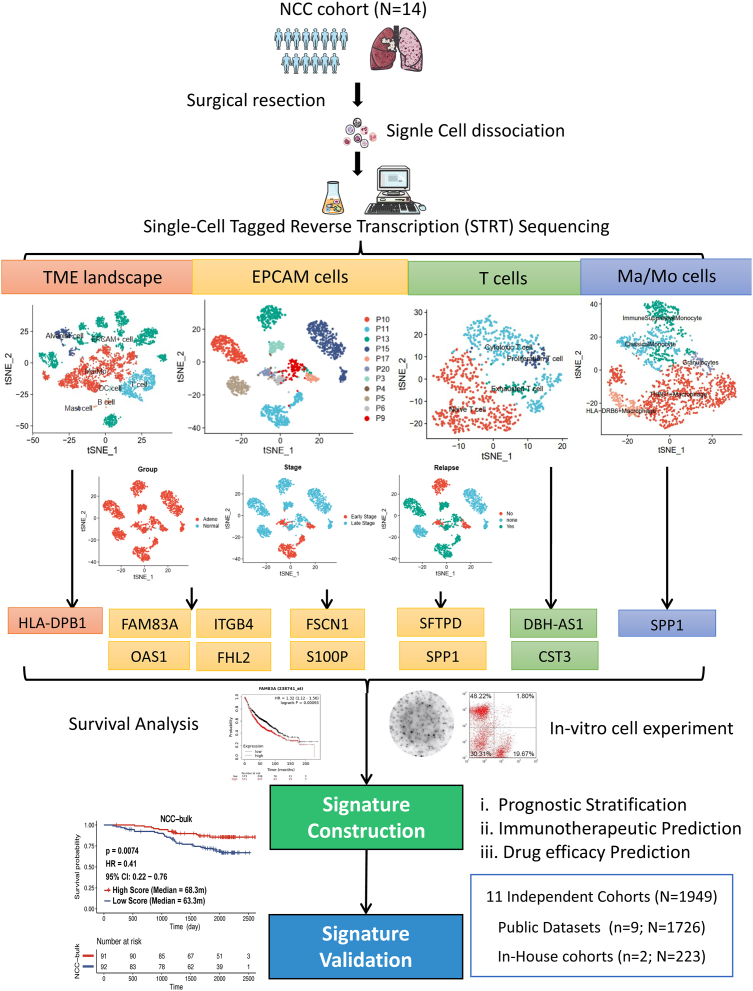
Summary
Background
The high heterogeneity of tumour and the complexity of tumour microenvironment (TME) greatly impacted the tumour development and the prognosis of cancer in the era of immunotherapy. In this study, we aimed to portray the single cell-characterised landscape of lung adenocarcinoma (LUAD), and develop an integrated signature incorporating both tumour heterogeneity and TME for prognosis stratification.
Methods
Single-cell tagged reverse transcription sequencing (STRT-seq) was performed on tumour tissues and matched normal tissues from 14 patients with LUAD for immune landscape depiction and candidate key genes selection for signature construction. Kaplan–Meier survival analyses and in-vitro cell experiments were conducted to confirm the gene functions. The transcriptomic profile of 1949 patients from 11 independent cohorts including nine public datasets and two in-house cohorts were obtained for validation.
Findings
We selected 11 key genes closely related to cell-to-cell interaction, tumour development, T cell phenotype transformation, and Ma/Mo cell distribution, including HLA-DPB1, FAM83A, ITGB4, OAS1, FHL2, S100P, FSCN1, SFTPD, SPP1, DBH-AS1, CST3, and established an integrated 11-gene signature, stratifying patients to High-Score or Low-Score group for better or worse prognosis. Moreover, the prognostically-predictive potency of the signature was validated by 11 independent cohorts, and the immunotherapeutic predictive potency was also validated by our in-house cohort treated by immunotherapy. Additionally, the in-vitro cell experiments and drug sensitivity prediction further confirmed the gene function and generalizability of this signature across the entire RNA profile spectrum.
Interpretation
This single cell-characterised 11-gene signature might offer insights for prognosis stratification and potential guidance for treatment selection.
Funding
Support for the study was provided by National key research and development project (2022YFC2505004, 2022YFC2505000 to Z.W. and J.W.), Beijing Natural Science Foundation (7242114 to J.X.), National Natural Science Foundation of China of China (82102886 to J.X., 81871889 and 82072586 to Z.W.), Beijing Nova Program (20220484119 to J.X.), NSFC general program (82272796 to J.W.), NSFC special program (82241229 to J.W.), CAMS Innovation Fund for Medical Sciences (2021-1-I2M-012, 2022-I2M-1-009 to Z.W. and J.W.), Beijing Natural Science Foundation (7212084 to Z.W.), CAMS Key lab of translational research on lung cancer (2018PT31035 to J.W.), Aiyou Foundation (KY201701 to J.W.). Medical Oncology Key Foundation of Cancer Hospital Chinese Academy of Medical Sciences (CICAMS-MOCP2022003 to J.X.)
Keywords: Lung adenocarcinoma, Single cell sequencing, Tumour heterogeneity, Immune microenvironment, Prognostic stratification
Research in context.
Evidence before this study
We searched from 1st Jan. 2006 to 30th Mar. 2023 with the keywords “lung cancer” (“prognosis” or “prognostic”), (“biomarker” or “signature”), (“immunotherapy” or “immune checkpoint inhibitor” or “ICI”) in PubMed and proceedings of international meetings, restricting the language to English, and found that the unsatisfactory long-term survival of cancer stemmed from the high heterogeneity of tumour, the complexity of tumour microenvironment (TME) and the uncomprehensive characterization provided by current prognostic and predictive indicators. Thus, it holds immense significance to keep exploring prognostic and predictive biomarkers that rely on integrated characterization incorporating both tumour heterogeneity and TME.
Added Value of this study
We provided a comprehensive portrayal of the single cell-characterised TME landscape in 14 patients with lung adenocarcinoma, and developed an integrated signature incorporating both tumour heterogeneity and TME based on high-precision single-cell transcriptome analysis, as validated by 1949 patients from 11 independent cohorts including nine public datasets and two in-house cohorts.
Implications of all the available evidence
This study underscored the importance of the current immunotherapy approach for advanced-stage tumour, highlighted the potential for future drug development by targeting TME components, and offered promising insights of integrated signature incorporating tumour heterogeneity and TME for prognostic stratification and treatment selections.
Introduction
Lung cancer stands as the leading cause of cancer-related death worldwide, with lung adenocarcinoma (LUAD) representing the predominant pathological subtype.1,2 Although various treatments, including immune checkpoint inhibitors (ICIs), have transformed the management of lung cancer and led to significantly improved clinical outcomes, challenges including the unsatisfactory 5-year survival rate and the limited proportion of patients experiencing durable response still existed. Such issues stemmed from the high heterogeneity of tumour, the complexity of tumour microenvironment (TME) and the uncomprehensive characterization provided by current prognostic and predictive indicators.3, 4, 5 Therefore, it holds immense significance to keep exploring prognostic and predictive biomarkers that rely on integrated characterization incorporating both tumour heterogeneity and TME.
Over the past decade, there has been growing recognition of the crucial role played by TME in the development of lung cancer. It is now understood that both tumour cells which exhibit significant heterogeneity, and the immune cells which infiltrate the TME, collectively influence the progression of cancer and shape the response to treatment.6, 7, 8, 9, 10, 11 Numerous prior studies have underscored the impact of both the quantity and nature of infiltrating cells in TME on the response to immunotherapy in lung cancer, particularly focusing on T lymphocytes and mononuclear macrophages. Notably, research by Zemin Zhang et al. has emphasized the important role of T cell migration and transformation in non-small cell lung cancer.12 Wu K et al. made a notable contribution by differentiating tumour-associated macrophages (TAMs) into M1-TAMs and M2-TAMs, elucidating their distinct roles in anti-tumour and tumour-promoting functions. Their work shed light on the significant involvement of macrophages in tumour genesis and development.13 Therefore, it becomes imperative to thoroughly characterize tumour cells, lymphocytes, mononuclear macrophages, and other relevant components in order to explore comprehensive biomarkers and future druggable targets in the era of immunotherapy.
In recent years, single-cell RNA sequencing has undergone rapid development, allowing for the clear distinction of cell origins and the comprehensive display of interactions between various cells. It has emerged as a crucial tool for characterizing the TME and identifying potential biomarkers across multiple cancer types.14 Previous studies have delved into the creation of prognostic or predictive biomarkers, either based on the overarching TME characteristics or specific components revealed through single-cell sequencing, albeit independently.15 For instance, Jiang A et al. developed a 6-gene prognostic model based on the overall TME characteristics in patients with LUAD using 10× scRNA-seq.16 Pang J et al. emphasized the crucial role of neutrophil-related genes, considering different neutrophil subsets with varying differentiation states and established a 6-gene prognostic model.17 Chen J et al. unveiled the inhibitory capability of naive-like B cells towards lung cancer cells, driven by the secretion of four factors known to negatively regulate cell growth and impact prognosis, thereby suggesting them as potential prognostic biomarkers.18 Additionally, Zemin Z et al. detailedly described the heterogeneity of infiltrated CD8+T cells and Treg cells, providing a new way to classify patients.12 However, to date, there have been no biomarkers that exhibiting both prognostic and predictive potential by combining considerations of both tumour heterogeneity and infiltrating immune cells.
Herein, we provided a comprehensive portrayal of the single cell-characterised TME landscape in LUAD, and developed an integrated signature incorporating both tumour heterogeneity and TME based on high-precision single-cell transcriptome analysis, aiming to offer insights for prognosis stratification and potential guidance for the selection of treatment strategies.
Methods
Experimental design
14 Patients with pathologically-diagnosed lung adenocarcinoma underwent surgical resections at the National Cancer Center/Cancer Hospital and Chinese Academy of Medical Sciences were prospectively recruited as NCC cohort from May 18th 2017 to Apr 9th 2018. All patients had signed informed consent. Tumour tissues and matched normal tissues were both collected from each patient for single-cell tagged reverse-transcription (STRT)-single cell analysis, with detailed methods as previously reported.19 Rigorous quality control measures were employed to ensure high cell viability for single-cell RNA-seq. This study received approval from the ethics committees of the National Cancer Center (NCC-22/250-3454, NCC-22/429-3631, NCC1798).
Dimension reduction and unsupervised clustering
By using STRT technology, we isolated single cells from 14 tumours and 14 adjacent tissues of 14 patients. Single-cell cDNA amplification was performed based on a modified STRT-seq protocol. The constructed libraries were sequenced on HiSeq 4000 system as paired-end 150-bp reads. After removing the low quality, poly A, TSO, and adapter contaminated reads, 6985 cells were obtained through filtering based on specific conditions, ensuring that the proportion of red blood cells was less than 2%, and the proportion of mitochondria was less than 10%. Then, we used R package Seurat 3.2.2 to create SeuratObject, and candidate cells filter by the following criteria: min.cells = 10 & nFeature_RNA >2000 & nFeature_RNA <12,000 & percent.mt < 10 & percent. HB < 2. Finally, we acquired 5543 cells in total for subsequent analysis. We generated normalized expression matrices using the log 2 (TPM/10 + 1) and function ScaleData in Seurat. To reduce the data dimension, we performed principal component analysis with top 2000 variable genes characterised by FindVariable function. The first 50 principal components were used as input to perform clustering based on shared nearest neighbor (SNN) algorithm at a resolution of 2. Further dimensionality reduction was performed by unsupervised t-distributed stochastic neighborhood embedding (t-SNE) analysis using RunTSNE function in Seurat. DimPlot function was applied to visualize the clustering results.
Cell–cell interaction analysis
We employed CellPhoneDB20 to identify significant ligand-receptor pairs within samples. Specific expression of a receptor by one cell type and a corresponding ligand by another cell type was identified to reflect the potential interaction between cell types. The interaction score was determined as the total mean of the average expression values of individual ligand-receptor partners within their respective interacting pairs of cell types. The expression of any complexes generated by CellPhoneDB was calculated as the sum of the expression values of their constituent genes.
Identification of differentially expressed genes and functional analysis
We employed the “FindAllMarkers” function within the Seurat package (v3.2.2) to identify differentially expressed genes (DEGs) across various groups. Significant DEGs were selected from the genes with P value ≤ 0.01 and average fold change (avglog FC) ≥ 0.5 after applying a logarithmic transformation, setting the stage for further analysis and visualization. To gain insights into the biological significance of these important DEGs, we conducted Gene Ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis. These analyses were performed using the Metascape platform (http://metascape.org). Additionally, we explored pathway enrichment comparisons across various combinations of the two clusters through Gene Set Enrichment Analysis (GSEA). For GSEA, we utilized a matrix encompassing all genes detected in our dataset. We employed the desktop tool available for download at http://software.broadinstitute.org/gsea/index.jsp to perform GSEA. We further conducted Gene Set Variation Analysis (GSVA) utilising the GSVA package 34. To determine differences between distinct cell groups, we utilized a linear model provided by the Limma package.
Cell developmental trajectory
We inferred the cell lineage trajectory of T cells and monocytes/macrophages (Ma/Mo)Ma/Mo cells using Monocle 2.21 To start, we utilized the ‘relative2abs’ function in Monocle2 to convert TPM values into normalized mRNA counts. Subsequently, we created an object with the ‘expression Family = negbinomial. size’ parameter, in accordance with the Monocle2 tutorial. To identify DEGs, we employed the differentialGeneTest function on each cluster, selecting relevant genes with q-value <1e-8 were assessed to order the cells for subsequent pseudotime analysis. Once the cell trajectories were constructed, we conducted an analysis of differentially expressed genes along the pseudotime, once again utilising the differentialGeneTest function. Subsequently, plot cell trajectory was performed to visualize cells order in pseudotime progression.
Simultaneous gene regulatory network analysis
To measure the difference between cell clusters based on transcription factors or their target genes, SCENIC, a new computational method used in the construction of regulatory networks and in the identification of different cell states from scRNA-seq data,22 was performed on all single cells, and the preferentially expressed regulons were calculated by the Limma package.23 Only regulons significantly upregulated or downregulated in at least one cluster, with adj. P-value <0.05, were involved in further analysis.
Public data acquisition and pre-processing
We conducted a comprehensive search in publicly available databases, including The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO), to access mRNA expression data and clinical information related to LUAD. TCGA, GEO databases were used to search mRNA expression and clinical information of LUAD. We identified a total of 9 public LUAD cohorts for validation. To ensure the reliability of our analysis and the relevance of our findings, we carefully curated these datasets. We excluded patients who lacked sufficient follow-up information from further consideration.
In-house patient cohorts
We incorporated two additional in-house datasets into our study. One dataset comprised 183 patients with stage I-III LUAD who had undergone surgery (NCC-bulk cohort). The second dataset consisted of 40 patients with advanced-stage lung cancer who received treatment with single anti-PD-(L)1 antibodies at our centere (NCC-ICIs cohort). For bulk RNA sequencing, we obtained Formalin-Fixed and Paraffin-Embedded (FFPE) samples from the NCC-bulk cohort and NCC-ICIs cohort. The RNA extraction, sequencing library construction, sequencing and FASTQ data quality control were performed in accordance with the protocol by Nick D.L. Owens et al.24
In-vitro cell experiments
The human lung cancer cell line H1650 (RRID:CVCL_B260) and PC-9 (RRID:CVCL_B260) were procured from the Cell Resource Centere at Peking Union Medical College, which serves as the headquarters of the National Infrastructure of Cell Line Resources (NSTI). Both cell lines were validated and recently assessed to ensure they were free of mycoplasma contamination.
H1650 and PC-9 cells were transfected with FAM83A, ITGB4, OAS1, FHL2, S100P, FSCN1 siRNAs and normal cell in parallel using Lipofectamine 3000. Following transfection for 24 h, we evaluated cell viability using the CellTiter 96 AQueous Non-Radioactive Cell Proliferation Assay (MTS), following the manufacturer's guidelines (Promega). Simultaneously, cell proliferation test and cell scratch test was conducted following previously-reported protocols.25
Full length of DBH-AS1, CST3, HLA-DPB1 and SFTPD were constructed into plasmid pCDNA3.1 vector and electroporated into T cells from Peripheral blood mononuclear cells (PBMCs) who volunteered for. Then enzyme linked immunospot assay (ELISPOT) test was conducted following protocols.26 Then the cells were subsequently subjected to flow cytometry analysis under Human anti-cytokine (CD3, CD4, CD8) detection antibodies (RRID:AB_400457 and AB_398476) using the BD FACSymphony™ A1 flow cytometer, supported by BD FACSDiva™ software, following manusfacturer's guidelines.27
SPP1 was constructed into plasmid pCDNA3.1 vector and electroporated into Ma/Mo cells from Peripheral blood mononuclear cells (PBMCs) who volunteered for. CD206 is a highly expressed co-stimulatory molecule in M2-type macrophages after polarization, so we applied it as a marker of macrophage polarization. Flow cytometry was then performed to sort the isolated macrophages and determine the number of macrophages expressing CD206 in both the experimental and control groups.
Signature construction
To analyse the gene signature within the context of a gene expression matrix E, we initiated a process involving gene binning. Initially, we categorized genes into 50 expression bins based on their average expression levels across the sample set. Ei,j denotes the expression value of gene i in sample j. The average expression of a gene i across a set of N samples is defined as ∑jEi,j/N.The Raw Score of a gene i in sample j is defined as: Raw Score i,j= (Ei,j−∑jEi,j)/N. Next, we introduced the concept of a gene signature S, comprised of K genes, with kb genes situated in bin b. We implemented random sampling of S-compatible signatures to facilitate normalization. A random signature was considered S-compatible if it consisted of a total of K genes, with kb genes residing in each bin (b). To compute the overall expression of a gene i in sample j, we employed the following formula: OEi,j = Raw Scorei,j–Random Scorei,j. The overall expression of a gene i in sample j is then defined as: OEi,j = Raw Scorei,j−Random Scorei,j. Our scoring formula is: Score = (OEDBH-AS1+OECST3+OEHLA-DP1+OESFTPD)–(OEFAM83A + OEITGB4+OEOAS1+OEFHL2+OES100P + OEFSCN1+OE SPP1).
Statistical analysis
For NCC cohort, the lockup date of follow-up was July 28th, 2022, with a median follow-up time of 52.4 [interquartile range (IQR): 39.8, 62.3]. The origin and start times for survival analysis were the same. The survival analysis was carried out using R function survfit from survival package. Kaplan–Meier survival curves were plotted, and log-rank test was performed using R function ggsurvplot from survminer package. Cox regression analysis was applied to determine the hazard ratio (HR)28,29 along with its corresponding 95% confidence interval (95% CI). Wilcoxon rank-sum test was used to compare groups of variables. Kruskal–Wallis test was used to compare three or more groups of variables. Fisher's exact test was used to compare categorical variables. Clinicopathological features associated with lung cancer and significantly associated with survival through univariable Cox regression analyses (with an alpha level <0.2) were included in the multivariable Cox regression analyses. A statistical significance threshold of P < 0.05 was used. Statistical analysis was performed using R (version 4.0.3).
Role of funders
This study was independently conducted by authors and the funders had no role in study design, collection, analysis, interpretation, manuscript writing and submission.
Results
The single cell-originated landscape and cell–cell mutual interaction of patients with lung adenocarcinoma
STRT seq was performed on tumour tissues and matched normal tissues from 14 patients with LUAD (NCC cohort). The clinicopathological characteristics of these patients were listed in Table 1 and the study flowchart was shown in Fig. 1. A total of 5543 cells were included for analysis, revealing the presence of seven major cell types based on their characteristic expression of typical cell markers. These cell types included epithelial cells, alveolar cells, and various immune cell populations, such as Ma/Mo, T lymphocytes, B lymphocytes, mast cells, and follicular dendritic cells (Figure S1a).
Table 1.
The clinicopathological characteristics of patients enrolled.
| NCC | TCGA | GSE31210 | GSE8894 | GSE68571 | GSE42127 | GSE4573 | GSE37745 | GSE30219 | GSE50081 | NCC-bulk | NCC-ICIs | NCC-ICIs | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | 14 | 522 | 226 | 138 | 86 | 176 | 130 | 172 | 106 | 170 | 183 | 40 | 22 |
| Cancer Type | LUAD | LUAD | LUAD | LUAD + LUSC | LUAD | LUAD + LUSC | LUAD | LUAD + LUSC | LUAD + LUSC | LUAD + LUSC | LUAD | Lung cancer | LUAD |
| Agea (years) | 58.3 ± 9.0 | 65.4 ± 10.0 | 59.3 ± 7.8 | 60.8 ± 9.5 | 63.8 ± 9.8 | 66.3 ± 9.7 | 67.5 ± 9.8 | 63.8 ± 9.2 | 61.5 ± 11.6 | 68.8 ± 9.4 | 60.5 ± 8.8 | 56.9 ± 9.2 | 55 ± 8.9 |
| Biological and Self-reported Sex | |||||||||||||
| Female | 8 (57%) | 280 (54%) | 121 (54%) | 34 (24%) | 51 (59%) | 83 (47%) | 48 (37%) | 80 (47%) | 20 (19%) | 80 (47%) | 105 (57%) | 10 (25%) | 9 (41%) |
| Male | 6 (43%) | 242 (46%) | 105 (46%) | 104 (75%) | 35 (41%) | 93 (53%) | 82 (63%) | 92 (53%) | 86 (81%) | 90 (53%) | 78 (43%) | 30 (75%) | 13 (59%) |
| TNM Stage | |||||||||||||
| I | 6 (43%) | 279 (53%) | 168 (74%) | – | 67 (78%) | 112 (64%) | 73 (56%) | 110 (64%) | 75 (71%) | 119 (70%) | 79 (43%) | – | – |
| II | 1 (7%) | 124 (24%) | 58 (26%) | – | 32 (18%) | 34 (26%) | 34 (20%) | 18 (17%) | 51 (30%) | 44 (24%) | – | – | |
| III | 6 (43%) | 85 (16%) | – | – | 19 (22%) | 26 (15%) | 23 (18%) | 23 (13%) | 4 (4%) | – | 60 (33%) | 5 (12.5%) | 4 (18%) |
| IV | 1 (7%) | 26 (5%) | – | – | – | 5 (3%) | – | 4 (2%) | 5 (5%) | – | – | 35 (87.5%) | 18 (82%) |
| Smoking | |||||||||||||
| Ever | 2 (14%) | 356 (68%) | 111 (49%) | – | 74 (86%) | – | 120 (92%) | – | – | 23 (18%) | 57 (31%) | 24 (60%) | 9 (41%) |
| Never | 12 (86%) | 166 (32%) | 115 (51%) | – | 9 (10%) | – | 4 (3%) | – | – | 92 (72%) | 126 (69%) | 16 (40%) | 13 (59%) |
| TP53 | |||||||||||||
| MUT | – | 247 (47%) | – | – | – | – | – | – | – | – | – | – | – |
| WT | – | 260 (50%) | – | – | – | – | – | – | – | – | – | – | – |
| EGFR | |||||||||||||
| 70 (13%) | 127 (56%) | – | – | – | – | – | – | – | – | – | – | ||
| WT | 2 (14%) | 437 (84%) | 68 (30%) | – | – | – | – | – | – | – | – | – | – |
| KRAS | |||||||||||||
| MUT | – | 140 (27%) | 20 (9%) | – | 39 (45%) | – | – | – | – | – | – | – | – |
| WT | – | 367 (70%) | 68 (30%) | – | 46 (54%) | – | – | – | – | – | – | – | – |
| ALK | |||||||||||||
| MUT | 1 (7%) | 23 (4%) | – | – | – | – | – | – | – | – | – | – | – |
| WT | 13 (93%) | 484 (93%) | – | – | – | – | – | – | – | – | – | – | – |
| Platform | – | Illumina-Hiseq | Affy.Plus 2 | Affy.Plus 2 | Affy.HuGeneFL | Illu.WG-6 V3 | Affy.U133A | Affy.Plus 2 | Affy.Plus 2 | Affy.Plus 2 | – | – | – |
| Reference | – | TCGA | Okayama et al., 2012 | Lee et al., 2008 | Beer et al., 2002 | Tang et al., 2013 | Raponi et al., 2006 | Botling et al., 2013 | Rousseaux et al., 2013 | Der et al., 2014 | – | – | – |
Abbreviations: LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; TP53, Tumour Protein P53; MUT, mutation; WT, wild type; EGFR, epidermal growth factor receptor; KRAS, kirsten rat sarcoma viral oncogene; ALK, anaplastic lymphoma kinase; TKI, tyrosine kinase inhibitor; N/A = Not Applicable; SD, standard deviation.
Mean (SD).
Fig. 1.
The overview of this study. 14 samples from the NCC cohort were analysed through STRT sequencing. By separately detecting differentially-expressed genes and key genes of TME landscape, EPCAM cells, T cells and Ma/Mo cells, 11 genes (HLA-DPB1, FAM83A, ITGB4, OAS1, FHL2, FSCN1, S100P, SFTPD, SPP1, DBH-AS1, CST3) were obtained to construct a prognostic and immunotherapeutic predictive signature with validation from public datasets and in-house cohorts. Abbreviations: STRT, Single-cell Tagged Reverse Transcription; EPCAM, epithelial cell adhesion molecule; Ma/Mo, macrophage and monocyte; TME, tumour microenvironment; HR, hazard ratio.
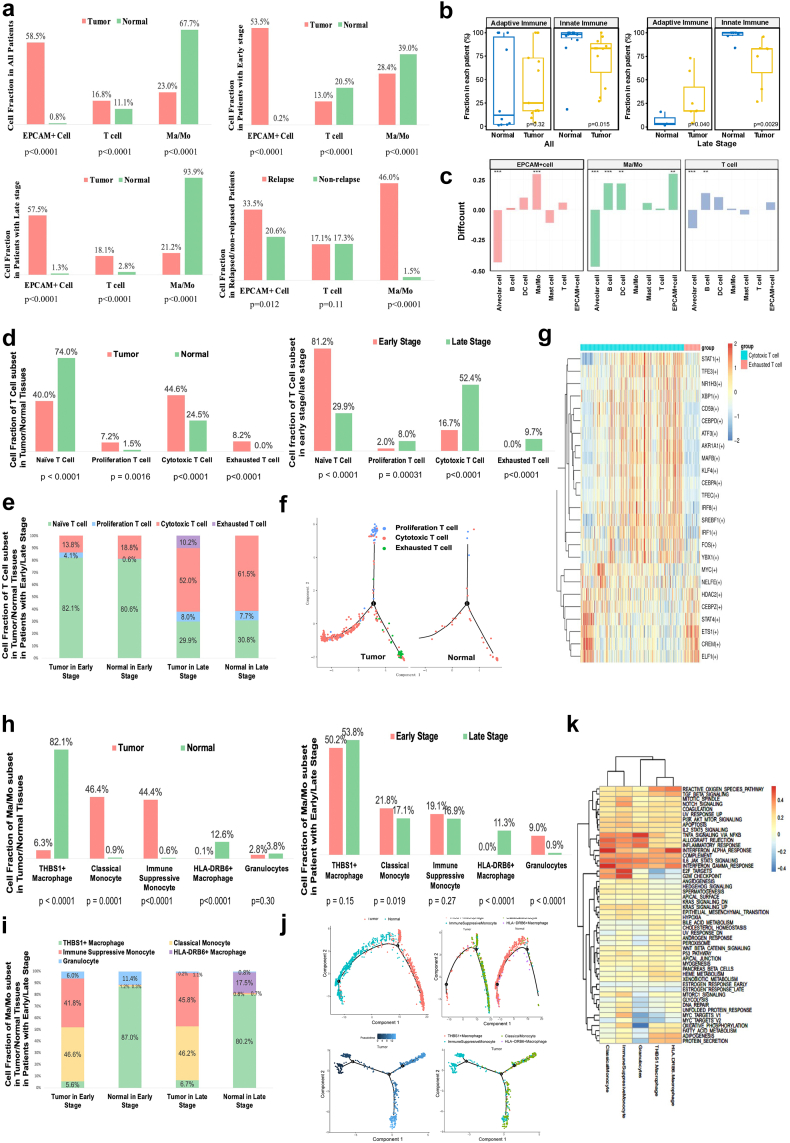
Generally, the most annotated cells were Ma/Mo cells, followed by EPCAM + epithelial cells, and T lymphocytes. Almost all EPCAM + epithelial cells were captured in tumour tissues compared to normal tissues, suggesting the captured EPCAM + epithelial cells mainly represented tumour cells in our study. In all patients, a higher abundance of T lymphocytes was observed in tumour tissues compared to normal tissues. However, inconsistent results could be observed across different clinical stages. Among patients in the early stages (Stage I-II), normal tissues exhibited a higher T cell count, whereas in patients with late stages (Stage III-IV), tumour tissues recruited more T cells, suggesting a progressive increase in T cell infiltration within tumour tissues as the tumour developed, underscoring the significance of T cell-targeted immunotherapy particularly for patients in advanced stages. In contrast, Ma/Mo cells were predominantly enriched in normal tissues, particularly in patients with advanced stages, highlighting the need for further exploration of the potential therapeutic strategies targeting Ma/Mo cells (Fig. 2a).
Fig. 2.
The subdivision analyses of EPCAM cells, T cells and Ma/Mo cells. (a) Cell fraction difference between tumour vs. normal, relapsed vs. non-relapsed in different patient groups. P values were determined by Fisher's exact test. (b) The differences in adaptive and innate immune infiltration between tumour tissue and normal tissue in patients with all stages and late stages separately. P values were determined by Wilcoxon rank-sum test. (c) The analysis of cellular interaction in EPCAM + cell, Ma/Mo cell and T cell with different type of cells. ∗∗P < 0.01; ∗∗∗P < 0.001. P values were determined by Fisher's exact test. (d) Cell fraction of T cell subset in tumour/normal tissues and early/late stage. P values were determined by Fisher's exact test. (e) Cell fraction of T cell subset in tumour/normal tissues in patients with early/late stage. (f) The T cell differentiation trajectory analysis between tumour/normal tissues. (g) The heatmap of differentially-expressed transcriptomic factors between cytotoxic T cells and exhausted T cells. (h) Cell fraction of Ma/Mo subset in tumour/normal tissues, and early and late stage. P values were determined by Fisher's exact test. (i) Cell fraction of Ma/Mo cell subset in tumour/normal tissues in patients with early/late stage. (j) The Ma/Mo cell differentiation trajectory analysis. (k) The heatmap of different functions in different Ma/Mo subsets. Abbreviations: EPCAM, epithelial cell adhesion molecule; Ma/Mo, macrophage and monocyte; DEGs, differentially expressed genes.
To provide a more comprehensive and direct illustration of the immune status, we conducted further analyses of innate and adaptive immune cell infiltration concerning different regions and stages. Our findings revealed that innate immune cell infiltration (Ma/Mo cells, Mast cells, Dendritic cells) was notably more pronounced in normal tissues, while in contrast, tumour tissues exhibited a higher level of adaptive immune invasion (T cells, B cells), particularly in patients with advanced stages when compared to normal tissues (Fig. 2b). However, there were no difference in immune status between tumour tissues and normal tissues in patients with early stage (Figure S1b). These results suggested that the significance of adaptive immunity increases progressively with the tumour development, while innate immunity primarily functions within the adjacent immune microenvironment.
Moreover, we focused on the difference between relapsed vs. non-relapsed patients. 12 out of 14 patients had complete follow-up information, and the other two patients were lost to follow-up due to their subjective withdrawl from the regular visit or on-line communication. Among the 12 patients, six patients experienced disease recurrence, with the median follow-up time of 52.4 [IQR 39.8, 62.3] months. Among the relapsed patients, the proportion of Ma/Mo cells was significantly higher compared to the non-relapsed ones, again emphasizing the potential significance of Ma/Mo cells in tumour progression (Fig. 2a).
To better illustrate the interaction between tumour cells and immune cells, we conducted a detailed analysis of cell-to-cell communication involving EPCAM + cells and other immune cells. Our findings revealed that the interactions between Ma/Mo cells and EPCAM + cells, as well as Ma/Mo cells and T cells, were more pronounced in tumour tissues compared to normal tissues, highlighting the close interplay between the immune system and malignant cells, as well as the intimate mutual interactions among various immune cell types within the tumour environment (Fig. 2c). Further ligand-receptor analyses revealed significant enrichment of AXL-GAS6 and CXCR3-CXCL9 interactions in the communication between tumour cells and macrophages in tumour tissues (Figure S1c–d). In the communication between T cells and macrophages, we observed significant enrichment of interactions involving HLA-DP1/TNFSF13B and LAIR1/LILRB4. Notably, among these ligands and receptors, HLA-DPB1 showed a consistent positive correlation with prognosis, as demonstrated consistently across TCGA and GSE 3141 dataset (Table S1).
The tempospatial distribution and the differential gene expression analyses between heterogenous subsets of EPCAM + cell
To identify candidate genes associated with tumour development, we conducted a subdivision analysis of EPCAM + cell subsets in 11 patients with adequate EPCAM + cell annotated, revealing substantial heterogeneity among individuals (Fig. 1).
We conducted DGE analysis on EPCAM + cells, comparing tumour tissues vs. adjacent normal tissues, early-stage vs. late-stage cases, and relapsed vs. non-relapsed patients. The immune-related functional enrichment analysis revealed significant interactions between EPCAM + cells and both T cells and Ma/Mo cells, underscoring their importance in the context of tumour development (Figure S2a–l). Among the DEGs, we identified 8 key genes consistently associated with prognosis across multiple datasets, including DEGs between tumour tissues vs. normal tissues (FAM83A, ITGB4, OAS1, FHL2), between early vs. late stage (S100P, FSCN1), and between relapsed vs. non-relapsed patients (SFTPD, SPP1) (Table S1). Of these 8 genes, SFTPD was identified as a protective gene, while the remaining 7 genes were associated with poorer prognoses based on survival analyses across multiple datasets.
The temporal/spatial distribution, the subset transformation analyses and the differential gene expression analyses between heterogenous subsets of T cells
Next, we subcategorized T cells into four traditional subsets: naïve T cells, proliferative T cells, cytotoxic T cells and exhausted T cells, and further investigated the critical factors associated with T cell subset transformation (Figure S3a).
We observed a significant enrichment of naïve T cells in adjacent normal tissues, while tumour tissues predominantly contained a higher proportion of proliferation T cells, cytotoxic T cells, and almost all exhausted T cells, suggesting that T cells infiltrating tumour tissues underwent a complete immune process of activation, proliferation, and exhaustion within the immune microenvironment (Fig. 2d). In patients with early stage, we observed a higher presence of naïve T cells and an absence of exhausted T cells. Conversely, in patients with late stage, there was an increased abundance of proliferative T cells, cytotoxic T cells and exhausted T cells, suggesting that T cell-mediated killing processes were more prevalent as the tumour progressed (Fig. 2d). In patients with early stage, we observed increased proliferation of T cells in tumour tissues compared to normal tissues. In contrast, in patients with late stage, exhausted T cells firstly appeared, and the fraction of cytotoxic T cells decreased in tumour tissues compared to normal tissues, again indicating that infiltrated T cells initiated to proliferate in the primary stage of tumourigenesis while generally exhausted as tumour developed (Fig. 2e). The further function analyses of these four subsets emphasized different hallmark function of different phase of T cells (Figure S3b–e).
To identify key factors associated with T cell transformation, we conducted T cell differentiation trajectory analysis, followed by DGE analysis and transcription factor analysis among various CD8+ T cell subsets. Our findings revealed a noticeable differentiation process from proliferative T cells to cytotoxic T cells and exhausted T cells within tumour tissues, emphasizing the importance of identifying key factors related to the transformation from cytotoxic T cells to exhausted T cells (Fig. 2f). Through DGE analysis, we identified several genes associated with the transformation from cytotoxic T cells to exhausted T cells. Based on following validation of survival analysis, we pinpointed two key genes, DBH-AS1 and CST3, that consistently correlated with a favourable prognosis across multiple datasets (Figure S3f, Table S1). The transcription factor analyses comparing cytotoxic and exhausted T cells revealed that cytotoxic T cells were more inclined to immune processes, while exhausted T cells were more linked to cell metabolism (Fig. 2g).
The temporal/spatial distribution and the differential gene expression analyses between heterogenous subsets of Ma/Mo cells
Given the high heterogeneity of Ma/Mo cells, we further characterised Ma/Mo cells using multiple specific markers, revealing the presence of five distinct subpopulations, namely classical monocytes, immunosuppressed monocytes, granulocytes, THBS1+ macrophages, and HLA-DRB6+ macrophages (Figure S4a).
We observed a significant increase in monocytes, including both classical monocytes and immunosuppressed monocytes, in tumour tissues compared to normal tissues, regardless of whether patients were in the early or late stage of the disease (Fig. 2h). This highlighted the consistent and underlying immune suppression status within tumour tissues and underscored the potential for future interventions targeting these monocytes, especially the immunosuppressed monocyte subset, to reshape the immune microenvironment. Interestingly, THBS1+ macrophages were predominantly found in normal adjacent tissues, irrespective of the patient's stage, indicating the stability of this subset in the peritumoural environment throughout tumour development (Fig. 2h). HLA-DRB6+ macrophages were predominantly observed in samples from patients with late stage, suggesting a gradual increase in the interaction of this macrophage subset with tumour cells as the disease progressed (Fig. 2i). Similarly, Ma/Mo cell pseudotime inference analysis exhibited a comparable distribution pattern, it revealed that immunosuppressive monocytes had a tendency to differentiate into classical monocytes, together constituting the two most prevalent subsets of Ma/Mos in tumour tissues, highlighting the importance of exploring factors that might promote this transformation in the future (Fig. 2j).
We next conducted analyses of the function and hallmark pathways among different Ma/Mo subsets, revealing distinct physiological processes emphasized by each subset (Fig. 2k, Figure S4a–c). In general, classical monocytes and HLA-DRB6+ macrophages exhibited a more active inflammatory response. Furthermore, the differential functional analysis comparing tumour and normal tissues indicated an inflammatory reaction in THBS + macrophages (Figure S4c and d).
To identify key genes related to prognosis, we conducted a DGE analysis of Ma/Mo cells comparing tumour tissues to normal tissues (Figure S4e and f), and identified SPP1 to be differentially expressed in tumour tissues as well as consistently associated with poorer prognosis in multiple datasets (Table S1), the same gene identified by DGE analysis in EPCAM + cells between relapsed/non-relapsed patients.
The in-vitro validation of candidate genes
To further validate the function of candidate genes, we conducted in-vitro cell experiments on the selected 11 key genes.
Overall, the selected 11 key genes were: gene related to immunocytes interactions (HLA-DPB1), genes specifically prognosis-related in tumour cells (differentially-expressed genes between tumour tissues/adjacent tissues [FAM83A, ITGB4, OAS1, FHL2] & early stage/late stage [S100P, FSCN1] & relapsed/non-relapsed [SFTPD, SPP1]), genes related to T cell transformation (DBH-AS1, CST3), and gene associated with Ma/Mo cell distribution (SPP1, the same gene as differentially-expressed genes of EPCAM + cells between relapsed/non-relapsed patients). Among them, HLA-DPB1, SFTPD, DBH-AS1 and CST3 were positively correlated with survival, while the rest demonstrated negative correlation with the survival (Table S1).
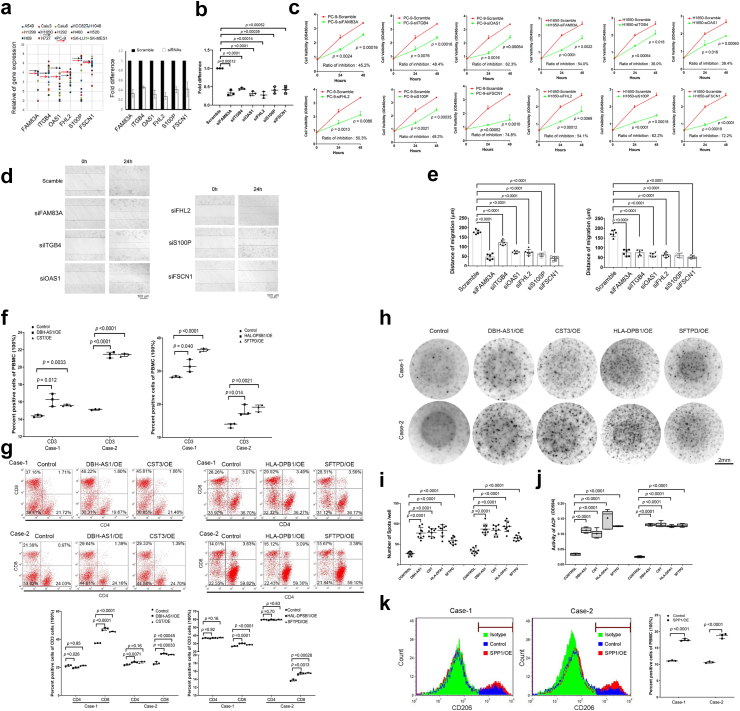
We conducted cell experiments using specific siRNAs for genes with different functions. After being transfected with FAM83A, ITGB4, OAS1, FHL2, S100P, and FSCN1 siRNAs, we observed a significant inhibition of cell proliferation and migration ability in H1650 and PC-9 cell lines (Fig. 3a–e). After being electroporated with DBH-AS1, CST3, HLA-DPB1, and SFTPD, peripheral blood mononuclear cells (PBMC) derived T cells with overexpression of these genes displayed significantly enhanced immune effects, as indicated by ELISPOT assays, as well as notably higher expression levels of CD3 and CD8, as demonstrated by flow cytometry (Fig. 3f–j). After being electroporated with SPP1, the PBMC-derived macrophages exhibited an enhanced M2 polarization (Fig. 3k). These in-vitro results all aligned with the clinical findings mentioned earlier, providing further validation of the prognostic capability of these candidate genes.
Fig. 3.
Results of In-vitro cell experiments. (a) The scatter plot shows the expression difference of multiple genes in multiple lung cancer cell lines. The black arrow represents H1650 and the red arrow represents PC9. (b) The fold change of the gene expression following multiple gene respectively knockdown in PC-9 cells was confirmed by RT-PCR. (c) The cell variability curve demonstrated a significant decrease in cell survival following genes knockdown in PC9 and H1650. (d) Wounding assay shows cells movement following genes knockdown after 24-h. Scale Bar = 100 μm. (e) The bar chart shows the distance of migration by the cells of genes knockdown in PC-9 and H1650. (f) The flow cytometry results revealed the presence of the percentage of CD3 cells in the peripheral blood mononuclear cells (PBMCs) of patients with overexpression of DBH-AS1, CST3, HLA-DPB1 and SFTPD. (g) The flow cytometry results the percentage of CD4 and CD8 in PBMCs of patients. (h) and (i) Elispots assay shows number of spots signature in DBH-AS1, CST3, HLA-DPB1 and SFTPD overexpressed cells of PBMCs. The horizontal lines denoted the mean quantity of spots per well. Scale Bar = 2 mm. (j) ELISA assay shows ACP activity in genes overexpressed cells of PBMCs. (k) The flow cytometry results revealed the presence of the percentage of CD206 cells in PBMCs with SPP1 overexpression. All experiments were conducted independently three times. The error bars refer to the standard deviation of the values. P values were determined by independent t-test. Two-tail paired t-test for b and c by mean with SD; One-way analysis of variance for e, i and j by mean with SD; Chi-square test for f, g and k by medium with 95% CI. Abbreviations: RT-PCR, reverse transcription-polymerase chain reaction; OE, overexpression model; siRNA, small interfering RNA; CI, confidence interval.
The prognostic signature development and validation
Given the crucial role of various cells and their interactions in the immune microenvironment, we subsequently combined the candidate genes to establish an integrated prognostic signature.
The 11-gene signature was established based on the positive or negative impact on survival: Score = (OEDBH-AS1 + OECST3 + OEHLA-DP1 + OESFTPD)−(OEFAM83A + OEITGB4 + OEOAS1 + OEFHL2 + OES100P + OEFSCN1 + OE SPP1), classifying High-Score and Low-Score subgroup.
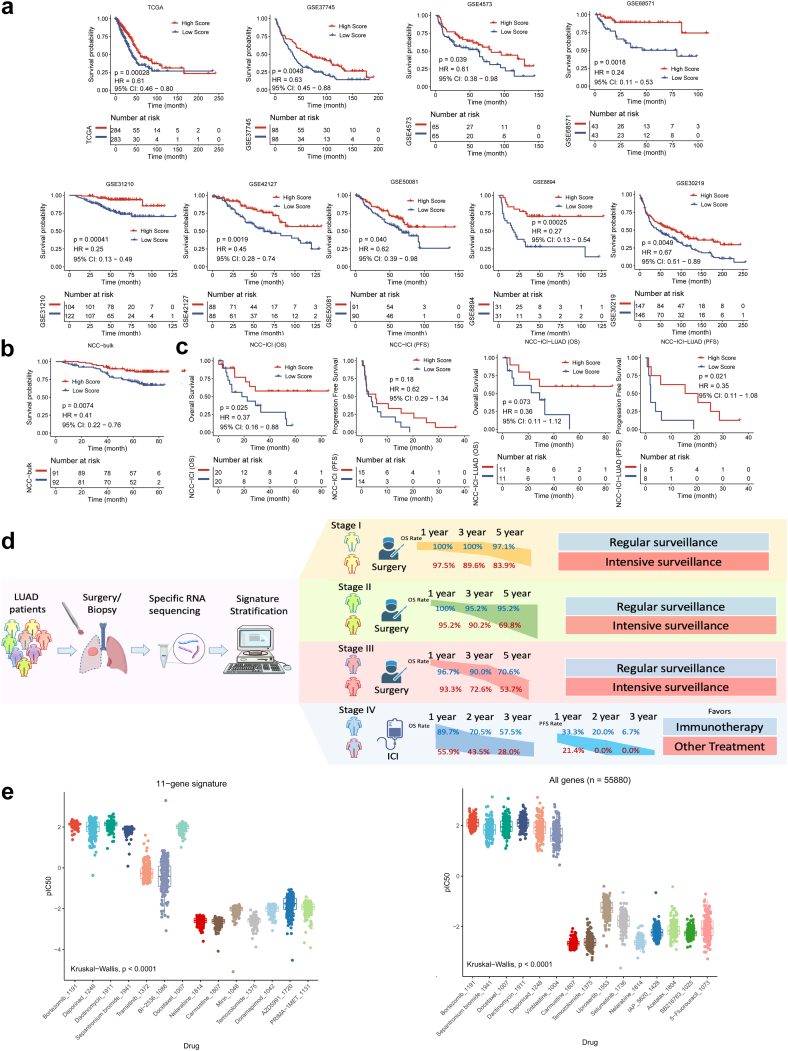
Next, we assessed the prognostic performance of the signature in nine public datasets and our in-house dataset (NCC-bulk cohort with 183 samples, with a median follow-up time of 66.3 [ IQR: 41.4, 72.2 ] months). The baseline clinicopathological characteristics of these datasets were summarized in Table 1. In TCGA and eight GSE cohorts, patients in the High-score group exhibited significantly prolonged overall survival (OS) compared to those in the Low Score group (Fig. 4a, Table S2). In our in-house validation NCC-bulk dataset, we also observed a significantly longer OS in patients in the High-Score subgroup compared to the Low-Score subgroup (median OS NA [not applicable] vs. NA, HR 0.41 [95% CI 0.22–0.76], P = 0.0074, log-rank test.) (Fig. 4b, Table S2).
Fig. 4.
The validation of the signature and the overallflow-diagram. (a) The OS stratified by High/Low score in TCGA dataset, GSE8894, GSE68571, GSE42127, GSE4573, GSE37745, GSE50081, GSE31210, GSE30219 datasets. P values were determined by log-rank test. (b) The OS stratified by High/Low score in NCC-bulk cohort. P values were determined by log-rank test. (c) The OS and PFS stratified by High/Low score in all patients and patients with LUAD in NCC-ICI cohort, respectively. P values were determined by log-rank test. (d) The clinical flow-diagram demonstrating different overall survival rate of different stages and suggestions for surveillance stratified by the signature. (e) Drug sensitivity prediction analysis of 183 samples to 198 drugs was predicted based on 11-gene signature and prediction based on all genes (n = 55,880) of the RNA-seq expression profile. P values were determined by Kruskal–Wallis test. Abbreviations: CI, confidence interval; DEGs, differentially expressed genes; HR, hazard ratio; ICI, immune checkpoint inhibitors; IC50, half maximal inhibitory concentration; OS, overall survival; PFS, progression-free survival; TCGA, The Cancer Genome Atlas.
To validate the signature's independence as a prognostic biomarker, we conducted multivariate analyses. The results indicated that, among important clinicopathological characteristics such as age, gender, TNM stage, smoking status, etc., the 11-gene scoring signature could function as an independent biomarker (Table 2).
Table 2.
The Multivariable Cox regression analyses of OS in separate cohort.
| Dataset | Factor | HR (95% CI) | P value | Dataset | Factor | HR | P value |
|---|---|---|---|---|---|---|---|
| TCGA | 11-gene signature (High Score vs. Low Score) | 0.5 (0.4–0.8) | 0.0046 | GSE68571 | 11-gene signature (High Score vs. Low Score) | 0.3 (0.1–0.8) | 0.021 |
| Age (years) | 1.0 (1.0–1.0) | 0.10 | Age (years) | 1.0 (1.0–1.1) | 0.34 | ||
| Sex (Male vs. Female) | 1.3 (0.8–1.9) | 0.27 | Sex (Male vs. Female) | 1.5 (0.6–3.7) | 0.38 | ||
| Stage (III-IV vs. I-II) | 2.6 (1.7–4.0) | <0.0001 | Stage (III vs. I) | 5.3 (2.1–13.4) | 0.00045 | ||
| Smoking status (Ever vs. Never) | 0.7 (0.4–1.2) | 0.15 | Smoking (Ever vs. Never) | 0.8 (0.2–4.0) | 0.81 | ||
| GSE31210 | 11-gene signature (High Score vs. Low Score) | 0.3 (0.1–0.8) | 0.017 | KRAS status (Mutated vs. Wild Type) | 1.5 (0.6–3.5) | 0.40 | |
| Age (years) | 1.0 (1.0–1.1) | 0.17 | GSE50081 | 11-gene signature (High Score vs. Low Score) | 0.6 (0.3–1.0) | 0.056 | |
| Sex (Male vs. Female) | 1.1 (0.5–2.7) | 0.82 | Age (years) | 1.0 (1.0–1.0) | 0.22 | ||
| Stage (II vs. I) | 3.2 (1.6–6.4) | 0.0010 | Sex (Male vs. Female) | 1.9 (1.1–3.2) | 0.020 | ||
| Smoking status (Ever vs. Never) | 1.2 (0.5–3.0) | 0.70 | Stage (II vs. I) | 1.9 (1.2–3.2) | 0.012 | ||
| GSE37745 | 11-gene signature (High Score vs. Low Score) | 0.6 (0.4–0.9) | 0.0055 | Smoking status (Ever vs. Never) | 0.8 (0.4–1.8) | 0.62 | |
| Age (years) | 1.0 (1.0–1.0) | 0.0056 | GSE30219 | 11-gene signature (High Score vs. Low Score) | 0.7 (0.5–1.0) | 0.048 | |
| Sex (Male vs. Female) | 1.0 (0.7–1.4) | 0.90 | Age (years) | 1.0 (1.0–1.1) | <0.0001 | ||
| Stage (III-IV vs. I-II) | 1.7 (1.1–2.6) | 0.012 | Sex (Male vs. Female) | 1.4 (0.9–2.3) | 0.14 | ||
| GSE4573 | 11-gene signature (High Score vs. Low Score) | 0.6 (0.4–1.0) | 0.043 | T Stage (T3-4 vs. T1-2) | 1.7 (1.1–2.6) | 0.014 | |
| Age (years) | 1.0 (1.0–1.0) | 0.15 | N Stage (N+ vs. N0) | 1.8 (1.3–2.7) | 0.0015 | ||
| Sex (Male vs. Female) | 1.3 (0.8–2.3) | 0.27 | M Stage (M+ vs. M0) | 3.2 (1.3–8.0) | 0.014 | ||
| Stage (III-IV vs. I-II) | 2.08 (1.1–3.7) | 0.029 | NCC-bulk | 11-gene signature (High Score vs. Low Score) | 0.5 (0.2–0.9) | 0.033 | |
| Smoking status (Ever vs. Never) | 0.5 (0.1–2.3) | 0.38 | Age (years) | 1.0 (1.0–1.1) | 0.25 | ||
| GSE42127 | 11-gene signature (High Score vs. Low Score) | 0.5 (0.3–0.9) | 0.013 | Sex (Male vs. Female) | 1.8 (0.6–5.0) | 0.27 | |
| Age (years) | 1.0 (1.0–1.1) | 0.033 | Stage (III-IV vs. I-II) | 3.2 (1.7–6.1) | 0.00039 | ||
| Sex (Male vs. Female) | 1.2 (0.7–2.1) | 0.52 | Smoking status (Ever vs. Never) | 1.6 (0.6–4.4) | 0.34 | ||
| Stage (III-IV vs. I-II) | 1.7 (1.0–3.1) | 0.072 |
The immunotherapeutic predictive potency of the signature
To evaluate the signature's predictive potency for immunotherapy, we conducted immune microenvironment analyses using CYBERSORT and XCELL, and also performed survival analyses in one in-house cohort, which comprised patients with lung cancer treated with immune checkpoint inhibitors, stratified by the model.
In the High-Score group, CD8+ T lymphocytes were the most commonly enriched cells, while in the Low-Score group, M0/M1 macrophages predominated. Additionally, the immune score, stroma score, and microenvironment score were generally higher in the High-Score group, indicating a more active immune microenvironment (Figure S5). Functional analyses revealed that the Low-Score group was linked to mitosis and cell proliferation, while the High-Score group showed associations with antigen presentation, membrane transport, and other immune reactions. These findings underscore the “hot” immune microenvironment in the High-Score group and suggest the model's potential predictive power for patients undergoing immunotherapy (Figure S6).
Therefore, we enrolled an in-house cohort of 40 patients with lung cancer treated with single immune checkpoint inhibitors (named NCC-ICI cohort, Table 1, with a median follow-up time of 19.7 [IQR: 8.5, 42.9] months). In 40 patients with OS results, patients in High-Score group harbored a significantly longer OS than those in Low-Score group (Median OS NA vs. 18.3 months, HR 0.37 [95% CI 0.16–0.88], P = 0.025, log-rank test). In 29 patients with progression-free survival (PFS) outcomes, patients in High-Score group had a longer PFS compared to those in Low-Score group (Median PFS 5.6 vs. 3.4 months, HR 0.62 [95% CI 0.29–1.34], P = 0.18, log-rank test), although not statistically significant, there was a clear trend (Fig. 4c). For the 22 patients with LUAD, patients in High-Score subgroup also exhibited a statistically higher OS and significantly higher PFS (median OS NA vs. 28.9 months, HR 0.36 [95% CI 0.11–1.12], P = 0.073, median PFS 17.9 vs. 2.0 months, HR [95% CI 0.11–1.08], P = 0.021, log-rank test) (Fig. 4c, Table S2). These results indicated the potential of this 11-gene signature to stratify beneficiaries from immunotherapeutic strategies, especially in patients with LUAD.
Thus, we drew a clinical flow-diagram to demonstrate the overall survival rate of patients with different stages stratified by this signature (stage I-III referenced on NCC-bulk cohort, and stage IV on NCC-ICI cohort), and made suggestions on clinical surveillance and treatment selection (Fig. 4c).
The prediction of drug sensitivity by the signature
To validate the representativeness of the 11 key genes, we conducted drug sensitivity prediction analyses using both all gene expression profiles and the 11 key genes. We obtained response data for 198 drugs from the Genomics of Drug Sensitivity in Cancer (GDSC) database. In the NCC cohort with 14 patients, bortezomib, dactinomycin, and daporinad were the most sensitive drugs, while carmustine and temozolomide were the most resistant drugs when considering all gene expression profiles (n = 20,317) (Figure S7). Notably, the drug prediction results based on the 11-gene signature were consistent with those based on all genes. We further validated these findings in the NCC-bulk cohort, where bortezomib, daporinad, dactinomycin, and docetaxel were identified as sensitive drugs, while nelarabine, carmustine, and temozolomide were insensitive. This consistency underscores the representativeness of these 11 genes across all RNA profiles (Fig. 4e).
Discussion
In this study, we systematically characterised the tumour heterogeneity and immune microenvironment landscape of LUAD using comprehensive single-cell analysis. We provided a detailed exploration of the spatial and temporal dynamics of key cell subsets, delved into the functions and intrinsic transformation modes of key immune cells (T cell and Ma/Mo cell subsets), and ultimately constructed a prognostic and immunotherapeutic predictive signature based on 11 key genes closely related to tumour development, immune cell interactions, T cell transformation, or Ma/Mo cell distribution. To the best of our knowledge, this study represented the frontier of a comprehensive single-cell-characterised signature combining both tumour heterogeneity and immune microenvironment for prognosis stratification.
Current immunotherapy, especially anti-PD-1/PD-L1 therapies, primarily relies on T cell-mediated adaptive immunity.33,34 We emphasized the importance of T cell-based treatment for patients with advanced stage due to the gradually increasing adaptive immune infiltration, particularly T cells, as tumours developed. Additionally, research has underscored the essential role of innate immune components in activating and modulating the adaptive anti-tumour immune response, leading to the exploration and development of therapeutic strategies targeting the innate immune system, including NK cells and macrophages.35 Our study focused on Ma/Mo cells and provided further evidence to support the consideration of these cells as potential therapeutic targets in future research.
With advances in cell-originated detection techniques and bioinformatics, we have gradually unveiled cell–cell interactions. For instance, granulocytes can influence the recruitment and activation of dendritic cells, thus impacting T cell responses,36 and macrophages were reported to interact closely with CD8+ T cells and CD4+ T cells.37,38 However, limited interpretation of mutual interactions has hindered the translation of these observations into clinical practice. In our study, we highlighted the close interactions between T cells and Ma/Mo cells, as well as between EPCAM + cells and Ma/Mo cells, highlighting the crucial and central role of Ma/Mo cells in the TME. We also identified a key interaction-related gene, HLA-DPB1, a transmembrane glycoprotein mainly found on the surfaces of lymphocytes and macrophages, which plays an important role in promoting T cell proliferation and cytotoxic reactions.39
Currently, the intrinsic transformation of cell subsets has been recognized as a promising and potential exploration hotspot, especially the T cell transformation, due to its vital importance in TME. Among all the T cell subsets, the cytotoxic T cell was the most important component to kill cancer cells, however the inevitable transformation to exhausted T cell limited the long-lasting anti-tumour effect, making it important to explore the transition pattern and dig out key factor for promoting the T cell exhaustion. Our study provided a visual representation of the T cell evolution process alongside tumour development and highlighted the active metabolic processes associated with exhausted T cells. Previous research has suggested that cholesterol may contribute to the dysfunction and exhaustion of CD8+ T cells,40 implying the potential of targeting metabolic pathways, such as reducing cholesterol levels, as a promising strategy to reverse T cell dysfunction. Additionally, we identified two key genes related to T cell transformation, DBH-AS1 and CST3, which could enhance T cell immune activity.41,42 These findings may provide insights for future research on T cell recovery.
As another important component in the immune landscape, Ma/Mo cells were demonstrated to interact closely with both tumour cells and T cells,37,38,43,44 also shown in our study, implying its essentiality in TME. In our study, we identified a key gene, SPP1, which was significantly related to both tumour development and macrophage polarization. SPP1 has been confirmed by previous studies to up-regulate PD-L1 mediating the polarization of macrophages and promote immune escape, and high expression of SPP1 can lead to low infiltration of CD8+ T cells and high infiltration of M2 macrophages. Other than the regulatory effect on M2 polarization and T cells, SPP1 was also reported to associate with the apoptosis of lung adenocarcinoma cells.45,46 Our finding again highlighted the importance and rationale for focusing on SPP1 for future drug development.
Recent studies have highlighted the concept of tumour niche integrity, where the components of the immune environment and tumour cells form an interactive and symbiotic entity.2,47 Current prognostic or predictive models were often based on individual components, lacking the comprehensive integration of different elements, which limited their representation of the integrated TME. In our study, we combined the key factors of heterogeneous tumourigenesis with essential microenvironment components to create an integrated signature based on single-cell characterization. This signature stratified patients into High-Score or Low-Score groups, effectively predicting prognosis, and was validated in both public databases and our in-house cohorts, indicating a more intensive surveillance of patients with Low-Score in clinical practice. Importantly, it also showed the potential to predict improved progression-free survival in our in-house cohort treated with immune checkpoint inhibitors. Patients in our immunotherapy cohort were treated with single-agent immunotherapy using PD-1 or PD-L1 inhibitors, including a variety of commonly used immunotherapy regimens (atezolizumab, Nivolumab, pembrolizumab, toripalimab, sintilimab, etc.). Our results suggest the selection of treatment in patients with Low-Score might have to turn to other therapeutic strategies.
Furthermore, in drug prediction analysis, we found that the response predictions using our 11-gene signature and all 55,880 genes from RNA-seq expression profiles significantly overlapped. This indicated the representativeness of the selected genes across all RNA profiles, underscoring the promising feasibility and potential practicality of our signature for specific RNA sequencing due to its representativeness and cost-effectiveness.
This study has certain limitations. Firstly, we used a previous generation single-cell sequencing technique, which identified fewer cells compared to newer techniques like 10× sequencing, leading to the cautious interpretation of our results. However, this technology allowed a measurement of broader range of gene expressions enabling the detection of long noncoding RNA (lncRNA); Moreover, the key genes we identified demonstrated the expected functionality in subsequent in vitro experiments, and their representative nature was confirmed in drug sensitivity analysis, further underscoring the high specificity of this technology. Secondly, the analyses involving other crucial immune cells, such as B cells, NK cells, etc. were limited, future research with higher depth and diversity could provide denser information. Thirdly, the unavoidable confounding factors, the measurement bias of sequencing data from different platforms, and the built-in selection bias in HRs48 may impact the viability of our model thus further standardization was warranted.
In conclusion, we provided a comprehensive view of the single cell-characterised TME landscape in LUAD, underscoring the importance of the current immunotherapy approach for advanced-stage tumour and highlighted the potential for future drug development by targeting TME components. Moreover, we developed an integrated signature consisted of 11 key genes incorporating both tumour heterogeneity and TME, as validated in multiple independent public datasets and our in-house cohorts, demonstrating its potential for prognostic stratification and treatment selections.
Contributors
Jiachen Xu: Conceptualization, methodology, investigation, data curation, formal analysis, writing–original draft, writing–review & editing, funding acquisition.
Yundi Zhang: Conceptualization, methodology, investigation, data curation, formal analysis, writing–original draft, writing–review & editing.
Man Li: Conceptualization, methodology, investigation, data curation, formal analysis, writing–review & editing.
Zhuo Shao: Conceptualization, methodology, investigation, visualization, data curation, formal analysis, writing–review & editing.
Yiting Dong: Data curation, writing–review & editing.
Qingqing Li: Methodology, investigation.
Hua Bai: Data curation, writing–review & editing.
Jianchun Duan: Data curation, writing–review & editing.
Jia Zhong: Data curation, writing–review & editing.
Rui Wan: Data curation, writing–review & editing.
Jing Bai: Methodology, visualization, data curation, formal analysis, writing–review & editing.
Xin Yi: investigation, data curation, formal analysis.
Fuchou Tang: Methodology, investigation, data curation.
Jie Wang: Conceptualization, methodology, investigation, data curation, supervision, writing–review & editing, funding acquisition.
Zhijie Wang: Conceptualization, methodology, investigation, supervision, writing–review & editing, funding acquisition.
Jiachen Xu, Yundi Zhang, Man Li and Zhuo Shao contributed equally as co-first authors.
Jie Wang and Zhijie Wang contributed equally as co-corresponding authors.
All authors read and approved the final version of the manuscript.
Jiachen Xu, Yundi Zhang, Man Li, Zhuo Shao, Jing Bai, Jie Wang and Zhijie Wang have verified the underlying data.
Data sharing statement
Data from publicly archive datasets are available from the Cancer Genome Atlas (TCGA), gene expression omnibus (GEO) database and OncoSG database (https://src.gisapps.org/OncoSG/), as publications cited in the manuscript. Data from in-house cohorts are available from the corresponding author on reasonable request.
Declaration of interests
The authors declare that they have no competing interests.
Acknowledgements
Support for the study was provided by National key research and development project (2022YFC2505004, 2022YFC2505000 to Z.W. and J.W.), Beijing Natural Science Foundation (7242114 to J.X.), National Natural Science Foundation of China (82102886 to J.X., 81871889 and 82072586 to Z.W.), Beijing Nova Program (20220484119 to J.X.), NSFC general program (82272796 to J.W.), NSFC special program (82241229 to J.W.), CAMS Innovation Fund for Medical Sciences (2021-1-I2M-012, 2022-I2M-1-009 to Z.W. and J.W.), Beijing Natural Science Foundation (7212084 to Z.W.), CAMS Key lab of translational research on lung cancer (2018PT31035 to J.W.), Aiyou Foundation (KY201701 to J.W.). Medical Oncology Key Foundation of Cancer Hospital Chinese Academy of Medical Sciences (CICAMS-MOCP2022003 to J.X.)
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2024.105092.
Contributor Information
Jie Wang, Email: zlhuxi@163.com.
Zhijie Wang, Email: wangzj@cicams.ac.cn.
Appendix A. Supplementary data
References
- 1.Sung H., Ferlay J., Siegel R.L., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Thai A.A., Solomon B.J., Sequist L.V., Gainor J.F., Heist R.S. Lung cancer. Lancet (London, England) 2021;398(10299):535–554. doi: 10.1016/S0140-6736(21)00312-3. [DOI] [PubMed] [Google Scholar]
- 3.Chansky K., Detterbeck F.C., Nicholson A.G., et al. The IASLC lung cancer staging project: external validation of the revision of the TNM stage groupings in the eighth edition of the TNM classification of lung cancer. J Thorac Oncol. 2017;12(7):1109–1121. doi: 10.1016/j.jtho.2017.04.011. [DOI] [PubMed] [Google Scholar]
- 4.Hua X., Zhao W., Pesatori A.C., et al. Genetic and epigenetic intratumor heterogeneity impacts prognosis of lung adenocarcinoma. Nat Commun. 2020;11(1):2459. doi: 10.1038/s41467-020-16295-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hussaini S., Chehade R., Boldt R.G., et al. Association between immune-related side effects and efficacy and benefit of immune checkpoint inhibitors–a systematic review and meta-analysis. Cancer Treat Rev. 2021;92 doi: 10.1016/j.ctrv.2020.102134. [DOI] [PubMed] [Google Scholar]
- 6.Barnes T.A., Amir E. HYPE or HOPE: the prognostic value of infiltrating immune cells in cancer. Br J Cancer. 2017;117(4):451–460. doi: 10.1038/bjc.2017.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Quail D.F., Joyce J.A. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013;19(11):1423–1437. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bense R.D., Sotiriou C., Piccart-Gebhart M.J., et al. Relevance of tumor-infiltrating immune cell composition and functionality for disease outcome in breast cancer. J Natl Cancer Inst. 2017;109(1) doi: 10.1093/jnci/djw192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fridman W.H., Zitvogel L., Sautes-Fridman C., Kroemer G. The immune contexture in cancer prognosis and treatment. Nat Rev Clin Oncol. 2017;14(12):717–734. doi: 10.1038/nrclinonc.2017.101. [DOI] [PubMed] [Google Scholar]
- 10.Hanahan D., Coussens L.M. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell. 2012;21(3):309–322. doi: 10.1016/j.ccr.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 11.Turley S.J., Cremasco V., Astarita J.L. Immunological hallmarks of stromal cells in the tumour microenvironment. Nat Rev Immunol. 2015;15(11):669–682. doi: 10.1038/nri3902. [DOI] [PubMed] [Google Scholar]
- 12.Guo X., Zhang Y., Zheng L., et al. Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nat Med. 2018;24(7):978–985. doi: 10.1038/s41591-018-0045-3. [DOI] [PubMed] [Google Scholar]
- 13.Wu K., Lin K., Li X., et al. Redefining tumor-associated macrophage subpopulations and functions in the tumor microenvironment. Front Immunol. 2020;11:1731. doi: 10.3389/fimmu.2020.01731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischer D.S., Fiedler A.K., Kernfeld E.M., et al. Inferring population dynamics from single-cell RNA-sequencing time series data. Nat Biotechnol. 2019;37(4):461–468. doi: 10.1038/s41587-019-0088-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liang L., Yu J., Li J., et al. Integration of scRNA-seq and bulk RNA-seq to analyse the heterogeneity of ovarian cancer immune cells and establish a molecular risk model. Front Oncol. 2021;11 doi: 10.3389/fonc.2021.711020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang A., Wang J., Liu N., et al. Integration of single-cell RNA sequencing and bulk RNA sequencing data to establish and validate a prognostic model for patients with lung adenocarcinoma. Front Genet. 2022;13 doi: 10.3389/fgene.2022.833797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pang J., Yu Q., Chen Y., Yuan H., Sheng M., Tang W. Integrating Single-cell RNA-seq to construct a Neutrophil prognostic model for predicting immune responses in non-small cell lung cancer. J Transl Med. 2022;20(1):531. doi: 10.1186/s12967-022-03723-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen J., Tan Y., Sun F., et al. Single-cell transcriptome and antigen-immunoglobin analysis reveals the diversity of B cells in non-small cell lung cancer. Genome Biol. 2020;21(1):152. doi: 10.1186/s13059-020-02064-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Q., Wang R., Yang Z., et al. Molecular profiling of human non-small cell lung cancer by single-cell RNA-seq. Genome Med. 2022;14(1):87. doi: 10.1186/s13073-022-01089-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Efremova M., Vento-Tormo M., Teichmann S.A., Vento-Tormo R. CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat Protoc. 2020;15(4):1484–1506. doi: 10.1038/s41596-020-0292-x. [DOI] [PubMed] [Google Scholar]
- 21.Qiu X., Mao Q., Tang Y., et al. Reversed graph embedding resolves complex single-cell trajectories. Nat Methods. 2017;14(10):979–982. doi: 10.1038/nmeth.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van de Sande B., Flerin C., Davie K., et al. A scalable SCENIC workflow for single-cell gene regulatory network analysis. Nat Protoc. 2020;15(7):2247–2276. doi: 10.1038/s41596-020-0336-2. [DOI] [PubMed] [Google Scholar]
- 23.Ritchie M.E., Phipson B., Wu D., et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7) doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Owens N.D.L., De Domenico E., Gilchrist M.J. An RNA-seq protocol for differential expression analysis. Cold Spring Harb Protoc. 2019;2019(6) doi: 10.1101/pdb.prot098368. [DOI] [PubMed] [Google Scholar]
- 25.Mao W., Wang K., Xu B., et al. ciRS-7 is a prognostic biomarker and potential gene therapy target for renal cell carcinoma. Mol Cancer. 2021;20(1):142. doi: 10.1186/s12943-021-01443-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malyguine A.M., Strobl S., Dunham K., Shurin M.R., Sayers T.J. ELISPOT assay for monitoring cytotoxic T lymphocytes (CTL) activity in cancer vaccine clinical trials. Cells. 2012;1(2):111–126. doi: 10.3390/cells1020111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spidlen J., Moore W., Parks D., et al. Data file standard for flow cytometry, version FCS 3.2. Cytometry A. 2021;99(1):100–102. doi: 10.1002/cyto.a.24225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greenland S., Mansournia M.A., Joffe M. To curb research misreporting, replace significance and confidence by compatibility: a Preventive Medicine Golden Jubilee article. Prev Med. 2022;164 doi: 10.1016/j.ypmed.2022.107127. [DOI] [PubMed] [Google Scholar]
- 29.Mansournia M.A., Nazemipour M., Etminan M. P-value, compatibility, and S-value. Glob Epidemiol. 2022;4 doi: 10.1016/j.gloepi.2022.100085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Altman D.G., Royston P. The cost of dichotomising continuous variables. BMJ. 2006;332(7549):1080. doi: 10.1136/bmj.332.7549.1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi Y., Au J.S., Thongprasert S., et al. A prospective, molecular epidemiology study of EGFR mutations in Asian patients with advanced non-small-cell lung cancer of adenocarcinoma histology (PIONEER) J Thorac Oncol. 2014;9(2):154–162. doi: 10.1097/JTO.0000000000000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi J.F., Wang L., Wu N., et al. Clinical characteristics and medical service utilization of lung cancer in China, 2005-2014: overall design and results from a multicenter retrospective epidemiologic survey. Lung Cancer. 2019;128:91–100. doi: 10.1016/j.lungcan.2018.11.031. [DOI] [PubMed] [Google Scholar]
- 33.Ribas A. Adaptive immune resistance: how cancer protects from immune attack. Cancer Discov. 2015;5(9):915–919. doi: 10.1158/2159-8290.CD-15-0563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang Q., Chen Y., Li X., et al. The role of PD-1/PD-L1 and application of immune-checkpoint inhibitors in human cancers. Front Immunol. 2022;13 doi: 10.3389/fimmu.2022.964442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ginefra P., Lorusso G., Vannini N. Innate immune cells and their contribution to T-cell-based immunotherapy. Int J Mol Sci. 2020;21(12) doi: 10.3390/ijms21124441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liew P.X., Kubes P. The neutrophil's role during health and disease. Physiol Rev. 2019;99(2):1223–1248. doi: 10.1152/physrev.00012.2018. [DOI] [PubMed] [Google Scholar]
- 37.Kersten K., Hu K.H., Combes A.J., et al. Spatiotemporal co-dependency between macrophages and exhausted CD8(+) T cells in cancer. Cancer Cell. 2022;40(6):624–638.e9. doi: 10.1016/j.ccell.2022.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mo Z., Liu D., Chen Y., et al. Single-cell transcriptomics reveals the role of Macrophage-Naive CD4 + T cell interaction in the immunosuppressive microenvironment of primary liver carcinoma. J Transl Med. 2022;20(1):466. doi: 10.1186/s12967-022-03675-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klobuch S., Hammon K., Vatter-Leising S., et al. HLA-DPB1 reactive T cell receptors for adoptive immunotherapy in allogeneic stem cell transplantation. Cells. 2020;9(5) doi: 10.3390/cells9051264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma X., Bi E., Lu Y., et al. Cholesterol induces CD8(+) T cell exhaustion in the tumor microenvironment. Cell Metab. 2019;30(1):143–156.e5. doi: 10.1016/j.cmet.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shi X.Y., Tao X.F., Wang G.W., et al. LncDBH-AS1 knockdown enhances proliferation of non-small cell lung cancer cells by activating the Wnt signaling pathway via the miR-155/AXIN1 axis. Eur Rev Med Pharmacol Sci. 2021;25(1):139–144. doi: 10.26355/eurrev_202101_24377. [DOI] [PubMed] [Google Scholar]
- 42.Sokol J.P., Neil J.R., Schiemann B.J., Schiemann W.P. The use of cystatin C to inhibit epithelial-mesenchymal transition and morphological transformation stimulated by transforming growth factor-beta. Breast Cancer Res. 2005;7(5):R844–R853. doi: 10.1186/bcr1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jakubzick C.V., Randolph G.J., Henson P.M. Monocyte differentiation and antigen-presenting functions. Nat Rev Immunol. 2017;17(6):349–362. doi: 10.1038/nri.2017.28. [DOI] [PubMed] [Google Scholar]
- 44.Xia Y., Rao L., Yao H., Wang Z., Ning P., Chen X. Engineering macrophages for cancer immunotherapy and drug delivery. Adv Mater. 2020;32(40) doi: 10.1002/adma.202002054. [DOI] [PubMed] [Google Scholar]
- 45.Zhang Y., Du W., Chen Z., Xiang C. Upregulation of PD-L1 by SPP1 mediates macrophage polarization and facilitates immune escape in lung adenocarcinoma. Exp Cell Res. 2017;359(2):449–457. doi: 10.1016/j.yexcr.2017.08.028. [DOI] [PubMed] [Google Scholar]
- 46.Dong B., Wu C., Huang L., Qi Y. Macrophage-related SPP1 as a potential biomarker for early lymph node metastasis in lung adenocarcinoma. Front Cell Dev Biol. 2021;9 doi: 10.3389/fcell.2021.739358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen X., Song E. The theory of tumor ecosystem. Cancer Commun. 2022;42(7):587–608. doi: 10.1002/cac2.12316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hernan M.A. The hazards of hazard ratios. Epidemiology. 2010;21(1):13–15. doi: 10.1097/EDE.0b013e3181c1ea43. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.