Abstract
Liver cancer accounts for 6% of all malignancies causing death worldwide, and hepatocellular carcinoma (HCC) is the most common histological type. HCC is a heterogeneous cancer, but how the tumour microenvironment (TME) of HCC contributes to the progression of HCC remains unclear. In this study, we investigated the immune microenvironment by multiomics analysis. The tumour immune infiltration characteristics of HCC were determined at the genomic, epigenetic, bulk transcriptome and single-cell levels by data from The Cancer Genome Atlas portal and the Gene Expression Omnibus (GEO). An epigenetic immune-related scoring system (EIRS) was developed to stratify patients with poor prognosis. SPP1, one gene in the EIRS system, was identified as an immune-related predictor of poor survival in HCC patients. Through receptor-ligand pair analysis in single-cell RNA-seq, SPP1 was indicated to mediate the crosstalk between HCC cells and macrophages via SPP1-CD44 and SPP1-PTGER4 association. In vitro experiments further validate SPP1 can trigger the polarization of macrophages to M2-phenotype tumour-associated macrophages (TAMs).
Supplementary information
The online version contains supplementary material at (doi:10.1007/s00262-021-02967-8).
Keywords: Hepatocellular carcinoma (HCC), Tumour microenvironment (TME), Prognosis, SPP1, Tumour-associated macrophages (TAMs), Crosstalk
Introduction
Liver cancer accounts for 6% of all cancers and 9% of all deaths from cancer [1]. Liver cancer is the sixth-most diagnosed cancer and the second-leading cause of cancer death worldwide. Hepatocellular carcinoma (HCC) accounts for approximately 75% of liver cancers. Although surgical resection helps to improve the survival of patients with HCC, recurrence rates after surgery remain high, leading to poor prognosis of patients with HCC. Current progress in immune checkpoint inhibitor (ICI)-based and neoantigen vaccine-based immunotherapy provides a promising strategy to improve the prognosis of HCC [2–5]. For ICI-based immunotherapy, the effect is largely dependent on the tumour microenvironment (TME) [6–11].
The TME is characterized by the interaction of various cell types, e.g. cancer-associated fibroblasts, tumour cells and immune cells [12]. The crosstalk of the different cell types in the TME not only has a strong association with tumour progression but also relates to therapeutic outcome. Uncovering the pre-existing antitumour adaptive immune reaction and boosting the CD8 + T cell-mediated immune response by regulating the abundance and function of cells in the TME are critical for improving the reaction to immunotherapy.
In the present study, we aimed to elucidate the TME and the crosstalk of immune cells in HCC through multiomics analysis. The immune infiltration characteristics and the underlying mechanism were analysed by bulk transcriptome, single-cell transcriptome, whole-exome sequencing (WES) and DNA methylation array analyses. An epigenetic immune-related scoring system (EIRS) was constructed for stratification of patients with poor prognosis and improvement of individualized treatment. SPP1, which is in the epigenetic immune-related signature, was found to be a key regulator from tumour cells contributing to M2 polarization. Thus, targeting SPP1 may be a promising way to regulate the TME and improve the response to immunotherapy in HCC.
Materials and methods
Data source
Single-cell transcriptome files of GSE125449 were downloaded from the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/). The clinical information, DNA methylation data and transcriptome data of The Cancer Genome Atlas (TCGA) LIHC cohort were downloaded via the UCSC Xena browser (https://xenabrowser.net/). R software (version 3.5.3) and Python (version 3.6) were used for all the analyses in the manuscript.
TME construction
The ESTIMATE algorithm was performed to obtain an estimate score, stromal score and immune score based on the transcriptome data [13]. Higher tumour purity, lower infiltration levels of stromal populations and lower infiltration levels of immune cell populations in tumours were associated with a lower estimate score, stromal score and immune score, respectively. The deconvolutional method was performed with the CIBERSORT algorithm to calculate the abundance of immune cell populations in TCGA-LIHC cohort by transcriptome data [14].
Functional enrichment analysis
The “GSVA” package was applied for single-sample GSEA (ssGSEA) analysis for each sample [15]. The hallmark and KEGG gene sets were used for the enrichment analysis and were downloaded from the Broad Institute. The “GSEAPY” package was used to perform GSEA. Gene ontology (GO) analysis was performed with the “clusterProfiler” package [16]. The results were plotted with the “ggplot2” and “pheatmap” packages [17]. The correlation between immune score and ssGSEA value was performed with Pearson’s coefficient analysis.
DNA methylation data analysis
The DNA methylation level of one gene can be represented by the β value (0–1 = hypomethylated to hypermethylated). The correlation between the gene expression level and DNA methylation level was calculated with Pearson’s coefficient analysis. The genes with an absolute r2 value less than -0.7 were considered significant.
WES data analysis
WES data of both the high-immunity cohort and low-immunity cohort were used to detect SNVs, SNPs and INDELs using VarScan2.3.9 software [18]. The co-occurrence and mutually exclusive mutations were calculated with the CoMEt algorithm. The “maftools” package was used for the visualization of the somatic mutations in TCGA-LIHC cohort [19]. For the differential mutation pattern between the high-immunity cohort and the low-immunity cohort, Fisher’s exact test was used to calculate the most differential mutations between the high-immunity cohort and the low-immunity cohort.
Differentially expressed gene (DEG) and differentially methylated gene (DMG) analyses
The DEG analysis and DMG analyses were applied with the “Limma” package [20]. An empirical Bayesian method was applied to estimate the fold change between the high-immunity cohort and the low-immunity cohort using moderated t tests. The adjusted p value for multiple testing was calculated using the Benjamin-Hochberg correction. The genes with an absolute log2 fold change greater than 1 and adjusted p value less than 0.05 were identified as DEGs.
Construction of the EIRS scoring system
The least absolute shrinkage and selection operator (LASSO) was applied to construct the epigenetic-related immune gene signature by TCGA-LIHC cohort transcriptome data [21]. The EIRS scoring system was built by including individual normalized gene expression values weighted by their LASSO Cox coefficients as follows: (The detailed code is in supplementary file 1).
Immunohistochemistry
The tissue sections were placed in a 63 °C oven (PH-070A) for one hour, and then deparaffinized in an automatic dyeing machine (LEICAST5020, LEICA). The antigen retrieval of the slides was carried out by the PT Link (Dako North America, Inc). After the repair was completed, the sections were naturally cooled to room temperature and washed in distilled water for 5 min. Primary antibody incubation was performed after deparaffinization and antigen retrieval. Those sections were rinsed in PBST before blocking with 5% bovine serum albumin (BSA) for 30 min and incubating with anti-osteopontin antibody (1:100, ab8448, Abcam) at 4 °C overnight. After that, the slides were rewarmed at room temperature for 45 min followed by washing three times in PBST buffer (5 min each time).
Immunohistochemical staining was performed by an automated staining platform (Autostainer Link 48, Dako North America, Inc) according to manufacturer’s instructions, and the appropriate programme was selected to run blocking, secondary antibody binding and diaminobenzidine (DAB) colour development programmes. After the slides were stained with hematoxylin (SLBT4555, Sigma-Aldrich) for 1 min, they were immersed in 0.25% hydrochloric acid alcohol (400 ml 70% alcohol + 1 ml concentrated hydrochloric acid) for no less than 2 s. The glass slides were rinsed thoroughly with tap water for more than 2 min and dried at room temperature for several hours and sealed with neutral resin. The slides were digitized using Aperio scanner (Aperio XT, LEICA) at 20 × magnification.
Immunofluorescence staining was carried out according to standard staining procedures. The slides were incubated with secondary antibody in the dark at room temperature for 50 min, followed by washing in PBS buffer. The autofluorescence quencher (Servicebio) was then added to it for 5 min and washed with running water for 20 min. After the slices were slightly dried, 4,6-diamidino-2-phenylindole (DAPI; Beyotime, Shanghai, China) was added and incubated at room temperature in the dark for 10 min. After the sections were washed with PBS buffer and dried, they were mounted with anti-fluorescence quenching mounting tablets (Servicebio, G1401). Thereafter, the slides were digitized using fluorescence microscope (NIKON DS-U3) at 20 × magnification.
Patient cohort and TMA
The commercialized TMA (panel HLivH180Su15) and clinical data were provided by Shanghai Outdo Biotech Co. Ltd. (Shanghai, China). The TMA comprised samples of 90 patients with stage I-III HCC were collected between 2007 and 2012. All tumour tissue samples were obtained from patients considered to be optimally debulked at the time of initial surgery. When categorized by American Joint Committee on Cancer (AJCC) 7th edition clinical staging, 63 samples were stage I, 25 were stage II and 2 were stage III. The mean age of the patients was 52 years (range 31–78 years), the median progression-free survival time was 42 months (range 4–56 months) and the median follow-up time was 66.5 months (range 5–104 months).
Single-cell analysis
The single-cell expression matrix was downloaded from the GEO database and was analysed by the “scanpy” package in Python [22]. Dimension reduction was performed with the uMAP method. The crosstalk between different cells and the ligand-receptor pair was inferred by cellphoneDB2 software [23].
Cell culture
The human HCC cell lines HepG2, Hep3B, Huh 7, SK and the human monocyte cell line THP-1 cells were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). The human HCC cell lines were grown routinely in Dulbecco’s modified Eagle’s medium (DMEM, HyClone, USA) supplemented with 10% foetal bovine serum (FBS, HyClone, USA), while THP-1 cells were cultured in RPMI-1640 (Invitrogen, CA, USA) containing 10% heat-inactivated FBS. The THP-1 cells were differentiated into M0 macrophages when incubated in phorbol 12-myristate 13-acetate (PMA, 0.01 mg/ml, Abcam, ab120297) for 18 h. All cell cultures were maintained in a humidified incubator supplied with 5% CO2 at 37 °C.
siRNA transfection
Human HCC cells were transfected with SPP1 siRNA (siRNAs for human SPP1: genOFFTM st-h-SPP1_002: GAACGACTCTGATGATGTA, genOFFTM st-h-SPP1_003: CCAAGTAAGTCCAACGAAA) and Negative Control siRNA, respectively. Transfection was performed using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions. 48 h after transfection, the cells were collected to extract RNA or protein identification, and the transfection efficiency was recorded. The specific two siRNA sequences are shown in Additional file.
RNA extraction and quantification
For quantitative RT-PCR (qPCR), total RNA content of cells was extracted using TRIzol (Invitrogen) according to the manufacturer’s instructions. After detecting the total concentration and purity of the extracted RNA, the collected RNA was reverse transcribed into complementary DNA (cDNA) using SYBR Green Master Mix (Vazyme, China). Each sample was subjected to qRT-PCR in triplicate to assess the expression level of SPP1 (forward primer: GAAGTTTCGCAGACCTGACAT, reverse primer: GTATGCACCATTCAACTCCTCG), and GAPDH was used as an internal reference. Relative mRNA quantification was calculated by the formula 2 − ΔΔCt method. The specific primers used for amplification are shown in Additional file.
Western blot analysis
After being washed twice by phosphate-buffered saline (PBS), cells were harvested and solubilized in cold radioimmunoprecipitation assay (RIPA) lysis buffer (Beyotime, China) containing phenylmethylsulfonyl fluoride (PMSF) and protease inhibitor cocktail. The concentration of total protein was determined by a bicinchoninic acid (BCA) protein assay kit (Vazyme, China). Equal amounts of each protein sample (30 mg) were separated by 4–20% precast polyacrylamide gels (Bio-Rad, Hercules, CA, USA) and transferred onto polyvinylidene fluoride (PVDF) membranes. After blocking with 5% bovine serum albumin in PBS at room temperature for 1 h, the membranes were incubated with primary rabbit antibodies to anti-osteopontin antibody (1:1000, ab8448, Abcam) or anti-GAPDH (1:2000, ab8245, Abcam) at 4 °C overnight. Subsequently, the membrane was washed with Tris-buffered saline with Tween 20 (TBST) three times (5 min each) and incubated with secondary anti-rabbit antibody to IgG (1:2000, ab97051, Abcam) at room temperature for 1 h. After washing, the protein signals on the membranes were visualized with enhanced chemiluminescence (ECL) substrate in a FluorChem Q imaging system.
Immunofluorescence
Cells were seeded on a coverslip in six-well plates. After 24 h, the cells were fixed with 4% paraformaldehyde for 30 min at room temperature. Then, the cells were washed 3 times for 5 min each in PBS. Thereafter, cells were permeabilized with 0.5% Triton X-100 for 5 min. After being washed for three times in PBS, cells were blocked with 3% BSA for 30 min and incubated with the primary antibody to anti-osteopontin antibody (1:1000, ab8448, Abcam) at 4 °C overnight. Subsequently, cells were washed three times in PBS and incubated with goat anti-rabbit secondary antibody in the dark at 37 °C for 30 min. After the final washing, cells were stained with DAPI and examined using a Zeiss LSM510 laser scanning confocal microscope (Carl Zeiss, Jena, Germany).
Wound-healing assay
Cells were seeded in six-well plates at a total of 105 cells per well for transfection. When cellular confluence reached ∼90% confluence, a 100 μl sterile pipette tip was used to create wounds in the monolayer cells. Wells were then gently washed with PBS to remove any cell fragments or free-floating cells, and 2 mL medium without serum was added. Photographs of both control and treated wells were taken with a phase-contrast microscopy at 0, 24 and 48 after the creation of wounds. Cell migratory activity of SPP1 silenced human HCC cells was measured by the rate of cells moving towards the scratched area, which was analysed by ImageJ software and calculated as follows: migration index = recovered wound area/original wound area.
Cell migration assay
Cell migration abilities were assayed by using transwell with a noncoated polycarbonate membrane (8.0 μm pore size, Corning Incorporated). Each well of the top chamber was filled with 200 μL serum-free DMEM medium containing 105 transfected cells per mL. The lower chambers contained 800 μL complete medium containing 10% FBS as a chemoattractant. The Transwell plate was then incubated at 37 °C for 24 h followed by 30 min of fixation with 4% paraformaldehyde. The chambers were then stained with 0.1% crystal violet for 30 min, washed gently by PBS. The cells remaining on the upper surface of the chamber were wiped off with cotton swabs. Finally, the remaining cells on the undersurface were photographed and counted by using a light microscope with10 × objective lens.
Co-culture assay
In order to explore the polarization effect of SPP1 gene on M0 macrophages, we designed a co-culture system. The co-culture groups were established between human HCC cells (105 cells per well) in transwell inserts (0.4 μm pore size, Corning Incorporated) and M0 macrophages (105 cells per well) in six-well plates. The co-cultures were incubated for 72 h in the humidified incubator supplied with 5% CO2 at 37 °C. Then, the relevant markers of M1 and M2 macrophages were detected using quantitative RT-PCR.
Colony formation assay
Cells were seeded in six-well plates (1000 cells per well) after transfection in complete medium for 7 days. The cells were then fixed with 4% paraformaldehyde for 30 min at room temperature and stained using 0.1% crystal violet for 30 min. Cells were washed with PBS, photographed and counted using Image J software to determine colony formation rates.
Statistical analysis
All experimental data were analysed using the GraphPad prism 9 software (GraphPad software, USA). Measurement data were expressed as mean ± standard error. A two tailed Student’s t test was applied to measure the significance of data analysis between two groups. A p value < 0.05 was considered significant.
Results
Construction of the TME in HCC
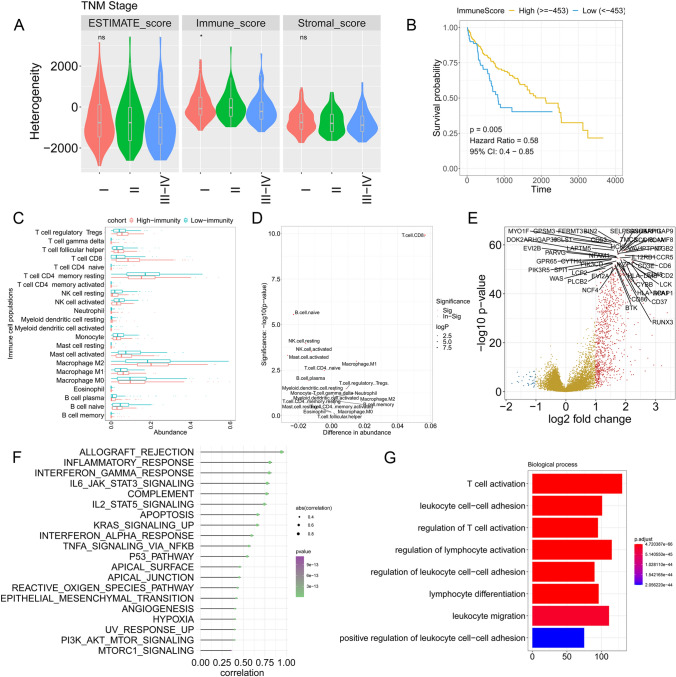
The workflow of this study is shown in Figure S1. The heterogeneity of HCC tumours was analysed by the ESTIMATE algorithm and is shown by the estimate score, immune score and stromal score. The immune score decreased with increased TNM stage (P < 0.05 by T test), whereas no significance was found in the estimated score and stromal score among the different TNM stages (Fig. 1a). Kaplan–Meier survival analysis demonstrated that the patients with high- (> = −453) and low-immune scores (< −453) (threshold selected by best cut-off method) exhibited significantly different outcomes (hazard ratio (HR) = 0.58, p = 0.005 by univariate Cox regression) (Fig. 1b). The immune cell populations were calculated by a deconvolutional algorithm by CIBERSORT (Fig. 1c). The results indicated that CD8+ T cells, M1 macrophages and several other immune cells were enriched in the high-immune group, while activated mast cells, activated NK cells and resting NK cells were more enriched in the low-immune group (Fig. 1d). DEG analysis was performed between the high- and low-immune groups, and the results revealed that immune-related molecules, such as LCK, RUNX3, LCP2 and CCR5, were upregulated in the high-immune group (Fig. 1e). ssGSEA was performed and showed that allograft rejection, inflammatory response, interferon-gamma response, IL6-JAK-STAT3 signalling and complement (Cor > 0.75) were the most significant hallmarks associated with the immune score (Fig. 1f). GO analysis was performed with the DEGs and indicated that T cell activation, leukocyte cell–cell adhesion, regulation of T cell activation and other enrichments were identified (Fig. 1g).
Fig. 1.
Construction of the TME in HCC. a Comparison of the distributions of immune scores between tumours in different TNM stages. b Kaplan–Meier curve showing the association between overall survival and immune scores in HCC patients. c Immune cell population distribution in the high- and low-immunity cohorts. d Statistical analysis of the immune cell population in the high- and low-immunity cohorts. e Volcano plot indicating the DEGs between the high- and low-immunity cohorts. f ssGSEA revealing the association between significant hallmarks and immune score. g GO analysis. The x axis indicates the overlapping numbers between each GO term and DEGs. The colour of the bars represents the adjusted p values (with FDR correction)
Identification of the immune infiltration-dependent expression profile and related pathways
Among the DEGs identified between the high- and low-immune groups, 31 chemotactic factors, such as CXC3 L1, CCL8 and CCL13, were significantly upregulated in the high-immune group and regulated the recruitment of a variety of immune cells to tumours (Fig. 2a). B7-CD28 and the TNF superfamily are involved in immune checkpoint-based immune therapy and tumour immunity. We then checked the molecules involved in B7-CD28, the TNF superfamily and other important molecules in TCGA-HCC cohort. The results indicated that CD274, PDCD1, ICOS, CTLA4, LAG3, HAVCR2 and other molecules were enriched in patients with high-immune scores (Fig. 2b). We also investigated the expression of metagenes involved in IgG, interferon, LCK, MHC-II and STAT1. As expected, genes associated with MHC-II, interferon, LCK and STAT1 were highly expressed in the high-immune group (Fig. 2c). MHC-II molecules (e.g. HLA-DMA, HLA-DMB, HLA-DRA and HLA-DPA1), as well as the activity of the involved pathways and hallmarks, were analysed in the HCC cohort. The results demonstrated that allograft rejection, antigen processing, antigen presentation, TCR signalling and other MHC-II-related pathways were upregulated with increased expression of MHC-II molecules in the high-immune group (Fig. 2d).
Fig. 2.
DEGs and related pathway changes in the immune infiltration-dependent status. a Heatmap displaying the expression changes of chemotactic factors. b Heatmap displaying the expression changes of genes involved in the B7-CD28 family and TNF family. c Metagenes related to IgG, interferon, LCK, MHC-II and STAT1. d Genes involved in MHC-II family and MHC-II-related pathway alterations
Comparisons of immune infiltration-dependent somatic mutations
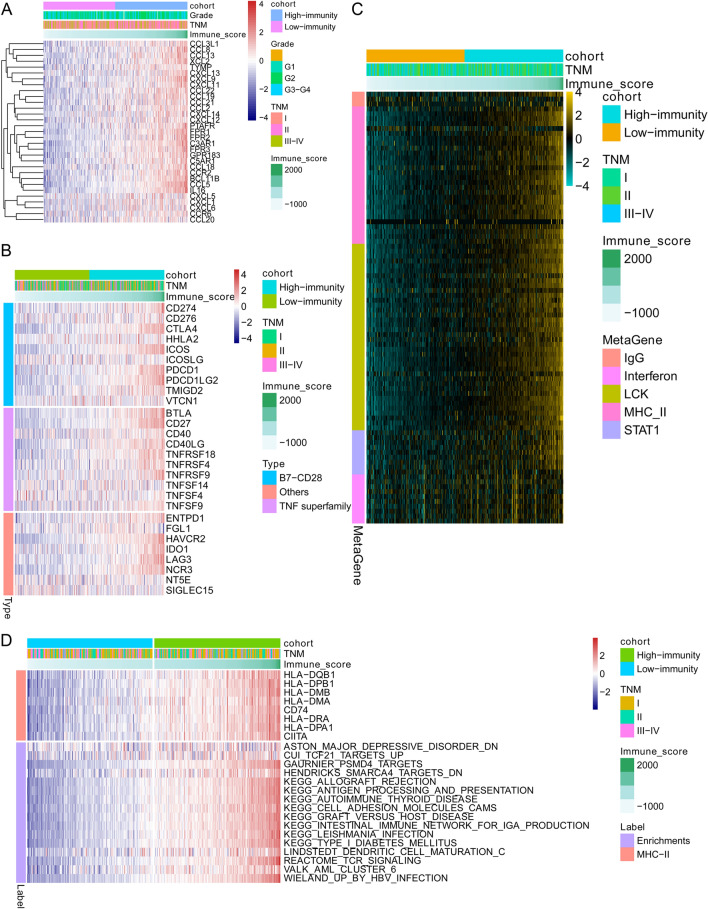
After depicting the difference in transcriptome levels in the above section, we then investigated the discrepancy between the high- and low-immunity groups at the genomic level. The WES data from TCGA were analysed by maftools in which the mutations were called by VarScan2. Missense mutations accounted for a large proportion of all mutation types in both the high-immunity and low-immunity groups (Fig. 3a, b). Furthermore, the most frequently mutated genes were TP53, CTNNB1 and TTN in both the high- and low-immunity groups, which indicated the importance of these three genes in the malignant progression of HCC rather than the regulation of immune infiltration. The somatic mutation types, including polymorphisms (SNPs), insertions (INSs) and deletions (DELs), showed no significance in the low-immunity and high-immunity groups (Fig. 3c). Thus, we further analysed the difference in mutation frequency between the two groups. Fisher’s exact test indicated a high frequency of ABCB5, AXIN1, FBN1 and ANK1 mutation frequency in the low-immunity group, as well as a high frequency of NUP214 mutation in the high-immunity group (Fig. 3d). Nevertheless, there was no significant difference in the prognosis associated with these gene mutations between the high- and low-immunity groups. Considering the top 10 most frequent mutations in the HCC cohort, the CTNNB1 mutation frequency was significantly higher in the low-immunity group than in the high-immunity group (Fig. 3e). In the next step, we analysed the co-interaction (both co-occurring and exclusive mutations) events of the 25 most frequently mutated genes in the low-immunity and high-immunity groups using the CoMEt algorithm. The co-interaction analysis demonstrated that different co-occurring and exclusive mutation events occurred between the high-immunity and low-immunity groups (Fig. 3f). For instance, the CTNNB1 gene was mutually exclusive to the AXIN1 gene in the high-immunity group, while CTNNB1 only showed one significant co-occurrence event with ARID2 in the low-immunity group.
Fig. 3.
Genomic landscape of HCC in the high-immunity and low-immunity cohorts. a, b OncoPlot showing the mutation distribution of the top 15 most frequently mutated genes in the high-immunity and low-immunity cohorts. The upper panel displays the mutation frequency of each HCC sample. The middle panel displays the types of mutations. The bar plot on the left indicates the frequency and mutation type of genes mutated in the low-immunity and high-immunity cohorts, respectively. The lower panel displays the clinical features (tumour stage, grade and immunity cohort) of each sample. The bottom panel shows the frequency and distribution of mutation types in each patient. c Mutation numbers of DELs, INSs and SNPs in the high- and low-immunity cohorts. d Forest plot displaying the top 3 most significantly differentially mutated genes between the two cohorts. e Comparison of the rate of the 10 most frequent gene mutations in the low- and high-immunity cohorts. f Heatmap displaying the mutually co-occurring and exclusive mutations of the top 20 frequently mutated genes in the high- and low-immunity groups. The colour and star symbol in each cell represent the statistical significance of the exclusivity or co-occurrence for each pair of genes
Elucidation of the DNA methylation pattern associated with the TIME of HCC and construction of an EIRS scoring system
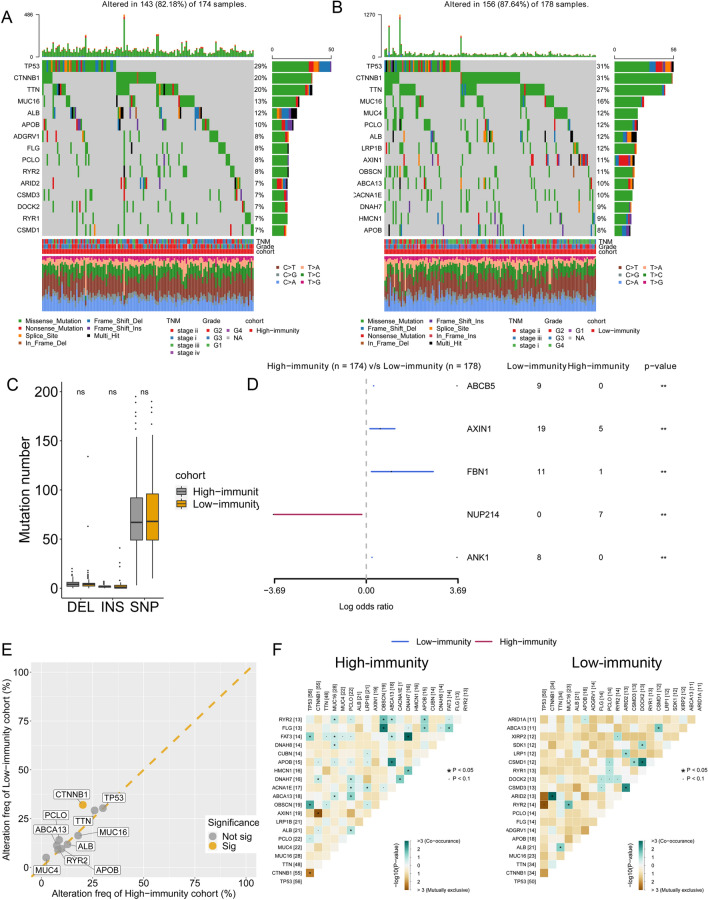
The alteration of DNA methylation status (hypermethylation in CpG islands and hypomethylation in CpG-poor regions) is associated with the malignant transformation and progression of HCC. Hence, we investigated the changed DNA methylation pattern in the high- and low-immunity groups using Illumina 450 k DNA methylation data from TCGA portal. The gene-level DNA methylation status was represented with the β value, and the different methylated genes (DMGs) were identified (Fig. 4a). CCR5, GPSM3, BIN2, SELPLG and other genes showed different methylation statuses between the high- and low-immunity groups. GO analysis was performed using the DMGs, and the results indicated that T cell activation, regulation of lymphocyte activation, positive regulation of cytokine production and other hallmarks were enriched (Fig. 4b). We then performed Pearson’s test on TCGA transcriptome data and DNA methylation data. Genes with Cor < −0.6 were considered to be regulated by DNA methylation. The Venn plot indicated that among the DNA methylation-regulated genes, 16 genes were differentially expressed and differentially methylated between the high- and low-immunity groups (Fig. 4c and Figure S2). A LASSO Cox regression model was performed to identify robust features among the 16 candidates. Cross-validation was applied to prevent overfitting, and the optimal log(λ) = − 4.2 was selected (Fig. 4d). Eleven genes (FERMT3, SPP1, MTHFD2, CYP3A4, PTPRCAP, LOC100233209, IFITM1, CXCR6, RIPK3, SPN and HLA-DMA) were retained with their nonzero individual coefficients (Fig. 4e). The distribution of coefficients of the 11 genes is demonstrated in Fig. 4e. Finally, the immune-related epigenetic risk score (EIRS) of the gene signature was established as follows:. Among the 11 genes, eight genes were significantly associated with the OS of HCC patients (Fig. 4f). Univariate Cox regression demonstrated that high expression of SPP1 was associated with poor prognosis, while high expression of the remaining seven genes showed favourable survival outcomes in HCC patients. With EIRS, HCC patients can be stratified into high- and low-risk groups. Kaplan–Meier survival analysis demonstrated that the patients with high (> = −453) and low EIRS (< −453) exhibited significantly different outcomes (hazard ratio (HR) = 3.63, p < 0.001 by univariate Cox regression) (Fig. 4g). To further explore the biological meaning underlying the EIRS scoring system, the associations of ICI expression level, immune cell populations and EIRS were calculated. Strikingly, PD-1 and PD-L1 expression levels were lower in the high-EIRS group (P < 0.001 and P < 0.01, respectively) (Fig. 4h). The association between EIRS and the immune cell populations was calculated with Pearson’s correlation analysis (Fig. 4i). The results indicated that high EIRS was correlated with a low CD8+ T cell population (R = −0.36, P < 0.001), while a positive association was found between EIRS and M0 macrophages (R = 0.42, P < 0.001) (Fig. 4j–k).
Fig. 4.
Construction of an epigenetic immune-related scoring system (EIRS). a Volcano plot showing the differentially methylated genes (DMGs). b GO analysis of DMGs. c Venn plot indicating the key immune-related genes. d LASSO Cox regression. e Coefficient for each gene in the EIRS signature in the LASSO Cox model. f Genes with a significant association with OS in the EIRS signature and the HR value. g Kaplan–Meier curve showing the association between overall survival and EIRS values in HCC patients. h PD1, PD-L1 and CTLA4 expression in the high-EIRS cohort and low-EIRS cohort. i Association between EIRS value and immune cell abundance. The colour and star symbol represent the statistical significance for Pearson’s coefficient. The cross symbol indicates insignificance. j Association of EIRS and CD8 + T cell. k Association of EIRS and M0 macrophages
SPP1-mediated interaction between HCC cells and macrophages
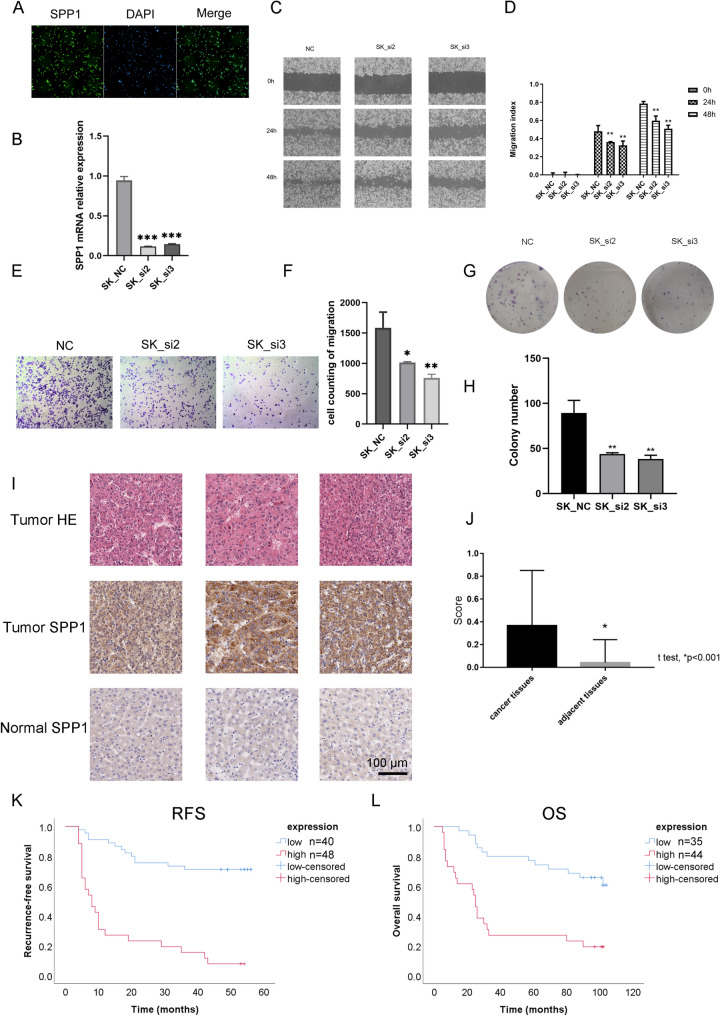
SPP1 was the only gene associated with poor prognosis in our epigenetic signature in the HCC cohort. We then focused on the mechanism of SPP1 in HCC patients. Through immunofluorescence (IF) staining, we confirmed that SPP1 was expressed in SK cells (Fig. 5a). siRNA was applied to knockdown the expression of SPP1 in SK cells (Fig. 5b). Quantification of the wound area revealed significantly lower closure of SPP1KD SK cells than control SK cells in the in vitro closure assay (Fig. 5c, d). The cell migration assay indicated that knockdown of SPP1 weakened the migration capacity of SK cells (Fig. 5e, f). In the colony formation assay, fewer colonies were found in SPP1KD SK cells than in control SK cells (Fig. 5g, h). The results above indicated a potential oncogenic role of SPP1 in HCC cells. We then measured the protein level of SPP1 in HCC using a tissue microarray chip. High protein levels of SPP1 were found in tumours compared with normal tissues (Fig. 5i, j). Kaplan–Meier survival analysis indicated that high SPP1 protein levels were associated with poor OS and RFS in this cohort, which indicated that SPP1 may be a potential biomarker for prognosis in HCC patients (Fig. 5k, l). Detailed information on this cohort is provided in supplemental table S1. Multivariate Cox regression confirmed that SPP1 was an independent risk factor for RFS (P < 0.001) and OS (P = 0.001) (Supplemental Tables S2-S3).
Fig. 5.
SPP1 acts as an immune-related prognostic factor in HCC. a Immunofluorescence staining of SPP1 in SK cells. b Relative mRNA level between SK cells and SPPKD SK cells. c Wound-healing assay of SK cells and SPPKD SK cells. d Quantification of the wound-healing assay between SK cells and SPPKD SK cells. e Migration assay of SK cells and SPPKD SK cells. f Quantification of the migration assay between SK cells and SPPKD SK cells. (E) Migration assay of SK cells and SPPKD SK cells. g Colony formation assay of SK cells and SPPKD SK cells. h Quantification of the colony formation assay between SK cells and SPPKD SK cells. i, j SPP1 expression in tumour and normal tissues according to IHC staining and HE staining. k Association of SPP1 protein and RFS in HCC patients. l Association of SPP1 protein and OS in HCC patients
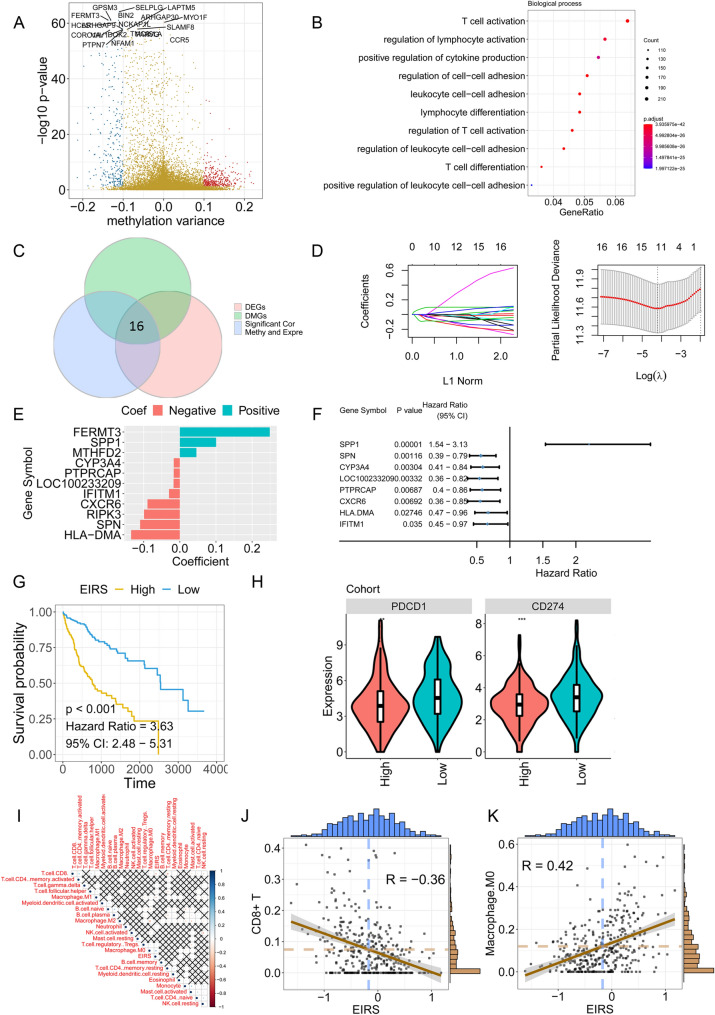
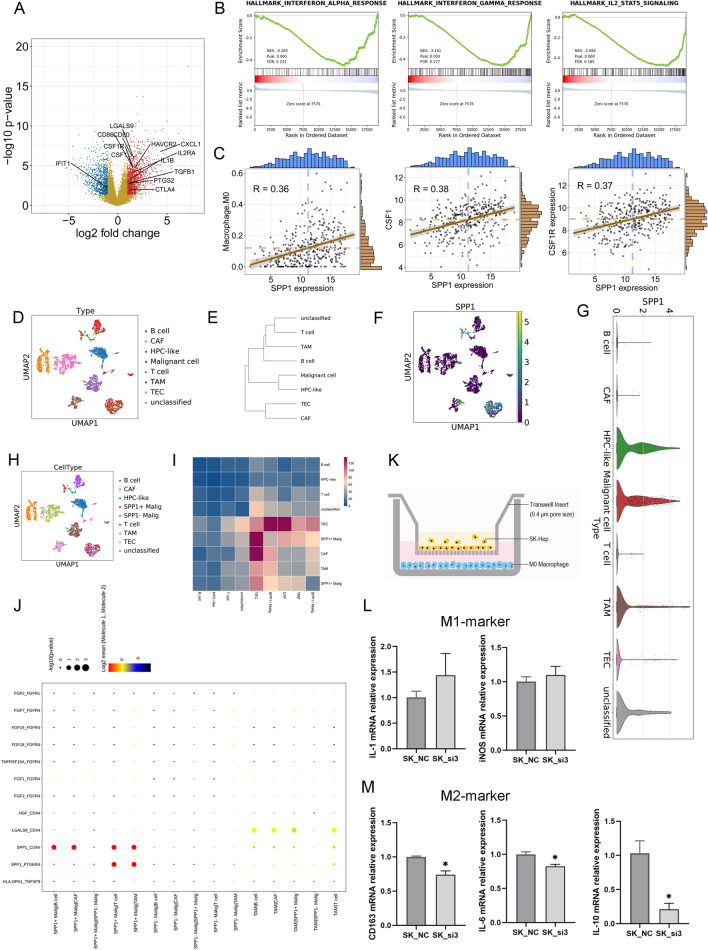
To illustrate the potential role of SPP1 in tumour immunity, we selected tumour samples with high SPP1 expression and tumour samples with low SPP1 expression from TCGA-HCC cohort and performed DEG analysis. The results showed that several genes, such as CTLA4, CSF1, CSF1R, IL1B, IL2RA, CXCL1 and some other molecules, were upregulated in samples with high SPP1 expression (Fig. 6a). GSEA validated that the interferon-alpha response, interferon-gamma response and IL2-STAT5 signalling were downregulated in samples with high SPP1 expression (Fig. 6b). SPP1 was identified as a key component in our epigenetic signature, which significantly correlated with M0 macrophages, and tumours with high SPP1 expression also showed upregulation of CSF1 and CSF1R expression. We then analysed the correlation between SPP1 expression and M0 macrophages. Significant associations were found between M0 macrophage populations and SPP1, CSF1 and CSF1R expression (Fig. 6c). Thus, these data suggested that SPP1 may play a potential role in regulating the function of macrophages.
Fig. 6.
Single-cell transcriptome analysis for tumour heterogeneity in HCC patients. a Volcano plot indicating the DEGs between the SPP1 high-expression and SPP1 low-expression groups. B GSEA between the SPP1 high-expression and SPP1 low-expression groups. c Association of M0 macrophage abundance with SPP1, CSF1 and CSF1R expression. d UMAP showing the distribution of various cell types. e Hierarchical clustering of cell types. f UMAP indicating SPP1 expression in different cell types. g UMAP showing the distribution of various cell types, including SPP1 + malignant cells and SPP1-malignant cells. h Histogram showing the expression level of SPP1. i Heatmap indicating the number of LR pairs between different cell types. j Heatmap indicating the ligand-receptor pairs between different cells
To further confirm the relationship between SPP1 and macrophages, we reanalysed the scRNA-seq data. The HCC tumours were composed of several cell types, such as malignant cells, cancer-associated fibroblasts (CAFs), tumour-associated macrophages (TAMs) and others (Fig. 6d). uMAP and unsupervised clustering showed the cluster of cell types from GSE125449 (Fig. 6e). We then analysed SPP1 expression at the single-cell transcriptome level. SPP1 was expressed mainly in malignant cells and TAMs (Fig. 6f). A proportion of malignant cells highly expressed SPP1 (Fig. 6g), and the malignant cells were then divided into SPP1 + cells and SPP- cells (Fig. 6h). The cell–cell interaction was investigated by CellphoneDB. SPP1 + malignant cells had a high number of ligand-receptor pairs with cancer-associated fibroblasts, TAM cells and tumour endothelial cells. Strong SPP1-CD44 interactions and SPP1-PTGER4 interactions were predicted between SPP1 + malignant cells and TAMs (Fig. 6i, j). IF staining suggested the co-localization of SPP1 and CD68 in HCC tissues (Figure S3). Hence, we applied non-contact co-culture of HCC cells and M0-macrophages (Fig. 6k). SPP1KD SK cells or SPP1Ctrl SK cells were co-cultured with M0-macrophages for 72 h and then macrophages were harvested for QPCR analysis. Results showed that M2 markers were downregulated in SPP1KD group while not significant change was found for M1 markers, which suggested the contribution of SPP1 in M2 polarization (Fig. 6l, m).
Discussion
The inter- and intratumour heterogeneity of liver cancer shapes cancer progression and affects the response to chemotherapy and immunotherapy [12, 24, 25]. Thus, identifying the immune contexture and the interaction between tumour cells and immune components can be beneficial to improve the effect of ICI-based and individual neoantigen vaccination-based therapy [24–28]. Nevertheless, the current understanding of the TME, liver cancer prognosis and therapeutic targets is still not clear. In this study, we performed multiomics analysis to analyse the heterogeneity of liver cancer and to elucidate the TME of liver cancer at both the single-cell level and bulk tissue level.
Using the ESTIMATE algorithm, an individual immune score was generated for liver cancer patients. As expected, the patients with high-immune scores showed a better prognosis than the patients with low-immune scores. The immune cell populations were identified by the CIBERSORT algorithm. CD8 + T cells, activated NK cells and M1 macrophages were enriched in patients with high-immune scores, which indicated a “hot” tumour status in patients with high-immune infiltration. Several of the most significant genes, such as RUNX3, BTK and CD37, were identified using DEG analysis, and these genes deserve further analysis of their roles in regulating antitumour immunity in liver cancer. The presence of the appropriate cocktail of chemotactic factors and their cellular sources is important for the establishment of a “hot” tumour microenvironment, which in turn can guide antitumour immunity and lead to better prognosis. Thus, we further analysed chemotactic factors in liver cancer. Thirty-one chemotactic factors were found to be highly expressed in patients with high-immune scores. CXCL9, CXCL10, CXCL11 and CXCL13 correlate positively with patient survival in a variety of cancers, while CXCL5 has been invariably associated with poor survival in multiple cancers [27–35]. In our analysis, highly expressed CXCL9, CXCL11, CXCL13 (favourable) and CXCL5 (unfavourable) were found in patients with high-immune infiltration, which indicated the need for a multifactor analysis for liver cancer patients. In a pan-cancer analysis, relapsing patients have been found to be associated with decreased expression or function of the interferon-γ signalling components, Janus kinase 1 (JAK1) and JAK2, as well as the antigen-presenting protein, β2-microglobulin (B2M), and MHC class II [12]. In patients with high-immune infiltration, MHC-II molecules and related genes and pathways were enriched. The better prognosis of “hot” tumour patients was related to the enrichment of these pathways.
We further investigated the mechanism behind the antitumour immune activities by exploring WES and DNA methylation data. Alterations of several key genes, such as TP53, CTNNB1 and TNN, were identified in patients with HCC. The mutation numbers regarding DELs, INSs and SNPs did not significantly differ between the low- and high-immunity groups. Several alterations in genes, including ABCB5, AXIN5, FBN1, NUP214 and ANK1, were identified to be significantly different. Interestingly, among the 10 most significantly altered genes, CTNNB1, the WNT pathway mediator, was found to be significantly highly mutated in the low-immunity cohort, which suggested the potential role of the WNT pathway in regulating antitumour immunity in HCC. Wnt ligands released by tumour cells in the TME facilitate the immune evasion of cancer cells, and aberrant Wnt signalling results in reduced immune cell infiltration [36]. These findings suggest that targeting Wnt may be a promising method to improve immunotherapy in HCC.
Additionally, we explored the DNA methylation level change between the low- and high-immunity cohorts. Not surprisingly, GO analysis demonstrated that canonical pathways, such as T cell activation and regulation of lymphocyte activation, were enriched. Among all the dysregulated DEGs and DMGs between the high- and low-immunity cohorts, we found that 16 genes were regulated by DNA methylation changes. An epigenetically regulated prognosis model EIRS scoring system was constructed and showed superior performance with regard to the OS of patients with HCC. High-expression levels of PD-1 and PD-L1 were found in patients with lower EIRS. Additionally, enriched CD8 + T cells were also correlated with low EIRS, which indicated that patients with lower EIRS may benefit more from ICI-based immunotherapy.
SPP1 was the only gene that contributed to poor prognosis in the EIRS scoring system. We further analysed the molecular function of SPP1 in HCC. SPP1 is a secretory acidic glycoprotein that belongs to the small integrin binding ligand N-linked glycoprotein (SIBLING) family. SIBLINGs can specifically bind and activate matrix metalloproteinases (MMPs), which play crucial roles in cancer progression [37]. SPP1 has been found to be highly expressed in various tumour types [36–40]. As expected, SPP1 was an oncogenic gene in HCC, and knockdown of this gene weakened the migration capacity and colony formation ability of SK cells. We also confirmed that SPP1 was highly expressed in HCC tumour tissues compared with normal tissues. Using a TMA, we confirmed the poor prognosis of patients with high SPP1 expression by IHC staining. DEG analysis between the SPP1 high-expression group and the SPP1 low-expression group revealed that several key immune coinhibitory molecules were dysregulated, and GSEA revealed that interferon-α, interferon-β and IL2-STAT5 signalling pathways were downregulated in the SPP1 high-expression group. Thus, we confirmed that SPP1 may be involved in reshaping the immune microenvironment and promoting immune escape of HCC cells.
It has been reported that SPP1 is highly expressed in human lung adenocarcinoma tissues and in TAMs. Lung cancer cells induce M2 polarization of macrophages through SPP1 [41]. SPP1 knockdown in macrophages mitigates lung cancer progression and activates T cells [41]. Another study has indicated that the expression of SPP1 is highly upregulated in both murine and human glioma-associated microglia/macrophages and is associated with poor prognosis in human glioblastoma multiforme [42]. Thus, we reanalysed the scRNA-seq data of HCC tissues to identify the expression level of SPP1 in different cells in the TME. The results indicated that SPP1 was highly expressed in HCC tumour cells and TAMs but expressed at low levels in other cell types in the TME. LR analysis of single-cell RNA-seq data indicated that SPP1/CD44 and SPP1/PTGER4 were paired as ligands and receptors between SPP1 + malignant cells and TAMs in HCC. The ligand pair relationship was also validated in other tumour types [41, 43, 44].
Thus, we demonstrated that the expression of SPP1 was regulated by a DNA methylation-controlling pattern in HCC tissues and that the expression of SPP1 was higher in HCC tissues than in normal cells. SPP1 may play several roles in HCC tumour progression, such as regulating migration capacity and colony formation ability. Importantly, SPP1 can also reshape the tumour microenvironment by acting as a ligand to interact with TAMs and contributing to M2 polarization.
There are some limitations to our study. First, the prognostic robustness and clinical usefulness of the EIRS scoring system need further validation in larger prospective trials. Second, we were not able to perform in vivo analysis for the validation of SPP1-CD44 pair from single-cell transcriptome data, which required further study.
In summary, we determined inter- and intratumour heterogeneity in HCC through multiomics analysis. By elucidating the TME of HCC, we identified the factors associated with the prognosis of HCC patients. A prognostic prediction model was constructed and demonstrated potential clinical value that may help develop individualized immunotherapeutic strategies for HCC patients. Finally, we confirmed the prognostic value of SPP1 and the potential role of HCC-secreted SPP1 in TME of HCC patients.
Supplementary information
Acknowledgements
We would like to thank Dr. Yanfang Wang for helpful discussions.
Author contributions
X.B. and Y.Z. supervised the project. L.L. and X.B. designed the workflow. X.B. performed the bioinformatic analysis. W.F. and P.Z. led the study. R.Z. and J.D. performed the wet experiment. X.B., R.Z. and L.L. wrote the first draft. X.D., X.Z., H.Z., X.Z., Q.F., Z.T., W.F. and P.Z. reviewed the manuscript. All authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Funding
This research was supported by the Major Scientific Project of Zhejiang Province (LY21H160029) and the National Natural Science Foundation of China Program (Grant No. 81602128).
Data availability
All presented data in this study are available from the corresponding author upon reasonable request. The public data used in this study can be found in TCGA and GEO database.
Declarations
Conflict of interest
The authors declare that there are no conflicts of interest.
Consent for publication
All authors reviewed and approved the manuscript.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Lulu Liu, Ruyi Zhang, Jingwen Deng have contributed equally to this work.
Contributor Information
Yi Zheng, Email: oncologist@zju.edu.cn.
Xuanwen Bao, Email: baoxuanwen@gmail.com.
References
- 1.Center MM, Jemal A. International trends in liver cancer incidence rates. Cancer Epidemiol Prev Biomark. 2011;20(11):2362–2368. doi: 10.1158/1055-9965.EPI-11-0643. [DOI] [PubMed] [Google Scholar]
- 2.Greten TF, Sangro B. Targets for immunotherapy of liver cancer. J Hepatol. 2018;68(1):157–166. doi: 10.1016/j.jhep.2017.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elsegood CL, Tirnitz-Parker JE, Olynyk JK, Yeoh GC. Immune checkpoint inhibition: prospects for prevention and therapy of hepatocellular carcinoma. Clini Trans Immunol. 2017;6(11):e161. doi: 10.1038/cti.2017.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu L, Jiang J, Zhan M, Zhang H, Wang QT, Sun SN, et al. (2020) Targeting neoantigens in hepatocellular carcinoma for immunotherapy: A futile strategy? Hepatology 73(1):414–421 [DOI] [PubMed]
- 5.Löffler MW, Mohr C, Bichmann L, Freudenmann LK, Walzer M, Schroeder CM, et al. Multi-omics discovery of exome-derived neoantigens in hepatocellular carcinoma. Genom Med. 2019;11(1):1–16. doi: 10.1186/s13073-019-0636-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bao X, Zhang H, Wu W, Cheng S, Dai X, Zhu X, et al. Analysis of the molecular nature associated with microsatellite status in colon cancer identifies clinical implications for immunotherapy. J Immunother Cancer. 2020;8(2):e001437. doi: 10.1136/jitc-2020-001437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bao X, Shi R, Zhao T, Wang Y, Anastasov N, Rosemann M, et al. Integrated analysis of single-cell RNA-seq and bulk RNA-seq unravels tumour heterogeneity plus M2-like tumour-associated macrophage infiltration and aggressiveness in TNBC. Cancer Immunol Immunother. 2020;70:189–202. doi: 10.1007/s00262-020-02669-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bao X, Shi R, Zhao T, Wang Y. Mast cell-based molecular subtypes and signature associated with clinical outcome in early-stage lung adenocarcinoma. Mol Oncol. 2020;14(5):917–932. doi: 10.1002/1878-0261.12670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bao X, Shi R, Zhao T, Wang Y (2020) Immune landscape and a novel immunotherapy-related gene signature associated with clinical outcome in early-stage lung adenocarcinoma. J Mol Med 98:805–818 [DOI] [PMC free article] [PubMed]
- 10.Dai X, Guo Y, Hu Y, Bao X, Zhu X, Fu Q, Zhang H, Tong Z, Liu L, Zheng Y, Zhao P, Fang W (2021) Immunotherapy for targeting cancer stem cells in hepatocellular carcinoma. Theranostics 11(7):3489–3501. 10.7150/thno.54648 [DOI] [PMC free article] [PubMed]
- 11.Bao X, Shi R, Zhao T, et al (2021) Integrated analysis of single-cell RNA-seq and bulk RNA-seq unravels tumour heterogeneity plus M2-like tumour-associated macrophage infiltration and aggressiveness in TNBC[J]. Cancer Immunol Immunotherapy 70(1):189–202 [DOI] [PMC free article] [PubMed]
- 12.Bruni D, Angell HK, Galon J. The immune contexture and Immunoscore in cancer prognosis and therapeutic efficacy. Nat Rev Cancer. 2020;20:662–680. doi: 10.1038/s41568-020-0285-7. [DOI] [PubMed] [Google Scholar]
- 13.Yoshihara K, Shahmoradgoli M, Martínez E, Vegesna R, Kim H, Torres-Garcia W, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4(1):1–11. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen B, Khodadoust MS, Liu CL, Newman AM, Alizadeh AA. Profiling tumor infiltrating immune cells with CIBERSORT. Cancer Systems Biology: Springer; 2018. pp. 243–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14(1):7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu G, Wang L-G, Han Y, He Q-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics J Integr Biol. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wickham H (2016) ggplot2: elegant graphics for data analysis: Springer
- 18.Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22(3):568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mayakonda A, Lin D-C, Assenov Y, Plass C, Koeffler HP. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018;28(11):1747–1756. doi: 10.1101/gr.239244.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smyth GK. Limma: linear models for microarray data. Bioinformatics and computational biology solutions using R and Bioconductor: Springer; 2005. pp. 397–420. [Google Scholar]
- 21.Hastie T, Qian J. Glmnet vignette. Retriev June. 2016;2014(9):1–30. [Google Scholar]
- 22.Wolf FA, Angerer P, Theis FJ. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018;19(1):15. doi: 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Efremova M, Vento-Tormo M, Teichmann SA, Vento-Tormo R. Cell PhoneDB v2. 0: inferring cell-cell communication from combined expression of multi-subunit receptor-ligand complexes. BioRxiv. 2019;563:680926. doi: 10.1038/s41596-020-0292-x. [DOI] [PubMed] [Google Scholar]
- 24.Shi R, Bao X, Sun J, Lu S, Belka C, Li M (2020) Tumor microenvironment characterization in head and neck squamous carcinoma reveals distinct genomic alterations and clinical outcomes. Clin Trans Med 10(5):e187 [DOI] [PMC free article] [PubMed]
- 25.Bao X, Shi R, Zhang K, Xin S, Li X, Zhao Y, et al. Immune landscape of invasive ductal carcinoma tumour microenvironment identifies a prognostic and immunotherapeutically relevant gene signature. Front Oncol. 2019;9:903. doi: 10.3389/fonc.2019.00903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pages F, Galon J, Dieu-Nosjean M, Tartour E, Sautes-Fridman C, Fridman W. Immune infiltration in human tumors: a prognostic factor that should not be ignored. Oncogene. 2010;29(8):1093–1102. doi: 10.1038/onc.2009.416. [DOI] [PubMed] [Google Scholar]
- 27.Galon J, Angell HK, Bedognetti D, Marincola FM. The continuum of cancer immunosurveillance: prognostic, predictive, and mechanistic signatures. Immunity. 2013;39(1):11–26. doi: 10.1016/j.immuni.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 28.Melero I, Rouzaut A, Motz GT, Coukos G. T-cell and NK-cell infiltration into solid tumors: a key limiting factor for efficacious cancer immunotherapy. Cancer Discov. 2014;4(5):522–526. doi: 10.1158/2159-8290.CD-13-0985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bronger H, Singer J, Windmüller C, Reuning U, Zech D, Delbridge C, et al. CXCL9 and CXCL10 predict survival and are regulated by cyclooxygenase inhibition in advanced serous ovarian cancer. Br J Cancer. 2016;115(5):553–563. doi: 10.1038/bjc.2016.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohta M, Tanaka F, Yamaguchi H, Sadanaga N, Inoue H, Mori M. The high expression of Fractalkine results in a better prognosis for colorectal cancer patients. Int J Oncol. 2005;26(1):41–47. [PubMed] [Google Scholar]
- 31.Ding Q, Lu P, Xia Y, Ding S, Fan Y, Li X, et al. CXCL9: evidence and contradictions for its role in tumor progression. Cancer Med. 2016;5(11):3246–3259. doi: 10.1002/cam4.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Chaisemartin L, Goc J, Damotte D, Validire P, Magdeleinat P, Alifano M, et al. Characterization of chemokines and adhesion molecules associated with T cell presence in tertiary lymphoid structures in human lung cancer. Can Res. 2011;71(20):6391–6399. doi: 10.1158/0008-5472.CAN-11-0952. [DOI] [PubMed] [Google Scholar]
- 33.Crotty S. T follicular helper cell differentiation, function, and roles in disease. Immunity. 2014;41(4):529–542. doi: 10.1016/j.immuni.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kowalczuk O, Burzykowski T, Niklinska WE, Kozlowski M, Chyczewski L, Niklinski J. CXCL5 as a potential novel prognostic factor in early stage non-small cell lung cancer: results of a study of expression levels of 23 genes. Tumor Biol. 2014;35(5):4619–4628. doi: 10.1007/s13277-014-1605-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou S-L, Dai Z, Zhou Z-J, Chen Q, Wang Z, Xiao Y-S, et al. CXCL5 contributes to tumor metastasis and recurrence of intrahepatic cholangiocarcinoma by recruiting infiltrative intratumoral neutrophils. Carcinogenesis. 2014;35(3):597–605. doi: 10.1093/carcin/bgt397. [DOI] [PubMed] [Google Scholar]
- 36.Luke JJ, Bao R, Sweis RF, Spranger S, Gajewski TF. WNT/β-catenin pathway activation correlates with immune exclusion across human cancers. Clin Cancer Res. 2019;25(10):3074–3083. doi: 10.1158/1078-0432.CCR-18-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fedarko NS, Jain A, Karadag A, Fisher LW. Three small integrin-binding ligand N-linked glycoproteins (SIBLINGs) bind and activate specific matrix metalloproteinases. FASEB J. 2004;18(6):734–736. doi: 10.1096/fj.03-0966fje. [DOI] [PubMed] [Google Scholar]
- 38.Dalla-Torre CA, Yoshimoto M, Lee C-H, Joshua AM, de Toledo SR, Petrilli AS, et al. Effects of THBS3, SPARC and SPP1 expression on biological behavior and survival in patients with osteosarcoma. BMC Cancer. 2006;6(1):1–10. doi: 10.1186/1471-2407-6-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sharon Y, Raz Y, Cohen N, Ben-Shmuel A, Schwartz H, Geiger T, et al. Tumor-derived osteopontin reprograms normal mammary fibroblasts to promote inflammation and tumor growth in breast cancer. Can Res. 2015;75(6):963–973. doi: 10.1158/0008-5472.CAN-14-1990. [DOI] [PubMed] [Google Scholar]
- 40.Anborgh PH, Caria LB, Chambers AF, Tuck AB, Stitt LW, Brackstone M. Role of plasma osteopontin as a biomarker in locally advanced breast cancer. Am J Transl Res. 2015;7(4):723. [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y, Du W, Chen Z, Xiang C. Upregulation of PD-L1 by SPP1 mediates macrophage polarization and facilitates immune escape in lung adenocarcinoma. Exp Cell Res. 2017;359(2):449–457. doi: 10.1016/j.yexcr.2017.08.028. [DOI] [PubMed] [Google Scholar]
- 42.Szulzewsky F, Pelz A, Feng X, Synowitz M, Markovic D, Langmann T, et al. Glioma-associated microglia/macrophages display an expression profile different from M1 and M2 polarization and highly express Gpnmb and Spp1. PLoS ONE. 2015;10(2):e0116644. doi: 10.1371/journal.pone.0116644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kijewska M, Kocyk M, Kloss M, Stepniak K, Korwek Z, Polakowska R, et al. The embryonic type of SPP1 transcriptional regulation is re-activated in glioblastoma. Oncotarget. 2017;8(10):16340. doi: 10.18632/oncotarget.14092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klement JD, Paschall AV, Redd PS, Ibrahim ML, Lu C, Yang D, et al. An osteopontin/CD44 immune checkpoint controls CD8+ T cell activation and tumor immune evasion. J Clin Investig. 2018;128(12):5549–5560. doi: 10.1172/JCI123360. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All presented data in this study are available from the corresponding author upon reasonable request. The public data used in this study can be found in TCGA and GEO database.