Abstract
Background
Porcine circovirus type 2 (PCV2) infection is ubiquitous around the world. Diagnosis of the porcine circovirus-associated disease requires clinic-pathological elements together with the quantification of viral loads. Furthermore, given pig farms in regions lacking access to sufficient laboratory equipment, developing diagnostic devices with high accuracy, accessibility, and affordability is a necessity.
Objectives
This study aims to investigate two newly developed diagnostic tools that may satisfy these criteria.
Methods
We collected 250 specimens, including 170 PCV2-positive and 80 PCV2-negative samples. The standard diagnosis and cycle threshold (Ct) values were determined by quantitative polymerase chain reaction (qPCR). Then, two point-of-care (POC) diagnostic platforms, convective polymerase chain reaction (cPCR, qualitative assay: positive or negative results are shown) and EZtargex (quantitative assay: Ct values are shown), were examined and analyzed.
Results
The sensitivity and specificity of cPCR were 88.23% and 100%, respectively; the sensitivity and specificity of EZtargex were 87.65% and 100%, respectively. These assays also showed excellent concordance compared with the qPCR assay (κ = 0.828 for cPCR and κ = 0.820 for EZtargex). The statistical analysis showed a great diagnostic power of the EZtargex assay to discriminate between samples with different levels of positivity.
Conclusions
The two point-of-care diagnostic platforms are accurate, rapid, convenient and require little training for PCV2 diagnosis. These POC platforms can discriminate viral loads to predict the clinical status of the animals. The current study provided evidence that these diagnostics were applicable with high sensitivity and specificity in the diagnosis of PCV2 infection in the field.
Keywords: Porcine circovirus, molecular diagnostics, point-of-care testing, point-of-care, pigs
INTRODUCTION
Porcine circovirus type 2 (PCV2) is a small, non-enveloped, circular, and single-stranded DNA virus that is classified within the genus Circovirus of the family Circoviridae [1]. PCV2 is one of the primary agents contributing to high mortality in porcine respiratory disease complex (PRDC) [2] and caused economic loss in the swine industry worldwide two decades ago. PCV2 vaccination is one of biggest success stories in pig medicine, with substantial improvements in the mortality rate and the average daily weight gain since PCV2 vaccination worldwide [3]. A previous study suggested that nationwide eradication by PCV2 vaccination alone would not be practical with current vaccines [4]. Similar to breakthrough infection of coronavirus disease 2019 (COVID-19) in humans, the presence of a leaky vaccine situation of PCV2 was also observed [5]. Both are characterized by an imperfect vaccine that reduces disease severity, but it does not provide full protection against infection, transmission and completely eliminated from the body [6].
Evidence from the field supports that the categories of lesion severity of porcine circovirus-associated disease (PCVAD or PCVD) were significantly correlated with the level of PCV2 load in the serum [7]. Therefore, diagnosis of PCVAD requires clinic-pathological elements together with the quantification of viral loads [8]. A high PCV2 load is a major feature of PCVAD-affected pigs. Therefore, PCV2 loads of < 103, 103 to 104, and > 104 DNA copies/μL of serum sample can be used to categorize pigs as subclinically infected, suspected, and PCVAD, respectively [7,9]. Therefore, the determination of viral loading and cycle threshold (Ct) values are critical for PCVAD diagnosis. Quantitative real-time polymerase chain reaction (PCR) platforms allow very sensitive DNA detection in clinical samples. However, these assays are technically more complex than rapid test or point-of-care (POC) platforms and require a much higher level of training, facilities and competency for accurate testing. Quantitative real-time PCR platform difficulty emphasizes the need for the development of rapid, simple, and convenient diagnostic tools in the field.
Several diagnostic tools are widely accepted and used for the diagnosis of PCV2, although they are usually time-consuming and costly. PCV2 was primarily determined by the restriction fragment length polymorphism (RFLP) assay for genotype differentiation [10] or detected viral DNA in tissues through immunohistochemistry (IHC) [11,12] and/or in situ hybridization (ISH) [13]. PCR-based diagnostic methods were also proposed, including real-time PCR [12], multiplex real-time PCR [14], loop-mediated isothermal amplification (LAMP) PCR [15,16], droplet digital PCR (ddPCR) [17], and SYBR TaqMan-based PCR [18] assays. Furthermore, commercial kits or antigens detecting PCV2 antibodies based on enzyme-linked immunosorbent assay (ELISA) were also available [19,20,21]. However, the detection of the PCV2 virus by IHC and ISH requires samples that are collected from tissues, which makes these diagnostic tools more time-consuming. PCR-base and ELISA diagnosis rely on more advanced laboratory equipment and well-trained manpower. These issues highlight the urgent demands for developing diagnostic tools for PCV2 that can furnish elaborate details and thus guide treatment strategies.
Although several commercial diagnostic tools have been established, rapid tools that can precisely detect PCV2 DNA are still needed. Furthermore, in undeveloped or developing countries/regions, in-house rapid tests are necessary for PCVAD diagnosis in the field. Certain methods known as POC tests have been more appropriate for fields since the global COVID-19 pandemic [22]. These devices are simple to use, require low levels of training, and immediately provide results. Similar to COVID-19, the development of new diagnostic tests for PCV2 remains needed; therefore, this study was designed to verify two novel POC tests, convective PCR (cPCR) and EZtargex, for the rapid and accurate identification of PCV2. The cPCR is a PCR-based diagnostic tool and EZtargex is a newly developed quantitative PCR (qPCR) assay. These two POC tests targeted the ORF1 gene of PCV2 for diagnosis. The detecting primer probe (TaqMan probe) is labelled with a fluorescent reporter. Therefore, when the specific gene is amplified, the fluorescent signal is generated as follows. Because cPCR is a qualitative assay, the plug-in fluorescent detecting system will then calculate the signal intensity and show the positive or negative results within 45 min. For the EZtargex assay, the fluorescent signal intensity will be recorded during every cycle, which fulfills the purpose of real-time quantification. During the automatic sample-to-answer procedure, the EZtargex device only takes 2.5 h for diagnosis, which significantly saves time and minimizes the errors from operators. The current study will prove and provide evidence that these tools are applicable with high sensitivity and specificity in detecting PCV2.
MATERIALS AND METHODS
Sample collection and classification
Clinical samples were collected from the Animal Disease Diagnostic Center, National Pingtung University of Science and Technology (NPUST). These samples were actively submitted by the owners for pathological and molecular diagnosis. No animal experiments were involved in the current study; therefore, formal approval was unnecessary for this research considered by the Institutional Animal Care and Use Committee of NPUST. According to previous studies, PCV2 loads of < 103, 103 to 104, and > 104 DNA copies/μL of serum sample can be used to categorize pigs as subclinically infected, suspected, and PCVAD, respectively [7,9]. Therefore, we assume that the criteria for weak, middle, and strong positivity were PCV2 viral loads less than 103, 103 to 104, and more than 104 DNA copies/μL for the serum samples, respectively.
DNA extraction and PCV2 qPCR
DNA extraction and qPCR for PCV2 were conducted as previously described [23]. Briefly, viral DNA from serum samples was extracted using a MagNA Pure LC total nucleic acid isolation kit (Roche Diagnostics GmbH, Germany) according to the manufacturer’s instructions. Extracted DNA was used as a template combined with 10 μM of each primer (Supplementary Table 1) and 10 μM of the probe using the 2X Probe Master. The samples were initially heated to 95°C for 10 min, followed by a 45-repeated cycling profile: 95°C for 20 sec, 60°C for 20 sec, and 72°C for 20 sec. The thermal cycles were run on a LightCycler® 96 System (Roche Diagnostics GmbH), which take more than 3 h.
cPCR and EZtargex
The cPCR was conducted using a commercial PCV2 cPCR kit (Schweitzer Biotech Company Ltd. [SBC], Taiwan) according to the instruction guidelines. The commercial kit contained three working reagents (reagents A, B, and C), dilution buffer, positive control, positive control reconstitution buffer, and negative control. Briefly, 10 μL of reagent A was added to 10 μL of serum in an Eppendorf tube. After gentle pipetting, the mixture was incubated at room temperature for 5 min. Then, the samples were heated to 95°C for 5 min, followed by the addition of 20 μL of dilution buffer. The well-prepared sample solution was then subjected to cPCR analysis. For the preparation of the positive control, 100 μL of reconstitution buffer was first added to the positive control tube. The reaction buffer was prepared by transferring all reagent B into reagent C. Then, 50 μL of reaction buffer was transferred into each cPCR tube. A total of 5 μL of sample solution, positive control, and negative control was added to the corresponding reaction tube. After gently mixing, caps were placed on cPCR tubes which were then shaken by hand or spin down to ensure no air bubbles were trapped in the tubes. The tubes were placed into the cPCR device (SBC) and the reaction program was started. The interpretation of results was shown as positive or negative. The positive results indicate that copies of the target nucleic acid of PCV2 were detected in an amount equal to or above the detection limit of the cPCR kit. Conversely, no copies of the target sequences of PCV2 were identified in the samples or the copy number of the targets was lower than the detection limit of the kit.
EZtargex (SBC) is an all-in-one device including a compact automated nucleic acid extraction system (magnetic bead-based) and a real-time PCR test. The samples were prepared according to the manufacturer’s instructions. Briefly, 200 μL of serum was added to the wells of the extraction cartridge for EZtargex which contained pre-filled buffer (SBC). The package of PCV2 lyophilized qPCR kit (SBC) includes one pack of lyophilized reagents, positive control, and negative control. The qPCR reagent tubes were placed into the transfer cartridges, and the transfer and extraction cartridges were then subjected to the process in the EZtargex device. After 150 min, the Ct values were shown on the screen.
Statistical analysis
Statistical analyses were performed with GraphPad Prism v8 (GraphPad Software, Inc., USA). Data are described as the median ± interquartile range (range from the 25th to the 75th percentile). Statistical difference was defined as a p value below 0.05. For some statistical analyses, the Ct value of negative samples was considered to be Ct > 40 [24]. To verify the differences between Ct values, the Mann–Whitney U test or Kruskal–Wallis test was employed to compare two or three groups. The sensitivity and specificity of cPCR and EZtargex were calculated by samples confirmed as PCV2-positive or PCV2-negative by qPCR methods. A kappa between 0 and 0.2 is considered slight, 0.2 to 0.4 fair, 0.4 to 0.6 moderate, 0.6 to 0.8 substantial, and > 0.8 almost perfect agreement [25]. Pearson’s correlation analysis was used to examine the relationship between Ct values and PCV2 viral loads.
RESULTS
Sample characteristics
In the current study, we analyzed 250 samples, including 170 PCV2-positive and 80 PCV2-negative samples. The 170 PCV2-positive samples were further classified into 60 weak-, 60 middle-, and 50 strong-positive groups determined by the qPCR assay. The negative controls were collected from 20 specific pathogen-free (SPF) pigs, and 60 negative samples were collected from the fields. The correlation analysis between viral loads and Ct values of qPCR was employed in the 170 PCV2-positive samples. As shown in Supplementary Fig. 1, the Ct values were positively associated with PCV2 viral loads (R2 = 0.9554, p < 0.0001).
Ideal sensitivity and specificity of cPCR and EZtargex assays
To elucidate the diagnostic power of cPCR and EZtargex, we compared the results with the qPCR assay. First, the cPCR assay was evaluated. The PCV2 samples with middle and strong positivity also showed positive results. The negative samples showed a consistent result in qPCR and cPCR assays. However, the weakly positive samples showed inconsistent outcomes. In the 60 weakly positive samples, 40 and 20 cPCR-diagnosed samples showed positivity and negativity, respectively. Based on these findings, cPCR exhibited 88.23% sensitivity and 100% specificity in the diagnosis of PCV2 DNA (Table 1). The perfect consistency of cPCR and qPCR assays was found (κ = 0.828, p < 0.0001), revealing a high agreement between these assays (Table 1).
Table 1. Statistical analysis of the cPCR assay compared with qPCR.
| qPCR | cPCR | Sensitivity | Specificity | p value | Kappa coefficient | |
|---|---|---|---|---|---|---|
| Positive (+) | Negative (−) | |||||
| Positive (+) | 150 | 20 | 88.23% | 100.00% | < 0.00001 | 0.828 |
| Negative (−) | 0 | 80 | ||||
cPCR, convective polymerase chain reaction; qPCR, quantitative polymerase chain reaction.
In the EZtargex analysis, similar findings were found. Consistent results were found in middle-positive, strong-positive, and negative samples diagnosed by the EZtargex and qPCR assays. Conversely, 60 weakly positive samples showed different results. Of these 60 samples, 39 samples were diagnosed as positive, and the resulting 21 samples showed negative results. The sensitivity and specificity for EZtargex were 87.65% and 100.00%, respectively (Table 2). The novel assay also showed a high concordance compared with the conventional qPCR assay (κ = 0.82, p < 0.0001) (Table 2). Taken together, these results indicate that the newly developed diagnostic tools cPCR and EZtargex harbored an ideal diagnostic power in PCV2 compared with the traditional assay.
Table 2. Statistical analysis of the EZtargex assay compared with qPCR.
| qPCR | EZtargex | Sensitivity | Specificity | p value | Kappa coefficient | |
|---|---|---|---|---|---|---|
| Positive (+) | Negative (−) | |||||
| Positive (+) | 149 | 21 | 87.65% | 100% | < 0.00001 | 0.820 |
| Negative (−) | 0 | 80 | ||||
qPCR, quantitative polymerase chain reaction.
An original median Ct value below 33.67 was considered a cPCR-positive sample
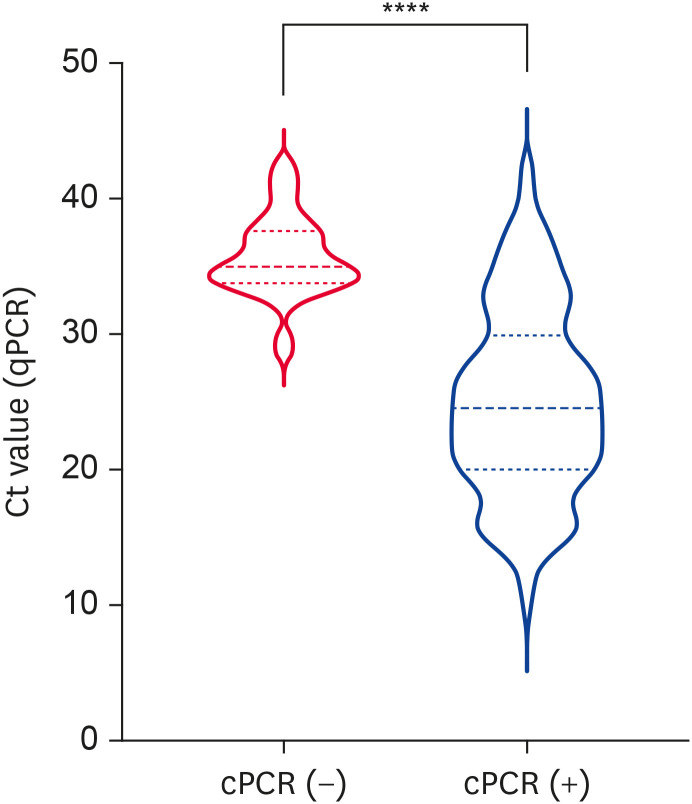
Our findings suggested that the cPCR assay exerted outstanding sensitivity and specificity; however, this qualitative assay only provided positive or negative results. Therefore, we analyzed the cPCR results from qPCR-positive samples (n = 170). The median Ct values of cPCR-negative samples (n = 20, 35.02 ± 3.84) were significantly higher than those of cPCR-positive samples (n = 150, 24.56 ± 9.88, p < 0.0001) (Fig. 1). Notably, two samples that had Ct values of 41.55 and 42.08 (determined by qPCR test) showed positive results in the cPCR assay, indicating the high sensitivity of the cPCR assay. Then, we further elucidate clinical samples with how many Ct values would be considered as PCV2-positivity in the cPCR assay. Based on the maximum value of J [26], an optimal cutoff value of < 33.67 with a sensitivity of 87.33% and a specificity of 90.00% was selected (Table 3). These results suggested that PCV2-positive samples with an original Ct value below 33.67 might be deemed positive in the cPCR assay.
Fig. 1. Comparisons of Ct values between cPCR-negative and cPCR-positive samples.
Data are described as the median ± interquartile range and were analyzed by the Mann–Whitney U test; p values < 0.05 were considered to indicate statistical significance.
cPCR, convective polymerase chain reaction; qPCR, quantitative polymerase chain reaction; Ct, cycle threshold.
****p < 0.0001.
Table 3. Sensitivity and specificity of cPCR for the differentiation of positive or negative results at different cutoff values.
| Cutoff Ct value | Sensitivity | 95% CI | Specificity | 95% CI |
|---|---|---|---|---|
| < 32.97 | 84.67 | 77.89%–90.02% | 90.00 | 68.30%–98.77% |
| < 33.01 | 85.33 | 78.64%–90.57% | 90.00 | 68.30%–98.77% |
| < 33.27 | 86.00 | 79.40%–91.12% | 90.00 | 68.30%–98.77% |
| < 33.58 | 86.67 | 80.16%–91.66% | 90.00 | 68.30%–98.77% |
| < 33.67 | 87.33 | 80.93%–92.20% | 90.00 | 68.30%–98.77% |
| < 33.70 | 87.33 | 80.93%–92.20% | 85.00 | 62.11%–96.79% |
| < 33.72 | 87.33 | 80.93%–92.20% | 80.00 | 56.34%–94.27% |
| < 33.83 | 87.33 | 80.93%–92.20% | 75.00 | 50.90%–91.34% |
| < 33.95 | 88.00 | 81.70%–92.73% | 75.00 | 50.90%–91.34% |
Positive or negative results were categorized by cPCR assay. The statistical analysis was performed based on the original Ct values determined by qPCR assay.
cPCR, convective polymerase chain reaction; Ct, cycle threshold; CI, confidence interval; qPCR, quantitative polymerase chain reaction.
Excellent performance of the EZtargex assay for distinguishing weak, middle, and strong positivity
Different from the cPCR assay, EZtargex is a quantitative assay, and thus Ct values were available for all samples. To compare the correlation between Ct values of qPCR assay and EZtargex, correlation analysis was carried out. The Ct values of EZtargex were positively associated with those of qPCR assay (R2 = 0.8670, p < 0.0001) (Fig. 2A). These results indicated that EZtargex was a reliable diagnostic tool as a qPCR assay. In the EZtargex assay, the median Ct values for samples with weak, middle, and strong positivity were 37.71 ± 5.96, 28.24 ± 4.88, and 22.58 ± 4.51, respectively (Fig. 2B). Furthermore, the median Ct values in weakly positive samples were significantly higher than those in middle- and strongly positive samples (p < 0.0001); a significant difference was also observed in samples with middle and strong positivity (p < 0.0001). These results indicated the ideal diagnostic performance of the EZtargex assay to discriminate between weak, middle, and strong positivity of clinical samples.
Fig. 2. Correlation analysis and comparisons of Ct values between pigs with weak, middle, and strong PCV2 infection. The criteria for weak, middle, and strong positivity were PCV2 viral loads less than 103, 103 to 104, and more than 104 DNA copies/μL for the serum samples, respectively.
(A) Pearson’s correlation analysis was performed to analyze the correlation between Ct values of qPCR and EZtargex (R2 = 0.8670, p < 0.0001). (B) Ct values of weak, middle, and strong PCV2-positive samples diagnosed by the EZtargex test; data are described as the median ± interquartile range and were analyzed by the Kruskal–Wallis test. p values < 0.05 were considered to indicate statistical significance.
Ct, cycle threshold; PCV2, porcine circovirus type 2; qPCR, quantitative polymerase chain reaction; R2, Pearson’s correlation coefficient.
****p < 0.0001.
Recommended cutoff Ct values in the EZtargex assay for defining weak, middle, and strong positivity
Additional diagnostic test parameters were employed to determine the cutoff Ct values for defining “weak or middle” and “middle or strong” positive samples in the EZtargex assay. Based on the maximum value of J [26], an optimal cutoff value of < 33.20 with a sensitivity of 96.67% and a specificity of 81.67% was selected (Table 4). These results indicate that at a Ct value below 33.20, samples were defined as middle positivity. An ideal cutoff Ct value of < 25.42 with a sensitivity of 92.00% and a specificity of 78.33% was selected (Table 5). Samples were regarded as strongly positive with a Ct value below 25.42.
Table 4. Sensitivity and specificity of EZtargex for the differentiation of weak or middle positivity at different cutoff values.
| Cutoff Ct value | Sensitivity | 95% CI | Specificity | 95% CI |
|---|---|---|---|---|
| < 32.58 | 93.33 | 83.80%–98.15% | 85.00 | 73.43%–92.90% |
| < 32.63 | 93.33 | 83.80%–98.15% | 83.33 | 71.48%–91.71% |
| < 32.81 | 95.00 | 86.08%–98.96% | 83.33 | 71.48%–91.71% |
| < 32.95 | 95.00 | 86.08%–98.96% | 81.67 | 69.56%–90.48% |
| < 33.20 | 96.67 | 88.47%–99.59% | 81.67 | 69.56%–90.48% |
| < 33.55 | 96.67 | 88.47%–99.59% | 80.00 | 67.67%–89.22% |
| < 33.68 | 98.33 | 91.06%–99.96% | 80.00 | 67.67%–89.22% |
| < 33.78 | 98.33 | 91.06%–99.96% | 78.33 | 65.80%–87.93% |
| < 33.94 | 98.33 | 91.06%–99.96% | 76.67 | 63.96%–86.62% |
PCV2 viral loads determined by qPCR less than 103 or between 103 to 104 DNA copies/μL were defined as weak or middle positivity. The statistical analysis was performed based on the original Ct values determined by EZtargex.
Ct, cycle threshold; CI, confidence interval; PCV2, porcine circovirus type 2; qPCR, quantitative polymerase chain reaction.
Table 5. Sensitivity and specificity of EZtargex for the differentiation of middle or strong positivity at different cutoff values.
| Cutoff Ct value | Sensitivity | 95% CI | Specificity | 95% CI |
|---|---|---|---|---|
| < 25.09 | 86.00 | 73.26%–94.18% | 80.00 | 67.67%–89.22% |
| < 25.14 | 88.00 | 75.69%–95.47% | 80.00 | 67.67%–89.22% |
| < 25.18 | 90.00 | 78.19%–96.67% | 80.00 | 67.67%–89.22% |
| < 25.23 | 90.00 | 78.19%–96.67% | 78.33 | 65.80%–87.93% |
| < 25.42 | 92.00 | 80.77%–97.78% | 78.33 | 65.80%–87.93% |
| < 25.69 | 92.00 | 80.77%–97.78% | 76.67 | 63.96%–86.62% |
| < 25.94 | 92.00 | 80.77%–97.78% | 75.00 | 62.14%–85.28% |
| < 26.13 | 92.00 | 80.77%–97.78% | 71.67 | 58.56%–82.55% |
| < 26.18 | 94.00 | 83.45%–98.75% | 71.67 | 58.56%–82.55% |
PCV2 viral loads determined by qPCR between 103 to 104 or above 104 DNA copies/μL were defined as middle or strong positivity. The statistical analysis was performed based on the original Ct values determined by EZtargex.
Ct, cycle threshold; CI, confidence interval; PCV2, porcine circovirus type 2; qPCR, quantitative polymerase chain reaction.
DISCUSSION
PCV2 is an important pathogen in numerous countries. Its global prevalence results in a seropositivity rate of 20%–80% in pigs coupled with a very high incidence rate of 60% [27]. These results revealed the clinical relevance of PCV2 infection, and a precise diagnosis should be immediately made on the farm. This study investigated two novel molecular assays that can accurately diagnose PCV2 infection in industrial animal areas. The currently used cPCR and EZtargex assays are rapid for clinical application and required low capacities compared with traditional qPCR assays. Our results suggested that these diagnostic techniques can be complemented with qPCR methods in clinical diagnostic laboratories.
To establish prevention and control strategies for PCV2, PCR assays are sensitive methods to detect viruses in animals [28]. Several different PCR-related methods have been employed, such as ddPCR [17], real-time PCR [12,14,18], and LAMP [15,16]. The ddPCR require an expensive thermocycler and are time-consuming and difficult to perform for inexperienced investigators. Furthermore, multiple steps expose the reactions to risks of cross-contamination [29]. The two TaqMan probe-based PCR testing platforms, cPCR and EZtargex, are simple to use and have several advantages compared to traditional PCR, ddPCR, and LAMP assay. In the present study, the authors not only evaluated the clinical efficacy of cPCR and EZtargex but also compared the results of these two methods with those obtained from qPCR assay. For the diagnosis of PCV2 through qPCR assay, the qPCR program took at least three hours, excluding the extraction of DNA, data analysis, well-trained manpower, and risks of contamination. The characteristics of cPCR and EZtargex used in the study were similar, and they were sensitive, specific (Tables 1 and 2), and comparatively easy to perform without requiring any specialized laboratory equipment. Most importantly, compared with qPCR assay, these two diagnostic tools are relatively rapid (cPCR for 1 h and EZtargex for 2.5 h) and user-friendly. The users without training can easily handle the diagnostic procedures, and the all-in-one devices can conduct the diagnostic process from whole blood or swab samples. The weakness of the cPCR only provided positive or negative results, the sensitivity and specificity remain convincing in PCV2 diagnosis when compared with the exact Ct values obtained by conventional assay. Furthermore, these two are more suitable for small-scale tests, in which testing of only one sample can be easily conducted, and further improvement was needed. The interface of the two methods is more user-friendly because of the straightforward design of instructions.
The sensitivity and specificity of these two assays were relatively high. When we compared the original Ct values from samples with weak, middle, and strong positivity (defined by qPCR test) in EZtargex assays, the median Ct values for these three groups were significantly different (Fig. 2B). These results indicate that EZtargex has a high potential to discriminate patients with different viral loads, which could provide a quantitative Ct value corresponding to clinical observation. Furthermore, two specimens that had Ct values of 41.55 and 42.08 (determined by qPCR test) showed positive results in the cPCR assay. The Ct values correspond to a viral load with as few as one copy number per microliter. Notably, no signal of the truly negative sample (determined by qPCR) was detected by cPCR or EZtargex, which revealed excellent specificity in PCV2 diagnosis. These results indicated that these assays had high sensitivity and specificity for PCV2 detection. The rapid diagnostic methods also offered easy-to-interpreted results.
The accuracy of a newly developed diagnostic tool is important, but not all. In human medicine, ASSURED criteria have been set by the World Health Organization for the development of clinical diagnostics, especially the diagnosis of infectious diseases [30]. The POC tests were expected to meet all the ASSURED criteria, being affordable, sensitive, specific, user-friendly, rapid, equipment-free or simple, and deliverable to end-users criteria [31]. As no test is perfect, ASSURED criteria therefore embody three key characteristics, namely, accuracy, accessibility, and affordability [30]. However, except for one previous study [32], reports from research in the development of novel diagnostic tools for PCV2 rarely present the characteristics that meet ASSURED criteria. Herein, in our study, we proposed two novel diagnostics that exhibited a high potential to meet several criteria. These two assays not only provide sensitive, specific, and rapid results but also harbor several characteristics that can be applied in regions without sufficient equipment. The cPCR, a hand-held device, is equipped with an internet (WiFi) and global positioning system (GPS), enabling PCV2 diagnosis anytime and anywhere. Although EZtargex is a laboratory-used device, it is a POC testing diagnostic, rendering it eligible for limited resource settings as well. Furthermore, the EZtargex system is equipped with USB ports for data transfer, and test results can be exported to a computer in the engineer mode for further analyses and creation of customized reports. Because of these advantages, these two assays are ideal choices for POC tests, not only for convenient, rapid, and accurate diagnosis but also for the nonrequirement of well-trained staff and unique equipment.
The current study has some limitations. First, the newly developed assays in the current study failed to distinguish subtypes of PCV2. Fortunately, the treatment strategies are similar among various subtypes of PCV2. Second, cPCR and EZtargex assays can handle 8 and 16 samples at a time. The limited sample numbers might restrict the application of a large-scale herd diagnosis. Last, the key obstacle preventing PCR-based tests from satisfying all the ASSURED criteria is the requirement of thermal cycles. Studies developing diagnostics that can meet the ASSURED criteria are highly expected in the future.
In conclusion, the two recently developed molecular tests are rapid and convenient for the detection of PCV2 on farms. These assays can directly detect PCV2 DNA from clinical samples in only 1–2.5 h. Therefore, these two diagnostic assays can provide vital information that can help expedite therapeutic decisions or preventive strategies on farms. These POC platforms can discriminate viral loads to predict the clinical status of the animals. We highly believe that in the future, these two assays may reduce the labor and time for PCV2 diagnosis and provide convenient and accurate on-demand test results.
Footnotes
Funding: This work was supported by Schweitzer Biotech Company Ltd., Taiwan.
Conflict of Interest: Schweitzer Biotech Company (SBC) Ltd. is the sponsor of the project. Hsieh WJ, Lin CC, and Ting JM. are employees of the SBC. However, the authors declare that the research was conducted with integrity, following all ethical guidelines. All other authors declare no competing interests.
- Conceptualization: Chiou MT, Lin CN.
- Data curation: Du MY, Ke CH, Lin CN.
- Formal analysis: Ke CH, Lin CN.
- Funding acquisition: Lin CN, Chiou MT.
- Investigation: Chiou MT, Lin CN.
- Methodology: Du MY.
- Project administration: Lin CN, Chiou MT.
- Resources: Chiou MT, Lin CN.
- Software: Ke CH.
- Supervision: Ting JM, Lin CN, Chiou MT.
- Validation: Ke CH, Hsieh WJ, Lin CC, Lin CN.
- Visualization: Ke CH, Lin CN.
- Writing - original draft: Ke CH.
- Writing - review & editing: Lin CN, Chiou MT.
SUPPLEMENTARY MATERIALS
Primers used in qPCR
Pearson’s correlation analysis was performed to analyse the correlation between PCV2 viral load and Ct values of qPCR (R2 = 0.9554, p < 0.0001).
References
- 1.Breitbart M, Delwart E, Rosario K, Segalés J, Varsani A Ictv Report Consortium. ICTV virus taxonomy profile: circoviridae. J Gen Virol. 2017;98(8):1997–1998. doi: 10.1099/jgv.0.000871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Segalés J. Porcine circovirus type 2 (PCV2) infections: clinical signs, pathology and laboratory diagnosis. Virus Res. 2012;164(1-2):10–19. doi: 10.1016/j.virusres.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 3.Kristensen CS, Baadsgaard NP, Toft N. A meta-analysis comparing the effect of PCV2 vaccines on average daily weight gain and mortality rate in pigs from weaning to slaughter. Prev Vet Med. 2011;98(4):250–258. doi: 10.1016/j.prevetmed.2010.11.015. [DOI] [PubMed] [Google Scholar]
- 4.Afghah Z, Webb B, Meng XJ, Ramamoorthy S. Ten years of PCV2 vaccines and vaccination: is eradication a possibility? Vet Microbiol. 2017;206:21–28. doi: 10.1016/j.vetmic.2016.10.002. [DOI] [PubMed] [Google Scholar]
- 5.Karuppannan AK, Opriessnig T. Porcine circovirus type 2 (PCV2) vaccines in the context of current molecular epidemiology. Viruses. 2017;9(5):99. doi: 10.3390/v9050099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin CN, Chan KR, Ooi EE, Chiou MT, Hoang M, Hsueh PR, et al. Animal Coronavirus diseases: parallels with COVID-19 in humans. Viruses. 2021;13(8):1507. doi: 10.3390/v13081507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olvera A, Sibila M, Calsamiglia M, Segalés J, Domingo M. Comparison of porcine circovirus type 2 load in serum quantified by a real time PCR in postweaning multisystemic wasting syndrome and porcine dermatitis and nephropathy syndrome naturally affected pigs. J Virol Methods. 2004;117(1):75–80. doi: 10.1016/j.jviromet.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 8.Segalés J, Sibila M. Revisiting porcine circovirus disease diagnostic criteria in the current Porcine circovirus 2 epidemiological context. Vet Sci. 2022;9(3):110. doi: 10.3390/vetsci9030110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Opriessnig T, Meng XJ, Halbur PG. Porcine circovirus type 2 associated disease: update on current terminology, clinical manifestations, pathogenesis, diagnosis, and intervention strategies. J Vet Diagn Invest. 2007;19(6):591–615. doi: 10.1177/104063870701900601. [DOI] [PubMed] [Google Scholar]
- 10.Ciacci-Zanella JR, Morés N. Diagnosis of post-weaning multisystemic wasting syndrome in pigs in Brazil caused by porcine circovirus type 2. Arq Bras Med Vet Zootec. 2003;55(5):522–527. [Google Scholar]
- 11.Chianini F, Majó N, Segalés J, Domínguez J, Domingo M. Immunohistochemical characterisation of PCV2 associate lesions in lymphoid and non-lymphoid tissues of pigs with natural postweaning multisystemic wasting syndrome (PMWS) Vet Immunol Immunopathol. 2003;94(1-2):63–75. doi: 10.1016/S0165-2427(03)00079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dela Cruz A, Palmieri C, Azul R, Legaspi C, Lola S, Barnes TS, et al. Detection of porcine circovirus type 2 (PCV2) in the Philippines and the complexity of PCV2-associated disease diagnosis. Trop Anim Health Prod. 2021;53(3):371. doi: 10.1007/s11250-021-02823-y. [DOI] [PubMed] [Google Scholar]
- 13.Kim D, Ha Y, Lee YH, Chae S, Lee K, Han K, et al. Comparative study of in situ hybridization and immunohistochemistry for the detection of porcine circovirus 2 in formalin-fixed, paraffin-embedded tissues. J Vet Med Sci. 2009;71(7):1001–1004. doi: 10.1292/jvms.71.1001. [DOI] [PubMed] [Google Scholar]
- 14.Wang HY, Song JK, Shin S, Kim H. Comparison of multiplex real-time PCR and PCR-reverse blot hybridization assays for the direct and rapid detection of porcine circovirus type 2 genotypes. Front Vet Sci. 2020;7:200. doi: 10.3389/fvets.2020.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou S, Han S, Shi J, Wu J, Yuan X, Cong X, et al. Loop-mediated isothermal amplification for detection of porcine circovirus type 2. Virol J. 2011;8(1):497. doi: 10.1186/1743-422X-8-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen HT, Zhang J, Sun DH, Chu YF, Cai XP, Liu XT, et al. Rapid detection of porcine circovirus type 2 by loop-mediated isothermal amplification. J Virol Methods. 2008;149(2):264–268. doi: 10.1016/j.jviromet.2008.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao S, Lin H, Chen S, Yang M, Yan Q, Wen C, et al. Sensitive detection of Porcine circovirus-2 by droplet digital polymerase chain reaction. J Vet Diagn Invest. 2015;27(6):784–788. doi: 10.1177/1040638715608358. [DOI] [PubMed] [Google Scholar]
- 18.Zhao K, Han F, Zou Y, Zhu L, Li C, Xu Y, et al. Rapid detection of porcine circovirus type 2 using a TaqMan-based real-time PCR. Virol J. 2010;7(1):374. doi: 10.1186/1743-422X-7-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nainys J, Lasickiene R, Petraityte-Burneikiene R, Dabrisius J, Lelesius R, Sereika V, et al. Generation in yeast of recombinant virus-like particles of porcine circovirus type 2 capsid protein and their use for a serologic assay and development of monoclonal antibodies. BMC Biotechnol. 2014;14(1):100. doi: 10.1186/s12896-014-0100-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nawagitgul P, Harms PA, Morozov I, Thacker BJ, Sorden SD, Lekcharoensuk C, et al. Modified indirect porcine circovirus (PCV) type 2-based and recombinant capsid protein (ORF2)-based enzyme-linked immunosorbent assays for detection of antibodies to PCV. Clin Diagn Lab Immunol. 2002;9(1):33–40. doi: 10.1128/CDLI.9.1.33-40.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shuai J, Li X, Chen N, Chen X, Fang W. Characterization and potential use of truncated PCV2 capsid protein and its polyclonal antibody for diagnosis of PCV2 infections. Wei Sheng Wu Hsueh Pao. 2008;48(1):85–90. [PubMed] [Google Scholar]
- 22.Iliescu FS, Ionescu AM, Gogianu L, Simion M, Dediu V, Chifiriuc MC, et al. Point-of-care testing-the key in the battle against SARS-CoV-2 pandemic. Micromachines (Basel) 2021;12(12):1464. doi: 10.3390/mi12121464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsai GT, Lin YC, Lin WH, Lin JH, Chiou MT, Liu HF, et al. Phylogeographic and genetic characterization of porcine circovirus type 2 in Taiwan from 2001-2017. Sci Rep. 2019;9(1):10782. doi: 10.1038/s41598-019-47209-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henriques AM, Duarte M, Barros SC, Fagulha T, Ramos F, Luís T, et al. Development and validation of a real-time PCR for the detection and quantification of porcine circovirus type 2. Virusdisease. 2018;29(3):355–361. doi: 10.1007/s13337-018-0476-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Audigé L. Veterinary epidemiologic research: I. Dohoo, W. Martin, H. Stryhn, Atlantic Veterinary College, Charlottetown, PE, Canada, 2003. Prev Vet Med. 2005;68:289–292. [Google Scholar]
- 26.Unal I. Defining an optimal cut-point value in ROC analysis: an alternative approach. Comput Math Methods Med. 2017;2017:3762651. doi: 10.1155/2017/3762651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X, He K, Wang W, Zhou Z, Yang G, Mao A, et al. Genetic variation of porcine circovirus type 2 isolate 201105ZJ. Agric Sci Tech. 2014;15(11):1860–1864. [Google Scholar]
- 28.Caprioli A, McNeilly F, McNair I, Lagan-Tregaskis P, Ellis J, Krakowka S, et al. PCR detection of porcine circovirus type 2 (PCV2) DNA in blood, tonsillar and faecal swabs from experimentally infected pigs. Res Vet Sci. 2006;81(2):287–292. doi: 10.1016/j.rvsc.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 29.Yuan H. In: Polymerase Chain Reaction for Biomedical Applications. Samadikuchaksaraei A, editor. Rijeka: IntechOpen; 2016. Chapter 4. Regulatory concern of polymerase chain reaction (pcr) carryover contamination. [Google Scholar]
- 30.Land KJ, Boeras DI, Chen XS, Ramsay AR, Peeling RW. REASSURED diagnostics to inform disease control strategies, strengthen health systems and improve patient outcomes. Nat Microbiol. 2019;4(1):46–54. doi: 10.1038/s41564-018-0295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Otoo JA, Schlappi TS. REASSURED multiplex diagnostics: a critical review and forecast. Biosensors (Basel) 2022;12(2):124. doi: 10.3390/bios12020124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang M, Wang A, Sun Y, Li Y, Chen Y, Zhou J, et al. Development of a gold nanoparticle-based immunochromatographic strip for rapid detection of porcine circovirus type 2. Microbiol Spectr. 2023;11(4):e0195322. doi: 10.1128/spectrum.01953-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers used in qPCR
Pearson’s correlation analysis was performed to analyse the correlation between PCV2 viral load and Ct values of qPCR (R2 = 0.9554, p < 0.0001).