Abstract
Background
Chimeric antigen receptor-modified T cells (CAR T-cells) have shown exhilarative clinical efficacy for hematological malignancies. However, a shared antigen pool between healthy and malignant T-cells remains a concept to be technically and clinically explored for CAR T-cell therapy in T-cell cancers. No guidelines for engineering CAR T-cells targeting self-expressed antigens are currently available.
Method
Based on anti-CD70 CAR (CAR-70) T-cells, we constructed CD70 knock-out and wild-type CAR (CAR-70KO and CAR-70WT) T-cells and evaluated their manufacturing and anti-tumor capability. Single-cell RNA sequencing and TCR sequencing were performed to further reveal the underlying differences between the two groups of CAR T-cells.
Results
Our data showed that the disruption of target genes in T-cells before CAR transduction advantaged the expansion and cell viability of CAR T-cells during manufacturing periods, as well as the degranulation, anti-tumor efficacy, and proliferation potency in response to tumor cells. Meanwhile, more naïve and central memory phenotype CAR+ T-cells, with higher TCR clonal diversity, remained in the final products in KO samples. Gene expression profiles revealed a higher activation and exhaustion level of CAR-70WT T-cells, while signaling transduction pathway analysis identified a higher level of the phosphorylation-related pathway in CAR-70KO T-cells.
Conclusion
This study evidenced that CD70 stimulation during manufacturing process induced early exhaustion of CAR-70 T-cells. Knocking-out CD70 in T-cells prevented the exhaustion and led to a better-quality CAR-70 T-cell product. Our research will contribute to good engineering CAR T-cells targeting self-expressed antigens.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00262-023-03475-7.
Keywords: CAR T-cell therapy, T-cell malignancy, CD70, Single-cell RNA sequencing, Single-cell TCR sequencing
Introduction
Chimeric antigen receptor (CAR) T-cell infusion is one of the most potent strategies for cancer therapy, which re-directs T-cells to antigens expressed on tumors and mediates the cytolysis [1, 2]. Besides B cells [3, 4], CAR T-cell therapy for T-cell malignancies is intensively studied, but research progressed slowly [5]. One of the obstacles is that shared target expression between healthy and malignant T-cells may induce fratricide of CAR-70 T-cells, hindering their ex vivo production and in vivo anti-tumor function [6, 7]. Dissection and then elimination of such fratricidal phenomenon are essential for developing efficient CAR T-cells targeting self-expressed antigens.
CD70 is expressed in many tumors, like peripheral T-cell lymphomas and clear cell renal carcinoma, offering an attractive therapeutic target for CAR T-cells [8–10]. More than ten clinical trials have been developed to investigate the efficacy of CAR-70 T-cells in cancer therapy (NCT05420519, NCT05518253, and NCT05420545). However, the manufacture processes for CAR-70 T-cells varied, with or without considering the potential fratricide of CAR T-cells induced by CD70 expression on the activated T-cells [11, 12]. Panowski et al. suggested that CAR-70 T-cells based on a subset of specific single-chain fragment variables (scFvs) can mask CD70 on themselves and therefore avoid the fratricide [11]. While identifying such fratricide-resistant binders was challenging, and no acknowledged guidelines were available. CD27, the receptor of CD70, is the natural binder of CD70. Sauer et al. reported that CAR-70 T-cells employing the structure of CD27 to recognize CD70 had an excellent anti-tumor performance without addressing the issue of CD70-mediated CAR T-cells fratricide [13].
In this study, we aimed to elucidate the effect of target expression in T-cells on the manufacture and function of CAR T-cells, hoping to provide a reference for the engineering of CAR T-cells targeting self-expressed antigens. By comparing CD70 wild-type CAR-70 (CAR-70WT) T-cells with CD70 knocked-out CAR-70 (CAR-70KO) T-cells through single-cell RNA (scRNA) and TCR (scTCR) sequencing and in vitro functional experiments, we showed that knocking-out CD70 in T-cells before CAR transduction benefited the manufacture and anti-tumor functionality of CAR-70 T-cells, possibly due to reduced antigen-stimulation-mediated early exhaustion. This project provides a comprehensive understanding of the antigen-dependent fratricidal phenomenon and its impact on CAR T-cells and contributes to the establishment of new protocols for the generation of CAR T-cells targeting self-expressed antigens.
Materials and methods
Cells lines and culture
THP-1 (acute monoblastic/monocytic leukemia), MV4-11 (acute monoblastic/monocytic leukemia), and Lenti-X™293 T (human embryonic kidney cell) were purchased from the American Type Culture Collection bank and verified to be negative for mycoplasma. THP-1/MV4-11 and Lenti-X™293 T cells were maintained in RPMI-1640 (Gibco, USA) and DMEM (Gibco, USA), respectively. Both mediums are supplemented with 10% fetal bovine serum (FBS) (Gibco, USA). Luciferase-expressing cell lines were established by lentivirus transduction and cultured in mediums supplemented with 1 µg/mL puromycin.
CAR-70 Construction, lentivirus production, CD70 KO, and CAR-70 T-cell production
The second-generation CAR was constructed as follows: Cusatuzumab-derived CD70- specific scFv fused to a CD8α hinge/transmembrane domain, followed by a 4-1BB co-stimulatory and a CD3ζ activation domain. A truncated human epidermal growth factor receptor (tEGFR) was incorporated following a 2A self-cleaving (T2A) sequence as a safety switcher and detection marker.
The lentivirus encoding CAR-70 was produced by Lenti-X™293 T cells using packaging plasmids (pMDlg/pRRE, pRSV-Rev, and pMD2.G) and polyethyleneimine transfection reagent (PEI, Invitrogen, USA). Supernatants containing the released virus were collected 48 h after transfection and concentrated through super centrifugation at 30,000 g for 2.5 h at 4 °C. The viral titer was measured by fluorescence tittering assay using HEK-293 cells. The virus was stored at -80 °C.
On day 0, T-cells were isolated from commercial peripheral blood mononuclear cells (PBMC) (Milestone® Biotechnologies, China) using anti-CD3 microbeads (Miltenyi Biotec, Germany), activated by Dynabeads™ Human T-Activator CD3/CD28 (Thermo Fisher Scientific, USA), and cultured in CTS medium (Gibco, USA) supplemented with 10% FBS, 200 IU IL-2 (Sigma-Aldrich, USA) and 100 µg/mL L-Glutamine (Thermo Fisher Scientific, USA). CD70 gene disruption was performed on day 1 by electroporation with Cas9 protein and small guide RNA targeting CD70 (Genscript Biotech Corporation, China) using the Celetrix system (Celetrix, USA). Eight hours later, the CD70 KO T-cells were transduced with lentivirus at a multiplicity of infection (MOI) of 5 to generate CAR-70KO T-cells. The cells continued to culture in fresh medium 24 h post-transduction at a density of 1 ~ 2 × 106 cells/mL.
CD107A degranulation assay
For degranulation in the absence of tumor cells, CAR-70 T-cells were cultured in 10% RPMI-1640 and incubated with 1:100 diluted PE/Cyanine7-conjugated CD107A antibody (BioLegend, USA) and 1:500 diluted Monesin (BioLegend, USA) for 4 h. The CD107A expression level on CD8+CAR+ T-cells was then detected by flow cytometry. For degranulation in response to tumor cells, CAR-70 T-cells were co-cultured with tumor cells at a ratio of 1:1 and the CD107A expression was measured under the same condition as described above.
Cytotoxicity assay
Tumor cell lines stably expressing firefly-luciferase were used to assess the cytotoxicity of CAR-70 T-cells. After 24-h co-incubation with tumor cells, luciferase activity (reflecting the amount of the remaining live cells) was quantified using Steady-Glo® Luciferase Assay System (Promega, USA) and Synergy H1 microplate reader (BioTek, USA). 2 × 104 tumor cells were added in triplicates. CAR+ T-cells were added according to the specified effector-to-target ratio. Percentage of Lysis = (Luminescencetumor cells only- LuminescenceCAR-70 T-cells + Tumor cell) / Luminescencetumor cells only.
Repetitive antigen stimulation assay
After three times washing by phosphate-buffered saline (PBS), mitomycin C (1 µg/mL) overnight-treated targets (MV4-11 and THP-1) were used to stimulate CAR-70 T-cells at an effector-to-target ratio of 3:1. A round of stimulation was defined as 3-day co-culture in a complete T-cell medium. The absolute number of CAR-positive cells before and after stimulation was obtained from total cell counting and CAR-70 positivity rate measurement. The expansion rate was calculated by dividing the number of CAR-positive cells after stimulation by the number before stimulation. After each round, a certain amount of CAR-70 T-cells from the last pool were picked and used for the next round of stimulation. The cumulative proliferation rate of CAR-positive cells was the product of the expansion rate in each round.
Flow cytometry
Samples were washed by washing buffer (PBS supplemented with 2% FBS) and then stained with antibodies at room temperature for 15 min in the dark. Anti-CD70-PE (clone:113–16, BioLegend, USA) was used for CD70 detection. For the degranulation assay, the staining panel was anti-EGFR-APC, anti-CD8-BV421, and anti-CD107A-PE/Cy7. The staining panel for memory T-cell subpopulation detection was anti-CD45RA-APC, anti-CCR7-BV421, anti-CD8-FITC, and anti-EFGR-PE. After twice washing, samples were ready for acquisition by MACS Quant Analyzer 10 (Miltenyi Biotec, USA). Data were analyzed by FlowJo (Version 10.0.7, USA).
scRNA-seq data analysis
CAR-70WT T-cells and CAR-70KO T-cells were used for scRNA sequencing on day 13 of the culture. The scRNA-seq library was constructed by 10X Genomics according to the manufacturer's protocol. For each sample, the expressed genes were sequenced using Novaseq 6000 System (Illumina, USA) with a paired-end 150-bp (PE150) reading strategy (CapitalBio Technology, China). Cell Ranger (version 4.0) aligns the reads of each sample's original gene expression matrix pipeline with the human reference genome (GRCh38). R package Seurat (version 4.0.6) [14] was used for the subsequent processing of the qualified gene expression matrix. Following the data filtering, cells with greater than 0.1% of gene expression and > 200 genes detection were selected. Data from cells with < 500 unique molecular identifiers (UMI), or gene numbers < 500 or > 7800, or a percentage of mitochondrial genes (UMI derived from mitochondria) > 15% were removed. The DoubletFinder method [15] was then used to remove technical artifact doublets from the data. The two samples data were integrated and corrected for the batch effect and then normalized by the NormalizeData function. FindVariableFeatures were used to calculate 5000 features with a high degree of cell-to-cell variation. Linear-transformation scaled data were generated by using the ScaleData function with default parameters, and the RunPCA function was utilized to reduce the dimensions of the dataset. The FindNeighbors and FindClusters functions were used to cluster cells, and the RunUMAP function was used for nonlinear dimensional reduction.
Identification of CAR+ T cells
According to the specific CAR sequence, cells can be divided into T cells expressing CAR-RNA (CAR+) or not (CAR−). The average depth of the CAR sequence in each sample was used to estimate the expression level of CAR. The level was normalized using the NormalizeData function of the Seurat software package (version 4.0).
Elimination of the cell-cycle effect
Cells in the same cell cycle may be clustered together although with different gene expression profiles. In order to remove the artifact of cell cycle on clustering, we implement the CellCycleScore function in Seurat to evaluate cell cycle status based on established G1/S and G2/M phase-specific genes [16] and then the regressout in Seurat's ScaleData function to remove the cell cycle effect.
Sequencing and analysis of V(D)J of TCR
The full-length cDNA fragment of TCR V(D)J was obtained by using the single cell V(D)J enrichment kit per the manufacturer's protocol (10X Genomics). Using GRCh38 as a reference, the gene quantification and TCR clonal allocation were calculated using CellRanger (v.4.0) vdj pipeline. The R package scRepertoire was used for TCR analysis of CAR+ cells [17]. Only cells with at least one productive TCRα chain (TRA) and one productive TCRβ chain (TRB) were further analyzed. A clonotype is defined as every unique TRA(s)-TRB(s) pair. The cloning degree is determined by the cell numbers with the same clonotype. The UMAP profiles of different clones were obtained from the combined information of scRNA-seq and scTCR-seq. In addition, the STARTRAC (Single T-cell Analysis by RNA-seq and TCR TRACking) algorithm [18] was used to analyze the differences in distribution preference and clone expansion of two CAR-T cells.
Evaluation of the cell subsets status
We normalized the expression of the genes using SCTransform function in the Seurat package. The status of cell subsets was assessed by the average expression level of specific gene sets. The cytotoxicity genes (PRF1, IFNG, GNLY, NKG7, GZMB, GZMA, KLRK1, KLRD1, CTSW, CST7), the cell cycle genes [16], the exhaustion genes (CTLA4, LAG3, PDCD1, CD160, ENTPD1, HAVCR2, TIGIT) and the apoptosis genes (ACIN1, BIRC3, CYCS, NFKB1, PARP1, BCL2A1, CDK1) were used to identify the cytotoxic, proliferative, exhaustion and apoptotic status, respectively. We used Wilcox test for statistical analysis.
DEGs identification and functional enrichment analysis
The FindMarkers function in the Seurat R package was used to analyze the differentially expressed genes (DEG). Remark the KO / WT sample as ident.1 / ident.2. Genes satisfying the following conditions were regarded as DEGs: |avg_log2FC|≥ 0.25, p ≤ 0.01, and expressed in > 10% of cells. CompareCluster functions in the clusterProfiler R package (v.4.0.5) were used for the enrichment. The pathway enrichment analysis was performed using the GO database and the KEGG database. For the associations among proteins enriched in the phosphorylation pathway, the analysis was conducted in the STRING database [19] and visualized by Cytoscape software [20].
Results
CD70 KO benefits the manufacturing of CAR-70 T-cells
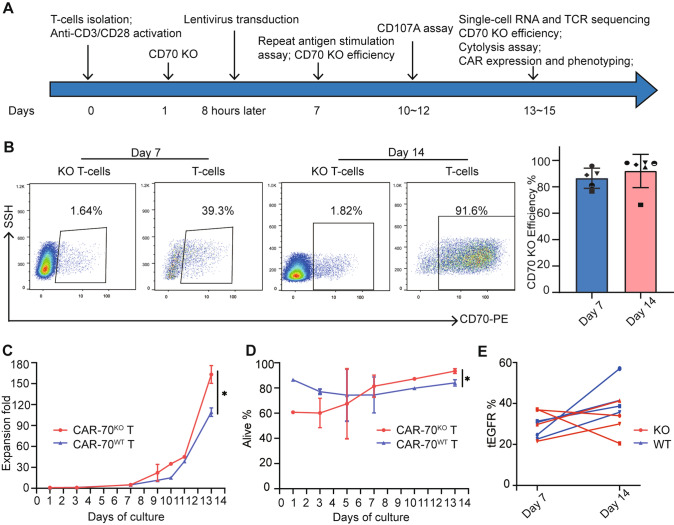
Given that CAR T-cells were commonly infused in patients after approximately two weeks of culture in the clinical practice [3, 21], the timeline of the study was intentionally crafted to simulate real-world clinical settings, as illustrated in Fig. 1A. CD70 expression in T-cells increased as the culture was prolonged. The CD70 knock-out (KO) efficiencies were defined as the reduction of CD70 expression in CD70 KO T-cells compared with that of parental wild-type (WT) T-cells, and ranged from 76.5% to 95.8% at day 7 and from 66.3% to 98.2% at day 14 (Fig. 1B). The expansion of CAR-70KO T-cells is more robust than that of CAR-70WT T-cells during the manufacturing culture (163.1 12.8 vs. 109.1 6.2, P = .037) (Fig. 1C), as well as the viability (93.4% 2.9% vs. 84.1% 2.5%, P = .013) (Fig. 1D). However, the proportion of CAR-positive (CAR+) cells in the final product (at day 14 of culture) in the CD70 KO group is slightly lower than that of the CD70 WT group (43.2% 9.5% vs. 31.5% 8.7%, P = .11). It was also found that the CAR expression consistently increased in WT groups, but not in KO groups, which may account for the higher CAR+ positivity in WT samples. (Fig. 1E). These data suggested that electroporation of the Cas9/sgRNA complex can efficiently knock out CD70 in T cells and benefit the proliferation and viability of the final product.
Fig. 1.
CD70 knock-out and production of CAR-70 T-cells. A Design of the study. B Typical flow cytometric dot plot of CD70 expression in WT T-cells and KO T-cells, and the calculated CD70 KO efficiency at day 7 (n = 5) and day 14 (n = 6) of the culture. C The cumulative proliferation of CAR-70WT T-cells and CAR-70KO T-cells at different days of culture (n = 3). D The viability of CAR-70WT T-cells and CAR-70KO T-cells at different days of culture (n = 3). E The positive rate of tEGFR (the proportion of CAR+ cells) at day 7 and day 14 of the culture. A pair of WT and KO samples were indicated by the same point shape. Paired t-test was used and *P < .05
CAR-70KO T-cells confer superior anti-tumor functionality than CAR-70WT T-cells
We then compared the anti-tumor functionality of CAR-70KO T-cells and CAR-70WT T-cells at day 13 ~ 15 of culture, the same as the time of CAR-T infusion in patients. In response to CD70 highly expressed THP-1 cell line, the expression of CD107A (lysosomal-associated membrane protein-1, a marker of T-cell’s degranulation) in both CAR T-cells specifically increased compared with the control T-cells. Moreover, the degranulation level in CAR-70KO T-cells was significantly higher than that of CAR-70WT T-cells (74.4% 2.1% vs. 65.9% 4.3%, P = .028) (Fig. 2A). The same phenomenon was observed upon stimulation of CD70 moderately expressed MV4-11 cell line (60.7% 4.1% vs. 49.9% 3.0%, P = .004) (Fig. 2B). Also, both CAR T-cells showed strong cytotoxicity against THP-1 (Fig. 2C) and MV4-11 (Fig. 2D) cell lines. The cytolysis of CAR-70KO T-cells for THP-1 was significantly superior to that of CAR-70WT T-cells in donor 1 (94.7% 2.3% vs. 89.3% 2.3%, P = .047) but comparable in donor 2 (98% 0% vs. 98% 1%, P > .99). However, improved cytotoxicity of CAR-70KO T-cells against MV4-11 was observed in both two donors (donor 1: 55.0% 8.2% vs 25.3% 6.7%, P = .008; donor 2: 85.7% 2.1% vs 50.7% 9.3%, P = .003). In the repetetive antigen stimulation assay, CAR-70KO T-cells exhibited better proliferation ability than CAR-70WT T-cells in response to both THP-1 (Fig. 2E) (cumulative expansion fold: 107.0 vs. 50.0 for donor 1; 1284.0 vs. 84.3 vs. for donor 2) and MV4-11 (Fig. 2F) (cumulative expansion fold: 83.5 vs. 45.1 for donor 1; 1140.0 vs. 139.0 for donor 2), implying better expansion of CAR T-cells in vivo upon stimulation of tumor cells. In general, CAR-70KO T-cells performed better than CAR-70WT T-cells in response to tumor cells.
Fig. 2.
Functional evaluation of CAR-70WT T-cells and CAR-70KO T-cells. A and B Typical dot plot and statistics of the CD107A expression in CD8+CAR+ T-cells after 4-h stimulation of THP-1 (CD70 + +) A and MV4-11 (CD70 +) B at an effector-to-target (E:T) ratio of 1:1 (n = 3, * P < .05, **P < .01, paired t-test). C and D the lysis percentage of THP-1 (C) and MV4-11 D after 24-h co-culture of CAR-70KO T-cells and CAR-70WT T-cells from two different donors (n = 3, * P < .05, **P < .01, ns, no significance, t-test). The E:T ratio for THP-1/MV4-11 was 1:1/3:1. E and F the cumulative expansion of CAR+-70WT T-cells and CAR+-70KO T-cells after repeated THP-1 E and MV4-11 F stimulation from 2 different donors
CAR-70KO T-cells possessed a higher proportion of naïve and central memory CAR+ cells
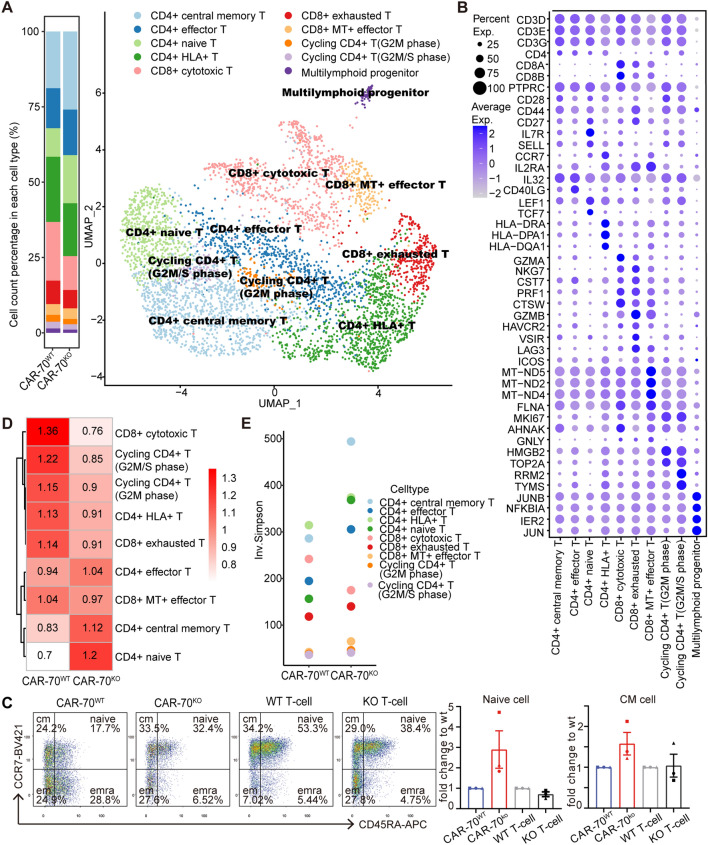
To determine the mechanism underlying the better function of CAR-70KO T-cells against tumor cells, we performed scRNA-seq and scTCR-seq for the final CAR-T products. After strict quality control, we obtained about 6k cells per sample, of which CAR+ cells accounted for 41.2% and 30.7% (Fig. S1A and B). Since the cell cycle significantly impacted the data, its fraction was regressed to leave biologically significant differences (Fig. S1C and D). As a result, 4,647 CAR+ cells with 20,177 genes were obtained for further analysis. CAR+ cells were clustered into ten subsets based on their marker genes (Fig. 3A, B), without a unique population in the KO or WT sample. We found that CD4+ central memory (25.9% vs. 18.8%) and CD4+ naïve (16.0% vs. 9.5%) subsets were predominantly enriched in the KO CAR+ pool when compared with the WT (Fig. 3A). Results from flow cytometry were consistent: the proportion of CD8− naïve T-cells (CD8−EGFR+CD45RA+CCR7+) and CD8− central memory T-cells (CD8−EGFR+CD45RA−CCR7+) in CAR-70KO T-cells is averagely 2.9 and 1.6 folds of that in CAR-70WT T-cells in three different donors (Fig. 3C). Moreover, our data revealed a decreasing trend in the proportion of CD8− naïve cells within KO T-cells when compared to WT T-cells (Fig. 3C). This indicates that the greater proportion of naïve CAR+ cells in KO samples was not a result of the lack of CD70 signaling but rather, it was likely induced by the CAR signaling triggered by CD70. By utilizing scTCR-seq, we examined the cloning of the ten subsets of CAR+ cells in the two samples. The result from the STARTRAC algorithm indicated that the TCRs of the CD4+ central memory and CD4+ naïve subsets were more abundant in the KO samples (Fig. 3D). Additionally, the clonal diversity of the two subsets was higher in the KO samples than in the WT samples (Fig. 3E). These data collectively suggested that knock-out CD70 in CAR-70 T-cells led to an enrichment of naïve and central memory CD4+CAR+ cells and increased TCR diversity.
Fig. 3.
CAR-70KO T cells possessed a superior phenotype. A UMAP diagram of CAR-T cells of different cell subsets according to marker gene expression (right). Each dot represents a cell, and different colors represent different subsets. The histogram shows the proportion of different cell subsets in each sample (left). B The bubble chart shows the expression ratio of typical marker genes of each cell subset, and cell types are determined by these genes. C The typical dot plot of flow cytometry of the memory phenotype of KO/WT CAR+CD8− T-cells, KO/WT CD8− T-cells, and the statistics of fold changes of naïve and central memory population of KO groups relative to WT groups (n = 3).The same point shape refers to a pair of WT and KO sample. D The STARTRAC-dist index estimated the cell type preference of WT and KO CAR-T samples. E Clone diversity of each cell type (Inv. Simpson index)
CD70 KO improved the functional status of CAR-70 T-cells
Next, we analyzed the functional status of CAR-70 T-cells, including the cytotoxicity, proliferation, exhaustion, and apoptosis, by evaluating the expression levels of the corresponding genes. The results showed that in the CAR+ pools, the KO sample had significantly lower cytotoxicity (P < 2.22e-16), proliferation (P < 2.22e-16), exhaustion (P < 2.22e-16), and apoptosis (P = .012) scores compared to the CAR-70WT sample (Fig. 4A–D). Meanwhile, in the absence of tumor cell stimulation, CD8+CAR-70KO T-cells had a higher degranulation level than the CD8+CAR-70WT T-cells (6.6% ± 1.9% vs. 4.0% ± 1.1%, P = .030) (Fig. 4E). These data collectively indicated the activation of CAR-70WT T-cells induced by CD70 stimulation, which led to their early exhaustion. The differentially expressed representative genes regarding cytotoxicity (e.g., CST7, CTSW and PRF1), proliferation (e.g., TMPO, MKI67 and TYMS), exhaustion (e.g., LAG3, HAVCR2 and CTLA4) and apoptosis (e.g., PARP1, ACIN1 and BIRC3) are summarized in Fig. 4F. In conclusion, compared to CAR-70KO T-cells, the higher transcriptional levels of cytotoxicity, proliferation, differentiation, and exhaustion in CAR-70WT T-cells implied the fratricide of CAR-70WT T-cells induced by the CD70 stimulation. Knock-out of CD70 prevented CAR-70 T-cells from pre-activation and early exhaustion.
Fig. 4.
The functional status of KO and WT CAR-T cells. A-D Violin plots of cytotoxicity score A, proliferation score B, exhaustion score C, and apoptosis score D. Wilcox test was used for A-D. E The CD107A expression level in CD8+CAR+ cells of KO groups and WT groups without tumor cell stimulation (n = 3, paired t-test). F The bubble chart shows the expression of typical genes used in the cell status score of two samples. Statistical significance: * P < .05, ** P < .01, *** P < .001; **** P < .0001, while ns indicates non-significance
CAR-70KO T-cells exhibited higher phosphorylation-related pathway
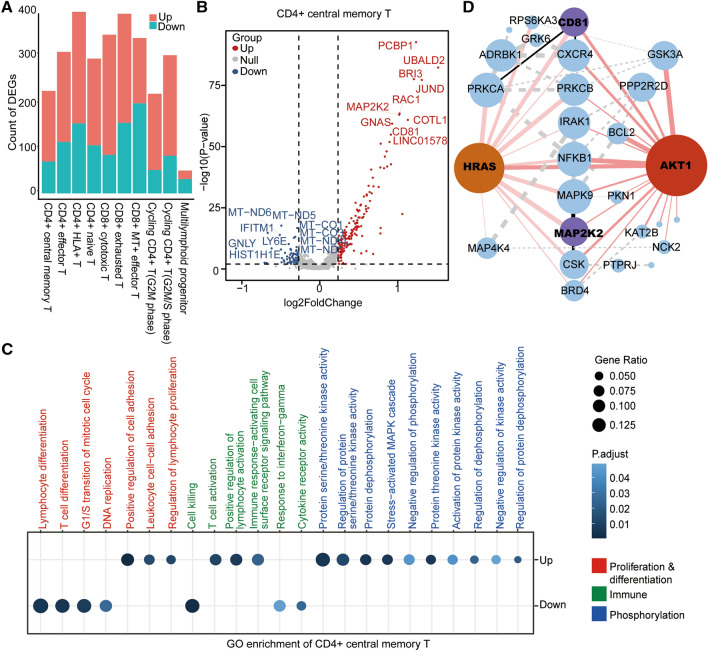
To delve deeper into the molecular mechanism underlying the difference between KO and WT samples, we analyzed the functional enrichment of the differentially expressed genes (DEGs). We first extracted the up-regulated and down-regulated genes in KO samples compared with WT samples according to the cell subsets (Fig. 5A) and identified 148/182 up-regulated and 67/101 down-regulated genes in CD4+ central memory/naïve T-cells (Fig. 5A). Moreover, 105 up-regulated and 15 down-regulated genes were found in the both subsets (Figure S2A). The DEGs were visualized in the volcano maps (Fig. 5B, Figure S2B). Seven out of the top-10 up-regulated and 6 out of the top-10 down-regulated genes overlapped in the central memory and naïve CAR+ T-cells, indicating similarity in the differences between KO and WT samples of the two subsets. Furthermore, we conducted the GO analysis of the DEGs in CD4+ central memory/naïve T-cells and identified 22 (15 up-regulated and 7 down-regulated) /27 (18 up-regulated and 9 down-regulated) functional terms, involving proliferation and differentiation, cell killing, and phosphorylation-related pathways (Fig. 5C, Figure S2C). In general, the KO group displayed reduced levels in proliferation, differentiation, and cell-killing pathways, implying a limited antigen-stimulation-mediated activation in CAR-70KO T-cells. However, the KO groups showed significantly enhanced activity in the phosphorylation-related pathway, which is closely associated with the functionality of CAR T-cells. We further analyzed the interactions among the phosphorylation-related genes and found that AKT1 (essential for the PI3K-Akt signaling pathway) and HRAS (vital for the RTK-Ras-MAPK pathway) were in the central position of the network maps (Fig. 5D). The genes, MAP2K2 and CD81, with essential roles in the phosphorylation network, were also among the top-10 up-regulated genes in KO groups (Fig. 5B, S2C), consistent with the altered phosphorylation level in the CAR-70KO T-cells. Additionally, the KEGG analysis generated similar results (Figure S2D, E).
Fig. 5.
The differential genes between KO and WT CAR-T and their functional enrichment. A The histogram shows the statistics of the number of differential genes in each cell subset under the threshold of | log2FC |≥ 0.25, min. pct = 0.1, p ≤ .01) (ko vs. wt). B Volcano map of up-regulated and down-regulated genes of CD4+ central memory T-cells, with the top-10, differentially expressed genes marked. C GO pathway with significantly enriched DEGs in KO CAR-T sample compared with WT CAR-T sample in CD4+ central memory T-cells (P ≤ .01). D A network diagram of the interactions of the phosphorylation-related proteins was constructed based on the phosphorylation-related pathways in Figure C. The size of the node represents the number of proteins that interacted, and the thickness of the connection line represents the interaction intensity. Red nodes represent the central proteins with the most interactions with the remaining proteins, and purple nodes represent the proteins in the top 10 with fold enrichment. Red lines indicate direct interaction with central proteins, black lines indicate direct interaction with the top 10 proteins, and dotted lines represent interactions between the remaining proteins
Discussion
The possible fratricide of CAR T-cells mediated by the shared antigens on healthy T-cells hindered its development in the therapy for T-cell malignancies [22, 23]. To address this issue, our study employed Cas9/sgRNA gene-editing technology [24] to knock-out CD70 before the transduction of the CAR lentivirus. Our results demonstrated that CAR-70KO T-cells had a better expansion, viability, and anti-tumor efficacy than CAR-70WT T-cells. Further analysis of scRNA-seq and scTCR-seq data supported the fratricide phenomenon of CAR-70WT T-cells. Thus, this study highlights the detrimental effects of antigen-induced fratricide on the manufacturing, function, and transcriptional status of CAR T-cells, and proposes a practical strategy to overcome the limitations imposed by shared antigen between healthy and malignant T-cells in CAR T-cell therapy. However, verification of this phenomenon in a tumor xenograft mouse model is absent in this study and is urgently needed.
CD70 is a pan-cancer target up-regulated in various T-cells malignancies and solid tumors, making it an attractive therapeutic target for CAR T-cells [10, 25–28]. However, this study suggested that CD70 expression in activated T-cells can lead to fratricide of CAR-70 T cells, interfering with their production and anti-tumor functions. Additionally, other reports have described the deficient expansion of the engineered CAR T-cells targeting another self-expressed antigen, like CD26 and CD7 [7, 29]. Moreover, antigen expression in CAR T-cells mediated by trogocytosis can even result in fratricide and exhaustion, leading to the depressed activity of CAR T-cells and subsequently the relapses of diseases [6]. Altogether, target expression in T-cells tends to disadvantage the production and function of CAR T-cells. To circumvent this issue, some researchers have selected target negative T-cells to produce CAR T-cells to avoid fratricide, like CD7-CARCD7- T-cells [30]. However, the number of these T-cells in peripheral blood is small, and activation-induced expression targets, like CD70, are unsuitable for this strategy. In the present study, CRISPR gene-editing technology was utilized to disrupt the CD70 gene in T-cells with good practicability and universality. Nevertheless, the effectiveness of gene editing and the risk of unpredicted events due to off-target effects are significant limitations [31]. Notably, CD70 was reported to play a role in T-cells’ priming, expansion, and differentiation by activating CD27 signals [32], which must be considered when knocking-out CD70 in T-cells. We found that after the depletion of CD70, the expression of CD27 in CAR+ T-cells and T-cells both increased compared to WT controls. While CD70 was undetectable at day 6 of culture in both CAR-70WT and CAR-70KO T-cells, indicative of the absence of CD27 signals in both CAR-70 T-cells (Supplementary Fig. 3). A model of CAR T-cells targeting antigens other than CD70 is required to investigate the role of CD70 on CAR T-cell functions.
Data from scRNA-seq and scTCR-seq indicated a higher proportion of both naïve and central memory CAR+ T-cells cells [18]. It was well established that T-cells with naïve or central memory phenotype have a greater potential for proliferation and long-term survival [18, 33]. Research on anti-CD19 CAR T-cells also indicated that a higher frequency of CD8+CD45RA+CCR7+CAR T-cells (which have a central memory phenotype) in the infused products is associated with improved expansion of CAR T-cells in vivo and more favorable clinical response [34]. In the study, a higher proportion of naïve and central memory CAR+ cells was observed in the KO sample, which may explain the more robust expansion of CAR-70KO T-cells in the repetitive antigen stimulation assay and suggest enhanced potency of survival and proliferation of CAR-70KO T-cells in vivo after repetitive encounters with the targets. Meanwhile, the TCR diversity of the naïve and central memory CAR+ subsets in the KO sample was higher than that of the WT sample, further supporting the greater proliferative and functional potential of naïve and central memory CAR+ cells in KO than those in the WT sample.
In the analysis of the functional status of CAR+ cells, we found that WT CAR+ cells had a higher cytotoxic score but exhibited weaker anti-tumor efficacy; had a higher proliferative score but expanded less after repeated antigen stimulation. Together with the higher score of exhaustion and apoptosis, these implied the fratricide of CAR T-cells in the WT sample, where CD70 stimulated the activation of CAR-70WT T-cells, followed by exhaustion and apoptosis, resulting in diminished anti-tumor functions. However, there was no direct evidence of CAR T-cells attacking each other. In vivo, the interaction between CAR T-cells and CD70-expressing tumor cells also led to similar problems of "fratricide" exhaustion after activation. Our data hinted that LAG3 but not PDCD1 was predominantly expressed in exhausted WT CAR+ cells. It is worth exploring whether blocking LAG3 signaling can inhibit CAR T-cells exhaustion or restore its function ex vivo and in vivo. Further understanding of CAR T-cells' exhaustion and tuning the balance between activation and exhaustion are urgently required to overcome the problems of self-targeting CAR T-cells fratricide and develop sustained well-functional CAR T-cells in vivo.
Interestingly, naïve and central memory CAR+ cells in the KO sample up-regulated phosphorylation-related pathways compared to the WT sample. The activation of T-cells involves the phosphorylation of sequential molecules, such as Lck and ZAP70 [35]. Previous research indicated that altering the phosphorylation of crucial molecules can modulate T-cell activation and proliferation [35]. Recent studies showed that larger-magnitude changes in protein phosphorylation upon CAR stimulation correlated with increased CAR T-cells effectiveness [36]. The up-regulation of the phosphorylation-related pathway in naïve and central memory CAR+ cells of the KO sample strongly implied improved function than that of the WT sample. MAP2K2 and CD81 were among the top 10 up-regulated DEGs in the KO sample and were concurrently present in the network of phosphorylation-related protein interaction. Meanwhile, AKT1 and HRAS are well acknowledged to positively influence the activation of T-cells, further supporting the high confidence of our results and indicating the vital role of MAP2K2 and CD81 in regulating phosphorylation signaling and CAR T-cells activation. Taken together, the increased phosphorylation pathways in CAR-70KO T-cells confirmed their improved function and offered insights into enhancing CAR T-cells potential.
In conclusion, this study on CAR-70 T-cells showed that CAR T-cells targeting antigens expressed in T-cells may undergo fratricide during the culture process, resulting in diminished expansion and anti-tumor function. Depletion of naïve and central memory subsets, as well as increased exhaustion level in CAR T-cells following target stimulation may account for the impaired anti-tumor function. By gene-editing technology, knocking out targets in T-cells can prevent CAR T-cells' fratricide and avail their function. However, more studies are necessary to comprehensively understand the phenomenon of CAR T-cells fratricide and develop strategies to address it.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We gratefully thank Wei Mu for the instructions for manufacturing the CAR T-cells.
Author contributions
A-YG, QL and XZ proposed the ideas, oversaw the whole project, and revised the manuscript. JC conducted the functional experiments and analyzed the related data. Y. Zhao and HH analyzed the data of scRNA and scTCR sequencing. LT and SD helped with the experiments and participated in the discussion. JC, Y. Zhao and Y. Zeng wrote the manuscript.
Funding
This work was supported by the National Key R&D Program of China (2021YFF0703704), National Natural Science Foundation of China (82270183, 32100527, and 82100241), Natural Sciences Foundation of Hubei Province of China (2020CFB790), and the Excellent Young Science Foundation Project of Tongji Hospital (No.2020YQ0012).
Availability of data and materials
Data were contained in the manuscript, and raw data for the scRNA-seq and scTCR-seq are available via HRA002418 at the National Genome Data Center (NGDC) (https://ngdc.cncb.ac.cn/).
Declarations
Conflict of interest
There is no competing interest for all the authors.
Ethics approval
Not applicable.
Consent for publication
Not applicable.
Consent to participate
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jiali Cheng and Yuyan Zhao have contributed equally to this work.
Contributor Information
An-Yuan Guo, Email: guoay@hust.edu.cn.
Qing Li, Email: Liqing54070907@163.com.
Xiaojian Zhu, Email: zhuxiaojian@hust.edu.cn.
References
- 1.Singh AK, McGuirk JP. CAR T cells: continuation in a revolution of immunotherapy. Lancet Oncol. 2020;21(3):e168–e178. doi: 10.1016/S1470-2045(19)30823-X. [DOI] [PubMed] [Google Scholar]
- 2.Lu J, Jiang G. The journey of CAR-T therapy in hematological malignancies. Mol Cancer. 2022;21(1):194. doi: 10.1186/s12943-022-01663-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang N, Hu X, Cao W, Li C, Xiao Y, Cao Y, et al. Efficacy and safety of CAR19/22 T-cell cocktail therapy in patients with refractory/relapsed B-cell malignancies. Blood. 2020;135(1):17–27. doi: 10.1182/blood.2019000017. [DOI] [PubMed] [Google Scholar]
- 4.Wang D, Wang J, Hu G, Wang W, Xiao Y, Cai H, et al. A phase 1 study of a novel fully human BCMA-targeting CAR (CT103A) in patients with relapsed/refractory multiple myeloma. Blood. 2021;137(21):2890–2901. doi: 10.1182/blood.2020008936. [DOI] [PubMed] [Google Scholar]
- 5.Safarzadeh Kozani P, Safarzadeh Kozani P, Rahbarizadeh F. CAR-T cell therapy in T-cell malignancies: Is success a low-hanging fruit? Stem Cell Res Ther. 2021;12(1):527. doi: 10.1186/s13287-021-02595-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hamieh M, Dobrin A, Cabriolu A, van der Stegen SJC, Giavridis T, Mansilla-Soto J, et al. CAR T cell trogocytosis and cooperative killing regulate tumour antigen escape. Nature. 2019;568(7750):112–116. doi: 10.1038/s41586-019-1054-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou S, Zhu X, Shen N, Li Q, Wang N, You Y, et al. T cells expressing CD26-specific chimeric antigen receptors exhibit extensive self-antigen-driven fratricide. Immunopharmacol Immunotoxicol. 2019;41(4):490–496. doi: 10.1080/08923973.2019.1637889. [DOI] [PubMed] [Google Scholar]
- 8.Jacobs J, Deschoolmeester V, Zwaenepoel K, Rolfo C, Silence K, Rottey S, et al. CD70: an emerging target in cancer immunotherapy. Pharmacol Ther. 2015;155:1–10. doi: 10.1016/j.pharmthera.2015.07.007. [DOI] [PubMed] [Google Scholar]
- 9.Flieswasser T, Camara-Clayette V, Danu A, Bosq J, Ribrag V, Zabrocki P, et al. Screening a broad range of solid and haematological tumour types for CD70 expression using a uniform IHC methodology as potential patient stratification method. Cancers. 2019;11(10):1611. doi: 10.3390/cancers11101611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park YP, Jin L, Bennett KB, Wang D, Fredenburg KM, Tseng JE, et al. CD70 as a target for chimeric antigen receptor T cells in head and neck squamous cell carcinoma. Oral Oncol. 2018;78:145–150. doi: 10.1016/j.oraloncology.2018.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Panowski SH, Srinivasan S, Tan N, Tacheva-Grigorova SK, Smith B, Mak YSL, et al. Preclinical development and evaluation of allogeneic CAR T cells targeting CD70 for the treatment of renal cell carcinoma. Can Res. 2022;82(14):2610–2624. doi: 10.1158/0008-5472.CAN-21-2931. [DOI] [PubMed] [Google Scholar]
- 12.Leick MB, Silva H, Scarfò I, Larson R, Choi BD, Bouffard AA, et al. Non-cleavable hinge enhances avidity and expansion of CAR-T cells for acute myeloid leukemia. Cancer Cell. 2022;40(5):494–508.e495. doi: 10.1016/j.ccell.2022.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sauer T, Parikh K, Sharma S, Omer B, Sedloev D, Chen Q, et al. CD70-specific CAR T cells have potent activity against acute myeloid leukemia without HSC toxicity. Blood. 2021;138(4):318–330. doi: 10.1182/blood.2020008221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36(5):411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McGinnis CS, Murrow LM, Gartner ZJ. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 2019;8(4):329–337.e324. doi: 10.1016/j.cels.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tirosh I, Izar B, Prakadan SM, Wadsworth MH, 2nd, Treacy D, Trombetta JJ, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352(6282):189–196. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Borcherding N, Bormann NL, Kraus G. scRepertoire: an R-based toolkit for single-cell immune receptor analysis. F1000Research. 2020;9:47. doi: 10.12688/f1000research.22139.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang L, Yu X, Zheng L, Zhang Y, Li Y, Fang Q, et al. Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature. 2018;564(7735):268–272. doi: 10.1038/s41586-018-0694-x. [DOI] [PubMed] [Google Scholar]
- 19.Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S, et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49(D1):D605–d612. doi: 10.1093/nar/gkaa1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neelapu SS, Locke FL, Bartlett NL, Lekakis LJ, Miklos DB, Jacobson CA, et al. Axicabtagene ciloleucel CAR T-cell therapy in refractory large B-cell lymphoma. N Engl J Med. 2017;377(26):2531–2544. doi: 10.1056/NEJMoa1707447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alcantara M, Tesio M, June CH, Houot R. CAR T-cells for T-cell malignancies: challenges in distinguishing between therapeutic, normal, and neoplastic T-cells. Leukemia. 2018;32(11):2307–2315. doi: 10.1038/s41375-018-0285-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Went P, Agostinelli C, Gallamini A, Piccaluga PP, Ascani S, Sabattini E, et al. Marker expression in peripheral T-cell lymphoma: a proposed clinical-pathologic prognostic score. J Clin Oncol. 2006;24(16):2472–2479. doi: 10.1200/JCO.2005.03.6327. [DOI] [PubMed] [Google Scholar]
- 24.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157(6):1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pal SK, Forero-Torres A, Thompson JA, Morris JC, Chhabra S, Hoimes CJ, et al. A phase 1 trial of SGN-CD70A in patients with CD70-positive, metastatic renal cell carcinoma. Cancer. 2019;125(7):1124–1132. doi: 10.1002/cncr.31912. [DOI] [PubMed] [Google Scholar]
- 26.Riether C, Pabst T, Höpner S, Bacher U, Hinterbrandner M, Banz Y, et al. Targeting CD70 with cusatuzumab eliminates acute myeloid leukemia stem cells in patients treated with hypomethylating agents. Nat Med. 2020;26(9):1459–1467. doi: 10.1038/s41591-020-0910-8. [DOI] [PubMed] [Google Scholar]
- 27.Deng W, Chen P, Lei W, Xu Y, Xu N, Pu JJ, et al. CD70-targeting CAR-T cells have potential activity against CD19-negative B-cell lymphoma. Cancer Commun (London, England) 2021;41(9):925–929. doi: 10.1002/cac2.12201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang M, Tang X, Zhang Z, Gu L, Wei H, Zhao S, et al. Tandem CAR-T cells targeting CD70 and B7–H3 exhibit potent preclinical activity against multiple solid tumors. Theranostics. 2020;10(17):7622–7634. doi: 10.7150/thno.43991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gomes-Silva D, Srinivasan M, Sharma S, Lee CM, Wagner DL, Davis TH, et al. CD7-edited T cells expressing a CD7-specific CAR for the therapy of T-cell malignancies. Blood. 2017;130(3):285–296. doi: 10.1182/blood-2017-01-761320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Freiwan A, Zoine J, Crawford JC, Vaidya A, Schattgen SA, Myers J, et al. Engineering naturally occurring CD7 negative T cells for the Immunotherapy of hematological malignancies. Blood. 2022;25:2684–2696. doi: 10.1182/blood.2021015020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang XH, Tee LY, Wang XG, Huang QS, Yang SH. Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol Ther Nucleic Acids. 2015;4(11):e264. doi: 10.1038/mtna.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Borst J, Hendriks J, Xiao Y. CD27 and CD70 in T cell and B cell activation. Curr Opin Immunol. 2005;17(3):275–281. doi: 10.1016/j.coi.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 33.McLellan AD, Ali Hosseini Rad SM. Chimeric antigen receptor T cell persistence and memory cell formation. Immunol Cell Biol. 2019;97(7):664–674. doi: 10.1111/imcb.12254. [DOI] [PubMed] [Google Scholar]
- 34.Xu Y, Zhang M, Ramos CA, Durett A, Liu E, Dakhova O, et al. Closely related T-memory stem cells correlate with in vivo expansion of CAR.CD19-T cells and are preserved by IL-7 and IL-15. Blood. 2014;123(24):3750–3759. doi: 10.1182/blood-2014-01-552174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gaud G, Lesourne R, Love PE. Regulatory mechanisms in T cell receptor signalling. Nat Rev Immunol. 2018;18(8):485–497. doi: 10.1038/s41577-018-0020-8. [DOI] [PubMed] [Google Scholar]
- 36.Salter AI, Ivey RG, Kennedy JJ, Voillet V, Rajan A, Alderman EJ, et al. Phosphoproteomic analysis of chimeric antigen receptor signaling reveals kinetic and quantitative differences that affect cell function. Sci Signal. 2018 doi: 10.1126/scisignal.aat6753. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data were contained in the manuscript, and raw data for the scRNA-seq and scTCR-seq are available via HRA002418 at the National Genome Data Center (NGDC) (https://ngdc.cncb.ac.cn/).