Abstract
Tumor microenvironment (TME) is a complex and dynamic evolving environment which facilitates tumor proliferation and progression. We aimed at investigating the characteristics of tumor microenvironment and its prognostic value in gliomas. Transcriptome data of 702 glioma samples from The Cancer Genome Atlas were included as training dataset, while 325 samples from Chinese Glioma Genome Atlas database and 268 samples from GSE16011 database were used to validate. We found that the infiltration of stromal and immune cell varied in gliomas of different grades and pathological types, and was associated with poor prognosis. Based on the gene expression profile, we constructed a TME-related signature (TMERS), which was closely related to clinical features and genomic variation of gliomas. In TMERS-high group, specific gene mutations and increased copy number alternations were observed. Kaplan–Meier survival and Cox regression analysis showed that TMERS was an independent prognostic indicator. Then we developed a nomogram prognostic model to predict 1-year, 3-year and 5-year survival of patients. Functional analysis confirmed that TMERS could reflect the status of glioma microenvironment, and immunological analysis showed that macrophages were significantly enriched in the TMERS-high group. We established a novel TME-related signature for predicting prognosis and provided new insights into immunotherapy.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00262-021-03058-4.
Keywords: Tumor microenvironment, Gliomas, Signature, Genetic, Survival
Introduction
Gliomas are the most common histological type of primary tumor of the central nervous system and originate from glial cells. Globally, the annual incidence of gliomas is 5–6 per 100,000 people [1]. Compared to lower-grade gliomas (LGG, WHO grade II and III), glioblastomas (GBM, WHO Grade IV) have a poor prognosis with a median overall survival of 14.4 months, despite maximum resection of the tumor combined with chemo- and radiotherapy [1, 2]. Tumor recurrence and treatment resistance are the main dilemma in the treatment of gliomas. Therefore, in-depth analysis of the mechanism of gliomagenesis and exploring effective treatments can better extend the survival of these patients.
Tumor microenvironment (TME) is a complex and dynamic evolving environment, which mainly includes various types of immune cells, stromal cells and the cytokines released by the cells. Tumor cells secrete various cytokines to reshape the surrounding microenvironment, which in turn facilitates tumor proliferation and progression [3]. Compared with malignant tumor cells, the genetic stability of normal cells in the TME determines the therapeutic stability of this target [4]. For example, the earliest immune-checkpoint blockade (ICB) therapy (CLTA-4 and PD-1 antibodies) have demonstrated pronounced clinical benefits by activating T cells [5]. However, a significant proportion of patients do not respond to ICB therapy [6]. In GBM, the therapeutic effectiveness of ICB has been unpredictable and uncommon, with only 8% of patients having a definite response [7, 8]. Therefore, a comprehensive understanding of TME is beneficial to improve the current therapies targeting dysfunctional cells in the TME.
Our study integrated 1295 transcriptome data across whole grade gliomas and comprehensively estimated the glioma microenvironment, including immune and stromal components. We screened differentially expressed genes representing TME and developed a TME-related signature (TMERS), which was associated with clinical and genetic properties of gliomas. Then we verified TMERS as a poor prognostic indicator and a prognostic nomogram model based on TMERS was constructed. TMERS could reflect the status of glioma microenvironment, and macrophages were significantly enriched in the TMERS-high group. Our research might contribute to the understanding of glioma TME and help guide the development of TME-targeted therapy for gliomas.
Methods
Samples and data collection
Totally, 1295 glioma samples from three datasets were collected: The Cancer Genome Atlas (TCGA), Chinese Glioma Genome Atlas (CGGA) and GSE16011. For TCGA training dataset (702 glioma samples), the RNA sequencing data, somatic mutation, copy-number alterations (CNAs) and clinicopathological information were downloaded online (http://cancergenome.nih.gov/). For CGGA validation dataset, we collected 325 RNA sequencing data generating with Illumina HiSeq 2,000 sequencing system [9]. Corresponding clinical information was also collected and the pathological reports were confirmed by two experienced neuropathologists according to WHO guidelines in 2016. The detection method of IDH mutation has been described in our previous study [10, 11]. Overall survival (OS) was defined from surgery to death or last follow-up [12]. The CGGA dataset was approved by the Beijing Tiantan Hospital Capital Medical University Institutional Review Board (IRB KY2013-017–01) and all patients signed an informed consent form [13]. Another validation dataset, GSE16011 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE16011), included RNA expression microarray data of 268 glioma patients and corresponding clinical information.
Estimate
The ESTIMATE algorithm was used to predict tumor purity, tumor invasion stroma/immune cells [14]. Using RNA sequencing data, ESTIMATE outputs three scores: stromal score, immune score and estimate score. Patients from TCGA dataset were divided into high or low score groups based on the median immune or stromal score. The differentially expressed genes (DEGs, p < 0.001 and foldchange > 5 or < 0.2) between two groups were defined as immune DEGs or stromal DEGs, respectively. Immune DEGs or stromal DEGs were eventually merged into TME-related genes.
Development of TME-related signature (TMERS)
Cox regression analysis was used to screen out TME-related genes associated with prognosis in TCGA dataset (p < 0.05). Least absolute shrinkage and selection operator (LASSO) regression algorithm [15] generated the Cox model with the minimum mean cross-validated error based on tenfold cross-validation. Our model included 20 genes and the corresponding LASSO Cox coefficients. TME-related signature (TMERS) of each patient was calculated using a linear combination of signature gene expression weighted by their LASSO Cox coefficients: TMERS = (exprgene1 x coefficientgene1) + (exprgene2 x coefficientgene2) + … + (exprgene20 x coefficientgene20). The coefficients from TCGA dataset were used to calculate TMERS for two validation datasets.
DAVID bioinformatics resources and Gene Set Enrichment Analysis (GSEA)
TMERS positively or negatively related genes (Pearson |R|> 0.6, p < 0.05) were input in DAVID functional annotation tool [16], and Gene ontology (GO) terms were selected to identify enriched biological themes. Gene set enrichment analysis (GSEA) was performed with R Package “fgsea” and top 15 pathways were selected. The number of permutations was 10,000 and the enrichment was significant when |NES|> 1 and adjusted p-value < 0.05.
Single-Cell RNA sequencing
We collected two samples of primary IDH-wildtype glioblastoma patients from Beijing Tiantan Hospital, Capital Medical University. Fresh glioma samples taken during the operation were immediately washed with phosphate-buffered saline (PBS), completely dissociated and enzymatically digested with trypsin–EDTA solution. We passed the dissociated cells through a cell strainer (BD), then suspended the cells in red blood cell lysis buffer (Solarbio) to lyse the red blood cells. After washing with PBS, the cell pellets were re-suspended for further analysis. According to the manufacture’s introduction, the cDNA libraries were constructed with Chromium Single Cell 3′ Library and Gel Bead kit v2 (120,267, 10x Genomics). The libraries were sequenced on Illumina HiSeq platform and the raw data was preprocessed with Cell Ranger pipeline (v3.0.2, 10x Genomics), mapped to the hg19 reference genome. Cells in gene-cell matrices with fewer than 200 transcripts and genes with fewer than two counts in two cells were filtered and removed. The matrix was then normalized such that the number of unique molecular identifiers (UMIs) in each cell was equal to the median UMI count across the dataset and log-transformed. Gene-barcode matrices were analyzed with the R package “Seurat” [17]. Based on QC metrics in the standard preprocessing workflow for scRNA-seq data, we filtered cells that had unique feature counts over 2000 or less than 200, UMI counts over 7000 and > 5% mitochondrial counts. To reduce the gene expression matrix to its most important features, we used principal component analysis (PCA) to decrease the dimensionality of the dataset. To visualize data in 2-D space, we passed the PCA-reduced data into UMAP (uniform manifold approximation and projection), a non-linear dimensional reduction method. We inferred and visualized copy number variation in cells with R packages “infercnv”.
Statistical analysis
Statistical analyses were mainly conducted with R software (v4.0.0, https://www.r-project.org/). Prognostic significance was evaluated with Kaplan–Meier survival curve by two-sided log-rank test and differences in clinical features were assessed by Student’s t test or Chi-square test. R packages “survival” were used for univariate and multivariate Cox regression analysis. R package “timeROC” was used to draw the one-, three- and five-year ROC curve [18]. A combination of independent prognostic indicators was used to construct the nomogram model with R package “rms”. Other R packages used in the study included ComplexHeatmap, gglpot2, pheatmap, Hmisc, corrgram and circlize. A p value < 0.05 was considered statistically significant.
Results
Tumor microenvironment (TME) characterization of gliomas
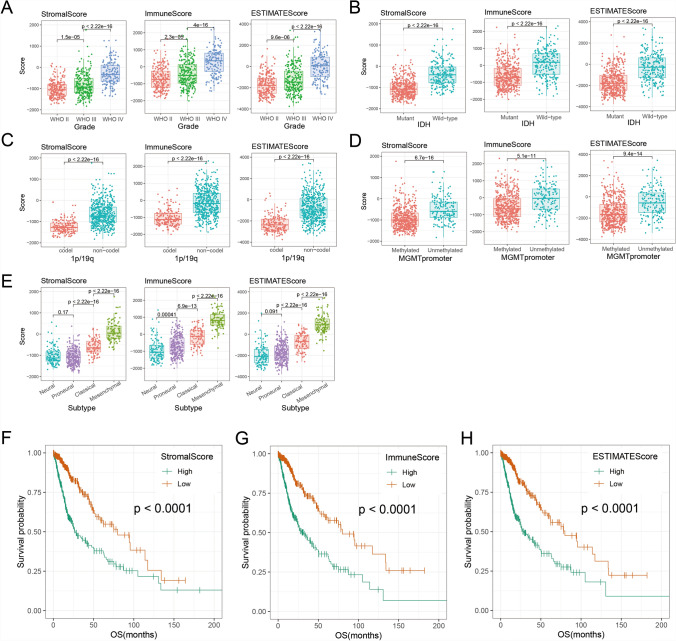
To investigate the tumor microenvironment (TME) of gliomas, ESTIMATE algorithm was performed to reflect the infiltration of stromal and immune cell. We included 702 RNA sequencing data of gliomas from TCGA dataset and calculated stromal score, immune score and estimate score. In Fig. 1A, three scores increased significantly in higher-grade glioma (p < 0.05). In addition, three scores were significantly higher in glioma with IDH wild-type, 1p/19q non-codeletion or MGMT promoter unmethylation (Fig. 1B–D, p < 0.05). In different molecular subtypes, three scores were significantly upregulated in classical and mesenchymal subtypes (Fig. 1E, p < 0.05). Furthermore, survival analysis revealed that patients with high score lived significantly shorter than patients with low score (Fig. 1F–H, p < 0.05). These results suggested that TME was associated with glioma malignancy and poor prognosis.
Fig. 1.
Evaluation of tumor microenvironmental status in gliomas with ESTIMATE algorithm. The stromal score, immune score and estimate score showed different distribution patterns in WHO grades (A), IDH mutation status (B), 1p/19q codeletion status (C), MGMT promoter methylation status (D) and molecular subtypes (E). The KM survival curve revealed that patients with high stromal score (F), immune score (G) or estimate score (H) lived significantly shorter than patients with low score (p < 0.0001, log-rank test)
Establishment of TMERS in gliomas
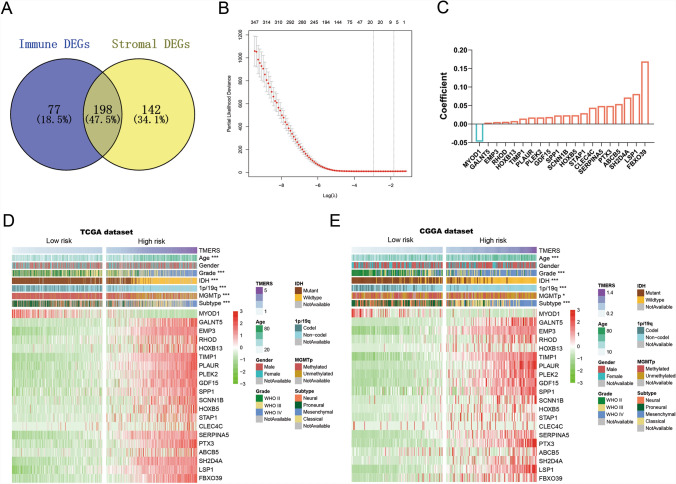
Considering the correlation between TME and glioma prognosis, we intended to construct a TME-related signature for prognosis prediction. In TCGA training dataset, we screened 340 stromal differentially expressed genes (DEGs) and 275 immune DEGs between high and low score gliomas (Fig. 2A, fold change > 5 or < 0.2, p < 0.001). Stromal and immune DEGs merged into 417 TME-related genes. Next, univariate Cox regression analysis further screened out 411 TME-related genes associated with overall survival (p < 0.05). Finally, the LASSO Cox regression algorithm was performed and 20 genes were selected with best prognostic value (Fig. 2B). A TME-related signature (TMERS) consisting of 20 genes was identified and the risk score was calculated using a linear combination of signature gene expression weighted by their regression coefficients (Fig. 2C). In CGGA and GSE16011 validation dataset, TMERS of each patient was calculated with the same regression coefficients from TCGA dataset. Based on the median TMERS, patients were divided into TMERS-low and TMERS-high groups, and we compared the clinical and molecular differences between two groups in TCGA dataset (Fig. 2D and Table 1). TMERS-high patients were significantly older than TMERS-low patients (p < 0.001) and more TMERS-high patients tend to be higher-grade (p < 0.001). Compared with TMERS-low patients at the molecular level, more patients were IDH wild-type, 1p/19q intact and MGMT promoter unmethylated in TMERS-high patients (p < 0.001). Meanwhile, more classical and mesenchymal subtypes were enriched in TMERS-high groups (p < 0.001). In CGGA and GSE16011 validation dataset, we observed consistent differences between TMERS-high and TMERS-low groups (Fig. 2E, Figure S1 and Table 1).
Fig. 2.
Identification of TME-related signature. A Venn diagram showed the differentially expressed genes (DEGs) of immune score and stroma score. B In LASSO regression analysis, partial likelihood deviance plot showed cross-validation for tuning parameter screening. C 20 genes were screened and corresponding regression coefficients in LASSO regression. Heatmaps showed the clinical and pathological differences between low risk and high risk patients in TCGA dataset (D) and CGGA dataset (E)
Table 1.
Correlation between TMERS and clinicopathological factors of glioma patients in the three datasets
| Characteristics | TCGA dataset | CGGA dataset | GSE16011 dataset | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Low-risk group (n = 351) | High-risk group (n = 351) | P-Value | Low-risk group (n = 162) | High-risk group (n = 163) | P-Value | Low-risk group (n = 134) | High-risk group (n = 134) | P-Value | |
| Age | |||||||||
| Mean(range) | 41 (17–74) | 54 (21–89) | < 0.001 | 39 (10–75) | 47 (8–81) | < 0.001 | 46 (15–81) | 56 (14–79) | < 0.001 |
| Gender | |||||||||
| Female | 126 | 129 | n.s | 65 | 57 | n.s | 46 | 42 | n.s |
| Male | 176 | 178 | 97 | 106 | 88 | 92 | |||
| NA | 49 | 44 | 0 | 0 | 0 | 0 | |||
| Grade | |||||||||
| II | 175 | 41 | < 0.001 | 92 | 13 | < 0.001 | 21 | 3 | < 0.001 |
| III | 127 | 114 | 39 | 37 | 64 | 21 | |||
| IV | 0 | 152 | 31 | 113 | 49 | 110 | |||
| NA | 49 | 44 | 0 | 0 | 0 | 0 | |||
| IDH status | |||||||||
| Mutant | 336 | 92 | < 0.001 | 141 | 35 | < 0.001 | 57 | 23 | < 0.001 |
| Wildtype | 5 | 229 | 21 | 128 | 51 | 85 | |||
| NA | 10 | 30 | 0 | 0 | 26 | 26 | |||
| 1p/19q status | |||||||||
| Codel | 150 | 19 | < 0.001 | 62 | 5 | < 0.001 | 39 | 7 | < 0.001 |
| Non-codel | 193 | 302 | 97 | 153 | 33 | 55 | |||
| NA | 8 | 30 | 3 | 5 | 62 | 72 | |||
| MGMT promoter | |||||||||
| Methylated | 317 | 160 | < 0.001 | 90 | 68 | < 0.05 | NA | NA | |
| Unmethylated | 26 | 136 | 62 | 87 | NA | NA | |||
| NA | 8 | 55 | 10 | 8 | NA | NA | |||
| TCGA subtype | |||||||||
| Proneural | 190 | 48 | < 0.001 | 85 | 17 | < 0.001 | 87 | 8 | < 0.001 |
| Neural | 74 | 37 | 65 | 16 | 17 | 9 | |||
| Classical | 0 | 86 | 11 | 63 | 22 | 36 | |||
| Mesenchymal | 0 | 95 | 1 | 67 | 8 | 81 | |||
| NA | 87 | 85 | 0 | 0 | 0 | 0 | |||
Associations of TMERS with pathological features in gliomas
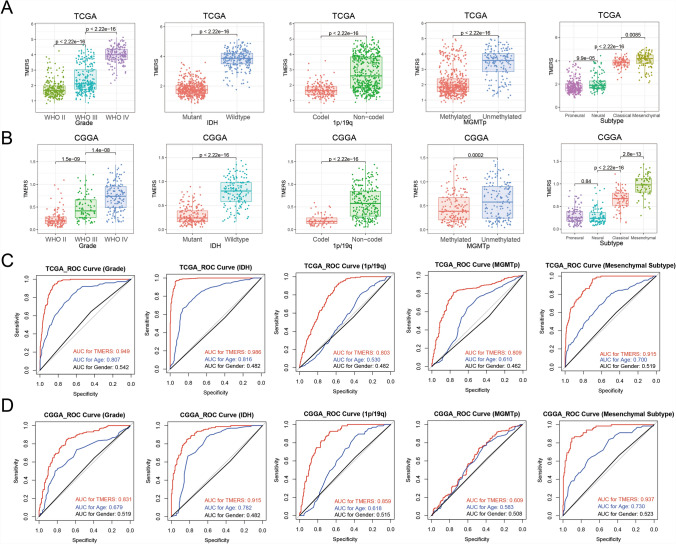
Due to heterogeneity of different pathological types of glioma, TMERS was analyzed according to WHO grade and molecular pathology (Fig. 3A). Compared with WHO grade II and grade III glioma, TMERS was the highest in grade IV glioma (p < 2.22e–16). Moreover, TMERS was significantly higher in patients with IDH wild-type, 1p/19q non-codeletion or MGMT promoter unmethylation (p < 2.22e–16). Among molecular subclasses of gliomas defined by TCGA network, TMERS was dramatically upregulated in mesenchymal subtype (p < 0.05). In two validation datasets, the distribution of TMERS was consistent with the above results (Fig. 3B and Figure S2A). Then receiver operating characteristic (ROC) curves were used to evaluate the diagnostic ability of TMERS to distinguish true states of pathological features in gliomas (Fig. 3C, D). Our TMERS showed excellent predictive ability in glioma grade (AUC 0.949 or 0.831 in TCGA or CGGA), IDH mutation status (AUC 0.986 or 0.915), 1p/19q co-deletion status (AUC 0.803 or 0.859), MGMT promoter methylation status (AUC 0.809 or 0.609) and mesenchymal subtype (AUC 0.915 or 0.937), which was better than age and gender. And in GSE16011 dataset, we observed similar results (Figure S2B). In brief, these findings emphasized that TMERS was closely associated with glioma malignancy and served as a biomarker for pathological features.
Fig. 3.
Associations between TMERS and pathological features. In TCGA dataset (A) and CGGA dataset (B), patients were grouped by WHO grade, IDH mutation status, 1p/19q codeletion status, MGMT promoter methylation status, TCGA molecular subtype. In TCGA dataset (C) and CGGA dataset (D), the ROC curve evaluated the predictive value of TMERS in pathological features
TMERS was associated with different genomic variations
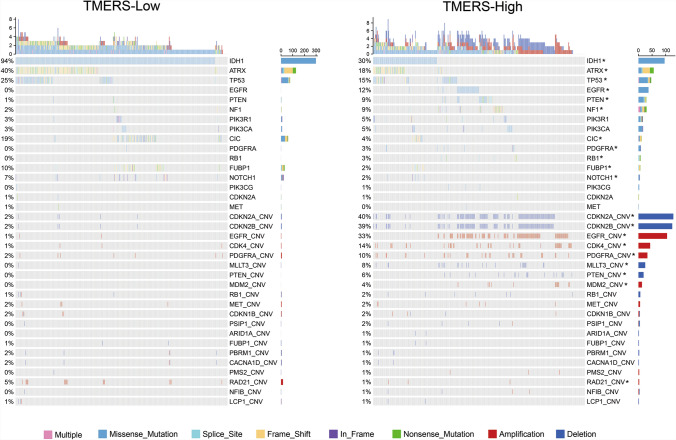
To better understand the molecular mechanisms of glioma microenvironment, we analyzed the difference in genomic alterations between TMERS-high and TMERS-low groups of TCGA dataset (Fig. 4). Mutations in IDH1, ATRX, TP53, CIC and FUBP1 were significantly enriched in TMERS-low group (p < 0.05, Chi-square test). Meanwhile, higher-frequency of mutations in EGFR, PTEN, NF1, PDGFRA, RB1 and NOTCH1 were found in TMERS-high group. GISTIC2.0 analysis showed distinct copy number alterations (CNA) between TMERS-high and TMERS-low groups. The most commonly deleted regions like CDKN2A/B, MLLT3, PTEN or amplified regions like EGFR, CDK4, PDGFRA, MDM2 were enriched in TMERS-high group (Fig. 4, p < 0.05, Chi-square test). These results suggested that gliomas with high TMERS tended to have more frequent genomic variations.
Fig. 4.
Genomic variation analysis based on TMERS grouping. Differential somatic mutations and copy number alternations analyses between TMERS-low and TMERS-high group. Chi-square test, *p < 0.05
Prognostic value of TMERS in gliomas
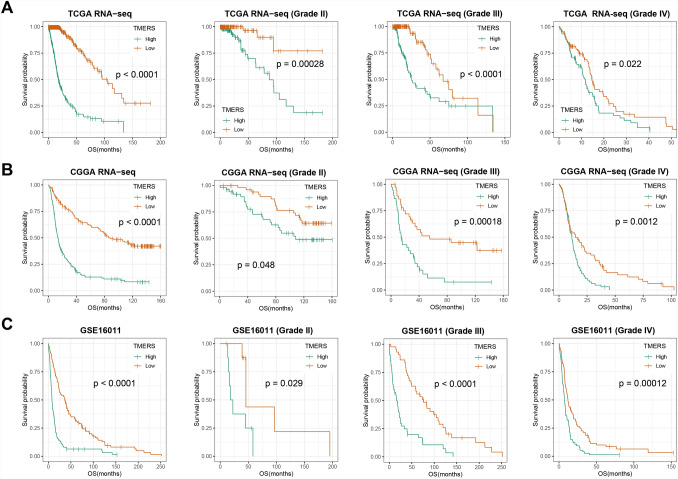
In consideration of correlation between TMERS and clinical and genetic features, we further assessed the prognostic value of TMERS. TMERS-high patients lived significantly shorter than TMERS-low patients in all three glioma datasets (Fig. 5, p < 0.0001). We then validated the prognostic value of TMERS in WHO grade II gliomas (p < 0.05), grade III gliomas (p < 0.05) and grade IV GBMs (p < 0.05). According to 2016 CNS tumor classification guidelines, gliomas were classified into five subtypes based on histopathology and IDH mutation and 1p/19q codeletion status. Overall survival of TMERS-high patients was significantly shorter than TMERS-low patients in LGG-IDHmut-codel (p < 0.05), LGG-IDHwt (p < 0.05) and GBM-IDHwt (p < 0.05, Figure S3). However, in LGG-IDHmut-non-codel and GBM-IDHmut, there was no significant difference in survival between the two groups. In addition, univariate and multivariate Cox regression analysis showed TMERS as an independent prognostic indicator by adjusting for clinicopathological variables (age, gender, grade, IDH mutation, 1p/19q codeletion and MGMT promoter methylation, Table 2).
Fig. 5.
Clinical prognostic value of TMERS. Kaplan–Meier survival analysis were performed in gliomas and different grades. A TCGA dataset, B CGGA dataset, C GSE16011 dataset
Table 2.
Variables related to OS in gliomas: univariate and multivariate analysis
| TCGA | Univariate Cox Regression | Multivariate Cox Regression | ||||
|---|---|---|---|---|---|---|
| HR | 95%CI | p Value | HR | 95%CI | p Value | |
| Age (≥ 45 vs. < 45) | 5.142 | 3.571–7.403 | < 2e–16* | 2.461 | 1.472–4.115 | 0.0006* |
| Gender (male vs. female) | 1.001 | 0.743–1.347 | 0.997 | |||
| Grade (GBM vs. LGG) | 9.576 | 6.835–13.420 | < 2e–16* | 1.392 | 0.860–2.252 | 0.178 |
| IDH (wild vs. mutant type) | 11.070 | 7.772–15.770 | < 2e–16* | 0.992 | 0.460–2.139 | 0.984 |
| 1p/19q (non-codel vs. codel) | 4.541 | 2.671–7.719 | 2.28e–08* | 1.420 | 0.741–2.724 | 0.291 |
| MGMT promoter (unmethylated vs. methylated) | 3.207 | 2.312–4.447 | 2.88e–12* | 1.199 | 0.818–1.756 | 0.352 |
| TMERS (high vs.low) | 3.629 | 3.083–4.273 | < 2e–16* | 2.714 | 1.873–3.933 | 1.32e–07* |
| CGGA | Univariate Cox Regression | Multivariate Cox Regression | ||||
|---|---|---|---|---|---|---|
| HR | 95%CI | p Value | HR | 95%CI | p Value | |
| Age (≥ 45 vs. < 45) | 2.006 | 1.532–2.627 | 4.2e–07* | 1.304 | 0.968–1.757 | 0.080 |
| Gender (male vs. female) | 0.998 | 0.759–1.312 | 0.988 | |||
| Grade (GBM vs. LGG) | 4.919 | 3.670–6.593 | < 2e–16* | 2.519 | 1.801–3.522 | 6.64e–08* |
| IDH (wild vs. mutant type) | 2.866 | 2.171–3.782 | 1.05e–13* | 0.669 | 0.442–1.013 | 0.058 |
| 1p/19q (non-codel vs. codel) | 5.877 | 3.602–9.588 | 1.33e–12* | 3.252 | 1.924–5.496 | 1.06e–05* |
| MGMT promoter (unmethylated vs. methylated) | 1.195 | 0.911–1.566 | 0.199 | |||
| TMERS (high vs.low) | 8.733 | 5.951–12.810 | < 2e–16* | 4.036 | 2.258–7.215 | 2.51e–06* |
| GSE16011 | Univariate Cox Regression | Multivariate Cox Regression | ||||
|---|---|---|---|---|---|---|
| HR | 95%CI | p Value | HR | 95%CI | p Value | |
| Age (≥ 45 vs. < 45) | 2.377 | 1.790–3.157 | 2.16e–09* | 2.224 | 1.387–3.568 | 0.0009* |
| Gender (male vs. female) | 1.066 | 0.811–1.401 | 0.647 | |||
| Grade (GBM vs. LGG) | 3.131 | 2.353–4.166 | 4.92e–15* | 1.105 | 0.634–1.927 | 0.725 |
| IDH (wild vs. mutant type) | 1.930 | 1.423–2.618 | 2.34e–05* | 1.615 | 0.993–2.626 | 0.053 |
| 1p/19q (non-codel vs. codel) | 2.445 | 1.645–3.633 | 9.68e–06* | 1.559 | 0.931–2.611 | 0.091 |
| TMERS (high vs.low) | 10.234 | 6.401–16.360 | < 2e–16* | 4.833 | 1.697–13.770 | 0.003* |
| HR, hazard ratio; CI, confidence interval; *Significant | ||||||
An individualized prognostic model based on TMERS
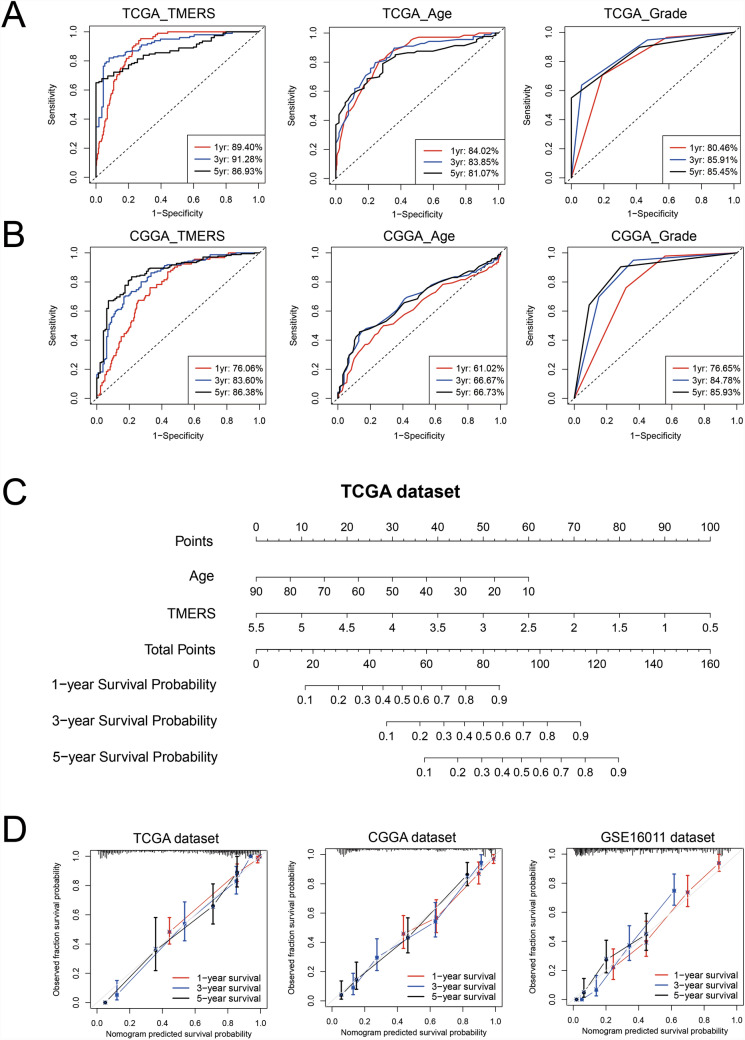
With the “timeROC” algorithm, 1-year, 3-year and 5-year ROC curves were used to evaluate the predictive efficacy of TMERS. The 1-year, 3-year and 5-year AUC of TMERS were 0.8940, 0.9128 and 0.8693, respectively, superior to age (0.8402, 0.8385 and 0.8107) and grade (0.8046, 0.8591 and 0.8545) (Fig. 6A). And in two validation datasets, the predictive efficacy of TMERS was also superior to that of age and grade (Fig. 6B and Figure S4). Next, we developed a nomogram prognostic model with independent prognostic indicators (age and TMERS) (Fig. 6C). The C-indices of our model were 0.872, 0.766 and 0.738 for TCGA, CGGA and GSE16011 datasets, respectively. The calibration curve also showed the satisfactory ability of nomogram model to predict 1-year, 3-year and 5-year survival in three datasets (Fig. 6D).
Fig. 6.
A clinical survival prediction model based on TMERS in gliomas. In TCGA dataset (A) and CGGA dataset (B), the timeROC curve evaluated the predictive value of TMERS, age and grade in 1-year, 3-year and 5-year overall survival. (C) A nomogram model integrating TMERS and age in TCGA dataset. (D) Calibration curves for the nomogram model predicting 1-year (red), 3-year (blue) and 5-year (black) survival in three datasets
TMERS related biological functions and microenvironment analysis
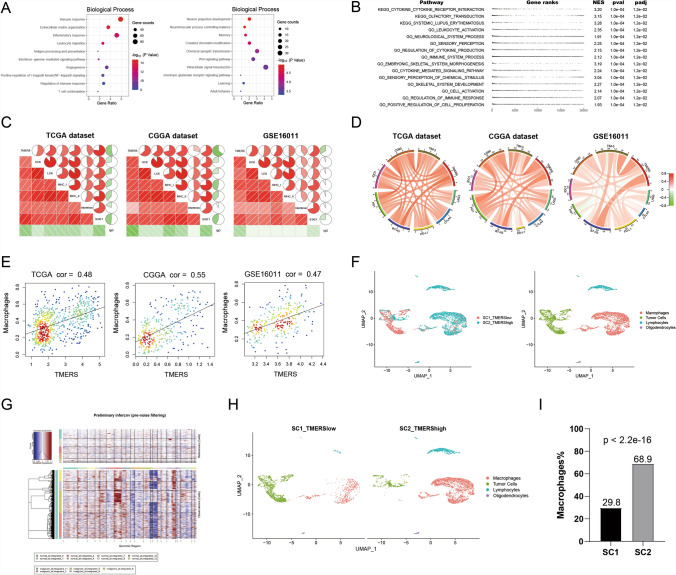
To reveal biological characteristics of TMERS, DAVID functional annotation tool was performed. First, genes related to TMERS (1528 positively and 882 negatively related genes, Pearson |R|> 0.6, p < 0.05) were screened with Pearson correlation analysis in TCGA dataset. In Gene Ontology analysis, positively related genes were enriched in the biological processes of “immune response”, “Extracellular matrix organization”, “Inflammatory response”, “Leukocyte migration”, “Angiogenesis” and so on (Fig. 7A). And negatively related genes were enriched in the biological processes of normal neural function, such as “Neuron projection development”, “Neuromuscular process controlling balance”, “Memory”, “Covalent chromatin modification”, and “Chemical synaptic transmission”. Meanwhile, GSEA analysis showed that pathways including “KEGG_cytokine_cytokine_receptor_interaction” (NES = 3.20, padj = 1.2e − 02), “GO_leukocyte_activation” (NES = 2.35, padj = 1.2e − 02) and “GO_immune_system_process” (NES = 2.12, padj = 1.2e − 02) were enriched in TMERS-high group (Fig. 7B).
Fig. 7.
Functional annotation and immunological analysis of TMERS. A Biological processes analysis of genes positively or negatively associated with TMERS using DAVID functional annotation tool. B Gene set enrichment analysis (GSEA) revealed the top 15 pathways which were enriched in TMERS-high group. C Correlation analysis of TMERS and inflammatory activities in three datasets. D Correlation analysis of TMERS and immune checkpoints in three datasets. E Correlation analysis of TMERS and macrophage proportions by CIBERSORT in three datasets. F Two single-cell RNA-sequencing samples (SC1 was TMERS-low and SC2 was TMERS-high) were integrated, and all cells were labeled as four clusters: macrophages (red), tumor cells (green), lymphocytes (blue), oligodendrocytes (purple). G The InferCNV heatmap showed single cell chromosome copy number amplification (red) or deletion (blue). The upper part was reference normal cells, the lower part was malignant cells. H SC1 (left) and SC2 (right) in the UMAP plot. I The proportion of macrophages in SC1 was significantly lower than that in SC2 (p < 2.2e-16, chi-squared test)
In order to understand the relationship between TMERS and inflammatory response, we analyzed seven inflammatory metagenes using the method described previously [19, 20]. As shown in Fig. 7C, TMERS was positively correlated with HCK, LCK, MHC-I, MHC-II, interferon and STAT1, while it showed a negative correlation with IgG in all three datasets. These findings indicated an enhanced inflammatory response involving macrophages, T lymphocytes and antigen-presenting cells, but not B lymphocyte responses in TMERS-high patients. Meanwhile, TMERS was involved in immune response, and we included immune checkpoints and immune cells for further study. As shown in Fig. 7D, immune checkpoints (LAG3, CTLA4, PD-L1, PD1, B7-H3, IDO1, CD80, TIM-3) were positively correlated with TMERS in three datasets, indicating immunosuppressive status in TMERS-high patients. To assess abundances of immune cell, we imputed mRNA expression data in CIBERSORT analytical tool and observed a positive correlation between TMERS and macrophage abundance (Fig. 7E). In order to better verify the macrophage abundance, single-cell RNA sequencing (scRNA-seq) was performed on two IDH-wildtype glioblastomas (SC1 was TMERS-low and SC2 was TMERS-high). All cells were clustered into four groups, including macrophages, tumor cells, lymphocytes and oligodendrocytes (Fig. 7F). The InferCNV algorithm showed significant chromosomal copy number variation in “malignant” cells, confirming our subgroup classification of cells (Fig. 7G). Separately, the proportion of macrophages in SC1 was significantly lower than that in SC2 (p < 2.2e-16, Fig. 7H, I). In addition to the difference in proportions, we further compared the differences in the phenotype of macrophages. We found that the expression of anti-inflammatory factors in the macrophages of SC2 was significantly increased, while the expression of pro-inflammatory factors in the macrophages of SC2 was significantly lower than that in SC1 (p < 0.05, Figure S5). These results revealed that macrophages in gliomas with high TMERS may promote tumor growth by restraining inflammation.
Discussion
TME is a crucial component of tumor. Recently, with the realization of the essential role of TME in the evolution of tumor, the previous tumor cell-centered treatment has gradually turned to the treatment of complex tumor systems. Some well-known targets in TME, such as PD-1 and CTLA-4, inhibit the activation of T cells in TME. Currently, antibodies targeting PD-1 or CTLA-4 have shown significant tumor-suppressive effects in some solid tumors, such as advanced melanoma [21, 22], non-small-cell lung cancer [23, 24], and so on. However, the efficacy of PD-1 or CTLA-4 blockade therapy was limited in gliomas [7, 8], and the current immunotherapy strategies for gliomas need to be improved urgently. Our study has integrated 1295 RNA sequencing data of whole grade gliomas and evaluated the characteristics of the glioma tumor microenvironment. Then we developed a TME-related signature (TMERS), which was significantly up-regulated in higher grade gliomas, IDH wild-type gliomas, 1p/19q intact gliomas, MGMT promoter unmethylated gliomas and mesenchymal subtype. These results revealed that TMERS was closely related to malignant progression of glioma.
Then based on TMERS grouping, we analyzed the genomic variation of gliomas. Mutations in IDH1, ATRX, TP53, CIC and FUBP1 were significantly enriched in TMERS-low group. IDH mutations are present in more than 80% of lower-grade gliomas and secondary glioblastomas [25]. IDH mutation is an early and stable mutation event [26], which plays an important role in maintaining the growth of glioma cells [27]. Mutations in ATRX and TP53 were reported to drive gliomas towards astrocytoma phenotype [28], while mutations in CIC and FUBP1 were driver events for oligodendrogliomas [29]. In TMERS-high group, higher-frequency of mutations in EGFR, PTEN, NF1, PDGFRA, RB1 and NOTCH1 was found. EGFR, PTEN, NF1 and PDGFRA mainly affected the RTK-RAS-PI3K pathway, which controlled the most basic cellular functions including cell cycle, proliferation, differentiation, migration, and survival [30]. Alterations in RTK-RAS-PI3K pathways were common in glioma malignant progression [31]. The protein encoded by RB1 gene could negatively regulate cell cycle and mutation in RB1 induced the occurrence of various cancers [32]. NOTCH1 gene belonged to the NOTCH family, which was involved in the Notch signaling pathway. Notch signaling pathway was associated with cancer cell survival and drug resistance [33]. In addition to the significant association between gene mutations and TMERS, more copy number alterations were found in TMERS-high group, including the most commonly deleted regions like CDKN2A/B, MLLT3, PTEN or amplified regions like EGFR, CDK4, PDGFRA, MDM2. Therefore, genomic variation might be the root cause of the TME differences in gliomas.
We further evaluated the prognostic value of TMERS by Kaplan–Meier survival curve and Cox regression analysis. Compared with the traditional indicators (age and grade), the ROC curve showed that TMERS had a better prediction effect on 1-year, 3-year and 5-year overall survival. Based on the superior prognosis and predictive value of TMERS, we combined TMERS with age to construct a nomogram model for survival prediction, implying the clinical practice value of TMERS.
Next, biological function analysis suggested that TMERS could effectively reflect the status of glioma tumor microenvironment, including immune and inflammatory response, extracellular matrix, angiogenesis and cytokines. We then included seven inflammatory metagenes and observed that TMERS was primarily associated with the enhanced inflammatory response involving macrophages, T lymphocytes and antigen-presenting cells, but not B lymphocytes. To better understand the relationship between TMERS and immune response, common immune checkpoint genes were selected. Correlation analysis showed a positive correlation between TMERS and immune checkpoints, indicating that TMERS reflected immunosuppression in gliomas. Immune cells were involved in the regulation of tumor immune response, and our study showed that macrophages mediated immune response were involved TMERS-high group of glioma patients. The majority of non-tumor cells in the microenvironment are macrophages, which support tumor survival and invasion. At present, the interaction between glioma cells and macrophages has been extensively studied. Peiwen Chen et al. found that LOX secreted by glioma cells functioned as a potential macrophage chemoattractant, while SPP1 secreted by macrophages could support the survival of glioma cells and stimulate angiogenesis. Inhibition of LOX expression significantly down-regulated macrophage infiltration and glioma progression [34]. Salvatore J Coniglio et al. revealed that CSF-1 secreted by glioma cell lines stimulated microglia infiltration, while EGF secreted by microglia stimulated glioma cell migration. Blocking the CSF-1R signaling pathway reduced microglial infiltration and glioma invasion [35]. Symbiotic interactions between glioma cells and macrophages offer new opportunities for targeted therapy.
In recent years, research focusing on the tumor microenvironment has become more and more intense, and TME-related signatures, including immune-related signatures, have emerged by a large margin. Qiu et al. constructed a prognostic microenvironment-related immune signature via ESTIMATE for glioma, but the prediction accuracy of this model (AUCs > 0.6) needs to be improved [36]. Zhang et al. built a immune-related gene signature that was effective in risk stratification in primary lower-grade gliomas, but did poorly in predicting survival in recurrent lower-grade gliomas [37]. In a recent study by Xia et al., immune-related lncRNAs were screened and eleven immune-related lncRNAs were adopted to construct the prognostic signature [38]. These studies have reflected the close correlation between tumor microenvironment (including immune components) and tumor development and clinical prognosis. In-depth study of the tumor microenvironment is conducive to the discovery of prognostic markers and the development of novel therapeutic targets.
Conclusions
In conclusion, we evaluated the characteristics of the glioma microenvironment and identified a TME-related signature (TMERS) that could predict the prognosis of glioma patients and reflect the status of glioma microenvironment. More prospective studies, including validation in immunized populations, could further explore the potential value of TMERS. TMERS may help us understand the glioma microenvironment and develop immunotherapy strategies for gliomas.
Supplementary Information
Below is the link to the electronic supplementary material.
Author contributions
QWW and YJZ directed and supervised the study. QWW performed most of the experiments and wrote the original draft. ZSB and TJ participated in study design, discussed, and interpreted the data.
Funding
This study was funded by Provincial Key R&D Program, Science and Technology Department of Zhejiang Province (Grant No. 2017C03018); Key Program of Administration of Traditional Chinese Medicine, Zhejiang Province (No.2018ZZ015); National Natural Science Foundation of China (NSFC)/Research Grants Council (RGC) Joint Research Scheme (81761168038); Beijing Municipal Administration of Hospitals’ Mission Plan (SML20180501); National Natural Science Foundation of China (81902337; 82002647; 81902528).
Availability of data and materials
The authors confirm the data that has been used in this work is available on reasonable request.
Declarations
Conflict of interest
The authors have no conflicts of interest to declare.
Ethical approval
The authors have no ethical conflicts to disclose.
Informed consent
All patients were provided written consent for the research or molecular diagnostic use of residual tumor tissues obtained during their surgical treatment.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Jiang T, Nam DH, Ram Z, et al. Clinical practice guidelines for the management of adult diffuse gliomas. Cancer Lett. 2021;499:60–72. doi: 10.1016/j.canlet.2020.10.050. [DOI] [PubMed] [Google Scholar]
- 2.Stupp R, Hegi ME, Mason WP, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459–466. doi: 10.1016/s1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 3.Hinshaw DC, Shevde LA. The Tumor Microenvironment Innately Modulates Cancer Progression. Can Res. 2019;79:4557–4566. doi: 10.1158/0008-5472.can-18-3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Binnewies M, Roberts EW, Kersten K, Chan V. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nature Med. 2018;24:541–550. doi: 10.1038/s41591-018-0014-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Topalian SL, Drake CG, Pardoll DM. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell. 2015;27:450–461. doi: 10.1016/j.ccell.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pitt JM, Vétizou M, Daillère R, et al. Resistance Mechanisms to Immune-Checkpoint Blockade in Cancer: Tumor-Intrinsic and -Extrinsic Factors. Immunity. 2016;44:1255–1269. doi: 10.1016/j.immuni.2016.06.001. [DOI] [PubMed] [Google Scholar]
- 7.Zhao J, Chen AX, Gartrell RD, et al. Immune and genomic correlates of response to anti-PD-1 immunotherapy in glioblastoma. Nature Med. 2019;25:462–469. doi: 10.1038/s41591-019-0349-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Filley AC, Henriquez M, Dey M. Recurrent glioma clinical trial, CheckMate-143: the game is not over yet. Oncotarget. 2017;8:91779–91794. doi: 10.18632/oncotarget.21586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao Z, Meng F, Wang W, Wang Z, Zhang C, Jiang T. Comprehensive RNA-seq transcriptomic profiling in the malignant progression of gliomas. Scientific data. 2017;4:170024. doi: 10.1038/sdata.2017.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Q, Wang Z, Bao Z, Zhang C, Wang Z, Jiang T. PABPC1 relevant bioinformatic profiling and prognostic value in gliomas. Future Oncol. 2020;16:4279–4288. doi: 10.2217/fon-2019-0268. [DOI] [PubMed] [Google Scholar]
- 11.Yan W, Zhang W, You G, et al. Correlation of IDH1 mutation with clinicopathologic factors and prognosis in primary glioblastoma: a report of 118 patients from China. PLoS ONE. 2012;7:e30339. doi: 10.1371/journal.pone.0030339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang QW, Wang YW, Wang ZL, Bao ZS. Clinical and molecular characterization of incidentally discovered lower-grade gliomas with enrichment of aerobic respiration. Onco Targets Therapy. 2020;13:9533–9542. doi: 10.2147/ott.s248623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang QW, Liu HJ, Zhao Z, Zhang Y, Wang Z, Jiang T, Bao ZS. Prognostic correlation of autophagy-related gene expression-based risk signature in patients with glioblastoma. Onco Targets Therapy. 2020;13:95–107. doi: 10.2147/ott.s238332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoshihara K, Shahmoradgoli M, Martínez E, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao J, Kwan PW, Shi D. Sparse kernel learning with LASSO and Bayesian inference algorithm Neural networks : the official journal of the International. Neural Network Society. 2010;23:257–264. doi: 10.1016/j.neunet.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 16.da Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 17.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Q, Wang Z, Li G, Zhang C, Bao Z, Wang Z, You G, Jiang T. Identification of IDH-mutant gliomas by a prognostic signature according to gene expression profiling. Aging. 2018;10:1977–1988. doi: 10.18632/aging.101521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Z, Zhang C, Liu X, et al. Molecular and clinical characterization of PD-L1 expression at transcriptional level via 976 samples of brain glioma. Oncoimmunology. 2016;5:e1196310. doi: 10.1080/2162402x.2016.1196310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rody A, Holtrich U, Pusztai L, et al. T-cell metagene predicts a favorable prognosis in estrogen receptor-negative and HER2-positive breast cancers. Breast cancer research : BCR. 2009;11:R15. doi: 10.1186/bcr2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Topalian SL, Sznol M, McDermott DF, et al. Survival, durable tumor remission, and long-term safety in patients with advanced melanoma receiving nivolumab. J Clin Oncol: Off J Am Soc Clin Oncol. 2014;32:1020–1030. doi: 10.1200/jco.2013.53.0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Larkin J, Chiarion-Sileni V, Gonzalez R, et al. Combined Nivolumab and Ipilimumab or monotherapy in untreated melanoma. N Engl J Med. 2015;373:23–34. doi: 10.1056/NEJMoa1504030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brahmer J, Reckamp KL, Baas P, et al. Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N Engl J Med. 2015;373:123–135. doi: 10.1056/NEJMoa1504627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garon EB, Rizvi NA, Hui R, et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med. 2015;372:2018–2028. doi: 10.1056/NEJMoa1501824. [DOI] [PubMed] [Google Scholar]
- 25.Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watanabe T, Nobusawa S, Kleihues P, Ohgaki H. IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am J Pathol. 2009;174:1149–1153. doi: 10.2353/ajpath.2009.080958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rohle D, Popovici-Muller J, Palaskas N, et al. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science. 2013;340:626–630. doi: 10.1126/science.1236062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu XY, Gerges N, Korshunov A, et al. Frequent ATRX mutations and loss of expression in adult diffuse astrocytic tumors carrying IDH1/IDH2 and TP53 mutations. Acta Neuropathol. 2012;124:615–625. doi: 10.1007/s00401-012-1031-3. [DOI] [PubMed] [Google Scholar]
- 29.Wesseling P, van den Bent M, Perry A. Oligodendroglioma: pathology, molecular mechanisms and markers. Acta Neuropathol. 2015;129:809–827. doi: 10.1007/s00401-015-1424-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103:211–225. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- 31.Bai H, Harmancı AS, Erson-Omay EZ, et al. Integrated genomic characterization of IDH1-mutant glioma malignant progression. Nat Genet. 2016;48:59–66. doi: 10.1038/ng.3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dyson NJ. RB1: a prototype tumor suppressor and an enigma. Genes Dev. 2016;30:1492–1502. doi: 10.1101/gad.282145.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aster JC, Pear WS, Blacklow SC. The Varied Roles of Notch in Cancer. Annu Rev Pathol. 2017;12:245–275. doi: 10.1146/annurev-pathol-052016-100127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen P, Zhao D, Li J, et al. Symbiotic Macrophage-Glioma Cell Interactions Reveal Synthetic Lethality in PTEN-Null Glioma. Cancer Cell. 2019;35:868–84.e6. doi: 10.1016/j.ccell.2019.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Coniglio SJ, Eugenin E, Dobrenis K, Stanley ER, West BL, Symons MH, Segall JE. Microglial stimulation of glioblastoma invasion involves epidermal growth factor receptor (EGFR) and colony stimulating factor 1 receptor (CSF-1R) signaling. Mol Med. 2012;18:519–527. doi: 10.2119/molmed.2011.00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qiu H, Li Y, Cheng S, Li J, He C, Li J. A prognostic microenvironment-related immune signature via estimate (promise model) predicts overall survival of patients with glioma. Front Oncol. 2020;10:580263. doi: 10.3389/fonc.2020.580263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang M, Wang X, Chen X, Zhang Q, Hong J. Novel immune-related gene signature for risk stratification and prognosis of survival in lower-grade glioma. Front Genet. 2020;11:363. doi: 10.3389/fgene.2020.00363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xia P, Li Q, Wu G. An immune-related lncRNA signature to predict survival in glioma patients. Cell Mol Neurobiol. 2021;41:365–375. doi: 10.1007/s10571-020-00857-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm the data that has been used in this work is available on reasonable request.