Abstract
Background
It is widely considered that pancreatic cancer (PC) is an immunosuppressive cancer. Immune-based therapies remain promising therapeutic strategies for PC. Overexpression of lipase H (LIPH) was reported to be related to immunity in cattle and has also been demonstrated to promote tumor progression in several tumors, but its role in pancreatic carcinogenesis remains unclear. Study on LIPH in PC might provide a new insight into the immunosuppression in PC.
Methods
The potential biological and clinical significance of LIPH was evaluated by bioinformatics analysis. We further investigated potential associations between the expression of LIPH and tumor immune infiltration using the CIBERSORT algorithm, the ESTIMAT algorithm, and single sample gene set enrichment analysis (ssGSEA).
Results
LIPH was significantly overexpressed in tumor tissues compared with normal tissues. LIPH overexpression correlated with tumor recurrence, advanced histologic grade, and poorer overall survival (OS). Four of the most common somatic mutation, including KRAS, TP53, CDKN2A, and SMAD4, in PC were all correlated with high LIPH expression. And high LIPH expression was significantly correlated with KRAS activation and SMAD4 inactivation. Besides, LIPH expression was involved in various biological pathways such as negative regulation of cell–cell adhesion, actin cytoskeleton, EMT, angiogenesis, and signaling by MST1. And LIPH overexpression caused high infiltration of TAMs, Treg cells, and Th2/Th1, but reduced the infiltration of CD8+ T cells and Th1 cells.
Conclusions
Our findings demonstrated that LIPH correlated with immune suppression or evasion and may function as a novel unfavorable prognostic biomarker in PC.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00262-021-03019-x.
Keywords: Pancreatic cancer (PC), Lipase H (LIPH), The Cancer Genome Atlas (TCGA), The Gene Expression Omnibus (GEO), Immune infiltration
Introduction
As one of the most aggressive malignancies, pancreatic cancer (PC) causes nearly 5% of all cancer-related deaths worldwide [1]. Poor survival is attributed to its high aggressiveness and chemotherapeutic resistance [2]. Presently, surgery remains the major therapy for PC, but only 20% of PC patients present with surgically resectable status, 80% among which die within 5 years [3]. PC is a highly immunosuppressive cancer and unique from an immunological perspective [4]. Immune-based therapies that recruit or enhance antitumor immune cells in the tumor microenvironment (TME) remain novel therapeutic strategies for PC [5]. In recent years, immune checkpoint inhibitors (ICIs) have been promising potent drugs in the treatment of several solid tumors, such as malignant melanoma, non-small cell lung cancer, hepatocellular carcinoma, triple negative breast cancer, and head-neck squamous cell carcinoma, but so far, lack of efficacy in advanced PC patients [6–12]. Therefore, a deeper understanding of the molecular mechanisms involved in immune suppression is required, which would helpful to develop immune-based therapies and improve prognosis in PC [13].
LIPH, also known as mPA-PLA1, is a protein coding gene that encodes a membrane-bound protease that can catalyze the production of 2-acyl lysophosphatidic acid (LPA) [14, 15]. Moreover, LPA was reported as a lipid mediator with diverse biological properties that include proliferation, migration, survival, and angiogenesis in multiple cancers [16]. Early studies about LIPH focused on its mutation and its correlation with hypotrichosis [17, 18]. Besides, Orozco-terWengel et al. reported that LIPH was related to immunity in cattle [19]. However, oncologic researches about LIPH have been conducted in only four cancers (e.g., breast cancer, lung cancer, papillary thyroid carcinoma, and esophageal adenocarcinoma) [14, 20–22]. The precise biological mechanism of LIPH in PC progression remains poorly understood.
In the present study, we systemically analyzed the expression pattern of LIPH and potential biological role in PC. For the first time, potential correlation between LIPH expression and immune cells infiltration levels in PC was investigated using the CIBERSORT algorithm, the ESTIMATE algorithm, and ssGSEA [23, 24].
Materials and methods
Data acquisition
The mRNA-sequencing data and clinical information of patients with PC were obtained from the Cancer Genome Atlas (TCGA, https://cancergenome.nih.gov/) database [13, 25]. Mutation data on KRAS, TP53, CDKN2A, and SMAD4 in TCGA PC dataset were obtained from cBioportal database (http://www.cbioportal.org/) [25]. Of the 177 PC cases in TCGA PC dataset, 171 were patients with OS > 1 month [25]. In addition, several GEO datasets, including GSE79668, GSE28735, GSE60979, and GSE62452, were selected for further analysis. All datasets are freely available as public resources. Consequently, local ethics approval was not required.
LIPH expression analysis
Differential expression analysis for LIPH was, respectively, performed in GSE60979 and GSE62452 datasets. Then, LIPH expression in PC was further validated in the Oncomine database (https://www.oncomine.org/resource/main.html) and Gene Expression Profiling Interactive Analysis (GEPIA; http://gepia.cancerpku.cn/index.html). And GSE28735 dataset (45 pairs of adjacent non-tumor tissues and pancreatic tumor) was used for paired differential expression analysis of LIPH. Moreover, the Human Protein Atlas database (http://proteinatlas.org/) was used to further validate the protein expression of LIPH and the expression of LIPH in cancer cell lines.
Survival analysis
Kaplan–Meier (KM) survival analysis was conducted to investigate the correlation between LIPH expression and OS of PC patients in TCGA PC dataset, GSE79668 dataset, and GSE62452 dataset. The optimal cutoff points were, respectively, obtained from the X-tile 3.6.1 software (Yale University, New Haven, CT, USA), and patients were, respectively, divided into low expression (low-Exp) and high expression (high-Exp) groups [13, 26].
Association between LIPH expression and somatic mutation
Early studies reviewed that KRAS, TP53, CDKN2A, and SMAD4 mutation are four of the most frequent genetic alterations for PC [25, 27]. Firstly, we evaluated the association between these mutation statuses and LIPH expression. Then, we also evaluated the association between LIPH expression and these four genes in TCGA PC dataset and GSE62452 dataset. Besides, we also assessed the association between the total mutational burden (TMB) and LIPH expression. In summary, we tried to preliminarily figure out whether somatic mutation level had an influence on the expression of LIPH in PC.
Functional enrichment analysis of LIPH expression
Co-expression genes of LIPH in TCGA PC dataset and the Broad Institute Cancer Cell Line Encyclopedia (CCLE) were separately screened out with the threshold of |Pearson correlated coefficient|> 0.6 and P < 0.05 [13]. Venny 2.1.0 (https://bioinfogp.cnb.csic.es/tools/venny/index.html) was used to figure out the overlapped co-expression genes between TCGA PC dataset and CCLE database. Then, the overlapped co-expression genes were imported in ConsensusPathDB (http://cpdb.molgen.mpg.de/) for functional enrichment analysis; P < 0.05 was considered statistically significant [13, 28].
Immune infiltration analysis for TCGA PC dataset
The CIBERSORT algorithm is an analysis tool that assesses specific immune cell fractions using gene expression data [23]. Therefore, the CIBERSORT algorithm was performed to evaluate the infiltration level of 22 immune cell types in TCGA PC dataset. The PC samples with a CIBERSORT output of P < 0.05 were included for further study. Subsequently, using R package estimate, the ESTIMATE algorithm was performed to generate an immune score and a tumor purity score [24]. A higher tumor purity score indicates a low level of immune cell infiltration in tumor tissue. PC samples with higher immune scores showed higher infiltration level of immune cells in tumor tissues [25]. Then, using R package gsva, ssGSEA was performed to assess the enrichment levels of immune-related terms in the cancer samples [29]. The following 29 immune-related terms were obtained: tumor-infiltrating lymphocyte (TIL), CD8+ T cells, regulatory T cells (Treg), cytolytic activity, type-2 T helper cells (Th2 cells), T cell co-stimulation, type-1 T helper cells (Th1 cells), T cell co-inhibition, checkpoint, natural killer cells (NK cells), tumor-associated macrophages (TAMs), antigen-presenting cell (APC) co-stimulation, major histocompatibility complex (MHC) class-1, antigen-presenting cell (APC) co-inhibition, follicular helper T cells (Tfh), type-1 IFN response, dendritic cells, parainflammation, plasmacytoid dendritic cells (pDCs), activated dendritic cells (aDCs), immature dendritic cells (iDCs), mast cells, B cells, neutrophils, inflammation-promoting, human leukocyte antigen (HLA), T helper cells, type-2 IFN response, and chemokine receptor (CCR) [30]. Using R package sparcl, TCGA PC dataset was divided into three clusters—immunity L, immunity M, and immunity H—according to the enrichment scores of 29 immune-related terms. Moreover, GSE62452 dataset was utilized to validate the immune infiltration of PC through the ESTIMATE algorithm and ssGSEA method.
Statistical analysis
All statistical analyses were performed using R software (http:///www.r-project.org/), GraphPad prism 8.0 software (San Diego, CA, USA), and SPSS 25.0 software (Chicago, IL, USA). The chi-square test or Fisher’s exact test and contingency analysis were used to assess the association between LIPH expression and clinicopathological features. Correlations were assessed using Pearson correlated coefficient. Group differences were evaluated by the Student’s t-test and expressed as mean ± SD. Statistical significance was defined by a value of P < 0.05.
Results
LIPH overexpression predicts poor prognosis in PC
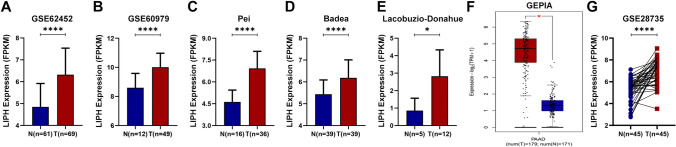
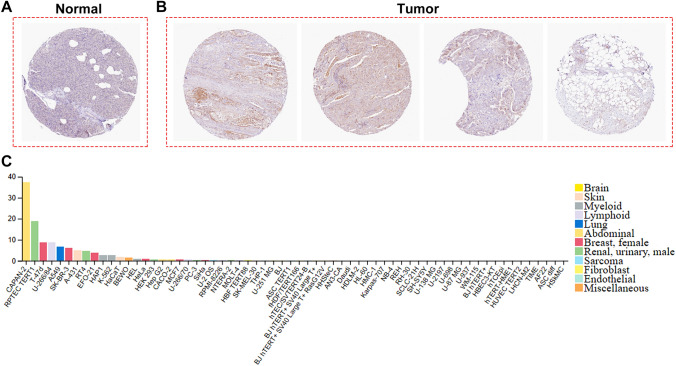
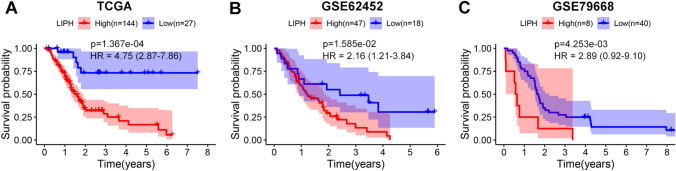
In GSE62452 and GSE60979 datasets, LIPH was overexpressed in PC tissues compared with normal pancreatic tissues (P < 0.0001) (Fig. 1A, B). Three studies in the Oncomine database showed that LIPH was significantly upregulated in PC tissues (P < 0.05) (Fig. 1C–E). And similar result was found in the GEPIA database (Fig. 1F). In GSE28735 dataset, LIPH was overexpressed in tumor tissues when compared with that in the adjacent non-tumor tissues (P < 0.0001) (Fig. 1G). We also used the Human Protein Atlas database to investigate the protein expression of LIPH and the expression of LIPH in cancer cell lines, and demonstrated that the protein expression of LIPH in PC tissues was significantly upregulated (Fig. 2A, B), and CAPAN-2 cells showed much higher LIPH expression compared with various other cancer cells (Fig. 2C). Of note, patients with lower expression of LIPH had a better survival than those with higher expression of LIPH (P < 0.05) (Fig. 3A–C). Taken together, the current study identified LIPH as an unfavorable prognostic factor for patient with PC.
Fig. 1.
Multiple databases demonstrated that LIPH was overexpressed in PC. A, B The expression of LIPH in PC was significantly upregulated in PC tissues compared with that in normal tissues in both the GSE62452 and the GSE60979 datasets. C–E Three studies in the Oncomine database demonstrated that LIPH was overexpression in PC tissues. F GEPIA database demonstrated that the expression of LIPH was significantly higher in PC tissues than that in normal tissues. G GSE28735 dataset demonstrated the expression LIPH was upregulated in PC tissues compared with that in the adjacent non-tumor tissues. (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001). PC, pancreatic cancer; LIPH, lipase H; N, normal; T, tumor
Fig. 2.
The Human Protein Atlas database analysis. A, B The Human Protein Atlas database was used to validate the protein expression of LIPH, which demonstrated that the protein expression of LIPH was significantly upregulated in tumor tissues. C The expression of LIPH in CAPAN-2 cells was much higher than that in various other cancer cells
Fig. 3.
KM survival analysis for LIPH through TCGA, GSE62425, and GSE79668 datasets. LIPH overexpression was significantly associated with poor survival in PC (P < 0.05). KM, Kaplan–Meier; HR, hazard ratio
Correlation between LIPH expression and the clinicopathological characteristics of PC
The correlation between LIPH expression and clinicopathological characteristics of PC is shown in Table 1. Higher expression of LIPH was significantly correlated with advanced histologic grade (P = 0.000008) and tumor recurrence (P = 0.001). These results indicated that LIPH overexpression was associated with tumor progression in PC.
Table 1.
Correlation of LIPH expression to clinicopathological features in PC
| Parameters | LIPH expression | P | |
|---|---|---|---|
| Low (n = 27) | High (n = 144) | ||
| Age | |||
| ≦60 | 9 (33.3%) | 48 (33.3%) | 1.00 |
| > 60 | 18 (66.7%) | 96 (66.7%) | |
| Gender | |||
| Female | 13 (48.1%) | 65 (45.1%) | 0.77 |
| Male | 14 (51.9%) | 79 (54.9%) | |
| AJCC stage | |||
| I–IIa | 9 (33.3%) | 38 (26.4%) | 0.33 |
| IIb–IV | 16 (59.3%) | 105 (72.9%) | |
| Unknown | 2 (7.4%) | 1 (0.7%) | |
| Histologic grade | |||
| G1 | 11 (40.7%) | 17 (11.8%) | 0.000008**** |
| G2 | 10 (37.0%) | 82 (56.9%) | |
| G3 | 3 (11.1%) | 44 (30.6%) | |
| G4 | 2 (7.4%) | 0 | |
| Unknown | 1 (3.7%) | 1 (0.7%) | |
| Recurrence | |||
| No | 18 (66.7%) | 48 (33.3%) | 0.001** |
| Yes | 9 (33.3%) | 96 (66.7%) | |
| Alcohol history | |||
| No | 10 (37.0%) | 52 (36.1%) | 0.99 |
| Yes | 15 (55.6%) | 82 (56.9%) | |
| Unknown | 2 (7.4%) | 10 (6.9%) | |
| Diabetes history | |||
| No | 16 (59.3%) | 89 (61.8%) | 0.77 |
| Yes | 5 (18.5%) | 31 (21.5%) | |
| Unknown | 6 (22.2%) | 24 (16.7%) | |
| Tumor size | |||
| < 4 | 17 (63.0%) | 73 (50.7%) | 0.21 |
| ≧4 | 10 (37.0%) | 58 (40.3%) | |
| Unknown | 0 | 13 (9.0%) | |
| Tumor site | |||
| Head | 17 (63.0%) | 116 (80.6%) | 0.07 |
| Body and tail | 21 (77.8%) | 6 (4.2%) | |
| Unknown | 7 (25.9%) | 4 2.8%) | |
Statistical significance was calculated by the chi-square test and Fisher’s extract test
Association between LIPH expression and somatic mutation
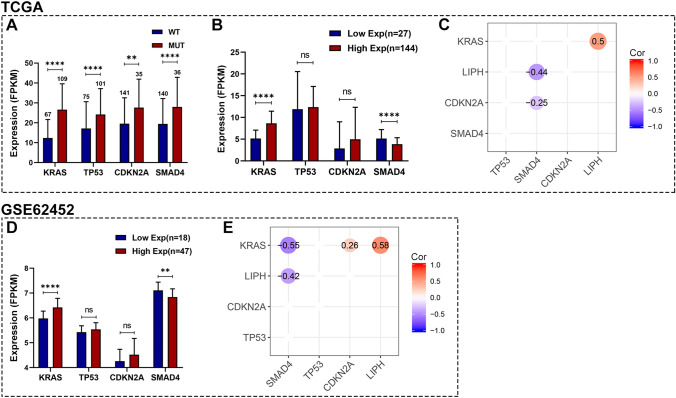
The mutation landscape of PC in TCGA PC dataset is shown in Figure S2, and consistently, KRAS, TP53, CDKN2A, and SMAD4 were four of the most frequent genetic alterations for PC. Our study showed that KRAS, TP53, CDKN2A, and SMAD4 mutation status were significantly correlated with LIPH overexpression (Fig. 4A). Differential expression analysis for these 4 genes between the high- and low-Exp LIPH groups in TCGA PC dataset revealed that KRAS was significantly upregulated in high-Exp group, while SMAD4 was significantly downregulated in high-Exp group (Fig. 4B). And correlation analyses demonstrated positive correlation between LIPH expression and KRAS expression (Cor = 0.5, P < 0.05), while negative correlation between LIPH expression and SMAD4 expression (Cor = − 0.44, P < 0.05) (Fig. 4C). Similar results were found in GSE62452 dataset (Figure D, E). Besides, the correlations among LIPH, KRAS, and SMAD4 were also validated in cBioportal database, which were consistent with our findings (Figure S3A-B). In addition, Figure S4 demonstrated that LIPH expression was significantly correlated with TMB. These results suggested that LIPH highly correlated with KRAS and SMAD4, and thereby promoted tumor progression through cooperating with KRAS and SMAD4.
Fig. 4.
Association between LIPH expression and somatic mutations. A KRAS, TP53, CDKN2A, and SMAD4 mutation status were significantly associated with higher expression of LIPH. B Differential expression analysis of KRAS, TP53, CDKN2A, and SMAD4 in high- and low-Exp groups in TCGA PC dataset. C Correlation matrix of KRAS, TP53, SMAD4, and CDKN2A in TCGA PC dataset. D Differential expression analysis of KRAS, TP53, CDKN2A, and SMAD4 in high- and low-Exp groups in the GSE62452 dataset. E Correlation matrix of KRAS, TP53, SMAD4, and CDKN2A in the GSE62452 dataset. (*P < 0.05; **P < 0.01;***P < 0.001; ****P < 0.0001). PC, pancreatic cancer; TCGA, the Cancer Genome Atlas; Exp, expression; Cor, Pearson correlated coefficient
Functional enrichment analysis of LIPH expression in PC
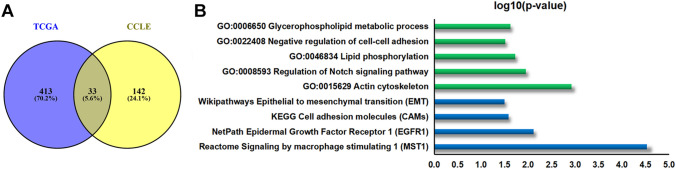
We further investigated the biological role of LIPH in PC. We conducted co-expression analysis (Pearson correlated coefficient|> 0.6, P < 0.05) for LIPH based on TCGA PC dataset and the CCLE database. Significantly overlapped co-expression genes (33 overlapped co-expression genes; see in Fig. 5A, Table 2) were imported in ConsensusPathDB and subjected to functional enrichment analysis (P < 0.05) (Fig. 5B). GO enrichment analysis showed that LIPH might play a vital role in negative regulation of cell–cell adhesion, regulation of Notch signaling pathway, actin cytoskeleton, lipid phosphorylation, and glycerophospholipid metabolic process (Fig. 5B). Moreover, pathway enrichment analysis revealed that LIPH may be important for the regulation of cell adhesion molecules, epithelial to mesenchymal transition (EMT), epidermal growth factor receptor 1 (EGFR1), and signaling by macrophage stimulating 1 (MST1) (Fig. 5B). These results implied that the expression of LIPH provided necessary support for tumorigenesis, progression, and immune infiltration in PC.
Fig. 5.
Co-expression analysis and functional enrichment analysis for LIPH. A Venn diagrams showing the co-expression genes of LIPH in TCGA dataset and CCLE database. B GO and pathway enrichment analysis for LIPH. TCGA, the Cancer Genome Atlas; CCLE, the Broad Institute Cancer Cell Line Encyclopedia; GO, gene oncology
Table 2.
Co-expression analysis for LIPH (33 overlapped co-expression genes between TCGA dataset and CCLE database)
| Gene | TCGA | CCLE | ||
|---|---|---|---|---|
| Cor | P | Cor | P | |
| TIMM22 | − 0.605 | 4.56E−19 | −0.617 | 1.73E−05 |
| ABCC3 | 0.699 | 2.56E−27 | 0.65 | 4.17E−06 |
| AGR2 | 0.668 | 2.93E−24 | 0.614 | 1.96E−05 |
| ARL14 | 0.705 | 6.94E−28 | 0.698 | 3.87E−07 |
| B3GNT3 | 0.703 | 1.07E−27 | 0.624 | 1.29E−05 |
| BCL2L15 | 0.775 | 1.19E−36 | 0.635 | 8.27E−06 |
| C1orf106 | 0.82 | 2.65E−44 | 0.67 | 1.66E−06 |
| CDH1 | 0.663 | 9.39E−24 | 0.641 | 6.42E−06 |
| CGN | 0.774 | 1.47E−36 | 0.644 | 5.61E−06 |
| CLDN4 | 0.68 | 2.10E−25 | 0.693 | 5.05E−07 |
| ELF3 | 0.722 | 8.13E−30 | 0.725 | 8.50E−08 |
| EPS8L3 | 0.719 | 1.88E−29 | 0.649 | 4.40E−06 |
| ERBB3 | 0.736 | 1.75E−31 | 0.742 | 2.82E−08 |
| GPR35 | 0.7 | 2.15E−27 | 0.645 | 5.34E−06 |
| HKDC1 | 0.744 | 1.72E−32 | 0.648 | 4.72E−06 |
| LGALS3 | 0.764 | 3.45E−35 | 0.611 | 2.18E−05 |
| LRRC1 | 0.736 | 1.98E−31 | 0.67 | 1.65E−06 |
| MST1R | 0.804 | 1.92E−41 | 0.658 | 2.90E−06 |
| PIK3C2B | 0.612 | 1.52E−19 | 0.617 | 1.71E−05 |
| PLEKHA7 | 0.648 | 2.00E−22 | 0.657 | 3.05E−06 |
| PLS1 | 0.796 | 4.54E−40 | 0.626 | 1.20E−05 |
| POF1B | 0.764 | 4.29E−35 | 0.667 | 1.94E−06 |
| RNF103 | 0.606 | 3.86E−19 | 0.653 | 3.77E−06 |
| SCNN1A | 0.651 | 1.09E−22 | 0.619 | 1.60E−05 |
| SPINT1 | 0.625 | 1.50E−20 | 0.608 | 2.52E−05 |
| ST14 | 0.729 | 1.36E−30 | 0.612 | 2.12E−05 |
| TJP3 | 0.783 | 6.46E−38 | 0.747 | 2.08E−08 |
| TMC5 | 0.756 | 4.44E−34 | 0.652 | 3.92E−06 |
| TMEM62 | 0.604 | 5.43E−19 | 0.622 | 1.42E−05 |
| TRIM31 | 0.665 | 5.67E−24 | 0.615 | 1.90E−05 |
| TSPAN15 | 0.782 | 9.99E−38 | 0.614 | 1.95E−05 |
| VAMP8 | 0.611 | 1.84E−19 | 0.654 | 3.50E−06 |
| LIPH | 1 | 0.00E+00 | 1 | 0.00E+00 |
Cor: Pearson correlated coefficient > 0.6 or <− 0.6, P < 0.05
LIPH overexpression correlates with immune suppression or invasion in PC
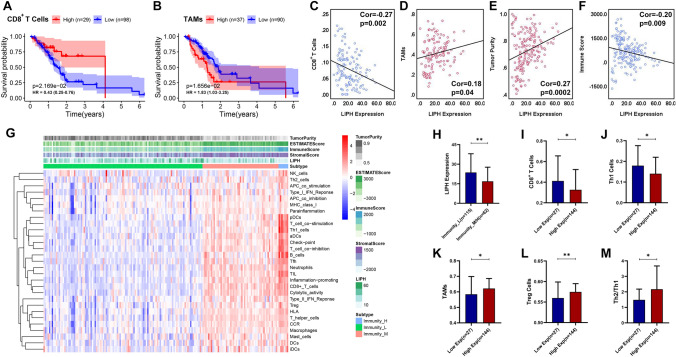
Through CIBERSORT algorithm, 127 PC samples with a CIBERSORT output P < 0.05 were involved in the assessment of 22 immune cells (ICs) types infiltration. And we found that low infiltration level of CD8 + T cells or high infiltration of TAMs significantly correlated with poor prognosis in PC (P < 0.05), consistent with previous studies (Fig. 6A, B). And our study showed that LIPH overexpression associated with lower infiltration level of CD8+ T cells (Cor = − 0.27, P < 0.01), but higher infiltration level of TAMs (Cor = 0.18, P < 0.05) (Fig. 6C, D). ESTIMATE algorithm showed that LIPH overexpression correlated with higher tumor purity (Cor = 0.27, P < 0.001), but lower immune score (Cor = − 0.20, P < 0.01) (Fig. 6E, F), a finding which was also found in GSE62452 dataset (Fig. 7A, B). Using ssGSEA method, the enrichment level of 29 immune-related terms was obtained and 177 PC samples in TCGA PC dataset were divided into immunity L (n = 115), immunity M (n = 55), and immunity H (n = 7) (Fig. 6G). We merged immunity M and immunity H into immunity M/H (n = 62) and found that the expression of LIPH in immunity M/H was much lower than that in immunity L (P < 0.05) (Fig. 6H). In TCGA PC dataset, ssGSEA analysis demonstrated that LIPH overexpression was significantly associated with low infiltration levels of CD8+ T cells and Th1 cells (Fig. 6I, J). In contrast, high infiltration levels of TAMs, Treg cells, and Th2/Th1 were significantly associated with high LIPH expression (P < 0.05) (Fig. 6K, M). Similar results were observed in the GSE62452 dataset (Fig. 7C–E). These results suggested that LIPH overexpression promoted immune suppression or invasion in PC.
Fig. 6.
Association between LIPH expression and the immune infiltration within tumors in TCGA dataset. A KM survival analysis showed that patients with lower infiltration levels of CD8+ T cells had a short OS than those with higher infiltration levels of CD8+ T cells (P < 0.05). B Patients with higher infiltration of macrophages had a shorter OS (P < 0.05) than those with lower infiltration of TAMs (P < 0.05). C LIPH expression negatively correlated with the infiltration level of CD8+ T cells (Cor = − 0.27, P < 0.01). D LIPH expression positively correlated with the infiltration level of TAMs (Cor = 0.18, P < 0.05) and E tumor purity (Cor = 0.27, P < 0.001). F LIPH expression negative correlated with immune score (Cor = − 0.20, P < 0.01). G Based on the ssGSEA analysis, the enrichment scores of 29 immune-related term were obtained and 177 PC samples in TCGA PC dataset were divided into the immunity L (n = 115), immunity M (n = 55), and immunity H (n = 7) groups. H LIPH expression was significantly upregulated in the immunity-L group compared with the immunity-M/H group. I LIPH overexpression was significantly associated with low infiltration levels of CD8+ T cells. J LIPH overexpression was significantly associated with low infiltration levels of Th1 cells, K high infiltration of TAMs, L Treg cells and M Th2/Th1. (*P < 0.05; **P < 0.01;***P < 0.001; ****P < 0.0001). TAMs, tumor-associated macrophages; Treg cells, regulatory T cells; Th1, type-1 T helper cells; type-2 T helper cells; ssGSEA, single sample gene set enrichment analysis; KM, Kaplan–Meier; OS, overall survival; Cor, pearson correlated coefficient
Fig. 7.
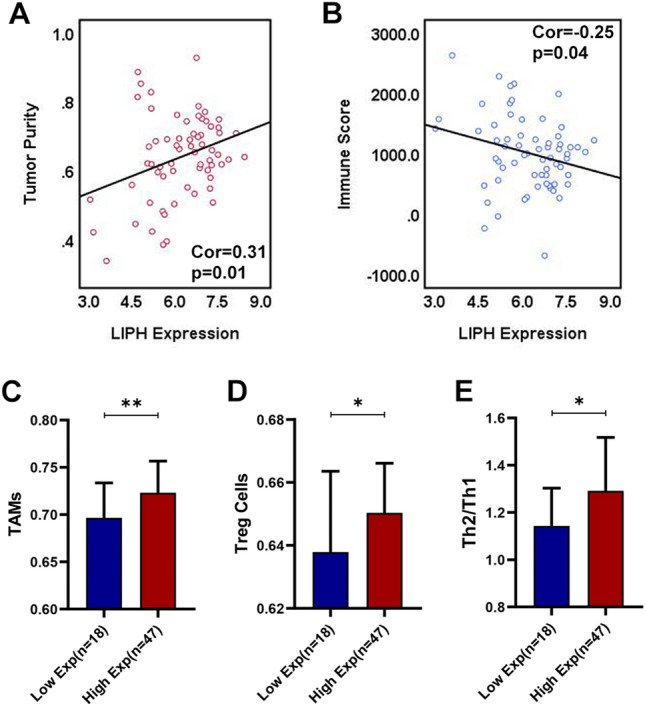
ESTIMATE algorithm and ssGSEA to validate the association between LIPH expression and immune infiltration within tumors in the GSE62452 dataset. A LIPH expression positively correlated with tumor purity (Cor = 0.31, P = 0.01) and B negative correlated with immune score (Cor = − 0.25, P = 0.04). C High LIPH expression was significantly associated with high infiltration of TAMs, D Treg cells and E Th2/Th1. (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001). TAMs, tumor-associated macrophages; Treg cells, regulatory T cells; Th1, type-1 T helper cells; type-2 T helper cells; ssGSEA, single sample gene set enrichment analysis; Exp, expression; Cor, Pearson correlated coefficient
Discussion
Pancreatic cancer (PC) is one of the most aggressive cancers with a very poor prognosis, and its incidence is rising every year [31]. It is consistently considered that PC is a “cold” tumor, which is highly immunosuppressive [32]. Understanding of the molecular mechanisms involved in PC immune suppression is believed to be helpful to develop novel immune-based therapies for PC [33]. The present study mainly investigated the role of LIPH during pancreatic carcinogenesis and immunosuppression. Consistent with previous studies about LIPH in other carcinomas [14, 20–22], multiple databases showed that LIPH was significantly upregulated in PC (Figs. 1, 2) and correlated with advanced histologic grade, tumor recurrence, and poor survival of patients with PC (Table 1, Fig. 3). Thus, LIPH could be a critical unfavorable prognostic factor for patients with PC.
Function enrichment analysis showed that LIPH overexpression in PC might take part in various biological processes such as negative regulation of cell–cell adhesion, actin cytoskeleton, EMT, angiogenesis, and signaling by MST1 (Fig. 5B). These findings suggested that LIPH overexpression in PC may promote tumor progression through affecting actin cytoskeleton, cell–cell adhesion, EMT, and angiogenesis. But further experimental studies should be conducted to elucidate the potential mechanisms of LIPH in pancreatic carcinogenesis.
Our study found that LIPH expression correlated with the immune infiltration level in PC. Interestingly, in 2015, Orozco-terWengel et al. reported that LIPH was related to immunity in cattle [19]. But we are the first to demonstrate the correlation between LIPH expression and immune infiltration within tumor. Higher LIPH expression was associated with higher tumor purity score and lower immune score (Fig. 6E, F). The expression of LIPH in immunity L was much higher than that in immunity M/H (P < 0.05) (Fig. 6H). In PC, there was a strong negative association between LIPH expression and the infiltration of CD8+ T cells, Th1 cells. In contrast, LIPH overexpression upregulated the infiltration level of TAMs, Treg cells, and Th2/Th1 in PC. TAMs and Tregs could induce an immunosuppressive tumor microenvironment (TME) through production of immune suppressive cytokines like transforming growth factor β (TGF-β), interleukin-10 (IL-10), and IL-35 [34–36]. These factors could antagonize the antitumor effects of CD8+ T cells and Th1 cells [37, 38]. These results suggested that LIPH played an important role in the formation of immunosuppressive TME in PC through upregulating the pro-tumor effects of TAMS and Treg cells, and downregulating the antitumor effect of CD8+ T cells and Th1, and therefore influenced PC prognosis.
Our study demonstrated that high infiltration of CD8+ T cells significantly prolonged survival for patients with PC (Fig. 6A, B). Fukunaga et al. also reported that increasing infiltration of CD8+ T cells was significantly correlated with prolonged survival in PC [39]. However, the infiltration of CD8+ T cells is usually rare in the TME of PC. A high number of tumor-associated immunosuppressive cells (e.g., TAMs) functions as a barrier to CD8+ T cells infiltration [40]. Moreover, our data demonstrated that the proportion of CD8+ T cells within tumors was much less than that of TAMs. In our study, high infiltration of TAMs within tumors was observed to correlate with poor prognosis in PC. Cui et al. reported that TAMs played large role in promoting tumor growth and inducing an immunosuppressive microenvironment [34]. Taken together, we suggested that the antitumor effect of CD8+ T cells in PC was impaired or overwhelmed by the pro-tumor effect of TAMs. Considering TAMs occupying the major proportion of infiltrating ICs within tumors, targeting TAMs could be a promising therapeutic strategy to complement current chemotherapeutic, anti-angiogenic, or ICI therapies.
Through ssGSEA analysis, the significant positive associations between LIPH expression and the infiltration of TAMs, Treg cells and Th2/Th1 in PC were observed in both TCGA PC dataset and GES62452 datasets. Of note, we found LIPH might be involved in the MST1 signaling pathway, which further supported that high expression of LIPH significantly associated with high infiltration of TAMs. Bayne et al. reported that KRAS mutated PC cells secret granulocyte–macrophage colony-stimulating factor, recruiting myeloid-derived suppressor cell and impairing the antitumor activity of CD8+ T cells [41]. Early studies have also reported that KRAS inhibits the expression of components of the antigen presentation pathway, allowing the evasion of TILs [42]. Our study demonstrated that KRAS mutation significantly upregulated the expression of LIPH, and LIPH overexpression was significantly correlated with KRAS activation (Cor = 0.5, P < 0.05 in TCGA PC dataset; Cor = 0.58, P < 0.05 in GSE62452 dataset; Cor = 0.51, P < 0.05 in cBioportal database) (Fig. 4C, E, and S3A). Taken together, we proposed that LIPH overexpression and KRAS activation cooperated with each other to induce immune suppression or evasion in PC.
Previous study by Bellone et al. showed that TGF-β is elevated in PC cell lines [43]. TGF-β is reported to suppress tumor formation through blocking cell cycle progression [44, 45]. But the tumor suppressive effect of TGF-β is often inhibited in PC due to the inactivation of TGF-β signaling mediator, SMAD4 [46]. In this study, we observed that LIPH was notably upregulated by SMAD4 mutation in PC. Furthermore, LIPH upregulation was significantly correlated with SMAD4 downregulation (Cor = − 0.44, P < 0.05 in the TCGA dataset; Cor = − 0.42, P < 0.05 in the GSE62452 dataset; Cor = − 0.43, P < 0.05 in cBioportal database) (Fig. 4A3, 4B2, and S1B). Thus, we postulated that there was a bidirectional regulation between LIPH and SMAD4. It has been reported that SMAD4 inactivation promotes KRAS-mediated tumor progression in PC [47]. Moreover, it was reviewed that KRAS mutation in PC increases TGF-β expression, promoting Treg cells recruitment and TAMs polarization and contributing to immunosuppression in the TME [48, 49]. TGF-β could also upregulate Treg cells through switch Th1/Th2 [50]. And the expression of LIPH was positively associated with the infiltration of Treg cells and Th2/Th1. Treg cells and Th2/Th1 have been reported to be strongly associated with poor prognosis and negatively correlated to the presence of CD8+ T cells in PC [51, 52]. In the study by Whiteside et al., Treg cells produce IL-10 and TGF-β, causing the impairment of CD8+ T cells [53, 54]. Bellone G et al. reported that the function of CD8+ T cells in PC patients is impaired when Th2 dominates in the TME [43]. Thus, higher infiltration of Treg cells and Th2/Th1 reflected an immunosuppressive status in the TME. Taken together, we proposed that LIPH overexpression upregulated the infiltration of pro-tumor immune cells, such as TAMs, Tregs, and Th2 cells, through KRAS-SMAD4-TGF-β signaling pathway.
Function enrichment analysis demonstrated that LIPH mediated the EMT signaling pathway and the cell–cell adhesion in PC (Fig. 5B). Li et al. reported that LIPH promoted the progression of papillary thyroid carcinoma through EMT signaling pathway [14]. It was reviewed that EMT played a critical role in tumor immunosuppression [55]. The activation of EMT impairs the therapeutic effects of ICIs [56, 57]. With respect to cell–cell adhesion in PC, it was reported that targeting focal adhesion kinase increases PC responsive to checkpoint immunotherapy [40]. Taken together, we supposed that targeting LIPH in PC also enhanced the effect of ICIs. All these results provided an initial understanding about the role of LIPH in immune suppression or evasion within tumor, which remains to be proved in future works. And targeting LIPH-induced EMT, TGF-β or cell–cell adhesion may be a potential effective therapeutic strategy in PC.
Our study has some limitations. Firstly, both the sample sizes of the TCGA PC dataset and the GSE62452 dataset were small. Our findings should be validated with a large sample size dataset in future study. Secondly, the potential function of LIPH in pancreatic carcinogenesis and immune infiltration has not been explored in vitro or in vivo. Furthermore, future works should be conducted to prove the hypothesis about the pathway of KRAS-SMAD4-LIPH-TGF-β in the immunosuppressive TME of PC.
Conclusions
In this study, we found that LIPH overexpression might promote tumor progression, and positively associated with the infiltration of TAMs, Treg cells, and Th2/Th1, while negatively associated with the infiltration of CD8+ T cells and Th1 cells for the first time. Therefore, we identified LIPH as a novel unfavorable prognostic biomarker correlated with immunosuppression, and a novel potential therapeutic target in PC. This study provides new insights into the tumor-immune microenvironment and immune-based therapies for PC.
Supplementary Information
Below is the link to the electronic supplementary material.
KM survival analysis for OS of PC patients in TCGA dataset according to the median of LIPH expression. KM, Kaplan-Meier; PC, pancreatic cancer; OS, overall survival; HR, hazard ratio (TIF 3189 kb)
The mutation landscape of PC in TCGA dataset. PC, pancreatic cancer (TIF 6564 kb)
Validation of the association among LIPH, KRAS, and SMAD4 using cBioportal database. (A) LIPH expression was positively correlated to KRAS expression (Cor=0.51, P<0.05). (B) LIPH expression was negatively correlated to SMAD4 expression (Cor=-0.43, P<0.05) (TIF 1823 kb)
Association between LIPH expression and TMB. TMB, the total mutational burden (TIF 299 kb)
Acknowledgements
This study was supported by High-level Hospital Construction Project (DFJH201921), Fundamental Research Funds for the Central Universities (y2syD2192230), and National Natural Science Foundation of China (Nos. 81672475, 81702783, 82072635 and 82072637).
Author contributions
HZ, BC, CZ, and BH conceived and designed the study. HZ, XC, and YW performed the data analysis. HZ, XC, BC, and SH wrote the paper. All authors read and approved the manuscript.
Data availability statement
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary.
Declarations
Conflict of interest
All authors declare that they have no conflicts of interest.
Ethical standards
All datasets are freely available as public resources. Therefore, local ethics approval was not needed.
Footnotes
Hongkai Zhuang, Xinming Chen and Ying Wang co-first authors.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Bo Chen, Email: chenbo@gdph.org.cn.
Chuanzhao Zhang, Email: 641703837@qq.com.
Baohua Hou, Email: 15917919681@163.com.
References
- 1.Kamisawa T, Wood LD, Itoi T, Takaori K. Pancreatic cancer. Lancet. 2016;388:73–85. doi: 10.1016/S0140-6736(16)00141-0. [DOI] [PubMed] [Google Scholar]
- 2.Lai E, Puzzoni M, Ziranu P, et al. New therapeutic targets in pancreatic cancer. Cancer Treat Rev. 2019;81:101926. doi: 10.1016/j.ctrv.2019.101926. [DOI] [PubMed] [Google Scholar]
- 3.Parkin A, Man J, Chou A, Nagrial AM, Samra J, Gill AJ, Timpson P, Pajic M. The evolving understanding of the molecular and therapeutic landscape of pancreatic ductal adenocarcinoma. Diseases. 2018 doi: 10.3390/diseases6040103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bazhin AV, Shevchenko I, Umansky V, Werner J, Karakhanova S. Two immune faces of pancreatic adenocarcinoma: possible implication for immunotherapy. Cancer Immunol Immunother CII. 2014;63:59–65. doi: 10.1007/s00262-013-1485-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elaileh A, Saharia A, Potter L, Baio F, Ghafel A, Abdelrahim M, Heyne K. Promising new treatments for pancreatic cancer in the era of targeted and immune therapies. Am J Cancer Res. 2019;9:1871–1888. [PMC free article] [PubMed] [Google Scholar]
- 6.Cervello M, Emma MR, Augello G, et al. New landscapes and horizons in hepatocellular carcinoma therapy. Aging. 2020 doi: 10.18632/aging.102777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schouwenburg MG, Suijkerbuijk KPM, Koornstra RHT, et al. Switching to immune checkpoint inhibitors upon response to targeted therapy; the road to long-term survival in advanced melanoma patients with highly elevated serum LDH? Cancers. 2019 doi: 10.3390/cancers11121940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ridolfi L, De Rosa F, Petracci E, et al. Anti-PD1 antibodies in patients aged >/= 75 years with metastatic melanoma: a retrospective multicentre study. J Geriatr Oncol. 2020 doi: 10.1016/j.jgo.2019.12.012. [DOI] [PubMed] [Google Scholar]
- 9.Fu Y, Liu S, Zeng S, Shen H. From bench to bed: the tumor immune microenvironment and current immunotherapeutic strategies for hepatocellular carcinoma. J Exp Clin Cancer Res: CR. 2019;38:396. doi: 10.1186/s13046-019-1396-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balachandran VP, Beatty GL, Dougan SK. Broadening the impact of immunotherapy to pancreatic cancer: challenges and opportunities. Gastroenterology. 2019;156:2056–2072. doi: 10.1053/j.gastro.2018.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li J, Yuan S, Norgard RJ, Yan F, Yamazoe T, Blanco A, Stanger BZ. Tumor cell-intrinsic USP22 suppresses antitumor immunity in pancreatic cancer. Cancer Immunol Res. 2019 doi: 10.1158/2326-6066.CIR-19-0661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hou YC, Chao YJ, Hsieh MH, Tung HL, Wang HC, Shan YS. Low CD8(+) T cell infiltration and high PD-L1 expression are associated with level of CD44(+)/CD133(+) cancer stem cells and predict an unfavorable prognosis in pancreatic cancer. Cancers. 2019 doi: 10.3390/cancers11040541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhuang H, Zhang C, Hou B. FAM83H overexpression predicts worse prognosis and correlates with less CD8(+) T cells infiltration and Ras-PI3K-Akt-mTOR signaling pathway in pancreatic cancer. Clin Transl Oncol. 2020;22:2244–2252. doi: 10.1007/s12094-020-02365-z. [DOI] [PubMed] [Google Scholar]
- 14.Li Y, Zhou X, Zhang Q, Chen E, Sun Y, Ye D, Wang O, Zhang X, Lyu J. Lipase member H is a downstream molecular target of hypoxia inducible factor-1alpha and promotes papillary thyroid carcinoma cell migration in BCPAP and KTC-1 cell lines. Cancer Manag Res. 2019;11:931–941. doi: 10.2147/CMAR.S183355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inoue A, Arima N, Ishiguro J, Prestwich GD, Arai H, Aoki J. LPA-producing enzyme PA-PLA(1)alpha regulates hair follicle development by modulating EGFR signalling. EMBO J. 2011;30:4248–4260. doi: 10.1038/emboj.2011.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin ME, Herr DR, Chun J. Lysophosphatidic acid (LPA) receptors: signaling properties and disease relevance. Prostaglandins Other Lipid Mediat. 2010;91:130–138. doi: 10.1016/j.prostaglandins.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mizukami Y, Hayashi R, Tsuruta D, Shimomura Y, Sugawara K. Novel splice site mutation in the LIPH gene in a patient with autosomal recessive woolly hair/hypotrichosis: case report and published work review. J Dermatol. 2018;45:613–617. doi: 10.1111/1346-8138.14257. [DOI] [PubMed] [Google Scholar]
- 18.Ahmad F, Sharif S, FurqanUbaid M, Shah K, Khan MN, Umair M, Azeem Z, Ahmad W. Novel sequence variants in the LIPH and LPAR6 genes underlies autosomal recessive woolly hair/hypotrichosis in consanguineous families. Congenit Anom. 2018;58:24–28. doi: 10.1111/cga.12226. [DOI] [PubMed] [Google Scholar]
- 19.Orozco-terWengel P, Barbato M, Nicolazzi E, et al. Revisiting demographic processes in cattle with genome-wide population genetic analysis. Front Genet. 2015;6:191. doi: 10.3389/fgene.2015.00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cui M, Jin H, Shi X, Qu G, Liu L, Ding X, Wang Y, Niu C. Lipase member H is a novel secreted protein associated with a poor prognosis for breast cancer patients. Tumour Biol. 2014;35:11461–11465. doi: 10.1007/s13277-014-2436-5. [DOI] [PubMed] [Google Scholar]
- 21.Seki Y, Yoshida Y, Ishimine H, et al. Lipase member H is a novel secreted protein selectively upregulated in human lung adenocarcinomas and bronchioloalveolar carcinomas. Biochem Biophys Res Commun. 2014;443:1141–1147. doi: 10.1016/j.bbrc.2013.12.106. [DOI] [PubMed] [Google Scholar]
- 22.Ishimine H, Zhou R, Sumitomo K, Ito Y, Seki Y, Yoshida Y, Kurisaki A. Lipase member H frequently overexpressed in human esophageal adenocarcinomas. Tumour Biol. 2016;37:2075–2081. doi: 10.1007/s13277-015-3985-y. [DOI] [PubMed] [Google Scholar]
- 23.Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, Diehn M, Alizadeh AA. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoshihara K, Shahmoradgoli M, Martinez E, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhuang H, Zhou Z, Zhang Z, Chen X, Ma Z, Huang S, Gong Y, Zhang C, Hou B. B3GNT3 overexpression promotes tumor progression and inhibits infiltration of CD8(+) T cells in pancreatic cancer. Aging. 2020;13:2310–2329. doi: 10.18632/aging.202255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Camp RL, Dolled-Filhart M, Rimm DL. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. 2004;10:7252–7259. doi: 10.1158/1078-0432.CCR-04-0713. [DOI] [PubMed] [Google Scholar]
- 27.Singh RR, Goldberg J, Varghese AM, Yu KH, Park W, O'Reilly EM. Genomic profiling in pancreatic ductal adenocarcinoma and a pathway towards therapy individualization: a scoping review. Cancer Treat Rev. 2019;75:27–38. doi: 10.1016/j.ctrv.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kamburov A, Stelzl U, Lehrach H, Herwig R. The ConsensusPathDB interaction database: 2013 update. Nucleic Acids Res. 2013;41:D793–800. doi: 10.1093/nar/gks1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bindea G, Mlecnik B, Tosolini M, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39:782–795. doi: 10.1016/j.immuni.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 31.Kong K, Guo M, Liu Y, Zheng J. Progress in animal models of pancreatic ductal adenocarcinoma. J Cancer. 2020;11:1555–1567. doi: 10.7150/jca.37529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi J, Xue J. Inflammation and development of pancreatic ductal adenocarcinoma. Chin Clin Oncol. 2019;8:19. doi: 10.21037/cco.2019.04.02. [DOI] [PubMed] [Google Scholar]
- 33.Li M, Li M, Yang Y, et al. Remodeling tumor immune microenvironment via targeted blockade of PI3K-gamma and CSF-1/CSF-1R pathways in tumor associated macrophages for pancreatic cancer therapy. J Controlled Release. 2020 doi: 10.1016/j.jconrel.2020.02.011. [DOI] [PubMed] [Google Scholar]
- 34.Cui R, Yue W, Lattime EC, Stein MN, Xu Q, Tan XL. Targeting tumor-associated macrophages to combat pancreatic cancer. Oncotarget. 2016;7:50735–50754. doi: 10.18632/oncotarget.9383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paluskievicz CM, Cao X, Abdi R, Zheng P, Liu Y, Bromberg JS. T Regulatory cells and priming the suppressive tumor microenvironment. Front Immunol. 2019;10:2453. doi: 10.3389/fimmu.2019.02453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ohue Y, Nishikawa H. Regulatory T (Treg) cells in cancer: Can Treg cells be a new therapeutic target? Cancer Sci. 2019;110:2080–2089. doi: 10.1111/cas.14069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sideras K, Braat H, Kwekkeboom J, van Eijck CH, Peppelenbosch MP, Sleijfer S, Bruno M. Role of the immune system in pancreatic cancer progression and immune modulating treatment strategies. Cancer Treat Rev. 2014;40:513–522. doi: 10.1016/j.ctrv.2013.11.005. [DOI] [PubMed] [Google Scholar]
- 38.Najafi S, Mirshafiey A. The role of T helper 17 and regulatory T cells in tumor microenvironment. Immunopharmacol Immunotoxicol. 2019;41:16–24. doi: 10.1080/08923973.2019.1566925. [DOI] [PubMed] [Google Scholar]
- 39.Fukunaga A, Miyamoto M, Cho Y, et al. CD8+ tumor-infiltrating lymphocytes together with CD4+ tumor-infiltrating lymphocytes and dendritic cells improve the prognosis of patients with pancreatic adenocarcinoma. Pancreas. 2004;28:e26–31. doi: 10.1097/00006676-200401000-00023. [DOI] [PubMed] [Google Scholar]
- 40.Jiang H, Hegde S, Knolhoff BL, et al. Targeting focal adhesion kinase renders pancreatic cancers responsive to checkpoint immunotherapy. Nat Med. 2016;22:851–860. doi: 10.1038/nm.4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bayne LJ, Beatty GL, Jhala N, Clark CE, Rhim AD, Stanger BZ, Vonderheide RH. Tumor-derived granulocyte-macrophage colony-stimulating factor regulates myeloid inflammation and T cell immunity in pancreatic cancer. Cancer Cell. 2012;21:822–835. doi: 10.1016/j.ccr.2012.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.El-Jawhari JJ, El-Sherbiny YM, Scott GB, et al. Blocking oncogenic RAS enhances tumour cell surface MHC class I expression but does not alter susceptibility to cytotoxic lymphocytes. Mol Immunol. 2014;58:160–168. doi: 10.1016/j.molimm.2013.11.020. [DOI] [PubMed] [Google Scholar]
- 43.Bellone G, Turletti A, Artusio E, Mareschi K, Carbone A, Tibaudi D, Robecchi A, Emanuelli G, Rodeck U. Tumor-associated transforming growth factor-beta and interleukin-10 contribute to a systemic Th2 immune phenotype in pancreatic carcinoma patients. Am J Pathol. 1999;155:537–547. doi: 10.1016/s0002-9440(10)65149-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen Y, Di C, Zhang X, et al. Transforming growth factor beta signaling pathway: a promising therapeutic target for cancer. J Cell Physiol. 2020;235:1903–1914. doi: 10.1002/jcp.29108. [DOI] [PubMed] [Google Scholar]
- 45.Fang P, Li X, Dai J, et al. Immune cell subset differentiation and tissue inflammation. J Hematol Oncol. 2018;11:97. doi: 10.1186/s13045-018-0637-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ahmed S, Bradshaw AD, Gera S, Dewan MZ, Xu R. The TGF-beta/Smad4 signaling pathway in pancreatic carcinogenesis and its clinical significance. J Clin Med. 2017 doi: 10.3390/jcm6010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leung L, Radulovich N, Zhu CQ, Wang D, To C, Ibrahimov E, Tsao MS. Loss of canonical Smad4 signaling promotes KRAS driven malignant transformation of human pancreatic duct epithelial cells and metastasis. PLoS ONE. 2013;8:e84366. doi: 10.1371/journal.pone.0084366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheng H, Fan K, Luo G, et al. Kras(G12D) mutation contributes to regulatory T cell conversion through activation of the MEK/ERK pathway in pancreatic cancer. Cancer Lett. 2019;446:103–111. doi: 10.1016/j.canlet.2019.01.013. [DOI] [PubMed] [Google Scholar]
- 49.Cullis J, Das S, Bar-Sagi D. Kras and tumor immunity: friend or foe? Cold Spring Harb Perspect Med. 2018 doi: 10.1101/cshperspect.a031849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Neuzillet C, Tijeras-Raballand A, Cohen R, Cros J, Faivre S, Raymond E, de Gramont A. Targeting the TGFbeta pathway for cancer therapy. Pharmacol Ther. 2015;147:22–31. doi: 10.1016/j.pharmthera.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 51.Protti MP, De Monte L. Cross-talk within the tumor microenvironment mediates Th2-type inflammation in pancreatic cancer. Oncoimmunology. 2012;1:89–91. doi: 10.4161/onci.1.1.17939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brunetto E, De Monte L, Balzano G, et al. The IL-1/IL-1 receptor axis and tumor cell released inflammasome adaptor ASC are key regulators of TSLP secretion by cancer associated fibroblasts in pancreatic cancer. J Immunother Cancer. 2019;7:45. doi: 10.1186/s40425-019-0521-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Whiteside TL. FOXP3+ Treg as a therapeutic target for promoting anti-tumor immunity. Expert Opin Ther Targets. 2018;22:353–363. doi: 10.1080/14728222.2018.1451514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Whiteside TL. Regulatory T cell subsets in human cancer: are they regulating for or against tumor progression? Cancer Immunol Immunother CII. 2014;63:67–72. doi: 10.1007/s00262-013-1490-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jiang Y, Zhan H. Communication between EMT and PD-L1 signaling: New insights into tumor immune evasion. Cancer Lett. 2020;468:72–81. doi: 10.1016/j.canlet.2019.10.013. [DOI] [PubMed] [Google Scholar]
- 56.Hugo W, Zaretsky JM, Sun L, et al. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell. 2017;168:542. doi: 10.1016/j.cell.2017.01.010. [DOI] [PubMed] [Google Scholar]
- 57.Soundararajan R, Fradette JJ, Konen JM, et al. Targeting the interplay between epithelial-to-mesenchymal-transition and the immune system for effective immunotherapy. Cancers. 2019 doi: 10.3390/cancers11050714. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
KM survival analysis for OS of PC patients in TCGA dataset according to the median of LIPH expression. KM, Kaplan-Meier; PC, pancreatic cancer; OS, overall survival; HR, hazard ratio (TIF 3189 kb)
The mutation landscape of PC in TCGA dataset. PC, pancreatic cancer (TIF 6564 kb)
Validation of the association among LIPH, KRAS, and SMAD4 using cBioportal database. (A) LIPH expression was positively correlated to KRAS expression (Cor=0.51, P<0.05). (B) LIPH expression was negatively correlated to SMAD4 expression (Cor=-0.43, P<0.05) (TIF 1823 kb)
Association between LIPH expression and TMB. TMB, the total mutational burden (TIF 299 kb)
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary.