Abstract
Background
Despite high expression of PD-L1, around half of advanced non-small cell lung cancer (NSCLC) will not experience tumor response with pembrolizumab. There is an need for a better understanding of the resistance mechanisms in this setting.
Methods
This bi-centric retrospective study included all consecutive patients with PDL1 ≥ 50% advanced NSCLC treated with pembrolizumab in first-line treatment between 2016 and 2020. We compared the clinical characteristics of patients with early progression (refractory) vs others. We performed a comprehensive gene expression profile screening by RNAseq capture on tumor samples.
Results
We included 46 patients. Twenty-two patients were refractory to pembrolizumab, mainly women, with poor performance status and lower albumin concentration. RNAseq analysis was performed on 19 samples. Hierarchical clustering allowed the identification of 3 clusters with various proportion of refractory tumors: intermediate (C1: 57%), high (C2: 71%) and low proportion (C3: 40%). Comparative analysis between C2 and C3 allowed the identification of overexpressed (n = 137) and underexpressed (n = 40) genes. Among the genes of interest, C2 exhibits higher activation of pathways associated with stemness phenotype (Hedgehog, Notch and Hippo pathways) and pathways associated with loss of PTEN and JAK2. In C2, genes associated with PD-1, toll-like receptor-9 (TLR-9), major histocompatibility complex (MHC) and interferon-γ pathways were underexpressed.
Conclusion
This study gives an overview of activated and downregulated pathways in high PD-L1 NSCLC refractory to pembrolizumab. These tumors showed activation of pathways associated with cancer stem cells, loss of PTEN and JAK2, and inhibition of both priming and effector phases of the immune response.
Keywords: Non-small cell lung cancer, PD-L1, Pembrolizumab, Resistance, RNAseq
Introduction
Non-small cell lung cancer (NSCLC) is the leading cause of cancer-related death in the world. Often diagnosed at an advanced stage, its treatment has been revolutionized by immune checkpoint inhibitors (ICIs). Programmed-death Ligand 1 (PD-L1) is a membranous protein mostly expressed by cancer cells. Its ligand, programmed death-1 (PD-1), is expressed on the surface of activated T cells. PD-L1–PD-1 interaction inhibits T cell activity and downregulates the immune response against cancer cells, inducing tumor proliferation. Pembrolizumab is a monoclonal antibody directed against PD-1, thus able to restore cytotoxic T-cells activity against NSCLC cells. Keynote-024 phase III trial has shown, in NSCLC tumors with high PD-L1 expression (≥ 50% in immunohistochemistry), that pembrolizumab given in first-line treatment was more effective than platinum-based chemotherapy, providing an overall response rate (ORR) of 45% and a median overall survival (OS) of 26,3 months vs. 13,4 months with chemotherapy [1]. However, around half of the high PD-L1 NSCLC patients will not experience tumor response with pembrolizumab, and 25% of these patients will have an acceleration of tumor growth, defining a hyperprogressive pattern [2]. The association of pembroliuzmab with chemotherapy is also an effective treatment in advanced NSCLC treatment, whatever the PD-L1 expression, with, however, higher toxicity events and similar ORR than pembrolizumab given alone [3, 4]. A better comprehension of primary resistance mechanisms is therefore urgently needed in order to improve our ICI treatment strategies. We aimed in this study to describe the clinical, molecular and gene expression patterns of NSCLC with high PD-L1 expression and primary resistance (refractory tumors) to pembrolizumab.
Material and methods
Patients and demographic comparisons
Consecutive patients with PDL1 ≥ 50%, EGFR/ALK/ROS1 wild-type, advanced NSCLC treated with first-line pembrolizumab at Hospital Ambroise Paré or Hospital André Mignot between 01/01/2016 and 30/06/2020 were retrospectively included. Patients received intravenous 200 mg pembrolizumab every 3 weeks, until progression or unacceptable toxicity. CT scan of brain, chest and abdomen was performed every 9 weeks for tumor response assessment. Patients with progression as best tumor response or death within the first month of treatment were considered as refractory patients. Other patients were considered as non-refractory patients. Censoring date was September 1, 2020. Statistical tests used to compare the two patients ‘groups (refractory vs others) were Chi square, Student and Fisher tests.
Molecular screening
Molecular screening was performed as part of the clinical practice for advanced NSCLC. All included patients had negative EGFR, ALK and ROS1 testing.
Gene expression profiling
RNA extraction
RNA was extracted from the tumor areas of formalin-fixed and paraffin-embedded tissue (FFPE) after selection by a pathologist, with the guidance of the hematoxylin–eosin-stained section. Extraction was performed as previously described, using the AS1480 Maxwell RSC Simply RNA tissue kit (Promega, USA) according to manufacturer’s instructions [13]. Each RNA sample extracted by Maxwell RSC was eluted with 50 μl of the Tris EDTA (TE) buffer. Then, RNA purity and concentrations were calculated using Multiscan GO reader, V.1.01.10 (ThermoFisher Scientific, France).
Sequencing
RNA-Seq libraries were performed with NEBNext® Ultra™ II Directional RNA Library Prep Kit for Illumina according to supplier recommendations (NEB). The capture was then performed on cDNA libraries with the Twist Human Core Exome + Custom IntegraGen Enrichment System according to supplier recommendations (Twist Bioscience). First of all, a RNA quality control was performed on Fragment Analyzer (AATI) with the RNA kit (DNF-489) to check the integrity of the RNA profile and to assess the RNA concentration. For degraded samples the DV200 (% of fragments > 200nt) was extracted to define the correct input for the process. The input was 15 ng for a DV200 > 70%, 30 ng for a DV200 between 50 and 70% and 75 ng for a DV200 between 30 and 50%. The process was not recommended for a DV200 < 30%. The protocol permitted to convert total RNA into a library of template molecules of known strand origin. Then a capture of the coding regions of the transcriptome was performed, and the resulting library was suitable for subsequent cluster generation and sequencing. Briefly, the RNA was fragmented into small pieces using divalent cations under elevated temperature. cDNA was generated from the cleaved RNA fragments using random priming during first and second strand synthesis, and sequencing adapters were ligated to the resulting double-stranded cDNA fragments and enriched by 7 PCR cycles. The coding regions of the transcriptome were then captured from this library using sequence-specific probes to create the final library. For that purpose 500 ng of purified libraries were hybridized to the twist oligo probe capture library for 16 h in a singleplex reaction. After hybridization, washing and elution, the eluted fraction was PCR-amplified with 8 cycles, purified and quantified by QPCR to obtain sufficient DNA template for downstream applications. Each eluted-enriched DNA sample was then sequenced on an Illumina NovaSeq as Paired End 100b reads. Image analysis and base calling were performed using Illumina Real Time Analysis (3.4.4) with default parameters.
Quantification of gene expression
STAR was used to obtain the number of reads associated to each gene in the Gencode v31 annotation (restricted to protein-coding genes, antisense and lincRNAs). Raw counts for each sample were imported into R statistical software. Extracted count matrix was normalized for library size and coding length of genes to compute FPKM expression levels.
Unsupervised analysis
The Bioconductor edgeR package was used to import raw counts into R statistical software and compute normalized log2 CPM (counts per millions of mapped reads) using the TMM (weighted trimmed mean of M-values) as normalization procedure. The normalized expression matrix from the 1000 most variant genes (based on standard deviation) was used to classify the samples according to their gene expression patterns using principal component analysis (PCA), hierarchical clustering and consensus clustering. PCA was performed by FactoMineR::PCA function with “ncp = 10, scale.unit = FALSE” parameters. Hierarchical clustering was performed by stats::hclust function (with distance and method). Consensus clustering was performed by ConsensusClusterPlus::ConsensusClusterPlus function to examine the stability of the clusters. We established consensus partitions of the data set in K clusters (for K = 2, 3,..., 8), on the basis of 1000 resampling iterations (80% of genes, 80% of sample) of hierarchical clustering, with distance and method. Then, the cumulative distribution functions (CDFs) of the consensus matrices were used to determine the optimal number of clusters (K = 3 for instance), considering both the shape of the functions and the area under the CDF curves. tSNE analysis was performed with the Bioconductor Rtsne package applied to the PCA object (theta = 0.0, perplexity = , max_iter = 1000).
Differential expression analysis
The Bioconductor edgeR package was used to import raw counts into R statistical software. Differential expression analysis was performed using the Bioconductor limma package and the voom transformation. To improve the statistical power of the analysis, only genes expressed in at least one sample (FPKM ≥) were considered. A q-value threshold of ≤0.05 and a minimum fold change of 1.2 were used to define differentially expressed genes.
Pathway enrichment analysis—ORA
Hypergeometric tests were used to identify gene sets from the MSigDB v7.4 database overrepresented among the lists of up- or downregulated genes, correcting for multiple testing with the Benjamini–Hochberg procedure. Gene sets from MSigDB v7.4 database were selected among the C2_curated, C5_GO, C6_oncogenic, Hallmark classes, keeping only gene sets defined by 10–500 genes.
Pathway enrichment analysis—GSEA
Gene list from the differential analysis was ordered by decreasing log2 fold change. Gene set enrichment analysis was performed by clusterProfiler::GSEA function using the fgsea algorithm. Gene sets from MSigDB v7.4 database were selected among the C2_curated, C5_GO, C6_oncogenic, C7_immunologic, Hallmark classes, keeping only gene sets defined by 10–500 genes.
Results
Clinical data
We included 46 consecutive patients who received at least one course of pembrolizumab as first line treatment for NSCLC (squamous cell subtype: n = 7; non-squamous cell subtype: n = 39). Sixteen patients (34.8%) had tumors with very high PD-L1 expression (≥ 90%).
Twenty-four patients (52%) had refractory tumors, whereas others (n = 22) had tumor response or stability. We compared the clinical characteristics of the patients between the two groups (Table 1). Patients with refractory tumors were more frequently women (58.3 vs. 27.3%, p = 0.034), with poorer performance status score (PS 2: 45.9 vs. 13.7%, p = 0.017), with lower albumin rate (p = 0.031).
Table 1.
Comparison of clinical data between refractory and non-refractory patients
| Characteristic | non-refractory patients (N = 22) | Refractory patients (n = 24) | p-value |
|---|---|---|---|
| Sex (female), n (%) | 6 (27.3) | 14 (58.3) | 0.034 |
| Age (years), median (range) | 66.8 (36–89) | 67.0 (40–89) | 0.94 |
| Comorbidity Score Charlson (range) | 10.2 (7–14) | 10.2 (7–15) | 0.97 |
| ECOG PS, n (%) | |||
| 0–1 | 19 (86.3) | 13 (54.1) | 0.017 |
| 2 | 3 (13.7) | 11 (45.9) | |
| Past or current smoker, n (%) | 21 (95.5) | 20 (83.3) | 0.35 |
| Tobacco (PY) | 42 | 46 | 0.61 |
| Albumin, g/l (mean) | 35.7 | 31.6 | 0.031 |
| CRP, mg/l (mean) | 45.1 | 52.1 | 0.73 |
| Neutrophils (G/l) (mean) | 6.52 | 7.57 | 0.25 |
| Lymphocytes (G/l) (mean) | 1.49 | 1.67 | 0.34 |
| Weight loss (% from baseline) (mean) | -5.9 | -7.5 | 0.53 |
| Pretreatment Proton pump inhibitor use, n (%) | 9 (40.9) | 10 (41.7) | 0.95 |
| Pretreatment Steroids, n (%)* | 2 (9) | 8 (33) | 0.074 |
| Antibiotics within 2 months before pembrolizumab, n (%) | 8 (36.3) | 11 (50.0) | 0.36 |
| Brain metastases, n (%) | 3 (13.6) | 2 (8.3) | 0.66 |
| Liver Metastases, n (%) | 3 (13.6) | 4 (16.7) | 1.00 |
*Maximum dose ≤ 10 mg per day for all patients
Other variables did not significantly differ between the two groups of patients. Notably, we did not show any statistical difference concerning comorbidity score, liver or brain metastasis prevalence, antibiotic use before the beginning of the treatment, or use of proton pomp inhibitor between the two groups.
Molecular profile
Molecular screening was available for 40 samples (87%): for all non-squamous cell carcinoma except three (n = 36), and for 4 squamous cell carcinomas, as part of clinical routine. KRAS mutations were observed in 15 samples (37.5%). Additional DNA next-generation sequencing was performed for 27 samples: STK11 mutation, V600 BRAF mutation and skip exon 14 MET mutation were observed respectively in one sample (3.7%) each, and MET amplification in two samples (7.4%).
Gene expression profile
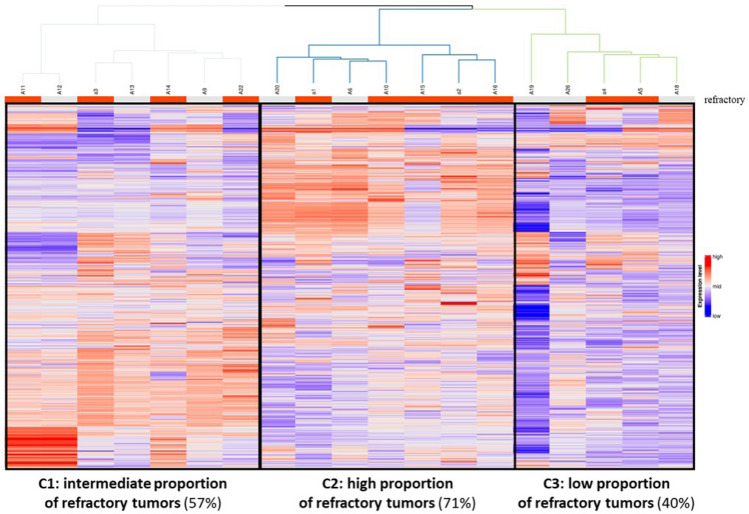
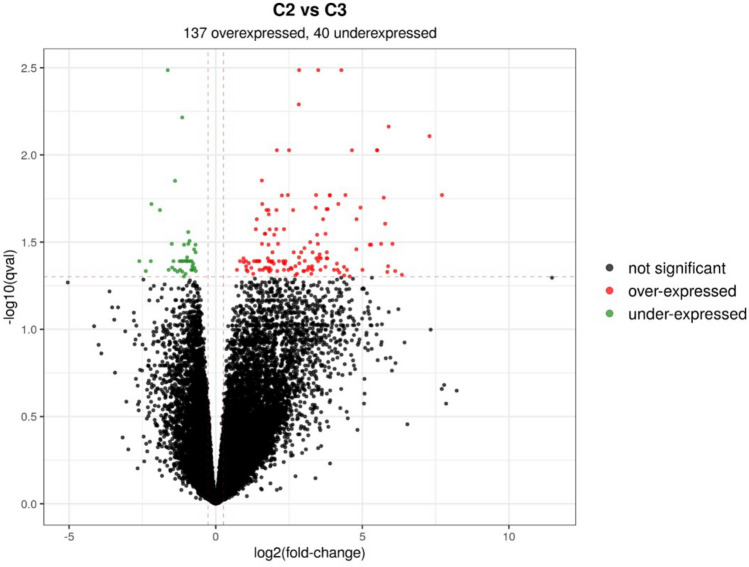
Twenty tumor samples were available with enough material for RNA extraction and RNAseq analysis. After quality control (Qc) and data normalization, one sample was excluded due to poor quality. Gene expression profiling was therefore feasible for 19 NSCLC samples (11 refractory samples, 8 samples with response to pembrolizumab), corresponding to 16 adenocarcinomas and 3 squamous cell carcinomas. Unsupervised hierarchical clustering allowed the separation of three different clusters of samples, with various proportions of refractory tumors: intermediate (57%) for C1, high for C2 (71%) and low for C3 (40%) (Fig. 1). Then we performed differential analysis between C2 (high proportion of refractory tumors) versus C3 (low proportion of refractory tumors). We observed a significant numbers of overexpressed (n = 137) and underexpressed (n = 40) genes in C2 in comparison of C3. Using GSEA enrichment analysis, we were able to determine several over- and underexpressed genes of interest in C2 (Fig. 2).
Fig. 1.
Hierarchical clustering according to RNAseq analysis (n = 19)
Fig. 2.
Differential analysis of gene expression between C2 and C3 clusters
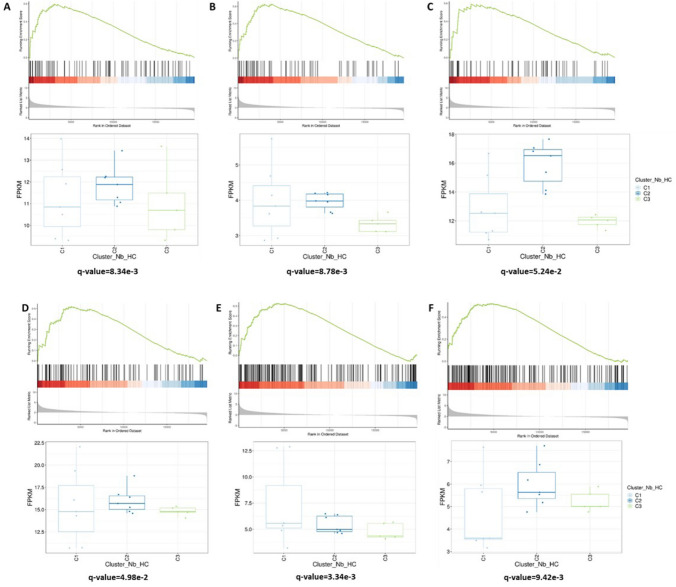
Among overexpressed genes (Fig. 3), stem cell-related pathways were activated in C2 (q-value = 8.34e-3). Notably, Hedgehog (q-value = 8.78e−3) and Notch (q-value = 5.24e-2) genes were overexpressed. We found also a significant activation of Hippo pathway (q-value = 4.98e-2) and pathways associated with downregulation of PTEN (q-value = 3.34e-3) and JAK2 (q-value = 9.42e-3).
Fig. 3.
Overexpressed genes of interest in C2 compared to C3 using GSEA enrichment analysis. a stemness maintenance-associated pathways; b Hedgehog signaling pathway; c Notch signaling pathway; d Hippo signaling pathway; e pathways associated with loss of PTEN; f pathways associated with loss of JAK2
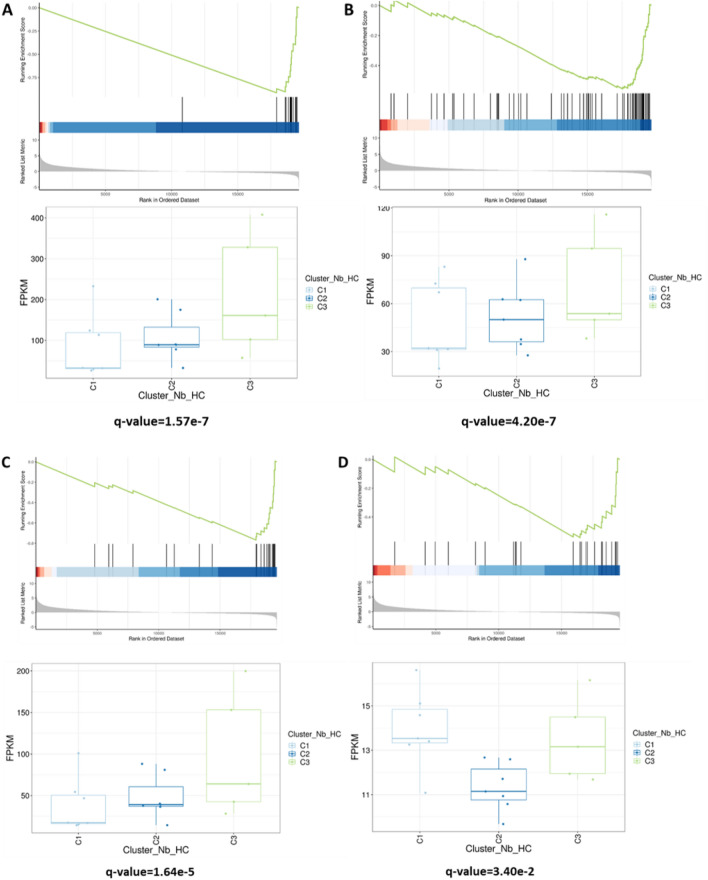
Among underexpressed genes in C2 (Fig. 4), pathways associated with interferon-gamma (IFN-γ) and major histocompatibility complex (MHC) were downregulated in C2 compared to C3 (q-value = 1.57e-7 and q-value = 4.20e-7, respectively). Genes related to PD-1 and toll-like receptor-9 (TLR-9) pathways were also underexpressed in C2 (q-value = 1.64e-5 and q-value = 3.40e-2, respectively).
Fig. 4.
Underexpressed genes of interest in C2 compared to C3 using GSEA enrichment analysis. a MHC-associated pathway; b pathways associated with response to IFN-γ; c PD-1-associated pathway; d toll-like receptor-9-associated pathway
Discussion
Our study gives an extensive overview of the pathways and phenotypes associated with early resistance to pembrolizumab in first-line treatment of high PD-L1 advanced NSCLC. To our knowledge, this is the first study that gives gene expression profiles of this specific NSCLC population.
Cancer stem cells (CSCs) are poorly differentiated cells, known to be insensitive to cytotoxic chemotherapy and radiotherapy [5]. Stemness phenotype seems to be also associated with ICI resistance [6–8]. Several pathways are closely associated with CSCs and stemness feature, as Hedgehog (Hh) and Notch pathways. Hh pathway is associated with epithelial–mesenchymal transition (EMT) [9] and with resistance to chemotherapy [10, 11]. Concerning ICI, in a medulloblastoma mice model, the activation of Hh pathway induced resistance to treatment [12]. In our study, Hh pathway was overexpressed in refractory tumors to pembrolizumab. We have shown recently that Hh pathway activation, reflected by Gli1 expression in cancer cells in IHC, was closely associated with tumor response and early progression with ICI given in monotherapy in advanced NSCLC [13]. Altogether, previous preclinical and translational results suggest that Hh pathway activation is associated with early progression and primary resistance to ICI. Notch pathway is another pathway associated with CSCs and resistance to cancer treatment [14]. We found an overexpression of Notch pathway in C2 cluster compared to C3 cluster. Notch signaling is also a master developmental signaling involved in the development and activation of immune cells. Several preclinical and clinical data suggested an interaction between Notch signaling, immune response and IO efficacy in solid tumors [15–17]. Recent studies provided evidence that high mutated NOTCH signaling in NSCLC patients treated with ICIs is related to inflammatory immune microenvironment and correlated with treatment outcomes and prognosis [17, 18]. Hippo pathway is a signaling cascade involved in tissue patterning and development. The pathway consists in a cascade of protein kinases activation, which finally leads to the phosphorylation and inactivation of cytoplasmic transcription factors Yes-associated protein (YAP) and the transcriptional coactivator with PDZ-binding motif (TAZ). YAP activation is frequently observed in many solid tumors and associated with chemoresistance [19]. Emerging data suggest also a role of Hippo signaling in resistance to ICIs. YAP acts indeed as a regulator of PD-L1 expression in tumor cells [20, 21].
Other overexpressed pathways in C2 cluster concerned pathways associated with downregulation of PTEN and JAK2. Loss of PTEN is a well-known resistance mechanism to ICIs, through upregulation of PI3-kinase (PI3K) and STAT3 activation, leading to the release of immunosuppressive cytokines and chemokines by the tumor (notably Il-6, Il-10 and VEGF) [22]. Loss of PTEN is associated with lower tumor lymphocyte infiltration and poor tumor response to ICIs in melanoma [23]. Silencing mutations of JAK2 induce loss of interferon-gamma (IFN-γ) receptor and loss of IFN-γ signaling in tumor cells and negate adaptive PD-L1 expression on cell surface [24]. In melanoma, JAK2 mutations are associated with secondary resistance to ICIs [25]. Similarly, we found in our study a downregulation of IFN- γ signaling pathway in NSCLC with early resistance to pembrolizumab, consistent with these results. Pathways associated with MHC protein complex were also downregulated in C2 cluster. Loss of MHC can occur notably through loss of beta-2 microglobuline (B2M) expression (through mutations or loss of expression). Loss of B2M has been found to be associated with poor response to ICIs, including NSCLC with secondary resistance to ICIs [26]. In a whole exome sequencing analysis on ctDNA of advanced NSCLC with secondary late resistance to ICIs, we have also shown a high rate of mutations of PTEN, HLA and B2M genes [27]. In the present study, we confirmed the frequent downregulation of these pathways in high PD-L1 NSCLC refractory to pembrolizumab. Finally, we observed in C2 a downregulation of pathways associated with both priming (TLR-9) and effector (PD-1) immune phases, suggesting that all the steps of the antitumor immune response are inhibited in refractory tumors.
In conclusion, our work is the first to our knowledge to give the gene expression profile of high PD-L1 advanced NSCLC refractory to pembrolizumab in first-line treatment. Stemness phenotype, with activation of Hh, Notch and Hippo pathways, along with downregulation of PTEN- and JAK-related pathways, are frequent in this population. Combined treatment strategies targeting these pathways are therefore needed in order to improve response to ICI in first-line treatment.
Author contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by JT, PTK and EGI. The first draft of the manuscript was written by JT and EGI, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Funding
The authors declare that no funds, grants, or other supports were received during the preparation of this manuscript.
Data Availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Declarations
Conflict of interest
EGL: MSD (advisory board). Other authors declare no potential conflicts of interest.
Ethics approval
No ethics approval was needed for retrospective studies according to French legislation in force at the beginning of the study. Non-opposition form for retrospective data collection was provided to all patients. None objected to data collection.
Consent to participate
Informed consent was obtained from all individual participants included in the study.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jamila Talb and Paul Takam Kamga are contribute equally.
References
- 1.Reck M, Rodríguez-Abreu D, Robinson AG, Hui R, Csőszi T, Fülöp A, Gottfried M, Peled N, Tafreshi A, Cuffe S, O'Brien M, Rao S, Hotta K, Leiby MA, Lubiniecki GM, Shentu Y, Rangwala R, Brahmer JR. Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N Engl J Med. 2016;375(19):1823–1833. doi: 10.1056/NEJMoa1606774. [DOI] [PubMed] [Google Scholar]
- 2.Champiat S, Dercle L, Ammari S, Massard C, Hollebecque A, Postel-Vinay S, Chaput N, Eggermont A, Marabelle A, Soria JC, Ferté C. Hyperprogressive disease is a new pattern of progression in cancer patients treated by Anti-PD-1/PD-L1. Clin Cancer Res. 2014;23(8):1920–1928. doi: 10.1158/1078-0432.CCR-16-1741. [DOI] [PubMed] [Google Scholar]
- 3.Gandhi L, Rodríguez-Abreu D, Gadgeel S, Esteban E, Felip E, De Angelis F, Domine M, Clingan P, Hochmair MJ, Powell SF, Cheng SY, Bischoff HG, Peled N, Grossi F, Jennens RR, Reck M, Hui R, Garon EB, Boyer M, Rubio-Viqueira B, Novello S, Kurata T, Gray JE, Vida J, Wei Z, Yang J, Raftopoulos H, Pietanza MC, Garassino MC. Pembrolizumab plus chemotherapy in metastatic non-small-cell lung cancer. N Engl J Med. 2018;378(22):2078–2092. doi: 10.1056/NEJMoa1801005. [DOI] [PubMed] [Google Scholar]
- 4.Paz-Ares L, Luft A, Vicente D, Tafreshi A, Gümüş M, Mazières J, Hermes B, Çay Şenler F, Csőszi T, Fülöp A, Rodríguez-Cid J, Wilson J, Sugawara S, Kato T, Lee KH, Cheng Y, Novello S, Halmos B, Li X, Lubiniecki GM, Piperdi B, Kowalski DM. Pembrolizumab plus chemotherapy for squamous non-small-cell lung cancer. N Engl J Med. 2018;379(21):2040–2051. doi: 10.1056/NEJMoa1810865. [DOI] [PubMed] [Google Scholar]
- 5.Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat Rev Cancer. 2005;5:275–284. doi: 10.1038/nrc1590. [DOI] [PubMed] [Google Scholar]
- 6.Maccalli C, Parmiani G, Ferrone S. Immunomodulating and immunoresistance properties of cancer-initiating cells: implications for the clinical success of immunotherapy. Immunol Invest. 2017;46:221–238. doi: 10.1080/08820139.2017.1280051. [DOI] [PubMed] [Google Scholar]
- 7.Chen L, Yang QC, Li YC, Yang LL, Liu JF, Li H, Xiao Y, Bu LL, Zhang WF, Sun ZJ. Targeting CMTM6 suppresses stem cell-like properties and enhances antitumor immunity in head and neck squamous cell carcinoma. Cancer Immunol Res. 2020;8:179–191. doi: 10.1158/2326-6066.CIR-19-0394. [DOI] [PubMed] [Google Scholar]
- 8.Liao TT, Lin CC, Jiang JK, Yang SH, Teng HW, Yang MH. Harnessing stemness and PD-L1 expression by AT-rich interaction domain-containing protein 3B in colorectal cancer. Theranostics. 2020;10:6095–6112. doi: 10.7150/thno.44147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giroux-Leprieur E, Costantini A, Ding VW, He B. Hedgehog signaling in lung cancer: from oncogenesis to cancer treatment resistance. Int J Mol Sci. 2018;19:2835. doi: 10.3390/ijms19092835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giroux Leprieur E, Vieira T, Antoine M, Rozensztajn N, Rabbe N, Ruppert AM, Lavole A, Cadranel J, Wislez M. Sonic Hedgehog pathway activation is associated with resistance to platinum-based chemotherapy in advanced non-small-cell lung carcinoma. Clin Lung Cancer. 2016;17:301–308. doi: 10.1016/j.cllc.2015.12.007. [DOI] [PubMed] [Google Scholar]
- 11.Giroux Leprieur E, Tolani B, Li H, Leguay F, Hoang NT, Acevedo LA, Jin JQ, Tseng HH, Yue D, Kim IJ, Wislez M, Wang C, Jablons DM, He B (2017) Membrane-bound full-length Sonic Hedgehog identifies cancer stem cells in human non-small cell lung cancer. Oncotarget 8:103744–103757. 10.18632/oncotarget.21781. [DOI] [PMC free article] [PubMed]
- 12.Pham CD, Flores C, Yang C, Pinheiro EM, Yearley JH, Sayour EJ, Pei Y, Moore C, McLendon RE, Huang J, Sampson JH, Wechsler-Reya R, Mitchell DA. Differential immune microenvironments and response to immune checkpoint blockade among molecular subtypes of murine medulloblastoma. Clin Cancer Res. 2016;22:582–595. doi: 10.1158/1078-0432.CCR-15-0713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mehlman C, Takam Kamga P, Costantini A, Julié C, Dumenil C, Dumoulin J, Ouaknine J, Giraud V, Chinet T, Emile JF, Giroux Leprieur E. Baseline Hedgehog pathway activation and increase of plasma Wnt1 protein are associated with resistance to immune checkpoint inhibitors in advanced non-small-cell lung cancer. Cancers (Basel) 2021;13:1107. doi: 10.3390/cancers13051107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Purow B. Notch inhibition as a promising new approach to cancer therapy. Adv Exp Med Biol. 2012;727:305–319. doi: 10.1007/978-1-4614-0899-4_23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ayaz F, Osborne BA. Non-canonical notch signaling in cancer and immunity. Front Oncol. 2014;4:345. doi: 10.3389/fonc.2014.00345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chung J, Maillard I. Notch signaling in alloreactive T cell immunity. Curr Top Microbiol Immunol. 2012;360:135–150. doi: 10.1007/82_2012_226. [DOI] [PubMed] [Google Scholar]
- 17.Li X, Wang Y, Li X, Feng G, Hu S, Bai Y. The impact of NOTCH pathway alteration on tumor microenvironment and clinical survival of immune checkpoint inhibitors in NSCLC. Front Immunol. 2021;12:638763. doi: 10.3389/fimmu.2021.638763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Z, Gu Y, Su X, Bai J, Guan W, Ma J, Luo J, He J, Zhang B, Geng M, Xia X, Guan Y, Shen C, Chen C. Co-occurring alteration of NOTCH and DDR pathways serves as novel predictor to efficacious immunotherapy in NSCLC. Front Oncol. 2021;11:659321. doi: 10.3389/fonc.2021.659321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang K, Qi HX, Hu ZM, Chang YN, Shi ZM, Han XH, Han YW, Zhang RX, Zhang Z, Chen T, Hong W. YAP and TAZ take center stage in cancer. Biochemistry. 2015;54:6555–6566. doi: 10.1021/acs.biochem.5b01014. [DOI] [PubMed] [Google Scholar]
- 20.Hsu PC, Yang CT, Jablons DM, You L. The role of Yes-Associated Protein (YAP) in Regulating Programmed Death-Ligand 1 (PD-L1) in thoracic cancer. Biomedicines. 2018;6:114. doi: 10.3390/biomedicines6040114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miao J, Hsu PC, Yang YL, Xu Z, Dai Y, Wang Y, Chan G, Huang Z, Hu B, Li H, Jablons DM, You L (2017) YAP regulates PD-L1 expression in human NSCLC cells. Oncotarget 8:114576–114587. 10.18632/oncotarget.23051. [DOI] [PMC free article] [PubMed]
- 22.Piro G, Carbone C, Carbognin L, Pilotto S, Ciccarese C, Iacovelli R, Milella M, Bria E, Tortora G. Revising PTEN in the era of immunotherapy: new perspectives for an old story. Cancers (Basel) 2019;11:1525. doi: 10.3390/cancers11101525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peng W, Chen JQ, Liu C, Malu S, Creasy C, Tetzlaff MT, Xu C, McKenzie JA, Zhang C, Liang X, Williams LJ, Deng W, Chen G, Mbofung R, Lazar AJ, Torres-Cabala CA, Cooper ZA, Chen PL, Tieu TN, Spranger S, Yu X, Bernatchez C, Forget MA, Haymaker C, Amaria R, McQuade JL, Glitza IC, Cascone T, Li HS, Kwong LN, Heffernan TP, Hu J, Bassett RL, Jr, Bosenberg MW, Woodman SE, Overwijk WW, Lizée G, Roszik J, Gajewski TF, Wargo JA, Gershenwald JE, Radvanyi L, Davies MA, Hwu P. Loss of PTEN promotes resistance to T cell-mediated immunotherapy. Cancer Discov. 2016;6:202–216. doi: 10.1158/2159-8290.CD-15-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalbasi A, Ribas A. Tumour-intrinsic resistance to immune checkpoint blockade. Nat Rev Immunol. 2020;20:25–39. doi: 10.1038/s41577-019-0218-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zaretsky JM, Garcia-Diaz A, Shin DS, Escuin-Ordinas H, Hugo W, Hu-Lieskovan S, Torrejon DY, Abril-Rodriguez G, Sandoval S, Barthly L, Saco J, Homet Moreno B, Mezzadra R, Chmielowski B, Ruchalski K, Shintaku IP, Sanchez PJ, Puig-Saus C, Cherry G, Seja E, Kong X, Pang J, Berent-Maoz B, Comin-Anduix B, Graeber TG, Tumeh PC, Schumacher TN, Lo RS, Ribas A. Mutations associated with acquired resistance to PD-1 blockade in melanoma. N Engl J Med. 2016;375:819–829. doi: 10.1056/NEJMoa1604958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gettinger S, Choi J, Hastings K, Truini A, Datar I, Sowell R, Wurtz A, Dong W, Cai G, Melnick MA, Du VY, Schlessinger J, Goldberg SB, Chiang A, Sanmamed MF, Melero I, Agorreta J, Montuenga LM, Lifton R, Ferrone S, Kavathas P, Rimm DL, Kaech SM, Schalper K, Herbst RS, Politi K. Impaired HLA Class I antigen processing and presentation as a mechanism of acquired resistance to immune checkpoint inhibitors in lung cancer. Cancer Discov. 2017;7:1420–1435. doi: 10.1158/2159-8290.CD-17-0593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giroux Leprieur E, Hélias-Rodzewicz Z, Takam Kamga P, Costantini A, Julie C, Corjon A, Dumenil C, Dumoulin J, Giraud V, Labrune S, Garinet S, Chinet T, Emile JF. Sequential ctDNA whole-exome sequencing in advanced lung adenocarcinoma with initial durable tumor response on immune checkpoint inhibitor and late progression. J Immunother Cancer. 2020;8:e000527. doi: 10.1136/jitc-2020-000527. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.