Abstract
With the use of a high-throughput biochemical DNA helicase assay as a screen, T157602, a 2-amino thiazole compound, was identified as a specific inhibitor of herpes simplex virus (HSV) DNA replication. T157602 inhibited reversibly the helicase activity of the HSV UL5-UL8-UL52 (UL5/8/52) helicase-primase complex with an IC50 (concentration of compound that yields 50% inhibition) of 5 μM. T157602 inhibited specifically the UL5/8/52 helicase and not several other helicases. The primase activity of the UL5/8/52 complex was also inhibited by T157602 (IC50 = 20 μM). T157602 inhibited HSV growth in a one-step viral growth assay (IC90 = 3 μM), and plaque formation was completely prevented at concentrations of 25 to 50 μM T157602. Vero, human foreskin fibroblast (HFF), and Jurkat cells could be propagated in the presence of T157602 at concentrations exceeding 100 μM with no obvious cytotoxic effects, indicating that the window between antiviral activity and cellular toxicity is at least 33-fold. Seven independently derived T157602-resistant mutant viruses (four HSV type 2 and three HSV type 1) carried single base pair mutations in the UL5 that resulted in single amino acid changes in the UL5 protein. Marker rescue experiments demonstrated that the UL5 gene from T157602-resistant viruses conferred resistance to T157602-sensitive wild-type viruses. Recombinant UL5/8/52 helicase-primase complex purified from baculoviruses expressing mutant UL5 protein showed complete resistance to T157602 in the in vitro helicase assay. T157602 and its analogs represent a novel class of specific and reversible anti-HSV agents eliciting their inhibitory effects on HSV replication by interacting with the UL5 helicase.
Herpes simplex virus types 1 and 2 (HSV-1 and HSV-2) each comprise at least 77 genes whose expression is tightly regulated (42). These genes are assigned to four kinetic classes, designated as α, β, γ1, and γ2 on the basis of the timing of and requirements for their expression (46). The five α genes, α0, α4, α22, α27, and α47, are expressed first in the absence of viral protein synthesis and are responsible for the regulated expression of the other viral genes. The β genes require functional α gene products for their expression and encode proteins and enzymes that are directly involved in DNA synthesis and nucleotide metabolism. The γ genes form the last set of viral genes to be expressed, with the γ2 class having viral DNA replication as a strict requirement for their expression.
The HSV genome contains three origins of replication (44, 45, 47, 48, 50, 54) and encodes seven viral proteins that are essential for DNA replication (34, 59). These include an origin binding protein (OBP) encoded by open reading frame (ORF) UL9 (14, 15, 17, 35), a DNA binding protein encoded by UL29 (40, 53, 54), a DNA polymerase encoded by ORF UL30 and its accessory factor encoded by UL42 (1, 4, 8, 18, 19, 21, 24, 37), and a heterotrimeric complex consisting of proteins encoded by ORFs UL5, UL8, and UL52, which include both 5′-to-3′ helicase activity and primase activity (10–12). Although extensively studied, the roles of the individual subunits of the helicase-primase complex and their specific interactions with each other have not been completely defined. However, several lines of evidence suggest that the UL5 gene encodes the helicase activity of the complex. Examination of the amino acid sequence of the UL5 protein revealed that it contains six conserved motifs that are found in many DNA and RNA helicases, two of these motifs defining an ATP binding site (20, 25, 32, 52, 61). Site-specific mutagenesis of amino acids within each of the six motifs revealed that all six are critical for the function of the UL5 protein as a helicase in transient replication assays (60, 61).
The observation that recombinant UL5, UL52, and UL8 proteins could be purified from baculovirus-infected insect cells as a complex that displays DNA-dependent ATPase, helicase, and primase activities that are identical to those produced during a herpesvirus infection allowed functional and biochemical analyses of the individual components of the complex (10, 13, 38). Although the UL5 protein alone contained the defining helicase amino acid sequence motifs, the UL5 protein does not display helicase activity in vitro in the absence of the UL52 protein. Purified UL5 protein has less than 1% of the ATPase activity of the complex UL5-UL8-UL52 (UL5/8/52) complex (2, 43). In addition, studies with recombinant herpesviruses carrying mutations in the UL5 gene that abolish helicase activity revealed that the UL5 protein could still form specific interactions with UL8 and UL52 proteins (60). These results indicate that the functional domains of UL5 protein required for helicase activity are separate from those involved in protein-protein interactions and that UL5 and UL52 must interact to yield efficient helicase activity. Further mutagenesis studies with the UL52 protein identified mutations that abolish the primase activity of the complex, while the helicase and ATPase activities are unaffected, suggesting that the UL52 protein is responsible for the primase activity of the complex (27). The third component of the helicase-primase complex, the UL8 protein, interacts with other viral replication proteins, including the OBP, the single-stranded DNA binding protein, and the viral DNA polymerase (30, 33). It has been postulated that the interaction of the UL8 protein with the OBP (encoded by the UL9 gene) may function to recruit helicase-primase complexes to initiation complexes at viral origins (30). The UL8 protein is also required for stimulation of primer synthesis by the UL52 protein and for stimulation of the helicase activity of the helicase-primase complex which is crucial to allow efficient unwinding of long stretches of duplex DNA (16, 43, 49). Additionally, UL8 appears to be required for efficient nuclear entry of the helicase-primase complex (1, 3, 31).
As the UL5, UL8, and UL52 gene products are essential for HSV replication and have not been exploited previously for antiviral drug discovery, they represent attractive targets for the development of novel anti-HSV agents. Current anti-HSV drugs include vidarabine (adenine arabinoside; Ara-A), foscarnet (phosphonoformic acid; PFA), and a wide variety of nucleoside analogs, the most clinically successful being acyclovir (ACV) and its analogs valacyclovir and famciclovir. ACV is phosphorylated by viral thymidine kinase (TK) to its monophosphate form, an event that occurs to a much lesser extent in uninfected cells. Subsequent phosphorylation events by cellular enzymes convert the ACV monophosphate to its triphosphate form. The ACV triphosphate derivative directly inhibits the DNA polymerase by competing as a substrate with dGTP. Because the ACV triphosphate lacks the 3′ hydroxyl group required to elongate the DNA chain, DNA replication is terminated. The triphosphorylated form of ACV is a much better substrate for the viral DNA polymerase than it is for the cellular DNA polymerase; thus, very little ACV triphosphate is incorporated into cellular DNA. Although ACV has proven to be safe and successful at reducing the duration, severity, and in some cases recurrence of HSV infections, eradication of the infection symptoms is far from complete and latent virus can reactivate frequently (55–58). In addition, primarily as a result of poor patient compliance with inconvenient ACV dosage regimens, virulent HSV strains resistant to ACV that contain mutations in either the viral TK or DNA polymerase gene have arisen (6, 7, 9, 26, 39). More potent and efficacious drugs that target other essential components of the virus replicative cycle would be invaluable as therapeutic agents to treat HSV and ACV-resistant HSV infections.
To identify novel inhibitors of the HSV helicase-primase enzyme, we developed a high-throughput in vitro helicase assay and screened >190,000 samples. Using this biochemical approach, we identified T157602, a 2-amino thiazole, as a specific inhibitor of HSV replication. By generating and analyzing T157602-resistant viruses, we further demonstrate genetically that the molecular target of T157602 is the UL5 component of the HSV helicase-primase complex.
MATERIALS AND METHODS
Cells and viruses.
Vero cells were used for high-titer virus stocks and viral DNA preparations as well as for virus titrations and viral yield determinations. Human foreskin fibroblasts (HFF cells) and Jurkat cells were used in the cytotoxic studies. Vero, HFF, and Jurkat cell lines were obtained from the American Type Culture Collection (Manassas, Va.) and were maintained in minimum essential medium (JRH Biosciences, Lenexa, Kans.) supplemented with 10% fetal bovine serum (JRH Biosciences), l-glutamine (2 mM), penicillin (100 U/ml), streptomycin (0.1 mg/ml), and pyruvate (1 mM). Rabbit skin cells were a gift from B. Roizman (University of Chicago, Chicago, Ill.). High Five cells (Invitrogen, Carlsbad, Calif.) were maintained in Ex-Cell 405 medium (JRH Biosciences) supplemented with penicillin (100 U/ml), streptomycin (100 μg/ml), and amphotericin B (0.25 μg/ml). Spodoptera frugiperda (SF9) cells were maintained as a suspension in TNM-FH medium (JRH Biosciences) supplemented with 10% heat-inactivated fetal bovine serum and antibiotic-antimycotic as described for High Five cells. The recombinant baculoviruses Autographa californica nuclear polyhedrosis virus (AcMNPV)/(HSV-1) UL5, AcMNPV/(HSV-1) UL8, and AcMNPV/(HSV-1) UL52 were gifts from R. Lehman (Stanford University, Stanford, Calif.). AcMNPV/(HSV-1) N-His UL8 was constructed by PCR amplification of total cellular DNA isolated from AcMNPV/(HSV-1) UL8-infected SF9 cells, using a 5′ PCR primer to place a nine-histidine tag in frame with the second amino acid of the coding sequence. Recombinant baculoviruses AcMNPV/(HSV-2)wt UL5 and AcMNPV/(HSV-2) R6(K355N) UL5 were constructed by PCR amplification of viral DNA obtained from HSV-2 wild-type and T157602-resistant mutant HSV-2 R6(K355N)-infected cells and subcloning of these products into pVL1392. Recombinant baculoviruses were made by cotransfection with BaculoGold linearized baculovirus DNA (Pharmingen, San Diego, Calif.). Routine methods were used in the propagation of these viruses (36). HSV-2(G) and HSV-1(MacIntyre) were used as the wild-type viruses; both were obtained from the American Type Culture Collection. Recombinant virus rHSV-2LUC contains a luciferase reporter gene in the glycoprotein C ORF. It was constructed by cotransfection of intact viral HSV-2(G) DNA with plasmid TP3 into Vero cells. Individual plaques were purified on Vero cells and screened for reporter gene by expression, luciferase activity measurement, and restriction enzyme analysis. To construct TP3, pBluescript (pBS; Stratagene, San Diego, Calif.) was digested with EcoRV and Ecl136 and religated to remove the BamHI site from the vector, generating ΔpBS. The 2,880-bp SalI fragment was excised from HSV-2(G) DNA and cloned into the SalI site of ΔpBS to generate ΔpBSSal. An oligonucleotide with AgeI and NcoI terminal restriction enzyme sites containing internal NheI and BamHI restriction sites was inserted at the NcoI site in the glycoprotein C ORF within ΔpBSSalI, generating ΔpBSSalIN. The luciferase gene was excised from pGL-2 (Promega, Madison, Wis.) by PCR with primers containing an internal NcoI site that corresponded to the luciferase ATG and terminal BamHI and HindIII sites. The PCR product was cloned into pBS at the HindIII and BamHI sites to generate pBSluc. The entire luciferase gene was excised from pBSluc as an NcoI-BamHI fragment and cloned into ΔpBSSalIN at the NcoI and BamHI sites, replacing a portion of the glycoprotein C coding sequence. The resulting plasmid, designated TP3, contained the luciferase gene under the control of the glycoprotein C gene promoter and the HSV-2 flanking sequences necessary for recombination into the viral genome. Intact HSV-2(G) viral DNA was cotransfected with plasmid TP3 into Vero cells, and the progeny of the transfection were screened for luciferase activity and by restriction enzyme analyses. The resulting recombinant virus, which contained a functional luciferase gene and was designated rHSV-2LUC, was plaque purified on Vero cells four times.
A recombinant consisting of cytomegalovirus (CMV) containing a luciferase gene was generated and designated rCMVLUC.
Purification procedure for the UL5/8/52 helicase-primase complex.
Forty-two Intergrid dishes (150 by 25 mm) of High Five (2 × 107) cells were coinfected with baculovirus recombinants AcMNPV/(HSV-1) UL52, AcMNPV/(HSV-1) N-His UL8, and either AcMNPV/(HSV-2)wt UL5 or AcMNPV/(HSV-2) R6(K355N) UL5. At 60 h postinfection, cells were dislodged from the plates and harvested from the cell culture medium by centrifugation for 10 min at 2,500 rpm in a Beckman GS-6KR centrifuge. All subsequent purification steps were performed at 4°C. The cells were resuspended in 160 ml of buffer A (20 mM HEPES [pH 7.9], 400 mM KCl, 5% glycerol, 15 mM imidazole, 0.1% Nonidet P-40, 1.5 mM MgCl2, 8 mM β-mercaptoethanol, 1 mM AEBSF [Boehringer Mannheim, Indianapolis, Ind.]), allowed to sit on ice for 15 min, and then sonicated for 2 min at a setting of 6 (Branson Sonifier 450). The homogenate was centrifuged for 30 min at 12,000 rpm in a Sorvall SS34 rotor. The supernatant was decanted and incubated for 2 h with 3 ml of equilibrated Ni2+-agarose beads (Qiagen, Chatsworth, Calif.). The beads were centrifuged at 3,000 rpm for 10 min, and the unbound material was removed. The beads were then washed twice with 10 volumes of buffer A and twice with 10 volumes of buffer B (buffer A containing 30 mM imidazole). The helicase-primase complex was eluted from the beads with 3 volumes of buffer C (buffer A containing 200 mM imidazole and 10% glycerol), frozen in liquid N2, and stored at −80°C. The proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Coomassie blue staining and were determined to be greater than 90% pure.
DNA helicase gel assays.
The synthetic oligonucleotide GO135 (5′-GCAGCAAGCGGTCCACGCTGGTTTG-3′), which is complementary to nucleotides 5902 to 5926 of M13mp18(+) DNA, was annealed to M13 DNA and 3′ end labeled with [α-32P]dCTP and Klenow enzyme. The substrate was phenol-chloroform extracted, and unincorporated nucleotides were removed by two sequential centrifugations through Sephadex G-50 QuickSpin columns (Boehringer Mannheim). Standard helicase reaction mixtures (30 μl) contained 20 mM HEPES (pH 7.6), 5% glycerol, 4 mM MgCl2, 100 μg of bovine serum albumin (BSA) per ml, 2 mM dithiothreitol (DTT), 10% dimethyl sulfoxide (DMSO) (or T-compound in DMSO), 10,000 cpm of M13/GO135 DNA (3 to 5 ng of M13 DNA), and sufficient UL5 [wild type or R6(K355N)]–UL8 N-His–UL52 enzyme complex to displace ≈70% of the radiolabeled oligomer (upper linear range of a standard dose-response curve). Reaction mixtures were preincubated at room temperature for 10 min, and ATP was added to a final concentration of 4 mM to start the reaction. The reaction mixtures were incubated for 1 h further at room temperature, and reactions were terminated by the addition of an equal volume of 1.2% sodium dodecyl sulfate–60 mM EDTA–10% glycerol–0.05% bromophenol blue. The products were separated on a nondenaturing 12% polyacrylamide gel in 0.5× TBE (25 mM Tris base, 25 mM boric acid, 0.5 mM EDTA), and the gel was dried and subjected to autoradiography. Dried gels were scanned for the percentage of displaced oligonucleotide on a Fuji FujixBAS1000 phosphorimager.
High-throughout helicase screening assay.
The high-throughput helicase screen was based on a labeled primer displacement assay, the details of which will be published elsewhere. Essentially, an α-33P-labeled primer was annealed to an M13 template, and pure, baculovirus-expressed UL5/8/52 HSV-1 helicase complex was added together with ATP and Mg2+. The extent to which the helicase enzyme complex could displace the labeled primer in the presence of the test compound was measured by counting the radioactivity that remained associated with the M13 substrate.
Primase assay.
Helicase primase enzyme (2 μg) was incubated in a buffer (50 mM NaCl, 50 mM Tris [pH 8], 4 mM MgCl2, 200 μg of BSA per ml, 10% glycerol, 1 mM DTT, 1 mM ATP, 1 mM GTP, 0.1 mM CTP, 25 fmol of [33P]UTP, 5% DMSO) containing the test drug together with 10 pmol of single-stranded oligonucleotide template. The reaction was allowed to proceed for 90 min at 30°C. The reaction products were separated on a nondenaturing 12% polyacrylamide gel in 0.5× TBE. The gel was dried and subjected to autoradiography, and dried gels were scanned for the primer products on a Fuji FujixBAS1000 phosphorimager.
ATPase assay.
Purified HSV helicase-primase complex (150 ng) was incubated with 20 mM HEPES (pH 7.6), 4 mM MgCl2, 4 mM ATP, 100 μg of BSA per ml, 5% glycerol, 2 mM DTT, and 500 ng of M13 DNA for 1 h at room temperature. The released inorganic phosphate was detected colorimetrically as described previously (28).
Purification and analyses of viral DNA.
Viral DNA was prepared from NaI gradients as described previously (51). Specific ORFs were cloned as PCR products either from the viral DNA or from plasmids containing EcoRI, BamHI, and HindIII viral DNA restriction fragments cloned in pGEM3Zf+. Clones were verified by restriction enzyme digestion. Southern (DNA) blotting and sequencing methods are described elsewhere (41).
Marker rescue.
NaI-purified intact viral DNA from either HSV-2(G) or rHSV-2LUC was cotransfected with 50, 100, or 200 μg of plasmid DNA cloned from the T157602-resistant viruses onto rabbit skin cells as described previously except that 50 μM T157602 compound was added to the transfection medium 2 h posttransfection. After 3 days, the progeny viruses were titered on Vero cells and overlayed with medium containing 50 μM T157602. Plaques were counted 2 to 3 days postinfection.
Viral yield determinations.
Vero cells were infected with either wild-type or T157602-resistant viruses at a multiplicity of infection (MOI) of 1 PFU/cell. After 1 h of exposure to virus, the cells were trypsinized and seeded into 96-well plates. The infected cells were incubated for a further 2 h before T157602 was added at concentrations ranging from 0.2 to 100 μM. After incubation for 24 h, the cell monolayers were frozen and thawed, and the viral yield was determined by serial dilution in 96-well plates and monitoring of cytopathic effect after 48 h.
Luciferase assays.
The 50% inhibitory concentration (IC50) for T157602 was determined by measuring the luciferase activity in cells infected with recombinant virus rHSV-2LUC at various concentrations of the drug T157602. A 25-cm2 flask of Vero cells was infected with rHSV-2LUC at an MOI of 0.1 PFU/cell. After 1 h of exposure to virus, the inoculum was removed, and the cells were transferred to 96-well plates and incubated for a further 2 h. The test drug was added at concentrations ranging from 0.1 to 100 μM, and the infected cells were incubated for a further 16 h at 37°C before luciferase expression was measured. To measure luciferase activity, the supernatant was removed from each well, and 50 μl of assay buffer (8 mM magnesium acetate, 30 mM Tricine [pH 7.8], 200 μM EDTA, 1.5 mM ATP, 0.5 mM luciferin, 1.5 mM coenzyme A, 0.1 M 2-mercaptoethanol, 10% Triton X-100) was added to each well. After 5 min, luminescence (a measure of the luciferase activity) was measured with a Lumicount luminometer (Packard).
Site-directed mutagenesis.
The 3.29-kbp XbaI-HindIII fragment that contained UL5 was excised from TP103 and cloned into pALTER-1 (Promega) at the XbaI and HindIII sites to generate plasmid TP201. Site-directed mutagenesis was performed on single-stranded DNA with an Altered Sites II in vitro mutagenesis system kit as instructed by the manufacturer (Promega). Mutagenic oligonucleotides were synthesized by Operon Technologies (Alameda, Calif.). All mutations in the final clones that were used for marker rescue experiments were verified by DNA sequence analysis using Sequenase (United States Biochemical Corp., Cleveland, Ohio) as instructed by the supplier.
RESULTS
Identification of a new class of drugs with anti-HSV activity.
A high-throughput helicase assay was used to screen >190,000 samples consisting of random pure chemicals and natural products for specific inhibitors of HSV-1 UL5/8/52 helicase-primase. This screen identified a single class of 2-amino thiazole compounds, represented by T157602. The structure of T157602 and a summary of its characteristics are shown in Fig. 1 and Table 1, respectively. Briefly, T157602 inhibited reversibly the HSV helicase activity with an IC50 of 5 μM. The inhibition was specific for HSV helicases, as several other unrelated helicases tested, including Escherichia coli DnaB and RecQ and HSV OBP, were unaffected. The inhibitory effects of T157602 on HSV helicase were found to be completely reversible upon removal of the drug. Competition assays showed that T157602 did not compete for either ATP or DNA binding.
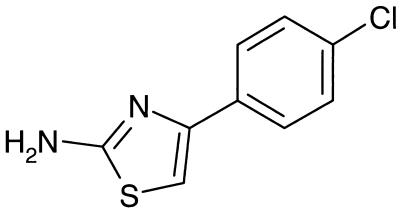
FIG. 1.
Molecular structure of the 2-amino thiazole compound T157602.
TABLE 1.
Assay characteristics of T157602
| Type of assay | IC50 (μM) |
|---|---|
| Biochemical | |
| HSV UL5/8/52 helicase (gel assay) | 5 |
| HSV UL5/8/52 primase | 20 |
| HSV UL5/8/52 ATPase | 10 |
| HSV OBP helicase | >100 |
| E. coli RecQ helicase | >100 |
| E. coli DnaB helicase | >100 |
| Viral | |
| rHSV-2LUC (luciferase) | 10 |
| rCMVLUC (luciferase) | >100 |
| HSV yield (one-step growth) | 0.8 |
| 3 (IC90) | |
| HSV plaque reduction | 25 (IC99.9) |
| Cytotoxicity | |
| Vero | >100 |
| HFF | >100 |
| Jurkat | >100 |
| CFU-GM | >75 |
| BFU-e | >75 |
The primase activity of the UL5/8/52 complex was also inhibited by T157602 with an IC50 of 20 μM (Table 1). In addition, T157602 inhibited viral growth in a one-step growth assay with an IC90 of 3 μM, and plaque formation was completely prevented at concentrations of 25 to 50 μM. By comparison, ACV prevented plaque formation at concentrations of 3 to 6 μM and inhibited viral growth with an IC90 of ≈0.8 μM (data not shown). Cells infected with the recombinant virus rHSV-2LUC, in which a luciferase gene under the control of the HSV-2 glycoprotein C promoter is expressed as a late (γ2) viral gene, showed reduced luciferase expression in the presence of T157602, with an IC50 of 10 μM. Vero, HFF, and Jurkat cells could be propagated in the presence of T157602 at a concentration exceeding 100 μM with no obvious cytotoxic effects, indicating that the window between antiviral activity and cellular toxicity is approximately 33-fold.
As a more sensitive cellular toxicity assay, the effects of T157602 on the growth of bone marrow cells in soft agar were assessed. As shown in Table 1, no obvious cytotoxic effect of T157602 was observed on either the colony-forming ability of granulocyte/macrophage precursors (CFU-GM) or the blast-forming ability of erythrocytes (BFU-e) at a T157602 concentration of 75 μM. In addition, the replication abilities of ACV-resistant strains of HSV and HSV clinical isolates were also decreased in tissue culture to levels similar to those of wild-type HSV strains in the presence of T157602 (data not shown). Taken together, these results suggest that T157602 and its analogs represent a novel class of specific and reversible anti-HSV agents that may elicit their effects by inhibiting viral replication through interactions with the viral helicase-primase complex.
Generation of T157602-resistant HSV strains.
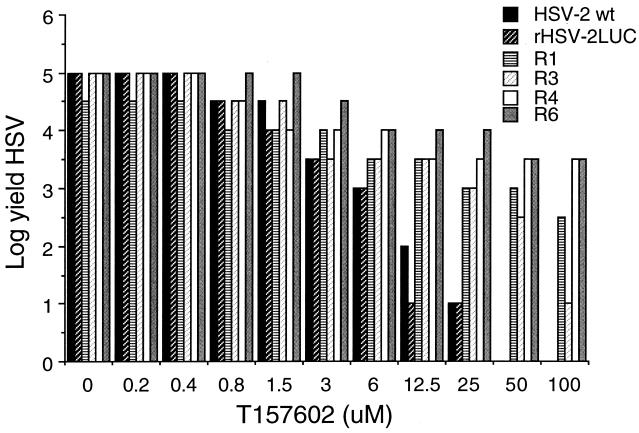
Viral replication can be inhibited by either interfering directly with virus-specific processes or incapacitating the host cell. If virus mutants resistant to an inhibitory compound can be selected, it is likely that the target of the inhibitor is a viral process. Drug targets can be identified by defining the gene in which the mutation conferring the resistant phenotype has occurred. To determine the molecular target of T157602 in the context of a viral infection, we selected resistant viruses in the presence of increasing concentrations of the T157602. Four independently derived T157602-resistant HSV-2 mutants (R1, R3, R4, and R6) were selected and plaque purified four times in the presence of 50 μM T157602. Three T157602-resistant HSV-1 mutants (R1, R2, and R3) were also isolated and plaque purified by a similar procedure. As shown in Fig. 2, the viral yields obtained from the resistant viruses derived from HSV-2 were 2 to 4 logs greater than those of wild-type virus in the presence of 50 μM T157602; in addition, no plaques were formed by wild-type viruses at this concentration (data not shown). All of the resistant HSV mutants grew to high titers in Vero cells, indicating that the mutations in the resistant isolates did not significantly impair their growth. The T157602-resistant mutants all showed relatively high levels of luciferase expression compared to the parental wild-type virus (data not shown), demonstrating that DNA replication-dependent HSV late gene expression was no longer inhibited by T157602. These results suggest that the target of T157602 is encoded by the viral genome.
FIG. 2.
Comparison of viral yields from cells infected with resistance mutant R1, R3, R4, or R6, rHSV-2LUC, or wild-type (wt) HSV-2(G) in the presence of increasing concentrations of T157602.
Single base pair mutations in the UL5 genes of T157602-resistant viruses.
Although the HSV helicase-primase complex comprises UL5, UL52, and UL8, we focused our initial genetic analysis on UL5 and UL52 because of their known enzymatic activities and our observations that the dimeric complex (UL5/52) is inhibited by T157602 with the same IC50. The 4,940-bp HSV-2 BamHI K fragment from R6, which contains the entire 3.2-kb UL52 ORF and flanking sequences, was cloned into pGEM3Zf+ to generate TP604. TP604 was sequenced completely, and no sequence differences from wild-type virus were identified. Moreover, this plasmid could not rescue the wild-type virus in the presence of 50 μM T157602, indicating that the resistance mutation was not in the UL52 gene.
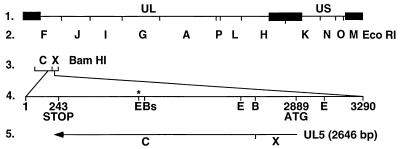
The UL5 gene was obtained as a single 3.2-kbp fragment by PCR amplification of viral DNA from each of the resistant HSV-2 strains (Fig. 3). The corresponding PCR products from T157602-sensitive HSV-2(G) and the parental HSV-2 recombinant (rHSV-2LUC) were also synthesized. The UL5 genes from R6, R4, R3, R1, rHSV-2LUC, and HSV-2(G) were sequenced, and single point mutations were identified in the resistant virus DNA (Fig. 4). T157602-resistant mutant viruses R6 and R4 carried the same mutation at lysine residue 355, where the third base pair of the codon was mutated from a G to a T, resulting in a lysine-to-asparagine substitution. This mutation also destroyed an EcoNI restriction site, allowing for quick identification of the point mutation within the viral DNA by restriction enzyme analysis. R1 contained a single mutation at methionine residue 354, where the second base of the codon was mutated from a T to a C, generating a threonine residue. This mutation destroyed a BspHI restriction site, which allowed rapid identification of the mutation. R3 carried a single mutation at glutamic acid residue 399, where the third base of the codon was mutated from a G to a T, resulting in an aspartic acid residue.
FIG. 3.
Schematic representation of the DNA sequence arrangement in the genomes of HSV-2 and the resistance mutants used in this study. Line 1, HSV-2(G) prototype orientation, with unique sequences denoted by a thin line and inverted repeats flanking the UL and US components denoted by filled rectangles. Line 2, EcoRI restriction fragments of HSV DNA designated alphabetically according to size in base pairs. Line 3, HSV BamHI restriction fragments C and X containing the UL5 ORF. Line 4, the 3,290-bp PCR-amplified fragment containing the entire 2,646-bp UL5 ORF. The start codon is represented by ATG at nucleotide 2889, and the stop codon is represented by STOP at nucleotide 243. The letters B, Bs, and E, represent restriction enzyme sites BamHI (bp 2590), BspHII (bp 1827), and EcoNI (bp 1824, 2556, and 3124), respectively. The asterisk indicates the EcoNI site at position 1824 that is destroyed in T157602-resistant mutants R6 and R4 and the BspHI site at position 1827 that is destroyed in mutant R1. Line 5, orientation of the 2,646-bp UL5 ORF and its relative position spanning BamHI fragments C and X on the viral genome.
FIG. 4.
Amino acid sequence of the HSV-2(G) UL5 ORF showing the positions and nature of the point mutations contained in the UL5 protein of the T157602-resistant viruses R1, R3, R4, and R6. Resistant virus R1 contains a single base pair mutation at methionine residue 354, where the second base of the codon was mutated from a T to a C, thus changing the methionine residue to a threonine residue. Resistant virus R3 carried a mutation at glutamic acid residue 399, where the third base of the codon was mutated from a G to a T, thus changing the glutamic acid residue to an aspartic acid residue. Resistant viruses R6 and R4 carried similar mutations at lysine residue 355, where the third base pair of the codon was mutated from a G to a T, thus resulting in lysine-to-asparagine substitution. The boxed regions indicate the six protein domains required for helicase activity of the UL5 protein.
Compared with HSV-2 UL5, the HSV-1 UL5 gene has been shown to contain one extra leucine at position 20 in the N-terminal region. Sequence analyses of the HSV-1 T157602-resistant mutants revealed that they also carried a single point mutation in the UL5 gene. Specifically, the mutation was a G-to-T substitution in the third base pair of codon 356, which resulted in a lysine-to-asparagine change at this position. This mutation in HSV-1 DNA also resulted in the destruction of an EcoNI site.
Conferring T157602 resistance upon drug-sensitive viruses with a resistant form of UL5.
To determine whether the UL5 genes carrying T157602-resistant mutations could transfer this resistant phenotype to wild-type drug-sensitive strains of HSV-2, we conducted a series of marker rescue experiments. The 3.2-kb PCR products containing the UL5 genes from wild-type and T157602-resistant R6, R1, and R3 viruses were cloned into the SmaI site of pGEM3Zf+ to generate plasmids TP106, TP103, TP108, and TP107, respectively. The existence of the appropriate point mutations in the UL5 genes was verified by restriction enzyme analysis and DNA sequencing.
Rabbit skin cells were cotransfected with intact viral DNA from rHSV-2LUC and plasmids containing UL5 genes from R6, R1, R3, or wild-type virus in the presence of 50 μM T157602. After 3 days, the progeny recombinant viruses were titrated on Vero cells in the presence of 50 μM T157602. The results from these marker rescue experiments are shown in Table 2. No plaques were observed in any of the dishes transfected with the wild-type plasmid when T157602 was present at a concentration of 50 μM. All of the mutant UL5 genes could rescue the wild-type viruses with titers ranging from 102 to 104 PFU/ml. Plasmids containing the R6 K355N and the R1 M354T mutations were the most efficient at rescuing the wild-type viruses in the presence of T157602. It is noteworthy that plaques formed by the R1-rescued viruses were significantly smaller than those generated by the R6-rescued viruses. The R3 E399D mutation was less effective at rescuing wild-type viruses, as evidenced by the fact that the virus titers were at least 1 log lower than those routinely obtained from marker rescue experiments. The plaques produced by the R3(E399D)-rescued virus also appeared to be smaller and took longer to develop than those of R6(K355N). In the absence of T157602, the plaque morphology of the R3(E399D) recombinant was no different from that of R6(K355N), and both viruses grew to the same titers. In addition, we compared the growth of R6 and R3 in the presence of different concentrations of T157602, using high-titered stocks of each virus (Fig. 2). These results indicate that the E399D-mutated UL5 is less resistant to T157602 than is either the R6(K355N) or the R1(M354T) virus.
TABLE 2.
Marker rescue of wild-type viruses with plasmids carrying mutations in the UL5 gene
| Parental virus DNA (1 μg) | Transfected plasmida | Amt of plasmid DNA transfected (ng) | T157602 concn (μM) | Titer of rescued viruses (PFU/ml) |
|---|---|---|---|---|
| rHSV-2LUC | —b | 4.2 × 103 | ||
| HSV-2G | — | 2.0 × 103 | ||
| rHSV-2LUC | — | 50 | 0 | |
| HSV-2G | — | 50 | 0 | |
| rHSV-2LUC | TP103 (R6) | 50 | 50 | 5.6 × 103 |
| TP103 | 100 | 50 | 9.5 × 103 | |
| TP103 | 200 | 50 | 9.4 × 103 | |
| TP106 (WT) | 50 | 50 | 0 | |
| TP106 | 100 | 50 | 0 | |
| TP106 | 200 | 50 | 0 | |
| TP106 | 100 | 8.7 × 103 | ||
| HSV-2G | TP103 (R6) | 50 | 50 | 1.0 × 103 |
| TP103 | 100 | 50 | 5.3 × 103 | |
| TP103 | 200 | 50 | 2.8 × 103 | |
| rHSV-2LUC | TP108 (R1) | 50 | 50 | 5.9 × 103 |
| TP108 | 100 | 50 | 8.5 × 103 | |
| TP108 | 200 | 50 | 5.2 × 103 | |
| TP107 (R3) | 50 | 50 | 1.4 × 102 | |
| TP107 | 100 | 50 | 2.0 × 102 | |
| TP107 | 200 | 50 | 0.8 × 102 | |
| TP630 (R6c) | 50 | 50 | 1.5 × 103 | |
| TP630 | 100 | 50 | 1.2 × 103 | |
| TP630 | 200 | 50 | 1.8 × 104 | |
| — | 50 | 0 | ||
| TP34(WTc) | 50 | 50 | 0 | |
| TP34 | 100 | 50 | 0 | |
| TP34 | 200 | 50 | 0 |
WT, wild type; R6c and WTc, cloned fragment viral DNA.
—, no plasmid, viral DNA only.
The UL5 gene of HSV-2 spans the two HSV-2 BamHI fragments C and X. As an additional control, the HSV-2 BamHI C restriction fragments from both the wild-type and R6 viruses were cloned directly from the viral DNA into pGEM3Zf+ to generate plasmids TP34 and TP630, respectively. Only the BamHI C fragment from R6 (designated TP630) containing the K355N mutation was able to rescue the wild-type viruses in the presence of 50 μM T157602 (Table 2).
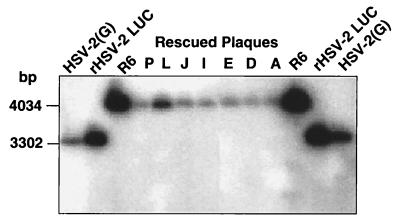
To ensure that the R6 K355N mutation had been transferred to the rescued viruses, we picked several plaques from the R6-rescued viruses [both rHSV-2LUC and HSV-2(G)] and subjected them to one more round of plaque purification. The DNA of the rescued plaques was then analyzed by Southern analysis for the presence of the R6 K355N mutation. As an additional control, the UL5 gene was obtained by PCR from the purified viral DNA from each of the rescued virus isolates and analyzed by digestion with EcoNI and sequencing. Shown in Fig. 5 is a representative Southern blot of an EcoNI digestion of DNA from wild-type, resistant mutant R6, and rescued wild-type virus plaque isolates probed with a portion of the UL5 gene. When the EcoNI site at UL5 position 355 has been destroyed as in R6, the 3,302-bp wild-type EcoNI fragment is lengthened by 732 bp to 4,034 bp. In all of the rescued recombinant virus isolates, the EcoNI fragment is the same size as that in R6, indicating that the rescued virus with a T157602-resistant phenotype has acquired the K355N mutation in UL5. The rescued viruses could be distinguished clearly from the original T157602-resistant virus R6 by the lack of the luciferase gene, verifying that there was no contamination of the rescued viruses with R6 (data not shown).
FIG. 5.
Computer-generated photograph of an autoradiographic image of a Southern blot analysis of the structures of HSV-2(G), rHSV-2LUC, resistant virus R6, and plaque isolates from drug-resistant viruses obtained by marker rescue with plasmid containing the R6 K355N point mutation in UL5. EcoNI digests of viral DNA were electrophoretically separated on 0.8% agarose gels, transferred to Hy-Bond nylon membranes (Amersham), and hybridized with a radiolabeled probe containing specific UL5 sequences. In resistant virus R6, the 3,302-bp wild-type EcoNI band containing UL5 sequences is shifted up by 732 bp to 4,034 bp. In the rescued recombinant virus isolates (P, L, J, I, E, D, and A), the EcoNI fragment is shifted to the R6 position. This indicates that the rescued virus with a T157602-resistant phenotype had acquired the K355N mutation in UL5, which destroys the EcoNI site at this position in the viral DNA.
T157602 resistance of a helicase-primase enzyme that carries the K355N mutation in an in vitro helicase assay.
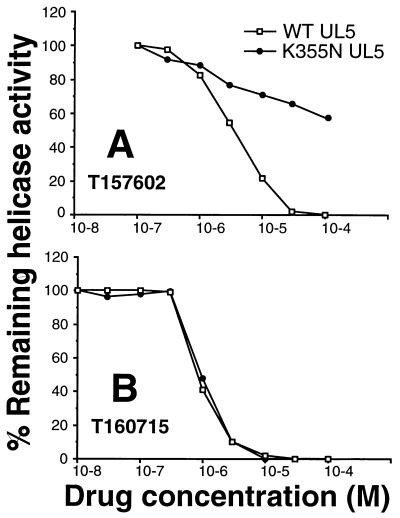
Helicase-primase enzyme complexes containing either the wild-type HSV-2 UL5 protein or the K355N UL5 protein cloned from resistant virus R6 DNA were purified from baculovirus-infected High Five cells and tested in an in vitro gel-based helicase assay. The specific activity of the helicase-primase complex containing the K355N UL5 protein was found to be similar to that of the wild-type UL5-containing complex (data not shown). As shown in Fig. 6A, the R6(K355N) mutant complex was found to be resistant to T157602, with an IC50 of >100 μM. A nonspecific inhibitor of helicase function (T160715) exhibited the same level of inhibition on both the mutant and wild-type enzyme complexes (Fig. 6B). T160715 was identified as a nonspecific inhibitor since it also inhibited HSV OBP and E. coli RecQ and DnaB helicases with IC50 values of approximately 10 μM (data not shown). In addition, the primase and ATPase activities of the complex were shown to be resistant to T157602 (data not shown). Thus, the mutant and wild-type complexes possess similar enzymatic activities but the mutant complex is selectively resistant to T157602. In summary, our biochemical data confirm the conclusion drawn from the genetic analysis, that the target of T157602 is the UL5 helicase protein.
FIG. 6.
Graphical representation of helicase activities from helicase gel assays with purified helicase-primase enzyme complexes containing either the wild-type HSV-2 UL5 or the K355N UL5 protein cloned from resistant virus R6 DNA. (A) Activity observed in both the mutant and wild-type (WT) helicase-primase complexes in the presence of various concentrations of T157602; (B) activity observed in the presence of a nonspecific inhibitor of helicase function T160715. The relative percentage of remaining activity is plotted versus the molar concentration of drug, with 100% being that activity obtained in the absence of drug.
Use of site-directed mutagenesis of the UL5 protein for identification of amino acids capable of conferring T157602 resistance.
To investigate which amino acids could substitute for lysine 355 and overcome drug sensitivity, we performed site-directed mutagenesis. As shown in Table 3, the K355N mutation was the best at producing a drug-resistant UL5 helicase, and this probably accounts for the fact that it was represented in four of the seven selected resistant mutants. Other residues that were relatively well tolerated at this position were threonine, serine, and histidine; however, in order to produce these amino acids at position 355, two bases of the lysine codon would have to mutate in tissue culture during the selection procedure, making it less likely that they would occur naturally. Large hydrophobic residues or residues with branched side chains also conferred resistance to the drug, but they were either less efficient at preventing drug interaction or less well tolerated in terms of helicase activity levels, as they consistently rescued wild-type viruses 2 to 3 logs less well than the more conservative substitutions that had shorter side chains. The methionine residue at position 354 could be changed to a cysteine residue, but valine, alanine, and leucine were not tolerated at this position.
TABLE 3.
Marker rescue of wild-type viruses with plasmids carrying mutations in the UL5 gene
| Plasmida | Site-specific mutation | Codon change | Titer of rescued viruses (avg PFU/ml) |
|---|---|---|---|
| TP201 | UL5 wild type | None | 0 |
| TP202 | 355K-N | AAG-AAT | 5.0 × 104 |
| TP227 | 355K-V | AAG-GTA | 0 |
| TP205 | 355K-T | AAG-ACG | 1.0 × 103 |
| TP206 | 355K-L | AAG-TTG | 3.0 × 102 |
| TP213 | 355K-C | AAG-TGC | 4.0 × 10 |
| TP212 | 355K-Q | AAG-CAG | 3.0 × 102 |
| TP214 | 355K-S | AAG-AGC | 1.4 × 104 |
| TP215 | 355K-I | AAG-ATA | 3.0 × 102 |
| TP216 | 355K-M | AAG-ATG | 0 |
| TP217 | 355K-G | AAG-GGG | 0 |
| TP218 | 355K-R | AAG-AGA | 2.0 × 102 |
| TP220 | 355K-D | AAG-GAC | 0 |
| TP221 | 355K-E | AAG-GAA | 0 |
| TP222 | 355K-H | AAG-CAC | 3.0 × 103 |
| TP223 | 355K-P | AAG-CCA | 0 |
| TP224 | 355K-W | AAG-TGG | 1.3 × 102 |
| TP219 | 355K-Y | AAG-TAC | 0 |
| TP208 | 354M-C | ATG-TGC | 3.0 × 103 |
| TP231 | 354M-L | ATG-TTG | 0 |
| TP209 | 354M-A | ATG-GCC | 0 |
| TP210 | 354M-V | ATG-GTC | 0 |
Transfections were performed in triplicate with 50, 100, or 200 ng of plasmid DNA and 1 μg of rHSV-2LUC DNA.
DISCUSSION
In this report, we describe an approach to the discovery of novel inhibitors of HSV replication for potential development as anti-HSV agents. Our rationale was to use a high-throughput biochemical helicase assay to screen libraries of synthetic and natural compounds for anti-HSV helicase activity. Using this strategy, we identified a 2-amino thiazole compound, designated T157602, and its analogs as a class of small molecules capable of specifically and effectively inhibiting HSV replication.
The results obtained from the genetic analysis and mapping studies are in agreement with the biochemical experiments inasmuch as both indicate that the target of T157602 is the UL5 component of the viral helicase-primase complex. The biochemical assay used in the screening process contained purified HSV helicase-primase complex, which comprised only three viral components UL5, UL8, and UL52. In order to elucidate the mechanism of action of T157602 in vivo, it was necessary to determine its target in the context of a viral infection. To decipher how inhibition of the viral helicase in vitro relates to the inhibition of viral replication in vivo, we generated T157602-resistant mutants, mapped their mutations on the viral genome, and performed marker rescue experiments. Using this strategy, we demonstrated that single point mutations within the UL5 gene were sufficient to confer resistance to T157602 to wild-type viruses. Moreover, biochemical assays with the mutated drug-resistant UL5 protein in the context of the helicase-primase complex showed that the helicase was resistant to T157602 in vitro, confirming the genetic findings that the in vivo target of T157602 is UL5.
Drug-resistant mutants arise frequently in passaged laboratory stocks of HSV. This phenomenon has been observed for mutations in the TK gene, which is not essential for viral replication, and in the essential viral DNA polymerase gene. Estimations of the mutation frequency for HSV in the presence of ACV, Ara-A or phosphonoacetic acid have been reported to be one in 10−5 to one in 10−3, and one source of this high mutation rate is the relative infidelity of the HSV DNA polymerase (5–7, 22, 23, 26). The frequency with which drug resistance mutations arose in the UL5 gene was approximately 1 in 10−7, which is lower than the frequencies of DNA polymerase point mutations. Of the three types of resistant HSV-2 viruses obtained, one had lysine 355 mutated to an asparagine (K355N), while a second had the neighboring methionine residue at position 354 changed to a threonine (M354T); the last resistant virus that we obtained had the glutamic acid at position 399 changed to an aspartic acid residue (E399D). Resistant viruses with the K355N mutation arose more frequently, as two independently derived HSV-2 and two HSV-1 T157602-resistant viruses (four of seven) contained this mutation. This result may be explained in part by the observation that resistant viruses carrying either the K355N or the M354T substitution were able to grow more efficiently than the E399D virus at lower MOIs (0.01 PFU/cell) and in the presence of high concentrations of T157602. It is noteworthy that all resistant mutants grew equally well in the absence of the drug. These data indicate that the R3(E399D) virus was less resistant to T157602 and that other functions of the UL5 protein were not impaired by this amino acid change.
What might be the mechanism of action of T157602? Interestingly, the drug inhibits all three activities of the enzyme complex; the genetic and biochemical evidence all points to UL5 as the target. Moreover, T157602 does not act simply to block binding of either the DNA substrate or the nucleotide cofactor. Preliminary fluorescence anisotropy experiments with T157602 suggest that it stabilizes the helicase-primase-DNA complex (43a). This finding suggests a model in which the drug traps the enzyme complex on the DNA substrate, effectively blocking all three of its enzymatic activities.
All of the T157602-resistant mutations mapped to a region of the UL5 protein that is outside the six motifs conserved in most helicases. Structural studies suggest that these protein domains come together in a three-dimensional structure to form the substrate and cofactor binding cleft (29, 60, 61). As the drug does not compete for binding with ATP or DNA, it presumably binds to a region other than the highly conserved cleft. Thus, the simplest interpretation of the data is that the mutations mark residues in UL5 that may be involved directly in interactions with T157602. All of these mutations (K355N, M354T, and E399D) are relatively conservative changes, and strikingly, all shorten the length of the amino acid side chain, perhaps resulting in reduced drug binding.
Although the mutations identified are outside the highly conserved general helicase motifs, they are found, nevertheless, within regions of the UL5 protein that are highly conserved among the UL5 homologs in other herpesviruses (Fig. 7). The K355 and E399 residues are found to be completely conserved across the nine herpesviruses, while residue 354 can be either an M or an L. Direct analysis of varicella-zoster virus and CMV replication revealed that these viruses were resistant to the inhibitory effects of T157602 (data not shown). Both of these members of the herpesvirus family have an L at position 354, but this difference is unlikely to be the sole reason for their resistance to T157602, because Epstein-Barr virus is also resistant to T157602 (25a) and because, like that of HSV, the UL5 gene of Epstein-Barr virus has an M at position 354. In addition, residue 354 was mutated by site-specific mutagenesis to an L residue, and this version of the UL5 protein was not able to rescue replication of wild-type viruses in the presence of T157602. Several scenarios might explain these observations. It is possible that the tertiary structure of the binding pocket is conserved, but its primary structure is not. It is also possible that certain nonconserved residues are involved in contacting T157602 or that the shape of the pocket in which T157602 binds is not well conserved.
FIG. 7.
Comparison of amino acid sequences of the UL5 protein in the region of the T157602 resistance mutations described in this report from various members of the herpesvirus family. Completely conserved residues are indicated by boxes; positions of amino acid mutations found in T157602-resistant viruses are indicated by arrows. HHV7, human herpesvirus 7; HHV6, human herpesvirus 6; HCMV, human cytomegalovirus; HSVSA, herpesvirus saimiri; EBV, Epstein-Barr virus; VZV, varicella-zoster virus; HSVEB, equine herpesvirus 1; HSV2, herpes simplex virus type 2; HSV1, herpes simplex virus type 1.
The results presented in this study demonstrate that the HSV helicase-primase enzyme is an attractive target for the development of new HSV therapeutics. Moreover, the approach of designing biochemical assays that measure essential HSV functions and are adaptable to a high-throughput screening format is an effective and invaluable strategy for the rapid identification of novel anti-HSV agents.
ACKNOWLEDGMENTS
We thank Lisa Marshall for invaluable assistance with the high-throughput screen and Keith Williamson for sequencing our constructs. We also thank Robert Lehman and Bernard Roizman for the gifts of recombinant baculoviruses and cell lines and Kelly La Marco for editing the manuscript.
REFERENCES
- 1.Bush M, Yager D R, Gao M, Weisshart K, Marcey A I, Coen D M, Knipe D. Correct intranuclear localization of herpes simplex virus DNA polymerase requires the viral ICP8 DNA binding protein. J Virol. 1991;65:1082–1089. doi: 10.1128/jvi.65.3.1082-1089.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Calder J M, Stow N D. Herpes simplex virus helicase-primase: the UL8 protein is not required for DNA dependent ATPase and DNA helicase activities. Nucleic Acids Res. 1990;18:3573–3578. doi: 10.1093/nar/18.12.3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calder J M, Stow E C, Stow N D. On the cellular localization of the components of the herpes simplex virus type 1 helicase-primase complex and the viral origin-binding protein. J Gen Virol. 1992;73:531–538. doi: 10.1099/0022-1317-73-3-531. [DOI] [PubMed] [Google Scholar]
- 4.Chartrand P, Crumpacker C S, Schaffer P A, Wilkie N M. Physical and genetic analysis of the herpes simplex DNA polymerase locus. Virology. 1980;103:311–326. doi: 10.1016/0042-6822(80)90190-7. [DOI] [PubMed] [Google Scholar]
- 5.Coen D M. General aspects of virus drug resistance with special reference to herpes simplex virus. J Antimicrob Chemother. 1986;18:1–10. doi: 10.1093/jac/18.supplement_b.1. [DOI] [PubMed] [Google Scholar]
- 6.Coen D M. Antiviral drug resistance. Ann N Y Acad Sci USA. 1990;616:224–237. doi: 10.1111/j.1749-6632.1990.tb17843.x. [DOI] [PubMed] [Google Scholar]
- 7.Coen D M. Acyclovir resistant, pathogenic herpes viruses. Trends Microbiol. 1994;2:481–485. doi: 10.1016/0966-842x(94)90652-1. [DOI] [PubMed] [Google Scholar]
- 8.Coen D M, Aschmann D P, Gelep P T, Retondo M J, Weller S K, Schaffer P A. Fine mapping and molecular cloning of mutations in the herpes simplex virus DNA polymerase locus. J Virol. 1984;49:236–247. doi: 10.1128/jvi.49.1.236-247.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coen D M, Kosz-Vnenchak M, Jacobsen J G, Leib D A, et al. Thymidine kinase negative herpes simplex virus mutants establish latency in mouse trigeminal ganglia but do not reactivate. Proc Natl Acad Sci USA. 1989;86:4736–4740. doi: 10.1073/pnas.86.12.4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crute J J, Lehman I R. Herpes simplex virus-1 DNA polymerase. Identification of an intrinsic 5′ to 3′ exonuclease with ribonucleaseH activity. J Biol Chem. 1989;264:19266–19270. [PubMed] [Google Scholar]
- 11.Crute J J, Lehman I R. Herpes simplex virus helicase:primase. Physical and catalytic properties. J Biol Chem. 1991;266:4484–4488. [PubMed] [Google Scholar]
- 12.Crute J J, Tsurumi L, Zhu L, Weller S K, Olivio P D, Mocarski E S, Challberg M D, Lehman I R. Herpes simplex virus 1 helicase-primase: a complex of three herpes encoded gene products. Proc Natl Acad Sci USA. 1989;86:2186–2189. doi: 10.1073/pnas.86.7.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dodson M S, Lehman I R. Overexpression and assembly of the herpes simplex virus type 1 helicase-primase in insect cells. J Biol Chem. 1991;264:20835–20838. [PubMed] [Google Scholar]
- 14.Elias P, Lehman I R. Interaction of origin binding protein with an origin of replication of herpes simplex type 1. Proc Natl Acad Sci USA. 1988;85:2959–2963. doi: 10.1073/pnas.85.9.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elias P, O’Donnell M E, Mocarski E S, Lehman I R. A DNA binding protein specific for an origin of replication of herpes simplex virus type 1. Proc Natl Acad Sci USA. 1986;86:6322–6326. doi: 10.1073/pnas.83.17.6322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Falkenberg M, Bushnell D A, Elias P. The UL8 subunit of the heterotrimeric herpes simplex virus type 1 helicase primase is required for the unwinding of single strand DNA binding protein (ICP8) coated DNA substrate. J Biol Chem. 1997;272:22766–22770. doi: 10.1074/jbc.272.36.22766. [DOI] [PubMed] [Google Scholar]
- 17.Fierer D S, Challberg M D. Purification and characterization of UL9, the herpes simplex virus type 1 origin-binding protein. J Virol. 1992;66:3986–3995. doi: 10.1128/jvi.66.7.3986-3995.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gallo M L, Dorsky D I, Crumpacker C S, Parris D S. The essential 65-kilodalton DNA binding protein of herpes simplex virus stimulates the virus-encoded DNA polymerase. J Virol. 1989;63:5023–5029. doi: 10.1128/jvi.63.12.5023-5029.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gallo M L, Jackwood D H, Murphy M, Marsden H S, Parris D S. Purification of the herpes simplex virus type 1 65-kilodalton DNA-binding protein: properties of the protein and evidence of its association with the virus-encoded DNA polymerase. J Virol. 1988;62:2874–2883. doi: 10.1128/jvi.62.8.2874-2883.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gorbalenya A E, Koonin E V, Donchenko A P, Blinov V M. A novel superfamily of nucleoside triphosphate binding motif containing proteins which are probably involved in duplex unwinding in DNA and RNA replication and recombination. FEBS Lett. 1988;235:16–24. doi: 10.1016/0014-5793(88)81226-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gottlieb J, Marcy A I, Coen D M, Challberg M D. The herpes simplex virus type 1 gene product: a subunit of DNA polymerase that functions to increase processivity. J Virol. 1990;64:5976–5987. doi: 10.1128/jvi.64.12.5976-5987.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hall J D, Coen D M, Fisher B L, Weisslitz M, Almy R E, Gelep P T, Schaffer P A. Generation of genetic diversity in herpes simplex virus: an antimutator phenotype maps to the DNA polymerase locus. Virology. 1984;132:26–37. doi: 10.1016/0042-6822(84)90088-6. [DOI] [PubMed] [Google Scholar]
- 23.Hall J D, Furman P A, St. Clair M H, Knopf C W. Reduced in vivo mutagenesis by mutant herpes simplex DNA polymerase involves improved nucleotide selection. Proc Natl Acad Sci USA. 1985;82:3889–3893. doi: 10.1073/pnas.82.11.3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hernandez T R, Lehman I R. Functional interactions between the herpes simplex-1 DNA polymerase and the UL42 protein. J Biol Chem. 1990;265:11227–11232. [PubMed] [Google Scholar]
- 25.Hodgeman T C. A new superfamily of replicative proteins. Nature (London) 1988;333:22–23. doi: 10.1038/333022b0. [DOI] [PubMed] [Google Scholar]
- 25a.Kern, E. Personal communication.
- 26.Kimberlin D W, Coen D M, Biron K K, Cohen J I, Lamb R A, McKinlay M, Emini E A, Whitley R J. Molecular mechanisms of antiviral resistance. Antiviral Res. 1995;26:369–401. doi: 10.1016/0166-3542(95)00027-j. [DOI] [PubMed] [Google Scholar]
- 27.Klinedinst D K, Challberg M D. Helicase-primase complex of herpes simplex virus type 1: a mutation in the UL52 subunit abolishes primase activity. J Virol. 1994;68:3693–3701. doi: 10.1128/jvi.68.6.3693-3701.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lanzetta P A, Alvarez L J, Reinach P S, Candia O A. An improved assay for nanomole amounts of inorganic phosphate. Anal Biochem. 1979;100:95–97. doi: 10.1016/0003-2697(79)90115-5. [DOI] [PubMed] [Google Scholar]
- 29.Marians K J. Helicase structures: a new twist on DNA unwinding. Structure. 1997;5:1129–1137. doi: 10.1016/s0969-2126(97)00263-3. [DOI] [PubMed] [Google Scholar]
- 30.Marsden H S, Cross A M, Francis G J, Patel A H, MacEachran K A, Murphy M, McVey G L, Haydon D, Abbotts A, Stow N D. The herpes simplex type 1 UL8 protein influences the intracellular localization of the UL52 but not the ICP8 or POL replication proteins in virus infected cells. J Gen Virol. 1996;77:2241–2249. doi: 10.1099/0022-1317-77-9-2241. [DOI] [PubMed] [Google Scholar]
- 31.Marsden H S, McLean G W, Barnard E C, Francis G J, MacEachran K, Murphy M, McVey G, Cross A, Abbotts A P, Stow N D. The catalytic subunit of the DNA polymerase of herpes simplex type 1 interacts specifically with the C terminus of the UL8 component of the viral helicase-primase complex. J Virol. 1997;71:6390–6397. doi: 10.1128/jvi.71.9.6390-6397.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McGeoch D J, Dalrymple M A, Dolan A, McNab D, Perry L J, Taylor P, Challberg M D. Structures of the herpes simplex type 1 genes required for virus DNA replication. J Virol. 1988;62:444–453. doi: 10.1128/jvi.62.2.444-453.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McLean G, Abbots A, Parry M E, Marsden H S, Stow N D. The herpes simplex virus type 1 origin binding protein interacts specifically with the viral UL8 protein. J Gen Virol. 1994;75:2699–2706. doi: 10.1099/0022-1317-75-10-2699. [DOI] [PubMed] [Google Scholar]
- 34.Olivo P D, Nelson N J, Challberg M D. Herpes simplex virus type 1 gene products required for DNA replication: identification and overexpression. J Virol. 1989;63:196–204. doi: 10.1128/jvi.63.1.196-204.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Olivo P D, Nelson N J, Challberg M D. Herpes simplex virus DNA replication: the UL9 gene encodes an origin binding protein. Proc Natl Acad Sci USA. 1988;85:5414–5418. doi: 10.1073/pnas.85.15.5414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O’Reilly D R, Miller L K, Luckow V A. Baculovirus expression vectors: a laboratory manual. W. H. New York, N.Y: Freeman & Co.; 1992. [Google Scholar]
- 37.Parris, D. S., C. A., L. Harr, A. Orr, M. C. Frame, M. Murphy, D. J. McGeoch, and H. S. Marsden. 1988. Identification of the gene encoding the 65-kilodalton DNA binding protein of herpes simplex type 1. J. Virol. 62:894–902. [DOI] [PMC free article] [PubMed]
- 38.Parry M E, Stow N D, Marsden H S. Purification and properties of the herpes simplex virus type 1 UL8 protein. J Gen Virol. 1993;74:607–612. doi: 10.1099/0022-1317-74-4-607. [DOI] [PubMed] [Google Scholar]
- 39.Pelosi E, Hicks K A, Sacks S L, Coen D M. Heterogeneity of a herpes simplex virus clinical isolate exhibiting resistance to ACV and foscarnet. Adv Exp Med Biol. 1992;312:151–158. doi: 10.1007/978-1-4615-3462-4_15. [DOI] [PubMed] [Google Scholar]
- 40.Powell K L, Littler E, Purifoy D J M. Nonstructural proteins of herpes simplex virus. II. Major virus-specific DNA-binding protein. J Virol. 1981;39:894–902. doi: 10.1128/jvi.39.3.894-902.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Purves F C, Roizman B. The UL13 gene of herpes simplex virus 1 encodes the functions for posttranslational processing associated with phosphorylation of the regulatory protein α22. Proc Natl Acad Sci USA. 1992;89:7310–7314. doi: 10.1073/pnas.89.16.7310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roizman B, Sears A E. Herpes simplex viruses and their replication. In: Fields B N, Howley P M, Chanock R M, Melnick J L, Monath T P, Roizman B, Straus S E, editors. Fields virology. 3rd ed. Vol. 2. New York, N.Y: Lippincott-Raven; 1996. pp. 2231–2295. [Google Scholar]
- 43.Sherman G, Gottlieb J, Challberg M D. The UL8 subunit of the herpes simplex helicase-primase complex is required for efficient primer utilization. J Virol. 1992;66:4884–4892. doi: 10.1128/jvi.66.8.4884-4892.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43a.Sivaraja, M. Unpublished data.
- 44.Spaete R, Frenkel N. The herpes simplex amplicon: a new eucaryotic defective virus cloning-amplifying vector. Cell. 1982;30:295–304. doi: 10.1016/0092-8674(82)90035-6. [DOI] [PubMed] [Google Scholar]
- 45.Spaete R R, Frenkel N. The herpes simplex virus amplicon: analyses of cis acting replication functions. Proc Natl Acad Sci USA. 1985;82:694–698. doi: 10.1073/pnas.82.3.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spector D, Purves F C, King R, Roizman B. Regulation of α and γ gene expression in cells infected with herpes simplex 1. New York, N.Y: Plenum; 1993. pp. 25–42. [Google Scholar]
- 47.Stow N D. Localization of an origin of DNA replication within the TRS/IRS repeated region of herpes simplex virus type 1. EMBO J. 1982;1:863–867. doi: 10.1002/j.1460-2075.1982.tb01261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stow N D, McMonagle E C. Characterization of the TRS/IRS origin of DNA replication of herpes simplex virus type 1. Virology. 1983;130:427–438. doi: 10.1016/0042-6822(83)90097-1. [DOI] [PubMed] [Google Scholar]
- 49.Tenney D J, Hurlburt W W, Micheletti P A, Bifano M, Hamatake R K. The UL8 component of the herpes simplex virus helicase-primase complex stimulates primer synthesis by a subassembly of the UL5 and UL52 components. J Biol Chem. 1993;269:5030–5035. [PubMed] [Google Scholar]
- 50.Vlazny D A, Frenkel N. Replication of herpes simplex virus DNA: localization of replication recognition signals within defective virus genomes. Proc Natl Acad Sci USA. 1981;78:742–746. doi: 10.1073/pnas.78.2.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Walboomers J M, Ter Schagget J. A new method for the isolation of herpes simplex virus type 2 DNA. Virology. 1976;74:256–258. doi: 10.1016/0042-6822(76)90151-3. [DOI] [PubMed] [Google Scholar]
- 52.Walker J E, Saraste M, Runswicke M J, Gay N J. Distantly related sequences in the A and B subunits of ATP synthetase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weller S K, Lee K J, Sabourin D J, Schaffer P A. Genetic analysis of temperature-sensitive mutants which define the gene for the major herpes simplex virus type 1 DNA-binding protein. J Virol. 1983;45:354–366. doi: 10.1128/jvi.45.1.354-366.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weller S K, Spadaro A, Schaffer J E, Murray A W, Maxam A M, Scahffer P A. Cloning, sequencing and functional analysis of oriL, a herpes simplex type 1 origin of DNA synthesis. Mol Cell Biol. 1985;5:930–942. doi: 10.1128/mcb.5.5.930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Whitley R J. Herpes simplex viruses. In: Fields B N, Howley P M, Chanock R M, Melnick J L, Monath T P, Roizman B, Straus S E, editors. Fields virology. 3rd ed. Vol. 2. New York, N.Y: Lippincott-Raven; 1996. pp. 2297–2342. [Google Scholar]
- 56.Whitley R J, Gnann J W. Acyclovir: a decade later. N Engl J Med. 1992;327:782–789. doi: 10.1056/NEJM199209103271108. [DOI] [PubMed] [Google Scholar]
- 57.Whitley R J, Gnann J W., Jr . The epidemiology and clinical manifestations of herpes simplex virus infections. In: Roizman B, Whitley R J, Lopez C, editors. The human herpesviruses. Vol. 3. New York, N.Y: Raven Press; 1993. pp. 69–105. [Google Scholar]
- 58.Whitley R J, Kern E R, Chatterjee S, Chou J, Roizman B. Replication, establishment of latency, and induced reactivation of herpes simplex virus γ-1 34.5 deletion mutants in rodent models. J Clin Invest. 1993;91:2837–2843. doi: 10.1172/JCI116527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu C A, Nelson N, McGeoch D J, Challberg M D. Identification of herpes simplex virus type 1 genes required for origin dependent DNA synthesis. J Virol. 1988;62:435–443. doi: 10.1128/jvi.62.2.435-443.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhu L, Weller S K. UL5, a protein required for HSV DNA synthesis: genetic analysis, overexpression in Escherichia coli, and generation of polyclonal antibodies. Virology. 1988;166:366–378. doi: 10.1016/0042-6822(88)90507-7. [DOI] [PubMed] [Google Scholar]
- 61.Zhu L, Weller S K. The six conserved helicase motifs of the UL5 gene product, a component of the herpes simplex virus type 1 helicase-primase, are essential for its function. J Virol. 1992;66:469–479. doi: 10.1128/jvi.66.1.469-479.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]