Abstract
We have used the yeast three-hybrid system (D. J. SenGupta, B. Zhang, B. Kraemer, P. Pochart, S. Fields, and M. Wickens, Proc. Natl. Acad. Sci. USA 93:8496–8501, 1996) to study binding of the human immunodeficiency virus type 1 (HIV-1) Gag protein to the HIV-1 RNA encapsidation signal (HIVΨ). Interaction of these elements results in the activation of a reporter gene in the yeast Saccharomyces cerevisiae. Using this system, we have shown that the HIV-1 Gag protein binds specifically to a 139-nucleotide fragment of the HIVΨ signal containing four stem-loop structures. Mutations in either the Gag protein or the encapsidation signal that have been shown previously to impair this interaction reduced the activation of the reporter gene. Interestingly, the nucleocapsid portion of Gag retained the RNA binding activity but lost its specificity compared to the full-length Gag. These results demonstrate the utility of this system and suggest that a variety of genetic analyses could be performed to study Gag-encapsidation signal interactions.
The retrovirus genomic RNA typically accounts for less than 1% of the total cytoplasmic mRNA, whereas it is the major RNA species found in the virion particle. This enrichment is thought to be mediated by the specific binding and encapsidation of the genomic RNA by the Gag precursor polyprotein (reviewed in reference 7). For the human immunodeficiency virus type 1 (HIV-1) RNA, the major encapsidation signal (Ψ) lies downstream of the primer binding site and extends into the 5′ portion of the gag gene. Computerized sequence analysis, chemical and RNase accessibility mapping, and mutational analyses have identified several stem-loop structures within the encapsidation region (5, 6, 14, 27, 28, 32, 35, 37, 44, 47). Four adjacent stem-loops are thought to mediate the binding to the HIV-1 Gag protein and contribute to the encapsidation of the viral RNA into the virion (8, 14, 15, 38, 39). The major protein component of the interaction is the nucleocapsid (NC) region of Gag, a basic domain containing two copies of a motif known as a Cys-His box (consensus sequence Cys-X2-Cys-X4-His-X4-Cys) or a zinc knuckle because the four conserved residues coordinate a zinc ion. Mutational analysis of the HIV-1 Cys-His boxes has demonstrated their importance in the binding and encapsidation of the HIV-1 RNA (2, 11, 18, 25, 26, 43, 45, 54, 55). Recently, the three-dimensional structure of an HIV-1 NC bound to RNA stem-loop 3 (SL3) was determined (17).
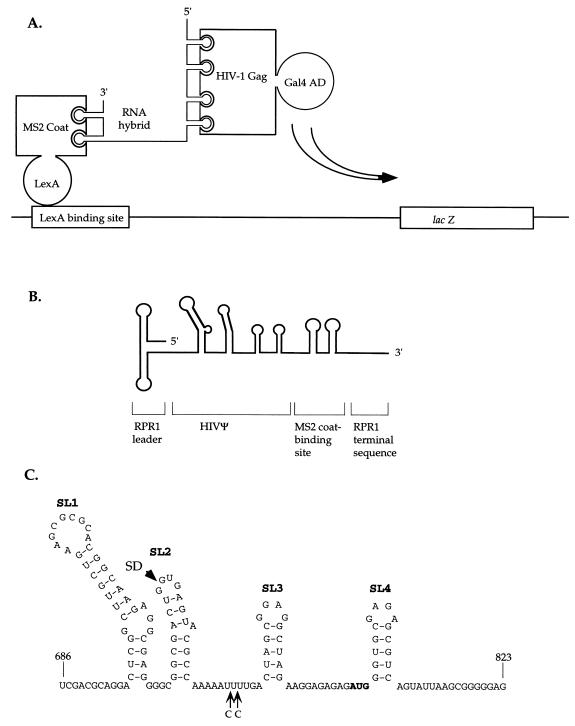
Here we report the use of the yeast three-hybrid system to detect and analyze the interaction between the HIV-1 Gag protein and the Ψ RNA. In this system, a hybrid RNA molecule bridges two fusion proteins, one containing a DNA-binding domain and the other containing a transcriptional activation domain, resulting in the transcriptional activation of a lacZ reporter downstream of the binding site for the DNA-binding domain (46). To apply the system to HIV-1, we designed constructs fusing the Gal4 activation domain (Gal4AD) to the HIV Gag as one protein partner and made use of a preexisting construct fusing LexA (containing a DNA-binding domain) to the phage MS2 coat protein as the other partner (46). To bridge the proteins, we prepared a plasmid encoding a fusion RNA with both the HIV-1 RNA encapsidation signal (HIVΨ) and the MS2 RNA-binding sites (Fig. 1A and B).
FIG. 1.
(A) General scheme of the three-hybrid system used in this study, adapted from reference 46. Binding of the RNA hybrid to the LexA-MS2 coat protein and to the Gal4AD-HIV Gag protein leads to formation of a complex which activates transcription of the lacZ gene. The yeast strain, L40-coat, used in this study constitutively expresses the LexA-MS2 coat fusion protein from an integrated gene (46). (B) Structure of the RNA hybrid. Shown from 5′ to 3′ are the stem-loop RPR1 leader (84 nt), linker (30 nt), the four stem-loops of the HIVΨ (139 nt), linker (23 nt), two stem-loop structures that bind the MS2-coat protein (60 nt), and the RPR1 3′ terminal sequence (41 nt). The structures have not been drawn to scale. (C) HIVΨ RNA sequence, adapted from reference 14. The alternative positions of the cytosine insertion are marked with arrows. The location of the HIV-1 splicing donor (SD) is marked with an arrowhead, and the translation start codon appears in bold letters. Each of the stem-loops is marked by a number (SL1 to SL4).
To prepare the Gal4AD-Gag fusion, the full-length HIV-1 gag sequence (1.7 kb), derived from the infectious molecular clone HXBC2 (19), was cloned into the expression vector pGADNOT (33). The resulting plasmid, pGADZX2, encodes a fusion protein with an N-terminal GAL4AD and a C-terminal HIV-1 Gag polyprotein (Gal4AD-HIV Gag). The LexA-MS2 coat protein fusion (46) was constitutively expressed from an integrated construct resident in Saccharomyces cerevisiae L40-coat. The RNA fusion was prepared by using the yeast RNA expression plasmid pIIIA/MS2-2 (53), containing an RNA polymerase III promoter and terminator from the S. cerevisiae RNase P RNA gene (RPR1) (24, 46). The encoded RNA is composed of the 5′ stem-loop leader sequence and the 3′ end of RPR1, separated by two stem-loop structures that bind the MS2 coat protein (4, 46). PCR was used to amplify a 138-nucleotide (nt) fragment of the HIVΨ (nt 686 to 823 [14] [Fig. 1C]) from plasmid pNLENV-1 (a derivative of pNL4-3 [1, 36]) and to introduce SmaI and SphI sites at the 5′ and the 3′ ends, respectively. The HIVΨ sequence contains a stretch of four consecutive thymidine residues which might serve as a termination signal for RNA polymerase III, preventing formation of the complete RNA (12, 22). To avoid this problem, PCR was used to change the sequence TTTT to either TCTTT or TTCTT (Fig. 1C) (the latter sequence is present in the HIV-1 MN isolate [14]). In subsequent tests, similar results were obtained with or without the inserted cytosines. These HIVΨ sequences were inserted between the SmaI and SphI sites of pIIIA/MS2-2. The resultant chimeric RNA, named HIVΨ-MS2, contains the HIVΨ sequences near the 5′ end and the MS2 sequences near the 3′ end (Fig. 1B).
To test for the ability of the RNA to successfully bridge the two protein fusions, we coexpressed different combinations of the components of this system in yeast strain L40-coat (Table 1). Expression of the Gal4AD-HIV Gag fusion protein together with the HIVΨ-MS2 hybrid RNA resulted in a strong activation of the lacZ reporter, as judged by 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) staining of the transformed colonies. Inversion of the Ψ sequence (HIVΨinv-MS2) abolished the lacZ activation. Yeast cotransformed with the HIVΨ-MS2 hybrid and Gal4AD showed no detectable lacZ expression, indicating that the Gal4AD by itself did not interact with either the HIVΨ hybrid RNA or the LexA-MS2 coat protein. When the Gal4AD-HIV Gag was coexpressed with an MS2 RNA lacking the HIVΨ sequence, only very low levels of β-galactosidase were detectable in some of the transformants. This low level of gene activation may be a result of the nonspecific RNA binding activity of retrovirus Gag proteins (reviewed in reference 7). However, the level of β-galactosidase activity seen with the HIVΨ-MS2 chimeric RNA was much higher than the level with the nonspecific RNA (a strong blue color that appeared in less then an hour versus a faint blue color after 6 h of incubation), allowing a clear distinction between specific and nonspecific interactions in this assay.
TABLE 1.
Transcriptional activation of lacZ by the interaction of Gag protein and Ψ RNA
| RNA hybrida | Fusion proteina | Activationb |
|---|---|---|
| HIVΨ-MS2 | Gal4AD-HIV Gag | +++ |
| HIVΨ-MS2 | Gal4AD | − |
| HIVΨinv-MS2 | Gal4AD-HIV Gag | − |
| MS2 | Gal4AD-HIV Gag | −/+ |
| MoMLVΨ-MS2 | Gal4AD-HIV Gag | −/+ |
| HaMSVΨ-MS2 | Gal4AD-HIV Gag | −/+ |
| IRE-MS2 | Gal4AD-IRP1 | +++ |
| IRE-MS2 | Gal4AD-HIV Gag | −/+ |
S. cerevisiae L40-coat, stably expressing the LexA-MS2 coat fusion protein, was transformed (23) with different combinations of plasmids encoding the indicated RNA hybrid and fusion protein. The transformants were selected for uracil and leucine prototrophy for 3 days.
Colonies from the above selection were replica plated onto nitrocellulose filters, frozen at −80°C, thawed, soaked in buffer containing X-Gal, incubated at 30°C, and assayed for the appearance of a blue color (13). +++, color seen in about 15 min, turning to strong blue in about 1 h. Quantitative β-galactosidase assay yielded 420 ± 100 U for the HIVΨ-MS2/Gal4AD-HIV Gag pair. −/+, color seen in about 6 h, remaining faint blue even after overnight incubation. Quantitative β-galactosidase assay of Gal4AD-HIV Gag in combination with either HaMSVΨ-MS2 or IRE-MS2 yielded 14 ± 3 and 7 ± 1 U of β-galactosidase, respectively. −, no detectable color observed after overnight incubation.
Additional control experiments were performed by analyzing the interactions between HIV-1 Gag protein and a stem-loop RNA containing the iron response element (IRE). When this element was fused to the MS2 coat-binding sequence and coexpressed with iron regulatory protein 1 fused to the Gal4AD (Gal4AD-IRP1), strong lacZ activation occurred (46) (Table 1). However, only very low activation occurred when the IRE-MS2 RNA was coexpressed with HIV Gag, demonstrating specificity of the Gag interaction with HIVΨ.
To further evaluate the specificity of the HIV Gag protein-Ψ signal interaction, we tested the encapsidation signals from two additional retroviruses, Moloney murine leukemia virus (MoMLV) and Harvey murine sarcoma virus (HaMSV). The MoMLVΨ region (positions 303 to 386 according to the MoMLV genome map [50]), amplified by PCR from plasmid pNCA (16), contains the two stem-loop structures thought to be important for packaging (20, 31, 41, 42). The HaMSVΨ region (positions 205 to 380 [49]), amplified by PCR from pHAMDR1/A (40), is sufficient to mediate encapsidation (49). Flanking SmaI (MoMLVΨ) or 5′ SmaI and 3′ SphI (HaMSVΨ) sites were also introduced to enable the cloning of the amplified sequences into plasmid pIIIA/MS2-2.
In yeast containing the murine virus RNAs and the HIV Gag fusions, the HIV Gag protein interacted very weakly with both the MoMLVΨ and the HaMSVΨ (Table 1), similar to the nonspecific interaction with the MS2-binding-site RNA alone. These results demonstrate that a specific binding of the HIV Gag protein to the HIVΨ sequence can be achieved in the three-hybrid system. Coexpression of a Gal4AD-MoMLV Gag fusion protein with either the MoMLVΨ or the HaMSVΨ failed to activate lacZ significantly (data not shown). The interaction of the MoMLV Gag with its encapsidation signal could not be detected in RNA gel mobility shift assays (8a). The reason for the poor RNA binding activity of the MoMLV Gag in these systems is not known.
To provide an additional means to assay reporter gene activation, we performed a quantitative β-galactosidase liquid assay (3) in triplicate on selected pairs of constructs in S. cerevisiae. Yeast cells in liquid cultures were collected by centrifugation and disrupted by using glass beads, and the β-galactosidase activity and total protein concentration of the lysates were determined. The extracts were mixed in buffer containing o-nitrophenyl-β-d-galactoside and incubated at 30°C, and the appearance of a yellow color was determined spectrophotometrically by measuring the optical density at a wavelength of 420 nm (OD420). The β-galactosidase units were calculated from the equation U = (1,000 × OD420)/(t × mg), where t is the reaction time in minutes and mg is the lysate total protein in milligrams that was added to the reaction. As shown in Table 1, β-galactosidase activity was about 40 times higher in yeast colonies that developed a dark blue color in 1 h (the HIVΨ-MS2/Gal4AD-HIV Gag pair) than in control yeast colonies that developed a faint blue color in 6 h (HaMSVΨ-MS2/Gal4AD-HIV Gag or IRE-MS2/Gal4AD-HIV Gag pairs) in the filter assay. These assays demonstrate a good correlation between the quantitative β-galactosidase liquid assay and the filter assay.
If the interaction of HIV Gag with HIVΨ reflects binding in vivo, the response should be affected by mutations in both Gag and Ψ. Previous studies suggested that the NC portion of the Gag polyprotein is sufficient for binding in vitro (reviewed in reference 7). We tested whether Gag protein lacking the NC domain can bind HIVΨ in the yeast assay. Plasmid Gal4AD-HIV MA-CA encodes the matrix (MA) and capsid (CA) portions of the HIV-1 Gag protein and lacks the p2-NC-p1-p6 C-terminal region. This construct was created by PCR amplification of the MA-CA sequence from pGADZX2, using the primers 5′GCGCGGGATCCTGGGTGCGAGAGCG3′ (in which the ATG start codon was changed to CTG) and 5′GCGCCGTCGACAACTCTTGCCTTATGG3′. The amplified fragment was cloned into pGADNOT, using the BamHI and SalI sites, downstream of the GAL4AD-encoding sequence and in the same reading frame. Yeast expressing the Gal4AD-HIV MA-CA fusion protein and the HIVΨ-MS2 hybrid showed no detectable lacZ expression (Table 2). Thus, the interaction detected in yeast required the NC domain, as expected.
TABLE 2.
Effects of mutations in Gag or in the Ψ on interaction in the three-hybrid system
| RNA hybrida | Fusion proteina | Activationb |
|---|---|---|
| HIVΨ-MS2 | Gal4AD-HIV Gag | +++ |
| HIVΨ-MS2 | Gal4AD-HIV MA-CA | − |
| HIVΨ-MS2 | Gal4AD-HIV Gag F16A | + |
| HIVΨΔSL1,3,4-MS2 | Gal4AD-HIV Gag | ++ |
| HIVΨΔSL3,4-MS2 | Gal4AD-HIV Gag | +++ |
| HIVΨΔSL1-MS2 | Gal4AD-HIV Gag | +++ |
As in Table 1. HIVΨΔSLx, HIV-1 encapsidation signal lacking SLx, where x denotes the stem-loop number(s) as depicted in Fig. 1C. The sequence of each junction created by each deletion is as follows: ΔSL1 (deletion of nt 697 to 730 [14]), AGGAGGGG; ΔSL3 (deletion of nt 766 to 779), TTGAAAGG; ΔSL4 (deletion of nt 793 to 806), GATGAGTA. The bold letters denote the bases flanking the newly generated junctions.
As in Table 1. +++, color seen in about 15 min, turning to strong blue in about 1 h. Quantitative β-galactosidase assay yielded 420 ± 100 U for the HIVΨ-MS2/Gal4AD-HIV Gag pair. ++, color seen in about 15 min, turning blue in about 1 h. +, color seen in about 3 h, remaining fainter than the wild-type Gag after an additional 4 h of incubation. Quantitative β-galactosidase assay yielded 275 ± 100 U for both the HIVΨ-MS2/Gal4AD-HIV Gag F16A and HIVΨΔSL1,3,4-MS2/Gal4AD-HIV Gag pairs. −, no detectable color observed after overnight incubation.
To explore the sensitivity of the system to alterations in Gag in more detail, a point mutation affecting the NC was examined. A single change of phenylalanine to alanine (F16A) in the amino-terminal Cys-His box in the NC portion of the HIV-1 Gag was shown to significantly impair the binding of Gag to its RNA packaging signal (18). Northwestern analysis revealed that this mutation reduced the binding in vitro to about 20% of that of the wild-type protein and reduced the viral RNA content in virions to about 15% of that in the wild-type virions (18). The gag sequences were excised from pGADZX2 and cloned into pUC19, using BamHI-SalI sites. An internal 0.5-kb ApaI-SpeI fragment was replaced with the same fragment derived from plasmid pT7Gag 3D, containing the isogenic HIV Gag with the F16A mutation. The mutated gag sequence was then exchanged for the wild-type gag in pGADZX2, using the BamHI-SalI sites, to form pGADZX2/F16A. The F16A change creates a new HhaI site, which was used to confirm the presence of this mutation.
As shown in Table 2, this mutation dramatically reduced the activation of the lacZ reporter in the filter assay. Yeast colonies expressing the wild-type Gag turned dark blue in about an hour, whereas yeast colonies expressing the mutated Gag stained only faintly blue after 3 h of incubation. The intensity of the staining increased over another 4 h of incubation but never reached the intensity of the colonies expressing the wild-type allele. This result demonstrates a good correlation between the ability of the mutated Gag to bind RNA in vitro and to activate the lacZ gene in the three-hybrid system.
To test for the regions of the viral RNA required for interaction with Gag in the three-hybrid system, several variants of the Ψ region were examined. Previous analyses revealed that stem-loops SL1, SL3, and SL4 in the HIVΨ each contribute to encapsidation (7, 15, 38). The ability of Gag to bind single versus multiple stem-loops is dependent on the assay used. RNA gel mobility shift assays (8) revealed that SL2, SL3, and SL4 each binds Gag poorly compared to the binding of two or three of these stem-loops in tandem. In contrast, filter binding assays showed that SL3, SL4, and SL1 each binds independently with about the same affinity as the four stem-loops together (Kd of ∼200 nM for SL1, SL3, or SL4; Kd of ∼400 nM for SL2 [14]). In another study, the affinity of the NC to SL3 was estimated to have a Kd of ∼100 nM (17). The differences in these various assays can be explained by the different assay conditions, the use of different proteins (full-length Gag versus NC portion), and the different flanking sequences of the tested RNA. All of these experiments, however, suggest that each of the four stem-loop structures has at least substantial ability to bind the HIV Gag and that there is considerable redundancy in the RNA for binding the protein.
We deleted various combination of the stem-loops from HIVΨ by overlapping PCR using mutated oligonucleotides (29). All mutant clones were sequenced to verify the existence of the desired mutations (Table 2). Each construct was used along with the wild-type Gal4AD-HIV Gag to cotransform yeast strain L40-coat, and the resulting colonies were stained for β-galactosidase activity. The HIVΨ construct lacking SL1, SL3, and SL4 but retaining SL2 and the intervening sequences (HIVΨΔSL1,3,4-MS2) gave a considerably weaker signal than that obtained with the intact Ψ region (Table 2). HIVΨ constructs lacking SL3 and SL4 (HIVΨΔSL3,4-MS2) or lacking SL1 (HIVΨΔSL1-MS2) showed signals equal to that of the wild type (Table 2). These results are in good agreement with the above studies in which only weak binding occurred with one stem-loop, and relatively strong binding to the Gag protein occurred when two or three stem-loops were present.
In the quantitative β-galactosidase liquid assay, the HIVΨΔSL1,3,4-MS2 mutation in the RNA and the F16A mutation in the Gag protein each reduced β-galactosidase activity 30 to 40% compared to the wild-type sequences (Table 2). Although this is a relatively low reduction, the result further demonstrates the correlation between the ability of Gag protein to bind the Ψ sequence and the level of the β-galactosidase activation. For both mutations, the filter assay served as a better tool than the liquid assay to distinguish them from the wild-type sequences. Furthermore, it demonstrates the ability to screen quickly and easily on plate lifts for mutations that affect the Gag-Ψ interaction.
The NC proteins exhibit both nonspecific (7, 21, 30, 48, 52) and sequence-specific RNA binding activities. In the context of the Gag polyprotein, the NC portion mediates the specific recognition of the encapsidation signal. However, after it has been cleaved from the Gag polyprotein, the NC can bind to and cover the RNA nonspecifically at a density of one NC molecule per six to seven nucleotides (7, 30, 52). Thus, the NC molecule detached from the Gag polyprotein may lose at least some of its specific RNA binding activity. We compared the RNA binding activity of the Gal4AD-Gag fusion to that of a GAL4AD-NC fusion. To prepare the Gal4AD-NC fusion, PCR was used (with oligonucleotides 5′GGGGATCCGAATACAGAAAGGCAATTTTAGG3′ and 5′TCTCTCGAGCTCTAATTAGCCTGTCTCTCAGTAC3′) to amplify the NC coding sequence from the infectious molecular clone NL4-3, to introduce a stop codon at the 3′ end, and to introduce BamHI and XhoI sites at the 5′ and 3′ ends, respectively. The amplified fragment was cloned into the BamHI and XhoI sites of pACT2 (Clontech). The resulting plasmid, pGAL4AD-HIV NC, encodes a fusion protein with an N-terminal GAL4AD and a C-terminal HIV-1 NC protein (GAL4AD-HIV NC).
Expression of the GAL4AD-HIV NC fusion protein together with the HIVΨ-MS2 hybrid RNA resulted in a strong activation of the lacZ reporter, similar to the activation by the Gal4AD-HIV Gag (Table 3). However, in contrast to the GAL4AD-HIV Gag, expression of the GAL4AD-HIV NC together with HaMSVΨ-MS2 or IRE-MS2 RNAs resulted in an equally strong lacZ expression (compare Table 1 to Table 3). This activation is RNA dependent, as yeast transformed with pGAL4AD-HIV NC but without the plasmid that encodes an RNA hybrid did not activate the reporter gene (Table 3). These results indicate that while the GAL4AD-HIV NC retained its RNA binding activity, it lost the specificity of the parental Gal4AD-HIV Gag. Thus, the three-hybrid system may reflect the in vivo differences in RNA binding specificity better than the in vitro gel shift assay, in which either full-length Gag or NC, fused to glutathione S-transferase, showed roughly the same RNA binding specificity (10).
TABLE 3.
Transcriptional activation of lacZ by the interaction of NC protein and Ψ RNA
| RNA hybrida | Fusion proteina | Activationb |
|---|---|---|
| HIVΨ-MS2 | Gal4AD-HIV Gag | +++ |
| HIVΨ-MS2 | Gal4AD-HIV NC | +++ |
| HaMSVΨ-MS2 | Gal4AD-HIV NC | +++ |
| IRE-MS2 | Gal4AD-HIV NC | +++ |
| —c | Gal4AD-HIV NC | − |
a,b As in Table 1, except that the transformants were selected for uracil and leucine prototrophy for 4 days due to lower growth rate. Yeast colonies transformed only with pGal4AD-HIV NC were selected for leucine prototrophy.
—, no plasmid encoding an RNA hybrid was introduced.
By the criteria available, the yeast three-hybrid system provides a measure of the interaction of HIV-1 Gag with its cognate RNA encapsidation signal that reflects the interactions seen in vitro and in vivo. The main advantage of the genetic system described here over traditional biochemical approaches is the ability to screen a large number of combinations of RNA or protein for the ability to interact. Mutant Gags or RNAs with lower or higher binding affinities could be isolated from randomly mutated libraries, using the β-galactosidase filter assay. In our hands, this assay was sensitive enough for detecting these kind of mutations. In principle, this strategy would allow the isolation of compensatory mutations in both the encapsidation signal and the Gag protein. In addition, this system could be useful for screening drugs, or libraries of random peptides or RNAs, for molecules that interfere with the authentic formation of the Gag-RNA complex. The yeast system offers the possibility of screening a library made from random fragments of the HIV-1 genome for other RNA sequences that bind to the HIV Gag protein (9, 10, 34, 39). Finally, the three-hybrid system could be useful for identifying new protein players in the process. The system was used recently to isolate cellular proteins that bind specifically to defined RNA sequences (51, 53). If portions of the HIVΨ sequence also serve as a binding target for cellular proteins, these proteins may be isolated by screening cDNA libraries with the HIVΨ sequence.
Acknowledgments
All of the three-hybrid components were generously provided by Marvin Wickens. We thank Arthur Bank for plasmid pHAMDR1/A, Bing Yuan for plasmid pGal4AD-HIV MA-CA, Gilda Tachedjian for plasmid pGAL4AD-HIV NC, and Jeremy Luban for plasmids pGADZX2 and pT7Gag 3D. We thank Jason Gonsky, Guangxia Gao, Marion Dorsch, Sergei Kuchin, and Marian Carlson for helpful discussions, and we thank Sharon Boast and Kenia de los Santos for technical assistance.
E.B. is an Associate and S.P.G. is an Investigator of the HHMI.
REFERENCES
- 1.Adachi A, Gendelman H E, Koenig S, Folks T, Willey R, Rabson A, Martin M A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aldovini A, Young R A. Mutations of RNA and protein sequences involved in human immunodeficiency virus type 1 packaging result in production of noninfectious virus. J Virol. 1990;64:1920–1926. doi: 10.1128/jvi.64.5.1920-1926.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1995. [Google Scholar]
- 4.Bardwell V J, Wickens M. Purification of RNA and RNA-protein complexes by an R17 coat protein affinity method. Nucleic Acids Res. 1990;18:6587–6594. doi: 10.1093/nar/18.22.6587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baudin F, Marquet R, Isel C, Darlix J L, Ehresmann B, Ehresmann C. Functional sites in the 5′ region of human immunodeficiency virus type 1 RNA form defined structural domains. J Mol Biol. 1993;229:382–397. doi: 10.1006/jmbi.1993.1041. [DOI] [PubMed] [Google Scholar]
- 6.Berglund J A, Charpentier B, Rosbash M. A high affinity binding site for the HIV-1 nucleocapsid protein. Nucleic Acids Res. 1997;25:1042–1049. doi: 10.1093/nar/25.5.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berkowitz R, Fisher J, Goff S P. RNA packaging. Curr Top Microbiol Immunol. 1996;214:177–218. doi: 10.1007/978-3-642-80145-7_6. [DOI] [PubMed] [Google Scholar]
- 8.Berkowitz R D, Goff S P. Analysis of binding elements in the human immunodeficiency virus type 1 genomic RNA and nucleocapsid protein. Virology. 1994;202:233–246. doi: 10.1006/viro.1994.1339. [DOI] [PubMed] [Google Scholar]
- 8a.Berkowitz, R. D., and S. P. Goff. Unpublished data.
- 9.Berkowitz R D, Hammarskjold M L, Helga M C, Rekosh D, Goff S P. 5′ regions of HIV-1 RNAs are not sufficient for encapsidation: implications for the HIV-1 packaging signal. Virology. 1995;212:718–723. doi: 10.1006/viro.1995.1530. [DOI] [PubMed] [Google Scholar]
- 10.Berkowitz R D, Luban J, Goff S P. Specific binding of human immunodeficiency virus type 1 Gag polyprotein and nucleocapsid protein to viral RNAs detected by RNA mobility shift assays. J Virol. 1993;67:7190–7200. doi: 10.1128/jvi.67.12.7190-7200.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berkowitz R D, Ohagen A, Hoglund S, Goff S P. Retroviral nucleocapsid domains mediate the specific recognition of genomic viral RNAs by chimeric Gag polyproteins during RNA packaging in vivo. J Virol. 1995;69:6445–6456. doi: 10.1128/jvi.69.10.6445-6456.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bogenhagen D F, Brown D D. Nucleotide sequences in Xenopus 5S DNA required for transcription termination. Cell. 1981;24:261–270. doi: 10.1016/0092-8674(81)90522-5. [DOI] [PubMed] [Google Scholar]
- 13.Breeden L, Nasmyth K. Regulation of the yeast HO gene. Cold Spring Harbor Symp Quant Biol. 1985;50:643–650. doi: 10.1101/sqb.1985.050.01.078. [DOI] [PubMed] [Google Scholar]
- 14.Clever J, Sassetti C, Parslow T G. RNA secondary structure and binding sites for gag gene products in the 5′ packaging signal of human immunodeficiency virus type 1. J Virol. 1995;69:2101–2109. doi: 10.1128/jvi.69.4.2101-2109.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clever J L, Parslow T G. Mutant human immunodeficiency virus type 1 genomes with defects in RNA dimerization or encapsidation. J Virol. 1997;71:3407–3414. doi: 10.1128/jvi.71.5.3407-3414.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colicelli J, Goff S P. Sequence and spacing requirements of a retrovirus integration site. J Mol Biol. 1988;199:47–59. doi: 10.1016/0022-2836(88)90378-6. [DOI] [PubMed] [Google Scholar]
- 17.De G R, Wu Z R, Stalling C C, Pappalardo L, Borer P N, Summers M F. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science. 1998;279:384–388. doi: 10.1126/science.279.5349.384. [DOI] [PubMed] [Google Scholar]
- 18.Dorfman T, Luban J, Goff S P, Haseltine W A, Gottlinger H G. Mapping of functionally important residues of a cysteine-histidine box in the human immunodeficiency virus type 1 nucleocapsid protein. J Virol. 1993;67:6159–6169. doi: 10.1128/jvi.67.10.6159-6169.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fisher A G, Collalti E, Ratner L, Gallo R C, Wong S F. A molecular clone of HTLV-III with biological activity. Nature. 1985;316:262–265. doi: 10.1038/316262a0. [DOI] [PubMed] [Google Scholar]
- 20.Fisher J, Goff S P. Mutational analysis of stem-loops in the RNA packaging signal. Virology. 1998;244:133–145. doi: 10.1006/viro.1998.9090. [DOI] [PubMed] [Google Scholar]
- 21.Fisher R J, Rein A, Fivash M, Urbaneja M A, Casas F J, Medaglia M, Henderson L E. Sequence-specific binding of human immunodeficiency virus type 1 nucleocapsid protein to short oligonucleotides. J Virol. 1998;72:1902–1909. doi: 10.1128/jvi.72.3.1902-1909.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geiduschek E P, Tocchini V G. Transcription by RNA polymerase III. Annu Rev Biochem. 1988;57:873–914. doi: 10.1146/annurev.bi.57.070188.004301. [DOI] [PubMed] [Google Scholar]
- 23.Gietz R D, Schiestl R H. Transforming yeast with DNA. Methods Mol Cell Biol. 1995;5:255–269. [Google Scholar]
- 24.Good P D, Engelke D R. Yeast expression vectors using RNA polymerase III promoters. Gene. 1994;151:209–214. doi: 10.1016/0378-1119(94)90658-0. [DOI] [PubMed] [Google Scholar]
- 25.Gorelick R J, Chabot D J, Rein A, Henderson L E, Arthur L O. The two zinc fingers in the human immunodeficiency virus type 1 nucleocapsid protein are not functionally equivalent. J Virol. 1993;67:4027–4036. doi: 10.1128/jvi.67.7.4027-4036.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gorelick R J, Nigida S J, Bess J J, Arthur L O, Henderson L E, Rein A. Noninfectious human immunodeficiency virus type 1 mutants deficient in genomic RNA. J Virol. 1990;64:3207–3211. doi: 10.1128/jvi.64.7.3207-3211.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harrison G P, Lever A M. The human immunodeficiency virus type 1 packaging signal and major splice donor region have a conserved stable secondary structure. J Virol. 1992;66:4144–4153. doi: 10.1128/jvi.66.7.4144-4153.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hayashi T, Shioda T, Iwakura Y, Shibuta H. RNA packaging signal of human immunodeficiency virus type 1. Virology. 1992;188:590–599. doi: 10.1016/0042-6822(92)90513-o. [DOI] [PubMed] [Google Scholar]
- 29.Higuchi R. Recombinant PCR. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols. A guide to methods and applications. New York, N.Y: Academic Press, Inc.; 1990. pp. 177–188. [Google Scholar]
- 30.Khan R, Giedroc D P. Nucleic acid binding properties of recombinant Zn2 HIV-1 nucleocapsid protein are modulated by COOH-terminal processing. J Biol Chem. 1994;269:22538–22546. [PubMed] [Google Scholar]
- 31.Konings D A, Nash M A, Maizel J V, Arlinghaus R B. Novel GACG-hairpin pair motif in the 5′ untranslated region of type C retroviruses related to murine leukemia virus. J Virol. 1992;66:632–640. doi: 10.1128/jvi.66.2.632-640.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laughrea M, Jette L, Mak J, Kleiman L, Liang C, Wainberg M A. Mutations in the kissing-loop hairpin of human immunodeficiency virus type 1 reduce viral infectivity as well as genomic RNA packaging and dimerization. J Virol. 1997;71:3397–3406. doi: 10.1128/jvi.71.5.3397-3406.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luban J, Bossolt K L, Franke E K, Kalpana G V, Goff S P. Human immunodeficiency virus type 1 Gag protein binds to cyclophilins A and B. Cell. 1993;73:1067–1078. doi: 10.1016/0092-8674(93)90637-6. [DOI] [PubMed] [Google Scholar]
- 34.Luban J, Goff S P. Binding of human immunodeficiency virus type 1 (HIV-1) RNA to recombinant HIV-1 Gag polyprotein. J Virol. 1991;65:3203–3212. doi: 10.1128/jvi.65.6.3203-3212.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luban J, Goff S P. Mutational analysis of cis-acting packaging signals in human immunodeficiency virus type 1 RNA. J Virol. 1994;68:3784–3793. doi: 10.1128/jvi.68.6.3784-3793.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maldarelli F, Martin M A, Strebel K. Identification of posttranscriptionally active inhibitory sequences in human immunodeficiency virus type 1 RNA: novel level of gene regulation. J Virol. 1991;65:5732–5743. doi: 10.1128/jvi.65.11.5732-5743.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McBride M S, Panganiban A T. The human immunodeficiency virus type 1 encapsidation site is a multipartite RNA element composed of functional hairpin structures. J Virol. 1996;70:2963–2973. doi: 10.1128/jvi.70.5.2963-2973.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McBride M S, Panganiban A T. Position dependence of functional hairpins important for human immunodeficiency virus type 1 RNA encapsidation in vivo. J Virol. 1997;71:2050–2058. doi: 10.1128/jvi.71.3.2050-2058.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McBride M S, Schwartz M D, Panganiban A T. Efficient encapsidation of human immunodeficiency virus type 1 vectors and further characterization of cis elements required for encapsidation. J Virol. 1997;71:4544–4554. doi: 10.1128/jvi.71.6.4544-4554.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Metz M Z, Best D M, Kane S E. Harvey murine sarcoma virus/MDR1 retroviral vectors: efficient virus production and foreign gene transduction using MDR1 as a selectable marker. Virology. 1995;208:634–643. doi: 10.1006/viro.1995.1194. [DOI] [PubMed] [Google Scholar]
- 41.Mougel M, Barklis E. A role for two hairpin structures as a core RNA encapsidation signal in murine leukemia virus virions. J Virol. 1997;71:8061–8065. doi: 10.1128/jvi.71.10.8061-8065.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mougel M, Zhang Y, Barklis E. cis-active structural motifs involved in specific encapsidation of Moloney murine leukemia virus RNA. J Virol. 1996;70:5043–5050. doi: 10.1128/jvi.70.8.5043-5050.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Poon D T, Wu J, Aldovini A. Charged amino acid residues of human immunodeficiency virus type 1 nucleocapsid p7 protein involved in RNA packaging and infectivity. J Virol. 1996;70:6607–6616. doi: 10.1128/jvi.70.10.6607-6616.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sakaguchi K, Zambrano N, Baldwin E T, Shapiro B A, Erickson J W, Omichinski J G, Clore G M, Gronenborn A M, Appella E. Identification of a binding site for the human immunodeficiency virus type 1 nucleocapsid protein. Proc Natl Acad Sci USA. 1993;90:5219–5223. doi: 10.1073/pnas.90.11.5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schwartz M D, Fiore D, Panganiban A T. Distinct functions and requirements for the Cys-His boxes of the human immunodeficiency virus type 1 nucleocapsid protein during RNA encapsidation and replication. J Virol. 1997;71:9295–9305. doi: 10.1128/jvi.71.12.9295-9305.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.SenGupta D J, Zhang B, Kraemer B, Pochart P, Fields S, Wickens M. A three-hybrid system to detect RNA-protein interactions in vivo. Proc Natl Acad Sci USA. 1996;93:8496–8501. doi: 10.1073/pnas.93.16.8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Skripkin E, Paillart J C, Marquet R, Ehresmann B, Ehresmann C. Identification of the primary site of the human immunodeficiency virus type 1 RNA dimerization in vitro. Proc Natl Acad Sci USA. 1994;91:4945–4949. doi: 10.1073/pnas.91.11.4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Surovoy A, Dannull J, Moelling K, Jung G. Conformational and nucleic acid binding studies on the synthetic nucleocapsid protein of HIV-1. J Mol Biol. 1993;229:94–104. doi: 10.1006/jmbi.1993.1011. [DOI] [PubMed] [Google Scholar]
- 49.Torrent C, Gabus C, Darlix J L. A small and efficient dimerization/packaging signal of rat VL30 RNA and its use in murine leukemia virus-VL30-derived vectors for gene transfer. J Virol. 1994;68:661–667. doi: 10.1128/jvi.68.2.661-667.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Van Beveren C, Coffin J, Hughes S. Restriction analysis of two genomes and restriction maps of representative retroviral proviruses and cellular oncogenes. In: Weiss R, Teich N, Varmus H, Coffin J, editors. RNA tumor viruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1984. pp. 559–1209. [Google Scholar]
- 51.Wang Z F, Whitfield M L, Ingledue T R, Dominski Z, Marzluff W F. The protein that binds the 3′ end of histone mRNA: a novel RNA-binding protein required for histone pre-mRNA processing. Genes Dev. 1996;10:3028–3040. doi: 10.1101/gad.10.23.3028. [DOI] [PubMed] [Google Scholar]
- 52.You J C, McHenry C S. HIV nucleocapsid protein. Expression in Escherichia coli, purification, and characterization. J Biol Chem. 1993;268:16519–16527. [PubMed] [Google Scholar]
- 53.Zhang B, Gallegos M, Puoti A, Durkin E, Fields S, Kimble J, Wickens M P. A conserved RNA-binding protein that regulates sexual fates in the C. elegans hermaphrodite germ line. Nature. 1997;390:477–484. doi: 10.1038/37297. [DOI] [PubMed] [Google Scholar]
- 54.Zhang Y, Barklis E. Nucleocapsid protein effects on the specificity of retrovirus RNA encapsidation. J Virol. 1995;69:5716–5722. doi: 10.1128/jvi.69.9.5716-5722.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang Y, Barklis E. Effects of nucleocapsid mutations on human immunodeficiency virus assembly and RNA encapsidation. J Virol. 1997;71:6765–6776. doi: 10.1128/jvi.71.9.6765-6776.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]