Abstract
Loss of vaccinia virus A18R gene function results in an aberrant transcription profile termed promiscuous transcription, defined as transcription within regions of the genome which are normally transcriptionally silent late during infection. Promiscuous transcription results in an increase in the intracellular concentration of double-stranded RNA, which in turn results in activation of the cellular 2-5A pathway and subsequent RNase L-catalyzed degradation of viral and cellular RNAs. One of three hypotheses could account for promiscuous transcription: (i) reactivation of early promoters late during infection, (ii) random transcription initiation, (iii) readthrough transcription from upstream promoters. Transcriptional analysis of several viral genes, presented here, argues strongly against the first two hypotheses. We have tested the readthrough hypothesis by conducting a detailed transcriptional analysis of a region of the vaccinia virus genome which contains three early genes (M1L, M2L, and K1L) positioned directly downstream of the intermediate gene, K2L. The results show that mutation of the A18R gene results in increased readthrough transcription of the M1L gene originating from the K2L intermediate promoter. A18R mutant infection of RNase L knockout mouse fibroblast (KO3) cells does not result in 2-5A pathway activation, yet the virus mutant is defective in late viral gene expression and remains temperature sensitive. These results demonstrate that the A18R gene product is a negative transcription elongation factor for postreplicative viral genes.
Vaccinia virus, the prototypical member of the orthopoxvirus family, is unique among DNA viruses in that it replicates in the cytoplasm of the infected cell (32). This replication strategy requires that the virus encode the majority of the enzymes necessary for macromolecular synthesis, including RNA polymerase, associated transcription factors, and enzymes needed for DNA replication. Thus, vaccinia virus has served as a useful model system for understanding the basic mechanisms of RNA and DNA metabolism.
Vaccinia virus gene expression is controlled primarily at the level of transcription initiation (32). Vaccinia virus genes are expressed in a cascade which is divided into three gene classes, early, intermediate, and late. All three classes are transcribed by the same virus-encoded multisubunit RNA polymerase. Initiation of early vaccinia virus transcription requires, in addition to the RNA polymerase, the early transcription factor vETF (11) and the RNA polymerase-associated protein RAP94 (1, 18). Early gene expression is initiated from the infecting virion immediately following infection and results in synthesis of the factors required for intermediate gene expression, which to date include the vaccinia virus capping enzyme (23, 49) and at least two additional factors: VITF-1, which is a 30-kDa subunit of RNA polymerase encoded by gene E4L (38), and VITF-2, which is a cellular protein (39). Expression of the intermediate genes A1L, A2L, G8R (27), the early gene H5R (29), and one other unidentified gene, VLTF-X (53), supplies the factors for transactivating late gene expression. Intermediate and late gene expression are coupled to DNA replication (27); that is, intermediate and late gene expression is abolished in the presence of DNA replication inhibitors such as hydroxyurea (HU) and cytosine-d-arabinoside. Many of the late viral proteins are components of the early transcription apparatus which is packaged in the virion for subsequent rounds of infection.
Termination of intermediate and late gene transcription is strikingly different from termination of early gene transcription. Early transcripts are homogeneous in size due to specific transcription initiation and termination signals. Termination is signaled by transcription of a highly specific sequence (T5NT) (58) and results in a factor-dependent dissociation of the ternary elongation complex 30 to 50 nucleotides (nt) downstream of the termination signal, followed by polyadenylation of the nascent mRNA 3′ ends. The ternary complex responsible for elongation and termination of early transcripts contains the heterodimeric viral capping enzyme and a DNA-dependent ATPase, NPH-I (19). Transcription initiated from intermediate and late promoters reads through early transcription termination signals and does not terminate at discrete sites. Thus, initiation at each intermediate or late promoter results in synthesis of a family of transcripts with homogeneous 5′ ends and heterogeneous 3′ ends (31). Since intermediate and late transcripts are heterogeneous in length, and since both DNA strands of the linear genome are utilized in transcription, significant amounts of double-stranded RNA (dsRNA) are formed late during infection.
Previous genetic experiments implicate the vaccinia virus A18R gene in regulation of viral transcription at late times during infection. The A18R protein is a DNA-dependent ATPase (5) and a DNA helicase with 3′-to-5′ directionality (44). Although the A18R protein is expressed during both early and late phases of infection and packaged in virions (43), the phenotype of A18R mutant infections is expressed only late during viral infection (35). Specifically, A18R mutant infections display an aberrant late transcription profile termed promiscuous transcription, characterized by transcription of regions of the viral genome that are normally transcriptionally silent late during infection, for example, the early gene D9R (4). Promiscuous transcription results in an increase in the intracellular concentrations of dsRNA which activates the cellular 2-5A pathway and hence the cellular RNase L, resulting ultimately in a global degradation of viral and host mRNA and rRNA and an abortion of protein synthesis (4, 12).
One of three possible models could account for the promiscuous transcription phenotype: (i) reactivation of early promoters late during infection, (ii) random, promoter-independent transcription initiation throughout the genome, or (iii) readthrough transcription from upstream gene promoters. Previous genetic experiments favor the last of these hypotheses. Specifically, mutation of the A18R gene compensates for null mutation of gene G2R (17), which itself is implicated in control of transcription elongation. By itself, a G2R null mutation results in synthesis of intermediate and late viral mRNAs that are truncated at their 3′ ends, implying that the wild-type (wt) G2R gene product normally serves as a positive transcription elongation factor (8). The fact that mutation of the A18R gene compensates for loss of G2R function implies that the A18R mutation may have the effect of restoring or extending transcription of the abnormally 3′-truncated mRNAs, an activity consistent with the readthrough hypothesis.
We present here a detailed transcriptional analysis of A18R mutant infections designed to distinguish among the three above-mentioned hypotheses. The results show that early promoters are not reactivated during A18R mutant infections and that random transcription of the viral genome does not occur, thus discrediting the first two hypotheses. The results show further that A18R mutant infections result in synthesis of longer than normal intermediate mRNAs which could account for promiscuous transcription, thus supporting the readthrough hypothesis. Last, the results show that A18R mutations are lethal even in the absence of 2-5A pathway activation, implying that transcriptional readthrough compromises downstream gene expression. Taken together, the data show that the wt A18R protein is a negative transcription elongation factor which either restricts intermediate and late transcription elongation or promotes termination.
MATERIALS AND METHODS
Cells and virus.
The continuous African green monkey kidney cell line BSC40 and conditions for cell culture have been previously described (15, 16). KO3 cells are immortalized fibroblasts established from an RNase L knockout mouse (60). They were grown in Dulbecco’s modified Eagle medium supplemented with 10% fetal bovine serum and antibiotics. Wild-type vaccinia virus strain WR, the gene A18R temperature-sensitive mutant virus Cts23, and the conditions for their growth, infection, and plaque titration have been described previously (15, 16).
Plasmids. (i) G-less cassettes.
PCFW10 (54), which contains the vaccinia virus 11K (gene F17R) late promoter placed upstream of the 375-nt G-less cassette in pC2AT19 (41), was obtained from Cynthia Wright (University of South Carolina). pVGFG (14) contains the vaccinia virus growth factor (VGF, gene C11R) early promoter (10) placed upstream of the 375-nt G-less cassette in pC2AT19. PG8G (14) contains the vaccinia virus gene G8R intermediate promoter (2) placed upstream of the 375-nt G-less cassette in pC2AT19. pK2G, which contains the vaccinia virus gene K2L promoter placed upstream of the 375-nt G-less cassette in pC2AT19, was constructed as follows. Two complementary K2L promoter-containing oligonucleotides were synthesized, 5′ phosphorylated, annealed, and ligated to pC2AT19 linearized with SacI and blunt ended with T4 DNA polymerase. The inserted oligonucleotide has the sequence 5′ AGTACTAACATAAAAATAAGGTTAATTATTAATACCATAAAATCAT 3′, where the plain text represents K2L promoter, the first two nucleotides of the translation initiation ATG are underlined, and the italic text represents a ScaI site introduced for ease of identification of the desired clone. pK4G, containing the vaccinia virus gene K4L promoter, was constructed in the same fashion as pK2G except that the sequence of the inserted oligonucleotide was 5′ AGTACTGAGTGAAGTGATATAGGATTATTCTTTTAACAAATAAAAT 3′. The inserts in both pK4G and pK2G were sequenced to confirm their identity.
(ii) Riboprobe template clones.
pGEM-VGF, pGEM-30K, and pGEM-11K, clones used for synthesis of riboprobes specific for the 5′ ends of standard early, intermediate, and late genes, respectively, were kindly provided by Bernard Moss (2). pGEM-M2L, which contains the vaccinia virus M2L coding sequence (lacking the 3′-terminal 27 nt), was provided by Richard Moyer.
Riboprobes.
Riboprobes were synthesized as described by the supplier (Promega), using as a template for T7 or SP6 RNA polymerase either linearized plasmid DNA or PCR products (8) obtained by amplification of desired regions of wt vaccinia virus DNA. In the latter case, the downstream primer contained the consensus T7 RNA polymerase promoter sequence, 5′ tgTAATACGACTCACTATA 3′, where uppercase letters represent the T7 RNA polymerase promoter and lowercase letters represent extra nucleotides necessary for efficient T7 RNA polymerase binding. Riboprobes synthesized from PCR-amplified vaccinia virus genomic DNA were purified on a 6% polyacrylamide–50% urea gel (Sequagel; National Diagnostics, Atlanta, Ga.) to eliminate potential nonspecific hybridization.
Isolation of RNA.
RNA was purified from infected cells essentially as described previously (4). Briefly, confluent BSC40 cells or KO3 cells (107 cells in 100-mm-diameter dishes) were infected with wt or mutant virus at a multiplicity of infection (MOI) of 15 at 31°C (permissive temperature) or 40°C (nonpermissive temperature). At various times postinfection, cells were lysed with a guanidine thiocyanate-containing buffer and total cellular RNA was purified by centrifugation through a CsCl cushion. Alternatively, total cellular RNA was purified by using RNeasy Total RNA purification columns as described by the supplier (Quiagen, Inc., Chatsworth, Calif.). Control experiments revealed that these two RNA preparations were indistinguishable in the analyses reported here (not shown).
Northern analysis.
Purified RNA was denatured in formamide and electrophoresed through 1.2% formaldehyde agarose gels as previously described (4). The RNAs were transferred to a GeneScreen membrane (New England Nuclear) and hybridized with antisense riboprobes as described by the manufacturer.
RNase protection analysis.
RNase protection assay was done as described previously (8). Briefly, 2 μg of total RNA was hybridized to 5 ng of [α-32P]CTP-labeled riboprobe, digested with RNase A and RNase T1, and analyzed on a 6% acrylamide–8 M urea gel. When necessary, conditions of probe excess were determined by titrating the probe versus a constant amount of cellular RNA.
Drug swap assay.
Vaccinia virus genes were identified as early, intermediate, or late genes by using a drug swap assay performed essentially as described by Baldick and Moss (3). Confluent BSC40 cells (107 cells in 100-mm-diameter dishes) were infected with wt or mutant virus at an MOI of 15, and viral DNA replication was blocked by addition of HU (Sigma Biochemical, St. Louis, Mo.) at a final concentration of 10 mM, added at the end of the 30-min adsorption period. At 3 h postinfection, HU was removed and replaced with cycloheximide (CHX) at a final concentration of 100 μg/ml. After various times, total RNA was purified from the cells as described above.
In vitro transcription.
Infected cell extracts for transcription were prepared by lysolethicin permeabilization of infected cell monolayers as described previously (14). Transcription was assayed by incubation of extracts with DNA templates containing vaccinia virus promoter-driven G-less cassettes in the presence of ATP, UTP, [α-32P]CTP, and 3′-O-Me-GTP at 30°C for 30 min. Labeled RNA products were analyzed by electrophoresis on 4% polyacrylamide urea gels.
RT-PCR analysis of vaccinia virus-expressed RNA.
RNA was extracted from virus-infected BSC40 cells and purified on Quiagen RNeasy Total RNA purification columns as described above. For reverse transcription (RT)-PCR, the eluted RNA was DNase treated and repurified as described in the protocol for the Invitrogen RNA kit. Purified DNA-free RNA (1 μg) was incubated at 42°C for 1 h with 200 U of Moloney murine leukemia virus reverse transcriptase (Promega) in a 20-μl reaction containing 50 mM Tris-HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2, 10 mM dithiothreitol, 1 mM deoxynucleoside triphosphates, 25 pmol of experimental primer (antisense to the M1L gene 5′ region), 6 pmol of control primer (antisense to the K2L 3′ region), and 20 U of RNasin. Control reactions lacking either reverse transcriptase or template RNA were performed. For PCR amplification, 2 μl of the 20-μl RT mixtures was added to 23 μl of PCR mixture consisting of 1× PCR buffer (Promega), 0.2 mM deoxynucleoside triphosphates, 1.5 mM MgCl2, 6 pmol of each experimental primer (antisense to the M1L 5′ region and sense to the K2L 3′ region), 1.25 pmol of each control primer (antisense to the K2L 3′ region and sense to the K2L 5′ region), 0.25 μCi of [α-32P]dCTP, and 1.25 U of Taq polymerase (Promega). Cycling conditions (determined empirically) were an initial denaturation step of 2 min at 94°C, followed by 30 cycles of 94°C for 30 s, 51°C for 30 s, and 72°C for 3 min, followed by a final extension at 72°C for 5 min. Seven microliters of the reaction was loaded on 0.8% nondenaturing agarose gels. The gels were dried, and the RT-PCR products were detected by autoradiography and quantified by analysis on a Molecular Dynamics PhosphorImager.
One-step growth experiment.
One-step growth experiments were performed as previously described (24). Briefly, cells were infected with wt or Cts23 virus at an MOI of 6 and incubated at 31 or 40°C. At various time postinfection, virus was harvested and the yield from each time point was quantified by plaque titration at 31°C.
Protein pulse-labeling analysis.
Pulse-labeling of proteins in virus-infected cells was done as described previously (15). Briefly, cells were infected with wt or Cts23 at an MOI of 15 or mock infected. At various times postinfection, cells were metabolically labeled with [35S]methionine for 15 min. Cells were lysed on the dishes by addition of sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) sample buffer, and solubilized proteins were analyzed by SDS-PAGE. The gels were Coomassie blue stained, dried, and autoradiographed.
RESULTS
Promiscuous transcription is context sensitive.
The phenotype of infection with A18R mutants is characterized by promiscuous transcription (originally called aberrant transcription) (4). Promiscuous transcription is defined as transcription within regions of the genome, for example, early genes D9R and G2R, which are normally transcriptionally silent late during wt infection. One of three hypotheses could account for promiscuous transcription: reinitiation at early promoters, promoter-independent random transcription throughout the genome, or readthrough transcription from upstream intermediate or late promoters. To distinguish among these hypotheses, we extended our analysis of transcription in A18R mutant-infected cells by analyzing transcription of representatives of each of the three viral gene classes: early (gene C11R), intermediate (gene G8R), and late (gene F17R).
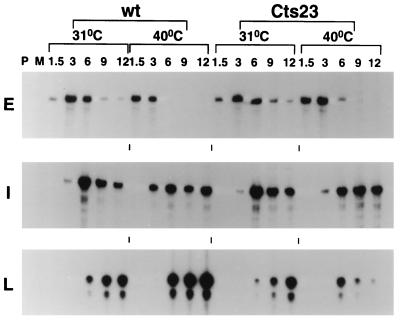
Promoter utilization in A18R mutant-infected BSC40 cells was assessed by RNase protection (Fig. 1). The riboprobes used contain antisense RNA sequence from both upstream and downstream of the previously determined mRNA 5′ ends. The sizes of the protected fragments shown in Fig. 1 correspond precisely to the mRNA 5′ ends previously determined for each gene tested (2). In the A18R mutant (Cts23) infection, protected fragments for early and intermediate promoters appear with similar kinetics and in similar amounts compared to the wt infection. The early C11R promoter turns on at 1.5-h postinfection, peaks at 3 h, and decreases at late times. The intermediate G8R promoter turns on at 3 h, peaks at 6 h, and stays on throughout the time course. Compared to wt infection, in the A18R mutant infection the late F17R promoter turns on at a similar time at 31 and 40°C (6 h postinfection), but the signal is reduced in intensity and decays prematurely at 40°C. Most importantly, the early promoter is not utilized at late times in the A18R mutant infection at 40°C, thereby contradicting the hypothesis that promiscuous transcription represents reactivation of early promoters late during infection. Significantly, we have also used RNase protection to measure the promoter activity of a promiscuously transcribed early gene, M2L, and found that this promoter is also not utilized at late times in an A18R mutant infection at 40°C (data not shown).
FIG. 1.
Promoter utilization in wt- and Cts23-infected BSC40 cells. BSC40 cells were infected with wt or Cts23 virus at an MOI of 15 and incubated at 31 or 40°C. Total RNA was extracted from infected cells at various times postinfection, indicated in hours above the lanes. RNA was hybridized to uniformly labeled antisense riboprobes specific for the 5′ end of an early (E), intermediate (I), or late (L) gene. After RNase digestion, the protected fragments were analyzed by denaturing PAGE and autoradiography. P, unhybridized probe digested with RNase; M, mock-infected cell RNA.
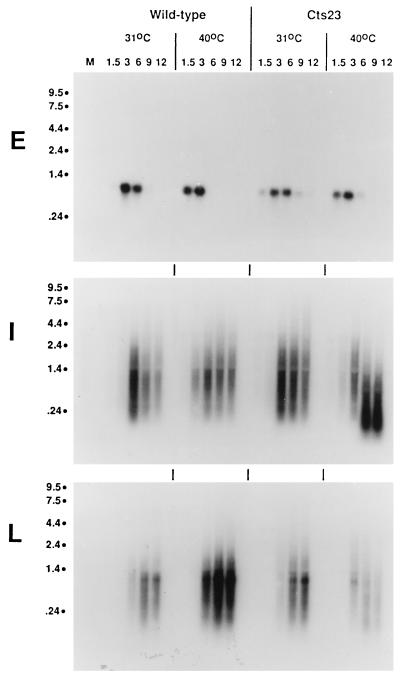
To determine whether random initiation occurs throughout the virus genome during A18R mutant infections, we performed Northern blot analysis using the same three standard early, intermediate, and late gene riboprobes (Fig. 2). The kinetics of mRNA synthesis observed are similar to those detected by RNase protection. Importantly, the early C11R mRNA signal disappears at late times, confirming that the early promoter does not reactivate late during infection. In Cts23-infected cells at 40°C at late times, some G8R transcripts are shorter than normal, indicating the expected RNase L-catalyzed breakdown of RNAs. Consistent with the RNase protection analysis described above, the late F17R gene seems to be poorly transcribed in A18R mutant infections at 40°C. Most importantly, promiscuous transcription is not observed with either the early or the late gene probe at late times postinfection, thereby contradicting the hypothesis that promiscuous transcription represents completely random initiation within untranscribed regions of the genome.
FIG. 2.
Northern blot analysis of RNA synthesized in wt- and Cts23-infected BSC40 cells. The total RNA was purified from infected BSC40 cells as described in the legend to Fig. 1. RNA was fractionated on formaldehyde-agarose gels, transferred to GeneScreen membranes, and probed with uniformly labeled antisense RNA riboprobes specific for an early (E), intermediate (I), or late (L) gene. Lanes M contain uninfected cell RNA. Sizes are denoted at the left in kilobases.
A compilation of all results obtained to date concerning transcription in A18R mutant-infected cells reveals that promiscuous transcription is context sensitive. Specifically, promiscuous transcription has been observed within the D9R and G2R genes (4) but not within the C11R and F17R genes (Fig. 2). Interestingly, inspection of the vaccinia virus genetic map (21) reveals that the C11R and F17R genes are unusual in that there are no other known promoters in the same transcriptional orientation within 14 kb upstream from either gene. By contrast, D9R and G2R each lie 3 to 5 kb downstream from intermediate or late promoters driving transcription of upstream genes. These observations provide further support for the hypothesis (referred to below as the readthrough hypothesis) that promiscuous transcription results from transcriptional readthrough from upstream intermediate or late gene promoters.
Promiscuous transcription in the region from K2L to M1L.
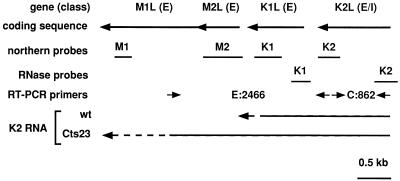
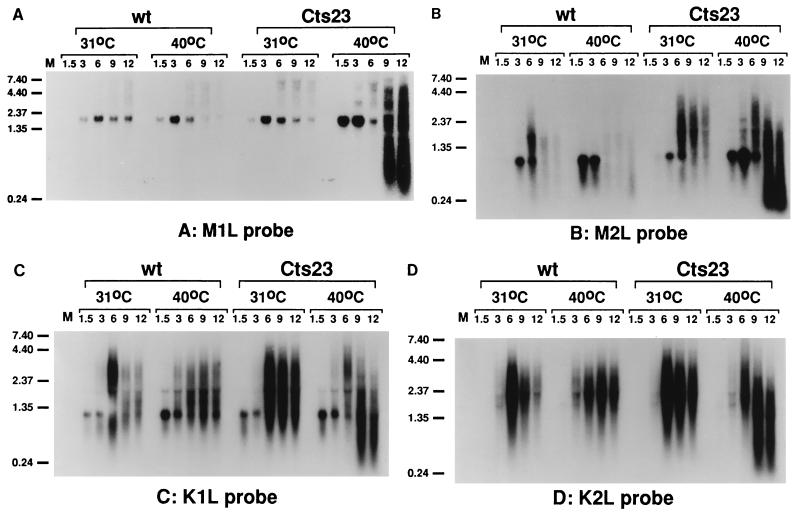
To test the readthrough hypothesis, we focused attention on a region spanning the vaccinia virus genes M1L through K2L (Fig. 3). Previous Northern blot analysis from other laboratories using exclusively early viral RNA shows that M1L, M2L, K1L, and K2L are each expressed early during infection. Furthermore, Northern blot analysis by Smith et al. (45) revealed transcription within the K2L region at late times. Whether any of the four genes contains intermediate or late promoters is unknown. Previous data from our lab showed that the M2L gene was promiscuously transcribed (6). We hypothesize that promiscuous transcription in the M2L region results from readthrough transcription from an upstream intermediate or late promoter. As an initial test of this hypothesis, we conducted Northern blot analysis within the region (Fig. 4), using antisense riboprobes specific for M1L, M2L, K1L, and K2L (Fig. 3). Northern blot analysis of M1L (Fig. 4A) and M2L (Fig. 4B) shows that early M1L transcripts (1.6 kb) and early M2L transcripts (800 bp) appear at 1.5 h postinfection and decease at late times in both wt- and Cts23-infected BSC40 cells. The sizes of the early transcripts are consistent with previous results from other laboratories (46, 47). At late times, the M1L and M2L genes are transcriptionally silent in wt infections at either 31 or 40°C. At late times in Cts23 infections at 31°C, the M1L gene is transcriptionally silent, while heterogeneous transcripts are observed within the M2L gene. Abundant transcription is observed in Cts23-infected cells at 40°C after 6 h postinfection in both the M1L and M2L genes. Some of these transcripts have a shorter than normal chain length resulting from RNA degradation catalyzed by RNase L. These results show that both the M1L and M2L genes are promiscuously transcribed in the absence of A18R activity. The low level of transcription observed at late times in Cts23-infected cells within the M2L gene may indicate that the A18R gene product is not fully active at permissive temperatures. Nevertheless, promiscuous transcription observed at 31°C in Cts23-infected cells is not sufficiently robust to either affect the M1L gene or activate RNase L. Northern blot analysis of the K1L gene (Fig. 4C) reveals a 1.1-kb early transcript, consistent with published experiments (46). Transcription is detected at late times in both wt and Cts23 infections at both temperatures. Some of the late transcripts in Cts23-infected cells at 40°C are shorter than normal, resulting from RNase L-catalyzed RNA degradation. Northern blot analysis of the K2L gene (Fig. 4D) reveals two barely detectable early transcripts of 1.5 and 2.1 kb, consistent with published experiments (45). At late times, the K2L gene is transcribed in both wt and Cts23 infections. Northern blot analysis of the K3L gene (data not shown) is very similar to the K1L analysis, indicating that the K3L gene is transcribed at both early and late times. Northern blot analysis of the K4L gene (data not shown) reveals transcription only at late times in both wt and Cts23 infections. Since intermediate and late vaccinia virus RNAs are normally heterogeneous in size and may read through into downstream genes, we cannot determine the origin of any of the late transcripts detected in this region from Northern blot analysis alone. Nevertheless, if the readthrough hypothesis is correct, we predict that promiscuous transcription of the M1L and M2L genes results from readthrough transcription originating from K1L, K2L, K3L, or K4L. Detailed analysis of transcription initiation is required to further investigate this hypothesis.
FIG. 3.
Diagram describing the M1L through K2L region of the vaccinia virus genome. From the top down, the drawing shows the identity and class (Fig. 4 to 6) of each gene, the coding sequence with arrows representing transcriptional orientation, the positions of Northern and RNase protection probes, the positions of primers used for RT-PCR (E:2466, experimental primer set, 2,466-bp product; C:862, control primer set, 862-bp product), and an interpretation of the transcriptional analysis in Fig. 4 to 8, showing extended readthrough transcription from the K2L promoter into the M1L gene in a Cts23 infection. E, early; I, intermediate.
FIG. 4.
Promiscuous transcription in the region from M1L through K2L in Cts23-infected BSC40 cells. The total RNA was purified from infected BSC40 cells as described in the legend to Fig. 1. Equal amounts of RNA were fractionated on formaldehyde-agarose gels, transferred to GeneScreen membranes, and probed with uniformly labeled antisense RNA riboprobes specific for the M1L (A), M2L (B), K1L (C), and K2L (D) genes. Lanes M contain uninfected cell RNA. Sizes are denoted at the left in kilobases.
Characterization of gene class in the region from M1L to K2L.
RNase protection analysis was used in an attempt to characterize the promoter type for each of the genes in the M1L–K2L region. Published S1 nuclease analysis shows that M1L contains an early promoter, consistent with Northern analysis (47). The published S1 nuclease analysis of the M1L gene also indicates the presence of a weak late promoter. This late promoter must be very weak, since late transcription occurs at only a very low level within the M1L gene, as revealed by Northern analysis. We did not attempt to analyze transcription of the M1L gene further. Antisense riboprobes for RNase protection analysis of the M2L, K1L, and K2L genes were designed to measure simultaneously both gene-specific transcription and upstream readthrough transcription. The viral sequence contained in each probe spans the 5′ end of each gene, so that protection of only a fraction of the viral sequences in the probe reveals a gene-specific 5′ end. Each probe also contained nonviral sequence, 6 nt at the 5′ end and 11 nt at the 3′ end, which permits differentiation between undigested full-length probe and digested probe in which all of the viral sequences are protected by upstream readthrough transcripts. Riboprobes specific for M2L, K1L, and K2L were used in RNase protection analysis of RNA extracted at different times after infection of BSC40 cells at 31 or 40°C with either wt virus or Cts23 (data not shown). Consistent with Northern blot analysis, the RNase protection analysis showed that each of these three genes contains an early promoter. The data showed further that neither M2L nor K1L contains a postreplicative promoter and therefore that any intermediate or late transcription through these genes represents readthrough from upstream genes. Last, the results showed that K2L contains a postreplicative promoter and is also transcribed by readthrough from an upstream postreplicative promoter. Detailed transcriptional analysis of the K1L and K2L 5′ regions is presented below.
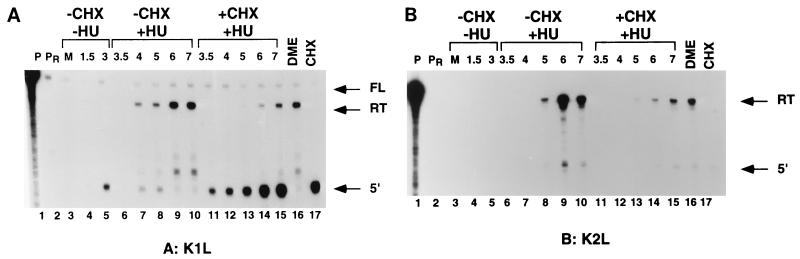
To further confirm the gene class of K1L, we performed RNase protection analysis on RNA purified from cells treated with a drug swap protocol designed to distinguish intermediate from late transcription. Vaccinia virus early, intermediate, and late RNAs can be distinguished by analysis of RNA from cells infected in the presence of the DNA replication inhibitor HU, followed by a shift to drug-free medium or medium supplemented with the protein synthesis inhibitor CHX (3). Since intermediate and late gene transcription is coupled to DNA replication, only early genes are expressed in the presence of HU. Early gene expression includes synthesis of intermediate transcription factors; thus, when the HU block is removed, intermediate and ultimately late transcription proceeds and early gene expression is shut off. If CHX is added at the time HU is removed, early transcription continues and additional transcription is limited to intermediate genes since synthesis of late transcription factors encoded by intermediate genes is inhibited. RNase protection analysis of the K1L and K2L 5′ regions by using RNA from the drug swap protocol is shown in Fig. 5. With the K1L-specific riboprobe (Fig. 5A), very weak 5′-protected fragments are detected in infected cells in drug-free medium at early times (lanes 4 and 5) and disappear at late times (lane 16). This 5′-protected fragment is also detected in cells infected in the presence of CHX only (lane 17). Lanes 6 to 10 show that the 5′-protected fragment is observed in the presence of HU but decreases when HU is removed and postreplicative gene expression is allowed to proceed in the absence of drug. Lanes 11 to 15 show that the 5′-protected fragment accumulates if, after removal of the HU block, late gene expression is inhibited by the addition of CHX. These results show that the 5′-protected fragment has the characteristics of an early transcript. While the drug swap experiments do not formally rule out the possibility that the K1L 5′-protected fragment is also expressed as an intermediate transcript, precise mapping of this 5′ end relative to a sequence ladder (not shown) shows that it originates approximately 20 nt upstream of the K1L translation start site, within a sequence that does not contain the requisite TAAA intermediate transcription initiation signal. We conclude that the K1L 5′-protected fragment represents an exclusively early transcript and therefore that the K1L gene contains an early promoter. RNase protection with the K1L riboprobe also reveals readthrough transcription. The readthrough transcripts are detected neither at early times (lanes 4 and 5) nor in the presence of CHX (lane 17). The readthrough transcripts are present in drug-free medium late during infection (lane 16); they appear after the HU block is removed without added CHX (lanes 7 to 10) and also following the drug swap (lanes 11 to 15). These results show that the readthrough transcript has the characteristics of an intermediate RNA. It is noteworthy that the signal representing readthrough transcription is more intense in the absence than in the presence of CHX following removal of HU (compare lanes 10 and 15), suggesting that these transcripts may have late as well as intermediate character.
FIG. 5.
Drug swap RNase protection analysis. BSC40 cells were infected with wt virus in the absence (lanes 3 to 5) or presence (lanes 6 to 15) of HU; 3 h postinfection, medium was removed and replaced with drug-free medium (lanes 6 to 10) or medium containing CHX (lanes 11 to 15). RNA was purified at various times postinfection, indicated above the lanes in hours. RNA was hybridized to uniformly labeled antisense riboprobes specific for the 5′ end of K1L (A) or K2L (B). After RNase digestion, the protected fragments were analyzed by denaturing PAGE and autoradiography. Lanes DME (lane 16) and CHX (lane 17) contain RNA extracted at 7 h postinfection from cells infected in drug-free medium and in the presence of CHX, respectively. P (lane 1), undigested probe; PR (lane 2), unhybridized probe digested with RNase; M, mock infection; FL, full-length probe; RT, readthrough transcript protected fragment; 5′, mRNA 5′-end-protected fragment.
RNase protection with the K2L riboprobe in the drug swap protocol (Fig. 5B) also reveals both a 5′-protected fragment and a readthrough transcript. Consistent with Northern blot analysis of the K2L gene, the 5′-protected fragment has characteristics consistent with a weak early transcript, notably a weak signal in the presence of CHX (lane 17). The K2L 5′-protected fragment also has characteristics of an intermediate transcript in that it is present at late times in the absence of inhibitors (lanes 8 to 10 and 16) and also following the drug swap (lanes 13 to 15). In this case, mapping of the K2L 5′ end (not shown) places it near the A4 stretch in the sequence TAAAATCATG proximal to the K2L translation start site (underlined). This sequence contains the requisite TAAA intermediate promoter initiation consensus, and thus the K2L gene may contain a compound early-intermediate promoter. The K2L readthrough transcript is absent early and in CHX-treated cells (lanes 4, 5, and 17) but is present late and during the drug swap (lanes 8 to 10 and 16) and thus has characteristics of an intermediate transcript. Sequence analysis and preliminary transcription analysis (not shown) indicate that readthrough into the K2L gene may arise from the K4L promoter. Once again it is noteworthy that the signals representing both the K2L 5′ end and the readthrough transcript are more intense in the absence than in the presence of CHX following removal of HU (compare lanes 10 and 15), suggesting that these transcripts may have late as well as intermediate character.
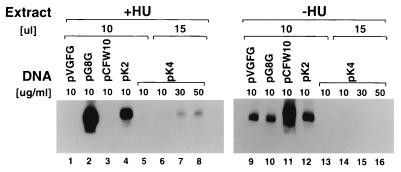
To confirm that K2L and K4L contain intermediate promoters, we analyzed these promoters in an in vitro transcription assay. In this assay, extracts from vaccinia virus-infected, HU-treated cells are used to transcribe a template containing an intermediate promoter fused to a G-less cassette. HU treatment prevents intermediate and late gene transcription and therefore prevents synthesis of late and early viral transcription factors (14). Thus, HU-treated extracts contain only early gene products, including intermediate gene transcription factors, and they are capable of initiating transcription at intermediate but not early or late vaccinia virus promoters. We cloned each of the candidate K2L and K4L promoter sequences upstream of the G-less cassette in pC2AT19. Results of in vitro transcription directed by these templates are shown in Fig. 6. Control experiments with standard early (pVGFG DNA, C11R promoter), intermediate (pG8G DNA, G8R promoter), and late (pCFW10 DNA, F17R promoter) templates (lanes 1 to 3 and 9 to 11) shows that early, intermediate, and late transcripts are detected with the extracts made in the absence of HU, (−HU extracts), while only intermediate transcripts are detected with extracts made in the presence of HU (+HU extracts). In addition, the intermediate template is transcribed more efficiently in the +HU extracts. In vitro transcription with the pK2 template shows that, like the G8R control, the K2L promoter is transcribed more efficiently with +HU extracts than with −HU extracts, confirming that the K2L promoter has intermediate character. In vitro transcription with the pK4 template shows that it is transcribed very weakly in a high concentration of +HU extract, suggesting that K4L contains a weak intermediate promoter. In summary, both the K2L and the K4L promoters have intermediate characteristics in the in vitro transcription analysis.
FIG. 6.
In vitro transcription of templates containing K2L and K4L promoter sequences. Transcription-competent extracts were made from cells infected with wt virus in the presence of 10 mM HU (+HU) or in the absence of drug (−HU). Transcription was done with 10 μl (lanes 1 to 5 and 9 to 13) or 15 μl (lanes 6 to 8 and 14 to 16) of extract. Reactions contained 10, 30, or 50 μg of pVGFG (early), pG8G (intermediate), pCFW10 (late), pK2, or pK4 DNA per ml, as indicated. Reaction products were separated on a 7% polyacrylamide gel, which was dried and autoradiographed.
The results from detailed transcription analysis of the M1L through K2L region are summarized in Fig. 3. We conclude that M1L, M2L, and K1L are exclusively early genes and that K2L contains a compound early-intermediate promoter. In addition, the K4L promoter also has intermediate character. Furthermore, intermediate transcription reads through from upstream genes into both the K1L and K2L genes. Most of the readthrough into K1L probably derives from the K2L intermediate promoter, while most of the readthrough into the K2L gene probably arises from the K4L promoter. We hypothesize that during an A18R mutant infection, transcription initiated from the K2L intermediate promoter reads through into the downstream early M1L and M2L genes, resulting in the promiscuous transcription phenotype.
RT-PCR analysis of the readthrough transcription from K2L into M1L.
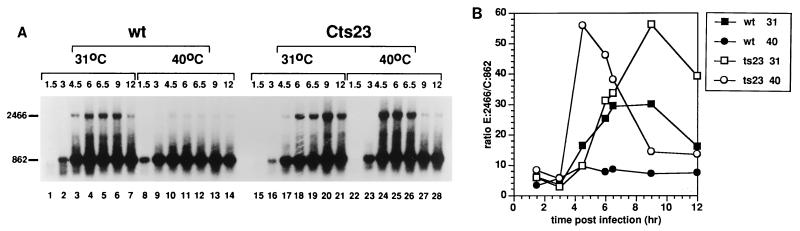
We used RT-PCR as a direct measure for transcription which extends from the K2L gene into M1L or M2L. Analysis of the size of nascent transcripts in A18R mutant infections is complicated by the fact that promiscuous transcription induces RNase L-catalyzed RNA breakdown. However, intermediate gene transcription begins at 3 h postinfection (Fig. 1 and 2) whereas RNA degradation is not evident until 7.5 h postinfection (reference 4 and data not shown). We therefore attempted to measure readthrough transcription by RT-PCR during the interval between the initiation of intermediate transcription and induction of RNase L (Fig. 7). RNA was extracted from BSC40 cells infected with wt or Cts23 at various times postinfection and analyzed by RT-PCR using two sets of primers simultaneously (Fig. 7A). The experimental primer set amplified a 2466-nt readthrough transcript (E:2466) extending from K2L to M1L, whereas the control primer set amplified an 862-nt transcript from within the K2L gene. Both transcripts were quantified by phosphorimage analysis, and the ratio of readthrough transcripts to control transcripts (E:2466/C:862) was determined. As expected, the 862-nt control transcript appears early after infection, persists throughout the experiment, and is present in similar amounts regardless of the virus or temperature used. Surprisingly, the long 2,466-nt transcript is observed in wt-infected cells at 31°C (lanes 1 to 7), even though the 2-5A pathway is not induced at this temperature. At 40°C, these readthrough transcripts are reduced in abundance in the wt infection (lanes 8 to 14). These results indicate that the incubation temperature affects the steady-state level of readthrough transcription and also activation of the 2-5A pathway in wt-infected cells. The amount of readthrough transcription observed in Cts23-infected cells at 31°C (lanes 15 to 21) is slightly increased relative to the wt infection at 31°C (lanes 1 to 7), consistent with the previous suggestion that the Cts23 mutant is slightly defective even under permissive conditions (Fig. 4B). Most importantly, at 40°C after induction of intermediate transcription but before induction of the 2-5A pathway, the transcripts which extend from K2L into M1L are much more abundant in the Cts23 infection (lanes 23 to 26) compared to the wt infection (lanes 8 to 12). At later times, the Cts23 readthrough transcripts disappear (lanes 27 and 28) due to activation of 2-5A pathway. (The 862-nt control transcript is presumably small enough to be a poor target for RNase L in vivo.) In summary, at 40°C, more readthrough transcription is detected in the Cts23 infection preceding RNA breakdown, which supports the hypothesis that the A18R mutation causes extended readthrough transcription from intermediate promoters.
FIG. 7.
Readthrough transcription of the M1L gene in Cts23-infected BSC40 cells detected by RT-PCR analysis. (A) Total RNA was purified from infected BSC40 cells as described in the legend to Fig. 1. RNA was DNase treated and analyzed by RT-PCR using primers extending from M1L into K2L, generating a 2,466-nt-long product (E:2466 in Fig. 3). Internal control primers measured a K2L RNA of 862 nt (C:862 in Fig. 3). (B) RT-PCR signals from panel A were quantified by phosphorimage analysis, and the ratio of E:2466 signal to C:862 signal was plotted as a function of time postinfection.
Readthrough transcription in the RNase L knockout cell line KO3.
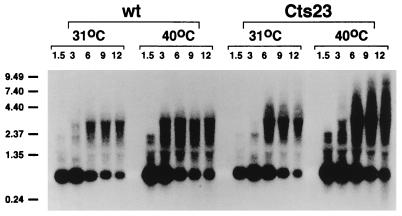
To prove conclusively that the A18R mutation causes readthrough transcription, we investigated the A18R mutant phenotype in mouse KO3 cells, which are derived from an RNase L knockout mouse and which therefore lack RNase L activity (60). Preliminary experiments demonstrated that infection of KO3 cells with Cts23 at 40°C does not cause rRNA breakdown (data not shown). Interestingly, Cts23 infection of normal control cells from the parental mouse at 40°C also revealed no rRNA breakdown, and therefore differences observed during vaccinia virus infection of KO3 cells compared to BSC40 cells cannot be attributed solely to a lack of RNase L. Nevertheless, KO3 cells provide a method for analyzing the A18R mutant phenotype in the absence of RNA degradation. In KO3 cells, we expect to observe abnormally long transcripts in A18R mutant infections in regions where promiscuous transcription occurs. Northern blot analysis using an M2L riboprobe was performed with RNAs extracted at various times from virus-infected KO3 (Fig. 8). The results show that the discrete 800-nt M2L early RNA is expressed similarly in Cts23- and wt-infected KO3 cells at both permissive and nonpermissive temperatures. In addition, larger heterogeneous readthrough transcripts appear at late times in both Cts23- and wt-infected KO3 cells at both temperatures. Most importantly, the readthrough transcripts observed in Cts23-infected KO3 cells at 40°C include a large population of transcripts longer than 4.4 kb which are not observed under any other condition of infection. This result is consistent with readthrough transcription from upstream genes into the M2L region in A18R mutant-infected KO3 cells. Northern blot analysis using M1L, K2L, D9R, and G2R riboprobes show the similar results (data not shown and reference 40).
FIG. 8.
Readthrough transcription of the M2L gene in Cts23-infected KO3 (RNase L−) cells. Total RNA was purified from infected KO3 cells as described in the legend to Fig. 1. RNA was analyzed by Northern blot analysis using a uniformly labeled antisense riboprobe specific for the M2L gene (Fig. 3). Sizes are denoted at the left in kilobases.
A18R mutant phenotype in KO3 cells.
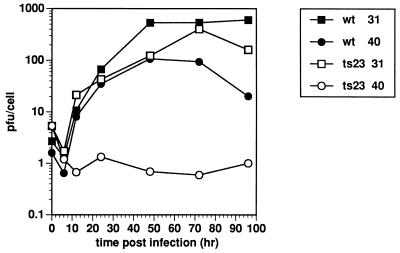
KO3 cells provide an opportunity to study whether the A18R gene is essential under circumstances where the 2-5A pathway is not activated. Cts23 is temperature sensitive on KO3 cells in a plaque assay (data not shown). A one-step growth experiment was done to quantify the growth of wt and Cts23 on KO3 cells (Fig. 9). The wt virus grown at 31 or 40°C shows a burst size of between 80 and 800 PFU per cell, with maximum yield occurring after 48 h postinfection. Growth of Cts23 at 31°C is identical to wt growth. Cts23 does not grow on KO3 cells at 40°C. This result shows that the A18R gene is essential even in the absence of 2-5A pathway induction.
FIG. 9.
One-step growth of wt and Cts23 in KO3 cells. KO3 cells were infected at an MOI of 6 with either wt or Cts23 and incubated at 31 or 40°C. Samples were taken at various times postinfection, and virus yields were determined by plaque titration at 31°C.
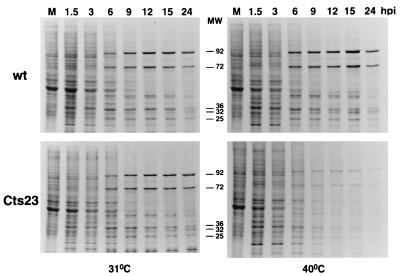
Protein pulse-labeling studies were done to examine the pattern of gene expression in wt- and Cts23-infected KO3 cells (Fig. 10). KO3 cells were infected with wt or Cts23; at various times postinfection, cells were pulse-labeled with [35S]methionine, and the labeled proteins were analyzed by SDS-PAGE and autoradiography. The profiles of protein synthesis observed in wt-infected cells at 31 and 40°C and in Cts23-infected cells at 31°C are identical and representative of the normal pattern of vaccinia virus gene expression observed on BSC40 cells. Specifically, early viral proteins are expressed concomitant with shutoff of host cell protein synthesis, followed by expression of intermediate and late viral proteins, which persists throughout the experiment. In a Cts23 infection at the nonpermissive temperature, host shutoff and early viral protein synthesis appear normal, and late protein synthesis is initiated at a normal time, but synthesis of late proteins is reduced in amount throughout the duration of the experiment. Thus, the A18R mutant virus is defective in late protein synthesis on KO3 cells.
FIG. 10.
Protein synthesis in wt- and Cts23-infected KO3 cells. KO3 cells were infected with wt or Cts23 at an MOI of 15, incubated at 31 or 40°C, and pulse-labeled for 15 min with [35S]methionine at the times postinfection indicated above the lanes in hours. Lane M, mock infection. Labeled proteins were electrophoresed on SDS–10% polyacrylamide gels and autoradiographed.
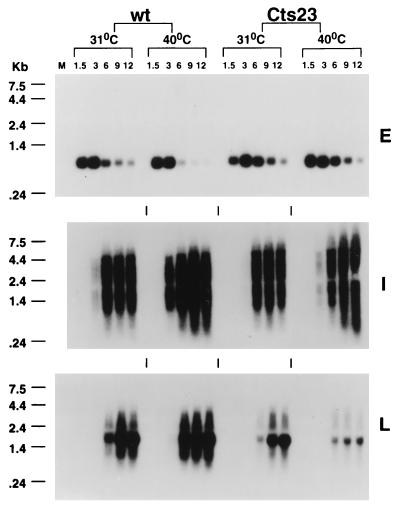
The defective late protein synthesis phenotype seen in Cts23-infected KO3 cells could be due to a deficiency in viral mRNA metabolism. To test this hypothesis, we used Northern blot analysis to determine kinetics of mRNA synthesis, as well as the size and quantity of the steady-state mRNAs synthesized. Total cellular RNA extracted from KO3 cells infected with wt or Cts23 was hybridized with antisense riboprobes specific for the early (C11R), intermediate (G8R), or late (F17R) gene (Fig. 11). Early C11R mRNAs in wt- and Cts23-infected cells are expressed at the same times postinfection, are expressed in the same quantities, and appear as discrete bands, regardless of the incubation temperature. Intermediate G8R mRNAs in wt- and Cts23-infected KO3 cells are expressed at the same time postinfection, are expressed in similar quantities, and appear as smears diagnostic of the expected 3′-end heterogeneity. In the Cts23 infection done at 40°C, there is an increase in intermediate transcripts larger than 4.4 kb, consistent with Northern analysis of the M2L gene (Fig. 8). At 31°C, late F17R mRNA synthesis in Cts23-infected KO3 cells is similar to that in the wt infection at 31°C. Under these conditions, the F17R mRNA appears as a characteristic smear, superimposed on a relatively discrete transcript peculiar to this late gene (Fig. 2). In Cts23-infected cells at 40°C, late F17R mRNA synthesis is initiated at the appropriate time postinfection, but the quantities of late mRNA are significantly decreased relative to all other conditions of infection. In summary, synthesis of steady-state late mRNA in A18R mutant-infected KO3 cells is reduced in quantity, consistent with the defective late protein synthesis phenotype described above.
FIG. 11.
Northern blot analysis of RNA synthesized in wt- and Cts23-infected KO3 cells. The total RNA was purified from infected KO3 cells as described in the legend to Fig. 1. RNA was analyzed by Northern blot analysis using uniformly labeled antisense RNA riboprobes specific for an early (E), intermediate (I), or late (L) gene. Lanes M contain uninfected cell RNA.
DISCUSSION
The experiments described here were done to refine our understanding of the effects of the vaccinia virus A18R gene on postreplicative viral transcription. Previous research had shown that mutations in the A18R gene cause promiscuous transcription, that is, transcription from regions of the genome which are normally transcriptionally silent late during infection. Our results discredit two possible explanations for promiscuous transcription. Specifically, the data show that (i) the early VGF and M2L promoters do not reactivate at late times postinfection and (ii) random transcription throughout the genome does not occur. Our detailed analysis of transcription within the M1L through K2L region of the viral genome provides positive support for the only remaining explanation for promiscuous transcription. Specifically, both RT-PCR analysis conducted in virus-infected BSC40 cells and Northern analysis conducted in RNase L knockout KO3 cells show that in A18R mutant infections transcription initiated from the K2L intermediate promoter yields longer than normal transcripts which read through into the downstream early M1L gene. In summary, these results show that late during a wt virus infection, the A18R gene product limits elongation by the viral RNA polymerase and thus has the properties of a negative transcription elongation factor.
In the course of our characterization of the A18R mutant, we have carried out a detailed transcription analysis of gene class in the region spanning the M1L and K2L genes which both confirms and extends previous analysis of individual genes within this region. Our results confirm that M1L, M2L, K1L, and K2L are expressed early during infection. While published Northern analysis of M1L transcription, like our own, shows little or no late transcription through this gene, the published S1 nuclease mapping indicates the presence of a very weak late M1L promoter a short distance upstream from the early M1L promoter (47). Our experiments do not address the existence of this late M1L promoter. Also consistent with our results, published Northern analysis of the K2L gene revealed late transcriptional activity (45) which we can now state represents both initiation from a complex early-intermediate promoter and readthrough from the upstream K4L promoter. Ours is the first transcriptional analysis of the K4L gene, and the results suggest that the K4L gene contains an intermediate promoter. Perhaps the most important outcome of this transcription analysis is the discovery of two new intermediate genes, K2L and K4L.
The discovery of two new intermediate genes, K2L and K4L, provides additional insight into intermediate promoter structure. Published analysis of the five known intermediate genes (2, 25, 50) shows that intermediate promoters are approximately 30 nt in length and contain two critical regions, an upstream 14-bp core element that is A/T rich, separated by 10 or 11 bp from a 4-bp initiator element which contains the sequence TAAA (Fig. 12). We have shown here that the 38-nt sequences upstream from both the K2L and K4L translation initiation codons have intermediate promoter activity in vitro and that in vivo, an mRNA 5′ end with characteristics of an intermediate RNA maps within 10 nt upstream of the K2L translation initiation codon. Importantly, we have not mapped with absolute precision the 5′ end of either the K2L or K4L in vivo mRNA. Nevertheless, inspection of the K2L and K4L upstream regions reveals sequence which matches precisely the initiator TAAA and which closely approximates the A/T-rich core. Closer inspection of these sequences, allowing for inclusion of 10 or 11 bp in the spacer region, reveals deviations from previously characterized critical residues in the G8R intermediate promoter which could both increase and decrease promoter activity (2). For example, both promoters contain a potentially inhibitory deviation from a consensus AAA in the region from −17 to −19, while both contain potentially stimulatory deviations from the G8R sequence in the −20 and −23 regions.
FIG. 12.
Intermediate gene upstream sequences. The upstream sequences from five intermediate genes are aligned relative to the critical TAAA initiator region. The G8R, A1L, A2L, and critical (Crit) sequences are from Baldick et al. (2). The critical sequence represents nucleotides in the G8R promoter in which two of three possible substitutions decrease activity by greater than or equal to 75%. Putative core and initiator regions in the K2L and K4L sequence are underlined. Translation initiation ATGs are italicized.
The discovery that K2L and K4L are intermediate genes provides some additional insight into the functional organization of the intermediate gene class. Zhang et al. (59) have provided evidence that the intermediate gene class contains a minimum of 13 genes. The five previously characterized intermediate genes, three late transcription factors (27), a virion RNA helicase (42), and a single-stranded DNA binding protein (37), all map within the central conserved region of the virus genome and are all implicated in nucleic acid metabolism. By contrast, while the function of K4L is unknown, K2L encodes SPI3, a serine protease inhibitor homolog which plays a role in virus-induced cell fusion (48), and both K2L and K4L map in the variable left terminus of the genome. Importantly, any vaccinia gene which has been previously classified as a late gene and not specifically tested for intermediate gene activity is potentially an intermediate gene. In summary, our observations support the idea that the intermediate gene class may in fact be relatively large and include genes with a wide variety of functions.
Our phenotypic analysis of the A18R mutant infection of KO3 cells provides fresh insight into the primary consequences of readthrough transcription on the viral infection. Interpretation of prior phenotypic analysis of A18R mutant infections, done exclusively on BSC40 cells, was complicated by the fact that readthrough transcription from converging promoters causes an accumulation of dsRNA, which triggers the 2-5A pathway, which in turn activates RNase L and causes a global degradation of mRNA and rRNA and a cessation of protein synthesis (4, 12, 35). Thus, it was unclear whether in the absence of RNase L activity, the A18R gene would be essential and whether readthrough transcription would have deleterious effects on the infection. Significantly, we have found that in RNase L knockout KO3 cells, the A18R mutant virus is temperature sensitive with respect to virus growth and that steady-state viral late mRNAs and late viral proteins are present in reduced amounts. Several possible explanations exist for the observed defect in late gene expression on KO3 cells. First, it is formally possible that the A18R gene product is directly involved in the initiation of late viral transcription. We feel that this possibility is unlikely since in vitro experiments from our lab (data not shown) and from other labs (29, 53) have failed to demonstrate any role of A18R in initiation of late viral transcription. Second, since readthrough transcription from converging intermediate promoters should still cause accumulation of dsRNA in KO3 cells, the dsRNA-dependent protein kinase pathway (30) may be activated at intermediate times, thus inhibiting synthesis of late viral transcription factors, which in turn could cause defective synthesis of late mRNAs. Third, it is possible that formation of dsRNA results in interference with translation of late transcription factors from intermediate mRNAs, also affecting late mRNA synthesis. Fourth, readthrough transcription could result in direct interference with initiation of transcription from downstream genes, a phenomenon previously documented in studies of transcription in mammalian cells (20). Unfortunately, the decrease in late mRNA synthesis in A18R mutant-infected cells has so far made it difficult to determine whether the A18R mutation affects readthrough transcription from late as well as intermediate promoters. In any case, the phenotypic analysis of A18R mutant infections on KO3 cells emphasizes the importance of restricting readthrough transcription from intermediate promoters during a normal vaccinia virus infection.
In both eucaryotic and procaryotic systems, a variety of negative transcription elongation factors have been identified. Virtually all of these factors have termination factor activity, defined experimentally as the release of nascent transcripts from a ternary elongation complex, and many are helicases and/or nucleic acid-dependent ATPases. The Escherichia coli factor Rho is the most extensively studied termination factor (26, 51, 52). Rho is an RNA-dependent ATPase and an RNA-DNA helicase which is thought to bind nascent RNA and to translocate in the 5′-to-3′ direction along the RNA in an ATP-dependent fashion, finally causing the dissociation of the ternary elongation complex by unknown mechanisms. Drosophila factor 2, a double-stranded DNA-dependent ATPase which lacks detectable helicase activity (55, 56), can cause the release of RNA polymerase II transcripts in an ATP-dependent manner (57). Recently it has been shown that in vaccinia virus, the ATP-dependent step in early transcription termination is catalyzed by a single-stranded DNA-dependent ATPase, the product of gene D11L, which also lacks detectable helicase activity (19). Thus, it is clear that transcription termination in several systems requires the participation of a factor which can bind single- or double-stranded RNA or DNA and hydrolyze ATP. The precise mechanism of action of these factors is not known; however, it seems reasonable that translocation or helicase activities of these factors within an elongation complex may destabilize the complex. We have shown here that the A18R protein is a negative transcription elongation factor. We have shown previously that the A18R protein, a member of the DExH helicase superfamily II (28), possesses both DNA-dependent ATPase and DNA helicase activities. The ATPase activity of A18R is stimulated by both single- and double-stranded DNA but not by RNA (5). The helicase activity is restricted exclusively to DNA-DNA hybrids, it is capable of separating only hybrids containing less than 25 bp, and it displays 3′-to-5′ directionality (44). Thus, based on prior biochemical analysis of the A18R protein, based on the A18R mutant analysis presented here, and by analogy with other known transcription termination factors, we propose that A18R serves as a termination factor for intermediate (and perhaps late) transcription in vivo.
Our experiments imply that 3′-end formation during postreplicative vaccinia virus transcription is a factor-mediated event but provide no information about potential cis-acting elements in either RNA or DNA that might be required for termination. In fact, termination of vaccinia virus postreplicative transcription resembles termination of transcription in metazoan cells in that it occurs at a large number of sites, generating extreme 3′-end heterogeneity (22, 31, 36). Thus, if specific nucleic acid sequences or structures mediate postreplicative vaccinia virus transcription termination, these elements must be both abundant and inefficient.
Prior genetic and biochemical experiments suggest that the A18R protein does not act alone but rather acts as part of a larger complex containing the viral RNA polymerase, the viral transcription factors G2R and H5R (9), and perhaps other factors as well. H5R is a 35-kDa DNA binding phosphoprotein (7, 33, 34) which has late transcription factor activity in vitro (29) and which interacts directly with G2R and either directly or indirectly with A18R. The precise role of H5R in stimulating late transcription has not been determined, and no virus mutants in H5R exist. G2R is a novel 26-kDa protein which interacts either directly or indirectly with A18R as well as undergoing a direct interaction with H5R. G2R mutants cause synthesis of 3′-truncated intermediate and late vaccinia virus RNAs (8) and also function as extragenic suppressors of A18R mutants (17). Thus, G2R behaves like a positive transcription elongation factor whose function serves to balance A18R activity. G2R could function independently of A18R, or it could be a positive regulator of A18R activity. Experiments with the antipoxvirus drug isatin-β-thiosemicarbazone (IBT) suggest that RNA polymerase interacts with A18R, since IBT induces promiscuous transcription (4), and IBT resistance maps to the second-largest subunit of the RNA polymerase (13). Biochemical experiments designed to elucidate the precise activities of A18R, G2R, and H5R in an elongating RNA polymerase complex are under way.
ACKNOWLEDGMENTS
We thank Jackie Lewis for technical support. We thank Carman Sancho for communication of unpublished data.
This work was funded by NIH grant AI 10894 to R.C.C. D.A.S. was supported in part by an NIH postdoctoral fellowship F32 AI 09252.
REFERENCES
- 1.Ahn B Y, Moss B. RNA polymerase-associated transcription specificity factor encoded by vaccinia virus. Proc Natl Acad Sci USA. 1992;89:3536–3540. doi: 10.1073/pnas.89.8.3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baldick C J, Jr, Keck J G, Moss B. Mutational analysis of the core, spacer, and initiator regions of vaccinia virus intermediate-class promoters. J Virol. 1992;66:4710–4719. doi: 10.1128/jvi.66.8.4710-4719.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baldick C J, Jr, Moss B. Characterization and temporal regulation of mRNAs encoded by vaccinia virus intermediate-stage genes. J Virol. 1993;67:3515–3527. doi: 10.1128/jvi.67.6.3515-3527.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bayliss C D, Condit R C. Temperature-sensitive mutants in the vaccinia virus A18R gene increase double-stranded RNA synthesis as a result of aberrant viral transcription. Virology. 1993;194:254–262. doi: 10.1006/viro.1993.1256. [DOI] [PubMed] [Google Scholar]
- 5.Bayliss C D, Condit R C. The vaccinia virus A18R gene product is a DNA-dependent ATPase. J Biol Chem. 1995;270:1550–1556. doi: 10.1074/jbc.270.4.1550. [DOI] [PubMed] [Google Scholar]
- 6.Bayliss, C. D., and R. C. Condit. 1993. Unpublished data.
- 7.Beaud G, Beaud R. Preferential virosomal location of underphosphorylated H5R protein synthesized in vaccinia virus-infected cells. J Gen Virol. 1997;78:3297–3302. doi: 10.1099/0022-1317-78-12-3297. [DOI] [PubMed] [Google Scholar]
- 8.Black E P, Condit R C. Phenotypic characterization of mutants in vaccinia virus gene G2R, a putative transcription elongation factor. J Virol. 1996;70:47–54. doi: 10.1128/jvi.70.1.47-54.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Black E P, Moussatche N, Condit R C. Characterization of the interactions among vaccinia virus transcription factors G2R, A18R, and H5R. Virology. 1998;245:313–322. doi: 10.1006/viro.1998.9166. [DOI] [PubMed] [Google Scholar]
- 10.Broyles S S, Li J, Moss B. Promoter DNA contacts made by the vaccinia virus early transcription factor. J Biol Chem. 1991;266:15539–15544. [PubMed] [Google Scholar]
- 11.Broyles S S, Yuen L, Shuman S, Moss B. Purification of a factor required for transcription of vaccinia virus early genes. J Biol Chem. 1988;263:10754–10760. [PubMed] [Google Scholar]
- 12.Cohrs R J, Condit R C, Pacha R F, Thompson C L, Sharma O K. Modulation of ppp(A2′p)nA-dependent RNase by a temperature-sensitive mutant of vaccinia virus. J Virol. 1989;63:948–951. doi: 10.1128/jvi.63.2.948-951.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Condit R C, Easterly R, Pacha R F, Fathi Z, Meis R J. A vaccinia virus isatin-β-thiosemicarbazone resistance mutation maps in the viral gene encoding the 132-kDa subunit of RNA polymerase. Virology. 1991;185:857–861. doi: 10.1016/0042-6822(91)90559-t. [DOI] [PubMed] [Google Scholar]
- 14.Condit R C, Lewis J I, Quinn M, Christen L M, Niles E G. Use of lysolecithin-permeabilized infected-cell extracts to investigate the in vitro biochemical phenotypes of poxvirus ts mutations altered in viral transcription activity. Virology. 1996;218:169–180. doi: 10.1006/viro.1996.0177. [DOI] [PubMed] [Google Scholar]
- 15.Condit R C, Motyczka A. Isolation and preliminary characterization of temperature-sensitive mutants of vaccinia virus. Virology. 1981;113:224–241. doi: 10.1016/0042-6822(81)90150-1. [DOI] [PubMed] [Google Scholar]
- 16.Condit R C, Motyczka A, Spizz G. Isolation, characterization, and physical mapping of temperature-sensitive mutants of vaccinia virus. Virology. 1983;128:429–443. doi: 10.1016/0042-6822(83)90268-4. [DOI] [PubMed] [Google Scholar]
- 17.Condit R C, Xiang Y, Lewis J I. Mutation of vaccinia virus gene G2R causes suppression of gene A18R ts mutants: implications for control of transcription. Virology. 1996;220:10–19. doi: 10.1006/viro.1996.0280. [DOI] [PubMed] [Google Scholar]
- 18.Deng L, Shuman S. A role for the H4 subunit of vaccinia RNA polymerase in transcription initiation at a viral early promoter. J Biol Chem. 1994;269:14323–14328. [PubMed] [Google Scholar]
- 19.Deng L, Shuman S. Vaccinia NPH-I, a DExH-box ATPase, is the energy coupling factor for mRNA transcription termination. Genes Dev. 1998;12:538–546. doi: 10.1101/gad.12.4.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eggermont J, Proudfoot N J. Poly(A) signals and transcriptional pause sites combine to prevent interference between RNA polymerase II promoters. EMBO J. 1993;12:2539–2548. doi: 10.1002/j.1460-2075.1993.tb05909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goebel S J, Johnson G P, Perkus M E, Davis S W, Winslow J P, Paoletti E. The complete DNA sequence of vaccinia virus. Virology. 1990;179:247–266. doi: 10.1016/0042-6822(90)90294-2. [DOI] [PubMed] [Google Scholar]
- 22.Hagenbuchle O, Wellauer P K, Cribbs D L, Schibler U. Termination of transcription in the mouse alpha-amylase gene Amy-2a occurs at multiple sites downstream of the polyadenylation site. Cell. 1984;38:737–744. doi: 10.1016/0092-8674(84)90269-1. [DOI] [PubMed] [Google Scholar]
- 23.Harris N, Rosales R, Moss B. Transcription initiation factor activity of vaccinia virus capping enzyme is independent of mRNA guanylylation. Proc Natl Acad Sci USA. 1993;90:2860–2864. doi: 10.1073/pnas.90.7.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hassett D E, Condit R C. Targeted construction of temperature-sensitive mutations in vaccinia virus by replacing clustered charged residues with alanine. Proc Natl Acad Sci USA. 1994;91:4554–4558. doi: 10.1073/pnas.91.10.4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirschmann P, Vos J C, Stunnenberg H G. Mutational analysis of a vaccinia virus intermediate promoter in vivo and in vitro. J Virol. 1990;64:6063–6069. doi: 10.1128/jvi.64.12.6063-6069.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin D J, Burgess R R, Richardson J P, Gross C A. Termination efficiency at rho-dependent terminators depends on kinetic coupling between RNA polymerase and rho. Proc Natl Acad Sci USA. 1992;89:1453–1457. doi: 10.1073/pnas.89.4.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keck J G, Baldick C J, Moss B. Role of DNA replication in vaccinia virus gene expression: a naked template is required for transcription of three late trans-activator genes. Cell. 1990;61:801–809. doi: 10.1016/0092-8674(90)90190-p. [DOI] [PubMed] [Google Scholar]
- 28.Koonin E V, Senkevich T G. Vaccinia virus encodes four putative DNA and/or RNA helicases distantly related to each other. J Gen Virol. 1992;73:989–993. doi: 10.1099/0022-1317-73-4-989. [DOI] [PubMed] [Google Scholar]
- 29.Kovacs G R, Moss B. The vaccinia virus H5R gene encodes late gene transcription factor 4: purification, cloning, and overexpression. J Virol. 1996;70:6796–6802. doi: 10.1128/jvi.70.10.6796-6802.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levin D, London I M. Regulation of protein synthesis: activation by double-stranded RNA of a protein kinase that phosphorylates eukaryotic initiation factor 2. Proc Natl Acad Sci USA. 1978;75:1121–1125. doi: 10.1073/pnas.75.3.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mahr A, Roberts B E. Arrangement of late RNAs transcribed from a 7.1-kilobase EcoRI vaccinia virus DNA fragment. J Virol. 1984;49:510–520. doi: 10.1128/jvi.49.2.510-520.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moss B. Poxviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J L, Monath T P, Roizman B, Strauss S E, editors. Fields virology. New York, N.Y: Lippincott-Raven; 1996. pp. 2637–2672. [Google Scholar]
- 33.Nowakowski M, Bauer W, Kates J. Characterization of a DNA-binding phosphoprotein from vaccinia virus replication complex. Virology. 1978;86:217–225. doi: 10.1016/0042-6822(78)90022-3. [DOI] [PubMed] [Google Scholar]
- 34.Nowakowski M, Kates J, Bauer W. Isolation of two DNA-binding proteins from the intracellular replication complex of vaccinia virus. Virology. 1978;84:260–267. doi: 10.1016/0042-6822(78)90246-5. [DOI] [PubMed] [Google Scholar]
- 35.Pacha R F, Condit R C. Characterization of a temperature-sensitive mutant of vaccinia virus reveals a novel function that prevents virus-induced breakdown of RNA. J Virol. 1985;56:395–403. doi: 10.1128/jvi.56.2.395-403.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Proudfoot N J. How RNA polymerase II terminates transcription in higher eukaryotes. Trends Biochem Sci. 1989;14:105–110. doi: 10.1016/0968-0004(89)90132-1. [DOI] [PubMed] [Google Scholar]
- 37.Rochester S C, Traktman P. Characterization of the single-stranded DNA binding protein encoded by the vaccinia virus I3 gene. J Virol. 1998;72:2917–2926. doi: 10.1128/jvi.72.4.2917-2926.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rosales R, Harris N, Ahn B Y, Moss B. Purification and identification of a vaccinia virus-encoded intermediate stage promoter-specific transcription factor that has homology to eukaryotic transcription factor SII (TFIIS) and an additional role as a viral RNA polymerase subunit. J Biol Chem. 1994;269:14260–14267. [PubMed] [Google Scholar]
- 39.Rosales R, Sutter G, Moss B. A cellular factor is required for transcription of vaccinia viral intermediate-stage genes. Proc Natl Acad Sci USA. 1994;91:3794–3798. doi: 10.1073/pnas.91.9.3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sancho, C., Y. Xiang, and R. C. Condit. 1997. Unpublished data.
- 41.Sawadogo M, Roeder R G. Factors involved in specific transcription by human RNA polymerase II: analysis by a rapid and quantitative in vitro assay. Proc Natl Acad Sci USA. 1985;82:4394–4398. doi: 10.1073/pnas.82.13.4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shuman S. Vaccinia virus RNA helicase: an essential enzyme related to the DE-H family of RNA-dependent NTPases. Proc Natl Acad Sci USA. 1992;89:10935–10939. doi: 10.1073/pnas.89.22.10935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simpson D A, Condit R C. The vaccinia virus A18R protein plays a role in viral transcription during both the early and the late phases of infection. J Virol. 1994;68:3642–3649. doi: 10.1128/jvi.68.6.3642-3649.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simpson D A, Condit R C. Vaccinia virus gene A18R encodes an essential DNA helicase. J Virol. 1995;69:6131–6139. doi: 10.1128/jvi.69.10.6131-6139.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith G L, Howard S T, Chan Y S. Vaccinia virus encodes a family of genes with homology to serine proteinase inhibitors. J Gen Virol. 1989;70:2333–2343. doi: 10.1099/0022-1317-70-9-2333. [DOI] [PubMed] [Google Scholar]
- 46.Smith K A, Stallard V, Roos J M, Hart C, Cormier N, Cohen L K, Roberts B E, Payne L G. Host range selection of vaccinia recombinants containing insertions of foreign genes into non-coding sequences. Vaccine. 1993;11:43–53. doi: 10.1016/0264-410x(93)90338-x. [DOI] [PubMed] [Google Scholar]
- 47.Tamin A, Villarreal E C, Weinrich S L, Hruby D E. Nucleotide sequence and molecular genetic analysis of the vaccinia virus HindIII N/M region encoding the genes responsible for resistance to alpha-amanitin. Virology. 1988;165:141–150. doi: 10.1016/0042-6822(88)90667-8. [DOI] [PubMed] [Google Scholar]
- 48.Turner P C, Moyer R W. Orthopoxvirus fusion inhibitor glycoprotein SPI-3 (open reading frame K2L) contains motifs characteristic of serine proteinase inhibitors that are not required for control of cell fusion. J Virol. 1995;69:5978–5987. doi: 10.1128/jvi.69.10.5978-5987.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vos J C, Sasker M, Stunnenberg H G. Vaccinia virus capping enzyme is a transcription initiation factor. EMBO J. 1991;10:2553–2558. doi: 10.1002/j.1460-2075.1991.tb07795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vos J C, Stunnenberg H G. Derepression of a novel class of vaccinia virus genes upon DNA replication. EMBO J. 1988;7:3487–3492. doi: 10.1002/j.1460-2075.1988.tb03224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Walstrom K M, Dozono J M, Robic S, von Hippel P H. Kinetics of the RNA-DNA helicase activity of Escherichia coli transcription termination factor rho. 1. Characterization and analysis of the reaction. Biochemistry. 1997;36:7980–7992. doi: 10.1021/bi963179s. [DOI] [PubMed] [Google Scholar]
- 52.Walstrom K M, Dozono J M, von Hippel P H. Kinetics of the RNA-DNA helicase activity of Escherichia coli transcription termination factor rho. 2. Processivity, ATP consumption, and RNA binding. Biochemistry. 1997;36:7993–8004. doi: 10.1021/bi963180r. [DOI] [PubMed] [Google Scholar]
- 53.Wright C F, Hubbs A E, Gunasinghe S K, Oswald B W. A vaccinia virus late transcription factor copurifies with a factor that binds to a viral late promoter and is complemented by extracts from uninfected HeLa cells. J Virol. 1998;72:1446–1451. doi: 10.1128/jvi.72.2.1446-1451.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wright C F, Moss B. Identification of factors specific for transcription of the late class of vaccinia virus genes. J Virol. 1989;63:4224–4233. doi: 10.1128/jvi.63.10.4224-4233.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xie Z, Price D. Unusual nucleic acid binding properties of factor 2, an RNA polymerase II transcript release factor. J Biol Chem. 1998;273:3771–3777. doi: 10.1074/jbc.273.6.3771. [DOI] [PubMed] [Google Scholar]
- 56.Xie Z, Price D. Drosophila factor 2, an RNA polymerase II transcript release factor, has DNA-dependent ATPase activity. J Biol Chem. 1998;272:31902–31907. doi: 10.1074/jbc.272.50.31902. [DOI] [PubMed] [Google Scholar]
- 57.Xie Z, Price D H. Purification of an RNA polymerase II transcript release factor from Drosophila. J Biol Chem. 1996;271:11043–11046. doi: 10.1074/jbc.271.19.11043. [DOI] [PubMed] [Google Scholar]
- 58.Yuen L, Moss B. Oligonucleotide sequence signaling transcriptional termination of vaccinia virus early genes. Proc Natl Acad Sci USA. 1987;84:6417–6421. doi: 10.1073/pnas.84.18.6417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang Y, Keck J G, Moss B. Transcription of viral late genes is dependent on expression of the viral intermediate gene G8R in cells infected with an inducible conditional-lethal mutant vaccinia virus. J Virol. 1992;66:6470–6479. doi: 10.1128/jvi.66.11.6470-6479.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou A, Paranjape J, Brown T L, Nie H, Naik S, Dong B, Chang A, Trapp B, Fairchild R, Colmenares C, Silverman R H. Interferon action and apoptosis are defective in mice devoid of 2′,5′-oligoadenylate-dependent RNase L. EMBO J. 1997;16:6355–6363. doi: 10.1093/emboj/16.21.6355. [DOI] [PMC free article] [PubMed] [Google Scholar]