Abstract
Tumour-associated macrophages (TAMs) support cancer cell survival and suppress anti-tumour immunity. Tumour infiltration by CD163pos TAMs is associated with poor outcome in several human malignancies, including multiple myeloma (MM). Signal transducer and activator of transcription 3 (STAT3) is over-activated in human cancers, and specifically within TAMs activation of STAT3 may induce an immunosuppressive (M2-like) phenotype. Therefore, STAT3-inhibition in TAMs may be a future therapeutic strategy.
We investigated TAM markers CD163, CD206, and activated STAT3 (pSTAT3) in patients with MGUS (n = 32) and MM (n = 45), as well as healthy controls (HCs, n = 13).
Blood levels of the macrophage biomarkers sCD163 and sCD206, and circulating cytokines, as well as bone marrow mRNA expression of CD163 and CD206, were generally increased in MGUS and MM patients, compared to HCs, but to highly similar levels. By immunohistochemistry, bone marrow levels of pSTAT3 were increased specifically within CD163pos cells in both MGUS and MM patients.
In conclusion, macrophage-related inflammatory changes, including activation of STAT3, were present already at the MGUS stage, at similar levels as in MM. Specific increase in pSTAT3 levels within CD163pos cells supports that the CD163 scavenger receptor may be a useful target for future delivery of STAT3-inhibitory drugs to TAMs in MM patients.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00262-021-02952-1.
Keywords: Bone marrow microenvironment, CD163, Macrophage, MGUS, Multiple myeloma, STAT3
Introduction
Multiple myeloma (MM) is the second most common hematology malignancy, in which proliferating clonal plasma cells expand within the bone marrow, causing anaemia, osteolytic bone lesions, and hypercalcaemia. Further, monoclonal protein deposition often leads to renal failure in MM patients. Despite major advances in treatment, MM remains incurable [1].
Virtually all cases of MM are preceded by the premalignant condition monoclonal gammopathy of undetermined significance (MGUS) [2], and studies showed that the monoclonal plasma cells (mPCs) in MGUS and MM were highly similar regarding mutations, translocations, deletions, gene expression, and immunophenotype [3–5]. These findings have prompted researchers to look for factors outside the malignant cells, thus in the bone marrow microenvironment, in search of the key factors that drive transitioning from MGUS to MM [3, 5, 6].
The bone marrow microenvironment (the “myeloma niche”) is known to be vital for MM cell survival, and the success of the “novel agents” is partly owing to their effects on MM supportive cells [7]. Therefore, the microenvironmental changes during transition from healthy to MGUS, and from MGUS to MM, are of particular interest. Along with the MM supportive bone marrow stromal cells (BMSCs), there has been a rising interest in the role of tumour-associated macrophages (TAMs) in MM pathobiology. In most human malignancies, infiltration by TAMs is associated with poor outcome [8, 9], due to TAM-mediated resistance to chemotherapy, suppression of anti-tumour immunity, increased angiogenesis, and ultimately metastasis [10–12].
The paradigm of macrophage-polarization comprises two extremes: M1-like (classically activated, pro-inflammatory) and M2-like (alternatively activated, anti-inflammatory) macrophages [13]. The activation state of TAMs mostly falls within the M2-spectrum, and TAMs are thought to perform tasks similar to those of M2-like macrophages in normal physiology—e.g. in wound healing [14, 15].
In patients with MM, high bone marrow infiltration by CD163pos TAMs has been associated with poor outcome [16–18]. In a recent study, MM bone marrow biopsies were investigated for inducible nitric oxide synthase (iNOS) and CD163 expression to identify M1-like and M2-like TAMs, respectively. High infiltration by CD163pos (M2) cells was associated with poor outcome, whereas high infiltration with iNOSpos (M1) cells was associated with longer survival [19]. These results may be explained, at least in part, by TAM-induced resistance to anti-myeloma treatment [20, 21], as well as immunosuppressive functions of M2-like TAMs [22, 23]. In line with this, we previously showed that higher serum levels of the M2-related macrophage-derived biomarkers soluble CD163 (sCD163) and soluble mannose receptor/CD206 (sCD206), at the time of MM diagnosis, were associated with poor outcome [24, 25].
Importantly, TAMs can be “re-programmed” from M2-like towards M1-like cells, which may restore anti-tumour immunity, and thus may be a strategy for novel anti-cancer therapy [12, 23, 26, 27]. Further, in murine models of MM, macrophages could be activated to kill MM cells in vivo by blocking the anti-phagocytic protein CD47 [28], or by M1-polarizing stimuli [29, 30]. One attractive target is inhibition of the signal transducer and activator of transcription 3 (STAT3) transcription factor within TAMs [12, 31]. In several solid tumours, STAT3 is over-activated within immune cells, which has been associated with cancer-promoting functions, including suppression of anti-tumour immunity [22, 32]. Thus, inhibition of STAT3 specifically within TAM may be able to restore anti-tumour immunity [31, 33, 34].
Strategies for re-programming of TAMs are now within reach of systemic therapy owing to the development of technologies for specific targeting of drugs to macrophages, e.g. by antibody targeting of the CD163 scavenger receptor, which is highly expressed by human TAMs [13, 35–37].
The aim of the present study was to characterize macrophages/TAMs in a prospective cohort of patients with newly diagnosed MGUS or MM, especially regarding expression of the TAM markers CD163 and CD206. Furthermore, we examined the expression of activated (phospho-Tyr-705)-STAT3 within the bone marrow of patients with MM and MGUS — including activated STAT3 localized within CD163pos TAMs, which may be a future therapeutic target.
Materials, subjects, and methods.
Patients and collected samples
In this prospective study, we included patients with a positive M-protein measurement, that were subsequently diagnosed with MGUS (n = 32) or MM (n = 45) at the Department of Hematology, Aarhus University Hospital, Aarhus, Denmark between 2011 and 2015. Included patients diagnosed with other conditions (e.g. CLL or plasmacytoma, n = 8) were excluded from the study. In addition, we included elderly volunteers to serve as healthy controls (HCs, n = 13).
At the time of diagnosis, we collected whole bone marrow aspirate for RNA purification using the PAXgene Bone Marrow RNA system (Qiagen, Hilden, Germany) according to the manufactures instructions, to stabilize bone marrow RNA levels at sample collection. Further, at diagnosis, serum and EDTA stabilised plasma samples were collected. All samples were stored at 80 °C. For immunohistochemistry (IHC), we used the Jamshidi biopsy taken for diagnostic purpose. For eight patients, this biopsy was not available, and the diagnostic bone marrow aspirate was used for IHC.
Data on basic patient characteristics along with clinical and laboratory data, as well as bone marrow mPC infiltration determined by a pathologist, were obtained from medical records. The study was approved by the Central Denmark Region Committee on Health Research Ethics (M-20100171) and conducted according to the declaration of Helsinki. All patients and control subjects gave informed written consent before inclusion.
Serum/plasma levels of sCD163, sCD206, and inflammatory cytokines
Serum concentrations of the macrophage activation biomarkers sCD163 and sCD206 were measured using our in-house enzyme-linked immunosorbent assays (ELISA) as described in detail previously [38, 39]. Samples were run on an automated BEP 2000 system (Siemens Healthcare Diagnostics, Munich, Germany). Plasma concentrations of cytokines were measured using electro-chemo-luminescence- based multiplex sandwich immunoassays according to the manufacturer instructions (Meso Scale Discovery, Rockville, MD): Interleukin-1 beta (IL-1β), IL-6, IL-10, IL-12p70, tumour necrosis factor alpha (TNFα), and interferon gamma (IFNγ).
Reverse transcription quantitative polymerase chain reaction (RT-qPCR)
Purified RNA was reverse transcribed to cDNA in a 20 μL reaction of 2.5 μM Oligo(dT) (DNA Technology, Risskov, Denmark), 1 mM dNTP mix (VWR International, Radnor, PA), 2.5 U/μL MulV reverse transcriptase enzyme, 1 U/μL RNase inhibitors, 1 × PCR buffer and 6.25 mM MgCl2 (all from Applied Biosystems, Thermo Fisher Scientific, Waltham, MA). In each reaction, the RNA input was 100 ng (measured using a NanoDrop 2000 spectrophotometer, Thermo Fisher Scientific).
Quantitative PCR was performed on a LightCycler 480 instrument (Roche, Basel, Switzerland) in a 10 μL reaction with primers, 480 SYBR Green I Master mix (Roche), and 1μL cDNA (generated as described above). Fifty qPCR cycles were run; denaturation at 95 °C for 10 s, 20 s at annealing temperature, and 72 °C for 5 s (see Supplemental table 1 for primer sequences, concentrations, and annealing temperatures). All samples were run in duplicates. For relative mRNA quantification by RT-qPCR, a standard curve (calibrator) was included in each run (The E-Method, recommended by Roche). This compensates for variation in reaction efficiency, both day-to-day and inter-run variation. All qPCR results were normalized to the household gene SDHA (succinate dehydrogenase complex, subunit A), which was identified as the most stable of five tested household genes (Actin, HMBS, GAPDH, SDHA, YWHAZ) using the NormFinder algorithm [40]. Primers were from Eurofins Genomics (Ebersberg, Germany).
Immunohistochemistry
Sections (3 µm) of formaldehyde-fixed paraffin-embedded bone marrow samples, as well as control sections from healthy organs (pancreas, liver, and pharyngeal tonsil—included on every slide, Supplemental Fig 2), were mounted on glass slides, incubated for 1 h at 60 °C, and stored at 4 °C until use. Staining was performed on the automated platform Benchmark XT (Ventana Medical Systems, Roche Group, Tucson, AZ). The following monoclonal antibodies were used: Mouse anti-human CD163 (clone Edhu-1, dilution 1:450, Bio-Rad, Oxford, the UK), rabbit anti-human phospho-(Y705)-STAT3 (clone D3A7, dilution 1:50, Cell Signaling Technologies, Danvers, MA), mouse anti-human CD206 (clone 5C11, dilution 1:200, LSBio, Seattle, WA).
Antigen retrieval was done using cell conditioning solution 2 (CC2, Ventana Medical Systems). For the CD163/pSTAT3 double staining, additional antigen retrieval was performed using Protease 3 (Ventana Medical Systems), and lastly, Ultra Wash (Ventana Medical Systems) was used to reduce non-specific staining.
Visualization of pSTAT3 was performed with OptiView DAB (3,3’-diaminobenzidine) IHC Detection Kit, CD206 by UltraView Universal DAB Detection Kit, and CD163 by UltraView Universal Alkaline Phosphatase (AP) Red Detection Kit (all from Ventana Medical Systems). Standard haematoxylin counterstaining was done for all sections. One section from each patient/control was examined for each staining. Staining protocols were validated by a hematopathologist, including comparisons between single and double stainings for the CD163/pSTAT3 protocol showing good concordance.
Digital image analysis
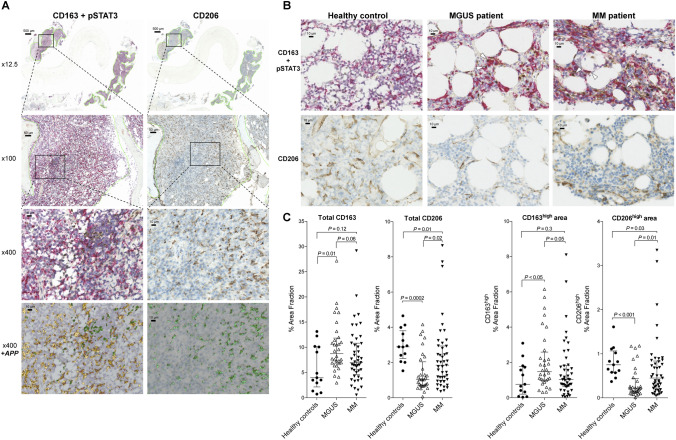
Slides were digitized using a Nanozoomer 2.0HT (Hamamatsu Photonics KK, Hamamatsu City, Japan) at a magnification of 20 × in the NDPI file format. Digitized slides were analysed using Visiopharm Visiomorph (VIS) software (Version 6, Visiopharm, Hørsholm, Denmark). Using VIS software, specific application protocol packages (APPs) were developed to analyse each staining (see Fig. 3a). The results were expressed as % Area Fraction (%AF), which is the positively stained area (µm2) as % of the total analysed area (µm2) within the biopsy. Thus, an AF of 8% for CD163 denotes that 8% of the analysed area stained positive for CD163. Tissue areas were identified using the “tissue detect” function in VIS, with subsequent manual adjustment to define the final used “region of interest” (ROI), which excluded areas with bone and cartilage tissues. Using the developed APPs, we were able to detect three staining intensities for CD163 and CD206, and two intensities for pSTAT3 (more intense staining is shown by darker colours in the + APP picture in Fig. 3a.) Due to some non-specific CD206 staining, we chose to exclude CD206low areas from further analyses (this did not change the statistical results.) To quantify CD163-associated pSTAT3 signal (a measure of activated STAT3 within CD163pos cells), we instructed the CD163/pSTAT3 APP to quantify the pSTAT3 positive area that was surrounded by CD163 positive area (> 50% of pSTAT3 area surrounded, Fig. 4c).
Fig. 3.
Bone marrow immunohistochemistry: Macrophage markers and STAT3 activation. Bone marrow Jamshidi biopsies from the time of diagnosis were double stained for CD163 and activated (phospho-Tyr-705)-STAT3 (pSTAT3), or single stained for CD206. CD163 was stained red (developed with red alkaline phosphatase), whereas pSTAT3 and CD206 positive areas were stained brown (developed with DAB). Using VIS software, we developed APPs (application protocol packages) to analyse the whole bone marrow biopsies by automated digital image analysis. a The two columns show stainings for CD163/pSTAT3 and CD206, respectively, at 12.5x, 100x, and 400 × magnifications. In the 12.5 × and 100 × images, the analysed “region of interest” is marked with green borders. 400 × pictures are shown with/without the respective APPs applied. Using digital image analysis, we detected three increasing staining intensities for CD163 (yellow/orange) and CD206 (green), and two intensities of pSTAT3 (green): More intense staining is shown with darker colours in the APP images (bottom). As described, to minimize impact of non-specific staining in the CD206 specimens, the CD206low areas were excluded from statistical analyses. b Representative pictures (× 400) of CD163/pSTAT3 (top) and CD206 (bottom) stainings from an HC, an MGUS, and an MM patient. Some MGUS and MM patients had high levels of pSTAT3 within CD163pos macrophages (white arrowheads). c % Area Fractions (%AF) for total CD163 and CD206 expression within the bone marrow (left), and areas with highest staining intensity are shown (right). For quantitative data on pSTAT3 expression, see Fig. 4. Lines and whiskers show median and IQR
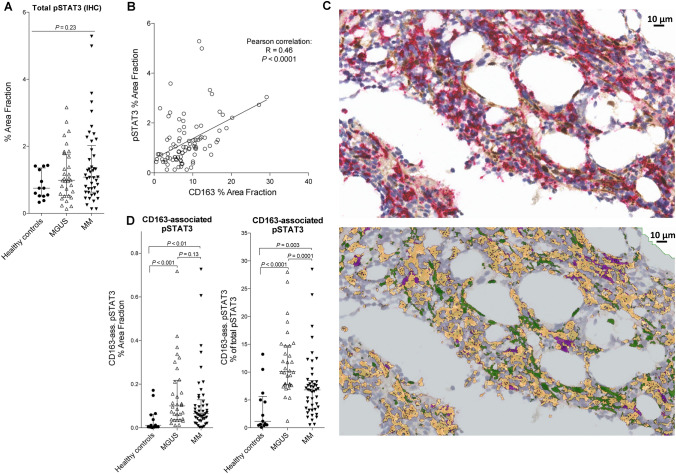
Fig. 4.
STAT3 activation within CD163pos macrophages is increased in MGUS and MM patients. a Quantitation of total pSTAT3% Area Fraction (%AF) showed high levels in some MGUS and MM patients, but with no overall statistically significant difference. b The bone marrow expression of CD163 and pSTAT3 showed positive correlation, with some patients having high levels of both markers (data from both HCs, MGUS, and MM patients included). c Using digital image analysis, we examined CD163-associated pSTAT3 as a measure of STAT3-activation level within CD163pos macrophages. Images show stainings from an MM patient without (top) and with the APP applied (bottom). In the APP picture, all CD163-positive staining is yellow, non-CD163-associated pSTAT3 is green, and CD163-associated pSTAT3 staining is purple (defined as pSTAT3 positive staining surrounded by ≥ 50% CD163 positive staining). d Quantitative data from HCs, MGUS, and MM patients showing total pSTAT3 that is CD163-associated (%AF of total bone marrow area, left), and also CD163-associated pSTAT3 %AF relative to total pSTAT3 %AF (measure of shift in STAT3 activity towards macrophages). Lines and whiskers show median and IQR
Statistical analyses
Statistical analyses were performed using STATA version 14 for Mac OS (StataCorp LP, TX). Graphs were made using Prism 5d for Mac OS (GraphPad software, Inc., La Jolla, CA). To test differences between patient groups, we used one-way analysis of variance (ANOVA) and t-test when appropriate. Gaussian distribution of data was assessed using Q-Q plots. Data with non-normal distribution were log-transformed using the natural logarithm to obtain a normal distribution before statistical analysis. For data with significant differences in standard deviations between groups Kruskal–Wallis and Mann–Whitney (rank-sum) tests were used. A P value ≤ 0.05 was considered statistically significant.
Results
Basic characteristics of the included patients and controls with paraclinical data from the time of diagnosis are shown in Table 1. MGUS and MM patients were slightly older than the HCs (P < 0.01). Eighty percent of the MM patients had treatment demanding disease at diagnosis. Patients with MM had lower haemoglobin (P < 0.0001), and levels of both serum M-protein and bone marrow mPC infiltration were higher in MM compared to MGUS (P < 0.0001) as expected.
Table 1.
Basic characteristics of the included patients and controls
| Healthy controls ( n = 13) | MGUS patients ( n = 32) | MM patients ( n = 45) | |||||
|---|---|---|---|---|---|---|---|
| n (%) | Median (IQR) | n (%) | Median (IQR) | n (%) | Median (IQR) | P | |
| Sex | |||||||
| Male | 7 (54) | 16 (50) | 25 (56) | 0.89 | |||
| Age | 13 (100) | 59 (57—63) | 32 (100) | 75 (61—79) | 45 (100) | 70 (64—75) | < 0.01 |
| M-protein subtype | |||||||
| IgA | 5 (16) | 9 (20) | |||||
| IgG | 26 (81) | 31 (69) | |||||
| LC only | 1 (3) | 5 (10) | |||||
| ISS stage | |||||||
| I | 8 (18) | ||||||
| II | 26 (58) | ||||||
| III | 11 (24) | ||||||
| Treatment demanding | 36 (80) | ||||||
| Bone lesions | |||||||
| No lesions | 17 (19) | ||||||
| Lesions in one area | 8 (19) | ||||||
| Several lesions | 18(42) | ||||||
| BM monoclonal PC (%) | 32 (100) | 0 (0—6) | 45 (100) | 45 (20—70) | < 0.0001 | ||
| M-protein (g/L) | 13 (100) | 0 (N.A.) | 32 (100) | 7.2 (3.6—10) | 44 (98) | 33.5 (20.5—48) | < 0.0001 |
| Creatinine (µmol/L) | 13 (100) | 72 (66—78) | 32 (100) | 77 (67—98) | 45 (100) | 78 (69—103) | 0.33 |
| CRP (mg/L) | 13 (100) | < 4 (N.A.) | 32 (100) | 2.8 (0.6—8.8) | 44 (98) | 3.9 (1.5—9.2) | 0.79 |
| Hb (mM) | 11 (85) | 8.8 (8.4—10.6) | 32 (100) | 8.4 (7.45—8.8) | 45 (100) | 6.9 (6.3—7.6) | < 0.0001 |
| Platelet count (10^9/L) | 11 (85) | 246 (225—250) | 32 (100) | 260 (209—313) | 45 (100) | 241 (181—323) | 0.68 |
| WBC count (10^9/L) | 11 (85) | 7.0 (5.7—7.8) | 32 (100) | 7.2 (5.8—8.6) | 45 (100) | 5.4 (4.6—7.6) | 0.02 |
P values by ANOVA. CRP was < 4 mg/L in all healthy controls, except one (6.3 mg/L); measured on standard CRP assay, whereas high-sensitivity CRP assay was used for MGUS and MM patients
LC light chain; ISS International Staging System; BM bone marrow; CRP C-reactive protein; Hb haemoglobin; WBC white blood cell
Increased levels of peripheral blood inflammatory markers in MGUS and MM patients
Previously, we showed in retrospective studies that higher serum levels of macrophage activation markers sCD163 and sCD206 were associated with poor overall survival in MM patients [24, 25].
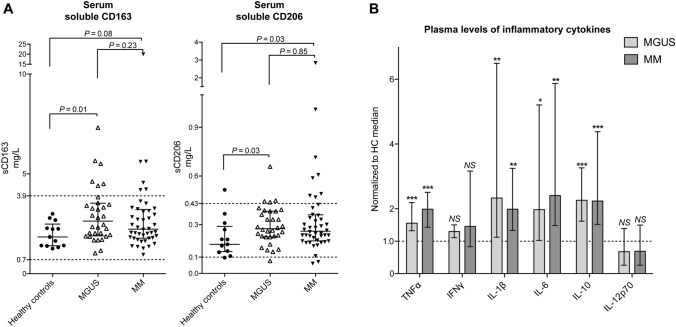
In the present prospective study, levels of sCD163 and sCD206 were slightly increased in both MGUS and MM compared to HCs (Fig. 1a, not statistically significant for sCD163 in MM vs. HCs). Interestingly, there were no significant differences between MGUS and MM patients regarding both sCD163 and sCD206 levels (P > 0.2). Further, levels of sCD163 and sCD206 were positively correlated in MGUS and MM patients (P < 0.05 and < 0.0005, respectively), whereas this was not seen in HCs (P = 0.2, Supplemental Fig 1a.)
Fig. 1.
Peripheral blood levels of macrophage activation markers and inflammatory cytokines. a Serum levels of macrophage-related inflammation markers sCD163 and sCD206, at the time of diagnosis, are shown for HCs, MGUS, and MM patients. Reference intervals are shown with dashed lines for sCD163 and sCD206. sCD163 (median and IQR); HCs: n = 13, 1.94 mg/L (1.37–2.47), MGUS: n = 32, 2.64 mg/L (1.92–3.51), MM: n = 44, 2.23 mg/L (1.79–3.20). sCD206 (median and IQR); HCs: n = 13, 0.18 mg/L (0.12–0.29), MGUS: n = 32, 0.28 mg/L (0.23–0.38), MM: n = 44, 0.26 mg/L (0.20–0.36). b Plasma levels of inflammatory cytokines at the time of diagnosis. Data are displayed as values in MGUS and MM patients, respectively, normalized to the median value for HCs for simplicity. Median values (and IQR) for the HC group (n = 13): TNFα: 1.58 pg/mL (1.47–1.92), IFNγ: 4.77 pg/mL (3.22–8.09), IL-1β: 0.06 pg/mL (0.01–0.08), IL-6: 0.61 pg/mL (0.40–0.85), IL-10: 0.21 pg/mL (0.19–0.27), IL-12p70: 0.10 pg/mL (0.05–0.19). TNFα was slightly higher in MM compared to MGUS patients (P = 0.05), otherwise there were no statistically significant differences between MGUS and MM patients (all P > 0.2). Statistical analyses were performed using raw data, not normalised values. Symbols above each column indicate statistical differences compared to the HC group: NS: P > 0.05, * P ≤ 0.05, ** P < 0.005, *** P < 0.0005. Box and whiskers show median and IQR
The examined cytokines showed a pattern similar to the macrophage activation markers, with significantly increased levels of TNFα, IL-1β, IL-6, IL-10 in MGUS and MM, compared to HCs, and the measured cytokine levels did not differ between MGUS and MM patients (all P > 0.2 except for TNFα (P = 0.05), Fig. 1b).
Increased gene expression of TAM markers CD163 and CD206 in MGUS and MM bone marrow
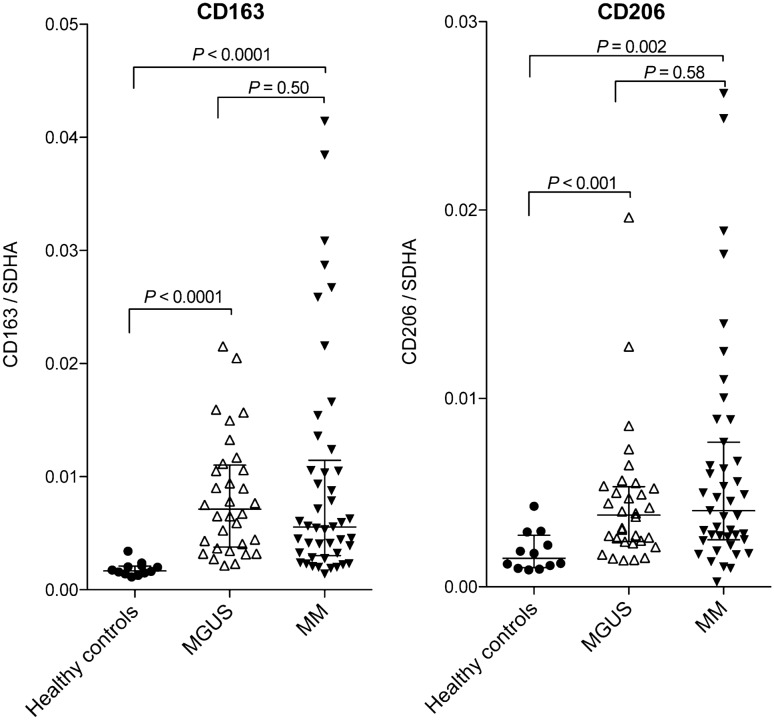
We investigated gene expression levels of CD163 and CD206 within the bone marrow microenvironment of HCs, MGUS, and MM patients by qPCR on RNA isolated from bone marrow aspirates. Both CD163 and CD206 mRNA expression were markedly higher in patients with MGUS (P < 0.001) and MM (P < 0.002) compared to HCs (Fig.2). Interestingly, CD163 and CD206 mRNA levels were increased to similar levels in MGUS and MM patients (P ≥ 0.50). Further, expression levels of CD163 and CD206 were positively correlated in both MGUS (P = 0.03) and MM patients (P < 0.0001), but not in HCs (P = 0.96, Supplemental Fig. 1b).
Fig. 2.
Bone marrow gene expression of CD163 and CD206 at diagnosis. Reverse transcriptase quantitative PCR (qPCR) was used to analyse mRNA expression in whole marrow aspirates from MGUS and MM patients at the time of diagnosis (n = 32 and 45, respectively) and HCs (n = 12). M2-like macrophage markers CD163 and CD206 were increased in both MGUS and MM patients, but with no difference between the two disease states. All qPCR results were normalized to housekeeping gene SDHA as described. Lines and whiskers show median and IQR. SDHA: Succinate dehydrogenase complex, subunit A
Bone marrow immunohistochemistry: Increased CD163 but decreased CD206 expression in MGUS patients
We also investigated protein levels of TAM markers CD163 and CD206 within the MGUS and MM bone marrow microenvironment using immunohistochemistry (IHC) with digital image analysis (Fig. 3a). Double stainings for CD163 and pSTAT3 and single stainings for CD206 were performed (Fig. 3b). Results are expressed as % Area Fraction (%AF) of positive staining relative to the total analysed area. Patients with MGUS had significantly higher CD163pos %AF compared to HC (P = 0.01), but only a tendency for higher CD163pos %AF was observed in MM patients (P = 0.12). Further, there was no significant difference in total CD163pos %AF between MGUS and MM (P = 0.06). For CD206, patients with both MGUS and MM had significantly lower %AF than HCs (P = 0.0002 and 0.01, respectively), and MM patients had slightly higher CD206%AF than in MGUS (P = 0.02).
As a measure of M2-like polarization of bone marrow macrophages, we quantified areas with “high expression” of CD163 or CD206 (the highest staining intensity as shown in Fig. 3a). This showed results similar to total CD163 and CD206 expression levels; compared to HCs, the CD163high area was significantly larger in MGUS (P < 0.05), but not in MM (P = 0.3), whereas the CD206high area was smaller in patients with MGUS compared to HCs (P < 0.001) and was larger in MM compared to MGUS patients (P = 0.01, Fig. 3c).
There was a positive correlation between CD163 and CD206 %AFs in MM patients (P = 0.03), but not for HCs (P = 0.92) or MGUS patients (P = 0.47, Supplemental Fig. 1c).
Increased bone marrow STAT3 activation within CD163pos cells in MGUS and MM patients
Bone marrow levels of activated STAT3 (by phosphorylation of Tyr-705, pSTAT3) and association with CD163pos macrophages were also examined by IHC. Overall, expression levels of pSTAT3 were not significantly higher in MGUS and MM patients, compared to HCs (P = 0.23, Fig. 4a).
However, there was a positive correlation between the expression levels of CD163 and pSTAT3 within the bone marrow biopsies (R = 0.46, P < 0.0001, Fig. 4b). Using digital image analysis, we then investigated the level of STAT3 activation specifically within CD163pos cells (quantified as CD163-associated pSTAT3 staining, Fig. 4c). Interestingly, both patients with MGUS and MM had significantly higher levels of pSTAT3 within CD163pos cells, compared to HCs (P < 0.001 and < 0.01, respectively, Fig.4d). Also, the relative amount of total pSTAT3 staining that was CD163-associated, was significantly higher in both MGUS (P < 0.0001) and MM patients (P = 0.003) compared to HCs, but also with markedly higher levels in MGUS compared to MM patients (P = 0.0001) —indicating a shift in the location of activated STAT3 towards macrophages in MM, and especially in the premalignant condition MGUS (Fig.4d).
Associations of TAM markers with MM pathophysiology and prognostic factors
There were no clear associations between the analysed TAM markers in the MM patients and bone marrow mPC infiltration, elevated LDH levels, or adverse cytogenetic profile (del(17p), t(4;14), or t(14;16), data not shown). Figure 5 shows CD163, CD206, and pSTAT3 data for the included MM patients, in relation to International Staging System (ISS) stage and bone disease severity at the time of diagnosis. Both serum sCD206 and CD206%AF tended to be higher in patients with ISS 3 (both P = 0.02), whereas pSTAT3 levels were lower in most patients with ISS 3 (P = 0.03). We also observed higher CD163 and CD206 mRNA expression in the bone marrow of patients with the most severe bone disease at diagnosis (P = 0.05 and 0.03, respectively).
Fig. 5.
Associations of TAM markers with prognostic factors in MM patients. Expression levels of M2/TAM markers CD163, CD206, and pSTAT3 are shown as stratified by ISS stage (a), or by the severity of bone disease (b) at diagnosis for included patients with MM. Shown P values are from ANOVA. NS: P > 0.05. Lines and whiskers show median and IQR. SDHA: Succinate dehydrogenase complex, subunit A
Discussion
Tumour-associated macrophages are abundant in tumours, including the myeloma bone marrow microenvironment, and have attracted increasing attention in recent years due to their tumour-promoting functions. Thus, TAMs are now considered important future therapeutic targets in cancers, including MM [12, 30, 41].
Here, we describe levels of circulating macrophage-related and general inflammatory markers, as well as bone marrow microenvironment levels of TAM markers CD163, CD206, and pSTAT3, covering the range from healthy to MGUS and to MM, in a prospective study.
The main findings of the study were increased levels of TAM markers, including activated STAT3 specifically within bone marrow CD163pos cells, in both MGUS and MM patients. Further, TAM markers were increased already at the MGUS stage, both in the bone marrow and in the circulation, to levels seen in MM, which was accompanied by increased systemic cytokine levels.
Statistical power of the study could have been increased by inclusion of more healthy controls. However, it is difficult to recruit healthy volunteers who are of relatively high age, and also willing to volunteer for bone marrow and blood sampling.
It is recognized that changes within the bone marrow microenvironment of MM patients establish a “niche” with MM-protective effects [6], and TAMs are known to play important roles in MM pathobiology [16, 17, 19–21, 30]. Interestingly, we observed that such microenvironmental changes seemed to be established already at the premalignant MGUS stage. Thus, we found significantly increased bone marrow levels of CD163 and CD206 mRNA in both MGUS and MM, but with no difference between the two disease states. Further, we found similarly increased blood levels of sCD163 and sCD206, as well as inflammatory cytokines, for MGUS and MM patients (not statistically significant for sCD163 in MM). The similarly increased cytokine levels correspond with recently published results [42]. Previously, we reported serum levels of sCD163 and sCD206 as independent prognostic biomarkers in MM patients [24, 25], but only one report with few patients has previously compared serum levels of sCD163 in patients with MGUS and MM with HCs, showing no significant difference [43].
In the present study, IHC analysis of bone marrow CD163 and CD206 expression showed a mixed picture, with increased expression of CD163 but decreased CD206 in MGUS, and no clear changes in MM. Further, healthy controls had the highest bone marrow CD206 levels. A previous study showed increased bone marrow CD206 expression, and no significant changes in CD163 expression, in MM patients vs. non-MM patient controls with “normal bone marrow” [41]. Thus, our CD206 IHC results are in contrast to this study. We are not aware of other studies that have compared CD206 bone marrow levels between MM and MGUS patients, and with healthy controls. Thus, further studies are needed.
Numerous studies have investigated tumour CD163 expression by IHC and demonstrated associations with clinical outcome [8]. However, few studies have investigated CD163 mRNA expression in human tumours, e.g. one study showed association between high CD163 mRNA levels and poor outcome in bladder cancer [44]. The increased CD163 and CD206 bone marrow mRNA levels in both MGUS and MM have not previously been described and should be investigated further as prognostic markers.
It has been shown that genetic abnormalities characteristic for MM mPCs is also present in mPCs from MGUS patients [3, 4], indicating that changes in the bone marrow microenvironment may drive the progression from MGUS to MM [5, 6]. Thus, our findings of highly similar levels of macrophage-related inflammation in MGUS and MM are interesting, indicating that TAMs of the myeloma microenvironment may be polarized already during MGUS.
In the bone marrow biopsies, there was a strong correlation between CD163 and activated STAT3 (pSTAT3) levels. This is not surprising since STAT3 activation is known to upregulate CD163 on human macrophages [45, 46]. Some might be concerned that the used CD163/pSTAT3 double staining would favour such an association, which should be taken into account. However, as described, our assay validation showed good concordance between single/double staining. Further, we observed significantly higher levels of STAT3 activation within CD163pos cells in the bone marrow of both MGUS and MM patients compared to HCs. Interestingly, the relative amount of pSTAT3 within CD163pos cells (% of total pSTAT3) was higher in MGUS compared to both HCs and MM. The lower levels of CD163-associated pSTAT3 in MM compared to MGUS patients may seem unexpected, however, this result could be impacted by the higher infiltration in MM of mPCs that may also be pSTAT3pos. Nevertheless, STAT3 signalling in TAMs and myeloid-derived suppressor cells can induce an immunosuppressive phenotype [22, 32, 47], and this may indicate a role for macrophage STAT3 over-activation in establishing the immunosuppressive “bone marrow niche” already during the MGUS stage. Such STAT3 activation in TAMs has been proposed as a future therapeutic target [33, 34, 36], since specific STAT3 inhibition can re-program TAMs from an M2-like (pro-tumour) to an M1-like (tumouricidal) phenotype [22, 31, 32]. Recently, M2-to-M1 re-programming of macrophages showed anti-myeloma effects in vivo [30]. However, specifically in MM, functional studies demonstrating anti-tumour effects of TAM-specific STAT3 inhibition are needed.
Thus, STAT3 inhibition specifically within TAMs may be an attractive anti-cancer therapeutic strategy in several malignancies [12, 31, 33, 34]. We recently showed that a novel CD163-targeted STAT3-inhibitory liposome drug was able to inhibit STAT3 specifically in CD163pos cells, with a pro-inflammatory (re-programming) effect [36]. The present findings of significantly elevated levels of CD163-associated pSTAT3, in both MGUS and MM patients, encourage future studies on CD163-targeted STAT3-inhibition as an anti-myeloma immunomodulatory drug.
In conclusion, our data demonstrated similar inflammatory changes in MGUS and MM patients, with similar bone marrow expression of CD163, and increased levels of circulating macrophage biomarkers and cytokines. The specific increase in pSTAT3 levels within CD163pos cells supports that the CD163 scavenger receptor may be a useful gateway for drug delivery of STAT3-inhibitors to TAMs in MM.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors wish to thank Kirsten Petersen, Lene Dabelstein, Helle Ryom, Christina Sønderskov, and Gitte Brix for excellent technical assistance.
Authors Contributions
MNA, NFA, MH, HJM contributed to conception and design. MNA, NFA, KLA, TP, BSS, HJM were involved in inclusion of patients and collection of data. MNA, NFA, KLA, AE, NA, MH, HJM contributed to analysis and interpretation of data. MNA was involved in drafting the article. NFA, KLA, AE, BSS, NA, TP, MH, HJM contributed to revising the manuscript critically. MNA, NFA, KLA, AE, BSS, NA, TP, MH, HJM were involved in approval of the version to be published.
Funding
The work was supported by the Department of Clinical Biochemistry, Aarhus University Hospital, Denmark; The Faculty of Health, Aarhus University, Denmark; The Danish Cancer Research Foundation; The Cancer Foundation (Denmark), The Memorial Foundation of Max and Inge Wørzner, and by The Memorial Foundation of Eva and Henry Frænkel.
Data availability
Data from the present study are available upon reasonable request to the corresponding author.
Declarations
Conflict of interest
HJM is a minority shareholder in DeLiver Pharma that holds IP protecting the use of CD163 drug targeting. All other authors declare no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Rajkumar SV, Dimopoulos MA, Palumbo A, et al. International Myeloma Working Group updated criteria for the diagnosis of multiple myeloma. Lancet Oncol. 2014;15:e538–e548. doi: 10.1016/S1470-2045(14)70442-5. [DOI] [PubMed] [Google Scholar]
- 2.Landgren O, Kyle RA, Pfeiffer RM, et al. Monoclonal gammopathy of undetermined significance (MGUS) consistently precedes multiple myeloma: a prospective study. Blood. 2009;113:5412–5417. doi: 10.1182/blood-2008-12-194241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bladé J, Rosinol L, Cibeira MT, de Larrea CF. Pathogenesis and progression of monoclonal gammopathy of undetermined significance. Leukemia. 2008;22:1651–1657. doi: 10.1038/leu.2008.203. [DOI] [PubMed] [Google Scholar]
- 4.Zhao S, Choi M, Heuck C, et al. Serial exome analysis of disease progression in premalignant gammopathies. Leukemia. 2014;28:1548–1552. doi: 10.1038/leu.2014.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dhodapkar MV. MGUS to myeloma: a mysterious gammopathy of underexplored significance. Blood. 2016;128:2599–2606. doi: 10.1182/blood-2016-09-692954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghobrial IM, Detappe A, Anderson KC, Steensma DP. The bone-marrow niche in MDS and MGUS: implications for AML and MM. Nat Rev Clin Oncol. 2018;15:219–233. doi: 10.1038/nrclinonc.2017.197. [DOI] [PubMed] [Google Scholar]
- 7.Podar K, Chauhan D, Anderson KC. Bone marrow microenvironment and the identification of new targets for myeloma therapy. Leukemia. 2008;23:10–24. doi: 10.1038/leu.2008.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Q-W, Liu L, Gong C-Y, et al. Prognostic significance of tumor-associated macrophages in solid tumor: a meta-analysis of the literature. PLoS ONE. 2012 doi: 10.1371/journal.pone.0050946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Komohara Y, Jinushi M, Takeya M. Clinical significance of macrophage heterogeneity in human malignant tumors. Cancer Sci. 2014;105:1–8. doi: 10.1111/cas.12314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qian B-Z, Pollard JW. Macrophage diversity enhances tumor progression and metastasis. Cell. 2010;141:39–51. doi: 10.1016/j.cell.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Noy R, Pollard JW. Tumor-Associated Macrophages: From Mechanisms to Therapy. Immunity. 2014;41:49–61. doi: 10.1016/j.immuni.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Komohara Y, Fujiwara Y, Ohnishi K, Takeya M. Tumor-associated macrophages: Potential therapeutic targets for anti-cancer therapy. Adv Drug Deliv Rev. 2016;99:180–185. doi: 10.1016/j.addr.2015.11.009. [DOI] [PubMed] [Google Scholar]
- 13.Biswas SK, Mantovani A. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat Immunol. 2010;11:889–896. doi: 10.1038/ni.1937. [DOI] [PubMed] [Google Scholar]
- 14.Wynn TA, Chawla A, Pollard JW. Macrophage biology in development, homeostasis and disease. Nature. 2013;496:445–455. doi: 10.1038/nature12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muliaditan T, Caron J, Okesola M, et al. Macrophages are exploited from an innate wound healing response to facilitate cancer metastasis. Nat Commun. 2018;9:2951. doi: 10.1038/s41467-018-05346-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suyanı E, Sucak GT, Akyürek N, et al. Tumor-associated macrophages as a prognostic parameter in multiple myeloma. Ann Hematol. 2013;92:669–677. doi: 10.1007/s00277-012-1652-6. [DOI] [PubMed] [Google Scholar]
- 17.Panchabhai S, Kelemen K, Ahmann G, et al. Tumor-associated macrophages and extracellular matrix metalloproteinase inducer in prognosis of multiple myeloma. Leukemia. 2016;30:951–954. doi: 10.1038/leu.2015.191. [DOI] [PubMed] [Google Scholar]
- 18.Wang H, Hu W-M, Xia Z-J, et al. High numbers of CD163+ tumor-associated macrophages correlate with poor prognosis in multiple myeloma patients receiving bortezomib-based regimens. J Cancer. 2019;10:3239–3245. doi: 10.7150/jca.30102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen X, Chen J, Zhang W, et al. Prognostic value of diametrically polarized tumor-associated macrophages in multiple myeloma. Oncotarget. 2017;8:112685–112696. doi: 10.18632/oncotarget.22340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng Y, Cai Z, Wang S, et al. Macrophages are an abundant component of myeloma microenvironment and protect myeloma cells from chemotherapy drug-induced apoptosis. Blood. 2009;114:3625–3628. doi: 10.1182/blood-2009-05-220285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng Y, Yang J, Qian J, et al. PSGL-1/selectin and ICAM-1/CD18 interactions are involved in macrophage-induced drug resistance in myeloma. Leukemia. 2012;27:702–710. doi: 10.1038/leu.2012.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer. 2009;9:798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quail DF, Joyce JA. Molecular Pathways: Deciphering Mechanisms of Resistance to Macrophage-Targeted Therapies. Clin Cancer Res. 2017;23:876–884. doi: 10.1158/1078-0432.CCR-16-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andersen MN, Abildgaard N, Maniecki MB, et al. Monocyte/macrophage-derived soluble CD163: a novel biomarker in multiple myeloma. Eur J Haematol. 2014;93:41–47. doi: 10.1111/ejh.12296. [DOI] [PubMed] [Google Scholar]
- 25.Andersen MN, Andersen NF, Rødgaard-Hansen S, et al. The novel biomarker of alternative macrophage activation, soluble mannose receptor (sMR/sCD206): Implications in multiple myeloma. Leuk Res. 2015;39:971–975. doi: 10.1016/j.leukres.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 26.Porcheray F, Viaud S, Rimaniol A-C, et al. Macrophage activation switching: an asset for the resolution of inflammation. Clin Exp Immunol. 2005;142:481–489. doi: 10.1111/j.1365-2249.2005.02934.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hagemann T, Lawrence T, McNeish I, et al. “Re-educating” tumor-associated macrophages by targeting NF- B. J Exp Med. 2008;205:1261–1268. doi: 10.1084/jem.20080108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim D, Wang J, Willingham SB, et al. Anti-CD47 antibodies promote phagocytosis and inhibit thegrowth of human myeloma cells. Leukemia. 2012;26:2538–2545. doi: 10.1038/leu.2012.141. [DOI] [PubMed] [Google Scholar]
- 29.Jensen JL, Rakhmilevich A, Heninger E, et al. Tumoricidal effects of macrophage-activating immunotherapy in a murine model of relapsed/refractory multiple myeloma. Cancer Immunol Res. 2015;3:881–890. doi: 10.1158/2326-6066.CIR-15-0025-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gutiérrez-González A, Martínez-Moreno M, Samaniego R, et al. Evaluation of the potential therapeutic benefits of macrophage reprogramming in multiple myeloma. Blood. 2016;128:2241–2252. doi: 10.1182/blood-2016-01-695395. [DOI] [PubMed] [Google Scholar]
- 31.Su Y-L, Banerjee S, White SV, Kortylewski M. STAT3 in tumor-associated myeloid cells: multitasking to disrupt immunity. Int J Mol Sci. 2018 doi: 10.3390/ijms19061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu H, Lee H, Herrmann A, et al. Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nat Rev Cancer. 2014;14:736–746. doi: 10.1038/nrc3818. [DOI] [PubMed] [Google Scholar]
- 33.Herrmann A, Kortylewski M, Kujawski M, et al. Targeting Stat3 in the myeloid compartment drastically improves the in vivo antitumor functions of adoptively transferred T cells. Cancer Res. 2010;70:7455–7464. doi: 10.1158/0008-5472.CAN-10-0736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dang W, Tang H, Cao H, et al. Strategy of STAT3β cell-specific expression in macrophages exhibits antitumor effects on mouse breast cancer. Gene Ther. 2015;22:977–983. doi: 10.1038/gt.2015.70. [DOI] [PubMed] [Google Scholar]
- 35.Etzerodt A, Maniecki MB, Graversen JH, et al. Efficient intracellular drug-targeting of macrophages using stealth liposomes directed to the hemoglobin scavenger receptor CD163. J Control Release. 2012;160:72–80. doi: 10.1016/j.jconrel.2012.01.034. [DOI] [PubMed] [Google Scholar]
- 36.Andersen MN, Etzerodt A, Graversen JH, et al. STAT3 inhibition specifically in human monocytes and macrophages by CD163-targeted corosolic acid-containing liposomes. Cancer Immunol Immunother. 2019;68:489–502. doi: 10.1007/s00262-019-02301-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Etzerodt A, Tsalkitzi K, Maniecki M, et al. Specific targeting of CD163 +TAMs mobilizes inflammatory monocytes and promotes T cell–mediated tumor regression. J Exp Med. 2019;216:2394–2411. doi: 10.1186/s40364-017-0106-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Møller HJ, Hald K, Moestrup SK. Characterization of an enzyme-linked immunosorbent assay for soluble CD163. Scand J Clin Lab Invest. 2002;62:293–299. doi: 10.1080/003655102760145852. [DOI] [PubMed] [Google Scholar]
- 39.Rødgaard-Hansen S, Rafique A, Christensen PA, et al. A soluble form of the macrophage-related mannose receptor (MR/CD206) is present in human serum and elevated in critical illness. Clin Chem Lab Med. 2014;52:453–461. doi: 10.1515/cclm-2013-0451. [DOI] [PubMed] [Google Scholar]
- 40.Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 41.De Beule N, De Veirman K, Maes K, et al. Tumour-associated macrophage-mediated survival of myeloma cells through STAT3 activation. J Pathol. 2017;241:534–546. doi: 10.1002/path.4860. [DOI] [PubMed] [Google Scholar]
- 42.Bosseboeuf A, Allain-Maillet S, Mennesson N, et al. Pro-inflammatory state in monoclonal gammopathy of undetermined significance and in multiple myeloma is characterized by low sialylation of pathogen-specific and other monoclonal immunoglobulins. Front Immunol. 2017;8:1347. doi: 10.3389/fimmu.2017.01347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kvorning SL, Nielsen MC, Andersen NF, et al. Circulating extracellular vesicle-associated CD163 and CD206 in multiple myeloma. Eur J Haematol. 2020;104:409–419. doi: 10.1111/ejh.13371. [DOI] [PubMed] [Google Scholar]
- 44.Maniecki MB, Etzerodt A, Ulhøi BP, et al. Tumor-promoting macrophages induce the expression of the macrophage-specific receptor CD163 in malignant cells. Int J Cancer. 2012;131:2320–2331. doi: 10.1002/ijc.27506. [DOI] [PubMed] [Google Scholar]
- 45.Hasita H, Komohara Y, Okabe H, et al. Significance of alternatively activated macrophages in patients with intrahepatic cholangiocarcinoma. Cancer Sci. 2010;101:1913–1919. doi: 10.1111/j.1349-7006.2010.01614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakamura R, Sene A, Santeford A, et al. IL10-driven STAT3 signalling in senescent macrophages promotes pathological eye angiogenesis. Nat Commun. 2015;6:7847. doi: 10.1038/ncomms8847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang J, De Veirman K, De Beule N, et al. The bone marrow microenvironment enhances multiple myeloma progression by exosome-mediated activation of myeloid-derived suppressor cells. Oncotarget. 2015;6:43992–44004. doi: 10.18632/oncotarget.6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data from the present study are available upon reasonable request to the corresponding author.