Abstract
The vpr sequences from six human immunodeficiency virus type 1 (HIV-1)-infected mother-infant pairs following perinatal transmission were analyzed. We found that 153 of the 166 clones analyzed from uncultured peripheral blood mononuclear cell DNA samples showed a 92.17% frequency of intact vpr open reading frames. There was a low degree of heterogeneity of vpr genes within mothers, within infants, and between epidemiologically linked mother-infant pairs. The distances between vpr sequences were greater in epidemiologically unlinked individuals than in epidemiologically linked mother-infant pairs. Moreover, the infants’ sequences displayed patterns similar to those seen in their mothers. The functional domains essential for Vpr activity, including virion incorporation, nuclear import, and cell cycle arrest and differentiation were highly conserved in most of the sequences. Phylogenetic analyses of 166 mother-infant pairs and 195 other available vpr sequences from HIV databases formed distinct clusters for each mother-infant pair and for other vpr sequences and grouped the six mother-infant pairs’ sequences with subtype B sequences. A high degree of conservation of intact and functional vpr supports the notion that vpr plays an important role in HIV-1 infection and replication in mother-infant isolates that are involved in perinatal transmission.
Mother-to-infant transmission of human immunodeficiency virus type 1 (HIV-1) occurs at an estimated rate of more than 30% and is one of the major causes of AIDS in children (1, 11, 17, 34, 43). In studies of the molecular characterization of HIV-1 from mothers and infants with perinatal transmission, we (2) and others (35, 42, 49) have shown a selective transmission of HIV-1 from mothers to infants, based on the sequence analyses of HIV-1 envelope variable regions. Selective transmission of HIV-1 has also been demonstrated in sexual transmission from transmitters to recipients, including a homogeneous sequence population present in the recipients (32, 48, 52, 53). Factors influencing maternal-fetal transmission of HIV-1 include advanced clinical stage of the mother, low CD4+ counts, maternal immune response to HIV-1 antigenemia, recent infection, high viral load in mothers, maternal disease progression, and HIV-1 heterogeneity in the env region of mothers (1, 2, 34, 43, 49). However, the viral determinants involved in perinatal or sexual transmission are not known, which makes it difficult to develop strategies for prevention and treatment of HIV-1 infection in children. Nonetheless, it is possible that several other regions of the HIV-1 genome including accessory and regulatory genes are one of the viral determinants associated with mother-to-infant transmission. Recently, we have shown that an intact and functional vif open reading frame was conserved in HIV-1-infected mother-infant pairs following perinatal transmission (50). Along with vif, HIV-1 encodes another accessory gene, namely, vpr, that is conserved among diverse members of the primate immunodeficiency viruses and confers selection for its function in vivo (15, 44, 46). We, therefore, sought to analyze vpr sequences from mother-infant isolates following perinatal transmission.
The vpr open reading frame encodes a 14-kDa virion- and cell-associated protein (6, 10, 27) that is dispensable for HIV-1 replication in T-cell lines (4, 9) but is required for efficient replication in primary monocytes/macrophages (4, 5, 7). Several possible roles for Vpr in HIV-1 replication have been suggested, including a modest transactivation of HIV-1 long terminal repeat (6), enhancement of the nuclear migration of the preintegration complex in newly infected nondividing cells (16), and inhibition of the establishment of chronic HIV-1 infection (36, 40). In addition, Vpr has been shown to arrest cells in the G2/M phase of the cell cycle (18, 40). Moreover, Vpr has been reported to be capable of inducing latent cells into high-level viral production (25). However, the role of Vpr in AIDS pathogenesis is not very well understood. Lang et al. (23) have shown that macaques infected with the simian immunodeficiency virus SIVmac239 defective in vpr progressed to AIDS slowly compared to SIVmac239 containing a wild-type vpr. In vitro studies have shown that generation of defective vpr genes results in viral persistence (20, 40) and loss of cytopathogenicity (20). While some studies have demonstrated an association between the presence of defective or mutated vpr quasispecies and long-term nonprogressors of HIV-1 infection (41, 47), others have shown a lack of correlation (8, 51). However, a complete analysis of vpr sequences following HIV-1 mother-infant transmission has not been performed. Mutations in the vpr gene may potentially affect mother-to-infant transmission of HIV-1, since this gene is essential for efficient viral replication in primary monocytes/macrophages (4, 5, 7) and the macrophage-tropic viruses are believed to be involved in transmission (31, 53).
To characterize the HIV-1 vpr isolates involved in mother-to-infant transmission, we have analyzed the vpr sequences from six infected mother-infant pairs following perinatal transmission. We show that the vpr open reading frame was conserved in most of the mother-infant pair sequences. The domains required for Vpr function were also present in most of the mother-infant pair sequences, suggesting selection for Vpr function in vivo. Taken together, these findings indicate that vpr is important for HIV-1 infection and replication following mother-to-infant transmission.
Patient population, sample collection, and clinical parameters.
This study was approved by the Human Subjects Committee of the University of Arizona, Tucson, Arizona, and the Institutional Review Board of the Children’s Hospital Medical Center, Cincinnati, Ohio, and written informed consent was obtained for participation in the study. Blood samples were collected from six HIV-1-infected mother-infant pairs, and the ages of the infants at the time of specimen collection were 6 weeks (infant A), 4.75 months (infant B), 14 months (infant C), 28 months (infant D), 34 months (infant E), and 1 week (infant F) following perinatal transmission. Mothers A, B, C, D, and F and infants A, B, and F were asymptomatic, whereas mother E and infants C, D, and E had symptomatic AIDS. Moreover, mother E and infants C, E, and F were on zidovudine, and infant D was on zalcitibine.
PCR amplification and cloning and sequencing of HIV-1 vpr genes.
The peripheral blood mononuclear cells (PBMC) were isolated by a single-step Ficoll-Paque procedure (Pharmacia-LKB) from whole-blood samples from HIV-1-infected mother-infant pairs. DNA was isolated by a modified version of the procedure described previously (2). TNE buffer (0.5 M Tris-HCl [pH 7.5], 0.1 M NaCl, 1 mM EDTA) (0.5 ml) was used. A two-step PCR amplification, first with outer primers VIF5 (5′TGGCAGCAATTTCACCGGTACTA, positions 4580 to 4602, sense) and VPR1 (5′CAACTTGGCAATGAAAGCAACAC, positions 5916 to 5939, antisense) and then with nested or inner primers VIF6 (5′TCAAGCAGGAATTTGGAATTCCC, positions 4633 to 4655, sense) and VPR2 (5′GGTACAAGCAGTTTAGGCTGACT, positions 5875 to 5898, antisense), was performed to amplify vpr sequences from infected patient PBMC DNA samples (2, 3). Equal amounts of HIV-1 PBMC DNA were used from the patients as determined by end point dilution (13). The PCRs were performed in a 25-μl reaction mixture containing 2.5 μl of 10× LA PCR buffer which consists of 25 mM TAPS [tris(hydroxymethyl)-methyl-amino propanesulfonic acid, sodium salt] (pH 9.3), 50 mM KCl, 2 mM MgCl2, 1 mM 2-mercaptoethanol, 400 μM dATP, 400 μM dCTP, 400 μM dGPT, and 400 μM TTP, 0.2 μM (each) outer primer, and 2.5 U of TaKaRa LA Taq polymerase (TaKaRa Biomedicals, Shiga, Japan). PCR was performed for 35 cycles, with 1 cycle consisting of 30 s at 95°C, 45 s at 50°C, and 3 min at 72°C. After the first round of PCR, 1 μl of the product was amplified for 35 cycles with the corresponding inner primers, with 1 cycle consisting of 30 s at 95°C, 45 s at 55°C, and 3 min at 72°C. We also included a known HIV-1 NL 4-3 sequence for PCR amplifications as a control to assess errors generated by TaKaRa LA Taq polymerase. The PCR products amplified by inner primer pair VIF6-VPR2 that yielded 1,246-bp fragments (not shown) were blunt ended by DNA polymerase I (Gibco-BRL, Gaithersburg, Md.), treated with T4 polynucleotide kinase (Gibco-BRL), and cloned into the SmaI site of pGem 3Zf (+) vector (Promega Corp., Madison, Wis.). The clones with the correct-size inserts were selected for DNA preparation followed by nucleotide sequencing of 9 to 19 clones from each patient according to Sequenase protocol (U.S. Biochemical Corp., Cleveland, Ohio).
Computer alignment and analysis of HIV-1 vpr sequences.
The nucleotide sequences of the vpr genes (288 bp) from the six mother-infant pairs were translated to the corresponding amino acid sequences (96 amino acids). Alignments were performed by hand, as one position contained a gap. Pairwise distances, defined as the percentages of mismatches between two aligned nucleotide sequences, were used to study the extent of genetic variability for sequences within an individual and between mother and infant. For intraindividual variability (within mothers’ and infants’ sequence sets), pairwise distances were calculated for all possible comparisons of pairs of sequences within the set. For interindividual variability (between mother-infant sets and between epidemiologically unlinked individual sets), each sequence from one set was compared with each sequence of the other set. The selection pressure was calculated as the ratio of nonsynonymous to synonymous substitutions (38) by comparing all possible pairs of sequences between mothers and their infants. The phylogenetic analysis was performed by using the software PHYLIP, version 3.5 (12). The tree was built from a distance matrix (function DNADIST) by using the neighbor-joining method (function NEIGHBOR). The robustness of the neighbor-joining tree was assessed by bootstrap resampling of the multiple alignments (function SEQBOOT). We traced two trees, one for 166 vpr sequences from 6 mother-infant pairs with HIV-1 NL 4-3 as a root and the second for 166 mother-infant pair vpr sequences and 195 outgroup sequences found in the HIV databases. These outgroup sequences were extracted by using Entrez version 6.04 (45) and handled by using Sequin version 2.25 (19).
Analysis of vpr sequences in mother-infant isolates.
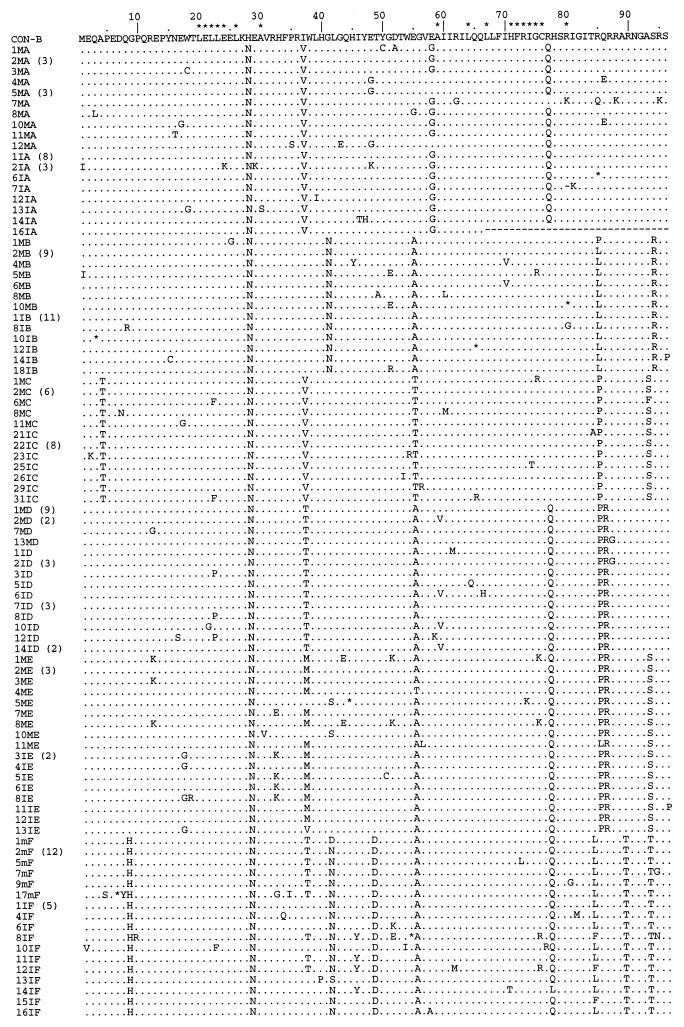
The alignment of multiple deduced amino acid sequences (96 amino acids) of 166 vpr clones from six mother-infant pairs and subtype B consensus sequence is shown in Fig. 1. The coding potential of the vpr open reading frame was maintained in most of the sequences in 47,808 bp sequenced. We analyzed 166 different vpr clones, and 153 clones contained intact vpr open reading frames, a 92.17% frequency of conservation of intact vpr open reading frames. The frequency of defective vpr genes in our six mother-infant pair sequences was 7.83%. We found that a total of seven clones contained stop codons. In addition, five clones (10IG, 5MB, 2IA, 8IA, and 10IA) lacked initiation codons, and one clone (16IA) had a 90-bp (30-amino-acid) deletion at the C terminus. Our vpr sequences displayed amino acid sequence patterns in each mother-infant pair that were not seen in epidemiologically unlinked pairs (Fig. 1). Interestingly, there were several amino acid motifs present in most of the mother-infant pair sequences, including an asparagine (N) at position 28, an alanine (A) or threonine (T) at position 55, and a glutamine (Q) or arginine (R) at position 77.
FIG. 1.
Alignment of multiple deduced amino acid sequences of the vpr gene of HIV-1 from six mother-infant pairs following perinatal transmission. In the six mother-infant pair sequence designations, the letters A to F indicate the mother-infant pairs A, B, C, D, E, and F, respectively, the letter M indicates the mother and the letter I indicates the infant in each mother-infant pair, and the numbers are the clone number. In the alignment, the top sequence (CON-B) is the consensus sequence of the clade or subtype B as defined elsewhere (37). In the sequences, amino acids identical to those in the CON-B sequence (.), gaps introduced to maximize alignment (−), and stop codons (asterisks) are shown. The number of identical clones is indicated in parentheses. Above the alignment, the asterisks indicate the amino acids essential for Vpr function, including virion incorporation, cytoskeleton function, nuclear transport, and cell cycle arrest (10, 28–30).
Comparison of vpr sequences of epidemiologically linked mother-infant isolates.
To determine the degree of variability of the vpr gene from six mother-infant pairs, we analyzed variation in nucleotide and amino acid sequences as shown in Table 1. The nucleotide sequences of the vpr gene within mothers (A, B, C, D, E, and F) differed by 2.2, 0.6, 0.8, 0.6, 2.5, and 0.7 (median values), respectively, ranging from 0 to 5.3%. The variability in sequences from the infants (A, B, C, D, E, and F) was very similar to the variability in the sequences from the mothers and differed by 0.8, 0.5, 1, 1.1, 1.5, and 1.5% (median values), respectively, ranging from 0 to 3.7%. Interestingly, the variability between mother-and-infant sets (epidemiologically linked pairs A, B, C, D, E, and F) was also on the same order of 2.4, 0.8, 1.2, 1.1, 2.9, and 1.6% (median values), respectively, ranging from 0 to 5.3%. For mother-infant pairs A, B, C, D, E, and F, the median values of amino acid sequence variability of vpr were as follows: within mothers, 3.5, 1.6, 1.3, 0.6, 3.5, and 1.1%, respectively; within infants, 2.1, 0.9, 2.2, 2.0, 2.1, and 3.8%, respectively; and between epidemiologically linked mother-infant pairs, 3.0, 1.3, 1.2, 1.3, 3.4, and 3.2%, respectively. Moreover, there was no difference in variability of vpr sequences with increasing infants’ age. We also determined whether low variability of the vpr gene from mother-infant pair isolates is due to the errors made by TaKaRa LA polymerase used in our study. We rarely found any errors made by TaKaRa LA polymerase when using a known sequence of HIV-1 NL 4-3 for PCR amplifications and DNA sequencing of the vpr gene. The median distributions of distances for sequences from the same mother and the same infant, between epidemiologically linked mother-infant pairs, and between two epidemiologically unlinked mothers were 1.1, 1.0, 1.8, and 5.8%, respectively (not shown). Thus, by using the sequence distances of a conserved region, such as vpr, we were able to easily differentiate the epidemiologically unlinked individuals from epidemiologically linked mother-infant pairs. The selective pressure on mother-infant pair vpr sequences was determined by calculating the ratios of nonsynonymous and synonymous substitutions and showed no evidence for positive selection pressure for change. Comparisons of infant sequences with mother sequences from pairs A, B, C, D, E, and F gave ratios of nonsynonymous to synonymous substitution, dn/ds, of 0.07, 0.08, 0.1, 0.2, 0.08, and 0.3, respectively. Thus, there was very little selection pressure (a ratio of <1) on vpr sequences to change.
TABLE 1.
Distances in the vpr sequences within mothers, within infants, and between mother-infant pairs
| Sequence | Pair | % Distance for sequencesa
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Within mothers
|
Within infants
|
Between mother-infant pairs
|
||||||||
| Min | Median | Max | Min | Median | Max | Min | Median | Max | ||
| Nucleotide | A | 0 | 2.2 | 5.3 | 0 | 0.8 | 2.5 | 0.3 | 2.4 | 5.3 |
| B | 0 | 0.6 | 1.9 | 0 | 0.5 | 1.6 | 0 | 0.8 | 2.5 | |
| C | 0 | 0.8 | 1.9 | 0 | 1.0 | 2.2 | 0.3 | 1.2 | 2.5 | |
| D | 0 | 0.6 | 1.6 | 0 | 1.1 | 2.8 | 0.3 | 1.1 | 2.5 | |
| E | 0.3 | 2.5 | 5.3 | 0.3 | 1.5 | 2.8 | 0.9 | 2.9 | 5.0 | |
| F | 0 | 0.7 | 2.2 | 0 | 1.5 | 3.7 | 0.6 | 1.6 | 4.0 | |
| Totalb | 0 | 1.1 | 5.3 | 0 | 1.0 | 3.7 | 0 | 1.5 | 5.3 | |
| Amino acid | A | 0 | 3.5 | 10.4 | 0 | 2.1 | 6.3 | 0 | 3.0 | 10.4 |
| B | 0 | 1.6 | 5.2 | 0 | 0.9 | 4.1 | 0 | 1.3 | 5.2 | |
| C | 0 | 1.3 | 4.2 | 0 | 2.2 | 4.2 | 0 | 1.2 | 4.2 | |
| D | 0 | 0.6 | 2.1 | 0 | 2.0 | 5.2 | 0 | 1.3 | 4.2 | |
| E | 0 | 3.5 | 8.3 | 0 | 2.1 | 4.2 | 0 | 3.4 | 7.3 | |
| F | 0 | 1.1 | 6.3 | 0 | 3.8 | 12.5 | 1.0 | 3.2 | 12.5 | |
| Total | 0 | 1.8 | 10.4 | 0 | 1.9 | 12.5 | 0 | 2.0 | 12.5 | |
Expressed as percent nucleotides (for nucleotide sequence) or percent amino acids (for amino acid sequence). Min, minimum; Max, maximum.
Calculated from all the pairs taken together.
Phylogenetic analysis of vpr sequences of mother-infant isolates.
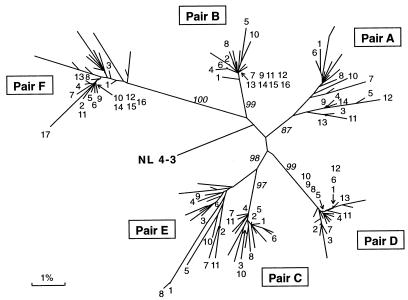
To determine the similarity relationships among the 166 vpr sequences from six mother-infant pairs and 195 other vpr sequences from infected individuals present in HIV databases, we performed phylogenetic analyses as shown in Fig. 2 and 3. The phylogenetic tree traced for 166 vpr sequences revealed that the six mother-infant pairs were well discriminated, separated, and confined within subtrees (Fig. 2), indicating the absence of PCR product cross-contamination (21, 24). These subtrees were equidistant from each other. The average distance between two sequences from different pairs (intersubtree distance) ranged from 5% between pairs C and E to 8.8% between pairs C and F. High bootstrap values further emphasized the separation between the subtrees as distinct clusters. Bootstrap analysis performed on resampling the data sets 100 times formed the same clusters of the sequences 87 to 100 times in the six mother-infant pairs. Furthermore, the six subtrees showed homogeneous vpr sequences in which some sequences were intermingled.
FIG. 2.
Phylogenetic analysis of vpr sequences from six mother-infant pairs (A, B, C, D, E, and F). The distances were calculated between the nucleotide sequences from the six mother-infant pairs. Each leaf of the tree represents one vpr sequence. The mother sequences in each pair are labeled with the number of the clones (Fig. 1), whereas the infant sequences are unlabeled. The tree was rooted by using the reference HIV-1 sequence, NL 4-3 (37). The numbers at branch points indicate the occurrence number of branches over 100 bootstrap resampling of the data sets. The mother-infant pairs formed a distinct cluster and are discriminated, separated, and confined within subtrees, indicating the absence of PCR product cross-contamination (21, 24).
FIG. 3.
Phylogenetic analysis of 166 mother-infant pair (A, B, C, D, E, and F) sequences and 195 other vpr sequences from HIV-1 databases. The sequences from Kuiken et al. (22) are labeled Dutch (samples from Amsterdam Cohorts) and Scott (from Scottish samples). The sequences from Michael et al. (33) were obtained from a long-term survivor, 3799, and two control patients CONTRL1 and CONTRL2. The sequences from Saksena et al. (41) were obtained from Australian patients Aus890 and Aus1149. The sequences from Ge et al. (14) and Wang et al. (47) were obtained from two long-term nonprogressors (LTNPs) and drug users (IVDUs). The other 45 sequences from the HIV databases belong to the following subtypes (accession numbers shown in parentheses): subtype B, MN (M17449), JRcsf (M38429), PNL4-3 (U26942), NY5 (M19921), clade B (U26546), PV22 (K02083), SF2 (K02007), HAN (U43141), D31 (U43096), WEAU (U21135), UK-Manchester (UK-Man) (U23487), F12 (Z11530), 89.6 (U39362), and C18MBC (U37270); subtype A, U455 (M62320), Z321 (U76035), IbNg (L39106), and 92UG027 (U51190); subtype C, 92BR025 (U52953) and ETH C2220 (U46016); subtype D, ELI (K03454), NDK (M27323), and 84ZR085 (U88822); subtype A/C recombinant, ZAM184 (U86780) and ZAM174 (U86768); subtype A/D recombinant, MAL (X04415); subtype A/E recombinants CM240 (U54771), 93TH253 (U51189), and 90CR402 (U51188); and subtype A/G recombinant, 92NG0003 (U88825) and 92NG083 (U88826). The largest, starlike cluster contained all subtype B sequences. Subtypes other than B are indicated in parentheses after the isolate names. The six mother-infant pair sequences clustered with subtype B sequences.
In the second phylogenetic analysis (Fig. 3), 166 vpr sequences from six mother-infant pairs and 195 other available vpr sequences were included. These additional vpr sequences from several independent studies analyzing vpr genes from infected individuals (14, 22, 41, 47, 51) and 48 other sequences belonged to two main HIV-1 groups, group M (M for major) and group O (O for outlier). Group M includes at least 10 distinct HIV-1 subtypes designated A to J, and we included subtypes A, B, C, and D and recombinants A/C, A/E, A/G, and A/D. The tree grouped the 361 sequences in a manner similar to that for the env, gag, and vif genes (2, 42, 49, 50). The largest, starlike cluster, contained all subtype B sequences, including the six mother-infant pair sequences. Although diverging independently from the center of the subtype B, subtype D (ELI, NDK, MAL, and 84ZR085) appeared as distant from subtype B sequences as two subtype B sequences. Subtype C, subtype A/C, and subtypes A, A/E, and A/G were more distant from subtype B and diverged in three separate lineages. In subtype B, sets of sequences from the same individuals and from each mother-infant pair were closely grouped in subtrees. Subtype B sequences from different individuals were shown approximately equidistant from the others as if arising from a common ancestor. The genetic distances among subtype B sequences, intra- and interindividual distances included, was 6% on average, ranging from 0 to 18%. This value did not change when subtype D was included. The average distance between subtypes A and C and recombinant sequences was 10%, ranging from 0 to 17%. We measured the global variability of subtype B by generating a consensus sequence, supposedly at the center of the subtree B, and comparing all subtype B sequences to it. We generated the consensus sequence from the most frequent nucleotide at each position of the alignment. The genetic distances between the 335 subtype B sequences and the consensus B sequence was 4%, ranging from 0.1 to 8%. When subtype D sequences were included in the calculation, the variability around the consensus B sequence was up to 10%. Subtypes A, C, and recombinants had genetic distances of 10 to 15% from the consensus B sequence, and group O had 21% genetic distance from the consensus B sequence on average.
Conservation of functional domains required for Vpr protein function in mother-infant isolates.
Three domains have been classified in the Vpr protein (37) that are involved in virion incorporation, oligomerization, nuclear transport, cell cycle arrest, and differentiation. The first domain includes amino acids 1 to 42 containing an oligomerization domain and a putative α-helix (amino acid residues 17 to 34) required for virion incorporation. The second domain encompasses an H(S/F)RIG motif related with cytoskeleton function (28), a leucine/isoleucine-rich sequence (LR motif from amino acids 60 to 81) important for nuclear localization (30), a conserved dipeptide (GC residues 75 and 76) and a potential α-helical motif (residues 46 to 74) postulated to play a role in virion incorporation and in the stability of Vpr (29). The third domain (residues 77 to 96) contains highly charged amino acids and is suggested to be involved in cell cycle arrest and differentiation (10). The NH2-terminal domain of the Vpr contains five negatively charged amino acids that are highly conserved and predicts an amphipathic α-helix (30, 31). Examination of vpr sequences from six mother-infant pairs showed that the five negatively charged amino acids at the same positions were highly conserved (Fig. 1). In addition, the HFRIG motif related with cytoskeleton function (25) was present in most of the sequences. Based on mutational analysis, several studies (10, 30) have identified specific domains in Vpr required for virion incorporation, nuclear import, and cell cycle arrest and differentiation. The two glutamic acids at positions 21 and 24, the hydrophobic polar leucines at positions 20, 22, 23, and 26, and the alanine at position 59 that are required for virion incorporation of Vpr (30) were highly conserved in most of the 166 vpr sequences analyzed. In six clones from pair D (2MD, 3MD, 6ID, 14ID, 15ID, and 16ID), alanine was substituted with valine. We then examined the nuclear transport properties of Vpr that comprises glutamic acid at positions 21 and 24, leucine at positions 20, 22, 23, 26, 67, and 68, and alanine at position 59 (30) and found them to be highly conserved in most of the mother-infant pairs’ vpr sequences analyzed (Fig. 1, pairs A to F). The cell cycle arrest properties of Vpr that require glutamic acid at positions 21 and 24, alanine at positions 30 and 59, leucine at positions 64 and 67, histidine at position 71, glycine at position 75, and cysteine at position 76 (30) as well as arginine at positions 73 and 80 (10) were conserved in mother-infant pairs’ vpr sequences (Fig. 1).
We have provided evidence for maintenance of an intact vpr open reading frame with functional domains conserved following mother-to-infant HIV-1 transmission. The vpr sequences directly derived from uncultured PBMC DNA of six mother-infant pairs revealed a 92.17% frequency of conserved vpr open reading frames. The functional domains required for Vpr function, including virion incorporation (10, 30), predicted α-helix formation (29), nuclear localization (10, 30), and cell cycle arrest and differentiation (10, 28, 30), were highly conserved in most of the vpr sequences from the mother-infant pairs. Our results also show a low degree of variability of vpr sequences following mother-to-infant transmission. However, the epidemiologically linked mother-infant pair vpr sequences were easily distinguishable from epidemiologically unlinked individuals. Taken together, these findings suggest that there is a selection mechanism in vivo that maintains intact and functional vpr open reading frames (15), making vpr necessary for HIV-1 infection and replication in mothers and infants during perinatal transmission.
The data presented here demonstrated that the coding potential of the vpr open reading frame was maintained in most of the sequences in 47,808 bp sequenced, except for seven sequences containing stop codons, five lacking initiation codons, and one having a deletion (Fig. 1). Our results are consistent with the earlier published limited analyses of vpr sequences from one infected patient (33), the C terminus of Vpr from several infected patients (48), and some long-term progressors (8, 52). In contrast, defective vpr genes clustering at the C terminus have been shown in some long-term nonprogressors (42, 48). The frequency of defective vpr genes in our six mother-infant pairs was 7.83% and was comparable to but lower than vif (10.2%) (50) and higher than those observed for gag (1.5%) (26) and nef (3.3%) (39) genes. Interestingly, the Vpr amino acid sequences of each mother-infant pair displayed a pattern that was not seen in epidemiologically unlinked pairs (Fig. 1), as also observed in the analyses of V3 region (2) and vif (50) sequences of the same mother-infant pairs. In addition, several amino acid motifs, beside the functional domains, were conserved in most of sequences at positions 28, 55, and 77, including an aspargine (N) at position 28 that is highly conserved in most of the macrophage-tropic but not in all lymphotropic clones and consensus clade B sequences (37).
Phylogenetic analysis performed on the vpr sequences from the six mother-infant pairs clearly demonstrated that the six pairs were well discriminated, separated, and confined within subtrees (Fig. 2), indicating the absence of PCR product cross-contamination (21, 24). In addition, these vpr sequences formed distinct clusters and grouped with subtype B sequences when subjected to a phylogenetic analysis with 195 vpr sequences of several subtypes present in HIV databases (Fig. 3). These results are consistent with the phylogenetic analyses of other genes such as vif (50) and env (2, 49). Our data also suggest that the low variability of vpr sequences was not due to errors made by TaKaRa polymerase but to persistence in vivo and was in agreement with those reported for infected individuals (22) and for vif (50) and gag (26, 32) genes. There was no evidence for a positive selection pressure for changes in vpr sequences by immune responses (38). In contrast, the env V3 region sequence from the same mother-infant pairs’ isolates showed a positive selection pressure for change (2), thus harboring variable sequence population containing several variants or genotypes (2, 35, 49). HIV-1 vpr (as vif [50] or gag [26]) evolves with little selection pressure for change, and the mothers’ variants persist in their infants. Evidence for this conclusion is provided by a low variability of and little selection pressure for change in mother-infant vpr sequences.
There seems to be a selection for Vpr function in mother-infant vpr sequences, consistent with data from a recent published report that suggested a selection of Vpr function in vivo (15). The functional domains required for Vpr function, including the two glutamic acids at positions 21 and 24, four leucines at positions 20, 22, 23, and 26, and an alanine at position 59 required for virion incorporation of Vpr (10, 30), were highly conserved, indicating that Vpr is required early in viral replication. The other functional domains, such as HFRIG related to cytoskeleton function (28), the glutamic acid at positions 21 and 24, leucine at positions 20, 22, 23, 26, 67, and 68, and alanine at position 59 required for nuclear transport (29), and the glutamic acid at positions 21 and 22, alanine at positions 30 and 59, leucine at positions 64 and 67, histidine at position 71, glycine at position 75 and cysteine at position 76 (30) as well as arginine at positions 73 and 80 (10) essential for cell cycle arrest, were also conserved in most of the 166 vpr sequences (Fig. 1). Mutational analysis of Vpr has revealed that these amino acids at the positions described above are critical for Vpr function (10, 28, 30). Our data on vpr sequences from six mother-infant pairs, the closest in vivo situation, is consistent with earlier reports on Vpr functional analysis performed in vitro (10, 30). Furthermore, we have recently shown a similar conservation of the functional domains for another accessory gene, vif, derived from the same mother-infant pairs following perinatal transmission (50).
Although maternal-fetal transmission of HIV-1 may be multifactorial in nature, identification of viral factors or determinants involved in maternal transmission may provide insights toward the development of strategies for prevention and treatment. Since vpr is conserved among primate immunodeficiency viruses (15, 44, 46) and required for efficient HIV-1 replication in primary monocytes/macrophages (4, 5, 7), it may have a role in transmission because macrophage-tropic isolates are believed to be involved in transmission (31, 53). The data presented here on 92.17% frequency of intact vpr open reading frames and conserved functional domains for Vpr activity support the notion that vpr is important for HIV-1 pathogenesis in maternal-fetal isolates and may be one of the viral determinants of perinatal transmission.
Nucleotide sequence accession numbers.
The 166 vpr sequences from six mother-infant pairs have been submitted to GenBank with accession no. AF042864 to AF043029.
Acknowledgments
We thank Raymond C. Baker, Children’s Hospital Medical Center, Cincinnati, Ohio, for providing blood samples from HIV-1-infected mother-infant pairs; Scott Martin and Ulrike Philippar for technical help; and John J. Marchalonis, Erik Matala, Tobias Hahn, and Mohammad Husain of the University of Arizona for reviewing the manuscript.
This work was supported in part by grants to N.A. from the National Institute of Allergy and Infectious Diseases (AI 40378) and the Arizona Disease Control Research Commission (9601).
REFERENCES
- 1.Ahmad N. Maternal-fetal transmission of human immunodeficiency virus. J Biomed Sci. 1996;2:238–250. doi: 10.1007/BF02253703. [DOI] [PubMed] [Google Scholar]
- 2.Ahmad N, Baroudy B M, Baker R C, Chappey C. Genetic analysis of human immunodeficiency virus type 1 envelope V3 region isolates from mothers and infants after perinatal transmission. J Virol. 1995;69:1001–1012. doi: 10.1128/jvi.69.2.1001-1012.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahmad N, Schiff G M, Baroudy B M. Detection of viremia by a one step polymerase chain reaction method in hepatitis C virus infection. Virus Res. 1993;30:303–315. doi: 10.1016/0168-1702(93)90098-8. [DOI] [PubMed] [Google Scholar]
- 4.Balliet J W, Kolson D L, Eiger G, Kim F M, Megann K A, Srinivasan A, Collman R C. Distinct effects in primary macrophages and lymphocytes of human immunodeficiency virus type 1 accessory genes vpr, vpu and nef: mutational analysis of a primary HIV-1 isolate. Virology. 1994;200:623–631. doi: 10.1006/viro.1994.1225. [DOI] [PubMed] [Google Scholar]
- 5.Ballota C, Lusso P, Crowley R, Gallo R C, Franchini G. Antisense phosphorothioate oligonucleotides targeted to the vpr gene inhibit human immunodeficiency virus type 1 replication in primary human macrophages. J Virol. 1993;67:4409–4414. doi: 10.1128/jvi.67.7.4409-4414.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen E A, Dehni G, Sodroski J G, Haseltine W A. Human immunodeficiency virus vpr product is a virion-associated regulatory protein. J Virol. 1990;64:3097–3099. doi: 10.1128/jvi.64.6.3097-3099.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Connor R L, Chen B K, Chen S, Landau N R. Vpr is required for efficient replication of human immunodeficiency virus type 1 in mononuclear phagocytes. Virology. 1995;206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 8.Cornelissen M, Kuiken C, Zorgdrager F, Hartman S, Goudsmit J. Gross defects in the vpr and vpu genes of HIV type 1 cannot explain the differences in RNA copy number in long-term asymptomatics and progressors. AIDS Res Hum Retroviruses. 1997;13:247–252. doi: 10.1089/aid.1997.13.247. [DOI] [PubMed] [Google Scholar]
- 9.Dedera D, Hu W, Vander Heyden N, Ratner L. Viral protein R of human immunodeficiency virus types 1 and 2 is dispensable for replication and cytopathogenicity in lymphoid cells. J Virol. 1989;63:3205–3208. doi: 10.1128/jvi.63.7.3205-3208.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DiMarzio P, Choe S, Elbright M, Knoblauch R, Landau N R. Mutational analysis of the cell cycle, nuclear localization, and virion packaging of human immunodeficiency virus type 1 Vpr. J Virol. 1995;69:3205–3208. doi: 10.1128/jvi.69.12.7909-7916.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.European Collaborative Study. Mother to child transmission of HIV-1. Lancet. 1988;ii:1039–1042. [PubMed] [Google Scholar]
- 12.Felsenstein J. PHYLIP phylogenetic inference package. Cladiatice. 1989;5:164–166. [Google Scholar]
- 13.Furtado M R, Kingsley L A, Wolinsky S M. Changes in the viral mRNA expression pattern correlate with a rapid rate of CD4+ T-cell number decline in human immunodeficiency virus type 1-infected individuals. J Virol. 1995;69:2092–2100. doi: 10.1128/jvi.69.4.2092-2100.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ge Y C, Wang B, Dwyer D E, Cunningham A L, Saxena N. Length polymorphism of the viral protein R of human immunodeficiency virus type 1 strains. AIDS Res Hum Retroviruses. 1996;12:351–354. doi: 10.1089/aid.1996.12.351. [DOI] [PubMed] [Google Scholar]
- 15.Goh W C, Rogel M E, Kinsey C M, Michael S C, Fultz P N, Nowak M A, Hahn B H, Emerman M. HIV-1 Vpr increases viral expression by manipulation of the cell cycle: a mechanism for selection of Vpr in vivo. Nat Med. 1998;4:65–71. doi: 10.1038/nm0198-065. [DOI] [PubMed] [Google Scholar]
- 16.Heinzinger N K, Bukrinsky M L, Haggerty S A, Ragland A M, Kewalramani V, Lee M A, Gendelman H E, Ratner L, Stevenson M, Emmereman M. The Vpr protein of human immunodeficiency virus type 1 influences nuclear localization of viral nucleic acids in non-dividing host cells. Proc Natl Acad Sci USA. 1994;91:7311–7315. doi: 10.1073/pnas.91.15.7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Italian Multiculture Study. Epidemiology, clinical features and prognostic factors of pediatric HIV infection. Lancet. 1988;ii:1043–1046. [PubMed] [Google Scholar]
- 18.Jowett J B M, Planelles V, Poon B, Shah N P, Chen M-L, Chen I S Y. The human immunodeficiency virus type 1 vpr gene arrests infected T cells in the G2 + M phase of the cell cycle. J Virol. 1995;69:6304–6313. doi: 10.1128/jvi.69.10.6304-6313.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kans J A. Conference Proceedings of the Cold Spring Harbor Laboratory Genome Mapping and Sequencing Meeting. 1996. Sequin; p. 114. [Google Scholar]
- 20.Kishi M, Zheng Y-H, Bahmani M K, Tokunaga K, Takahahi H, Kakinuma M, Lai P K, Nonoyama M, Luftig R B, Ikuta K. Naturally occurring accessory gene mutations lead to persistent human immunodeficiency virus type 1 infection of CD4-positive T cells. J Virol. 1995;69:7507–7518. doi: 10.1128/jvi.69.12.7507-7518.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Korber B T M, Learn G, Mulins J I, Hahn B H, Wolinsky S M. Protecting HIV databases. Nature. 1995;378:242–243. doi: 10.1038/378242a0. [DOI] [PubMed] [Google Scholar]
- 22.Kuiken C L, Cornelissen M T E, Zorgdrager F, Hartman S, Gibbs A J, Goudsmit J. Consistent risk group-associated differences in human immunodeficiency virus type 1 vpr, vpu and V3 sequences despite independent evolution. J Gen Virol. 1996;77:783–792. doi: 10.1099/0022-1317-77-4-783. [DOI] [PubMed] [Google Scholar]
- 23.Lang S M, Weeger M, Stahl-Hening C, Coulibaly C, Hunsmann G, Muller J, Muller-Hermelink H, Fuchs D, Wachter H, Daniel M M, Desrosiers R C, Fleckenstein B. Importance of vpr for infection of rhesus monkeys with simian immunodeficiency virus. J Virol. 1993;67:902–912. doi: 10.1128/jvi.67.2.902-912.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Learn G H, Jr, Korber B T M, Foley B, Hahn B H, Wolinsky S M, Mullins J I. Maintaining the integrity of human immunodeficiency virus sequence databases. J Virol. 1996;70:5720–5730. doi: 10.1128/jvi.70.8.5720-5730.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levy D N, Fernandes L S, Williams W V, Weiner D B. Induction of cell differentiation by human immunodeficiency virus 1 vpr. Cell. 1993;72:541–550. doi: 10.1016/0092-8674(93)90073-y. [DOI] [PubMed] [Google Scholar]
- 26.Louwagie J, McCutchan F E, Peeters M, Brennan T P, Sanders-Buell E, Eddy E, van der Groen G, Fransen K, Gershy-Damet G-M, Deleys R, Burke D S. Phylogenetic analysis of gag genes from 70 international HIV-1 isolates provides evidence of multiple genotypes. AIDS. 1993;7:769–780. doi: 10.1097/00002030-199306000-00003. [DOI] [PubMed] [Google Scholar]
- 27.Lu Y L, Spearman P, Ratner L. Human immunodeficiency virus type 1 viral protein R localization in infected cells and virions. J Virol. 1993;67:6542–6550. doi: 10.1128/jvi.67.11.6542-6550.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Macreadie I G, Castelli L A, Hewish D R, Kirkpatrick A, Ward A C, Azad A A. A domain of human immunodeficiency virus type 1 Vpr containing repeated H(S/F)RIG amino acid motifs causes growth arrest and structural defects. Proc Natl Acad Sci USA. 1995;92:2770–2774. doi: 10.1073/pnas.92.7.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mahalingam S, Khan S A, Jabbar M A, Monken C E, Collman R G, Srinivasan A. Identification of residues in the N-terminal acidic domain of HIV-1 Vpr essential for virion incorporation. Virology. 1995;207:297–302. doi: 10.1006/viro.1995.1081. [DOI] [PubMed] [Google Scholar]
- 30.Mahalingam S, Ayyavoo V, Patel M, Kieber-Emmons T, Weiner D B. Nuclear import, virion incorporation, and cell cycle arrest/differentiation are mediated by distinct functional domains of human immunodeficiency virus type 1 Vpr. J Virol. 1997;71:6339–6347. doi: 10.1128/jvi.71.9.6339-6347.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matala, E., V. R. K. Yedavalli, and N. Ahmad. Unpublished data.
- 32.McNearny T, Hornickora Z, Markham R, Birdnell A, Arnes M, Saah A, Ratner L. Relationship of human immunodeficiency virus type 1 sequence heterogeneity to stage of disease. Proc Natl Acad Sci USA. 1992;89:10247–10251. doi: 10.1073/pnas.89.21.10247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Michael N L, Chang G, D’Arcy L A, Ehrenberg P K, Mariani R, Busch M P, Brix D L, Schwartz D H. Defective accessory genes in a human immunodeficiency virus type 1-infected long-term survivor lacking recoverable virus. J Virol. 1995;69:4228–4236. doi: 10.1128/jvi.69.7.4228-4236.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mok J Q, Giaquinto C, DeRossi A, Gruch-Worner I, Ades A E, Pekham C S. Infants born to mothers seropositive for human immunodeficiency virus—preliminary findings from a multiculture European study. Lancet. 1987;i:1164–1168. doi: 10.1016/s0140-6736(87)92142-8. [DOI] [PubMed] [Google Scholar]
- 35.Mulder-Kampinga G A, Simonon A, Kuiken C L, Dekker J, Scherpbier H J, van de Perre P, Boer K, Goudsmit J. Similarity in env and gag genes between genomic RNAs of human immunodeficiency virus type 1 (HIV-1) from mother and infant is unrelated to time of HIV-1 RNA positivity in the child. J Virol. 1995;69:2285–2296. doi: 10.1128/jvi.69.4.2285-2296.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mustafa F, Robinson H L. Context-dependent role of human immunodeficiency virus type 1 auxillary genes in the establishment of chronic virus producers. J Virol. 1993;67:6909–6915. doi: 10.1128/jvi.67.11.6909-6915.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Myers G, Korber B, Hahn B H, Jeang K-T, Mellors J W, McCutchan F E, Henderson L E, Pavlakis G N. Human retroviruses and AIDS database. Los Alamos, N.Mex: Theoretical Biology, Los Alamos National Laboratory; 1995. [Google Scholar]
- 38.Nei M, Gojobori T. Simple methods for estimating the number of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 39.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rogel M E, Lu L I, Emerman M. The human immunodeficiency virus type 1 vpr gene prevents cell proliferation during chronic infection. J Virol. 1995;69:882–888. doi: 10.1128/jvi.69.2.882-888.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saksena N K, Ge Y C, Wang B, Xiang S H, Dwyer D E, Randle C, Palasanthiran P, Ziegler J, Cunningham A L. An HIV-1 infected long-term non-progressor (LTNP): molecular analysis of HIV-1 strains in the vpr and nef genes. Ann Acad Med Singapore. 1996;25:848–854. [PubMed] [Google Scholar]
- 42.Scarlatti G, Leitner T, Hapi E, Wahlberg J, Marchisi P, Clerici-Schoeller M A, Wigzell H, Fenyo E M, Albert J, Uhlen M, Rossi P. Comparison of variable region 3 sequences of human immunodeficiency virus type 1 from infected children with RNA/DNA sequences of virus population of their mothers. Proc Natl Acad Sci USA. 1993;90:1721–1725. doi: 10.1073/pnas.90.5.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scott G B, Fischl M A, Khmas N, Fletcher M A, Dickinson G M, Levine R S, Parks W P. Mothers of infants with acquired immunodeficiency syndrome: evidence for both symptomatic and asymptomatic carriers. JAMA. 1985;253:363–366. [PubMed] [Google Scholar]
- 44.Sharp P M, Balles E, Stevenson M, Emerman M, Hahn B H. Gene acquisition in HIV and SIV. Nature. 1996;383:586–587. doi: 10.1038/383586a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shuler G D, Epstein J A, Ohkawa H, Kans J A. Entrez: molecular biology database and retrieval system. Methods Enzymol. 1996;266:141–162. doi: 10.1016/s0076-6879(96)66012-1. [DOI] [PubMed] [Google Scholar]
- 46.Stivahtis G L, Soares M A, Vodicka M A, Hahn B H, Emerman M. Conservation and host specificity of Vpr-mediated cell cycle arrest suggest a fundamental role in primate lentivirus evolution and biology. J Virol. 1997;71:4331–4338. doi: 10.1128/jvi.71.6.4331-4338.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang B, Ge Y C, Palasanthiran P, Xiang S-H, Ziegler J, Dwyer D E, Randle C, Dowton D, Cunningham A, Saxena N K. Gene defects clustered at the C-terminus of the vpr gene of HIV-1 in long-term nonprogressing mother and child pair: in vivo evolution of vpr quasispecies in blood and plasma. Virology. 1996;223:224–232. doi: 10.1006/viro.1996.0471. [DOI] [PubMed] [Google Scholar]
- 48.Wolfs T, Zwart G, Bakker M, Goudsmit J. HIV-1 genomic RNA diversification following sexual and parental virus transmission. Virology. 1992;189:103–110. doi: 10.1016/0042-6822(92)90685-i. [DOI] [PubMed] [Google Scholar]
- 49.Wolinsky S M, Wike C M, Korber B T M, Hutto C, Parks W P, Rosenblum L L, Kuntsman K J, Furtado M R, Munoz I L. Selective transmission of human immunodeficiency virus type 1 variants from mother to infants. Science. 1992;255:1134–1137. doi: 10.1126/science.1546316. [DOI] [PubMed] [Google Scholar]
- 50.Yedavalli V R K, Chappey C, Matala E, Ahmad N. Conservation of an intact vif gene of human immunodeficiency virus type 1 during maternal-fetal transmission. J Virol. 1998;72:1092–1102. doi: 10.1128/jvi.72.2.1092-1102.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang L, Huang Y, Yuan H, Tuttleton S, Ho D D. Genetic characterization of vif, vpr, and vpu sequences from long-term survivors of human immunodeficiency virus type 1 infection. Virology. 1997;228:340–349. doi: 10.1006/viro.1996.8378. [DOI] [PubMed] [Google Scholar]
- 52.Zhang L Q, MacKenzie P, Cleland A, Holmes E C, Leigh Brown A J, Simmonds P. Selection for specific sequences in the external envelope protein of human immunodeficiency virus type 1 upon primary infection. J Virol. 1993;67:3345–3356. doi: 10.1128/jvi.67.6.3345-3356.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu T, Mo H, Wang N, Nam D S, Cao Y, Koup R A, Ho D D. Genotypic and phenotypic characterization of HIV-1 in patients with primary infection. Science. 1993;261:1179–1181. doi: 10.1126/science.8356453. [DOI] [PubMed] [Google Scholar]