Abstract
Whereas DNA viruses are known to be abundant, diverse, and commonly key ecosystem players, RNA viruses are insufficiently studied outside disease settings. In this study, we analyzed ≈28 terabases of Global Ocean RNA sequences to expand Earth’s RNA virus catalogs and their taxonomy, investigate their evolutionary origins, and assess their marine biogeography from pole to pole. Using new approaches to optimize discovery and classification, we identified RNA viruses that necessitate substantive revisions of taxonomy (doubling phyla and adding >50% new classes) and evolutionary understanding. “Species”-rank abundance determination revealed that viruses of the new phyla “Taraviricota,” a missing link in early RNA virus evolution, and “Arctiviricota” are widespread and dominant in the oceans. These efforts provide foundational knowledge critical to integrating RNA viruses into ecological and epidemiological models.
RNA viruses of 47 of 103 established families included in the riboviriad (with RNA genomes) kingdom Orthornavirae [orthornavirans; encoding an RNAdirected RNA polymerase (RdRp) for replication] have been studied deeply and mechanistically for their roles in human, live-stock, and plant diseases (1–3). The remaining viruses are less well studied because they infect less economically critical but nevertheless ecologically essential organisms, such as in-vertebrates, fungi, protists, and bacteria. Not surprisingly, virus discovery efforts, largely by using environmental RNA sequencing, have recently forced drastic changes in our understanding of orthornaviran diversity and evolution (4–7). Specifically, these studies have expanded diversity within known orthornaviran groups (4–6), revealed altered genome architecture among viruses with broad host ranges (4), and posited large host range jumps as driving much of orthornaviran evolution (8, 9).
Because the gene encoding RdRp is ancient, thought to be among the first genes of the peptide-RNA world (10–12), it serves as a deep evolutionary gene marker and is often used to understand orthornaviran origins and more generally to explore the origins of life (7, 12–15). Recently, RdRp-inferred orthornaviran evolutionary relationships resolved five major branches (7), which were subsequently recognized by the International Committee on Taxonomy of Viruses (ICTV) as five phyla (16, 17). This five-branch phylogenetic structure that underpins current orthornaviran megataxonomy was hypothesized to be stable, and the question of whether phylum-rank diversity was saturated was opened (5, 17). Beyond taxonomy, the evolutionary origins of orthornavirans, because of challenges in deep phylogenetic inferences (18), remain contentious, puzzling, and complex (19–21). Also problematic is that environmental surveys lack scalable and systematic approaches to taxonomically classify new data and assess their impact on our understanding of orthornaviran evolution
In this study, we update several key analytics and apply these to ≈28 terabases (Tb) of Global Ocean RNA metatranscriptome sequences to identify and characterize previously unknown RNA viruses and use them to (i) test hypotheses about orthornaviran megataxonomy stability and evolutionary origins and (ii) establish baseline planetary-scale ocean biogeographic context.
Marine RNA viruses double known orthornaviran phyla from 5 to 10
Given how little RNA virus diversity is explored in the Global Ocean (tables S1 and S2), we sought to leverage systematically collected and globally distributed Tara Oceans resources (table S3). These include RNA-sequencing data from 771 metatranscriptomes (table S4 for sample metadata) that span 10 organismal size fractions (fig. S1), three ocean layers, and 121 locations distributed throughout the world’s five oceans and include ≈6 Tb of new sequencing data from 143 metatranscriptomes obtained throughout the Arctic Ocean (Fig. 1A and table S4). To maximize our inferences from these metatranscriptomes, we developed and/or improved and benchmarked methods for the identification, classification, and organization of the orthornaviran genome–derived sequence space.
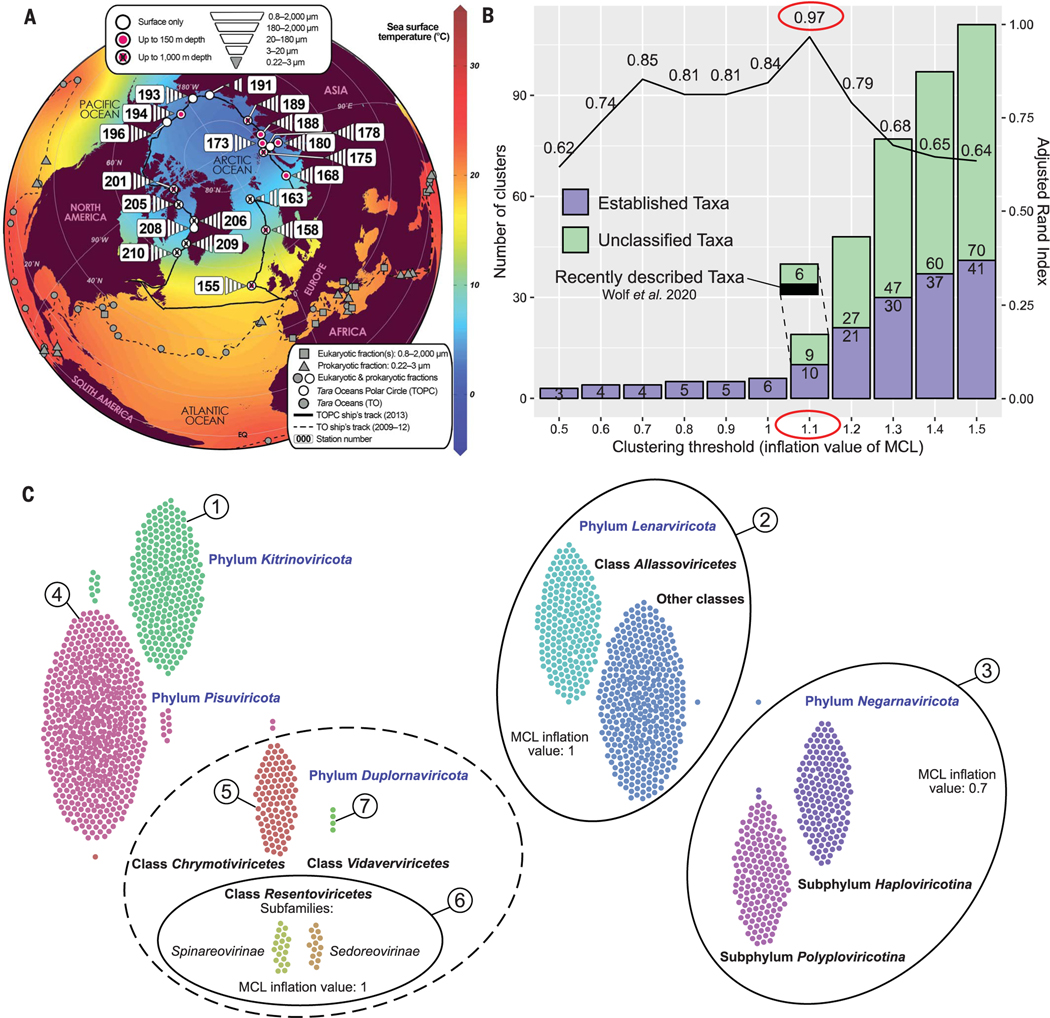
Fig. 1. Establishment of RdRp domain megaclusters.
(A) Arctic projection of the Global Ocean highlighting the new size-fractionated metatranscriptomes described here (white polygons). Gray symbols indicate previously published metatranscriptomes, whereas numbered stations indicate circumpolar Arctic Ocean data. Sea surface temperature gridding was done by using the weighted-average method in Ocean Data View (43) from the in situ temperature measurements collected during Tara expeditions. TO, Tara Oceans; TOPC, Tara Oceans Polar Circle. (B) Percent agreement (line) of our network-guided and phylogeny-based megataxonomy at different clustering thresholds (materials and methods). Stacked bars represent the number of taxonomic clusters of near-complete RdRp domains (at least 90% of the domain) (materials and methods) at these different clustering thresholds. Only sequences representing established taxa (violet) were used for calculating the agreement percentage. At an inflation value of 1.1, three (black box) of the nine unclassified clusters have been recently described by Wolf et al. (5), bringing the number of new major taxa in our study to six. (C) Swarm plot of the 10 ICTV-established taxa emerging at an inflation value 1.1 in the Markov Clustering Algorithm (MCL) analysis [from (A)]. Solid lines encompass taxa that were exclusively joined at a lower inflation value, as indicated within each ellipse. The dashed line encompasses the three established duplornaviricot classes, which were never exclusively joined at lower inflation values. Dots that have the same color but are not part of their swarm represent discrepancies from GenBank taxonomy (aligned vertically with the cluster that recruited them in the network). The resultant seven clusters (numbered) along with the six new clusters from our study (A) were used to build the 13 individual phylogenetic trees in Fig. 2A. Phylum Kitrinoviricota encompasses two of the three recently described unclassified megaclusters (A) at an MCL inflation value of 1. The third megacluster represents viruses with permuted motifs in the RdRp domain (“permutotetra-like” and “birna-like” viruses) and hence was excluded from phylogenetic analyses.
We first searched our Global Ocean data for nucleic acids that encode RdRps, which are specific to orthornavirans and have no known relationship to cellular RdRps (22) or DNA-directed RNA polymerases (23). Given notoriously divergent RdRp sequences, we maximized RdRp identification by means of an iterative search-and-update hidden Markov model (HMM) approach that we improved and automated in our work (supplementary materials, materials and methods, and fig. S2). This approach identified 44,779 RdRp-encoding contigs (after removing 134 false positives) (materials and methods and fig. S2C) (details per contig are available in table S5), a ≈26-fold improvement over standard BLAST (Basic Local Alignment Search Tool)–based approaches (fig. S2G). Of these 44,779 contigs, 6686 encoded complete or near-complete RdRp domain sequences (≥90% completeness) (materials and methods).
Because the oceans are vastly undersampled for orthornavirans, we sought to assess how these new data compared with the current five-branch understanding of orthornaviran megataxonomy (7). This introduced our second major analytical challenge because although this phylogeny-based unified framework was groundbreaking, RdRp phylogenies are complex and require a manual and stepwise approach for construction, including a laborious iterative process of multiple sequence alignments, manual refinement, tree building, and representative selections to establish the global phylogeny. We worried that as seen in the literature (7, 24), subjectivity in the iterative manual curation step could lead to varied perspectives on orthornaviran evolutionary inferences. Thus, to mitigate these concerns, we developed and benchmarked a scalable, network-based, iterative clustering approach to assess RdRp diversity; once performed, it nearly completely recapitulated the previously established phylogeny-based ICTV-accepted taxonomy (7, 17) at the phylum and class ranks (97% agreement) (Fig. 1, B and C, and materials and methods).
With this approach, we then evaluated the Global Ocean data to classify the subset with complete or nearly complete RdRp domains and assess their novelty. Joint analysis of 111,760 complete or nearly complete RdRp domain sequences from all available (terrestrial and oceanic) viruses—6686 from our dataset, 101,819 from GenBank [release 233; only 3850 established species (25), indicating high species-rank redundancy] (materials and methods), and 3255 fromcoastal ocean RNAviromes (5)—revealed 19 “megaclusters” (Fig. 1B and table S6). Whereas our dataset represents only ≈6% of the total sequences in this analysis, our data covered vast diversity across the RNA orthovirosphere as follows (Fig. 2 and fig. S3): 13 of the 19 megaclusters from our analysis were known previously; together they compose the five ICTV-recognized phyla of the orthornaviran megataxonomy (17), with oceanrepresentative viruses for all five established phyla, all 20 established classes, and 49 of 103 established families (Fig. 2 and figs. S3 and S4). Although “known” at these taxon ranks, virtually all (99.7%) of the ocean viruses that could be evaluated represent new species (determined from whole-genome or contig information as described later) (table S5) that substantially augment undersampled taxa, because as much as 70% of sequences for some families were ocean derived (fig. S4A and table S7).
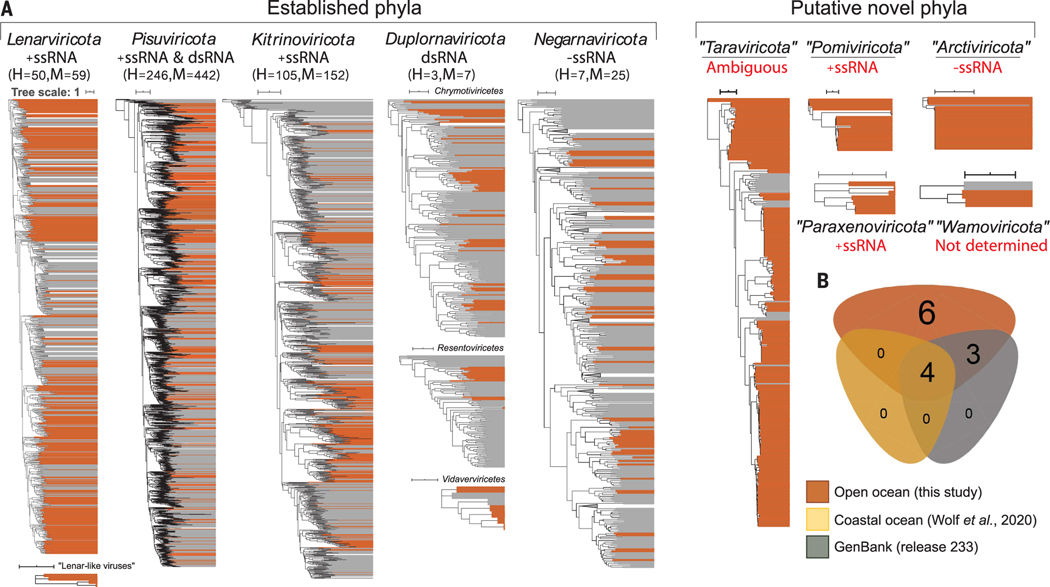
Fig. 2. Phylum- and class-rank RdRp-based phylogenetic analyses showing the taxonomic diversity of Global Ocean orthornavirans.
(A) Thirteen maximum-likelihood phylogenetic trees encompassing the 19 megaclusters that emerged from network analyses of near-complete RdRp sequences (details in Fig. 1). Brown color indicates virus sequences discovered in this study, whereas gray indicates previously known reference sequences. The scale bar indicates one amino acid substitution per site. Classes were merged into a unified phylum-ranked tree only if the results from both phylogeny and network-guided clustering analysis were in agreement (materials and methods). Sequences were preclustered at 50% identity, and clades supported by 100% bootstrap values were collapsed. Genome strandedness (red text) for the new phyla was inferred in this study (as described in fig. S8 and materials and methods). A conservative estimate of the number of new complete or high-quality (H) and medium-quality (M) genomes retrieved in this study is indicated with parentheses. Underlined new phyla are supported by long- and short-read assemblies, whereas the remainder were supported by multiple independent assemblies from short-read assemblies (domain motifs are available in table S10). (B) Euler diagram of the shared, well-resolved phylum-or class-rank clusters of the near-complete RdRp domains across all available data from GenBank, a prior coastal ocean survey, and this study. Established megataxa represented in all datasets are Lenarviricota, Pisuviricota, Kitrinoviricota, and Duplornaviricota; Chrymotiviricetes. Established megataxa represented in our dataset and GenBank are Duplornaviricota; Vidaverviricetes, Duplornaviricota; Resentoviricetes, and Negarnaviricota. Unestablished megataxa inferred in this study are “Taraviricota,”“Pomiviricota,” “Paraxenoviricota,”“Arctiviricota,”“Wamoviricota,” and “lenar-like viruses.” In all analyses, RdRp domain clusters with permuted motifs (“permutotetra-like” and “birna-like” viruses) were excluded.
Beyond these more established taxa of the five-phylum system, 6 of the 19 megaclusters from our analysis were new (hereafter indicated with double quotation marks) and dominated by Global Ocean RdRps (Fig. 2A and data S1 and S2) (explanations for the suggested names are provided in the supplementary materials, materials and methods). In the current orthornaviran megataxonomic framework (17), these six clusters would correspond to five new phyla, which we suggest to call “Arctiviricota,” “Paraxenoviricota,” “Pomiviricota,” “Taraviricota” [includes the 22 previously identified “quenyaviruses” (24) with near-complete RdRp domains], and “Wamoviricota,” as well as a new lenarviricot class, which we refer to here as “lenar-like viruses.” Manual sequence inspection revealed that three of seven canonical RdRp motifs (26) are missing from members of this class-rank megataxon. Cluster-specific phylogenetic analyses (data S3) revealed that some virus groups were well represented in the oceans and else-where (such as ICTV-recognized pisuviricots), whereas others were primarily (“taraviricots”) orexclusively(“pomiviricots,” “paraxenoviricots,” “arctiviricots,” and “lenar-like viruses”) oceanic (Fig. 2A).
To further assess the validity of our RdRp-inferred five new phyla, we evaluated phylogenetic (primary sequences) (Fig. 3A) and three-dimensional (3D) alignment (predicted and resolved tertiary structures) (Fig. 3B, fig. S5, and table S8) analyses of the RdRp domain, as well as other genomic features for which data were available (such as domain enrichments outside the RdRp, available for 7 of the 10 phyla) (table S9). In all cases, the network-derived clusters were supported by the phylogenetic and 3D-structure network information and contained features (statistically significant enrichment of domains outside the RdRp) (complete list is provided in table S9) that are consistent with variation observed at the established phylum rank. Marine representatives from established families have genome organizations similar to those from nonmarine taxa, whereas virus contigs of new phyla and classes were poorly annotated beyond the RdRp domains (figs. S6 and S7 and table S9). Together, these findings further suggest that the Global Ocean sequences add five phyla to the five already established as well as increase the number of known orthornaviran classes >50% by adding at least 11 classes (figs. S3 and S7) within previously established phyla. This expands the current megataxonomic framework beyond a stable five-phylum structure (5, 17) and invites further exploration of its sequence space.
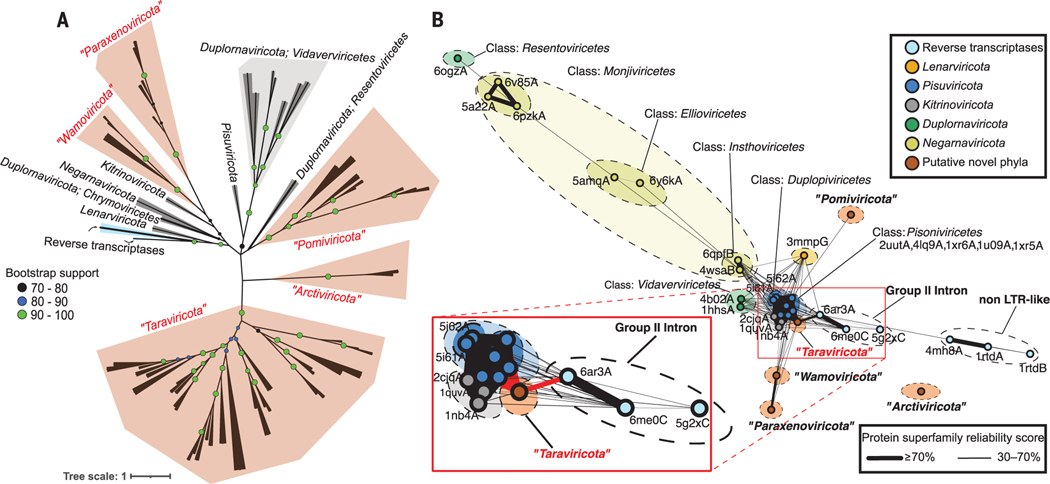
Fig. 3. Global RdRp-based phylogeny and network analyses inferring the early evolutionary history of orthornavirans.
(A) Maximum-likelihood phylogenetic tree of RdRp domain sequences with RT sequences (cyan). The gray branches and polygons represent established megataxa, whereas the brown polygons represent megataxa inferred here. Each branch represents either a consensus or an individual sequence from a megataxon (materials and methods). Nodes in each branch represent bootstrap support. The scale bar indicates one amino acid substitution per site. (B) Three-dimensional structure similarity network of predicted (brown) and experimentally resolved (other colors; labeled with accession numbers) RdRp and RT protein domain structures. Each node represents a different structure, and the edges represent the reliability scores, for each connected pair, that they belong to the same protein superfamily (materials and methods). (Inset) The probability of “taraviricot” RdRps belonging to the same superfamily as group II–intron RTs and pisuviricot RdRps is 75 and 98%, respectively. In all analyses, RdRp domain clusters with permuted motifs (“permutotetra-like” and “birna-like” viruses) were excluded. LTR, long terminal repeat.
Marine RNA viruses revise the early evolution of orthornaviran megataxa
RdRp domain–based phylogeny has been used to infer deep orthornaviran evolutionary history (7), with different opinions on its robustness for this purpose (21, 24, 27) owing to the challenges of assigning homology in highly divergent primary sequences (28, 29). The deepest parts of the RdRp phylogenetic tree are controversial (21, 27) because only 55 of 441 sites showed an alignment homogeneity score ≥0.3 (as compared with 128 or more such sites for more broadly accepted phyla) (27). Although controversial and challenging, we interpret current literature to suggest that RdRp primary-sequence inferences lack confidence for interphyla relationships (7, 21, 24, 27) but do suggest most phyla appear monophyletic (27). Given the extensive, new orthornaviran diversity, we revisited these deep evolutionary inferences using primary sequence–inferred phylogeny but also other features such as RdRp 3D structures and network-based clusters, other genomic domains, and whole-genome characteristics.
First, we assessed the monophyletic origin of double-stranded RNA (dsRNA) viruses of Duplornaviricota, which is one of the five orthornaviran phyla thought to have more recently evolved from positive-sense single-stranded RNA (+ssRNA) viruses (7). Previously, all viruses in Duplornaviricota were placed in a single phylum with three classes because Duplornaviricota and Negarnaviricota were strongly monophyletic [Duplornaviricota and Negarnaviricota are labeled as branches 4 and 5, respectively, in (7, 17)]. However, reexamination of alignment homogeneity from previous work (27) suggests that these taxa are polyphyletic because (i) only 72 sites within the duplornaviricot sequence alignment showed homogeneity ≥0.3 as compared with at least 128 sites for sequences from the other phylaand (ii) Duplornaviricota showed a paraphyletic relationship with respect to Negarnaviricota (7), which hinted toward accommodating Duplornaviricota taxonomically by at least three phyla (7, 17). Our global phylogenetic tree also suggests, with strong support, that these dsRNA viruses are polyphyletic (Fig. 3A). The Duplornaviricota polyphyly we observed is further supported by (i) the lack of strong duplornaviricot intertaxon connections in our 3D structure network (Fig. 3B), (ii) the absence of a homogeneous cluster encompassing these taxa that are emerging from our iterative clustering approach (Fig. 1), and (iii) differential extraneous-to-RdRp domain enrichment across these taxa (table S9). Hence, the grouping of all dsRNA viruses (apart from the class Duplopiviricetes) into one phylum (Duplornaviricota), as established currently(7), appears incorrect. Instead, we suggest—as the ICTV has done for +ssRNA viruses that were recently split into three phyla [Lenarviricota, Pisuviricota, and Kitrinoviricota; also supported by our data (Figs. 2 and 3)] (7)—that Duplornaviricota represent three different phyla along the lines of the currently recognized classes. If ultimately ICTV approved, this would expand currently known diversity to a total of 12 phyla.
The second deep evolutionary orthornaviran inference we assessed was the proposition that negative-sense single-stranded RNA(−ssRNA) viruses (phylum Negarnaviricota) evolved from the dsRNA duplornaviricots, which is considered a low-confidence link in the literature (7, 17, 27). Our global phylogenetic tree also indicates a last common ancestor of negarnaviricots and one of the dsRNA virus “classes,” but we found the well-supported sister taxon to be the dsRNA “class” Chrymotiviricetes (Fig. 3A), as opposed to the prior observed “class” Resentoviricetes (7). Because such deep evolutionary phylogenetic inferences are prone to long branch attraction artefacts, we evaluated other lines of evidence. This revealed that these prior proposed relationships were not supported in (i) our 3D structure network (only Resentoviricetes was connected, and only weakly, to Negarnaviricota) (Fig. 3B) or (ii) our iterative primary sequence–based clustering approach (the two taxa never formed a homogeneous cluster) (Fig. 1). Additionally, domain enrichment analysis (table S9, section B) showed that negarnaviricots did not share any domains with dsDNA viruses but did share a virus-capping methyltransferase domain (Pfam: PF14314) with >50 viruses classified in Pisuviricota and Kitrinoviricota (table S9). When we examined the suggested phyla for their “strandedness” (materials and methods and fig. S8), which helps identify the virus genome type (+ssRNA, −ssRNA, or dsRNA), “Arctiviricota” emerged as −ssRNA. Both phylogenetic (Fig. 3A) and 3D structure network (Fig. 3B) analyses suggest that “arctiviricots” evolved independently from negarnaviricots (and dsRNA viruses) and represent a second −ssRNA phylum and further polyphyly within the orthornavirans. These findings argue that all orthornaviran genome types (+ssRNA, −ssRNA, and dsRNA viruses) have multiple evolutionary origins.
Third, we revisited the RdRp primary sequence–inferred hypothesis that considers orthornavirans monophyletic and assumes reverse transcriptases (RTs) of retroelements as the root of the global RdRp tree (7). In that scenario, lenarviricots (some of which infect bacteria and carry capsid proteins) are a sister group to the remaining orthornavirans, and retroelements appear more likely (and parsimoniously) to be ancestral to orthornavirans (7), arguing against the emergence of virus RdRp in the peptide-RNA world (12, 30). Instead, our RdRp phylogeny revealed lenarviricot RdRps sharing ancestry with RTs (well supported) (Fig. 3A and data S4), which (assuming amonophyletic origin of orthornavirans) suggests a capsidless RNA replicon as the ancestor of both retroelements and RNA viruses and agrees with the thinking that virus RdRps were part of the earlier peptide-RNA world. Lenarviricota harbors the short (<5 kb) capsidless RNA replicons (mitovirids that carry only an RdRp, infect eukaryotes, and replicate in host mitochondria).
An alternative scenario, however, was inferred from 3D structure analyses, which are often considered more informative than primary sequence information for deep evolutionary inferences (31). These analyses suggest, with high calculated probability (materials and methods), that viruses from our suggested phylum “Taraviricota” represent a missing link between retroelements (riboviriad pararnavirans) and orthornavirans (Fig. 3B). If true, this implies that “Taraviricota” RdRp represents the capsidless RNA replicon ancestor of retroelements and orthornaviran RdRps—potentially the RdRp replicon postulated to have originated from junctions of proto-tRNAs (11, 12). To evaluate this scenario further, we examined genomic information of “taraviricots” as follows.
First, similar to mitovirids (phylum Lenarviricota), all but four of the marine “taraviricots” that were recovered from short- (n = 220) or long-read (n = 32) assemblies (Fig. 2A) have short genomes (<3.4 kb) (fig. S7) and encode only RdRp. Noother well-sampled(>10viruses) phylum in our dataset showed such a feature, which we interpret to be due to either short virus genome length or consistent genome segmentation [ “quenyaviruses” always encode RdRp on its own segment (24)]. If the former is true—that most “taraviricots” have short genomes—it implies that orthornavirans evolved from an RdRp-only ancestor through gene gains (and potential later losses) (7). If the latter is true, then genome segmentation in orthornavirans evolved early and potentially contributed to an accelerated early diversification of orthornavirans (Fig. 3A, “Taraviricota”). Genome segmentation is not common among lenarviricots, and many of its non-segmented lineages encode single jelly-roll capsid proteins that were hypothesized (although, notably, unparsimoniously) to be horizontally transferred from viruses of other phyla (7). Both of these observations support our alternative 3D structure–inferred scenario presented here.
Second, of the four marine “taraviricots” encoding more than just RdRp, two encoded only a putative phospholipase [Pfam, PF11618 (CL14603) or PF02230 (CL0028); not found in any other orthornaviran (table S9)]. This observation suggests that at least some “taraviricots” ancestrally or currently infect a cell wall–deficient prokaryotic host or the mitochondria of eukaryotes (sensu mitovirids). Although this link is still speculative, we interpret this finding—together with “taraviricots” overwhelmingly encoding just the RdRp on very short genomes and/or potential consistent genome segmentation and their 3D structure resemblance to multiple orthornaviran types (+ssRNA and dsRNA) and RTs—to provide a parsimonious scenario for “Taraviricota” as an early basal lineage from which other orthornaviran phyla have subsequently evolved.
Collectively, we sought to reevaluate deep evolutionary inferences using multiple data types beyond primary sequence, and these analyses suggest (i) polyphyletic origins of dsRNA “phylum” Duplornaviricota (splitting it into three different phyla) and −ssRNA phyla (Negarnaviricota and “Arctiviricota”) and (ii) an ancient presence of “taraviricots” on Earth, with a potential important role in the orthornaviran and pararnaviran evolution.
Abundance and biogeography of orthornaviran “species”
Given this extensive, new orthornaviran diversity, we next sought to biogeographically contextualize it globally, at least for the oceans. Such analyses are possible because of two major advances: (i) systematic Tara Oceans’ global sampling (table S4) and (ii) a recent consensus approach (32) that establishes virus operational taxonomic units (vOTUs; a species-rank approximation) by evaluating genomic sequence space for discontinuities. Applying this approach to our whole-genome and contig data revealed such a discontinuity, although at different cutoffs supported by our sensitivity analyses (fig. S9 and materials and methods). The empirically derived vOTU definition suggested from these analyses was 90% average nucleotide identity over 80% coverage of the smaller contig and ≥1 kb in length. Dereplicating our 44,779 virus contigs at this cutoff revealed 5504 vOTUs (vOTU contig length range of 1001 to 25,584 nucleotides, with a median of 1958) (table S5). Of these 5504 vOTUs, a subset (n = 624) is related enough to known complete virus genomes that we can estimate their completeness—433 high-quality or complete genomes (belonging to 188 vOTUs), 719 medium-quality genomes (belonging to 246 additional vOTUs), and 807 low-quality genomes (belonging to 190 additional vOTUs)—whereas the remainder (n = 4880) are so divergent from reference genomes that their completeness cannot be estimated by using available approaches (table S5). Virtually all of these vOTUs (n = 5485; 99.7%), including those with at least medium-quality genomes (n = 430; 99.6%), belong to new species (table S5). Additionally, to compare our methods with those that rely on just the RdRp domain sequences for vOTU construction [for example, (33)], we examined a range of clustering and contig length cutoffs (materials and methods) and found general and robust agreement for contigs ≥1 kb in length (at least 93% agreement) (fig. S9 and materials and methods). Hence, our vOTU definition both respects RdRp-inferred relationships among individual contigs in a cluster and expands on them by including genomic information to resolve ambiguity in RdRp-based identity cutoffs (fig. S9).
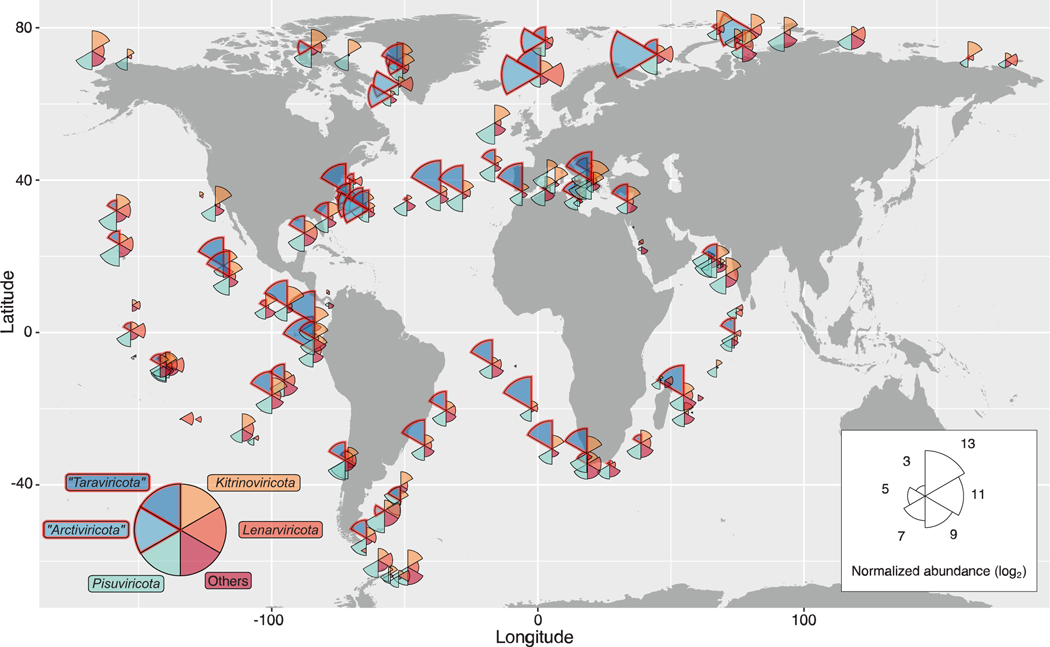
Given this robustness, we quantified vOTUs by means of read mapping to assess abundance and global biogeography across the 771 Global Ocean meta transcriptomes (materials and methods). This revealed three phyla—Pisuviricota, Kirinoviricota, and “Taraviricota”—as collectively abundant and widespread (fig. S10). The first two phyla include “picorna-like” and “tombus-like” viruses commonly found in site-focused surveys (34, 35), whereas the third phylum (“Taraviricota”) consists of at least 220 previously unknown viruses (with near-complete RdRp domain sequences) described here. This phylum’s vOTUs were, on average, the most abundant across most temperate and tropical waters (Fig. 4). This finding suggests ecological importance for these previously overlooked viruses and provides broader context for previously described viruses (“quenyaviruses”) that were found to be abundant in some arthropods and other animals (24) and are now more clearly recognized as members of the most abundant ocean orthornaviran phylum. Although with more restricted geographic range, vOTUs belonging to the −ssRNA phylum “Arctiviricota” were, on average, the most abundant across most of the Atlantic Arctic waters (Fig. 4). None of the other −ssRNA viruses (negarnaviricots) showed similar patterns in any area of the ocean, suggesting a specific ecological footprint for the “arctiviricots” described here. Although the biogeographic data shown here represent relative abundances of a mixture of abundances derived from genomes and transcripts, the relative abundances of “Taraviricota” and “Arctiviricota” are likely mostly derived from their genomes (fig. S8). Together, these data provide an orthornaviran-wide, systematically sampled, and large-scale complement to prior RNA virus diversity studies in the ocean (24, 33–35).
Fig. 4. Biogeography of orthornaviran megataxa.
Global map showing the distribution and average relative abundance (on a log2 scale) of vOTUs inferred in this study per phylum. The position and color of the wedges are fixed for the same megataxon across the Global Ocean. Wedge lengths are proportional to the average abundance in the sample as well as across the global dataset. Biogeography per size fraction is provided in fig. S11.
Last, having established this environmental context and vast ocean-derived orthornaviran diversity, we sought to identify their hosts. Unfortunately, host identification for environmental RNA virus contigs is challenging, which limits us to reporting only domain-rank hosts for the new megataxa from multiple analytical approaches that include preestablished host linkages to previously known RNA virus taxa, abundance-based co-occurrence networks, and screening of endogenous virus elements (materials and methods). Results from this effort revealed that viruses of “Taraviricota,” “Arctiviricota,” “Pomiviricota,” “Wamoviricota,” and eight of the new classes are associated with eukaryotes (table S11), whereas only pisuviricot class 27 viruses likely infect prokaryotes (table S12). The latter finding of infecting prokaryotes is rare but not unknown for RNA viruses and is supported by a statistically significant signal of Shine-Dalgarno motifs (table S12 and materials and methods) and one of the representative virus genomes encoding a putative preprotein translocase subunit SecY of the bacterial type II secretion system (fig. S7). The remaining new megataxa (one phylum and two classes) could not be associated with hosts. Together, these findings suggest that eukaryotes remain the main hosts of orthornavirans but suggest addition of our new pisuviricot class 27 to known RNA phage groups alongside levivirids (phylum Lenarviricota), cystovirids (phylum Duplornaviricota), and potentially (36) picobirnavirids (phylum Pisuviricota).
Conclusions
Although clear population- and genomere-solved approaches have been developed for dsDNA viruses and revealed the existence of hundreds of thousands of distinct dsDNA virus species in the oceans alone (37), few parallel studies for RNA viruses exist—despite urgent needs (38) and suggestions that our understanding of the virosphere will increase with the study of microbial eukaryotes (4, 5). Our study and several prior studies (4, 5, 39) confirm this suggestion and are now reshaping our understanding of RNA virus diversity and evolution, with thousands of previously unknown RNA virus species presented in this study alone. Although documentation of such RNA virus diversity might now be scalable to that observed in nature, several challenges need to be addressed. These include (i) identifying hosts for previously undiscovered viruses, (ii) scalably improving genome completeness in survey approaches, and (iii) directly capturing RNA virus particles from environmental samples to assess their diversity in a targeted manner and complement the host metatranscriptomic sequence space–based abundance calculations presented in this study. Although challenges remain, the global and systematic effort presented here provides critical information and resources, an analytical roadmap, and foundational advances to feed the predictive models that are needed to assess RNA virus ecosystem, eco-evolutionary, and epidemiological impacts.
Supplementary Material
ACKNOWLEDGMENTS
We thank Y. I. Wolf (National Center for Biotechnology Information, U.S. National Library of Medicine, National Institutes of Health) for advice and guidance in analyzing RdRp sequences and A. Crane (Integrated Research Facility at Fort Detrick, National Institute of Allergy and Infectious Diseases, National Institutes of Health) for critically editing the manuscript. Tara Oceans would not exist without the leadership of the Tara Expeditions Foundation and the continuous support of 23 institutes. The extensive Tara Oceans expeditionary support is detailed in the supplementary text.
Funding:
The virus-specific work presented here was supported in part through the following: Gordon and Betty Moore Foundation (award #3790); US National Science Foundation (awards OCE#1829831, ABI#1759874, and DBI#2022070); The Ohio Supercomputer and Ohio State University’s Center of Microbiome Science; Ramon-Areces Foundation Postdoctoral Fellowship to G.D.-H.; Laulima Government Solutions, LLC prime contract with the U.S. National Institute of Allergy and Infectious Diseases (NIAID—Contract No. HHSN272201800013C); SNSF project (grant 205321_184955 to S.S.); and France Génomique for funding for the sequencing (ANR-10-INBS-09) (P.W.).
Footnotes
Competing interests: The authors declare that they have no competing interests.
Data and materials availability:
The authors declare that all data reported here are fully and freely available from the date of publication without restrictions and that all of the analyses, publications, and ownership of data are free from legal entanglement or restriction by the various nations whose waters were sampled during the Tara Oceans expeditions. This article is contribution number 129 of Tara Oceans. Newly generated raw sequence reads for the 143 eukaryote-size fraction metatranscriptomes from the Arctic Ocean are available at ENA/SRA under BioProjectID PRJEB9738 and PRJEB9739. Processed data are publicly available through iVirus (40), including all metatranscriptome assemblies, RNA virus contigs and vOTUs, RdRp sequences and clusters, and HMM profiles. Multiple sequence alignments and phylogenetic trees are publicly available through DRYAD (41), whereas the HMM pipeline developed in this work can be accessed through Zenodo (42). In addition, scripts used to generate figures are uploaded to the MAVERICKlab bitbucket page (https://bitbucket.org/MAVERICLab/global-rna-virus-evolution-2021).
REFERENCES AND NOTES
- 1.Woolhouse MEJ, Brierley L, Sci. Data 5, 180017 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scholthof K-BG et al. , Mol. Plant Pathol. 12, 938–954 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brun A, Methods Mol. Biol. 1349, 1–24 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shi M. et al. , Nature 540, 539–543 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Wolf YI et al. , Nat. Microbiol. 5, 1262–1270 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi M. et al. , Nature 556, 197–202 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Wolf YI et al. , mBio 9, e02329–e18 (2018). [Google Scholar]
- 8.Krupovic M, Dolja VV, Koonin EV, Biol. Direct 10, 12 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dolja VV, Koonin EV, Virus Res. 244, 36–52 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carter CW Jr., Life 5, 294–320 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chatterjee S, Yadav S, Life 9, 25 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pereira Dos Santos A Jr., José MV, Torres de Farias S, Biosystems 206, 104442 (2021). [DOI] [PubMed] [Google Scholar]
- 13.de Farias ST, Dos Santos AP Jr., Rêgo TG, José MV,Front. Genet. 8, 125 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forterre P, Virus Res. 117, 5–16 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Forterre P, Prangishvili D, Res. Microbiol. 160, 466–472 (2009). [DOI] [PubMed] [Google Scholar]
- 16.Kuhn JH et al. , Nature 566, 318–320 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koonin EV et al. , Microbiol. Mol. Biol. Rev. 84, e00061–e19 (2020). [Google Scholar]
- 18.Whitfield JB, Lockhart PJ, Trends Ecol. Evol. 22, 258–265 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Krupovic M, Dolja VV, Koonin EV, Nat. Rev. Microbiol. 18, 661–670 (2020). [DOI] [PubMed] [Google Scholar]
- 20.Kristensen DM, Mushegian AR, Dolja VV, Koonin EV,Trends Microbiol. 18, 11–19 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holmes EC, Duchêne S, mBio 10, 1–2 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burroughs AM, Ando Y, Aravind L, Wiley Interdiscip. Rev. RNA 5, 141–181 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iyer LM, Koonin EV, Aravind L, BMC Struct. Biol. 3, 1 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Obbard DJ, Shi M, Roberts KE, Longdon B, Dennis AB,Virus Evol. 6, vez061 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dance A, Nature 595, 22–25 (2021). [DOI] [PubMed] [Google Scholar]
- 26.te Velthuis AJW, Cell. Mol. Life Sci. 71, 4403–4420 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wolf YI et al. , mBio 10, e00542–e19 (2019). [Google Scholar]
- 28.Rost B, Protein Eng. 12, 85–94 (1999). [DOI] [PubMed] [Google Scholar]
- 29.Zanotto PM, Gibbs MJ, Gould EA, Holmes EC, Virol J. 70, 6083–6096 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Farias ST, Rêgo TG, José MV, Life (Basel) 6, 15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Illergård K, Ardell DH, Elofsson A, Proteins 77, 499–508 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Roux S. et al. , Nat. Biotechnol. 37, 29–37 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gustavsen JA, Winget DM, Tian X, Suttle CA,Front. Microbiol. 5, 703 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Culley AI, Lang AS, Suttle CA, Science 312, 1795–1798 (2006). [DOI] [PubMed] [Google Scholar]
- 35.Culley A, Virus Res. 244, 84–89 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Ghosh S, Malik YS, Front. Vet. Sci. 7, 615293 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gregory AC et al. , Cell 177, 1109–1123.e14 (2019). [Google Scholar]
- 38.French RK, Holmes EC, Trends Microbiol. 28, 165–175 (2020). [DOI] [PubMed] [Google Scholar]
- 39.Li C-X et al. , eLife 4, e05378 (2015). [Google Scholar]
- 40.Zayed AA, Wainaina JM, Dominguez-Huerta G,Cryptic and abundant marine viruses at the evolutionary origins of Earth’s RNA virome. CyVerse Data Commons (2021). [DOI] [PMC free article] [PubMed]
- 41.Wainaina JM, Zayed AA, Dominguez-Huerta G, Bolduc B,Sullivan MB, Supporting trees and alignments for the publication: Cryptic and abundant marine viruses at the evolutionary origins of Earth’s RNA virome. DRYAD; (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guo J, rdrpsearch, Version 0.1.Zenodo (2021).
- 43.Schlitzer R, Ocean Data View (2018); https://odv.awi.de/.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all data reported here are fully and freely available from the date of publication without restrictions and that all of the analyses, publications, and ownership of data are free from legal entanglement or restriction by the various nations whose waters were sampled during the Tara Oceans expeditions. This article is contribution number 129 of Tara Oceans. Newly generated raw sequence reads for the 143 eukaryote-size fraction metatranscriptomes from the Arctic Ocean are available at ENA/SRA under BioProjectID PRJEB9738 and PRJEB9739. Processed data are publicly available through iVirus (40), including all metatranscriptome assemblies, RNA virus contigs and vOTUs, RdRp sequences and clusters, and HMM profiles. Multiple sequence alignments and phylogenetic trees are publicly available through DRYAD (41), whereas the HMM pipeline developed in this work can be accessed through Zenodo (42). In addition, scripts used to generate figures are uploaded to the MAVERICKlab bitbucket page (https://bitbucket.org/MAVERICLab/global-rna-virus-evolution-2021).