Abstract
Bacterial flora has clinical significance for the host. The metabolic environment created by this flora influences immunotherapy in urothelial carcinoma. However, there are no reports on the clinical significance of bacterial flora in the host bloodstream. We aimed to clarify the correlation between extracellular vesicle (EV)-derived blood microflora information and tumor immunological status in urothelial carcinoma (UC) patients. Serum samples were collected from 20 healthy donors, 50 patients with localized UC, and 31 patients with metastatic UC (mUC) who had undergone pembrolizumab treatment. Bacterial DNA in EVs was extracted from each sample. Metagenomic sequencing was performed after amplification of the V1–V2 region of the bacterial 16S rRNA gene. Using the matched tumor tissue and serum samples, we revealed that the smaller amount of peripheral EVs carrying Firmicutes DNA was significantly correlated with the higher number of infiltrating T cells within tumor tissues (CD3; p = 0.015, CD4; p = 0.039, CD8; p = 0.0084) and the higher expression of activation markers on their surface (ICOS on both CD4; p = 0.0013 and CD8 T cells; p = 0.016 and 4-1BB on CD4 T cells; p = 0.016). In terms of circulating metabolic information, l-Ser and l-Pro levels, which play important roles in T cell expansion and proliferation, were significantly higher in the Firmicutes-low group (p = 0.010). All of the patients with higher Firmicutes abundance had disease progression without any clinical response (p = 0.026) and significantly inferior prognosis for pembrolizumab therapy (p = 0.035). This is the first study on the importance of peripheral bacterial EVs in cancer patients treated with cancer immunotherapy.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00262-022-03213-5.
Keywords: Immunotherapy, Firmicutes, Extracellular vesicles, Urothelial carcinoma, Bloodstream, Microbiome
Introduction
Urothelial carcinoma (UC) is the ninth most frequently diagnosed cancer worldwide, of which the highest incidence is observed in men. Up to 10–15% of initially diagnosed patients have metastatic UC (mUC), with a 5-year survival of 15% worldwide [1]. Although the advancements in immune checkpoint inhibitors (ICIs), such as pembrolizumab and avelumab, have revolutionized the management of mUCs, only 20–30% of mUC patients achieve significant response to ICIs therapy. We previously classified the tumor immunological status based on the expression of surface markers in UC patients [2], suggesting the clinical need for predictive biomarkers of immunological status in tumor microenvironment that could discern mUC patients who could benefit from ICI therapy.
Extracellular vesicles (EVs) are released by all three domains of life (eukaryotes, bacteria, and archaea) and represent an evolutionarily conserved mechanism for intercellular communication [3]. Although several reports suggest that bacteria-derived EVs can enter the bloodstream [4, 5], their physiological and clinical significance remains unclear.
Bacterial flora exists in various parts of the human body, including the intestine, and is thought to play a role in the etiology of various diseases, including multiple sclerosis, bowel disease, male infertility, and uterine-related diseases [6–9]. In the field of clinical oncology, it has become clear that the metabolic environment created by the intestinal microbiota influences carcinogenesis [10], cancer progression [11], and multidisciplinary treatment, including immunotherapy, such as ICIs, in various cancer types, including UC [12–15]. Moreover, organ-specific microflora exists for different cancer types [16, 17]; although it remains unclear if this associated bacterial content arises from lysed bacterial cells or bacterial EVs. Components of cancer-specific microflora has also been reported in the bloodstream, which may be useful in cancer diagnosis [18]. However, no reports have clarified the clinical significance of bacterial flora components in the bloodstream of the host. It is, therefore, necessary to identify more accurate diagnostic markers by combining information on multiple bacterial species for diagnosis and cancer staging of UC patients, as has been accomplished for other cancer types [18]. Therefore, in this study, we aimed to investigate the correlation of EV-derived blood microflora information and tumor immunological status, and evaluate its clinical significance and impact on the host.
Materials and methods
Clinical specimens
The healthy donor and urothelial cancer specimens were obtained from patients undergoing primary resection at the Osaka University Medical Hospital, Japan. Histological diagnosis was established with standard hematoxylin and eosin-stained sections by two senior pathologists experienced in UC diagnosis. Tumors were staged according to the seventh TNM staging system [19]. Written informed consent was obtained from each patient, and the study was approved by the ethics review board of both the Osaka University Medical Hospital (#13397–2, #14069–3). The pathological information of clinical serum samples used in Figs. 1, 2, 3, 4 is shown in Supplementary Tables S1.
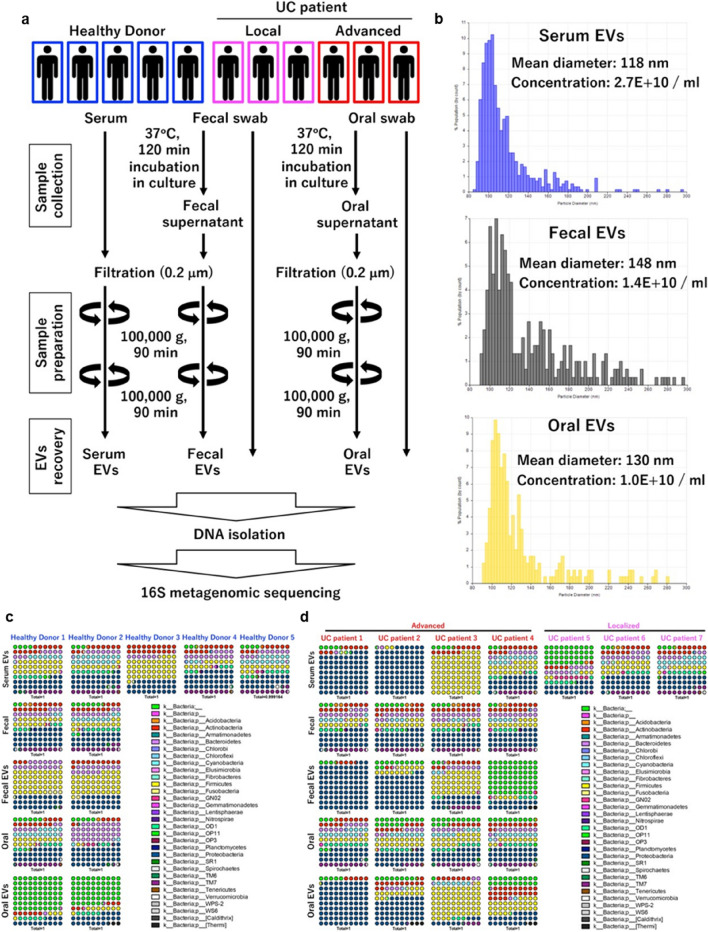
Fig. 1.
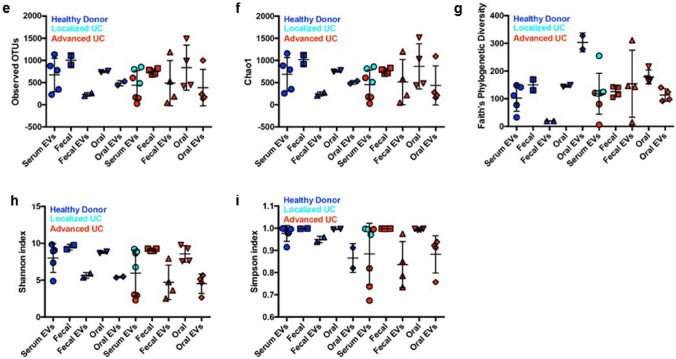
Serum EVs enriched with bacterial 16S rRNA genes. a Experimental design for EV isolation and subsequent 16S metagenomic sequencing analysis. b Representative results of nanoparticle analysis. Serum EVs enriched with bacterial 16S rRNA genes with high diversity. Percent dot plot of phylogenetic composition of common bacterial taxa (> 0.1% abundance) at the phylum level in each sample in healthy donors (c) or urothelial cancer patients (d). e Observed OTUs. Diversity analysis using Chao1 (f), Faith’s Phylogenetic Diversity (g), Shannon index (h), Simpson index (i)
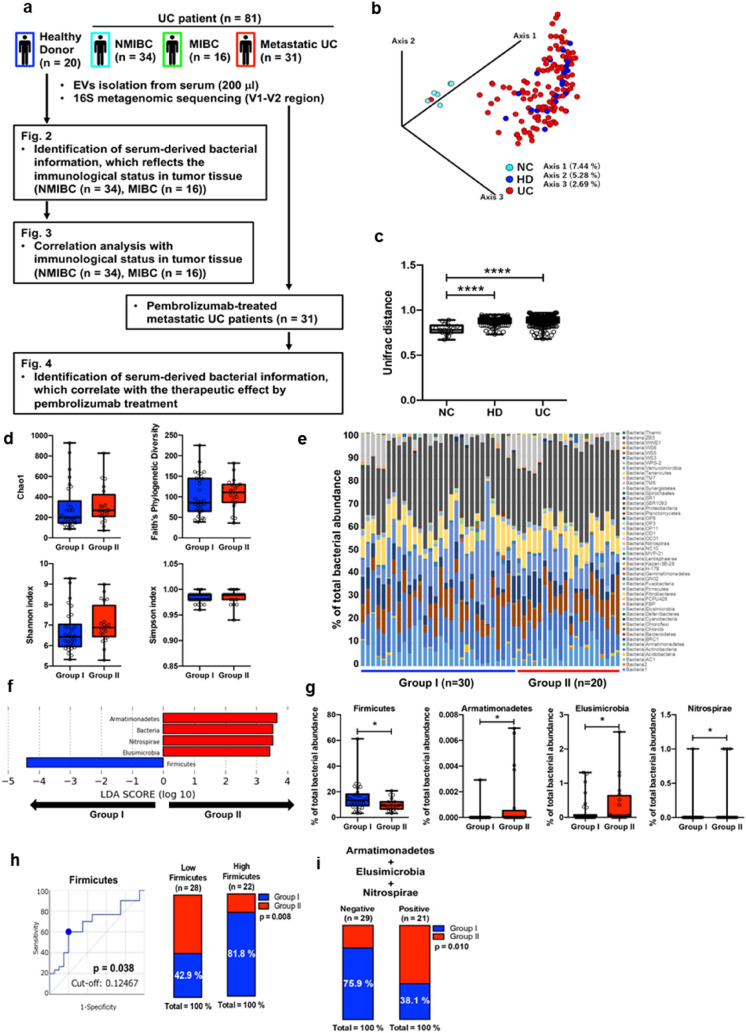
Fig. 2.
16S metagenomic sequencing of peripheral EVs in UC patients. a Schematic design of the study. b β diversity plots for 20 healthy donors (HD), 81 urothelial carcinoma patients (UC), and six negative controls (NC) (unweighted). c β diversity analysis using UniFrac distance. Kruskal–Wallis test followed by Dunn’s multiple comparisons test. ****p < 0.0001. Exact p values: p < 0.0001 (NC vs. HD), p < 0.0001 (NC vs. UC), p = 0.0862 (HD vs. UC). Immunological classification of peripheral bacterial information. d α diversity analysis based on immunological classification (Group I: 30, Group II: 20). e Taxa bar plots of UC peripheral EVs based on immunological classification. f Distinctive bacterial phylum in UC patients and the percent of each phylum abundance depending on immunological classification. g Mann–Whitney test (Two-tailed). *p < 0.05. Exact p values: p = 0.038 (Firmicutes), p = 0.012 (Armatimonadetes), p = 0.043 (Elusimicrobia), p = 0.028 (Nitrospirae). h ROC analysis based on Firmicutes abundance in UC (left panel). Cumulative bar chart based on abundance of peripheral Firmicutes (right panel). i Cumulative bar chart based on positivity of peripheral AEN (Armatimonadetes, Elusimicrobia and Nitrospirae). Fisher’s exact probability test
Fig. 3.
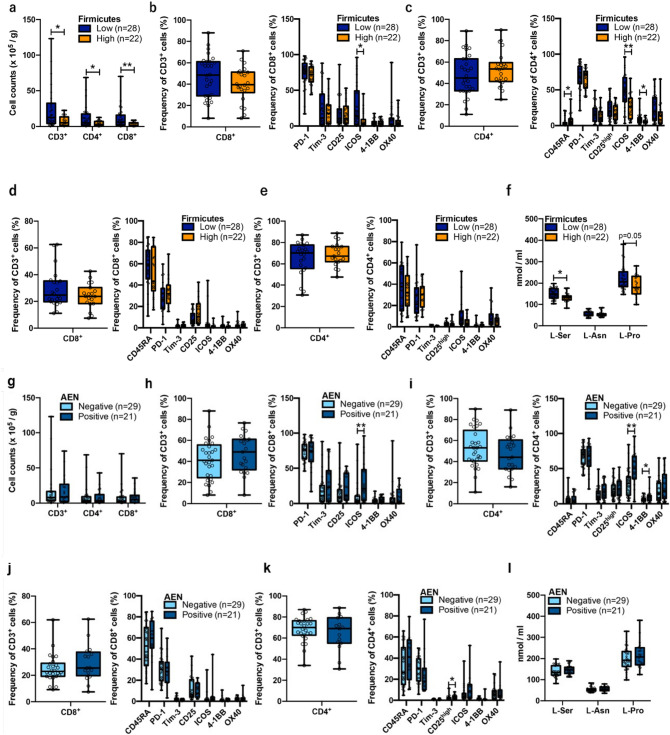
Peripheral Firmicutes phylum abundance correlated with the number and expression of activation surface markers of tumor tissue-infiltrating T cells. Number of tumor tissue-infiltrated T cells, frequency of CD8+ or CD4+ cells, and surface activation marker expression were analyzed based on peripheral Firmicutes abundance (a–e). a Number of tumor tissue-infiltrating T cells. b Frequency of tumor tissue-infiltrating CD8+ cells (left panel). Expression of immune checkpoint markers in CD8+ cells (right panel). c Frequency of tumor tissue-infiltrating CD4+ cells (left panel). Expression of immune checkpoint markers in CD4+ cells (right panel). d Frequency of peripheral CD8+ cells (left panel). Expression of immune checkpoint markers in CD8+ cells (right panel). e Frequency of peripheral CD4+ cells (left panel). Expression of immune checkpoint markers in CD4+ cells (right panel). Peripheral levels of L-amino acids based on peripheral Firmicutes abundance (f). Correlation analysis between peripheral AEN abundance and the number of tumor tissue-infiltrating T cells and myeloid cells and expression of activation markers on their surface. g Number of tumor tissue-infiltrated T cells, frequency of CD8+ or CD4+ cells, and surface activation marker expression were analyzed based on the presence/absence of AEN. h Frequency of tumor tissue‒infiltrating CD8+ cells (left panel). Expression of immune checkpoint markers in CD8+ cells (right panel). i Frequency of tumor tissue‒infiltrating CD4+ cells (left panel). Expression of immune checkpoint markers in CD4+ cells (right panel). (j) Frequency of peripheral CD8+ cells (left panel). Expression of immune checkpoint markers in CD8+ cells (right panel). k Frequency of peripheral CD4+ cells (left panel). Expression of immune checkpoint markers in CD4+ cells (right panel). l Peripheral levels of L-amino acids based on the presence/absence of AEN. Mann–Whitney test (Two-tailed). Mann–Whitney test (Two-tailed). *p < 0.05; **p < 0.01. Exact p values for (a): p = 0.015 (CD3+), p = 0.039 (CD4+), p = 0.0084 (CD8+); for (b): p = 0.24 (PD-1), p = 0.97 (Tim-3), p = 0.84 (CD25), p = 0.016 (ICOS), p = 0.66 (4-1BB), p = 0.71 (OX40); for (c): p = 0.017 (CD45RA), p = 0.60 (PD-1), p = 0.56 (Tim-3), p = 0.45 (CD25high), p = 0.0013 (ICOS), p = 0.016 (4-1BB), p = 0.084 (OX40); for (f): p = 0.010 (L-Ser), p = 0.75 (l-Asn), p = 0.051 (l-Pro). Expression of immune checkpoint markers in CD8+ cells (right panel). *p < 0.05; **p < 0.01. Exact p values for (h): p = 0.86 (PD-1), p = 0.24 (Tim-3), p = 0.35 (CD25), p = 0.0022 (ICOS), p = 0.10 (4-1BB), p = 0.36 (OX40); for (i): p = 0.93 (CD45RA), p = 0.74 (PD-1), p = 0.14 (Tim-3), p = 0.49 (CD25high), p = 0.0022 (ICOS), p = 0.039 (4-1BB), p = 0.12 (OX40); for (k): p = 0.27 (CD45RA), p = 0.070 (PD-1), p = 0.56 (Tim-3), p = 0.028 (CD25high), p = 0.18 (ICOS), p = 0.30 (4-1BB), p = 0.93 (OX40)
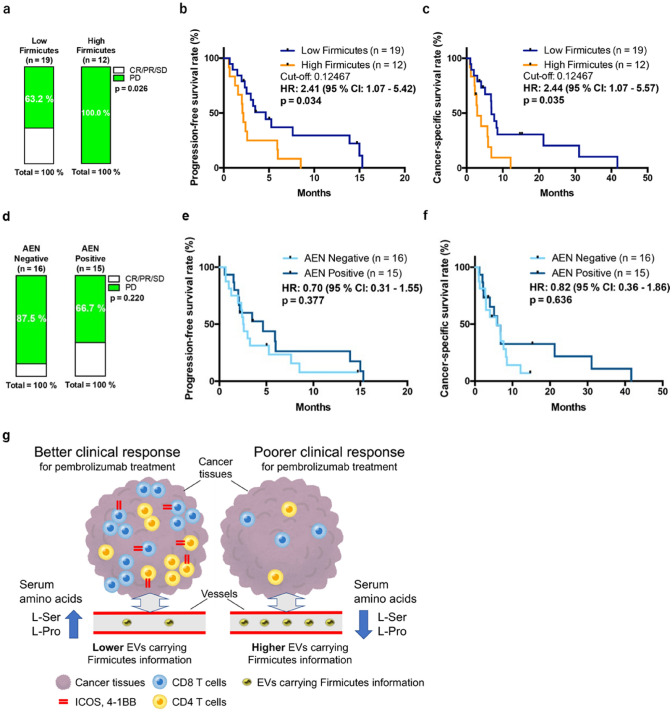
Fig. 4.
Peripheral Firmicutes abundance could predict the response and prognosis of metastatic urothelial carcinoma patients treated with pembrolizumab. a Cumulative bar chart based on abundance of peripheral Firmicutes. Fisher’s exact probability test. Progression-free survival ratio (b) and cancer-specific survival ratio (c) based on peripheral Firmicutes abundance. Peripheral AEN could not predict the response and prognosis of metastatic urothelial carcinoma patients treated with pembrolizumab. d Cumulative bar chart based on the presence/absence of AEN. Fisher’s exact probability test. Progression-free survival ratio (e) and cancer-specific survival ratio (f) based on the presence/absence of peripheral AEN (g) Graphical abstract representing the primary findings of this study
Sample preparation
Clinical specimens were collected from seven UC patients (local [n = 3], advanced [n = 4]) who were TURBT at Osaka University Hospital (Osaka, Japan) and five healthy individuals for Fig. 1. Meanwhile, 50 patients with localized UC (muscle invasive bladder cancer [MIBC], n = 16; non-MIBC [NMIBC], n = 34) who had undergone TURBT and were included in our previous study to evaluate the local immune status of UC [2], as well as 31 metastatic UC patients who had undergone treatment with pembrolizumab (anti-PD-1 antibody) were included in our analysis shown in Figs. 2, 3, 4. Healthy individuals were defined as those without a current malignant disease or medical history of urinary cancer. All UC patients were histologically diagnosed.
Serum samples
Whole blood (2.0–7.0 mL) was collected directly into Venoject II tubes (TERUMO, Tokyo, Japan). Within three hours after collection, all blood samples were centrifuged at 1200 × g for 15 min; the serum (supernatant) was collected and stored at – 80 °C. Serum samples were subsequently centrifuged at 2000 × g for 30 min and filtered with 0.2 µm syringe filter (KURABO, Osaka, Japan). Next, EVs were isolated according to the following protocol. Serum EVs were isolated using ultracentrifugation (Fig. 1a) or Exosome Isolation Kit (serum) (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions.
Fecal and oral samples
Fecal and oral swab samples were obtained with FLOQSwabs (COPAN Diagnostics, Murrieta, CA, USA). Swabbed samples were immediately immersed in 1 mL of DMEM medium (FUJIFILM Wako) without fetal calf serum and incubated at 37 °C for 2 h. The swab-immersed medium, containing EVs released by fecal (fecal EVs) or oral (oral EVs) bacteria, was then centrifuged at 2000 × g, for 30 min, and the supernatants were filtered through a 0.2-µm syringe filter (KURABO) and subjected to ultracentrifugation for recovery of EVs (Fig. 1c).
Circulating lymphocytes and tissue-infiltrating lymphocytes (TILs) were extracted as previously described [2].
Isolation of DNA
Serum EVs, fecal EVs, and oral EVs
DNA was isolated from EV samples using the QIAamp® Circulating Nucleic Acid Kit (QIAGEN, Hilden, Germany) according to the manufacturer’s protocol.
Fecal and oral swab: DNA was isolated from swab samples using the DNeasy PowerSoil Pro Kit (QIAGEN) according to the manufacturer’s protocol.
16S rRNA gene PCR with whole blood
Whole blood (5.0 mL) was collected directly into EDTA tubes. Then, 16S rRNA was analyzed via PCR using the following primer sets [20]: JRP1 forward primer: 5ʹ-CTCCTACGGGAGGCAGCAG-3ʹ and JRP3 reverse primer: 5ʹ-ACATGCTCCACCGCTTGTG-3ʹ (expected amplicon size: 600–650 bp). The cycling protocol was as follows: denaturation for 1 min at 94 °C, annealing for 30 s at 62 °C, and extension for 40 s at 72 °C for 30 cycles. Amplicons were resolved by agarose gel electrophoresis and detected by SAFELOOK™ Green Nucleic Acid Stain (FUJIFILM Wako, Osaka, Japan).
16S metagenomic sequencing
V1–V2 region of the bacterial 16S rRNA gene was PCR-amplified with primers 27Fmod (5'-AGRGTTTGATYMTGGCTCAG-3') and 338R (5'-TGCTGCCTCCCGTAGGAGT-3'), and sequencing of the 16S rRNA was performed on a MiSeq platform (Illumina, San Diego, CA). QIIME version 2.202002 was used to process all raw sequencing data. Linear discriminant analysis effect size (LEfSe, [21]) was used to elucidate bacterial genus classification taxa that were associated with malignancy of UC and immunological classification of T cells. LEfSe was calculated using the Galaxy web platform, and only linear discriminant analysis (LDA) scores ≥ 2 were listed.
Nanoparticle measurement
The size and concentration of EVs were determined using qNano Gold (Izon Science, Christchurch, New Zealand). Data were analyzed with Izon Control Suite Software (V3.3.2.2001).
Immunological classification of T cells
Lymphocytes including TILs were stained with multiple extracellular antibodies as previously described [2]. The positively stained cells were defined above the negative region for isotype control. Immunological classification was defined based on surface immune checkpoint molecules in our previous study [2]. In the current study, we used only 50 of our previous 70 samples that could pair off with serum EVs samples in the present study.
Measurement of amino acid concentration
Concentration of l-serine and l-proline was quantified using the micro-2D-HPLC. In brief, amino acids were labeled with fluorescence (4-fluoro-7-nitro-2,1,3-benzoxadiazole), and l-amino acids were quantified via chiral selectivity. Fluorescence-labeled amino acids were separated by reversed-phase column, followed by chiral separation using a chiral-selective column. Detection of fluorescence-labeled amino acids was carried out at 530 nm with an excitation of 470 nm using two photomultiplier tubes. Detailed methods were previously described [22].
Survival analysis of mUC patients treated with pembrolizumab
Cancer-specific survival (CSS) time was calculated from the date of initiation of pembrolizumab until death or the date of the patient’s last follow-up visit. Progression-free survival (PFS) time was calculated from the date of initiation of pembrolizumab until radiological and clinical disease progression or death. The CSS and PFS rates were calculated using Kaplan–Meier method. We also used the Cox regression model to calculate the hazard ratios in univariate and multivariate analysis.
Statistical analysis
Statistical analyses were performed using JMP Pro software (v.14.0.0; SAS Institute), and quantification was performed using GraphPad Prism software (GraphPad Prism 6.0, GraphPad software). Differences between the values were statistically analyzed using the Mann–Whitney test or Kruskal–Wallis test followed by Dunn’s multiple comparisons test. A p value < 0.05 was considered statistically significant. The patient characteristics were compared using the Mann–Whitney U test and χ2-test. Univariate analysis was performed with the Mann–Whitney U test. Multiple regression analyses were performed to assess the relative contributions of factors (age, sex, BMI, smoking status, NLR, PLR, MLR, and FAN score [23]) and the Firmicutes or Armatimonadetes, Elusimicrobia, and Nitrospirae (AEN) abundance in serum EVs. Probability analysis was performed using Fisher’s exact probability test. Significant predictors of urothelial carcinoma were identified by logistic regression analysis. The optimal cut-off value for each EV protein was determined from the receiver operating characteristic (ROC) curve using Youden index.
Results
Identification of bacterial information from serum-derived EVs in healthy donors and UC patients
First, to evaluate the existence of bacterial EVs in circulation and their source, we first extracted DNA from the oral cavity, feces, and blood from two healthy donors and four patients with advanced UC. DNA was also extracted from the blood of three healthy donors and three patients with localized UC. Extracted DNA was used for 16S rRNA metagenomic analysis to compare the expression profiles (Fig. 1a, b). We found that the bacterial diversity in both oral and fecal samples was reduced in EV samples compared to samples collected by normal collection methods. Moreover, no significant correlation was identified between the bacterial information contained in each sample and clinical background information (Fig. 1c–i). The bacterial information in fecal and oral EVs were reflected in serum EVs, particularly in a proportion of the advanced UC patients, suggesting that serum EVs harbor a unique cancer-specific bacterial signature within some UC patients (Fig. 1c, d).
Next, we used archived serum samples collected from 20 healthy donors, those collected immediately before treatment from 50 patient with localized UC who had undergone TURBT and those collected from 31 metastatic UC patients who had undergone treatment with pembrolizumab (Supplementary Table S1; Fig. 2a). Six negative control samples from sterilized PBS were set in each experiment for EV isolation and DNA extraction, and the 16 s rRNA sequencing results of all 107 samples were evaluated. As a result, we clarified that the flora information isolated from negative controls differed significantly from that derived from the sera of both healthy donors and UC patients (p < 0.001; Fig. 2b, c).
Bacterial information from serum-derived EVs reflects the immunological condition within the tumor microenvironment
We attempted to identify the correlation between bacterial flora in the bloodstream and immunological status within the tumor microenvironment using 50 matched serum and tumor tissue samples (MIBC, n = 16; NMIBC n = 34: identical cohort as in Fig. 2) selected from our previously collected samples (Supplementary Figs. 1a–c) [2]. No difference was detected in the number of immune cells infiltrating the tissues between two groups, classified based on the expression of surface markers (Supplementary Fig. 1d). As previously reported, this immunological classification is clearly divided into two groups, i.e., Group I, in which ratio of CD4 is significantly higher, and Group II, in which ratio of CD8 and various immune checkpoint molecules expressed significantly higher (Supplementary Figs. 1e, f) [2]. However, the immunological status of peripheral blood cells has no such features (Supplementary Figs. 1 g, h).
There were no significant differences in the α-diversity in the bloodstream according to the immunological classification (Fig. 2d). Thus, we attempted to identify specific bacterial information for immunological classification (Fig. 2e). LEfSe analysis based on phylum information showed that the Nitrospirae, Elusimicrobia, and Armatimonadetes phyla were significantly upregulated in Group II, and the Firmicutes phylum was significantly upregulated in Group I (Fig. 2f, g). In contrast, there was no significant difference in the Bacteroidetes phylum abundance between the two groups, and the Firmicutes/Bacteroidetes (F/B) ratio, which is also known to be significantly correlated with obesity [24, 25] and aging [26] with respect to intestinal bacteria, tended to be higher in Group I (p = 0.089; Supplementary Figs. 2a–c).
We then set a cut-off value using ROC curve analysis to divide the groups for prediction of immune classification. The best cut-off value for Firmicutes to predict Group I was set at 0.12467 (Fig. 2h); 81.8% of the cases with higher abundance of Firmicutes (≧ 0.12467) belonged to Group I, whereas 42.9% of the cases with lower abundance of Firmicutes belonged to Group I (p = 0.008; Fig. 2h). When the best cut-off for the F/B ratio to predict Group I was set at 1.2165, 75% of the cases with higher F/B ratio belonged to Group I, and 46.2% of cases with lower F/B ratio belonged to Group I (p = 0.048; Supplementary Fig. 2d). AEN, which were upregulated in Group II, yielded positive results in 6 (12.0%), 18 (36.0%), and 8 (16.0%) patients, respectively, and the percentage of Group II cases was significantly higher in all cases (Supplementary Figs. 3a‒c). Additionally, when the presence of any of these three phyla (AEN) was considered positive, 60.9% of the AEN-positive cases belonged to Group II (Fig. 2i).
Using the above classification, we evaluated the clinical characteristics and local immune status of UC patients depending on the Firmicutes and AEN abundance. Although no clinical characteristics significantly correlated with Firmicutes and AEN abundance, Firmicutes abundance tended to be higher in older patients (age ≥ 71, p = 0.091; Table 1). We also found that the proportion of CD3, CD4, and CD8 T cells per weight of tumor tissue was significantly higher in the Firmicutes-low group than in the Firmicutes-high group (Fig. 3a). In addition, although the percentages of tissue-infiltrating CD8 and CD4 T cells did not change, inducible costimulatory molecule (ICOS), a member of the CD28/CTLA4 family, was expressed at significantly higher levels on the surface of CD8 and CD4 T cells in the Firmicutes-low group than Firmicutes-high group (p = 0.016 and p = 0.0013, respectively). Additionally, 4-1BB, a member of the tumor necrosis factor receptor superfamily T cell costimulatory receptor, was also significantly upregulated on CD4 T cells in the Firmicutes-low group. Meanwhile, CD4+CD45RA+ cells, a naive CD4+ fraction, were significantly upregulated in the Firmicutes-high group (p = 0.017) (Fig. 3b, c). The expression of T cell surface molecules and abundance of Firmicutes in the bloodstream were not significantly correlated with CD4 or CD8 T cells (Fig. 3d, e).
Table 1.
Univariate logistic regression analysis to identify a correlation between clinical characteristics and the abundance of peripheral EVs containing Firmicutes and presence of AEN in patients with localized UC
| Characteristics | Firmicutes phylum | AEN expression | ||||
|---|---|---|---|---|---|---|
| OR | 95% CI | P | OR | 95% CI | P | |
| Age (< 71 vs. ≥ 71) | 2.705 | 0.854–8.568 | 0.091 | 1.641 | 0.529–5.093 | 0.391 |
| Sex (male vs. female) | 1.022 | 0.239–4.368 | 0.976 | 0.639 | 0.140–2.912 | 0.563 |
| BMI (< 23.9 vs. ≥ 23.9) | 1.000 | 0.327–3.055 | 1.000 | 0.434 | 0.138–1.371 | 0.155 |
| Smoking status | ||||||
| (never vs. current/former) | 0.519 | 0.167–1.611 | 0.257 | 0.434 | 0.138–1.371 | 0.155 |
| pT stage (Ta vs. T1 and more) | 1.132 | 0.357–3.587 | 0.833 | 2.031 | 0.614–6.710 | 0.246 |
| Tumor grade (2 vs. 3) | 1.444 | 0.468–4.460 | 0.523 | 0.642 | 0.207–1.989 | 0.442 |
| NLR (< 2.39 vs. ≥ 2.39) | 0.519 | 0.167–1.611 | 0.257 | 1.179 | 0.383–3.627 | 0.775 |
| PLR (< 0.81 vs. ≥ 0.81) | 0.722 | 0.235–2.215 | 0.569 | 1.179 | 0.383–3.627 | 0.775 |
| MLR (< 0.23 vs. ≥ 0.23) | 0.539 | 0.174–1.671 | 0.285 | 1.244 | 0.402–3.853 | 0.705 |
| FAN score (0, 1 vs. 2, 3) | 0.900 | 0.212–3.822 | 0.886 | 1.143 | 0.279–4.683 | 0.853 |
OR, odds ratio; CI, confidence interval; BMI, body mass index; NLR, neutrophil-to-lymphocyte ratio; PLR, platelet-to-lymphocyte ratio; MLR, monocyte-to-lymphocyte ratio
Next, to assess the metabolic information associated with T cells in blood, we evaluated the correlation between Firmicutes abundance and concentrations of l-serine (l-Ser, [27]), l-asparagine (l-Asn, [28]), and l-proline (l-Pro [29],), which play important roles in T cell expansion and proliferation. l-Ser and l-Pro levels were higher in the Firmicutes-low group (p = 0.010) (Fig. 3f).
We conducted further analysis based on AEN abundance and found no significant correlation with the number of tumor-infiltrating T cells (Fig. 3g). ICOS was highly expressed on CD8 and CD4 T cells in the AEN-positive group. In addition, 4-1BB+CD4 T cells were significantly upregulated in the AEN-positive group (Fig. 3h, i). Furthermore, unlike that for Firmicutes abundance, there was no significant correlation between AEN abundance and T cell surface molecule expression in the blood nor l-Ser or l-Pro concentration (Fig. 3j–l). We also compared the expression profile of cells in the Firmicutes- or AEN-positive groups with that of myeloid cells in tumor tissues, and found no significant differences, including that for PD-L1 [30] or CCR2 [31] expression, which is associated with immunosuppression (Supplementary Figs. 4a, b). These results together indicate that peripheral EV-derived bacterial information significantly reflects the immune status of T cells, but not myeloid cells, within the tumor microenvironment.
Serum-derived EVs carrying Firmicutes represent potential predictive and prognostic markers of mUC patients treated with pembrolizumab
Finally, we investigated whether peripheral EV-derived bacterial information could predict response and prognosis for ICI therapy using 31 mUC samples from patients who had undergone treatment with pembrolizumab. Among the 31 patients treated with pembrolizumab, none of the 12 patients classified in the Firmicutes-high group, stratified using the cut-off value of 0.12467, could achieve a therapeutic response higher than stable disease (SD). All seven patients who achieved a therapeutic response higher than SD belonged to the Firmicutes-low group (Fig. 4a). Additionally, patients in the Firmicutes-high group had significantly lower progression-free survival (PFS) (hazard ratio [HR]: 2.41, 95% confidence interval [CI]: 1.07–5.42, p = 0.034) and cancer-specific survival (CSS) (HR: 2.44, 95% CI: 1.07–5.57, p = 0.035) than those in the Firmicutes-low group (Fig. 4b, c). When the F/B ratio was stratified using the cut-off value of 1.2165, only one (6.2%) of the 16 patients with higher F/B ratio showed a therapeutic effect of SD or higher, and six (40%) of the 15 patients with a lower F/B ratio showed therapeutic effect of SD or higher (p = 0.037; Supplementary Fig. 5a). Patients with a higher F/B ratio had significantly lower PFS (HR: 2.96, 95% CI: 1.26–6.96p = 0.013) and CSS (HR: 3.29, 95% CI: 1.38–7.86, p = 0.007) than those with higher F/B ratios (Supplementary Figs. S5b, c). In contrast, 33.3% of the 15 AEN-positive patients and 12.5% of the 16 AEN-negative patients showed therapeutic effect higher than SD. Although the therapeutic effect was higher in AEN-positive patients, there was no significant difference between the two groups (AEN-positive vs -negative, Fig. 4d). In terms of prognosis, some AEN-positive patients achieved long-term survival though there was no significant difference in PFS (HR: 0.70, 95% CI: 0.31–1.55, p = 0.377) or CSS (HR: 0.82, 95% CI: 0.36–1.86, p = 0.636; Fig. 4e, f). According to multivariate analysis, peripheral EVs carrying Firmicutes, but not AEN, significantly correlated with PFS (HR: 3.906, 95% CI: 1.436–10.625, p = 0.008) and CSS (HR: 4.593, 95% CI: 1.697–12.428, p = 0.003) (Table 2).
Table 2.
Univariate and multivariate analyses with Cox regression analysis for progression-free survival (A) and cancer-specific survival (B) of 31 metastatic UC patients treated with pembrolizumab
| Characteristic | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | |
| (A) | ||||||
| Age (< 73 vs. ≥ 73) | 1.026 | 0.460–2.289 | 0.950 | 0.822 | 0.320–2.114 | 0.684 |
| Sex (male vs. female) | 2.137 | 0.934–4.888 | 0.072 | 1.029 | 0.410–2.581 | 0.951 |
| BMI (< 20.2 vs. ≥ 20.2) | 0.880 | 0.403–1.923 | 0.749 | |||
| Smoking status (never vs. current/former) | 0.842 | 0.350–2.028 | 0.702 | |||
| Firmicutes abundance (low vs. high) | 2.408 | 1.071–5.417 | 0.034 | 3.906 | 1.436–10.625 | 0.008 |
| AEN presence (negative vs. positive) | 0.698 | 0.314–1.550 | 0.377 | 0.484 | 0.197–1.189 | 0.114 |
| NLR (< 5.0 vs. ≥ 5.0) | 1.353 | 0.598–3.061 | 0.468 | |||
| PLR (< 1.50 vs. ≥ 1.50) | 1.424 | 0.640–3.165 | 0.386 | |||
| MLR (< 0.52 vs. ≥ 0.52) | 1.658 | 0.740–3.713 | 0.219 | |||
| FAN score (0, 1 vs. 2, 3) | 2.601 | 1.137–5.951 | 0.024 | 3.820 | 1.354–10.775 | 0.011 |
| (B) | ||||||
| Age (< 73 vs. ≥ 73) | 0.675 | 0.301–1.512 | 0.339 | 0.541 | 0.208–1.404 | 0.207 |
| Sex (male vs. female) | 3.051 | 1.243–7.491 | 0.015 | 1.841 | 0.713–4.757 | 0.207 |
| BMI (< 20.2 vs. ≥ 20.2) | 0.648 | 0.293–1.435 | 0.285 | |||
| Smoking status (never vs. current/former) | 1.071 | 0.442–2.594 | 0.879 | |||
| Firmicutes abundance (low vs. high) | 2.920 | 1.251–6.815 | 0.013 | 4.593 | 1.697–12.428 | 0.003 |
| AEN presence (negative vs. positive) | 0.652 | 0.281–1.513 | 0.320 | 0.643 | 0.268–1.540 | 0.321 |
| NLR (< 5.0 vs. ≥ 5.0) | 1.666 | 0.725–3.830 | 0.229 | |||
| PLR (< 1.50 vs. ≥ 1.50) | 1.144 | 0.511–2.562 | 0.744 | |||
| MLR (< 0.52 vs. ≥ 0.52) | 1.564 | 0.705–3.468 | 0.271 | |||
| FAN score (0, 1 vs. 2, 3) | 2.259 | 0.999–5.109 | 0.050 | 2.328 | 0.898–6.040 | 0.082 |
HR, hazard ratio; CI, confidence interval; BMI, body mass index; NLR, neutrophil-to-lymphocyte ratio; PLR, platelet-to-lymphocyte ratio; MLR, monocyte-to-lymphocyte ratio
Bold values represent the statistically significant
Discussion
The present study is the first, to our knowledge, to demonstrate the importance of bacterial information in peripheral EVs in cancer patients (Fig. 4g). We found that aging tended to be associated with increased blood Firmicutes abundance, which correlated with a decrease in the abundance, and activation of markers such as ICOS and 4–1 BB on CD4 and CD8 tissue-infiltrating T cells. In previous reports, an increase in the F/B ratio was closely correlated with aging [24] and obesity [5, 25]. Although the underlying molecular mechanism requires further clarification, these results suggest that higher peripheral abundance of Firmicutes leads to senescence of the immune system.
In patients with malignant melanoma, bacterial peptides within cancer cells are presented on HLA molecules and activate local immune cells [16]. In addition, we recently clarified that Helicobacter pylori infection increases ICOS expression on CD4 regulatory T cells in gastric and colorectal cancer patients [32, 33]. Moreover, the presence of tumor-infiltrating cytotoxic CD4 T cells is reportedly important for predicting the efficacy of ICI treatment in patients with metastatic UC [34]. These results support our findings that the peripheral abundance of Firmicutes could reflect the local immune status and serve as a predictor of the efficacy and prognosis of ICI treatment.
We previously showed that an increase in Tim-3+ T cells in peripheral blood mononuclear cells (PBMCs) before and after ICI treatment, but not in either time period independently, correlated with treatment responses [35]. Interestingly, the present study did not show a significant correlation between the expression of markers on PBMCs and the abundance of Firmicutes or AEN information in the bloodstream, which supports the notion that predicting the therapeutic effect of ICI by evaluating PBMC samples before treatment was more difficult than comparing PBMC samples before and after ICI treatment.
Although the evaluation of bacterial flora in stool has great potential for facilitating the implementation of treatments, such as stool transplantation [36] and oral administration of bacteria [37], it is limited by defecation conditions [38]. Furthermore, it is difficult to monitor bacteria in the mucus secreted from the mucosa [39]. In comparison, the evaluation of bacterial flora information contained in blood-derived EVs using the methods described in our research is highly versatile as it does not involve complicated ultracentrifugation methods and allows for stable blood sample collection. In addition, this method makes it possible to perform various analyses on stored samples.
In the present study, we used operational taxonomic units (OTUs) to analyze bacterial information. Future studies are required to evaluate bacterial information with amplicon sequence variant (AVE), which enables higher resolution.
In general, the average size of bacteria is approximately 0.4 to 2 μm in diameter and 0.5 to 5 μm in length [40]. In contrast, the mean diameter of EVs secreted from oral and fecal flora was smaller than 200 nm (Fig. 1b), which enable their passage through a 200-nm filter (but not bacteria). Moreover, it has been reported that bacterial EVs in the gut lumen can gain access to the circulatory system [41]. Therefore, we believe that we have analyzed the bacterial-derived EVs which originate from the fecal and oral flora.
This study has several limitations. Considering that this is a pilot study with a relatively small population size, further large-scale and multi-institutional studies with an independent cohort are warranted to confirm our findings. Second, we could not clarify the source of circulating EVs with bacteria DNA. Since our present data showed that serum EVs contained several bacterial components, which represent the gut and oral bacterial flora (Bacteroidetes, Firmicutes, Actinobacteria, and Proteobacteria phylum), we postulate that each localized bacterial flora releases EVs into the bloodstream. Third, although we identified a strong correlation between the Firmicutes abundance in serum EVs and the immune status within tumor tissues, we could not clarify the underlying mechanism; hence, further investigation is warranted.
In conclusion, information on Firmicutes abundance derived from EVs in the blood can reflect the local immune status of the tumor microenvironment and could be used to predict the efficacy and prognosis of UC immunotherapy.
Supplementary Information
Below is the link to the electronic supplementary material.
Authors contributions
Conceptualization was done by KJ, AK, and TS, Methodology was done by KJ, AK, TK, DM, TK, MM, AY, TU, GY, KO, YK, ET, and MU; Investigation was done by KJ, AK, and TS; Visualization was done by KJ and AK; Funding acquisition was don by KJ, AK, HW, and NN; Project administration was done by AK; Supervision was done by KT, HW, and NN; Writing—original draft were done by KJ and AK; Writing—review and editing were done by KJ, AK, TS, TK, MM, KT, HW, and NN. All authors read and approved the final manuscript.
Funding
Japan Society for the Promotion of Science Grants-in-Aid for Scientific Research (C) (19K09709 awarded to AK); and the Suzuki Urological Foundation in Japan (awarded to AK).
Data availability
The data generated in this study are available within the article and its supplementary data files.
Declarations
Conflict of interest
The authors declare that they have no conflicting interest.
Ethical approval
The study was approved by the ethics review board of both the Osaka University Medical Hospital (#13397–2, #14069–3).
Consent to participate
Informed consent was obtained from all individual participants included in the study.
Consent to publish
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Kentaro Jingushi, Atsunari Kawashima and Takuro Saito have contributed equally to this work.
References
- 1.Nadal R, Bellmunt J. Management of metastatic bladder cancer. Cancer Treat Rev. 2019;76:10–21. doi: 10.1016/j.ctrv.2019.04.002. [DOI] [PubMed] [Google Scholar]
- 2.Kawashima A, Kanazawa T, Jingushi K, et al. Phenotypic analysis of tumor tissue-infiltrating lymphocytes in tumor microenvironment of bladder cancer and upper urinary tract carcinoma. Clin Cancer. 2019;17:114–124. doi: 10.1016/j.clgc.2018.11.004. [DOI] [PubMed] [Google Scholar]
- 3.Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Sci. 2020;367:6478. doi: 10.1126/science.aau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stentz R, Carvalho AL, Jones EJ, Carding SR. Fantastic voyage: the journey of intestinal microbiota-derived microvesicles through the body. Biochem Soc Trans. 2018;46:1021–1027. doi: 10.1042/BST20180114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tulkens J, De Wever O, Hendrix A. Analyzing bacterial extracellular vesicles in human body fluids by orthogonal biophysical separation and biochemical characterization. Nat Protoc. 2020;15:40–67. doi: 10.1038/s41596-019-0236-5. [DOI] [PubMed] [Google Scholar]
- 6.Chen C, Song X, Wei W, et al. The microbiota continuum along the female reproductive tract and its relation to uterine-related diseases. Nat Commun. 2017;8:875. doi: 10.1038/s41467-017-00901-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lundy SD, Sangwan N, Parekh NV, et al. Functional and taxonomic dysbiosis of the gut, urine, and semen microbiomes in male infertility. Eur Urol. 2021;79:826–836. doi: 10.1016/j.eururo.2021.01.014. [DOI] [PubMed] [Google Scholar]
- 8.Miyauchi E, Kim SW, Suda W, et al. Gut microorganisms act together to exacerbate inflammation in spinal cords. Nat. 2020;585:102–106. doi: 10.1038/s41586-020-2634-9. [DOI] [PubMed] [Google Scholar]
- 9.Park SY, Hwang BO, Lim M, et al. Oral-gut microbiome axis in gastrointestinal disease and cancer. Cancers (Basel) 2021;13:2124. doi: 10.3390/cancers13092124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saito T, Nishikawa H, Wada H, et al. Two FOXP3(+)CD4(+) T cell subpopulations distinctly control the prognosis of colorectal cancers. Nat Med. 2016;22:679–684. doi: 10.1038/nm.4086. [DOI] [PubMed] [Google Scholar]
- 11.Matsushita M, Fujita K, Hayashi T, et al. Gut microbiota-derived short-chain fatty acids promote prostate cancer growth via IGF1 signaling. Cancer Res. 2021;81:4014–4026. doi: 10.1158/0008-5472.CAN-20-4090. [DOI] [PubMed] [Google Scholar]
- 12.Hopkins AM, Kichenadasse G, Karapetis CS, Rowland A, Sorich MJ. Concomitant antibiotic use and survival in urothelial carcinoma treated with atezolizumab. Eur Urol. 2020;78:540–543. doi: 10.1016/j.eururo.2020.06.061. [DOI] [PubMed] [Google Scholar]
- 13.Mager LF, Burkhard R, Pett N, et al. Microbiome-derived inosine modulates response to checkpoint inhibitor immunotherapy. Sci. 2020;369:1481–1489. doi: 10.1126/science.abc3421. [DOI] [PubMed] [Google Scholar]
- 14.Pederzoli F, Bandini M, Raggi D, et al. Is there a detrimental effect of antibiotic therapy in patients with muscle-invasive bladder cancer treated with neoadjuvant pembrolizumab? Eur Urol. 2021;80:319–322. doi: 10.1016/j.eururo.2021.05.018. [DOI] [PubMed] [Google Scholar]
- 15.Routy B, Le Chatelier E, Derosa L, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Sci. 2018;359:91–97. doi: 10.1126/science.aan3706. [DOI] [PubMed] [Google Scholar]
- 16.Kalaora S, Nagler A, Nejman D, et al. Identification of bacteria-derived HLA-bound peptides in melanoma. Nat. 2021;592:138–143. doi: 10.1038/s41586-021-03368-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nejman D, Livyatan I, Fuks G, et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Sci. 2020;368:973–980. doi: 10.1126/science.aay9189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poore GD, Kopylova E, Zhu Q, et al. Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nat. 2020;579:567–574. doi: 10.1038/s41586-020-2095-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edge SB, Committee American Joint, on Cancer, AJCC cancer staging manual. 7. New York: Springer; 2010. [Google Scholar]
- 20.Moriyama K, Ando C, Tashiro K, et al. Polymerase chain reaction detection of bacterial 16S rRNA gene in human blood. Microbiol Immunol. 2008;7:375–382. doi: 10.1111/j.1348-0421.2008.00048.x. [DOI] [PubMed] [Google Scholar]
- 21.Afgan E, Baker D, Batut B, et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018;46:W537–W544. doi: 10.1093/nar/gky379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hesaka A, Sakai S, Hamase K, Ikeda T, Matsui R, Mita M, Horio M, Isaka Y, Kimura T. D-Serine reflects kidney function and diseases. Sci Rep. 2019;9:5104. doi: 10.1038/s41598-019-41608-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawashima A, Yamamoto Y, Sato M, et al. FAN score comprising fibrosis-4 index, albumin-bilirubin score and neutrophil-lymphocyte ratio is a prognostic marker of urothelial carcinoma patients treated with pembrolizumab. Sci Rep. 2021;11:21199. doi: 10.1038/s41598-021-00509-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: Human gut microbes associated with obesity. Nat. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 25.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nat. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 26.Liang D, Leung RK, Guan W, Au WW. Involvement of gut microbiome in human health and disease: Brief overview, knowledge gaps and research opportunities. Gut Pathog. 2018;10:3. doi: 10.1186/s13099-018-0230-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma EH, Bantug G, Griss T, et al. Serine is an essential metabolite for effector T cell expansion. Cell Metab. 2017;25:482. doi: 10.1016/j.cmet.2017.01.014. [DOI] [PubMed] [Google Scholar]
- 28.Wu J, Li G, Li L, Li D, Dong Z, Jiang P. Asparagine enhances LCK signalling to potentiate CD8(+) T-cell activation and anti-tumour responses. Nat Cell Biol. 2021;23:75–86. doi: 10.1038/s41556-020-00615-4. [DOI] [PubMed] [Google Scholar]
- 29.Chu TL, Guan Q, Nguan CY, Du C. Halofuginone suppresses T cell proliferation by blocking proline uptake and inducing cell apoptosis. Int Immunopharmacol. 2013;16:414–423. doi: 10.1016/j.intimp.2013.04.031. [DOI] [PubMed] [Google Scholar]
- 30.Boussiotis VA. Molecular and biochemical aspects of the PD-1 checkpoint pathway. N Engl J Med. 2016;375:1767–1778. doi: 10.1056/NEJMra1514296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Flores-Toro JA, Luo D, Gopinath A, et al. CCR2 inhibition reduces tumor myeloid cells and unmasks a checkpoint inhibitor effect to slow progression of resistant murine gliomas. Proc Natl Acad Sci USA. 2020;117:1129–1138. doi: 10.1073/pnas.1910856117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagase H, Takeoka T, Urakawa S, et al. ICOS(+) Foxp3(+) TILs in gastric cancer are prognostic markers and effector regulatory T cells associated with helicobacter pylori. Int J Cancer. 2017;140:686–695. doi: 10.1002/ijc.30475. [DOI] [PubMed] [Google Scholar]
- 33.Urakawa S, Yamasaki M, Makino T, et al. The impact of ICOS(+) regulatory T cells and Helicobacter pylori infection on the prognosis of patients with gastric and colorectal cancer: potential prognostic benefit of pre-operative eradication therapy. Cancer Immunol Immunother. 2021;70:443–452. doi: 10.1007/s00262-020-02696-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oh DY, Kwek SS, Raju SS, et al. Intratumoral CD4(+) T cells mediate anti-tumor cytotoxicity in human bladder cancer. Cell. 2020;181:1612–25.e13. doi: 10.1016/j.cell.2020.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kato R, Yamasaki M, Urakawa S, et al. Increased Tim-3(+) T cells in PBMCs during nivolumab therapy correlate with responses and prognosis of advanced esophageal squamous cell carcinoma patients. Cancer Immunol Immunother. 2018;67:1673–1683. doi: 10.1007/s00262-018-2225-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ianiro G, Rossi E, Thomas AM, et al. Faecal microbiota transplantation for the treatment of diarrhoea induced by tyrosine-kinase inhibitors in patients with metastatic renal cell carcinoma. Nat Commun. 2020;11:4333. doi: 10.1038/s41467-020-18127-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li S, Jiang W, Zheng C, et al. Oral delivery of bacteria: basic principles and biomedical applications. J Control Release. 2020;327:801–833. doi: 10.1016/j.jconrel.2020.09.011. [DOI] [PubMed] [Google Scholar]
- 38.David LA, Materna AC, Friedman J, Campos-Baptista MI, Blackburn MC, Perrotta A, Erdman SE, Alm EJ. Host lifestyle affects human microbiota on daily timescales. Gen Biol. 2014;15:R89. doi: 10.1186/gb-2014-15-7-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Desai MS, Seekatz AM, Koropatkin NM, et al. A dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell. 2016;167:1339–53.e21. doi: 10.1016/j.cell.2016.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marshall WF, Young KD, Swaffer M, et al. What determines cell size? BMC Biol. 2012;10:101. doi: 10.1186/1741-7007-10-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chronopoulos A, Kalluri R. Emerging role of bacterial extracellular vesicles in cancer. Oncogene. 2020;39:6951–6960. doi: 10.1038/s41388-020-01509-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in this study are available within the article and its supplementary data files.