst the multitude of other infections affecting millions10.
Furthermore, the challenge of isolating N. fowleri appears to persist when it is present in low quantities in surface water, in contrast to situations where the pathogen is abundant, such as in the cooling waters of industrial facilities. Were it not for the growth of this pathogen in temperate climate zones within industrial cooling waters, this disease might conceivably be relegated exclusively to tropical regions10.
Biology and dissemination of N. fowleri
Life cycle
N. fowleri is a microscopic organism with a complex life cycle consisting of three distinct morphological stages: the trophozoite, the flagellate, and the cyst (Fig. 3)14. Each stage serves specific functions in the amoeba’s survival, reproduction, and environmental adaptation.
Figure 3.
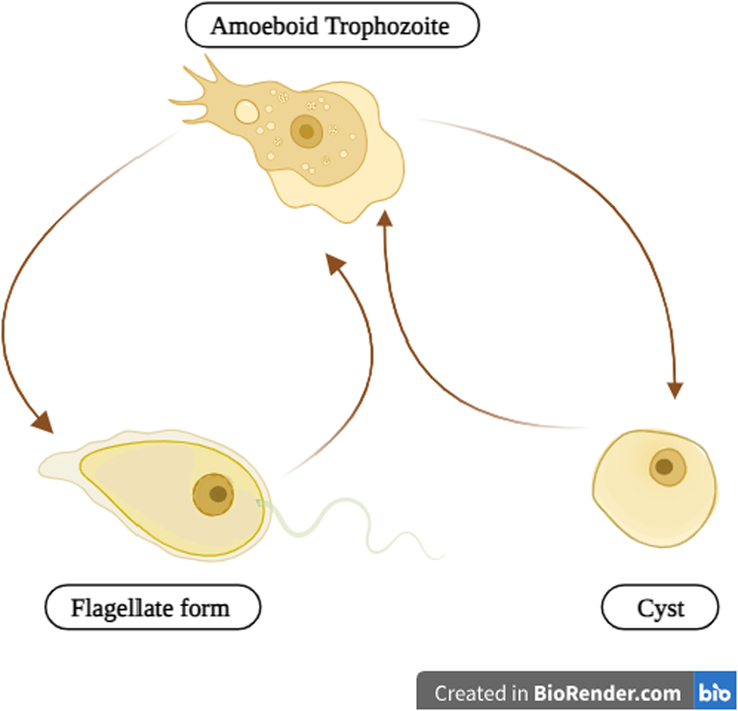
The life cycle of Naegleria fowleri.
The trophozoite stage is the most critical and versatile phase in N. fowleri’s life cycle. This stage is typically the infective phase when environmental conditions are conducive to its survival1,15. Trophozoites are amoeboid in shape and fall within a size range of 10–25 μm in diameter16. These single-celled organisms are uninucleate, meaning they possess a single prominent nucleus within their cell17,18. One of the most remarkable features of trophozoites is their ability to adapt to changing conditions. They exhibit a high degree of plasticity in size and shape, continually undergoing morphological changes. This adaptability allows them to navigate and thrive in diverse aquatic environments, ranging from freshwater bodies like rivers and lakes to poorly maintained swimming pools17,18.
Within this trophozoite stage, N. fowleri reproduces through binary fission, a process in which the nuclear membrane remains intact during cell division, known as ‘promitosis’17,18. This mode of reproduction enables rapid multiplication when conditions are favorable. The optimal temperature range for the growth of trophozoites lies between 35°C and 46°C. Within this temperature range, trophozoites are highly active and can feed on bacteria, which constitutes their primary source of nutrition in free-living conditions. Their amoeboid shape and motility aid in the pursuit and engulfing of bacterial prey. However, when these amoebae find themselves within a host’s tissues, their feeding behavior shifts toward phagocytosis of red and white blood cells. This shift in feeding behavior can lead to tissue destruction, a hallmark of N. fowleri infections18.
Trophozoites can transform into a flagellate form in response to specific environmental cues, such as changes in ionic concentration, exposure to distilled water, or nutrient deprivation19. The flagellate stage is characterized by two flagella at the broader end of the cell. These flagella endow the amoebae with remarkable motility, allowing them to disperse effectively within their surroundings. While in the flagellate stage, N. fowleri can survive from 27°C to 37°C18,19. This adaptability to different temperature ranges facilitates the dispersion of amoebae within both soil and aquatic habitats. It is important to note that the flagellate stage is nonreproductive and nonfeeding, primarily serving as a means of dispersal. This phase allows the amoeba to seek more favorable conditions and potentially locate new sources of nutrients. However, when conditions favorable for the trophozoite stage return, N. fowleri can revert to its amoeboid form; this reversibility showcases the amoeba’s remarkable ability to adjust its morphology in response to its environment, ensuring its continued survival18,19.
When environmental conditions become significantly adverse, trophozoites of N. fowleri may encyst. Encystation is a protective mechanism that occurs under conditions such as food deprivation, overcrowding, desiccation (lack of moisture), the accumulation of waste products, or exposure to cold temperatures below 10°C. The cyst form of N. fowleri is characterized by a thick, double-walled shell that provides protection and durability. The amoeba’s metabolism slows down during encystation, becoming nonreproductive and nonfeeding. This state of dormancy allows N. fowleri to endure adverse environmental conditions until conditions become more favorable20. The cysts of N. fowleri can persist in the environment for extended periods, waiting for the right conditions to return. Once these conditions improve, cysts can undergo excystation, returning to the trophozoite form21.
Each stage plays a unique and crucial role in the amoeba’s survival, reproduction, and adaptation to changing environmental conditions. Trophozoites are infective and actively feeding; flagellates are motile but nonreproductive, and cysts serve as a protective dormant form1. The amoeba’s ability to transition between these stages demonstrates its remarkable adaptability in response to its surroundings. Understanding these stages is essential for recognizing and managing the risks associated with N. fowleri in aquatic environments and its potential to cause severe human infections.
Transmission of N. fowleri
N. fowleri inhabits various environments, including freshwater bodies, soil, thermal effluents from power plants, geothermal wells, and inadequately chlorinated recreational and tap water sources22. The primary route of N. fowleri infection involves infiltrating the nasal mucosa, typically occurring during activities such as swimming in contaminated water or using improperly sterilized equipment for sinus irrigation (Fig. 4). Once within the nasal passages, N. fowleri trophozoites, in their active feeding stage, can traverse the nasal mucosa and gain access to the olfactory nerves22,23. This migration along the olfactory nerves provides the critical pathway for N. fowleri to reach the brain.
Figure 4.
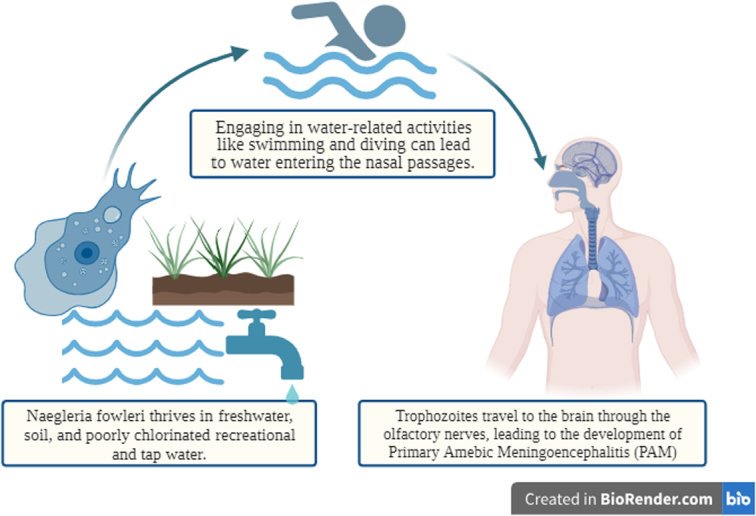
Transmission of Naegleria fowleri.
As N. fowleri trophozoites make their way along the olfactory nerves, they eventually reach the central nervous system (CNS). Although the precise mechanism by which the amebae invade the CNS remains partially understood, it is postulated that the trophozoites utilize the olfactory nerves as a conduit to travel along the cribriform plate and enter the subarachnoid space of the brain, where CSF circulates23. This migration culminates in the onset of PAM, a grave and rapidly progressing brain infection24.
Upon successful brain infiltration, N. fowleri initiates its pathogenic effects, which can have catastrophic consequences. These voracious trophozoites display a strong appetite for host cells and essential nutrients, primarily targeting erythrocytes (red blood cells) and nerve cells within the CNS. The presence of N. fowleri trophozoites within the brain triggers an inflammatory response involving the activation of immune cells and the release of pro-inflammatory molecules. This inflammation contributes to the hallmark symptoms of PAM, including a severe headache, fever, and neck stiffness. It is crucial to emphasize that consuming water contaminated with N. fowleri does not lead to infection, as stomach acid effectively neutralizes N. fowleri trophozoites when ingested23.
A comprehensive and multifaceted approach is imperative to effectively mitigate the transmission of N. fowleri and reduce the risk of PAM. These strategies encompass several key components, each pivotal in minimizing the threat of N. fowleri infection. The government-ensured structures for wastewater surveillance systems can also serve as an early warning system25. Firstly, avoiding contaminated water sources demands proactive measures such as educating the public about associated risks, conducting regular water quality monitoring to identify high-risk areas, implementing advisory systems to communicate potential dangers, and promoting responsible behavioral practices during aquatic activities. Secondly, safeguarding nasal passages is crucial, which involves using protective measures like nose clips or plugs to prevent contaminated water from infiltrating the nasal passages and practicing appropriate diving techniques to minimize the risk of forceful water entry. Additionally, using sterile water for sinus irrigation, achieved through boiling and cooling, is paramount in preventing N. fowleri introduction through this route. Lastly, public health initiatives must encompass awareness campaigns that educate the populace about N. fowleri’s risks, symptoms, and preventive measures, alongside promoting early diagnosis through swift medical evaluation for individuals displaying symptoms indicative of PAM, including severe headache, fever, nausea, vomiting, and a stiff neck20. By diligently adhering to these multifaceted strategies, we can collectively strive to significantly reduce the incidence of N. fowleri transmission and safeguard public health.
PAM
Clinical progression
Upon infiltrating the CNS, N. fowleri initiates a series of devastating events, culminating in a rapidly progressive, hemorrhagic, and necrotizing form of meningoencephalitis referred to as PAM. Symptoms typically emerge within a relatively narrow timeframe, 1–9 days following exposure, with a median onset of around 5 days. However, the insidious nature of this infection lies in its initial presentation, often manifesting as subtle alterations in sensory perception, particularly related to taste and smell. Though potentially unsettling, these early symptoms may not immediately raise concern or prompt recognition of the severe threat they present. As the disease advances, the clinical profile of PAM rapidly parallels that of bacterial meningitis. Patients commonly present with elevated body temperatures, severe frontal headaches, sensitivity to light (photophobia), and signs of meningismus, including neck stiffness. Nausea and vomiting frequently accompany these symptoms, adding to the distress experienced by affected individuals9. At this stage, the striking resemblance to bacterial meningitis can obscure the true nature of the infection, potentially leading to delays in diagnosis and the initiation of treatment.
The full extent of the horrors inflicted by PAM becomes strikingly evident as the disease relentlessly targets the CNS. The onset of cerebral edema, characterized by swelling of brain tissue in response to the amoebic invasion, triggers a dramatic decline in neurological function. Confusion sets in, often accompanied by visual hallucinations that further disorient afflicted individuals. Whether focal or generalized, seizures emerge as a distressing hallmark of the disease, characterized by violent and uncontrollable convulsions that compound the suffering of individuals affected by PAM. Ultimately, as the amoebae continue their assault on brain tissue and edema worsens, patients frequently descend into a coma, marking a grave turning point during the illness. The prognosis for individuals afflicted with PAM is exceedingly bleak. Death typically occurs within a median duration of ~5 days from the emergence of symptoms8,26,27. This swift progression toward a fatal outcome underscores the utmost importance of speedy diagnosis and the immediate commencement of treatment.
Factors contributing to the mortality rate
This disease is sporadic and highly lethal, with a staggering 98% mortality rate (Fig. 5)28. The high fatality rate can be attributed to several factors, primarily stemming from the unique nature of the disease. One key challenge is the remarkably nonsuggestive symptomology during the early stages of the disease. This subtleness, often characterized by symptoms like changes in sensory perception, makes it difficult to promptly recognize the infection’s severity, leading to a delayed diagnosis. Moreover, the diagnostic process itself presents a significant hurdle. Confirming the presence of N. fowleri necessitates microbial culture of the CSF, which can be time-consuming and may not yield timely results29. This diagnostic delay further contributes to the high mortality rate.
Figure 5.
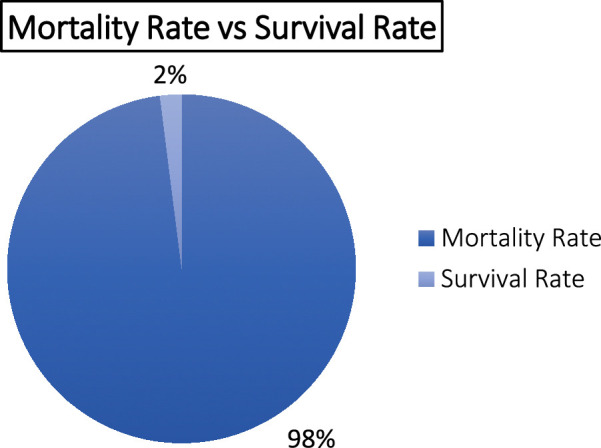
The mortality and survival rate of primary amoebic meningoencephalitis (PAM)
Another formidable aspect of the disease is its rapid late-stage propagation. The parasite demonstrates an alarming ability to traverse the nerves of the olfactory system swiftly, infiltrating multiple regions of the brain simultaneously. This includes the vulnerable medulla, a critical part of the brainstem regulating vital autonomic functions. This rapid dissemination through the nervous system compounds the challenge of treating PAM effectively. For the rare survivors of these infections, complications can vary significantly and depend on the extent of CNS involvement. Respiratory failure is one of the gravest complications, occurring when the infection spreads to the brainstem, where it destroys the autonomic nerve cells of the medulla oblongata29. This devastating consequence underscores the critical importance of early detection and treatment, as preventing the progression of the disease to this stage is paramount for any chance of survival and a positive outcome.
Current diagnostic approaches
Laboratory diagnosis
CSF analysis
Diagnosing N. fowleri infection requires meticulously examining CSF samples. Working with fresh, unrefrigerated CSF specimens is crucial to ensure accurate results. A valuable tool for visualizing actively moving N. fowleri trophozoites is a wet mount prepared from freshly centrifuged CSF sediment17. These trophozoites, typically measuring between 15 and 30 µm, exhibit rapid and sinuous movement characterized by the utilization of eruptive pseudopods. Various staining methods, including Hematoxylin and Eosin (H&E), Periodic Acid-Schiff (PAS), Trichrome, Giemsa, or Wright–Giemsa stains, can effectively identify Naegleria within CSF smears or cultures. It is essential to avoid using Gram staining as it may potentially damage the amebae during heat fixation. Stained CSF smears reveal trophozoites with the characteristic Naegleria morphology, including a nucleus featuring a large, centrally located, and densely staining nucleolus. Following the identification of amebae in the CSF, the diagnosis of PAM is confirmed through polymerase chain reaction (PCR), ensuring diagnostic accuracy and facilitating prompt medical intervention30.
Patients with PAM often exhibit leukocytosis with a left shift, signifying an elevated white blood cell count in the bloodstream, with a notable presence of immature neutrophils indicative of an ongoing infection. In addition, PAM is characterized by a markedly high CSF opening pressure upon initial examination, reflecting increased intracranial pressure (ICP). The CSF analysis typically reveals a significantly elevated white cell count ranging from 300 to 26 000 cells per millimeter, with a prevalence of polymorphonuclear cells, indicating a robust inflammatory response within the CNS. Furthermore, the presence of red blood cells in the CSF is familiar, and as the disease progresses, the CSF may become hemorrhagic. Another distinctive feature is hypoglycorrhachia, signifying reduced glucose levels in the CSF, characteristic of PAM. Lastly, elevated protein levels in the CSF serve as an additional marker, reflecting the ongoing immune response and inflammation within the CNS17.
Brain tissue
N. fowleri diagnosis can also be achieved through a comprehensive examination of brain biopsy or autopsy specimens. Microscopic examination of these specimens, stained with H&E, PAS, trichrome, Giemsa, or Wright–Giemsa, can reveal trophozoites displaying characteristic N. fowleri morphology. These amoeboid trophozoites exhibit a size range of 10–35 µm, with a more common diameter of 10–15 µm when rounded. Their cytoplasm appears granular and contains numerous vacuoles, while the single nucleus is notably large, featuring a large and densely staining karyosome. It is worth noting that N. fowleri does not form cysts within human tissues30.
Immunostaining techniques
Immunohistochemical (IHC) staining and indirect immunofluorescent (IIF) staining are advanced laboratory techniques employed to detect the presence of N. fowleri in formalin-fixed tissue or CSF30. These methods rely on specific antibodies that selectively target and bind to N. fowleri antigens, enabling their identification through microscopic examination.
In the IHC staining process, tissue sections or CSF samples are first prepared and preserved in formalin to fix the biological material. Specific antibodies, designed to recognize and bind to N. fowleri antigens, are then applied to the samples. These antibodies form a particular complex with the target antigens if N. fowleri is present within the tissue or CSF. Once the antibody–antigen complexes are formed, they can be visualized under a microscope30. The presence of these complexes within the tissue or CSF sample indicates the presence of N. fowleri. IHC staining offers a precise and reliable method for confirming the infection within formalin-fixed specimens.
Similarly, IIF staining utilizes specific antibodies for N. fowleri. In this technique, the antibodies are labeled with fluorescent markers, such as fluorophores, that emit distinctive fluorescence when exposed to specific wavelengths of light. The labeled antibodies are applied to the tissue or CSF samples, and if N. fowleri is present, the antibodies will bind to the antigens on the pathogen’s surface30.
Under fluorescent microscopy, fluorescence within the samples indicates the binding of the labeled antibodies to N. fowleri. This method provides a visual and highly sensitive means of detecting the pathogen within the specimen30. IHC and IIF staining techniques enhance the diagnostic capabilities for N. fowleri infection, especially when dealing with formalin-fixed tissues or CSF samples. Their high specificity and ability to target specific antigens make them invaluable tools in confirming the presence of this pathogenic amoeba.
Serology
Serologic testing for N. fowleri, which involves using indirect immunofluorescent antibody (IFA) assays to measure patient serum antibody titers, has limited diagnostic value. This limitation stems from the rapid and aggressive nature of PAM, with most patients succumbing to the disease before their immune systems can generate significant antibodies. Consequently, serologic testing is often impractical for timely diagnosis, and alternative diagnostic methods, such as direct microscopic examination or molecular assays, are generally more reliable in clinical practice30.
Culture
The culture procedure is a method employed to detect free-living amoebae in clinical and environmental samples. This process involves introducing the sample into mammalian cell cultures and monitoring them for signs of cytopathogenicity or growth on Escherichia coli bacterial lawns. When growing on an E. coli lawn, the sample is placed on a growth plate covered with bacteria that can serve as a food source for N. fowleri. The initial screening step is carried out by incubating the plate at a higher temperature (108°F/42°C), which is lethal to most free-living amoebae but favors the growth of thermophilic amoebae like N. fowleri. During this initial screening, tracks created by an amoeba as it moves across the plate while consuming bacteria become visible. If no amoebae are observed on the plate incubated at a higher temperature, it indicates the absence of N. fowleri. However, suppose thermophilic amoebae are incubated at a higher temperature on the plate. In that case, further specific testing is required to confirm the presence of N. fowleri, as other thermophilic free-living amoebae might also be present30.
Naegleria can be definitively identified in cultures from clinical specimens using various staining methods such as H&E, PAS, trichrome, Giemsa, or Wright–Giemsa stains. Stained cultures may reveal trophozoites with morphological characteristics typical of N. fowleri, as previously described. In culture, these trophozoites may measure more than 40 µm. It is important to note that a negative culture result does not conclusively rule out the presence of free-living amoebae, and additional testing should be considered30.
Imaging techniques
Computed tomography (CT) scanning and magnetic resonance imaging (MRI)
In cases where clinical signs suggest focal CNS involvement or elevated ICP, it is advisable to perform head CT scanning or MRI before conducting a lumbar puncture. In PAM, these imaging studies may reveal nonspecific findings, such as the obliteration of the cisterns around the midbrain and subarachnoid space31.
Limitations
CT and MRI scans are typically utilized to visualize structural anomalies within the brain, such as tumors, vascular problems, or injuries. They are not intended to detect microscopic pathogens like N. fowleri directly. In advanced PAM, where neurological complications have developed, these imaging modalities can be supplementary diagnostic tools to assess brain damage and guide treatment decisions. However, it is crucial to emphasize that CT and MRI scans are not the primary methods for diagnosing N. fowleri infections.
Genomic profiling techniques for early detection
The standard procedure for diagnosing PAM in a laboratory typically involves culturing1,32 and confirming the presence of N. fowleri in CSF32. In addition to this, a flagellation test (FT) is commonly employed as a supplementary diagnostic tool for detecting N. fowleri. However, in practical application, FT often requires further verification using alternative diagnostic methods such as enzyme-linked immunosorbent assay (ELISA) or PCR due to the potential for false-positive and false-negative outcomes1,33,34. ELISA-based diagnostic procedures, while widely utilized, often yield retrospective and post-mortem diagnoses, which may not be timely for effective treatment. Conversely, RFLP (restriction fragment length polymorphism) is a diagnostic technique specific to species within the Naegleria genus based on distinct restriction patterns35–37. A DNA probe-based detection method, introduced in the mid-1990s, provides an alternative to these traditional approaches38. Nevertheless, these methods possess limitations, including time-intensive culturing, expensive RFLP, delayed diagnosis through ELISA, or insufficient outcomes from the FT.
PCR as a molecular diagnostic tool for N. fowleri
PCR diagnostic methods offer significant advantages by overcoming many of these issues. They are valuable for diagnosing both clinical and environmental samples. PCR allows DNA isolation from samples without prior cultivation, streamlining the diagnostic process. Moreover, PCR not only detects N. fowleri but also facilitates the differentiation of other Naegleria species within the genus. Specific PCR tests for N. fowleri are highly recommended to confirm suspected infectious agents in clinical samples. They offer improved sensitivity, quicker results, and the ability to identify various Naegleria species, making them a valuable tool in diagnosing PAM and related conditions39.
Presently, the laboratory detection of PAM using PCR encompasses conventional PCR techniques40–43, nested PCR approaches39,44,45, multiplex PCR procedures46, and real-time PCR techniques47–52.
Conventional PCR
In conventional PCR, a genetic analysis of various Naegleria species was conducted using PCR primers designed for telomeric and simple repeat sequences. This study demonstrated the ability to identify Naegleria isolates at species and subspecies levels with a single PCR assay, resulting in characteristic DNA banding patterns, often called fingerprints41. These fingerprints were generated using oligonucleotide primers specific for variable eukaryotic DNA regions. These repetitive sequences, consisting of short GC (guanine–cytosine)-rich units repeated in tandem, ranged from 0.1 to 20 kbp in length53. Notably, these identical sequences were employed in genetic analyses of humans for purposes such as paternity disputes and forensics53,54. In the case of the Naegleria subspecies, distinct fingerprints allowed discrimination based on specific genetic characteristics. This described a way to simplify complex genome typing using short oligonucleotides, carefully chosen to reduce the complexity of the generated fingerprints. This approach extends beyond Naegleria species and can be used to discriminate pathogenic amoebae, such as N. fowleri, from nonpathogenic ones. While some isolates showed limited variability based on geographic origin, the method remained highly specific. Furthermore, it was noted that the inter-repeat PCR banding pattern did not correlate with the degree of pathogenicity in isolates. This versatile DNA typing approach applies to other lower eukaryotic organisms and has potential applications in epidemiological studies and the isolation of species-specific DNA probes41.
Nested PCR
In a significant 2002 study, researchers introduced a complementary DNA (cDNA) clone named Mp2Cl5 isolated from N. fowleri. This clone had a remarkable ability – it could recognize DNA from the pathogenic N. fowleri and a closely related nonpathogenic species known as Naegleria lovaniensis. To make Mp2Cl5 more versatile and practical for their research objectives, they employed the XbaI restriction enzyme to cleave it into two distinct fragments, creatively named Mp2Cl5.G and Mp2Cl5.P. These researchers then conducted experiments, evaluating these fragments against DNA samples from various Naegleria species and even another free-living amoeba called Acanthamoeba. Astonishingly, the results unveiled that one of these fragments, Mp2Cl5.P, exhibited an extraordinary level of specificity for N. fowleri. Empowered by the cDNA probe Mp2Cl5.P sequence, they strategically designed primers to develop PCR assays. These specialized assays were meticulously crafted to distinguish N. fowleri from other free-living amoebae. While the initial standard PCR assay proved to be highly specific, selectively amplifying N. fowleri DNA, it fell short in sensitivity, only capable of detecting DNA quantities equal to or exceeding 0.5 ng. In response to this sensitivity limitation, they proposed an innovative solution – the nested PCR assay. This novel approach not only maintained a high level of specificity but also achieved exceptional sensitivity. Remarkably, the nested PCR assay could pinpoint the presence of N. fowleri DNA in quantities as minuscule as 5 pg. Furthermore, both the standard PCR and nested PCR assays developed in this study demonstrated their efficacy in detecting pathogenic N. fowleri strains from diverse geographical regions, including the United States and Australia45.
A study examined the nested PCR assay’s feasibility to detect pathogenic N. fowleri in water samples. Remarkably, they discovered that a suspension of intact N. fowleri amoebae in water could serve as a source of genomic DNA (gDNA), eliminating the need for time-consuming and meticulous gDNA extraction and purification. This streamlined approach, complemented by a brief incubation step during the initial PCR, endowed the technique with exceptional sensitivity, allowing the detection of as few as five N. fowleri amoebae. Furthermore, this breakthrough eliminated the need for cumbersome gDNA extraction and purification steps, enhancing the assay’s efficiency. Notably, it paved the way for a rapid DNA extraction-free, filter-based method that demonstrated its effectiveness in detecting protozoa in various contexts, whether from food or clinical specimens, through nested PCR40.
By employing this nested PCR assay, even minute numbers of N. fowleri amoebae in contaminated water sources could be promptly identified. Notably, the assay proved resilient against potential river and lake water interference, although it could not discern between live and deceased amoebae. Nonetheless, it stood as a compassionate qualitative tool for detecting the presence of N. fowleri at levels as low as five organisms. This swift and cost-effective testing approach opens the door to assessing a significant number of samples to ascertain whether N. fowleri is present. Positive samples can be analyzed through more labor-intensive quantitative counting procedures41,42.
Furthermore, the clinical potential of this test is promising, with the potential for early detection of pathogenic N. fowleri in CSF or nasal swabs. Such a clinical application could be pivotal in preventing the misdiagnosis of PAM as bacterial meningitis, which, if left unaddressed, can lead to fatal outcomes45.
Multiplex PCR
A multiplex PCR assay was meticulously developed to enable the simultaneous detection of N. fowleri and other Naegleria species present in the environment. This innovative technique also distinguished between N. fowleri isolates with different internal transcribed spacer (ITS) lengths46. When the conserved and species-specific primers were combined, the expected outcome was amplifying four distinct fragments for N. fowleri. Notably, the higher and lower fragments corresponded to the extension of the conserved and specific primers, respectively. In contrast, the two intermediate fragments arose from the interaction between the clear and conserved primers. Interestingly, when equal concentrations of conserved and species-specific primers were employed, the most minor stained bands on the gel often exhibited lower intensity than the others. To achieve uniform amplification product intensities, it was necessary to use higher concentrations of species-specific primers compared to nonspecific primers46.
This multiplex PCR approach could discern various variants of N. fowleri characterized by ITS1 length polymorphism. For instance, the short-ITS variant yielded all four expected bands, featuring a 42-bp ITS1. Conversely, the long-ITS variant, boasting an ITS1 length of 86 bp, produced only three bands due to the comigration of the two intermediate fragments measuring 388 and 376 bp in the gel. Significantly, fungal contaminants alongside N. fowleri did not compromise the test’s species specificity. Furthermore, to rigorously assess the specificity of the N. fowleri PCR, DNA from Acanthamoeba, Hartmannella, Nuclearia, Vahlkampfia, and Willaertia isolates was scrutinized. These isolates were frequently encountered as contaminants in culture plates during monitoring efforts. Encouragingly, no species-specific PCR product was observed for any of the tested strains. When employing either the conserved or the multiple primers, a single fragment was discernible with Hartmannella, Vahlkampfia, and Willaertia isolates, each featuring a distinct fragment size. This underlines the multiplex PCR’s capability to identify the prevalent competitors coexisting with thermotolerant Naegleria species46. This breakthrough enhances our ability to detect Naegleria species accurately. It provides valuable insights into the broader spectrum of amoebae present in the sample sites, some of which are known to harbor pathogenic bacteria55–57.
Real-time PCR (qPCR)
Real-time PCR-based diagnostic techniques offer significant advantages, primarily in speed and the ability to monitor the amplification process as it happens50. This quantitative real-time PCR (qPCR) has consistently demonstrated superior reliability, robustness, sensitivity, and speed compared to traditional endpoint PCR58. qPCR has the remarkable capacity to detect and quantify suspected pathogens, including microorganisms that may evade detection or easy identification through conventional cultivation methods efficiently and precisely. This rapid and highly accurate molecular method accelerates result availability, enabling swift action to address potential contamination issues59.
In the research conducted by Behets et al.47, their methodology involved primers and a probe designed based on the Mp2Cl5 gene, utilizing a single FAM-labeled probe. A previous study examined the reactivity of Mp2Cl5 with four species from the Naegleria genus and four species from the Acanthamoeba genus, conclusively demonstrating specificity exclusively for N. fowleri 45. In the real-time PCR-based diagnostic approach, multiplex PCR is employed, with the detection of N. fowleri facilitated by a HEX (hexachlorofluorescein)-labeled probe48.
Furthermore, Robinson et al.49 in 2006 detailed a real-time PCR procedure for N. fowleri detection, utilizing primers designed for noncoding spacers ITS1 and ITS2, complemented by the intercalating dye SYT09. Another study harnessed a LightCycler instrument for amplification, with primers tailored for detecting the ribosomal small-unit SSU (18s) rRNA gene and employing SYBR Green 1 labeling60.
In essence, this real-time PCR assay serves as a compassionate surveillance tool, enabling the rapid detection of an organism that poses a significant risk to human health.
Different approaches for gene silencing and editing in N. fowleri
RNA interference (RNAi)
In the pursuit of gene manipulation within N. fowleri, RNAi technology was explored. Initial efforts focused on utilizing laboratory-synthesized double-stranded RNA (dsRNA) molecules directed toward specific genes deemed pivotal for the amoeba’s proliferation. These genes encompassed NfEF1a, NfTUB, and NfENO, alongside control genes such as eYFP and NfCPI. Introduction of the dsRNA into N. fowleri was accomplished through a transfection method, followed by an evaluation of cell viability using microscopy and growth assays. Remarkably, no substantial disparities in growth rates emerged between the targeted genes and control genes, signifying an absence of impact on amoeba growth stemming from our gene silencing endeavors.
Additionally, experimentation involving dsRNA generated by bacteria (E. coli HT115) was undertaken, wherein dsRNA production was induced and then exposed to the amoebae. Once again, no discernible variances in cell growth or gene expression were observed when comparing experimental targets to control groups. These findings collectively indicate that the RNAi approaches did not yield the intended impact on gene function61,62.
CRISPR-Cas9 gene editing
Inspired by gene-editing techniques employed in previous studies, CRISPR-Cas9 technology was applied to disrupt gene expression in N. fowleri 63. The recombinant Staphylococcus aureus Cas9 protein was paired with a specific guide RNA (sgRNA) directed toward a predicted gene, forming a ribonucleoprotein (RNP) complex. This RNP complex was introduced into N. fowleri trophozoites alongside a repair template containing eYFP64, utilizing the Amaxa nucleofection technique, which had proven effective in related organisms. gDNA was extracted 2 days later, and PCR analysis indicated successful gene editing had occurred, with enhanced yellow fluorescent protein (eYFP) integration into the target gene. Nevertheless, the relatively low abundance of PCR products suggested our gene-editing efficiency was limited, and the modified cells did not exhibit detectable fluorescence. After 6 days of culture, eYFP became undetectable, likely due to prolonged culturing resulting in the loss of the limited number of modified cells62.
Potential benefits for the field
Developing genetic tools for N. fowleri represents a significant advancement, offering the potential to manipulate gene expression in these amoebae. RNAi and CRISPR/Cas9 technology could provide valuable insights into gene function. RNAi may enable wide-scale genetic screening, allowing us to explore specific phenotypes across the entire genome. Additionally, using expression plasmids for transgenesis, including fluorescent reporter genes, will facilitate studies related to protein localization and function within these amoebae62.
The need for noninvasive diagnostic biomarkers
Rationale for noninvasive diagnostic methods
N. fowleri is a deadly infection with a mortality rate exceeding 95% despite advancements in antimicrobial treatment and supportive care. Initially, symptoms of PAM are indistinguishable from bacterial meningitis. Therefore, a swift diagnosis can potentially reduce the overall death rate65. Identifying the specific type of free-living amoeba involved is equally essential to characterize and identify the potentially distinctive disease progression, risk factors, organ distribution, route of infection, and disease severity. Though slight morphologic differences among amoebae are visible on routine histologic staining, these distinctions can be subtle, especially in cases with low infection intensity. Identifying cysts or multinucleated trophozoites can be challenging and may require advanced magnification and visualization techniques to confirm the pathogen66. Obtaining a thorough patient history that considers freshwater exposure can enhance the chances of arriving at a likely diagnosis. Regardless of transmissibility, accurately measuring the infection burden remains challenging due to the need for more conclusive diagnostic methods, leading to underreporting cases. The initial vague symptoms of PAM, such as fever and headache, which mimic bacterial meningitis, often lead to delayed or incorrect diagnoses, compounded by the rarity of the infection and limitations in diagnostic tools. For instance, observing N. fowleri directly in a sample can be influenced by the operator’s skill and may be less effective with untrained personnel. Additionally, PCR techniques for analysis may be limited in specific centers, particularly in rural and district hospitals4. Hence, the key to diagnosis rests on clinical suspicion, and a definitive gold standard test for diagnosing N. fowleri has yet to be established.
Advantages and potential of biomarkers in early detection
Efforts to achieve a prompt diagnosis and implement an aggressive treatment approach, ultimately improving the patient’s prognosis, are pivotal areas where advancements can significantly reduce mortality rates67,68. An examination of CSF after the onset of neurological symptoms enhances the chances of making an accurate diagnosis. The utilization of neuroimaging techniques, such as MRI and CT, aids in better visualization of affected regions of the brain4. CSF sample results may help to rule out the viral causes of CNS involvement. Low CSF glucose levels and raised levels of CSF proteins eliminate viral causes. When the laboratory has the necessary skills, it is possible to directly examine the CSF sample using light microscopy to identify motile amoebae. This is important because host cells like leukocytes and macrophages can resemble amoeba trophozoites, which could result in incorrect interpretations if not carefully examined. Therefore, diagnosing PAM is more reliably achieved using molecular techniques, as advised by the US CDC (United States Centers for Disease Control and Prevention), on a CSF sample or brain tissue containing N. fowleri. In a case report by Huang et al., N. fowleri was also detected in the bloodstream, albeit with lower sequence copy numbers in CSF. This finding suggests that N. fowleri could spread beyond the CNS, in line with a study of autopsies conducted by the CDC in the United States from 2009 to 2012. Besides the CNS, N. fowleri was found in the lungs, kidneys, liver, spleen, and other organs in four cases reported in the literature. This leads us to speculate that N. fowleri may enter the bloodstream through a compromised blood–brain barrier and subsequently reach other tissues and organs via the bloodstream; therefore, potential biomarkers could be present, though this requires validation68.
Overview of various potential biomarkers
Indirect immunofluorescence from tissue has been widely used to distinguish between Balamuthia mandr illaris, N. fowleri, and Acanthamoeba spp. in various species, and our review of published cases found that it was the most employed method66. The debate surrounding whether an exudate obtained from the olfactory region, either through modified transcritical devices or conventional methods, could be an alternative to CSF for getting amoeba in PAM for a definitive real-time PCR-based diagnosis requires further validation67. Matrix-assisted laser desorption ionization–time-of-flight mass spectrometry (MALDI-TOF MS), a practical and rapidly advancing application of mass spectrometry for microorganism identification and strain differentiation, has gained attention in clinical microbiology as a fast and effective method. It provides characteristic protein patterns (unique biomarker fingerprints) from entire organisms, facilitating the identification of bacteria, viruses, protozoa, and fungi. This technique is considered a revolution in routine microbial identification and is a rapid, reproducible, high-throughput method for identifying Naegleria isolates69.
Furthermore, the use of metagenomic next-generation sequencing (mNGS) can be considered a vital diagnostic tool for the rapid and accurate detection of unique, rare, unexpected, or challenging-to-detect pathogens, including PAM and other life-threatening infectious conditions68. In diagnosing PAM itself, there have been likely unrecognized cases leading to underreporting to health authorities. Drawing upon the extensive observations from the cases we have analyzed, we suggest several key traits that can aid healthcare professionals when faced with patients displaying comparable symptoms. The illness typically manifests suddenly with fever, accompanied by headache and vomiting. Meningeal symptoms after exposure to freshwater should trigger immediate testing for a confirmed diagnosis. Although essential blood parameters such as complete blood counts and tests for renal and liver function may not display notable variations, general infective markers such as ESR (erythrocyte sedimentation rate) and CRP (C-reactive protein) will consistently be elevated. Therefore, it is crucial to interpret the initial results promptly and meticulously to approach an accurate diagnosis4.
Genomic characterization of N. fowleri
Various genotyping systems have been proposed for N. fowleri, including those targeting genes like 12S rRNA, 16S rRNA, ITS1, and 5.8S Rrna70. The most widely recognized system relies on ITS1 length and a 5.8S rDNA sequence transition, categorizing N. fowleri into eight genotypes (T1–T8). Geographically, Europe has seen up to seven genotypes (T2–T8), and the USA and mainland Asia (excluding Japan) have reported three (T1–T3) and two (T2–T3) genotypes, respectively. Oceania and Japan have predominantly one genotype (T5). Notably, in Thailand, PAM cases are linked to at least three genotypes (T2, T3, and T4), with T3 being the most prevalent N. fowleri genotype observed to date10.
Methods used for genomic sequencing and key genomic features
The genome of N. fowleri was sequenced using Illumina HiSeq 2000 and Roche 454 GS FLX technologies, resulting in a 30-Mb genome sequence, including RNA sequencing data. The assembly comprised 1729 contigs with an N50 of 38 128 bp and L50 of 212.
Utilizing Oxford Nanopore Technology (ONT) enhanced the draft genome of the pathogenic amoeba N. fowleri ATCC 30894, achieving a highly contiguous reference with 83 contigs. This ONT-based assembly substantially reduced the contig count (90 vs. 1729) and boosted the N50 by 18-fold (717 491 vs. 38 128 bp). Additional refinement with signal-level raw data and high-quality Illumina reads further improved the assembly quality. The final genome assembly exhibited a comparable number of complete BUSCOs to other sequenced Naegleria species, indicating similar quality and completeness. The genomic size measured 29.5 Mb, with a genomic content of 36.9. This high-quality reference genome supports precise gene prediction and serves as a valuable resource for downstream experiments71.
The N. fowleri TY isolate responsible for a PAM case in a Virginia patient was sequenced using HiSeq Illumina and PacBio technologies, with genome assembly validation through optical mapping data. The resultant haploid assembly comprised 37 chromosomes, with a total size of 27.9 Mb (27 994 426 bp). Individual chromosome sizes ranged from 1 206 962 to 537 351 bp72.
Importance of high-quality genomic data for accurate comparative analysis
The N. fowleri TY genome72, refined through optical mapping and Illumina polishing of PacBio-assembled contigs, stands as the highest-quality reference to date for this human-pathogenic amoeba. With 37 contigs, it approximates a complete set of chromosomes, though karyotyping studies are required for confirmation. Comparatively, the ONT-sequenced NF30894 genome71, comprising 90 contigs, boasts a larger genome size (29.54 Mb) but a lower N50 value (717 491 vs. 756 811 bp) than TY. TY exhibited 215 complete BUSCO genes, whereas NF30894 had 208 complete BUSCO genes and more fragmented ones. The TY genome’s fewer protein-coding genes (9,405) may be a more realistic estimate due to superior genome scaffolding via optical mapping and RNAseq data incorporation. In contrast, NF30894’s gene prediction lacked transcriptomic data, contributing to its larger contig count and gene count. BUSCO analysis indicated a greater number of complete genes in TY (220) compared to NF30894 (219) and NF30863 (210)73, suggesting TY’s gene prediction may be more accurate due to optical mapping scaffolding. TY’s genome, scaffolded using optical mapping, likely provides a realistic estimate of N. fowleri genes74 (Table 1).
Table 1.
Comparison of sequenced Naegleria genomes
| N. fowleri ATCC 3089471 | N. fowleri ATCC 3086373 | N. lovaniensis 75 | N. gruberi 76 | |
|---|---|---|---|---|
| GeneBank accession | VFQX00000000 | AWXF00000000 | PYSW00000000 | ACER00000000 |
| Genome size (Mb) | 29.5 | 27.7 | 30.2 | 40.9 |
| GC content (%) | 36.9 | 37 | 37 | 35 |
| Repeat content (%) | 6 | 2.5 | 3.5 | 5.1 |
| Number of contigs (scaffolds) | 90 | 1729 (574) | 111 | 1977 (784) |
| N50 (bp) | 717 491 | 38 128 | 658 530 | 159 679 |
| L50 | 18 | 212 | 21 | 68 |
| Number of predicted genes | 13 925 | 17 252 (based on RNAseq data) | 15 195 | 16 620 |
Comparative genomics
Genetic diversity
Naegleria species are thermophilic amebae that are commonly found in freshwater and soil all around the world75,76. There have been over 40 Naegleria species identified by studying the genetic differences in the (ITS sequences of their ribosomal DNA and the mitochondrial small subunit (mtSSU) rRNA gene74. The rDNA in Naegleria is located on a circular plasmid in each cell, with ~4000 copies. The ITS and 5.8S rDNA sequences are located between SSU and LSU rDNA and are of particular interest in Naegleria studies77. There are eight types of N. fowleri based on differences in ITS1 length and a single nucleotide difference in the 5.8S rDNA sequence, but only four have been identified in patients so far10. The distribution of these types around the world is uneven, with seven types being detected in Europe and only one (type 5) being found in the West Pacific, which was also observed twice in Europe. Two of the three American types are also present in Europe, while type 1 has only been detected in the USA and not in Europe10. All N. fowleri strains have identical ITS2 sequences, which distinguishes them from other Naegleria species. The types of N. fowleri are differentiated by the length of ITS1 and a T to C transition at position 31 in the 5.8S rDNA sequence10. The difference between type 1/2 and type 3/4 is only the T to C transition since the ITS1 lengths are identical (42 bp and 86 bp, respectively)10. However, type 5 does not have the same ITS1 type.
Comparison of N. fowleri and N. lovaniensis
N. lovaniensis is the closest relative to N. fowleri, differing in the nucleotide sequence in the SSU rDNA sequence by 16 bp (0.8%) only78. Both species differ by only one nucleotide at the 31st location in the 5.8S rDNA sequence. Some strains of N. fowleri have the same sequence as N. lovaniensis, while others have a C to T transition. Additionally, the M1 and M2 sequences in the ITS1 of N. lovaniensis are like those in type 1 and 2 of N. fowleri. The M2 sequences are the same in both species, but the M1 sequence of N. lovaniensis differs by 4 bp transitions and 1 bp deletion compared to the M1 sequence in N. fowleri 78. To understand the differences in virulence between closely related species, pathogenic N. fowleri and nonpathogenic N. lovaniensis, a comparative analysis of their phenotypic traits was conducted79. This research involved exploring various factors such as growth rate and their susceptibility to complement-mediated lysis, both in contact-dependent and contact-independent contexts. The growth rates of N. fowleri and N. lovaniensis were compared by monitoring the density of amoebas at various time points. N. fowleri exhibited a generation time of 1.5 h, whereas N. lovaniensis had a generation time of 6.5 h (FIG: 01)79. After 72 h of growth, light micrographs were captured to illustrate the contrasting growth patterns between the two species, revealing a disparity in the number of amoebae79. Contact-independent cytotoxicity assays were employed to explore whether there were distinctions in the substances released by N. fowleri and N. lovaniensis in response to the presence of human nasal epithelial cells. The evaluation of the integrity of the target cells using a crystal violet staining technique revealed a 10% variation following a 4.5-h incubation period79.
Shared gene families and genetic similarities
To distinguish N. fowleri genes that differ from those in Naegleria gruberi and N. lovaniensis, the proteins of each species (N. fowleri TY, N. lovaniensis 76-15-250, and N. gruberi) were initially classified using Neptune, CD-HIT, and OrthoMCL. From each of these tools, the unique genes and clusters specific to the genus were extracted. As a result, the genus formed a total of 11 530 protein clusters and 7511 orthologous clusters. Among them, 5749 protein clusters were found to be shared by all Naegleria species. Furthermore, there were 1246 clusters that were commonly shared between N. fowleri and N. lovaniensis, whereas N. fowleri exhibited only 255 shared protein clusters with N. gruberi (FIG: 02). A total of 404 N. fowleri genes were identified as unique in comparison to N. gruberi and N. lovaniensis 74.
Phylogenomic relationship
The PHYLO-WIN software was utilized for the phylogenetic analysis, which was based on the ITS and 5.8s sequences. During the analysis, the global gap was eliminated80. The neighbor-joining (NJ) algorithm was employed to reconstruct the phylogenetic tree81. To assess the reliability of each node, the bootstrap procedure was applied82. Additionally, a parsimony analysis using the DOLMIX program in the PHYLIP package was conducted to investigate the phylogenetic relationships within N. fowleri 83. The phylogenetic tree, generated using the NJ algorithm, was constructed based on the ITS and 5.8s sequences. The tree revealed four main clusters (FIG: 03). The first cluster consisted of all N. fowleri strains and N. lovaniensis. Additionally, Naegleria morganensis NG236, along with the isolates NG427 and NG334 formed a sister group to the first cluster. In the second main cluster, N. gruberi diverged with Naegleria australiensis, Naegleria clarki, Naegleria italica, and Naegleria galeacystis and was referred to as the gruberi–australiensis cluster. The third cluster exclusively included Naegleria jumiesoni and Naegleria andersoni. Naegleria pussurdi and N. gruberi NG260 formed the fourth cluster. The reliability of these clusters was well-supported, with over 90% bootstrap support83.
Differential adhesion and interaction with extracellular matrix (ECM)
A crucial stage in the infection process by N. fowleri involves its interaction with the host’s basement membrane, which is a complex layer comprising specialized ECM glycoproteins and proteoglycans. This membrane serves the purpose of separating the epithelium from the underlying stromal tissues84. During its journey to the brain, N. fowleri needs to traverse the epithelial layer and meet specific ECM elements. These include laminin 1, a significant constituent of the basement membrane85; collagen I, which is a prevalent ECM component in connective tissues86; and fibronectin, an adhesive glycoprotein found in both connective tissues and the bloodstream87. A comparison was made between the adhesive and invasive characteristics of thermotolerant pathogenic N. fowleri and a thermotolerant nonpathogenic species, N. lovaniensis. The findings revealed differences in their ability to adhere to ECM components, with N. fowleri exhibiting a notably higher level of adhesion88.
Advanced microscopic imaging for characterization
N. gruberi, a microbial eukaryote that exists independently and is quite distinct in its evolutionary lineage from animals, yeast, and plants, can be commonly located in various soil and freshwater environments around the world, whether they are oxygen-rich or have low oxygen levels89,90. In 2010, researchers published the genome of N. gruberi, unveiling a remarkably intricate collection of cytoskeletal, sexual, signaling, and metabolic elements, along with a highly comprehensive membrane trafficking system (MTS)91. Naegleria belongs to the Excavata supergroup, which also includes parasitic organisms of significance such as trypanosomatids, Trichomonas vaginalis, and Giardia intestinalis, as well as the anaerobic and amitochondriate Monocercomonoides sp., a commensal organism found in the guts of rodents. Consequently, N. gruberi remains among the limited number of free-living excavates whose complete genome is accessible to the public92.
Electron microscopy for ultrastructural analysis
Scanning electron microscopy (SEM) showed variations in the structure of N. fowleri in comparison to N. lovaniensis. N. fowleri displayed an extended appearance characterized by the presence of focal adhesion-like extensions. Western immunoblots detected two protein species that reacted with an anti-integrin antibody. Among these, the larger one, a 70 kDa protein resembling integrin, was present in higher quantities in N. fowleri in comparison to N. lovaniensis 88. Additionally, when these amoebae were exposed to ECM components, they displayed lamellipodia and morphological characteristics resembling focal adhesions. These characteristics were absent when N. fowleri adhered to uncoated glass surfaces. Conversely, N. lovaniensis maintained an elongated shape when placed on any type of substrate. Notably, focal adhesion-like structures were not observed in N. lovaniensis when it was placed on any of the substrates88. Electron microscopy of the infected mouse brain has previously shown amoebae in the process of engulfing collagen I fibrils during the invasion, as documented by Martinez et al. in 197393. It was observed that N. fowleri displayed a seemingly quicker invasion of collagen I and30 atrigel constructs compared to N. lovaniensis. This apparent enhanced invasion capability may be attributed to increased motility79. For instance, a study that compared the movement patterns of Naegleria species within agarose found a correlation between their locomotive ability and pathogenic potential94. Furthermore, it is worth noting that N. fowleri, unlike N. lovaniensis, has demonstrated heightened motility when in proximity to rat B103 nerve cells, as reported by Cline and Marciano-Cabral in 198695. This suggests that N. fowleri selectively responds to neurotropic factors, setting it apart from N. lovaniensis and other free-living amoebae that lack the potential to cause neurological pathogenesis79,95.
Molecular differences
One unique characteristic of Naegleria is the absence of a visibly distinguishable Golgi organelle, which is, in fact, a defining trait of the broader taxonomic group to which Naegleria belongs92. The sole supporting evidence for the existence of this organelle in Naegleria has been the bioinformatic predictions of Golgi-associated proteins identified during the genome project. According to Fritz-Laylin et al.91, 2010 study, they tackled the task of identifying and visualizing the Golgi structure in Naegleria by employing a multidisciplinary approach. They presented the first molecular and cellular proof supporting the presence of a punctate Golgi structure in N. gruberi 91. Naegleria possesses the genetic coding and expresses the machinery associated with Golgi-related membrane trafficking. Traditionally, Naegleria, along with other heteroloboseans, has been conventionally described as lacking a visible stacked Golgi structure92. Through an examination of a publicly available transcriptome for N. gruberi, we were able to identify expression data for 37 out of 67 genes96. We observed expressed transcripts for all 66 Golgi-associated MTS genes in N. fowleri. Every N. gruberi sequence was found to have a counterpart in N. fowleri, except for three specific paralogues unique to N. gruberi (Vps53A, Ykt6B, and Vps45B)96.
Confocal microscopy for three-dimensional visualization
Herman et al.96. conducted localization experiments and captured images using confocal microscopy to gain a better understanding of the Naegleria Golgi’s ultrastructure. Employing the same concentrations of antibodies as in previous experiments, they visualized the localization of COPB in various cells. The 3D representations of multiple cell sections revealed a clearly defined tubular arrangement, which they confirmed through 24 separate experiments involving different antibody concentrations. To delve even deeper into the subcellular localization of NgCOPB, they employed transmission electron microscopy, followed by immuno-gold electron microscopy. These methods provided strong evidence for the specific localization of this protein within distinct membrane organelles, each ranging from 1 to 4 μm in length. This localization contrasted with its presence in the cytosol, nucleus, larger membrane organelles, and membrane vesicles96.
Immunofluorescence staining
One study conducted a comparison of two immunofluorescence staining protocols97. To visualize the amoebae, a specific monoclonal antibody, 5D12 (provided by Indicia Biotechnology), was employed98,99. This antibody was diluted to varying concentrations in a solution of PBS (containing 8 g of NaCl, 0.2 g of KCl, 1.44 g of Na2HPO4, and 0.24 g of KH2PO4 per liter, pH 7.2) supplemented with 0.1% Tween 20 and 2% bovine serum albumin (reagents sourced from Sigma Chemical Company). The antibody was subsequently linked with either biotin or HRP (horseradish peroxidase) and then detected using streptavidin linked with RPE-Cy5 or fluorescein isothiocyanate (FITC)-conjugated tyramide, respectively97. The RPE-Cy5 method proved to be the most effective procedure, enabling the identification of both trophozoite and cyst forms in the water samples97.
Advanced staining techniques for characterization
In another study, seven different staining methods were examined to identify the most suitable color and contrast for staining the developmental stages of free-living pathogenic Acanthamoeba and Naegleria species. The acid-fast bacilli stain (AFB) resulted in a blue color but lacked contrast. The trichrome-eosin and modified Field’s stains exhibited a variety of color contrasts. Giemsa, iron-hematoxylin, modified AFB, and Gram stains all yielded a single color, which effectively distinguished components like the nucleus, nucleolus, cytoplasm, food vacuoles, and water vacuoles100. These staining methods revealed that Naegleria trophozoites possess a single motile protruding structure known as the lobopodium or bluntly eruptive pseudopodium, which enables them to move actively in a unidirectional manner21. The process of enflagellation, which involves the formation of flagella, can be induced in these trophozoites by various factors, including nutrient depletion, temperature changes, the growth phase, and agitation in culture101,102. The ability of Naegleria trophozoites to undergo enflagellation is a significant characteristic used to distinguish different Naegleria species100. However, it is important to note that certain Naegleria species, such as N. chilensis and N. indonesiensis, are incapable of enflagellation33. Upon staining, the cysts of Naegleria are seen as uniformly spherical, possessing smooth and thin double walls. They tend to cluster closely together, forming colonies on the agar surface103,104. The application of staining to the amoebae yielded a detailed depiction of their cellular organelles. The motile organelles became readily apparent after the staining process100.
Out of the seven staining methods evaluated, trichrome-eosin and iron-hematoxylin stains consistently exhibited good color contrast across all three stages (trophozoite, cyst, and flagellate). Giemsa and Gram stains effectively stained the trophozoite and flagellate stages, while modified Field’s and modified AFB stains exclusively stained the trophozoite stage. The original AFB stain did not provide satisfactory color contrast at any of the stages, and the modified Field’s stain was ineffective for staining the flagellate stage of Naegleria 100.
Integration of biomarkers, genomic profiling, and microscopic imaging
PAM is a rare infectious disease caused by the protozoan N. fowleri, and it is associated with a mortality rate exceeding 95%. For clinicians, it is crucial to swiftly and accurately identify the responsible pathogen to guide their decisions regarding treatment strategies in a clinical setting105. In patients with PAM, the initial signs typically include headaches, nausea, alterations in the sense of smell and taste, fever, backache, and vomiting. These symptoms can advance to more severe manifestations such as confusion, hallucinations, diminished attention span, and seizures. When these symptoms manifest alongside a history of swimming or exposure of the nasal passages to contaminated water, there should be a strong suspicion of PAM. The involvement of the CNS is often revealed through CT scans in affected patients106,107. For laboratory diagnosis, it is recommended to screen CSF for the presence of N. fowleri 108,109. A distinctive morphological characteristic of N. fowleri is the presence of food cups and flagellates, which can be observed through microscopic examination106,110. When exposed to distilled water at 37°C, Naegleria can transition from the trophozoite form to the flagellate form, exhibiting two flagella107,111–113. On the other hand, when cultured on non-nutrient agar lacking essential nutrients, it can transform into cysts. These processes, known as encystation and enflagellation, are crucial for identifying N. fowleri.
Microscopic identification is often combined with staining techniques, such as Gram staining and Wright–Giemsa staining114. Alternatively, various other methods can be employed, including indirect immunofluorescence staining, flow cytometry, immune phosphate staining, ELISA, and PCR assays19,111.
PCR – a rapid and sensitive diagnostic method
PCR is frequently the preferred method due to its sensitivity and rapidity in diagnosing PAM by detecting N. fowleri in the nasal mucosa. Given that nasal mucosa samples can be easily collected, and PCR is a quick detection technique, employing a primer mix capable of detecting the genetic material of N. fowleri in nasal mucosa could offer a fast, sensitive, specific, and noninvasive approach for PAM detection115. Next-generation sequencing (NGS) has also been applied to detect N. fowleri 105.
Untargeted metabolomics method
Another recently developed method for N. fowleri detection involves ‘untargeted metabolomics methods’. This approach includes quenching the parasite using a mixture of methanol and ammonium bicarbonate, followed by hot methanol extractions to isolate the cell metabolites116.
These metabolites are then analyzed using liquid chromatography–mass spectrometry. The resulting data is compared to existing libraries and subjected to analysis through bioinformatics tools116. Further examination of the obtained peaks, using these peaks to identify molecules, and assessing their properties through bioinformatics software such as Pepstats, leads to the identification of specific biomarkers for N. fowleri 69.
Next-generation sequencing (NGS)
In a research report, the first-ever documented case of PAM in mainland China was reported. Next-generation sequencing (NGS) was employed for swift diagnosis, and it played a crucial role in guiding the selection of appropriate medications for treatment105.
Early detection using CSF wet mount sample
In another report from India, a case involving a 6-month-old boy was documented. The child was brought to the pediatric emergency room in a state of status epilepticus, accompanied by a history of high-grade fever lasting for 3 days, as well as lethargy, poor feeding, and altered consciousness over the past 24 h117. Examination of the CSF sample using a wet mount revealed a substantial number of actively moving trophozoites, a finding further confirmed by Giemsa staining, which indicated the likely presence of Naegleria species117.
This case underscores the importance of maintaining a high level of suspicion when dealing with infants who present with symptoms resembling those of pyogenic meningitis, particularly when a CSF gram stain fails to detect bacteria. In cases where amoebic infection is suspected, it is advisable to conduct a wet mount preparation to identify motile trophozoites early on118, facilitating the prompt detection of N. fowleri infection and the implementation of appropriate intervention measures117.
Real-time PCR as a confirmatory test
In Turkey, a case was encountered of an 18-year-old male patient whose family reported that he had recently visited a hot spring. His complaint of headaches began ~2–3 days after he returned from the trip. To determine the presence of the pathogen, a CSF sample was collected from the patient and subjected to a thorough examination. This examination involved direct microscopic observation, real-time PCR analysis, and sequence analysis. The CSF sample was carefully introduced into distilled water, as there was a possibility of trophozoites transforming into an intermediate form. It was then incubated at 37°C for 1–2 h. During direct microscopic examination, pear-shaped nonpermanent flagellated forms were observed, further raising suspicion. To confirm the diagnosis, molecular typing was conducted. This study presents a comprehensive case of N. fowleri infection in Turkey, where the causative agent was successfully isolated and confirmed using real-time PCR119.
The CSF wet mount is a rapid and immediate diagnostic method commonly employed before a patient’s death. Additionally, PCR testing, brain tissue immunofluorescent analysis (IF), and next-generation sequencing (NGS) were utilized but are more time-consuming120.
Conclusion
The presented study highlights various laboratory diagnostic approaches, including CSF analysis, brain tissue examination, immunostaining techniques, and culture methods for the detection of N. fowleri in clinical samples. Additionally, it discusses the limitations of CT and MRI in diagnosing N. fowleri infections.
The genomic profiling techniques, particularly PCR-based methods, are instrumental in the accurate and timely diagnosis of N. fowleri infections. These techniques, such as conventional PCR, nested PCR, multiplex PCR, and real-time PCR, offer improved sensitivity, specificity, and speed, making them valuable tools for early detection and differentiation of N. fowleri strains.
The research emphasizes the importance of early detection in enhancing patient outcomes and public health measures. Furthermore, it discusses the potential of advanced microscopic imaging techniques to characterize N. fowleri’s morphology and behavior at different infection stages, contributing to a deeper understanding of its life cycle and pathogenic mechanisms.
Ethical approval
Not applicable.
Consent
Informed consent was not required for this review.
Sources of funding
Not applicable.
Author contribution
All authors contributed equally.
Conflicts of interest disclosure
There are no conflicts of interest.
Research registration unique identifying number (UIN)
Registry used: nil.
Unique identifying number or registration ID: nil.
Hyperlink to your specific registration: nil.
Guarantor
Nahid Raufi.
Data availability statement
Not applicable.
Provenance and peer review
Not commissioned, externally peer-reviewed.
Acknowledgements
Not applicable.
Footnotes
Sponsorships or competing interests that may be relevant to content are disclosed at the end of this article.
Contributor Information
Ayesha Shaukat, Email: ayeshaa.skt@gmail.com.
Nawal Khaliq, Email: nawmkm@gmail.com.
Rumaisa Riaz, Email: riazrumaisa@gmail.com.
Rabbia Munsab, Email: rabbiamunsab2956@gmail.com.
Tayyaba Ashraf, Email: tayyabakhatri1203@gmail.com.
Nahid Raufi, Email: nahidraufi99@outlook.com.
Hafsa Shah, Email: alizeroman@gmail.com.
References
- 1.Visvesvara GS, Moura H, Schuster FL. Pathogenic and opportunistic free-living amoebae: Acanthamoeba spp., Balamuthia mandrillaris, Naegleria fowleri, and Sappinia diploidea. FEMS Immunol Med Microbiol 2007;50:1–26. [DOI] [PubMed] [Google Scholar]
- 2.Yoder JS, Straif-Bourgeois S, Roy SL, et al. Primary amebic meningoencephalitis deaths associated with sinus irrigation using contaminated tap water. Clin Infect Dis 2012;55:e79–e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zain A, Ashfaq F, Azhar M. Lahore’s encounter with Naegleria fowleri: unveiling health hazards and heightened concerns. Health Sci Rep 2023;6:e1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahmad Zamzuri M, Ammar I, Abd Majid FN, et al. Systematic review of brain-eating amoeba: a decade update. Int J EnvironRes Public Health 2023;20:3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baig AM, Khan NA. Novel chemotherapeutic strategies in the management of primary amoebic meningoencephalitis due to Naegleria fowleri. CNS Neurosci Ther 2014;20:289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shakoor S, Beg MA, Mahmood SF, et al. Primary amebic meningoencephalitis caused by Naegleria fowleri, Karachi, Pakistan. Emerg Infect Dis 2011;17:258–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cooper AM, Aouthmany S, Shah K, et al. Killer amoebas: primary amoebic meningoencephalitis in a changing climate. JAAPA 2019;32:30–35. [DOI] [PubMed] [Google Scholar]
- 8.Cope JR, Ali IK. Primary amebic meningoencephalitis: what have we learned in the last 5 years? Curr Infect Dis Rep 2016;18:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gompf SG, Garcia C. Lethal encounters: the evolving spectrum of amoebic meningoencephalitis. IDCases 2019;15:e00524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Jonckheere JF. Origin and evolution of the worldwide distributed pathogenic amoeboflagellate Naegleria fowleri. Infect Genet Evol 2011;117:1520–1528. [DOI] [PubMed] [Google Scholar]
- 11.De Jonckheere JF. Seasonal primary amebic meningoencephalitis (PAM) in the south: summertime is PAM time. J La State Med Soc 2012;164:148–50; 152–5. [PubMed] [Google Scholar]
- 12.Maciver SK, Piñero JE, Lorenzo-Morales J. Is Naegleria fowleri an emerging parasite? Trends Parasitol 2020;36:19–28. [DOI] [PubMed] [Google Scholar]
- 13.GRIFFIN JL. The pathogenic amoeboflagellate Naegleria fowleri: environmental isolations, competitors, ecologic interactions, and the flagellate-empty habitat hypothesis. J Protozool 1983;30:403–409. [DOI] [PubMed] [Google Scholar]
- 14.Martínez AJ, Visvesvara GS. Pathogenic and opportunistic free-living amebas: Naegleria fowleri, Acanthamoeba spp. and Balamuthia mandrillaris. In: Gillespie SH, Pearson RD, editors. Principles and Practise of Clinical Parasitology. Wiley; 2001. pp. 269–85.
- 15.Sohn HJ, Kim JH, Kim K, et al. De novo transcriptome profiling of Naegleria fowleri trophozoites and cysts via RNA sequencing. Pathogens 2023;12:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trabelsi H, Dendana F, Sellami A, et al. Pathogenic free-living amoebae: epidemiology and clinical review. Pathol Biol 2012;60:399–405. [DOI] [PubMed] [Google Scholar]
- 17.Pana A, Vijayan V, Anilkumar AC. Amebic Meningoencephalitis. StatPearls; 2023. [PubMed] [Google Scholar]
- 18.Abo El-Maaty DA, Hamza RS. Primary amoebic meningoencephalitis caused by Naegleria fowleri. PUJ 2012;5:93–104. [Google Scholar]
- 19.Siddiqui R, Ali IKM, Cope JR, et al. Biology and pathogenesis of Naegleria fowleri. Acta Trop 2016;164:375–394. [DOI] [PubMed] [Google Scholar]
- 20.Nadeem A, Malik IA, Afridi EK, et al. Naegleria fowleri outbreak in Pakistan: unveiling the crisis and path to recovery. Front Public Health 2023;11:1266400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marciano-Cabral F. Biology of Naegleria spp. Microbiol Rev 1988;52:114–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.CDC . DPDx - Free Living Amebic Infections. Accessed 11 October 2023. https://www.cdc.gov/dpdx/freelivingamebic/index.html
- 23.Schuster FL, Visvesvara GS. Free-living amoebae as opportunistic and non-opportunistic pathogens of humans and animals. Int J Parasitol 2004;34:1001–1027. [DOI] [PubMed] [Google Scholar]
- 24.Marciano-Cabral F, Cabral GA. The immune response to Naegleria fowleri amebae and pathogenesis of infection. FEMS Immunol Med Microbiol 2007;51:243–259. [DOI] [PubMed] [Google Scholar]
- 25.Marfani WB, Vohra LI, Sahito AM, et al. Wastewater surveillance in Pakistan: preventing future epidemics. Ann Med Surg 2022;81:104495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baig AM. Primary amoebic meningoencephalitis: neurochemotaxis and neurotropic preferences of Naegleria fowleri. ACS Chem Neurosci 2016;7:1026–1029. [DOI] [PubMed] [Google Scholar]
- 27.John DT. Primary amebic meningoencephalitis and the biology of Naegleria fowleri. Annu Rev Microbiol 1982;36:101–123. [DOI] [PubMed] [Google Scholar]
- 28.Sarfraz MR, Tariq H, Rehman S, et al. Naegleria fowleri – the brain-eating amoeba: an emerging threat in Pakistan. Acta Biomed 2023;94:e2023024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.WikiDoc . Primary amoebic meningoencephalitis natural history, complications and prognosis - wikidoc. Accessed 11 October 2023. https://www.wikidoc.org/index.php/Primary_amoebic_meningoencephalitis_natural_history,_complications_and_prognosis
- 30.CDC . Diagnosis | Naegleria fowleri | CDC. Accessed 11 October 2023. https://www.cdc.gov/parasites/naegleria/diagnosis-hcp.html
- 31.da Rocha-Azevedo B, Tanowitz HB, Marciano-Cabral F. Diagnosis of infections caused by pathogenic free-living amoebae. Interdiscip Perspect Infect Dis 2009;2009:251406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schuster FL. Cultivation of pathogenic and opportunistic free-living amebas. Clin Microbiol Rev 2002;15:342–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Jonckheere JF, Brown S, Dobson PJ, et al. The amoeba-to-flagellate transformation test is not reliable for the diagnosis of the genus Naegleria. Description of three new Naegleria spp. Protist 2001;152:115–121. [DOI] [PubMed] [Google Scholar]
- 34.Behets J, Seghi F, Declerck P, et al. Detection of Naegleria spp. and Naegleria fowleri: a comparison of flagellation tests, ELISA and PCR. Water Sci Technol 2003;47:117–22. [PubMed] [Google Scholar]
- 35.De Jonckheere JF. Characterization of Naegleria species by restriction endonuclease digestion of whole-cell DNA. Mol Biochem Parasitol 1987;24:55–66. [DOI] [PubMed] [Google Scholar]
- 36.McLaughlin GL, Brandt FH, Visvesvara GS. Restriction fragment length polymorphisms of the DNA of selected Naegleria and Acanthamoeba amebae. J Clin Microbiol 1988;26:1655–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.MILLIGAN SM, BAND RN. Restriction endonuclease analysis of mitochondrial DNA as an aid in the taxonomy of Naegleria and Vahlkampfia. J Protozool 1988;35:198–204. [DOI] [PubMed] [Google Scholar]
- 38.Sparagano O. Detection of Naegleria fowleri cysts in environmental samples by using a DNA probe. FEMS Microbiol Lett 1993;112:349–351. [DOI] [PubMed] [Google Scholar]
- 39.Maclean RC, Richardson DJ, LePardo R, et al. The identification of Naegleria fowleri from water and soil samples by nested PCR. Parasitol Res 2004;93:211–217. [DOI] [PubMed] [Google Scholar]
- 40.Sparagano O. Differentiation of Naegleria fowleri and other naegleriae by polymerase chain reaction and hybridization methods. FEMS Microbiol Lett 1993; 110:325–330. [DOI] [PubMed] [Google Scholar]
- 41.Van Belkum A, De Jonckheere J, Quint WGV. Genotyping Naegleria spp. and Naegleria fowleri isolates by interrepeat polymerase chain reaction. J Clin Microbiol 1992;30:2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sheehan KB, Fagg JA, Ferris MJ, et al. PCR detection and analysis of the free-living amoeba Naegleria in hot springs in Yellowstone and Grand Teton National Parks. Appl Environ Microbiol 2003;69:5914–5918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McLaughlin GL, Vodkin MH, Huizinga HW. Amplification of repetitive DNA for the specific detection of Naegleria fowleri. J Clin Microbiol 1991;29:227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marciano-Cabral F, MacLean R, Mensah A, et al. Identification of Naegleria fowleri in domestic water sources by nested PCR. Appl Environ Microbiol 2003;69:5864–5869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Réveiller FL, Cabanes PA, Marciano-Cabral F. Development of a nested PCR assay to detect the pathogenic free-living amoeba Naegleria fowleri. Parasitol Res 2002;88:443–450. [DOI] [PubMed] [Google Scholar]
- 46.Pélandakis M, Pernin P. Use of multiplex PCR and PCR restriction enzyme analysis for detection and exploration of the variability in the free-living amoeba Naegleria in the environment. Appl Environ Microbiol 2002;68:2061–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Behets J, Declerck P, Delaedt Y, et al. A duplex real-time PCR assay for the quantitative detection of Naegleria fowleri in water samples. Water Res 2007;41:118–126. [DOI] [PubMed] [Google Scholar]
- 48.Qvarnstrom Y, Visvesvara GS, Sriram R, et al. Multiplex real-time PCR assay for simultaneous detection of Acanthamoeba spp., Balamuthia mandrillaris, and Naegleria fowleri. J Clin Microbiol 2006;44:3589–3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robinson BS, Monis PT, Dobson PJ. Rapid, sensitive, and discriminating identification of Naegleria spp. by real-time PCR and melting-curve analysis. Appl Environ Microbiol 2006;72:5857–5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maďarová L, Trnková K, Feiková S, et al. A real-time PCR diagnostic method for detection of Naegleria fowleri. Exp Parasitol 2010;126:37–41. [DOI] [PubMed] [Google Scholar]
- 51.Mull BJ, Narayanan J, Hill VR. Improved method for the detection and quantification of Naegleria fowleri in water and sediment using immunomagnetic separation and real-time PCR. J Parasitol Res 2013;2013:608367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Puzon GJ, Lancaster JA, Wylie JT, et al. Rapid detection of Naegleria fowleri in water distribution pipeline biofilms and drinking water samples. Environ Sci Technol 2009;43:6691–6696. [DOI] [PubMed] [Google Scholar]
- 53.Jeffreys AJ, Wilson V, Thein SL. Hypervariable “minisatellite” regions in human DNA. Nature 1985;314:67–73. [DOI] [PubMed] [Google Scholar]
- 54.Nakamura Y, Leppert M, O’Connell P, et al. Variable number of tandem repeat (VNTR) markers for human gene mapping. Science 1987;235:1616–1622. [DOI] [PubMed] [Google Scholar]
- 55.Brown MRW, Barker J. Unexplored reservoirs of pathogenic bacteria: protozoa and biofilms. Trends Microbiol 1999;7:46–50. [DOI] [PubMed] [Google Scholar]
- 56.Moffat JF, Tompkins LS. A quantitative model of intracellular growth of Legionella pneumophila in Acanthamoeba castellanii. Infect Immun 1992;60:296–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wadowsky RM, Wilson TM, Kapp NJ, et al. Multiplication of Legionella spp. in tap water containing Hartmannella vermiformis. Appl Environ Microbiol 1991;57:1950–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mackay IM. Real-time PCR in the microbiology laboratory. Clin Microbiol Infect 2004;10:190–212. [DOI] [PubMed] [Google Scholar]
- 59.Bej AK. Molecular based methods for the detection of microbial pathogens in the environment. J Microbiol Methods 2003;53:139–140. [DOI] [PubMed] [Google Scholar]
- 60.Schild M, Gianinazzi C, Gottstein B, et al. PCR-based diagnosis of Naegleria sp. infection in formalin-fixed and paraffin-embedded brain sections. J Clin Microbiol 2007;45:564–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lê HG, Ham AJ, Kang JM, et al. A novel cysteine protease inhibitor of Naegleria fowleri that is specifically expressed during encystation and at mature cysts. Pathogens 2021;10:388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Milanes J. Glycolytic inhibitors as leads for drug discovery in the pathogenic free-living amoebae [dissertation]. TigerPrints; 2023. https://tigerprints.clemson.edu/all_dissertations
- 63.Medeiros LCS, South L, Peng D, et al. Rapid, selection-free, high-efficiency genome editing in Protozoan parasites using CRISPR-Cas9 ribonucleoproteins. mBio 2017;8:e01788–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Faktorová D, Kaur B, Valach M, et al. Targeted integration by homologous recombination enables in situ tagging and replacement of genes in the marine microeukaryote Diplonema papillatum. Environ Microbiol 2020;22:3660–3670. [DOI] [PubMed] [Google Scholar]
- 65.Che X, He Z, Tung TH, et al. Diagnosis of primary amoebic meningoencephalitis by metagenomic next-generation sequencing: a case report. Open Life Sci 2023;18:20220579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Niedringhaus KD, Gordon M, Yabsley MJ, et al. Fatal balamuthosis in a Siberian tiger and a literature review of detection options for free-living amoebic infections in animals. J Vet Diagn Invest 2023;35:311–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Baig AM. Proposals for amendments in the diagnosis and treatment of encephalitis caused by free-living amoebae. Infect Disord Drug Targets 2020;20:115–121. [DOI] [PubMed] [Google Scholar]
- 68.Huang S, Liang X, Han Y, et al. A pediatric case of primary amoebic meningoencephalitis due to Naegleria fowleri diagnosed by next-generation sequencing of cerebrospinal fluid and blood samples. BMC Infect Dis 2021;21:1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Moura H, Izquierdo F, Woolfitt AR, et al. Detection of biomarkers of pathogenic Naegleria fowleri through mass spectrometry and proteomics. J Eukaryot Microbiol 2015;62:12–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.De Jonckheere JF. Sequence variation in the ribosomal internal transcribed spacers, including the 5.8S rDNA, of Naegleria spp. Protist 1998;149:221–228. [DOI] [PubMed] [Google Scholar]
- 71.Liechti N, Schürch N, Bruggmann R, et al. Nanopore sequencing improves the draft genome of the human pathogenic amoeba Naegleria fowleri. Sci Rep 2019;9:16040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ali IKM, Kelley A, Joseph SJ, et al. Draft chromosome sequences of a clinical isolate of the free-living ameba Naegleria fowleri. Microbiol Resour Announc 2021;10:e01034–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zysset-Burri DC, Müller N, Beuret C, et al. Genome-wide identification of pathogenicity factors of the free-living amoeba Naegleria fowleri. BMC Genomics 2014;15:496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Joseph SJ, Park S, Kelley A, et al. Comparative genomic and transcriptomic analysis of Naegleria fowleri clinical and environmental isolates. mSphere 2021;6:e0063721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Martinez AJ. Free-living amebas: infection of the central nervous system. Mt Sinai J Med 1993;60:271–278. [PubMed] [Google Scholar]
- 76.Martinez AJ, Visvesvara GS. Free-living, amphizoic and opportunistic amebas. Brain Pathol 1997;7:583–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.De Jonckheere JF. What do we know by now about the genus Naegleria? Exp Parasitol 2014;145(suppl):S2–S9. [DOI] [PubMed] [Google Scholar]
- 78.Jonckheere D. A century of research on the amoeboflagellate genus Naegleria. Acta Protozool 2002;41:309–342. [Google Scholar]
- 79.Jamerson M. A comparative study of a pathogenic versus a nonpathogenic Naegleria species [dissertation]. Medicine and Health Sciences Commons; 2011. https://scholarscompass.vcu.edu/etd/249
- 80.Galtier N, Gouy M, Gautier C. SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Bioinformatics 1996;12:543–548. [DOI] [PubMed] [Google Scholar]
- 81.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 1987;4:406–425. [DOI] [PubMed] [Google Scholar]
- 82.Felsenstein J. Confidence limits on phylogenies: an approach using the Bootstrap. Evolution 1985;39:783–791. [DOI] [PubMed] [Google Scholar]
- 83.Pélandakis M, Serre S, Pernin P. Analysis of the 5.8S rRNA gene and the internal transcribed spacers in Naegleria spp. and in N. fowleri. J Eukaryot Microbiol 2000;47:116–121. [DOI] [PubMed] [Google Scholar]
- 84.LeBleu VS, MacDonald B, Kalluri R. Structure and function of basement membranes. Exp Biol Med (Maywood) 2007;232:1121–1129. [DOI] [PubMed] [Google Scholar]
- 85.Yurchenco PD, Schittny JC. Molecular architecture of basement membranes. FASEB J 1990;4:1577–1590. [DOI] [PubMed] [Google Scholar]
- 86.Nimni ME. Collagen: structure, function, and metabolism in normal and fibrotic tissues. Semin Arthritis Rheum 1983;13:1–86. [DOI] [PubMed] [Google Scholar]
- 87.Hynes RO, Yamada KM. Fibronectins: multifunctional modular glycoproteins. J Cell Biol 1982;95(2 Pt 1):369–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jamerson M, da Rocha-Azevedo B, Cabral GA, et al. Pathogenic Naegleria fowleri and non-pathogenic Naegleria lovaniensis exhibit differential adhesion to, and invasion of, extracellular matrix proteins. Microbiology (Reading) 2012;158(Pt 3):791–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fulton C. Naegleria: a research partner for cell and developmental biology. J Eukaryot Microbiol 1993;40:520–532. [Google Scholar]
- 90.Fulton C. Axenic cultivation of Naegleria gruberi. Requirement for methionine. Exp Cell Res 1974;88:365–370. [DOI] [PubMed] [Google Scholar]
- 91.Fritz-Laylin LK, Prochnik SE, Ginger ML, et al. The genome of Naegleria gruberi illuminates early eukaryotic versatility. Cell 2010;140:631–642. [DOI] [PubMed] [Google Scholar]
- 92.Adl SM, Simpson AGB, Lane CE, et al. The revised classification of eukaryotes. J Eukaryot Microbiol 2012;59:429–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Martinez J, Duma RJ, Nelson EC, et al. Experimental naegleria meningoencephalitis in mice. Penetration of the olfactory mucosal epithelium by Naegleria and pathologic changes produced: a light and electron microscope study. Lab Invest 1973;29:121–33. [PubMed] [Google Scholar]
- 94.Thong YH, Ferrante A. Migration patterns of pathogenic and nonpathogenic Naegleria spp. Infect Immun 1986;51:177–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cline M, Carchman R, Marciano‐Cabral F. Movement of Naegleria fowleri stimulated by mammalian cells in vitro. J Protozool 1986;33:10–13. [DOI] [PubMed] [Google Scholar]
- 96.Herman EK, Yiangou L, Cantoni DM, et al. Identification and characterisation of a cryptic Golgi complex in Naegleria gruberi. J Cell Sci 2018;131:jcs213306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pougnard C, Catala P, Drocourt JL, et al. Rapid detection and enumeration of Naegleria fowleri in surface waters by solid-phase cytometry. Appl Environ Microbiol 2002;68:3102–3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Réveiller FL, Marciano-Cabral F, Pernin P, et al. Species specificity of a monoclonal antibody produced to Naegleria fowleri and partial characterization of its antigenic determinant. Parasitol Res 2000;86:634–641. [DOI] [PubMed] [Google Scholar]
- 99.Sparagano O, Drouet E, Brebant R, et al. Use of monoclonal antibodies to distinguish pathogenic Naegleria fowleri (cysts, trophozoites, or flagellate forms) from other Naegleria species. J Clin Microbiol 1993;31:2758–2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ithoi I, Ahmad AF, Mak JW, et al. Morphological characteristics of developmental stages of Acanthamoeba and Naegleria species before and after staining by various techniques. Southeast Asian J Trop Med Public Health 2011;42:1327–38. [PubMed] [Google Scholar]
- 101.Fulton C. Intracellular regulation of cell shape and motility in Naegleria. First insights and a working hypothesis. J Supramol Struct 1977;6:13–43. [DOI] [PubMed] [Google Scholar]
- 102.Woodworth TW, John DT, Bradley SG. Biological factors affecting enflagellation of Naegleria fowleri. J Bacteriol 1982;152:803–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Init I, Lau YL, Arin Fadzlun A, et al. Detection of free living amoebae, Acanthamoeba and Naegleria, in swimming pools, Malaysia. Trop Biomed 2010;27:566–77. [PubMed] [Google Scholar]
- 104.Carter RF. Description of a Naegleria sp. isolated from two cases of primary amoebic meningo-encephalitis, and of the experimental pathological changes induced by it. J Pathol 1970;100:217–244. [DOI] [PubMed] [Google Scholar]
- 105.Wang Q, Li J, Ji J, et al. A case of Naegleria fowleri related primary amoebic meningoencephalitis in China diagnosed by next-generation sequencing. BMC Infect Dis 2018;18:349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Linam WM, Ahmed M, Cope JR, et al. Successful treatment of an adolescent with Naegleria fowleri primary amebic meningoencephalitis. Pediatrics 2015;135:e744–e748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lam AH, De Silva M, Procopis P, et al. Primary amoebic (Naegleria) meningoencephalitis. J Comput Assist Tomogr 1982;6:620–623. [DOI] [PubMed] [Google Scholar]
- 108.Reyes-Batlle M, Wagner C, López-Arencibia A, et al. Isolation and molecular characterization of a Naegleria strain from a recreational water fountain in Tenerife, Canary Islands, Spain. Acta Parasitol 2017;62:265–268. [DOI] [PubMed] [Google Scholar]
- 109.da Rocha-Azevedo B, Tanowitz HB, Marciano-Cabral F. Diagnosis of infections caused by pathogenic free-living amoebae. Interdiscip Perspect Infect Dis 2009;2009:251406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jamerson M, Schmoyer JA, Park J, et al. Identification of Naegleria fowleri proteins linked to primary amoebic meningoencephalitis. Microbiology (Reading) 2017;163:322–332. [DOI] [PubMed] [Google Scholar]
- 111.Bellini NK, Santos TM, da Silva MTA, et al. The therapeutic strategies against Naegleria fowleri. Exp Parasitol 2018;187:1–11. [DOI] [PubMed] [Google Scholar]
- 112.El-Badry AA, Aufy SM, El-Wakil ES, et al. First identification of Naegleria species and Vahlkampfia ciguana in Nile water, Cairo, Egypt: seasonal morphology and phylogenetic analysis. J Microbiol Immunol Infect 2020;53:259–265. [DOI] [PubMed] [Google Scholar]
- 113.Benterki MS, Ayachi A, Bennoune O, et al. Meningoencephalitis due to the amoeboflagellate Naegleria fowleri in ruminants in Algeria. Parasite 2016;23:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Capewell LG, Harris AM, Yoder JS, et al. Diagnosis, clinical course, and treatment of primary amoebic meningoencephalitis in the United States, 1937–2013. J Pediatric Infect Dis Soc 2015;4:e68–e75. [DOI] [PubMed] [Google Scholar]
- 115.Mungroo MR, Khan NA, Siddiqui R. Naegleria fowleri: diagnosis, treatment options and pathogenesis. Expert Opin Orphan Drugs 2019;7:67–80. [Google Scholar]
- 116.Yu Z, Miller HC, Puzon GJ, et al. Development of untargeted metabolomics methods for the rapid detection of pathogenic Naegleria fowleri. Environ Sci Technol 2017;51:4210–4219. [DOI] [PubMed] [Google Scholar]
- 117.Hebbar S, Bairy I, Bhaskaranand N, et al. Fatal case of Naegleria fowleri meningo-encephalitis in an infant: case report. Ann Trop Paediatr 2005;25:223–226. [DOI] [PubMed] [Google Scholar]
- 118.Shenoy S, Wilson G, Prashanth HV, et al. Primary meningoencephalitis by Naegleria fowleri: first reported case from Mangalore, South India. J Clin Microbiol 2002;40:309–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Oncel K, Karaagac L, Dagci H, et al. Real-time PCR confirmation of a fatal case of primary amoebic meningoencephalitis in Turkey caused by Naegleria fowleri or brain-eating amoeba. Acta Parasitol 2022;67:697–704. [DOI] [PubMed] [Google Scholar]
- 120.Zhou W, Ouyang Y, Zhang D, et al. Case Report and Literature Review: Bacterial meningoencephalitis or not? Naegleria fowleri related primary amoebic meningoencephalitis in China. Front Pediatr 2022;10:785735. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.