Abstract
B7-H3, an important co-inhibitor, is abnormally highly expressed in a variety of malignancies. The antibodies targeting B7-H3 have exhibited beneficial therapeutic effects in clinical trials. Therefore, discovery of the regulatory factors in B7-H3 expression may provide new strategies for tumor therapy. Here, we investigated the splicing factors involved in the splicing of B7-H3. By individual knockdown of the splicing factors in colorectal cancer (CRC) cells, we found that B7-H3 expression was markedly inhibited by SRSF3 and SRSF8, especially SRSF3. Then we found that both SRSF3 and B7-H3 were highly expressed in CRC tissues. Moreover, high-expression of either SRSF3 or B7-H3 was significantly correlated with poor prognosis of patients. The expression of B7-H3 mRNA and protein were evidently reduced by SRSF3 silence, but were enhanced by overexpression of SRSF3 in both HCT-116 and HCT-8 cells. The results from the RNA immunoprecipitation (RIP) assays demonstrated that SRSF3 protein directly binds to B7-H3 mRNA. In addition, we constructed a minigene recombinant plasmid for expressing B7-H3 exons 3–6. We found that SRSF3 contributed to the retention of B7-H3 exon 4. These findings demonstrate that SRSF3 involves in the splicing of B7-H3 by directly binding to its exon 4 and/or 6. It may provide novel insights into the regulatory mechanisms of B7-H3 expression and potential strategies for the treatment of CRC.
Electronic supplementary material
The online version of this article (10.1007/s00262-020-02683-9) contains supplementary material, which is available to authorized users.
Keywords: Colorectal cancer, Splicing, B7-H3, SRSF3
Introduction
CRC is one of the most common gastrointestinal tumors. There were 1.8 million new cases of CRC worldwide in 2018, and 881,000 deaths, accounting for about 10% of the total number of cancer deaths in the world [1]. CRC has become the third most common cancer in the world, and the fatality rate ranks the second. The incidence of CRC is closely related to factors such as age, environment, and living habits. For instance, 90% of patients with CRC are over 50 years of age, and the incidence of urban residents is much higher than that of rural residents [2, 3]. With the economic development and improvement of living standards, people’s dietary structure has been improved, leading to increased incidence of CRC and number of deaths, especially the young patients [4]. The high morbidity and mortality of CRC have seriously threatened human health. The main treatments for CRC include surgery, chemotherapy, and radiation therapy. However, the outcomes are still not ideal for the patients at advanced stage, eg. metastatic CRC [5–7]. In recent years, immunotherapy has become an emerging therapy applied in the treatment of CRC [8]. Immunotherapy is applied to activate the immune cells in the body and enhance the anti-tumor immune response, as well as remove residual tumor lesions and inhibit tumor growth.
T cell activation requires the stimulation of dual signals generated from the TCR-antigen peptide-MHC molecular complex and the co-stimulatory molecules, in which the B7/CD28 family immune-checkpoint molecules play critical roles [9, 10]. The ligands expressed on the surface of antigen-presenting cells or tumor cells can bind to co-stimulatory or co-inhibitory receptors on the surface of T cells, and promote or hinder the activation and proliferation of T cells, thereby regulating the immune response of T cells [11–13].
B7-H3 (CD276) is a member of the B7/CD28 family [14]. In the beginning, B7-H3 was observed to exhibit a co-stimulatory effect on the proliferation of CD4+ and CD8+ T cells [15]. Recently, numerous studies have shown that B7-H3 is a co-inhibitory molecule, which can inhibit the activation and proliferation of T cells, reduce the secretion of IL-2, IFN-γ and other cytokines, and promote immune escape of tumor cells [16]. There are two splicing isoforms of B7-H3, in which only 2Ig B7-H3 is expressed in mice, while 4Ig B7-H3 is the main expression form in humans. B7-H3 mRNA is widely expressed in a variety of tissues and cells, such as heart, spleen, thymus, pancreas, prostate, small intestine, and colon, but the expression level of B7-H3 protein is low in the normal tissues [17]. A number of studies have shown that B7-H3 protein is abnormally highly expressed in CRC, prostate cancer, breast cancer, melanoma and other malignant tumors, and is closely related to the poor prognosis and clinical outcome of cancer patients [18, 19]. Studies have demonstrated that B7-H3 expression in CRC is significantly related to TNM stage, cancer metastasis, and poor patient prognosis. In addition, B7-H3 can promote tumor angiogenesis through the NF-κB/VEGFA signaling pathway, or up-regulate the expression of Smad1 via the PI3K-Akt pathway to promote epithelial-mesenchymal transition of CRC cells, or induce the proliferation, metastasis and invasion of tumor cells through the signaling pathways including PI3K/AKT/STAT3 and JAK2/STAT3 [18, 20]. Therefore, targeting B7-H3 can not only enhance the anti-tumor immunity, but also inhibit tumor angiogenesis, which is expected to become a new target for cancer immunotherapy. Indeed, antibodies targeting B7-H3 have entered into clinical trials, for instance, Enoblituzumab is at clinical phase III.
Alternative splicing is an important mechanism for regulating gene expression and generating proteomic diversity. Approximately 90–95% of human genes undergo alternative splicing after transcription to produce alternative splice isomers [21, 22]. The types of alternative splicing include exon skipping, intron retention, mutually exclusive exons, variable 5′/ 3′ splicing sites, etc. [23]. There are multiple splicing sites on the pre-mRNA [24]. Alternative splicing occurs in aspects of cell biology, including proliferation, differentiation, cell cycle, metabolism, apoptosis, migration, invasion, and angiogenesis [25]. Strict gene splicing is critical to maintain the homeostasis of body. However, abnormal splicing of key genes can promote the occurrence of cancers, such as VEGFA, TP53, BCL-X, and PKM [26]. Alternative splicing is catalyzed by a ribonucleoprotein complex called spliceosome, which can bind to versatile splicing sites on targets. The spliceosome usually consists of five small nuclear ribonucleoprotein particles (U1, U2, U4, U5, and U6), hnRNP proteins, SR proteins, and SF proteins. Among them, serine/arginine-rich proteins (SRp) and nuclear heterogeneous ribonucleoproteins (hnRNP) are highly conserved proteins [27, 28]. Studies have revealed that several SR proteins and hnRNP proteins are highly expressed in CRC, lung cancer, and breast cancer [29–31]. Moreover, high-expression of these proteins are closely related to the occurrence and development of cancers.
In this study, we investigated the splicing factors participated in the splicing of B7-H3. We first utilized the TCGA data to analyze the correlation between splicing factors and B7-H3. Then we discovered that SRSF3 was involved in the expression of B7-H3 by individual knockdown of splicing factors. Consequently, we employed experiments, such as lose- and gain-of-function, RIP, and minigene reporter, to figure out that SRSF3 regulated B7-H3 splicing through direct binding to B7-H3 exon 4.
Materials and methods
Cell lines and cell culture
Human CRC cell lines HCT-116, HCT-8, and Lovo cells were cultured in DMEM medium (Hyclone, USA) containing 10% fetal bovine serum (FBS; Gibco); SW480, SW620, Caco-2, and DLD-1 cells were cultured in RPMI 1640 medium (Hyclone) containing 10% FBS. The cell lines were purchased from ATCC (American Type Culture Collection). All cells were cultured in an incubator (Thermo) with constant temperature of 37 °C and 5% CO2.
Tissue samples
The colorectal tumor and adjacent tissues were collected from the First Affiliated Hospital of Soochow University. The human CRC tissue chips (No.HcolH180su12) were provided by Outdo Biotech (Shanghai). All the tissues were stained with hematoxylin–eosin and confirmed by pathologists. All tissue-derived patients did not receive chemotherapy or radiotherapy before surgery. This study was approved by the ethics committee of Soochow University, and all patients signed the informed consent.
Transfection of siRNAs and plasmids
siRNAs targeting splicing factors were synthesized by Genepharma (Suzhou). The Flag-SRSF3 overexpression vector containing SRSF3 coding sequence and a flag tag was synthesized by GENEWIZ (Suzhou). The siRNAs and vectors were transfected into CRC cells using lipofectamine 2000 (Invitrogen). The cells were collected after 48 h for RNA analysis or 78 h for protein detection.
Reverse transcription PCR (RT-PCR) and quantitative PCR (qPCR)
Total cellular RNA was extracted by RNAiso Plus reagent (TaKaRa), and was stored in RNase-free tubes. RNA concentration and integrity was measured using a NanoDrop ND-2000 spectrophotometer (Thermo). About 1000 ng RNA were reversely transcripted into cDNA with random primers (TaKaRa) and RT-Kit (Thermo). To detect the expression of 2Ig- and 4Ig-B7-H3, the cDNA fragment containing exons 3–6 was amplified by PCR using premix Taq™ (TaKaRa) and primers (5′-TGG CAT GGG TGT GCA TGT G-3′ and 5′-CCA CCA GCA GTG CAA TGA G-3; GENEWIZ). To measure the expression level of mRNA, qPCR was performed on CFX96 Touch™ real-time PCR system (Bio-Rad) using SYBR Green Supermix (Bio-Rad) and primers (Table S1). GAPDH or β-actin was used as an endogenous quantitative control. The data were analyzed using the quantification technique 2−ΔΔCq method.
Immunohistochemistry (IHC)
IHC staining was performed by Outdo Biotech (Shanghai) on human CRC tissue chips (No.HcolH180su12). Briefly, the tissue chips were deparaffinized in xylene, hydrated in ethyl alcohol and washed in tap water. The adjacent sections were stained with B7-H3 (#376769; Santa Cruz) or SRSF3 (#398541; Santa Cruz) antibody and diaminobenzidine in an Envision System (Dako). Slides were viewed and imaged on a microscope system (Olympus). Two pathologists performed an independent review of the IHC results. The stain strength was scored at 0–3, and the stain prevalence was scored at 0 (negative), 1 (1–25%), 2 (26–50%), 3 (51–75%), or 4 (76–100%). The sample with a product of stain strength and stain prevalence > 6 is classified as high-expression and ≤ 6 as low-expression.
Western blotting (WB)
Cells or tissues were lysed in RIPA buffer (Beyotime) with protease inhibitor, phosphatase inhibitor and EDTA for 1 h on ice. After centrifugation at 12,000g, 4 °C for 20 min, the protein concentration was determined using BCA Protein Assay Kit (Thermo). The proteins were separated on 10% SDS-PAGE and then transferred onto PVDF membranes (Millipore). After being blocked with 5% skimmed milk in TBST for 1.5 h at room temperature, the membranes were washed with TBST for three times and incubated with antibodies targeting SRSF3 (#398541; Santa Cruz), B7-H3 (#376769; Santa Cruz), DDDDK (#166355; Santa Cruz), GAPDH (#47724; Santa Cruz), and β-actin (Beyotime) overnight at 4 °C. The membranes were washed, incubated with a horseradish peroxidase-conjugated secondary antibodies (Beyotime) diluted at 1:2500 for 1.5 h at room temperature. Finally, blots were visualized with enhanced chemiluminescence reaction (Millipore) in ChemiDoc™ MP Imaging System (Bio-Rad).
RIP
HCT-116 cells were transfected with Flag-SRSF3/pcDNA3.1 vectors. Forty-eight hours later, the cells were washed twice with 5 ml ice-cold PBS, and were lysed in IP buffer with phosphatase inhibitor, protease inhibitor, EDTA, DTT, and RNase inhibitor on ice for 30 min, and then were collected by centrifugation at 12,000g, 4 °C for 15 min. The supernatant of cell lysis were incubated with 2 μg anti-DDDDK at 4° C for 12 h; then 40 μl protein-A/G beads were added and incubated for 2 h. The beads were collected by centrifugation and were washed with IP wash buffer for five times. The beads were divided into two parts: one was added with 2 × SDS loading buffer for WB detection of the immunoprecipitated proteins; the other was resuspended with RNAiso Plus to extract the immunoprecipitated RNA for qPCR assays.
Minigene reporter assay
The minigene recombinant vectors were constructed by cloning a genomic DNA fragment containing B7-H3 exons 3–6 and 100-bp flanking sequence into the pcDNA3.1 (+) plasmid. The minigene vectors were synthesized by GENEWIZ (Suzhou). First the minigene vectors were transfected into HCT-116 cells to test the transcription of the minigene vectors. To investigate the effect of SRSF3 on the transcription of the minigene vectors, the minigene vectors were co-transfected with SRSF3 siRNA or expression vector into HCT-116 cells. The transcripts were determined by RT-PCR with primers (FP1, 5′-AGC ATC CGG GAT TTC GGC A-3′; FP2, 5′-GCA CAG CTC TGT CAC CAT CAC-3′; RP1, 5′-TGA GCT GTG CCA GGC TGA A-3′; RP2, 5′-TCA TGC TGG GCT TCG AGT AGG-3′).
Results
SRSF3 involved in the expression of B7-H3
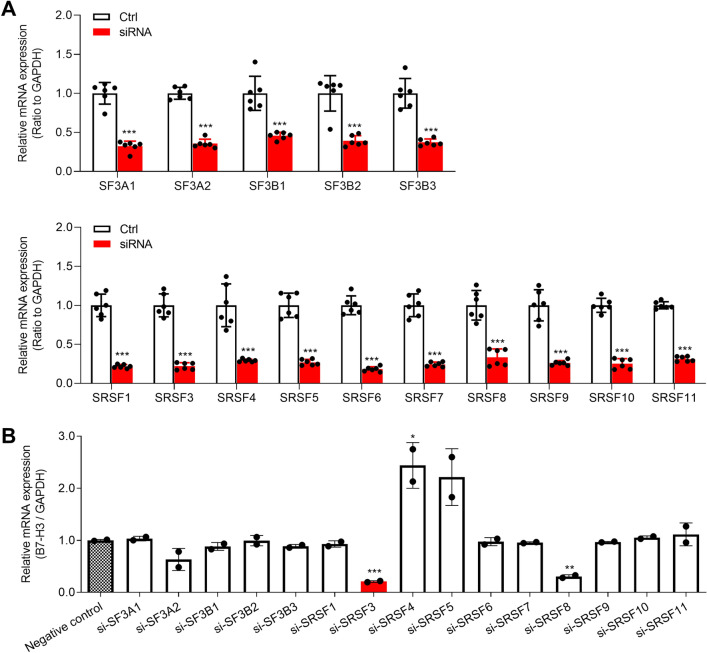
To investigate the splicing factors involved in the splicing of B7-H3, we first utilized TCGA data to analyze the correlation of B7-H3 expression with the SF- and SR-family splicing factors using ChIPBase v2.0 (https://rna.sysu.edu.cn/chipbase/). Several splicing factors were significantly related to B7-H3 expression in TCGA samples (Table S2). Then we individually silenced the splicing factors in HCT-116 cells and measured the expression of B7-H3. The results demonstrated that the expression of splicing factors were significantly inhibited by siRNAs (Fig. 1a). Moreover, the expression of B7-H3 mRNA were evidently suppressed by the silence of SRSF3 or SRSF8, especially SRSF3 (Fig. 1b), suggesting that SRSF3 might be involved in the splicing of B7-H3.
Fig. 1.
The effects of splicing factors on the expression of B7-H3 mRNA. a The efficiencies of siRNA knockdown were tested in HCT-116 cells. b The changes in B7-H3 expression upon silence of splicing factors in HCT-116 cells. The cells were transfected with 50 nM each siRNA. Data represent mean ± SD. Significance was assessed by t test. ***P < 0.001; **P < 0.01; *P < 0.05
Both B7-H3 and SRSF3 were highly expressed in CRC
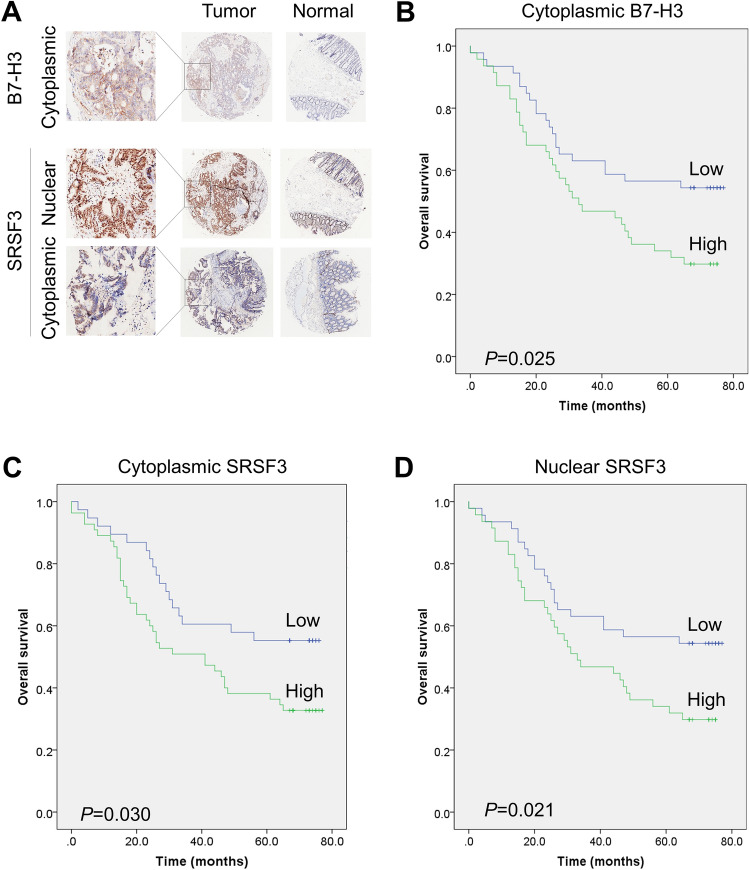
To explore the roles of B7-H3 and SRSF3 in CRC, we detected the expression of B7-H3 and SRSF3 in 87 pairs of CRC tissues and adjacent normal tissues using IHC method. The results showed that B7-H3 was highly expressed in the cytoplasm of cancer cells of 43 CRC tissues, but was low or not expressed in the adjacent tissues (Fig. 2a and Table S3). B7-H3 expression was significantly correlated with the age (P = 0.029; Table S3) and short overall survival (P = 0.025; Fig. 2b) of CRC patients. We also found that SRSF3 was highly expressed in the cytoplasm of cancer cells of 52 CRC tissues, but was lowly expressed in the adjacent tissues (Table S4). The expression of cytoplasmic SRSF3 was closely related to lymph node metastasis (P = 0.007) and TNM stage (P = 0.017), as well as the expression of B7-H3 (P = 0.001), p53 (P = 0.018), and Ki67 (P < 0.001). Furthermore, high-expression of cytoplasmic SRSF3 was significantly related to the poor prognosis of CRC patients (P = 0.030; Fig. 2c). Additionally, SRSF3 was highly expressed in the nucleus of cancer cells of 61 CRC tissues and 8 adjacent tissues, and was low expressed in the cellular nucleus of the other tissues (Table S5). The expression of nuclear SRSF3 was markedly related to lymph node metastasis (P < 0.001), TNM stage (P < 0.001), and the expression of Ki67 (P = 0.004). High-expression of nuclear SRSF3 was also apparently correlated with short overall survival of patients (P = 0.021; Fig. 2d). Further analysis demonstrated that the tumor stage, lymph node metastasis, TNM stage, MSH6, and MSH2 were significantly related to the overall survival of the patients (Table 1).
Fig. 2.
The expression and clinical significance of B7-H3 and SRSF3 in CRC. a IHC stains of cytoplasmic B7-H3 and cytoplasmic and nuclear SRSF3 in the tumor and normal tissues. b–d The significant correlation between poor prognosis of CRC patients and the highly expressed B7-H3 in cytoplasm (b) or SRSF3 in cytoplasm (c) or nucleus (d)
Table 1.
The relationship between the factors and overall survival of CRC patients
| Variables | HR | 95% CI | P value |
|---|---|---|---|
| Cytoplasmic B7-H3 | 1.844 | 1.066–3.190 | 0.025 |
| Cytoplasmic SRSF3 | 1.865 | 1.049–3.316 | 0.030 |
| Nuclear SRSF3 | 2.196 | 1.104–4.370 | 0.021 |
| Sex | 0.803 | 0.471–1.370 | 0.421 |
| Age | 1.230 | 0.720–2.102 | 0.448 |
| Grade | 3.714 | 2.155–6.401 | 0.000 |
| T stage | 1.697 | 0.977–2.949 | 0.061 |
| N stage | 2.523 | 1.696–3.754 | 0.000 |
| M stage | 2.372 | 0.846–6.647 | 0.101 |
| TNM stage | 2.190 | 1.465–3.273 | 0.000 |
| MSH6 | 0.106 | 0.042–0.267 | 0.000 |
| MSH2 | 0.212 | 0.120–0.372 | 0.000 |
| Ki67 | 0.745 | 0.426–1.304 | 0.302 |
| p53 | 0.975 | 0.572–1.664 | 0.927 |
SRSF3 expression was positively correlated with B7-H3 in CRC
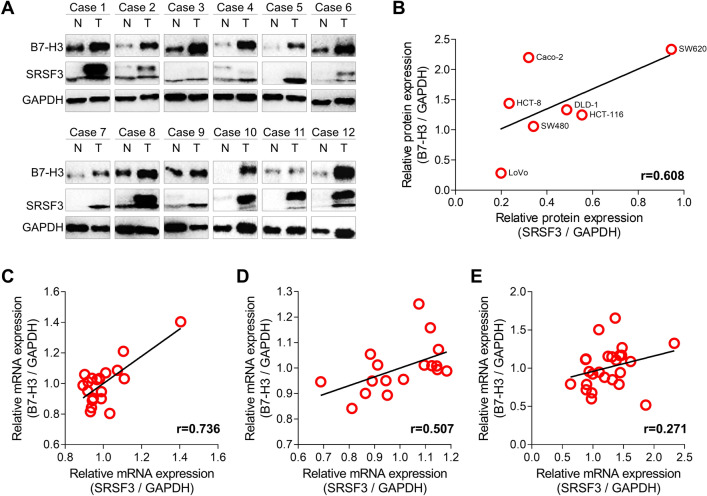
To further investigate the relationship between B7-H3 and SRSF3, we employed WB to detect the expression of B7-H3 and SRSF3 in 12 pairs of tumor and adjacent normal tissues. We found that both B7-H3 and SRSF3 were highly expressed in 9/12 tumor tissues (Fig. 3a). Also, we measured the expression of B7-H3 and SRSF3 proteins in seven CRC cell lines including HCT-116, HCT-8, Caco-2, DLD-1, SW480, SW620, and Lovo. We observed a significantly positive relationship between the expression of B7-H3 and SRSF3 (r = 0.608; Figs. 3b and S1). Moreover, the gene microarray data (GEO IDs: GDS4718, GDS4382, and GSE41657) [32–34] demonstrated that the mRNA expression of both B7-H3 and SRSF3 were significantly upregulated in the carcinoma tissues as compared with the normal tissues (Fig. S2A-C), and the expression of B7-H3 mRNA was positively correlated with SRSF3 in the carcinoma tissues (Fig. 3c–e).
Fig. 3.
The relationship between the expression of B7-H3 and SRSF3 in CRC. a WB results of B7-H3 and SRSF3 in colorectal normal (N) and tumor (T) tissues. b Positive correlation between the expression of B7-H3 and SRSF3 proteins in CRC cell lines. c–e Positive correlation between the expression of B7-H3 and SRSF3 mRNA in CRC tissues. The gene microarray data were collected from GEO Datasets with IDs of GDS4718, GDS4382, and GSE41657, respectively
SRSF3 positively regulated B7-H3 expression in CRC
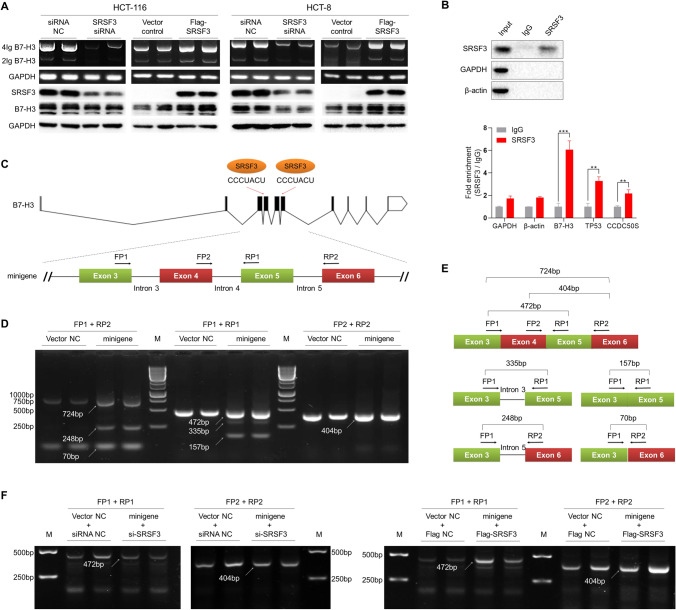
To test the effect of SRSF3 on B7-H3 expression, we silenced or overexpressed SRSR3 expression in HCT-116 and HCT-8 cells by transfecting with siRNA and expression vector, respectively, and then detected the expression of B7-H3 mRNA by RT-PCR or protein by WB. The results demonstrated that the expression level of SRSF3 protein was obviously attenuated by siRNA but was elevated by the expression vector in both HCT-116 and HCT-8 cells (Fig. 4a). Moreover, the expressions of B7-H3 mRNA and protein were markedly suppressed by SRSF3 siRNA in HCT-116 and HCT-8 cells (Fig. 4a). Meanwhile, the expressions of B7-H3 mRNA and protein were apparently enhanced by SRSF3 expression vector in HCT-116 and HCT-8 cells (Fig. 4a). However, two bands were observed in the B7-H3 blots. Further investigation showed that only one truncated band was detected in the blots of PNGase F-digested B7-H3, indicating that the two bands in the B7-H3 blots were originated from multiple glycoforms of B7-H3 (Fig. S4).
Fig. 4.
The effect of SRSF3 on B7-H3 splicing in CRC. a The effect of SRSF3 on the expression of B7-H3 mRNA and protein in HCT-116 and HCT-8 cells. The cells were transfected with 50 nM SRSF3 siRNA or 200 ng Flag-SRSF3 vectors. b RIP analysis of the binding of SRSF3 protein to the mRNA of B7-H3, TP53, and CCDC50S. The expression B7-H3, TP53, and CCDC50S mRNA were determined by qPCR assays. Data represent mean ± SD. Significance was assessed by t test. **P < 0.01; ***P < 0.001. c The schematic diagram of B7-H3 mRNA and minigene. The B7-H3 mRNA contains ten exons. Two binding-sites of SRSF3 were predicted on the exons 4 and 6. The exons 3–6 were constructed into pcDNA3.1 vectors. Two forward primers (FP1 and FP2) and two reverse primers (RP1 and RP2) were designed to amplify the transcripts of minigene. d The gel electrophorgrams of PCR amplicons of the transcripts in HCT-116 cells. The cells were transfected with 200 ng minigene vectors for 48 h. e The schematic diagram and sizes of transcripts in figure d. f The gel electrophorgrams of PCR amplicons of the minigene transcripts in HCT-116 cells. The cells were co-transfected with 200 ng minigene vectors and 50 nM SRSF3 siRNA or 200 ng Flag-SRSF3 vectors for 48 h. NC, negative control; M, DNA marker; bp, base pair
SRSF3 directly bond to B7-H3 mRNA
To investigate whether SRSF3 is directly involved in the splicing of B7-H3 mRNA, RIP experiments were performed to analyze the binding of SRSF3 protein to B7-H3 mRNA in CRC cells. Since the TP53 and CCDC50S genes have been identified as direct targets of SRSF3 [35, 36], we used the TP53 and CCDC50S mRNA as positive controls. We first lysed the cells and mixed the cell lysis solution with SRSF3 antibody on magnetic beads. An irrelevant IgG was used as negative control. Next, the beads/antibody/SRSF3/mRNA complexes were pulled down for determination of SRSF3 protein, B7-H3, CCDC50S, and TP53 mRNA. The WB results demonstrated that SRSF3 protein was specifically pulled down by the beads (Fig. 4b). Moreover, the results of qPCR assays showed that B7-H3, TP53, and CCDC50S mRNAs were also pulled down by the beads (Figs. 4b and S3). These findings indicate that SRSF3 protein directly binds to B7-H3 mRNA.
SRSF3 regulated the splicing of B7-H3 mRNA
To further explore the regulatory role of SRSF3 on B7-H3 mRNA, we used the online software RBPmap (https://rbpmap.technion.ac.il) to predict the binding-sites of SRSF3 on B7-H3. We found there were two binding-sites (CCCUACU) at the 5′-terminal of exons 4 and 6 (Fig. 4c). Thus, we constructed a minigene recombinant plasmid by cloning the DNA fragment containing exons 3–6 and 100-bp flanking sequence into pcDNA3.1 (+) vector (Fig. 4c). Then the minigene recombinant plasmids were transfected into HCT-116 cells, and RT-PCR assays were performed with primers on exons 3–6 (Fig. 4c) to detect the transcripts. Three amplicons (724-, 248-, and 70-bp) were produced by FP1 and RP2, three amplicons (472-, 335-, and 157-bp) were produced by FP1 and RP1, and an amplicon with the size of 404 bp was produced by FP2 and RP2 (Fig. 4d). Furthermore, the PCR products with the lengths of 724 bp, 472 bp, 404 bp, 335 bp, 248 bp, and 157 bp were sequenced. Based on the sizes and sequences of the PCR products, we speculated that at least five kinds of transcripts were presented in the cells transfected with the minigene recombinant plasmids, including the skipping of exon 4 and/or 5 and the retention of intron 3 or 5 (Fig. 4e). Next, we co-transfected the minigene plasmids with SRSF3 siRNA or expression plasmid into HCT-116 cells, and then detected the transcripts in the cells by RT-PCR. The results demonstrated that the amount of 472- and 404-bp products were reduced by knockdown of SRSF3, but were elevated by overexpression of SRSF3 (Fig. 4f).
Discussion
In this study, we discovered that both B7-H3 and SRSF3 were highly expressed in CRC tissues and were positively correlated with the poor prognosis of patients. SRSF3 positively regulated the expression of B7-H3 mRNA and protein in CRC cells. Further investigation demonstrated that SRSF3 involved in the splicing of B7-H3 through directly binding to B7-H3 mRNA.
In recent years, immunotherapy has been widely used in the treatment of various cancers including CRC due to its advantages of less side effects, obvious therapeutic efficacy, and ability to treat metastatic lesions. In particular, the successful treatment with immune-checkpoint inhibitors in melanoma has promoted the research of immunotherapy in CRC [37]. Currently, US FDA has approved the antibody drugs, such as ipilimumab, pembrolizumab, and nivolumab, for the treatment of CRC. However, the patients with pMMR/MSI-L phenotype accounting for 95% of CRC cases cannot benefit from these drugs [38]. Thus the inhibitors of TIM3, LAG3, and TIGIT, as well as the agonists of CD137, ICOS, CD40L, and CD27 are contending clinical trials for the treatment of CRC [37, 39]. B7-H3, a crucial member of immune-checkpoint molecules, has also been identified as a potential immunotherapeutic target for CRC. Numerous studies have shown that B7-H3 is evidently overexpressed in CRC and is closely related to the prognosis of patients [17]. In this study, we also found that B7-H3 was markedly overexpressed in CRC tissues and cell lines. Moreover, high-expression of B7-H3 is closely related to tumor TNM stage, lymph node metastasis, and overall survival of patients.
Up to now, several studies have reported the regulatory mechanisms of B7-H3 expression in tumors. At the transcriptional level, BRD4 promoted the expression of B7-H3 mRNA in pancreatic cancer [40]. An immunoglobulin transcript ILT4 induced B7-H3 expression through PI3K/AKT/mTOR signaling in non-small cell lung cancer and consequently resulted in immunosuppression of tumors [41]. At the post-transcriptional level, in addition to miR-29 [42], we previously identified miR-143 as an inhibitor of B7-H3 expression by directly binding to its 3′-UTR in CRC [43]. Moreover, both in vivo and in vitro experiments have confirmed that these miRNAs exert anti-tumor activities by suppressing B7-H3 expression. In addition, other studies have shown that the expression of B7-H3 in tumors is positively correlated with the density of tumor infiltrating FOXP3+ regulatory T cells, and is induced by CD40-activated dendritic cells or CagA oncoprotein of Helicobacter pylori in gastric cancer [44–46]. However, little is known about the splicing process of B7-H3 expression.
To investigate the splicing process of B7-H3, we first utilized the TCGA data to analyze the correlation between the expression of splicing factors and B7-H3. By individually silenced the splicing factors in HCT-116 cells, we found that the expression of B7-H3 mRNA was dramatically reduced by SRSF3, suggesting that SRSF3 might be involved in B7-H3 splicing. Thereafter, we provided solid evidences to support the conclusion that B7-H3 is spliced by SRSF3 in CRC. First, both B7-H3 and SRSF3 were highly expressed in CRC tissues and cell lines. High-expression of B7-H3 and SRSF3 was positively associated with poor prognosis of CRC patients. Moreover, the expression of B7-H3 protein in CRC tissues and cell lines was significantly positively related to SRSF3. This was also supported by the published data (GEO IDs: GDS4718, GDS4382, and GSE41657). Second, the expressions of B7-H3 mRNA and protein were evidently suppressed by knockdown of SRSF3, but were markedly enhanced by overexpression of SRSF3 in both HCT-116 and HCT-8 cells. Third, the RIP results demonstrated that SRSF3 protein directly bond to B7-H3 mRNA. Final, minigene reporter experiments were performed to further confirm that SRSF3 participated in the splicing of B7-H3 mRNA by binding to exon 4 and/or 6.
More and more studies have demonstrated that dysregulation of splicing factors leads to abnormal splicing of genes and consequent occurrence of various diseases. For instance, SRSF3, an important member of the SR protein family, is significantly upregulated in various cancers including CRC [31, 47, 48]. SRSF3 has been identified as a key regulator in the splicing of TP53, CD44, RAC1, FOXM1, KLF6, HER2, MDM4, MAP4K4, and PKM [47–49], and is closely related to the proliferation, migration, and invasion of tumor cells [35]. In this study, we found that SRSF3 was overexpressed in the cytoplasm and nucleus of CRC cells and was significantly correlated with lymph node metastasis, TNM stage, and the expression of B7-H3, p53, and Ki67, as well as poor prognosis of CRC patients. Moreover, SRSF3 participated in the splicing of B7-H3, which broadened our understanding of the immune regulatory roles of SRSF3.
Although a positive correlation was observed between B7-H3 and SRSF4, but not SRSF5 and SRSF8 in TCGA colon cancer samples, B7-H3 mRNA was apparently downregulated by SRSF8 silence and upregulated by knockdown of SRSF4 and SRSF5. However, the regulatory mechanisms mediated by these splicing factors are still required to be further investigated. For instance, whether or not there is a synergic effect of SRSF8 and SRSF3 on the splicing of B7-H3. Two isoforms of B7-H3, 2Ig B7-H3 and 4Ig B7-H3, have been detected in CRC cells, but the splicing factors involved in the alternative splicing of B7-H3 are still unknown. In addition, the effects of SRSF3 on the proliferation, migration, and invasion of tumor cells are also worthy to be further studied. Nevertheless, we discovered that a splicing factor SRSF3 involved in the splicing of B7-H3 mRNA in CRC cells. Mechanistic studies demonstrated that SRSF3 spliced B7-H3 mRNA by directly binding to its exon 4 and/or 6. These findings provide novel insights into the regulatory mechanisms of B7-H3 expression and regulatory roles of SRSF3 in tumor immune evasion, which offers potential therapeutic strategies for the treatment of CRC.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Author contributions
WPW and CWC conceived the idea and directed the study. ZCX, CYS, LFC, CWC, and WPW designed the experiments and wrote the manuscript. ZCX, CYS, LFC, MY, FYM, and ZYW performed experiments. ZCX, CYS, and LFC analyzed the data. ZCX, CYS, LFC, CWC, and WPW generated figures. CWC and WPW critically revised the manuscript for important intellectual content.
Funding
This research was supported by the National Natural Science Foundation of China (No. 81773044), Science and Technology Special Project of Clinical Medicine in Jiangsu Province (BL2014046), Social Development Project of Jiangsu Province (BE2019657), Qinglan Project of Jiangsu Province, and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Compliance with ethical standards
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the ethics committee of Soochow University (No.IRB-29-20120512H) and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Chunxia Zhang, Yinshuang Chen and Fuchao Li contributed equally to this work.
Contributor Information
Weichang Chen, Email: weichangchen@126.com.
Weipeng Wang, Email: wangweipeng@suda.edu.cn.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Gessani S, Belardelli F. Immune dysfunctions and immunotherapy in colorectal cancer: the role of dendritic cells. Cancers. 2019 doi: 10.3390/cancers11101491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thanikachalam K, Khan G. Colorectal cancer and nutrition. Nutrients. 2019 doi: 10.3390/nu11010164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bailey CE, Hu CY, You YN, Bednarski BK, Rodriguez-Bigas MA, Skibber JM, Cantor SB, Chang GJ. Increasing disparities in the age-related incidences of colon and rectal cancers in the United States, 1975–2010. JAMA Surg. 2015;150:17–22. doi: 10.1001/jamasurg.2014.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Messersmith WA. NCCN guidelines updates: management of metastatic colorectal cancer. J Natl Comprehensive Cancer Netw. 2019;17:599–601. doi: 10.6004/jnccn.2019.5014. [DOI] [PubMed] [Google Scholar]
- 6.Spartalis C, Schmidt EM, Elmasry M, Schulz GB, Kirchner T, Horst D. In vivo effects of chemotherapy on oncogenic pathways in colorectal cancer. Cancer Sci. 2019;110:2529–2539. doi: 10.1111/cas.14077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Torring ML, Falborg AZ, Jensen H, Neal RD, Weller D, Reguilon I, Menon U, Vedsted P. Advanced-stage cancer and time to diagnosis: an international cancer benchmarking partnership (ICBP) cross-sectional study. Eur J Cancer Care. 2019;28:e13100. doi: 10.1111/ecc.13100. [DOI] [PubMed] [Google Scholar]
- 8.Jiao Q, Ren Y, Ariston Gabrie AN, Wang Q, Wang Y, Du L, Liu X, Wang C, Wang YS. Advances of immune checkpoints in colorectal cancer treatment. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2020;123:109745. doi: 10.1016/j.biopha.2019.109745. [DOI] [PubMed] [Google Scholar]
- 9.Janakiram M, Shah UA, Liu W, Zhao A, Schoenberg MP, Zang X. The third group of the B7-CD28 immune checkpoint family: HHLA2, TMIGD2, B7x, and B7–H3. Immunol Rev. 2017;276:26–39. doi: 10.1111/imr.12521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pollizzi KN, Powell JD. Integrating canonical and metabolic signalling programmes in the regulation of T cell responses. Nat Rev Immunol. 2014;14:435–446. doi: 10.1038/nri3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Driessens G, Kline J, Gajewski TF. Costimulatory and coinhibitory receptors in anti-tumor immunity. Immunol Rev. 2009;229:126–44. doi: 10.1111/j.1600-065X.2009.00771.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seliger B, Quandt D. The expression, function, and clinical relevance of B7 family members in cancer. Cancer Immunol Immunother. 2012;61:1327–1341. doi: 10.1007/s00262-012-1293-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen L, Flies DB. Molecular mechanisms of T cell co-stimulation and co-inhibition. Nat Rev Immunol. 2013;13:227–42. doi: 10.1038/nri3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Picarda E, Ohaegbulam KC, Zang X. Molecular pathways: targeting B7–H3 (CD276) for human cancer immunotherapy. Clin Cancer Res. 2016;22:3425–3431. doi: 10.1158/1078-0432.Ccr-15-2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chapoval AI, Ni J, Lau JS, et al. B7–H3: a co stimulatory molecule for T cell activation and IFN-gamma production. Nat Immunol. 2001;2:269–274. doi: 10.1038/85339. [DOI] [PubMed] [Google Scholar]
- 16.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12:252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang T, Wang F, Wu JY, et al. Clinical correlation of B7–H3 and B3GALT4 with the prognosis of colorectal cancer. World J Gastroenterol. 2018;24:3538–3546. doi: 10.3748/wjg.v24.i31.3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang B, Zhang T, Liu F, Sun Z, Shi H, Hua D, Yang C. The co-stimulatory molecule B7–H3 promotes the epithelial-mesenchymal transition in colorectal cancer. Oncotarget. 2016;7:31755–31771. doi: 10.18632/oncotarget.9035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zang X, Thompson RH, Al-Ahmadie HA, Serio AM, Reuter VE, Eastham JA, Scardino PT, Sharma P, Allison JP. B7–H3 and B7x are highly expressed in human prostate cancer and associated with disease spread and poor outcome. Proc Natl Acad Sci USA. 2007;104:19458–19463. doi: 10.1073/pnas.0709802104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li G, Quan Y, Che F, Wang L. B7–H3 in tumors: friend or foe for tumor immunity? Cancer Chemother Pharmacol. 2018;81:245–253. doi: 10.1007/s00280-017-3508-1. [DOI] [PubMed] [Google Scholar]
- 21.Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song X, Wan X, Huang T, et al. SRSF3-regulated RNA alternative splicing promotes glioblastoma tumorigenicity by affecting multiple cellular processes. Can Res. 2019;79:5288–5301. doi: 10.1158/0008-5472.Can-19-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee Y, Rio DC. Mechanisms and regulation of alternative pre-mRNA splicing. Annu Rev Biochem. 2015;84:291–323. doi: 10.1146/annurev-biochem-060614-034316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He C, Zhou F, Zuo Z, Cheng H, Zhou R. A global view of cancer-specific transcript variants by subtractive transcriptome-wide analysis. PLoS ONE. 2009;4:e4732. doi: 10.1371/journal.pone.0004732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang J, Manley JL. Misregulation of pre-mRNA alternative splicing in cancer. Cancer Discov. 2013;3:1228–1237. doi: 10.1158/2159-8290.Cd-13-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koedoot E, Wolters L, van de Water B, Devedec SEL Splicing regulatory factors in breast cancer hallmarks and disease progression. Oncotarget. 2019;10:6021–6037. doi: 10.18632/oncotarget.27215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen J, Weiss WA. Alternative splicing in cancer: implications for biology and therapy. Oncogene. 2015;34:1–14. doi: 10.1038/onc.2013.570. [DOI] [PubMed] [Google Scholar]
- 29.Cohen-Eliav M, Golan-Gerstl R, Siegfried Z, Andersen CL, Thorsen K, Orntoft TF, Mu D, Karni R. The splicing factor SRSF6 is amplified and is an oncoprotein in lung and colon cancers. J Pathol. 2013;229:630–639. doi: 10.1002/path.4129. [DOI] [PubMed] [Google Scholar]
- 30.Das S, Anczukow O, Akerman M, Krainer AR. Oncogenic splicing factor SRSF1 is a critical transcriptional target of MYC. Cell Rep. 2012;1:110–117. doi: 10.1016/j.celrep.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park WC, Kim HR, Kang DB, et al. Comparative expression patterns and diagnostic efficacies of SR splicing factors and HNRNPA1 in gastric and colorectal cancer. BMC Cancer. 2016;16:358. doi: 10.1186/s12885-016-2387-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi X, Zhang Y, Cao B, et al. Genes involved in the transition from normal epithelium to intraepithelial neoplasia are associated with colorectal cancer patient survival. Biochem Biophys Res Commun. 2013;435:282–288. doi: 10.1016/j.bbrc.2013.04.063. [DOI] [PubMed] [Google Scholar]
- 33.Tsukamoto S, Ishikawa T, Iida S, Ishiguro M, Mogushi K, Mizushima H, Uetake H, Tanaka H, Sugihara K. Clinical significance of osteoprotegerin expression in human colorectal cancer. Clin Cancer Res. 2011;17:2444–2450. doi: 10.1158/1078-0432.CCR-10-2884. [DOI] [PubMed] [Google Scholar]
- 34.Khamas A, Ishikawa T, Shimokawa K, et al. Screening for epigenetically masked genes in colorectal cancer Using 5-Aza-2'-deoxycytidine, microarray and gene expression profile. Cancer Genomics Proteomics. 2012;9:67–75. [PubMed] [Google Scholar]
- 35.Kurokawa K, Akaike Y, Masuda K, Kuwano Y, Nishida K, Yamagishi N, Kajita K, Tanahashi T, Rokutan K. Downregulation of serine/arginine-rich splicing factor 3 induces G1 cell cycle arrest and apoptosis in colon cancer cells. Oncogene. 2014;33:1407–1417. doi: 10.1038/onc.2013.86. [DOI] [PubMed] [Google Scholar]
- 36.Wang H, Zhang CZ, Lu SX, et al. A coiled-coil domain containing 50 splice variant is modulated by serine/arginine-rich splicing factor 3 and promotes hepatocellular carcinoma in mice by the Ras signaling pathway. Hepatology. 2019;69:179–195. doi: 10.1002/hep.30147. [DOI] [PubMed] [Google Scholar]
- 37.Ganesh K, Stadler ZK, Cercek A, Mendelsohn RB, Shia J, Segal NH, Diaz LA., Jr Immunotherapy in colorectal cancer: rationale, challenges and potential. Nature reviews. Gastroenterol Hepatol. 2019;16:361–375. doi: 10.1038/s41575-019-0126-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kreidieh M, Mukherji D, Temraz S, Shamseddine A. Expanding the scope of immunotherapy in colorectal cancer: current clinical approaches and future directions. BioMed Res Int. 2020;2020:9037217. doi: 10.1155/2020/9037217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Emambux S, Tachon G, Junca A, Tougeron D. Results and challenges of immune checkpoint inhibitors in colorectal cancer. Expert Opin Biol Ther. 2018;18:561–573. doi: 10.1080/14712598.2018.1445222. [DOI] [PubMed] [Google Scholar]
- 40.Zhao J, Meng Z, Xie C, et al. B7–H3 is regulated by BRD4 and promotes TLR4 expression in pancreatic ductal adenocarcinoma. Int J Biochem Cell Biol. 2019;108:84–91. doi: 10.1016/j.biocel.2019.01.011. [DOI] [PubMed] [Google Scholar]
- 41.Zhang P, Yu S, Li H, et al. ILT4 drives B7–H3 expression via PI3K/AKT/mTOR signalling and ILT4/B7-H3 co-expression correlates with poor prognosis in non-small cell lung cancer. FEBS Lett. 2015;589:2248–2256. doi: 10.1016/j.febslet.2015.06.037. [DOI] [PubMed] [Google Scholar]
- 42.Purvis IJ, Avilala J, Guda MR, Venkataraman S, Vibhakar R, Tsung AJ, Velpula KK, Asuthkar S. Role of MYC-miR-29-B7-H3 in medulloblastoma growth and angiogenesis. J Clin Med. 2019 doi: 10.3390/jcm8081158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou X, Mao Y, Zhu J, et al. TGF-beta1 promotes colorectal cancer immune escape by elevating B7–H3 and B7–H4 via the miR-155/miR-143 axis. Oncotarget. 2016;7:67196–67211. doi: 10.18632/oncotarget.11950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen C, Zhu YB, Qu QX, Ge Y, Huang JA, Wang Y, Zhang XG (2009) CD40-activated apoptotic tumor cell-pulsed dendritic cell could potentially elicit antitumor immune response: involvement of up-regulation of B7–H3 expression. J Immunother 32:29–35. 10.1097/CJI.0b013e31818c8816 [DOI] [PubMed]
- 45.Inamura K, Amori G, Yuasa T, Yamamoto S, Yonese J, Ishikawa Y. Relationship of B7–H3 expression in tumor cells and tumor vasculature with FOXP3+ regulatory T cells in renal cell carcinoma. Cancer Manag Res. 2019;11:7021–7030. doi: 10.2147/CMAR.S209205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lina TT, Gonzalez J, Pinchuk IV, Beswick EJ, Reyes VE. Helicobacter pylori elicits B7H3 expression on gastric epithelial cells: implications in local T cell regulation and subset development during infection. Clin Oncol Res. 2019 doi: 10.31487/j.cor.2019.05.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuranaga Y, Sugito N, Shinohara H, Tsujino T, Taniguchi K, Komura K, Ito Y, Soga T, Akao Y. SRSF3, a splicer of the PKM gene, regulates cell growth and maintenance of cancer-specific energy metabolism in colon cancer cells. Int J Mol Sci. 2018 doi: 10.3390/ijms19103012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin JC, Lee YC, Tan TH, Liang YC, Chuang HC, Fann YC, Johnson KR, Lin YJ. RBM4-SRSF3-MAP4K4 splicing cascade modulates the metastatic signature of colorectal cancer cell. Biochimica et biophysica acta. Mol Cell Res. 2018;1865:259–272. doi: 10.1016/j.bbamcr.2017.11.005. [DOI] [PubMed] [Google Scholar]
- 49.Corbo C, Orrù S, Salvatore F. SRp20: An overview of its role in human diseases. Biochem Biophys Res Commun. 2013;436(1):1–5. doi: 10.1016/j.bbrc.2013.05.027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.