Abstract
In cells infected with the herpes simplex virus 1 (HSV-1) recombinant R3616 lacking both copies of the γ134.5 gene, the double-stranded protein kinase R (PKR) is activated, eIF-2α is phosphorylated, and protein synthesis is shut off. Although PKR is also activated in cells infected with the wild-type virus, the product of the γ134.5 gene, infected-cell protein 34.5 (ICP34.5), binds protein phosphatase 1α and redirects it to dephosphorylate eIF-2α, thus enabling sustained protein synthesis. Serial passage in human cells of a mutant lacking the γ134.5 gene yields second-site, compensatory mutants lacking various domains of the α47 gene situated next to the US11 gene (I. Mohr and Y. Gluzman, EMBO J. 15:4759–4766, 1996). We report the construction of two recombinant viruses: R5103, lacking the γ134.5, US8, -9, -10, and -11, and α47 (US12) genes; and R5104, derived from R5103 and carrying a chimeric DNA fragment containing the US10 gene and the promoter of the α47 gene fused to the coding domain of the US11 gene. R5104 exhibited a protein synthesis profile similar to that of wild-type virus, whereas protein synthesis was shut off in cells infected with R5103 virus. Studies on the wild-type parent and mutant viruses showed the following: (i) PKR was activated in cells infected with parent or mutant virus but not in mock-infected cells, consistent with earlier studies; (ii) lysates of R3616, R5103, and R5104 virus-infected cells lacked the phosphatase activity specific for eIF-2α characteristic of wild-type virus-infected cells; and (iii) lysates of R3616 and R5103, which lacked the second-site compensatory mutation, contained an activity which phosphorylated eIF-2α in vitro, whereas lysates of mock-infected cells or cells infected with HSV-1(F) or R5104 did not phosphorylate eIF-2α. We conclude that in contrast to wild-type virus-infected cells, which preclude the shutoff of protein synthesis by causing rapid dephosphorylation of eIF-2α, in cells infected with γ134.5− virus carrying the compensatory mutation, eIF-2α is not phosphorylated. The activity made apparent by the second-site mutation may represent a more ancient mechanism evolved to preclude the shutoff of protein synthesis.
Cells infected with a variety of viruses synthesize complementary RNA either because RNA viruses require a RNA template for the synthesis of complementary strands or because of overlapping transcription of genes encoded on both strands of DNA viruses. The consequence of annealing of complementary RNAs is activation of a double-stranded RNA-dependent protein kinase R (PKR), phosphorylation of the α subunit of the translation initiation factor 2 (eIF-2α), and total shutoff of protein synthesis. Viruses have evolved a variety of mechanisms to block the shutoff of protein synthesis. These mechanisms include degradation of PKR (poliovirus), proteins which block the binding of double-stranded RNA to PKR (influenza virus NS1 protein), production of short double-stranded RNA that binds but fails to activate PKR (adenovirus VaIRNA), and proteins which block the phosphorylation of eIF-2α (vaccinia virus K3L) (2, 3, 8, 16, 19, 21, 23). Herpes simplex viruses 1 and 2 (HSV-1 and HSV-2) are especially vulnerable to the shutoff of protein synthesis inasmuch as viral genes are located on both strands of the DNA and signals at the end of transcriptional units are not entirely effective in terminating transcription. Earlier studies have shown that fully half of the HSV-1 genome is represented in double-stranded RNA prepared by self-annealing of RNA extracted from infected cells. The melting temperature of the RNA was consistent with duplexes containing few mismatches and therefore not due to double-stranded stems arising from secondary structures of mRNA (15, 17). HSV evolved a gene, γ134.5, whose product, infected-cell protein 34.5 (ICP34.5), precludes the shutoff of protein synthesis by activated PKR (5–7). The γ134.5 gene maps in the inverted repeat sequences ab and b′a′ flanking the unique long (UL) sequence (Fig. 1) and therefore is present in two copies per genome (1, 4, 27, 29). Unlike the gene products of other viruses, ICP34.5 blocks the shutoff of protein synthesis by interacting with protein phosphatase 1 and redirecting its activity to dephosphorylate eIF-2α (14). In cells infected with wild-type virus or the genetically engineered virus from which the γ134.5 genes had been deleted, PKR is activated, eIF-2α is phosphorylated, and protein synthesis is shut off in cells infected with the γ134.5− virus (7).
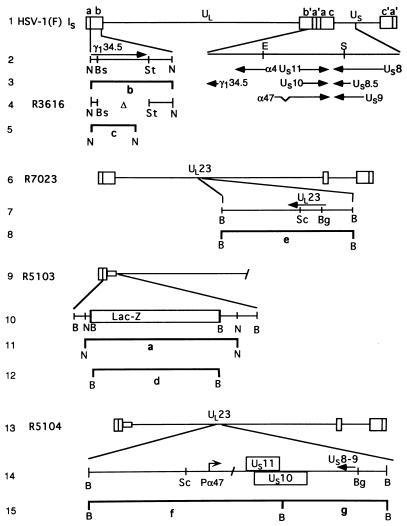
FIG. 1.
Schematic representation of the DNA sequence arrangements of the wild-type and recombinant viruses used in this study. Line 1, schematic representation of the wild-type HSV-1 genome. The genome consists of two covalently linked components, L and S, each consisting of unique sequences (UL and US) flanked by inverted repeats. The arrangement shown is the IS isoform in which the S component is inverted relative to the prototypic orientation of the L component. The inverted repeat sequences designated ab and b′a′ flanking the UL sequence are 9 kb in size, whereas the repeat sequences ac and c′a′ flanking the US sequence are each 6.3 kb in size. Line 2, expansion of specific domains of the genome showing gene arrangements within the expanded region. Line 4, schematic representation of one of two γ134.5 coding domains in recombinant R3616 in which the sequences between the BstEII and StuI restriction endonuclease sites had been deleted. Line 6, representation of a portion of the genome of the recombinant R7023 shown in the IS arrangement. Line 7, schematic representation of the wild-type UL23 gene encoding thymidine kinase and key restriction endonuclease sites present in R7023. The domain encoding the genes US8 through US12 as well as the reiterated sequences a′ and ac and the portion of the b′ sequence encoding one of the copies of the γ134.5, ORF O, and ORF P genes are absent. R7023 was the parent of R5103 schematically represented in line 9. In R5103, the coding domain between the BstEII and StuI restriction sites in the remaining copy of the γ134.5 gene was replaced with the E. coli lacZ gene represented in line 10. Line 13, schematic representation of the recombinant virus R5104 constructed by homologous recombination between plasmid pRB4999 and R5103 DNA. In the resulting recombinant, R5104 (line 14), the UL23 gene was disrupted by the insertion of US10 and US11 driven by the α47 promoter. Lines 3, 5, 8, 11, 12, and 15 represent the predicted bands produced by restriction endonuclease digestion of viral DNAs and are shown as reference for the bands shown in Fig. 2. Abbreviations: B, BamHI; N, NcoI; Bs, BstEII; St, StuI; Sc, SacI; Bg, BglII.
Mohr and Gluzman (24) reported that serial passage of γ134.5− virus in human cells yielded a series of mutants capable of sustained protein synthesis. The compensatory mutation was mapped to a domain located at the junction between the unique short (US) sequence and the inverted repeat sequence ca flanking it (Fig. 1). Thus, all of the characterized mutants contained a deletion between the US11 gene and the inverted repeat sequence. Analyses of one second-site compensatory mutation revealed that the deletion brought the α47 promoter 5′ to the coding sequence of the US11 gene, converting the latter to an early gene (13).
In this report, we show that insertion of a DNA fragment carrying the US10 gene and the US11 gene driven by the α47 promoter into the genome of a virus (R5103) lacking γ134.5, US8, US9, US10, US11, and US12 (α47) genes yielded a recombinant virus (R5104) which exhibited the capacity for sustained protein synthesis similar to that of the wild-type parent virus. We also report that PKR was activated by R5103, the parent virus by R5104, the virus whose capacity for sustained protein synthesis was restored, and by R3616, the virus lacking the γ134.5 genes. Whereas cells infected with wild-type parent virus exhibited activated phosphatase activity specific for eIF-2α, the cells infected with R5103 or R5104 lacked this activity. Finally, lysates of cells infected with R5103 but not with R5104 phosphorylated eIF-2α in vitro. The key conclusion is that although PKR is activated, the second-site compensatory gene created or activated in R5104 blocks the phosphorylation of eIF-2α by the activated PKR.
Relevant to this report are the function and distribution of the γ134.5 gene among the members of herpesvirus family. The γ134.5 gene encodes two functions. The first, mapping throughout the coding domain of the gene, enables the replication and spread of the virus in the central nervous system (4). The second, mapping in the carboxyl-terminal domain, enables the interaction of ICP34.5 with protein phosphatase 1α and precludes the premature termination of protein synthesis described above (5, 6, 12, 14). The carboxyl-terminal domain of ICP34.5 is homologous to the corresponding domain of a conserved mammalian protein known as GADD34, an acronym for growth arrest and DNA damage (12, 30). GADD proteins are expressed during differentiation, serum deprivation, or repair of damaged DNA. The sequence encoding the carboxyl-terminal domain of the murine homolog of GADD34 can substitute for the corresponding γ134.5 in blocking the premature shutoff of protein synthesis, but the recombinants carrying the chimeric gene are avirulent (12).
The γ134.5 gene is encoded by HSV-1, HSV-2, and the simian B virus but has not been found in other herpesvirus genomes (6). Since it is expected that all herpesviruses have evolved mechanism to preclude shutoff of protein synthesis by activated PKR, it could be expected that viruses lacking the γ134.5 gene have evolved alternative mechanisms to deal with this host response to infection. Our interest in the second-site compensatory mutation first reported by Mohr and Gluzman (24) stems from the possibility that GADD34 performs a similar function in uninfected cells under conditions of stress, i.e., that in the course of its evolution HSV acquired the carboxyl-terminal domain of GADD34 but retained in a modified form a more ancient mechanism for precluding shutoff of protein synthesis that is perhaps more widespread among the members of the herpesvirus family.
MATERIALS AND METHODS
Plasmids.
Standard methods (22) were used for all plasmid constructions described here. The HSV DNA in plasmid pRB4999 consisted of a BamHI Q fragment carrying in the BglII restriction endonuclease site a chimeric gene consisting of the promoter of the α47 gene juxtaposed to the US11 gene and the adjacent US10 gene (Fig. 1, line 14). This was constructed as follows. In the first step, plasmid pRB421, encoding the coding domains of US11, US10, and α47 within the 3.2-kb EcoRI-SalI fragment, was digested with BstEII and NruI, deleting the 720-bp α47 coding domain and the US11 promoter (26a). The new plasmid, pRB4028, contains the US10 gene and the α47 promoter juxtaposed to the US11 coding sequence. Next, a plasmid encoding the BamHI Q fragment, pRB3982, containing an XbaI-KpnI polylinker inserted between the BglII and SacI sites of UL23, was digested with the restriction endonuclease BglII. The 2.5-kb EcoRI-SalI fragment of pRB4028 and the BglII-digested pRB3982 were blunt ended with Klenow enzyme and then ligated. In plasmid pRB849, the Escherichia coli lacZ gene replaced the γ134.5 open reading frame (ORF) contained in the BstEII-StuI fragment of BamHI S (Fig. 1, line 10) (6a). Plasmid pRB4974, containing the 1.8-kb NcoI fragment encoding the γ134.5 coding sequence, was reported elsewhere (18).
Cells and viruses.
The Vero, HeLa, and SK-N-SH cell lines were obtained from the American Type Culture Collection. The 143 thymidine kinase-minus (143TK−) and the rabbit skin cells were originally obtained from Carlo Croce and J. McClaren, respectively. The cell lines were propagated in Dulbecco modified Eagle medium supplemented with 5% newborn calf serum (Vero and rabbit skin cells), 10% (SK-N-SH cells), or 5% (HeLa cells) fetal bovine serum, and 100 μg of bromodeoxyuridine per ml (143TK− cells). HSV-1(F) is the prototype strain used in this laboratory (9). The recombinant virus R3616, described elsewhere (4) in detail, lacks 1,000 bp in the domain of both copies of the HSV-1(F) γ134.5 gene.
The recombinant virus R7023 (Fig. 1, line 6), described elsewhere (20), lacks the genes US8 through US12 (α47), contains single copies of the γ134.5 ORF O, ORF P, α4, and α0 genes, and maintains the remainder of the S component frozen in an inverted orientation (IS and ISL) (20). Recombinant virus R5103 (Fig. 1, line 9) was isolated from among the progeny of cotransfection of rabbit skin cells with intact R7023 DNA plasmid pRB849 on the basis of expression of β-galactosidase (25). In the recombinant R5103, the gene encoding β-galactosidase replaced the remaining copy of the γ134.5 gene. This virus therefore lacks the native US8 through US12, γ134.5, ORF P, and ORF O genes. The recombinant R5104 (Fig. 1, line 13) was isolated from the progeny of transfection of 143TK− cells maintained in medium containing bromodeoxyuridine and transfected with intact R5103 DNA and pRB4999 by selection against thymidine kinase. R5104 therefore lacks the native US8 through US12, UL23, UL24, γ134.5, ORF P, and ORF O genes. The insert restored the US10 gene as well as the US11 gene fused to the α47 promoter and included the 3′ 186 nucleotides of the US9 gene.
Analyses of viral DNA.
Digestion of viral DNAs with appropriate enzymes, electrophoretic separation in agarose gels, transfer to Zeta Probe membranes by capillary transfer, and hybridization using radiolabeled DNA probes were done as previously described (12, 28).
Determination of eIF-2α kinase and phosphatase activities.
Generation of cytoplasmic (S10) fractions from HeLa cells 7 h after infection and measurements of phosphatase activity were done as described elsewhere (7, 12). Briefly, eIF-2α-specific phosphatase activity of infected-cell lysates was measured by incubating radiolabeled eIF-2 [1.0 pmol, 1.0 pmol eIF-2(α32P), and 0.7 pmol eIF-2(β32P) at 0.6 Ci/mmol] with 6 μl of infected-cell S10 fractions supplemented with 4.5 μl of TKM buffer (10 mM Tris-HCl [pH 7.5], 20 mM KCl, 2.25 mM MgCl2) and 1.0 mM ATP in a final volume of 12.5 μl at 34°C. Aliquots (6 μl) were removed at 60 and 150 s for all recombinant viruses and at 20 and 60 s after incubation for HSV-1(F), denatured in sodium dodecyl sulfate (SDS), and electrophoretically separated on a 7% denaturing polyacrylamide gel that was stained, dried, and autoradiographed. The eIF-2α band was cut from each gel lane, and its Cerenkov radioactivity was measured.
To determine eIF-2α kinase activity of infected-cell lysates, the equivalent of 0.2 μl of each lysate was incubated with 5 pmol of purified eIF-2 and 0.04 mM [γ-32P]ATP (25 Ci/mmol) in 10 μl of TKM buffer for 1 min at 34°C. Reactions were terminated and electrophoretically separated on a 7% denaturing polyacrylamide gel (11). Following electrophoretic separation, the gel was silver stained, dried, and autoradiographed as described above and elsewhere (14).
In vivo kinase assay and in vivo protein synthesis.
Replicate 25-cm2 flask cultures of HeLa cells were exposed to 1 ml of medium 199V (medium 199 supplemented with 1% calf serum) containing 10 PFU of recombinant gE− viruses (R7023, R5103, and R5104) per cell and then incubated at 13 h after infection first in Eagle minimal essential medium lacking phosphate and 1 h later in the same medium but supplemented with 200 μCi of [32P]orthophosphate (carrier free; New England Nuclear). At 18 h after infection, the cells were rinsed once with phosphate-buffered saline lacking Ca and Mg (PBS-A) and then transferred in ice-cold PBS-A to an Eppendorf tube, pelleted by centrifugation at 4°C, and lysed in 100 μl of radioimmunoprecipitation assay buffer (PBS-A containing 0.1% SDS, 0.5% deoxycholate, 1% Nonidet P-40, and 2 mM sodium benzamidine) (7, 26). The lysates were shaken for 30 min at 4°C with 25 μl of protein A conjugated to Sepharose beads and centrifuged, and the supernatant fluids were reacted at 4°C with 1.5 μg of rabbit polyclonal anti-PKR immunoglobulin (K-17; Santa Cruz Research) for 3.5 h. The immunoprecipitate mixture was then incubated with protein A-Sepharose for 1 h, centrifuged, and washed three times with cold radioimmunoprecipitation assay buffer. Samples were resuspended in disruption buffer (50 mM Tris-HCl [pH 7.0], 2% SDS, 700 mM β-mercaptoethanol, 2.75% sucrose), boiled for 3 min, electrophoretically separated on 0.1% SDS-containing 10% (vol/vol) polyacrylamide gels cross-linked with N,N′-diallyltartardiamide (DATD), electrically transferred to a nitrocellulose sheet, and exposed to Kodak XAR5 film at −70°C.
In vivo protein synthesis.
Protein labeling experiments were done as previously described (5, 26). Briefly, infected SK-N-SH cells in 25-cm2 flasks were incubated for 30 min at 13.5 h after infection in medium 199V lacking methionine and then in 1 ml of the same medium supplemented with 50 μCi of [35S]methionine. After 1 h of labeling, the cells were rinsed twice with PBS-A and then scraped in 1 ml of ice-cold PBS-A, pelleted, solubilized in disruption buffer, boiled, electrophoretically separated on a 12% (vol/vol) polyacrylamide gel cross-linked with DATD, electrically transferred to a nitrocellulose sheet, and subjected to autoradiography.
RESULTS
Genotypes of the recombinants derived for this study.
The DNA sequence arrangements of recombinant viruses R7023 and R3616 were described elsewhere and are shown schematically in Fig. 1 (4, 5, 20). R3617, the parent of R3616, lacks a 500-nucleotide stretch in the sequence of the tk gene (4, 5). The recombinant virus R5103 was constructed by homologous recombination and contained the E. coli lacZ gene in place of the remaining copy of the γ134.5 gene (25). Hybridization of electrophoretically separated, immobilized, NcoI-digested viral DNA with a nick-translated probe from plasmid pRB4974 revealed that the 1.8-kb NcoI fragment present in HSV-1(F) and R7023 DNA (Fig. 2A, band B) shifted to 4.0 kb in R5103 and R5104 viral DNAs carrying the lacZ gene (Fig. 2A, band A). The evidence that the lacZ gene was responsible for the altered electrophoretic mobility of the BamHI S fragment in R5103 and R5104 is shown in Fig. 2B. Specifically, the 389-bp lac operon sequence present in plasmid pGEM 3Zf(+) hybridized with the 3.2-kb BamHI fragment (Fig. 2B, band D) in R5103 and R5104 but failed to hybridize with any fragments derived from wild-type, R7023, or R3616 virus.
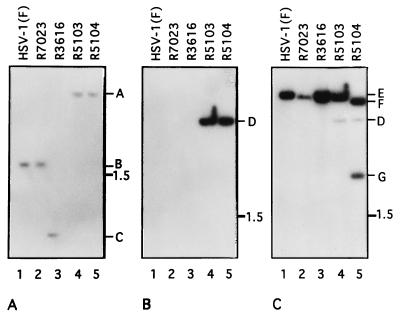
FIG. 2.
Autoradiographic images of electrophoretically separated digests of viral DNAs hybridized with specific probes. (A) Electrophoretically separated NcoI digests hybridized with 32P-labeled nick-translated probe of the wild-type NcoI fragment encoding the γ134.5 gene cloned as pRB4974. Both wild-type HSV-1(F) and recombinant virus R7023 contained the wild-type γ134.5 gene and the radioactive probe hybridized with a 1.8-kb NcoI fragment (band B); in recombinant virus R3616, the NcoI fragment (band C) was approximately 800 bp smaller since it lacked the sequence between the BstEII and StuI sites encoding the γ134.5 gene. In recombinant viruses R5103 and R5104, the 1-kb γ134.5 sequence between BstEII and StuI was replaced by the 3.2-kb E. coli lacZ gene, yielding a 4-kb DNA fragment (band A). (B) Electrophoretically separated BamHI digests probed with the labeled pGEM 3Zf(+) vector containing approximately 400 bp of the lacZ sequence. The lacZ sequences in recombinant viruses R5103 and R5104 formed single 3.2-kb bands (band D). The probe failed to hybridize with digests of HSV-1(F), R7023, or R3616 DNA. (C) Autoradiogram of the electrophoretically separated fragments shown in panel B but stripped and hybridized with labeled plasmid pRB3982 carrying the BamHI Q DNA fragment in a pUC-9 vector. The probe hybridized with wild-type 3.58-kb BamHI Q fragments (band E) in digests of HSV-1(F), R7023, R3616, or R5103 consistent with the expected size of the wild-type BamHI Q fragment and with an additional band (band D) in digests of recombinants R5103 and R5104 corresponding to the hybridization of the lacZ sequence in the probe with the gene inserted in place of the γ134.5 gene. In lane 5, two R5104 bands of 3.4 kb (band F) and 2.1 kb (band G) hybridized with the BamHI Q fragment. The insertion of the α47-US11 chimeric gene into BglII site of the BamHI Q fragment introduced an additional BamHI site from the US11 gene as shown schematically in Fig. 1, line 15.
The recombinant virus R5104 was constructed by transfection of intact R5103 DNA and a DNA fragment containing the native US10 gene and the ORF of US11 fused to the α47 promoter inserted into the BglII site of the BamHI Q fragment. Consequently, hybridization of electrophoretically separated BamHI-digested viral DNA with a radioactive probe prepared from plasmid pRB3982 detected a 3.58-kb BamHI Q fragment (Fig. 1, line 8; Fig. 2C, band E) in wild-type, R7023, R3616, and R5103 viruses, whereas in R5104 the probe hybridized with 3.43- and 2.18-kb fragments (Fig. 2C, bands F and G). In addition, the 389-bp lac operon sequence from the vector hybridized with the 3.2-kb lacZ fragment in the DNA of viruses R5103 and R5104 (Fig. 2C, band D), as shown in Fig. 2B.
The ability to synthesize proteins late in infection was restored in a γ134.5− recombinant by the insertion of a DNA fragment containing US10 and of the US11 gene driven by the α47 promoter.
The objective of this series of experiments was to characterize the phenotypes of the recombinant viruses generated for these studies and described in detail in Materials and Methods. Replicate cultures of HeLa cells were mock infected or exposed to 10 PFU of HSV-1(F), R7023, R3616, R3617, R5103, or R5104. The cultures were labeled with [35S]methionine at 13.5 h after infection for 1 h and processed for autoradiography as described in Materials and Methods. The results (Fig. 3) were as follows.
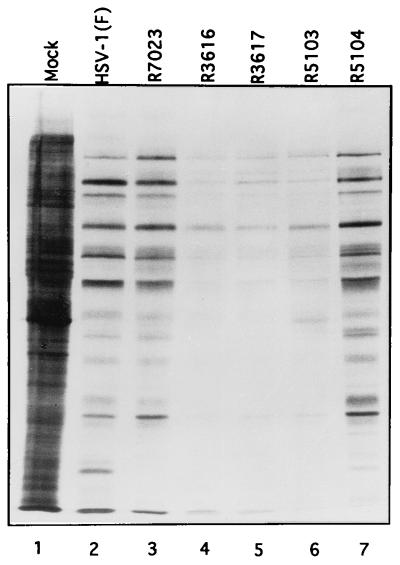
FIG. 3.
Autoradiographic image of electrophoretically separated [35S]methionine-labeled proteins prepared from lysates of SK-N-SH cells mock infected or virus infected. Replicate cultures of SK-N-SH cells were mock infected or infected with 10 PFU of HSV-1(F), R7023, R3616, R3617, R5103, or R5104. At 13.5 h after infection the medium was replaced with medium 199V lacking methionine but supplemented with 50 μCi of [35S]methionine (specific activity, >1,000 Ci/mmol; Amersham) for 1 h. After labeling, the infected cells were rinsed, harvested, solubilized in an SDS-containing buffer, and electrophoretically separated on a denaturing 12% polyacrylamide gel cross-linked with DATD. The electrophoretically separated proteins were transferred to a nitrocellulose sheet and subjected to autoradiography.
(i) The level of protein synthesis in cells infected with HSV-1(F) (Fig. 3, lane 2) could not be differentiated from that of cells infected with R7023 or R5104 (lane 3 or 7, respectively). In contrast, there was a significant decrease in the accumulation of labeled proteins in cells infected with R3616, R3617, and R5103 (lanes 4 to 6, respectively). Each of the latter three recombinants lacks the γ134.5 gene.
(ii) Restoration of protein synthesis was readily apparent in cells infected with the recombinant virus R5104 (Fig. 3, lane 7). The higher accumulation of labeled proteins was not due to the absence of the tk gene since this gene was also absent in R3617 (lane 5). We may conclude therefore that the restoration of protein synthesis seen in cells infected with R5104 was due to the insertion of a DNA fragment containing the US10 gene under its own promoter and the US11 gene driven by the α47 promoter. The phenotype of R5104 resembles that of the serially passaged γ134.5− virus of Mohr and Gluzman reported elsewhere (24).
PKR phosphorylation status at late time points after infection.
The purpose of this series of experiments was twofold. First, earlier studies led to the conclusion that in infected cells PKR was activated irrespective of the presence or absence of a functional γ134.5 gene, and therefore the factors which determined whether protein synthesis was shut off acted subsequent to the activation of PKR (7). It was of interest, therefore, to verify that PKR was also activated in cells infected with R5103 and R5104, since both lack the γ134.5 gene but exhibit very different protein synthesis profiles. Second, earlier studies were based on precipitation of PKR from lysates of mock-infected or infected cells labeled in vitro by the addition of labeled ATP (7, 13). It was of interest to determine whether PKR was labeled in the infected cell.
In this experiment, HeLa cells mock infected or infected with R7023, R5103, or R5104 were labeled with [32P]orthophosphate and lysed, and the PKR was precipitated with the anti-PKR rabbit polyclonal antibody. The precipitate was electrophoretically separated on a polyacrylamide gel and subjected to autoradiography as described in Materials and Methods. The recombinant R7023 served as the parent virus and represents the phenotype of the wild-type virus since the accumulation of labeled proteins late in infection could not be differentiated from that of the wild-type parent HSV-1(F). The results (Fig. 4) indicate the presence of several labeled protein bands brought down by the anti-PKR antibody from lysates of infected cells. These include PKR (apparent Mr of approximately 67,000), two labeled bands estimated to have apparent Mrs of 75,000 and 200,000, and a less intense, broad band with an apparent Mr of approximately 90,000. The studies described here extend the results reported earlier (7, 13) in that PKR was labeled not only in vitro but also in vivo. These results reinforce the conclusion that PKR is activated in all infected cells irrespective of the level of protein synthesis in these cells.
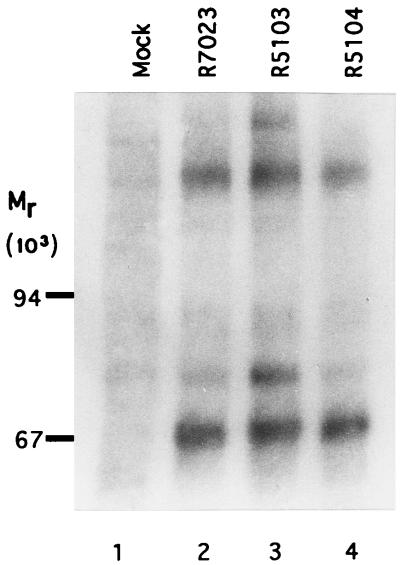
FIG. 4.
Autoradiographic image of PKR electrophoretically separated from immune complexes derived from labeled infected HeLa cells. Replicate HeLa cell cultures were mock infected or infected with R7023, R5103, or R5104 as described in Materials and Methods. At 18 h after infection, the cells were harvested and lysed, and immune precipitates obtained with the anti-PKR antibody (K-17; Santa Cruz Research) were electrophoretically separated on a 12% polyacrylamide gel, transferred to a nitrocellulose sheet, and subjected to autoradiography. The positions of the Mr-200,000, -90,000, and -67,000 proteins determined from the molecular weight markers (Pharmacia) are indicated on the left. The heavily labeled bands with an apparent Mr of 67,000 were identified as PKR (7).
The eIF-2α-specific phosphatase activity in cells infected with R5104 cannot be differentiated from that of other viruses lacking the γ134.5 gene.
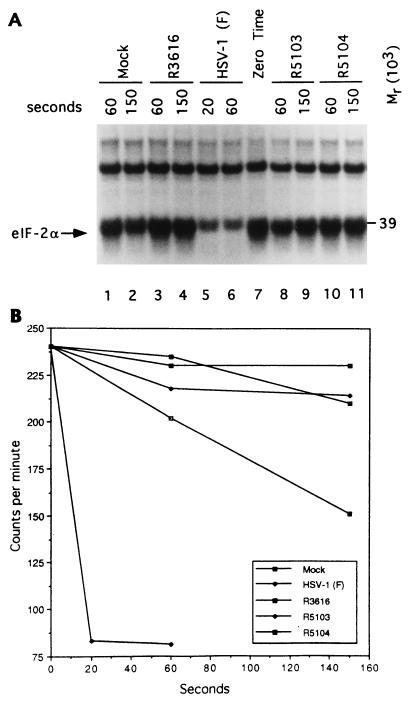
In light of the evidence presented here that PKR was activated but protein synthesis was not shut off in cells infected with R5104, the question arose as to whether eIF-2α is selectively dephosphorylated in a fashion similar to that shown to occur in wild-type-infected cells. In this series of experiments, purified eIF-2 phosphorylated in vitro was reacted with the S10 fraction prepared from lysates of mock-infected cells, cells infected with HSV-1(F), or cells infected with recombinant virus R3616, R5103, or R5104. The results were consistent with the previously reported results that the lysates of cells infected with HSV-1(F) contain a potent phosphatase activity which rapidly dephosphorylates the α subunit of eIF-2 but has no effect on the β subunit or the adventitious Mr-39,000 protein (Fig. 5A, lanes 5 to 7) and that this activity is absent in cells infected with R3616 (lanes 3, 4, and 7) or in mock-infected cells (lanes 1, 2, and 7). The new and significant result is that this activity was also absent in cells infected with R5104. As shown in Fig. 5A, lanes 7 to 11, the rates of dephosphorylation of eIF-2α by lysates of cells infected with R5103 and R5104 cannot be differentiated from those obtained with lysates of cells infected with R3616. The α subunit of eIF-2 migrates closely to the Mr-39,000 phosphoprotein present in the purified eIF-2 fraction. Earlier studies have shown that this protein is unrelated to the eIF-2 and plays no role in protein synthesis (10). Attempts to improve the separation of the Mr-39,000 protein from eIF-2α for autoradiographic analyses have been unsuccessful (10). In additional experiments, purified eIF-2 was electrophoretically separated after reaction with infected-cell lysates and the eIF-2α protein band, and its radioactivity was measured. The results shown in Fig. 5B indicate that only the wild-type-infected lysate rapidly dephosphorylated eIF-2α.
FIG. 5.
Dephosphorylation of eIF-2α by lysates of mock-infected and infected cells. (A) Autoradiographic image of purified, in vitro-labeled eIF-2α reacted with lysates of mock-infected cells or cells infected with wild-type or recombinant viruses for time intervals shown and then electrophoretically separated in a denaturing gel. The reaction was terminated by mixing with buffer containing SDS; the reaction times are indicated above the image. The arrow identifies the α subunit of eIF-2 and the Mr-39,000 protein which copurifies with eIF-2. The slowly migrating unlabeled band represents the β subunit of eIF-2. (B) Radioactivity contained in individual bands as a function of time of exposure to infected cell lysates.
The significant conclusion is that the compensatory mutation which arises in mutants lacking the γ134.5 genes enables continuous protein synthesis by a mechanism different from that observed in wild-type-infected cells. Cells infected with wild-type virus contain an activity which is capable of dephosphorylating eIF-2α, whereas in cells infected with second-site mutants of viruses lacking the γ134.5 gene, this activity was absent. Since protein synthesis continued, it could be predicted that in these cells eIF-2α was not phosphorylated even though PKR was activated.
eIF-2α is not phosphorylated in cells infected with R5104 recombinant virus.
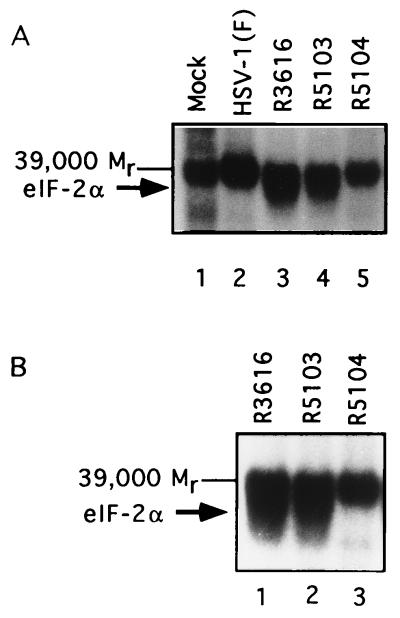
The experiments described above suggested that even though PKR was activated, eIF-2α was not phosphorylated in cells infected with the R5104 recombinant virus. To test this hypothesis, we analyzed the ability of lysates of mock- or virus-infected cells to phosphorylate eIF-2α in the presence of [γ32P]ATP. The results (Fig. 6) show the following: (i) the Mr-39,000 protein was labeled to approximately the same level by all of the lysates tested, and (ii) the α subunit was labeled by lysates of cells infected with R3616 and R5103 but not by the HSV-1(F)- or R5104-infected cell lysates.
FIG. 6.
(A) Autoradiographic image of electrophoretically separated purified eIF-2 samples after reaction in the presence of [γ32P]ATP with S10 fractions from cells mock-infected or infected with HSV-1(F), R7023, R3616, R5103, or R5104. The arrows indicate the position of the α subunit of eIF-2 and the Mr-39,000 protein unrelated to eIF-2. (B) Autoradiographic image of electrophoretically separated proteins contained in the eIF-2 kinase reactions carried out as described above. The samples were electrophoretically separated and run on a longer gel to increase the separation of the Mr-39,000 phosphoprotein from eIF-2α. The arrows indicate the position of the α subunit of eIF-2 and the unrelated Mr-39,000 phosphoprotein present in the purified eIF-2 samples.
Whereas the failure of eIF-2α to be labeled by HSV-1(F) could be attributed to the highly efficient phosphatase activity mobilized by ICP34.5 and directed to eIF-2α, as shown in Fig. 5A, lanes 5 and 6, no such phosphatase activity was present in lysates of cells infected with R5104. We conclude from these analyses that the second-site mutation resulted in a block in the phosphorylation of eIF-2α, enabling protein synthesis to continue even in the absence of the γ134.5 gene product.
DISCUSSION
The double-stranded RNA-activated PKR pathway is an important mechanism by which cells can respond to infectious agents to restrict their multiplication and thereby reduce their ability to spread throughout the body. Viruses, in turn, have evolved numerous strategies for subverting this host response to infection. HSV strains are no exception. Earlier studies have shown that PKR is activated in cells infected with wild-type and mutant HSV-1, but that eIF-2α is phosphorylated concomitant with total shutoff of protein synthesis in cells infected with mutants lacking both copies of the γ134.5 gene (7). Subsequent studies have shown that γ134.5 protein binds to protein phosphatase 1α and redirects its activity to dephosphorylate eIF-2α (14). The domain of the γ134.5 protein which binds protein phosphatase 1α is highly homologous to and in fact replaceable by the corresponding domain of the GADD34 gene (5, 12). In this respect, wild-type HSV-1 differs from other viruses in that its main armamentarium to counter host response to infection was to capture a fragment of a cellular gene to dephosphorylate eIF-2α rather than to evolve mechanisms to prevent the activation of PKR or the phosphorylation of eIF-2α.
The study described in this report stemmed from two considerations. First, the γ134.5 gene is not highly conserved among herpesviruses, yet all herpesviruses could be expected to trigger the activation of PKR and, in turn, evolve mechanisms to preclude the shutoff of protein synthesis by the activated kinase (6). In principle, herpesviruses must have evolved alternative mechanisms for blocking this response to infection. Second, Mohr and Gluzman reported that serial passage of γ134.5− virus in cell culture resulted in the selection of mutants which did not induce the shutoff of protein synthesis characteristic of the γ134.5− viruses (24). Analyses of the mutants derived by Mohr and Gluzman led to two key observations. Thus, they reported that a characteristic of the compensatory mutants is a variable-size deletion encompassing all or most of the coding domain of the α47 gene. This deletion brought the promoter of the α47 gene in juxtaposition to the US11 coding domain, resulting in a change in the kinetics of expression of US11 from that of a γ2 gene to a gene expressed early in infection (24). Moreover, subsequent studies have shown that PKR is activated in cells infected with these compensatory mutants (13). This result suggested that HSV-1 encodes a secondary, cryptic mechanism to block the shutoff of protein synthesis by the activated PKR and that this mechanism was activated by the compensatory mutation discovered by Mohr and Gluzman (24).
In this report, we showed the following.
(i) We have unambiguously localized the secondary, compensatory mutation of Mohr and Gluzman (24) by showing that protein synthesis is shut off in cells infected with the R5103 mutant lacking the γ134.5, US11, and α47 genes but not in cells infected with a mutant carrying an insertion containing as its key element the native US10 gene and the US11 gene driven by the α47 promoter. We should stress that extensive serial passages of R5103 mutant in human SK-N-SH cells failed to yield second-site mutants capable of sustained protein synthesis in infected cells (6a).
(ii) We have demonstrated that in cells infected with the R5104 mutant, in which protein synthesis was restored, the level of phosphatase activity specific for eIF-2α was significantly lower than that present in cells infected with wild-type virus. The significance of this finding stems from the consideration that since there was no increase in phosphatase activity yet protein synthesis was unaffected even though PKR was phosphorylated, it could be expected that the activated PKR did not phosphorylate eIF-2α. This was in fact the case. The conclusion to be drawn from the study described in this report is that the secondary mutation leading to sustained protein synthesis results in the expression of a factor which precludes the phosphorylation of eIF-2α.
Two issues confront us. First, although we have not excluded the expression of hitherto silent ORFs or subtle changes in the US10 gene, the only novel factor immediately apparent in cells infected with the mutant of Mohr and Gluzman (24) derived by serial passage or reconstructed in the study reported here is the expression of US11 as an early gene. It is conceivable that US11 made early blocks PKR from phosphorylating eIF-2α. We should note, however, that US11 is an abundant tegument protein brought into cells early in infection and one which would be available to block the phosphorylation of eIF-2α if this were one of its functions. The role of US11 protein remains unresolved.
The second issue that confronts us is why in the course of its evolution HSV abandoned the process revealed by the second-site mutations in favor of the γ134.5 protein. If US11 protein plays a role in precluding the phosphorylation of eIF-2α, it is conceivable, for example, that in its original composition, US11 was less efficient than the modern version of the γ134.5 protein, that after the acquisition of the γ134.5 gene the US11 gene evolved additional functions, and that expression of US11 late in the replicative cycle confers maximal benefits to the replication of HSV-1. At least some of the issues raised here are amenable to future investigation.
As noted in the introduction, many diverse families of viruses are subject to repression by activated PKR and in turn have evolved mechanisms to alleviate the repression. It is a reflection of the importance of this host response that viruses have evolved diverse mechanisms for dealing with this host response. In some instances, as in the case of vaccinia virus, this host response is blocked by the products of two genes acting by different pathways (3, 8). In the case of HSV-1, while a second and entirely different mechanism appears to exist, it is effective only secondary to mutation. Inasmuch as a mutation rendered this mechanism for abating the host response cryptic but did not fully eliminate it, our results suggest that the evolution of the γ134.5 gene may be a relatively recent phenomenon.
ACKNOWLEDGMENTS
We thank Alice P. W. Poon for careful reading of the manuscript and Suzanne Hessefort and Annette Olin for technical assistance.
This study was aided by Public Health Service grants from the National Cancer Institute (CA47451) to B.R. and the National Heart, Lung, and Blood Institute (HL30121) to M.G. Grant support was also provided to K.A.C. by the Pediatric Scientist Development Program of the National Institute of Child Health and Human Development administered by the Association of Medical School Pediatric Department Chairmen Inc.
REFERENCES
- 1.Ackermann M, Chou J, Sarmiento M, Lerner R A, Roizman B. Identification by antibody to a synthetic peptide of protein specified by a diploid gene located in the terminal repeats of the L component of herpes simplex virus genome. J Virol. 1986;58:843–850. doi: 10.1128/jvi.58.3.843-850.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Black T L, Barber G N, Katze M G. Degradation of the interferon-induced 68,000-Mr protein kinase by poliovirus requires RNA. J Virol. 1993;67:791–800. doi: 10.1128/jvi.67.2.791-800.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carroll K, Elroy-Stein O, Moss B, Jagus R. Recombinant vaccinia virus K3L gene product prevents activation of double-stranded RNA-dependent, initiation factor 2α-specific protein kinase. J Biol Chem. 1993;268:12837–12842. [PubMed] [Google Scholar]
- 3a.Cassady, K., and B. Roizman. Unpublished results.
- 4.Chou J, Kern E R, Whitley R J, Roizman B. Mapping of herpes simplex virus-1 neurovirulence to γ134.5, a gene nonessential for growth in cell culture. Science. 1990;250:1262–1266. doi: 10.1126/science.2173860. [DOI] [PubMed] [Google Scholar]
- 5.Chou J, Roizman B. The γ134.5 gene of herpes simplex virus 1 precludes neuroblastoma cells from triggering the total shutoff of protein synthesis characteristic of programmed cell death in neuronal cells. Proc Natl Acad Sci USA. 1992;89:3266–3270. doi: 10.1073/pnas.89.8.3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chou J, Roizman B. Herpes simplex virus 1 γ134.5 gene function, which blocks the host response to infection maps in the homologous domain of the genes expressed during growth arrest and DNA damage. Proc Natl Acad Sci USA. 1994;91:5247–5251. doi: 10.1073/pnas.91.12.5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6a.Chou, J., and B. Roizman. Unpublished results.
- 7.Chou J, Chen J-J, Gross M, Roizman B. Association of a Mr 90,000 phosphoprotein with protein kinase PKR in cells exhibiting enhanced phosphorylation of translation initiation factor eIF-2α and premature shutoff of protein synthesis after infection with γ134.5− mutants of herpes simplex virus 1. Proc Natl Acad Sci USA. 1995;92:10516–10520. doi: 10.1073/pnas.92.23.10516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davies M V, Chang H W, Jacobs B L, Kaufman R J. The E3L and K3L vaccinia virus gene products stimulate translation through inhibition of the double-stranded RNA-dependent protein kinase by different mechanisms. J Virol. 1993;67:1688–1692. doi: 10.1128/jvi.67.3.1688-1692.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ejercito P M, Kieff E D, Roizman B. Characterization of herpes simplex virus strains differing in their effect on social behavior of cells. J Gen Virol. 1968;2:357–364. doi: 10.1099/0022-1317-2-3-357. [DOI] [PubMed] [Google Scholar]
- 10.Gross M, Kaplansky D A. Identification of a Mr = 39,000 phosphoprotein in highly purified preparations of rabbit reticulocyte eIF-2 that is distinct from the Mr = 35,000 subunit phosphorylated by the hemin-controlled translational repressor. J Biol Chem. 1980;255:6270–6375. [PubMed] [Google Scholar]
- 11.Gross M, Kaplansky D A. Differential effect of Mn2+ on the hemin-controlled translational repressor and the double-stranded RNA-activated inhibitor. Biochim Biophys Acta. 1983;740:255–263. doi: 10.1016/0167-4781(83)90134-3. [DOI] [PubMed] [Google Scholar]
- 12.He B, Chou J, Lieberman D A, Hoffman B, Roizman B. The carboxy terminus of murine MyD116 gene substitutes for the corresponding domain of the γ134.5 gene of herpes simplex virus to preclude the premature shutoff of total protein synthesis in infected human cells. J Virol. 1996;70:84–90. doi: 10.1128/jvi.70.1.84-90.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He B, Chou J, Brandimarti R, Mohr I, Gluzman Y, Roizman B. Suppression of the phenotype of γ134.5− herpes simplex virus 1: failure of activated RNA-dependent protein kinase to shut off protein synthesis is associated with a deletion in the domain of the α47 gene. J Virol. 1997;71:6049–6054. doi: 10.1128/jvi.71.8.6049-6054.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He B, Gross M, Roizman M. The γ134.5 protein of herpes simplex virus 1 complexes with protein phosphatase 1α to dephosphorylate the α subunit of eukaryotic translation initiation factor 2 and preclude the shutoff of protein synthesis by double-stranded RNA-activated protein kinase. Proc Natl Acad Sci USA. 1997;94:843–848. doi: 10.1073/pnas.94.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jacquemont B, Roizman B. RNA synthesis in cells infected with herpes simplex virus. X. Properties of viral symmetrical transcripts and double-stranded RNA prepared from them. J Virol. 1975;15:707–713. doi: 10.1128/jvi.15.4.707-713.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Katze M. Regulation of interferon-induced PKR: can viruses cope? Trends Microbiol. 1995;3:75–78. doi: 10.1016/s0966-842x(00)88880-0. [DOI] [PubMed] [Google Scholar]
- 17.Kozak M, Roizman B. RNA synthesis in cells infected with herpes simplex virus. IX. Evidence for accumulation of abundant symmetrical transcripts in nuclei. J Virol. 1975;15:36–40. doi: 10.1128/jvi.15.1.36-40.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lagunoff M, Randall G, Roizman B. Phenotypic properties of herpes simplex virus 1 containing a derepressed open reading frame P gene. J Virol. 1996;70:1810–1817. doi: 10.1128/jvi.70.3.1810-1817.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee T G, Tang N, Thompson S, Miller J, Katze M G. The 58,000-dalton cellular inhibitor of the interferon-inducible double-stranded RNA-activated protein kinase (PKR) is a member of the tetratricopeptide repeat. Mol Cell Biol. 1994;14:2331–2342. doi: 10.1128/mcb.14.4.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Longnecker R, Roizman B. Generation of an inverting herpes simplex virus 1 mutant lacking the L-S junction a sequences, an origin of DNA synthesis and several genes including those specifying glycoprotein E and the α47 gene. J Virol. 1986;65:583–591. doi: 10.1128/jvi.58.2.583-591.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu Y, Wambach M, Katze M G, Krug R M. Binding of the influenza virus NS1 protein to the protein kinase that phosphorylates the eIF-2 translation initiation factor. Virology. 1995;214:222–228. doi: 10.1006/viro.1995.9937. [DOI] [PubMed] [Google Scholar]
- 22.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 23.Mathews M B, Schenk T. Adenovirus virus-associated RNA and translation control. J Virol. 1991;65:5657–5662. doi: 10.1128/jvi.65.11.5657-5662.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mohr I, Gluzman Y. A herpesvirus genetic element which affects translation in the absence of viral GADD34 function. EMBO J. 1996;15:4759–4766. [PMC free article] [PubMed] [Google Scholar]
- 25.Post L E, Roizman B. A generalized technique for deletion of specific genes in large genomes: α gene α22 of herpes simplex virus 1 is not essential for growth. Cell. 1981;25:227–232. doi: 10.1016/0092-8674(81)90247-6. [DOI] [PubMed] [Google Scholar]
- 26.Purves F C, Spector D, Roizman B. UL34, the target of the herpes simplex virus US3 protein kinase is a membrane protein which in its unphosphorylated state associates with novel phosphoproteins. J Virol. 1992;66:4295–4303. doi: 10.1128/jvi.66.7.4295-4303.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26a.Roller, R., and B. Roizman. Unpublished results.
- 27.Sheldrick P, Berthelot N. Inverted repetitions in the chromosome of herpes simplex virus. Cold Spring Harbor Symp Quant Biol. 1975;39:667–678. doi: 10.1101/sqb.1974.039.01.080. [DOI] [PubMed] [Google Scholar]
- 28.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 29.Wadsworth S, Jacob R J, Roizman B. Anatomy of herpes simplex virus DNA. II. Size composition, and arrangement of inverted terminal repetitions. J Virol. 1975;15:1487–1497. doi: 10.1128/jvi.15.6.1487-1497.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhan Q, Lord K A, Alamo I, Jr, Hollander M C, Carrier F, Ron D, Kohn K W, Hoffman B, Liebermann D A, Fornace A J., Jr The gadd and MyD genes define a novel set of mammalian genes encoding acidic proteins that synergistically suppress growth. Mol Cell Biol. 1994;14:2361–2371. doi: 10.1128/mcb.14.4.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]