Abstract
Purpose
Identification of reliable postoperative indicators for accurately evaluating prognosis of clear cell renal cell carcinoma (ccRCC) patients remains an important clinical issue. This study determined the prognostic value of UBR5 expression in ccRCC patients by combining with CD163+ tumor-associated macrophages (TAMs) and the established clinical parameters.
Methods
The expression of UBR5 was analyzed in ccRCC patients from TCGA databases. A total of 310 ccRCC patients were randomly divided into the training and validation cohorts at a 3:2 or 1:1 ratio, and immunohistochemistry (IHC) and statistical analyses were performed to examine the prognostic value of UBR5 and CD163+ TAMs.
Results
UBR5 expression was commonly downregulated in human ccRCC specimens, which was associated with TNM stage, SSIGN, WHO/ISUP Grading and poor prognosis of ccRCC patients. In addition, UBR5 expression was negatively correlated with CD163 expression (a TAM marker) in ccRCC tissues, and combining expressions of UBR5 and CD163 better predicted worse overall survival and progression-free survival of ccRCC patients. Even after multivariable adjustment, UBR5, CD163, TNM stage and SSIGN appeared to be independent risk factors. By time-dependent c-index analysis, the integration of intratumoral UBR5 and CD163 achieved higher c-index value than UBR5, CD163, TNM stage or SSIGN alone in predicting ccRCC patients’ prognosis. Moreover, the incorporation of both UBR5 and CD163 into the clinical indicators TNM stage or SSIGN exhibited highest c-index value.
Conclusions
Integrating intratumoral UBR5 and CD163+ TAMs with the current clinical parameters achieves better accuracy in predicting ccRCC patients’ postoperative prognosis.
Electronic supplementary material
The online version of this article (10.1007/s00262-021-02885-9) contains supplementary material, which is available to authorized users.
Keywords: Clear cell renal cell carcinoma, UBR5, CD163, Tumor-associated macrophages, Prognosis
Introduction
Kidney cancer in the USA is the 6th and 8th most frequent cancer type in men and women, respectively [1]. And clear cell renal cell carcinoma (ccRCC) is the most common type of kidney cancer. Even though surgical resection such as radical or partial nephrectomy affords a curative treatment for early-stage RCC, postoperative recurrence can occur in approximately 40% of RCC with high-risk features [2]. In the clinic, the specific surgical procedures or monitoring and follow-up of ccRCC patients after surgery depend largely on clinicopathological parameters including TNM stage [3]. In addition, the stage-scoring system SSIGN (for stage, size, grade and necrosis) classifies ccRCC patients into subgroups with different risks, which is helpful for predicting postoperative prognosis [4]. Moreover, oncogenes, tumor suppressive genes and cancer stem-like cell (CSC) markers can be applied to predict postoperative prognoses of patients [5, 6]. However, these previous researches mainly focused on the characteristics of tumor cells.
Recently, many studies have suggested that indicators in the tumor microenvironment (TME) may more accurately predict postoperative outcomes of cancer patients [7]. TME has garnered much attention by scientists and clinicians due to its vital role in facilitating tumor progression [8]. Stromal markers in TME are associated with risk classification of cancer patients [9]. A growing number of studies including ours have reported that TAMs, one of the important components of TME, constitute a special niche with tumors through their interaction and promote the progression and drug resistance of tumors [10]. Little has been reported about whether integrating intratumoral markers and TAMs may achieve better predictive value for postoperative prognosis of ccRCC patients. Since CD163+ TAMs are important based on prognostic associations in cancers [11] and a preliminary bioinformatics analysis of TCGA-derived gene-associated survival data that we performed suggested that genomic alterations of a novel molecule, ubiquitin–protein ligase E3 component n-recognin 5 (UBR5), may be linked with poor ccRCC patient prognosis. Thus, in the present study, we comprehensively investigated whether combining with UBR5, CD163+ TAMs and the current clinical parameters could serve as a useful biomarker for postoperative prognosis of ccRCC patients.
UBR5, one of the 29 members of the HECT-domain E3 ubiquitin–protein ligase family, has been reported to be necessary for development and maintenance of pluripotency [12]. UBR5 exhibits a crucial role in homeostasis of ubiquitin-mediated signaling after DNA damage [13]. UBR5 has been implicated in various types of cancers [14]. Data from the Cancer Genome Atlas (TCGA) indicate that UBR5 gene amplification is a familiar alteration in various types of malignant tumors. Studies have shown that UBR5 promotes the growth and progression of malignant tumors including colon cancer, breast cancer, ovarian cancer [15–17]. On the other hand, UBR5 missense mutations also occur in some cancers such as mantle cell lymphoma with recurrent UBR5 mutations [18]. However, the prognostic value of UBR5 in ccRCC remains unknown.
Materials and methods
Collection and analysis of public databases
Datasets from the Cancer Genome Atlas (TCGA) were downloaded from National Cancer Institute GDC Data Portal (https://portal.gdc.cancer.gov) using Firebrowse R package. We selected KIRC dataset from all TCGA cohorts and filtered out 533 samples from ccRCC patients with bar codes and gene mRNA expression profiles.
Patients and specimens
A total of 310 ccRCC patients who underwent either nephron-sparing surgery or radical nephrectomy at Changhai Hospital between 2010 and 2014 were retrospectively recruited in our study. Patients’ clinicopathologic data for age, gender, WHO/ISUP Grading, TNM stage, SSIGN and clinical outcomes were collected. Paired tumor tissues and adjacent tissues of 310 ccRCC patients were obtained. Another 60 paired ccRCC specimens were used for real-time PCR analysis and 16 paired ccRCC samples for Western blot analysis. This study followed the recommendations for prognostic studies of tumor biomarkers (REMARK) [19]. All experiments were approved by the institutional ethical review boards from the hospital, and all written informed consents were obtained from the ccRCC patients. The clinical characteristics of the ccRCC patients are summarized in Supplementary Table S1.
Real-time polymerase chain reaction (real-time PCR)
The real-time PCR assay was performed as we reported previously [10]. The primer sequences used in the study are listed as follows: UBR5 (forward primer, 5`-CCAGACAGATTGGAATTGGGTAA-3`, and reverse primer, 5`-CATGGAGAGTCGCTTGTCCT-3`), GAPDH (forward primer, 5′-GGAAGGTGAAGGTCGGAGT-3′, and reverse primer, 5′-CCTGGAAGATGGTGATGGG-3′). All results were normalized to the expression of GAPDH and fold change relative to the mean value was determined by 2−△△Ct.
Western blot analysis
Western blot was done as we previously reported [10]. The following primary antibodies were used: rabbit anti-UBR5 antibody (NB100-1591, Novus Biologicals, Littleton, CO, USA) and rabbit anti-GAPDH antibody (#2118S, Cell Signaling Technology, Danvers, MA, USA).
Immunohistochemistry (IHC)
IHC assay was carried out as described in our previous study [20]. The following primary antibodies were used: rabbit anti-UBR5 antibody (NB100-1591, Novus Biologicals) and rabbit anti-CD163 (#93498S, Cell Signaling Technology). UBR5 staining was scored by an H-score which was generated as a score of 0–3 intensity multiplied by the percentage of positive cells (range 0-300) as described in our previous study [20]. For CD163+ TAMs, the counts of all cells were defined as the number of nucleated stained cells per field and are presented as the density (cells/mm2). The scores of each patient were calculated by two independent and experienced pathologists in double blind way.
Cell culture
ccRCC cell lines used in the present study were obtained from the Cell Bank of the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China) in 2018. HK-2 cells were maintained in DMEM (Gibco, Waltham, MA, USA). ACHN cells were cultured in MEM (Gibco), and 786-O, 769-P and OS-RC-2 cells were maintained in RPMI-1640 medium (Gibco). The culture media of all cell lines were supplemented with fetal bovine serum (FBS, 10%, Gibco) and 1% penicillin/streptomycin (Gibco). ccRCC cell lines were cultured at 37 °C in 5% Co2. Sunitinib-resistant 786-O and 769-P cell lines (786-O-SR and 769-P-SR) were established as described in our previous study [3], and the established 786-O-SR and 769-P-SR cell lines were maintained in RPMI-1640 medium with 10% (v/v) FBS and 10 µM sunitinib.
Statistical analysis
Numerical data were expressed as the mean ± S.D. Two-tailed Student’s t test or Wilcoxon test was conducted for continuous variables. Chi squared test or Fisher’s exact test was conducted for categorical variables. Survival curves were plotted using Kaplan–Meier analysis and compared via log-rank test. Difference was considered significant at p < 0.05. Prognostic accuracy of the UBR5 classifier and other prognostic indicators was indicated by Harrell’s concordance index using “rms” package(c-index). All the statistical analyses were performed using R software (version 3.5.2).
Results
UBR5 expression negatively associates with tumor stage and sunitinib resistance of ccRCC
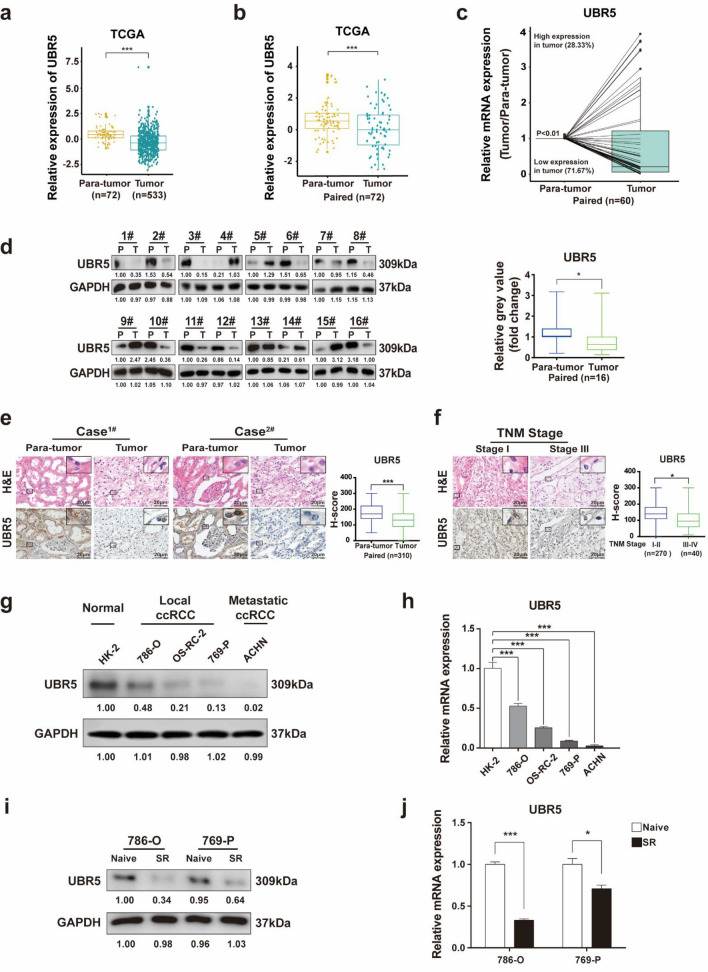
UBR5 expression was first analyzed in datasets from the Cancer Genome Atlas (TCGA), which showed that UBR5 was decreased in ccRCC samples (Fig. 1a, b). In addition, the mRNA and protein expressions of UBR5 were lower in ccRCC specimens than that in adjacent renal samples (Fig. 1c, d). Consistent with the above results, a majority of ccRCC displayed decreased UBR5 expression as examined immunohistochemistry (IHC), compared with paired adjacent tissues (Fig. 1e). These results indicate that UBR5 is usually downregulated in human ccRCC specimens.
Fig. 1.
Low UBR5 expression negatively associates with tumor stage and sunitinib resistance of ccRCC. a The expression of UBR5 in human ccRCC tissues (n = 533) and normal renal tissues (n = 72) from TCGA datasets was compared. b The expression of UBR5 was analyzed the same way as described above between tumor tissues (n = 72) and paired normal tissues (n = 72) in human ccRCC from TCGA datasets. c Real-time PCR was performed to assess the mRNA expression of UBR5 in tumor samples and their paired adjacent renal tissues from ccRCC patients (n = 60). d Western blot analysis was performed to measure the protein expression of UBR5 in ccRCC specimens and their matched adjacent renal tissues (n = 16). e Representative images of hematoxylin and eosin (H&E) staining and immunohistochemistry (IHC) staining for UBR5 in ccRCC tissues and the adjacent tissues of tissue microarrays are presented (n = 310; scale bar = 20 μm). f Representative images of H&E and IHC staining for UBR5 in ccRCC tissues with different TNM stages are shown (scale bar = 20 μm). g–h Western blot (g) or Real-time PCR (h) was performed to analyze the protein expression of UBR5 in a normal cell line (HK-2), local and metastatic ccRCC cell lines. i–j Western blot (i) or real-time PCR (j) was applied to detect the expression of UBR5 in 786-O-SR and 769-P-SR cells. All p values are defined as: *p < 0.05; **p < 0.01 and ***p < 0.001 (SR: sunitinib-resistant)
Given the decreased expression of UBR5 in ccRCC, we postulated that UBR5 might be negatively associated with clinicopathological features of ccRCC. According to the IHC assay, UBR5 expression was lower in ccRCC with TNM stage III or IV than that in stage I or II (Fig. 1f). In addition, higher UBR5 expression was observed in the metastatic ccRCC cell line ACHN than that in local ccRCC cell lines (Fig. 1g, h). Furthermore, the protein and mRNA expressions of UBR5 were both lower in sunitinib-resistant ccRCC cell lines (786-O–SR and 769-P-SR, as we described previously [3, 20]) than that in naïve ccRCC cells (Fig. 1i, j). These data demonstrate that decreased UBR5 expression in specimens is correlated with tumor stage and sunitinib resistance of ccRCC.
Low UBR5 expression is predictive of unfavorable clinicopathological characteristics and poor postoperative prognosis of ccRCC patients
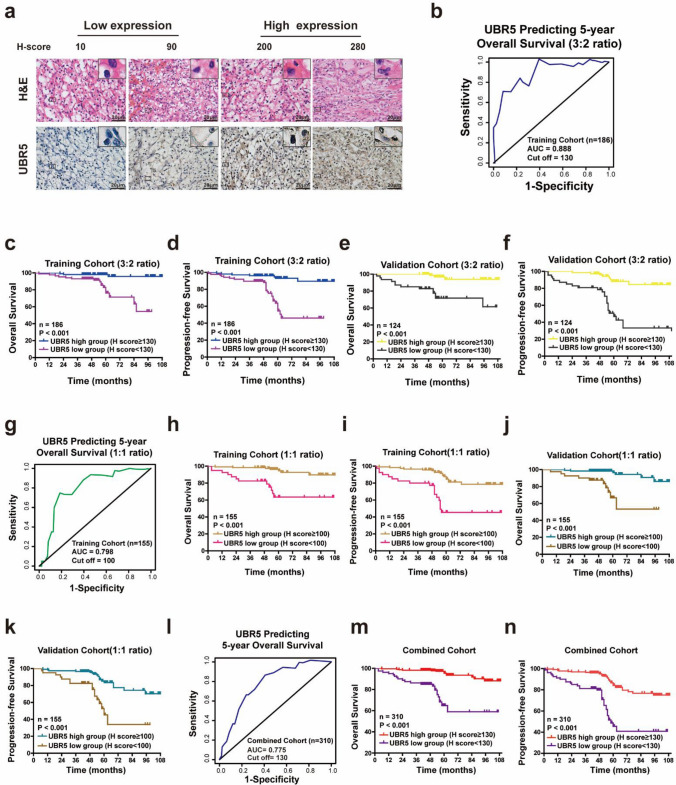
To determine the relationship between UBR5 expression and clinicopathological features or prognosis of ccRCC patients, specimens from 310 ccRCC patients were stochastically divided into the training cohort (n = 186) and validation cohort (n = 124) at a 3:2 ratio. First, IHC was performed to determine the expression of UBR5 (Fig. 2a), and the optimal cutoff value for dividing ccRCC patients was determined by time-dependent ROC analysis in the training cohort, which showed that the best cutoff value was 130 (Fig. 2b). By using this cutoff value, ccRCC patients in the training cohort were divided into UBR5high and UBR5low groups. As shown in Table 1, low expression of UBR5 predicted unfavorable clinicopathological features such as higher WHO/ISUP Grading, TNM stage and SSIGN score. Furthermore, Kaplan–Meier survival analysis demonstrated that UBR5low subgroup presented worse OS and progression-free survival (PFS), compared with that of UBR5high (Fig. 2c, d). We then employed the validation cohort to corroborate above findings using the cutoff value derived from the training cohort (Fig. 2e, f; Supplementary Table S2).
Fig. 2.
Low UBR5 expression is predictive of unfavorable clinicopathological characteristics and poor postoperative prognosis of ccRCC patients. a Representative images of H&E and IHC staining of UBR5 in ccRCC specimens from the training cohort are presented (scale bar = 20 µm). b A time-dependent receiver operating characteristics (ROC) analysis was used to determine the optimum cutoff value of UBR5 to predict 5-year overall survival (OS) in the training cohort (according to a 3:2 ratio). c–f Kaplan–Meier curves for OS and progression-free survival (PFS) of ccRCC patients were analyzed based on the UBR5 expression in the randomized training cohort (c, d) and the validation cohort (e, f) (3:2 ratio). g A time-dependent ROC analysis was used to calculate the optimum cutoff value of UBR5 to predict 5-year OS in the training cohort (1:1 ratio). h–k Kaplan–Meier curves for OS and PFS of ccRCC patients were analyzed according to UBR5 expression in the randomized training cohort (h, i) and validation cohort (j, k) according to a 1:1 ratio. l A time-dependent ROC analysis was used to examine the optimum cutoff value of UBR5 to predict 5-year OS in the combined cohort. m–n Kaplan–Meier curves for OS and PFS of ccRCC patients were analyzed according to UBR5 expression in the combined cohort. All p values are defined as: *p < 0.05; **p < 0.01 and ***p < 0.001
Table 1.
Correlation between UBR5 expression and clinicopathologic characteristics of patients with clear cell renal cell carcinoma in the training cohort (n = 186) (3:2 ratio)
| Characteristic | UBR5 | Sum (186) | p** value | |
|---|---|---|---|---|
| Low expression* (n = 86) | High expression* (n = 100) | |||
| Age | 0.620 | |||
| <60 | 52 | 64 | 116 | |
| ≥60 | 34 | 36 | 70 | |
| Gender | 0.890 | |||
| Male | 61 | 70 | 131 | |
| Female | 25 | 30 | 55 | |
| WHO/ISUP Grading | 0.013 | |||
| I–II | 59 | 84 | 143 | |
| III–IV | 27 | 16 | 43 | |
| TNM stage | 0.021 | |||
| I–II | 68 | 91 | 159 | |
| III | 18 | 9 | 27 | |
| SSIGN | 0.028 | |||
| 0-4 | 75 | 96 | 171 | |
| ≥5 | 11 | 4 | 15 | |
*UBR5Low expression: H-scores of UBR5 < 130; UBR5High expression: H-scores of UBR5 ≥ 130
**Statistical significance was calculated by Chi squared test or Fisher’s exact test for categorical/binary measures
P-value for this item is less than 0.05 and there is a statistical difference between the corresponding two groups of data
To validate above results, the ccRCC patients were also stochastically divided into the training cohort (n = 155) and validation cohort (n = 155) at a 1:1 ratio. As shown in Supplementary Tables S3-4, low expression level of UBR5 predicted unfavorable clinicopathological features and worse prognosis of ccRCC patients in the training and validation cohorts (Fig. 2g–k). Moreover, the similar findings were confirmed in the combined cohort (Supplementary Table S5, Fig. 2l–n). Thus, UBR5 expression negatively correlates with disease progression and short survival of ccRCC patients.
Combining UBR5 and CD163+ TAMs better predicts prognosis of ccRCC patients
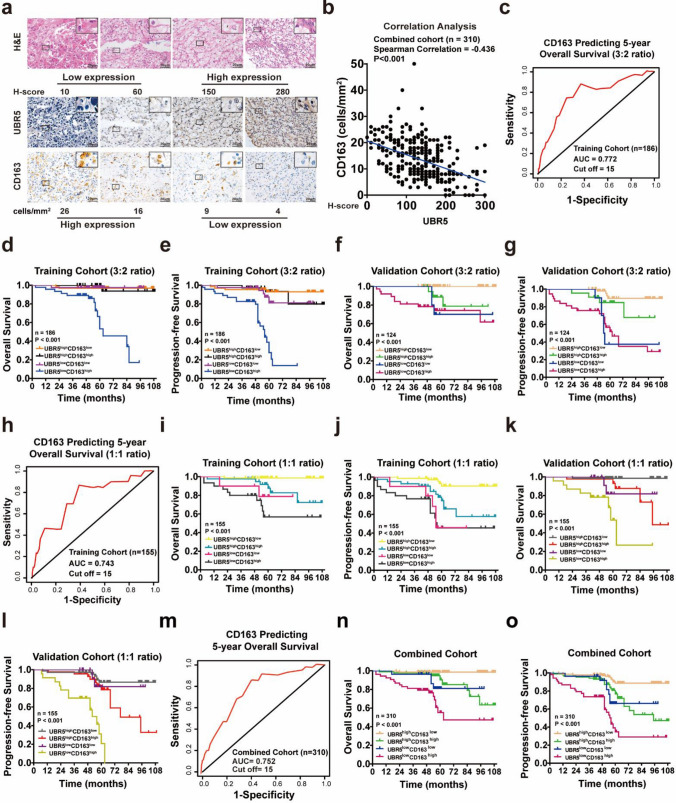
Given others and our previous studies have demonstrated that combining expressions of intratumoral markers and TAMs better predicts prognosis of tumor patients [10], we next sought to determine the predictive value of concomitant expression of UBR5 and CD163+ TAMs in ccRCC patients (Supplementary Fig. S1). First, the correlation analysis demonstrated that UBR5 expression was negatively correlated with CD163 in ccRCC (Fig. 3a, b). Secondly, the ccRCC patients were first stochastically divided into the training cohort (n = 186) and the validation cohort (n = 124) at a 3:2 ratio, and time-dependent ROC analysis of CD163 in the training cohort exhibited 15 (Fig. 3c). According to the optimal cutoff of UBR5 (Fig. 2b) and CD163 (Fig. 3c), ccRCC patients in the training cohort were divided into four groups. As shown in Table 2 and Supplementary Table S6, UBR5lowCD163 high group presented unfavorable clinicopathological features and worst prognosis (Fig. 3d–g). These findings were confirmed in the training cohort and validation cohort, which were randomly divided at a 1:1 ratio (Fig. 3h–l; Supplementary Tables S7-8). Moreover, data from the combined cohort also verified the above results (Fig. 3m-o; Supplementary Table S9). Thus, combining low UBR5 expression and more infiltration of CD163+ TAMs predicts poor prognosis of ccRCC patients.
Fig. 3.
Combining UBR5 and CD163+ TAMs better predicts prognosis of ccRCC patients. a Representative images of H&E and IHC staining of UBR5 and CD163 in ccRCC specimens are shown (n = 310; scale bar = 20 µm). b The correlation analysis between UBR5 and CD163 in ccRCC specimens is shown. c A time-dependent ROC analysis was applied to calculate the optimum cutoff value of CD163 to predict 5-year OS in the training cohort (3:2 ratio). d–g Kaplan–Meier curves for OS and PFS of ccRCC patients were analyzed based on the expressions of UBR5 and CD163 in the training cohort (d, e) and the validation cohort (f, g) (3:2 ratio). h A time-dependent ROC analysis was used to examine the optimum cutoff value of CD163 to predict 5-year OS in the training cohort (1:1 ratio). i–l Kaplan–Meier curves for OS and PFS of ccRCC patients were analyzed according to expressions of UBR5 and CD163 in the randomized training cohort (i, j) and validation cohort (k, l) (1:1 ratio). m A time-dependent ROC analysis was used to examine the optimum cutoff value of CD163 to predict 5-year OS in the combined cohort. n–o Kaplan–Meier curves for OS and PFS of ccRCC patients were analyzed based on the expressions of UBR5 and CD163 in the combined cohort. All p values are defined as: *p < 0.05; **p < 0.01 and ***p < 0.001
Table 2.
Correlation between expressions of UBR5, CD163 and clinicopathologic characteristics of patients with clear cell renal cell carcinoma in the training cohort (n = 186) (3:2 ratio)
| Characteristic | UBR5/CD163 expression* | |||||
|---|---|---|---|---|---|---|
| UBR5high CD163low (n = 65) |
UBR5high CD163high (n = 35) |
UBR5low CD163low (n = 40) |
UBR5low CD163high (n = 46) |
Sum (186) | p** value | |
| Age | 0.239 | |||||
| <60 | 44 | 20 | 28 | 24 | 116 | |
| ≥60 | 21 | 15 | 12 | 22 | 70 | |
| Gender | 0.194 | |||||
| Male | 41 | 29 | 27 | 34 | 131 | |
| Female | 24 | 6 | 13 | 12 | 55 | |
| WHO/ISUP Grading | 0.022 | |||||
| I–II | 58 | 26 | 29 | 30 | 143 | |
| III–IV | 7 | 9 | 11 | 16 | 43 | |
| TNM stage | 0.003 | |||||
| I–II | 64 | 27 | 32 | 36 | 159 | |
| III | 1 | 8 | 8 | 10 | 27 | |
| SSIGN | 0.004* | |||||
| 0-4 | 63 | 33 | 39 | 36 | 171 | |
| ≥5 | 2 | 2 | 1 | 10 | 15 | |
*UBR5low expression: H-scores of UBR5 < 130; UBR5high expression: H-scores of UBR5 ≥ 130; CD163low expression: Density of CD163 < 15 cells/mm2; CD163high expression: Density of CD163 ≥ 15 cells/mm2
**Statistical significance was calculated by Chi squared test or Fisher’s exact test for categorical/binary measures and ANOVA for continuous measures
P-value for this item is less than 0.05 and there is a statistical difference between the corresponding two groups of data
Combining UBR5, CD163+ TAMs and the currently established indicators yields superior prognostic accuracy in predicting ccRCC patients’ postoperative prognosis
To appraise the clinical significance of UBR5 and CD163+ TAMs in postoperative ccRCC patients, univariate and multivariate Cox regression analyses were employed, which showed that UBR5, CD163, TNM stage and SSIGN respectively appeared to be an independent risk factor in the training and validation cohorts (whether according to 3:2 ratio or 1:1 ratio) or in the combined cohort (Supplementary Tables S10-15). Next, by using time-dependent concordance index (C-index) analysis, the higher c-index value of combining UBR5 and CD163 was observed in OS and PFS of ccRCC patients, compared with that of UBR5, CD163 or other currently molecular markers [21, 22] such as EZH2 or CXCR4 alone (Table 3; Supplementary Tables S14-19; Supplementary Fig. S2). In addition, combining UBR5 and CD163 exhibited higher c-index value than TNM stage or SSIGN score alone (Table 3; Supplementary Tables S16-17). Moreover, the incorporation of both UBR5 and CD163 into the clinical indicators TNM stage and SSIGN score exhibited highest c-index value than other groups (Table 3 and Supplementary Tables S16-17). Taken together, better prognostic accuracy could be completed by combining UBR5 and CD163+ TAMs with the current clinical indicators in predicting OS and PFS of ccRCC patients.
Table 3.
C-index analysis of the prognostic accuracy of UBR5, CD163 and other variables for overall survival and progression-free survival in the training and validation cohorts (3:2 ratio)
| C-index (95% CI) | Overall survival | Progression-free survival | ||
|---|---|---|---|---|
| Training cohort (n = 186) | Validation cohort (n = 124) | Training cohort (n = 186) | Validation cohort (n = 124) | |
| TNM stage | 0.677 (0.634–0.72) | 0.684 (0.646–0.722) | 0.639 (0.61–0.669) | 0.647 (0.622–0.672) |
| SSIGN | 0.676 (0.644–0.709) | 0.679 (0.65–0.707) | 0.682 (0.663–0.701) | 0.63 (0.612–0.648) |
| UBR5 | 0.705 (0.643–0.768) | 0.757 (0.694–0.82) | 0.701 (0.656–0.745) | 0.727 (0.682–0.771) |
| CD163 | 0.683 (0.621–0.745) | 0.698 (0.633–0.764) | 0.675 (0.631–0.719) | 0.646 (0.6–0.693) |
| UBR5 + CD163 | 0.767 (0.697–0.837) | 0.803 (0.732–0.874) | 0.767 (0.717–0.816) | 0.749 (0.699–0.8) |
| UBR5 + CD163 + TNM stage | 0.804 (0.734–0.875) | 0.851 (0.779–0.923) | 0.801 (0.751–0.851) | 0.802 (0.751–0.853) |
| UBR5 + CD163 + SSIGN | 0.801 (0.73–0.871) | 0.852 (0.78–0.923) | 0.84 (0.79–0.89) | 0.791 (0.74–0.842) |
Discussion
Though many prognostic biomarkers have been demonstrated to reflect the prognosis of postoperative ccRCC patients [23], they may not be applicable to clinical practice. The reason for this “dilemma” is that these markers only focus on the features of tumors but neglect the microenvironment. Our present study integrates UBR5 and CD163+ TAMs into a prognostic model with the clinical parameters, which exhibits the superiority in predicting ccRCC patients’ prognosis.
The biological role of UBR5 as either an oncogene or a tumor suppressor is based on the tumor type. UBR5 has been reported to exert oncogenic role in gallbladder, colorectal, breast and gastric cancers [14]. Additionally, colorectal cancer patients with high UBR5 mRNA levels, UBR5 gene amplification or high nuclear UBR5 protein levels had poor prognosis [24]. In contrast, loss-of-function mutations of UBR5 were found in mantle cell lymphoma [18], which suggests that UBR5 could play a tumor suppressive role in malignant cancers. Moreover, UBR5-mediated ubiquitination of oncogene SOX2 in esophageal cancer also suggests that UBR5 has a tumor suppressive role [25]. Meanwhile, data from TCGA have shown that there is a significant advantage for overall survival of breast cancer patients with normal UBR5 expression in contrast to that with UBR5 mutations [16]. Our present study demonstrates that UBR5 expression is lower in ccRCC specimens, and low UBR5 expression is associated with poor prognosis of ccRCC. The underlying mechanism for the apparently conflicting results in different cancers is presently unknown. Given its role in DNA damage repair and in the maintenance of cellular homeostasis [13], it is possible that sufficient UBR5 may be required for normal renal cell homeostasis and an insufficiency causes malignant changes in the kidney. Thus, the UBR5 down-expression or its genetic variants may indicate certain underlying abnormal development at the cellular or organismic level.
A large number of prognostic indicators including molecular and genomic prognostic factors including VEGF, p53 and mutations of PBRM1, BAP1 have been reported for ccRCC patients [26, 27], Additionally, integrated prognostic scoring systems such as SSIGN score have been used to predict ccRCC patients’ prognosis due to the limited prognostic value of TNM staging system. However, these indicators are based on ccRCC tissues and reflect only tumor-intrinsic properties. Given the trend that integrated multiple biomarkers might exhibit superiority in predicating postoperative prognosis of tumor patients [28], our present study demonstrates that combining intratumoral UBR5 expression, CD163+ TAMs and the established clinical indicators TNM stage or SSIGN exhibit superior accuracy in predicting ccRCC patients’ prognosis.
Some limitations remain in our study. Only postoperative ccRCC patients were employed in the present study, which could not determine whether the UBR5-CD163+ TAMs-TNM stage (or SSIGN) classifier serves as a potential TKI or immunotherapy response/resistance indicator for ccRCC patients. Our future studies will involve more ccRCC patients who have received TKI or immunotherapy, and enroll more ccRCC patients who have therapy resistance. Meanwhile, CD163+ TAMs in Classical Hodgkin Lymphoma was correlated with the immunosuppression of T lymphocytes [29], which may be involved in the observed unfavorable clinicopathological features of ccRCC patients associated with the high expression of CD163. In addition, Song et al. [30] found that UBR5 drove breast cancer cell growth particularly through inhibiting the cytotoxic response mediated by CD8+ T lymphocytes, the major subset of leukocytes in the tumor microenvironment. This suggests that UBR5 plays a profound role in the interaction between tumor cells and different kinds of immune cells. Thus, we will further examined the effects of UBR5 on the interaction of ccRCC cells and other immunocytes in future studies.
Conclusions
In the present study, we have demonstrated that integrating UBR5 expression and CD163+ TAMs with the existing clinical indicators presents obvious advantages compared to using these markers individually in predicting the postoperative value of ccRCC patients.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by the Top-level Clinical Discipline Project of Shanghai Pudong (PWYgf2018-03), National Natural Science Foundation of China (Nos. 81773154, 81772747, 81974391), Shanghai Natural Science Foundation (20ZR1449600), Pudong New Area Science and technology development fund special fund for people’s livelihood Research (medical and health) (PKJ2019-Y19), the Program of Shanghai Academic/Technology Research Leader (No. 19XD1405100), the Shanghai “Rising Stars of Medical Talent” Youth Development Program: Outstanding Youth Medical Talents (Xin-gang Cui), Meng Chao Talent Training Program-Cultivation of Leading Talents Reserve (Xin-gang Cui), and the Shanghai Medical Guidance (Chinese and Western Medicine) Science and Technology Support Project (No. 17411960200). We thank Dr. Aiping Zhang (Department of Urinary Surgery, Gongli Hospital, Shanghai, China) for her assistance in providing the clinical samples of ccRCC patients. We also thank Shanghai BioGenius biotech. Co., Ltd (China) for his BioGenius Cloud Computing Service and bioinformatics analysis.
Authors’ contributions
Chao Wang, Dong Zhuo, Jingcun Zheng, Xiaojing Ma and Xingang Cui designed the study; Chao Wang wrote the manuscript; Chao Wang, TianYu Hong, Guang Peng, Yuning Wang and Yongwei Yu analyzed the data, performed the experiments and statistical analysis; Jing Zhang provided the ccRCC patient samples; and Zhuo Dong, Jingcun Zheng, Xiaojing Ma and Xingang Cui supervised the study and reviewed the manuscript.
Declarations
Conflict of interest
All authors have no conflicts of interest to disclose. All authors have contributed significantly, and all authors are in agreement with the content of the manuscript.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Chao Wang, TianYu Hong, Yuning Wang, Guang Peng, and Yongwei Yu contributed equally to the study.
Contributor Information
Dong Zhuo, Email: whzhuo2008@sina.com.
Jingcun Zheng, Email: zhengjingc@sina.com.
Xiaojing Ma, Email: xim2002@med.cornell.edu.
Xingang Cui, Email: cuixingang@smmu.edu.cn.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics. CA Cancer J Clin. 2020;70(1):7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 2.Ridyard DG, Buller DM, Ristau BT. The current state of adjuvant therapy following surgery for high-risk renal cell carcinoma. Eur Urol Focus. 2019;5(6):935–938. doi: 10.1016/j.euf.2019.03.020. [DOI] [PubMed] [Google Scholar]
- 3.Wang C, Li Y, Chu C, et al. Gankyrin is a novel biomarker for disease progression and prognosis of patients with renal cell carcinoma. EBioMedicine. 2019;39:255–264. doi: 10.1016/j.ebiom.2018.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang M, Li X, Zhang J, et al. AHNAK2 is a novel prognostic marker and oncogenic protein for clear cell renal cell carcinoma. Theranostics. 2017;7(5):1100–1113. doi: 10.7150/thno.18198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li X, Ma X, Chen L, et al. Prognostic value of CD44 expression in renal cell carcinoma: a systematic review and meta-analysis. Sci Rep. 2015;5(1):13157. doi: 10.1038/srep13157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morihiro T, Kuroda S, Kanaya N, et al. PD-L1 expression combined with microsatellite instability/CD8 + tumor infiltrating lymphocytes as a useful prognostic biomarker in gastric cancer. Sci Rep. 2019;9(9):2045–2322. doi: 10.1038/s41598-019-41177-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fu H, Zhu Y, Wang Y, Liu Z, et al. Identification and validation of stromal immunotype predict survival and benefit from adjuvant chemotherapy in patients with muscle invasive bladder cancer. Clin Cancer Res. 2018;24(13):3069–3078. doi: 10.1158/1078-0432.CCR-17-2687. [DOI] [PubMed] [Google Scholar]
- 8.Becht E, de Reynies A, Giraldo NA, et al. Immune and stromal classification of colorectal cancer is associated with molecular subtypes and relevant for precision immunotherapy. Clin Cancer Res. 2016;22(16):4057–4066. doi: 10.1158/1078-0432.CCR-15-2879. [DOI] [PubMed] [Google Scholar]
- 9.Chittezhath M, Dhillon MK, Lim JY, et al. Molecular profiling reveals a tumor-promoting phenotype of monocytes and macrophages in human cancer progression. Immunity. 2014;41(5):815–829. doi: 10.1016/j.immuni.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 10.Huang H, Wang C, Liu F, et al. Reciprocal network between cancer stem-like cells and macrophages facilitates the progression and androgen deprivation therapy resistance of prostate cancer. Clin Cancer Res. 2018;24(18):4612–4626. doi: 10.1158/1078-0432.CCR-18-0461. [DOI] [PubMed] [Google Scholar]
- 11.Shi Y, Ping Y, Zhou W, et al. Tumour-associated macrophages secrete pleiotrophin to promote PTPRZ1 signalling in glioblastoma stem cells for tumour growth. Nat Commun. 2017;8:15080. doi: 10.1038/ncomms15080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koyuncu S, Saez I, Lee HJ, et al. The ubiquitin ligase UBR5 suppresses proteostasis collapse in pluripotent stem cells from Huntington’s disease patients. Nat Commun. 2018;9(1):2886. doi: 10.1038/s41467-018-05320-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li CG, Mahon C, Sweeney N, et al. PPARγ interaction with UBR5/ATMIN promotes DNA repair to maintain endothelial homeostasis. Cell Rep. 2019;26(5):1333. doi: 10.1016/j.celrep.2019.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shearer RF, Iconomou M, Watts CKW, Saunders DN. Functional roles of the E3 ubiquitin ligase UBR5 in cancer. Mol Cancer Res. 2015;13(12):1523–1532. doi: 10.1158/1541-7786.MCR-15-0383. [DOI] [PubMed] [Google Scholar]
- 15.Ji SQ, Zhang YX, Yang BH. UBR5 promotes cell proliferation and inhibits apoptosis in colon cancer by destablizing P21. Pharmazie. 2017;72(7):408. doi: 10.1691/ph.2017.7433. [DOI] [PubMed] [Google Scholar]
- 16.Liao L, Song M, Li X, et al. E3 ubiquitin ligase UBR5 drives the growth and metastasis of triple-negative breast cancer. Cancer Res. 2017;77(8):2090–2101. doi: 10.1158/0008-5472.CAN-16-2409. [DOI] [PubMed] [Google Scholar]
- 17.Matsuura K, Huang N, Cocce K, Zhang L, Kornbluth S. Downregulation of the proapoptotic protein MOAP-1 by the UBR5 ubiquitin ligase and its role in ovarian cancer resistance to cisplatin. Oncogene. 2017;36(12):1698–1706. doi: 10.1038/onc.2016.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meissner B, Kridel R, Lim RS, et al. The E3 ubiquitin ligase UBR5 is recurrently mutated in mantle cell lymphoma. Blood. 2013;121(16):3161–3164. doi: 10.1182/blood-2013-01-478834. [DOI] [PubMed] [Google Scholar]
- 19.McShane LM, Altman DG, Sauerbrei W, et al. Reporting recommendations for tumor marker prognostic studies. J Clin Oncol. 2005;23(36):9067–9072. doi: 10.1200/JCO.2004.01.0454. [DOI] [PubMed] [Google Scholar]
- 20.Wang C, Wang Y, Hong T, et al. Blocking the autocrine regulatory loop of Gankyrin/STAT3/CCL24/CCR3 impairs the progression and pazopanib resistance of clear cell renal cell carcinoma. Cell Death Dis. 2020;11(2):117. doi: 10.1038/s41419-020-2306-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ho TH, Kapur P, Eckel-Passow JE, et al. Multicenter validation of enhancer of zeste homolog 2 expression as an independent prognostic marker in localized clear cell renal cell carcinoma. J Clin Oncol. 2017;35(32):3706–3713. doi: 10.1200/JCO.2017.73.3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen D, Gassenmaier M, Maruschke M, et al. Expression and prognostic significance of a comprehensive epithelial-mesenchymal transition gene set in renal cell carcinoma. J Urol. 2014;191(2):479–486. doi: 10.1016/j.juro.2013.08.052. [DOI] [PubMed] [Google Scholar]
- 23.Wu Q, Yang L, Liu H, et al. Elevated expression of N-acetylgalactosaminyltransferase 10 predicts poor survival and early recurrence of patients with clear-cell renal cell carcinoma. Ann Surg Oncol. 2015;22(7):2446–2453. doi: 10.1245/s10434-014-4236-y. [DOI] [PubMed] [Google Scholar]
- 24.Xie Z, Liang H, Wang J, et al. Significance of the E3 ubiquitin protein UBR5 as an oncogene and a prognostic biomarker in colorectal cancer. Oncotarget. 2017;8(64):108079–108092. doi: 10.18632/oncotarget.22531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Z, Kang L, Zhang H, et al. AKT drives SOX2 overexpression and cancer cell stemness in esophageal cancer by protecting SOX2 from UBR5-mediated degradation. Oncogene. 2019;38:5250–5264. doi: 10.1038/s41388-019-0790-x. [DOI] [PubMed] [Google Scholar]
- 26.Park JS, Pierorazio PM, Lee JH, et al. Gene expression analysis of aggressive clinical T1 stage clear cell renal cell carcinoma for identifying potential diagnostic and prognostic biomarkers. Cancers (Basel) 2020;12(1):222. doi: 10.3390/cancers12010222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wierzbicki PM, Klacz J, Kotulak-Chrzaszcz A, et al. Prognostic significance of VHL, HIF1A, HIF2A, VEGFA and p53 expression in patients with clearcell renal cell carcinoma treated with sunitinib as firstline treatment. Int J Oncol. 2019;55(2):371–390. doi: 10.3892/ijo.2019.4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Graham J, Dudani S, Heng DYC. Prognostication in kidney cancer: recent advances and future directions. J Clin Oncol. 2018;36(36):3567–3573. doi: 10.1200/JCO.2018.79.0147. [DOI] [PubMed] [Google Scholar]
- 29.Jones K, Vari F, Keane C, Crooks P, Nourse JP, Seymour LA, Gottlieb D, Ritchie D, Gill D, Gandhi MK. Serum CD163 and TARC as disease response biomarkers in classical Hodgkin lymphoma. Clin Cancer Res. 2013;19(3):731–742. doi: 10.1158/1078-0432.CCR-12-2693. [DOI] [PubMed] [Google Scholar]
- 30.Song M, Wang C, Wang H, et al. Targeting ubiquitin protein ligase E3 component N-recognin 5 in cancer cells induces a CD8 + T cell mediated immune response. Oncoimmunology. 2020;9(1):1746148. doi: 10.1080/2162402X.2020.1746148. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.