Abstract
The production of adenosine by CD73 on cancer cells in the tumor microenvironment is a recognized immunosuppressive mechanism contributing to immune evasion in many solid tumors. While NK cells have been purported to overexpress CD73 under certain conditions, this phenomenon has remained elusive and unclear. We have found that while NK cells are able to upregulate expression of CD73 on their surface when exposed to CD73+ cancer cells, this upregulation is not universal, nor is it often substantial. Rather, our data point to the extent of CD73 expression on NK cells to be both cancer-specific and environmentally-driven, and largely limited in intensity. We found that NK cell overexpression of CD73 responds to the level of CD73 on cancer cells and is enhanced in hypoxia. Interestingly, human CD73+ NK cells appear hyperfunctional in vitro compared to CD73− NK cells, suggesting that CD73 expression could be a bystander of NK cell activation. In addition, glioblastoma patient data show that tumor-infiltrating NK cells express CD73 variably, depending on donor, and present lower expression of CD16, alongside patient-specific changes in CEACAM1, CXCR3 and TIM-3, suggesting some functional changes in NK cell responses associated with expression of CD73 on NK cells in vivo. Taken together, our study is the first to show that while NK cells are largely resistant to the upregulation of CD73, CD73 expression is inducible on NK cells in response to CD73 on cancer cells, and these cells are associated with distinct functional signatures.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00262-022-03219-z.
Keywords: NK cells, CD73, Tumor microenvironment, Immunometabolism, Adenosine
Introduction
Solid tumors are commonly hypoxic, and this hypoxia enhances extracellular adenosine accumulation due to the metabolism of ATP though ectoenzymes CD39 and CD73 present on the tumor cells [1, 2]. CD73 is linked to the plasma membrane on cancer cells through a glycosylphosphatidylinositol (GPI) linker but can also exist as a catalytically active soluble form through shedding from the membrane either proteolytically or by phosphatidylinositol-specific phospholipases [3, 4]. The overexpression of CD73 is prevalent in many cancers and accompanies worse survival in solid tumor patients since CD73 can contribute to metastasis and immune evasion [5–7].
In hypoxic environments, CD73 expression is induced through the binding of HIF-1 to the NT5E gene in the cancer cells [8]. HIF-1 is a heterodimer, consisting of HIF-1α and HIF-1β domains. Under normal conditions, HIF-1α is rapidly degraded; however, under hypoxic conditions, HIF-1α dimerizes with HIF-1β to trigger signaling and expression [9]. HIF-1α has also been shown to increase intracellular adenosine through the inhibition of adenosine kinase [10]. This elevated concentration of intracellular adenosine can then be transported outside of the cell to inhibit immune cell function. The accumulation of adenosine through both increases in intracellular concentrations and CD73 activity have been reported to impair the cytotoxic ability, proliferation, and maturation of tumor-infiltrating natural killer (NK) cells [11, 12].
In healthy tissues and peripheral blood, NK cells do not normally express CD73. However, evidence has begun to emerge that under certain conditions NK cells may acquire CD73 expression and anti-inflammatory responses to produce adenosine and IL-10 [13, 14]. It has also been reported that tumor-infiltrating NK cells isolated from various tumors have had increases in CD73 expression [15]. More recently, the presence of CD73+ NK cells in the tumor microenvironment of breast cancer patients was found to correlate to a larger tumor size. These tumor-infiltrating CD73+ NK cells were shown to express higher amounts of multiple immune checkpoints compared to non-CD73+-expressing, tumor-infiltrating and peripheral blood NK cells. CD73 expression was also induced on peripheral blood NK cells after culture with tumor cells, and this increase was linked to engagement with the 4-1BB ligand that initiates actin polymerization-dependent exocytosis and eventually IL-10 and TGF-β production through STAT3 [16]. Although these studies have shown that tumor-infiltrating NK cells are able to express CD73, this occurrence is not fully understood and may be tumor-specific. Conflicting data [17] on the extent of expression of CD73 on tumor NK cells have, moreover, suggested this may not be a universal, but rather a defined, environmentally-driven process.
Here, we report that NK cells, while largely resistant to the overexpression of CD73, are able to upregulate its surface expression in response to CD73+ cancer cells under specific conditions, and that CD73 expression on cancer cells is a likely prerequisite for induction of CD73 expression on NK cells. This phenomenon was seen on both peripheral blood human NK cells as well as NK-92 cells, though it was more pronounced on NK-92 cells, while glioblastoma (GBM) patient data show that this is not universal. Despite the lower induction of CD73 on primary human NK cells, CD73+ primary human NK cells were associated with hyperfunction against cancer targets in terms of degranulation, suggesting a role for CD73+ as a possible consequence of NK cell activation or maturation. Patient-derived CD73+ NK cells, on the other hand, presented upregulated inhibitory and downregulated activating NK cell receptors, evidencing the induction of an inhibitory phenotype associated with CD73+ expression in vivo. This is the first report of the induction of CD73 on NK cells specifically in response to CD73+ cancer cells, as well as the characterization of the functional identity of CD73+ NK cells, which will have implications for the use of these cells in adoptive transfer settings.
Materials and methods
Reagents and cell lines
Commercial cell lines used for cocultures were NK-92, K562, PC3, A549, and U87 (ATCC). GBM43 cells were provided by Dr. Karen E. Pollok (Indiana University School of Medicine, Indianapolis, IN) and were derived from xenograft tissue provided by Dr. Jann Sarkaria (Mayo Clinic, Rochester MN) according to described protocols [18, 19]. The identity of the GBM43 cell line was confirmed by DNA fingerprint analysis for species and baseline short-tandem repeats (IDEXX BioResearch, Columbia, MO) and was found to be 100% human [20]. K562 and U87 were maintained in IMDM (Gibco, Thermo Fisher Scientific, Waltham, MA) supplemented with 10% FBS (Corning, Corning, NY) and 1% penicillin/streptomycin (Gibco, Thermo Fisher Scientific, Waltham, MA), PC3 and A549 were cultured in DMEM (Gibco) supplemented with 10% FBS and 1% penicillin/streptomycin, and GBM43 was grown in DMEM without sodium pyruvate (Gibco, Thermo Fisher Scientific, Waltham, MA) supplemented with 10% FBS and 1% HEPES (Fisher Scientific, Waltham, MA). NK92 cells were maintained in RPMI (Gibco, Thermo Fisher Scientific, Waltham, MA), with 10% FBS, 1% penicillin/streptomycin, 100 IU/ml IL-2 (Akron Biotech, Boca Raton, FL), and 0.1 mM 2-mercaptoethanol. Cobalt chloride hexahydrate (Sigma-Aldrich, St. Louis, MO) was used for generating hypoxic conditions.
Patient samples
Samples of whole blood and resected GBM tumors were collected from GBM surgical patients by Indiana University Health ECRO Biorepository according to HIPAA and approved IRB protocol #1011004282 (04-093). Sample processing of fresh whole blood to purify peripheral blood mononuclear cell (PBMCs) was done by diluting the blood with an equal volume of 2% FBS in 1X PBS before density gradient separation using Lymphoprep (Stem Cell Technologies, Vancouver, Canada). NK cells were then analyzed using flow cytometry.
Human glioblastoma tissue was stored in Tissue Storage Buffer (Miltenyi Biotec, Gaithersburg, MD) until ready to process, no longer than 24 h after resection. Tissue was washed once with 1X PBS and was minced into small pieces using a scalpel. Tissue was suspended in digestion mix (1 mg/ml Collagenase IV (Worthington Biochemical, Lakewood, NJ) and 400 µg/ml DNase I (Worthington Biochemical, Lakewood, NJ) in 1X PBS) and incubated for 1 h at 37 °C with agitation. After enzymatic digestion, the mixture was diluted with an equal volume of cold 1X PBS, filtered through a 70-µm nylon cell strainer (Corning, Corning, NY), and centrifuged. Pellet was resuspended in ice cold 1X PBS and strained a second time though a 40-µm nylon cell strainer (Corning, Corning, NY) and then centrifuged and resuspended in 2% FBS, 1X PBS. The cell suspension was carefully layered on top of Lymphoprep (Stem Cell Technologies, Vancouver, Canada) and centrifuged at 800 g for 30 min at room temperature with no brake. Interface was collected and washed with FACS buffer. NK cells were further isolated from the tumor tissue by negative selection using the EasySep™ Direct Human NK Cell Isolation Kit (StemCell Technologies, Vancouver, Canada).
Peripheral blood NK cell isolation
Blood samples were obtained from normal healthy donor volunteers who gave written consent through Purdue University’s IRB protocol (#1804020540). Natural killer (NK) cells were isolated from whole blood by negative selection using the EasySep™ Direct Human NK Cell Isolation Kit to > 80–90% purity (StemCell Technologies, Vancouver, CA). Cells were cultured at a density of 1 × 106 – 1.5 × 106 cells/ml in OpTmizer (Thermo Fisher, Waltham, MA) with the addition of the expansion supplement following the manufacturer’s instructions along with 5% hAB serum (Valley Biomedical, Winchester, VA), 0.2 mM L-glutamine (Gibco, Thermo Fisher Scientific, Waltham, MA), 500 IU/ml hIL-2 (Akron Biotech, Boca Raton, FL), 10 ng/ml hIL-15 (Gold Bio, St. Louis, MO), and 25 ng/ml hIL-21 (Gold Bio, St. Louis, MO).
CRISPR knockout of CD73 on GBM43 cells
CD73 knockout of patient-derived GBM43 cells was carried out via CRISPR/Cas9 genome editing using the Gene Knockout Kit v2 from Synthego (Redwood City, CA) according to the manufacturer’s instructions. Briefly, the sgRNA and cas9 were mixed with the Lipofectamine™ CRISPRMAX™ Cas9 Transfection Reagent (Thermo Fisher, Waltham, MA) according to the manufacturer’s instructions. Next, this complex was incubated for 10 min (RT) prior to incubating with cancer cells plating in a 24-well plate (37 °C, 5% CO2). Following CRISPR/Cas9 genome editing, cells were expanded and sorted to > 99% knockout purity (CD73) using fluorescence-activated cell sorting (BD Aria III cell sorter). CD73 KO GBM43 cells were placed in culture and expanded under the same conditions as WT GBM43 cells prior to being used in co-culture assays.
Cancer cell co-cultures for determination of expression of CD73 on NK cells
For co-culture studies to determine expression of CD73 on NK cells, adherent cancer cell lines were attached to the plate for at least four hours before co-incubation with NK cells. Next, the NK cells (effector cells) were incubated with the cancer cells (target cells) at a ratio of effector/target (E:T) of 2.5:1 and 10:1 in 48-well plates by removing the cancer cell media, washing the cancer cells once with 1X PBS, and replacing with NK cell culture media and NK cells. The NK cells were kept at a density around 1 × 106 cells/ml. Controls with primary cells or NK-92 only were also cultured. After co-culture for 6 h at 37 °C, 5% CO2, the supernatant with NK cells was collected, and the cancer cells were washed with 1X PBS, trypsinized, and added to the NK cell supernatant. K562 cells were collected at the same time as the NK cells. All cells were then stained and visualized using flow cytometry.
Induction of hypoxia
For hypoxic experiments, A549 and GBM43 cells were attached to the plate for at least four hours before co-incubation. One hour before co-incubation, cancer cell media was replaced with NK cell culture media containing 250 µM cobalt chloride and the cancer cells were left to rest in the incubator at 37 °C, 5% CO2. NK cells were then added into the wells with the cancer cells for 6 h before staining and flow cytometry.
Phenotypic analysis by flow cytometry
All antibodies for flow cytometric analysis were from Biolegend (San Diego, CA) unless otherwise stated. After co-incubation experiments, both cancer and NK cells were collected and washed with FACS buffer (1X PBS, 5% FBS) and then stained for 30 min at 4 °C with manufacturer-recommended amounts of antibodies, CD56 (PE-Cy5.5, clone: CMSSB, eBioscience, Thermo Fisher Scientific, Waltham, MA), CD3 (PE-CY7, Clone: UCHT1), and CD73 (APC, Clone: 82). After staining, cells were washed with FACS buffer and Sytox™ Green Dead Cell Stain (Thermo Fisher, Waltham, MA) was added for cell viability. For co-incubation studies with PC3, Sytox™ Blue Dead Cell Stain (Thermo Fisher, Waltham, MA) was used. Gating was performed using both dead cell staining and CD3− CD56+ (peripheral blood human NK) or CD56 + (NK92). Representation of the gating strategy is shown in Figure S1, while representative histogram for induction of CD73 expression is shown in Figure S2.
For analysis of patient tumor data, manufacturer-recommended amounts of antibodies CD56 (APC Fire 750, clone: 5.1H11), CD3 (FITC, clone HIT3a), CD14 (FITC, clone: M5E2), CD19 (FITC, clone: SJ25C1), CD16 (BUV395, clone: 3G8), CEACAM-1 (PE, clone: 283340), R&D Systems), CXCR3 (PECy7, Clone: G025H7), NKp46 (BV785, clone: 9E2), Tim-3 (APC, clone: F38-2E2) and CD73 (BV605, clone: AD2) were used. Sytox™ Blue Dead Cell Stain was also used for cell viability. Gating was performed using dead cell staining, CD3− CD14− CD19− to remove T-cells, B-cells, and macrophages, and CD56+NKp46+ for peripheral blood human NK cells. All cell populations were analyzed on the BD Fortessa (BD Biosciences, Franklin Lakes, NJ) and evaluated with FlowJo software (Tree Star, Ashland, OR).
For analysis of patient whole blood data, recommended amounts of antibodies CD56 (PE-Cy5.5, clone: CMSSB), CD3 (PE-CY6, Clone: UCHT1), and CD73 (APC, Clone: 82 or BV605, Clone: AD2) were used. When not indicated, the same antibodies were used as for patient tumor samples. Sytox™ Green or Blue Dead Cell Stain was also used for cell viability.
To phenotype CD73+ NK cells, after a 2.5:1 co-incubation with U87 WT cells, all cells were stained with antibodies prior to analysis by flow cytometry. Antibodies used for phenotyping included CD56 (FITC or PE, clone:5.1H11), CD3 (PE-Cy7, clone: UCHT1), CD73 (APC, clone:82), TIGIT (PerCP-Cy5.5, clone: A15153G), CD57 (BV605, clone:QA17A04), NKG2D (BV605, clone: 1D11), PD-1 (BV510, clone: NAT105), Tim-3 (BV711, clone: F38-2E2), and NKG2A (FITC, clone: 1D11). The Live/Dead Fixable Violet Stain (Thermo Fisher, Waltham, MA) was used to distinguish live cells. For CD107a and IFN-γ expression analysis, NK cells were co-cultured for 6 h with U87 WT cells in media containing CD107a-FITC (clone: H4A3) at an E:T ratio of 2.5:1. Brefeldin A and monensin were added after 1 h of co-culture. After a total of 6 h, NK cells were harvested, stained for CD56/CD3 surface markers, fixed, permeabilized, and stained for intracellular cytokines (IFN-γ-PerCP-Cy5.5 clone: BD.B3), before being analyzed on the BD Fortessa (BD Biosciences, Franklin Lakes, NJ).
HIF-1α expression by Western Blot
To determine the concentration of cobalt chloride needed to induce hypoxia through HIF-1α in A549 and GBM43 cells, cells were placed in normoxic, 100 µM, or 250 µM cobalt chloride for 2 h or 6 h before western blot analysis. Nuclear cell extracts were then prepared from normoxic or hypoxic cells scraped from dishes into ice cold hypotonic solution (20 mM Tris–HCl, 10 mM NaCl, and 3 mM MgCl2) containing HALT protease inhibitor (Thermo Fisher, Waltham, MA) for 15 min. Nonidet P-40 (10%) was then added to cell extracts and centrifuged to remove the cytoplasmic fraction. The nuclear pellet was resuspended in Cell Extraction Buffer (Thermo Fisher, Waltham, MA) with HALT protease inhibitor for 30 min on ice with incremental vortexing. Extracts were centrifuged and supernatant was collected for analysis. Protein was quantified using the Quant-iT™ Protein Quantitation Kit (Thermo Fisher, Waltham, MA). A total of 100 µg of protein was loaded onto a 4–15% SDS/polyacrylamide gel, and after electrophoresis, it was blotted onto nitrocellulose membranes. The primary mouse anti-human HIF-1α (BD, Franklin Lakes, NJ, clone: 54) was used at a 1:250 dilution, and the anti-lamin B1 antibody (Biolegend, San Diego, CA, Clone: 5G8-D3-H7) was used at a 1:500 dilution in OneBlock Western-CL Blocking Buffer (Prometheus, Genesee Scientific, San Diego, CA). The HRP goat anti-mouse antibody (BD, Franklin Lakes, NJ) was used at a 1:1000 dilution, and the signal was analyzed by chemiluminescence (SuperSignal West Pico PLUS substrate, Thermo Fisher, Waltham, MA). Under these conditions, HIF1-α appeared as a band around 100 kDa with degraded protein around 40–60 kDa. Blots were analyzed using Image Studio Lite (LI-COR Biotechnology, Lincoln, NE).
Data sharing
For original data, please contact sandro@purdue.edu.
Statistical analysis
Prism 9 (Graphpad Software, San Diego, CA) was used for all statistical analysis with a p < 0.05 (*) considered to be significant. Normality of data was tested with the Shapiro–Wilk test. Ordinary one-way analysis-of-variance tests, Brown–Forsythe tests, or Kruskal–Wallis tests were used for multiple-group comparisons. To compare unpaired sample groups, the Dunnett’s multiple comparison test, Dunnett’s T3 multiple comparison test, and the Dunn’s multiple comparison test were used. Equal variances for all ANOVA-based tests were checked with the Brown–Forsythe test.
Results
Cancer cells express different levels of CD73, while patient-derived NK cells express limited CD73
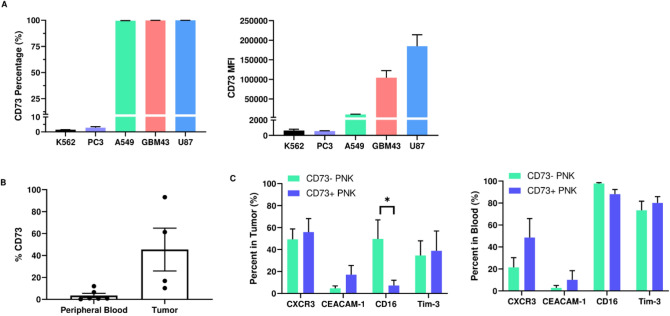
We found CD73 to be expressed on A549, GBM43 and U87MG cells with almost 100% of each cell population expressing the ectoenzyme. Despite the broad and consistent expression of CD73 on the three cancer cell lines, its expression intensity on these cell lines differs, with A549 cells displaying the lowest CD73 MFI, followed by GBM43 cells, and U87 cells with the highest CD73 MFI (Fig. 1A). PC3 cells, on the other hand, express very little CD73 (Fig. 1A). Similarly, leukemic cell line K562 does not express CD73.
Fig. 1.
Expression of CD73 on cancer cells and NK cells isolated from peripheral blood and tumors of GBM patients. A Percentage (left) and MFI (right) of CD73 expression on leukemia (K562), prostate (PC3), lung adenocarcinoma (A549), patient-derived glioblastoma (GBM43) and commercial glioblastoma (U87) cells; B expression of CD73 on NK cells freshly isolated from the peripheral blood and tumors of GBM patients (unpaired t-test); C phenotyping of CD73− and CD73+ NK cells isolated from GBM patient tumors and peripheral blood (unpaired t-test). *p < 0.05
Human NK cells from peripheral blood have also been reported to express low levels of CD73 [12]. NK cells extracted from the peripheral blood of GBM patients showed low expression of CD73, ranging from about 1–12%, depending on donor (Fig. 1B). On the other hand, NK cells extracted from tumors of GBM patients had a slightly higher expression of CD73, ranging from about 13–93%, depending on donor (Fig. 1B), but this was not significantly higher than CD73 expression on NK cells from patient blood.
We further analyzed how the phenotype of human NK cells extracted from tumors of GBM patients correlates to their CD73 expression, and found that CD73+ NK cells are characterized by significantly lower expression of activating receptor CD16, alongside patient-specific changes in inhibitory receptor CEACAM1 and chemokine receptor CXCR3, suggesting an altered activation phenotype and inhibitory state displayed by these cells (Fig. 1C). Specifically, CEACAM1 and CXCR3 expression was either increased or unchanged, depending on patient. Conversely, CD73+ NK cells from the blood of patients showed no changes in receptor expression compared to CD73− NK cells. For comparison, we also quantified expression of these receptors on these patient-derived NK cells stratified as CD56bright and CD56dim and found upregulated CXCR3 and downregulated CD16, as expected, on patient-derived NK cells (Figure S3).
NK cells acquire CD73 in response to high CD73-expressing cancer cells
The percentage of human peripheral blood-derived NK cells expressing CD73 did not significantly change upon co-culture with either PC3, A549 or GBM43 cells under normoxic conditions (Fig. 2A–C). However, peripheral blood human NK cells cultured with GBM43 cells did have a slight increase in CD73 at the E:T 2.5:1 ratio as indicated by a fold change of about 1.6. In these situations, the MFI remained consistent except in the case of A549 co-cultures, where it showed a slight decrease (0.8-fold change; Fig. 2B). In co-culture with U87 cells, on the other hand, CD73 expression on NK cells increased at an E:T 2.5:1, from 2.7% on peripheral blood-derived NK cells alone to 10% on NK cells after co-culture, with a fold change of about 3.7. (Fig. 2D). Additionally, the MFI increased by a fold change of about 1.33. Despite the increase in percentage of CD73-expressing NK cells, the overall MFI levels remained low before and after co-culture.
Fig. 2.
Induction of expression of CD73 on human peripheral blood-derived NK cells under normoxic conditions upon co-culture with cancer cells. Percentage (%, left panels), MFI (center panels) and fold change over peripheral blood NK cells alone (right panels) of CD73 expression on peripheral blood-derived human NK cells in co-culture with A PC3 [Brown–Forsythe (%), one-way ANOVA (MFI), no statistics since fold-change values were under one (fold-change)]; B A549 [Brown–Forsythe (%), one-way ANOVA (MFI), no statistics since fold-change values were under one (fold-change)]; C GBM43 [Kruskal–Wallis test (%), one-way ANOVA (MFI), one-way ANOVA (fold-change]; D U87 cells [one-way ANOVA (%), Kruskal–Wallis test (MFI), one-way ANOVA (fold-change)]. Co-cultures were carried out by co-incubating NK cells and cancer cells for 6 h at 37 °C + 5% CO2 at E:T of 2.5:1 and 10:1. Expression of CD73 was determined by flow cytometry. *p < 0.05
Expression of CD73 on NK-92 cells followed a similar pattern to that of peripheral blood NK cells, but these cells showed a broader acquisition of CD73 expression. While no change in CD73 expression was observed after co-culture with either K562, A549 or PC3 cells (Fig. 3A–C)—the cancer cells with the lowest levels of CD73—expression of CD73 on NK-92 cells changed after co-culture with both GBM43 and U87 cells (Fig. 3D, E). In the case of co-cultures with GBM43 cells, CD73 expression on NK-92 cells increased from 1.5 to 43%, while the MFI increased by a fold change of 1.8 (Fig. 3D) for the E:T 2.5:1 ratio. Co-culture with U87 cells induced a significant increase in CD73 expression on NK-92 cells at both E:T ratios, with an increase from 1% to 18% at the 2.5:1 ratio and from 1% to 6.8% at the 10:1 ratio. The MFI also increased with a fold change of 1.36 at the 2.5:1 ratio (Fig. 3E). Similarly to human peripheral blood-derived NK cells, however, overall MFI levels for CD73 expression remained low before and after co-culture.
Fig. 3.
Induction of expression of CD73 on NK-92 cells under normoxic conditions upon co-culture with cancer cells. Percentage (%, left panels), MFI (center panels) and fold change over peripheral blood NK cells alone (right panels) of CD73 expression on NK-92 cells in co-culture with A K562 [Kruskal–Wallis test (%), Kruskal–Wallis test (MFI), Kruskal–Wallis test (fold-change)]; B PC3 [one-way ANOVA (%), one-way ANOVA (MFI), one-way ANOVA (fold-change)]; C A549 [Brown–Forsythe (%), one-way ANOVA (MFI) one-way ANOVA (fold-change)]; D GBM43 [Kruskal–Wallis test (%), one-way ANOVA (MFI), Kruskal–Wallis (fold-change)]; E U87 cells [one-way ANOVA (%), one-way ANOVA (MFI), Kruskal–Wallis (fold-change)]. Co-cultures were carried out by co-incubating NK cells and cancer cells for 6 h at 37 °C + 5% CO2 at E:T of 2.5:1 and 10:1. Expression of CD73 was determined by flow cytometry. *p < 0.05
Hypoxia induces expression of CD73 on NK cells
Next, we sought to establish whether hypoxic conditions, such as those likely encountered by tumor-infiltrating NK cells in the microenvironment of solid tumors, can affect CD73 expression on them. Since we had seen no change in expression of CD73 on human peripheral blood-derived NK cells upon co-culture with A549 or GBM43 cells, we used these cells to check for CD73 expression after co-culture with human NK cells under hypoxic conditions. Induction of hypoxia was verified by Western blot via overexpression of HIF-1α (Figure S4). Under hypoxia, the percentage of CD73+ human NK cells increased after co-culture with both A549 and GBM43 cells. With A549 cells, CD73 expression increased from 2.8 to 12.4% at the 2.5:1 ratio, but the MFI remained unchanged (Fig. 4A). Similarly, co-cultures with GBM43 cells under hypoxia induced an increase in CD73 expression on human NK cells from 5.9 to 14.8% at the 2.5:1 ratio. In this case, the MFI also remained unchanged (Fig. 4B). Chemically-induced hypoxic conditions were also shown to upregulate expression of CD73 on cancer cells (Figure S5).
Fig. 4.
Induction of expression of CD73 on human peripheral blood-derived NK cells under hypoxic conditions upon co-culture with cancer cells. Percentage (%, left panels), MFI (center panels) and fold change over peripheral blood NK cells alone (right panels) of CD73 expression on peripheral blood-derived human NK cells in co-culture under hypoxia with A A549 [Brown–Forsythe (%), Kruskal–Wallis test (MFI), one-way ANOVA (fold-change)] and B GBM43 cells [one-way ANOVA (%), Brown–Forsythe (MFI), Kruskal–Wallis test (fold-change)]. Hypoxia was induced by treating cancer cells with NK cell media containing 250 µM cobalt chloride for one hour before co-culture with NK cells at 37 °C, 5% CO2 for 6 h at E:T of 2.5:1 and 10:1. C Histogram plots of co-cultures of PNK cells over 6 h at 37 °C under hypoxic conditions with GBM43 cells. Expression of CD73 was determined by flow cytometry. *p < 0.05
We performed similar studies under hypoxic conditions with NK-92 cells, which had previously shown an increase in CD73 expression following co-culture with GBM43 cells, but not A549 cells. When co-cultured with A549 cells, expression of CD73 on NK-92 cells increased from 1 to 9.4%, while the MFI increased by 1.25-fold at the 2.5:1 ratio (Fig. 5A). A more significant increase in CD73 expression was observed upon co-culture with GBM43 cells, where CD73+ NK-92 cells increased in percentage from 0.6 to 57% at the 2.5:1 ratio and 0.6% to 22.8% at the 10:1 ratio, while the MFI for overall CD73 expression increased by a 2.6 fold change at the 2.5:1 ratio (Fig. 5B). These increases were higher than those observed on NK-92 cells in co-culture with GBM43 cells under normoxic conditions.
Fig. 5.
Induction of expression of CD73 on NK-92 cells under hypoxic conditions upon co-culture with cancer cells. Percentage (%, left panels), MFI (center panels) and fold change over NK-92 cells alone (right panels) of CD73 expression on NK-92 cells in co-culture under hypoxia with A A549 [one-way ANOVA (%), Kruskal–Wallis test (MFI), Kruskal–Wallis test (fold-change)] and B GBM43 cells [one-way ANOVA (%), Kruskal–Wallis test (MFI), Kruskal–Wallis test (fold-change)]. Hypoxia was induced by treating cancer cells with NK-92 cell media containing 250 µM cobalt chloride for 1 h before co-culture with NK cells at 37 °C, 5% CO2 for 6 h at E:T of 2.5:1 and 10:1. Expression of CD73 was determined by flow cytometry. *p < 0.05
CD73 on cancer cells is a prerequisite for CD73 expression on NK cells
We next generated CD73 knockout (KO) patient-derived GBM43 cells using CRISPR/Cas9 gene editing in order to further validate whether the presence of CD73 is a prerequisite for acquisition of CD73 expression by NK cells, and subjected these cells to the same normoxic co-culture conditions with NK cells as previously. Because GBM43 cells had previously induced high CD73 expression on NK-92 cells, but not peripheral blood-derived NK cells, we decided to use NK-92 cells for these assays. Upon co-culture with CD73 KO cancer cells, we saw no induction of CD73 expression on NK cells at any of the E:T ratios tested (Fig. 6A, B). This contrasted with the significant expression induced in the presence of WT GBM43 cells, indicating that the presence of CD73 might be necessary for expression of CD73 on NK cells.
Fig. 6.
Effect of CD73 knockout (KO) on cancer cells on expression of CD73 on NK cells. A Induction of CD73 expression on NK cells after co-culture with CD73 KO GBM43 cells at E:T 2.5:1 and 10:1 [one-way ANOVA (%), Brown–Forsythe (MFI)]. B Histogram plots of co-cultures of NK-92 cells over 6 h at 37 °C under normoxic conditions with CD73 CRISPR KO GBM cells
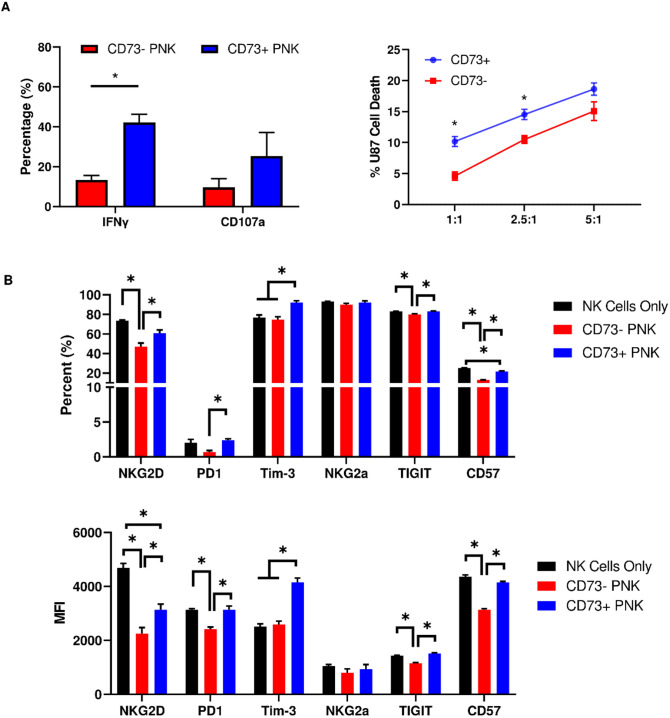
CD73+ NK cells are hyperfunctional compared to CD73− NK cells against cancer targets
To understand the functional significance of CD73 expression on NK cells, we evaluated cytokine activity through IFN-γ and degranulation through CD107a production. CD73+ NK cells showed higher IFN-γ production (Fig. 7A), and although degranulation by CD73+ NK cells was also higher, it was not significant. CD73+ NK cells were shown to be more cytotoxic than CD73− NK cells, as measured by a higher killing capacity both before (Fig. 7A) and after resting (Figure S6). We then phenotyped CD73+ NK cells and found them to express higher NKG2D, PD-1, TIM-3, TIGIT, and CD57 compared to CD73− NK cells after co-culture with cancer cells (Fig. 7B). However, only NKG2D, TIM-3 and CD57 expression differed from CD73+ NK cells compared to CD73− NK cells not co-incubated with cancer cells.
Fig. 7.
Functional and phenotypic characterization of CD73+ NK cells. (A, left) Production of IFN-γ and CD107a from CD73+ and CD73− NK cells at an E:T ratio of 2.5:1 (unpaired t-test); (A, right) killing of U87MG cells by sorted CD73+ and CD73− NK cells at three E:T ratios (unpaired t-test); B percentage (top) and MFI (bottom) of expression of receptors on CD73+ and CD73− NK cells in co-culture with U87 cells an E:T ratio of 2.5:1 [one-way ANOVA (%), one-way ANOVA (MFI)] *p < 0.05
In summary, while both human peripheral blood-derived NK cells and NK-92 cells increased their expression of CD73 following co-culture with CD73+ cancer cells, this depended on the presence and levels of CD73 expression on cancer cells, as well as the presence of hypoxia. Generally, the higher the CD73 expression on cancer cells, the higher the likelihood that CD73 expression on NK cells would increase, while CD73 expression on cancer cells was required for induction of CD73 on NK cells. Overall, it should be noted the expression of CD73 on NK cells is and remains low even after co-culture and appears to be induced most potently only in the presence of cancer cells that express substantial CD73. Functional analysis of CD73 reveals a potentially hyperfunctional subset of NK cells associated with higher receptor expression levels compared to CD73− NK cells in contact with cancer cells.
Discussion
In the microenvironment of solid tumors, adenosine is produced primarily by the ectoenzyme CD73 from the hydrolysis of precursor ATP to AMP through the action of CD39; however, a non-canonical pathway of adenosine production, independent of CD39, involving CD38 is also active [2, 11, 21]. CD73, by producing adenosine, causes serious immunosuppression of NK cell effector functions [12]. Though peripheral blood-derived NK cells do not express CD73 on their surface, some reports have suggested that NK cells are able to induce expression of CD73 on their surface under specific conditions. One such condition includes exposure to mesenchymal stem cells [22], which increased expression of CD73 on NK cells from ~ 1 to 5–10% upon co-culture. Higher expression of CD73 on NK cells was also observed on NK cells isolated from human breast cancer tumors [23], gastrointestinal stromal tumors [24] and from mouse melanoma (SM1WT1) tumors [15]. More recently, induction of CD73 expression on NK cells in the tumor microenvironment was associated with an increase in expression of inhibitory receptors, including PD-1, LAG-3 and PD-L1, alongside an upregulation of regulatory-like functions which include a heightened production of IL-10 [16]. Although upregulation was demonstrated in a co-culture setting, results were pooled from multiple cancer cell co-cultures, thus preventing analysis isolated to one specific cancer type.
Despite evidence suggesting that NK cells are able to acquire some CD73 expression, conflicting reports of such expression on NK cells extracted from patient samples obfuscate firm conclusions. For instance, Perrot et al. [17] reported no significant change in CD73 expression on NK cells isolated from peripheral blood or tumors.
Based on these conflicting results, we sought to investigate if the induction of CD73 expression on NK cells is a conserved process that occurs consistently upon contact with different types of cancer cells. We found this not to be the case. Generally, our study indicated that NK cells are largely resistant to the upregulation of CD73, and that this occurs under specific conditions. Interestingly, we observed that NK cells upregulate CD73 primarily after contact with cancer cells that have a high expression of CD73. In other words, the higher the CD73 expression on cancer cells, the more likely it was for NK cells to upregulate CD73 on their surface. Accordingly, there was no change in expression of CD73 on NK cells in co-culture with either K562 cells or PC3 cells, both of which express low to no CD73. The same pattern of behavior occurred on both human peripheral blood NK cells as well as on NK-92 cells, with NK-92 cells displaying more intense responses to cancer cell co-culture in terms of their ability to upregulate CD73. This heightened response on NK-92 cells may be related to the fact that NK-92 cells are sourced from a cancer cell line, or that primary NK cells are generally more adverse to the uptake of exogenous material, though the exact reason is not known. Further, CRISPR knockout of CD73 on cancer cells completely abrogated the increase in CD73 expression that occurred on NK cells in the presence of WT cancer cells, suggesting that the presence of CD73 is required for the upregulation of CD73 on NK cells that come in contact with them.
This was true under hypoxic conditions as well, which yielded an additional increase in CD73 expression on NK cells by upregulating HIF1-α, a driver of CD73 expression. Generally, however, expression of NK cells after co-culture remained low, with limited change in CD73 expression intensity, even in cases where the percentage of CD73+ NK cells increased significantly. Nonetheless, the ability for NK cells to acquire CD73 expression was consistent, despite the low levels at which this occurred with human NK cells in vitro.
The mechanism of CD73 expression in contact with cancer cells is unclear. Recent reports suggest that NK cells transport intracellularly expressed CD73 to the cell surface via exocytosis, upon contact with cancer cells or fibroblasts in a 4-1BB-dependent manner [16]. Our data suggest that CD73 expression levels on cancer cells themselves drive the strength of CD73 expression on NK cells. While HIF1-α is involved in this process, normoxic data suggest this not to be the only mechanism at play. Trogocytosis is a mechanism that has been suggested as being involved in upregulation of receptors on NK cells and may be playing a role in acquisition of CD73 by NK cells. Shedding of CD73 from cancer cells could also be involved in this process. The phenomenon was also reversible, as NK cells appeared to lose some expression of CD73 they had acquired upon removal of cancer cells, such as by being placed in culture (data not shown). Interestingly, the functional changes in terms of higher killing ability these cells displayed upon uptake of CD73+ were maintained after resting in culture following sorting (Figure S6). What is clear, however, was that this process was not universal, but rather cancer cell-specific.
The hyperfunctionality of CD73+ NK cells against cancer targets correlated to higher expression of functional markers of NK cell activation, such as IFN-γ and NKG2D, higher levels of receptors TIM-3, PD-1, and TIGIT, and slightly higher senescence marker CD57 compared to CD73− NK cells upon co-culture with cancer targets. In addition, CD73+ NK cells were more cytototic against cancer targets than CD73− NK cells. This suggests that CD73 could be associated with NK cell activation, possibly maturation, and that the upregulation of CD73 may correlate with a hyperfunctional state of activation on NK cells. Though TIM-3 has been thought of as a marker of NK cell dysfunction, studies have shown that in vitro, it can also be associated with functional and potentially cytotoxic NK cells [1, 2]. Additionally, in our studies both populations of NK cells after co-culture with cancer targets showed downregulated NKG2D and upregulated TIM-3. Patient data also indicated that CD73 expression on NK cells isolated from human GBM tumors upregulate CD73 only to a certain extent. These data largely agree with evidence by others that CD73 expression on NK cells in tumors is not a significant phenotypic feature of these cells in the TME. Based on our in vitro data, which showed that hypoxia is able to enhance CD73 expression on NK cells, the lack of a substantial increase in CD73 expression on NK cells from some patient tumors could be explained by the dynamic and heterogeneous nature of tumor-experienced NK cells. We hypothesize, based on data from our and other groups, that CD73 expression on NK cells is an environmentally-driven process. NK cells extracted from tumors represent a diverse population of spatially distinct cells: some peripheral, others more infiltrative. Since CD73 expression appears to require distinct functional triggers, it is likely that NK cells more peripheral to the tumor change their expression of CD73 either very little, or not at all. NK cells that do acquire expression of CD73 in vivo, however, appear to undergo other functional changes to a more inhibitory, or “dysfunctional” phenotype. In our case, significantly lower expression of CD16 observed in patient samples occurred alongside CD73 upregulation on patient tumor-infiltrating NK cells. This was accompanied by a number of patients displaying enhanced expression of the inhibitory receptor CEACAM1. It is possible that these cells, like recent studies suggest, adopt an “inhibitory” and/or “dysfunctional” role in the tumor microenvironment with implications in tumor progression and metastasis. Acquisition of an inhibitory identity might be the dominant phenomenon that is associated with NK cell dysfunction in vivo.
Taken together, our data showed that CD73 expression on NK cells is not a universal process, but instead occurs in response to specific triggers in the tumor microenvironment and correlates to the expression of CD73 on cancer cells. We also show that presence of high levels of CD73 on cancer cells is a requirement for induction of CD73 expression on NK cells. The functional changes that these NK cells undergo are indicative of a role in CD73 driving NK cell function and could have implications in the immunotherapeutic targeting of CD73 [3] and role in the progression of tumor growth. Additionally, our study provides new implications for the possible role and function of CD73+ NK cells in adoptive cell therapy [25].
Supplementary Information
Below is the link to the electronic supplementary material.
Author contributions
AMC and SM developed, planned the experiments, and wrote the manuscript. AMC developed the methodology, analyzed the data, and designed the figures. AMC, TND, JW, KBL, PV, and MGA performed experiments. VS and ACG provided patient samples. SM provided funding. All authors provided feedback and agreed to the published version of the manuscript.
Funding
The funding statement should be changed to: This work was supported by the V Foundation for Cancer Research (Grant #D2019-039), a Lilly Graduate Fellowship and a Migliaccio/Pfizer Graduate Fellowship to Andrea Chambers, and a McKeehan Graduate Fellowship to Kyle Lupo. The authors also gratefully acknowledge the support of the Biological Evaluation Shared Resource and the Flow Cytometry Shared Resource, with support from the Purdue Center for Cancer Research, NIH grant P30 CA023168, the IU Simon Cancer Center NIH grant P30 CA082709, and the Walther Cancer Foundation.
Declarations
Conflict of interest
The authors declare no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Antonioli L, Blandizzi C, et al. Anti-CD73 immunotherapy: a viable way to reprogram the tumor microenvironment. Oncoimmunology. 2016;5(9):e1216292. doi: 10.1080/2162402X.2016.1216292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allard B, et al. The ectonuucleotidases CD39 and CD73: novel checkpoint inhibitor targets. Immunol Rev. 2017;276(1):121–144. doi: 10.1111/imr.12528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang B. CD73: a novel target for cancer immunotherapy. Cancer Res. 2010;70(16):6406–6411. doi: 10.1158/0008-5472.CAN-10-1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heuts DP, Weisenborn MJ, et al. Crystal structure of a soluble form of human CD73 with ecto-5’-nucleotidase activity. ChemBioChem. 2012;13(16):2384–2391. doi: 10.1002/cbic.201200426. [DOI] [PubMed] [Google Scholar]
- 5.Wang R, Zhang Y, et al. Prognositic value of CD73-adenosinergic pathway in solid tumor: a meta-analysis and systematic review. Oncotarget. 2017;8(34):57327–57336. doi: 10.18632/oncotarget.16905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stagg J, et al. Anti-CD73 antibody therapy inhibits breast tumor growth and metastasis. Proc Natl Acad Sci. 2010;107(4):1547–1552. doi: 10.1073/pnas.0908801107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loi S, et al. CD73 promotes anthracycline resistance and poor prognosis in triple negative breast cancer. Proc Natl Acad Sci. 2013;110(27):11091–11096. doi: 10.1073/pnas.1222251110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Synnestvedt K, et al. Ecto-5’-nucleotidase (CD73) regulation by hypoxia-inducible factor-1 mediates permeability changes in intestinal epithelia. J Clin Investig. 2002;110(7):993–1002. doi: 10.1172/JCI0215337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zagzag D, et al. Expression of hypoxia-inducible factor 1alpha in brain tumors: association with angiogenesis, invasion, and progression. Cancer. 2000;88(11):2606–2618. doi: 10.1002/1097-0142(20000601)88:11<2606::AID-CNCR25>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 10.Decking UK, et al. Hypoxia-induced inhibition of adenosine kinase potentiates cardiac adenosine release. Circ Res. 1997;81(2):154–164. doi: 10.1161/01.RES.81.2.154. [DOI] [PubMed] [Google Scholar]
- 11.Chambers AM, Matosevic S. Immunometabolic dysfunction of natural killer cells mediated by the hypoxia-CD73 axis in solid tumors. Front Mol Biosci. 2019;6:60. doi: 10.3389/fmolb.2019.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chambers AM, et al. Adenosinergic signaling alters natural killer cell functional responses. Front Immunol. 2018;30:9. doi: 10.3389/fimmu.2018.02533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morandi F, et al. CD56brightCD16− NK cells produce adenosine through a CD38-mediated pathway and act as regulatory cells inhibiting autologous CD4+ T cell proliferation. J Immunol. 2015;195(3):965–972. doi: 10.4049/jimmunol.1500591. [DOI] [PubMed] [Google Scholar]
- 14.Vivier E, Ugolini S. Regulatory natural killer cells: new players in the IL-10 anti-inflammatory response. Cell Host Microbe. 2009;6(6):493–495. doi: 10.1016/j.chom.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 15.Young A, et al. A2AR adenosine signaling suppresses natural killer cell maturation in the tumor microenvironment. Can Res. 2018;78(4):1003–1016. doi: 10.1158/0008-5472.CAN-17-2826. [DOI] [PubMed] [Google Scholar]
- 16.Neo SY, et al. CD73 immune checkpoint defines regulatory NK cells within the tumor microenvironment. J Clin Investig. 2020;130(3):1185–1198. doi: 10.1172/JCI128895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perrot I, et al. Preclinical development of humanized CD39 and CD73 blocking antibodies targeting the ATP/adenosine immune checkpoint pathway for cancer immunotherapy. Cancer Res. 2018;78:2718–2718. doi: 10.1158/1538-7445.AM2018-2718. [DOI] [Google Scholar]
- 18.Sarkaria JN, et al. Use of an orthotopic xenograft model for assessing the effect of epidermal growth factor amplification on glioblastoma radiation response. Clin Cancer Res. 2006;12:2264–2271. doi: 10.1158/1078-0432.CCR-05-2510. [DOI] [PubMed] [Google Scholar]
- 19.Carlson BL, Pokorny JL, Schroeder MA, Sarkaria JN. Establishment, maintenance, and in vitro and in vivo applications of primary human glioblastoma multiforme (GBM) xenograft models for translational biology studies and drug discovery. Curr Protoc Pharmacol. 2011;14:14–16. doi: 10.1002/0471141755.ph1416s52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang H, et al. Combination therapy in a xenograft model of glioblastoma: enhancement of the antitumor activity of temozolomide by and MDM2 antagonist. J Neurosurg. 2017;126:446–459. doi: 10.3171/2016.1.JNS152513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen S, et al. CD73: an emerging checkpoint for cancer immunotherapy. Immunotherapy. 2019;11:983–997. doi: 10.2217/imt-2018-0200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chatterjee D, et al. Natural killer cells acquire CD73 expression upon exposure to mesenchymal stem cells. Blood. 2014;123:594–595. doi: 10.1182/blood-2013-09-524827. [DOI] [PubMed] [Google Scholar]
- 23.Buisseret L, et al. Abstract 3361: CD73 expression on tumor-infiltrating breast cancer leukocytes. Cancer Res. 2015;75:3361–3361. doi: 10.1158/1538-7445.AM2015-3361. [DOI] [Google Scholar]
- 24.Vijayan D, et al. Selective activation of anti-CD73 mechanisms in control of primary tumors and metastases. Oncoimmunology. 2017;6:e1312044. doi: 10.1080/2162402X.2017.1312044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang J, Lupo KB, Chambers AM, Matosevic S. Purinergic targeting enhances immunotherapy of CD73+ solid tumors with piggyBac-engineered chimeric antigen receptor natural killer cells. J Immunother Cancer. 2018;6:136. doi: 10.1186/s40425-018-0441-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.