Abstract
Ductal carcinoma in situ (DCIS) represents pre-invasive breast carcinoma. In untreated cases, 25–60% DCIS progress to invasive ductal carcinoma (IDC). The challenge lies in distinguishing between non-progressive and progressive DCIS, often resulting in over- or under-treatment in many cases. With increasing screen-detected DCIS in these years, the nature of DCIS has aroused worldwide attention. A deeper understanding of the biological nature of DCIS and the molecular journey of the DCIS-IDC transition is crucial for more effective clinical management. Here, we reviewed the key signaling pathways in breast cancer that may contribute to DCIS initiation and progression. We also explored the molecular features of DCIS and IDC, shedding light on the progression of DCIS through both inherent changes within tumor cells and alterations in the tumor microenvironment. In addition, valuable research tools utilized in studying DCIS including preclinical models and newer advanced technologies such as single-cell sequencing, spatial transcriptomics and artificial intelligence, have been systematically summarized. Further, we thoroughly discussed the clinical advancements in DCIS and IDC, including prognostic biomarkers and clinical managements, with the aim of facilitating more personalized treatment strategies in the future. Research on DCIS has already yielded significant insights into breast carcinogenesis and will continue to pave the way for practical clinical applications.
Subject terms: Breast cancer, Breast cancer
Introduction
Ductal carcinoma in situ (DCIS) is a stage 0 breast cancer characterized by the abnormal proliferation of epithelial cells within the ductal-lobular system of the breast. It accounts for approximately 20% cases of newly diagnosed breast cancer.1–3 Based on architectural patterns, DCIS has been classified into comedo, papillary, solid, cribriform, and micropapillary subtypes.4 Based on the histopathologic nuclear features, DCIS can present as a spectrum, with low-grade (I), intermediate-grade (II), and high-grade (III) lesions; the latter are associated with a higher likelihood of invasive ductal carcinoma (IDC).5,6 DCIS is commonly considered as a direct precursor to IDC. Studies have reported that approximately 25–60% of untreated DCIS cases progressed to IDC within 9–24 years of follow-up, based on the limited sample size statistics.7–10 However, the natural history and definite etiology of these two disease classifications remain poorly understood. Nevertheless, the development of novel technologies has offered new insights into these lesions. In this context, discoveries related to the initiation and progression of DCIS and IDC are essential for further investigation into their origin and clinical management.
DCIS is often categorized as non-invasive or pre-invasive stage of breast cancer. Nonetheless, our understanding of the underlying causes of DCIS as well as how it progressed to be invasive is limited.11,12 Notably, similarities and differences have been observed between DCIS and IDC. Over the years, the rapid increase in the incidence of DCIS has accompanied the widespread adoption of mammography (mostly based on screening-detected calcifications).13–15 In this context, mammographic calcification is more frequently detected in DCIS than in IDC. Once IDC presents with calcifications on mammography, it is more likely to be associated with synchronous high-grade DCIS.16 The histologic characteristic that primarily distinguishes between DCIS and IDC is that DCIS tumor cells remain confined to the mammary ductal-lobular system without invading the surrounding parenchyma and the myoepithelial layer and basement membrane are intact, while IDC tumor cells have escaped the myoepithelial layer and spread into surrounding tissues.17 Based on the molecular features, some studies have categorized DCIS into four intrinsic subtypes similar to those of IDC; these include luminal A, luminal B, human epidermal growth factor receptor 2 (HER2/ERBB2)-positive, and basal-like subtypes. However, there is a variation in prevalence, as the HER2-positive subtype is more commonly observed in DCIS than in IDC (approximately 35% versus 15–20%, respectively).18–20 Notably, different intrinsic subtypes of DCIS have been reported to be associated with distinct tumor microenvironments (TMEs) and evolutionary pathways compared to IDC.21 Studies have shown that DCIS and IDC share certain risk factors that contribute to their incidence; these include age, family history, breast density, and hormone therapy.22 However, it is commonly considered that the prognosis of DCIS is superior to IDC. DCIS is not considered a life-threatening disease and is linked to a high rate of overall survival and a normal life expectancy.23 After treatments, the overall recurrence rate of DCIS is approximately 20%, in which 50% are in situ recurrence while another 50% invasive recurrence.24 It is therefore essential to identify the initiation of these lesions and the relationships between them; in particular, the molecular events underlying progression from DCIS to IDC warrant further investigation.
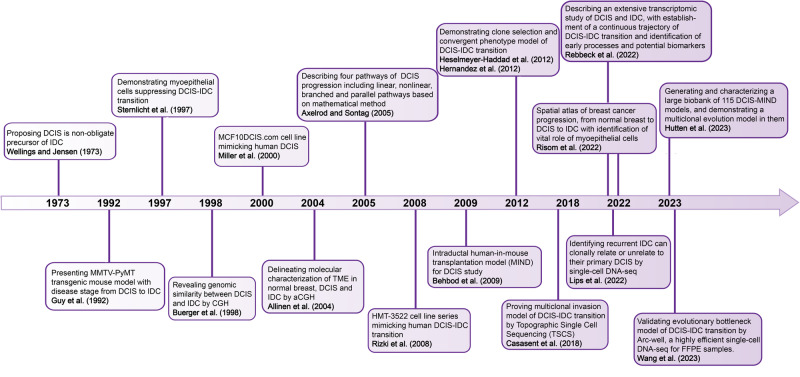
Although the epidemiological and clinicopathological characteristics indicate the progression from DCIS to IDC, the underlying biological mechanisms remain obscure. Here, we review the highlights of DCIS-IDC studies (Fig. 1). Since DCIS was termed as the precursor of IDC in 1973,25 studies on DCIS-IDC transition have been rapidly evolving since the establishment of DCIS cell lines and in vivo models. Recent advancements in technology, such as single-cell sequencing, spatial transcriptomics, and artificial intelligence, have provided deeper insights into the molecular changes occurring during the transition from DCIS to IDC. These studies have illuminated the alterations in both tumor-intrinsic features and the surrounding microenvironment. However, different studies relied on a variety of the sample populations and diverse techniques, making it necessary to systematically review all these related studies to get a clear picture of the known precise biological mechanisms underlying DCIS-IDC transition.26 Moreover, the clinical and pathological markers that are currently used to predict prognosis mostly rely on a combination of factors including patient age, surgical margins, tumor size, and nuclear grade, however, all these factors fail to predict prognosis independently with high confidence.27–30 Thus, the question whether current treatment is overly aggressive for indolent DCIS or insufficient for progressive DCIS remains unanswered. This ongoing debate highlights the need for further research and a better understanding of the natural history and biology of DCIS in order to develop personalized and targeted management approaches for each individual patient.
Fig. 1.
Highlights of DCIS-IDC research. Since DCIS was termed as the precursor of IDC in 1973, research on the transition from DCIS to IDC has rapidly advanced. This progress has been fueled by the establishment of in vitro DCIS cell lines and in vivo animal models. In recent years, technological advancements have offered deeper insights into the changes occurring in both tumor cells and the microenvironment during the transition from DCIS to IDC
This review highlights the pivotal signaling pathways in breast cancer and subsequently delves into the molecular distinctions between DCIS and IDC. It offers valuable insights into the biology of the transition from DCIS to IDC, shedding light on notable alterations occurring in tumor cells and the surrounding microenvironment. Essential experimental models and advanced technologies that are used for studies on DCIS-IDC have also been discussed. In addition, the recent clinical advances in DCIS and IDC, including prognostic biomarkers and advanced treatments have been described. Finally, this review aimed to identify the biology of the origin and progression of DCIS in order to better individualize treatments for DCIS with variable malignant potential.
Key signaling pathways in breast cancer
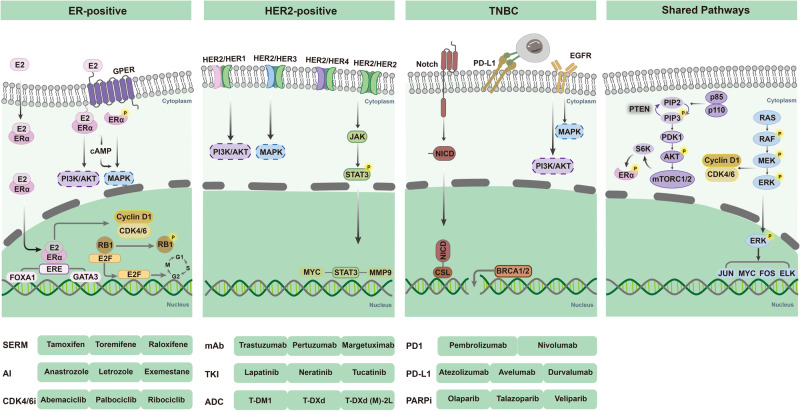
Why DCIS attracts much attention although itself is not invasive? It is probably because that DCIS can progress to IDC. IDC represents 60–75% of invasive breast cancer cases and breast cancer remains a leading cause of cancer-related mortality in women.31 Breast cancer is heterogeneous. IDC of different subtypes shared some common oncogenic pathways, as well as being perturbed by dominant pathways distinctively. The most important pathways acknowledged in breast cancer including estrogen receptor (ER) pathway,32,33 HER2 signaling pathway,34,35 Phosphoinositide 3-kinase/protein kinase B/mammalian target of rapamycin (PI3K/AKT/mTOR) pathway,36 mitogen-activated protein kinases (MAPK) pathway,36,37 and cyclin D1/cyclin-dependent kinase 4/6/retinoblastoma protein (cyclin D1/CDK4/6/RB1) pathway (Fig. 2). Metastasis retains a significant problem of breast cancer, with 20–30% of patients in early-stage breast cancer still die of metastatic disease.38 The metastatic cascade is admitted as a multistep process39 and the route of disseminated breast cancer cell to metastatic success or failure is decided by tumor cell intrinsic factors such as genetic/epigenetic plasticity, stemness, epithelial–mesenchymal transition and the tumor cell extrinsic factors especially including the metastatic microenvironment and the anti-cancer drug action.40–42
Fig. 2.
Key signaling pathways in breast cancer. Breast cancer is a heterogeneous disease characterized by diverse subtypes. The development and progression of breast cancer result from the influence of subtype-specific and shared signaling pathways, as well as the intricate crosstalk between them. Inhibitors that target these key signaling pathways have led to improvement in the prognosis of breast cancer. E2 estradiol, ER estrogen receptor, ERE estrogen response element, GPER G protein-coupled ER, HER2 human epidermal growth factor receptor 2, TNBC triple-negative breast cancer, EGFR epidermal growth factor receptor, NICD Notch intracellular domain, SERM selective ER modulator, AI aromatase inhibitor, CDK4/6i cyclin-dependent kinase 4/6 inhibitor, mAb monoclonal antibody, TKI tyrosine kinase inhibitor, ADC antibody–drug conjugates, PARPi Poly (ADP‑ribose) polymerase inhibitor
Estrogen receptor pathway
Hormone exposure is served as the primary risk factor for sporadic breast cancer.43 The two main steroid hormones involved in breast cancer are estrogen and progesterone, which are linked to the growth and proliferation of breast cells. Higher hormone exposure increases the risk of breast cancer, including shorter menstrual cycle, early menarche, and late menopause.44 Estrogen is regarded as a major promoter of breast cancer, especially for ER-positive breast cancer subtype, and the most common type in human is 17β-estradiol (E2).45 The binding of E2 to estrogen receptors (ERs, mostly ERα, encoded by the ESR1 gene) stimulates the classical genomic ER signaling pathway, which ultimately contributes to the increase of breast cancer cells’ proliferation and decrease of their apoptosis.46 More specifically, when activated by E2 binding, the estrogen-ER dimer translocates to the cell nucleus and engages with coregulator proteins and specific DNA sequences known as estrogen responsive elements (EREs). These interactions further modulate the expression of downstream genes such as GATA3 and FOXA1 that participate in breast carcer initiation, progression, and metastasis (Fig. 2).47,48 On the other hand, a nongenomic ER signaling pathway involves a membrane-anchored G protein-coupled estrogen receptor (GPER). Activation of GPER triggers signaling cascades such as PI3K/AKT and Ras/MAPK, which in turn regulate the transcription of genes involved in breast cancer development.45,49
HER2 signaling pathway
HER2, encoded by the ERBB2 gene, is one of the four members of epidermal growth factor receptor (EGFR) family, which includes EGFR (HER1), HER2, HER3, and HER4.43,50 HER2 is enriched in approximately 15–20% of breast cancers, which is correlated with a highly aggressive phenotype and unfavorable prognosis.50,51 Although HER2 has no specific ligand, it forms homodimers or heterodimers with HER1, HER3, or HER4 to initiate downstream signaling pathways (Fig. 2).52–54 In breast cancer, activation of HER2 signaling further triggers various downstream signalings, such as PI3K/AKT, MAPK, and JAK/STAT signaling pathways, all of which leading to cancer cell proliferation, survival, adhesion and metastasis.50,55 Targeting HER2 is proved to have great efficacy in the treatment of HER2-positive breast cancer patients, which profoundly benefits their overall survival.
PI3K/AKT/mTOR pathway
The phosphatidylinositol 3-kinases (PI3Ks) are a group of intracellular kinases that are categorized into three classes (class I, II and III). Among them, class I PI3Ks composed of a regulatory (p85) and a catalytic (p110) subunit are the most commonly studied and definitely implicated in oncogenesis.41,56 Mutations commonly occur in the p110α subunit (encoded by PIK3CA), among all breast cancer subtypes, but the mutation frequency is especially higher in ER-positive breast cancers. Approximately, mutations in PIK3CA are observed in 40% of ER-positive breast cancers, 25% of HER2-positive breast cancers, and 9% of TNBC.56 PIK3CA mutations lead to the activation of PI3K, which activates downstream targets such as protein kinase B (AKT) and mammalian target of rapamycin (mTOR). The excessive activation of the PI3K/AKT/mTOR pathway is strongly linked to uncontrolled breast cancer development.57 Phosphatase and tensin homolog (PTEN) and inositol polyphosphate 4-phosphatase type II (INPP4B) are two crucial negative regulators of the PI3K pathway (Fig. 2). In breast cancer, there is often a decrease in the expression of PTEN and INPP4B, further enhancing the activation of the PI3K/AKT/mTOR pathway,58–60 and dysregulation of this signaling pathway is recognized as a mechanism of resistance to endocrine and anti-HER2-targeted treatment.61–63 Consequently, targeting PI3K/AKT/mTOR pathway has emerged as a promising approach for precise therapeutic intervention in breast cancer.
MAPK pathway
Mitogen-activated protein kinases (MAPKs) are phosphoproteins stimulated by mitogens and play a vital role in controlling various cellular processes such as cell proliferation, stress adaptation, differentiation, and apoptosis. Mammalian cells have three major branches of the MAPK signaling pathways: the extracellular signal-regulated kinases (ERK), the c-Jun N-terminal kinases (JNK), and the p38 MAPKs (Fig. 2). MAPKs are strongly related to breast cancer prognosis, which participate in hormone receptor modulation, response to growth factors and targeted therapies.64–66 The MAPK pathway is activated in approximately 50% of breast cancers.67 RAS mutation, which frequently observed in tumors such as pancreatic cancer and colorectal cancer,68 leading to constitutive activation of ERK1/2, is not considered as the main cause of MAPK signaling activation in breast cancer, as RAS mutation occurs in less than 5% of breast cancer cases. Contrarily, MAPK signaling activation in breast cancer is usually considered as a result of constitutive upstream signaling, such as the ER signaling pathway and HER2 overexpression. In ER-positive breast cancers, estrogen can stimulate growth factors such as transforming growth factor beta (TGF-β), insulin-like growth factor type 1 (IGF-1), that ultimately activate MAPK pathway.69 The activated MAPKs can also phosphorylate ER, either through direct or indirect signaling pathways, resulting in an sustainedly enhanced transcriptional efficiency of the receptor.70
Cyclin D1/CDK4/6/RB1 pathway
In breast cancer, the cyclin D1/CDK4/6/RB1 complex plays a crucial role in cell proliferation mediated by ER signaling.71 Particularly, the presence of estrogen in ER-positive breast cancers induces the expression of cyclin D1(encoded by CCDN1), resulting in the activation of CDK4/6. CDK4/6 activity consequently leads to the hyperphosphorylation of RB1, facilitating the progression of the cell cycle and promoting cellular proliferation (Fig. 2).72,73 What’s more, increased MAPK and PI3K/AKT pathways can also drive CCDN1 transcription that finally activate cyclin D1/CDK4/6/RB1 pathway.74 CDK4/6 inhibitors are undeniably considered as one of the most significant advancements in breast cancer treatments in the last two decades.75 Different from ER-positive breast cancer, TNBC often exhibits a loss of RB1 expression, which consequently renders them unresponsive to CDK4/6 inhibitors.76,77
Molecular features of DCIS and IDC
A traditional theory presumes that DCIS and IDC are derived from mammary ducts, while lobular carcinoma in situ and invasive lobular carcinoma arise from lobules,78 although in Wellings’ research, they found that most early-stage breast carcinomas including ductal and lobular types arise from the same structure, namely, the terminal duct lobular unit.25,79 The ductal structure of terminal duct lobular unit consists of two cell layers, namely, the epithelial cell layer within the lumen and the myoepithelial cells (MECs) layer (surrounded by basement membrane). From a morphological perspective, some DCIS can be visually distinguished from IDC on H&E slides, which is characterized by neoplastic proliferation within the terminal duct lobular unit and invariable encasement by the basement membrane,80 while some other small DCIS need to be differentiated from early IDC by a combination of myoepithelial markers such as cytokeratin 5/6 (CK5/6), tumor protein p63 (P63), calponin, α‐smooth muscle actin, etc.81–83 To date, numerous studies have explored the genomic events in DCIS and compare them with those of IDC, they have put insights into the distinctions between synchronous DCIS and IDC, pure DCIS and DCIS with synchronous IDC, and primary DCIS and recurrent DCIS or IDC (Table 1).
Table 1.
Comparison of molecular features between DCIS and IDC
| Author | Year | Samples | Analyses | Methods | Highlights |
|---|---|---|---|---|---|
| a. Synchronous DCIS VS Synchronous IDC | |||||
| Moelans et al.90 | 2011 | 39 synchronous DCIS-IDC | DNA | MLPA | No significant differences were found in copy numbers of 21 genes in synchronous DCIS and IDC, except BIRC5 being more prevalent in DCIS but without significance. |
| Hernandez et al.84 | 2012 | 13 synchronous DCIS-IDC | DNA | aCGH, Sequenom MassARRAY | The genomic profiles of synchronous DCIS-IDC were largely similar, but PIK3CA mutations were limited to the DCIS component in 2/13 cases, and 1/13 case exhibited a higher frequency of PIK3CA mutations in DCIS compared to IDC. |
| Johnson et al.91 | 2012 | 21 synchronous DCIS-IDC | DNA | MIP array | 83% of the genome was shared in synchronous DCIS-IDC, with recurrent losses at 3q, 6q, 8p, 11q and gains at 5q, 16p, 19q and 20 observed in IDC but not in DCIS. Additionally, amplification of CCND1 and MYC was more pronounced in IDC, while loss of 17p11.2 was specific to DCIS. |
| Casasent et al.92 | 2018 | 10 synchronous DCIS-IDC | DNA | TCSC | A direct genomic lineage existed between synchronous DCIS and IDC, with most mutations and CNAs evolved within the ducts prior to invasion. |
| Pareja et al.86 | 2020 | 27 synchronous DCIS-IDC | DNA | WES, MSK-IMPACT | Genetic alterations were similar between synchronous DCIS and IDC, the most overlapped mutations include TP53, PIK3CA and GATA3. |
| Fortunato et al.87 | 2021 | 53 synchronous DCIS-IDC | DNA | DNA-seq | Statistically significant differences were identified that IDC harbored increased mutations and higher genetic divergence than synchronous DCIS. |
| b. Primary DCIS VS Recurrent DCIS/IDC | |||||
|---|---|---|---|---|---|
| Waldman et al.95 | 2000 | 18 primary DCIS, 18 recurrent DCIS | DNA | CGH | DCIS recurrences were clonally related to their primary DCIS in 17/18 cases. The most common chromosomal alterations shared between primary and recurrent DCIS were gains involving chromosome 17q and losses involving chromosomes 8p and 17p. |
| Gorringe et al.96 | 2015 | 8 primary DCIS, 6 recurrent DCIS, 1 IDC, 1 mixed DCIS | DNA | SNP arrays | There was no significant difference in fraction genome alterations between primarily DCIS and matched recurrent tumors, in which a large variation in the copy number altered base pairs showing the same gain or loss event ranging from 11 to 58%. |
| Trinh et al.94 | 2021 | 6 primary DCIS, 6 recurrent IDC | DNA, mRNA |
WES, RNA-Seq |
Amongst pure DCIS and matched recurrent IDC, frequent changes were observed including 1q, 8q, 16p, 17 amplification and loss of 11q and 16q. |
| Lips et al.97 | 2022 | 129 primary DCIS, 34 recurrent DCIS, 95 recurrent IDC | DNA |
WES, Single-cell sequencing |
Most of the recurrent IDC or DCIS were found clonally related to primary DCIS. The most common shared mutations between primary DCIS and recurrent IDC occur in TP53 and PIK3CA, and 1q and 8p11 gain were more common in recurrent IDC compared to primary DCIS, while 3p21 loss was more common in primary DCIS compared to recurrent IDC. |
| Wang et al.98 | 2023 | 10 primary DCIS, 10 recurrent DCIS/IDC | DNA | Arc-well | Seven samples with evolutionary bottlenecks were found, in which common CNA events of persistent subclones showed increased chr3q (PIK3CA), chr8p (MYC, CCNE2) and 20q (ZNF217, AURKA) gains in recurrent tumors comparing to primary DCIS. |
| c. Pure DCIS VS DCIS with synchronous IDC | |||||
|---|---|---|---|---|---|
| Iakovlev et al.85 | 2008 | 6 pure DCIS, 17 DCIS with synchronous IDC | DNA | aCGH | Pure DCIS exhibited a higher degree of genomic complexity than DCIS with synchronous IDC, in which the gain on 17q22-q24.2 was less common while the gain at 17q12-21.2 was more common in pure DCIS. |
| Zhou et al.99 | 2009 | 32 pure DCIS, 48 DCIS with synchronous IDC | DNA | DNA sequencing | TP53 mutation frequency was found slightly lower in pure DCIS (15.6%) than synchronous DCIS (20.8%). |
| Miron et al.100 | 2010 | 43 pure DCIS, 31 DCIS with synchronous IDC | DNA | Sanger sequencing | PIK3CA mutations were less common in pure DCIS (5%) than DCIS with synchronous IDC (16%). |
| Sakr et al.101 | 2014 | 89 pure DCIS, 119 DCIS with synchronous IDC | DNA | Sequenom MassARRAY | PIK3CA hotspot mutations and pAKT expression were more prevalent in ER + /HER2- DCIS with synchronous IDC, while INPP4B loss of expression was more frequent in ER-/HER2 + DCIS with synchronous IDC than pure DCIS. |
| Afghahi et al.102 | 2015 | 120 pure DCIS, 151 DCIS with synchronous IDC | DNA | FISH | Pure DCIS had lower frequencies of CNAs at three common chromosomal loci 1q, 8q24 and 11q13 than DCIS with synchronous IDC |
| Kim et al.103 | 2015 | 6 pure DCIS, 5 DCIS with synchronous IDC | DNA |
aCGH, WES |
Gains of PIK3CA, CDK12, MLF1, EVI1, SOX2, TFRC, ERG and MTCP1, and losses of PIK3R1, APC, FGFR2, PDGFRB, CD74, ITK, EBF1, RANBP17, TLX3, NPM1, NR4A3, IL6ST and MAP2K4 were more frequent in DCIS with synchronous than pure DCIS |
| Lin et al.106 | 2019 | 65 pure DCIS, 60 DCIS with synchronous IDC | DNA | Targeted sequencing | The mutations of PIK3CA kinase domain were found more frequent in pure DCIS. |
| Bergholtz et al.104 | 2020 | 10 pure DCIS, 13 DCIS with synchronous IDC | DNA | Targeted sequencing | A lower frequency of TP53, PIK3CA, and ERBB2 mutations was found in pure DCIS. |
| Rebbeck et al.108 | 2022 | A whole of >2,000 ductal lesions from 145 patients | mRNA | RNA-seq | CAMK2N1, MNX1, ADCY5, HOXC11 and ANKRD22 were found reduced expression in DCIS with synchronous than pure DCIS. |
DCIS ductal carcinoma in situ, IDC invasive ductal carcinoma, CGH comparative genomic hybridization, MLPA multiplex ligation-dependent probe amplification, MIP molecular inversion probe, WES whole-exome sequencing, MSK-IMPACT MSK Integrated Mutation Profiling of Actionable Cancer Targets, TCSC Topographic Single Cell Sequencing, IHC immunohistochemistry, FISH fluorescence in situ hybridization, aCGH array-comparative genomic hybridization, CNAs copy number aberrations, Arc-well Archival nanowell sequencing
Comparison of synchronous DCIS and IDC
Synchronous DCIS and IDC are frequently found in patients who are diagnosed with invasive breast carcinoma. To identify the progressive markers, a number of studies have explored genomic profiles of synchronous DCIS and IDC which were usually micro-dissected in the same tumor sample. Most of these studies concluded that synchronous DCIS and IDC share high levels of genomic concordance, which is reflected by their copy number profiles or copy number aberrations (CNAs) (Table 1).84–89 Moelans et al. described no significant difference in copy numbers of 21 genes in synchronous DCIS and IDC.90 Hernandez et al. demonstrated a largely similar genomic profiles of synchronous DCIS and IDC,84 and Johnson et al. further depicted 83% of the genome in synchronous DCIS and IDC were shared.91 In a recent whole-exome sequencing study, Pareja et al. evaluated 27 formalin-fixed paraffin-embedded (FFPE) samples of synchronous DCIS and IDC, finding that the most frequent gains and losses in both synchronous DCIS and IDC were on 1q, 16p, 5q, 6q, and 8p, and the most overlapped mutations in genes included tumor protein p53 (TP53), phosphatidylinositol-4, 5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA), and GATA binding protein 3,86 which have been considered to play pivotal roles in breast cancer initiation and progression. In essence, given the notable genetic similarity observed between synchronous DCIS and IDC, it may be inferred that synchronous DCIS has fundamentally invasive potential, wherein invasive modifications have already occurred as early as DCIS stage. However, the limited findings of the differences between synchronous DCIS and IDC may attribute to the bottleneck of high-resolution techniques especially at single-cell level. Most recently, by using spatially-resolved single-cell DNA sequencing, Casasent et al. reported a direct genomic lineage between synchronous DCIS and IDC, with most mutations and copy number aberrations evolved within the ducts prior to invasion.92
However, notably, studies have also found differences between synchronous DCIS and IDC. Fortunato et al. analyzed single nucleotide variants in synchronous DCIS and IDC lesions and found significantly increased mutations and genetic divergence in IDC than synchronous DCIS.87 A meta analyses performed by Rane et al. found that DCIS displays more frequent losses on 5q31.1–5q35.3, 6q25.3–6q26, and 13q32.3–13q33.1 and gains on 11p12 in synchronous DCIS than synchronous IDC.88
In essence, given the notable genetic resemblance observed between synchronous DCIS and IDC, it can be inferred that synchronous DCIS is fundamentally invasive, wherein invasive modifications have already taken place. Although slight disparities between these two conditions have been documented, there is limited consensus regarding the genomic alterations that signify transitions. The critical events associated with the transition from DCIS to IDC may occur in earlier lesions such as pure DCIS or possibly even in lesions preceding DCIS.
Comparison of primary DCIS and recurrent DCIS or IDC
Recurrent DCIS or IDC refers to subsequent carcinoma lesions that develop after a primary diagnosis of DCIS. Notably, up to 20% of patients with DCIS develop recurrent DCIS or IDC despite treatment and approximately half of these are IDC.4,93 Owing to difficulties in collecting samples from primary DCIS and their matched recurrent tumors, related data of genomics, transcriptomics, and proteomics are still limited. Recurrence is considered as the DCIS progression and comparison studies of primary pure DCIS and recurrent DCIS or IDC have been launched by the aim of exploring the biological factors and molecular features leading to DCIS progression, however limited significant alternations were founded between primary DCIS and recurrent DCIS or IDC at genetic level. Previous studies by using conventional techniques such as comparative genomic hybridization, single nucleotide polymorphism arrays or whole-exome sequencing, consistently found that primary DCIS and their recurrent tumors were genetically related.94–96 In recent important study by Lips et al. whole-exome and single-cell sequencing were conducted among 129 primary DCIS, 34 matched recurrent DCIS and 95 matched recurrent IDC. Most of the recurrent IDC or DCIS were found clonally related to primary DCIS. When comparing primary DCIS and recurrent IDC, the numbers of shared and private mutations were found to be highly variable for different matched tumor pairs, with most common shared mutations occurring in TP53 and PIK3CA. Besides, 1q and 8p11 gain were more common in recurrent IDC compared to primary DCIS, while 3p21 loss was more common in primary DCIS compared to recurrent IDC. When comparing primary DCIS and DCIS recurrence, whole-exome sequencing and copy number profiling data revealed 29/34 cases were related, suggesting the DCIS recurrence as the residual DCIS which was not detected by imaging preoperatively.97 Another recent important study by Wang et al. conducted a high-throughput single-cell DNA sequencing (Arc-well) in 10 paired archival FFPE samples of primary DCIS and recurrent tumors.98 Evolutionary analysis indicated that majority of DCIS cases in the cohort went through an evolutionary bottleneck. Specific chromosome aberrations were identified in the persistent subclones across the primary DCIS and recurrent tumors, which were considered to be closely related to DCIS recurrence. In the seven samples with evolutionary bottlenecks, increased chr3q (PIK3CA), chr8p (MYC, CCNE2) and 20q (ZNF217, AURKA) gains were found in the persistent subclones in recurrent tumors comparing to primary DCIS.
In summary, genomic investigations have demonstrated a close genetic connection between recurrent DCIS or IDC and their primary pure DCIS counterparts. Only minimal alternations were noted between primary DCIS and their recurrence. It indicates that genomic changes responsible for recurrence may occur as an early occurrence in primary DCIS. Based on the numerous genomic studies, landscape of transcriptomics and proteomics is required to further discover the molecular features occurring in DCIS recurrence. Besides, the influence of microenvironment in DCIS recurrence needs to be further investigated too.
Comparison between pure DCIS and DCIS with synchronous IDC
Studies also interested in distinguishing the molecular features between pure DCIS and DCIS from patients diagnosed with co-occurring IDC, which refer to DCIS with synchronous IDC here. Many studies observed that pure DCIS exhibited lower genomic instability compared to DCIS with synchronous IDC.99–104 For example, in Afghahi et al.’s study with large sample size, pure DCIS was found to have lower frequencies of CNAs at three common chromosomal loci 1q, 8q24, and 11q13 than DCIS with synchronous IDC.102 However, there are some dissenting voices. For example, a previous study by Iakovlev et al. with limited sample size, demonstrated that pure DCIS exhibited a higher degree of genomic complexity than DCIS with synchronous IDC, in which the gain on 17q22-q24.2 was less common while the gain at 17q12-21.2 was more common in pure DCIS.85 Mutations of driver genes such as TP53 and PIK3CA have been identified with significant differences between pure DCIS and DCIS with synchronous IDC.99,105–107 For example, in Zhou et al.’s study, TP53 mutations were less common in pure DCIS than IDC with synchronous IDC.99 In Bergholtz et al.’s study, a lower frequency of TP53, PIK3CA, and ERBB2 mutations was found in pure DCIS comparing to DCIS with synchronous IDC. It should be noted that mutations in specific regions of driver genes should be paid more attentions to.107 Earlier studies by Miron et al. and Sakr et al. typically found more PIK3CA mutations in DCIS with synchronous IDC than pure DCIS.100,103 Contrarily, a recent study by Lin et al., by targeted exon sequencing in the kinase domain of PIK3CA, discovered that mutations in this specific domain was more frequent in pure DCIS than those in DCIS with synchronous IDC.106 The discrepancy may attribute to that Lin et al.‘s study focusing on a more precise mutation region in PIK3CA kinase domain, while earlier studies looked at a combination of regions in PIK3CA termed as “hotspot mutations”.
Intriguingly, Rebbeck et al. recently conducted a comprehensive transcriptomic study involving over 2,000 micro-dissected ductal lesions from 145 patients.108 In the study, they compared pure DCIS with concurrent DCIS which refer to DCIS with synchronous IDC and successfully identified some genes that were potentially responsible for DCIS progression, such as CAMK2N1, MNX1, ADCY5, HOXC11, and ANKRD22, which exhibited reduced expression in concurrent DCIS.
In brief, pure DCIS exhibited fewer genetic events and less frequent mutations in driver genes than DCIS with synchronous IDC, suggesting that concurrent DCIS is probably in a more progressive stage rather than pure DCIS in. Emerging studies of transcriptome in pure DCIS and DCIS with synchronous decipher distinguished related genes expression, although these studies are very limited. However, if molecular features derived from the comparisons between pure DCIS and DCIS with synchronous IDC are considered as to be responsible for DCIS progression, it should be admitted first that DCIS is the precursor of IDC.
Progression from DCIS to IDC
Although the origin and evolution of breast cancer has been long discussed, the mechanisms involved remain unclear. In regard to tumor heterogeneity, two distinct but not mutually exclusive models were proposed, namely, the clonal evolution and cancer stem cell models.109–112 In the cancer stem cell model, tumor-initiating cells originate from a small fraction of stem or progenitor cells which can undergo self-renewal. In the clonal evolution model, any breast epithelial cell has tumorigenic potential and tumor progression is promoted by cumulative genetic and epigenetic changes.
However, as a key stage in breast carcinogenesis, the evolution from DCIS to IDC remains largely obscure. The major questions in this regard are: (1) does IDC originate from DCIS? and (2) if so, how does DCIS progress during the DCIS-IDC transition? The four proposed models of DCIS-IDC transition may answer the former question. The theories regarding DCIS progression have been broadly divided into two categories, namely, genetic and non-genetic. The former posits the crucial role of the neoplastic cell itself while the latter emphasizes on non-genetic factors, particularly the TME.
Proposed models of DCIS progression
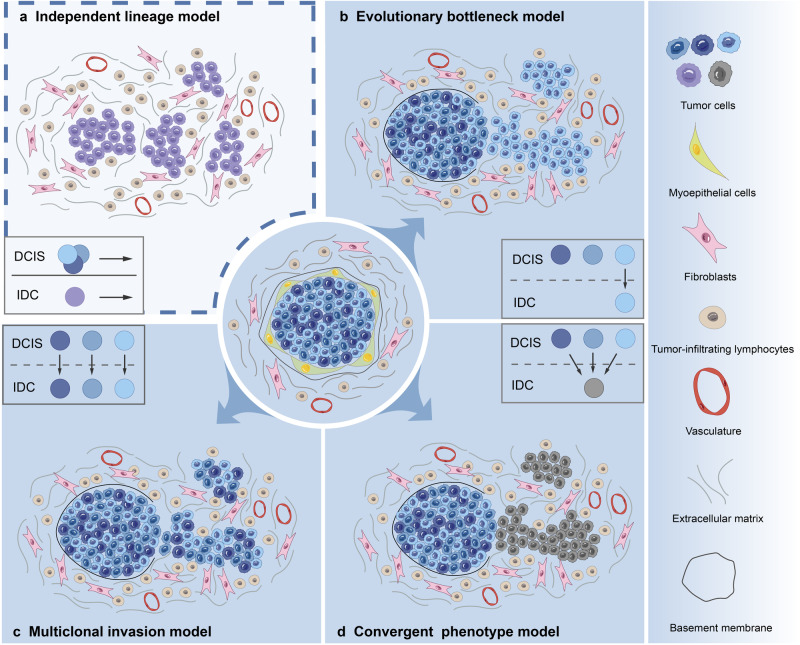
Four different but complementary models have been proposed for DCIS progression, including the independent lineage, evolutionary bottleneck, multiclonal invasion, and convergent phenotype models (Fig. 3).113–115 The independent lineage model and the other three direct lineage models differ on whether DCIS is a precursor of IDC. Naturally, only up to 60% of DCIS was observed to progress to IDC if left untreated, while at least the left 40% of DCIS remained indolent and never progressed. Thus, the existence of indolent DCIS provided evidences for independent lineage model that DCIS and IDC were of independent origin.7 Moreover, independent lineage model opposes the precursor theory based on the identification of discordant markers between DCIS and IDC. Contrarily, Wellings and Jensen et al. suggested that ductal breast carcinomas undergo continuous histological progression from atypical ductal hyperplasia (ADH) to DCIS and subsequently to IDC.25 The gradual histological continuity, similar intrinsic subtypes, and general genetic similarity between DCIS and adjacent IDC further validates the theory.116–118 However, low-grade and high-grade DCIS are supposed to individually progress to IDC via different pathways.78,119 In addition to partially explaining whether DCIS is a precursor of IDC, these models illustrate the transition from DCIS to IDC in terms of changes in the tumor cell. Extensive research using sequencing has clearly demonstrated intratumor heterogeneity in DCIS and IDC; this heralds a new era in investigations on DCIS progression.
Fig. 3.
Proposed models of DCIS progression. a Independent lineage model, which presumes that DCIS and IDC derive from two distinct normal epithelial cells which share no overlapping CNAs or mutations. b Evolutionary bottleneck model, which presumes that a specific clone within DCIS is selected and it evolves into IDC. c Multiclonal invasion model, which presumes that multiple clones escape and co-migrate to invasive regions to generate IDC. d Convergent phenotype model, which presumes that subclones of different genotypes within DCIS can all give rise to an invasive phenotype to establish IDC
Independent lineage model
The independent lineage model hypothesizes that DCIS and IDC derive from two distinct normal epithelial cells (Fig. 3a).92 It presumes that DCIS and IDC arise from and progress through two independent cell lineages which never share any CNAs or mutations. The model opposes the opinion that DCIS is the precursor of IDC which has been generally accepted in recent years. In support of this theory, Studies results of discordant targeted gene or protein markers and CNAs in DCIS and IDC supported the theory of independent lineage model.91,100,120,121 In this context, Sontag et al. used a mathematical approach and concluded that DCIS and IDC develop in parallel pathways.122 Johnson et al. consistently identified IDC-specific gains and losses on chromosomal regions to be restricted to IDC; this was not observed in DCIS.91 Moelan et al.’s study found the observed methylation of CDKN2A and CHFR to only exist in DCIS.123 The independent model is also supported by the fact that no clonal relatedness is observed between certain synchronous DCIS-IDC lesions located in different quadrants of the breast.86,120 In Pareja et al.’s study, clonal relatedness was not observed between DCIS and IDC foci arising from different mammary quadrants, but was present among lesions in the same quadrant.86 In Lip et al.’s recent study, when investigating clonal relationships in 95 paired cases of DCIS and invasive recurrence by advanced single-cell sequencing, they only found 75% (71 out of 95) of the tumor pairs exhibited clear clonal connections while there still remained 18% (17/95) of which displayed no discernible relationship.97
Summarily, when earlier studies defined the independent lineage model according to a few discordant markers in DCIS and IDC, which general concordance of other markers might be neglected, was limited by the sample size. Recent extensive genomic analysis of finding a certain proportion of discernible relationships in DCIS and invasive recurrence partially supported the probability of the independent model.
Evolutionary bottleneck model
The heterogeneity of breast cancer creates hindrances to targeted therapy and leads to drug resistance. Nevertheless, it facilitates the study of tumor evolution. Previous studies on breast cancer progression have demonstrated a phenomenon of evolutionary bottlenecks in metastases compared with the primary tumor.124 Subpopulations of tumor cells from the primary site are enriched in metastases, with or without new mutations that are acquired during the process of metastasis.125 Here, the evolutionary bottleneck model for DCIS-IDC transition hypothesizes that during the transition, only a small proportion of DCIS tumor cells with specific genetic events are selected to form a single clone, which subsequently breaks the evolutionary bottleneck and evolves into IDC (Fig. 3b).113–115 The evolutionary bottleneck model emphasizes the existence of clonal selection and decreased clonal diversity from DCIS to IDC transition.
Despite the general genetic similarities between synchronous DCIS and IDC, clear differences exist between the two components. In terms of PIK3CA mutations in synchronous DCIS and IDC, some studies reported that these mutations were restricted to DCIS,84,91,100 while others identified these mutations in IDC but not in DCIS.100,101 It may be speculated that the DCIS-IDC transition obeys the: (1) independent lineage model (based on the discordance in PIK3CA mutations between DCIS and IDC), (2) evolutionary bottleneck model, with the selected clone demonstrating clonal shifts of gain or loss PIK3CA mutations in IDC, and (3) evolutionary bottleneck model, with the selected clone having PIK3CA mutations subsequently developing into the dominant clone in IDC. Besides, based on a small sample size, Doebar et al.’s study selected a subset of 92 invasive tumor-specific variants from 4 synchronous DCIS and IDC lesions, of which 52 variants overlapped between DCIS and IDC lesions, while the other 40 were only restricted to IDC.126
In brief, it suggests that clonal selection probably occur during the transition from DCIS to IDC. However, the selected subclones that harbor specific genetic events in DCIS may vary between different patients and new genetic events may even be acquired after DCIS evolving into IDC. In conjunction, these observations support an evolutionary model in which the transition from DCIS to IDC occurs as a result of clonal selection and may obey the rules of Darwinian evolution.113
Multiclonal invasion model
The multiclonal invasion model differs from the evolutionary bottleneck model in that it refers to multiple subclones escaping and co-migrating to invasive regions to generate IDC, while the evolutionary bottleneck model mainly refers to the dominant subclones selection during the evolution from DCIS to IDC (Fig. 3c).86,114,115 Two scenarios are proposed for the multiclonal invasion model. In one scenario, multiple subclones form a relationship of mutual cooperation and even cooperate with the TME. In another scenario, these multiple subclones have different identities and may be considered as “leader” and “follower” subclones, and once the leader subclones break through the basement membrane, their followers join them.115 In either scenario, more than one clone may be detected in both DCIS and IDC. Previous studies have mostly examined the model based on genomic evidence, demonstrating highly concordant CNAs and mutations in synchronous DCIS-IDC.84,91,92,127–130 However, these studies did not conduct a direct clonal analysis of DCIS and IDC. Recently emerging technologies actualized the tracing of clonal evolution during DCIS progression. In Casasent et al.’s study, a novel technology known as topographic single-cell sequencing was used for analyzing the evolution from DCIS to IDC.92 In each matched sample of synchronous DCIS and IDC lesions, they clustered several major clonal tumor subpopulations with highly concordant copy number profiles, which were indicative of stable clonal expansion. Their data showed that in addition to existing in synchronous DCIS and IDC lesions, these subclones shared a common origin in the ducts. By performing deep-exome sequencing in micro-dissected DCIS and IDC, high concordance of nonsynonymous mutations (>87%) were further identified between synchronous DCIS and IDC lesions. These results therefore demonstrated the co-migration of multiple subclones in DCIS to IDC transition, which is in complete contrast to the theory of the bottleneck model. Notably, the shared origin of these subclones from a common ancestor also opposed the independent lineage model.
The multiclonal invasion model may imply that the DCIS-IDC transition: (1) is decided by multiple cancerous cells rather than a specific cell population, and (2) is influenced by noncancerous factors such as TME changes especially in the context of as high genomic concordance between DCIS and IDC.
Convergent phenotype model
Another direct lineage model of DCIS progression is the convergent phenotype model.131 The model describes that subclones of different genotypes within DCIS may all give rise to an invasive phenotype to establish IDC, with concordant genomic profiles between the DCIS and related IDC (Fig. 3d).113,114 This suggests that discordant genotypic tumor cells may undergo potential similar or complementary alterations and finally gain the same invasive phenotype. In particular, the invasive phenotype of IDC may be determined by various combinations of multiple distinct genomic aberrations in DCIS.113 This may also explain the negative findings from previous genomic comparisons between DCIS and IDC. Intriguingly, Yates et al. reported two distinct PTEN driver mutations appeared in different regions of multifocal DCIS, both of which parallelly evolved into PTEN-null IDC.132 Convergent evolution may occur despite genetic divergence acquired during DCIS progression, supporting the presence of mutational diversity in DCIS.
Accumulating evidence suggests that DCIS doesn’t progress with a predetermined pattern in patients, in which both independent and direct lineage models can be observed in different study populations. Thus, the natural pattern of DCIS progression still remains enigmatic. Moreover, microenvironment of the DCIS probably influences the DCIS progression, which may lead to different DCIS progression models. Advanced technologies such as single-cell sequencing and spatial transcriptomics have provided deeper insights into the subclonal dynamics in DCIS progression. During years of follow-up, one or more subclones can persist in the primary lesion and subsequently progress to IDC. These discoveries allow further exploration of prognostic biomarkers for progressive DCIS and corresponding active treatment. The independent DCIS progression model poses new challenges in identifying tumor-intrinsic prognostic biomarkers. In brief, models for DCIS progression remain theoretical, and further research is urgently needed to understand the natural molecular feature of DCIS progression process and the clinical significance relatively.
Role of the TME in DCIS progression
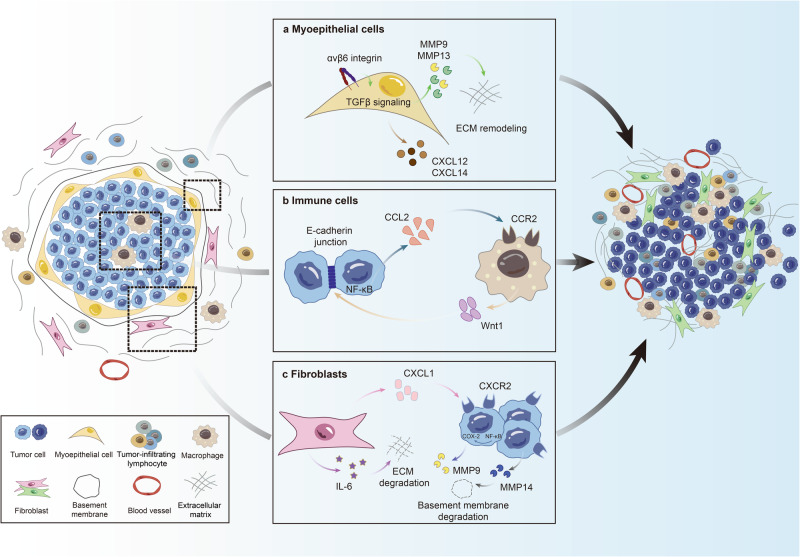
Since limited difference have been identified in tumor cells of DCIS and IDC at the genomic or transcriptomic level, many studies have been putting insight into their neighborhood----microenvironment. Emerging evidence suggests that considerable changes in the microenvironment pave the way for DCIS progression.133 Notably, different intrinsic subtypes of DCIS have been reported to be associated with distinct tumor microenvironment (TME) and evolutionary pathways to IDC.21 In breast cancer, the TME refers to all components surrounding to the cancerous cells in the tumor; it is mainly composed of MECs, immune cells, fibroblasts, extracellular matrix, and blood vessels.134–136 Numerous studies have demonstrated that the DCIS-IDC transition is not solely triggered by intrinsic changes in tumor cells; it is also regulated largely by the TME (Fig. 4).137,138
Fig. 4.
Role of microenvironment in DCIS progression. Tumor-microenvironment crosstalk may facilitate DCIS progression. The environment consists of a range of components including MECs, immune cells, fibroblasts, blood vessels, basement membrane and ECM. a MECs: Upregulation of αvβ6 integrin expressed in MECs activates TGFβ signaling, resulting in upregulation of MMP9 and MMP13 and ECM remodeling. Moreover, increased secretion of CXCL12 and CXCL14 from MECs promotes DCIS tumor cell invasion. b Immune cells: DCIS cell-derived CCL2 recruit macrophages into the tumor, driving increased Wnt-1 secretion from macrophages, contributing to myoepithelium disruption and the breakdown of E-cadherin junctions. c Fibroblasts: Increased secretion of CXCL1 and IL-6 from CAFs drives activation of NF-κB and COX-2 in DCIS cells, which induced and upregulation of MMP9 and MMP14 from DCIS cells, resulting in ECM remodeling and basement membrane degradation. DCIS ductal carcinoma in situ, MECs myoepithelial cells, ECM extracellular matrix, TGFβ transforming growth factor-beta, MMP9 matrix metalloproteinase 9, CXCL12 C-X-C motif chemokine ligand 12, CCL2 C-C motif ligand 2, IL6 interleukin-6, CAFs tumor-associated fibroblasts, NF-κB nuclear factor-kappaB, COX-2 cyclooxygenase 2
MECs
In breast ducts, the MECs are located between the epithelium and basement membrane and act as a physical barrier between the epithelium and surrounding stroma.139 The MEC layer remains intact in normal ducts, some breast benign lesions, and DCIS. On progression of DCIS, they play a significant role as a physical gatekeeper morphologically, although in some studies they were regarded as an active tumor suppressive factor.140
Normal MECs are regarded as natural tumor suppressors due to their anti-tumorigenic, anti-angiogenic, and anti-invasive functions.141–145 In this context, they express tumor suppressor proteins including P63, tumor protein p73, laminin 1, and maspin.80,146,147 However, DCIS-associated MECs differ from normal MECs. Evidence suggests that DCIS-associated MECs fail to polarize luminal epithelial cells.148 Discordant markers have been identified between normal and DCIS-associated MECs on immunohistochemical analysis; these include P63, calponin, CD10, and αvβ6 integrin.147,149–152 Notably, Ding et al. found that the functions of normal MECs are partly maintained via an interactive network involving p63 and TCF7 which accomplished by regulation of extracellular matrix proteins and cell adhesion. They further observed a significant decrease of p63 + TCF7+ MECs in MCF10DCIS models compared to normal MECs which correlate with invasive progression.152 Thus, the downregulation of tumor suppressor markers and upregulation of pro-invasive markers in DCIS-associated MECs indicate the possibility of achievement of an invasive phenotype, despite the presence of morphological similarities with normal MECs. It is therefore speculated that the progression from DCIS to IDC may be partly attributed to alterations in MECs.
Previous, some in vivo and in vitro studies have demonstrated that the progression from DCIS to IDC is regulated by loss of MEC integrity and function.140,147,153–155 Emerging evidence supports the hypothesis that MECs which are conventionally regarded as natural tumor suppressors may act as a promoter during DCIS progression. Upregulation of αvβ6 integrin which expressed in MECs may promote DCIS progression via activation of the transforming growth factor-beta (TGFβ) signaling pathway.147,156 The activation of TGFβ signaling has been found to induce the upregulation of matrix metallopeptidase (MMP), specifically MMP9 and MMP13, which play crucial roles in driving the progression of DCIS by contributing to the remodeling of the extracellular matrix and facilitating the invasive properties of DCIS cells.157,158 These findings were further supported by clinical observations that elevated MMP13 expression in myoepithelial cells associated with high-grade DCIS cases in Gibson et al.’s study.158 In addition, myoepithelial cells in DCIS were found to contribute to tumor-promoting effects through increased expression of C-X-C motif chemokines, such as CXCL12 and CXCL14. Both of which have been shown to promote the migration and invasion of DCIS tumor cells.159
Immune cells
The immune system is accepted to be a vital component of the TME. A single-cell atlas of breast cancer has revealed the presence of diverse immune cells in the TME including T cells, myeloid cells, macrophages, natural killer cells, and B cells, with T and myeloid cells being the most abundant.160,161 Immune cells may suppress tumor growth and metastasis by immune surveillance or drive tumor growth by immunosuppression.162 Within the DCIS, the proportion of T cells, B cells, macrophages, and Tregs stands significantly elevated in comparison to the neighboring normal tissue. While within the IDC, the proportion of all T cells (including helper, cytotoxic, and regulatory subtypes), B cells, macrophages, and PD-L1+ immune cells experiences a noteworthy increase in contrast to the adjacent DCIS.163,164
Tumor-infiltrating lymphocytes (TILs) have been broadly considered as significant components of the TME. They refer to a cluster of T and B cells that migrate into the tumor and stroma during tumor progression. Previous studies have focused on a subset of TILs mostly having CD3, CD4, CD8, and FOXP3 markers.165–169 A higher density of TILs in IDC has been found to be associated with a generally favorable prognosis and better response to adjuvant therapy in clinical trials.170,171 In particular, CD8 + T, CD4 + T helper, and CD20 + B cells always indicate good prognosis, while CD4 + FOXP3+ regulatory T cells are proven to drive tumor growth.172,173 TILs in DCIS has also been proved to play a potential role in prognostic significance.164,166 In comparison to pure DCIS, Toss et al. found that DCIS with synchronous IDC had significantly increased levels of CD8+, CD20+, FOXP3+, PD1+ and PDL1+ cells.166 Campbell et al. observed an increased abundance of CD8+, CD4+, CD20+, and FOXP3+ TILs in high-grade DCIS than low-grade DCIS.174 Thompson et al. also found increased numbers of CD3+ CD8+, CD4+, CD20+, and FOXP3+ TILs in grade II and III DCIS.175 What’s more, DCIS tissue is characterized by a higher abundance of CD4+ helper T cells compared to CD8+ cytotoxic T cells,175–177 the opposite is observed in IDC tissue, where cytotoxic T cells outnumber helper T cells.164 This discrepancy suggests that IDC exhibits a more pronounced immune-activated environment. There are hypothesis that: (1) greater immune cell activation around the disrupted MEC layer may subsequently trigger further disruption of this layer and result in basement membrane degradation, thereby favoring DCIS progression, or (2) MEC layer disruption and a highly reactive immune system may serve as protective factors against subsequent progression and recurrence.178
Macrophages were observed to infiltrate in DCIS. A study by Linde et al. using the MMTV-HER2 model showed that CD206hi macrophages could be drawn to DCIS via NF-κB-driven C-C motif ligand 2 (CCL2) production. The interaction of which subsequently led to heightened Wnt-1 secretion, resulting in myoepithelial disruption and the breakdown of E-cadherin junctions.179 More recently, a clinical study found that a high macrophage density in the stroma around DCIS was linked to less favorable outcomes. Specifically, DCIS with a greater density of CD163+ macrophages was indicative of recurrence and ipsilateral invasive recurrence.180
In brief, although numerous studies have demonstrated the crucial role of immune cells in DCIS progression, none of the cells could independently predict progression or recurrence alone. The potential mechanisms of DCIS-IDC transition that are regulated by the immune system and cooperate with other factors therefore warrant further evaluation.
Fibroblasts
Fibroblasts are the predominant component in the stroma, that produce extracellular matrix and cytokines and respond to the immune system. Fibroblasts also correlate with the polarity and proliferation of epithelium.172 Cancer-associated fibroblasts (CAFs) have been proven to play an important role in breast cancer progression. CAFs extracted from IDC differ considerably from those in the normal breast,181 and activated fibroblasts (myofibroblasts which express α-smooth muscle actin) have been found in large numbers in IDC. Fibroblasts are proven to promote the DCIS-IDC transition, while MECs suppress progression.140
Cytokines, proteases, and growth factors produced by CAFs have been found to facilitate tumor progression, including stromal cell-derived factor 1,181,182 TGF-β1,140,182 and hepatocyte growth factor.183,184 Studies by Hu et al. demonstrated that CAFs promoted DCIS invasion, primarily through triggering NF-κB and COX-2, which leads to an increase in MMP9 and MMP14 expressions,185 and further resulted in extracellular matrix (ECM) remodeling and basement membrane degradation respectively.140 Utilizing a 3D in vitro model, Osuala et al. delved into the role of interleukin 6 (IL-6) in the DCIS-IDC transition and discovered that CAFs-derived IL-6 initiated DCIS progression partly through cathepsin B-mediated ECM degradation.186 Moreover, Sameni et al. demonstrated an interplay between CAFs and myoepithelial cells; CAFs-derived IL-6 enhanced DCIS progression and invasion, while this could be attenuated by myoepithelial cells, partly due to inhibition of CAFs-mediated proteolysis of the extracellular matrix through inhibiting the production of CAFs-derived IL-6.187 In Bernard et al.’s study by using MMTV-PyVmT mouse model, they observed that CAFs-derived CXCL1 expression was more pronounced in IDC than in DCIS. CXCL1 produced by CAFs furthered DCIS progression via the CXCR2 receptor and the subsequent activation of various signaling cascades, including the MAPK, NF-κB, Akt, and Stat3 pathways.188
Actually, TME is an ecosystem in which progression from DCIS to IDC depends on an evolving spatial distribution and function of multiple cell types, rather than on any single cell subset. The crosstalk between tumor cells and TME components including MECs, immune cells, and fibroblasts synergistically drive DCIS progression. In a recent study, Risom et al. utilized advanced techniques named multiplexed ion beam imaging by time of flight (MIBI-TOF) to construct a spatial cellular map of the progression from DCIS to IDC.178 They identified that the myoepithelial layer in DCIS showed less phenotypic diversity and higher proliferation compared to normal tissue. Interestingly, this was accompanied by an increase of stromal CD4 + T cells and mast cells, which subsequently decreased in IDC. Along with the loss of myoepithelium, a greater number of proliferating CAFs and densely aligned fibrillar collagen was found in IDC, however, the potential regulatory mechanisms remain to be discovered. Current advanced techniques may allow investigation of the TME as an integral ecosystem rather than as one of the single-cell subtypes; they may further reveal spatial information and intrinsic interactive mechanisms in the TME.
Tools for DCIS-IDC research
As the underlying mechanisms of DCIS-IDC transition remain unclear, valuable tools including preclinical models and advanced new technologies are urgently needed for DCIS-IDC research. Both in vitro and in vivo preclinical models have their own advantages and disadvantages, and they should therefore be selected carefully as appropriate. Emerging advanced technologies, such as single-cell sequencing and spatial transcriptomics, have revolutionized our understanding of cancer biology. These powerful tools have provided new insights into DCIS-IDC transition.
In vitro and in vivo models for research on DCIS progression
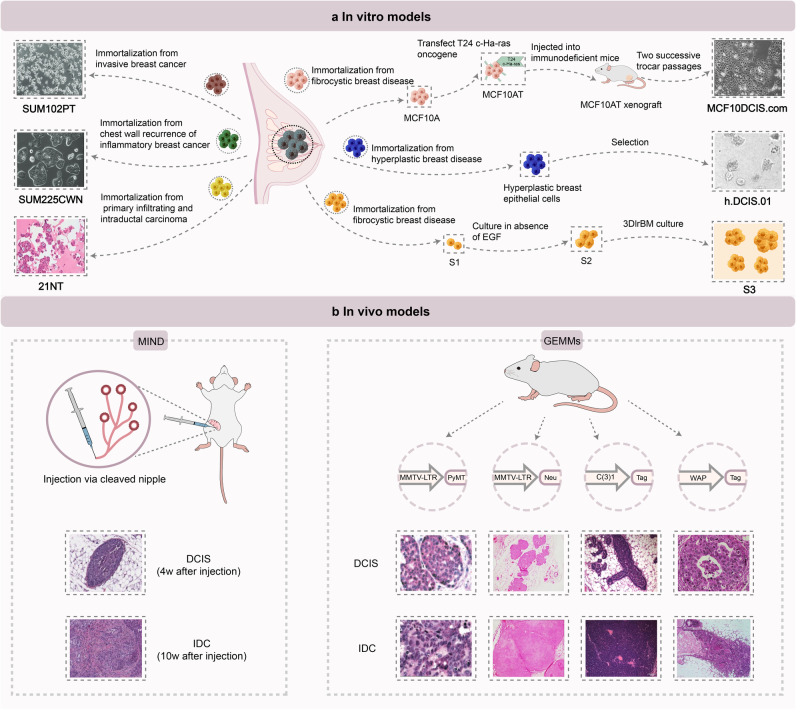
The study of DCIS-IDC transition has been hindered by lacking of suitable model systems and techniques that recapitulate human DCIS and their progression to IDC. In vitro models mainly include early 2-dimensional (2D) and modified 3D culture models, which allow the study of specific molecular pathways or evaluation of drug efficacy during DCIS progression by controlling experimental conditions. The widely used models including xenografts and genetically engineered mouse models (GEMMs) are suited for studying the biology process of DCIS in vivo. As different models have their own strengths and weakness, it is essential that researchers should choose the appropriate model based on the study aim (Fig. 5).
Fig. 5.
In vitro and in vivo models for DCIS progression research. a In vitro models, in which DCIS cell lines that include the MCF10 series,189 HMT-3522 series,191 21Tseries,193 SUM225CWN,189 SUM102PT,189 and h.DCIS.01197 authentically mimic human DCIS progression for 2D and 3D culture in vitro. b In vivo models, in which MIND models149 and GEMMs including MMTV-PyMT,233 MMTV-Neu,240 C3(1)/Tag,245 and WAP-T247 mouse are available for studying the biology of DCIS in vivo
DCIS cell lines
A few DCIS cell lines have been established and utilized in in vitro and in vivo models for the study of DCIS; these include MCF10DCIS.com from the MCF10 series,189,190 HMT-3522 series,191 21Tseries,192,193 SUM225CWN,194 SUM102PT,195 and h.DCIS.01 (Fig. 5a).196,197
Among these, the MCF10 progression series is regarded as one of the most widely used cell lines in functional studies on DCIS. It was derived from MCF10A, a spontaneously immortalized normal breast epithelial cell line derived from benign fibrocystic breast disease.198 A range of cell lines derived from MCF10A provide indispensable model systems that mimic breast cancer progression; these include MCF10AT (proliferation stage),199 MCF10DCIS.com (DCIS stage),190 and MCF10CA (invasive stage).200,201 MCF10DCIS.com is frequently utilized for mimicking human DCIS. Lee et al. used the DCIS.com cell line for constructing in vitro and in vivo models to investigate genes that may regulate DCIS progression. They found that CSTA, FAT1, and DST function as strongly suppressors of DCIS-IDC transition; they also obtained similar results when using the SUM225 and h.DCIS.01 DCIS cell lines.202 Maguire et al. observed driver mutations in TP53 and PIK3CA during transformation of the MCF10 progression series (from proliferation stage MCF10AT to invasive stage MCF10CA); this was similar to the findings in primary DCIS and IDC tissues.203 These results suggest that the MCF10 series represents a good model for DCIS research. In this context, these DCIS cell lines have been used frequently in both in vivo and in vitro studies on DCIS.
2D culture models
Prior to the use of 3D and in vivo models, conventional 2D cell culture models were the mainstay for breast cancer research.204 Studies on DCIS-IDC often use the 2D model to validate specific regulators or pathways that modulate DCIS progression. Compared 3D model, the 2D model offers the advantages of reduced costs, faster results, and easier use.205 However, it fails to reflect the natural construction of the tumor which reduces the potential to translate results into 3D, in vivo models, and clinical practice.206,207
MCFDCIS cells are widely used for imitating DCIS lesions in 2D models. To explore the role of peroxisome proliferator-activated receptor γ agonists in DCIS progression, Ory et al. used efatutazone, which could activate this pathway in a 2D culture model of the MCFDCIS cell line; the MCFDCIS cells showed an increase in the expression of luminal markers, similar to differentiating MCFDCIS cells. The authors concluded that peroxisome proliferator-activated receptor γ-induced differentiation of MCFDCIS cells could delay the progression from DCIS to IDC.208 However, studies have shown that results achieved in 2D models may differ from those obtained in 3D and in vivo models. Li et al. investigated the changes in sensitivity to mitogen-activated protein kinase inhibitors during breast cancer progression; they used the MCF10 cell line series and compared the results between 2D and 3D culture models.209 They found the results between these two models to be discordant, with increased sensitivity to these inhibitors in the 3D than in the 2D model. Similarly, Hu et al. identified certain differences between MCFDCIS 2D culture and xenograft models in terms of luminal and myoepithelial markers.140 Thus, as the 2D model has certain limitations, studies on DCIS need to employ 3D culture and in vivo models for further investigation.
3D culture models
Unlike the oversimplified 2D models, 3D models attempt to recapitulate the native tumor microenvironment in vitro by building a 3D culture environment that supports cell-cell and cell-matrix interactions.210,211 Emerging evidence suggests that the DCIS-IDC transition partially results from interactions between DCIS cells and their TME,139,140,186 and that 3D culture models provide a significant in vitro model by co-culturing tumor cells with stromal cells.187,211–215 Carter et al. developed a novel 3D culture model that could recapture DCIS using a native physiological bilayer arrangement of myoepithelial and luminal cells; this differed from the traditional use of cell lines to form spheroids.212
In summary, 3D models may partly recapitulate the complex TME by co-cultivating multiple cell types and may better mimic the architecture of DCIS in vitro.
Mouse-intraductal (MIND) model
The MIND model is a widely used xenograft model,149,216–220 which was first developed by Behbod et al.216 Two DCIS cell lines or a patient-derived DCIS line were transplanted into the mammary ducts of immunocompromised mice (Fig. 5b). Two DCIS cell lines, namely, MCF10DCIS.com (ER/ progesterone receptor/ HER2-negative) and SUM225 (HER2-positive) formed a basal and HER2-positive subtype xenograft model, respectively; both formed DCIS-like lesions during tumor formation and slowly developed to IDC.216 Valdez et al. identified the reproducible growth of patient-derived DCIS in NOD-SCID IL2rγ mice using the MIND model.217 In another study, the researchers found that only a proportion of the patient-derived DCIS (54%) line progressed to IDC over a median follow-up of nine months.218
Studies using MIND models to investigate the DCIS-IDC transition have reported on the underlying mechanisms of DCIS progression by using candidate promoters or suppressors.140,149,202,219 For instance, Elsarraj et al. targeted the B cell lymphoma-9 in a MIND model and found that it acted as a promoter of DCIS.219 Compared to other DCIS xenografts (such as xenotransplants of DCIS cell lines or patient-derived DCIS into the mammary fat pad), MIND models better mimic human DCIS lesions with recapitulation of the initial ductal environment.152,221–223 However, all these models are limited by the failure to explore the immune effect in DCIS progression (due to their immunodeficient hosts); they do not therefore fully mimic the natural evolution of human DCIS. The artificial construction of DCIS models using human DCIS cells and immunodeficient mice may therefore bias experimental results.
Mouse mammary tumor virus-polyoma middle tumor-antigen (MMTV-PyMT) model
Genetically engineered mouse model (GEMMs) are mostly utilized to elucidate the underlying mechanisms of breast cancer biology.224–227 Most transgenic models are constructed by targeting tissue-specific oncogenes in mice through tissue-specific promoters.228,229 The mammary-specific promoters that are commonly used to produce GEMMs include mouse mammary tumor virus long terminal repeat (MMTV-LTR) and whey acidic protein (WAP) and C(3)1.229–231 The MMTV-PyMT model is one of the most widely used GEMMs for breast cancer research. Notably, the MMTV is an important virus that can cause breast cancer in mice. The MMTV-PyMT is a transgenic model, which is generated by overexpression of the PyMT oncogene that is driven by the MMTV-LTR promoter (Fig. 5b).232 The MMTV-PyMT model has been demonstrated to undergo four stages during tumor progression (hyperplasia, adenoma/mammary intraepithelial neoplasia, DCIS, and IDC); this is similar to the course of DCIS progression in humans.233 The entire course lasts for 10–14 weeks; it generates multifocal tumors and leads to the rapid development of lung metastases. The similarity of pathological and molecular features between MMTV-PyMT and human samples make it a faithful model for research on DCIS progression.227
The lack of a natural immune environment in xenograft models makes it impossible to perform experiments pertaining to the immune response for the DCIS initiation and progression, however, the MMTV-PyMT model can compensate for this limitation. Martinez et al. crossed MMTV-PyMT with Foxp3DTR knock in mice to establish the regulatory T cell ablation model. They found that regulatory T cell ablation at the DCIS stage resulted in a more aggressive phenotype that promoted DCIS progression.234 Boyle and colleagues developed a MMTV-PyMT, C-C chemokine receptor 6-null mouse model and found that the chemokine receptor, CCR6, could promote DCIS initiation and progression by mediating pro-tumorigenic macrophages in the tumor microenvironment.235
In summary, the MMTV-PyMT model faithfully reproduces the natural progression of breast cancer with spontaneous tumor initiation. Although the MMTV-PyMT model compensates for weaknesses of the MIND model based on an intact immune system, this model does not authentically mimic human DCIS progression owing to the lack of actual human-derived cells and human tissue microenvironment.204
MMTV-neu model
MMTV-neu is another GEMM used for modeling breast cancer progression. The prominent ErbB2/HER2/Neu oncogene is associated with breast cancer initiation.236 Reports suggest that ErbB2 is overexpressed in nearly 15–20% of breast cancers.237 The MMTV-neu model is especially designed for mimicking ErbB2-amplified breast cancer progression.
Notably, the MMTV-neu model was constructed by ErbB2 oncogene overexpression in mice and was driven by the MMTV promoter (Fig. 5b).238 Studies have validated that the model can generate pre-invasive disease stage that exhibit histological similarities to human DCIS.228,236 Based on this characteristic, Lezzi et al. developed a dynamic mouse model (Balb-NeuT mice) for studying breast cancer progression in vivo. This transgenic model was induced by expressing the r-neuT oncogene driven by MMTV-LTR promoter in BALB/c mice; it underwent transformation from atypical hyperplasia to DCIS, became invasive, and subsequently metastasized.239 By using this model, Hosseini et al. found that nearly 80% of metastatic lesions had developed by dissemination from very early lesions (DCIS) rather than those in later stages.240 Similarly, Harper et al. found early disseminated cancer cells to be associated with HER2 upregulation on using MMTV-neu mice.241
Thus, the MMTV-neu model may be an ideal transgenic model for the study of HER2-positive DCIS. However, it remains unclear whether progression of hyperplasia in the MMTV-neu model can replicate the progression of human HER2-positive DCIS.
C3(1)/Tag and WAP-T model
The SV40 large T-antigen (Tag) is an efficient inducer of tumor formation; this is achieved by inactivation of tumor suppressor proteins such as p53 and retinoblastoma protein.242 Tag overexpression can lead to formation of breast and prostate cancers that histologically resemble the diseases found in humans.243 Similar to the MMTV-PyMT model, the C3(1)/Tag model can mimic human DCIS progression in mice. Green et al. found that the C3(1)/Tag model showed ductal atypia at an age of 8 weeks; it then progressed to DCIS at approximately 12 weeks and to IDC at 16 weeks.244 A recent study that used single-cell sequencing showed the existence of pre-DCIS, DCIS, and IDC-like lesions in the C3(1)/Tag model and further demonstrated that DCIS progression was associated with microenvironmental changes.245 Comparison of conserved gene expression between the C3(1)/Tag model and human breast cancers revealed that it represented the basal-like subtype found in humans.245,246 Another set of transgenic mice, namely, WAP-T mice, were introduced by Schulze-Garg et al. in 2000; they were driven by the SV40 large-T antigen and were induced by the whey acidic protein promoter (Fig. 5b). These mice also develop typical DCIS and IDC-like lesions that morphologically resemble those found in humans.247
New technologies for research on DCIS progression
Single-cell sequencing
Single-cell sequencing is a rapidly developing tool.248 Bulk sequencing only delineates average biological information of bulk cell populations;249 in contrast, single-cell sequencing provides an insight into the single cell level and has revolutionized understanding on tumor biology.
In recent years, single-cell sequencing has made considerable advances in cancer research, with the first single-cell ribonucleic acid (RNA) sequencing (scRNA-seq) study performed in 2009,250 the first single-cell deoxyribonucleic acid sequencing conducted in 2011,251 and the first single-cell exome sequencing completed in 2012.252,253 As intratumor heterogeneity is considerably common in breast cancer,254–256 the application of single-cell sequencing is particularly appropriate for breast cancer research.115,257 Studies that analyzed clone evolution in breast cancer using single nucleus sequencing,251 in which the whole-genome and exome single-cell sequencing258 have revealed the clonal relationships among subpopulations.251 A recent study by Wang et al. introduced a high-throughput single-cell DNA sequencing technique----Arc-well, specifically designed for the analysis of archival FFPE samples.98 While most previous genomic studies of DCIS have been limited to single time-point samples, either utilizing synchronous DCIS-IDC samples or mismatched pairs of DCIS and IDC, Arc-well made the analysis of FFPE sample possible. Arc-well is the first method capable of sequencing FFPE tissue that have been stored for decades, facilitating gene copy number analysis in thousands of single cells. By using this method, the research team conducted a systematic study on primary DCIS and their matched recurrent DCIS or IDC samples. Through this investigation, they unveiled an evolutionary bottleneck model of DCIS progression, providing crucial insights for the treatment of primary DCIS.
Single-cell sequencing has also been used to investigate the role of rare populations in breast cancer progression that could not be detected by prior technologies.259 This indicates that it has the potential to identify a rare population of cells in DCIS which may play a pivotal role in promoting DCIS-IDC transition. However, single-cell sequencing has certain limitations; single cells isolated from bulk tissues lose their spatial information, and the process of isolation may influence the cell status (such as causing dissociation-induced gene expression).260 In brief, single-cell sequencing has provided a detailed overview of individual cells. However, emerging powerful tools such as multi-omics offer more information than genomic or transcriptomic evaluation261 which is expected to bring remarkable benefits for future research on DCIS-IDC transition.
Spatial transcriptomics
As scRNA-seq fails to capture in situ spatial information and reflect intercellular communication within tissues, spatial transcriptomics may compensate for these limitations by providing a comprehensive atlas of tissue structure. To date, three main technologies have been developed for detecting spatial transcriptomic information; these include fluorescence in situ hybridization-based, in situ RNA sequencing-based, and spatial barcoding technologies.262,263
In addition to scRNA-seq, spatial transcriptomics can be a potent tool for investigating the heterogeneity of cancers.264–267 Ståhl and colleagues first introduced the in-situ spatial labeling technology and applied it to breast cancer with synchronous DCIS-IDC components; they found considerable spatial intratumoral heterogeneity among different ductal regions of DCIS. Gene expression was found to be divergent between these regions, probably indicating the different subclones that contributed to DCIS progression.268
Spatial transcriptomics can profile spatial information in tissues, but lacks of complete single-cell resolution similar to scRNA-seq. Recent studies have attempted to integrate these two methods to clearly delineate a single cell spatial atlas.264,266,269,270 Wei et al. developed a novel computational method, namely, CellTrek to integrate single cell and spatial data;269 they further applied CellTrek to two DCIS samples. In one sample, they identified three main tumor subclones; however, the different subclones were mapped to distinct ductal regions, implying extensive spatial intratumor heterogeneity in DCIS. In another sample with synchronous IDC components, they succeeded to map the spatial tumor-immune microenvironment and demonstrated the presence of tertiary lymphoid structures.
In addition to improving understanding on breast cancer biology, spatial transcriptomics can provide new insights into clinical diagnosis of DCIS and IDC and identify predictive markers for DCIS recurrence and treatment.271,272 In the study by Yoosuf et al., signatures generated from spatial transcriptomic data of expert-defined DCIS and IDC tissue sections offered highly accurate diagnoses of DCIS and IDC.272 In summary, spatial transcriptomics can effectively characterize the gene expression profiles of different cells while retaining the corresponding spatial information, and has considerable potential for providing a detailed spatial map of intercellular communication. However, more advanced technologies are needed for modification.
Artificial intelligence
The utilization of artificial intelligence (AI) has presented a viable solution for revolutionizing the interpretation and analysis of histological images. AI possesses the remarkable capability to learn and identify distinctive patterns and interconnections within biological tissues, thereby facilitating a significant transformation in this field.273 In DCIS-related studies, utilization of digital image analysis can help pathologists to histologically distinguish DCIS from IDC and differentiate well between different grades of DCIS.274,275 Specially designed AI model has been applied to evaluate future breast cancer risk, including the risk of developing DCIS and IDC.276 Recently, AI has been rapidly developed in accurately identifying DCIS among other pre-invasive breast lesions as well as in determining the prognostic outcome of DCIS.273 Diagnostic disagreements between DCIS and pre-invasive lesions like usual ductal hyperplasia (UDH) and ADH is common in pathologists.277 A study performed by Hayward et al. assessed the efficiency of machine learning in differentiating DCIS from atypia, the diagnosis accuracy of which was higher than that of the pathologists while specificity of the feature classification was similar to pathologists.278 Moreover, Yamamoto et al. applied a machine learning system to classify four pre-invasive stages of normal tissues, UDH, low and high-grade DCIS solely relying on morphological distinctions in myoepithelial cells, without the need for any information regarding tumor cells.279 Given that the distinct composition of the stroma surrounding DCIS compared to normal breast tissue and IDC,280 Bejnordi et al. used a deep learning algorithms to distinguish different grades of DCIS and found that the amount of tumor-associated stroma generally increased with higher lesion grade. Interestingly, DCIS with synchronous IDC possessed higher amounts of tumor-associated stroma than pure DCIS.281 Klimov et al. conducted an investigation of developing a risk classifier using machine learning-based image analysis on H&E staining of DCIS. Their analysis primarily focused on capturing the spatial relationships between various components of the DCIS, including normal and cancer ducts, lymphocyte region, stroma, and blood vessels. Interestingly, this innovative approach demonstrated remarkable success in accurately predicting the 10-year risk of DCIS recurrence.282
Studies to date have exhibited promising advances in applying AI in identifying novel features of DCIS and DCIS progression, which surpassing the dependence of traditional clinicopathological variables. However, it is essential to subject these methods to further testing in extensive patient cohorts with long-term follow-up. Only through rigorous evaluation, the clinical utility of these AI techniques in guiding DCIS management can be ascertained.
Clinical advancements in DCIS and IDC
Prognostic biomarkers of DCIS
Approximately 20% of DCIS cases are at risk of future recurrence despite current treatments.283 DCIS recurrence is associated with numerous clinical and pathological features.14,284,285 In terms of nuclear features, DCIS is classified into three grades, namely, low, intermediate, and high.14 Clinically, high-grade DCIS has been demonstrated to possess a significantly higher risk of progression to IDC and subsequent DCIS recurrence than low or intermediate grade DCIS.7,286,287 In addition to these well-established risk factors, certain prognostic biomarkers that can predict progression or recurrence and guide treatments are under evaluation; however, they are rarely used in clinical practice.
The ER is a well-established prognostic biomarker and predictor of treatment response in IDC. However, its prognostic and predictive role in DCIS remains unclear. Several studies have reported a significantly lower risk of recurrence in ER-positive DCIS28,288–292 compared to ER-negative cases; however numerous studies have found no significant association.293,294 Based on the UK/ANZ DCIS trial, a recent study by Thorat et al. used the “clonal method” that scored ER status by clonality of ER expression to analyze the relationship between ER expression and subsequent recurrent events.290 The investigators distinguished ER-multi-clonal DCIS (labeled as ER-positive DCIS but showed presence of at least one carcinoma in situ with a complete lack of ER expression) from conventionally classified ER-positive DCIS which lacked complete ER expression in one or more DCIS ducts. Unexpectedly, a similar recurrence risk was found in ER-multi-clonal DCIS and ER-negative DCIS; this was higher than that of ER-completely positive DCIS. This study provided a novel method for determining the ER status more accurately, and avoiding misclassification; this may facilitate evaluation of the actual prognostic value of ER. In this context, a robust association has been found between ER-positvie status and other prognostic factors including lower nuclear grade, smaller tumor size, HER2-negative status, and the absence of comedo necrosis.293 Notably, ER expression is higher in low-grade DCIS but lower in high-grade DCIS.14
HER2 status is commonly assessed in IDC and is considered an independent prognostic factor for poor outcomes.295 However, the predictive value of HER2 for DCIS progression and recurrence remains controversial. Several studies have shown that HER2-positive DCIS is associated with a higher risk of relapse. In clinical trials, HER2-positive DCIS tends to recur in situ, while HER2-negative DCIS has a lower recurrence rate.19,292,296–298 Based on these findings, it was supposed that the majority of IDC cases may arise from HER2-negative DCIS (or the HER2-negative subclone of DCIS in cases of heterogeneous HER2 expression). Moreover, HER2 overexpression is more common in pure DCIS than in DCIS with synchronous IDC, suggesting that HER2 expression may not be associated with progression from DCIS to IDC.20,299,300 To summarize, HER2-positive DCIS is more likely to relapse, whereas HER2-negative DCIS has a lower recurrence rate. HER2 expression in DCIS is correlated with other poor prognosis indicators such as higher nuclear grade, larger tumor size, ER-negative status, etc.19,298,301,302 However, the exact role of HER2 in DCIS initiation and DCIS-IDC transition still remains unclear.
Cyclooxygenase 2 (COX-2) expression is regarded as a potential biomarker in DCIS for the prediction of recurrence; it is also of value as a therapeutic target. COX-2 is highly expressed in both DCIS and IDC, with no significant differences in expression; notably, it is considered to play an important role in early breast cancer carcinogenesis.303,304 However, recent studies have demonstrated that COX-2 could be a promising marker for predicting progression from DCIS to IDC.305 It was further found that high COX-2 expression in DCIS with large adipocytes predicted a high risk of subsequent IDC.306 Other studies have reported that COX-2-positive DCIS may either recur as DCIS or progress to IDC.307,308 Notably, COX-2 overexpression in DCIS is also related to the expression of markers indicative of poor prognosis, such as higher nuclear grade, ER-negative status, and HER2-positive status.14,304
Ki67 is a predictor of cellular proliferation; high Ki67 expression in DCIS has been demonstrated to be associated with an increased risk of recurrence as either DCIS or IDC.93,309–311 An increased risk of subsequent progression to IDC has also been identified in DCIS with combined expression of p16, COX-2, and Ki67.292,312
In addition to the above potential protein biomarkers, gene expression detected at the mRNA level also shows prognostic value. The Oncotype DX DCIS score® (Genomic Health) multi-gene panel is the only commercially available tool with a clinically validated signature for prognostication in DCIS. It is a 12-gene panel that includes 7 and 5 cancer-related and reference genes, respectively, and classifies DCIS into low-, intermediate-, and high-risk categories.313,314 In addition to predicting the risk of recurrence, it helps to guide treatments. However, the Oncotype DX DCIS score® only predicts the 10-year risk of recurrence after breast-conserving surgery without radiotherapy for primary DCIS; this limits its clinical application to a certain extent.315,316 As it has only been validated by the ECOG-ACRIN E5194 trial in a low-risk DCIS patient cohort which met strict criteria, this novel prognostic tool has not been widely adopted.
In summary, the biomarkers discussed previously have not demonstrated good performance as independent predictors of DCIS recurrence and progression. Using a combination of two or more markers may offer better prognostication than an individual marker in certain cases. Most markers correlate with the well-established clinicopathological factors that include age, margin status, tumor size, nuclear grade, and presence of comedo necrosis.27,28,317 However, inadequate validation and their uncertain prognostic value have prevented their widespread adoption in clinical practice. The integration of emerging biomarkers and initial clinicopathological factors is expected to offer more reliable stratification of DCIS (based on the risk of recurrence or progression to IDC) in the future.
Treatment for DCIS
Owing to uncertainties regarding the risk of evolution to IDC or recurrence, current therapeutic approaches for DCIS remain controversial (especially in terms of over- or under-treatment). The primary objective of treatment is to prevent both, potentially progressive DCIS from progressing to malignant IDC and future recurrence. Patients with DCIS commonly undergo surgery and adjuvant therapy. However, there are inaccuracies in DCIS risk stratification and the treatment of DCIS remains controversial. Here, we have summarized all current treatments and related clinical trials for DCIS in Table 2.
Table 2.
Current treatments and relevant clinical trials for DCIS
| Trial name | Start year | Participant | Median follow-up (year) | DCIS feature | Control arm | Experimental arm | All new breast events (%) | Status | |
|---|---|---|---|---|---|---|---|---|---|
|
Surgery, radiation therapy |
NSABP B-17 | 1985 | 818 | 10.7 | / | BCS | BCS + RT | 35.1 VS 17.7 | Completed |
| EORTC 10853 | 1986 | 1010 | 10.5 | / | BCS | BCS + RT | 27 VS 15 | Completed | |
| SweDCIS | 1987 | 1067 | 8.0 | / | BCS | BCS + RT | 32 VS 20 | Completed | |
| UK/ANZ DCIS | 1990 | 1701 | 12.7 | / | BCS (±TAM) | BCS + RT (±TAM) | 21.7 VS 9.6 | No longer recruiting | |
| RTOG 9804 | 1999 | 636 | 7.2 | Good risk DCIS | BCS (±TAM) | BCS + RT (±TAM) | 6.7 VS 0.9 | Completed | |
| Endocrine therapy | NSABP B-24 | 1991 | 1804 | 7.0 | ER-positive DCIS | BCS + RT+placebo | BCS + RT + TAM | 31 VS 20 | Completed |
| NSABP B-35 | 2003 | 3104 | 9.0 | HR-positive DCIS | BCS + RT + TAM | BCS + RT + ANA | 7.9 VS 5.8 | Completed | |
| IBIS II DCIS | 2005 | 2980 | 7.2 | HR-positive DCIS | BCS + RT + TAM | BCS + RT + ANA | 5 VS 5 | Completed | |
| Active surveillance | LORIS | 2014 | 181 | / | Low risk DCIS | Surgery | AS | / | No longer recruiting |
| COMET | 2017 | 997 | / | Low risk DCIS | GCC | AS | / | Active, not recruiting | |
| LORD | 2017 | 2500a | / | Low risk DCIS | GCC | AS | / | Recruiting | |
| LORETTA | 2017 | 340a | / |
ER-positive, Low risk DCIS |
/ | TAM alone | / | Recruiting |
DCIS ductal carcinoma in situ, HR hormone receptor, ER estrogen receptor, BCS breast-conserving surgery, RT radiation therapy, TAM tamoxifen, ANA anastrozole, GCC guideline concordant care, AS active surveillance
aEstimated participant
Surgery and radiation therapy
Mastectomy and breast-conserving surgery are the most two common surgical approaches for DCIS; breast-conserving surgery followed by radiation therapy has been regarded as an acceptable surrogate for mastectomy.318–320 Mastectomy may be advised based on risk factors including tumor size, type of DCIS, patient age and preference, and recurrence risk.321 For radiation therapy, whole-breast radiation therapy is recommended in a majority of DCIS cases after breast-conserving surgery with the aim of reducing recurrence.322
In the clinic, mastectomy is advised for patients with multicentric DCIS or DCIS with microcalcifications scattered along the breast ducts. It remains unclear whether breast-conserving surgery followed by radiation therapy can be advised in these cases.323–325 By recruiting all kinds of DCIS, four randomized clinical trials including NSABP B-17, EORTC 10853, SweDCIS, and UK/ANZ DCIS (Table 2) have reported a reduction of local recurrence rate by approximately 50% following breast-conserving surgery plus adjuvant radiotherapy compared to breast-conserving surgery alone326–330 The survival benefits of radiation therapy following breast-conserving surgery have been further confirmed. In the RTOG 9804 trial (Table 2), which compared the impact of radiation therapy versus that of observation alone after breast-conserving surgery in good-risk DCIS, the risk of 15-year cumulative overall ipsilateral breast recurrence was found to be 7.1% in the radiation therapy group versus 15.1% in the observation group; the corresponding rates of invasive recurrence were found to be 5.4% versus 9.5%, respectively.330,331
Endocrine therapy
In ER-positive DCIS, systemic endocrine therapy can reduce both ipsilateral recurrence and contralateral breast cancer, albeit without a significant impact on overall survival.322,332–334 In this context, the NSABP B-24 and UK/ANZ DCIS trials (Table 2) evaluated the impact of tamoxifen treatment on recurrence and survival in DCIS.326,329 In the NSABP B-24 trial, patients underwent breast-conserving surgery and radiation therapy; this was followed by treatment with either tamoxifen or placebo. The tamoxifen group demonstrated a relatively lower (by 32%) incidence of ipsilateral IDC recurrence compared to the placebo group.326 In the UK/ANZ DCIS trial, patients were recruited into a 2 × 2 factorial trial of radiation therapy, tamoxifen, or both treatments after breast-conserving surgery.329 The tamoxifen group showed a reduced incidence of both, recurrent ipsilateral DCIS and contralateral new tumors; however, the incidence of recurrent ipsilateral IDC did not differ significantly. Notably, tamoxifen plus radiation therapy failed to show benefit compared with radiation therapy alone. The different results from these two trials may be explained by the younger average age of the participants in the UK/ANZ DCIS trial. Unfortunately, the ER status was not considered in either trial; however, further retrospective analysis of data from the NSABP B-24 and UK/ANZ DCIS trial identified the benefit from tamoxifen to be confined to the ER-positive DCIS group. By contrast, no tamoxifen benefit was found in ER-negative DCIS group.335,336
The IBIS-II DCIS and NSABP B-35 trials (Table 2) compared the effect of anastrozole (a non-steroidal aromatase inhibitor) with that of tamoxifen in hormone receptor-positive postmenopausal DCIS patients.337,338 They found that anastrozole could be a comparable surrogate for tamoxifen, as it showed a significant improvement in the breast cancer-free interval in postmenopausal population who were younger than 60 years. Both trials found a similar incidence of adverse events with anastrozole or tamoxifen. Despite its effectiveness in reducing recurrent events in hormone receptor-positive DCIS, the side effects of endocrine therapy should be taken into consideration on an individual basis.
Other treatments
In order to avoid overtreatment, active surveillance has been tested as an option in low-risk DCIS. Active surveillance refers to the use of regular mammography or other imaging examinations, with intervention only in cases where IDC is detected. Four ongoing prospective randomized trials are comparing active surveillance with standard treatment for low-risk DCIS; these include the LORIS from the UK, LORD from the Netherlands, COMET from the USA, and LORETTA from Japan (Table 2).339–342 A computational risk analysis conducted by Ryser et al. compared the disease-specific mortality between active surveillance and usual treatment for DCIS, they concluded that active surveillance could be a viable option for patients with DCIS, especially in older individuals.343
The efficacy of anti-HER2-targeted therapy has been established in treating HER2-positive IDC. However, the effectiveness of anti-HER2 therapy in reducing recurrence and improving survival in DCIS remains unclear. A clinical trial previously found that anti-HER2 dendritic cell vaccination could induce a tumor-specific T cell reaction, resulting in improved outcomes in patients with HER2-positive DCIS.344 Although HER2 receptor-targeted therapy is the standard in HER2-positive IDC, its role in HER2-positive DCIS needs further validation. The first prospective randomized phase III clinical trial (NSABP B-43) on high-risk HER2-positive DCIS evaluated whether trastuzumab (a HER2 inhibitor) and radiation therapy could reduce recurrence compared to radiation therapy alone (Table 2);345,346 the combination failed to meet the target of 36% reduction in ipsilateral breast cancer recurrence and showed a statistically non-significant reduction of 19%. Thus, from current clinical trial results, the use of adjuvant trastuzumab was not supported.
In summary, the current treatment and management approaches for DCIS often lead to under- or over-treatment due to a lack of precise risk stratification for patients. Moving forward, there is a need to implement personalized treatment strategies for both non-progressive and progressive DCIS, ensuring that patients receive appropriate and tailored care (Fig. 6).
Fig. 6.
Future management of non-progressive and progressive DCIS. Historically, distinguishing between progressive and non-progressive DCIS in women was challenging, resulting in overtreatment with limited benefits. It is crucial to find a solution to this problem. Advanced technologies, such as experimental models, multi-omics, single-cell sequencing, spatial transcriptomics, artificial intelligence and other emerging technologies, can facilitate the development of a risk stratification system for personalized treatment. Tailoring treatments for non-progressive DCIS differently from progressive DCIS is essential to alleviate the unnecessary burdens associated with overtreatment
Treatment for IDC
Treatment for IDC varies by their molecular subtypes, that include ER-positive, HER2-positive, and TNBC. Typically, a multimodal strategy is employed, combining surgery, radiation therapy, and systemic therapies. Surgical intervention plays a crucial role in the treatment of IDC. Mastectomy and breast-conserving surgery are the two widely used surgical approaches.347 Postoperative radiation therapy reduces the recurrence and enhances the survival rates of patients with lymph node involvement and/or who undergo breast-conserving therapy.348 Both surgery and radiation therapy are localized treatments for IDC. Nevertheless, systemic therapies, such as endocrine therapy, targeted therapies, chemotherapy, and immunotherapy, have made significant progress in recent years, significantly improving the prognosis of breast cancer. Table 3 presents the representative relevant phase 3 clinical trials of drugs for breast cancers. These drugs have revolutionized the disease trajectory for all subtypes of IDC.
Table 3.
Phase 3 randomized clinical trials of drugs for breast cancer
| Drug | Trial | Participant | Arms | POM | Status | Reference |
|---|---|---|---|---|---|---|
| a. ER-positive breast cancer | ||||||
| Aromatase inhibitor | ||||||
| Anastrozole | ATAC | 9366 | Anastrozole vs. tamoxifen vs. anastrozole + tamoxifen | DFS | Completed | ATAC Trialists’ Group.351 |
| Anastrozole | TARGET | 668 | Anastrozole vs. tamoxifen | TTP, OR | / | Bonneterre et al.385 |
| Anastrozole | The North American | 353 | Anastrozole vs. tamoxifen | OR, CR, PR, TTP | / | Nabholtz et al.386 |
| Letrozole | / | 907 | Letrozole vs. tamoxifen | TTP | / | Mouridsen et al.387 |
| Exemestane | SOFT and TEXT | 5738 | Exemestane + OFS vs. tamoxifen + OFS | DFS | Active, not recruiting | Pagani et al.388 |
| CDK4/6 inhibitor | ||||||
| Abemaciclib | MONARCH 2 | 669 | Abemaciclib + fulvestrant vs. fulvestrant | PFS | Active, not recruiting | Sledge et al.389 |
| Abemaciclib | MONARCH 3 | 493 | Abemaciclib + anastrozole or letrozole vs. anastrozole or letrozole | PFS | Active, not recruiting | Goetz et al.356 |
| Palbociclib | PENELOPE-B | 1250 | Palbociclib (1 yr) + ET vs. ET | iDFS | Completed | Loibl et al.390 |
| Palbociclib | PALLAS | 5760 | Palbociclib (2 yrs) + ET vs. ET | iDFS | Active, not recruiting | Mayer et al.391 |
| Palbociclib | monarchE | 5637 | Abemaciclib + ET vs. ET | iDFS | Active, not recruiting | Johnston et al.392 |
| Palbociclib | PALOMA-3 | 521 | Palbociclib+ fulvestrant vs. fulvestrant | PFS | Completed | Cristofanilli et al.393 |
| Ribociclib | MONALEESA-2 | 668 | Ribociclib+ letrozole vs. letrozole | PFS | Completed | Hortobagyi et al.394 |
| Ribociclib | MONALEESA-3 | 726 | Ribociclib+ fulvestrant vs. fulvestrant | PFS | Completed | Slamon et al.395 |
| Ribociclib | MONALEESA-7 | 672 | Ribociclib+ ET vs. ET | PFS | Completed | Lu et al.396 |
| b. HER2-positive breast cancer | ||||||
|---|---|---|---|---|---|---|
| Monoclonal antibody | ||||||
| Trastuzumab | HERA | 5099 | Trastuzumab (2 yrs) vs. trastuzumab (1 yr) vs. observation | DFS | Completed | Piccart-Gebhart et al.397 |
| Trastuzumab | NSABP B-31/N9831 | 3351 | Trastuzumab+ chemo. vs. chemo. | DFS | Completed | Romond et al.398 |
| Trastuzumab | BCIRG 006 | 3222 | AC → T vs. AC → TH vs. TCH | DFS | Completed | Slamon et al.399 |
| Pertuzumab | CLEOPATRA | 808 | Pertuzumab + trastuzumab + docetaxel vs. trastuzumab + docetaxel | PFS | Completed | Swain et al.400 |
| Pertuzumab | APHINITY | 4804 | Pertuzumab vs. placebo | iDFS | Active, not recruiting | von Minckwitz et al.362 |
| Margetuximab | SOPHIA | 536 | Margetuximab vs. trastuzumab | PFS | Completed | Rugo et al.401 |
| Tyrosine kinase inhibitor | ||||||
| Lapatinib | NeoALTTO | 455 | Lapatinib vs. trastuzumab vs. lapatinib + trastuzumab | pCR | Completed | Baselga et al.402 |
| Lapatinib | GeparQuinto | 620 | Lapatinib+ chemo. vs. trastuzumab+ chemo. | pCR | Completed | Untch et al.403 |
| Lapatinib | NSABP B-41 | 529 | Lapatinib+ chemo. vs. trastuzumab+ chemo. vs. lapatinib + trastuzumab+ chemo. | pCR | / | Robidoux et al.404 |
| Lapatinib | ALTTO | 8381 | Trastuzumab vs. lapatinib vs. trastuzumab followed by lapatinib vs. trastuzumab + lapatinib | DFS | Completed | Piccart-Gebhart et al.405 |
| Neratinib | NALA | 621 | Neratinib+ capecitabine vs. lapatinib + capecitabine | PFS | Complete | Saura et al.406 |
| Neratinib | ExteNET | 2840 | Neratinib vs. placebo | iDFS | Complete | Chan et al.407 |
| Tucatinib | HER2CLIMB | 612 | Tucatinib + capecitabine + trastuzumab vs. placebo + capecitabine + trastuzumab | PFS | Complete | Murthy et al.408 |
| Antibody–drug conjugates | ||||||
| T-DM1 | EMILIA | 991 | T-DM1 vs. lapatinib + capecitabine | PFS | Complete | Verma et al.365 |
| T-DM1 | KATHERINE | 1486 | T-DM1 vs. trastuzumab | iDFS | Active, not recruiting | von Minckwitz et al.409 |
| T-DXd | DESTINY-Breast03 | 524 | T-DXd vs. T-DM1 | PFS | Active, not recruiting | Cortés et al.366 |
| c. TNBC | ||||||
|---|---|---|---|---|---|---|
| PD1 inhibitor | ||||||
| Pembrolizumab | SWOG 1418 | 1155 | Pembrolizumab vs. observation | iDFS | Active, not recruiting | / |
| Pembrolizumab | KEYNOTE-119 | 622 | pembrolizumab vs. chemo. | OS | Completed | Winer et al.410 |
| Pembrolizumab | KEYNOTE-355 | 847 | Pembrolizumab + chemo. vs. chemo. | AEs, PFS, OS | Active, not recruiting | Cortes et al.411 |
| Pembrolizumab | KEYNOTE-522 | 1174 | Pembrolizumab + chemo. vs. chemo. | pCR, EFS | Active, not recruiting | Schmid et al.412 |
| PD-L1 inhibitor | ||||||
| Atezolizumab | IMpassion031 | 333 | Atezolizumab + chemo. vs. chemo. | pCR | Completed | Mittendorf et al.413 |
| Atezolizumab | IMpassion130 | 902 | Atezolizumab + nab-paclitaxel vs. nab-paclitaxel | PFS, OS | Completed | Schmid et al.383 |
| Atezolizumab | IMpassion132 | 572 | Atezolizumabe + chemo. vs. chemo. | OS | Active, not recruiting | / |
| Avelumab | A-Brave | 474 | Avelumab vs. observation | DFS | Active, not recruiting | / |
| PARP inhibitor | ||||||
| Olaparib | OlympiA | 1836 | Olaparib vs. placebo | iDFS | Active, not recruiting | Geyer et al.414 |
| Olaparib | OlympiAD | 302 | Olaparib vs. TPC | PFS | Active, not recruiting | Robson et al.415 |
| Veliparib | BROCADE3 | 509 | Veliparib+ carboplatin–paclitaxel vs. carboplatin–paclitaxel | PFS | Active, not recruiting | Diéras et al.416 |
| Talazoparib | EMBRACA | 431 | Talazoparib vs. TPC | PFS | Completed | Litton et al.417 |
ER estrogen receptor, HER2 human epidermal growth factor receptor 2, TNBC triple-negative breast cancer, POM primary outcome measures, DFS disease-free survival, TTP time to progression, OR objective response, CR complete response, PR partial response, OFS ovarian functional suppression, PFS progression free survival, AEs adverse event, iDFS invasive disease-free survival, pCR pathologic complete response, OS overall survival, EFS event-free survival, AC → T doxorubicin + cyclophosphamide followed by docetaxel, AC → TH AC followed by docetaxel + Herceptin, TCH docetaxel + carboplatin + Herceptin, TPC physician’s choice
ER-positive IDC
Endocrine treatment strategies encompass various approaches, including reducing estrogen production, modulating ER signaling, and antagonizing the ER itself.349 Among these strategies, the selective ER modulator tamoxifen and aromatase inhibitors (AIs), which inhibit estradiol synthesis, have long been the cornerstone of treatment for ER-positive breast cancer (Table 3).350 AIs such as anastrozole, letrozole, fadrozole (available in Japan only), were specifically developed for postmenopausal women. Numerous clinical trials have investigated the efficacy of these drugs in treating postmenopausal patients.350 For instance, the ATAC trial evaluated postmenopausal women with early-stage HR-positive breast cancer, randomizing them to receive either anastrozole or tamoxifen. After a median follow-up of 10 years, anastrozole demonstrated superiority over tamoxifen in terms of disease-free survival (DFS), time to recurrence and distant recurrence, although no significant difference was observed in overall survival (OS).351 Another meta-analysis conducted by the Early Breast Cancer Trialists’ Collaborative Group (EBCTCG) analyzed the benefits of 5-year treatment with an aromatase inhibitor or tamoxifen in 31,920 postmenopausal women with ER-positive early breast cancer. The analysis revealed that compared to tamoxifen, 5 years of treatment with an aromatase inhibitor reduced the 10-year breast cancer mortality by 15%.352 In addition, several randomized trials have aimed to evaluate the efficacy of ovarian function suppression (OFS) and AIs in premenopausal women. The SOFT and TEXT trials compared 5-year treatment with exemestane plus OFS versus tamoxifen plus OFS in premenopausal women with ER-positive early breast cancer. After a median follow-up of 13 years, the combined analysis demonstrated that exemestane plus OFS significantly improved DFS and distant recurrence-free interval, although there was no significant difference in OS compared to the tamoxifen plus OFS regimen.353
In the past decade, the significance of utilizing combination therapy in the treatment of ER-positive disease in the metastatic setting has become increasingly evident. Extensive research on the crosstalk between ER signaling and cyclin D1/CDK46 has paved the way for numerous groundbreaking studies. Consequently, the development and approval of CDK4/6 inhibitors have revolutionized the treatment approach for HR-positive metastatic breast cancer (Table 3). Several phase III trials, utilizing CDK4/6 inhibitors such as palbociclib, ribociclib, and abemaciclib in combination with endocrine therapy, have demonstrated remarkable improvements in progression-free survival (PFS) and OS for patients with hormone receptor-positive, HER2-negative metastatic breast cancer.354–356
Furthermore, the activation of the PI3K/AKT/mTOR pathway has been identified in ER-positive breast cancer and is often associated with resistance to endocrine therapy. Targeting this pathway has emerged as a potential therapeutic strategy to overcome resistance.357 Currently, multiple clinical trials are underway to evaluate the efficacy and safety of various inhibitors, including pan-class I PI3K inhibitors, selective PI3Kα inhibitors, AKT inhibitors, and mTOR inhibitors.349
HER2-positive IDC
The identification of HER2 as a therapeutic target marked a significant breakthrough in breast cancer treatment. Following the introduction of the first HER2-targeted drug, trastuzumab, in 1990, the survival rates for HER2-positive breast cancer improved dramatically. Previously, the prognosis for HER2-positive breast cancer was similarly gloomy as that for TNBC. Trastuzumab, a humanized monoclonal antibody, binds to the extracellular domain of HER2 and exerts its effects by inhibiting dimerization, suppressing intracellular HER2 signaling through pathways such as MAPK and PI3K/AKT/mTOR, and facilitating antibody-dependent cell-mediated cytotoxicity (ADCC).358 Currently, the standard-of-care treatment for HER2-positive breast cancer involves neoadjuvant and adjuvant chemotherapy in combination with anti-HER2 therapy. Pertuzumab, another humanized anti-HER2 monoclonal antibody, with a distinct binding site from trastuzumab, prevents the heterodimerization of HER2 with HER1, HER3, and HER4, ultimately leading to the attenuation of intracellular HER2 signaling.359,360 Clinical trials have demonstrated noteworthy outcomes by combining these two monoclonal antibodies with chemotherapy for the treatment of HER2-positive breast cancer. In the CLEOPATRA trial, the combination of pertuzumab, trastuzumab, along with docetaxel, resulted in a median increase of 15.7 months in OS compared to the trastuzumab plus docetaxel only (Table 3).361 In another trial, APHINITY, observed similar improvement in invasive DFS in patients who received pertuzumab plus trastuzumab and chemotherapy compared to those who did not receive pertuzumab.362
Antibody–drug conjugates (ADCs) drugs have been designed to deliver the cytotoxic effects of chemotherapy specifically to tumor cells (Table 3).363 Trastuzumab emtansine (T-DM1) was the first ADC developed for targeting HER2, which comprises trastuzumab linked to the tubulin-binding agent DM1 via a stable thioether linker.364 T-DM1 has demonstrated efficacy in women with HER2-positive advanced-stage breast cancer who were previously treated with trastuzumab and a taxane.365 Recently, a novel ADC, T-DXd, involving a humanized HER2 antibody with the same sequence as trastuzumab conjugated to deruxtecan (DXd), has shown great activity, including in cases refractory to T-DM1.366,367
Other HER2-targeted therapies like tyrosine kinase inhibitors (TKIs), are small molecules that target the intracellular catalytic kinase domain of HER2 (Table 3). Lapatinib, a reversible inhibitor of HER1 and HER2, overcomes trastuzumab resistance in HER2-positive breast cancer.368 Neratinib, an irreversible panHER TKI that targets HER1, HER2 and HER4,369 improves the 2-year invasive DFS rate compared to placebo when administered after chemotherapy and adjuvant therapy with trastuzumab to women with HER2-positive breast cancer.370 Ongoing clinical trials are evaluating pyrotinib, another oral irreversible pan-HER TKI targeting HER1, HER2, and HER4 for the treatment of HER2-positive metastatic breast cancer.371 In addition, tucatinib, a HER2-specific TKI capable of crossing the blood-brain barrier, may exhibit activity in patients with brain metastases.372
TNBC
In TNBC, conventional chemotherapy is standard of care based on an anthracycline and a taxane. However, the toxicity of chemotherapy is a burden on patients and sometimes lack of effectiveness.373 More recently, with improvements in understanding of TNBC biology and an increasing appreciation of the potential of personalized therapy strategies, a paradigm shift in the treatment of TNBC is under way. Targeted drugs that intended for BRCA1/2 mutations, intracellular signaling pathways, immune checkpoint have brought great opportunities to improve the prognosis of TNBC patients.374
BRCA1/2 gene mutations have been detected in approximately 15–20% of TNBC patients.31 These genes play a significant role in the repair of double-stranded DNA through homologous recombination. Tumor cells carrying mutations in BRCA1/2 genes exhibit impaired DNA repair due to deficiencies in homologous recombination repair.375,376 Exploiting this vulnerability, PARP inhibitors are utilized to disrupt DNA damage repair, leading to the buildup of excessive DNA damage and consequent elimination of tumor cells in BRCA1/2-deficient TNBC.377 Recent clinical trial data have demonstrated that PARP inhibitors (such as olaparib or talazoparib) improved PFS and enhanced quality of life in patients with BRCA-mutated breast cancer when compared to single-agent chemotherapy (Table 3).378,379
Immune checkpoint targeting, particularly the programmed death receptor (PD-1) and its ligand PD-L1, has been reported as effective in treating various types of tumors, including melanoma, non-small cell lung cancer, renal cell carcinoma.380–382 As TNBC shows heightened genomic instability with abundant immune cell infiltration compared to other breast cancer subtypes, immune checkpoint inhibitors (ICIs) such as pembrolizumab, atezolizumab have demonstrated benefit for TNBC patients (Table 3). Recently, data from clinical trials indicated that the combination of ICIs (especially for PD-1 inhibitor) with chemotherapy in TNBC patients, particularly in those with PD-L1 overexpression, resulted in a higher percentage of pathologic complete response (pCR) and prolonged PFS compared to chemotherapy alone.383,384
Ongoing research is exploring additional targeted therapies to inhibit intracellular signaling pathways such as PI3K/AKT/mTOR, EGFR, Notch signaling, and STAT3 signaling, aiming to evaluate their safety and effectiveness in TNBC patients.374 However, the current therapeutic options for TNBC are limited compared to ER-positive and HER2-positive breast cancer due to the absence of well-defined targets and the inherent heterogeneity of TNBC itself. As a result, effectively treating TNBC remains a significant challenge that needs to be addressed.
Conclusion and perspective
The progress and treatment of IDC are advancing rapidly. However, the available treatment options for DCIS remain limited, mainly due to a lack of comprehensive understanding of its underlying mechanisms. While several models, such as the independent lineage model, evolutionary bottleneck model, multiclonal invasion model, and convergent phenotype model, have been proposed in DCIS, there are still more unanswered questions and puzzles that warrant further exploration.
Obviously, in the field of DCIS-IDC research, bottlenecks also persist. Distinguishing between DCIS and early-stage IDC remains a challenge with the present imaging techniques. In addition, predicting which DCIS will transition to IDC versus those that will remain indolent is still a major challenge. As DCIS detection has become more prevalent with advanced screening techniques, there’s a risk of overtreating lesions that might never progress to invasive cancer. While the current treatment recommendations for DCIS are varied, the optimal strategies for management to prevent progression to IDC remain elusive. Furthermore, the biological intricacies governing the shift from DCIS to IDC are yet to be fully unraveled, and the inherent heterogeneity of DCIS further complicate research efforts. Currently, there are already a certain number of tools available for the study of DCIS, such as cells and animal models. However, these tools are still far from sufficient for extensive research. Of course, the use of high-throughput sequencing methods like single-cell sequencing can further uncover the biological characteristics of DCIS.
In the past, we used to focus primarily on the DCIS tumor itself. However, we are now gradually shifting our perspective and recognizing the importance of the surrounding microenvironment in influencing DCIS cells, such as myoepithelial cells, immune cells, fibroblasts. Extensive research has been conducted on the microenvironment of IDC, and drugs targeting the microenvironment to inhibit tumors have emerged. For example, anti-angiogenic drugs like VEGF/VEGFR inhibitors (Bevacizumab, Apatinib) and immune checkpoint inhibitors (Pembrolizumab, Atezolizumab) have demonstrated promising anti-tumor effects.374 In the case of DCIS, we are eagerly awaiting research on potential targets for anti-tumor effects specifically focused on the microenvironment.
In clinical practice, there is a great concern for aggressive DCIS because although DCIS is generally considered to have a good prognosis, some patients may experience recurrence or even progression to invasive disease. We aim to identify aggressive DCIS to ensure that they receive adequate treatment, while distinguishing non-aggressive DCIS to spare patients from unnecessary treatments. Prognostic indicators and appropriate treatment strategies are needed, aligning with the concept of precision medicine. As patients’ expectations for survival increase, individualized and precise treatment becomes crucial. A fundamental cornerstone for implementing precision treatment for DCIS patients lies in gaining a clear and in-depth understanding of the underlying biological mechanisms of DCIS and its progression to IDC.
The field of DCIS to IDC research is evolving, with both challenges to overcome and promising avenues to explore. Continued collaboration and innovation in the field will be crucial for improving patient outcomes and understanding the biology of disease progression.
Acknowledgements
This study was supported by grants from the National Natural Science Foundation of China (Grant No. 82172344 to J.Z.), the Natural Science Foundation of Zhejiang Province (Grant No. LY21H160039 to J.Z.), the funding of Medical Science and Technology Project of Zhejiang Province (Grant No. 2022RC174 to J.Z.), the Natural Science Foundation of Zhejiang Province (Grant No. LGF21H030010 to B.L.).
Author contributions
J.Z., S.Z., and S.Z.Z. proposed the topic and main idea. J.W. and J.Z. wrote the original manuscript and drew the figures. J.W., B.L., and M.L. were responsible for collecting data and making tables. J.W., J.H., and K.Z. were responsible for the literature search; S.Z.Z. and J.Z. commented on and revised the manuscript. All authors have read and approved the article.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Jing Wang, Baizhou Li, Meng Luo.
Contributor Information
Suzhan Zhang, Email: Zrsj@zju.edu.cn.
Jiaojiao Zhou, Email: zhoujj@zju.edu.cn.
References
- 1.DeSantis CE, et al. Breast cancer statistics, 2019. CA Cancer J. Clin. 2019;69:438–451. doi: 10.3322/caac.21583. [DOI] [PubMed] [Google Scholar]
- 2.Giaquinto AN, et al. Breast cancer statistics, 2022. CA Cancer J. Clin. 2022;72:524–541. doi: 10.3322/caac.21754. [DOI] [PubMed] [Google Scholar]
- 3.Badve SS, Gökmen-Polar Y. Ductal carcinoma in situ of breast: update 2019. Pathology. 2019;51:563–569. doi: 10.1016/j.pathol.2019.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shaaban AM, et al. Pathological features of 11,337 patients with primary ductal carcinoma in situ (DCIS) and subsequent events: results from the UK Sloane Project. Br. J. Cancer. 2021;124:1009–1017. doi: 10.1038/s41416-020-01152-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Groen EJ, et al. Prognostic value of histopathological DCIS features in a large-scale international interrater reliability study. Breast Cancer Res. Treat. 2020;183:759–770. doi: 10.1007/s10549-020-05816-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maxwell AJ, et al. Unresected screen-detected ductal carcinoma in situ: outcomes of 311 women in the Forget-Me-Not 2 study. Breast Edinb. Scotl. 2022;61:145–155. doi: 10.1016/j.breast.2022.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanders ME, Schuyler PA, Dupont WD, Page DL. The natural history of low-grade ductal carcinoma in situ of the breast in women treated by biopsy only revealed over 30 years of long-term follow-up. Cancer. 2005;103:2481–2484. doi: 10.1002/cncr.21069. [DOI] [PubMed] [Google Scholar]
- 8.Collins LC, et al. Outcome of patients with ductal carcinoma in situ untreated after diagnostic biopsy: results from the Nurses’ Health Study. Cancer. 2005;103:1778–1784. doi: 10.1002/cncr.20979. [DOI] [PubMed] [Google Scholar]
- 9.Page DL, Dupont WD, Rogers LW, Landenberger M. Intraductal carcinoma of the breast: follow-up after biopsy only. Cancer. 1982;49:751–758. doi: 10.1002/1097-0142(19820215)49:4<751::AID-CNCR2820490426>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 10.Betsill WL, Rosen PP, Lieberman PH, Robbins GF. Intraductal carcinoma: long-term follow-up after treatment by biopsy alone. JAMA. 1978;239:1863–1867. doi: 10.1001/jama.1978.03280450035020. [DOI] [PubMed] [Google Scholar]
- 11.Fu F, Gilmore RC, Jacobs LK. Ductal carcinoma in situ. Surg. Clin. North Am. 2018;98:725–745. doi: 10.1016/j.suc.2018.03.007. [DOI] [PubMed] [Google Scholar]
- 12.Nguyen, D. L., Greenwood, H. I., Rahbar, H. & Grimm, L. J. Evolving treatment paradigms for low-risk ductal carcinoma in situ: imaging needs. AJR Am. J. Roentgenol. 10.2214/AJR.23.30503 (2023). [DOI] [PubMed]
- 13.DeSantis CE, Ma J, Goding Sauer A, Newman LA, Jemal A. Breast cancer statistics, 2017, racial disparity in mortality by state. CA Cancer J. Clin. 2017;67:439–448. doi: 10.3322/caac.21412. [DOI] [PubMed] [Google Scholar]
- 14.Sanati S. Morphologic and molecular features of breast ductal carcinoma in situ. Am. J. Pathol. 2019;189:946–955. doi: 10.1016/j.ajpath.2018.07.031. [DOI] [PubMed] [Google Scholar]
- 15.Grimm LJ, Rahbar H, Abdelmalak M, Hall AH, Ryser MD. Ductal carcinoma in situ: state-of-the-art review. Radiology. 2022;302:246–255. doi: 10.1148/radiol.211839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gajdos C, et al. Mammographic appearance of nonpalpable breast cancer reflects pathologic characteristics. Ann. Surg. 2002;235:246–251. doi: 10.1097/00000658-200202000-00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farante G, et al. Advances and controversies in management of breast ductal carcinoma in situ (DCIS) Eur. J. Surg. Oncol. 2022;48:736–741. doi: 10.1016/j.ejso.2021.10.030. [DOI] [PubMed] [Google Scholar]
- 18.Yu K-D, et al. Different distribution of breast cancer subtypes in breast ductal carcinoma in situ (DCIS), DCIS with microinvasion, and DCIS with invasion component. Ann. Surg. Oncol. 2011;18:1342–1348. doi: 10.1245/s10434-010-1407-3. [DOI] [PubMed] [Google Scholar]
- 19.Thorat MA, et al. Prognostic and predictive value of HER2 expression in ductal carcinoma in situ: results from the UK/ANZ DCIS randomized trial. Clin. Cancer Res. 2021;27:5317–5324. doi: 10.1158/1078-0432.CCR-21-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Latta EK, Tjan S, Parkes RK, O’Malley FP. The role of HER2/neu overexpression/amplification in the progression of ductal carcinoma in situ to invasive carcinoma of the breast. Mod. Pathol. 2002;15:1318–1325. doi: 10.1097/01.MP.0000038462.62634.B1. [DOI] [PubMed] [Google Scholar]
- 21.Sadeghalvad M, Mohammadi-Motlagh H-R, Rezaei N. Immune microenvironment in different molecular subtypes of ductal breast carcinoma. Breast Cancer Res. Treat. 2021;185:261–279. doi: 10.1007/s10549-020-05954-2. [DOI] [PubMed] [Google Scholar]
- 22.Virnig BA, Tuttle TM, Shamliyan T, Kane RL. Ductal carcinoma in situ of the breast: a systematic review of incidence, treatment, and outcomes. JNCI J. Natl Cancer Inst. 2010;102:170–178. doi: 10.1093/jnci/djp482. [DOI] [PubMed] [Google Scholar]
- 23.Worni M, et al. Trends in treatment patterns and outcomes for ductal carcinoma in situ. J. Natl Cancer Inst. 2015;107:djv263. doi: 10.1093/jnci/djv263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Bockstal MR, Agahozo MC, Koppert LB, van Deurzen CHM. A retrospective alternative for active surveillance trials for ductal carcinoma in situ of the breast. Int. J. Cancer. 2020;146:1189–1197. doi: 10.1002/ijc.32362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wellings SR, Jensen HM. On the origin and progression of ductal carcinoma in the human breast. J. Natl Cancer Inst. 1973;50:1111–1118. doi: 10.1093/jnci/50.5.1111. [DOI] [PubMed] [Google Scholar]
- 26.Doebar SC, et al. Gene expression differences between ductal carcinoma in situ with and without progression to invasive breast cancer. Am. J. Pathol. 2017;187:1648–1655. doi: 10.1016/j.ajpath.2017.03.012. [DOI] [PubMed] [Google Scholar]
- 27.Collins LC, et al. Risk factors for non-invasive and invasive local recurrence in patients with ductal carcinoma in situ. Breast Cancer Res. Treat. 2013;139:453–460. doi: 10.1007/s10549-013-2539-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang S-Y, Shamliyan T, Virnig BA, Kane R. Tumor characteristics as predictors of local recurrence after treatment of ductal carcinoma in situ: a meta-analysis. Breast Cancer Res. Treat. 2011;127:1–14. doi: 10.1007/s10549-011-1387-4. [DOI] [PubMed] [Google Scholar]
- 29.Atakpa EC, Thorat MA, Cuzick J, Brentnall AR. Mammographic density, endocrine therapy and breast cancer risk: a prognostic and predictive biomarker review. Cochrane Database Syst. Rev. 2021;10:CD013091. doi: 10.1002/14651858.CD013091.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knowlton CA, Jimenez RB, Moran MS. DCIS: risk assessment in the molecular era. Semin. Radiat. Oncol. 2022;32:189–197. doi: 10.1016/j.semradonc.2022.01.005. [DOI] [PubMed] [Google Scholar]
- 31.Nolan E, Lindeman GJ, Visvader JE. Deciphering breast cancer: from biology to the clinic. Cell. 2023;186:1708–1728. doi: 10.1016/j.cell.2023.01.040. [DOI] [PubMed] [Google Scholar]
- 32.Hua H, Zhang H, Kong Q, Jiang Y. Mechanisms for estrogen receptor expression in human cancer. Exp. Hematol. Oncol. 2018;7:24. doi: 10.1186/s40164-018-0116-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clusan L, Le Goff P, Flouriot G, Pakdel F. A closer look at estrogen receptor mutations in breast cancer and their implications for estrogen and antiestrogen responses. Int. J. Mol. Sci. 2021;22:756. doi: 10.3390/ijms22020756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swain SM, Shastry M, Hamilton E. Targeting HER2-positive breast cancer: advances and future directions. Nat. Rev. Drug Discov. 2023;22:101–126. doi: 10.1038/s41573-022-00579-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang Y, et al. HER2-driven breast tumorigenesis relies upon interactions of the estrogen receptor with coactivator MED1. Cancer Res. 2018;78:422–435. doi: 10.1158/0008-5472.CAN-17-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoxhaj G, Manning BD. The PI3K-AKT network at the interface of oncogenic signalling and cancer metabolism. Nat. Rev. Cancer. 2020;20:74–88. doi: 10.1038/s41568-019-0216-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Butti R, et al. Receptor tyrosine kinases (RTKs) in breast cancer: signaling, therapeutic implications and challenges. Mol. Cancer. 2018;17:34. doi: 10.1186/s12943-018-0797-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Riggio AI, Varley KE, Welm AL. The lingering mysteries of metastatic recurrence in breast cancer. Br. J. Cancer. 2021;124:13–26. doi: 10.1038/s41416-020-01161-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin D, et al. Circulating tumor cells: biology and clinical significance. Signal Transduct. Target. Ther. 2021;6:404. doi: 10.1038/s41392-021-00817-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buyuk B, Jin S, Ye K. Epithelial-to-mesenchymal transition signaling pathways responsible for breast cancer metastasis. Cell. Mol. Bioeng. 2022;15:1–13. doi: 10.1007/s12195-021-00694-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miricescu D, et al. PI3K/AKT/mTOR signaling pathway in breast cancer: from molecular landscape to clinical aspects. Int. J. Mol. Sci. 2020;22:173. doi: 10.3390/ijms22010173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feng Y, et al. Breast cancer development and progression: risk factors, cancer stem cells, signaling pathways, genomics, and molecular pathogenesis. Genes Dis. 2018;5:77–106. doi: 10.1016/j.gendis.2018.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harbeck N, et al. Breast cancer. Nat. Rev. Dis. Prim. 2019;5:66. doi: 10.1038/s41572-019-0111-2. [DOI] [PubMed] [Google Scholar]
- 44.Hankinson SE, Colditz GA, Willett WC. Towards an integrated model for breast cancer etiology: the lifelong interplay of genes, lifestyle, and hormones. Breast Cancer Res. BCR. 2004;6:213–218. doi: 10.1186/bcr921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moisand A, et al. Hormone receptor signaling and breast cancer resistance to anti-tumor immunity. Int. J. Mol. Sci. 2023;24:15048. doi: 10.3390/ijms242015048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yager, J. D. Estrogen carcinogenesis in breast cancer. N. Engl. J. Med.10.1056/NEJMra050776 (2006). [DOI] [PubMed]
- 47.Brufsky AM, Dickler MN. Estrogen receptor-positive breast cancer: exploiting signaling pathways implicated in endocrine resistance. Oncologist. 2018;23:528–539. doi: 10.1634/theoncologist.2017-0423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nagini S. Breast cancer: current molecular therapeutic targets and new players. Anticancer Agents Med. Chem. 2017;17:152–163. doi: 10.2174/1871520616666160502122724. [DOI] [PubMed] [Google Scholar]
- 49.Clusan L, Ferrière F, Flouriot G, Pakdel F. A basic review on estrogen receptor signaling pathways in breast cancer. Int. J. Mol. Sci. 2023;24:6834. doi: 10.3390/ijms24076834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Drago JZ, Ferraro E, Abuhadra N, Modi S. Beyond HER2: targeting the erbb receptor family in breast cancer. Cancer Treat. Rev. 2022;109:102436. doi: 10.1016/j.ctrv.2022.102436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eroles P, Bosch A, Alejandro Pérez-Fidalgo J, Lluch A. Molecular biology in breast cancer: intrinsic subtypes and signaling pathways. Cancer Treat. Rev. 2012;38:698–707. doi: 10.1016/j.ctrv.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 52.Friedlaender A, et al. EGFR and HER2 exon 20 insertions in solid tumours: from biology to treatment. Nat. Rev. Clin. Oncol. 2022;19:51–69. doi: 10.1038/s41571-021-00558-1. [DOI] [PubMed] [Google Scholar]
- 53.Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat. Rev. Mol. Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 54.Yarden Y. Biology of HER2 and its importance in breast cancer. Oncology. 2001;61(Suppl 2):1–13. doi: 10.1159/000055396. [DOI] [PubMed] [Google Scholar]
- 55.Hynes NE, MacDonald G. ErbB receptors and signaling pathways in cancer. Curr. Opin. Cell Biol. 2009;21:177–184. doi: 10.1016/j.ceb.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 56.Cerma K, et al. Targeting PI3K/AKT/mTOR pathway in breast cancer: from biology to clinical challenges. Biomedicines. 2023;11:109. doi: 10.3390/biomedicines11010109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat. Rev. Genet. 2006;7:606–619. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- 58.Pascual J, Turner NC. Targeting the PI3-kinase pathway in triple-negative breast cancer. Ann. Oncol. 2019;30:1051–1060. doi: 10.1093/annonc/mdz133. [DOI] [PubMed] [Google Scholar]
- 59.Bose S, Wang SI, Terry MB, Hibshoosh H, Parsons R. Allelic loss of chromosome 10q23 is associated with tumor progression in breast carcinomas. Oncogene. 1998;17:123–127. doi: 10.1038/sj.onc.1201940. [DOI] [PubMed] [Google Scholar]
- 60.Garcia JM, et al. Allelic loss of the PTEN region (10q23) in breast carcinomas of poor pathophenotype. Breast Cancer Res. Treat. 1999;57:237–243. doi: 10.1023/A:1006273516976. [DOI] [PubMed] [Google Scholar]
- 61.Razavi P, et al. The genomic landscape of endocrine-resistant advanced breast cancers. Cancer Cell. 2018;34:427–438.e6. doi: 10.1016/j.ccell.2018.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ali S, Coombes RC. Endocrine-responsive breast cancer and strategies for combating resistance. Nat. Rev. Cancer. 2002;2:101–112. doi: 10.1038/nrc721. [DOI] [PubMed] [Google Scholar]
- 63.Fujimoto Y, et al. Combination treatment with a PI3K/Akt/mTOR pathway inhibitor overcomes resistance to anti-HER2 therapy in PIK3CA-mutant HER2-positive breast cancer cells. Sci. Rep. 2020;10:21762. doi: 10.1038/s41598-020-78646-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ahmad DAJ, et al. Clinicopathological and prognostic significance of mitogen-activated protein kinases (MAPK) in breast cancers. Breast Cancer Res. Treat. 2016;159:457–467. doi: 10.1007/s10549-016-3967-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Haagenson KK, Wu GS. The role of MAP kinases and MAP kinase phosphatase-1 in resistance to breast cancer treatment. Cancer Metastasis Rev. 2010;29:143–149. doi: 10.1007/s10555-010-9208-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Janczur Velloso F, et al. The crossroads of breast cancer progression: insights into the modulation of major signaling pathways. OncoTargets Ther. 2017;10:5491–5524. doi: 10.2147/OTT.S142154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.von Lintig FC, et al. Ras activation in human breast cancer. Breast Cancer Res. Treat. 2000;62:51–62. doi: 10.1023/A:1006491619920. [DOI] [PubMed] [Google Scholar]
- 68.Hobbs, G. A., Der, C. J. & Rossman, K. L. RAS isoforms and mutations in cancer at a glance. J. Cell Sci. 10.1242/jcs.182873 (2016). [DOI] [PMC free article] [PubMed]
- 69.Santen RJ, et al. The role of mitogen-activated protein (MAP) kinase in breast cancer. J. Steroid Biochem. Mol. Biol. 2002;80:239–256. doi: 10.1016/S0960-0760(01)00189-3. [DOI] [PubMed] [Google Scholar]
- 70.Kato S, et al. Activation of the estrogen receptor through phosphorylation by mitogen-activated protein kinase. Science. 1995;270:1491–1494. doi: 10.1126/science.270.5241.1491. [DOI] [PubMed] [Google Scholar]
- 71.Pandey K, et al. Molecular mechanisms of resistance to CDK4/6 inhibitors in breast cancer: a review. Int. J. Cancer. 2019;145:1179–1188. doi: 10.1002/ijc.32020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Roberts PJ, et al. Multiple roles of cyclin-dependent kinase 4/6 inhibitors in cancer therapy. J. Natl Cancer Inst. 2012;104:476–487. doi: 10.1093/jnci/djs002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thangavel C, et al. Therapeutically activating RB: reestablishing cell cycle control in endocrine therapy-resistant breast cancer. Endocr. Relat. Cancer. 2011;18:333–345. doi: 10.1530/ERC-10-0262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Goel S, Bergholz JS, Zhao JJ. Targeting cyclin-dependent kinases 4 and 6 in cancer. Nat. Rev. Cancer. 2022;22:356–372. doi: 10.1038/s41568-022-00456-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Spring LM, et al. Cyclin-dependent kinase 4 and 6 inhibitors for hormone receptor-positive breast cancer: past, present, and future. Lancet. 2020;395:817–827. doi: 10.1016/S0140-6736(20)30165-3. [DOI] [PubMed] [Google Scholar]
- 76.Bosco EE, Knudsen ES. RB in breast cancer: at the crossroads of tumorigenesis and treatment. Cell Cycle Georget. Tex. 2007;6:667–671. doi: 10.4161/cc.6.6.3988. [DOI] [PubMed] [Google Scholar]
- 77.Treré D, et al. High prevalence of retinoblastoma protein loss in triple-negative breast cancers and its association with a good prognosis in patients treated with adjuvant chemotherapy. Ann. Oncol. 2009;20:1818–1823. doi: 10.1093/annonc/mdp209. [DOI] [PubMed] [Google Scholar]
- 78.Lopez-Garcia MA, Geyer FC, Lacroix-Triki M, Marchió C, Reis-Filho JS. Breast cancer precursors revisited: molecular features and progression pathways: molecular evolution of breast cancer. Histopathology. 2010;57:171–192. doi: 10.1111/j.1365-2559.2010.03568.x. [DOI] [PubMed] [Google Scholar]
- 79.Wellings SR, Jensen HM, Marcum RG. An atlas of subgross pathology of the human breast with special reference to possible precancerous lesions. J. Natl Cancer Inst. 1975;55:231–273. [PubMed] [Google Scholar]
- 80.Rakha EA, et al. Invasion in breast lesions: the role of the epithelial-stroma barrier. Histopathology. 2018;72:1075–1083. doi: 10.1111/his.13446. [DOI] [PubMed] [Google Scholar]
- 81.Wilson GM, Dinh P, Pathmanathan N, Graham JD. Ductal carcinoma in situ: molecular changes accompanying disease progression. J. Mammary Gland Biol. Neoplasia. 2022;27:101–131. doi: 10.1007/s10911-022-09517-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mitchell E, et al. Loss of myoepithelial calponin-1 characterizes high-risk ductal carcinoma in situ cases, which are further stratified by T cell composition. Mol. Carcinog. 2020;59:701–712. doi: 10.1002/mc.23171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bonacho T, Rodrigues F, Liberal J. Immunohistochemistry for diagnosis and prognosis of breast cancer: a review. Biotech. Histochem. 2020;95:71–91. doi: 10.1080/10520295.2019.1651901. [DOI] [PubMed] [Google Scholar]
- 84.Hernandez L, et al. Genomic and mutational profiling of ductal carcinomas in situ and matched adjacent invasive breast cancers reveals intra-tumour genetic heterogeneity and clonal selection. J. Pathol. 2012;227:42–52. doi: 10.1002/path.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Iakovlev VV, et al. Genomic differences between pure ductal carcinoma in situ of the breast and that associated with invasive disease: a calibrated aCGH study. Clin. Cancer Res. 2008;14:4446–4454. doi: 10.1158/1078-0432.CCR-07-4960. [DOI] [PubMed] [Google Scholar]
- 86.Pareja F, et al. Whole-exome sequencing analysis of the progression from non–low-grade ductal carcinoma in situ to invasive ductal carcinoma. Clin. Cancer Res. 2020;26:3682–3693. doi: 10.1158/1078-0432.CCR-19-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fortunato A, et al. A new method to accurately identify single nucleotide variants using small FFPE breast samples. Brief. Bioinform. 2021;22:bbab221. doi: 10.1093/bib/bbab221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rane SU, Mirza H, Grigoriadis A, Pinder SE. Selection and evolution in the genomic landscape of copy number alterations in ductal carcinoma in situ (DCIS) and its progression to invasive carcinoma of ductal/no special type: a meta-analysis. Breast Cancer Res. Treat. 2015;153:101–121. doi: 10.1007/s10549-015-3509-x. [DOI] [PubMed] [Google Scholar]
- 89.Buerger H, et al. Comparative genomic hybridization of ductal carcinoma in situ of the breast-evidence of multiple genetic pathways. J. Pathol. 1999;187:396–402. doi: 10.1002/(SICI)1096-9896(199903)187:4<396::AID-PATH286>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 90.Moelans CB, de Wegers RA, Monsuurs HN, Maess AHJ, van Diest PJ. Molecular differences between ductal carcinoma in situ and adjacent invasive breast carcinoma: a multiplex ligation-dependent probe amplification study. Cell. Oncol. Dordr. 2011;34:475–482. doi: 10.1007/s13402-011-0043-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Johnson CE, et al. Identification of copy number alterations associated with the progression of DCIS to invasive ductal carcinoma. Breast Cancer Res. Treat. 2012;133:889–898. doi: 10.1007/s10549-011-1835-1. [DOI] [PubMed] [Google Scholar]
- 92.Casasent AK, et al. Multiclonal invasion in breast tumors identified by topographic single cell sequencing. Cell. 2018;172:205–217.e12. doi: 10.1016/j.cell.2017.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Williams KE, et al. Molecular phenotypes of DCIS predict overall and invasive recurrence. Ann. Oncol. 2015;26:1019–1025. doi: 10.1093/annonc/mdv062. [DOI] [PubMed] [Google Scholar]
- 94.Trinh A, et al. Genomic alterations during the in situ to invasive ductal breast carcinoma transition shaped by the immune system. Mol. Cancer Res. 2021;19:623–635. doi: 10.1158/1541-7786.MCR-20-0949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Waldman FM, et al. Chromosomal alterations in ductal carcinomas in situ and their in situ recurrences. J. Natl Cancer Inst. 2000;92:313–320. doi: 10.1093/jnci/92.4.313. [DOI] [PubMed] [Google Scholar]
- 96.Gorringe KL, et al. Copy number analysis of ductal carcinoma in situ with and without recurrence. Mod. Pathol. 2015;28:1174–1184. doi: 10.1038/modpathol.2015.75. [DOI] [PubMed] [Google Scholar]
- 97.Lips EH, et al. Genomic analysis defines clonal relationships of ductal carcinoma in situ and recurrent invasive breast cancer. Nat. Genet. 2022;54:850–860. doi: 10.1038/s41588-022-01082-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang, K. et al. Archival single-cell genomics reveals persistent subclones during DCIS progression. Cell10.1016/j.cell.2023.07.024 (2023). [DOI] [PubMed]
- 99.Zhou W, et al. Full sequencing of TP53 identifies identical mutations within in situ and invasive components in breast cancer suggesting clonal evolution. Mol. Oncol. 2009;3:214–219. doi: 10.1016/j.molonc.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Miron A, et al. PIK3CA mutations in in situ and invasive breast carcinomas. Cancer Res. 2010;70:5674–5678. doi: 10.1158/0008-5472.CAN-08-2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sakr RA, et al. PI3K pathway activation in high-grade ductal carcinoma in situ-implications for progression to invasive breast carcinoma. Clin. Cancer Res. 2014;20:2326–2337. doi: 10.1158/1078-0432.CCR-13-2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Afghahi A, et al. Chromosomal copy number alterations for associations of ductal carcinoma in situ with invasive breast cancer. Breast Cancer Res. 2015;17:108. doi: 10.1186/s13058-015-0623-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kim SY, et al. Genomic differences between pure ductal carcinoma in situ and synchronous ductal carcinoma in situ with invasive breast cancer. Oncotarget. 2015;6:7597–7607. doi: 10.18632/oncotarget.3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bergholtz H, et al. Contrasting DCIS and invasive breast cancer by subtype suggests basal-like DCIS as distinct lesions. NPJ Breast Cancer. 2020;6:26. doi: 10.1038/s41523-020-0167-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pang J-MB, et al. Breast ductal carcinoma in situ carry mutational driver events representative of invasive breast cancer. Mod. Pathol. 2017;30:952–963. doi: 10.1038/modpathol.2017.21. [DOI] [PubMed] [Google Scholar]
- 106.Lin C-Y, et al. Genomic landscape of ductal carcinoma in situ and association with progression. Breast Cancer Res. Treat. 2019;178:307–316. doi: 10.1007/s10549-019-05401-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bergholtz H, et al. Comparable cancer-relevant mutation profiles in synchronous ductal carcinoma in situ and invasive breast cancer. Cancer Rep. 2020;3:e1248. doi: 10.1002/cnr2.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rebbeck CA, et al. Gene expression signatures of individual ductal carcinoma in situ lesions identify processes and biomarkers associated with progression towards invasive ductal carcinoma. Nat. Commun. 2022;13:3399. doi: 10.1038/s41467-022-30573-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bombonati A, Sgroi DC. The molecular pathology of breast cancer progression. J. Pathol. 2011;223:308–318. doi: 10.1002/path.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Butti R, Gunasekaran VP, Kumar TVS, Banerjee P, Kundu GC. Breast cancer stem cells: biology and therapeutic implications. Int. J. Biochem. Cell Biol. 2019;107:38–52. doi: 10.1016/j.biocel.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 111.Polyak K. Breast cancer: origins and evolution. J. Clin. Invest. 2007;117:3155–3163. doi: 10.1172/JCI33295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhang M, Lee AV, Rosen JM. The cellular origin and evolution of breast cancer. Cold Spring Harb. Perspect. Med. 2017;7:a027128. doi: 10.1101/cshperspect.a027128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Cowell CF, et al. Progression from ductal carcinoma in situ to invasive breast cancer: Revisited. Mol. Oncol. 2013;7:859–869. doi: 10.1016/j.molonc.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.van der Borden CL, Stoffers S, Lips EH, Wesseling J. Avoiding overtreatment of ductal carcinoma in situ. Trends Cancer. 2019;5:391–393. doi: 10.1016/j.trecan.2019.05.005. [DOI] [PubMed] [Google Scholar]
- 115.Casasent AK, Edgerton M, Navin NE. Genome evolution in ductal carcinoma in situ: invasion of the clones: DCIS genome evolution. J. Pathol. 2017;241:208–218. doi: 10.1002/path.4840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Allred DC, et al. Ductal carcinoma in situ and the emergence of diversity during breast cancer evolution. Clin. Cancer Res. 2008;14:370–378. doi: 10.1158/1078-0432.CCR-07-1127. [DOI] [PubMed] [Google Scholar]
- 117.Gao Y, Niu Y, Wang X, Wei L, Lu S. Genetic changes at specific stages of breast cancer progression detected by comparative genomic hybridization. J. Mol. Med. 2009;87:145–152. doi: 10.1007/s00109-008-0408-1. [DOI] [PubMed] [Google Scholar]
- 118.Ma X-J, et al. Gene expression profiles of human breast cancer progression. Proc. Natl Acad. Sci. USA. 2003;100:5974–5979. doi: 10.1073/pnas.0931261100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Simpson PT, Reis-Filho JS, Gale T, Lakhani SR. Molecular evolution of breast cancer: Molecular evolution of breast cancer. J. Pathol. 2005;205:248–254. doi: 10.1002/path.1691. [DOI] [PubMed] [Google Scholar]
- 120.Foschini MP, et al. Genetic clonal mapping of in situ and invasive ductal carcinoma indicates the field cancerization phenomenon in the breast. Hum. Pathol. 2013;44:1310–1319. doi: 10.1016/j.humpath.2012.09.022. [DOI] [PubMed] [Google Scholar]
- 121.Hagio K, et al. Genetic heterogeneity during breast cancer progression in young patients. Breast Edinb. Scotl. 2021;60:206–213. doi: 10.1016/j.breast.2021.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sontag L, Axelrod DE. Evaluation of pathways for progression of heterogeneous breast tumors. J. Theor. Biol. 2005;232:179–189. doi: 10.1016/j.jtbi.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 123.Moelans CB, Verschuur-Maes AH, van Diest PJ. Frequent promoter hypermethylation of BRCA2, CDH13, MSH6, PAX5, PAX6 and WT1 in ductal carcinoma in situ and invasive breast cancer. J. Pathol. 2011;225:222–231. doi: 10.1002/path.2930. [DOI] [PubMed] [Google Scholar]
- 124.Ding L, et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464:999–1005. doi: 10.1038/nature08989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yap TA, Gerlinger M, Futreal PA, Pusztai L, Swanton C. Intratumor heterogeneity: seeing the wood for the trees. Sci. Transl. Med. 2012;4:127ps10. doi: 10.1126/scitranslmed.3003854. [DOI] [PubMed] [Google Scholar]
- 126.Doebar SC, et al. Progression of ductal carcinoma in situ to invasive breast cancer: comparative genomic sequencing. Virchows Arch. 2019;474:247–251. doi: 10.1007/s00428-018-2463-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Martelotto LG, et al. Whole-genome single-cell copy number profiling from formalin-fixed paraffin-embedded samples. Nat. Med. 2017;23:376–385. doi: 10.1038/nm.4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Oikawa M, et al. A novel diagnostic method targeting genomic instability in intracystic tumors of the breast. Breast Cancer. 2015;22:529–535. doi: 10.1007/s12282-013-0516-9. [DOI] [PubMed] [Google Scholar]
- 129.Liao S, et al. Differential copy number aberrations in novel candidate genes associated with progression from in situ to invasive ductal carcinoma of the breast. Genes. Chromosomes Cancer. 2012;51:1067–1078. doi: 10.1002/gcc.21991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Krøigård AB, et al. Clonal expansion and linear genome evolution through breast cancer progression from pre-invasive stages to asynchronous metastasis. Oncotarget. 2015;6:5634–5649. doi: 10.18632/oncotarget.3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Casasent, A. K. et al. Learning to distinguish progressive and non-progressive ductal carcinoma in situ. Nat. Rev. Cancer10.1038/s41568-022-00512-y (2022). [DOI] [PubMed]
- 132.Yates LR, et al. Subclonal diversification of primary breast cancer revealed by multiregion sequencing. Nat. Med. 2015;21:751–759. doi: 10.1038/nm.3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Gibson SV, et al. Everybody needs good neighbours: the progressive DCIS microenvironment. Trends Cancer. 2023;9:326–338. doi: 10.1016/j.trecan.2023.01.002. [DOI] [PubMed] [Google Scholar]
- 134.Arendt LM, Rudnick JA, Keller PJ, Kuperwasser C. Stroma in breast development and disease. Semin. Cell Dev. Biol. 2010;21:11–18. doi: 10.1016/j.semcdb.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Polyak K, Kalluri R. The role of the microenvironment in mammary gland development and cancer. Cold Spring Harb. Perspect. Biol. 2010;2:a003244. doi: 10.1101/cshperspect.a003244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Soysal SD, Tzankov A, Muenst SE. Role of the tumor microenvironment in breast cancer. Pathobiol. J. Immunopathol. Mol. Cell. Biol. 2015;82:142–152. doi: 10.1159/000430499. [DOI] [PubMed] [Google Scholar]
- 137.Man Y, et al. Cell clusters overlying focally disrupted mammary myoepithelial cell layers and adjacent cells within the same duct display different immunohistochemical and genetic features: implications for tumor progression and invasion. Breast Cancer Res. 2003;5:R231–R241. doi: 10.1186/bcr653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Man Y-G, et al. A subset of in situ breast tumor cell clusters lacks expression of proliferation and progression related markers but shows signs of stromal and vascular invasion. Cancer Detect. Prev. 2005;29:323–331. doi: 10.1016/j.cdp.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 139.Schnitt SJ. The transition from ductal carcinoma in situ to invasive breast cancer: the other side of the coin. Breast Cancer Res. 2009;11:101. doi: 10.1186/bcr2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hu M, et al. Regulation of in situ to invasive breast carcinoma transition. Cancer Cell. 2008;13:394–406. doi: 10.1016/j.ccr.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lakhani SR, O’Hare MJ. The mammary myoepithelial cell-Cinderella or ugly sister? Breast Cancer Res. 2001;3:1–4. doi: 10.1186/bcr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Deugnier M-A, Teulière J, Faraldo MM, Thiery JP, Glukhova MA. The importance of being a myoepithelial cell. Breast Cancer Res. 2002;4:224–230. doi: 10.1186/bcr459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Sternlicht MD, Kedeshian P, Shao ZM, Safarians S, Barsky SH. The human myoepithelial cell is a natural tumor suppressor. Clin. Cancer Res. 1997;3:1949–1958. [PubMed] [Google Scholar]
- 144.Jones JL, Shaw JA, Pringle JH, Walker RA. Primary breast myoepithelial cells exert an invasion-suppressor effect on breast cancer cells via paracrine down-regulation of MMP expression in fibroblasts and tumour cells. J. Pathol. 2003;201:562–572. doi: 10.1002/path.1483. [DOI] [PubMed] [Google Scholar]
- 145.Nguyen M, et al. The human myoepithelial cell displays a multifaceted anti-angiogenic phenotype. Oncogene. 2000;19:3449–3459. doi: 10.1038/sj.onc.1203677. [DOI] [PubMed] [Google Scholar]
- 146.Zhang RR, et al. A subset of morphologically distinct mammary myoepithelial cells lacks corresponding immunophenotypic markers. Breast Cancer Res. 2003;5:R151–R156. doi: 10.1186/bcr635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Allen MD, Marshall JF, Jones JL. αvβ6 expression in myoepithelial cells: a novel marker for predicting DCIS progression with therapeutic potential. Cancer Res. 2014;74:5942–5947. doi: 10.1158/0008-5472.CAN-14-1841. [DOI] [PubMed] [Google Scholar]
- 148.Gudjonsson T, et al. Normal and tumor-derived myoepithelial cells differ in their ability to interact with luminal breast epithelial cells for polarity and basement membrane deposition. J. Cell Sci. 2002;115:39–50. doi: 10.1242/jcs.115.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Russell TD, et al. Myoepithelial cell differentiation markers in ductal carcinoma in situ progression. Am. J. Pathol. 2015;185:3076–3089. doi: 10.1016/j.ajpath.2015.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Knudsen ES, et al. Progression of ductal carcinoma in situ to invasive breast cancer is associated with gene expression programs of EMT and myoepithelia. Breast Cancer Res. Treat. 2012;133:1009–1024. doi: 10.1007/s10549-011-1894-3. [DOI] [PubMed] [Google Scholar]
- 151.Hilson JB, Schnitt SJ, Collins LC. Phenotypic alterations in ductal carcinoma in situ-associated myoepithelial cells: biologic and diagnostic implications. Am. J. Surg. Pathol. 2009;33:227–232. doi: 10.1097/PAS.0b013e318180431d. [DOI] [PubMed] [Google Scholar]
- 152.Ding L, et al. Perturbed myoepithelial cell differentiation in BRCA mutation carriers and in ductal carcinoma in situ. Nat. Commun. 2019;10:4182. doi: 10.1038/s41467-019-12125-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Sarper M, et al. Loss of MMP-8 in ductal carcinoma in situ (DCIS)-associated myoepithelial cells contributes to tumour promotion through altered adhesive and proteolytic function. Breast Cancer Res. 2017;19:33. doi: 10.1186/s13058-017-0822-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Lo P-K, et al. Tumor-associated myoepithelial cells promote the invasive progression of ductal carcinoma in situ through activation of TGFβ signaling. J. Biol. Chem. 2017;292:11466–11484. doi: 10.1074/jbc.M117.775080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Aguiar FN, Cirqueira CS, Bacchi CE, Carvalho FM. Morphologic, molecular and microenvironment factors associated with stromal invasion in breast ductal carcinoma in situ: role of myoepithelial cells. Breast Dis. 2015;35:249–252. doi: 10.3233/BD-150416. [DOI] [PubMed] [Google Scholar]
- 156.Allen MD, et al. Altered microenvironment promotes progression of preinvasive breast cancer: myoepithelial expression of αvβ6 integrin in DCIS identifies high-risk patients and predicts recurrence. Clin. Cancer Res. 2014;20:344–357. doi: 10.1158/1078-0432.CCR-13-1504. [DOI] [PubMed] [Google Scholar]
- 157.Hayward M-K, et al. Mechanostimulation of breast myoepithelial cells induces functional changes associated with DCIS progression to invasion. NPJ Breast Cancer. 2022;8:109. doi: 10.1038/s41523-022-00464-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Gibson SV, et al. TGFβ-mediated MMP13 secretion drives myoepithelial cell dependent breast cancer progression. Npj Breast Cancer. 2023;9:9. doi: 10.1038/s41523-023-00513-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Allinen M, et al. Molecular characterization of the tumor microenvironment in breast cancer. Cancer Cell. 2004;6:17–32. doi: 10.1016/j.ccr.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 160.Wagner J, et al. A single-cell atlas of the tumor and immune ecosystem of human breast cancer. Cell. 2019;177:1330–1345.e18. doi: 10.1016/j.cell.2019.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Azizi E, et al. Single-cell map of diverse immune phenotypes in the breast tumor microenvironment. Cell. 2018;174:1293–1308.e36. doi: 10.1016/j.cell.2018.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Swann JB, Smyth MJ. Immune surveillance of tumors. J. Clin. Invest. 2007;117:1137–1146. doi: 10.1172/JCI31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Lal A, et al. FOXP3-positive regulatory T lymphocytes and epithelial FOXP3 expression in synchronous normal, ductal carcinoma in situ, and invasive cancer of the breast. Breast Cancer Res. Treat. 2013;139:381–390. doi: 10.1007/s10549-013-2556-4. [DOI] [PubMed] [Google Scholar]
- 164.Kim M, et al. Immune microenvironment in ductal carcinoma in situ: a comparison with invasive carcinoma of the breast. Breast Cancer Res. 2020;22:32. doi: 10.1186/s13058-020-01267-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Chung YR, Kim HJ, Jang MH, Park SY. Prognostic value of tumor infiltrating lymphocyte subsets in breast cancer depends on hormone receptor status. Breast Cancer Res. Treat. 2017;161:409–420. doi: 10.1007/s10549-016-4072-9. [DOI] [PubMed] [Google Scholar]
- 166.Toss MS, et al. The prognostic significance of immune microenvironment in breast ductal carcinoma in situ. Br. J. Cancer. 2020;122:1496–1506. doi: 10.1038/s41416-020-0797-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Denkert C, et al. Tumor-associated lymphocytes as an independent predictor of response to neoadjuvant chemotherapy in breast cancer. J. Clin. Oncol. 2010;28:105–113. doi: 10.1200/JCO.2009.23.7370. [DOI] [PubMed] [Google Scholar]
- 168.Loi S, et al. Prognostic and predictive value of tumor-infiltrating lymphocytes in a phase III randomized adjuvant breast cancer trial in node-positive breast cancer comparing the addition of docetaxel to doxorubicin with doxorubicin-based chemotherapy: BIG 02-98. J. Clin. Oncol. 2013;31:860–867. doi: 10.1200/JCO.2011.41.0902. [DOI] [PubMed] [Google Scholar]
- 169.Mahmoud SMA, et al. Tumor-infiltrating CD8+ lymphocytes predict clinical outcome in breast cancer. J. Clin. Oncol. 2011;29:1949–1955. doi: 10.1200/JCO.2010.30.5037. [DOI] [PubMed] [Google Scholar]
- 170.Loi S, et al. Tumor infiltrating lymphocytes are prognostic in triple negative breast cancer and predictive for trastuzumab benefit in early breast cancer: results from the FinHER trial. Ann. Oncol. 2014;25:1544–1550. doi: 10.1093/annonc/mdu112. [DOI] [PubMed] [Google Scholar]
- 171.Kotoula V, et al. Tumors with high-density tumor infiltrating lymphocytes constitute a favorable entity in breast cancer: a pooled analysis of four prospective adjuvant trials. Oncotarget. 2016;7:5074–5087. doi: 10.18632/oncotarget.6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Tower H, Ruppert M, Britt K. The immune microenvironment of breast cancer progression. Cancers. 2019;11:1375. doi: 10.3390/cancers11091375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Niwińska A, Olszewski WP. The role of stromal immune microenvironment in the progression of ductal carcinoma in situ (DCIS) to invasive breast cancer. Breast Cancer Res. 2021;23:118. doi: 10.1186/s13058-021-01494-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Campbell MJ, et al. Characterizing the immune microenvironment in high-risk ductal carcinoma in situ of the breast. Breast Cancer Res. Treat. 2017;161:17–28. doi: 10.1007/s10549-016-4036-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Thompson E, et al. The immune microenvironment of breast ductal carcinoma in situ. Mod. Pathol. 2016;29:249–258. doi: 10.1038/modpathol.2015.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Gil Del Alcazar CR, et al. Immune escape in breast cancer during in situ to invasive carcinoma transition. Cancer Discov. 2017;7:1098–1115. doi: 10.1158/2159-8290.CD-17-0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Lv S, et al. Functional CD3+CD8+PD1- T cell accumulation and PD-L1 expression increases during tumor invasion in DCIS of the breast. Clin. Breast Cancer. 2019;19:e617–e623. doi: 10.1016/j.clbc.2019.04.001. [DOI] [PubMed] [Google Scholar]
- 178.Risom T, et al. Transition to invasive breast cancer is associated with progressive changes in the structure and composition of tumor stroma. Cell. 2022;185:299–310.e18. doi: 10.1016/j.cell.2021.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Linde N, et al. Macrophages orchestrate breast cancer early dissemination and metastasis. Nat. Commun. 2018;9:21. doi: 10.1038/s41467-017-02481-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Chen X-Y, et al. Higher density of stromal M2 macrophages in breast ductal carcinoma in situ predicts recurrence. Virchows Arch. 2020;476:825–833. doi: 10.1007/s00428-019-02735-1. [DOI] [PubMed] [Google Scholar]
- 181.Orimo A, et al. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell. 2005;121:335–348. doi: 10.1016/j.cell.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 182.Kojima Y, et al. Autocrine TGF-beta and stromal cell-derived factor-1 (SDF-1) signaling drives the evolution of tumor-promoting mammary stromal myofibroblasts. Proc. Natl Acad. Sci. USA. 2010;107:20009–20014. doi: 10.1073/pnas.1013805107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Sung KE, et al. Understanding the impact of 2D and 3D fibroblast cultures on in vitro breast cancer models. PLoS ONE. 2013;8:e76373. doi: 10.1371/journal.pone.0076373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Jedeszko C, Victor BC, Podgorski I, Sloane BF. Fibroblast hepatocyte growth factor promotes invasion of human mammary ductal carcinoma in situ. Cancer Res. 2009;69:9148–9155. doi: 10.1158/0008-5472.CAN-09-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Hu M, et al. Role of COX-2 in epithelial-stromal cell interactions and progression of ductal carcinoma in situ of the breast. Proc. Natl Acad. Sci. USA. 2009;106:3372–3377. doi: 10.1073/pnas.0813306106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Osuala KO, et al. Il-6 signaling between ductal carcinoma in situ cells and carcinoma-associated fibroblasts mediates tumor cell growth and migration. BMC Cancer. 2015;15:584. doi: 10.1186/s12885-015-1576-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Sameni M, et al. Pathomimetic avatars reveal divergent roles of microenvironment in invasive transition of ductal carcinoma in situ. Breast Cancer Res. BCR. 2017;19:56. doi: 10.1186/s13058-017-0847-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Bernard S, et al. CXCL1 derived from mammary fibroblasts promotes progression of mammary lesions to invasive carcinoma through CXCR2 dependent mechanisms. J. Mammary Gland Biol. Neoplasia. 2018;23:249–267. doi: 10.1007/s10911-018-9407-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Barnabas N, Cohen D. Phenotypic and molecular characterization of MCF10DCIS and SUM breast cancer cell lines. Int. J. Breast Cancer. 2013;2013:872743. doi: 10.1155/2013/872743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Miller FR, Santner SJ, Tait L, Dawson PJ. MCF10DCIS.com xenograft model of human comedo ductal carcinoma in situ. J. Natl Cancer Inst. 2000;92:1185–1186. doi: 10.1093/jnci/92.14.1185a. [DOI] [PubMed] [Google Scholar]
- 191.Rizki A, et al. A human breast cell model of preinvasive to invasive transition. Cancer Res. 2008;68:1378–1387. doi: 10.1158/0008-5472.CAN-07-2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Band V, et al. Tumor progression in four mammary epithelial cell lines derived from the same patient. Cancer Res. 1990;50:7351–7357. [PubMed] [Google Scholar]
- 193.Souter LH, et al. Human 21T breast epithelial cell lines mimic breast cancer progression in vivo and in vitro and show stage-specific gene expression patterns. Lab. Investig. 2010;90:1247–1258. doi: 10.1038/labinvest.2010.97. [DOI] [PubMed] [Google Scholar]
- 194.Forozan F, et al. Molecular cytogenetic analysis of 11 new breast cancer cell lines. Br. J. Cancer. 1999;81:1328–1334. doi: 10.1038/sj.bjc.6695007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Sartor CI, Dziubinski ML, Yu CL, Jove R, Ethier SP. Role of epidermal growth factor receptor and STAT-3 activation in autonomous proliferation of SUM-102PT human breast cancer cells. Cancer Res. 1997;57:978–987. [PubMed] [Google Scholar]
- 196.Brock EJ, Ji K, Shah S, Mattingly RR, Sloane BF. In vitro models for studying invasive transitions of ductal carcinoma in situ. J. Mammary Gland Biol. Neoplasia. 2019;24:1–15. doi: 10.1007/s10911-018-9405-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Fang WB, et al. Expression of CCL2/CCR2 signaling proteins in breast carcinoma cells is associated with invasive progression. Sci. Rep. 2021;11:8708. doi: 10.1038/s41598-021-88229-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Soule HD, et al. Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res. 1990;50:6075–6086. [PubMed] [Google Scholar]
- 199.Dawson PJ, Wolman SR, Tait L, Heppner GH, Miller FR. MCF10AT: a model for the evolution of cancer from proliferative breast disease. Am. J. Pathol. 1996;148:313–319. [PMC free article] [PubMed] [Google Scholar]
- 200.Santner SJ, et al. Malignant MCF10CA1 cell lines derived from premalignant human breast epithelial MCF10AT cells. Breast Cancer Res. Treat. 2001;65:101–110. doi: 10.1023/A:1006461422273. [DOI] [PubMed] [Google Scholar]
- 201.Strickland LB, Dawson PJ, Santner SJ, Miller FR. Progression of premalignant MCF10AT generates heterogeneous malignant variants with characteristic histologic types and immunohistochemical markers. Breast Cancer Res. Treat. 2000;64:235–240. doi: 10.1023/A:1026562720218. [DOI] [PubMed] [Google Scholar]
- 202.Lee S, et al. Differentially expressed genes regulating the progression of ductal carcinoma in situ to invasive breast cancer. Cancer Res, 2012;72:4574–4586. doi: 10.1158/0008-5472.CAN-12-0636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Maguire SL, et al. Three-dimensional modelling identifies novel genetic dependencies associated with breast cancer progression in the isogenic MCF10 model. J. Pathol. 2016;240:315–328. doi: 10.1002/path.4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Bahcecioglu G, Basara G, Ellis BW, Ren X, Zorlutuna P. Breast cancer models: engineering the tumor microenvironment. Acta Biomater. 2020;106:1–21. doi: 10.1016/j.actbio.2020.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Duval K, et al. Modeling physiological events in 2D vs. 3D cell culture. Physiology. 2017;32:266–277. doi: 10.1152/physiol.00036.2016. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 206.Edmondson R, Broglie JJ, Adcock AF, Yang L. Three-dimensional cell culture systems and their applications in drug discovery and cell-based biosensors. Assay. Drug Dev. Technol. 2014;12:207–218. doi: 10.1089/adt.2014.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Katt ME, Placone AL, Wong AD, Xu ZS, Searson PC. In vitro tumor models: advantages, disadvantages, variables, and selecting the right platform. Front. Bioeng. Biotechnol. 2016;4:12. doi: 10.3389/fbioe.2016.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Ory V, et al. The PPARγ agonist efatutazone delays invasive progression and induces differentiation of ductal carcinoma in situ. Breast Cancer Res. Treat. 2018;169:47–57. doi: 10.1007/s10549-017-4649-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Li Q, Chow AB, Mattingly RR. Three-dimensional overlay culture models of human breast cancer reveal a critical sensitivity to mitogen-activated protein kinase kinase inhibitors. J. Pharmacol. Exp. Ther. 2010;332:821–828. doi: 10.1124/jpet.109.160390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Kim JB, Stein R, O’Hare MJ. Three-dimensional in vitro tissue culture models of breast cancer—a review. Breast Cancer Res. Treat. 2004;85:281–291. doi: 10.1023/B:BREA.0000025418.88785.2b. [DOI] [PubMed] [Google Scholar]
- 211.Huerta-Reyes M, Aguilar-Rojas A. Three‑dimensional models to study breast cancer (Review) Int. J. Oncol. 2021;58:331–343. doi: 10.3892/ijo.2021.5176. [DOI] [PubMed] [Google Scholar]
- 212.Carter EP, Gopsill JA, Gomm JJ, Jones JL, Grose RP. A 3D in vitro model of the human breast duct: a method to unravel myoepithelial-luminal interactions in the progression of breast cancer. Breast Cancer Res. 2017;19:50. doi: 10.1186/s13058-017-0843-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Choi Y, et al. A microengineered pathophysiological model of early-stage breast cancer. Lab. Chip. 2015;15:3350–3357. doi: 10.1039/C5LC00514K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Bischel LL, Beebe DJ, Sung KE. Microfluidic model of ductal carcinoma in situ with 3D, organotypic structure. BMC Cancer. 2015;15:12. doi: 10.1186/s12885-015-1007-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Asghar W, et al. Engineering cancer microenvironments for in vitro 3-D tumor models. Mater. Today. 2015;18:539–553. doi: 10.1016/j.mattod.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Behbod F, et al. An intraductal human-in-mouse transplantation model mimics the subtypes of ductal carcinoma in situ. Breast Cancer Res. 2009;11:R66. doi: 10.1186/bcr2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Valdez KE, et al. Human primary ductal carcinoma in situ (DCIS) subtype-specific pathology is preserved in a mouse intraductal (MIND) xenograft model. J. Pathol. 2011;225:565–573. doi: 10.1002/path.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Hong Y, et al. Mouse‐INtraDuctal (MIND): an in vivo model for studying the underlying mechanisms of DCIS malignancy. J. Pathol. 2022;256:186–201. doi: 10.1002/path.5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Elsarraj HS, et al. BCL9/STAT3 regulation of transcriptional enhancer networks promote DCIS progression. NPJ Breast Cancer. 2020;6:12. doi: 10.1038/s41523-020-0157-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Dobrolecki LE, et al. Patient-derived xenograft (PDX) models in basic and translational breast cancer research. Cancer Metastasis Rev. 2016;35:547–573. doi: 10.1007/s10555-016-9653-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Espina V, et al. Malignant precursor cells pre-exist in human breast DCIS and require autophagy for survival. PloS One. 2010;5:e10240. doi: 10.1371/journal.pone.0010240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Deome KB, Faulkin LJ, Bern HA, Blair PB. Development of mammary tumors from hyperplastic alveolar nodules transplanted into gland-free mammary fat pads of female C3H mice. Cancer Res. 1959;19:515–520. [PubMed] [Google Scholar]
- 223.Lyons TR, et al. Postpartum mammary gland involution drives progression of ductal carcinoma in situ through collagen and COX-2. Nat. Med. 2011;17:1109–1115. doi: 10.1038/nm.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Holen I, Speirs V, Morrissey B, Blyth K. In vivo models in breast cancer research: progress, challenges and future directions. Dis. Model. Mech. 2017;10:359–371. doi: 10.1242/dmm.028274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Manning HC, Buck JR, Cook RS. Mouse models of breast cancer: platforms for discovering precision imaging diagnostics and future cancer medicine. J. Nucl. Med. 2016;57(Suppl 1):60S–68SS. doi: 10.2967/jnumed.115.157917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Gengenbacher N, Singhal M, Augustin HG. Preclinical mouse solid tumour models: status quo, challenges and perspectives. Nat. Rev. Cancer. 2017;17:751–765. doi: 10.1038/nrc.2017.92. [DOI] [PubMed] [Google Scholar]
- 227.Attalla S, Taifour T, Bui T, Muller W. Insights from transgenic mouse models of PyMT-induced breast cancer: recapitulating human breast cancer progression in vivo. Oncogene. 2021;40:475–491. doi: 10.1038/s41388-020-01560-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Behbod F, Gomes AM, Machado HL. Modeling human ductal carcinoma in situ in the mouse. J. Mammary Gland Biol. Neoplasia. 2018;23:269–278. doi: 10.1007/s10911-018-9408-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Park MK, Lee CH, Lee H. Mouse models of breast cancer in preclinical research. Lab. Anim. Res. 2018;34:160–165. doi: 10.5625/lar.2018.34.4.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Taneja P, et al. MMTV mouse models and the diagnostic values of MMTV-like sequences in human breast cancer. Expert Rev. Mol. Diagn. 2009;9:423–440. doi: 10.1586/erm.09.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Zeng L, et al. Breast cancer animal models and applications. Zool. Res. 2020;41:477–494. doi: 10.24272/j.issn.2095-8137.2020.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Guy, C. T., Cardiff, R. D. & Muller, W. J. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol. Cell Biol. 12, 8 (1992). [DOI] [PMC free article] [PubMed]
- 233.Lin EY, et al. Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. Am. J. Pathol. 2003;163:2113–2126. doi: 10.1016/S0002-9440(10)63568-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Martinez LM, et al. Regulatory T cells control the switch from in situ to invasive breast cancer. Front. Immunol. 2019;10:1942. doi: 10.3389/fimmu.2019.01942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Boyle ST, Faulkner JW, McColl SR, Kochetkova M. The chemokine receptor CCR6 facilitates the onset of mammary neoplasia in the MMTV-PyMT mouse model via recruitment of tumor-promoting macrophages. Mol. Cancer. 2015;14:115. doi: 10.1186/s12943-015-0394-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Ursini-Siegel J, Schade B, Cardiff RD, Muller WJ. Insights from transgenic mouse models of ERBB2-induced breast cancer. Nat. Rev. Cancer. 2007;7:389–397. doi: 10.1038/nrc2127. [DOI] [PubMed] [Google Scholar]
- 237.Loibl S, Gianni L. HER2-positive breast cancer. Lancet. 2017;389:2415–2429. doi: 10.1016/S0140-6736(16)32417-5. [DOI] [PubMed] [Google Scholar]
- 238.Andrechek ER, et al. Amplification of the neu/erbB-2 oncogene in a mouse model of mammary tumorigenesis. Proc. Natl Acad. Sci. USA. 2000;97:3444–3449. doi: 10.1073/pnas.97.7.3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Iezzi, M. et al. BALB-neuT female mice as a dynamic model of mammary cancer. Transl. Anim. Models Drug Discov. Dev. 10.2174/978160805469511201010139 (2012).
- 240.Hosseini H, et al. Early dissemination seeds metastasis in breast cancer. Nature. 2016;540:552–558. doi: 10.1038/nature20785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Harper KL, et al. Mechanism of early dissemination and metastasis in Her2+ mammary cancer. Nature. 2016;540:588–592. doi: 10.1038/nature20609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Butel JS, Lednicky JA. Cell and molecular biology of simian virus 40: implications for human infections and disease. J. Natl Cancer Inst. 1999;91:119–134. doi: 10.1093/jnci/91.2.119. [DOI] [PubMed] [Google Scholar]
- 243.Maroulakou IG, Anver M, Garrett L, Green JE. Prostate and mammary adenocarcinoma in transgenic mice carrying a rat C3(1) simian virus 40 large tumor antigen fusion gene. Proc. Natl Acad. Sci. USA. 1994;91:11236–11240. doi: 10.1073/pnas.91.23.11236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Green JE, et al. The C3(1)/SV40 T-antigen transgenic mouse model of mammary cancer: ductal epithelial cell targeting with multistage progression to carcinoma. Oncogene. 2000;19:1020–1027. doi: 10.1038/sj.onc.1203280. [DOI] [PubMed] [Google Scholar]
- 245.Thennavan A, Garcia-Recio S, Liu S, He X, Perou CM. Molecular signatures of in situ to invasive progression for basal-like breast cancers: An integrated mouse model and human DCIS study. NPJ Breast Cancer. 2022;8:83. doi: 10.1038/s41523-022-00450-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Herschkowitz JI, et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007;8:R76. doi: 10.1186/gb-2007-8-5-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Schulze-Garg C, Löhler J, Gocht A, Deppert W. A transgenic mouse model for the ductal carcinoma in situ (DCIS) of the mammary gland. Oncogene. 2000;19:1028–1037. doi: 10.1038/sj.onc.1203281. [DOI] [PubMed] [Google Scholar]
- 248.Kaur H, et al. Next-generation sequencing: a powerful tool for the discovery of molecular markers in breast ductal carcinoma in situ. Expert Rev. Mol. Diagn. 2013;13:151–165. doi: 10.1586/erm.13.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Stegle O, Teichmann SA, Marioni JC. Computational and analytical challenges in single-cell transcriptomics. Nat. Rev. Genet. 2015;16:133–145. doi: 10.1038/nrg3833. [DOI] [PubMed] [Google Scholar]
- 250.Tang F, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods. 2009;6:377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 251.Navin N, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Hou Y, et al. Single-cell exome sequencing and monoclonal evolution of a JAK2-negative myeloproliferative neoplasm. Cell. 2012;148:873–885. doi: 10.1016/j.cell.2012.02.028. [DOI] [PubMed] [Google Scholar]
- 253.Xu X, et al. Single-cell exome sequencing reveals single-nucleotide mutation characteristics of a kidney tumor. Cell. 2012;148:886–895. doi: 10.1016/j.cell.2012.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Park SY, Gönen M, Kim HJ, Michor F, Polyak K. Cellular and genetic diversity in the progression of in situ human breast carcinomas to an invasive phenotype. J. Clin. Invest. 2010;120:636–644. doi: 10.1172/JCI40724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Farabegoli F, et al. Clone heterogeneity in diploid and aneuploid breast carcinomas as detected by FISH. Cytometry. 2001;46:50–56. doi: 10.1002/1097-0320(20010215)46:1<50::AID-CYTO1037>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 256.Torres L, et al. Intratumor genomic heterogeneity in breast cancer with clonal divergence between primary carcinomas and lymph node metastases. Breast Cancer Res. Treat. 2007;102:143–155. doi: 10.1007/s10549-006-9317-6. [DOI] [PubMed] [Google Scholar]
- 257.Navin NE. The first five years of single-cell cancer genomics and beyond. Genome Res. 2015;25:1499–1507. doi: 10.1101/gr.191098.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Wang Y, et al. Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature. 2014;512:155–160. doi: 10.1038/nature13600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Kim C, et al. Chemoresistance evolution in triple-negative breast cancer delineated by single-cell sequencing. Cell. 2018;173:879–893.e13. doi: 10.1016/j.cell.2018.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.van den Brink SC, et al. Single-cell sequencing reveals dissociation-induced gene expression in tissue subpopulations. Nat. Methods. 2017;14:935–936. doi: 10.1038/nmeth.4437. [DOI] [PubMed] [Google Scholar]
- 261.Lei Y, et al. Applications of single-cell sequencing in cancer research: progress and perspectives. J. Hematol. Oncol. J. Hematol. Oncol. 2021;14:91. doi: 10.1186/s13045-021-01105-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Dries R, et al. Advances in spatial transcriptomic data analysis. Genome Res. 2021;31:1706–1718. doi: 10.1101/gr.275224.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Burgess DJ. Spatial transcriptomics coming of age. Nat. Rev. Genet. 2019;20:317. doi: 10.1038/s41576-019-0129-z. [DOI] [PubMed] [Google Scholar]
- 264.Moncada R, et al. Integrating microarray-based spatial transcriptomics and single-cell RNA-seq reveals tissue architecture in pancreatic ductal adenocarcinomas. Nat. Biotechnol. 2020;38:333–342. doi: 10.1038/s41587-019-0392-8. [DOI] [PubMed] [Google Scholar]
- 265.Thrane K, Eriksson H, Maaskola J, Hansson J, Lundeberg J. Spatially resolved transcriptomics enables dissection of genetic heterogeneity in stage III cutaneous malignant melanoma. Cancer Res. 2018;78:5970–5979. doi: 10.1158/0008-5472.CAN-18-0747. [DOI] [PubMed] [Google Scholar]
- 266.Wu SZ, et al. A single-cell and spatially resolved atlas of human breast cancers. Nat. Genet. 2021;53:1334–1347. doi: 10.1038/s41588-021-00911-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Longo SK, Guo MG, Ji AL, Khavari PA. Integrating single-cell and spatial transcriptomics to elucidate intercellular tissue dynamics. Nat. Rev. Genet. 2021;22:627–644. doi: 10.1038/s41576-021-00370-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Ståhl PL, et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science. 2016;353:78–82. doi: 10.1126/science.aaf2403. [DOI] [PubMed] [Google Scholar]
- 269.Wei R, et al. Spatial charting of single-cell transcriptomes in tissues. Nat. Biotechnol. 2022;40:1190–1199. doi: 10.1038/s41587-022-01233-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Liu S-Q, et al. Single-cell and spatially resolved analysis uncovers cell heterogeneity of breast cancer. J. Hematol. Oncol. 2022;15:19. doi: 10.1186/s13045-022-01236-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Nagasawa S, et al. Genomic profiling reveals heterogeneous populations of ductal carcinoma in situ of the breast. Commun. Biol. 2021;4:438. doi: 10.1038/s42003-021-01959-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Yoosuf N, Navarro JF, Salmén F, Ståhl PL, Daub CO. Identification and transfer of spatial transcriptomics signatures for cancer diagnosis. Breast Cancer Res. 2020;22:6. doi: 10.1186/s13058-019-1242-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Hayward M-K, Weaver VM. Improving DCIS diagnosis and predictive outcome by applying artificial intelligence. Biochim. Biophys. Acta Rev. Cancer. 2021;1876:188555. doi: 10.1016/j.bbcan.2021.188555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Hayward M-K, et al. Derivation of a nuclear heterogeneity image index to grade DCIS. Comput. Struct. Biotechnol. J. 2020;18:4063–4070. doi: 10.1016/j.csbj.2020.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Sandbank J, et al. Validation and real-world clinical application of an artificial intelligence algorithm for breast cancer detection in biopsies. NPJ Breast Cancer. 2022;8:129. doi: 10.1038/s41523-022-00496-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Damiani C, et al. Evaluation of an AI model to assess future breast cancer risk. Radiology. 2023;307:e222679. doi: 10.1148/radiol.222679. [DOI] [PubMed] [Google Scholar]
- 277.Katayama, A. et al. Atypia in breast pathology: what pathologists need to know. Pathology) 54, 20–31 (2022). [DOI] [PubMed]
- 278.Mercan E, et al. Assessment of machine learning of breast pathology structures for automated differentiation of breast cancer and high-risk proliferative lesions. JAMA Netw. Open. 2019;2:e198777. doi: 10.1001/jamanetworkopen.2019.8777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Yamamoto Y, et al. Quantitative diagnosis of breast tumors by morphometric classification of microenvironmental myoepithelial cells using a machine learning approach. Sci. Rep. 2017;7:46732. doi: 10.1038/srep46732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Acerbi I, et al. Human breast cancer invasion and aggression correlates with ECM stiffening and immune cell infiltration. Integr. Biol. 2015;7:1120–1134. doi: 10.1039/c5ib00040h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Bejnordi BE, et al. Using deep convolutional neural networks to identify and classify tumor-associated stroma in diagnostic breast biopsies. Mod. Pathol. 2018;31:1502–1512. doi: 10.1038/s41379-018-0073-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Klimov S, et al. A whole slide image-based machine learning approach to predict ductal carcinoma in situ (DCIS) recurrence risk. Breast Cancer Res. 2019;21:83. doi: 10.1186/s13058-019-1165-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Donker M, et al. Breast-conserving treatment with or without radiotherapy in ductal carcinoma In Situ: 15-year recurrence rates and outcome after a recurrence, from the EORTC 10853 randomized phase III trial. J. Clin. Oncol. 2013;31:4054–4059. doi: 10.1200/JCO.2013.49.5077. [DOI] [PubMed] [Google Scholar]
- 284.Benson JR, Wishart GC. Predictors of recurrence for ductal carcinoma in situ after breast-conserving surgery. Lancet Oncol. 2013;14:e348–e357. doi: 10.1016/S1470-2045(13)70135-9. [DOI] [PubMed] [Google Scholar]
- 285.Rakovitch E, et al. Refined estimates of local recurrence risks by DCIS score adjusting for clinicopathological features: a combined analysis of ECOG-ACRIN E5194 and Ontario DCIS cohort studies. Breast Cancer Res. Treat. 2018;169:359–369. doi: 10.1007/s10549-018-4693-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Hughes LL, et al. Local excision alone without irradiation for ductal carcinoma in situ of the breast: a trial of the Eastern Cooperative Oncology Group. J. Clin. Oncol. 2009;27:5319–5324. doi: 10.1200/JCO.2009.21.8560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Solin LJ, et al. Surgical excision without radiation for ductal carcinoma in situ of the breast: 12-year results from the ECOG-ACRIN E5194 study. J. Clin. Oncol. 2015;33:3938–3944. doi: 10.1200/JCO.2015.60.8588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Roka S, et al. High nuclear grade and negative estrogen receptor are significant risk factors for recurrence in DCIS. Eur. J. Surg. Oncol. 2004;30:243–247. doi: 10.1016/j.ejso.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 289.Provenzano E, et al. Biological markers that predict clinical recurrence in ductal carcinoma in situ of the breast. Eur. J. Cancer. 2003;39:622–630. doi: 10.1016/S0959-8049(02)00666-4. [DOI] [PubMed] [Google Scholar]
- 290.Thorat MA, et al. Prognostic value of ER and PgR expression and the impact of multi-clonal expression for recurrence in ductal carcinoma in situ: results from the UK/ANZ DCIS trial. Clin. Cancer Res. 2021;27:2861–2867. doi: 10.1158/1078-0432.CCR-20-4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Ringberg A, et al. Cell biological factors in ductal carcinoma in situ (DCIS) of the breast-relationship to ipsilateral local recurrence and histopathological characteristics. Eur. J. Cancer. 2001;37:1514–1522. doi: 10.1016/S0959-8049(01)00165-4. [DOI] [PubMed] [Google Scholar]
- 292.Kerlikowske K, et al. Biomarker expression and risk of subsequent tumors after initial ductal carcinoma in situ diagnosis. J. Natl Cancer Inst. 2010;102:627–637. doi: 10.1093/jnci/djq101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Miligy IM, et al. The clinical significance of oestrogen receptor expression in breast ductal carcinoma in situ. Br. J. Cancer. 2020;123:1513–1520. doi: 10.1038/s41416-020-1023-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Lari SA, Kuerer HM. Biological markers in DCIS and risk of breast recurrence: a systematic review. J. Cancer. 2011;2:232–261. doi: 10.7150/jca.2.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Nofech-Mozes S, Spayne J, Rakovitch E, Hanna W. Prognostic and predictive molecular markers in DCIS: a review. Adv. Anat. Pathol. 2005;12:256–264. doi: 10.1097/01.pap.0000184177.65919.5e. [DOI] [PubMed] [Google Scholar]
- 296.Curigliano G, et al. Risk of subsequent in situ and invasive breast cancer in human epidermal growth factor receptor 2-positive ductal carcinoma in situ. Ann. Oncol. 2015;26:682–687. doi: 10.1093/annonc/mdv013. [DOI] [PubMed] [Google Scholar]
- 297.Rakovitch E, et al. HER2/neu and Ki-67 expression predict non-invasive recurrence following breast-conserving therapy for ductal carcinoma in situ. Br. J. Cancer. 2012;106:1160–1165. doi: 10.1038/bjc.2012.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Miligy IM, et al. The clinical and biological significance of HER2 over-expression in breast ductal carcinoma in situ: a large study from a single institution. Br. J. Cancer. 2019;120:1075–1082. doi: 10.1038/s41416-019-0436-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Lambein K, et al. Comparison of HER2 amplification status among breast cancer subgroups offers new insights in pathways of breast cancer progression. Virchows Arch. 2017;471:575–587. doi: 10.1007/s00428-017-2161-8. [DOI] [PubMed] [Google Scholar]
- 300.Hoque A, et al. Her-2/neu gene amplification in ductal carcinoma in situ of the breast. Cancer Epidemiol. Biomark. Prev. 2002;11:587–590. [PubMed] [Google Scholar]
- 301.Van Bockstal M, et al. Histopathological characterization of ductal carcinoma in situ (DCIS) of the breast according to HER2 amplification status and molecular subtype. Virchows Arch. 2014;465:275–289. doi: 10.1007/s00428-014-1609-3. [DOI] [PubMed] [Google Scholar]
- 302.Borgquist S, et al. The prognostic role of HER2 expression in ductal breast carcinoma in situ (DCIS); a population-based cohort study. BMC Cancer. 2015;15:468. doi: 10.1186/s12885-015-1479-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Glover JA, Hughes CM, Cantwell MM, Murray LJ. A systematic review to establish the frequency of cyclooxygenase-2 expression in normal breast epithelium, ductal carcinoma in situ, microinvasive carcinoma of the breast and invasive breast cancer. Br. J. Cancer. 2011;105:13–17. doi: 10.1038/bjc.2011.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Boland GP, Butt IS, Prasad R, Knox WF, Bundred NJ. COX-2 expression is associated with an aggressive phenotype in ductal carcinoma in situ. Br. J. Cancer. 2004;90:423–429. doi: 10.1038/sj.bjc.6601534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Visser LL, et al. Clinicopathological risk factors for an invasive breast cancer recurrence after ductal carcinoma in situ—a nested case-control study. Clin. Cancer Res. 2018;24:3593–3601. doi: 10.1158/1078-0432.CCR-18-0201. [DOI] [PubMed] [Google Scholar]
- 306.Almekinders MMM, et al. Breast adipocyte size associates with ipsilateral invasive breast cancer risk after ductal carcinoma in situ. NPJ Breast Cancer. 2021;7:31. doi: 10.1038/s41523-021-00232-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Generali D, et al. COX-2 expression is predictive for early relapse and aromatase inhibitor resistance in patients with ductal carcinoma in situ of the breast, and is a target for treatment. Br. J. Cancer. 2014;111:46–54. doi: 10.1038/bjc.2014.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Barnes N, Haywood P, Flint P, Knox WF, Bundred NJ. Survivin expression in in situ and invasive breast cancer relates to COX-2 expression and DCIS recurrence. Br. J. Cancer. 2006;94:253–258. doi: 10.1038/sj.bjc.6602932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Davis JE, et al. Her2 and Ki67 biomarkers predict recurrence of ductal carcinoma in situ. Appl. Immunohistochem. Mol. Morphol. 2016;24:20–25. doi: 10.1097/PAI.0000000000000223. [DOI] [PubMed] [Google Scholar]
- 310.Poulakaki N, et al. Hormonal receptor status, Ki-67 and HER2 expression: prognostic value in the recurrence of ductal carcinoma in situ of the breast? Breast Edinb. Scotl. 2016;25:57–61. doi: 10.1016/j.breast.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 311.Poulakaki N, et al. Ki-67 expression as a factor predicting recurrence of ductal carcinoma in situ of the breast: a systematic review and meta-analysis. Clin. Breast Cancer. 2018;18:157–167.e6. doi: 10.1016/j.clbc.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 312.Perez AA, Balabram D, Rocha RM, da Silva Souza Á, Gobbi H. Co-expression of p16, Ki67 and COX-2 is associated with basal phenotype in high-grade ductal carcinoma in situ of the breast. J. Histochem. Cytochem. 2015;63:408–416. doi: 10.1369/0022155415576540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Nofech-Mozes S, Hanna W, Rakovitch E. Molecular evaluation of breast ductal carcinoma in situ with oncotype DX DCIS. Am. J. Pathol. 2019;189:975–980. doi: 10.1016/j.ajpath.2018.12.003. [DOI] [PubMed] [Google Scholar]
- 314.Oncotype DX DCIS score predicts recurrence. Cancer Discov. 5, OF3 (2015). [DOI] [PubMed]
- 315.Solin LJ, et al. A multigene expression assay to predict local recurrence risk for ductal carcinoma in situ of the breast. J. Natl Cancer Inst. 2013;105:701–710. doi: 10.1093/jnci/djt067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Salvatorelli L, et al. Ductal carcinoma in situ of the breast: an update with emphasis on radiological and morphological features as predictive prognostic factors. Cancers. 2020;12:609. doi: 10.3390/cancers12030609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Bijker N, et al. Risk factors for recurrence and metastasis after breast-conserving therapy for ductal carcinoma-in-situ: analysis of European Organization for Research and Treatment of Cancer Trial 10853. J. Clin. Oncol. 2001;19:2263–2271. doi: 10.1200/JCO.2001.19.8.2263. [DOI] [PubMed] [Google Scholar]
- 318.Barrio AV, Van Zee KJ. Controversies in the treatment of ductal carcinoma in situ. Annu. Rev. Med. 2017;68:197–211. doi: 10.1146/annurev-med-050715-104920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Fisher B, et al. Lumpectomy and radiation therapy for the treatment of intraductal breast cancer: findings from National Surgical Adjuvant Breast and Bowel Project B-17. J. Clin. Oncol. 1998;16:441–452. doi: 10.1200/JCO.1998.16.2.441. [DOI] [PubMed] [Google Scholar]
- 320.Fisher B, et al. Lumpectomy compared with lumpectomy and radiation therapy for the treatment of intraductal breast cancer. N. Engl. J. Med. 1993;328:1581–1586. doi: 10.1056/NEJM199306033282201. [DOI] [PubMed] [Google Scholar]
- 321.Sue GR, Lannin DR, Au AF, Narayan D, Chagpar AB. Factors associated with decision to pursue mastectomy and breast reconstruction for treatment of ductal carcinoma in situ of the breast. Am. J. Surg. 2013;206:682–685. doi: 10.1016/j.amjsurg.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 322.Cardoso F, et al. Early breast cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2019;30:1194–1220. doi: 10.1093/annonc/mdz173. [DOI] [PubMed] [Google Scholar]
- 323.Curigliano G, et al. De-escalating and escalating treatments for early-stage breast cancer: the St. Gallen International Expert Consensus Conference on the Primary Therapy of Early Breast Cancer 2017. Ann. Oncol. 2017;28:1700–1712. doi: 10.1093/annonc/mdx308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Burstein HJ, Polyak K, Wong JS, Lester SC, Kaelin CM. Ductal carcinoma in situ of the breast. N. Engl. J. Med. 2004;350:1430–1441. doi: 10.1056/NEJMra031301. [DOI] [PubMed] [Google Scholar]
- 325.Martínez-Pérez C, et al. Current treatment trends and the need for better predictive tools in the management of ductal carcinoma in situ of the breast. Cancer Treat. Rev. 2017;55:163–172. doi: 10.1016/j.ctrv.2017.03.009. [DOI] [PubMed] [Google Scholar]
- 326.Wapnir IL, et al. Long-term outcomes of invasive ipsilateral breast tumor recurrences after lumpectomy in NSABP B-17 and B-24 randomized clinical trials for DCIS. J. Natl Cancer Inst. 2011;103:478–488. doi: 10.1093/jnci/djr027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.EORTC Breast Cancer Cooperative Group. et al. Breast-conserving treatment with or without radiotherapy in ductal carcinoma-in-situ: ten-year results of European Organisation for Research and Treatment of Cancer randomized phase III trial 10853-a study by the EORTC Breast Cancer Cooperative Group and EORTC Radiotherapy Group. J. Clin. Oncol. 2006;24:3381–3387. doi: 10.1200/JCO.2006.06.1366. [DOI] [PubMed] [Google Scholar]
- 328.Wärnberg F, et al. Effect of radiotherapy after breast-conserving surgery for ductal carcinoma in situ: 20 years follow-up in the randomized SweDCIS Trial. J. Clin. Oncol. 2014;32:3613–3618. doi: 10.1200/JCO.2014.56.2595. [DOI] [PubMed] [Google Scholar]
- 329.Cuzick J, et al. Effect of tamoxifen and radiotherapy in women with locally excised ductal carcinoma in situ: long-term results from the UK/ANZ DCIS trial. Lancet Oncol. 2011;12:21–29. doi: 10.1016/S1470-2045(10)70266-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.McCormick B, et al. RTOG 9804: a prospective randomized trial for good-risk ductal carcinoma in situ comparing radiotherapy with observation. J. Clin. Oncol. 2015;33:709–715. doi: 10.1200/JCO.2014.57.9029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.McCormick B, et al. Randomized phase III trial evaluating radiation following surgical excision for good-risk ductal carcinoma in situ: long-term report from NRG Oncology/RTOG 9804. J. Clin. Oncol. 2021;39:3574–3582. doi: 10.1200/JCO.21.01083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Staley H, McCallum I, Bruce J. Postoperative Tamoxifen for ductal carcinoma in situ: cochrane systematic review and meta-analysis. Breast Edinb. Scotl. 2014;23:546–551. doi: 10.1016/j.breast.2014.06.015. [DOI] [PubMed] [Google Scholar]
- 333.Staley H, McCallum I, Bruce J. Postoperative tamoxifen for ductal carcinoma in situ. Cochrane Database Syst. Rev. 2012;10:CD007847. doi: 10.1002/14651858.CD007847.pub2. [DOI] [PubMed] [Google Scholar]
- 334.van Seijen M, et al. Ductal carcinoma in situ: to treat or not to treat, that is the question. Br. J. Cancer. 2019;121:285–292. doi: 10.1038/s41416-019-0478-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Allred DC, et al. Adjuvant tamoxifen reduces subsequent breast cancer in women with estrogen receptor-positive ductal carcinoma in situ: a study based on NSABP protocol B-24. J. Clin. Oncol. 2012;30:1268–1273. doi: 10.1200/JCO.2010.34.0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Morrow M. Refining the use of endocrine therapy for ductal carcinoma in situ. J. Clin. Oncol. 2012;30:1249–1251. doi: 10.1200/JCO.2011.40.5514. [DOI] [PubMed] [Google Scholar]
- 337.Forbes JF, et al. Anastrozole versus tamoxifen for the prevention of locoregional and contralateral breast cancer in postmenopausal women with locally excised ductal carcinoma in situ (IBIS-II DCIS): a double-blind, randomised controlled trial. Lancet. 2016;387:866–873. doi: 10.1016/S0140-6736(15)01129-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Margolese RG, et al. Anastrozole versus tamoxifen in postmenopausal women with ductal carcinoma in situ undergoing lumpectomy plus radiotherapy (NSABP B-35): a randomised, double-blind, phase 3 clinical trial. Lancet. 2016;387:849–856. doi: 10.1016/S0140-6736(15)01168-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Francis A, et al. Addressing overtreatment of screen detected DCIS; the LORIS trial. Eur. J. Cancer. 2015;51:2296–2303. doi: 10.1016/j.ejca.2015.07.017. [DOI] [PubMed] [Google Scholar]
- 340.Elshof LE, et al. Feasibility of a prospective, randomised, open-label, international multicentre, phase III, non-inferiority trial to assess the safety of active surveillance for low risk ductal carcinoma in situ—The LORD study. Eur. J. Cancer. 2015;51:1497–1510. doi: 10.1016/j.ejca.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 341.Hwang ES, et al. The COMET (Comparison of Operative versus Monitoring and Endocrine Therapy) trial: a phase III randomised controlled clinical trial for low-risk ductal carcinoma in situ (DCIS) BMJ Open. 2019;9:e026797. doi: 10.1136/bmjopen-2018-026797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Kanbayashi C, Iwata H. Current approach and future perspective for ductal carcinoma in situ of the breast. Jpn. J. Clin. Oncol. 2017;47:671–677. doi: 10.1093/jjco/hyx059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Ryser MD, et al. Outcomes of active surveillance for ductal carcinoma in situ: a computational risk analysis. J. Natl Cancer Inst. 2016;108:djv372. doi: 10.1093/jnci/djv372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Lowenfeld L, et al. Dendritic cell vaccination enhances immune responses and induces regression of HER2pos DCIS independent of route: results of randomized selection design trial. Clin. Cancer Res. 2017;23:2961–2971. doi: 10.1158/1078-0432.CCR-16-1924. [DOI] [PubMed] [Google Scholar]
- 345.Siziopikou KP, et al. Preliminary results of centralized HER2 testing in ductal carcinoma in situ (DCIS): NSABP B-43. Breast Cancer Res. Treat. 2013;142:415–421. doi: 10.1007/s10549-013-2755-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Cobleigh MA, et al. Comparison of radiation with or without concurrent trastuzumab for HER2-positive ductal carcinoma in situ resected by lumpectomy: a phase III clinical trial. J. Clin. Oncol. 2021;39:2367–2374. doi: 10.1200/JCO.20.02824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Gu J, Delisle M, Engler-Stringer R, Groot G. Mastectomy versus breast-conservation therapy: an examination of how individual, clinicopathologic, and physician factors influence decision-making. Curr. Oncol. Tor. Ont. 2019;26:e522–e534. doi: 10.3747/co.26.5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.EBCTCG (Early Breast Cancer Trialists’ Collaborative Group). et al. Effect of radiotherapy after mastectomy and axillary surgery on 10-year recurrence and 20-year breast cancer mortality: meta-analysis of individual patient data for 8135 women in 22 randomised trials. Lancet 383, 2127–2135 (2014). [DOI] [PMC free article] [PubMed]
- 349.Huppert LA, Gumusay O, Idossa D, Rugo HS. Systemic therapy for hormone receptor‐positive/human epidermal growth factor receptor 2‐negative early stage and metastatic breast cancer. CA Cancer J. Clin. 2023;73:480–515. doi: 10.3322/caac.21777. [DOI] [PubMed] [Google Scholar]
- 350.Burstein HJ. Systemic therapy for estrogen receptor-positive, HER2-negative breast cancer. N. Engl. J. Med. 2020;383:2557–2570. doi: 10.1056/NEJMra1307118. [DOI] [PubMed] [Google Scholar]
- 351.Baum M, et al. Anastrozole alone or in combination with tamoxifen versus tamoxifen alone for adjuvant treatment of postmenopausal women with early breast cancer: first results of the ATAC randomised trial. Lancet. 2002;359:2131–2139. doi: 10.1016/S0140-6736(02)09088-8. [DOI] [PubMed] [Google Scholar]
- 352.Early Breast Cancer Trialists’ Collaborative Group (EBCTCG). Aromatase inhibitors versus tamoxifen in early breast cancer: patient-level meta-analysis of the randomised trials. Lancet 386, 1341–1352 (2015).. [DOI] [PubMed]
- 353.Pagani O, et al. Adjuvant exemestane with ovarian suppression in premenopausal breast cancer: long-term follow-up of the combined TEXT and SOFT trials. J. Clin. Oncol. 2023;41:1376–1382. doi: 10.1200/JCO.22.01064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Hortobagyi GN, et al. Ribociclib as first-line therapy for HR-positive, advanced breast cancer. N. Engl. J. Med. 2016;375:1738–1748. doi: 10.1056/NEJMoa1609709. [DOI] [PubMed] [Google Scholar]
- 355.Finn RS, et al. Palbociclib and letrozole in advanced breast cancer. N. Engl. J. Med. 2016;375:1925–1936. doi: 10.1056/NEJMoa1607303. [DOI] [PubMed] [Google Scholar]
- 356.Goetz MP, et al. MONARCH 3: abemaciclib as initial therapy for advanced breast. Cancer J. Clin. Oncol. 2017;35:3638–3646. doi: 10.1200/JCO.2017.75.6155. [DOI] [PubMed] [Google Scholar]
- 357.du Rusquec P, Blonz C, Frenel JS, Campone M. Targeting the PI3K/Akt/mTOR pathway in estrogen-receptor positive HER2 negative advanced breast cancer. Ther. Adv. Med. Oncol. 2020;12:1758835920940939. doi: 10.1177/1758835920940939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Valabrega G, Montemurro F, Aglietta M. Trastuzumab: mechanism of action, resistance and future perspectives in HER2-overexpressing breast cancer. Ann. Oncol. 2007;18:977–984. doi: 10.1093/annonc/mdl475. [DOI] [PubMed] [Google Scholar]
- 359.Oh D-Y, Bang Y-J. HER2-targeted therapies—a role beyond breast cancer. Nat. Rev. Clin. Oncol. 2020;17:33–48. doi: 10.1038/s41571-019-0268-3. [DOI] [PubMed] [Google Scholar]
- 360.Ishii K, Morii N, Yamashiro H. Pertuzumab in the treatment of HER2-positive breast cancer: an evidence-based review of its safety, efficacy, and place in therapy. Core Evid. 2019;14:51–70. doi: 10.2147/CE.S217848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Baselga J, et al. Pertuzumab plus trastuzumab plus docetaxel for metastatic breast cancer. N. Engl. J. Med. 2012;366:109–119. doi: 10.1056/NEJMoa1113216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Von Minckwitz G, et al. Adjuvant pertuzumab and trastuzumab in early HER2-positive breast cancer. N. Engl. J. Med. 2017;377:122–131. doi: 10.1056/NEJMoa1703643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Beck A, Goetsch L, Dumontet C, Corvaïa N. Strategies and challenges for the next generation of antibody-drug conjugates. Nat. Rev. Drug Discov. 2017;16:315–337. doi: 10.1038/nrd.2016.268. [DOI] [PubMed] [Google Scholar]
- 364.Lewis Phillips GD, et al. Targeting HER2-positive breast cancer with trastuzumab-DM1, an antibody-cytotoxic drug conjugate. Cancer Res. 2008;68:9280–9290. doi: 10.1158/0008-5472.CAN-08-1776. [DOI] [PubMed] [Google Scholar]
- 365.Verma S, et al. Trastuzumab emtansine for HER2-positive advanced breast cancer. N. Engl. J. Med. 2012;367:1783–1791. doi: 10.1056/NEJMoa1209124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Cortés J, et al. Trastuzumab deruxtecan versus trastuzumab emtansine for breast cancer. N. Engl. J. Med. 2022;386:1143–1154. doi: 10.1056/NEJMoa2115022. [DOI] [PubMed] [Google Scholar]
- 367.Modi S, et al. Trastuzumab deruxtecan in previously treated HER2-positive breast cancer. N. Engl. J. Med. 2020;382:610–621. doi: 10.1056/NEJMoa1914510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Konecny GE, et al. Activity of the dual kinase inhibitor lapatinib (GW572016) against HER-2-overexpressing and trastuzumab-treated breast cancer cells. Cancer Res. 2006;66:1630–1639. doi: 10.1158/0008-5472.CAN-05-1182. [DOI] [PubMed] [Google Scholar]
- 369.Collins DM, et al. Preclinical characteristics of the irreversible Pan-HER kinase inhibitor neratinib compared with lapatinib: implications for the treatment of HER2-positive and HER2-mutated breast cancer. Cancers. 2019;11:737. doi: 10.3390/cancers11060737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Martin M, et al. Neratinib after trastuzumab-based adjuvant therapy in HER2-positive breast cancer (ExteNET): 5-year analysis of a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2017;18:1688–1700. doi: 10.1016/S1470-2045(17)30717-9. [DOI] [PubMed] [Google Scholar]
- 371.B X, et al. Pyrotinib plus capecitabine versus lapatinib plus capecitabine for the treatment of HER2-positive metastatic breast cancer (PHOEBE): a multicentre, open-label, randomised, controlled, phase 3 trial. Lancet Oncol. 2021;22:351–360. doi: 10.1016/S1470-2045(20)30702-6. [DOI] [PubMed] [Google Scholar]
- 372.Lin NU, et al. Tucatinib vs placebo, both in combination with trastuzumab and capecitabine, for previously treated ERBB2 (HER2)-positive metastatic breast cancer in patients with brain metastases: updated exploratory analysis of the HER2CLIMB Randomized Clinical Trial. JAMA Oncol. 2023;9:197. doi: 10.1001/jamaoncol.2022.5610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Leon-Ferre, R. A. & Goetz, M. P. Advances in systemic therapies for triple negative breast cancer. BMJ10.1136/bmj-2022-071674 (2023). [DOI] [PubMed]
- 374.Zhu S, et al. Recent advances in targeted strategies for triple-negative breast cancer. J. Hematol. Oncol. 2023;16:100. doi: 10.1186/s13045-023-01497-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Li T, et al. Targeting PARP for the optimal immunotherapy efficiency in gynecologic malignancies. Biomed. Pharmacother. Biomed. Pharmacother. 2023;162:114712. doi: 10.1016/j.biopha.2023.114712. [DOI] [PubMed] [Google Scholar]
- 376.Yi M, et al. Advances and perspectives of PARP inhibitors. Exp. Hematol. Oncol. 2019;8:29. doi: 10.1186/s40164-019-0154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Luo L, Keyomarsi K. PARP inhibitors as single agents and in combination therapy: the most promising treatment strategies in clinical trials for BRCA-mutant ovarian and triple-negative breast cancers. Expert Opin. Investig. Drugs. 2022;31:607–631. doi: 10.1080/13543784.2022.2067527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Litton JK, et al. Talazoparib in patients with advanced breast cancer and a germline BRCA mutation. N. Engl. J. Med. 2018;379:753–763. doi: 10.1056/NEJMoa1802905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Robson M, et al. Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. N. Engl. J. Med. 2017;377:523–533. doi: 10.1056/NEJMoa1706450. [DOI] [PubMed] [Google Scholar]
- 380.Motzer RJ, et al. Nivolumab versus everolimus in advanced renal-cell carcinoma. N. Engl. J. Med. 2015;373:1803–1813. doi: 10.1056/NEJMoa1510665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Valsecchi ME. Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N. Engl. J. Med. 2015;373:1270. doi: 10.1056/NEJMc1509660. [DOI] [PubMed] [Google Scholar]
- 382.Antonia SJ, et al. Durvalumab after chemoradiotherapy in stage III non-small-cell lung cancer. N. Engl. J. Med. 2017;377:1919–1929. doi: 10.1056/NEJMoa1709937. [DOI] [PubMed] [Google Scholar]
- 383.Schmid P, et al. Atezolizumab and Nab-paclitaxel in advanced triple-negative breast cancer. N. Engl. J. Med. 2018;379:2108–2121. doi: 10.1056/NEJMoa1809615. [DOI] [PubMed] [Google Scholar]
- 384.Schmid P, et al. Pembrolizumab for early triple-negative breast cancer. N. Engl. J. Med. 2020;382:810–821. doi: 10.1056/NEJMoa1910549. [DOI] [PubMed] [Google Scholar]
- 385.Bonneterre J, et al. Anastrozole versus tamoxifen as first-line therapy for advanced breast cancer in 668 postmenopausal women: results of the tamoxifen or arimidex randomized group efficacy and tolerability study. J. Clin. Oncol. 2000;18:3748–3757. doi: 10.1200/JCO.2000.18.22.3748. [DOI] [PubMed] [Google Scholar]
- 386.Nabholtz JM, et al. Anastrozole is superior to tamoxifen as first-line therapy for advanced breast cancer in postmenopausal women: results of a North American multicenter randomized trial. J. Clin. Oncol. 2000;18:3758–3767. doi: 10.1200/JCO.2000.18.22.3758. [DOI] [PubMed] [Google Scholar]
- 387.Mouridsen H, et al. Superior efficacy of letrozole versus tamoxifen as first-line therapy for postmenopausal women with advanced breast cancer: results of a phase III study of the international letrozole breast cancer group. J. Clin. Oncol. 2001;19:2596–2606. doi: 10.1200/JCO.2001.19.10.2596. [DOI] [PubMed] [Google Scholar]
- 388.Pagani, O. et al. Adjuvant exemestane with ovarian suppression in premenopausal breast cancer. N. Engl. J. Med.10.1056/NEJMoa1404037 (2014). [DOI] [PMC free article] [PubMed]
- 389.Sledge GW, et al. The effect of abemaciclib plus fulvestrant on overall survival in hormone receptor-positive, ERBB2-negative breast cancer that progressed on endocrine therapy-MONARCH 2: a randomized clinical trial. JAMA Oncol. 2020;6:116–124. doi: 10.1001/jamaoncol.2019.4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Loibl S, et al. Palbociclib for residual high-risk invasive HR-positive and HER2-negative early breast cancer—the penelope-B trial. J. Clin. Oncol. 2021;39:1518–1530. doi: 10.1200/JCO.20.03639. [DOI] [PubMed] [Google Scholar]
- 391.Mayer EL, et al. Palbociclib with adjuvant endocrine therapy in early breast cancer (PALLAS): interim analysis of a multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2021;22:212–222. doi: 10.1016/S1470-2045(20)30642-2. [DOI] [PubMed] [Google Scholar]
- 392.Johnston SRD, et al. Abemaciclib plus endocrine therapy for hormone receptor-positive, HER2-negative, node-positive, high-risk early breast cancer (monarchE): results from a preplanned interim analysis of a randomised, open-label, phase 3 trial. Lancet Oncol. 2023;24:77–90. doi: 10.1016/S1470-2045(22)00694-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Cristofanilli M, et al. Fulvestrant plus palbociclib versus fulvestrant plus placebo for treatment of hormone-receptor-positive, HER2-negative metastatic breast cancer that progressed on previous endocrine therapy (PALOMA-3): final analysis of the multicentre, double-blind, phase 3 randomised controlled trial. Lancet Oncol. 2016;17:425–439. doi: 10.1016/S1470-2045(15)00613-0. [DOI] [PubMed] [Google Scholar]
- 394.Hortobagyi GN, et al. Overall survival with ribociclib plus letrozole in advanced breast cancer. N. Engl. J. Med. 2022;386:942–950. doi: 10.1056/NEJMoa2114663. [DOI] [PubMed] [Google Scholar]
- 395.Slamon DJ, et al. Ribociclib plus fulvestrant for postmenopausal women with hormone receptor-positive, human epidermal growth factor receptor 2-negative advanced breast cancer in the phase III randomized MONALEESA-3 trial: updated overall survival. Ann. Oncol. 2021;32:1015–1024. doi: 10.1016/j.annonc.2021.05.353. [DOI] [PubMed] [Google Scholar]
- 396.Lu Y-S, et al. Updated overall survival of ribociclib plus endocrine therapy versus endocrine therapy alone in pre- and perimenopausal patients with HR+/HER2- advanced breast cancer in MONALEESA-7: a phase III randomized clinical trial. Clin. Cancer Res. 2022;28:851–859. doi: 10.1158/1078-0432.CCR-21-3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Piccart-Gebhart MJ, et al. Trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer. N. Engl. J. Med. 2005;353:1659–1672. doi: 10.1056/NEJMoa052306. [DOI] [PubMed] [Google Scholar]
- 398.Romond EH, et al. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N. Engl. J. Med. 2005;353:1673–1684. doi: 10.1056/NEJMoa052122. [DOI] [PubMed] [Google Scholar]
- 399.Slamon D, et al. Adjuvant trastuzumab in HER2-positive breast cancer. N. Engl. J. Med. 2011;365:1273–1283. doi: 10.1056/NEJMoa0910383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Swain SM, et al. Pertuzumab, trastuzumab, and docetaxel in HER2-positive metastatic breast cancer. N. Engl. J. Med. 2015;372:724–734. doi: 10.1056/NEJMoa1413513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Rugo HS, et al. Efficacy of margetuximab vs trastuzumab in patients with pretreated ERBB2-positive advanced breast cancer: a phase 3 randomized clinical trial. JAMA Oncol. 2021;7:573–584. doi: 10.1001/jamaoncol.2020.7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Baselga J, et al. Lapatinib with trastuzumab for HER2-positive early breast cancer (NeoALTTO): a randomised, open-label, multicentre, phase 3 trial. Lancet. 2012;379:633–640. doi: 10.1016/S0140-6736(11)61847-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Untch M, et al. Lapatinib versus trastuzumab in combination with neoadjuvant anthracycline-taxane-based chemotherapy (GeparQuinto, GBG 44): a randomised phase 3 trial. Lancet Oncol. 2012;13:135–144. doi: 10.1016/S1470-2045(11)70397-7. [DOI] [PubMed] [Google Scholar]
- 404.Robidoux A, et al. Lapatinib as a component of neoadjuvant therapy for HER2-positive operable breast cancer (NSABP protocol B-41): an open-label, randomised phase 3 trial. Lancet Oncol. 2013;14:1183–1192. doi: 10.1016/S1470-2045(13)70411-X. [DOI] [PubMed] [Google Scholar]
- 405.Piccart-Gebhart M, et al. Adjuvant lapatinib and trastuzumab for early human epidermal growth factor receptor 2-positive breast cancer: results from the randomized phase III adjuvant lapatinib and/or trastuzumab treatment optimization trial. J. Clin. Oncol. 2016;34:1034–1042. doi: 10.1200/JCO.2015.62.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Saura C, et al. Neratinib plus capecitabine versus lapatinib plus capecitabine in HER2-positive metastatic breast cancer previously treated with ≥2 HER2-directed regimens: phase III NALA trial. J. Clin. Oncol. 2020;38:3138–3149. doi: 10.1200/JCO.20.00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Chan A, et al. Final efficacy results of neratinib in her2-positive hormone receptor-positive early-stage breast cancer from the phase III ExteNET trial. Clin. Breast Cancer. 2021;21:80–91.e7. doi: 10.1016/j.clbc.2020.09.014. [DOI] [PubMed] [Google Scholar]
- 408.Murthy RK, et al. Tucatinib, trastuzumab, and capecitabine for HER2-positive metastatic breast cancer. N. Engl. J. Med. 2020;382:597–609. doi: 10.1056/NEJMoa1914609. [DOI] [PubMed] [Google Scholar]
- 409.von Minckwitz G, et al. Trastuzumab emtansine for residual invasive HER2-positive breast cancer. N. Engl. J. Med. 2019;380:617–628. doi: 10.1056/NEJMoa1814017. [DOI] [PubMed] [Google Scholar]
- 410.Winer EP, et al. Pembrolizumab versus investigator-choice chemotherapy for metastatic triple-negative breast cancer (KEYNOTE-119): a randomised, open-label, phase 3 trial. Lancet Oncol. 2021;22:499–511. doi: 10.1016/S1470-2045(20)30754-3. [DOI] [PubMed] [Google Scholar]
- 411.Cortes J, et al. Pembrolizumab plus chemotherapy versus placebo plus chemotherapy for previously untreated locally recurrent inoperable or metastatic triple-negative breast cancer (KEYNOTE-355): a randomised, placebo-controlled, double-blind, phase 3 clinical trial. Lancet. 2020;396:1817–1828. doi: 10.1016/S0140-6736(20)32531-9. [DOI] [PubMed] [Google Scholar]
- 412.Schmid P, et al. Event-free survival with pembrolizumab in early triple-negative breast cancer. N. Engl. J. Med. 2022;386:556–567. doi: 10.1056/NEJMoa2112651. [DOI] [PubMed] [Google Scholar]
- 413.Mittendorf EA, et al. Neoadjuvant atezolizumab in combination with sequential nab-paclitaxel and anthracycline-based chemotherapy versus placebo and chemotherapy in patients with early-stage triple-negative breast cancer (IMpassion031): a randomised, double-blind, phase 3 trial. Lancet. 2020;396:1090–1100. doi: 10.1016/S0140-6736(20)31953-X. [DOI] [PubMed] [Google Scholar]
- 414.Geyer CE, et al. Overall survival in the OlympiA phase III trial of adjuvant olaparib in patients with germline pathogenic variants in BRCA1/2 and high-risk, early breast cancer. Ann. Oncol. 2022;33:1250–1268. doi: 10.1016/j.annonc.2022.09.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Robson ME, et al. OlympiAD final overall survival and tolerability results: Olaparib versus chemotherapy treatment of physician’s choice in patients with a germline BRCA mutation and HER2-negative metastatic breast cancer. Ann. Oncol. 2019;30:558–566. doi: 10.1093/annonc/mdz012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Diéras V, et al. Veliparib with carboplatin and paclitaxel in BRCA-mutated advanced breast cancer (BROCADE3): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2020;21:1269–1282. doi: 10.1016/S1470-2045(20)30447-2. [DOI] [PubMed] [Google Scholar]
- 417.Litton JK, et al. Talazoparib versus chemotherapy in patients with germline BRCA1/2-mutated HER2-negative advanced breast cancer: final overall survival results from the EMBRACA trial. Ann. Oncol. 2020;31:1526–1535. doi: 10.1016/j.annonc.2020.08.2098. [DOI] [PMC free article] [PubMed] [Google Scholar]