Abstract
Recent developments in cancer immunotherapy promise better outcomes for cancer patients, although clinical trials for difficult to treat cancers such as malignant brain cancer present special challenges, showing little response to first generation immunotherapies. Reasons for differences in immunotherapy response in some cancer types are likely due to the nature of tumor microenvironment, which harbors multiple cell types which interact with tumor cells to establish immunosuppression. The cell types which appear to hold the key in regulating tumor immunosuppression are the tumor-infiltrating immune cells. The current standard treatment for difficult to treat cancer, including the most malignant brain cancer, glioblastoma, continues to offer a bleak outlook for patients. Immune-profiling and correlation with pathological and clinical data will lead to a deeper understanding of the tumor immune microenvironment and contribute toward the selection, optimization and development of novel precision immunotherapies. Here, we review the current understanding of the tumor microenvironmental landscape in glioblastoma with a focus on next-generation technologies including multiplex immunofluorescence and computational approaches to map the brain tumor microenvironment to decipher the role of the immune system in this lethal malignancy.
Keywords: Tumor microenvironment, Immune cells, Glioblastoma, Glioma, Multiplex immunohistochemistry, Computational deconvolution
Introduction
The tumor immune microenvironment as an anti-cancer target
The tumor microenvironment comprises a heterogeneous milieu of cells including immune cells, termed the immune contexture, which has been shown to be intimately involved in carcinogenesis and tumor progression [1, 2]. Furthermore, the temporal–spatial dynamics of phenotypic densities of the tumor immune microenvironment is associated with an improved prognosis and therapeutic response in a range of solid tumors [3–7]. However, in tumor types that have been historically considered “immune privileged”, such as prostate cancer and glioblastoma (GBM), the response rates to immunotherapies have been disappointing [8, 9], emphasizing the need to comprehensively phenotype the tumor immune microenvironment of these tumor types beyond immune-checkpoint expression and T cell infiltration.
Inhibition of immune checkpoints has revolutionized the field of cancer immunotherapy, rejuvenated interest in the dynamics between malignant cells and the immune system and are now considered as first-line therapy in stage III and IV non-small lung cancer [10]. However, akin to targeted therapies, only a specific subset of patients are eligible, and indeed, about 20% of eligible patients derive benefit from these relatively expensive therapies. In those patients who are eligible, a subset exhibit de novo resistance, while others develop resistance over the course of treatment [11–13]. De novo resistance at the onset of treatment cannot be predicted based on PD-L1 expression status alone, highlighting the unmet clinical need for the identification and validation of new biomarkers to enhance precision immuno-oncology [14].
Tumor immunoscore
Traditional tumor staging by pathologists report a patient’s tumor burden based on many histopathological hallmarks, including the presence of cancer cells in draining and regional lymph nodes, evidence for metastases, mitotic index, the extent of invasion of primary tumor cells into surrounding healthy tissue and the level of tumor-specific biomarker expression. However, the recognition that clinical outcomes varied significantly among patients with the same diagnosis led to the notion that non-tumor cell components of the tumor should be considered. Given the abundance of intra-tumor immune cells in many cancer types, inclusion of immunological biomarkers has been examined as a tool for the prediction of prognosis and response to therapy. Accumulating data, collected from large cohorts of cancer patients, have demonstrated the importance of immune classification in prognosis and increasingly, prediction of response to specific immunotherapies. Measurement of immune cell number and type has been termed the ‘immunoscore’ and is increasingly being used for some cancer types, including colon cancer, in combination with traditional pathological classification [1, 15]. Immunoscore typically measures the extent of T-cell infiltration but research suggests that the measurement of multiple immune cell types will improve the understanding of tumor biology to enhance prognostic and predictive pathology. By focusing on glioblastoma (GBM) as an example of a highly heterogeneous and treatment resistant cancer, we review the properties of the tumor microenvironment and discuss the mechanisms which contribute to the establishment of immunosuppression and how immunotherapies are evolving to tackle these incurable diseases. We also describe key technologies being used to decipher the complex cellular and molecular interactions which play a role in tumor development and immunosuppression.
Cancer immunotherapy
Recent developments in novel immunotherapy approaches, including immune-checkpoint inhibitors, targeting the T-cell receptor, programmed cell death 1 (PD-1) [16], and cognate transmembrane ligands expressed in cancer cells, PD-1 ligand, programmed death ligand-1 (PD-L1) [16] and -2 (PD-L2) [17], have shown efficacy in GBM preclinical animal models [18, 19] but limited efficacy in human clinical trials [20, 21]. For example, a phase III randomized clinical trial using a PD-1 monoclonal antibody, Nivolumab, showed a non-significant effect compared to the anti-angiogenic drug, bevacizumab [22]. Another immunotherapy which has shown promising results in several cancers, uses genetically engineered T cells expressing antigen-specific chimeric antigen receptors (CAR-T) which are infused into a patient and is referred to as CAR-T adoptive transfer. To date, CAR-T therapies show some efficacy in blood cancers but little or no efficacy in various solid tumors tested [23, 24]. In GBM, CAR-T targeting the human epidermal growth factor receptor-2 (HER2) [25], epidermal growth factor receptor variant III (EGFRvIII) [26, 27] and interleukin-13 receptor α2 (IL-13Rα2) [28] have shown promise in preclinical animal models but failed in human clinical trials, where failure was attributed to the lack of sufficient CAR T-cell expansion and loss of target antigen, in vivo [25–28].
Failure of specific treatments is due to many factors. A long-standing issue in brain diseases treatment relying on systemically delivered therapeutics is the presence of the blood brain barrier (BBB), a unique feature of the central nervous system (CNS) where the blood vessel endothelial cells form robust tight junctions with neural cells. Another likely reason for the limited efficacy of immunotherapy for GBM is the immunosuppressive tumor microenvironment, first recognized as immune tolerance in cancer patients receiving radiation therapy [29]. Moreover, like most tumor types, the GBM microenvironment is dynamic which changes during tumor development and undergoes significant changes in response to therapy [30]. At a cellular level, much of the adaptation can be seen as changes in intra-tumor immune cell composition. Histopathological examination of GBM tissue using immune-specific antibodies has revealed the presence of diverse immune cell types. However, an understanding of immune cell composition and correlation with specific pathological, patient, and clinical data is only just beginning. As more sophisticated reagents and tools are developed to examine the GBM tissue microenvironment, a deeper understanding of the role of the brain tumor immune microenvironment in GBM development and immunosuppression will ultimately contribute toward the optimization of precision immunotherapies. The convergence of the development of highly specific antibodies targeting immune cell antigens, new generation multi-spectral microscopes techniques and machine learning is now being applied to investigating and understanding the immune cell composition in the GBM immune microenvironment.
The brain tumor microenvironment
The central nervous system (CNS) is not as immune privileged as previously thought, although difficulties remain for the integration of immunotherapies into the treatment paradigm for CNS cancers. Therapeutic interventions such as vaccines, monoclonal antibodies and targeted therapies have had little success in treating a range of brain diseases, including neurodegenerative disease and cancer [31–33]. Primary brain malignancies are characterized by the development of highly proliferative, invasive tumors that spread but are almost universally restricted to the brain and spine. The deadliest and most prevalent primary brain tumor in adults is glioblastoma (GBM), classified as a grade IV astrocytoma [34], which are both classified as gliomas, a broader reference to the group of brain cancers enriched in glial cells. GBM incidence is 3 per 100,000 people, and although patients of any age are affected, almost 80% of patients are over 55 years [35]. The current standard treatment for GBM patients employs maximal safe tumor resection, followed by radiotherapy and concomitant administration of temozolomide (TMZ), a DNA alkylating agent, resulting in a mean overall survival of 14.6 months [36]. Despite continual development of new anti-cancer therapies for non-brain cancers, including quantum leap advances such as immunotherapy, meaningful advances in brain cancer treatment have been limited, with only incremental improvement in the overall survival of GBM patients, over many decades. Significant advances in treating this disease require research aimed at a deeper understanding of the mechanisms involved in regulating brain cancer biology, including brain tumor immunity.
Gliomas are one of the most immunosuppressive solid tumors due in part to lymphopenia driven by bone marrow suppression [37]. GBM tumors are immunologically quiet, and exhibit low tumor mutational burden (TMB) [38, 39], few tumor-infiltrating T cells (TILS) [33], low expression of immune-checkpoint inhibitors, PD-1/PD-L1 [33], and a high density of immunosuppressive tumor-associated macrophages [40]. The standard treatment for GBM, which uses radiation therapy and alkylating chemotherapy, confounds this immunosuppression. In addition, the use of potent immunosuppressive drugs such as corticosteroids, for the management of peritumoral edema following surgery, interferes with the efficacy of immunotherapies and needs to be considered in this context. In addition to the hurdles for stimulating an effective immune response against GBM, robust immune activation within the intracranial space poses clinical safety risks, including complications of cytokine release syndrome, which can trigger robust systemic inflammation and autoimmune encephalitis. This proposes that any immunotherapy needs to establish a narrow, highly controlled response, balancing pro- and anti-inflammatory activation.
The GBM tumor microenvironment, GBM subtypes and relevance to diagnosis and therapy
Treatment of these tumors, thus, requires a clear and concise understanding of the patient’s tumor, which in the context of fast-developing technologies, pathologists and oncologists can leverage, to enable accurate diagnoses and selection of combination therapies tailored towards the specific patient tumor microenvironment [41]. DNA sequencing and gene expression analysis of GBM and low-grade glioma (LGG) via several landmark cancer genome atlas (TCGA) studies [42] has revealed that GBM can be classified into four distinct subtypes, proneural (PN), neural (N), classical (CL) and mesenchymal (MES). Patients with proneural and neural enriched tumor subtypes have a superior survival compared to the classical and mesenchymal subtypes [43, 44]. Data from the cancer genome atlas (TCGA) molecular signatures were applied to GBM samples only, the N and MES subtype had a similar survival rate, while the PN subtype remained the most prognostically significant [43].
A number of studies have identified that mesenchymal GBM is associated with an increased density of tumor-associated macrophages, T cells (CD3), cytotoxic T cells (CD8), T-helper cells (CD4) and FoxP3+ T-regulatory cells (Tregs) [30, 42, 45]. Paradoxically, the patients with mesenchymal GBM subtype tumors show no survival advantage exhibit nor benefit from immunotherapy [46]. With no apparent prognostic benefit from an increased immune cell infiltration, it is likely that the tumor cytokine and chemokine milieu and infiltrating immune cells, especially macrophages, have immuno-regulatory and immunosuppressive functions to enable the proliferation and invasion of the tumor cells into adjacent normal tissue [47].
The tumor microenvironment in recurrent GBM
GBM tumors are characterized by complex molecular and cellular heterogeneity in both tumor cells and the microenvironment. GBM cells undergo clonal selection processes during therapy, followed by treatment-induced mutations, manifested in recurrent GBM [48] and the resistant surviving cells ultimately contribute to recurrence. In most GBM patients, recurrence occurs within 6–8 months of primary therapy and the prognosis is poor. Surgery for recurrent GBM is challenging, since the recurrent tumor often occurs in multiple brain regions and while treatment with irradiation is possible, the risk of neuro-toxicity is high [49], highlighting the difficulties in managing relapsed GBM patients. Inhibiting tumor angiogenesis by targeting the vascular endothelial growth factor-A (VEGF-A) with bevacizumab has resulted in little improvement in patient overall survival, but some improvement in progression-free survival [32, 50, 51]. Recent trials have focused on using bevacizumab in combination with other drugs, including immunomodulatory drugs, exemplified by a study testing bevacizumab in combination with granulocyte–macrophage colony-stimulating factor (GM-CSF) and cyclophosphamide which resulted in survival benefit with minimal toxicity [52].
Comparing the immune cell profile between primary and recurrent GBM tumors can provide an insight into how treatment influences various immune cell related parameters including degree of immune cell infiltration, density and spatial distribution within the tumor microenvironment. A higher proportion of activated CD4+ T cells were present in recurrent GBM [53], whereas another study showed that TIL infiltration is highly variable and that the immune cell profile correlates with GBM subtype, rather than whether they are primary or recurrent tumors [54]. These studies compared primary and recurrent GBM samples from different patients. Due to patient to patient differences, the ideal comparison of tumor microenvironment changes, will be to compare the primary and recurrent brain cancer tumors from the same patients. Transcriptomic comparison of both the primary and recurrent GBM tumor-associated immune microenvironment [55], using the computational algorithm, CIBERSORT [56], showed GBM type-specific immune cell profile changes. By contrast, primary PN GBM exhibited higher fractions of several immune cell types compared with recurrent PN GBM, suggesting no net immune infiltration in PN GBM, upon recurrence [55].
The conclusions drawn from these studies must be considered with caution, given that the studies compare primary and recurrent GBM samples from different patients and the samples are unmatched, therefore not accounting for patient to patient variability. Further research investigating the tumor microenvironment in matched primary and recurrent GBM tumor samples from the same patients will more accurately determine the evolution of the GBM tumor microenvironment.
One of the complications in understanding the mechanisms involved in the evolution of the GBM microenvironment is that, typically, all patients with recurrent GBM will have had surgery, radiotherapy and temozolomide chemotherapy; treatment which will have an impact on the tumor microenvironment, including inflammation and genetic hypermutation [55, 57, 58]. Thus, investigating the therapeutic impact on the GBM microenvironment has begun to reveal specific cellular and molecular changes which occur in response to radiation and anti-cancer drugs.
Therapeutic impact on the GBM immune microenvironment
Standard therapy in GBM patients has been reported to cause lymphopenia, which is characterized by reduced T- and B-cell numbers, although this is not the case for all patients. Following surgery, radiation and chemotherapy, 96 patients with high-grade glioma (HGGs) experienced a drop in CD4+ cells, indicative of immunosuppression and lymphopenia [59]. However, other studies show that although lymphopenia was observed at early stages following radiotherapy and/or TMZ administration, a gradual normalization of the immune system occurred over time [60, 61]. The current data, therefore, suggest that although lymphopenia occurs at early stages following treatment, there is a gradual increase in TIL infiltration over time, providing the opportunity for immunotherapy, including T-cell mediated immunotherapy.
Aside from causing lymphopenia, TMZ contributes to hypermutation. In GBM, 17% of recurrent cases exhibit hypermutation after TMZ treatment, compared with no hypermutation in untreated patient tumor tissue [57]. Hypermutated recurrent GBM tumors exhibit CD8+ T-cell enrichment [55], likely due to mutation-induced immunogenicity. Radiation also enhances T-cell infiltration in GBM [62]. Treatment with bevacizumab for patients with recurrent GBM is also associated with increased levels of CD11b+ tumor-associated macrophages and correlates with poor patient outcome [63]. Tumor-associated macrophage infiltration may be associated with activation of pro-inflammatory signals by bevacizumab, which has been reported following intraocular bevacizumab injection for the treatment of ocular choroidal neovascularization and in cancer cell lines [64, 65].
Mapping the tumor immune microenvironment
Understanding the functional relationship of tumor and immune cells in the brain tumor immune microenvironment requires analytical methods which can identify tumor cells and the different immune cell types, and their spatial relationship. For example, the cell type on which PD-L1 is expressed is associated with immune-checkpoint inhibitor response [66, 67]; thus, PD-L1 expression on macrophages could be used as a biomarker to predict the efficacy of anti-PD-L1 therapy, thereby emphasizing the clinical need for assays that can differentiate between tumor cells, immune cells and the localization of immune-checkpoint expression.
To better understand the cellular and molecular mechanisms underlying the tissue pathology, contextual information through mapping tumor cell composition can be captured through multiplex labeling of biomarkers identifying individual cell types. Use of sophisticated image analysis software can help decipher complex spatial relationships and reveal functional mechanisms at play, to complement genomic information. To accomplish this level of tumor tissue analysis, the development and validation of optimized, informative assays, that go beyond “global tissue biomarker profiles” to “local tissue biomarker patterns” that will enable the visualization and interpretation of intra- and inter-heterogeneity is required. Furthermore, the application of artificial intelligence, convolutional neural networks and machine learning is also to quantify and spatially resolve the relationships between multiple cell types in the tumor. These high-dimensional, data-driven approaches will address the issue of the limited tissue availability, particularly in brain cancer, where biopsy collection is typically only possible during surgery.
In situ evaluation of the tumor immune microenvironment: immunohistochemistry
Since the introduction of immunohistochemistry (IHC) in the 1970s, this technique has revolutionized both diagnostic and translational histopathology. Immunohistochemistry remains a popular methodology for the detection, visualization of tissue proteins in a compartmentalized manner and can be used for patient stratification in diagnostic, therapeutic and/or genetic contexts [68].
Spatial mapping of tumor and tumor-associated cells has largely relied on immunohistochemistry (IHC). Although IHC allows localization of cells in tissue, the number of antibodies identifying different cell types on single tumor tissue sections is limited. With the development of more fluorophores, primary detection antibodies raised in different host species and microscopes with the capacity for high-resolution imaging, it is now possible to routinely perform multiplex IHC on single tissue sections. High-throughput automated tissue staining and image scanning and acquisition, combined with sophisticated image analysis software is now possible. Several integrated platforms are established and, here, we will describe our experience using these to map the brain tumor immune microenvironment.
Multiplex immunohistochemistry
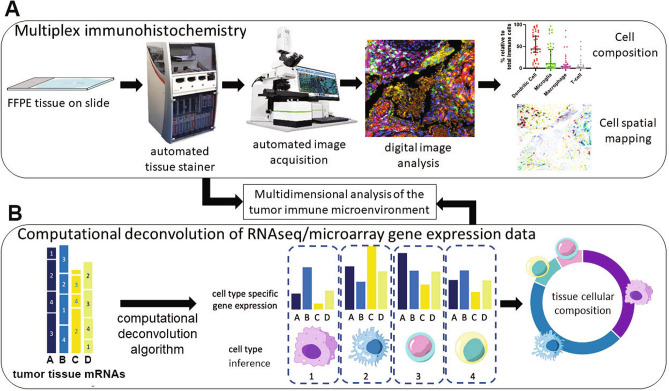
Multiplexing is possible, usually up to three antibodies, typically requiring iterative/repeated labeling. Recent advances and use of automated tissue stainers have allowed faster and higher throughput multiplex immunohistochemical analysis of tissue. Multiple iterative labeling, by sequential multiplex labeling increases the number of antibodies which can be used on a single tissue section. This type of technique has been used to characterize the tumor microenvironment in central nervous system lymphoma and link specific tumor immune cells with immune-checkpoint expression [69]. Opal is one of the recently developed methods allowing robust high-throughput automated multiplex immunohistochemical analysis of formalin-fixed paraffin-embedded (FFPE) tissue [70] (Fig. 1). The technique relies on sequential multiplexing technology using reactive fluorophores that covalently label the target epitope coupled with signal amplification and no cross-reactivity, resulting in minimal or no background signal. Using a microscope and multispectral camera, up to seven individual discrete colors can be detected. An example of the immune and tumor cell types which can be identified in GBM tumor tissue, using multiplex IHC, is shown in Fig. 2a. Cell phenotyping, spatial plotting and proximity analysis of each immune cell subset in relation to tumor cells can build an immune cell infiltration profile for specific tumor regions and histopathological markers in the tumors, including blood vessels, core and peripheral tumor zones [71–73]. Biomarkers for specific tumor characteristics, including hypoxia and cell signaling activation can provide further information on the biological relationship between specific cell types and other biomarkers.
Fig. 1.
Mapping of the tumor immune microenvironment. a Multiplex Immunohistochemistry (mIHC). The process flow for multiplex immunohistochemical staining and analysis of tumor tissue using Opal™ technology. Formalin-fixed paraffin-embedded (FFPE) tissue is stained in an automated tissue processor and stainer, where multiple predetermined primary antibodies are used in a series of incubation and washing steps, followed by signal amplification. The slides are scanned on a multispectral microscope and images are acquired. The images are then analyzed using morphometric software to enumerate specific cell types and spatial proximity analysis to determine the physical location in two dimensions and the location of any cell type with any other cell type or histopathological biomarker. b Computational deconvolution applied to gene expression data. Illustration of the deconvolution of heterogeneous mRNAs derived from tumor tissue expression analysis, typically RNA-sequencing. From the left, gene expression of genes A, B, C and D represents all mRNAs expressed in the tumor bulk, including all cell types 1, 2, 3, 4. Following computational deconvolution, the cell types present in the tumor sample can be inferred and the proportion of each cell type in the tissue can be estimated, based on the input of cell gene expression matrices from previous experiments (e.g., single-cell mRNA sequencing). Data from mIHC analysis and computational deconvolution can be combined to map the cellular content and physical distribution of cells in the tumor microenvironment
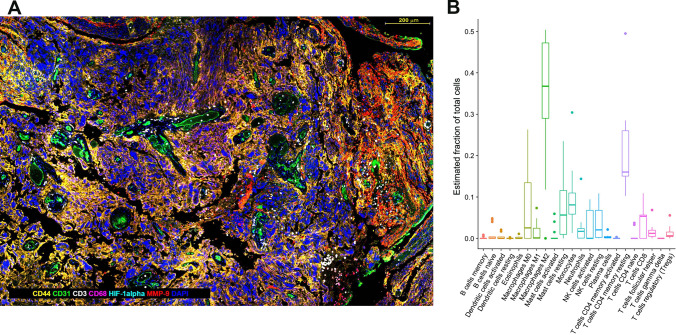
Fig. 2.
a Multiplex IHC using opal immunohistochemistry. GBM (WHO grade IV astrocytoma) tissue was stained with antibodies against CD44 labels tumor cells; CD31 labels endothelial cells (blood vessels); CD68, labels macrophages; HIF-1alpha, labels hypoxic cells; MMP-9 labels matrix metalloproteinase-9-producing cells; DAPI labels cell nuclei. Software-aided image analysis allows all single- and multiplex-labeled cells to be identified and counted, and the relative distances between cell types and histopathological hallmarks to be measured. Scale bar is 200 μm. b Immune cell composition in GBM inferred by computational deconvolution using CIBERSORT. GBM RNAseq data from the 168 GBM patient samples (The Cancer Genome Atlas (TCGA) [35]), was analyzed using the CIBERSORT algorithm to estimate the number of immune cells in GBM. Each data point represents an individual patient tumor sample
While multiplex immunohistochemistry is a significant advance in high-resolution tissue biomarker analysis, spatial transcriptomics to identify gene expression at single-cell level can help identify dynamic interactions between tumor and immune cells, which can help reveal functional changes in these cells. The development of spatial transcriptomic analytical methods highlights the rapid advances in technology which can be used to examine tumor tissue at high resolution. Preliminary data using spatial transcriptomics demonstrates the spatial heterogeneity of T-cell exhaustion in GBM, which correlates with GBM subtype-specific marker expression [74]. One of the limitations of current spatial transcriptomics is the low resolution [75]. However, by combining and integrating spatial transcriptomic data with multiplex immunohistochemistry data, a deeper understanding of the functional interactions between cells in the GBM microenvironment can be achieved. An example of integrating spatial gene expression data and single-cell sequencing has shown a distinct spatial relationship between tumor cells, macrophages, dendritic cells and inflammatory fibroblasts in pancreatic ductal adenocarcinoma [76]. Another example combining multiple analytical approaches to investigate dynamic changes of the tumor microenvironment in response to experimental immunotherapies in breast cancer mouse models used single-cell mass cytometry, single-cell sequencing and computational scaffold mapping to construct an interaction map of cells [77].
Computational deconvolution of gene expression data
In contrast to in situ tissue analysis such as IHC, gene expression profiling of large tumor sample numbers is possible using RNA-sequencing (RNA-seq) data. The Cancer Genome Atlas (TCGA) glioma and GBM cohort have genomic data from about 1100 patient tumors, with associated clinical information for each sample, including treatment, tumor grade and patient survival. How can the cellular composition from bulk tumor sample mRNA mixtures be interrogated? Computational tools have been developed to deconvolute mRNA sequence data, using gene signatures, where each gene signature, of between 10 and 20 genes, identifies specific cell types. Combined with statistical analyses, the cellular composition of the tissue from which the mRNA was derived can be inferred (Fig. 1b).
Computational tools have been developed to estimate cell-type composition using bulk gene expression data. The ability to perform computational deconvolution of complex mRNA mixtures is possible due to advances in cell prediction accuracy, achieved by combining pre-existing computational frameworks with machine learning algorithms. Machine learning, which is a form of artificial intelligence, allows analysis of specific tasks without explicit instructions, relying on patterns and inference to predict for example, cell identity. One of the most widely used computational deconvolution tools currently being used is CIBERSORT [56]. CIBERSORT uses an input matrix of reference gene expression signatures to deconvolve mRNA mixtures to calculate the relative proportions of specific cell types, combined with a machine learning approach. CIBERSORT is considered to be one of the most robust methods for resolving closely related cell subsets in tissues with complex mixtures of unknown cell types [56], a feature especially important in solid cancers, including brain tumors, where analysis is performed on a highly heterogeneous tissue.
To investigate the tumor immune microenvironment, the CIBERSORT algorithm uses pre-defined immune cell-specific gene expression signature matrices, derived from 22 different immune cell types, including B-cell, T-cell, dendritic cell and monocyte cell subtypes, as well as various stromal cell types. The analytical capacity of computational interrogation of gene expression data is exemplified in a landmark study investigating more than 10,000 tumors, across 33 different cancer types, where CIBERSORT was one of the key tools used to investigate the cancer immune landscape [78]. Other studies have defined activation-specific gene signatures in immune cells to investigate the role of immune cell subtype-specific antitumor mechanisms, including research focusing on growth factor-induced natural killer (NK) cell activation in GBM [79]. An example analysis of TCGA GBM RNAseq data using CIBERSORT (Fig. 2b), shows that 22 cell types can be identified and the proportions of each can be predicted revealing that M2 macrophages and resting CD4 memory T cells are the most abundant cell types.
Since computational approaches rely on pre-determined gene expression matrices to predict cellular identity, they are sensitive to experimental noise due to unknown cell mixture content and closely related cell types, limiting their utility for tumor immune microenvironment assessment. Using multiple computational tools and combining analysis with direct cell measurement using multiplex immunohistochemistry can provide a deeper characterization of the brain tumor immune microenvironment. For example, computational deconvolution analysis used in combination with single-cell genomic analysis examined the glioma immune microenvironment to show that NF1 gene deficiency correlates with increased tumor-associated macrophage number and that tumor-promoting M2 macrophages correlate with short-term relapse [30]. This study also demonstrated that recurrent GBM tumors exhibit a decrease in monocytes and a subtype-dependent increase in macrophages/microglia cells. The complexity of the tumor microenvironment can be further deciphered by integrating immunohistological evaluation, which has demonstrated the diversity of brain tumor immune microenvironment, particularly between primary and secondary GBMs, and brain metastases from different non-CNS primaries [80].
Another popular cell phenotyping computational tool developed recently is xCell [81], which performs cell-type enrichment analysis from gene expression data for 64 immune and stromal cell types, and has the ability to distinguish closely related cell types. The ability to identify more cell types comes with the trade-off of more noise and reduced certainty with respect to cell identification, although xCell has similar power to CIBERSORT, in defining immune cells based on gene expression profile (unpublished data); noting that CIBERSORT does not identify basophils, whereas xCell does, so algorithm selection should consider such differences. Both CIBERSORT and xCell have been used to investigate the tumor immune landscape from RNAseq data from 671 GBM patients, showing comparable results with respect to immune cell composition and that pro-tumorigenic M2 macrophages and resting CD4+ memory T cells are the most abundant cell types [82]. These computational tools are robust, as the same cell types were shown to be the most abundant in GBM from an independent analysis our laboratory has conducted (Fig. 2b).
Perspectives and future directions
Combining direct tissue analysis with computational analysis offers a powerful approach toward a better understanding of the nature of the brain tumor immune microenvironment and the clinical criteria associated with specific immune cell subtypes at diagnosis, during the disease progression, and during treatment. Ultimately, a deeper understanding of the tumor microenvironment and mechanisms regulating immunosuppression will provide opportunities for both optimal selection of existing immunotherapies and the development of novel brain cancer immunotherapies. Finally, if multidimensional analyses and technologies enabling these are not adopted, precision cancer immunotherapy, especially for difficult-to-treat cancers, will not be realized and attempts at re-purposing immunotherapies will continue to “bring owls to Athens”, in other words, be an exercise in futility.
Author contributions
SW, RAH and TM conceptualized the review. SW, YF, SM provided data for and prepared the figures. All authors contributed to the writing and editing of the review. All authors approved the final version.
Funding
We acknowledge the support of the CASS Foundation (Grant no. 28534), The Royal Melbourne Hospital Neuroscience Foundation, and the Brain Foundation, Australia. SW was supported by the Australia Awards Scholarship from the Department of Foreign Affairs and Trade of Australia. PKD was supported by a NHMRC Senior Research Fellowship (APP1136680).
Compliance with ethical standards
Conflict of interest
RAH has ongoing research collaborations with Definiens GmbH and Ultivue Inc.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Samuel S. Widodo and Ryan A. Hutchinson contributed equally.
References
- 1.Mlecnik B, Bindea G, Kirilovsky A, Angell HK, Obenauf AC, Tosolini M, et al. The tumor microenvironment and Immunoscore are critical determinants of dissemination to distant metastasis. Sci Transl Med. 2016;8:327ra26. doi: 10.1126/scitranslmed.aad6352. [DOI] [PubMed] [Google Scholar]
- 2.Mascaux C, Angelova M, Vasaturo A, Beane J, Hijazi K, Anthoine G, et al. Immune evasion before tumour invasion in early lung squamous carcinogenesis. Nature. 2019;571:570–575. doi: 10.1038/s41586-019-1330-0. [DOI] [PubMed] [Google Scholar]
- 3.Galon J. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006 doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 4.Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJM, Robert L, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515:568–571. doi: 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mlecnik B, Van den Eynde M, Bindea G, Church SE, Vasaturo A, Fredriksen T, et al. Comprehensive intrametastatic immune quantification and major impact of immunoscore on survival. J Natl Cancer Inst. 2018 doi: 10.1093/jnci/djx123. [DOI] [PubMed] [Google Scholar]
- 6.Van den Eynde M, Mlecnik B, Bindea G, Fredriksen T, Church SE, Lafontaine L, et al. The link between the multiverse of immune microenvironments in metastases and the survival of colorectal cancer patients. Cancer Cell. 2018;34(1012–1026):e3. doi: 10.1016/j.ccell.2018.11.003. [DOI] [PubMed] [Google Scholar]
- 7.Yang S, Liu T, Nan H, Wang Y, Chen H, Zhang X, et al. Comprehensive analysis of prognostic immune-related genes in the tumor microenvironment of cutaneous melanoma. J Cell Physiol. 2020;235:1025–1035. doi: 10.1002/jcp.29018. [DOI] [PubMed] [Google Scholar]
- 8.Beer TM, Kwon ED, Drake CG, Fizazi K, Logothetis C, Gravis G, et al. Randomized, double-blind, phase III trial of ipilimumab versus placebo in asymptomatic or minimally symptomatic patients with metastatic chemotherapy-naive castration-resistant prostate cancer. J Clin Oncol. 2017;35:40–47. doi: 10.1200/JCO.2016.69.1584. [DOI] [PubMed] [Google Scholar]
- 9.Reardon DA, Brandes AA, Omuro A, Mulholland P, Lim M, Wick A, et al. Effect of nivolumab vs bevacizumab in patients with recurrent glioblastoma: the checkmate 143 phase 3 randomized clinical trial. JAMA Oncol. 2020 doi: 10.1001/jamaoncol.2020.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gadgeel SM, Stevenson JP, Langer CJ, Gandhi L, Borghaei H, Patnaik A, et al. Pembrolizumab and platinum-based chemotherapy as first-line therapy for advanced non-small-cell lung cancer: phase 1 cohorts from the KEYNOTE-021 study. Lung Cancer. 2018;125:273–281. doi: 10.1016/j.lungcan.2018.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Straussman R, Morikawa T, Shee K, Barzily-Rokni M, Qian ZR, Du J, et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature. 2012;487:500–504. doi: 10.1038/nature11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson TR, Fridlyand J, Yan Y, Penuel E, Burton L, Chan E, et al. Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature. 2012;487:505–509. doi: 10.1038/nature11249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Syn NL, Teng MWL, Mok TSK, Soo RA. De-novo and acquired resistance to immune checkpoint targeting. Lancet Oncol. 2017;18:e731–e741. doi: 10.1016/S1470-2045(17)30607-1. [DOI] [PubMed] [Google Scholar]
- 14.De Gramont A, Watson S, Ellis LM, Rodón J, Tabernero J, De Gramont A, et al. Pragmatic issues in biomarker evaluation for targeted therapies in cancer. Nat Rev Clin Oncol. 2015;12:197. doi: 10.1038/nrclinonc.2014.202. [DOI] [PubMed] [Google Scholar]
- 15.Galon J, Pagès F, Marincola FM, Angell HK, Thurin M, Lugli A, et al. Cancer classification using the immunoscore: a worldwide task force. J Transl Med. 2012;10:205. doi: 10.1186/1479-5876-10-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishida Y, Agata Y, Shibahara K, Honjo T. Induced expression of PD-1, a novel member of the immunoglobulin gene superfamily, upon programmed cell death. EMBO J. 1992 doi: 10.1002/j.1460-2075.1992.tb05481.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Latchman Y, Wood CR, Chernova T, Chaudhary D, Borde M, Chernova I, et al. PD-L2 is a second ligand for PD-1 and inhibits T cell activation. Nat Immunol. 2001;2:261. doi: 10.1038/85330. [DOI] [PubMed] [Google Scholar]
- 18.Wainwright DA, Chang AL, Dey M, Balyasnikova IV, Kim CK, Tobias A, et al. Durable therapeutic efficacy utilizing combinatorial blockade against IDO, CTLA-4, and PD-L1 in mice with brain tumors. Clin Cancer Res. 2014;20:5290–5301. doi: 10.1158/1078-0432.CCR-14-0514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zeng J, See AP, Phallen J, Jackson CM, Belcaid Z, Ruzevick J, et al. Anti-PD-1 blockade and stereotactic radiation produce long-term survival in mice with intracranial gliomas. Int J Radiat Oncol Biol Phys. 2013;86:343–349. doi: 10.1016/j.ijrobp.2012.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schalper KA, Rodriguez-Ruiz ME, Diez-Valle R, López-Janeiro A, Porciuncula A, Idoate MA, et al. Neoadjuvant nivolumab modifies the tumor immune microenvironment in resectable glioblastoma. Nat Med. 2019;25:470–476. doi: 10.1038/s41591-018-0339-5. [DOI] [PubMed] [Google Scholar]
- 21.Cloughesy TF, Mochizuki AY, Orpilla JR, Hugo W, Lee AH, Davidson TB, et al. Neoadjuvant anti-PD-1 immunotherapy promotes a survival benefit with intratumoral and systemic immune responses in recurrent glioblastoma. Nat Med. 2019;25:477–486. doi: 10.1038/s41591-018-0337-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Omuro A, Vlahovic G, Lim M, Sahebjam S, Baehring J, Cloughesy T, et al. Nivolumab with or without ipilimumab in patients with recurrent glioblastoma: results from exploratory phase I cohorts of CheckMate 143. Neuro Oncol. 2018;20:674–686. doi: 10.1093/neuonc/nox208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davenport AJ, Cross RS, Watson KA, Liao Y, Shi W, Prince HM, et al. Chimeric antigen receptor T cells form nonclassical and potent immune synapses driving rapid cytotoxicity. Proc Natl Acad Sci USA. 2018;115:E2068–E2076. doi: 10.1073/pnas.1716266115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mardiana S, Lai J, House IG, Beavis PA, Darcy PK. Switching on the green light for chimeric antigen receptor T-cell therapy. Clin Transl Immunol. 2019;8:e1046. doi: 10.1002/cti2.1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ahmed N, Brawley V, Hegde M, Bielamowicz K, Kalra M, Landi D, et al. HER2-specific chimeric antigen receptor-modified virus-specific T cells for progressive glioblastoma: a phase 1 dose-escalation trial. JAMA Oncol. 2017;3:1094–1101. doi: 10.1001/jamaoncol.2017.0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O’Rourke DM, Nasrallah MP, Desai A, Melenhorst JJ, Mansfield K, Morrissette JJD, et al. A single dose of peripherally infused EGFRvIII-directed CAR T cells mediates antigen loss and induces adaptive resistance in patients with recurrent glioblastoma. Sci Transl Med. 2017 doi: 10.1126/scitranslmed.aaa0984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goff SL, Morgan RA, Yang JC, Sherry RM, Robbins PF, Restifo NP, et al. Pilot trial of adoptive transfer of chimeric antigen receptor-transduced T cells targeting EGFRvIII in patients with glioblastoma. J Immunother. 2019;42:126–135. doi: 10.1097/CJI.0000000000000260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brown CE, Alizadeh D, Starr R, Weng L, Wagner JR, Naranjo A, et al. Regression of glioblastoma after chimeric antigen receptor T-cell therapy. N Engl J Med. 2016;375:2561–2569. doi: 10.1056/NEJMoa1610497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Craver LF. Tolerance to whole body irradiation of patients with advanced cancer. At Energy Biophys Biol Med. 1948;1:148. [PubMed] [Google Scholar]
- 30.Wang Q, Hu B, Hu X, Kim H, Squatrito M, Scarpace L, et al. Tumor evolution of glioma-intrinsic gene expression subtypes associates with immunological changes in the microenvironment. Cancer Cell. 2018;33:152. doi: 10.1016/j.ccell.2017.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holmes C, Boche D, Wilkinson D, Yadegarfar G, Hopkins V, Bayer A, et al. Long-term effects of Aβ42 immunisation in Alzheimer’s disease: follow-up of a randomised, placebo-controlled phase I trial. Lancet. 2008;372:216–223. doi: 10.1016/S0140-6736(08)61075-2. [DOI] [PubMed] [Google Scholar]
- 32.Gilbert MR, Dignam JJ, Armstrong TS, Wefel JS, Blumenthal DT, Vogelbaum MA, et al. A randomized trial of bevacizumab for newly diagnosed glioblastoma. N Engl J Med. 2014;370:699–708. doi: 10.1056/NEJMoa1308573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berghoff AS, Kiesel B, Widhalm G, Rajky O, Ricken G, Wöhrer A, et al. Programmed death ligand 1 expression and tumor-infiltrating lymphocytes in glioblastoma. Neuro Oncol. 2015;17:1064–1075. doi: 10.1093/neuonc/nou307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 35.DeAngelis LM. Brain tumors. N Engl J Med. 2001;344:114–123. doi: 10.1056/NEJM200101113440207. [DOI] [PubMed] [Google Scholar]
- 36.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJB, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 37.Chongsathidkiet P, Jackson C, Koyama S, Loebel F, Cui X, Farber SH, et al. Sequestration of T cells in bone marrow in the setting of glioblastoma and other intracranial tumors. Nat Med. 2018;24:1459–1468. doi: 10.1038/s41591-018-0135-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SAJR, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hodges TR, Ott M, Xiu J, Gatalica Z, Swensen J, Zhou S, et al. Mutational burden, immune checkpoint expression, and mismatch repair in glioma: implications for immune checkpoint immunotherapy. Neuro Oncol. 2017;19:1047–1057. doi: 10.1093/neuonc/nox026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Darmanis S, Sloan SA, Croote D, Mignardi M, Chernikova S, Samghababi P, et al. Single-cell RNA-Seq analysis of infiltrating neoplastic cells at the migrating front of human glioblastoma. Cell Rep. 2017;21:1399–1410. doi: 10.1016/j.celrep.2017.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smyth MJ, Ngiow SF, Ribas A, Teng MWL. Combination cancer immunotherapies tailored to the tumour microenvironment. Nat Rev Clin Oncol. 2016;13:143–158. doi: 10.1038/nrclinonc.2015.209. [DOI] [PubMed] [Google Scholar]
- 42.Verhaak RGW, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin N, Yan W, Gao K, Wang Y, Zhang J, You Y. Prevalence and clinicopathologic characteristics of the molecular subtypes in malignant glioma: a multi-institutional analysis of 941 cases. PLoS One. 2014;9:e94871. doi: 10.1371/journal.pone.0094871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Teo W-Y, Sekar K, Seshachalam P, Shen J, Chow W-Y, Lau CC, et al. Relevance of a TCGA-derived glioblastoma subtype gene-classifier among patient populations. Sci Rep. 2019;9:7442. doi: 10.1038/s41598-019-43173-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaffes I, Szulzewsky F, Chen Z, Herting CJ, Gabanic B, Velázquez Vega JE, et al. Human mesenchymal glioblastomas are characterized by an increased immune cell presence compared to proneural and classical tumors. Oncoimmunology. 2019;8:e1655360. doi: 10.1080/2162402X.2019.1655360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao J, Chen AX, Gartrell RD, Silverman AM, Aparicio L, Chu T, et al. Immune and genomic correlates of response to anti-PD-1 immunotherapy in glioblastoma. Nat Med. 2019;25:462–469. doi: 10.1038/s41591-019-0349-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.House IG, Savas P, Lai J, Chen AXY, Oliver AJ, Teo ZL, et al. Macrophage-derived CXCL9 and CXCL10 are required for antitumor immune responses following immune checkpoint blockade. Clin Cancer Res. 2020;26:487–504. doi: 10.1158/1078-0432.CCR-19-1868. [DOI] [PubMed] [Google Scholar]
- 48.Kim J, Lee I-H, Cho HJ, Park C-K, Jung Y-S, Kim Y, et al. Spatiotemporal evolution of the primary glioblastoma genome. Cancer Cell. 2015;28:318–328. doi: 10.1016/j.ccell.2015.07.013. [DOI] [PubMed] [Google Scholar]
- 49.van Linde ME, Brahm CG, de Witt Hamer PC, Reijneveld JC, Bruynzeel AME, Vandertop WP, et al. Treatment outcome of patients with recurrent glioblastoma multiforme: a retrospective multicenter analysis. J Neurooncol. 2017;135:183–192. doi: 10.1007/s11060-017-2564-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Verhoeff JJC, Lavini C, van Linde ME, Stalpers LJA, Majoie CBLM, Reijneveld JC, et al. Bevacizumab and dose-intense temozolomide in recurrent high-grade glioma. Ann Oncol. 2010;21:1723–1727. doi: 10.1093/annonc/mdp591. [DOI] [PubMed] [Google Scholar]
- 51.Chinot OL, Wick W, Mason W, Henriksson R, Saran F, Nishikawa R, et al. Bevacizumab plus radiotherapy-temozolomide for newly diagnosed glioblastoma. N Engl J Med. 2014;370:709–722. doi: 10.1056/NEJMoa1308345. [DOI] [PubMed] [Google Scholar]
- 52.Bota DA, Chung J, Dandekar M, Carrillo JA, Kong X-T, Fu BD, et al. Phase II study of ERC1671 plus bevacizumab versus bevacizumab plus placebo in recurrent glioblastoma: interim results and correlations with CD4+ T-lymphocyte counts. CNS Oncol. 2018;7:CNS22. doi: 10.2217/cns-2018-0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mohme M, Schliffke S, Maire CL, Rünger A, Glau L, Mende KC, et al. Immunophenotyping of newly diagnosed and recurrent glioblastoma defines distinct immune exhaustion profiles in peripheral and tumor-infiltrating lymphocytes. Clin Cancer Res. 2018;24:4187–4200. doi: 10.1158/1078-0432.CCR-17-2617. [DOI] [PubMed] [Google Scholar]
- 54.Rahman M, Kresak J, Yang C, Huang J, Hiser W, Kubilis P, et al. Analysis of immunobiologic markers in primary and recurrent glioblastoma. J Neurooncol. 2018;137:249–257. doi: 10.1007/s11060-017-2732-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang Q, Hu B, Hu X, Kim H, Squatrito M, Scarpace L, et al. Tumor evolution of glioma-intrinsic gene expression subtypes associates with immunological changes in the microenvironment. Cancer Cell. 2017;32:42–56.e6. doi: 10.1016/j.ccell.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang J, Cazzato E, Ladewig E, Frattini V, Rosenbloom DIS, Zairis S, et al. Clonal evolution of glioblastoma under therapy. Nat Genet. 2016;48:768–776. doi: 10.1038/ng.3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Daniel P, Sabri S, Chaddad A, Meehan B, Jean-Claude B, Rak J, et al. Temozolomide induced hypermutation in glioma: evolutionary mechanisms and therapeutic opportunities. Front Oncol. 2019;9:41. doi: 10.3389/fonc.2019.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grossman SA, Ye X, Lesser G, Sloan A, Carraway H, Desideri S, et al. Immunosuppression in patients with high-grade gliomas treated with radiation and temozolomide. Clin Cancer Res. 2011;17:5473–5480. doi: 10.1158/1078-0432.CCR-11-0774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sampson JH, Aldape KD, Archer GE, Coan A, Desjardins A, Friedman AH, et al. Greater chemotherapy-induced lymphopenia enhances tumor-specific immune responses that eliminate EGFRvIII-expressing tumor cells in patients with glioblastoma. Neuro Oncol. 2011;13:324–333. doi: 10.1093/neuonc/noq157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rühle PF, Wunderlich R, Deloch L, Fournier C, Maier A, Klein G, et al. Modulation of the peripheral immune system after low-dose radon spa therapy: detailed longitudinal immune monitoring of patients within the RAD-ON01 study. Autoimmunity. 2017;50:133–140. doi: 10.1080/08916934.2017.1284819. [DOI] [PubMed] [Google Scholar]
- 62.Deng L, Liang H, Burnette B, Beckett M, Darga T, Weichselbaum RR, et al. Irradiation and anti-PD-L1 treatment synergistically promote antitumor immunity in mice. J Clin Investig. 2014;124:687–695. doi: 10.1172/JCI67313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lu-Emerson C, Snuderl M, Kirkpatrick ND, Goveia J, Davidson C, Huang Y, et al. Increase in tumor-associated macrophages after antiangiogenic therapy is associated with poor survival among patients with recurrent glioblastoma. Neuro Oncol. 2013;15:1079–1087. doi: 10.1093/neuonc/not082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bakri SJ, Larson TA, Edwards AO. Intraocular inflammation following intravitreal injection of bevacizumab. Graefes Arch Clin Exp Ophthalmol. 2008;246:779–781. doi: 10.1007/s00417-007-0754-7. [DOI] [PubMed] [Google Scholar]
- 65.EL-Hajjar L, Jalaleddine N, Shaito A, Zibara K, Kazan JM, El-Saghir J, et al. Bevacizumab induces inflammation in MDA-MB-231 breast cancer cell line and in a mouse model. Cell Signal. 2019;53:400–412. doi: 10.1016/j.cellsig.2018.11.007. [DOI] [PubMed] [Google Scholar]
- 66.Lin H, Wei S, Hurt EM, Green MD, Zhao L, Vatan L, et al. Host expression of PD-L1 determines efficacy of PD-L1 pathway blockade-mediated tumor regression. J Clin Investig. 2018;128:805–815. doi: 10.1172/JCI96113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tang H, Liang Y, Anders RA, Taube JM, Qiu X, Mulgaonkar A, et al. PD-L1 on host cells is essential for PD-L1 blockade-mediated tumor regression. J Clin Investig. 2018;128:580–588. doi: 10.1172/JCI96061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hutchinson RA, Adams RA, McArt DG, Salto-Tellez M, Jasani B, Hamilton PW. Epidermal growth factor receptor immunohistochemistry: new opportunities in metastatic colorectal cancer. J Transl Med. 2015;13:217. doi: 10.1186/s12967-015-0531-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Marcelis L, Antoranz A, Delsupehe A-M, Biesemans P, Ferreiro JF, Debackere K, et al. In-depth characterization of the tumor microenvironment in central nervous system lymphoma reveals implications for immune-checkpoint therapy. Cancer Immunol Immunother. 2020;69:1751–1766. doi: 10.1007/s00262-020-02575-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stack EC, Wang C, Roman KA, Hoyt CC. Multiplexed immunohistochemistry, imaging, and quantitation: a review, with an assessment of Tyramide signal amplification, multispectral imaging and multiplex analysis. Methods. 2014;70:46–58. doi: 10.1016/j.ymeth.2014.08.016. [DOI] [PubMed] [Google Scholar]
- 71.Lazarus J, Maj T, Joshua Smith J, Lanfranca MP, Rao A, D’Angelica MI, et al. Spatial and phenotypic immune profiling of metastatic colon cancer. JCI Insight. 2018 doi: 10.1172/jci.insight.121932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Halse H, Colebatch AJ, Petrone P, Henderson MA, Mills JK, Snow H, et al. Multiplex immunohistochemistry accurately defines the immune context of metastatic melanoma. Sci Rep. 2018;8:11158. doi: 10.1038/s41598-018-28944-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gorris MAJ, Halilovic A, Rabold K, van Duffelen A, Wickramasinghe IN, Verweij D, et al. Eight-color multiplex immunohistochemistry for simultaneous detection of multiple immune checkpoint molecules within the tumor microenvironment. J Immunol. 2018;200:347–354. doi: 10.4049/jimmunol.1701262. [DOI] [PubMed] [Google Scholar]
- 74.Ravi VM, Neidert N, Will P, Joseph K, Maier JP, Kückelhaus J et al (2020) Lineage and spatial mapping of glioblastoma-associated immunity. bioRxiv. 10.1101/2020.06.01.121467
- 75.Asp M, Bergenstråhle J, Lundeberg J. Spatially resolved transcriptomes—next generation tools for tissue exploration. BioEssays. 2020;42:e1900221. doi: 10.1002/bies.201900221. [DOI] [PubMed] [Google Scholar]
- 76.Moncada R, Barkley D, Wagner F, Chiodin M, Devlin JC, Baron M, et al. Integrating microarray-based spatial transcriptomics and single-cell RNA-seq reveals tissue architecture in pancreatic ductal adenocarcinomas. Nat Biotechnol. 2020;38:333–342. doi: 10.1038/s41587-019-0392-8. [DOI] [PubMed] [Google Scholar]
- 77.Spitzer MH, Carmi Y, Reticker-Flynn NE, Kwek SS, Madhireddy D, Martins MM, et al. Systemic immunity is required for effective cancer immunotherapy. Cell. 2017;168(487–502):e15. doi: 10.1016/j.cell.2016.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou Yang T-H, et al. The immune landscape of cancer. Immunity. 2018;48(812–830):e14. doi: 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Barrow AD, Edeling MA, Trifonov V, Luo J, Goyal P, Bohl B, et al. Natural killer cells control tumor growth by sensing a growth factor. Cell. 2018;172(534–548):e19. doi: 10.1016/j.cell.2017.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Klemm F, Maas RR, Bowman RL, Kornete M, Soukup K, Nassiri S, et al. Interrogation of the microenvironmental landscape in brain tumors reveals disease-specific alterations of immune cells. Cell. 2020;181(1643–1660):e17. doi: 10.1016/j.cell.2020.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18:220. doi: 10.1186/s13059-017-1349-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gusev Y, Bhuvaneshwar K, Madhavan S (2018) Exploration of the immune cell landscape in brain cancer utilizing gene expression and copy number data. bioRxiv. 10.1101/490599