Abstract
The microRNA let-7d has been reported to be a tumor suppressor in renal cell carcinoma (RCC). Tumor-associated macrophages (TAM) are M2-polarized macrophages that can enhance tumor growth and angiogenesis in many human cancers. However, the role of let-7d in TAM-associated RCC progression remains elusive. First, we observed a strongly inverse correlation between let-7d expression and microvessel density in RCC tissues. Furthermore, the proliferation, migration, and tube formation of HUVECs were significantly inhibited by conditioned medium from a coculture system of the phorbol myristate acetate pretreated human THP-1 macrophages and let-7d-overexpressing RCC cells. Moreover, the proportion of M2 macrophages was significantly lower in the group that was cocultured with let-7d-overexpressing RCC cells. Subcutaneous xenografts formed by the injection of let-7d-overexpressing RCC cells together with THP-1 cells resulted in a significant decrease in the M2 macrophage ratio and microvessel density compared with those formed by the injection of control RCC cells with THP-1 cells. In silico and experimental analysis revealed interleukin-10 (IL-10) and IL-13 as let-7d target genes. Importantly, the addition of IL-10 and IL-13 counteracted the inhibitory effects of the conditioned medium from the coculture system with let-7d-overexpressing RCC cells in vitro. Additionally, overexpression of IL-10 and IL-13 reversed the effects of let-7d on macrophage M2 polarization and tumor angiogenesis in vivo. Finally, the expression of IL-10 and IL-13 were inversely correlated with the expression of let-7d in RCC clinical specimens. These results suggest that let-7d may inhibit intratumoral macrophage M2 polarization and subsequent tumor angiogenesis by targeting IL-10 and IL-13.
Electronic supplementary material
The online version of this article (10.1007/s00262-020-02791-6) contains supplementary material, which is available to authorized users.
Keywords: microRNA, Let-7d, Macrophage, Angiogenesis, IL-10, IL-13
Introduction
Tumor angiogenesis has been indicated as a promising therapeutic target for advanced renal cell carcinoma (RCC). Targeted drugs that inhibit angiogenesis, such as sunitinib and sorafenib, have an established role in the clinical treatment of RCC [1]. Angiogenesis was a complex process that involves multiple steps, including endothelial cell proliferation and migration, matrix degradation and capillary sprouting, all of which are regulated by countervailing factors that either induce or oppose angiogenesis. The well-known inducers of angiogenesis include factors such as vascular endothelial growth factor (VEGF), basic fibroblast growth factor (bFGF), placental growth factor (PIGF) and platelet-derived growth factor (PDGF) [2].
It has long been recognized that the function of cells belonging to the monocyte-macrophage lineage can be profoundly affected by exposure to different microenvironmental signals [3]. Macrophage activation in response to lipopolysaccharide (LPS) and interferon-γ (IFN-γ) leads to the M1 phenotype, and these M1 macrophages exhibit elevated expression of tumor necrosis factor α (TNF-α) and increased ability to kill pathogens and cells [4]. In contrast, the M2 activation of macrophages can be induced by stimuli such as macrophage colony stimulating factor (MCSF), interleukin-4 (IL-4), IL-13, or IL-10 [5]. M2 macrophages exhibit reduced secretion of pro-inflammatory cytokines, such as TNF α, IL-1β, and IL-6, and produce high levels of anti-inflammatory cytokines, such as transforming growth factor-β (TGF-β) [6]. Tumor-associated macrophages (TAMs) represent a major population of the leukocytes that infiltrate tumors, and TAMs are driven by tumor- and T cell-derived cytokines to undergo polarization toward the M2 phenotype [7]. TAMs are known to promote tumor progression and metastasis by enhancing tumor growth, angiogenesis, and suppression of adaptive immunity [8]. A high density of tumor-infiltrating TAMs has been demonstrated to be associated with a poor prognosis in various types of cancer, including RCC [9, 10].
Previously, we found that microRNA let-7d expression was significantly downregulated in RCC. High densities of macrophage infiltration were observed in RCC and were inversely correlated with let-7d expression [11]. Considering the important role of TAMs in tumor angiogenesis, the current study was designed to investigate whether let-7d functionally participates in macrophage polarization and, thus, plays a role in tumor angiogenesis in RCC.
Materials and methods
Clinical samples
Histologically confirmed RCC tissues and paired adjacent normal tissue were collected from 80 patients who underwent radical nephrectomy at Peking University First Hospital between 2012 and 2013; after collection, the tissues were snap-frozen in liquid nitrogen. The Ethics Committee of Peking University First Hospital approved this study. Written informed consent was obtained from all the patients. The clinicopathological features of these patients can be seen in our previous study [11].
Cell lines and reagents
Human RCC cell lines 769P and 786O were purchased from ATCC (MA, USA). The human acute monocytic leukemia cell line THP-1 and human umbilical vein endothelial cells (HUVECs) were obtained from the China Center for Type Culture Collection (Wuhan, China). Pri-let-7d lentivirus stably transfected 786O and 769P cell lines overexpressing let-7d, as well as the corresponding vehicle-transfected control cells, were established as previously described [11]. The transformed human embryonic kidney 293FT cells were purchased from Invitrogen. The 786O and 769P cells were maintained in RPMI 1640 medium. The HUVECs were maintained in DMEM/F12 (50:50, v/v) medium. The 293FT cells were routinely cultured in DMEM. All the culture media were supplemented with 10% (v/v) fetal bovine serum (FBS), 100 U/ml penicillin sodium and 100 μg/ml streptomycin sulfate. Phorbol-12-myristate-13-acetate (PMA) was purchased from Sigma-Aldrich. THP-1 cells were seeded at 2 × 105 cells/cm2 and treated with PMA (200 nM) for 48 h to differentiate the cells into resting macrophages (M0 cells) as previously described [12].
In vivo assay
Approval of the animal studies was obtained from the Ethics Committee of Tsinghua Changgung Hospital. Specific pathogen-free (SPF) 5-week-old female BALB/C-nu/nu nude mice (Vital River Laboratory Animals, Beijing, China) were used in this study and maintained in accordance with institutional guidelines for the use of laboratory animals. A total of 3 × 106 786O cells stably transfected with lentivirus and 3 × 106 THP-1 cells treated with PMA (M0) were subcutaneously injected into the right flank of each mouse in a 1:1 mixture of Matrigel (BD Biosciences, San Jose, CA). Five mice were included in each group. After 7 weeks, the mice were euthanized, and the tumors were dissected, weighed and prepared for RNA isolation.
RNA extraction and quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted using the miRNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. To analyze the mature miRNA, 1 μg total RNA was polyadenylated to add a polyA tail with polyA polymerase (NEB, Beverly, MA, USA) and then reverse-transcribed with an oligo-dT adapter primer into cDNA for qRT-PCR quantification [11]. Quantitative RT-PCR was performed using the SYBR Select Master Mix (Life) in a final volume of 10 μL on an ABI7500 Fast PCR machine (Applied Biosystems, Foster City, CA, USA). U6 and GAPDH were selected as the endogenous references for miRNA and mRNA, respectively. Relative quantification (RQ) was calculated based on the 2−ΔCt method, where ΔCt = Ct (target) − Ct (reference). The fold change was calculated by the 2−ΔΔct method. For a list of all the primers, see Supporting Information, Table S1.
Lentiviral vector construction, virus packaging and cell infection
All the constructs were generated by standard DNA recombination techniques as previously described [11]. Briefly, the open reading frames of the IL-10 and IL-13 cDNAs were obtained by RT–PCR and cloned into the vector pcDNA3.1. All of the constructs were verified by sequencing. The sequences of the primers are listed in Table S1. Lentiviruses were produced using the ViraPower™ Packaging Mix (Invitrogen) in 293FT packaging cells. The virus-infected cells were selected with 5 μg/ml blasticidin + 500 μg/ml G418 (Invitrogen). The antibiotic-resistant clones were pooled and used for the subsequent assays.
In vitro cell coculture and conditioned medium preparation
RCC cells were seeded in 6-well plates at a density of 1 × 105 cells/well in serum-containing medium. Twenty-four hours later, the medium was replaced with serum-free RPMI 1640 medium, and the cells were incubated for an additional 48 h. The conditioned medium was collected and centrifuged at 3000×g for 5 min to remove the cells or cell debris. Transwell cell culture inserts (Corning Incorporated, Corning, NY) with 12-mm polyester (PET) membranes (pore size: 0.4 μM, density: 1.6 × 106 pores/cm2) were used for the noncontact coculture of RCC cells and THP-1 cells in 6-well plates, as previously described [13]. Briefly, 2 × 105 PMA-pretreated TPH-1 cells were cocultured with RCC cells in serum-free RPMI 1640 medium at 37 °C in an atmosphere containing 5% CO2. Twenty-four hours later, the culture medium from the lower compartments was collected, centrifuged and used as conditioned medium. In the in vitro rescue experiment, human recombinant IL-10 and IL-13 (R&D Systems, Minneapolis, MN) were added to the lower chamber of the coculture system at a concentration of 100 ng/mL and incubated for 24 h before collecting the conditioned medium.
In vitro cell proliferation, migration, and tube formation assay
Cell proliferation was evaluated by a Cell Counting Kit-8 (CCK-8, Dojindo, Kumamoto, Japan) at various time points for up to four days according to the manufacturer’s instructions. The cell migration assays were performed using a Boyden chamber as previously described [11]. Five high-power fields (magnification, 200×) per well were randomly selected and manually counted. The Fibrin In Vitro Angiogenesis Assay (Millipore) was used to analyze new blood vessel formation according to the manufacturer’s instructions. The enclosed networks in five random fields per well were measured by ImageJ software, and the average value was calculated. The experiment was repeated in triplicate.
Immunohistochemistry (IHC)
Paraffin-embedded tissues were analyzed using immunohistochemical staining with the following primaryantibodies: anti-CD68 antibody (DAKO, Carpinteria, CA), anti-CD204 antibody (Bioss, Woburn, MA), anti-CD31 antibody (DAKO, Carpinteria, CA). The microvessel density (MVD) was evaluated as previously described [14]. CD31 was used as a vascular endothelial cell marker. Any morphologically identifiable vessels with a lumen and CD31-positive endothelial cell clusters that were clearly separate from the adjacent microvessels in the selected field were all counted as individual vessels. Two pathologists who were blinded to the patients' clinical data independently examined all the slides.
ELISA
Cytokine secretion in the culture medium was assayed using an ELISA kit according to the manufacturer’s protocol [PIGF (R&D Systems, Minneapolis, MN), TNF-α, TGF-β and IL-6 (RayBiotech, Norcross, GA)].
Western blot analysis
Western blotting was performed as previously described [11]. The primary antibodies used in this study were anti-IL-10 antibody, anti-IL-13 antibody (Abcam, Cambridge, UK) and anti-GAPDH antibody (CST, Danvers, MA) as an internal loading control. The blotted membranes were then scanned using GeneSnap (Syngene, Cambridge, UK) acquisition software.
Flow cytometry
Cells were centrifuged at 1500 rpm for 5 min, washed with PBS and filtered through a 100-μm mesh for flow cytometry. The cells were subsequently stained with an antibody against an M2 macrophage surface markers (PE-labeled anti-human CD204 (R&D Systems, Minneapolis, MN)) for 30 min at 4 °C. Isotype control antibodies (Biolegend) were used as negative controls. The cells were analyzed using a flow cytometer (FAC-SCalibur, BD Biosciences). The collected data were processed by the CellQuest program (BD Biosciences).
Dual-luciferase activity assay
The 3′-untranslated regions (UTRs) of human IL-10 and IL-13 containing putative binding sites of let-7d and the corresponding mutant binding sites were chemically synthesized and inserted immediately downstream of the firefly luciferase cDNA in the pGL3-control vector (Promega, Madison, WI) by GenePharma (Shanghai, China) to generate the pGL3–IL-10, pGL3–IL-13, pGL3–IL-10–Mut, and pGL3–IL-13–Mut constructs. The luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega) as previously described [11].
Statistical analysis
All the data are presented as the mean ± SD of at least three independent experiments and were analyzed using SPSS 20.0 statistical software (IBM, Chicago, IL, USA). The significance of the difference between two groups was determined using double-sided Student’s t-test unless otherwise specified. Correlations were evaluated using two-tailed Spearman’s test. p < 0.05 was considered statistically significant.
Results
The conditioned medium from let-7d-overexpressing RCC cells had no effects on the proliferation, migration and tube formation of HUVECs
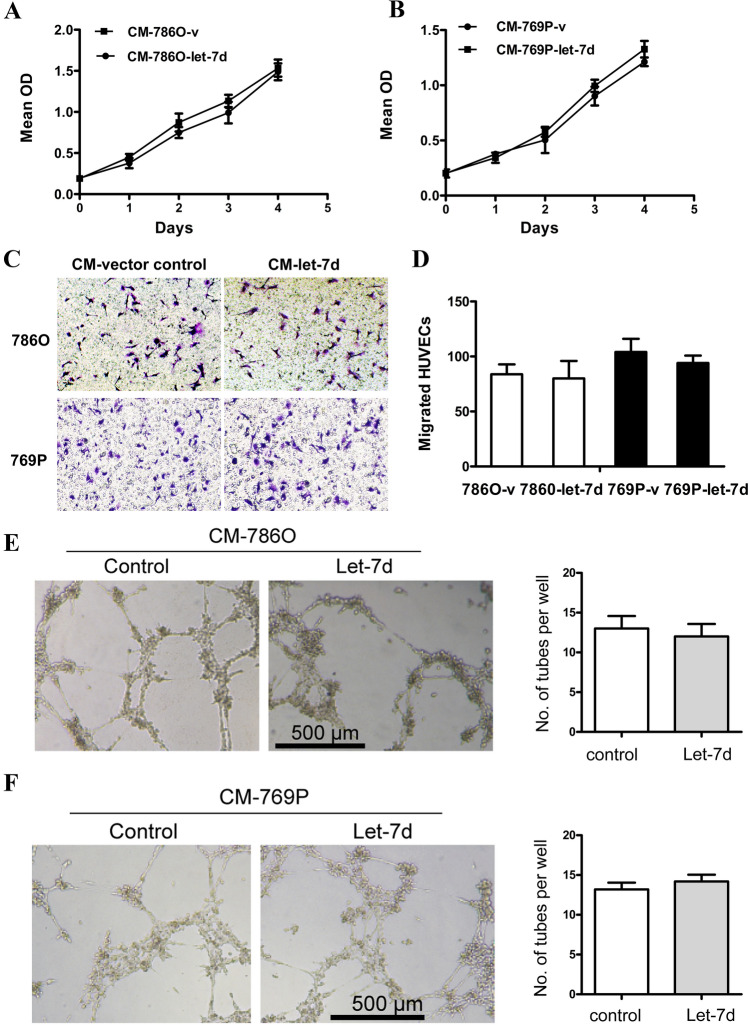
Previously we have demonstrated that let-7d expression was significantly downregulated in RCC tissues. By analyzing the TCGA database of RCC (KIRC, KIRP, KICH), the same result was found (Fig. S1A). Furthermore, let-7d expression in patients with lymphatic metastasis was significantly lower than those without metastasis (Fig. S1B). Low expression of let-7d was also associated with worse overall survival of ccRCC patients (Fig. S1C). To investigate the role of let-7d expression in RCC cells on tumor angiogenesis, we evaluated the association between let-7d expression and microvessel density (MVD) in 80 RCC tissues from our institute. An inverse correlation between let-7d expression and MVD was observed (Spearman’s r = − 0.40, p < 0.001) (Fig. S1C). Next, we used conditioned medium from the RCC cell lines stably transfected with pri–let-7d lentivirus and measured its effects on the proliferation, migration and tube formation of the HUVECs in vitro. Interestingly, as shown in Fig. 1, the proliferation, migration and tube formation of the HUVECs cultured with conditioned medium from the lenti–let-7d-transfected 786O and 769P cells were not significantly different from those of the HUVECs cultured with conditioned medium from the vehicle-transfected control cells. These results demonstrated that let-7d in RCC cells did not influence the proliferation, migration and tube formation of HUVECs in vitro. Considering the inverse relationship between let-7d expression and MVD in tumor tissues, our data suggested that let-7d expression in RCC cells might play an indirect role in tumor angiogenesis.
Fig. 1.
The conditioned medium from let-7d-overexpressing RCC cells had no effects on the proliferation, migration and tube formation of HUVECs. a, b The cell proliferation assay showed that conditioned medium from let-7d transfected 786O and 769P cells had no significant influence on proliferation of HUVEC cells compared with that from vehicle control transfected cells. The data represent the mean ± SD of three independent experiments with triplicates of each group. c, d Boyden chamber assays were performed, showing that HUVEC cell migration was not significantly different in conditioned medium from let-7d-overexpressing 786O and 769P cells compared with that in conditioned medium from respective control cells. Original magnification:×20. e, f Tube formation assay showed that the number of tubes formed by HUVEC cells in conditioned medium from let-7d-overexpressing 786O and 769P cells was not significantly different from those in conditioned medium from respective control cells. Bar, 500 μm. The quantification results of migrated cells and number of tubes per well were plotted in the right. Results were displayed as the mean ± SD of three independent experiments with five random fields counted for each chamber/well. Different numbers between two groups were analyzed by two-tailed Student’s t test. *p < 0.05
The conditioned medium from THP-1 cells cocultured with let-7d-overexpressing RCC cells inhibited the proliferation, migration, and tube formation of HUVECs
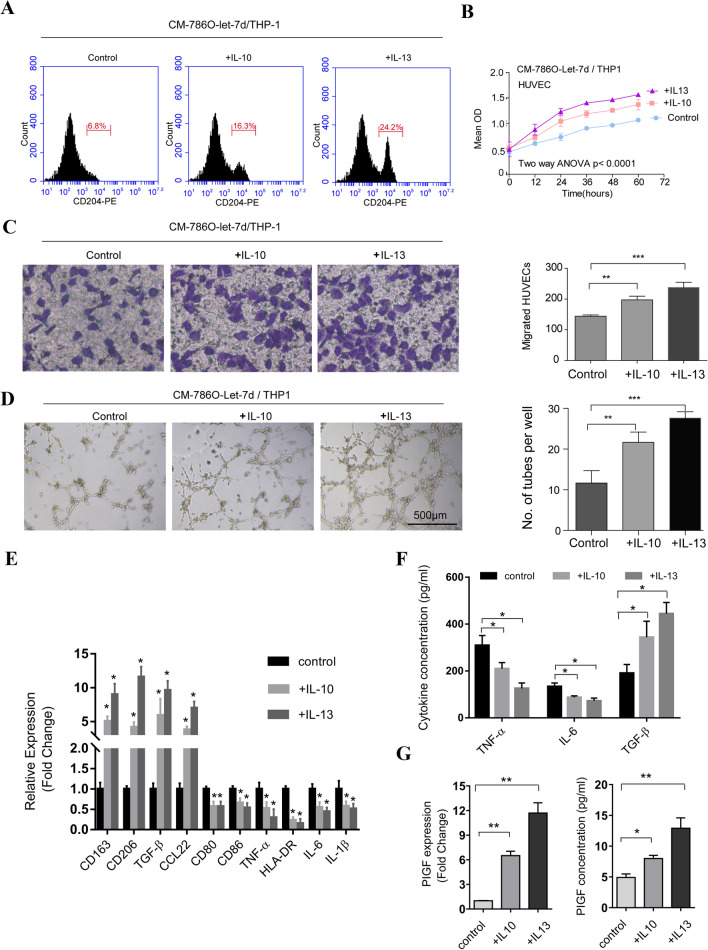
Increasing evidence has demonstrated that the interplay between cancer cells and neoplastic stromal cells is of great importance to tumor initiation and progression [15]. To investigate whether macrophages were involved in let-7d-related tumor angiogenesis in RCC, we employed a noncontact cell coculture system. The PMA-treated THP-1 cells (M0) were cultured alone or cocultured with let-7d-overexpressing or control RCC cells. The culture medium was then collected from the M0 cells and used as conditioned medium. As shown in Fig. 2, the proliferation, migration and tube formation of HUVECs cultured with conditioned medium from M0 cells cocultured with control RCC cells were significantly increased compared with those of HUVECs cultured with conditioned medium from M0 cells cultured alone or cocultured with let-7d-overexpressing RCC cells. These data suggested that let-7d expression in RCC cells could influence the HUVECs through the THP-1 macrophages, indicating the important role of macrophages in let-7d-related tumor angiogenesis in RCC.
Fig. 2.
The conditioned medium from THP-1 cells cocultured with let-7d-overexpressing RCC cells inhibited the proliferation, migration, and tube formation of HUVEC cells. a, b The cell proliferation assay showed that the proliferation of HUVEC cells in conditioned medium from coculture system with vector control-transfected 786O and 769P cells was significantly higher than those in conditioned medium from M0 cells cultured alone or coculture system with let-7d-transfected cells. The data represented the mean ± SD of three independent experiments with triplicates of each group. c, d Boyden chamber assay was performed, showing that HUVEC migration in conditioned medium from coculture system with vector control-transfected 786O and 769P cells was significantly higher than those in conditioned medium from M0 cells cultured alone or coculture system with let-7d-transfected cells. Original magnification: ×20. e, f Tube formation assay showed that the number of tubes formed by HUVEC cells in conditioned medium from coculture system with vector control transfected 786O and 769P cells was significantly higher than those in conditioned medium from M0 cells cultured alone or coculture system with let-7d-transfected cells. Bar, 500 μm. The quantification results of migrated cells and number of tubes are plotted in the right. Results were displayed as the mean ± SD of three independent experiments with five random fields counted for each chamber/well. Different numbers between three groups were analyzed by one-way ANOVA followed by the Bonferroni-Holm procedure. *p < 0.05
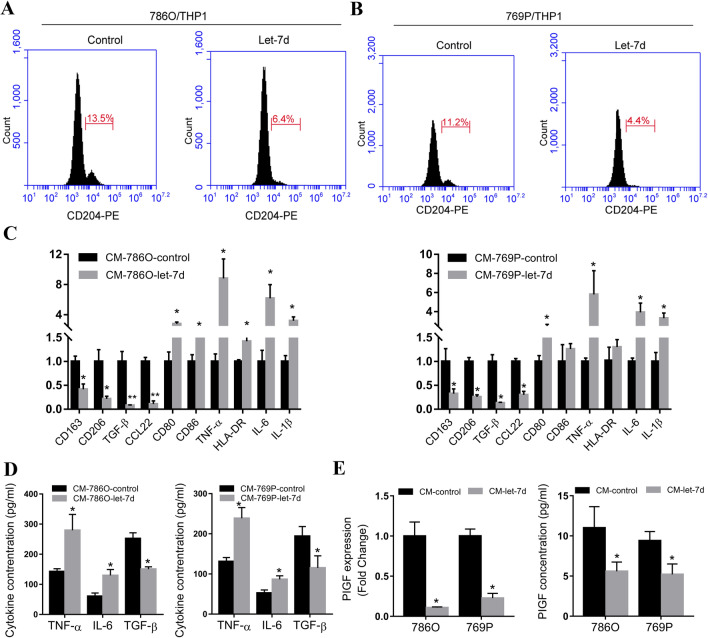
Let-7d overexpression in RCC cells inhibited the M2 polarization of THP-1 cells cocultured with RCC cells
Macrophages that infiltrate tumor tissue can polarize toward the M2 phenotype, and these M2 macrophages are oriented toward promoting tumor growth and facilitating angiogenesis [8]. To investigate whether the M0 macrophages cocultured with RCC cells polarized into the M2 phenotype and therefore affected the process of angiogenesis, we employed flow cytometry analysis to evaluate the percentage of M2 macrophages among the M0 macrophages cocultured with let-7d-overexpressing RCC cells. CD204 was used as an M2 surface macrophage marker [16]. As demonstrated by flow cytometry, the percentage of CD204-positive cells was lower in the M0 cells cocultured with let-7d-overexpressing RCC cells than in those cocultured with the corresponding control RCC cells (Fig. 3a, b). The mRNA expression levels of other M2 markers, including CD163, CD206, TGF-β and CCL22 were also significantly lower in the M0 cells cocultured with let-7d-overexpressing RCC cells compared with the M0 cells cocultured with control RCC cells. However, the mRNA expression levels of M1 markers inclding CD80, CD86, TNF-α, HLA-DR, IL-6 and IL-1β became significantly higher (Fig. 3c). Consistently, the protein levels of the proinflammatory cytokine TNF-α and IL-6 were increased, while the anti-inflammatory cytokine TGF-β was decreased in conditioned medium from M0 cells cocultured with let-7d-overexpressing RCC cells (Fig. 3d). Taking the surface marker and cytokine profiles together, these results showed that let-7d overexpression inhibited the RCC cell coculture-induced M2 polarization of the PMA-treated THP-1 macrophages. Since M2 macrophages have been reported to promote tumor angiogenesis, we evaluated whether let-7d expression in cocultured RCC cells influenced the angiogenic cytokine secretion of macrophages. Quantitative RT–PCR screening identified that among angiogenic factors including VEGF, PDGF, PIGF, bFGF and MMP-9 (Fig. S2), only the expression level of PIGF was significantly decreased in the M0 macrophages cocultured with let-7d-overexpressing RCC cells (Fig. 3e). As shown by ELISA, the protein level of PIGF in conditioned medium from the coculture system with let-7d-overexpressing RCC cells was also decreased. These results indicated that let-7d expression in RCC cells inhibited the M2 polarization of the THP-1 macrophages cocultured with RCC cells and therefore affected the process of angiogenesis.
Fig. 3.
Let-7d overexpression in RCC cells inhibited the M2 polarization of THP-1 cells cocultured with RCC cells. a, b Flow cytometry analysis showed that the percentage of CD204 (marker for M2 macrophage) positive cells were lower in M0 (THP-1 treated with 200 nM PMA for 48 h) cells cocultured with let-7d-overexpressing RCC cells, as compared with those cocultured with respective control RCC cells. c Real-time RT-PCR results showed that M0 cells cocultured with let-7d-overexpressing 786O cells showed decreased M2 markers, including CD163, CD206, TGF-β and CCL22, as well as increased M1 marker, including CD80, CD86, TNF-α, HLA-DR, IL-6 and IL-1β. The expression levels of CD86 and HLA-DR remain unchanged in M0 cells cocultured with let-7d-overexpressing 769P cells. The mRNA expression was normalized to GAPDH mRNA. The data represented the mean ± SD of three independent RT–PCRs (RNAs were from the same set). d The protein levels of pro-inflammatory cytokine TNF-α and IL-6 as well as anti-inflammatory cytokine TGF-β in conditioned medium from M0 cells cocultured with let-7d-overexpressing or control RCC cells were analyzed by ELISA assay. e The expression of PIGF was significantly decreased in M0 macrophages cocultured with let-7d-overexpressing RCC cells, compared with those cocultured with control RCC cells, at mRNA level and protein level as detected by real time RT–PCR (left) and ELISA assay (right). The data in d and e represented the mean ± SD of three independent experiments with triplicates of each sample. Student’s t test. *p < 0.05,**p < 0.01
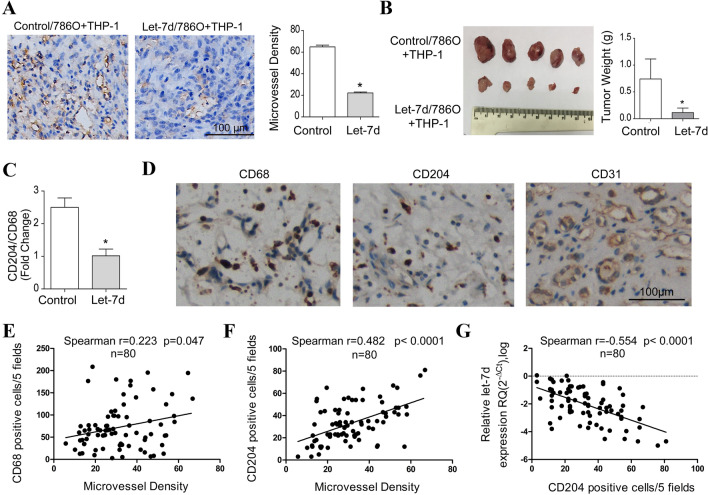
Let-7d overexpression inhibited macrophage M2 polarization and tumor angiogenesis in vivo
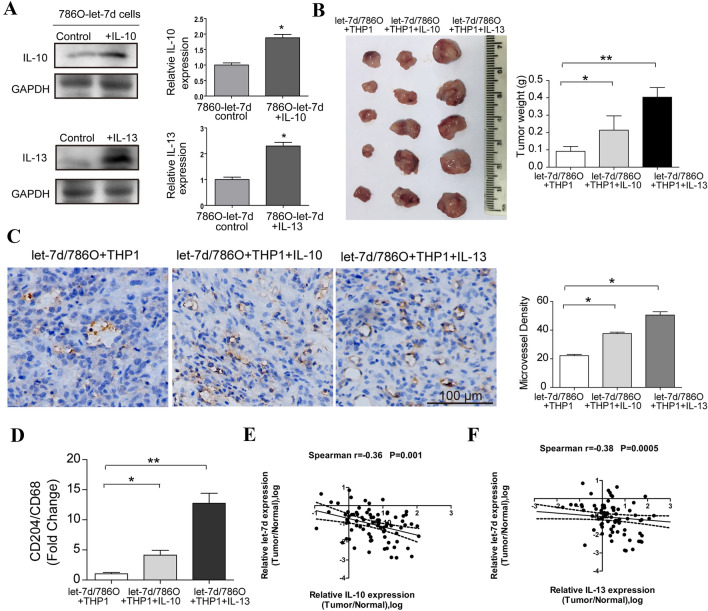
Next, we used a subcutaneous xenograft model that was established by inoculating 786O cells together with M0 macrophages into nude mice to investigate the influence of let-7d overexpression on macrophage polarization and tumor angiogenesis in vivo. As shown in Fig. 4a, the microvessel density in the let-7d-overexpressing tumors was significantly decreased compared with that in the control tumors. Consistently, the tumor growth of let-7d-overexpressing groups was also significantly inhibited (Fig. 4b). To investigate the phenotypic differentiation of the macrophages, we employed qRT-PCR to analyze the mRNA expression level of the M2 phenotype marker CD204 and the panmacrophage marker CD68. The ratio of the CD204/CD68 mRNA level in the let-7d-overexpressing group was significantly lower than that in the control group (Fig. 4c). Next we investigated the correlations between the let-7d, CD204 and MVD in clinical samples. Data from TCGA RCC dataset revealed that the expression of CD68 and CD204 were all upregulated in tumor tissues. High expression of CD68 and CD204 were associated with advanced pathological stage and poor overall survival (Fig. S3A–F). The mRNA expression levels of CD68 and CD204 were also positively correlated with the expression of CD31, respectively (Fig. S3G-H). Furthermore, as demonstrated by IHC staining in 80 RCC tissues from our institute, both CD68 and CD204 positive cell counts were correlated with MVD, but CD204 had a stronger correlationship (Spearman r 0.482 vs. 0.223, Fig. 4d–f). Additionally, a significant inverse correlation was observed between the expression of let-7d and CD204 positive cell couts in the same set of RCC samples (Fig. 4g). Taken together, these data suggested that let-7d overexpression inhibited macrophage M2 polarization, tumor growth and angiogenesis.
Fig. 4.
Let-7d overexpression inhibited macrophage M2 polarization and tumor angiogenesis in vivo. a Representative microphotograph of IHC staining shows the microvessels (CD31 positive) in subcutaneous xenografts formed by inoculating let-7d-overexpressing 786O cells or control 786O cells together with M0 macrophages. Bar, 100 μm. Three random sections for each xenograft were subjected to IHC staining and calculation of MVD. Quantitative data of the mean MVD in each group were compared. b Photograph of subcutaneous xenografts formed by inoculating let-7d-overexpressing 786O cells or control 786O cells together with M0 macrophages. Quantitative data of the mean tumor weight were shown in the right. c Real-time RT-PCR quantification of relative CD204 expression to CD68 expression in let-7d-overexpressing groups was significantly lower than that in the control groups. The mRNA expression was normalized to GAPDH mRNA. Data represented the mean ± SD of five mice. p values in a–c were obtained by two-tailed Student’s t-test. d IHC staining of CD68, CD204 and CD31 (MVD). Bar, 100 μm. The correlation between mean CD68 (e), CD204 (f) positive cell counts and MVD in each tumor sample were analyzed with r (Spearman) and p-values indicated (n = 80). (g) A linear regression and correlation among data from qRT-PCR of let-7d and the CD204 positive cell counts in 80 RCC tissues samples were shown with r (Spearman) and p-values indicated. Five independent areas with the highest numbers of discrete microvessels, CD68 or CD204 positive cells were selected and manually counted under a microscope using a 20 × objective lens.The average positive cell counts and vessel counts for each tumor tissue were used for the statistical analysis. Expression status of let-7d was shown in a log scale
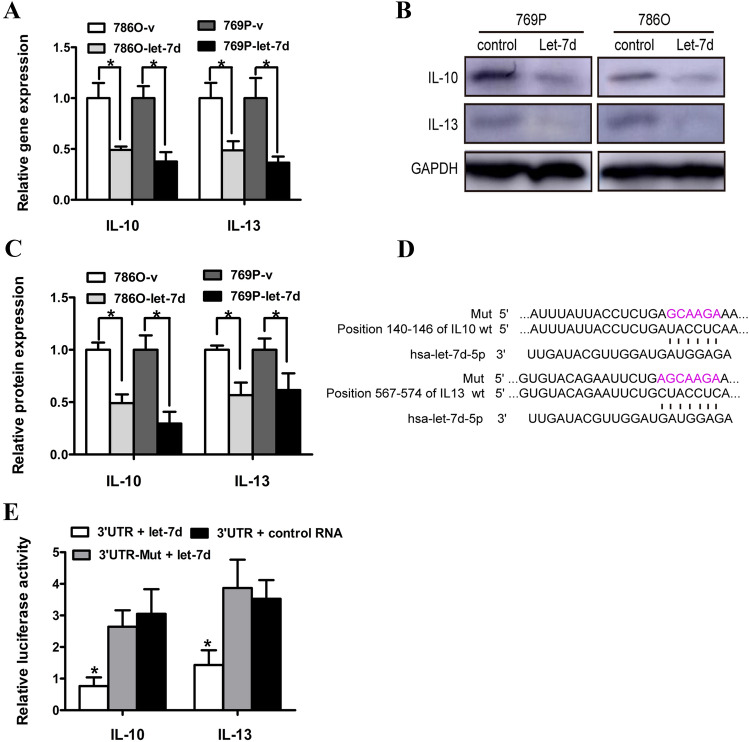
IL-13 and IL-10 were direct target genes of let-7d in RCC cells
To investigate the potential mechanisms involved in the suppressive effects of the overexpression of let-7d on the M2 polarization of macrophages cocultured with RCC cells, we first performed in silico screening using three computational algorithms for mRNA target prediction—miRanda, TargetScan, and PicTar. A list of predicted candidate let-7d target genes was obtained; among which we tried to identify genes that were potentially involved in macrophage polarization through data mining of the Gene Expression Omnibus database (GEO; accession number: GSE781; GSE15641) [17, 18]. Quantitative RT–PCR analysis confirmed that the expression of IL-13 and IL-10 was significantly decreased in the pri–let-7d lentivirus-transfected RCC cells compared with the control cells (Fig. 5a). The protein levels of IL-13 and IL-10 also decreased after let-7d overexpression, as demonstrated by western blot assay (Fig. 5b, c).
Fig. 5.
IL-13 and IL-10 were direct target genes of let-7d in RCC cells. a The expression of endogenous IL-10 and IL-13 was inhibited in the let-7d-overexpressing 786O and 769P cells, compared with the control at mRNA level as detected by real time RT–PCR; The data represented the mean ± SD of three independent RT–PCRs (RNAs were from the same set). b Representative Western blot results showed that the protein level of IL-10 and IL-13 were also downregulated following lenti–pri–let-7d infection. GAPDH served as an internal loading control. The corresponding densitometry of each band was presented in a bar graph in c. d Sequence alignment of human let-7d seed sequence with the 3′-UTRs of IL-10 and IL-13. The mutated sequence in the putative let-7d binding sites for each gene was shown above each gene set (Mut). e Luciferase activity of various reporter plasmids. Statistical significance was obtained using one way ANOVA. Data presented in a, c, e were the mean ± SD of three independent experiments.*p < 0.05
To further investigate whether these genes are bona fide targets of let-7d, luciferase reporters containing the wild-type and mutant (Mut) sequences of the putative let-7d binding site within the 3′-UTRs of the two genes were created (Fig. 5d). As shown in Fig. 5e, the luciferase activities were significantly decreased in reporters containing the wild-type 3′-UTRs of these two genes, but the luciferase activities in the corresponding mutant constructs were not decreased. Thus, these results suggested that IL-13 and IL-10 were direct targets of let-7d.
Rescue of IL-10 and IL-13 eliminated the effects of let-7d
To confirm that IL-10 and IL-13 are required for the effects of let-7d on macrophage M2 polarization and tumor angiogenesis, we performed rescue experiments. The addition of 100 ng/ml IL-10 or IL-13 rescued the M2 polarization of the cocultured THP-1 macrophages respectively, as demonstrated by flow cytometry analysis (Fig. 6a). Consistently, while the growth, migration and tube formation of HUVECs decreased upon exposure to conditioned medium from the coculture system with let-7d-overexpressing RCC cells, the addition of IL-10 or IL-13 completely eliminated this effect (Fig. 6b–d). Furthermore, the polarization markers, cytokine profiles and the expression levels of PIGF in conditioned medium from the THP-1 macrophages cocultured with let-7d-overexpressing RCC cells were also reversed in the presence of IL-10 or IL-13 respectively (Fig. 6e–h). Next, we separately introduced expression cassettes without the 3′-UTRs of these genes into let-7d-overexpressing 786O cells to evaluate the effects of let-7d in vivo. The let-7d-mediated suppression of macrophage M2 polarization, tumor growth, and angiogenesis in the subcutaneous xenograft model were reversed when the let-7d-overexpressing 786O cells were infected with IL-10- and IL-13-expressing lentiviruses (Fig. 7a–d). These data demonstrated that let-7d expression in RCC cells impeded macrophage M2 polarization and tumor angiogenesis by directly targeting IL-10 and IL-13.
Fig. 6.
Rescue of IL-10 and IL-13 eliminated the effects of let-7d in vitro. a Flow cytometry analysis showed that the percentage of CD204 positive cells was restored in the presence of purified IL-10 or IL-13 (100 ng/ml) respectively, as compared with control (PBS phosphate buffer solution). b The cell proliferation assay showed that the proliferation of HUVEC cells in conditioned medium from coculture system with let-7d-overexpressing RCC cells in the presence of 100 ng/ml IL-10 or IL-13 was significantly increased compared with that in the presence of control. The data represented the mean ± SD of three independent experiments with triplicates of each sample. *p < 0.05. Statistical significance was obtained using two way ANOVA. c Represented micrographs and quantitative data of a Boyden chamber assay showed that the migration of HUVEC cells was significantly restored in the conditioned medium from M0 macrophages cocultured with let-7d-overexpressing RCC cells in the presence of 100 ng/ml IL-10 or IL-13. Original magnification: ×40. d Photograph showed that the tubes formed by HUVEC cells following the addition of IL-10 or IL-13 in the coculture system. Bar, 500 μm. Histogram displayed the quantitative data of the number tubes formed by HUVEC cells in conditioned medium added with IL-10, IL-13 or control respectively. (c-d) The data presented are the mean ± SD of three independent experiments with five random fields counted for each chamber/well. e Real-time RT-PCR results showed that the expression of CD163, CD206, TGF-β and CCL22 were significantly increased and the expression of CD80, CD86, TNF-α, HLA-DR, IL-6 and IL-1β were significantly decreased in M0 cells cocultured with let-7d-overexpressing 786O after addition of 100 ng/ml IL-10 or IL-13 in the coculture system. The mRNA expression was normalized to GAPDH mRNA. The data represented the mean ± SD of three independent RT–PCRs (RNAs were from the same set). f ELISA assay showed decreased protein levels of TNF-α and IL-6 as well as increased protein level of TGF-β in conditioned medium from M0 cells cocultured with let-7d-overexpressing 786O after addition of 100 ng/ml IL-10 or IL-13 in the coculture system. g, h The expression of PIGF was significantly decreased in conditioned medium from M0 macrophages cocultured with let-7d-overexpressing 786O cells in the presence of control, compared with those in the presence of 100 ng/ml IL-10 or IL-13, at mRNA level (g) and protein level (h) as detected by real time RT–PCR and ELISA assay respectively. The data represented the mean ± SD of three independent experiments with triplicates of each sample. One-way ANOVA followed by the Bonferroni–Holm procedure. *p < 0.05, **p < 0.01, ***p < 0.001
Fig. 7.
Rescue of IL-10 and IL-13 overcame the effects of let-7d in vivo. Let-7d expression inversely correlated with the IL-10 and IL-13 mRNA levels in RCC tissues. a Western blot results demonstrated that the protein levels of IL-10 and IL-13 were increased in the let-7d-overexpressing 7860 cells infected with lentivirus carrying the expression cassette without the 3′-UTRs of IL-10 or IL-13 respectively. The corresponding densitometry of each band was presented in a bar graph. The data represented the mean ± SD of three independent experiments. b Photograph of subcutaneous xenografts from the three groups. Quantitative data of the mean tumor weight were shown in the right. c Representative graphs of IHC staining of MVD (CD31 positive) in xenografts formed by inoculating M0 macrophages together with let-7d-overexpressing 786O cells infected with IL-10, IL-13 expressing or vector control lentiviruses. Bar, 100 μm. Three random sections for each xenograft were subjected to IHC staining and calculation of MVD. Quantitative data of the mean MVD in each group were compared in the right. d Real-time RT-PCR quantification of relative CD204 expression to CD68 expression in xenografts formed by inoculating M0 macrophages together with let-7d-overexpressing 786O cells infected with IL-10, IL-13 expressing or vector control lentiviruses. The mRNA expression was normalized to GAPDH mRNA. Data in b, c and d represented the mean ± SD of five mice. p values were obtained by one-way ANOVA followed by the Bonferroni–Holm procedure. e, f The relationship between let-7d expression and IL-10 (e) and IL-13 (f) mRNA levels in RCC samples (80 cases). Statistically significant inverse correlations were observed between let-7d and IL-10 and between let-7d and IL-13 mRNA levels using the two-tailed Spearman’s test. *p < 0.05, **p < 0.01
Let-7d expression inversely correlated with the IL-10 and IL-13 mRNA levels in RCC tissues
We then analyzed the expression of let-7d and the mRNA levels of IL-10 and IL-13 by qRT-PCR in the same set of 80 RCC tissues. As shown in Fig. 7e, f, statistically significant inverse correlations were observed between the expression of let-7d and the mRNA levels of IL-10 and IL-13 in the RCC tissues. These data supported the premise that let-7d downregulation increased the mRNA expression levels of IL-10 and IL-13 in clinical specimens of human RCC.
Discussion
Tumor-associated macrophages (TAMs) or M2 macrophages were major components of the RCC tumor microenvironment and contributed to malignant cancer progression by orchestrating various aspects of cancer, such as tumor cell proliferation, invasion, metastasis, angiogenesis and immunosuppression [19]. In RCC, infiltrating TAMs increased the cancer stem cell-like populations, and the number of TAMs was highly correlated with the density of tumor microvessels [20, 21]. TAMs significantly contributed to tumor angiogenesis by producing a wide array of growth factors, and by stimulating several crucial signaling pathways [22]. The reported M2 or TAM markers in the tumor microenvironment included CD68, CD163, CD206, CD204 and CD209 [23, 24]. However, there were some discrepancies in the published literature regarding these markers. Some studies used CD68 or CD163 as a marker of TAMs and demonstrated that increased numbers of CD68- or CD163-positive cells in RCC were associated with poor prognosis [25, 26]. Some observations using an immunohistochemical approach have observed that CD163 could not be considered a reliable M2 marker when used alone, and CD68 was used as a panmacrophage marker [27, 28]. CD204 has been used as an M2 marker in many studies and has been validated to be associated with tumor aggressiveness and unfavorable prognosis in many cancers [29, 30]. In our study, we used CD204 as a marker of M2 macrophages and found that after coculturing with RCC cells, the proportion of CD204-positive macrophages was significantly increased, and the cytokine profiles shifted to profiles that were characteristic of the M2 phenotype. Furthermore, we found that CD204 expression is inversely correlated with let-7d expression in RCC tissues, indicating that the CD204 antigen might be a better marker of the M2 phenotype in RCC tissues.
Interleukin (IL)-10, which was originally called cytokine synthesis inhibitory factor produced by type 2 helper T-lymphocytes (Th2), has been described as a potentially immunosuppressive cytokine [31]. In RCC, IL-10 has been suggested to play a critical role in generating an immunosuppressive tumor microenvironment and to contribute to reduced tumor-infiltrating T cell effector functions [32]. Accumulating studies have suggested that this immunosuppressive function of IL-10 may be partly related to the M2 polarization of macrophages. IL-10 can polarize tumor-infiltrating macrophages toward the M2 phenotype, and M2-polarized macrophages, in turn, secrete more IL-10 [33]. In RCC, bone morphogenetic protein-6 (BMP-6) was reported to increase the secretion of IL-10 in tumor-infiltrating macrophages, polarize these macrophages toward the M2 phenotype and consequently promote RCC progression. Furthermore, patients with elevated IL-10 serum levels had worse outcomes after surgical treatment of RCC [26]. In our study, IL-10 was identified as a target gene of let-7d and was inversely correlated with the expression of let-7d. Furthermore, the restoration of IL-10 alone in let-7d-overexpressing RCC cells increased tumor growth and angiogenesis in vivo, indicating the important role of IL-10 in RCC progression.
IL-13 is a Th2 cytokine that is secreted by a wide variety of different cell types, including Th2-polarized T cells, monocytes/macrophages, granulocytes and cancer cells [34]. IL-13 has been reported to be an anti-inflammatory and immunoregulatory cytokine that can suppress tumor immunosurveillance through the activation of interleukin 4 receptor α (IL-4Rα) and its downstream transcriptional factor STAT6 in natural killer T cells (NKT cells) [35]. Moreover, a significant elevation in the plasma levels of IL-13 has been observed in patients with pancreatic, esophageal and gastric cancer [36]. High expression of IL-13 was associated with increased recurrence and reduced survival in localized clear cell RCC, as demonstrated by immunohistochemistry staining [37]. Many studies have validated that the IL-13 secreted by tumor cells or tumor stromal cells can polarize tumor-infiltrating macrophages toward the M2 phenotype, which can be tumor-trophic [38, 39]. Our study suggests that let-7d can simultaneously target two potent macrophage stimuli, IL-10 and IL-13, by which macrophages can be polarized toward the M2 phenotype and promote tumor angiogenesis, suggesting the potential role of let-7d as a target for immunotherapy of RCC.
In summary, our results indicate that let-7d can inhibit macrophage M2 polarization and tumor angiogenesis, at least in part through the direct destabilization of the mRNAs of IL-10 and IL-13. These findings may improve our understanding of the mechanisms involved in the progression of RCC and the interaction of RCC cells with tumor stromal cells, contributing to the discovery of better prognostic markers and therapeutic targets of RCC.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors thank Yunfei Fan for the support and help for technical assistance.
Abbreviations
- ccRCC
Clear cell renal cell carcinoma
- EGF
Epidermal growth factor
- FGF
Fibroblast growth factor
- HUVEC
Human umbilical vein endothelial cell
- KICH
Kidney chromophobe
- KIRC
Kidney renal clear cell carcinoma
- KIRP
Kidney renal papillary cell carcinoma
- MVD
Microvessel density
- PDGF
Platelet-derived growth factor
- PMA
Phorbol myristate acetate
- PIGF
Placental growth factor
- qRT–PCR
Quantitative reverse transcription–polymerase chain reaction
- RCC
Renal cell carcinoma
- TAM
Tumor-associated macrophage
- TCGA
The cancer genome atlas
- TGF-β
Transforming growth factor-β
- TNF-α
Tumor necrosis factor α
- 3′-UTR
3′-Untranslated regions
- VEGF
Vascular endothelial growth factor
Author contributions
Design: BS, HH, YG, WZ, GZ and LZ Experimental operation: BSu, HH, WH, XL, JY and JY. Acquisition of data and Analysis (acquired and managed patients’ information and statistical analysis): BS, HH, YG, XL and GZ. Writing, review, and/or revision of the manuscript: BS, HH, GZ and LZ. All authors read and approved the submitted manuscript.
Funding
This study was supported by Grants from the National Natural Science Foundation (Grant No.8170101445), the Beijing Natural Science Foundation (No.7182191) and the Capital Health Research and Development of Special (code: 2016-1-2241).
Data availability
All data supporting the conclusions of this article are included in the article.
Compliance with ethical standards
Conflict of interest
The authors declare no conflict of interest.
Ethics approval and consent to participate
All Procedures performed in studies involving human participants or animals were reviewed and granted by the Ethics Committee of Peking University First Hospital and the Ethics Committee of Tsinghua Changgung Hospital, Beijing, China. Informed consent was obtained from all the patients.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Boxing Su, Haibo Han, and Yanqing Gong contribute equally to this work.
Change history
2/14/2023
A Correction to this paper has been published: 10.1007/s00262-023-03389-4
Contributor Information
Boxing Su, Email: 05202154@163.com.
Gang Zhang, Email: haibian76@163.com.
Liqun Zhou, Email: zhoulqmail@china.com.
References
- 1.McDermott DF, Huseni MA, Atkins MB, et al. Clinical activity and molecular correlates of response to atezolizumab alone or in combination with bevacizumab versus sunitinib in renal cell carcinoma. Nat Med. 2018;24(6):749–757. doi: 10.1038/s41591-018-0053-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Viallard C, Larrivee B. Tumor angiogenesis and vascular normalization: alternative therapeutic targets. Angiogenesis. 2017;20(4):409–426. doi: 10.1007/s10456-017-9562-9. [DOI] [PubMed] [Google Scholar]
- 3.Mantovani A, Sozzani S, Locati M, et al. Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 2002;23(11):549–555. doi: 10.1016/S1471-4906(02)02302-5. [DOI] [PubMed] [Google Scholar]
- 4.Lu G, Zhang R, Geng S, et al. Myeloid cell-derived inducible nitric oxide synthase suppresses M1 macrophage polarization. Nat Commun. 2015;6:6676. doi: 10.1038/ncomms7676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murray PJ, Allen JE, Biswas SK, et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 2014;41(1):14–20. doi: 10.1016/j.immuni.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tjiu JW, Chen JS, Shun CT, et al. Tumor-associated macrophage-induced invasion and angiogenesis of human basal cell carcinoma cells by cyclooxygenase-2 induction. J Invest Dermatol. 2009;129(4):1016–1025. doi: 10.1038/jid.2008.310. [DOI] [PubMed] [Google Scholar]
- 7.Erreni M, Mantovani A, Allavena P. Tumor-associated Macrophages (TAM) and Inflammation in Colorectal Cancer. Cancer Microenviron. 2011;4(2):141–154. doi: 10.1007/s12307-010-0052-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pollard JW. Tumour-educated macrophages promote tumour progression and metastasis. Nat Rev Cancer. 2004;4(1):71–78. doi: 10.1038/nrc1256. [DOI] [PubMed] [Google Scholar]
- 9.Qian BZ, Pollard JW. Macrophage diversity enhances tumor progression and metastasis. Cell. 2010;141(1):39–51. doi: 10.1016/j.cell.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu L, Zhu Y, Chen L, et al. Prognostic value of diametrically polarized tumor-associated macrophages in renal cell carcinoma. Ann Surg Oncol. 2014;21(9):3142–3150. doi: 10.1245/s10434-014-3601-1. [DOI] [PubMed] [Google Scholar]
- 11.Su B, Zhao W, Shi B, et al. Let-7d suppresses growth, metastasis, and tumor macrophage infiltration in renal cell carcinoma by targeting COL3A1 and CCL7. Mol Cancer. 2014;13:206. doi: 10.1186/1476-4598-13-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miyake M, Hori S, Morizawa Y, et al. CXCL1-mediated interaction of cancer cells with tumor-associated macrophages and cancer-associated fibroblasts promotes tumor progression in human bladder cancer. Neoplasia. 2016;18(10):636–646. doi: 10.1016/j.neo.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Desai N, Ludgin J, Goldberg J, et al. Development of a xeno-free non-contact co-culture system for derivation and maintenance of embryonic stem cells using a novel human endometrial cell line. J Assist Reprod Genet. 2013;30(5):609–615. doi: 10.1007/s10815-013-9977-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang D, Stockard CR, Harkins L, et al. Immunohistochemistry in the evaluation of neovascularization in tumor xenografts. Biotech Histochem. 2008;83(3–4):179–189. doi: 10.1080/10520290802451085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Denton AE, Roberts EW, Fearon DT. Stromal cells in the tumor microenvironment. Adv Exp Med Biol. 2018;1060:99–114. doi: 10.1007/978-3-319-78127-3_6. [DOI] [PubMed] [Google Scholar]
- 16.Tuna B, Yorukoglu K, Unlu M, et al. Association of mast cells with microvessel density in renal cell carcinomas. Eur Urol. 2006;50(3):530–534. doi: 10.1016/j.eururo.2005.12.040. [DOI] [PubMed] [Google Scholar]
- 17.Jones J, Otu H, Spentzos D, et al. Gene signatures of progression and metastasis in renal cell cancer. Clin Cancer Res. 2005;11(16):5730–5739. doi: 10.1158/1078-0432.CCR-04-2225. [DOI] [PubMed] [Google Scholar]
- 18.Li W, Zhu W, Che J, et al. Microarray profiling of human renal cell carcinoma: identification for potential biomarkers and critical pathways. Kidney Blood Press Res. 2013;37(4–5):506–513. doi: 10.1159/000355726. [DOI] [PubMed] [Google Scholar]
- 19.Chevrier S, Levine JH, Zanotelli V, et al. An immune atlas of clear cell renal cell carcinoma. Cell. 2017;169(4):736–749. doi: 10.1016/j.cell.2017.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang Z, Xie H, He D, et al. Infiltrating macrophages increase RCC epithelial mesenchymal transition (EMT) and stem cell-like populations via AKT and mTOR signaling. Oncotarget. 2016;7(28):44478–44491. doi: 10.18632/oncotarget.9873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Toge H, Inagaki T, Kojimoto Y, et al. Angiogenesis in renal cell carcinoma: the role of tumor-associated macrophages. Int J Urol. 2009;16(10):801–807. doi: 10.1111/j.1442-2042.2009.02377.x. [DOI] [PubMed] [Google Scholar]
- 22.Fischer C, Jonckx B, Mazzone M, et al. Anti-PlGF inhibits growth of VEGF(R)-inhibitor-resistant tumors without affecting healthy vessels. Cell. 2007;131(3):463–475. doi: 10.1016/j.cell.2007.08.038. [DOI] [PubMed] [Google Scholar]
- 23.Tan HY, Wang N, Man K, et al. Autophagy-induced RelB/p52 activation mediates tumour-associated macrophage repolarisation and suppression of hepatocellular carcinoma by natural compound baicalin. Cell Death Dis. 2015;6:e1942. doi: 10.1038/cddis.2015.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jin H, He Y, Zhao P, et al. Targeting lipid metabolism to overcome EMT-associated drug resistance via integrin beta3/FAK pathway and tumor-associated macrophage repolarization using legumain-activatable delivery. Theranostics. 2019;9(1):265–278. doi: 10.7150/thno.27246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohba K, Miyata Y, Matsuo T, et al. High expression of Twist is associated with tumor aggressiveness and poor prognosis in patients with renal cell carcinoma. Int J Clin Exp Pathol. 2014;7(6):3158–3165. [PMC free article] [PubMed] [Google Scholar]
- 26.Lee JH, Lee GT, Woo SH, et al. BMP-6 in renal cell carcinoma promotes tumor proliferation through IL-10-dependent M2 polarization of tumor-associated macrophages. Cancer Res. 2013;73(12):3604–3614. doi: 10.1158/0008-5472.CAN-12-4563. [DOI] [PubMed] [Google Scholar]
- 27.Barros MH, Hauck F, Dreyer JH, et al. Macrophage polarisation: an immunohistochemical approach for identifying M1 and M2 macrophages. PLoS ONE. 2013;8(11):e80908. doi: 10.1371/journal.pone.0080908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rakaee M, Busund LR, Jamaly S, et al. Prognostic value of macrophage phenotypes in resectable non-small cell lung cancer assessed by multiplex immunohistochemistry. Neoplasia. 2019;21(3):282–293. doi: 10.1016/j.neo.2019.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ichimura T, Morikawa T, Kawai T, et al. Prognostic significance of CD204-positive macrophages in upper urinary tract cancer. Ann Surg Oncol. 2014;21(6):2105–2112. doi: 10.1245/s10434-014-3503-2. [DOI] [PubMed] [Google Scholar]
- 30.Kawachi A, Yoshida H, Kitano S, et al. Tumor-associated CD204(+) M2 macrophages are unfavorable prognostic indicators in uterine cervical adenocarcinoma. Cancer Sci. 2018;109(3):863–870. doi: 10.1111/cas.13476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wittke F, Hoffmann R, Buer J, et al. Interleukin 10 (IL-10): an immunosuppressive factor and independent predictor in patients with metastatic renal cell carcinoma. Br J Cancer. 1999;79(7–8):1182–1184. doi: 10.1038/sj.bjc.6690189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Daurkin I, Eruslanov E, Stoffs T, et al. Tumor-associated macrophages mediate immunosuppression in the renal cancer microenvironment by activating the 15-lipoxygenase-2 pathway. Cancer Res. 2011;71(20):6400–6409. doi: 10.1158/0008-5472.CAN-11-1261. [DOI] [PubMed] [Google Scholar]
- 33.Roszer T. Understanding the mysterious M2 macrophage through activation markers and effector mechanisms. Mediators Inflamm. 2015;2015:816460. doi: 10.1155/2015/816460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hallett MA, Venmar KT, Fingleton B, et al. Cytokine stimulation of epithelial cancer cells: the similar and divergent functions of IL-4 and IL-13. Cancer Res. 2012;72(24):6338–6343. doi: 10.1158/0008-5472.CAN-12-3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Terabe M, Matsui S, Noben-Trauth N, et al. NKT cell-mediated repression of tumor immunosurveillance by IL-13 and the IL-4R-STAT6 pathway. Nat Immunol. 2000;1(6):515–520. doi: 10.1038/82771. [DOI] [PubMed] [Google Scholar]
- 36.Gabitass RF, Annels NE, Stocken DD, et al. Elevated myeloid-derived suppressor cells in pancreatic, esophageal and gastric cancer are an independent prognostic factor and are associated with significant elevation of the Th2 cytokine interleukin-13. Cancer Immunol Immunother. 2011;60(10):1419–1430. doi: 10.1007/s00262-011-1028-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang Y, Xu L, An H, et al. Expression of IL-4 and IL-13 predicts recurrence and survival in localized clear-cell renal cell carcinoma. Int J Clin Exp Pathol. 2015;8(2):1594–1603. [PMC free article] [PubMed] [Google Scholar]
- 38.Satoh T, Takeuchi O, Vandenbon A, et al. The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat Immunol. 2010;11(10):936–944. doi: 10.1038/ni.1920. [DOI] [PubMed] [Google Scholar]
- 39.Gao S, Zhou J, Liu N, et al. Curcumin induces M2 macrophage polarization by secretion IL-4 and/or IL-13. J Mol Cell Cardiol. 2015;85:131–139. doi: 10.1016/j.yjmcc.2015.04.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the conclusions of this article are included in the article.