Abstract
This study examines the manufacturing, characterization, and biological evaluation of platinum nanoparticles, which were synthesized by Enterobacter cloacae and coated with Bovine Serum Albumin (BSA) and Resveratrol (RSV). The formation of PtNPs was confirmed with the change of color from dark yellow to black, which was due to the bioreduction of platinum chloride by E. cloacae. BSA and RSV functionalization enhanced these nanoparticles' biocompatibility and therapeutic potential. TGA, SEM, XRD, and FTIR were employed for characterization, where PtNPs and drug conjugation-related functional groups were studied by FTIR. XRD confirmed the crystalline nature of PtNPs and Pt-BSA-RSV NPs, while TGA and SEM showed thermal stability and post-drug coating morphological changes. Designed composite was also found to be biocompatible in nature in hemolytic testing, indicating their potential in Biomedical applications. After confirmation of PtNPs based nanocaompsite synthesis, they were examined for anti-bacterial, anti-oxidant, anti-inflammatory, and anti-cancer properties. Pt-BSA-RSV NPs showed higher concentration-dependent DPPH scavenging activity, which measured antioxidant capability. Enzyme inhibition tests demonstrated considerable anti-inflammatory activity against COX-2 and 15-LOX enzymes. In in vitro anticancer studies, Pt-BSA-RSV NPs effectively killed human ovarian cancer cells. This phenomenon was demonstrated to be facilitated by the acidic environment of cancer, as the drug release assay confirmed the release of RSV from the NP formulation in the acidic environment. Finally, Molecular docking also demonstrated that RSV has strong potential as an anti-oxidant, antibacterial, anti-inflammatory, and anticancer agent. Overall, in silico and in vitro investigations in the current study showed good medicinal applications for designed nanocomposites, however, further in-vivo experiments must be conducted to validate our findings.
Keywords: Bovine serum albumin, Enterobacter cloacae, Platinum nanoparticles, Resveratrol, Drug discovery
Subject terms: Materials science, Nanoscience and technology
Introduction
Liposomes, metal, polymer, albumin, solid fat, and lipid nanocarriers carry pharmaceutical particles. These nanoparticles have high surface-to-volume ratio, biocompatibility, and surface modifiability, which makes them excellent for drug delivery1–3. Important biological usage of platinum nanoparticles (PtNPs) is owing to their Biocompatibility, low toxicity, durability, cost-effectiveness, and manufacturing simplicity make them successful. PtNPs have been reported for various Biological applications including; anti-inflammatory, anti-bacterial, anti-diabetic, and wound-healing4–7. Furthermore, in vitro and in vivo studies have shown anti-cancer effects of PtNPs8. They destroy cells and alleviate oxidative stress by scavenging free radicals9,10. Modifying the surface of NPs is a promising approach to increase their therapeutic potential. Addition of Albumin is one such modification for PtNPs which helps the drug transportation more efficiently and effectively11,12. Albumin or other similar proteins increase nanoparticle dispersion, targeting, and toxicity13.
Researchers coupled bioactive chemicals to nanoparticles to achieve the object of targeted drug delivery14. This method is particularly important in anti-cancer drug discovery as it increases the concentration of nanoparticles at the site of tumor thereby assisting them in targeted drug discovery15. Increased nanoparticle uptake by cancer cells may lower drug use16. Nanoparticles with organic and bioactive compounds are also becoming increasingly popular17. Furthermore, loading NPs with therapeutics is also a recent way of increasing therapeutic potential of NPs. For this purpose, the use of natural compounds instead of synthetic ones is recommended because natural compounds have fewer side side-effects than synthetic ones18. Resveratrol (RSV) is one such natural compound that is recognized for regulating blood sugar, cholesterol, inflammation, antioxidants, and cancer prevention19.
There are multiple ways of synthesizing NPs, one such method is producing them via biological organisms including Bacteria. Bacterial nanoparticle production is a novel and ecologically friendly alternative to plant-based methods20. Chemical conjugation or physical adsorption of drugs onto nanoparticles is crucial to drug delivery21. Physical adsorption is straightforward, but nanoparticles often release their constituent before reaching their target22. Chemical conjugation provides a more reliable solution23.
Once the NPs are developed, conjugation drugs on them can be challenging. This requires strong amide, ester, hydrazine, and imine linkages24. The amine groups of medicines and the carbonyl groups of nanoparticles generate these connections25. This tight bond keeps drugs connected to nanoparticles until they reach their goal26. This technique relies on chemical links to respond to environmental changes and drug release27. The pH-sensitivity of imine, amide, and hydrazine connections is advantageous28. Acidic conditions accelerate connection breakdown in cancer cells29. Cancer cells produce an acidic environment, therefore pH-sensitive nanoparticles containing medicines may target medication delivery30. This ensures proper drug delivery, boosting efficacy and reducing negative effects31.
Pharmaceutical research increasingly incorporates computational tools like molecular docking to develop and enhance therapies. A therapeutic candidate ligand and a target protein interactions could be investigated through molecular docking experiments32. Examining the ligand's binding affinity, orientation, and possible interactions with the protein's active site may predict effective binding33, this helps in reduced dependency on wet lab experiments and overall reduction in cost of drug discovery Therefore, in this research work molecular docking approach was used to investigate the Resveratrol's interactions with therapeutic targets of different medical conditions.
In the current research work, PtNPs were synthesized from E. cloacae from a urinary tract infection patient. We next created amide linkages to load RSV onto BSA-conjugated PtNPs and release it into cancer cells' acidic environment. We also tested them against different other biological properties, such as; anti-bacterial, anti-oxidant, and anti-inflammatory properties. Overall, this research work examins the biocompatibility and therapeutic potential of the Pt-BSA-RSV-NPs formulation using both in vitro and in silico approaches of drug discovery.
Results and discussion
Phylogenetic analysis and PtNPs and Pt-BSA-RSV NPs synthesis
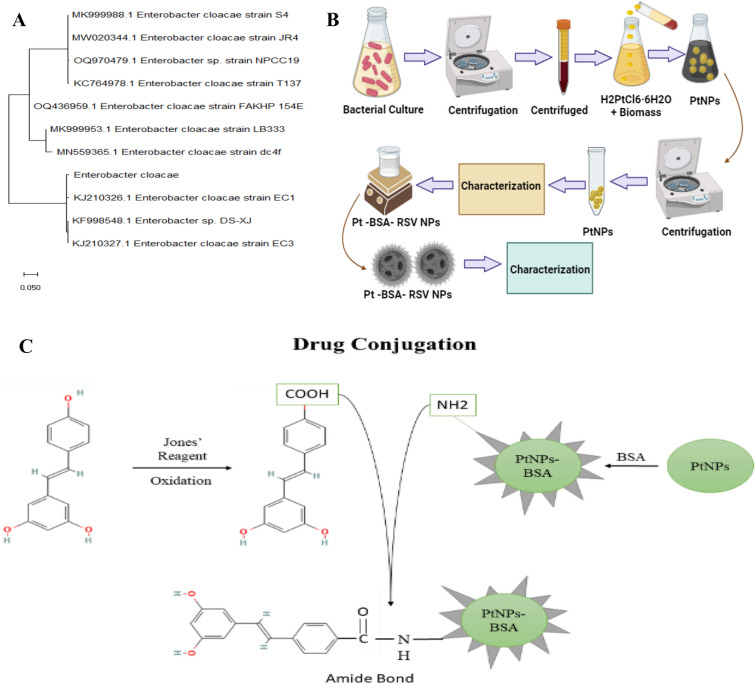
Platinum nanoparticles (PtNPs) made from E. cloacae from a urinary tract infection patient are an exciting blend of nanotechnology and biotechnology. Using phylogenetic analysis, E. cloacae was identified by comparing its ribosomal DNA (rDNA) to the databases of known bacterial genomes as shown in Fig. 1A. This approach is conventional in microbiology for species classification34. Once identified, PtNPs were made from the strains of E. cloacae to achieve the objective of synthesizing NPs using green technology, thus using an environmentally safe and sustainable approach to synthesis NPs35. The green synthesis of NPs is popular because of its cost effectiveness, large-scale manufacturing, and flexibility to change nanoparticles properties via bacterial growth conditions36,37. Bovine serum albumin (BSA) coating improves nanoparticles biocompatibility, stability, and cellular absorption37. This method improves nanoparticles for targeted drug delivery and medical use, therefore, developed PtNPs were further coated with BSA. Resveratrol (RSV) was then added to nanoparticles to boost their potential to fight cancer, inflammation, oxidative stress, microbial infections, and neurological diseases. This is largely due to the combine effect of NPs and RSV. RSV acts as an anticancer agent through antioxidant activity, anti-inflammatory effects, apoptosis induction, inhibition of angiogenesis, cell cycle arrest, interference with signaling pathways, and potential hormonal effects. It also possesses strong potential to be used in targeted drug delivery38. The schematic diagram of how PtNPs were synthesized and modified is shown in Fig. 1B, while the chemistry behind the conjugation of RSV with the BSA coated PtNPs is demonstrated in Fig. 1C.
Figure 1.
Phylogenetic analysis of identified bacterial strain, obtained from the patient of urinary tract infection (A), schematic synthesis and characterization of PtNPs coated with BSA and RSV (B), chemistry behind conjugation of RSV with PtNPs-BSA (C).
FTIR analysis
FTIR peaks were seen at 3350, 1650, 1450, 1390, 1070, 850, and 600 cm−1 for RSV, PtNPs, and Pt-BSA-RSV-NPs (Fig. 2). This peak is commonly associated with the stretching variations of either alcohol (O–H) and phenol or amine (N–H). It might also be due to proteins or polysaccharides in the bacterial biomass utilized to make PtNPs39. Peaks were also seen at 1650 cm−1 which frequently indicates amide bond formation, being induced by C=O stretching in proteins or peptides or C=C stretching in alkenes. Given drug conjugation, the Pt-BSA-RSV NPs sample's peak at this wavenumber may imply an amide connection between the drug and the BSA-PtNPs. This functionalization may increase PtNPs' drug delivery capabilities since the medicament binds selectively to nanoparticles. Peaks at 1450 and 1390 cm−1 suggest C–H bending vibrations in potentially alkanes and aromatic compounds. At 1070 cm−3 and 850 cm−3, stretching vibrations like C–O and C–Cl may be present in ethers or halo compounds40. The bands at 600 cm−1 and 470 cm−1 in Pt-BSA-RSV NPs suggest good nanoparticle-biomolecule bonding41. These wavenumbers also reveal Pt interactions, likely with BSA during drug conjugation.
Figure 2.
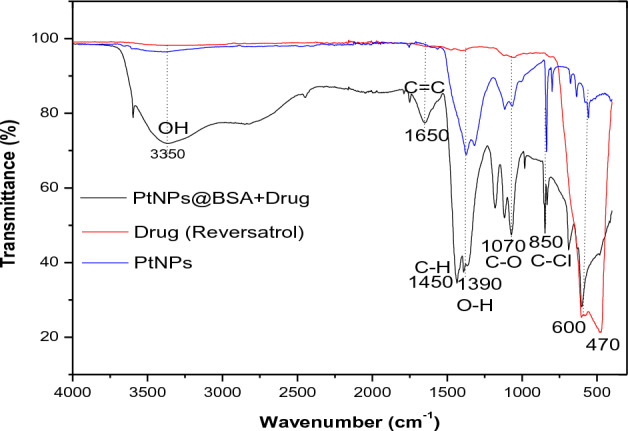
FTIR spectra of RSV, PtNPs and Pt-BSA-RSV-NPs.
XRD analysis
The X-ray diffraction (XRD) analysis confirmed the crystalline nature of both PtNPs and Pt-BSA-RSV NPs. The XRD analysis revealed the presence of peaks at specific angles (20.4, 21.7, 24.3, 30.4, 31.4, 36.7, and 44.4°) corresponding to PtNPs. However, a slight change in intensity and peaks was observed for BSA-coated drug-loaded PtNPs, as depicted in Fig. 3. The observed variations in the 2 theta values of the drug loaded NPs could potentially be attributed to the drug's presence on the surface of the PtNPs.
Figure 3.
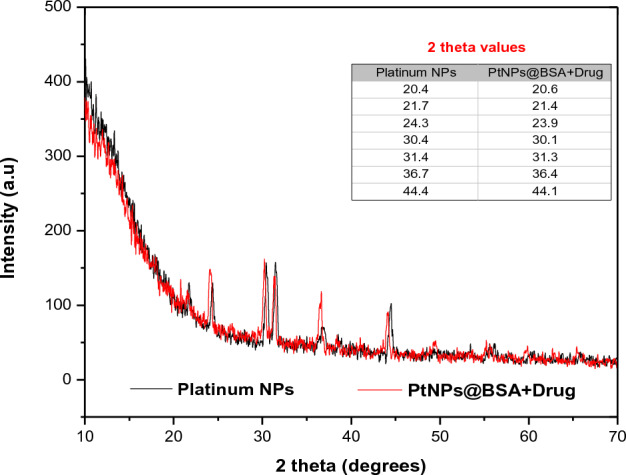
XRD spectra of the non-modified PtNPs and Pt-BSA-RSV NPs.
TGA analysis
The thermogravimetric analysis (TGA) shows a correlation between increase in temperature and weight loss of PtNPs and Pt-BSA-RSV NP. Figure 4A illustrates a decrease in weight of 0.3 mg as the temperature rises from 50 to 600 °C of PtNPs, while only 0.13 mg weight reduction was observed with the same change in temperature for drug coated Pt-NPs (Fig. 4B), which clearly indicates that the coating of BSA and RSV somehow stabilises the PtNPs, allowing them to withstand high temperatures.
Figure 4.
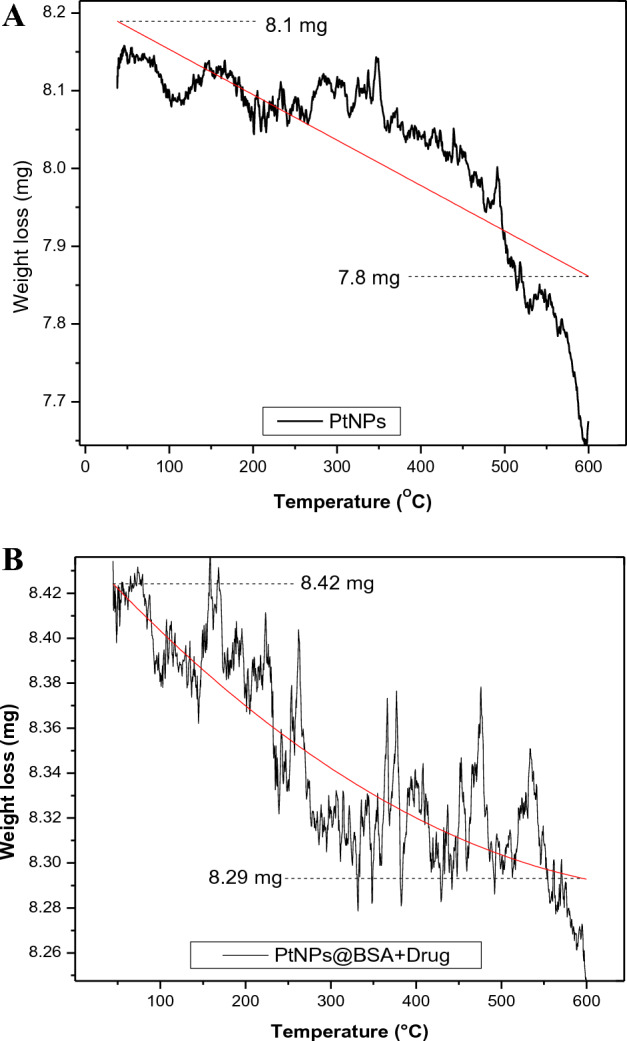
(A) TGA analysis of PtNPs and Pt-BSA-RSV NPs. (B) TGA analysis of PtNPs and Pt-BSA-RSV NPs at different temperatures.
SEM analysis
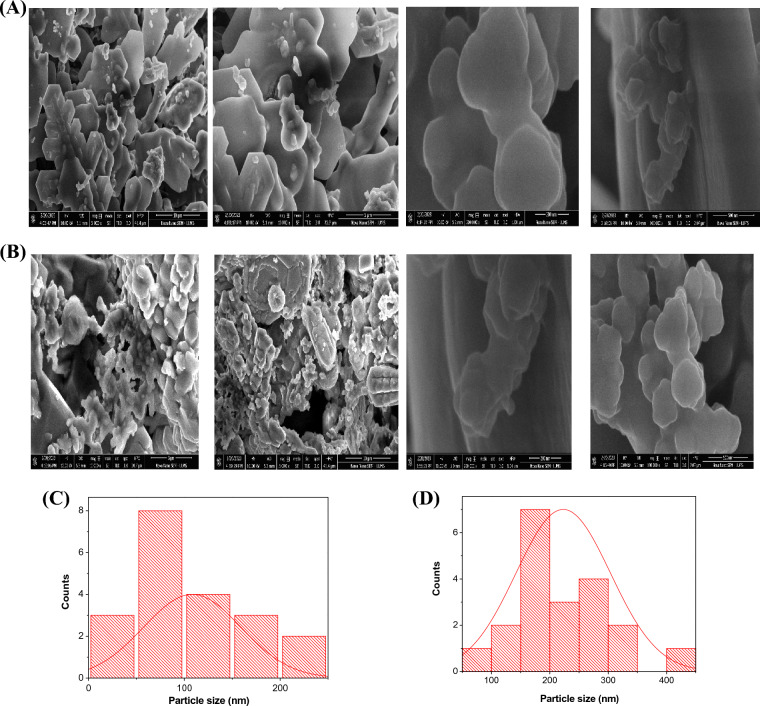
The Scanning Electron Microscope (SEM) images revealed the aggregation of PtNPs, as depicted in Fig. 5A. The particles exhibited a range of size, from 35.5 to 205.7 nm, with an average particle size of 108.2 nm, as determined by the ImageJ analysis (Fig. 5C). The drug coating had an impact on the size of PtNPs, with a slight increase in the size of the aggregates observed in Fig. 5B. Figure 5D shows that the smallest particle size observed was 96.8 nm, while the largest particle size was measured to 405 nm in diameter. The average particle size was determined to be 222.9 nm using ImageJ analysis.
Figure 5.
SEM images at different magnification of PtNPs (A) and Pt-BSA-RSV NPs (B). Particle size distribution of PtNPs (C), and Pt-BSA-RSV NPs (D).
RVS encapsulation efficiency
By creating a standard curve with absorbance values, measured at 280 nm for different RSV concentrations, the following equation Y = 0.9761x + 0.021 was computed.
This equation was obtained with a correlation coefficient (R2) of 0.91. Through the use of an equation developed from the RSV calibration curve and the absorption rate of the supernatant containing RSV, the encapsulation efficiency a proxy for the formulation's effectiveness was calculated to be around 95.91%.
Drugs release
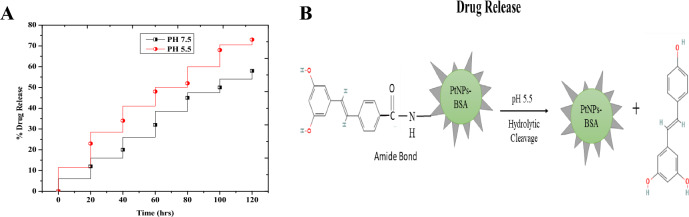
We studied the regulated release of RSV in an environment that replicated the endosomal pH seen in cancer cells and the physiological pH observed in healthy cells at 37 °C. When the pH was adjusted to 5.5 or 7.4, as shown in Fig. 6A, no initial or abrupt burst of drug release was observed. Instead of which, only 12.23% of the RSV was released in the first 20 h at pH 7.5. A total of 120 h were needed for the release of 58.89% of the RSV. The drug's slack attachment to the nanoparticle surface was the cause of this prolonged release. On the other hand, there was very little drug release when the pH was kept at a neutral level (pH 7.5). This resulted from an amide bond, which is a stable chemical link between Pt-BSA and RSV at neutral pH. On the other hand, we saw a more notable drug release pattern at a pH of 5.5. The drug release increased quickly, reaching 73.21% over the course of 48 h, with roughly 23.26% released in the first 20 h. These findings unequivocally show that the pH level affects the rate of RSV release. Over the course of 120 h, there is a noticeable change in drug release between pH 5.5 and 7.5, suggesting that the release of drug is pH-dependent (Fig. 6B).
Figure 6.
Drug release profiles of Pt-BSA-RSV-NPs under different pH conditions (pH 5.5 and pH 7.5) at a temperature of 37°C (A), Illustration of the drug release profiles (B).
Biological activity
Antibacterial activity
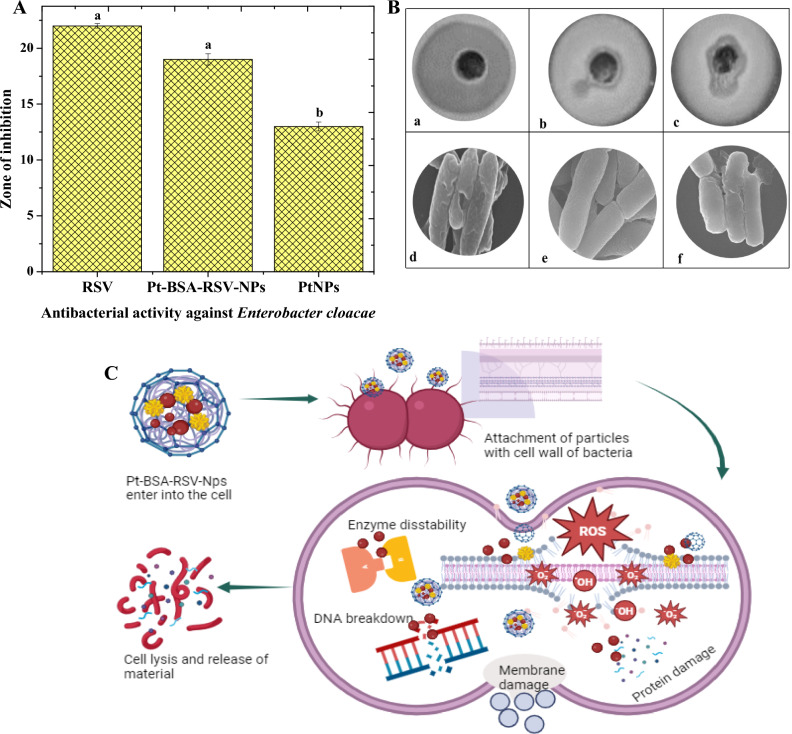
The antibacterial efficacy of PtNPs, RSV, and Pt-BSA-RSV NPs against E. cloacae were examined in this work. The test organism was the bacteria, which was initially recovered from the UTI patient. The main goal of this experiment was to find out the effectiveness of NPs against bacteria in vitro. Following a 24-h incubation period of test substances with the bacterial strain, the outcomes were noted. The zones of inhibition, or places where bacterial growth was inhibited surrounding the wells, were used to evaluate each tested chemical’s efficacy. These zones varied in size for each nanocomposite, suggesting different antibacterial activities. The disk diffusion experiment, a typical antibacterial test, confirmed these findings, as seen in the Fig. 7A–C.
Figure 7.
Graphical representation of inhibition zones for Enterobacter cloacae (A) Different letters are assigned to means when they are statistically significant (p-value < 0.05), while identical letters denote changes that are not significant. Pt-BSA-RSV-NPs activity against Enterobacter cloacae., Pt-BSA-RSV-NPs (a), RSV (b), and PtNPs (c) the SEM images of respective (d–f). (B) Schematic antibacterial mechanism of Pt-BSA-RSV-NPs (C).
RSV polyphenol is known to have antibacterial properties. Its antimicrobial impact may come from damaging bacterial cell membranes, increasing permeability and inducing cellular component loss. RSV also disrupts bacterial metabolism and critical enzyme functioning, which may kill bacterial cells42. Similarly, PtNP’s antibacterial properties come from their physicochemical properties and wide surface area. PtNPs cling to bacteria, rupturing their cell membranes and causes structural damage. When PtNPs engage with bacterial cells, they may generate ROS, causing oxidative stress which eventually leads to bacterial death43. Our proposed nanocomposite (Pt-BSA-RSV NPs) combines antibacterial capabilities of both PtNPs and RSV, where RSV may disrupt cell membranes and intracellular processes, increasing synergistic antibacterial effectiveness, whereas platinum coating increases bacterial cell contact44.
Antioxidant activities
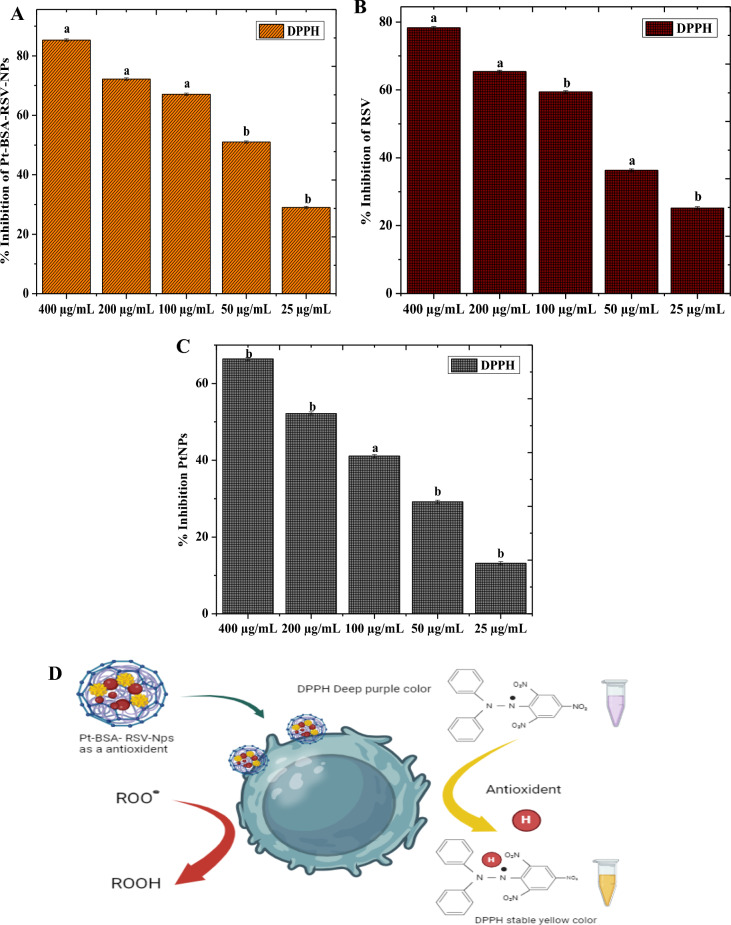
To examine their antioxidant capabilities, RSV, PtNPs, Pt-BSA-RSV NPs were tested for DPPH scavenging activity. Various concentrations (400, 200, 100, 50, and 25 µg/mL) were tested. Figure 8A–D shows that Pt-BSA-RSV NPs nanocomposites demonstrated strong antioxidant properties at high dosage of 400 µg/mL, indicating significant antioxidant potential. However, the nanocomposites' DPPH scavenging activity decreased at the lowest concentration (25 µg/mL), showing a concentration-dependent antioxidant activity. This pattern shows how nanoparticles concentration impacts antioxidant potential.
Figure 8.
Antioxidant activity of Pt-BSA-RSV-NPs (A), RSV (B), and PtNPs (C), different letters are assigned to means when they are statistically significant (p-value < 0.05), while identical letters denote changes that are not significant. Schematic antioxidant mechanism of Pt-BSA-RSV-NPs (D).
In the current study, Pt-BSA-RSV NPs nanocomposite combines RSV with PtNPs, where RSV is an antioxidant-rich natural phenol. Its antioxidant effect comes from free radical scavenging property. By donating a hydrogen atom, RSV may stabilize hydroxyl and peroxyl radicals and thereby prevents cell damage. RSV also chelates metal ions, preventing Fenton free radical formation45. Similarly, PtNPs may also act asantioxidants by breaking down Reactive Oxygen Species (ROS), for example, Hydrogen peroxide and superoxide anions, which are most likely to be converted into oxygen and water by PtNPs. This catalytic activity reduces cell oxidative stress46. Overall, platinum core in the nanocomposite may catalyse the antioxidant action, whereas RSV donates hydrogen and scavenges free radicals, while BSA coating enhances nanoparticle stability and bioavailability. Overall, the interaction between RSV and the PtNP’s surface may improve antioxidant capacity synergistically41,47, hence explaining the better anti-oxidant activity of Pt-BSA-RSV NPs composite as compared to the individual RSV or PtNPs.
Anti-inflammatory activities
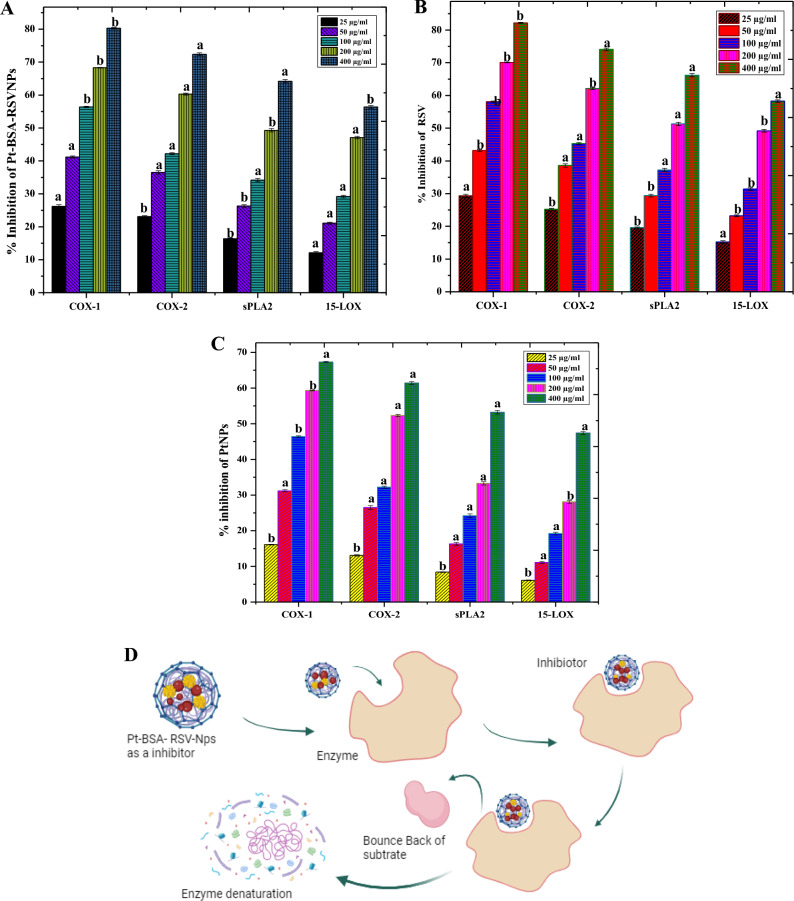
We examined the anti-inflammatory effects of RSV, PtNPs, and Pt-BSA-RSV NPs on COX-2, COX-1, sPLA2, and 15-LOX, enzymes that are known to initiate inflammation. Figure 9A–D shows that the best inhibition results were seen against 15-LOX, followed by sPLA2, COX-1, and COX-2. Furthermore, Pt-BSA-RSV NPs demonstrated many anti-inflammatory mechanisms. These include particular effects on COX-1 and COX-2 and targeting eicosanoid-producing lipoxygenases and phospholipase A2 which eventually reduces two key inflammatory mediators, namely; prostanoids and leukotrienes48.
Figure 9.
Anti-inflammatory activities of Pt-BSA-RSV-NPs (A), RSV (B), and PtNPs (C). Different letters are assigned to means when they are statistically significant (p-value < 0.05), while identical letters denote changes that are not significant. Schematic anti-inflammatory mechanism of Pt-BSA-RSV-NPs (D).
RSV is reported to possess anti-inflammatory potential by targeting several signalling pathways. It inhibits pro-inflammatory mediator-synthesising enzymes including LOX and COX. RSV also inhibits NF-κB activity, a transcription factor crucial for regulating inflammatory cytokines. By blocking these pathways, RSV reduces inflammatory mediator and cytokine production49. Organic compounds like RSV are more anti-inflammatory than PtNPs. PtNPs may inhibit cell inflammation by inhibiting macrophages reducing pro-inflammatory cytokines, and activating immune cell responses. Furthermore, PtNPs also reduce cell oxidative stress, which eventually alleviates inflammation50.
As both RSV and PtNPs have anti-inflammation potential, therefore, combining them in the form a nanocomposite makes them ever stronger anti-inflammatory substance. The developed Pt-BSA-RSV NPs works by combining the effects of both PtNPs and RSV, where PtNPs may reduce oxidative stress and alleviate immune cell function, while RSV may directly block generation of inflammation mediators. The BSA coating may increase nanoparticle bioavailability and stability, delivering active chemicals to inflammatory regions more efficiently51. Hence, synergistic effect of RSV and PtNPs may be larger than their individual anti-inflammatory effects. These nanocomposites may have dose-dependent anti-inflammatory effects, depending on concentration. This variation emphasizes the need of altering concentration levels to maximize treatment efficacy and minimize side effects4.
Invitro anticancer activity
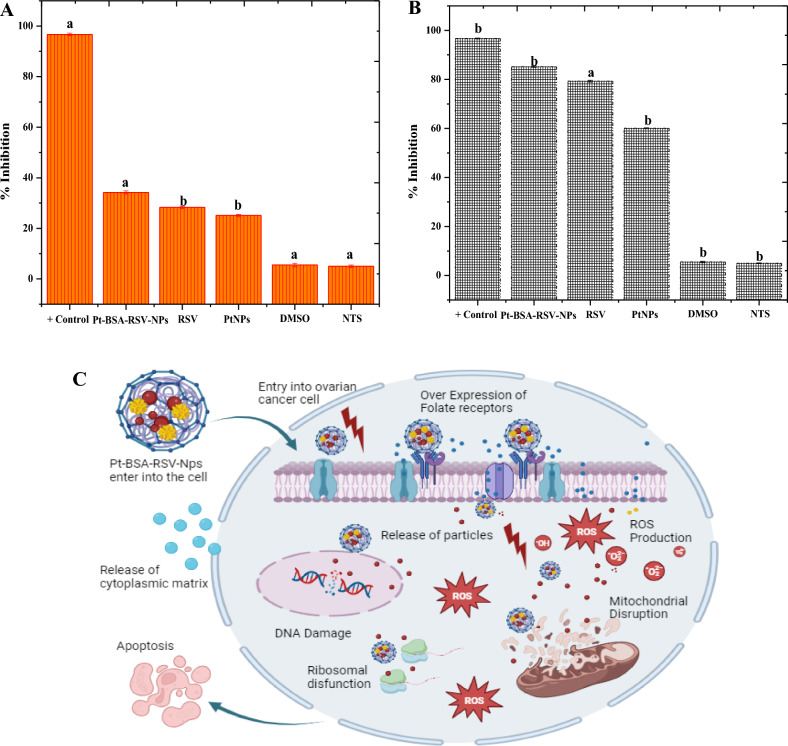
For this study The normal cell and ovarian cancer cells were purchased from Khyber Medical University Pakistan. This research also investigated the cytotoxic effects of RSV, PtNPs, and Pt-BSA-RSV NPs' on Normal cells and human ovarian cancer cells. Compared to the control groups, these test substances non-significantly reduce normal cell and also Significantly reduced ovarian cancer cell viability after 24 h. Phase-contrast microscopy indicated considerable cell death in ovarian cancer cells treated with RSV and Pt-BSA-RSV NPs with Pt-BSA-RSV NPs causing significant cytotoxicity in human ovarian cancer (Fig. 10A–C).
Figure 10.
In vitro cytotoxicity of Pt-BSA-RSV-NPs, RSV and PtNPs against normal cells (A), In vitro anticancer activity of Pt-BSA-RSV-NPs, RSV and PtNPs (B), Different letters are assigned to means when they are statistically significant (p-value < 0.05), while identical letters denote changes that are not significant. Schematic anticancer mechanism of Pt-BSA-RSV-NPs (C).
RSV may promote cancer cell apoptosis by activating signalling pathways such the p53 pathway, which regulates cell development and death52. RSV may also blocks angiogenesis in cancer cells, inhibit them from growing, and reduces inflammation and oxidative stress, all of which eventually hinders proliferation of cancer cells53–55.
PtNPs, like other platinum-based chemotherapy medicines, interact with DNA to fight cancer. PtNPs may form platinum–DNA adducts with the DNA of cancer cells, promoting severe damage to DNA which prompts death of cancer cells56. They may increase ROS production in cancer cells, causing oxidative stress and apoptosis57. Due to their small size, cancer cells may readily absorb PtNPs, which may increase possibility of targeted drug delivery56.
Combining PtNPs with RSV in a nanocomposite boost their anti-cancer properties, where BSA coating may help target cancer cells more accurately and release therapeutic components by enhancing nanoparticle stability and bioavailability. By acting as a carrier in nanoparticles, permitting prolonged drug release, improving targeted drug administration, offering biocompatibility, and maybe having direct antioxidant effects, BSA functions as an anticancer agent58. Similar to our study, Iqbal et. al in also showed that albumin coated NPs, loaded with drug, have been cancer specific, by taking the advantage of ph responsiveness59. Similarly, other recently published studies have also demonstrated that NPs based formulations or devices are better than the drug only to treat cancer, however, the conjugation between two can really increases the efficiency of both60–62.
Overall, the proposed nanocomposite in this study holds a strong potential as a novel medication to treat cancer18.
In-vitro biocompatibility study
In this study, we evaluated the hemolytic effects of RSV, PtNPs, and Pt-BSA-RSV NPs on human red blood cells (RBCs). We used 1 mL samples of fresh human RBCs, drawn into EDTA tubes from healthy individuals with their consent. the RBCs were cultured in a buffer solution designed to imitate biological circumstances. The basic premise is that a spectrophotometer can measure hemoglobin released into the medium when the RBC ruptures. Table 1 shows that hemolysis levels over 25% are hemolytic and below 10% are non-hemolytic. Pt-BSA-RSV NPs had excellent hemocompatibility at increasing concentrations, except for a modest hemolytic response at 400 µg/ml having no apparent activity at the studied levels. Biocompatibility makes Pt-BSA-RSV NPs promising candidates for biological applications. The constituents of this nanocomposites, are also biocompatible in nature. RSV occurs naturally in berries, grapes, and other plants. Biocompatibility is due to its natural nature and well-established antioxidant and anti-inflammatory properties, which may lower biological system oxidative stress and inflammation. This reduces unfavourable reactions in biological settings63. PtNPs, or platinum nanoparticles, are biocompatible depending on their size, shape, and surface. Small, biocompatible PtNPs have minimal toxicity and good cellular absorption. Functionalizations or surface coatings may improve PtNP compatibility with biological systems and reduce their risk64.
Table 1.
% hemolytic activity of Pt-BSA-RSV-NPs, RSV and PtNPs.
| Conc (µg/mL) | Pt-BSA-RSV NPs % Hemolysis |
RSV % Hemolysis |
PtNPs % Hemolysis |
|---|---|---|---|
| 400 | 2.61 ± 0.21 | 4.78 ± 0.62 | 3.45 ± 0.34 |
| 200 | 1.52 ± 0.23 | 3.19 ± 0.17 | 2.93 ± 0.51 |
| 100 | 0.85 ± 0.51 | 2.36 ± 0.21 | 1.92 ± 0.72 |
| 50 | 0.32 ± 0.31 | 1.20 ± 0.46 | 0.95 ± 0.69 |
Different letters are assigned to means when they are statistically significant (p-value < 0.05), while identical letters denote changes that are not significant.
In several aspects, Pt-BSA-RSV NPs increase biocompatibility. Nanoparticle stability and biocompatibility are typically improved using bovine serum albumin (BSA), which is intrinsically biocompatible. BSA-coated nanoparticles reduce cytotoxicity by creating a medically appropriate surface65. RSV and PtNPs may be released under controlled settings because to their structure, decreasing cell-damaging local concentrations66. Modifying Pt-BSA-RSV NPs' design allows tailored dispersion, reducing active component exposure to non-target cells and improving biocompatibility67. Their physical, chemical, and biological interactions affect this compounds’ in-vitro biocompatibility. Pt-BSA-RSV NPs are ideal for biological applications due to BSA's stabilizing effect, PtNPs' engineering adaptability, and RSV's intrinsic properties. This compatibility ensures safe and effective use in a biological setting, which is crucial for therapeutic use.
Findings from in silico experiments
Assessment of drug-like properties of resveratrol
The first phase of our computational experiments was focused on evaluating the drug-like properties of RSV. As illustrated in Table 2, RSV exhibits all the requisite drug-like properties without any violations from the five established rules of drug-likeness. Furthermore, an examination of its physiochemical and medicinal chemistry-related attributes revealed that RSV possesses properties considered acceptable for a "lead drug compound."
Table 2.
The attributes related to physiochemical characteristics, medicinal chemistry, and drug-likeness of Resveratrol were acquired through analysis using SwissADME.
| PubChem ID | Physiochemical properties | Medicinal chemistry | Drug likeliness (violations of rules) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular weight (g/mol) | Topological polar surface area (Å2) | No. of rotatable bonds | Hydrogen bond donor | Hydrogen bond acceptor | Pains alert | Synthetic accessibility | Lipinski rule of 5 | Veber rule | Egan rule | Muegge rule | Ghose rule | |
| 445,154 | 228.24 | 60.69 | 2 | 3 | 3 | 0 | 2.02 | 0 | 0 | 0 | 0 | 0 |
Molecular docking
The field of drug design has undergone a profound revolution, primarily driven by the integration of in silico analysis and bioinformatics. This transformative shift has significantly reduced both cost and time associated with drug discovery process. A considerable volume of research publications now highlights drugs and their associated targets, elucidated through the application of bioinformatics tools and software68. One such Bioinformatics method, widely used in computer-aided drug discovery, is molecular docking analysis which involves computationally predicting the binding interactions between a small molecule (ligand) and a target protein. It helps to assess the likelihood and strength of binding, aiding in the identification and optimization of potential drug candidates69. This approach plays a crucial role in rational drug design by providing insights into the molecular mechanisms underlying ligand–protein interactions. Therefore, in this study molecular docking analysis was conducted to get in-depth insight into the binding pattern of RSV into the active sites of therapeutic target proteins. For this purpose, we used MOE software which has been employed in many previously published studies70. MOE incorporates a scoring function that relies on essential factors such as hydrogen bonding, van der Waals forces, and other interactions, however, Hydrogen bonding is the key element which plays a critical role in discerning robust docked complexes. A higher count indicates more effective binding between the ligand and the target protein71. The Molecular docking investigations were conducted as the second and final step between RSV and P-Cs and the target proteins associated with various medical conditions previously tested in in-vitro experiments.
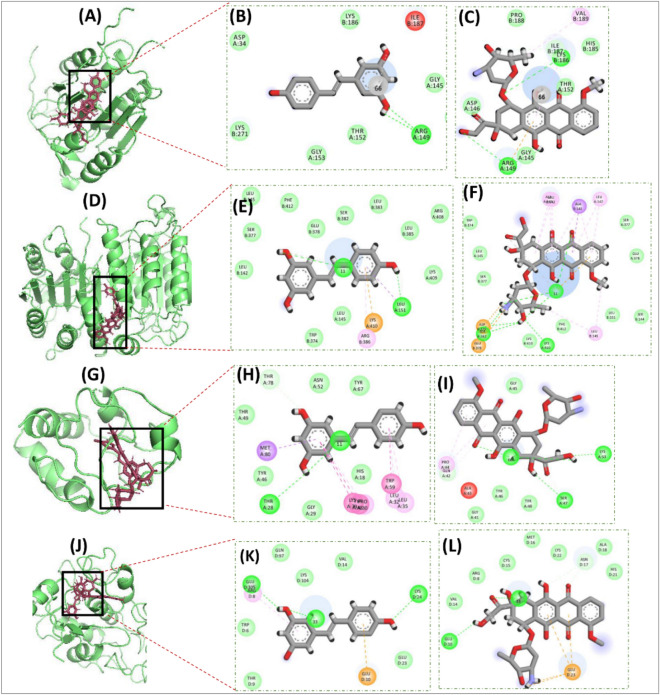
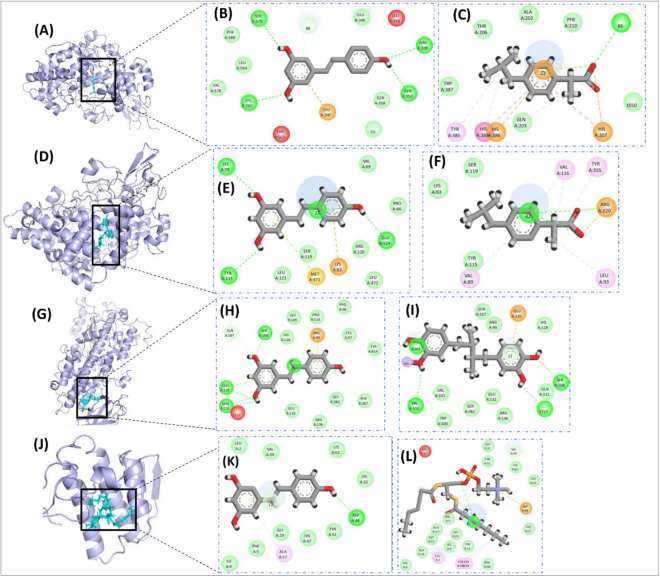
The outcomes are summarized in Table 3, highlighting that RSV demonstrates either better or comparable binding affinities, in comparison to the P-Cs, as indicated by docking score, RMSD values, and the number of hydrogen bonds formed. Additionally, surface images and 2-dimensional interactions of the ligand-receptor docked complexes are presented in Figs. 11, 12 for all the docking experiments. The antibacterial and anti-oxidant, related docking results are shown in Fig. 10, and if we visualize these results with Table 3 it is found that the interactions patterns for both RSV and P-Cs are similar. For the anti-bacterial activity, the docking protocols demonstrated that the number of Hydrogen Bonds (H-bond) was four and two for RSV and P–C, respectively, while the docking scores were also comparable (Fig. 11A–C). In the docking experiments of anti-oxidant activity, although the number of H-bonds developed by RSV was half as compared to P–C, i.e., six vs. twelve, yet, the docking scores of both were quite to close each other, showing that the interactions between Resveratrol are as strong as the P–C (Fig. 11D–F).
Table 3.
The outcomes of molecular docking investigations involving Resveratrol and positive controls (P–C) with various proteins to evaluate their potential in anti-Alzheimer’s disease, anti-oxidant, anti-bacterial, anti-cancer, and anti-inflammatory activities.
| Activity | Protein | Compound/drug | Docking score | RMSD | No. of hydrogen bond/s |
|---|---|---|---|---|---|
| Anti-cancer |
Caspase 3 6CKZ |
Resveratrol | − 16.7715 | 1.5104 | 2 |
| Doxorubicin (P–C) | − 8.8226 | 3.5228 | 2 | ||
|
Caspase 9 2AR9 |
Resveratrol | − 9.0870 | 1.2976 | 2 | |
| Doxorubicin (P–C) | − 14.2174 | 1.4186 | 7 | ||
|
Cytochrome-c 3zcf |
Resveratrol | − 10.6409 | 0.8256 | 1 | |
| Doxorubicin (P–C) | − 7.5731 | 2.0056 | 3 | ||
|
Folate receptors 4LRH |
Resveratrol | − 10.8684 | 1.8123 | 3 | |
| Doxorubicin (P–C) | − 11.9663 | 3.0038 | 2 | ||
| Anti inflammatory |
COX-1 6y3c |
Resveratrol | − 8.9569 | 0.9365 | 4 |
| Ibuprofin (P–C) | − 7.729646 | 1.584653 | 1 | ||
|
COX-2 1CX2 |
Resveratrol | − 10.39164 | 2.550156 | 2 | |
| Ibuprofn (P–C) | − 10.02839 | 1.839417 | 1 | ||
|
LOX5 1LOX |
Resveratrol | − 12.1118 | 0.8684 | 5 | |
| nordihydroguaiaretic acid (P–C) | − 12.0765 | 2.1353 | 3 | ||
|
Spla2 1DCY |
Resveratrol | − 10.43802 | 1.031119 | 1 | |
| Diheptanoyl thio-PC (P–C) | − 11.61481 | 2.263655 | 2 | ||
| Anti oxidant |
Myeloperoxidase enzymatic protein 1DNU |
Resveratrol | − 9.959191 | 1.871706 | 6 |
| Ascorbic acid (P–C) | − 10.5597 | 1.566998 | 12 | ||
| Anti-bacteria |
Thymidylate kinase 4GQQ |
Resveratrol | − 10.7747 | 2.2617 | 4 |
| Oxacillin (P–C) | − 12.4463 | 1.6937 | 2 |
The docking score reflects the binding energy, while the number of hydrogen bonds indicates the hydrogen bonds formed in the docked complex between the ligand and protein.
Figure 11.
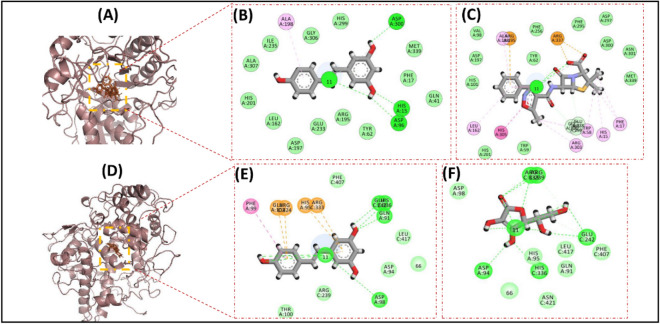
Images depicting interactions from docking experiments associated with the anti-bacteria and anti-oxidant activity of Resveratrol and positive controls. the surface images showcase the ligands docked with thymidylate kinase (A) for antibacterial, myeloperoxidase enzymatic protein (D) for anti-oxidant activities. The detailed amino acid level binding interactions are also shown for Resveratrol with thymidylate kinase (B), and myeloperoxidase enzymatic protein (E). Similar interaction images are also demonstrated for positive controls for thymidylate kinase (C), myeloperoxidase enzymatic protein (F).
Figure 12.
Images depicting interactions from docking experiments associated with the anti-cancer activity of Resveratrol and positive controls. The surface images showcase the ligands docked with Caspase 3 (A), Caspase 9 (D), Cytochrome-c (G), and Folate Receptor (J). The detailed amino acid level Binding interactions are also shown for Resveratrol with Caspase 3 (B), Caspase 9 (E), Cytochrome-c (H), and Folate Receptor (K) and for Doxorubicin as a positive control for Caspase 3 (C), Caspase 9 (F), Cytochrome-c (I), and Folate Receptor (L).
The molecular docking results for anti-cancer activities represented that the RSV docking score is better than P–C for both Caspase 3 and Cytochrome-c. In comparison, the docking scores of P–C for Caspase 9 and Folate receptors was better than the RSV, yet, it was comparable. Similarly, the number of H-bonds formation was also similar for test compound and P–C in all four cases (Fig. 12A–L).
Results of anti-inflammation activity for molecular docking experiments also demonstrated that the RSV has strong binding affinity for the all four receptor proteins, considered in molecular docking experiments. Figure 13A–I and Table 3 depicts that the number of H-bonds as well the docking function score of RSV is lower, hence better, than the P–C for COX-1, COX-2, and LOX5. Whereas, for Spla2, RSV found to forming only one H-bond with the active site residues of Spla2, as compared to P–C which formed two H-bonds (Fig. 13J–L). Manifestly, the docking score of RSV was also reported to be − 10.44 as compared to P–C whose score was − 11.62, although the score for Resveratrol was higher, yet comparable, with that of P–C (Table 3). Overall, the molecular docking results demonstrated that RSV develops multiple interactions (including hydrogen bonds) with target proteins, and these interactions are either stronger or comparable to the positive controls. Therefore, these results further indicate the therapeutic potential of Resveratrol with evidence from the molecular level interactions.
Figure 13.
Images depicting interactions from docking experiments associated with the anti-inflammation related activity of Resveratrol and positive controls. The surface images showcase the ligands docked with COX-2 (A), COX-1 (D), LOX-5 (G), and spLA2 (J). The detailed amino acid level Binding interactions are also shown for Resveratrol with COX-2 (B), COX-1 (E), LOX-5 (H), and spLA2 (K) and for Doxorubicin as a positive control for COX-2 (C), COX-1 (F), LOX-5 (I), and spLA2 (L).
Conclusion
This work effectively showcase the creative fusion of biotechnology and nanotechnology in the synthesis of platinum nanoparticles (PtNPs) by E. cloacae, which were then augmented by coatings of Resveratrol (RSV) and bovine serum albumin (BSA). In addition to supporting environmental sustainability, this green synthesis technique opens the door for the scalable and affordable manufacturing of nanoparticles with specific properties. The FTIR, XRD, TGA, and SEM extensive evaluation confirmed the effective production and functionalization of the produced nanoparticles and gave a detailed insight into their physicochemical characteristics. The PtNPs and Pt-BSA-RSV NPs shown strong antibacterial, antioxidant, anti-inflammatory, and anticancer effects, according to the biological activity evaluations. The BSA-RSV coated nanoparticles improved performance highlights the synergy that results from combining the special qualities of each component. The therapeutic potential of these nanocomposites was further confirmed by the in vitro experiments and molecular docking analyses, particularly in the areas of anti-inflammatory and cancer therapies. This work not only makes a substantial contribution to the area of nanomedicine, but it also serves as an example of how biological organisms and nanomaterials might be combined to create novel therapeutic agents. The results represent a major advancement in the search for novel and efficient medical treatments by opening up new options for the use of nanoparticles in medicine, notably in targeted drug delivery and precision medicine.
Methodology
Selection of bacterial specie and preparation of biomass
This study employed E. cloacae, which was isolated from a patient with a urinary tract infection. Informed consent was obtained from the participants. PCR, Sanger sequencing, and phylogenetic analysis revealed that the isolated specie is E. cloacae. E. cloacae was grown in a 500-mL Erlenmeyer flask with 300 mL of 5% nitrate broth. A rotary shaker shook the flask at 27 °C for 72 h at 200 rpm. This incubation was followed by 20 min of 6000 rpm centrifugation to remove microorganisms. The obtained pellet was washed with sterile phosphate buffer saline solution with pH 7.2. Washed pellet was then Re-suspended in 20 ml of triple-distilled water to produce ready-made biomass suspension.
Synthesis of platinum nanoparticles
To make platinum nanoparticles, 500 ml of distilled water was added into the one liter conical flask, followed by the addition of 0.0019 M platinum chloride solution (H2PtCl6·6H2O). Sonicating the solution ensured the full dissolution. 20 ml of ready-made biomass suspension, prepared in the last step, was then added into the mixture. The contact of platinum ions and bacterial cell wall was allowed for 48 h. This time produced a substantial black insoluble deposit at the conical flask's bottom. The condensed precipitates were washed with the distilled water after centrifuging the flask at neutralize pH. After air-drying, the precipitates were cleaned with acetone. The precipitates were heated to 500 °C for three hours to remove biomass-derived organic components and leaving behind purecrystalline platinum nanoparticles.
Physicochemical characterization of PtNPs
A thin layer of PtNPs was created for FTIR measurement by drop-casting an ethanol-based NP dispersion onto an IR-transparent substrate. FTIR spectra were obtained in the 4000–400 cm−1 region in order to examine distinctive peaks and functional groups. The PtNPs were subjected to a controlled temperature rise during thermogravimetric analysis (TGA), and the weight loss was tracked as a function of temperature. The PtNPs powder was subjected to XRD analysis utilizing Cu Kα radiation and a specific grade sample container. The sample was scanned across a variety of 2θ angles, such as 10–80°, to assess its crystal structure, phase purity, and average crystallite size. PtNPs were dissolved in ethanol, the suspension was drop-cast onto a glass slide, a small layer of conductive material was applied, and the morphology and size were observed using a high-resolution SEM.
Synthesis of Pt-BSA-RSV-NPs
The Pt-BSA-RSV Nano-formulation was synthesized by following methodology. First, sodium chloride (NaCl) was used to dissolve bovine serum albumin (BSA), which was agitated at 500 rpm for two hours at pH 8.3. Next, dissolved platinum nanoparticles (PtNPs) in distilled water were dropwise combined with the BSA solution. After that, the mixture was stirred for two more hours at 500 rpm. Resveratrol (RSV) was modified through oxidation, purification of oxidized RSV, and activation of the carboxylic group before loading. After dropwise addition of modified RSV to the BSA-containing solution, the mixture became turbid. After adding 30 µl of an 8% glutaraldehyde solution, the mixture was stirred and left at room temperature for a full day. After incubation, the solution was centrifuged at 13,000 rpm for 20 min. Encapsulated RSV was recovered from the supernatant to assess how much was encapsulated. After that, the nanoparticle precipitate was washed three times with distilled water to remove any leftover impurities. The washed precipitate dried after centrifugation again.
RSV encapsulation analysis using indirect approaches
In this work, the encapsulation efficiency of RSV was evaluated indirectly by the measurement of the sample adsorption rate at certain wavelengths through the Epoch-Biotech spectrophotometer. By making standard RSV solutions at different dosages, a workable curve was created. The formula for calculating the Encapsulation Efficiency (EE) was as follow
In vitro drug release study
The research examined the drug RSV release from Pt-BSA-RSV-NPs in two distinct pH settings: phosphate buffers with a pH of 5.5 and 7.4. First, a 250 mL conical flask containing a 75 mL dissolving media was filled with a precisely determined quantity of the conjugated NPs, which included 10.0 mg of drug, or 23.64% of the drug conjugated to the NPs. After that, these flasks were shaken vigorously in an incubator for 120 h at a speed of 75 rpm. Throughout the course of these 120 h, 5 mL of the solution was taken out of the flask at predetermined intervals, and 5 mL of new buffer was introduced back to keep the conditions of a "sink" in place. The cumulative drug release percentage was determined by using the following formula:
Here, “Total Drug" is the total drug in the conjugated NPs, and “Drug Released" is the drug released at a particular period. This approach tracks drug release from NPs across time and pH settings without affecting the sink state.
Characterization of Pt-BSA-RSV-NPs
FTIR spectra were used to identify functional groups present in prepared nanocomposite. In the first step, 2 mg of NPs and 200 mg of potassium bromide (KBr) were compressed to produce standard disks. After then, FTIR spectra were recorded between 400 and 4000 cm−1. For complete characterization, TGA, AFM, and XRD analysis were performed. The NPs' crystal structure and phase composition was revealed by XRD. The SEM was utilized to examine NPs’ morphology. NPs' thermal stability and breakdown behaviour was revealed by TGA. The combined efforts of these techniques helped us to characterize the designed Pt-BSA-RSV-NPs'.
In-vitro biological applications
Disc diffusion assay
The disc diffusion test identified E. cloacae strains for in-vitro antibacterial testing of RSV, PtNPs, and Pt-BSA-RSV-NPs. In this experiment, 100 μL quantities of standardized broth cultures of the bacterial strains (1 × 105 CFU/mL) were applied on culture plates, and sterile bent glass rods were used to create a uniform bacterial layer. Following sterilization, 400 μg/mL of RSV, PtNPs, and Pt-BSA-RSV-NPs were added to different 6 mm wells. The inhibition zones were determined after the incubation period of 24 h. The antibacterial effectiveness of the nanocomposites was confirmed by the notable difference we saw in the inhibitory zones between RSV, PtNPs, and Pt-BSA-RSV-NPs and control samples.
Collection of microorganisms and SEM imaging
After treatment, the microorganisms were collected and the excess fluid was promptly drained off by centrifuging them in PBS. The next day, they were fixed at 4 °C. Following the fixation process, the bacteria underwent three PBS cleanings prior to being dried using gradient concentration of ethanol solutions from 30 to 100%. After spreading out on silicon wafers, the bacteria were dried and coated with gold for field emission scanning electron microscopy (FE-SEM) imaging.
Antioxidant assays
Antioxidant experiments examined RSV, PtNPs, and Pt-BSA-RSV-NPs' free radical scavenging abilities. To evaluate the antioxidant capacity of test samples, RSV, PtNPs, and Pt-BSA-RSV-NPs were used at doses from 25 to 400 µg/mL. Each well on a 96-well plate received 10 µl of RSV, PtNPs, and Pt-BSA-RSV-NPs, followed by addition of 90 µl of DPPH (2,2-diphenyl-1-picrylhydrazyl) solution. In this experiment, the used negative and positive control was DMSO and ascorbic acid, respectively. Next, the microplate reader measured the mixture's absorbance at 515 nm and free radical scavenging potential was calculated by using the following equation:
where sample absorbance is "Abs" and negative control absorbance is "Abc."
Anti-inflammatory activities
The anti-inflammatory effects of the nanoparticles were examined by evaluating their capacity to obstruct pathways linked to the synthesis of prostaglandin E2 (PGE2), an inflammatory mediator, from arachidonic acid.
COX-2 and COX-1 inhibitory activity
Using particular test kits, the inhibitory activity of RSV, PtNPs, and Pt-BSA-RSV-NPs against COX-1 and COX-2 was assessed (Ovine Kit 701050 France for COX-1 and Human Kit 701050 France for COX-2). The positive control was ibuprofen at a concentration of 10 mM, whereas the substrate was arachidonic acid at a concentration of 1.1 mM. The kit's instructions were followed in order to quantify the peroxidase components of both COX enzymes. To guarantee precision, the experiment was carried out in a 96-well plate in triplicate. Using a Synergy II reader, the absorbance of N, N, N/, and N/-tetramethyl-p-phenylenediamine at 590 nm was determined.
Activity of inhibition for 15-LOX
Using the Cayman France 760700 kits, the capacity of RSV, PtNPs, and Pt-BSA-RSV-NPs to suppress the activity of 15-LOX was examined. Nordihydroguaiaretic acid (NDGA) was used as the positive control at a concentration of 100 mM, while arachidonic acid functioned as the substrate. Hydro-peroxides were produced as a result of lipooxygenation throughout the process, and their concentration was measured using a 15-lipoxygenase standard in a pH 7.4 Tris–HCl buffer. RSV, PtNPs, Pt-BSA-RSV-NPs, and the enzyme were combined in a 96-well plate, and the mixture was incubated for five minutes. They were incubated for five more minutes after adding the substrate, and then for fifteen minutes after adding the chromogen. Using a Synergy II reader (BioTek Instruments, Colmar, France), the absorbance at 590 nm was determined.
Activity of inhibition for secretory phospholipase A2 (sPLA2)
The ability of RSV, PtNPs, and Pt-BSA-RSV-NPs to reduce sPLA2 was examined using an assay kit (10004883, Cayman Chem. Co., Interchim, Montluçon, France). The positive control and substrate were diheptanoyl thio-PC at 1.44 mM and 100 mM thiotheramide-PC, respectively. Free thiols are released upon cleavage of the diheptanoyl thio-PC ester, and they were quantified in an aqueous solution at 420 nm using a Synergy II reader and DTNB (5,5'-dithio-bis-2-nitrobenzoic acid) in a 96-well microplate. The following formula was used to get the percentage inhibition:
In-vitro anti-cancer potential against human ovarian cancerous cells
The McCoy's 5a and Eagle Minimum Essential Medium growth media, together with 10% calf serum and non-essential amino acids, were used to cultivate human ovarian cancer cells. These cultivated cells were kept at 37 °C in a humidified environment with 5% CO2 under normal conditions. Following their cultivation, the cells were planted onto 96-well plates and allowed to incubate for 48 h. RSV, PtNPs, and Pt-BSA-RSV-NPs were applied to human ovarian cancer cells at a fixed concentration of 400 µg/mL. Untreated cells were used as a control which also received RSV, PtNPs, and Pt-BSA-RSV-NPs treatment (drug control). For the cytotoxicity test, we also tested the cells to the well-known anticancer drug doxorubicin at a stock concentration of 100 mM. The treated plates underwent a further 48–72 h of incubation. Next, the MTT test was run on the plates to evaluate the vitality of the cells. 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide, a yellow tetrazole, was produced at a concentration of 5 mg/mL in order to identify the presence of MTT. After adding 100 mL of MTT solution to each well, the mixture was allowed to sit at room temperature for four hours. Formazan crystals became purple, and dimethyl sulfoxide was used to dissolve them (DMSO). After that, the crystals' absorbance at 620 nm was measured using an ELISA plate reader with numerous wells to quantify them. Following equation was used to determine the experimental samples' optical density (OD) values and percentage viability.
The experimental sample (E) and the blank, untreated sample (B) are represented in the equation above.
Biocompatibility studies
The biogenic RSV, PtNPs, and Pt-BSA-RSV-NPs' compatibility with human red blood cells was investigated in a biocompatibility research in human Reb Blood Cells (hRBCs). In EDTA tubes, 1 ml of blood was drawn from willing, healthy subjects, and the RBCs were separated by centrifugation. Next, PBS was used to wash these RBC pellets. After a second cycle of centrifugation, 200 mL of RBCs and 9.8 mL of pH 7.2 PBS were gently mixed to create a suspension. The erythrocyte solution was then mixed with various amounts of RSV, PtNPs, and Pt-BSA-RSV-NPs, and incubated for one hour at 35 °C to allow the reaction. Following incubation, the resultant mixture was centrifuged and then moved to a well plate so that hemoglobin release could be measured using the standard absorption peak at 450 nm. Following equation was used to quantify hemolysis.
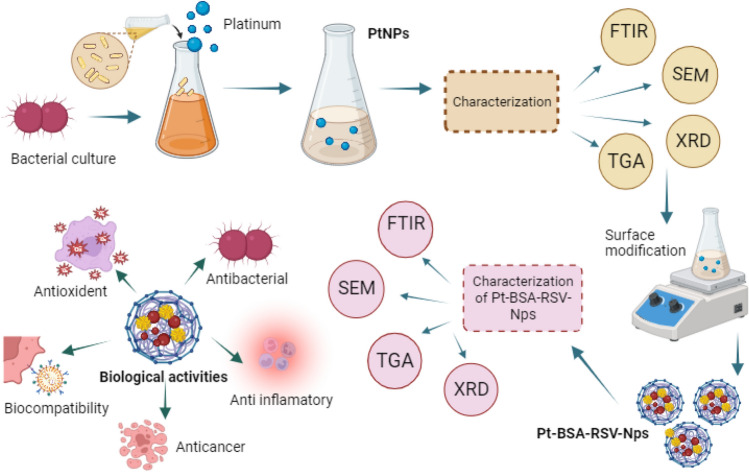
The over all methodology is shown in Scheme 1. The methodology starts with a bacterial culture used to synthesize PtNPs, followed by their characterization through FTIR, SEM, TGA, and XRD analysis. The PtNPs were then subjected to surface modification to create Pt-BSA-RSV-NPs, which were subsequently evaluated for their biological activities, including antioxidant, antibacterial, anti-inflammatory, and anticancer properties.
Scheme 1.
The overall methodology of this paper.
Insilco analysis
Assessment of drug-like properties
In this study, in silico experiments were conducted to evaluate various drug-like properties of RSV. The SwissADME online tool was employed to assess drug-likeness, pharmacokinetics, and medicinal chemistry-related characteristics. This tool utilizes five different rules, namely Lipinski Rule of 5, Muegge Rule, Veber Rule, Ghose Rule, and Egan Rule, to analyze the drug-likeness of test compounds. Additionally, pharmacokinetics-related properties, such as the number of hydrogen bond acceptors, the number of hydrogen bond donors, and the total number of rotatable bonds, were estimated. Synthetic Accessibility and PAINS Alert were also predicted by SwissADME to assess medicinal chemistry-related properties of small compounds.
Molecular docking
Molecular docking experiments were carried out to determine the binding potential of RSV with therapeutic targets associated with anti-cancer, anti-inflammatory, anti-bacterial, and anti-oxidant activities. The 3D structures of Resveratrol and Positive Controls (P–C), identical to those used in wet-lab experiments, were obtained in SDF format from the PubChem database. Oxacillin served as a P–C for anti-bacterial activity-related docking protocols, as no P–C was employed in wet-lab anti-bacterial experiments. Libraries of Resveratrol and P-Cs were established using the Molecular Operating Environment (MOE) software for diverse docking experiments. Ligands were prepared for molecular docking by protonation and energy minimization using the Pronate3D algorithm and AMBER99 force-field, respectively.
For target proteins, tertiary structures were retrieved from the Protein Data Bank (PDB). Proteins tested in wet lab experiments were considered for anti-cancer and anti-inflammatory docking protocols, while for anti-oxidant and anti-bacterial experiments, Myeloperoxidase Enzymatic Protein (MEP) and Thymidylate kinase were chosen after thorough literature review, as effects of Resveratrol was not tested on any specific proteins in wet-lab experiments. MEP contributes to the generation of free radical species, including reactive oxidants, whereas the Thymidylate Kinase plays a role in bacterial DNA biosynthesis. Consequently, both MEP and Thymidylate Kinase are considered viable therapeutic targets for antibacterial and antioxidant activities. The proteins and their corresponding PDB IDs are listed in Table 3. The downloaded PDB structures underwent the same preparation steps in MOE as the ligands.
Finally, the MOE software was employed for molecular docking, utilizing parameters such as Refinement: force field, Rescoring: London dg, Placement: Triangle matcher, and Retain: 10. This methodology aligns with approaches employed in prior published studies. Finally, Discovery studio (2021) and Pymol v2.5.7 software were utilized to visualize the docking results and to get a deep insight into the interaction patterns.
Ethics approval and consent to participate
All experiments and sampling procedures were ethically performed following standard protocol approved by Ethics Committee of Department of Physical Chemistry and Technology of Polymers, Silesian University of Technology, under reference No. Dir-SUT/EB/IV/00514. This study complies with the approved standard guidelines and regulations.
Acknowledgements
Authors wish to thank Researchers Supporting Project Number (RSP2024R346) at King Saud University Riyadh Saudi Arabia for financial support.
Author contributions
S.F, conceptualized the study. M.H.T curated the data., S.F conducted the investigation. S.F and Abdullah, contributed to the methodology. S.F, and M.H.T were responsible for project administration. S.F, provided the necessary resources. S.F, R.U.K and S.Z developed the software. S.F provided supervision., S.F and A.U.R carried out the validation. S.F, and K.U were involved in data visualization. S.F wrote the original draft, R.U. Z.N, and A.B participated in the review and editing of the manuscript. All authors reviewed and approved the final manuscript.
Funding
The research work is supported by researchers supporting project number (RSP2024R346) at King Saud University Riyadh Saudi Arabia.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article has been retracted. Please see the retraction notice for more detail: https://doi.org/10.1038/s41598-024-74394-5"
Change history
10/29/2024
This article has been retracted. Please see the Retraction Notice for more detail: 10.1038/s41598-024-74394-5
References
- 1.Soares, S., Sousa, J., Pais, A. & Vitorino, C. Nanomedicine: Principles, properties, and regulatory issues. Front. Chem.6, 360 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amoabediny, G. et al. Overview of preparation methods of polymeric and lipid-based (niosome, solid lipid, liposome) nanoparticles: A comprehensive review. Int. J. Polym. Mater. Polym. Biomater.67(6), 383–400 (2018). [Google Scholar]
- 3.Jain, A. K. & Thareja, S. In vitro and in vivo characterization of pharmaceutical nanocarriers used for drug delivery. Artif. Cells Nanomed. Biotechnol.47(1), 524–539 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Pedone, D., Moglianetti, M., De Luca, E., Bardi, G. & Pompa, P. P. Platinum nanoparticles in nanobiomedicine. Chem. Soc. Rev.46(16), 4951–4975 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Jeyaraj, M., Gurunathan, S., Qasim, M., Kang, M. H. & Kim, J. H. A comprehensive review on the synthesis, characterization, and biomedical application of platinum nanoparticles. Nanomaterials9(12), 1719 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jan, H. et al. A detailed review on biosynthesis of platinum nanoparticles (PtNPs), their potential antimicrobial and biomedical applications. J. Saudi Chem. Soc.25(8), 101297 (2021). [Google Scholar]
- 7.Fahmy, S. A., Preis, E., Bakowsky, U. & Azzazy, H. M. E. S. Platinum nanoparticles: Green synthesis and biomedical applications. Molecules25(21), 4981 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yogesh, B., Vineeta, B., Rammesh, N. & Saili, P. Biosynthesized platinum nanoparticles inhibit the proliferation of human lung-cancer cells in vitro and delay the growth of a human lung-tumor xenograft in vivo: In vitro and in vivo anticancer activity of bio-Pt NPs. J. Pharmacopuncture19(2), 114 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhattacharya, S. et al. Investigating in vitro and in vivo anti-tumor activity of Curvularia-based platinum nanoparticles. J. Environ. Pathol. Toxicol. Oncol.41(3), 13–32 (2022). [DOI] [PubMed] [Google Scholar]
- 10.Fani, A. et al. Green synthesis of a novel PtFe2O4@ Ag nanocomposite: Implications for cytotoxicity, gene expression and anti-cancer studies in gastric cancer cell line. J. Clust. Sci.34(1), 535–546 (2023). [Google Scholar]
- 11.Hosaka, H., Haruki, R., Yamada, K., Böttcher, C. & Komatsu, T. Hemoglobin–albumin cluster incorporating a Pt nanoparticle: Artificial O2 carrier with antioxidant activities. PLoS ONE9(10), e110541 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pourmadadi, M., Ghaemi, A., Shaghaghi, M., Rahdar, A. & Pandey, S. Cabazitaxel-nano delivery systems as a cutting-edge for cancer therapy. J. Drug Deliv. Sci. Technol.82, 104338 (2023). [Google Scholar]
- 13.Bhushan, B., Khanadeev, V., Khlebtsov, B., Khlebtsov, N. & Gopinath, P. Impact of albumin based approaches in nanomedicine: Imaging, targeting and drug delivery. Adv. Colloid Interface Sci.246, 13–39 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Martínez-Ballesta, M., Gil-Izquierdo, Á., García-Viguera, C. & Domínguez-Perles, R. Nanoparticles and controlled delivery for bioactive compounds: Outlining challenges for new “smart-foods” for health. Foods7(5), 72 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang, X. et al. A folate receptor-targeting nanoparticle minimizes drug resistance in a human cancer model. ACS Nano5(8), 6184–6194 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vlashi, E., Kelderhouse, L. E., Sturgis, J. E. & Low, P. S. Effect of folate-targeted nanoparticle size on their rates of penetration into solid tumors. ACS Nano7(10), 8573–8582 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Ezhilarasi, P. N., Karthik, P., Chhanwal, N. & Anandharamakrishnan, C. Nanoencapsulation techniques for food bioactive components: A review. Food Bioprocess Technol.6, 628–647 (2013). [Google Scholar]
- 18.Santos, A. C. et al. Targeting cancer via resveratrol-loaded nanoparticles administration: Focusing on in vivo evidence. AAPS J.21, 1–16 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Chung, I. M. et al. Resveratrol nanoparticles: A promising therapeutic advancement over native resveratrol. Processes8(4), 458 (2020). [Google Scholar]
- 20.Iravani, S. Bacteria in nanoparticle synthesis: Current status and future prospects. Int. Sch. Res. Notices2014, 2014 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mehanny, M., Hathout, R. M., Geneidi, A. S. & Mansour, S. Studying the effect of physically-adsorbed coating polymers on the cytotoxic activity of optimized bisdemethoxycurcumin loaded-PLGA nanoparticles. J. Biomed. Mater. Res. Part A105(5), 1433–1445 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Ossama, M., Hathout, R. M., Attia, D. A. & Mortada, N. D. Augmented cytotoxicity using the physical adsorption of Poloxamer 188 on allicin-loaded gelatin nanoparticles. J. Pharm. Pharmacol.73(5), 664–672 (2021). [DOI] [PubMed] [Google Scholar]
- 23.Morachis, J. M., Mahmoud, E. A. & Almutairi, A. Physical and chemical strategies for therapeutic delivery by using polymeric nanoparticles. Pharmacol. Rev.64(3), 505–519 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vigderman, L. & Zubarev, E. R. Therapeutic platforms based on gold nanoparticles and their covalent conjugates with drug molecules. Adv. Drug Deliv. Rev.65(5), 663–676 (2013). [DOI] [PubMed] [Google Scholar]
- 25.El-Boubbou, K. Magnetic iron oxide nanoparticles as drug carriers: Preparation, conjugation and delivery. Nanomedicine13(8), 929–952 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Huang, P. et al. Combination of small molecule prodrug and nanodrug delivery: Amphiphilic drug–drug conjugate for cancer therapy. J. Am. Chem. Soc.136(33), 11748–11756 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Feng, X. et al. Conjugated polymer nanoparticles for drug delivery and imaging. ACS Appl. Materi. Interfaces2(8), 2429–2435 (2010). [DOI] [PubMed] [Google Scholar]
- 28.Wang, Q. et al. Doxorubicin and adjudin co-loaded pH-sensitive nanoparticles for the treatment of drug-resistant cancer. Acta Biomater.94, 469–481 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Lu, D. et al. A pH-sensitive nano drug delivery system derived from pullulan/doxorubicin conjugate. J. Biomed. Mater. Res. Part B89(1), 177–183 (2009). [DOI] [PubMed] [Google Scholar]
- 30.Du, J. Z., Du, X. J., Mao, C. Q. & Wang, J. Tailor-made dual pH-sensitive polymer–doxorubicin nanoparticles for efficient anticancer drug delivery. J. Am. Chem. Soc.133(44), 17560–17563 (2011). [DOI] [PubMed] [Google Scholar]
- 31.Hu, R., Zheng, H., Cao, J., Davoudi, Z. & Wang, Q. Synthesis and in vitro characterization of carboxymethyl chitosan-CBA-doxorubicin conjugate nanoparticles as pH-sensitive drug delivery systems. J. Biomed. Nanotechnol.13(9), 1097–1105 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Abdelsattar, A. S., Dawoud, A. & Helal, M. A. Interaction of nanoparticles with biological macromolecules: A review of molecular docking studies. Nanotoxicology15(1), 66–95 (2021). [DOI] [PubMed] [Google Scholar]
- 33.Chibber, S. & Ahmad, I. Molecular docking, a tool to determine interaction of CuO and TiO2 nanoparticles with human serum albumin. Biochem. Biophys. Rep.6, 63–67 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu, W. Y., Wong, C. F., Chung, K. M. K., Jiang, J. W. & Leung, F. C. C. Comparative genome analysis of Enterobacter cloacae. PLoS ONE8(9), e74487 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Faisal, S. et al. Biofabrication of silver nanoparticles employing biomolecules of Paraclostridium benzoelyticum strain: Its characterization and their in-vitro antibacterial, anti-aging, anti-cancer and other biomedical applications. Microsc. Res. Tech.2023, 1–10 (2023). [DOI] [PubMed] [Google Scholar]
- 36.Faisal, S. et al. Paraclostridium benzoelyticum bacterium-mediated zinc oxide nanoparticles and their in vivo multiple biological applications. Oxid. Med. Cell. Longev.2022, 1–15 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nosrati, H., Sefidi, N., Sharafi, A., Danafar, H. & Manjili, H. K. Bovine serum albumin (BSA) coated iron oxide magnetic nanoparticles as biocompatible carriers for curcumin-anticancer drug. Bioorganic Chem.76, 501–509 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Jang, M. et al. Cancer chemopreventive activity of resveratrol, a natural product derived from grapes. Science275(5297), 218–220 (1997). [DOI] [PubMed] [Google Scholar]
- 39.Zafar, S. et al. Development of iron nanoparticles (FeNPs) using biomass of enterobacter: Its characterization, antimicrobial, anti-Alzheimer’s, and enzyme inhibition potential. Micromachines13(8), 1259 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Agrawal, A., Kapoor, N., Sirohi, A., Kumar, P., Dixit, R., & Kumar, L. K. G. P. Green synthesis, optimization, and characterization of zinc oxide nanoparticle using Lantana camara L. leaf extract 1–8 (2023).
- 41.Yu, S. et al. Biocompatible bovine serum albumin stabilized platinum nanoparticles for the oxidation of morin. New J. Chem.43(22), 8774–8780 (2019). [Google Scholar]
- 42.Bostanghadiri, N. et al. Comprehensive review on the antimicrobial potency of the plant polyphenol resveratrol. Biomed. Pharmacother.95, 1588–1595 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Hashimoto, M., Yanagiuchi, H., Kitagawa, H. & Honda, Y. Inhibitory effect of platinum nanoparticles on biofilm formation of oral bacteria. Nano Biomed.9(2), 77–82 (2017). [Google Scholar]
- 44.Mattio, L. M. et al. Antimicrobial activity of resveratrol-derived monomers and dimers against foodborne pathogens. Sci. Rep.9(1), 19525 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hunyadi, A. The mechanism(s) of action of antioxidants: From scavenging reactive oxygen/nitrogen species to redox signaling and the generation of bioactive secondary metabolites. Med. Res. Rev.39(6), 2505–2533 (2019). [DOI] [PubMed] [Google Scholar]
- 46.Moglianetti, M. et al. Platinum nanozymes recover cellular ROS homeostasis in an oxidative stress-mediated disease model. Nanoscale8(6), 3739–3752 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Younis, F. A. et al. Preparation, physicochemical characterization, and bioactivity evaluation of berberine-entrapped albumin nanoparticles. Sci. Rep.12(1), 17431 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rudrapal, M. et al. Dual synergistic inhibition of COX and LOX by potential chemicals from Indian daily spices investigated through detailed computational studies. Sci. Rep.13(1), 8656 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baur, J. A. & Sinclair, D. A. Therapeutic potential of resveratrol: The in vivo evidence. Nat. Rev. Drug Discov.5(6), 493–506 (2006). [DOI] [PubMed] [Google Scholar]
- 50.Rehman, M. U., Yoshihisa, Y., Miyamoto, Y. & Shimizu, T. The anti-inflammatory effects of platinum nanoparticles on the lipopolysaccharide-induced inflammatory response in RAW 264.7 macrophages. Inflamm. Res.61, 1177–1185 (2012). [DOI] [PubMed] [Google Scholar]
- 51.Aires, A. et al. BSA-coated magnetic nanoparticles for improved therapeutic properties. J. Mater. Chem. B3(30), 6239–6247 (2015). [DOI] [PubMed] [Google Scholar]
- 52.Kuo, P. L., Chiang, L. C. & Lin, C. C. Resveratrol-induced apoptosis is mediated by p53-dependent pathway in Hep G2 cells. Life Sci.72(1), 23–34 (2002). [DOI] [PubMed] [Google Scholar]
- 53.Berman, A. Y., Motechin, R. A., Wiesenfeld, M. Y. & Holz, M. K. The therapeutic potential of resveratrol: A review of clinical trials. NPJ Precis. Oncol.1(1), 35 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cao, Y. U., Fu, Z. D., Wang, F., Liu, H. Y. & Han, R. U. Anti-angiogenic activity of resveratrol, a natural compound from medicinal plants. J. Asian Nat. Prod. Res.7(3), 205–213 (2005). [DOI] [PubMed] [Google Scholar]
- 55.Rodríguez-Enríquez, S. et al. Resveratrol inhibits cancer cell proliferation by impairing oxidative phosphorylation and inducing oxidative stress. Toxicol. Appl. Pharmacol.370, 65–77 (2019). [DOI] [PubMed] [Google Scholar]
- 56.Li, F. et al. A Nuclease-mimetic platinum nanozyme induces concurrent DNA platination and oxidative cleavage to overcome cancer drug resistance. Nat. Commun.13(1), 7361 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Almarzoug, M. H., Ali, D., Alarifi, S., Alkahtani, S. & Alhadheq, A. M. Platinum nanoparticles induced genotoxicity and apoptotic activity in human normal and cancer hepatic cells via oxidative stress-mediated Bax/Bcl-2 and caspase-3 expression. Environ. Toxicol.35(9), 930–941 (2020). [DOI] [PubMed] [Google Scholar]
- 58.Solanki, R., Rostamabadi, H., Patel, S. & Jafari, S. M. Anticancer nano-delivery systems based on bovine serum albumin nanoparticles: A critical review. Int. J. Biol. Macromol.193, 528–540 (2021). [DOI] [PubMed] [Google Scholar]
- 59.Iqbal, H. et al. pH-responsive albumin-coated biopolymeric nanoparticles with lapatinib for targeted breast cancer therapy. Biomater. Adv.139, 213039 (2022). [DOI] [PubMed] [Google Scholar]
- 60.Aiqbal, H. et al. Breast cancer inhibition by biosynthesized titanium dioxide nanoparticles is comparable to free doxorubicin but appeared safer in BALB/c mice. Materials14(12), 3155 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang, Y. et al. Albumin-based nanodevices for breast cancer diagnosis and therapy. J. Drug Deliv. Sci. Technol.79, 104072 (2023). [Google Scholar]
- 62.Iqbal, H. et al. Serum protein-based nanoparticles for cancer diagnosis and treatment. J. Control. Release329, 997–1022 (2021). [DOI] [PubMed] [Google Scholar]
- 63.Hu, C. et al. Platinum-based nanocomposite Pt@ BSA as an efficient electrochemical biosensing interface for rapid and ultrasensitive determination of folate receptor-positive tumor cells. ACS Appl. Bio Mater.5(6), 3038–3048 (2022). [DOI] [PubMed] [Google Scholar]
- 64.Gulino, M., Santos, S. D. & Pêgo, A. P. Biocompatibility of platinum nanoparticles in brain ex vivo models in physiological and pathological conditions. Front. Neurosci.15, 787518 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Montero, N. et al. Biocompatibility studies of intravenously administered ionic-crosslinked chitosan-BSA nanoparticles as vehicles for antitumour drugs. Int. J. Pharm.554, 337–351 (2019). [DOI] [PubMed] [Google Scholar]
- 66.Rosli, N. A., Teow, Y. H. & Mahmoudi, E. Current approaches for the exploration of antimicrobial activities of nanoparticles. Sci. Technol. Adv. Mater.22(1), 885–907 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Naito, M., Yokoyama, T., Hosokawa, K., & Nogi, K. (Eds.). Nanoparticle Technology Handbook. (Elsevier, 2018).
- 68.Faisal, S. et al. Exploring the antibacterial, antidiabetic, and anticancer potential of mentha arvensis extract through in-silico and in-vitro analysis. BMC Complement. Med. Ther.23(1), 267 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tariq, M. H., Akram, M. & Sharif, Y. Insilico screening of bioactive phytochemicals against spike protein of COVID-19. Pak. J. Zool54(1), 433–438 (2022). [Google Scholar]
- 70.Pantsar, T. & Poso, A. Binding affinity via docking: Fact and fiction. Molecules23(8), 1899 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Morris, C. J. & Corte, D. D. Using molecular docking and molecular dynamics to investigate protein–ligand interactions. Mod. Phys. Lett. B35(08), 2130002 (2021). [Google Scholar]