Abstract
Background
Although immunotherapy with immune checkpoint inhibitors (ICIs) has become a standard therapeutic strategy in colorectal cancer (CRC) exhibiting microsatellite instability-high, limited patients benefit from this new approach. To increase the efficacy of ICIs in CRC patients, it is crucial to control the function of immunosuppressive cells in the tumor microenvironment. M2-tumor-associated macrophages (TAMs) are key immunosuppressive cells and promote tumor growth, angiogenesis, and epithelial-mesenchymal transition. In the present study, we focused on the VEGF signaling pathway in M2-TAMs to control their inhibitory function.
Methods
We evaluated the population of M2-TAMs, the VEGF receptor 2 (VEGFR2) expression on M2-TAMs, and the correlation between HIF-1α-positive cells and VEGFR2 expression levels on M2-TAMs in CRC using the analysis of The Cancer Genome Atlas colorectal adenocarcinoma dataset (n = 592), the flow cytometry of freshly resected surgical specimens of CRC (n = 20), and the immunofluorescence staining of formalin-fixed paraffin-embedded whole tissue samples of CRC (n = 20). Furthermore, we performed a functional assay of M2 macrophages through the VEGF/VEGFR2 signaling pathway in vitro.
Results
The population of M2-TAMs and their VEGFR2 expression significantly increased in the tumor compared to the normal mucosa in the CRC patients. HIF1-α-positive cells significantly correlated with the VEGFR2 expression level of M2-TAMs. M2 macrophages induced by cytokines in vitro produced TGF-β1 through the VEGF/VEGFR2 signaling pathway.
Conclusions
Our results suggest that anti-VEGFR2 therapy may have therapeutic potential to control the immune inhibitory functions of M2-TAMs in CRC, resulting in enhanced efficacy of immunotherapy with ICIs.
Electronic supplementary material
The online version of this article (10.1007/s00262-020-02676-8) contains supplementary material, which is available to authorized users.
Keywords: M2-tumor-associated macrophages (M2-TAMs), Colorectal cancer (CRC), VEGF receptor 2 (VEGFR2), TGF-β
Introduction
Colorectal cancer (CRC) has the third highest incidence and fourth highest mortality rate for males of all ages, as well as the second highest incidence and third highest mortality rate for females of all ages [1]. Patients with advanced CRC undergo multidisciplinary treatment such as surgical resection combined with chemotherapy and/or radiotherapy [2, 3]. Recently, although immunotherapy with immune checkpoint inhibitors (ICIs) has become a standard therapeutic strategy in CRC exhibiting microsatellite instability-high, its response rate is reported to be still limited at 30–55% [4, 5].
To increase the efficacy of this new treatment, the critical points are not only the accumulation of CTLs but also the inhibition of immunosuppressive cells, including MDSCs, Treg cells, and tumor-associated macrophages (TAMs), in the tumor microenvironment. The prevalence of them associated with tumor progression, metastasis, poor tumor differentiation, and poor survival [6–10].
TAMs can be categorized into inflammatory M1-TAMs (classically activated) or immunosuppressive M2-TAMs (alternatively activated) [11]. Especially, M2-TAMs promote tumor growth, angiogenesis, and epithelial-mesenchymal transition (EMT), and interfere with the antitumor function of immune cells, resulting in the suppression of antitumor immune responses in the tumor microenvironment [8–10, 12, 13]. Therefore, we focused on M2-TAMs in the present study to overcome the immune escape mechanism in the tumor microenvironment.
Recent reports on TAMs have suggested that they are polarized toward the M2 phenotype within the tumor microenvironment [14–16], and we have also previously reported that the distribution of M2-TAMs tended to increase in the tumor compared to the normal mucosa in CRC patients [17]. Since VEGF receptor 2 (VEGFR2) is expressed on macrophages [18], it is possible that anti-VEGFR2 therapy may control TAMs, especially M2-TAMs, in CRC.
In the present study, we evaluated the polarization of M1-/M2-TAMs and the VEGFR2 expression on M1-/M2-TAMs using freshly isolated samples, formalin-fixed paraffin-embedded (FFPE) tissue samples, and the public dataset from The Cancer Genome Atlas (TCGA) in CRC patients. Furthermore, we performed a functional assay of M2 macrophages through the VEGF/VEGFR2 signaling pathway in vitro.
Materials and methods
TCGA dataset analysis
The mRNA expression z-scores of genes (RNA-Seq V2 RSEM normalized, RNA-Seq data) were obtained from TCGA colorectal adenocarcinoma (PanCancer Atlas) dataset (n = 592) through cBioPortal (https://www.cbioportal.org/) [19, 20]. In the present study, we evaluated the mRNA expression levels of kinase insert domain receptor (KDR) (encoding VEGFR2) and hypoxia-inducible factor (HIF)1A. We utilized multi-gene expression signatures, including M1-TAMs signature (CD11c, CD40, CD64a, CD64b, CD64c, and CD80) and M2-TAMs signature (CD23, CD36, CD150, CD163, CD200, CD204, CD206, and CD209) [12]. The MRC1L1 gene expression was excluded because it was not available in TCGA dataset. The signature score was calculated by averaging the expression levels of included genes in M1-TAMs and M2-TAMs signature [21–23].
Patients
The inclusion criteria were patients who had undergone surgery for CRC at Fukushima Medical University Hospital between June 2018 and June 2019. The exclusion criteria were the preoperative treatment with a self-expanding metal stent and the tumor diameter of 3 cm or less because the stent induced local inflammation in the tumor and we need a 1–2 cm3 samples for flow cytometry. All procedures were conducted in accordance with the Helsinki Declaration and were approved by the Institutional Ethical Committee of Fukushima Medical University School of Medicine (Reference Nos. 2289 and 29316).
Freshly isolated clinical samples
Resected flesh samples of the normal mucosa and the tumor, sized 1–2 cm3, were immediately digested by the gentleMACS Octo Dissociator with Heaters (Miltenyi Biotec, Bergish Gladbach, Germany) using the Tumor Dissociation Kit (Miltenyi Biotec) according to the manufacturer’s protocol. Briefly, the samples were cut into 2–3 mm pieces and put in a C-tube (Miltenyi Biotec) with 5 ml of RPMI 1640 (Sigma‐Aldrich, St. Louis, MO) and enzymes H, R, and A in the Tumor Dissociation Kit. The samples were then dissociated by the gentleMACS Octo Dissociator with Heaters at 37 °C for 1 h, according to manufacturer’s recommendation. The collected cells were then filtered using Falcon® 70 µm Cell Strainer (Corning, Corning city, NY, USA), before being used for flow cytometric analysis.
Flow cytometry
The cells were stained according to the manufacturer’s preparation protocol for each antibody (Supplementary Table S1). To exclude the dead cells, 7-AAD (BD Biosciences, San Diego, CA, USA) was used, and the isotype-matched immunoglobulin served as a negative control. TAMs were detected by the gating used in our previous paper (M1-TAMs; CD14+CD11c+CD163−, M2-TAMs; CD14+CD11c−CD163+) (Fig. 1a) [17]. All staining was measured using a BD FACSCanto II flow cytometer (BD Biosciences) and data were analyzed using FlowJo software, version 10.3.0 (FlowJo, Ashland, OR, USA).
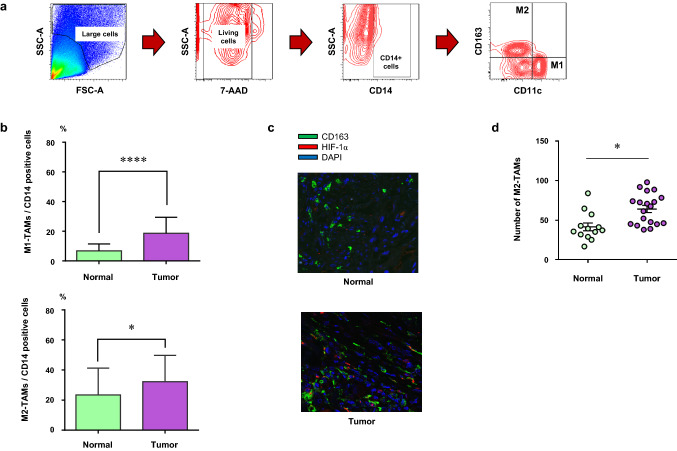
Fig. 1.
The number of M2-TAMs increased in the tumor. a The gating method of flow cytometry for freshly resected surgical specimens to detect M1-TAMs (CD14+CD11c+CD163−) and M2-TAMs (CD14+CD11c−CD163+). b The summary of population of M1- or M2-TAMs in CD14-positive cells in the normal mucosa (normal) and the tumor (tumor) by flow cytometric analysis (M1-TAMs; CD14+CD11c+CD163−, M2-TAMs; CD14+CD11c−CD163+). c Representative images showing the immunofluorescence staining of M2-TAMs in the normal mucosa (normal) and the tumor (tumor) samples. Green staining; CD163, red staining; HIF-1α, blue staining; DAPI. d The number of M2-TAMs in the normal mucosa (normal) and the tumor (tumor). *p < 0.05, ****p < 0.0001
Immunofluorescence staining
Four μm-thick sections were deparaffinized and rehydrated, and the sections were then incubated with epitope retrieval solution (pH9.0, Dako, Glostrup, Denmark) for 20 min at 100 °C. Thereafter, the sections were stained with anti-human CD163 antibody (clone 10D6, NCL-L-CD163, 1:200; Leica biosystems, Newcastle upon Tyne, UK) to detect M2-TAMs and anti-human HIF-1α antibody (polyclonal, ab82832, 1:100; abcam, Cambridge, UK) at 4 °C overnight. The sections were then incubated for 1 h with Alexa Fluor 488-conjugated anti-mouse (polyclonal, A-21202, 1:200; Thermo Fisher Scientific, Waltham, MA, USA) and Alexa Fluor 555-conjugated anti-rabbit secondary antibodies (polyclonal, A-31572, 1:200; Thermo Fisher Scientific). Nuclei were stained with DAPI (D9542, 40 ng/ml; Sigma-Aldrich) for 10 min. Finally, slides were mounted using ProLong Glass antifade reagent (Thermo Fisher Scientific), and images were obtained using an Olympus FV1000-D confocal microscope (Olympus, Tokyo, Japan). Green staining; CD163, red staining; HIF-1α, blue staining; DAPI. Counting of HIF-1α+ (red staining alone and/or merge around blue staining), CD163+ (green staining alone and/or merge around blue staining), and HIF-1α+CD163+ (green and red localized staining or merge around blue staining) cells was performed in four fields (× 400) of the hot spots of HIF-1α+ cells.
Induction of M1/M2 macrophages
CD14+ monocytes were isolated from peripheral blood mononuclear cells from healthy volunteers by magnetic labeling using a Pan Monocyte Isolation Kit (Miltenyi Biotec) according to the manufacturer’s instructions. The CD14+ monocytes were cultured in AIM-V (Fisher Scientific, Waltham, MA) with 25 ng/ml GM-CSF (215-GM-010; R&D systems, Minneapolis, MN, USA) to get M1 macrophages or 50 ng/ml M-CSF (216-MC-005; R&D systems) to get M2 macrophages for 5 days [24]. After 5 days, we cultured M1 or M2 macrophages with the addition of M1 or M2 cytokines (Table 1) for 48 h in hypoxia condition [24]. For hypoxia exposure, the cells were cultured with 100 μm cobalt chloride (Sigma-Aldrich) in a modulator incubator chamber (Billups-Rothenberg, Del Mar, CA, USA) with 1% O2, 5% CO2 and 94% N2 [25, 26].
Table 1.
Cytokines for the generation of M1/M2 macrophages used in the present study
| Cytokine | Company | Catalog number | Concentration (ng/ml) |
|---|---|---|---|
| M1 cytokines | |||
| IFN-γ | R&D systems (Minneapolis, MN, USA) | 285-IF-100 | 50 |
| LPS | Sigma-Aldrich (St. Louis, MO, USA) | L2630-10MG | 20 |
| M2 cytokines | |||
| IL-4 | Pepro Tech (Rocky Hill, NJ, USA) | 200-04 | 20 |
| IL-10 | Pepro Tech | 200-10 | 20 |
| IL-13 | Pepro Tech | 200-13 | 20 |
| TGF-β | Pepro Tech | 100-21 | 20 |
Cell treatment with VEGF and selective VEGFR2 inhibitor (small molecule)
We treated M1/M2 macrophages with 100 ng/ml of Recombinant Human VEGF 165 Protein (R&D systems) with/without 1 µm of selective VEGFR2 inhibitor (small molecule) (ZM323881, R&D systems). Each dose was recommended according to the manufacturer’s instructions. DMSO (Sigma-Aldrich) was used as a vehicle and negative control. The treated cells were analyzed by Western blotting after 2 h incubation and the culture supernatants were collected to measure the concentration of TGF-β1 after 48 h incubation.
Western blotting
Samples were prepared and stained with antibodies, and the protein signals were visualized as previously described [27, 28]. The primary antibodies and matched secondary antibodies used in the present study are presented in Supplementary Table S2.
ELISA
The TGF-β release in the culture supernatant of induced M2 macrophages was quantified with TGF beta-1 Human ELISA Kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol and the absorbance was measured using a Benchmark microplate spectrophotometer (Bio-Rad Laboratories, Nazareth, Belgium).
Statistical analysis
Correlations between two groups were evaluated using the Pearson correlation coefficient. A paired-two-tailed Student’s t test was used to compare two groups and one-way ANOVA with Turkey post hoc test was used to compare multiple groups. All statistical analyses were conducted using Graphpad Prism 6 software (Graphpad Software Inc., La Jolla, CA, USA). All p values were two-sided, and p < 0.05 was considered statistically significant.
Results
The number of M2-TAMs increased in the tumor
We evaluated the population of TAMs in CD14-positive cells in the tumor microenvironment of CRC by flow cytometry using the freshly resected surgical specimens (Fig. 1a) and immunofluorescence staining using FFPE whole tissue samples. The characteristics of patients and tumors are presented in Supplementary Table S3 (Stage I; 6, Stage II; 7, Stage III; 4, Stage IV; 3). The population of both M1- and M2-TAMs significantly increased in the tumor in comparison to the normal mucosa (p < 0.0001 and p = 0.02, respectively), with the population of M2-TAMs in the tumor being the highest (Fig. 1b). Representative images of immunofluorescence staining are presented in Fig. 1c. Immunofluorescence staining analysis also revealed that the number of M2-TAMs, CD163+ cells, significantly increased in the tumor (p = 0.0178) (Fig. 1d).
VEGFR2 was expressed on M2-TAMs in CRC patients
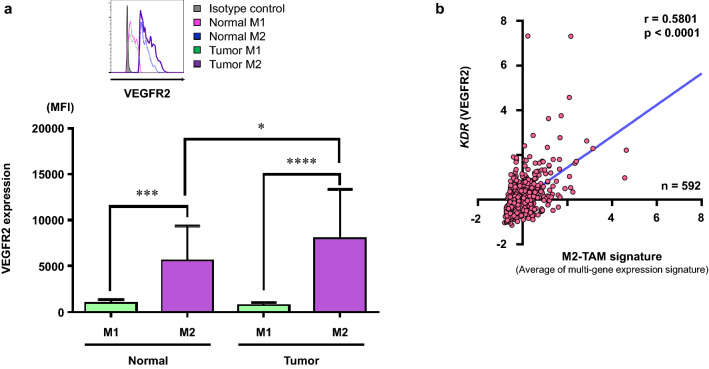
We assessed the expression of VEGFR2 on M1- and M2-TAMs in the tumor microenvironment of CRC by flow cytometry using the freshly resected surgical specimens. The gating method to detect the M1- and M2-TAMs is presented in Fig. 1a. A representative histogram of VEGFR2 expression is presented in Fig. 2a (upper). The summarized data of the 20 samples from CRC patients show that VEGFR2 expression on the M2-TAMs significantly increased in comparison to that on M1-TAMs in both the normal mucosa and the tumor, and, furthermore, VEGFR2 expression on M2-TAMs in the tumor was significantly higher than that in the normal mucosa (Fig. 2a). Next, we analyzed the correlation of VEGFR2 (KDR) and M2-TAMs signature using the TCGA colorectal adenocarcinoma (PanCancer Atlas) dataset, consisting of 592 samples. There was a significant correlation between mRNA expression of VEGFR2 (KDR) and M2-TAMs signature (CD23, CD36, CD163, CD200, CD209, MRC1, MSR1, and SLAMF1), which was calculated by averaging the expression levels of included genes (Fig. 2b). There was also a significant correlation between mRNA expression of VEGFR2 and M1-TAMs signature (CD40, CD80, FCGR1A, FCGR1B, FCGR1C, and ITGAX) (r = 0.4759, p < 0.0001).
Fig. 2.
VEGFR2 expression on M2-TAMs. a Summarized data for VEGFR2 expression on M1- and M2-TAMs in the normal mucosa (normal) and the tumor (tumor) by flow cytometric analysis (n = 20) (M1-TAMs; CD14+CD11c+CD163−, M2-TAMs; CD14+CD11c−CD163+). Representative histogram of VEGFR2 expression on M1- and M2-TAMs (upper). b mRNA expression z-scores in 592 samples with colorectal adenocarcinoma were obtained from TCGA (PanCancer Atlas) dataset. We utilized multi-gene expression signatures for M2-TAMs signature and the signature score was calculated by averaging the expression levels of included genes. Correlation of mRNA levels of KDR (VEGFR2) with M2-TAMs signature. *p < 0.05, ***p < 0.001, ****p < 0.0001
HIF1-α+ M2-TAMs increased in the tumor and HIF1-α+ cells significantly correlated with VEGFR2 expression of M2-TAMs
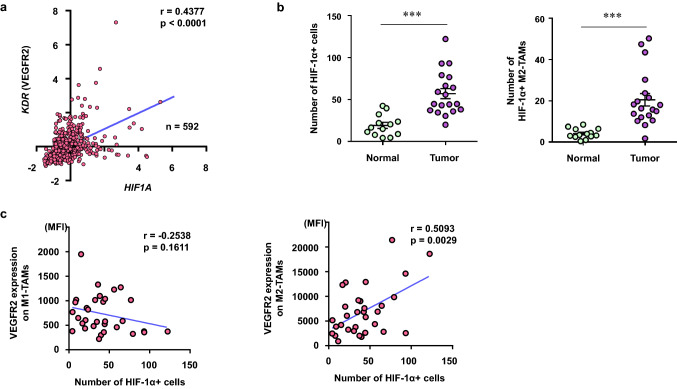
We then assessed the correlation between VEGFR2 and HIF1A using the TCGA colorectal adenocarcinoma (PanCancer Atlas) dataset. The mRNA expression of VEGFR2 was significantly correlated with that of HIF1A (Fig. 3a). Moreover, immunofluorescence staining analysis revealed that the number of HIF-1α+ cells and HIF-1α+ M2-TAMs significantly increased in the tumor in comparison to the normal mucosa (p = 0.0002, p = 0.0009, respectively) (Fig. 3b).
Fig. 3.
HIF1-α+ M2-TAMs increased in the tumor. a mRNA expression z-scores in 592 samples with colorectal adenocarcinoma were obtained from TCGA (PanCancer Atlas) dataset. Correlation of KDR (VEGFR2) with HIF1A. b The number of HIF-1α+ cells (left) and HIF-1α+ M2-TAMs (right) in the normal mucosa (normal) and the tumor (tumor). c The summary of correlation of VEGFR2 expression on M1-TAMs (CD14+ CD11c+ CD163−) and M2-TAMs (CD14+CD11c−CD163+) from flow cytometric analysis with the number of HIF-1α+ cells from immunofluorescence staining analysis in each patient. ***p < 0.001
Subsequently, we evaluated the correlation between the number of HIF-1α+ cells from the immunofluorescence staining analysis and the expression level of VEGFR2 on M1- or M2-TAMs from the flow cytometric analysis in each patient, respectively. There was a significant positive correlation between the number of HIF-1α+ cells and the VEGFR2 expression on M2-TAMs, but not with that on M1-TAMs (Fig. 3c).
M2 macrophages produced TGF-β1 through the VEGF/VEGFR2 signaling pathway
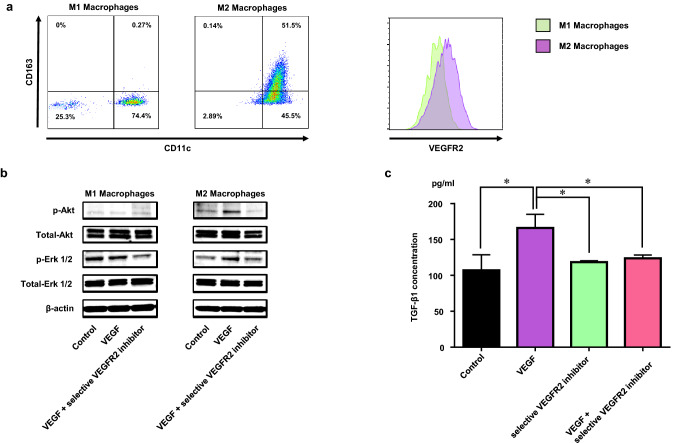
We generated M1 and M2 macrophages in vitro, as described in the “Materials and methods”. We evaluated their VEGFR2 expression using flow cytometry and confirmed the VEGFR2 expression on M2 macrophages (Fig. 4a). Next, we treated M1 and M2 macrophages with VEGF and/or selective VEGFR2 inhibitor (small molecule), as described in the “Materials and methods”. VEGF-signal-related molecules were activated by VEGF stimulation in the M2 but not the M1 macrophages, since p-Akt and p-Erk1/2 increased by stimulation with VEGF in the M2 macrophages only (Fig. 4b). This activation was canceled with the treatment of selective VEGFR2 inhibitor (small molecule) (Fig. 4b). Furthermore, we treated M2 macrophages in the same way and culture supernatants were collected to measure the concentration of TGF-β1. Production of TGF-β1 significantly increased in M2 macrophages by stimulation with VEGF and this TGF-β1 release was completely canceled by selective VEGFR2 inhibitor (small molecule) treatment (Fig. 4c).
Fig. 4.
M2 macrophages expressing VEGFR2 produced TGF-β1 through the VEGF/VEGFR2 signaling pathway. a VEGFR2 expression on induced M1 and M2 macrophages by flow cytometric analysis. b Western blot analysis showing the VEGF signaling molecules in induced M1 and M2 macrophages after stimulation by VEGF with/without selective VEGFR2 inhibitor (small molecule). c ELISA analysis showing the TGF-β1 secretion from induced M2 macrophages after stimulation by VEGF with/without selective VEGFR2 inhibitor (small molecule). *p < 0.05
Discussion
In the present study, our results clearly indicate that the population of M2-TAMs and the VEGFR2 expression on M2-TAMs significantly increased in the tumor compared to the normal mucosa of CRC patients. Furthermore, the results of the in vitro assay indicate that the inhibitory function of M2 macrophages is partially dependent on the VEGF/VEGFR2 signaling pathway.
Although the cell surface markers of M2-TAMs include CD163, CD204, and CD206, we used CD163 to detect M2-TAMs in our previous papers, in which we reported that TAMs may be shifted from M1- to M2-TAMs in the tumor microenvironment of CRC [12, 17]. Etzerodt A et al. indicated in a mouse model that CD163-expressing TAMs play a dominant role in suppressing anti-tumor immunity in melanomas resistant to anti-PD-1 therapy [29]. Furthermore, CD163 expression on TAMs was a particularly strong indicator of poor prognosis in several human cancers [30]. Therefore, we used CD163 to detect M2-TAMs in the present study as well. We found that the population of M2-TAMs significantly increased in the tumor compared to the normal mucosa in CRC patients (Fig. 1b, d); these results were in line with those of our previous studies [12, 17]. Of note, VEGFR2 was expressed on M2-TAMs, and the expression level of VEGFR2 was the highest in the M2-TAMs in the tumor (Fig. 2a). The analysis of TCGA colorectal adenocarcinoma dataset also showed a significant positive correlation between mRNA expression of VEGFR2 and M2-TAMs signature (Fig. 2b). These results suggest that the VEGFR2 expression on M2-TAMs plays an important role in anti-tumor response within the tumor microenvironment, and VEGFR2 on M2-TAMs is targetable by anti-VEGFR2 therapy.
The circumstance around tumor is usually hypoxic condition and our results also showed that the number of HIF-1α+ cells significantly increased in the tumor in comparison to the normal mucosa (Fig. 3b). Laoui et al. indicated that M2-like macrophages accumulate in hypoxic areas in the tumor microenvironment [31]. In line with this study, our results indicate that the number of HIF-1α+ M2-TAMs significantly increased in the tumor in comparison to the normal mucosa (Fig. 3b). Interestingly, although mRNA expression of VEGFR2 was significantly correlated with that of HIF1A (Fig. 3a), there was a significant positive correlation between the number of HIF-1α+ cells and the VEGFR2 expression on M2-TAMs, but not that on M1-TAMs (Fig. 3c). These results suggest that hypoxia may induce VEGFR2 expression on M2-TAMs only. Colegio OR et al. indicated that lactic acid produced by tumor cells induced the M2-like polarization of TAMs, and this effect of lactic acid was mediated by HIF-1α [32]. It is possible that polarized M2-TAMs induced by lactic acid express VEGFR2 in the tumor microenvironment. However, further study is needed to elucidate this mechanism.
It is well known that IL-10 and TGF-β are key immunosuppressive cytokines [33]. TAMs secrete IL-10 as well as TGF-β, which suppress the function of CD8+ T cells and DCs, and stimulate the amplification of Treg cells [16, 34–37]. Additionally, TGF-β is a key driver of metastasis, because TGF-β triggers the EMT; therefore, the inhibition of TGF-β is very important both to enhance the efficacy of immunotherapy and to inhibit metastasis [16, 34]. In the tumor microenvironment, VEGF is produced by tumor cells, endothelial cells, stromal cell, DCs, MDSCs, Treg cells, and M2-TAMs [38]. Furthermore, the present study show that M2 macrophages produced TGF-β1 though the VEGF/VEGFR2 signaling pathway (Fig. 4b). These previous studies and our results suggest that M2-TAMs suppressed the anti-tumor immune response in paracrine and autocrine VEGF signaling through VEGFR2 in the tumor microenvironment of CRC. It is possible that anti-VEGFR2 therapy have a potential to control the inhibitory function of M2-TAMs.
Hyper progressive disease (HPD) is a phenomenon, which is the acceleration of tumor growth in patients treated with ICIs, and its reported frequency ranges between 9 and 29% in patients with different types of cancer [39]. Lo Russo et al. recently suggested that non-small cell lung cancer patients with HPD showed the infiltration of clustered M2-TAMs expressing CD163+CD33+PD-L1+ in the pretreatment tissue samples [39]. In the present study, we also found that M2-TAMs are predominantly accumulated in the tumor microenvironment of CRC. Since our results indicate that VEGFR2 was abundantly expressed on M2-TAMs, the inhibition of VEGF-signaling by anti-VEGFR2 inhibitors or anti-VEGF antibodies in M2-TAMs may help in controlling their inhibitory function for anti-tumor immune response. Recent studies have confirmed that M2-TAMs express PD-1 as well as PD-L1 [39, 40]. It is possible that the accumulation of M2-TAMs may steal the anti-PD-1 mAb from CTL in the tumor microenvironment, resulting in the attenuation of efficacy of anti-PD-1 therapy. Taken together with the results of these previous reports and of the present study suggest that M2-TAMs may be involved in HPD, especially in ICIs targeted for PD-1/PD-L1 axis.
In conclusion, our results indicate that anti-VEGFR2 therapy may have therapeutic potential to control the immune inhibitory functions of M2-TAMs in CRC, resulting in an enhancement of the efficacy of immunotherapy with ICIs.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Abbreviations
- CRC
Colorectal cancer
- EMT
Epithelial-mesenchymal transition
- FFPE
Formalin-fixed paraffin-embedded
- HIF
Hypoxia-inducible factor
- HPD
Hyper progressive disease
- ICIs
Immune checkpoint inhibitors
- KDR
Kinase insert domain receptor
- TAMs
Tumor-associated macrophages
- TCGA
The Cancer Genome Atlas
- VEGFR2
VEGF receptor 2
Author contributions
KM and KK contributed to the study conception and design. HO, WS, SF, HE, MS, ZS, TM, and SO contributed to the acquisition of patient samples. AKTM and SN performed flow cytometry and in vitro assay. AKTM, SN, and KS performed immunofluorescence staining. AKTM, KM, SN, HO, KS, WS, SF, HE, MS, ZS, TM, SO, and KK performed analysis and interpretation of results. AKTM, KM, and KK drafted the manuscript. All the authors are aware of and agree to the contents of the paper, as well as them being listed as authors on the paper.
Funding
No relevant funding.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This study was conducted in accordance with the ethical principles of the 1964 Declaration of Helsinki and its later amendments and was approved by the Fukushima Medical University Research Ethics Committee (Reference Nos. 2289 and 29316).
Informed consent
Written informed consent was obtained from all patients included in the study for the use of their specimens and clinical data for research and publication prior to collecting the specimens at Fukushima Medical University Hospital.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Cercek A, Roxburgh CSD, Strombom P, Smith JJ, Temple LKF, Nash GM, Guillem JG, Paty PB, Yaeger R, Stadler ZK, Seier K, Gonen M, Segal NH, Reidy DL, Varghese A, Shia J, Vakiani E, Wu AJ, Crane CH, Gollub MJ, Garcia-Aguilar J, Saltz LB, Weiser MR. Adoption of total neoadjuvant therapy for locally advanced rectal cancer. JAMA Oncol. 2018 doi: 10.1001/jamaoncol.2018.0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gollins S, Sebag-Montefiore D. Neoadjuvant treatment strategies for locally advanced rectal cancer. Clin Oncol. 2016;28(2):146–151. doi: 10.1016/j.clon.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 4.Overman MJ, Lonardi S, Wong KYM, Lenz HJ, Gelsomino F, Aglietta M, Morse MA, Van Cutsem E, McDermott R, Hill A, Sawyer MB, Hendlisz A, Neyns B, Svrcek M, Moss RA, Ledeine JM, Cao ZA, Kamble S, Kopetz S, Andre T. Durable clinical benefit with nivolumab plus ipilimumab in DNA mismatch repair-deficient/microsatellite instability-high metastatic colorectal cancer. J Clin Oncol. 2018;36(8):773–779. doi: 10.1200/jco.2017.76.9901. [DOI] [PubMed] [Google Scholar]
- 5.Overman MJ, McDermott R, Leach JL, Lonardi S, Lenz HJ, Morse MA, Desai J, Hill A, Axelson M, Moss RA, Goldberg MV, Cao ZA, Ledeine JM, Maglinte GA, Kopetz S, Andre T. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): an open-label, multicentre, phase 2 study. Lancet Oncol. 2017;18(9):1182–1191. doi: 10.1016/S1470-2045(17)30422-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shibuya KC, Goel VK, Xiong W, Sham JG, Pollack SM, Leahy AM, Whiting SH, Yeh MM, Yee C, Riddell SR, Pillarisetty VG. Pancreatic ductal adenocarcinoma contains an effector and regulatory immune cell infiltrate that is altered by multimodal neoadjuvant treatment. PLoS ONE. 2014;9(5):e96565. doi: 10.1371/journal.pone.0096565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Monte L, Reni M, Tassi E, Clavenna D, Papa I, Recalde H, Braga M, Di Carlo V, Doglioni C, Protti MP. Intratumor T helper type 2 cell infiltrate correlates with cancer-associated fibroblast thymic stromal lymphopoietin production and reduced survival in pancreatic cancer. J Exp Med. 2011;208(3):469–478. doi: 10.1084/jem.20101876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qian BZ, Pollard JW. Macrophage diversity enhances tumor progression and metastasis. Cell. 2010;141(1):39–51. doi: 10.1016/j.cell.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Condeelis J, Pollard JW. Macrophages: obligate partners for tumor cell migration, invasion, and metastasis. Cell. 2006;124(2):263–266. doi: 10.1016/j.cell.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 10.Pollard JW. Tumour-educated macrophages promote tumour progression and metastasis. Nat Rev Cancer. 2004;4(1):71–78. doi: 10.1038/nrc1256. [DOI] [PubMed] [Google Scholar]
- 11.Yang L, Zhang Y. Tumor-associated macrophages: from basic research to clinical application. J Hematol Oncol. 2017;10(1):58. doi: 10.1186/s13045-017-0430-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakajima S, Koh V, Kua LF, So J, Davide L, Lim KS, Petersen SH, Yong WP, Shabbir A, Kono K. Accumulation of CD11c+CD163+ adipose tissue macrophages through upregulation of intracellular 11beta-HSD1 in human obesity. J Immunol. 2016;197(9):3735–3745. doi: 10.4049/jimmunol.1600895. [DOI] [PubMed] [Google Scholar]
- 13.Hinshaw DC, Shevde LA. The tumor microenvironment innately modulates cancer progression. Can Res. 2019;79(18):4557–4566. doi: 10.1158/0008-5472.Can-18-3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mantovani A, Sica A. Macrophages, innate immunity and cancer: balance, tolerance, and diversity. Curr Opin Immunol. 2010;22(2):231–237. doi: 10.1016/j.coi.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 15.Biswas SK, Gangi L, Paul S, Schioppa T, Saccani A, Sironi M, Bottazzi B, Doni A, Vincenzo B, Pasqualini F, Vago L, Nebuloni M, Mantovani A, Sica A. A distinct and unique transcriptional program expressed by tumor-associated macrophages (defective NF-kappaB and enhanced IRF-3/STAT1 activation) Blood. 2006;107(5):2112–2122. doi: 10.1182/blood-2005-01-0428. [DOI] [PubMed] [Google Scholar]
- 16.Batlle E, Massague J. Transforming growth factor-beta signaling in immunity and cancer. Immunity. 2019;50(4):924–940. doi: 10.1016/j.immuni.2019.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kikuchi T, Mimura K, Ashizawa M, Okayama H, Endo E, Saito K, Sakamoto W, Fujita S, Endo H, Saito M, Momma T, Saze Z, Ohki S, Shimada K, Yoshimura K, Tsunoda T, Kono K. Characterization of tumor-infiltrating immune cells in relation to microbiota in colorectal cancers. Cancer Immunol Immunother. 2020;69(1):23–32. doi: 10.1007/s00262-019-02433-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wheeler KC, Jena MK, Pradhan BS, Nayak N, Das S, Hsu CD, Wheeler DS, Chen K, Nayak NR. VEGF may contribute to macrophage recruitment and M2 polarization in the decidua. PLoS ONE. 2018;13(1):e0191040. doi: 10.1371/journal.pone.0191040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.Cd-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakayama Y, Mimura K, Tamaki T, Shiraishi K, Kua LF, Koh V, Ohmori M, Kimura A, Inoue S, Okayama H, Suzuki Y, Nakazawa T, Ichikawa D, Kono K. PhosphoSTAT1 expression as a potential biomarker for antiPD1/antiPDL1 immunotherapy for breast cancer. Int J Oncol. 2019;54(6):2030–2038. doi: 10.3892/ijo.2019.4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kikuchi T, Mimura K, Okayama H, Nakayama Y, Saito K, Yamada L, Endo E, Sakamoto W, Fujita S, Endo H, Saito M, Momma T, Saze Z, Ohki S, Kono K. A subset of patients with MSS/MSI-low-colorectal cancer showed increased CD8(+) TILs together with up-regulated IFN-gamma. Oncol Lett. 2019;18(6):5977–5985. doi: 10.3892/ol.2019.10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakayama Y, Mimura K, Kua LF, Okayama H, Min AKT, Saito K, Hanayama H, Watanabe Y, Saito M, Momma T, Saze Z, Ohki S, Suzuki Y, Ichikawa D, Yong WP, Kono K. Immune suppression caused by PD-L2 expression on tumor cells in gastric cancer. Gastric Cancer. 2020 doi: 10.1007/s10120-020-01079-z. [DOI] [PubMed] [Google Scholar]
- 24.Zarif JC, Hernandez JR, Verdone JE, Campbell SP, Drake CG, Pienta KJ. A phased strategy to differentiate human CD14+monocytes into classically and alternatively activated macrophages and dendritic cells. Biotechniques. 2016;61(1):33–41. doi: 10.2144/000114435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Onozawa H, Saito M, Saito K, Kanke Y, Watanabe Y, Hayase S, Sakamoto W, Ishigame T, Momma T, Ohki S, Takenoshita S. Annexin A1 is involved in resistance to 5-FU in colon cancer cells. Oncol Rep. 2017;37(1):235–240. doi: 10.3892/or.2016.5234. [DOI] [PubMed] [Google Scholar]
- 26.Okano M, Kumamoto K, Saito M, Onozawa H, Saito K, Abe N, Ohtake T, Takenoshita S. Upregulated Annexin A1 promotes cellular invasion in triple-negative breast cancer. Oncol Rep. 2015;33(3):1064–1070. doi: 10.3892/or.2015.3720. [DOI] [PubMed] [Google Scholar]
- 27.Thar Min AK, Okayama H, Saito M, Ashizawa M, Aoto K, Nakajima T, Saito K, Hayase S, Sakamoto W, Tada T, Hanayama H, Saze Z, Momma T, Ohki S, Sato Y, Motoyama S, Mimura K, Kono K. Epithelial-mesenchymal transition-converted tumor cells can induce T-cell apoptosis through upregulation of programmed death ligand 1 expression in esophageal squamous cell carcinoma. Cancer Med. 2018 doi: 10.1002/cam4.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ashizawa M, Okayama H, Ishigame T, Thar Min AK, Saito K, Ujiie D, Murakami Y, Kikuchi T, Nakayama Y, Noda M, Tada T, Endo H, Fujita S, Sakamoto W, Saito M, Saze Z, Momma T, Ohki S, Mimura K, Kono K. miRNA-148a-3p regulates immunosuppression in DNA mismatch repair-deficient colorectal cancer by targeting PD-L1. Mol Cancer Res. 2019;17(6):1403–1413. doi: 10.1158/1541-7786.Mcr-18-0831. [DOI] [PubMed] [Google Scholar]
- 29.Etzerodt A, Tsalkitzi K, Maniecki M, Damsky W, Delfini M, Baudoin E, Moulin M, Bosenberg M, Graversen JH, Auphan-Anezin N, Moestrup SK, Lawrence T. Specific targeting of CD163(+) TAMs mobilizes inflammatory monocytes and promotes T cell-mediated tumor regression. J Exp Med. 2019;216(10):2394–2411. doi: 10.1084/jem.20182124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Komohara Y, Jinushi M, Takeya M. Clinical significance of macrophage heterogeneity in human malignant tumors. Cancer Sci. 2014;105(1):1–8. doi: 10.1111/cas.12314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laoui D, Van Overmeire E, Di Conza G, Aldeni C, Keirsse J, Morias Y, Movahedi K, Houbracken I, Schouppe E, Elkrim Y, Karroum O, Jordan B, Carmeliet P, Gysemans C, De Baetselier P, Mazzone M, Van Ginderachter JA. Tumor hypoxia does not drive differentiation of tumor-associated macrophages but rather fine-tunes the M2-like macrophage population. Can Res. 2014;74(1):24–30. doi: 10.1158/0008-5472.Can-13-1196. [DOI] [PubMed] [Google Scholar]
- 32.Colegio OR, Chu NQ, Szabo AL, Chu T, Rhebergen AM, Jairam V, Cyrus N, Brokowski CE, Eisenbarth SC, Phillips GM, Cline GW, Phillips AJ, Medzhitov R. Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature. 2014;513(7519):559–563. doi: 10.1038/nature13490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mantovani A, Marchesi F, Malesci A, Laghi L, Allavena P. Tumour-associated macrophages as treatment targets in oncology. Nat Rev Clin Oncol. 2017;14(7):399–416. doi: 10.1038/nrclinonc.2016.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Massague J. TGFbeta in cancer. Cell. 2008;134(2):215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wrzesinski SH, Wan YY, Flavell RA. Transforming growth factor-beta and the immune response: implications for anticancer therapy. Clin Cancer Res. 2007;13(18 Pt 1):5262–5270. doi: 10.1158/1078-0432.Ccr-07-1157. [DOI] [PubMed] [Google Scholar]
- 36.Flavell RA, Sanjabi S, Wrzesinski SH, Licona-Limon P. The polarization of immune cells in the tumour environment by TGFbeta. Nat Rev Immunol. 2010;10(8):554–567. doi: 10.1038/nri2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moore KW, de Waal MR, Coffman RL, O'Garra A. Interleukin-10 and the interleukin-10 receptor. Annu Rev Immunol. 2001;19:683–765. doi: 10.1146/annurev.immunol.19.1.683. [DOI] [PubMed] [Google Scholar]
- 38.Fukumura D, Kloepper J, Amoozgar Z, Duda DG, Jain RK. Enhancing cancer immunotherapy using antiangiogenics: opportunities and challenges. Nat Rev Clin Oncol. 2018;15(5):325–340. doi: 10.1038/nrclinonc.2018.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lo Russo G, Moro M, Sommariva M, Cancila V, Boeri M, Centonze G, Ferro S, Ganzinelli M, Gasparini P, Huber V, Milione M, Porcu L, Proto C, Pruneri G, Signorelli D, Sangaletti S, Sfondrini L, Storti C, Tassi E, Bardelli A, Marsoni S, Torri V, Tripodo C, Colombo MP, Anichini A, Rivoltini L, Balsari A, Sozzi G, Garassino MC. Antibody-Fc/FcR interaction on macrophages as a mechanism for hyperprogressive disease in non-small cell lung cancer subsequent to PD-1/PD-L1 blockade. Clin Cancer Res. 2019;25(3):989–999. doi: 10.1158/1078-0432.Ccr-18-1390. [DOI] [PubMed] [Google Scholar]
- 40.Gordon SR, Maute RL, Dulken BW, Hutter G, George BM, McCracken MN, Gupta R, Tsai JM, Sinha R, Corey D, Ring AM, Connolly AJ, Weissman IL. PD-1 expression by tumour-associated macrophages inhibits phagocytosis and tumour immunity. Nature. 2017;545(7655):495–499. doi: 10.1038/nature22396. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.