Abstract
New series of escape mutants of human respiratory syncytial virus were prepared with monoclonal antibodies specific for the fusion (F) protein. Sequence changes selected in the escape mutants identified two new antigenic sites (V and VI) recognized by neutralizing antibodies and a group-specific site (I) in the F1 chain of the F molecule. The new epitopes, and previously identified antigenic sites, were incorporated into a refined prediction of secondary-structure motifs to generate a detailed antigenic map of the F glycoprotein.
Human respiratory syncytial virus (HRSV) is the major cause of severe respiratory infections in very young children (reviewed in reference 10). Viral isolates have been classified into two antigenic groups (A and B) on the basis of differences in reactivity with panels of monoclonal antibodies (1, 27). Like other paramyxoviruses, HRSV encodes two major surface glycoproteins (G and F), which are incorporated in the virus particle. The attachment protein (G) mediates virus binding to the cell receptor (21), and the fusion protein (F) promotes fusion of the viral and cell membranes, allowing penetration of the viral ribonucleoprotein into the cell cytoplasm (43). The F protein also promotes fusion of the membranes of infected cells with those of adjacent cells, leading to the formation of syncytia.
Antibodies directed against either G or F neutralize virus infectivity. Furthermore, experimental animals immunized with vaccinia virus recombinants expressing either antigen are protected against challenge with live HRSV (29, 37). However, whereas the immune response against the F protein protects the animals against viruses of both antigenic groups, the G protein induces a homotypic response protective only against viruses of the same antigenic group. These results reflect the extensive antigenic and genetic divergence in the G protein between group-A and group-B viruses (16), in contrast to the high degree of conservation of the F glycoprotein (17).
The F glycoprotein is synthesized as an inactive precursor (F0) (14) that is cotranslationally modified by the addition of N-linked carbohydrates in the endoplasmic reticulum, where it assembles into a homooligomer (probably a tetramer) (9). The F0 precursor is cleaved by trypsin-like proteases into two chains (F2 N-terminal to F1) before reaching the cell surface. The two chains remain disulfide linked. The F protein also contains palmitate (9).
Several laboratories have reported the isolation of monoclonal antibodies directed against the F protein that neutralize virus infectivity and/or inhibit membrane fusion. Virus-binding competition between antibodies identified three to four antigenic sites in the F molecule (4, 12). Some epitopes have been mapped by testing the reactivities of antibodies with synthetic peptides (5, 41) or protein fragments expressed in bacteria (26). This approach, however, is not applicable to epitopes that require the native conformation of the protein for antibody binding. As an alternative, we have isolated and sequenced HRSV escape mutants, resistant to certain anti-F antibodies, in order to identify amino acid residues that are essential for epitope integrity (2, 23). In this way, two major antigenic sites (II and IV) were located in the F protein primary structure (2, 23), and some of their epitopes were further characterized with synthetic peptides (24).
Identification of new antigenic sites recognized by neutralizing anti-F antibodies.
To expand our view of the antigenic sites in the F molecule, 12 neutralizing anti-F monoclonal antibodies, previously isolated against the WV4843 strain (antigenic group B) (30), were used to select escape mutants. Those antibodies cross-reacted and neutralized the Long strain (antigenic group A). Since the Long strain had been used in our previous studies of epitope mapping, we decided to use this virus for selecting new mutants.
The selection procedure involved passaging the virus in the presence of antibodies, as was done previously (2, 12). Although escape mutants could be selected after 4 to 5 passages with antibody 47F (which was done as a control), as previously reported (2), only four mutants resistant to antibody 7.936 and three mutants resistant to antibody 9.432 were selected after 12 to 20 passages. This might reflect more stringent structural or functional constraints in the new epitopes than in previously identified antigenic sites.
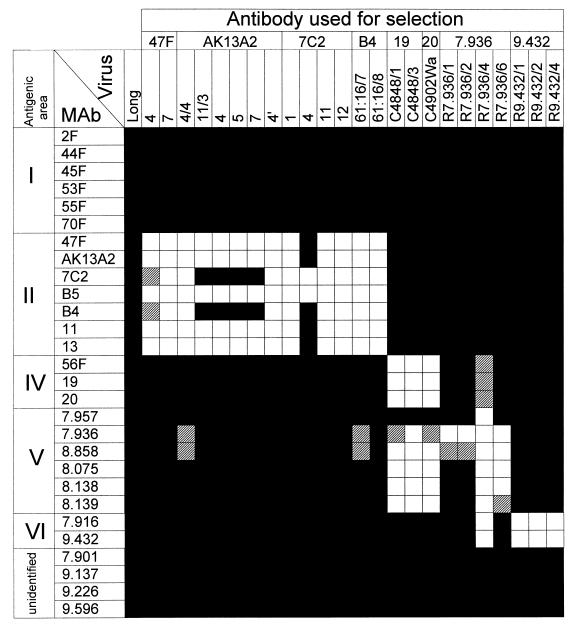
The reactivities of the new escape mutants with anti-F specific monoclonal antibodies are shown in Fig. 1. For comparison, previously described mutants and antibodies were included in the same assay. Antibody 7.957 and those below it on Fig. 1 reacted with mutants resistant to antibodies 47F, AK13A2, 7C2, and B4 of antigenic site II, indicating that their epitopes lie outside this region of the F molecule. Antibodies 7.936, 8.858, 8.075, 8.138, and 8.139 did not react with previously described mutants resistant to antibodies 19 and 20. These mutants had a single amino acid change (R429S) (2) that ablated reactivity with antibodies recognizing epitopes of antigenic site IV (see Table 1). In contrast, three mutants selected with antibody 7.936 (R7.936/1, R7.936/2, and R7.936/6) reacted normally with antibodies 56F, 19, and 20, and a fourth mutant (R7.936/4) reacted partially with these antibodies. These results indicated that epitopes 7.957, 7.936, 8.858, 8.075, 8.138, and 8.139, which were lost in some or all of the mutants resistant to antibody 7.936, identified a new antigenic site (V) that overlapped partially with site IV. At least three different classes of epitopes could be distinguished in antigenic site V by the reactivity of antibodies with escape mutants (Fig. 1): (i) epitope 7.957, (ii) epitopes 7.936 and 8.858, and (iii) epitopes 8.075, 8.138, and 8.139.
FIG. 1.
Reactivities of anti-F monoclonal antibodies with escape mutants of the Long strain. Each monoclonal antibody (MAb) was tested with an enzyme-linked immunosorbent assay (ELISA) using as the antigen extracts of HEp-2 cells (12) infected with the different escape mutants listed at the top. Antibodies included in antigenic areas I, II, and IV were classified previously by their competition for antigen binding (12) and reactivities with escape mutants (2). Mutants resistant to antibodies 47F, AK13A2, 7C2, B4, 19, and 20 have been described previously (2). Antibody 7.957 and those below it on the chart have also been described elsewhere (30). Viruses resistant to antibodies 7.936 and 9.432 are new to this study. Absorbance results: ▪, >75% of the value obtained with Long-infected cell extracts; ▨, between 25 and 75%; □, <25%.
TABLE 1.
Sequence changes in the F proteins of escape mutants resistant to antibodies from antigenic sites IV, V, and VIa
| Virus | Nucleotide
|
Amino acid
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 323 | 1298 | 1308 | 1311 | 1320 | 1353 | 104 | 429 | 432 | 433 | 436 | 447 | |
| Long | A | C | T | A | C | T | N | R | I | K | S | V |
| R7.936/1 | C | A | ||||||||||
| R7.936/2 | C | A | ||||||||||
| R7.936/4 | C | T | ||||||||||
| R7.936/6 | C | T | ||||||||||
| R9.432/1 | T | F | ||||||||||
| R9.432/2 | G | T | D | F | ||||||||
| R9.432/4 | T | F | ||||||||||
| C4848fb | A | S | ||||||||||
The three escape mutants selected with antibody 9.432 (R9.432/1, R9.432/2, and R9.432/4) lost reactivity only with this antibody and with antibody 7.916. Thus, epitopes 7.916 and 9.432 were included in a new antigenic site (VI). However, these epitopes were also lost in mutant R.7936/4, indicating partial overlapping of the newly identified antigenic sites V and VI of the F molecule. Finally, four antibodies (7.901, 9.137, 9.226, and 9.596) reacted with all the escape mutants in Fig. 1 and thus recognized epitopes of the F molecule as yet unidentified.
To locate the amino acid changes that were selected in the new escape mutants, poly(A) RNA extracted from cells infected with the different mutants was used to amplify the F gene by reverse transcription-PCR (RT-PCR). The amplified DNAs were cloned into pGEM-4 and sequenced by the dideoxy method with F-specific primers. Regions including the changes described below were confirmed by direct sequencing of the PCR product.
The sequence changes detected in the different mutants compared with the parental Long virus are listed in Table 1. Mutants R7.936/1 and R7.932/2 each had a single-nucleotide change at position 1353 (T to C) that led to the amino acid substitution V447A. The other two mutants (R7.936/4 and R7.936/6) resistant to antibody 7.936 had single-nucleotide substitutions at positions 1311 (A to C) and 1308 (T to C) that were translated into amino acid changes K433T and I432T, respectively. All these changes placed residues essential for epitopes of antigenic site V in a segment of 16 amino acids (residues 432 to 447) of the F1 chain. This segment is near residue 429, which was changed (R429S) in mutants resistant to antibodies 19 and 20 of site IV, and overlaps partially with a peptide (amino acids 422 to 438) that reacted to high titers of antibody 19 (2). These results are in agreement with the partial overlapping of profiles of reactivity with antibodies of antigenic sites IV and V observed in Fig. 1.
It is interesting that the drastic change (K433T) selected in mutant R7.936/4 ablated reactivity with all the antibodies from sites V and VI and partially inhibited reactivity with antibodies from site IV (Fig. 1). In contrast, a more conservative change at the adjacent position 432 (I432T), selected in mutant R7.936/6, influenced epitopes of site V only. Finally, the conservative change V447A (selected in mutants R7.936/1 and R7.936/2) eliminated only two of the epitopes included in antigenic site V.
Sequencing of the cDNA clones derived from escape mutants R9.432/1, R9.432/2, and R9.432/4 revealed the same nucleotide substitution at position 1320 (C to T), which resulted in the amino acid change S436F (Table 1). Mutant R9.432/2, however, had a second nucleotide substitution at position 323 (A to G), which resulted in the amino acid change N104D in the F2 chain. Although this change was also present in the RT-PCR product used for cDNA cloning, chimeric F proteins expressing only the N104D change reacted normally with antibody 9.432 (data not shown). Thus, it is likely that the substitution N104D represents an adventitious mutation incorporated during the selection process of mutant R9.432/2. It is worth emphasizing that residue 436 is included in the F gene segment (encoding residues 432 to 447) where mutations in mutants selected with antibodies from site V were located. Nonetheless, the three escape mutants selected with antibody 9.432 reacted normally with antibodies from site V (Fig. 1). In contrast, antibodies 7.916 and 9.432, included in site VI, did not react with mutant R7.936/4, which had a drastic change (K433T) only 3 residues away from amino acid 436.
Location of a group-specific antigenic site in the F protein primary structure.
In contrast to antibodies from other antigenic sites, some of the antibodies included previously in antigenic area I reacted exclusively with viruses from antigenic group A (12). Therefore, it was of interest to identify the residues of the F protein required for the integrity of these group-specific epitopes. Although antibodies from area I reduced virus infectivity by less than 1 log unit, two escape mutants could be obtained after the Long strain was passaged five to six times in the presence of antibody 55F (R55F/9 and R55F/11). The reactivities of these mutants with antibodies representative of previously identified antigenic sites are shown in Fig. 2.
FIG. 2.
Characterization of escape mutants selected with antibody 55F. Extracts of HEp-2 cells infected with either Long, CH18537, R55F/9, or R55F/11 were used in an ELISA. The results are presented as in Fig. 1. The amino acid at position 389 is indicated at the bottom for each virus.
Mutant R55F/9 lost reactivity with all the antibodies from site I but retained reactivity with antibodies from sites II and IV. Mutant R55F/11 showed a similar profile of reactivity, except that it reacted with antibody 44F (Fig. 2). Thus, at least two different epitopes could be distinguished in antigenic area I. The reactivity of virus CH18537 (a representative strain of antigenic group B) with antibodies resembled that of mutant R55F/9 (Fig. 2). cDNA copies of the F gene from viruses R55F/9 and R55F/11 were cloned in pGEM-4 and sequenced. Single-nucleotide substitutions were found at position 1179 in both viruses, C to T in mutant R55F/9 and C to A in mutant R55F/11. Those substitutions were translated into amino acid changes P389L and P389H, respectively (Fig. 2).
Amino acid 389 (P) is unchanged in the F sequence available for a second HRSV strain (A2) from antigenic group A, but it is changed in other pneumoviruses. Thus, it is S in an HRSV strain from antigenic group B (17), it is T or A in bovine respiratory syncytial virus (31), it is T in pneumonia virus of mice (8), and it is S in turkey rhinotracheitis virus (45). It is worth noting that amino acid changes in residue 389 can have different effects on antibody binding. Thus, the replacement P389H did not eliminate reactivity with antibody 44F, whereas the replacement P389S (in strain CH18537) or P389L (in the mutant R55F/9) ablated that epitope (Fig. 2). Residue 389 is located in the middle of the cysteine-rich region of the F1 chain (see Fig. 4).
FIG. 4.
Prediction of secondary-structure motifs of the F protein. (A) Linear diagram of the F protein polypeptide of the Long strain (22), showing cysteine residues (•), potential N-glycosylation sites ( ), site of proteolytic processing (↓), amino acid changes in escape mutants reported here and in previous publications (▾) and hydrophobic regions (▪). S-S, disulfide bridge. (B) Predicted secondary-structure elements of the F protein: α-helices (cylinders), β-sheets (rectangles), loops (turns). Other symbols are as explained for panel A. The segment between residues 255 and 275 (F255-275), predicted to fold in a helix-loop-helix structure, is boxed. F1h and F2h, amphipathic α-helices in the F1 and F2 chains, respectively. The diagram is not drawn to scale. The predictions were made with published sequences of pneumovirus F proteins and by using the following methods available on Worldwide Web servers: the PHD method (33, 34) (http://www.embl-heidelberg.de/predictprotein/predictprotei.html), the DSC method (18) (http://bonsai.lif.icnt.uk/bmm/dsc_form_align.html), and the PREDATOR method (11) (http://embl-heidelberg.de/predator/predator_info.html), which use multiple aligned protein sequences as input to estimate the probability of secondary-structure motifs and solvent accessibility; and the SOPMA method (13) (http://www.ibcp.fr/serv_pred.html), the nnPREDICT method (19) (http://www.cmpharm.ucsf.edu/~nomi/nnpredict.html), and the PSA method (38) (http://bmerc-www-bu.edu/psa/), which use single protein sequences as input. The final secondary-structure model was a consensus of the information obtained by the different methods. Details of the prediction studies are available from the authors upon request.
), site of proteolytic processing (↓), amino acid changes in escape mutants reported here and in previous publications (▾) and hydrophobic regions (▪). S-S, disulfide bridge. (B) Predicted secondary-structure elements of the F protein: α-helices (cylinders), β-sheets (rectangles), loops (turns). Other symbols are as explained for panel A. The segment between residues 255 and 275 (F255-275), predicted to fold in a helix-loop-helix structure, is boxed. F1h and F2h, amphipathic α-helices in the F1 and F2 chains, respectively. The diagram is not drawn to scale. The predictions were made with published sequences of pneumovirus F proteins and by using the following methods available on Worldwide Web servers: the PHD method (33, 34) (http://www.embl-heidelberg.de/predictprotein/predictprotei.html), the DSC method (18) (http://bonsai.lif.icnt.uk/bmm/dsc_form_align.html), and the PREDATOR method (11) (http://embl-heidelberg.de/predator/predator_info.html), which use multiple aligned protein sequences as input to estimate the probability of secondary-structure motifs and solvent accessibility; and the SOPMA method (13) (http://www.ibcp.fr/serv_pred.html), the nnPREDICT method (19) (http://www.cmpharm.ucsf.edu/~nomi/nnpredict.html), and the PSA method (38) (http://bmerc-www-bu.edu/psa/), which use single protein sequences as input. The final secondary-structure model was a consensus of the information obtained by the different methods. Details of the prediction studies are available from the authors upon request.
Escape mutants doubly resistant to antibodies from antigenic sites II and IV.
Until now only mutants resistant to individual anti-F antibodies have been reported in the literature (2, 4, 12). These viruses frequently displayed miniplaque phenotypes, indicative of functional alterations in the F molecule (4). This may be related to the unusual amino acid changes selected in the escape mutants at positions which are unchanged in HRSV isolates. Thus, it was of interest to know whether or not viruses doubly resistant to antibodies recognizing nonoverlapping antigenic sites could be isolated, and the effect of the accumulated mutations on F antigenicity.
The mutant C4848f, which is resistant to antibody 19 (2) (antigenic site IV), was chosen for selecting viruses also resistant to antibody 47F (antigenic site II). Four mutants (RRA3, RRA6, RRA9, and RRA10) were obtained after four passages in the presence of antibody 47F. The double mutants were impaired in cell membrane fusion, as denoted by a very small syncytium phenotype (data not shown). Those viruses lack reactivity with antibodies from sites IV and V, as does the parental C4848f virus, and in addition they did not react with most antibodies from antigenic site II (Fig. 3). Mutants RRA3 and RRA9 could be distinguished from RRA6 and RRA10 by their reactivity with antibodies 7C2 and B4. The reactivity profiles of the double mutants were those expected from the reactivities of mutants resistant to individual antibodies, and no major antigenic reorganization of the F protein was observed.
FIG. 3.
Characterization of double escape mutants. Extracts of HEp-2 cells infected with the viruses listed at the top were used in an ELISA. The results are presented as in Fig. 1. Amino acids at positions 262, 275, and 429 are indicated at the bottom for each virus.
The F genes of viruses doubly resistant to antibodies 19 and 47F were amplified by RT-PCR and sequenced. The four mutants had the original substitution at nucleotide 1298 (C to A) that resulted in the amino acid change R429S and distinguished the parental C4848f virus from the Long strain (Table 1). Mutants RRA6 and RRA10 had, in addition, a transversion at nucleotide 797 (A to T) that resulted in the amino acid change N262Y (Fig. 3). Single mutants with the change N262Y had been selected previously with antibody 47F or AK13A2, and they showed a reactivity profile with antibodies from site II identical to that of RRA6 and RRA10 (2). The other two double mutants shown in Fig. 3 (RRA3 and RRA9) had a transition at position 837 (C to T) that resulted in the amino acid change S275F. This change had not been observed previously among escape mutants of the Long strain, but it had been found in mutants of the A2 strain (10). It is interesting that the change S275F did not affect epitope B4, which is eliminated by an amino acid change at residue 262, 268, or 272 (2). B4 is the only epitope, among those included in site II, which is efficiently reproduced in a synthetic peptide spanning residues 255 to 275 (2, 24).
In conclusion, the changes selected in the double mutants were also found among those selected in single mutants, indicating that amino acid changes in individual antigenic sites of the F protein did not influence the changes selected in a separate site. Nevertheless, viruses with changes in sites II and IV had a clear impairment for syncytium formation (data not shown). This may reflect structural and/or functional constraints of the F protein that prevent accumulation of sequence changes and may explain the high degree of sequence conservation among the F protein genes of HRSV strains (17).
Structural considerations about the antigenic organization of the F molecule.
Figure 4A is a diagram of the primary structure of the F protein of the Long strain, showing the locations of amino acid changes selected in escape mutants described in this work and in previous publications (2, 23). To gain further insight into the structural requirements of the F protein epitopes, a model of secondary-structure motifs was built by using published sequences of different pneumoviruses (Fig. 4). This prediction is in good agreement with, and refines the structural elements proposed by Chambers et al. (8) on the basis of the comparison of 5 sequences of paramyxovirus fusion proteins. The F protein spikes of HRSV particles represent, at least, F1-F2 homodimers that are observed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis without heating and reduction (3, 9, 25). However, homotetramers have been observed after chemical cross-linking (9); presumably, these represent less-stable association between two dimers. Intermonomer associations appear to involve the F1 chain, which would form the tetramer core. By analogy with Sendai virus (15), a single disulfide bridge between cysteines 69 and 212 was proposed to link the F1 and F2 chains.
The F2 chain of 139 amino acids is the most divergent segment of the F molecule both between and within the antigenic groups of HRSV (17, 22). It is likely to be located outside the internal core, as indicated by high scores of solvent accessibility throughout its sequence (data not shown). F2 has the majority of potential sites for N glycosylation. Estimates of carbohydrate content suggest that most, if not all, of these sites are actually used. A region of turns and β-sheets is predicted at the N-terminal end before the amphipathic α-helix, which precedes a stretch of 28 hydrophilic amino acids that is present in HRSV but absent in other pneumoviruses (Fig. 4B) (8). The C-terminal end of F2 has the sequence KKRKRR, which is presumably the site of proteolytic processing of the F0 precursor.
The N-terminal end of the F1 chain has 19 contiguous hydrophobic amino acids that form the fusion peptide (Fig. 4). This is probably buried in the F-protein tetramer. Not only the hydrophobic character of this peptide but also certain amino acids are conserved among pneumoviruses, suggesting that they are required for fusion activity. The N-terminal one-third of F1 is rich in α-helices, including a long heptad repeat, which might be involved in coiled-coil interactions, and an amphipathic α-helix (Fig. 4B). The relevance of both the fusion peptide and the heptad repeat region for fusion activity has been demonstrated in Newcastle disease virus (NDV) by site-directed mutagenesis (35). The segment between residues 255 and 275 is predicted to fold in a helix-loop-helix structure, which is precisely the conformation adopted by a 21-residue peptide in 30% trifluoroethanol, as determined by two-dimensional nuclear magnetic resonance (39).
The cluster of cysteines in the middle of the F1 chain is larger in pneumoviruses than in other paramyxoviruses, which lack the last 4 cysteines. Nevertheless, the overall structure of this region is likely to be similar in all those viruses. Multiple intrachain cysteine loops have been found in the Sendai virus F protein (15), and several β-sheets are predicted in HRSV (Fig. 4B). Thus, a set of alternating loops and β-sheets should confer a globular conformation on this region of the F molecule, which might have an external location in the tetramer, as indicated by high hydrophilicity values (data not shown). Following the cysteine cluster there is a second heptad repeat, in close proximity to the membrane, which may participate in intra- or intermonomer coiled-coils.
The N-terminal segment of the F1 chain, up to the cysteine-rich region, has structural similarities with the HA2 subunit of influenza virus hemagglutinin (HA), another viral glycoprotein with membrane fusion activity and a known three-dimensional structure (44). This resemblance is reflected in a high α-helix content, the presence of fusion peptides, and resistance to trypsin digestion (23, 36). The relevance of this region for F protein assembly is inferred from the phenotype of mutants with mutations at positions 236 to 237 that are retained in the endoplasmic reticulum as a result of protein misfolding (25). The epitopes of antigenic site II are located in this protease-resistant region of F1. Previous studies indicated that a peptide spanning residues 215 to 275 could reproduce the bizarre local conformation of that region of the F molecule. Most antibodies included in site II (24) recognized that peptide.
The other region that included epitopes recognized by neutralizing antibodies (sites IV, V, and VI) is located near the C-terminal end of the cysteine-rich region and not far from the heptad repeat adjacent to the membrane. Thus, neutralizing antibodies bind to both sides of the cysteine cluster. It is likely that those antibodies would inhibit conformational changes of the F1 chain associated with the fusion process (20). These changes would be analogous to the structural reorganization of the HA2 subunit of influenza virus HA during fusion of the virus and cell membranes (6, 7). In this case, the HA2 chain folds back at an acid pH (6), bringing in close proximity the fusion peptide, inserted in the cell membrane, and the transmembrane anchor, inserted in the viral membrane. This notion of direct apposition of the two membranes before fusion would require that the cysteine-rich region of the F1 chain have flexible behavior. This is in agreement with the marginal effect that antibodies binding to site I (located in the middle of the cysteine cluster) have in virus neutralization. Antigenic sites equivalent to those of HRSV F protein have been described in Sendai virus (32), NDV (28, 40), and parainfluenza virus type 3 (42). Thus, virus neutralization afforded by anti-F antibodies may take place via a general mechanism in paramyxoviruses.
Acknowledgments
J.A.L. and R.B. contributed equally to this work.
We thank P. Coppe for antibody AK13A2, M. Trudel for antibody 7C2, and G. Taylor for antibodies B5, B4, 11, 13, 19, and 20.
This work was partially funded by grant PM96-0025 from the Dirección General de Enseñanza Superior (to J.A.M.) and by a grant from the Comisión Sectorial de Investigación Científica (to J.A.). A grant from the Agencia Española de Colaboración Internacional funded travel expenses between Madrid and Montevideo. J.A.L. was the recipient of a postdoctoral fellowship from the Instituto de Salud Carlos III.
REFERENCES
- 1.Anderson L J, Heirholzer J C, Tson C, Hendry R M, Fernie B N, Stone Y, McIntosh K. Antigenic characterization of respiratory syncytial virus strains with monoclonal antibodies. J Infect Dis. 1985;151:626–633. doi: 10.1093/infdis/151.4.626. [DOI] [PubMed] [Google Scholar]
- 2.Arbiza J, Taylor G, López J A, Furze J, Wyld S, Whyte P, Stott E J, Wertz G, Sullender W, Trudel M, Melero J A. Characterization of two antigenic sites recognized by neutralizing monoclonal antibodies directed against the fusion glycoprotein of human respiratory syncytial virus. J Gen Virol. 1992;73:2225–2234. doi: 10.1099/0022-1317-73-9-2225. [DOI] [PubMed] [Google Scholar]
- 3.Arumugham R G, Hildreth S W, Paradiso P R. Evidence that the fusion protein of respiratory syncytial virus exists as a dimer in the native form. Arch Virol. 1989;106:327–334. doi: 10.1007/BF01313961. [DOI] [PubMed] [Google Scholar]
- 4.Beeler J A, van Wyke Coelingh K. Neutralization epitopes of the F glycoprotein of respiratory syncytial virus: effect of mutation upon fusion function. J Virol. 1989;63:2941–2950. doi: 10.1128/jvi.63.7.2941-2950.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bourgeois C, Corvaisier C, Bour J B, Kohli E, Pothier P. Use of synthetic peptides to locate neutralizing antigenic domains on the fusion protein of respiratory syncytial virus. J Gen Virol. 1991;72:1051–1058. doi: 10.1099/0022-1317-72-5-1051. [DOI] [PubMed] [Google Scholar]
- 6.Bullough P A, Hughson F M, Skehel J J, Wiley D C. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 7.Carr C M, Kim P S. A spring-loaded mechanism for the conformational change of influenza haemagglutinin. Cell. 1993;73:823–832. doi: 10.1016/0092-8674(93)90260-w. [DOI] [PubMed] [Google Scholar]
- 8.Chambers P, Pringle C R, Easton A J. Sequence analysis of the gene encoding the fusion glycoprotein of pneumonia virus of mice suggests possible conserved secondary structure elements in paramyxovirus fusion glycoproteins. J Gen Virol. 1992;73:1717–1724. doi: 10.1099/0022-1317-73-7-1717. [DOI] [PubMed] [Google Scholar]
- 9.Collins P L, Mottet G. Post-translational processing and oligomerization of the fusion glycoprotein of human respiratory syncytial virus. J Gen Virol. 1991;72:3095–3101. doi: 10.1099/0022-1317-72-12-3095. [DOI] [PubMed] [Google Scholar]
- 10.Collins P L, McIntosh K, Chanock R M. Respiratory syncytial virus. In: Fields B N, Knipe D M, Howley P W, et al., editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1313–1351. [Google Scholar]
- 11.Frishman D, Argos P. Seventy-five percent accuracy in protein secondary structure prediction. Proteins Struct Funct Genet. 1997;27:329–335. doi: 10.1002/(sici)1097-0134(199703)27:3<329::aid-prot1>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 12.García-Barreno B, Palomo C, Peñas C, Delgado T, Perez-Breña P, Melero J A. Marked differences in the antigenic structure of human respiratory syncytial virus F and G glycoproteins. J Virol. 1989;63:925–932. doi: 10.1128/jvi.63.2.925-932.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geourjon C, Deleage C. SOPMA: a self-optimized method for protein secondary structure prediction. Protein Eng. 1994;7:157–164. doi: 10.1093/protein/7.2.157. [DOI] [PubMed] [Google Scholar]
- 14.Gruber C, Levine S. Respiratory syncytial virus polypeptides. III. The envelope-associated proteins. J Gen Virol. 1983;64:825–832. doi: 10.1099/0022-1317-64-4-825. [DOI] [PubMed] [Google Scholar]
- 15.Iwata S, Schmidt A C, Titani K, Suzuki M, Kido H, Goth B, Hamaguchi M, Nagai Y. Assignment of disulfide bridges in the fusion glycoprotein of Sendai virus. J Virol. 1994;68:3200–3206. doi: 10.1128/jvi.68.5.3200-3206.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson P R, Spriggs M K, Olmsted R A, Collins P L. The G glycoprotein of human respiratory syncytial viruses of subgroups A and B: extensive sequence divergence between antigenically related proteins. Proc Natl Acad Sci USA. 1987;84:5625–5629. doi: 10.1073/pnas.84.16.5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson P R, Collins P L. The fusion glycoprotein of human respiratory syncytial virus of subgroups A and B: sequence conservation provides a structural basis for antigenic relatedness. J Gen Virol. 1988;69:2623–2628. doi: 10.1099/0022-1317-69-10-2623. [DOI] [PubMed] [Google Scholar]
- 18.King R D, Saqi M, Sayle R, Sternberg M J E. DSC: public domain protein secondary structure prediction. CABIOS. 1997;13:473–474. doi: 10.1093/bioinformatics/13.4.473. [DOI] [PubMed] [Google Scholar]
- 19.Kneller A G, Cohen F E, Landridge R. Improvements in protein secondary structure prediction by an enhanced neural network. J Mol Biol. 1990;214:171–182. doi: 10.1016/0022-2836(90)90154-E. [DOI] [PubMed] [Google Scholar]
- 20.Lamb R A. Paramyxovirus fusion. A hypothesis for changes. Virology. 1993;197:1–11. doi: 10.1006/viro.1993.1561. [DOI] [PubMed] [Google Scholar]
- 21.Levine S, Klaiber-Franco R, Paradiso P R. Demonstration that glycoprotein G is the attachment protein of respiratory syncytial virus. J Gen Virol. 1987;68:2521–2524. doi: 10.1099/0022-1317-68-9-2521. [DOI] [PubMed] [Google Scholar]
- 22.López J A, Villanueva N, Melero J A, Portela A. Nucleotide sequence of the fusion and phosphoprotein genes of human respiratory syncytial (RS) virus Long strain: evidence of subtype genetic heterogeneity. Virus Res. 1986;10:249–262. doi: 10.1016/0168-1702(88)90020-2. [DOI] [PubMed] [Google Scholar]
- 23.López J A, Peñas C, García-Barreno B, Melero J A, Portela A. Location of a highly conserved neutralizing epitope in the F glycoprotein of human respiratory syncytial virus. J Virol. 1990;64:927–930. doi: 10.1128/jvi.64.2.927-930.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.López J A, Andreu D, Carreño C, Whyte P, Taylor G, Melero J A. Conformational constraints of conserved neutralizing epitopes from a major antigenic area of human respiratory syncytial virus fusion glycoprotein. J Gen Virol. 1993;74:2567–2577. doi: 10.1099/0022-1317-74-12-2567. [DOI] [PubMed] [Google Scholar]
- 25.López J A, Bustos R, Portela A, García-Barreno B, Melero J A. A point mutation in the F1 subunit of human respiratory syncytial virus F glycoprotein blocks its cell surface transport at an early stage of the exocytic pathway. J Gen Virol. 1996;77:649–660. doi: 10.1099/0022-1317-77-4-649. [DOI] [PubMed] [Google Scholar]
- 26.Martín-Gallardo A, Flen K A, Hu B T, Farly J F, Seid R, Collins P L, Hildreth S W, Paradiso P R. Expression of the F glycoprotein gene from human respiratory syncytial virus in Escherichia coli: mapping of a fusion inhibiting epitope. Virology. 1991;184:428–432. doi: 10.1016/0042-6822(91)90863-7. [DOI] [PubMed] [Google Scholar]
- 27.Mufson M A, Örvell C, Rafnar B, Norrby E. Two distinct subtypes of human respiratory syncytial virus. J Gen Virol. 1985;66:2111–2124. doi: 10.1099/0022-1317-66-10-2111. [DOI] [PubMed] [Google Scholar]
- 28.Neyt C, Geliebter J, Slaoui M, Morales D, Meulemans G, Burny A. Mutations located on both F1 and F2 subunits of the Newcastle disease virus fusion protein confer resistance to neutralization with monoclonal antibodies. J Virol. 1989;63:952–954. doi: 10.1128/jvi.63.2.952-954.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Olmsted R A, Elango N, Prince G, Murphy B R, Johnson P R, Moss B, Chanock R M, Collins P L. Expression of the F glycoprotein of respiratory syncytial virus by a recombinant vaccinia virus: comparison of the individual contribution of the F and G glycoproteins to host immunity. Proc Natl Acad Sci USA. 1986;83:7462–7466. doi: 10.1073/pnas.83.19.7462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Örvell C, Norrby E, Mufson M. Preparation and characterization of monoclonal antibodies directed against five structural components of human respiratory syncytial virus subgroup B. J Gen Virol. 1987;68:3125–3135. doi: 10.1099/0022-1317-68-12-3125. [DOI] [PubMed] [Google Scholar]
- 31.Pastey M K, Samal S K. Structure and sequence comparison of bovine respiratory syncytial virus fusion protein. Virus Res. 1993;29:195–202. doi: 10.1016/0168-1702(93)90059-v. [DOI] [PubMed] [Google Scholar]
- 32.Portner A, Scroggs R A, Naeve C W. The fusion glycoprotein of Sendai virus: sequence analysis of an epitope involved in fusion and virus neutralization. Virology. 1987;157:556–559. doi: 10.1016/0042-6822(87)90301-1. [DOI] [PubMed] [Google Scholar]
- 33.Rost B, Sander C. Combining evolutionary information and neural networks to predict secondary structure. Proteins Struct Funct Genet. 1994;19:55–72. doi: 10.1002/prot.340190108. [DOI] [PubMed] [Google Scholar]
- 34.Rost B, Sander C. Conservation and prediction of solvent accessibility in protein families. Proteins Struct Funct Genet. 1994;20:216–226. doi: 10.1002/prot.340200303. [DOI] [PubMed] [Google Scholar]
- 35.Sergel-Germano T, McQuain C, Morrison T. Mutations in the fusion peptide and heptad repeat regions of the Newcastle disease virus fusion protein block fusion. J Virol. 1994;68:7654–7658. doi: 10.1128/jvi.68.11.7654-7658.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skehel J J, Bayley P M, Brown E R, Martin S R, Waterfield M D, White J M, Wilson I A, Wiley D C. Changes in conformation of influenza virus haemagglutinin at the pH optimum of virus-mediated membrane fusion. Proc Natl Acad Sci USA. 1982;79:968–972. doi: 10.1073/pnas.79.4.968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stott E J, Taylor G, Ball L A, Anderson K, Young K-Y, King A M Q, Wertz G W. Immune and histopathological responses in animals vaccinated with recombinant vaccinia viruses that express individual genes of human respiratory syncytial virus. J Virol. 1987;61:3855–3861. doi: 10.1128/jvi.61.12.3855-3861.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stultz C M, White J B, Smith T F. Structural analysis based on state space modeling. Protein Sci. 1993;2:305–314. doi: 10.1002/pro.5560020302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Toiron C, López J A, Rivas G, Andreu D, Melero J A, Bruix M. Conformational studies of a short linear peptide corresponding to a major conserved neutralizing epitope of human respiratory syncytial virus fusion glycoprotein. Biopolymers. 1996;39:537–548. doi: 10.1002/(sici)1097-0282(199610)39:4<537::aid-bip6>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 40.Toyoda T, Gotoh B, Sakaguchi T, Kida H, Nagai Y. Identification of amino acids relevant to three antigenic determinants on the fusion protein of Newcastle disease virus that are involved in fusion inhibition and neutralization. J Virol. 1988;62:4427–4430. doi: 10.1128/jvi.62.11.4427-4430.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trudel M, Nadon F, Seguin C, Dionne C, Lacroix M. Identification of a synthetic peptide as part of a major neutralization epitope of respiratory syncytial virus. J Gen Virol. 1987;68:2273–2280. doi: 10.1099/0022-1317-68-9-2273. [DOI] [PubMed] [Google Scholar]
- 42.van Wyke Coelingh K, Tierney E L. Identification of amino acids recognized by syncytium-inhibiting and neutralizing monoclonal antibodies to the human parainfluenza type 3 virus fusion protein. J Virol. 1989;63:3755–3760. doi: 10.1128/jvi.63.9.3755-3760.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walsh E E, Hruska J. Monoclonal antibodies to respiratory syncytial virus proteins: identification of the fusion protein. J Virol. 1983;47:171–177. doi: 10.1128/jvi.47.1.171-177.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilson I A, Skehel J J, Wiley D C. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3Å resolution. Nature. 1981;289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 45.Yu Q, Davis P J, Barrett T, Binns M M, Boursnell M E G, Cavanagh D. Deduced amino acid sequence of the fusion glycoprotein of turkey rhinotracheitis virus has a greater identity with that of human respiratory syncytial virus, a pneumovirus, than that of paramyxoviruses and morbilliviruses. J Gen Virol. 1991;72:75–81. doi: 10.1099/0022-1317-72-1-75. [DOI] [PubMed] [Google Scholar]