Abstract
Background
Tumor heterogeneity plays essential roles in developing cancer therapies, including therapies for breast cancer (BC). In addition, it is also very important to understand the relationships between tumor microenvironments and the systematic immune environment.
Methods
Here, we performed single-cell, VDJ sequencing and spatial transcriptome analyses on tumor and adjacent normal tissue as well as axillar lymph nodes (LNs) and peripheral blood mononuclear cells (PBMCs) from 8 BC patients.
Results
We found that myeloid cells exhibited environment-dependent plasticity, where a group of macrophages with both M1 and M2 signatures possessed high tumor specificity spatially and was associated with worse patient survival. Cytotoxic T cells in tumor sites evolved in a separate path from those in the circulatory system. T cell receptor (TCR) repertoires in metastatic LNs showed significant higher consistency with TCRs in tumor than those in nonmetastatic LNs and PBMCs, suggesting the existence of common neo-antigens across metastatic LNs and primary tumor cites. In addition, the immune environment in metastatic LNs had transformed into a tumor-like status, where pro-inflammatory macrophages and exhausted T cells were upregulated, accompanied by a decrease in B cells and neutrophils. Finally, cell interactions showed that cancer-associated fibroblasts (CAFs) contributed most to shaping the immune-suppressive microenvironment, while CD8+ cells were the most signal-responsive cells.
Conclusions
This study revealed the cell structures of both micro- and macroenvironments, revealed how different cells diverged in related contexts as well as their prognostic capacities, and displayed a landscape of cell interactions with spatial information.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00262-022-03278-2.
Keywords: Breast cancer, Single-cell and spatial sequencing, Immune and stromal cell heterogeneity, Metastatic lymph nodes, Circulating immune system
Background
Tumor heterogeneity exists in all cancers. How tumor heterogeneity affects or is affected by the immune system has been a hot research topic in recent years, especially when single-cell sequencing technology was developed for cell classifications at high resolution. Generally, BC, as a molecularly diverse disease, is categorized into four subtypes (luminal A, luminal B, triple-negative (TNBC) and HER2+) based on hormone receptivity and the expression of epidermal growth factor receptor 2 (HER2), which are associated with specific histology, corresponding therapies and prognoses [1]. Nevertheless, these subtypes can be more deeply subdivided. For example, in 2011, Lehmann et al. subdivided TNBCs and corresponding cell lines into 6 groups [2], and Teschendorff et al. identified at least four distinct subtypes of estrogen receptor (ER)-negative BC [3]. Evidence has also shown that multiple subtypes can be present within a tumor [4] and even transform to each other [5]. Consistently, BC stem cells were reported to be heterogeneous [6–8] as sources for the dynamic evolution of cancer cells. All these findings confirmed the plasticity of BC cells and reflected ultrahigh heterogeneity.
Despite its high heterogeneity, BC is not traditionally considered a highly immunogenic tumor type, especially compared with melanoma and lung cancer. The causes of such immune silence are believed to derive from mechanisms downregulating immune recognition and promoting immunosuppression [9]. However, in recent decades, it has been reported that the BC tumor microenvironment, which includes a wide range of cells of both the innate and adaptive immune systems, has varying degrees of clinical relevance. TNBC and HER2 + are characterized by high tumor mutation burden (TMB) and are the most frequently infiltrated by tumor-infiltrating lymphocytes (TILs) compared to other subtypes. The abundance of TILs was a robust prognostic factor in TNBC and HER2 + patients treated with neoadjuvant therapy [10]. TILs in luminal-like subtypes were reported to be positively associated with unfavorable clinical features, such as high Ki67 levels, low ER expression and even shorter overall survival rates [10, 11]. For TIL subtypes, CD8+ T cell abundance is positively correlated with longer survival in patients undergoing surgery [12]. Furthermore, tissue-resident CD8+ T cells contribute to immunosurveillance and prolong BC patient survival [13]. Higher ratios of cytotoxic and regulatory T cells were reported to be associated with increased pathologic complete response (pCR) and better prognosis in patients receiving neoadjuvant treatment. All these observations emphasize the importance of the immune system in BC evolution and responses to related therapies.
Studying tumor heterogeneity and microenvironments inquires high-resolution technologies. Single-cell sequencing (SCS), as an advanced technology, sheds light on such studies in multiple cancers, including BC. In 2017, Woosung et al. first revealed the comprehensive tumor and immune profiles in primary BC at the single-cell RNA level, although their cell number was very limited [14]. Elham et al. reported a model of continuous T cell activation and macrophage polarization [15], revealing a dynamic pattern of the immune landscape. Peter et al. found that CD8+ tissue-resident T cells played key roles in the antitumor community and contributed to prolonging patient survival [13]. Recently, another single-cell study revealed T cell and macrophage trajectories during anti-PD1 treatment, highlighting immune responsiveness during immune checkpoint blockade [16]. These technologies also applied on the studies investigating metastasis in BC [17–19] which mostly focused on how cancer stem cells initiate and propagate metastatic tumors. Little is known about how the microenvironment altered in LNs when metastasis occurs.
Currently, our knowledge about BC is increasing dramatically. However, the tumor microenvironment has not been fully investigated, especially in the related macroenvironment and axillary LNs with or without metastasis. Our knowledge of the interactions among tumors, multiple cell types and the spatial connections of these cells is still rudimentary. The important role of the spatial distribution of cells and tumor microenvironments in the progression of BC remains elusive. In this study, we performed single-cell, VDJ and spatial transcriptome analyses on BC patients with the aim of unveiling tumor microenvironments as well as the systematic immune environment and the relationships among all cell types within.
Methods
Sample collection and clinical information
All procedures performed in this study were in accordance with the Declaration of Helsinki (as revised in 2013). Eight BC patients (B1-8) were recruited for this study. All patients were treatment-naïve. For patient B1-4, we collected samples from tumor, adjacent normal tissue, peripheral blood and axillar lymph node, where cell suspensions were obtained without sorting. For patient B5-7, we collected samples from tumor and adjacent normal tissue, where cell suspensions were obtained by CD45− sorting. Patients were placed in four groups according to their immunohistochemical results on ER, progesterone receptor (PR) and HER2. For patient B8, tumor and adjacent normal tissues were collected for spatial transcriptome analysis. Patient information details are listed in the supplementary file.
Single-cell suspension preparation, RNA library construction and Illumina sequencing
Tumor tissues, adjacent normal tissue and axillary lymph nodes obtained from resection surgeries were immediately sent to undergo the process of single-cell suspension preparation. We followed the protocol in a published article [20]. To sort CD45− cells in B5-7 patients, we used a CD45 MicroBeads kit from Milteny (130-045-801).
Single-cell 5’ RNA and T cell V(D)J libraries were generated by strictly following the user guide of Chromium Single-Cell V(D)J Reagent Kits (CG000086 Rev J) from 10X Genomics. For all libraries, the target cell number was 10,000.
The sequencing platform we used was Illumina Nova 6000 with pair end 150. Sequencing depth was set as recommended by 10X protocol, which was approximately 100G for one RNA library and 15G for one VDJ library.
Data processing
Expression matrixes were generated by Cellranger (v6.1.0) with default parameters. Cells were removed if they satisfied the following conditions: (1) RNA counts were less than 600; (2) RNA counts were greater than 98% of cells; (3) mitochondrial gene expression percentages were more than 15%. We applied the SCTransform normalization method [21] using 5000 genes as a highly variable gene set and integrated data from different batches by Seurat [22] (v4.0.4). Thirty principal components were selected for cell clustering and UMAP visualization. The resolution was set to 1.5. Cells were aligned to BLUEPRINT and ENCODE using SingleR [23] (v1.6.1). Based on the alignments and well-known cell markers, clusters were assigned to the corresponding cell groups. Then, we retrieved cell members from each cell group to perform subgroup analysis. Normalization, data integration and cell clustering were rerun for each cell group. The SCTransform and CCA methods [22] were applied to integrate nonepithelial cells in different batches. For epithelial cells and tumor cells, we employed Harmony [24] (v0.1.0) for data integration. If a subgroup highly expressed marker genes that should be uniquely expressed on other cell groups, we considered it to be a mixed cell type generated by doublets. Such subgroups were removed.
Pseudotime analysis and gene module calculation
We employed three methods to analyze cell differentiation. (1) Monocle3 [25] (v1.0.0) was used to build the pseudotime trajectory on UMAP. (2) A diffusion map algorithm [26] (package destiny v3.2.0) was also used to compute cell differentiation as supplementary evidence. (3) Directions of cell development were estimated by Velocity [27] (package velocyto.R v0.6).
Weighted gene coexpression network analysis (WGCNA) [28] was performed to retrieve coexpression gene modules (module eigengenes, MEs). Gene module scores were also obtained by the WGCNA package (v1.69) in R.
Gene set enrichment analysis and gene expression score calculation
Gene sets were obtained from MSigDB [29]. Gene set enrichment analysis between two cell groups was performed by GSEA [29]. For customized gene set comparisons among multiple cell groups, we calculated the scores of gene sets using the AUCell package [30] (v1.10.0) in R. This method was also applied to bulk RNA-seq data to calculate corresponding scores. Single gene expression score calculation was also based on the gene set constructed from the top 30 correlated genes, including the target gene itself.
Single-cell alignments to published transcriptome data
Raw count matrices of reference data were obtained from Gene Expression Omnibus (GEO), Molecular Taxonomy of BC International Consortium (METABRIC) [31] and The Cancer Genome Atlas (TCGA). RNA counts of the single-cell transcriptome were aligned to reference data by SingleR [23] (v1.6.1). The automatic annotation labels were obtained, as well as probability scores for each label. Higher probability scores represented better alignment and higher consistency.
Calculation of clonotype Morisita–Horn (MH) similarities between two T cell subgroups
MH similarities [32] were used to estimate the clonotypic similarities between the two αβ T cell subgroups. The calculation was based on the formula:
where xi is the number of cells within i clonotype among the total X cells from one group, yi is the number of cells within i clonotype among the total Y cells from the other group, and S is the number of unique clonotypes in both cell groups.
Cell interaction network construction
We randomly selected 20 k cells from the total cells to investigate cell group-to-group interactions. CellChat [33] (v1.1.0) was employed with built-in curated databases of cell surface molecule interactions. The normalized “SCT” gene count matrix was used as input to construct interaction networks.
Survival analysis
Survival analysis was based on two cohorts: METABRIC (n = 1904) and TCGA (n = 1102). Marker gene set scores were calculated using AUCell (v1.10.0). Overall survival rates were calculated using the “survival” (v3.2-11) and “survminer” (v0.4.8) packages in R.
Spatial transcriptome analysis
Spatial transcriptome technology from 10X Genomics Visium was employed to reveal the spatial heterogeneity of BCs. Space Ranger (v1.3.1) was used to generate the expression matrix. Published data [34] were also downloaded in this study. For each spot, cell group normalized counts were estimated by relative marker gene expression normalized by the average RNA counts of these marker genes in the corresponding cell type from SCS data. The normalized counts reflected the cell group relative abundance. The location correlations (colocations) of the two cell types were estimated by Spearman correlations between the log e values of their normalized counts. For high-throughput pairwise colocation estimation, cell group scores were calculated by cell2location [35] (v0.7).
Method of statistical analysis
The Wilcoxon signed-rank test was used to determine the statistical significance of data with two levels. P values of 0.05 or less were considered statistically significant. Statistical calculations were performed with R.
Results
Single-cell transcriptome analysis revealed predominant cell types in BC patients
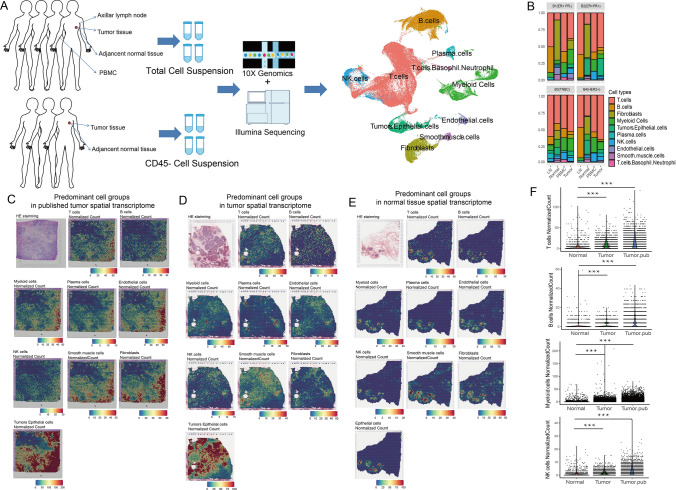
A thorough understanding of immune responses to cancer must encompass all immune cell lineages across the peripheral immune system in addition to the tumor microenvironment. In this study, we obtained 118,191 effective cells from tumor, adjacent normal tissue, PBMC and axillar LNs in 4 BC patients and 31,704 effective CD45− cells from tumor and normal tissue in 3 BC patients (Table S1). As shown in Fig. 1A, we observed multiple predominant cell clusters, including immune cells, epithelial cells, smooth muscle cells and endothelial cells, as well as fibroblasts. Top markers were shown in Table S2. Cell proportions are shown in Fig. 1B.
Fig. 1.
Single-cell and spatial transcriptome analyses revealed predominant cell types in BC patients. A Workflow of single-cell experiments. The UMAP plot shows the predominant cell types. B Histogram of predominant cell types in four patients from whom total cells were collected. Spatial normalized counts based on predominant cell type signature gene expression in the spatial transcriptome from published data (C), B8 patient tumor tissue (D) and adjacent normal tissue (E). F Scatter and violin plots comparing the immune infiltrations between tumor and normal tissue spatial transcriptome
Spatial analysis was also performed. All predominant cell types could be aligned into the spatial transcriptome using corresponding cell markers (Fig. 1C–E). Generally, spatial colocations could be observed across all cell types, suggesting a tidal interaction with each other. Immune infiltration was higher in tumor tissue than in normal tissue (Fig. 1F). Notably, tumor and epithelial cells were present in the highest abundance in the spatial transcriptome, while they were not highly obtained in the single-cell transcriptome, probably due to the single-cell preparation. Thus, an additional three BC patients were recruited, from whom CD45− cells were sorted using magnetic beads to enrich tumor and epithelial cells.
Myeloid cells exhibited microenvironment-dependent plasticity and different distributions between metastatic and nonmetastatic LNs
Hematopoietic dysregulation is common in cancers. Such dysregulation prominently expands monocytes in the periphery of patients, which traffic to the tumor microenvironment and play immune-suppressive roles [36, 37]. Comparing myeloid cells across different tissues (Fig. 2A and B, Table S3), we observed highly distinct phenotypes, indicating environment-dependent plasticity.
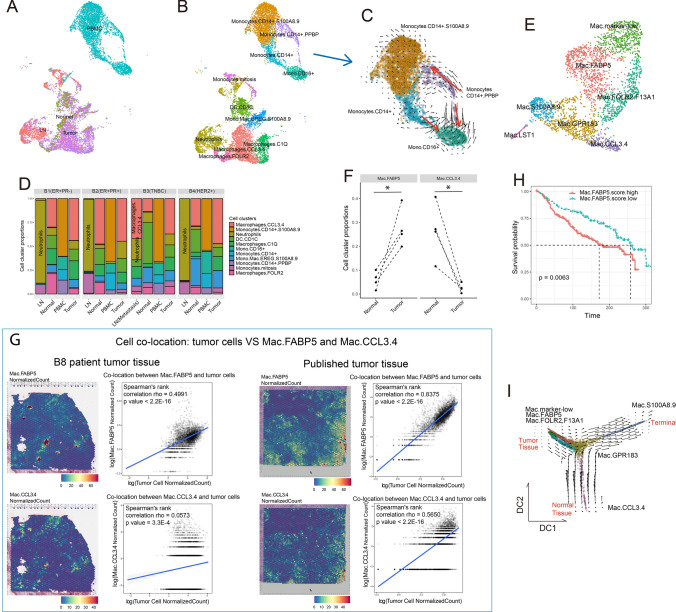
Fig. 2.
Myeloid cells exhibited microenvironment-dependent plasticity and showed clinical outcome relevance. A UMAP visualization of myeloid cell distributions based on histologic origin. B UMAP visualization of myeloid cell subtypes. C Velocity plot of the UMAP of monocytes from PBMCs. D Histogram of myeloid subtype compositions in the four patients. E UMAP visualization of LN- and tissue-infiltrated macrophage subtypes. F Paired dot plot showing cell cluster proportions between normal and tumor tissue in the four patients. G Spatial heatmaps and dotplots showing the cell colocations of tumor cells and the two groups of macrophages in tumor tissue. Cell colocation scores were estimated by Spearman correlation of normalized scores of corresponding cell groups. H Survival plot of patients from METABRIC based on Mac.FABP5 signature gene scores. I Velocity plot on the diffusion map of LN- and tissue-infiltrated macrophages
Myeloid cells were completely separated by their locations, where the differentiation path was not observed from cycling to tissue infiltration. In PBMCs, myeloid cells were primarily monocytes, which diverged along inflammatory (classical) and patrolling (nonclassical) switching paths (Fig. 2C) according to CD14 and FCGR3A (CD16) expression levels (Fig. S1), supporting the “monocyte continuum” theory [38]. Classical monocytes, also known as inflammatory monocytes, highly expressed CCR2 (Fig. S1) and were reported to be precursors of tissue-resident macrophages [39]. Additionally, we noticed that a group of macrophages highly expressed the growth factor gene PPBP, a potent cytokine and activator of neutrophils, and mostly turned into CCR2+ monocytes at high velocity (Fig. 2C), which might have recently differentiated from common myeloid precursors.
Additionally, we noticed that nonmetastatic LNs enriched a group of neutrophils (Fig. 2D), which possessed very limited marker genes and showed low total RNA abundance (Fig. S2A and B). However in the metastatic LN from B3 patient, neutrophils reduced and inflammatory macrophages infiltrated (Macrophages.CCL3.4, a group highly expressed M1 signatures as shown in Fig. S2C), suggesting the antitumor functions of macrophages in metastatic LNs.
Macrophage heterogeneity correlated with tumor-related spatial specificity and clinical outcomes
By retrieving macrophages only in LN and solid tissue (Fig. 2E, Table S4) and mapping them to the expression profiles of inactivated monocytes and tumor-associated macrophages (TAMs, GSE117970) (Fig. S3A and B), we found that Mac.CCL3.4 might play antitumor functions by expressing high M1 but low M2 signatures (Fig. S3C). Remarkably, Mac.FABP5 expressed relatively high signatures of both M1 and M2. Mac.CCL3.4 and Mac.FABP5 showed a reciprocal relationship, where Mac.CCL3.4 decreased and Mac.FABP5 increased from normal to tumor tissues (Fig. 2F and S3D). More importantly, Mac.FABP5 showed higher spatial colocalization with tumor cells than Mac.CCL3.4 (Fig. 2G). Such evidence suggested that macrophage polarization was heavily dependent on the environment, where Mac.CCL3.4 was tissue-resident and Mac.FABP5 was tumor-associated. CCL3 and CCL4 proteins are known as pro-inflammatory chemokines [40], while FABP5 played roles in fatty acid metabolism and was reported to facilitate tumor growth in many cancers [41, 42]. Using WGCNA, we located the gene module correlating with Mac.FABP5 (Fig. S4A and B). High expression of this gene module predicted worse survival (Fig. 2H).
By combining RNA velocity and pseudotime analysis, we also uncovered the macrophage evolution path (Fig. 2I and S4C). Mac.marker-low was the initial and neutrophil-like type, which developed into Mac.FABP5 and Mac.FOLR2.F13A, a group of macrophage reported to be associated with T cell infiltration [43], in tumor tissue. Then, both clusters transformed into Mac.GPR183 (GPR183 was reported to promote macrophage migration [44]), which could also originate from Mac.CCL3.4, a cluster of normal tissue-resident macrophages. Finally, Mac.GPR183 evolved into Mac.S100A8.9, the terminal of macrophage polarization. In previous studies, S100A8/A9 expressions on macrophages were report to promote tumor growth through several mechanisms [45, 46].
Cytotoxic T cells in tumor sites evolved in a separate path from those in the circulatory system
In total, we obtained 18 subtypes in cytotoxic cells, including CD8+ T cells and natural killer (NK) cells (Fig. 3A and S5A, Table S5). Cells could be grouped by tissue types in UMAP (Fig. 3B). Most innate lymphoid cells (ILCs), including NK cells, natural killer T cells (NKT) and gamma delta T cells (GDTs), were observed in PBMCs, while CD8+ effector memory (EM) cells were more widely distributed in tumor and normal tissues than in LNs and PBMCs, except for metastatic LNs in B3 patients (Fig. S5B).
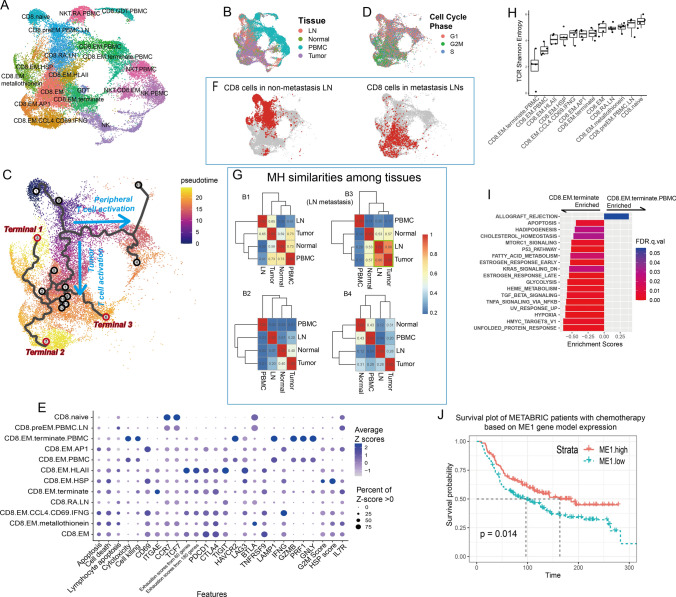
Fig. 3.
Cytotoxic cell heterogeneity. A UMAP visualization of cytotoxic cell subgroups. B UMAP visualization of cytotoxic cells labeled by histologic origin. C Pseudotime trajectories on UMAP. D UMAP visualization of cytotoxic cells labeled by cell cycle phases. E Dot plot of features from CD8 + T cell subgroups. Pathway signature genes were collected from REACTOME. Gene scores were calculated based on the expression of the top 30 positively correlated genes. F UMAP visualization of CD8 + T cells in nonmetastatic and metastatic LNs. G Heatmaps of CD8 + cell MH similarities among tissues. H Boxplot of TCR Shannon entropy in each CD8 + subgroup. I Hallmark pathway enrichment plot comparing two CD8 + terminal effector cell groups. J Survival plot of METABRIC patients with chemotherapy based on ME1 gene model expression
For CD8+ T cells, we observed two separated lineages constructed by pseudotime analysis (Fig. 3C). One evolved along the activation of peripheral T cells, while the other evolved along tumor-infiltrating T cells. T cells along the tumor infiltration path exhibited extremely high proliferation rates, where most effector cells were in the G2 or M phase of the cell cycle (Fig. 3D and S6A), especially in the CD8.EM.AP1 subtype. Three terminals were observed in this path (Fig. 3C). Terminal 1 highly expressed heat shock protein (HSP) genes, suggesting that they were undergoing stress stimulation, which was possibly caused by the preparation of the cell suspension. Terminal 2 was observed on cells highly expressing the AP1 family, mitosis and effector genes such as IFNG (Fig. 3E), suggesting that they were activated T cells, which perform killing functions and undergo cell proliferation. Based on their RNA velocity, these cells were turned into CD8.EMs (Fig. S6B), whose checkpoint inhibitor genes were upregulated accompanied by downregulated IFNG. Terminal 3, mostly occupied by CD8.EM.terminate cells, represented terminal effectors at the end of T cell activation. Compared to CD8.EM cells, they upregulated cytotoxic genes such as GZMB and GNLY (Figs. 3E and S6C) and enriched the inflammatory response pathway (Fig. S6D). The hypoxia pathway was also enriched in these cells, which further demonstrated their deeper tumor infiltration [47, 48] compared to the nonterminal effector cells. Notably, these cells had higher expression of ITGAE (Fig. 3E), indicating that a considerable portion of CD8.EM.terminate cells were tissue-resident memory T cells (Trms), which were described as highly exhausted in many tumors. However, these cells were not the most exhausted subtype. Instead, a group of EMs (CD8.EM.HLAII) with high expression of HLA II genes and possibly to be antigen-presenting T cells [49] were among the most exhausted (Fig. 3E).
CD8+ TCR tracking confirmed common neo-antigens existence across tumor and metastatic LNs
TCR is helpful in tracking clonotype expansion. We noticed that CD8+ cells in metastatic LNs were mostly effectors and undergoing T cell exhaustion, while in nonmetastatic LNs they were less activated (Fig. 3F). TCR clonotypes of CD8+ cells from the metastatic LN were in higher similarity (MH = 0.86) with CD8+ cells in the corresponding tumor, when compared to those between nonmetastatic LNs and tumors (MH = (0.65, 0.20, 0.28), Figs. 3G and S7A). These results indicated that tumor cells in the primary site and metastatic LNs provoked very similar clonotypes of cytotoxic T cells, suggesting that common neo-antigens existed across primary and metastatic sites, and targeting the those antigens could be helpful to tumor clearance in both primary and metastatic sites in adaptive immune therapy.
Similar to clonotypes in nonmetastatic LNs, PBMC clonotypes also showed low overlap with tumor-infiltrating T cells, except for B1 patients (Figs. 3G and S7A). Focusing on the highest four shared clonotypes in PBMCs and tumors of B1 (Fig. S7B–D), we found that three out of four were activated along the PBMC path and then ended up in terminal effectors in tumors, providing evidence that T cells could be activated in peripheral blood possibly by circulating tumor antigens, then infiltrate into the tumor site and perform effector functions.
Great differences existed between T cells in PBMCs and in tumor tissue. Tumor T cells, including CD8.EM and CD8.EM.terminate, were expanding, resulting in a lower TCR diversity than recently active and naïve cells (Fig. 3H). However, their diversities were much lower than those of effectors in PBMCs. The effector cytokine genes and cell-killing abilities in tumor CD8+ T cells were also significantly lower than those in PBMCs and even lower than those in recently activated T cells in LNs (Fig. 3E). Pathway enrichment analysis showed that although some antitumor pathways were observed in tumor effectors compared to PBMC T cells, they mostly enriched dysfunctional signaling pathways, such as hypoxia (response to a low oxygen environment and enhancing T cell infiltration [47, 48]), TGFB and MTORC1 signaling (exert prometastatic effects [50] and T cell autophagy-induced immunosenescence [51]) and apoptosis (Fig. 3I).
Activated CD8+ T cell showed high tumor specificity and their stemness correlated with survival
By WGCNA, we found three gene modules (Fig. S8A) that were significantly correlated with CD8+ T cell differentiation. ME1 correlated with tumor CD8+ cell activation, while ME2 and ME3 correlated with peripheral activation and CD8+ T cell stemness, respectively (Fig. S8B–D). Consistently, the spatial transcriptome indicated that CD8+ cells with ME1 signatures were more highly enriched and showed higher colocalization with tumor cells than cells with ME2 and ME3 (Fig. S8F–H). Overall, ME3 signatures were significantly associated with clinical outcomes (Fig. S8E), indicating that the accumulation of naïve CD8+ T cells suppressed tumor growth. However, in patients with chemotherapy, ME1 predicted improved patient survival (Fig. 3J), indicating that infiltrating CD8+ T cell responses to antigens released by chemotherapy could help to maintain long-term tumor resistance after surgery.
CD4+ T cells in metastatic LNs showed high consistency to those in tumors
CD4+ subtypes also showed tissue preference (Figs. 4A, B and S9A, Table S6). CD4+ Th1/17 cells could be observed in both normal and tumor tissue performing antitumor functions, while Treg cells were highly enriched in tumor-induced immune suppression. Meanwhile, follicular helper T cells (TFHs) and naïve cells were mostly observed in LNs and PBMCs.
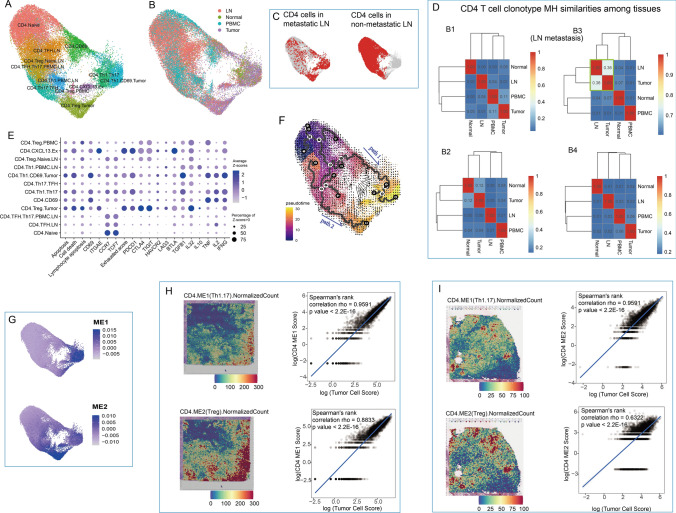
Fig. 4.
CD4+ T cell heterogeneity. A UMAP visualization of CD4+ T cell subgroups. B UMAP visualization of CD4+ T cells labeled by histologic origin. C UMAP visualization of CD4+ T cells in metastatic and nonmetastatic LNs. D Heatmaps of CD4 T cell clonotype MH similarities among tissues in B1-4 patients. E Dot plot of features from CD4 T cell subgroups. Pathway signature genes were collected from REACTOME. Gene scores were calculated based on expressions from top 30 positively correlated genes. F Cell differentiation path on UMAP combined by RNA velocity and pseudotime trajectories. G UMAP visualization of two gene model scores generated by WGCNA. Cell colocations between tumor cells and two groups of CD4 cells in tumor tissue in published data (H) and B8 patient (I). Cell colocation scores were estimated by Spearmen correlation of normalized scores of corresponding cell groups
Similar to CD8+ cells, CD4+ cells in metastatic LNs displayed significant phenotypic differences to those in nonmetastatic LNs (Fig. 4C). In metastatic LNs, CD4+ cells majorly differentiated into effector regulatory T cells (Tregs) and Th1/17, which showed high consistency to CD4+ cells in tumor. In line with this, TCR clonotypes in metastatic LNs were in higher similarity to tumor compared to those in nonmetastatic LNs (Fig. 4D), also providing evidence of common antigen existence across tumor sites and metastasis LNs.
In general, CD4+ cells did not evolve at fast rates based on RNA velocity. In line with this observation, CD4+ cells did not show fast clonotype expansions compared to CD8+ cells, thus displaying a relatively high TCR diversity (Fig. S9B), which is also consistent with previous studies [20, 52]. Tregs in PBMCs have extremely low TCR diversity due to their very limited cell number. The second lowest diversity was observed in a group of exhausted cells (Fig. S9C). Notably, in addition to exhaustion markers, they also highly expressed ITGAE (Fig. 4E), a tissue-resident cell marker, and CXCL13, an exhaustion marker [27] correlating with improved survival and immune cell tumor localization [53].
In CD4+ cell activation, two separate paths were observed (Fig. 4F), connecting the circulating system and solid tissues. One represented Th1.Th17 differentiation, while the other represented Treg activation, suggesting that in the circulatory system, CD4+ T cells already diverged before they infiltrated into tumors. In addition, by WGCNA, we obtained two gene modules (Fig. 4G), among which ME2 was Treg-specific and showed a negative correlation with overall survival, as expected [54] (Fig. S9D). ME2 also showed higher expression than ME1 but less tumor cell colocalization (Fig. 4H and I), suggesting that Th1.17 cells were more tumor-specific, while Tregs were more broadly distributed and with higher abundance.
Only a small proportion of B cells infiltrated the tumor to perform antitumor functions but also showed exhausted status
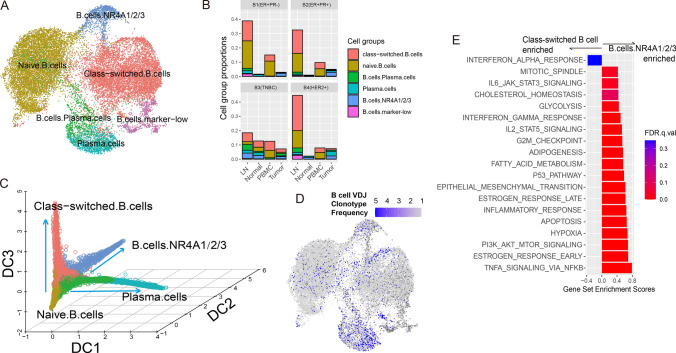
We obtained 6 subtypes of B cells (Fig. 5A, Table S7), most of which were found in LNs except the metastasis subtype in B3 patients (Fig. 5B). The most predominant subtypes were naïve and class-switched B cells, which could be identified by the expression of the IG family (Fig. S10). As shown in Fig. 5C, three B cell differentiation paths were observed, originating from naïve B cells to plasma cells (diffusion component 1, DC1), tumor-infiltrated exhausted B cells (DC2) and class-switched B cells (DC3). Of all subtypes, only plasma cells showed relatively high clonotype expansion (Fig. 5D).
Fig. 5.
B cell heterogeneity. A UMAP visualization of B cell subgroups. B Proportion B cell subgroups under total cells in related tissues from four patients. C Diffusion map 3D visualization showing B cell differentiation paths. D UMAP visualization of B cell clonotype frequency. E Hallmark pathway enrichment plot comparing two B cell subgroups
B.cells.NR4A1/2/3 were mostly tumor-infiltrating B cells. They highly expressed CD83 (Fig. S10), an activated B cell marker, which was reported to promote toxoid-stimulated B cell proliferation, dendritic cell-mediated T cell proliferation and the expression of IFNG and IL17A [55]. Gene set enrichment analysis (Fig. 5E) revealed that compared to class-switched B cells, B.cells.NR4A1/2/3 enriched the inflammatory response and performed antitumor activities through TNFA signaling, IL2 signaling and the IFN-γ response. However, high enrichment of hypoxia and apoptosis pathways in these subtypes demonstrated their exhaustion status. Although this subtype did not show prognostic relevance, the expression of the IG family predicted patient survival (Fig. S11), suggesting that the contribution of B cells to the antitumor community [56, 57] might be related to their expression levels of specific antigens rather than cell abundance.
Fibroblast heterogeneities correlated with patient survival
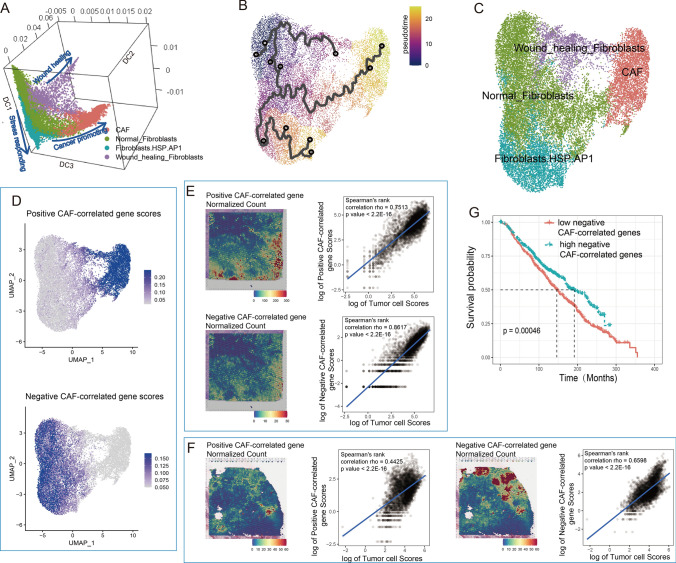
Fibroblasts were the most abundant nonimmune cells we found in both tumor and normal tissues. Based on their differentiation path (Fig. 6A and B), we clustered them into 4 subgroups: normal fibroblasts, cancer-associated fibroblasts (CAFs), wound-healing fibroblasts and stress-responsive fibroblasts that highly expressed heat shock proteins and AP-1 family genes (Fig. 6C, Table S8).
Fig. 6.
Fibroblast heterogeneity. A UMAP visualization of fibroblast subgroups. B Diffusion map 3D visualization showing fibroblast differentiation paths. C Pseudotime trajectory of fibroblast differentiation. D UMAP visualization of expression scores from positively and negatively correlated genes along with CAF development. Spatial heatmaps and dotplots showed cell colocations between tumor cells and two groups of CD4 + cells in tumor tissue in published data (E) and B8 patient (F). Cell colocation scores were estimated by Spearman correlation of normalized scores of corresponding cell groups. G Survival plot of METABRIC patients based on negative CAF-correlated gene scores
CAFs were found to be highly enriched in tumor tissue and showed extremely high similarity to those in published data (GSE29270, Fig. S12A and B). They widely expressed collagens and MMP11 (Fig. S12C), a gene recently found to hinder CD8+ T cell infiltration and significantly shorten survival [58, 59], as well as a series of genes maintaining CAF activities and modulating the tumor EMT process, such as FN1 [60], SPARC [61], and CTHRC1 [62].
Normal fibroblasts could be the origin of CAFs. They were observed in both tumor and adjacent normal tissue (Fig. S12A) and highly expressed antitumor genes, such as CCL2, TNFAIP6 and IRF1, as well as TAC1, an inducer of vasodilators and secretagogs with antimicrobial properties. However, the cell proliferation-suppressive gene BTG2 was upregulated in these cells, indicating their quiescent status and the limited pro-inflammatory functions.
The other two branches of fibroblast development were wound-healing and stress-responding fibroblasts (Fig. 6B and C). The former highly expressed extracellular matrix organization- and wound-healing-related genes, such as CCN5, TNXB, IGFBP5 and IGFBP6, representing regular responses to tissue damage. The latter highly expressed heat shock protein genes and AP1 family genes, exhibiting stress responses possibly caused by experimental treatment.
By WGCNA, we identified a gene module that was significantly correlated with developmental changes from normal fibroblasts to CAFs (Fig. 6D). Combined with the tumor spatial transcriptome (Fig. 6E and F), we found that CAFs showed less colocalization trends than normal fibroblasts, suggesting a wider distribution and less tumor specificity and inferring that CAFs might have wider communications with nontumor cells and play extensive roles in forming a suppressive microenvironment. Consistently, negatively correlated genes in this model predicted better overall survival (Fig. 6G), confirming the contribution of CAFs to tumor progression and normal fibroblasts to tumor suppression.
Cell interactions shaped an immunodeficient microenvironment and influenced the circulatory system
Before cell interaction analysis, we identified tumor cells with abnormal copy number alterations (CNAs) in chromatin (Fig. S13A). By comparing these cells to METABRIC transcriptome data, we found high consistency between our cancer type annotations and claudin subtype classifications from METABRIC (Fig. S13B), confirming that single-cell transcriptome signatures were in line with bulk RNA sequencing data and providing typical features from different BC subtypes (Fig. S13C and D).
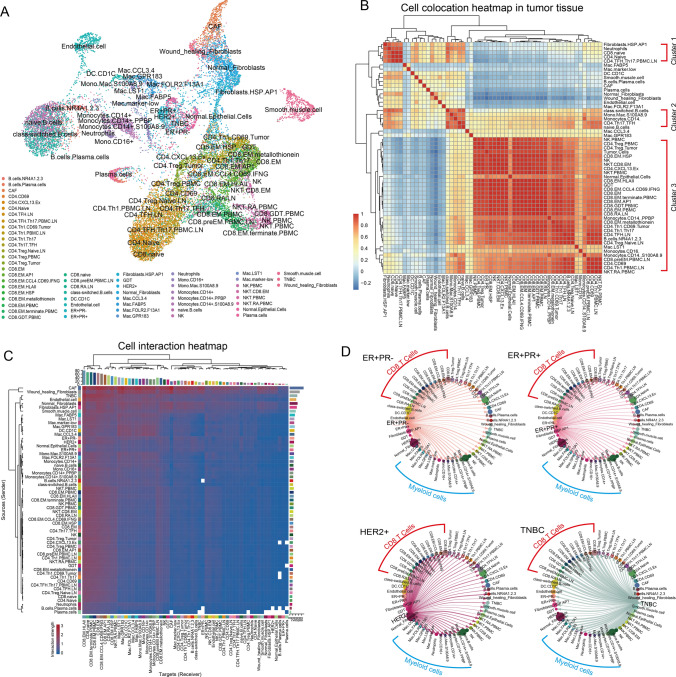
With 59 groups of cells we identified (Fig. 7A), the landscape of tumor microenvironment was constructed by cell colocations and interactions (Fig. 7B and C). Three major clusters could be observed in the heatmap of cell colocations (Fig. 7B). Cluster 1 could be represented by naïve T cells, neutrophils and TFHs, which helped the maturation of T cells. Cluster 2 could be represented by naïve B cells, class-switched B cells, TFHs, and monocytes, which were related to the maturation of B cells. Cluster 3 mostly occupied by NK cells, T cells and tumor cells, representing the terminal immune responses against the cancer.
Fig. 7.
Cell interactions. A UMAP visualization of all cell groups annotated in this research. B Heatmap of cell-type colocations. C Cell interaction heatmap in tumor tissue. Rows represent the senders (ligand genes), and columns represent the receivers (receptor genes). D Cell interactions sent from tumor cells in different BC subtypes. Dot size represents the cell numbers of each cell group. The width of the directed lines represents the interaction numbers observed between cell groups from the nodes of each end
From the cell interaction network, CAFs were found to contribute the most to outgoing roles (Fig. S14A and B), sending a series of signaling molecules to trigger biological responses in other cells. CAFs also did not show high colocation with other cell types (Fig. 7B), suggesting they widely distributed in tumor site. Most of the molecules CAF sent were reported to promote tumor growth and shorten survival, such as CXCL12-CXCR4 signaling targeting CD8+ effector cells to attenuate their infiltration [63], LAMA4-CD44 targeting CD4+ cells to inhibit Th1 and Th17 proliferation but favor Treg induction [64], PTN-NCL signaling targeting tumor cells to mediate their growth and angiogenesis [65], and THY1-ITGB2 signaling myeloid cells that secrete exosomes to tumor cells facilitating metastasis [66] (Fig. S14C). Conversely, CAFs could inhibit apoptosis of CAFs themselves through FN1/COL1A1-SDC1 signaling and maintain fibroblast stemness by PERIOSTIN signaling, consolidating normal fibroblast to CAF transaction. However, we also found some signaling pathways that may help tumor clearance in CAFs, such as ANXA1-PFR1 signaling targeting myeloid cells to facilitate the phagocytic uptake of dead-cell antigens, thus enhancing their immunogenicity [67].
Tumor cells were found to have impacts on all type of cells especially the CD8+ T cells and myeloid cells mechanically and spatially (Fig. 7B and C). TNBC cells, as the BC subtype with the highest immunogenicity, were found to be the most involved in cell interactions and served primarily outgoing roles compared to the others (Figs. 7C and S14B). In TNBC, tumor cells highly interacted with CD8+ T cells and myeloid cells (Figs. 7C and S15A). TNBC tumor cells could recruit CD8+ T cells and myeloid cells through the CX3CL1-CX3CR1 axis, facilitating immune infiltration [68]; TNBC tumor cells expressed higher levels of HLA-I molecules, resulting in higher antigen-presenting capability. Conversely, TNBC tumor cells helped transfer myeloid cells into suppressive cells via JAG1-NOTCH2 [69] and IL34-CSF1R [70]. MIF, an immunosuppressive factor [71], was observed to be expressed at higher levels in TNBC tumor cells targeting both CD8+ T cells and myeloid cells. Collectively, these observations explained the double faces of TNBC: higher immunogenicity and immune infiltration but also a more suppressive microenvironment.
Unsurprisingly, there were many other cells that helped construct an immune-suppressive microenvironment, facilitated tumor growth and even affected the circulating immune system. For example, through VEGF signaling, a group of cell types mediate angiogenesis and the immunosuppressive tumor microenvironment [72], including macrophages, DCs and smooth muscle cells (Fig. S15B). Wound-healing fibroblasts expressed CSF1 to stimulate both tissue-resident and cycling myeloid cells into suppressive cells (Fig. S15C). M2-like macrophages Mac.FABP5 expressed LGALS9 targeting both micro- and macroenvironmental immune cells by cell–cell interactions or exosomal delivery to modulate systematic immunodeficiency [73, 74] (Fig. S15D).
T cells showed high colocation with tumor cells and also were the most signal responder cells, especially CD8+ cells (Figs. 7B, S14B and S16A). For example, B cells targeted both CD8+ cells by CD22 and enhanced the interactions through a positive feedback loop, which was associated with better prognosis [75]. Through SPP1 signaling, macrophages control CD8+ cell activation and tumor immune evasion [76]. Through SELE signaling, endothelial cells mediate the accumulation of CD8+ T cells at sites of inflammation. In CD4 + cells, except for LAMININ and CD22 signaling, as mentioned above, FASL-FAS, a cell death signaling pathway, was observed in NK cells in PBMCs targeting Tregs in tumors (Fig. S16B), suggesting that PBMC NK cells might help suppress the activities of Tregs in tumors. Thus, facilitating NK-cell infiltration could help relieve immune suppression.
In general, cell subtypes were highly interactive with each other. Fibroblasts, macrophages and tumor cells were the most active contributors to forming an immune-suppressive microenvironment and affecting the macroenvironment, while CD8+ T cells were the most interactive responders affected by the microenvironment to reduce cytotoxicity.
Discussion
In this study, we performed single-cell and spatial transcriptome analyses on tumor microenvironments and systematic macroenvironments and found heterogeneities and tidal connections in all types of cells from both solid tissue and the circulating system in BC. BCs are considered to possess low immunogenicity and mutation burdens. In such a context, immune infiltration and activation are considerable key factors in defining disease etiology and treatment responses. Here, we identified multiple cell types in high resolution with spatial information and discovered how these cells evolved in different environments in BC. It is important to elucidate the evolutionary differences between immune cells in the circulatory system and the tumor microenvironment. The response for targeted and immune therapies would be different for cells in different niches. Understanding such heterogeneity could help to predict responses and side effects of related therapies.
With spatial information, we investigated tumor spatial specificity for each cell group. Activated CD8+ cells showed higher tumor spatial specificity than naïve CD8+ cells, consistent with their antitumor function. Interestingly, we found some cell groups, which induced immune deficiency, had less tumor spatial specificity, for example Tregs and CAFs. These results suggested they could have board effects on other cells especially immune cells to construct immune suppressive environment. On the contrary, a TAM-like cell group Mac.FABP5 possessing both M1 and M2 signature, showed higher tumor spatial specificity than pro-inflammatory macrophages, indicating these cells might be induced by tumor cells directly. Such analyses improved our understanding of how tumor cells interact with others and how tumor microenvironment established.
LNs are essential in connecting solid tissue with the systematic immune environment. LN metastasis has shown high clinical outcome relevance in BC. Recently, K. Xu et al. identified cancer stem cells evolved into metastatic clusters and infiltrated into lymph nodes [17]. However, how microenvironment changes in metastatic LNs is less elucidated. It was reported only in colorectal cancer that metastatic LNs showed significantly lower TCR diversity and higher frequencies of tumor-shared TCRs than nonmetastatic LNs [77], which was highly consistent with our observation in BC. In addition, we observed that the functions of T cells in metastatic LNs also showed high similarities to those of tumor-infiltrating T cells. Concomitantly, pro-inflammatory macrophages were significantly upregulated in metastatic LNs, accompanied by a decrease in B cells and neutrophils. The microenvironment of metastatic LNs was changed toward a tumor-like status. Given that LNs are considered a tissue to activate antitumor immunity, where antigen-specific effector T cells are generated, this observation supported the high possibility that TCR-sharing T cells in metastatic LNs and tumor sites were tumor antigen-specific. Previous studies showed that infiltration of total CD8+ T cells was confounded by the presence of bystander T cells [78, 79]. Considering TCR clonotypes with co-occurrence in both tumor site and metastatic LNs could be a useful way to distinguish the true effector cytotoxic T cells from the bystanders, which is important for developing personalized TCR-T therapies.
There are two prevailing and mutually inclusive theories describing tumor genesis [80]: somatic mutation theory and tissue organization field theory. The former is emphasized by tumor cells and their driver mutations that cause gain or loss of tumor-related genes. The latter describes cancer as a systematic disease that is caused by the dynamic and reciprocal relationship between cancer cells and their microenvironment, where stromal cells mostly contribute to restraining or promoting tumorigenesis. Here, we confirmed that CAFs send most molecular signals to other cells, acting as a primary key regulator in forming the tumor microenvironment. To date, there are no specific markers that can clearly distinguish CAFs from normal, resting fibroblasts [81]. Analogously, we observed a continuous path for fibroblasts differentiating into CAFs, where no clear edge could clearly separate CAFs from other fibroblasts. In addition, wound-healing fibroblasts are also involved in tumor progression, while normal fibroblasts might contribute to antitumor function. So far, CAF targeted therapy has not been clinically successful, in part due to a lack of understanding of CAF heterogeneity and function [82]. Our data gained insight into the heterogeneities of tumor fibroblasts and revealed related differentiation paths as well as their communication with tumor cells and immune microenvironments, which could help us to understand and develop CAF targeted therapy.
Conclusion
This study revealed the cell structures of both micro- and macroenvironments, revealed how different cells diverged in related contexts as well as their capacities for prognosis and provided useful information for developing targeted and immune therapies for BC.
Supplementary Information
Below is the link to the electronic supplementary material.
Authors' contributions
XM constructed sequencing libraries, performed data analysis and wrote the manuscript. DZ collected the samples. KL, BZ, SY, ChulingZ and ChunguoZ prepared single cell suspensions. JG, FL and SF helped to revise the manuscript. LZ, PC and GY helped to communicate with patients. GC and WL supervised this study.
Funding
This work was funded by grants from the National Natural Science Foundation of China (81972335, 82103347), Basic and Applied Basic Research Fund of Guangdong Province (2019A1515110677, 2019A1515110676) and Foshan City Climbing Peak Plan (2019A004), Medical Engineering Technology Research and Development Center of Immune Repertoire in Foshan.
Availability of data and materials
All data generated in this study have been uploaded to Genome Sequence Archive (GSA, https://ngdc.cncb.ac.cn/) with accession number HRA002051 and OMIX001111 under the project of PRJCA008495. Supplementary files were listed as below: Supplementary patient information file: Supplementary Table file: Table S1–S8; Supplementary Figure file: Figure S1–S16.
Declarations
Competing interests
The authors declare no potential conflict of interest.
Ethics approval and consent to participate
This study was approved by the independent ethics committee at The First People’s Hospital of Foshan. All patients provided written informed consent to participate in the study.
Consent for publication
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xiaofan Mao, Dan Zhou, Kairong Lin, and Beiying Zhang have contributed equally to this work.
Contributor Information
Guoqiang Chen, Email: 13929981788@139.com.
Wei Luo, Email: luowei_421@163.com.
References
- 1.Watkins EJ. Overview of breast cancer. JAAPA: Off J Am Acad Phys Assist. 2019;32:13–17. doi: 10.1097/01.JAA.0000580524.95733.3d. [DOI] [PubMed] [Google Scholar]
- 2.Lehmann BD, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Investig. 2011;121:2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Teschendorff AE, Miremadi A, Pinder SE, Ellis IO, Caldas C. An immune response gene expression module identifies a good prognosis subtype in estrogen receptor negative breast cancer. Genome Biol. 2007;8:R157. doi: 10.1186/gb-2007-8-8-r157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jordan NV, et al. HER2 expression identifies dynamic functional states within circulating breast cancer cells. Nature. 2016;537:102–106. doi: 10.1038/nature19328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roarty K, Pfefferle AD, Creighton CJ, Perou CM, Rosen JM. Ror2-mediated alternative Wnt signaling regulates cell fate and adhesion during mammary tumor progression. Oncogene. 2017;36:5958–5968. doi: 10.1038/onc.2017.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu S, et al. Breast cancer stem cells transition between epithelial and mesenchymal states reflective of their normal counterparts. Stem Cell Rep. 2014;2:78–91. doi: 10.1016/j.stemcr.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Visvader JE, Stingl J. Mammary stem cells and the differentiation hierarchy: current status and perspectives. Genes Dev. 2014;28:1143–1158. doi: 10.1101/gad.242511.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wahl GM, Spike BT. Cell state plasticity, stem cells, EMT, and the generation of intra-tumoral heterogeneity. NPJ Breast Cancer. 2017;3:14. doi: 10.1038/s41523-017-0012-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bates JP, Derakhshandeh R, Jones L, Webb TJ. Mechanisms of immune evasion in breast cancer. BMC Cancer. 2018;18:556. doi: 10.1186/s12885-018-4441-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denkert C, et al. Tumour-infiltrating lymphocytes and prognosis in different subtypes of breast cancer: a pooled analysis of 3771 patients treated with neoadjuvant therapy. Lancet Oncol. 2018;19:40–50. doi: 10.1016/S1470-2045(17)30904-X. [DOI] [PubMed] [Google Scholar]
- 11.Criscitiello C, et al. Tumor-infiltrating lymphocytes (TILs) in ER+/HER2− breast cancer. Breast Cancer Res Treat. 2020;183:347–354. doi: 10.1007/s10549-020-05771-7. [DOI] [PubMed] [Google Scholar]
- 12.Mahmoud SM, et al. Tumor-infiltrating CD8+ lymphocytes predict clinical outcome in breast cancer. J Clin Oncol: Off J Am Soc Clin Oncol. 2011;29:1949–1955. doi: 10.1200/JCO.2010.30.5037. [DOI] [PubMed] [Google Scholar]
- 13.Savas P, et al. Single-cell profiling of breast cancer T cells reveals a tissue-resident memory subset associated with improved prognosis. Nat Med. 2018;24:986–993. doi: 10.1038/s41591-018-0078-7. [DOI] [PubMed] [Google Scholar]
- 14.Chung W, et al. Single-cell RNA-seq enables comprehensive tumour and immune cell profiling in primary breast cancer. Nat Commun. 2017;8:15081. doi: 10.1038/ncomms15081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Azizi E, et al. Single-cell map of diverse immune phenotypes in the breast tumor microenvironment. Cell. 2018;174:1293–1308. doi: 10.1016/j.cell.2018.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bassez A, et al. A single-cell map of intratumoral changes during anti-PD1 treatment of patients with breast cancer. Nat Med. 2021;27:820–832. doi: 10.1038/s41591-021-01323-8. [DOI] [PubMed] [Google Scholar]
- 17.Xu K, et al. Single-cell RNA sequencing reveals cell heterogeneity and transcriptome profile of breast cancer lymph node metastasis. Oncogenesis. 2021;10:66. doi: 10.1038/s41389-021-00355-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lawson DA, et al. Single-cell analysis reveals a stem-cell program in human metastatic breast cancer cells. Nature. 2015;526:131–135. doi: 10.1038/nature15260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davis RT, et al. Transcriptional diversity and bioenergetic shift in human breast cancer metastasis revealed by single-cell RNA sequencing. Nat Cell Biol. 2020;22:310–320. doi: 10.1038/s41556-020-0477-0. [DOI] [PubMed] [Google Scholar]
- 20.Mao X, et al. Single-cell transcriptome analysis revealed the heterogeneity and microenvironment of gastrointestinal stromal tumors. Cancer Sci. 2021;112:1262–1274. doi: 10.1111/cas.14795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hafemeister C, Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019;20:296. doi: 10.1186/s13059-019-1874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stuart T, et al. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aran D, et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol. 2019;20:163–172. doi: 10.1038/s41590-018-0276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Korsunsky I, et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods. 2019;16:1289–1296. doi: 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cao J, et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature. 2019;566:496–502. doi: 10.1038/s41586-019-0969-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haghverdi L, Buttner M, Wolf FA, Buettner F, Theis FJ. Diffusion pseudotime robustly reconstructs lineage branching. Nat Methods. 2016;13:845–848. doi: 10.1038/nmeth.3971. [DOI] [PubMed] [Google Scholar]
- 27.La Manno G, et al. RNA velocity of single cells. Nature. 2018;560:494–498. doi: 10.1038/s41586-018-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005 doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- 29.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aibar S, et al. SCENIC: single-cell regulatory network inference and clustering. Nat Methods. 2017;14:1083–1086. doi: 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Curtis C, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hindley JP, et al. Analysis of the T-cell receptor repertoires of tumor-infiltrating conventional and regulatory T cells reveals no evidence for conversion in carcinogen-induced tumors. Can Res. 2011;71:736–746. doi: 10.1158/0008-5472.CAN-10-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin S, et al. Inference and analysis of cell–cell communication using cell chat. Nat Commun. 2021;12:1088. doi: 10.1038/s41467-021-21246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He B, et al. Integrating spatial gene expression and breast tumour morphology via deep learning. Nat Biomed Eng. 2020;4:827–834. doi: 10.1038/s41551-020-0578-x. [DOI] [PubMed] [Google Scholar]
- 35.Kleshchevnikov V, et al. Cell 2location maps fine-grained cell types in spatial transcriptomics. Nat Biotechnol. 2022;40:661–671. doi: 10.1038/s41587-021-01139-4. [DOI] [PubMed] [Google Scholar]
- 36.Gabrilovich DI, Ostrand-Rosenberg S, Bronte V. Coordinated regulation of myeloid cells by tumours. Nat Rev Immunol. 2012;12:253–268. doi: 10.1038/nri3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cane S, et al. The Endless Saga of Monocyte Diversity. Front Immunol. 2019;10:1786. doi: 10.3389/fimmu.2019.01786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bain CC, et al. Constant replenishment from circulating monocytes maintains the macrophage pool in the intestine of adult mice. Nat Immunol. 2014;15:929–937. doi: 10.1038/ni.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guilliams M, Mildner A, Yona S. Developmental and functional heterogeneity of monocytes. Immunity. 2018;49:595–613. doi: 10.1016/j.immuni.2018.10.005. [DOI] [PubMed] [Google Scholar]
- 40.Korbecki J, Grochans S, Gutowska I, Barczak K, Baranowska-Bosiacka I. CC chemokines in a tumor: a review of pro-cancer and anti-cancer properties of receptors CCR5, CCR6, CCR7, CCR8, CCR9, and CCR10 ligands. Int J Mol Sci. 2020;21:7619. doi: 10.3390/ijms21207619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garcia KA, et al. Fatty acid binding protein 5 regulates lipogenesis and tumor growth in lung adenocarcinoma. Life Sci. 2022;301:120621. doi: 10.1016/j.lfs.2022.120621. [DOI] [PubMed] [Google Scholar]
- 42.Wang W, et al. Downregulation of FABP5 suppresses the proliferation and induces the apoptosis of gastric cancer cells through the hippo signaling pathway. DNA Cell Biol. 2021;40:1076–1086. doi: 10.1089/dna.2021.0370. [DOI] [PubMed] [Google Scholar]
- 43.(2022) FOLR2+ macrophages are associated with T-cell infiltration and improved prognosis. Cancer Discov 12:1407. 10.1158/2159-8290.CD-RW2022-058 [DOI] [PubMed]
- 44.Preuss I, et al. Transcriptional regulation and functional characterization of the oxysterol/EBI2 system in primary human macrophages. Biochem Biophys Res Commun. 2014;446:663–668. doi: 10.1016/j.bbrc.2014.01.069. [DOI] [PubMed] [Google Scholar]
- 45.Li S, et al. S100A8 promotes epithelial-mesenchymal transition and metastasis under TGF-beta/USF2 axis in colorectal cancer. Cancer Commun. 2021;41:154–170. doi: 10.1002/cac2.12130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zha H, et al. S100A8 facilitates the migration of colorectal cancer cells through regulating macrophages in the inflammatory microenvironment. Oncol Rep. 2016;36:279–290. doi: 10.3892/or.2016.4790. [DOI] [PubMed] [Google Scholar]
- 47.Kim JH, et al. Functions of human liver CD69(+)CD103(-)CD8(+) T cells depend on HIF-2alpha activity in healthy and pathologic livers. J Hepatol. 2020;72:1170–1181. doi: 10.1016/j.jhep.2020.01.010. [DOI] [PubMed] [Google Scholar]
- 48.Palazon A, et al. An HIF-1alpha/VEGF-A axis in cytotoxic T cells regulates tumor progression. Cancer Cell. 2017;32:669–683. doi: 10.1016/j.ccell.2017.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pichler WJ, Wyss-Coray T. T cells as antigen-presenting cells. Immunol Today. 1994;15:312–315. doi: 10.1016/0167-5699(94)90078-7. [DOI] [PubMed] [Google Scholar]
- 50.Nieszporek A, Skrzypek K, Adamek G, Majka M. Molecular mechanisms of epithelial to mesenchymal transition in tumor metastasis. Acta Biochim Pol. 2019;66:509–520. doi: 10.18388/abp.2019_2899. [DOI] [PubMed] [Google Scholar]
- 51.Macian F. Autophagy in T cell function and aging. Front Cell Dev Biol. 2019;7:213. doi: 10.3389/fcell.2019.00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang L, et al. Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature. 2018;564:268–272. doi: 10.1038/s41586-018-0694-x. [DOI] [PubMed] [Google Scholar]
- 53.Rubio AJ, Porter T, Zhong X. Duality of B cell-CXCL13 axis in tumor immunology. Front Immunol. 2020;11:521110. doi: 10.3389/fimmu.2020.521110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ohue Y, Nishikawa H. Regulatory T (Treg) cells in cancer: can Treg cells be a new therapeutic target? Cancer Sci. 2019;110:2080–2089. doi: 10.1111/cas.14069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wong KY, et al. CD83 antibody inhibits human B cell responses to antigen as well as dendritic cell-mediated CD4 T cell responses. J Immunol. 2018;200:3383–3396. doi: 10.4049/jimmunol.1700064. [DOI] [PubMed] [Google Scholar]
- 56.Hollern DP, et al. B cells and T follicular helper cells mediate response to checkpoint inhibitors in high mutation burden mouse models of breast cancer. Cell. 2019;179:1191–1206. doi: 10.1016/j.cell.2019.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Garaud S, et al. Tumor infiltrating B-cells signal functional humoral immune responses in breast cancer. JCI insight. 2019;4:e129641. doi: 10.1172/jci.insight.129641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim HS, Kim MG, Min KW, Jung US, Kim DH. High MMP-11 expression associated with low CD8+ T cells decreases the survival rate in patients with breast cancer. PLoS ONE. 2021;16:e0252052. doi: 10.1371/journal.pone.0252052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Eiro N, et al. MMP11 expression in intratumoral inflammatory cells in breast cancer. Histopathology. 2019;75:916–930. doi: 10.1111/his.13956. [DOI] [PubMed] [Google Scholar]
- 60.Ji J, et al. Fibronectin 1 inhibits the apoptosis of human trophoblasts by activating the PI3K/Akt signaling pathway. Int J Mol Med. 2020;46:1908–1922. doi: 10.3892/ijmm.2020.4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ma J, et al. SPARC enhances 5-FU chemosensitivity in gastric cancer by modulating epithelial-mesenchymal transition and apoptosis. Biochem Biophys Res Commun. 2021;558:134–140. doi: 10.1016/j.bbrc.2021.04.009. [DOI] [PubMed] [Google Scholar]
- 62.Lv Y, et al. CTHRC1 overexpression promotes ectopic endometrial stromal cell proliferation, migration and invasion via activation of the Wnt/beta-catenin pathway. Reprod Biomed Online. 2020;40:26–32. doi: 10.1016/j.rbmo.2019.10.001. [DOI] [PubMed] [Google Scholar]
- 63.Murdamoothoo D, et al. Tenascin-C immobilizes infiltrating T lymphocytes through CXCL12 promoting breast cancer progression. EMBO Mol Med. 2021;13:e13270. doi: 10.15252/emmm.202013270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Simon T, et al. Differential regulation of T-cell immunity and tolerance by stromal laminin expressed in the lymph node. Transplantation. 2019;103:2075–2089. doi: 10.1097/TP.0000000000002774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Koutsioumpa M, et al. Pleiotrophin expression and role in physiological angiogenesis in vivo: potential involvement of nucleolin. Vascular cell. 2012;4:4. doi: 10.1186/2045-824X-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu J, et al. M2 macrophage-derived exosomes facilitate HCC metastasis by transferring alphaM beta2 integrin to tumor cells. Hepatology. 2021;73:1365–1380. doi: 10.1002/hep.31432. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 67.Baracco EE, et al. Contribution of annexin A1 to anticancer immunosurveillance. Oncoimmunology. 2019;8:e1647760. doi: 10.1080/2162402X.2019.1647760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Korbecki J, et al. Fractalkine/CX3CL1 in neoplastic processes. Int J Mol Sci. 2020;21:3723. doi: 10.3390/ijms21103723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Meurette O, Mehlen P. Notch signaling in the tumor microenvironment. Cancer Cell. 2018;34:536–548. doi: 10.1016/j.ccell.2018.07.009. [DOI] [PubMed] [Google Scholar]
- 70.Stanley ER, Chitu V. CSF-1 receptor signaling in myeloid cells. Cold Spring Harb Perspect Biol. 2014;6:a021857. doi: 10.1101/cshperspect.a021857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.de Azevedo RA, et al. MIF inhibition as a strategy for overcoming resistance to immune checkpoint blockade therapy in melanoma. Oncoimmunology. 2020;9:1846915. doi: 10.1080/2162402X.2020.1846915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tamura R, et al. The role of vascular endothelial growth factor in the hypoxic and immunosuppressive tumor microenvironment: perspectives for therapeutic implications. Med Oncol. 2019;37:2. doi: 10.1007/s12032-019-1329-2. [DOI] [PubMed] [Google Scholar]
- 73.Wang M, et al. Exosomal LGALS9 in the cerebrospinal fluid of glioblastoma patients suppressed dendritic cell antigen presentation and cytotoxic T-cell immunity. Cell Death Dis. 2020;11:896. doi: 10.1038/s41419-020-03042-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wu C, et al. Galectin-9-CD44 interaction enhances stability and function of adaptive regulatory T cells. Immunity. 2014;41:270–282. doi: 10.1016/j.immuni.2014.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ren X, Ji Y, Jiang X, Qi X. Down-regulation of siglec-2 (CD22) predicts worse overall survival from HBV-related early-stage hepatocellular carcinoma: a preliminary analysis from Gene Expression Omnibus. Biosci Rep. 2018;38:BSR20181423. doi: 10.1042/BSR20181423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Klement JD, et al. An osteopontin/CD44 immune checkpoint controls CD8+ T cell activation and tumor immune evasion. J Clin Investig. 2018;128:5549–5560. doi: 10.1172/JCI123360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Matsuda T, et al. TCR sequencing analysis of cancer tissues and tumor draining lymph nodes in colorectal cancer patients. Oncoimmunology. 2019;8:e1588085. doi: 10.1080/2162402X.2019.1588085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Simoni Y, et al. Bystander CD8(+) T cells are abundant and phenotypically distinct in human tumour infiltrates. Nature. 2018;557:575–579. doi: 10.1038/s41586-018-0130-2. [DOI] [PubMed] [Google Scholar]
- 79.Scheper W, et al. Low and variable tumor reactivity of the intratumoral TCR repertoire in human cancers. Nat Med. 2019;25:89–94. doi: 10.1038/s41591-018-0266-5. [DOI] [PubMed] [Google Scholar]
- 80.Alexander J, Cukierman E. Cancer associated fibroblast: mediators of tumorigenesis. Matrix Biol: J Int Soc Matrix Biol. 2020;91–92:19–34. doi: 10.1016/j.matbio.2020.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Houthuijzen JM, Jonkers J. Cancer-associated fibroblasts as key regulators of the breast cancer tumor microenvironment. Cancer Metastasis Rev. 2018;37:577–597. doi: 10.1007/s10555-018-9768-3. [DOI] [PubMed] [Google Scholar]
- 82.Jenkins BH, Buckingham JF, Hanley CJ, Thomas GJ. Targeting cancer-associated fibroblasts: challenges, opportunities and future directions. Pharmacol Ther. 2022;240:108231. doi: 10.1016/j.pharmthera.2022.108231. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated in this study have been uploaded to Genome Sequence Archive (GSA, https://ngdc.cncb.ac.cn/) with accession number HRA002051 and OMIX001111 under the project of PRJCA008495. Supplementary files were listed as below: Supplementary patient information file: Supplementary Table file: Table S1–S8; Supplementary Figure file: Figure S1–S16.