Abstract
Autographa californica nuclear polyhedrosis virus (AcNPV) encodes a 168-amino-acid polypeptide that contains the signature motif of the superfamily of protein phosphatases that act via a covalent cysteinyl phosphate intermediate. The sequence of the AcNPV phosphatase is similar to that of the RNA triphosphatase domain of the metazoan cellular mRNA capping enzyme. Here, we show that the purified recombinant AcNPV protein is an RNA 5′-triphosphatase that hydrolyzes the γ-phosphate of triphosphate-terminated poly(A); it also hydrolyzes ATP to ADP and GTP to GDP. The phosphatase sediments as two discrete components in a glycerol gradient: a 9.5S oligomer and 2.5S putative monomer. The 2.5S form of the enzyme releases 32Pi from 1 μM γ-32P-labeled triphosphate-terminated poly(A) with a turnover number of 52 min−1 and converts ATP to ADP with Vmax of 8 min−1 and Km of 25 μM ATP. The 9.5S oligomeric form of the enzyme displays an initial pre-steady-state burst of ADP and Pi formation, which is proportional to and stoichiometric with the enzyme, followed by a slower steady-state rate of product formation (approximately 1/10 of the steady-state rate of the 2.5S enzyme). We surmise that the oligomeric enzyme is subject to a rate-limiting step other than reaction chemistry and that this step is either distinct from or slower than the rate-limiting step for the 2.5S enzyme. Replacing the presumptive active site nucleophile Cys-119 by alanine abrogates RNA triphosphatase and ATPase activity. Our findings raise the possibility that baculoviruses encode enzymes that cap the 5′ ends of viral transcripts synthesized at late times postinfection by a virus-encoded RNA polymerase.
The m7GpppN cap structure is required for efficient translation of eukaryotic mRNAs. Eukaryotic viruses have evolved diverse, often exotic strategies to contend with the capping problem (1, 3, 21, 23, 24). RNA viruses that utilize virus-encoded RNA-dependent RNA polymerases to synthesize their mRNAs either steal the caps from cellular transcripts (e.g., influenza virus), encode their own enzymes that specifically cap and methylate the plus-strand transcripts (e.g., vesicular stomatitis virus, reovirus, and Sindbis virus), or else circumvent the need for a cap by including cis-acting RNA elements in the plus-strand transcript that promote cap-independent translation (e.g., poliovirus and other picornaviruses). DNA viruses employ somewhat less adventurous strategies. The mRNAs of retroviruses and most nuclear DNA viruses (e.g., papovaviruses, adenoviruses, and herpesviruses) are transcribed by cellular RNA polymerase II and are capped by the cellular capping and methylating enzymes. However, vaccinia virus and other poxviruses, which replicate entirely in the cytoplasm, encode and encapsidate their own DNA-dependent RNA polymerase and mRNA capping apparatus (16). African swine fever virus, which has a cytoplasmic replication phase, also encodes and encapsidates its own RNA polymerase and capping enzyme (19). Chlorella virus PBCV-1 encodes a capping enzyme (11) but appears not to encode its own RNA polymerase.
The biochemistry of cap synthesis was initially delineated for vaccinia virus and reovirus mRNAs and subsequently shown to apply to all eukaryotic cells (23). Capping entails three enzymatic reactions: (i) the triphosphate end of the primary transcript is hydrolyzed to a diphosphate by RNA triphosphatase, (ii) the diphosphate end is capped with GMP by GTP:RNA guanylyltransferase, and (iii) the GpppN structure is methylated by S-adenosylmethionine:RNA-(guanine N7)-methyltransferase. Vaccinia virus mRNA capping enzyme is a multifunctional protein that catalyzes all three reactions (29). The triphosphatase, guanylyltransferase, and methyltransferase active sites are arranged in a modular fashion within a single 95-kDa polypeptide (23). Metazoan species encode a two-component capping system consisting of a bifunctional triphosphatase-guanylyltransferase polypeptide and a monofunctional methyltransferase polypeptide (10, 15, 27, 30–32). Yeasts encode a three-component system in which the triphosphatase, guanylyltransferase, and methyltransferase reactions are catalyzed by separate gene products (14, 22, 28). The guanylyltransferase and methyltransferase domains are conserved between DNA viruses, fungi, and metazoans (14, 25, 30–32). In contrast, the triphosphatase components are structurally and mechanistically divergent.
Mammalian and nematode capping enzymes consist of an N-terminal RNA triphosphatase domain and a C-terminal guanylyltransferase domain (10, 15, 27, 31, 32). In the mouse enzyme, the two catalytic domains are autonomous and nonoverlapping (10, 32). The metazoan RNA triphosphatase domains contain an (I/V)HCxAGxGR(S/T)G signature motif initially described for the protein tyrosine phosphatase/dual-specificity protein phosphatase enzyme family (Fig. 1). These proteins catalyze phosphoryl transfer from a protein phosphomonoester substrate to the thiolate of a cysteine on the enzyme to form a covalent phosphocysteine intermediate (5). The intermediate is then attacked by water to liberate phosphate. The cysteine within the signature motif is the active site of phosphoryl transfer. The phosphatase signature motif has been encountered in scores of polypeptides that have been identified during genomic sequencing efforts, and such gene products are routinely designated as protein phosphatases. The discovery that Caenorhabditis elegans and mammalian RNA-specific triphosphatases contain the same phosphatase active site throws to the wind any presumptions regarding the substrate specificity of these putative protein phosphatases. Indeed, Takagi et al. (27) found that the C. elegans RNA triphosphatase domain was unable to hydrolyze phosphotyrosine- or phosphoserine-containing peptide substrates.
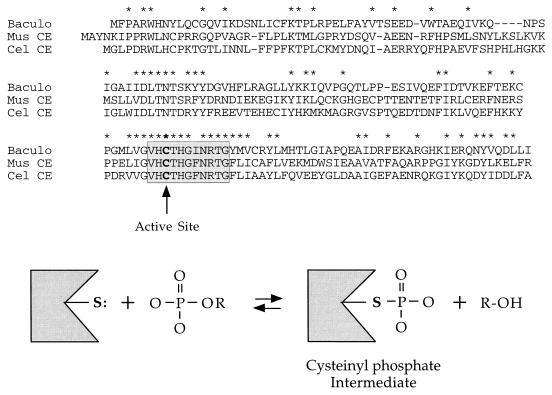
FIG. 1.
AcNPV phosphatase resembles the RNA triphosphatase domains of metazoan capping enzymes. The amino acid sequence of the 168-amino-acid AcNPV-encoded phosphatase (Baculo) is aligned with the N-terminal RNA triphosphatase domains of mouse capping enzyme (Mus CE) and C. elegans capping enzyme (Cel CE). Gaps in the sequences are indicated by dashes; amino acids conserved in all three proteins are denoted by asterisks. The protein phosphatase signature motif is highlighted in the shaded box. The active-site cysteine is in boldface. The presumptive reaction pathway involving formation of a cysteinyl phosphate intermediate is shown.
Takagi et al. (27) and Wang et al. (31) noted sequence similarity between the triphosphatase domain of metazoan capping enzyme and a “protein phosphatase” encoded by Autographa californica nuclear polyhedrosis virus (AcNPV) (9, 12, 20). The similarity extends well beyond the signature motif (Fig. 1). This extent of conservation raised the possibility that the baculovirus gene product is an RNA triphosphatase. We addressed this issue by expressing the AcNPV protein in bacteria, purifying the enzyme, and characterizing it biochemically. We show that the baculovirus protein is indeed an RNA triphosphatase (and a nucleoside triphosphatase [NTPase]) and that its activity requires Cys-119, the conserved active site nucleophile. The observed rates of Pi release from triphosphate-terminated RNA and ATP by the baculovirus phosphatase are much greater than the previously reported rates of hydrolysis of phosphoserine- and phosphotyrosine-containing phosphopeptides.
MATERIALS AND METHODS
T7-based vector for expression of AcNPV RNA triphosphatase in bacteria.
The AcNPV phosphatase open reading frame was amplified by PCR from a plasmid template containing a viral genomic DNA fragment (a gift of Linda Guarino). Oligonucleotide primers complementary to 5′ and 3′ ends of the gene were designed to introduce NdeI and BamHI restriction sites, respectively. The sequence of the 5′ flanking primer was 5′-GAAAGCACGTGTCATATGTTTCCCGCGCG, and that of the 3′ flanking primer was 5′-TGAAAATAGGATCCAGTATTTGTTAAAGAATGC. PCR was carried out with Taq polymerase (Boehringer). The PCR product was digested with NdeI and BamHI and then inserted between the NdeI and BamHI sites of the T7-based expression plasmid pET16b (Novagen). The resulting plasmid, pET-RTP, was transformed into Escherichia coli BL21(DE3).
Mutation of Cys-119 to alanine.
The codon for the conserved phosphatase active-site cysteine (Cys-119) was changed to an alanine codon by two-stage PCR overlap extension using mutagenic primers in the first-stage amplification. The second-stage PCR product was digested with NdeI and BamHI and then inserted into pET16b to yield plasmid pET-RTP-C119A. The insert was sequenced to confirm the presence of the C119A mutation.
Expression and purification of recombinant proteins.
One-liter cultures of E. coli BL21(DE3)/pET-RTP or BL21(DE3)/pET-RTP-C119A were grown at 37°C in Luria-Bertani medium containing 0.1 mg of ampicillin per ml until the A600 reached 0.7. The cultures were chilled for 30 min on ice, then adjusted to 2% ethanol, and incubated at 17°C for 15 h with continuous shaking. Cells were harvested by centrifugation, and the pellets were stored at −80°C. All subsequent procedures were performed at 4°C. Thawed bacteria were resuspended in 30 ml of buffer B (50 mM Tris HCl [pH 7.5], 150 mM NaCl, 10% sucrose) and then mixed with 10 ml of buffer B containing 8 mg of lysozyme. The samples were incubated on ice for 30 min, then adjusted to 0.1% Triton X-100, and incubated for an additional 30 min. The lysates were sonicated to reduce viscosity and then separated into soluble and insoluble fractions by centrifugation for 20 min at 18,000 rpm in a Sorvall SS34 rotor. The soluble fractions were mixed with 1.5 ml of Ni-nitrilotriacetic acid-agarose resin (Qiagen) that had been equilibrated with buffer C (20 mM Tris [pH 8.0], 300 mM NaCl, 10% glycerol, 0.1% Triton X-100), and the suspension was mixed by continuous rotation for 1 h. The Ni-agarose resin was recovered by centrifugation, and the supernatant was removed. The resin was resuspended in 20 ml of buffer C, and the slurry was poured into a column. The packed column was washed with 40 ml of buffer C and then eluted stepwise with 6-ml aliquots of buffer C containing 5, 10, 25, 50, 250, and 500 mM imidazole. The 23-kDa baculovirus polypeptide eluted at 250 mM imidazole, as judged by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie blue staining. The 250 mM imidazole fraction was diluted 20-fold in buffer A (50 mM Tris HCl [pH 8.0], 2 mM dithiothreitol, 1 mM EDTA, 10% glycerol, 0.1% Triton X-100) and then applied to a 2-ml phosphocellulose column that had been equilibrated in buffer A. The column was eluted stepwise with buffer A containing 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, and 1.0 M NaCl. The baculovirus protein was recovered in the 0.2 to 0.5 M NaCl steps.
Sedimentation analysis.
An aliquot (0.2 ml; 50 μg of protein) of the 0.3 M NaCl phosphocellulose eluate fraction was applied to a 4.8-ml 15 to 30% glycerol gradient containing either 0.3 or 1.0 M NaCl in buffer A. The gradients were centrifuged for 18 h at 50,000 rpm in a Beckman SW50 rotor at 4°C. Fractions (0.15 ml) were collected from the bottom of the tube. Protein standards—bovine catalase, bovine serum albumin (BSA), and bovine cytochrome c—were sedimented in a parallel gradient. Protein concentrations were determined by the Bio-Rad dye-binding assay with BSA as the standard.
RNA triphosphatase assay.
γ-32P-labeled triphosphate-terminated poly(A) was synthesized as described previously (26). RNA triphosphatase reaction mixtures (10 μl) containing 50 mM Tris HCl (pH 7.5), 5 mM dithiothreitol, 10 pmol of [γ-32P]poly(A), and enzyme as specified were incubated for 15 min at 30°C. The reactions were halted by adding 1 μl of 1 M formic acid. Aliquots (5 μl) were spotted onto a polyethyleneimine (PEI)-cellulose thin-layer chromatography (TLC) plate. The TLC plate was developed with 0.75 M potassium phosphate (pH 4.3). 32Pi and [γ-32P]poly(A) were visualized by autoradiographic exposure of the plate. The extent of release of 32Pi from [γ-32P]poly(A) was quantitated by scanning the plate with a Fuji BAS1000 phosphorimager.
ATPase assay.
ATPase activity was assayed as the release of 32Pi from [γ-32P]ATP or the conversion of [α-32P]ATP to [α-32P]ADP. Reaction mixtures (10 μl) containing 50 mM Tris HCl (pH 7.5), 5 mM dithiothreitol, and nucleotides and enzyme as specified were incubated at 30°C. The reaction was terminated by adding 1 μl of 1 M formic acid. Aliquots (5 μl) were spotted onto PEI-cellulose TLC plates, which were developed with 0.5 M LiCl–1 M formic acid. The reaction products were visualized by autoradiographic exposure, and activity was quantitated by scanning the TLC plates with a phosphorimager.
RESULTS
Purification of AcNPV RNA triphosphatase.
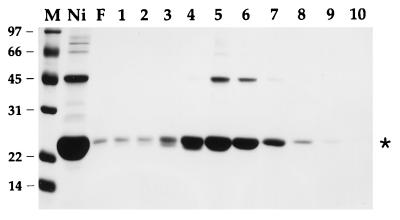
The AcNPV open reading frame encoding the phosphatase-like protein was PCR amplified from a subgenomic plasmid and cloned into a T7 RNA polymerase-based vector pET16b so as to place the gene in frame with an N-terminal leader encoding a 21-amino-acid peptide with 10 tandem histidines. The expression plasmid was introduced into E. coli BL21(DE3), a strain that contains the T7 RNA polymerase gene. A prominent 23-kDa polypeptide was detectable by SDS-PAGE in soluble bacterial extracts (not shown). Initial purification of the His-tagged fusion protein was achieved by adsorption to Ni-agarose and elution with 250 mM imidazole; the eluate was highly enriched with respect to the 23-kDa polypeptide (Fig. 2, lane Ni). The 23-kDa protein was retained on a column of phosphocellulose and eluted with 200 to 400 mM NaCl (Fig. 2, lanes 4 to 6).
FIG. 2.
Phosphocellulose chromatography. Aliquots of the Ni-agarose 250 mM imidazole eluate fraction (lane Ni), the phosphocellulose flowthrough fraction (lane F), and the phosphocellulose NaCl eluate fractions (lanes 1 and 2, 50 mM; lane 3, 100 mM; lane 4, 200 mM; lane 5, 300 mM, lane 6, 400 mM; lane 7, 500 mM; lane 8, 600 mM; lane 9, 700 mM; lane 10, 1.0 M NaCl) were analyzed by SDS-PAGE. The Coomassie blue-stained gel is shown. The positions and sizes (in kilodaltons) of marker proteins (lane M) are indicated at the left. The asterisk indicates the position of the baculovirus polypeptide.
RNA triphosphatase activity was assayed by the liberation of 32Pi from γ-32P-labeled triphosphate-terminated poly(A). The phosphocellulose preparation was active as an RNA triphosphatase. The extent of γ-phosphate hydrolysis during a 15-min incubation at 30°C was proportional to input protein (Fig. 3B). In the linear range of enzyme dependence, 350 fmol of 32Pi was released per fmol of 23-kDa protein.
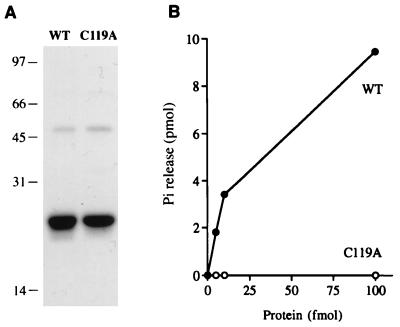
FIG. 3.
RNA triphosphatase activity. (A) Aliquots (3 μg of protein) of the 300 mM NaCl phosphocellulose eluate fraction of wild-type (WT) AcNPV phosphatase and the C119A mutant were electrophoresed through a 10% polyacrylamide gel containing 0.1% SDS. Polypeptides were visualized by staining with Coomassie blue dye. The positions and sizes (in kilodaltons) of marker proteins are indicated at the left. (B) RNA triphosphatase reaction mixtures containing 10 pmol of [γ-32P]poly(A) and either wild-type (WT) or C119A protein as specified were incubated for 15 min at 30°C. The extent of 32Pi release from poly(A) is plotted as function of input enzyme.
To confirm that the observed RNA triphosphatase activity was indeed catalyzed by the baculovirus polypeptide, we expressed in bacteria a mutated version in which the presumptive active-site cysteine moiety (Cys-119) was replaced by alanine. The C119A protein was isolated by Ni-agarose and phosphocellulose column chromatography. The purity of the C119A preparation was identical to that of the wild-type protein, as judged by SDS-PAGE (Fig. 3A). The C119A protein was unable to hydrolyze the γ-phosphate of [γ-32P]poly(A) (Fig. 3B). No activity was detectable with addition of up to 1,000 fmol of C119A protein (not shown). We calculate that the C119A mutation elicited at least a 10−3 decrement in specific activity. The inference from these observations is that the recombinant wild-type protein has intrinsic RNA triphosphatase activity and that it acts according to the predicted mechanism whereby the conserved cysteine serves as the active site nucleophile in phosphoryl transfer.
Characterization of AcNPV RNA triphosphatase.
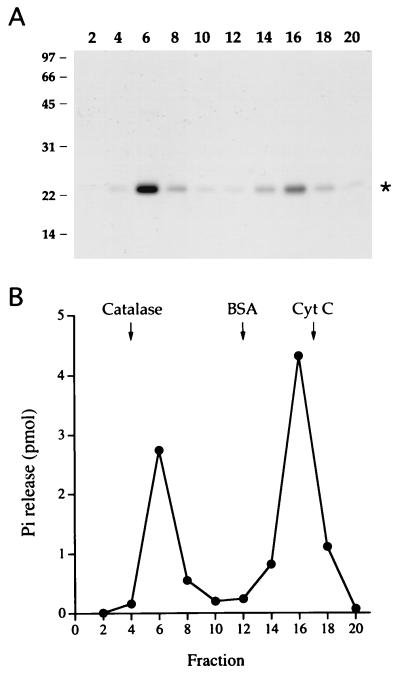
The native structure of AcNPV RNA triphosphatase was analyzed by sedimentation through a 15 to 30% glycerol gradient containing 0.3 M NaCl. The 23-kDa AcNPV polypeptide sedimented as two discrete peaks: a heavy component peaking in fraction 6 and a light component peaking in fraction 16 (Fig. 4A). There were also two discrete peaks of RNA triphosphatase activity that corresponded to the two peaks of the 23-kDa polypeptide (Fig. 4B). We calculated sedimentation coefficients of 9.5S and 2.5S for the heavy and light triphosphatase components, respectively, by comparison to marker proteins (catalase, 11.2S, 248 kDa; BSA, 4.4S, 66 kDa; and cytochrome c, 1.9S, 13 kDa) sedimented in a parallel gradient (Fig. 4B) and by cosedimentation of the markers in the same gradient as the AcNPV protein (not shown). We infer from comparison to the standards that the light component is a monomer of the 23-kDa polypeptide and that the heavy component is a stable oligomer. The sedimentation coefficient of 9.5S (∼200 kDa) suggests an octamer of the 23-kDa polypeptide, assuming that the oligomer is globular in shape. The same bimodal sedimentation behavior was also observed when the AcNPV RNA triphosphatase was centrifuged in a glycerol gradient containing 1 M NaCl (not shown). We conclude that the oligomerization state of the protein is not acutely dependent on ionic strength. The catalytically inactive C119A protein also sedimented in a bimodal fashion (data not shown). We verified that the 23-kDa polypeptides detected in fractions 6 and 16 were indeed the His-tagged AcNPV protein by subjecting both species to automated Edman sequencing after transferring the proteins from an SDS-gel to a polyvinylidene difluoride membrane. The experimentally determined sequence, GHHHHH, corresponded exactly to residues 2 to 7 of the N-terminal leader peptide; apparently, the recombinant protein suffered removal of the initiating methionine.
FIG. 4.
Sedimentation analysis. A sample of the phosphocellulose preparation of wild-type AcNPV phosphatase was sedimented in a 15 to 30% glycerol gradient containing 0.3 M NaCl as described in Materials and Methods. Fractions were collected from the bottoms of the tubes. (A) Aliquots (20 μl) of the indicated glycerol gradient fractions were analyzed by SDS-PAGE. A Coomassie blue-stained gel is shown. The positions and sizes (in kilodaltons) of coelectrophoresed marker polypeptides are indicated at the left. (B) RNA triphosphatase reaction mixtures containing 10 pmol of [γ-32P]poly(A) and 1 μl of the indicated glycerol gradient fractions were incubated for 15 min at 30°C. The peaks of marker proteins catalase, BSA, and cytochrome c (Cyt C), which were centrifuged in a parallel gradient, are indicated by arrows.
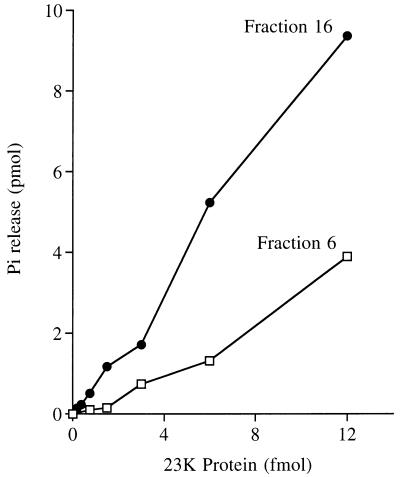
The activities of the heavy and light glycerol gradient fractions were characterized in parallel. The extent of γ-phosphate hydrolysis by the heavy component during a 15-min incubation at 30°C varied with input protein (Fig. 5). In the linear range of enzyme dependence, glycerol gradient fraction 6 released 280 fmol of 32Pi per fmol of 23-kDa polypeptide (∼0.3 s−1). The activity of the light component was also linear with respect to input protein; 780 fmol of 32Pi was released per fmol of the 23-kDa polypeptide (∼0.9 s−1). The specific activity of the light component was ∼3-fold higher than that of the heavy component. The activity of both the heavy and light fractions was optimal in the absence of a divalent cation and was unaffected by EDTA. Inclusion of magnesium elicited a concentration-dependent inhibition of the RNA triphosphatase activity of both enzyme fractions. 32Pi release was inhibited by 50 to 60% at 1 mM MgCl2 and by 90% at 5 mM MgCl2.
FIG. 5.
Specific activity of the 9.5S and 2.5S forms of AcNPV RNA triphosphatase. Reaction mixtures containing 10 pmol of [γ-32P]poly(A) and glycerol gradient fraction 6 or 16 as specified were incubated for 15 min at 30°C. The extent of 32Pi release from poly(A) is plotted as function of input enzyme (expressed as femtomoles of the 23-kDa protomer).
Hydrolysis of ATP.
The heavy and light forms of the AcNPV phosphatase also utilized ATP as a substrate. ATPase activity was detected either by the release of 32Pi from [γ-32P]ATP or the conversion of [α-32P]ATP to [α-32P]ADP. 32Pi release from 100 μM [γ-32P]ATP during a 15-min reaction at 30°C was proportional to input enzyme; the specific ATPase activity of the 2.5S fraction was fivefold higher than that of the 9.5S fraction (not shown).
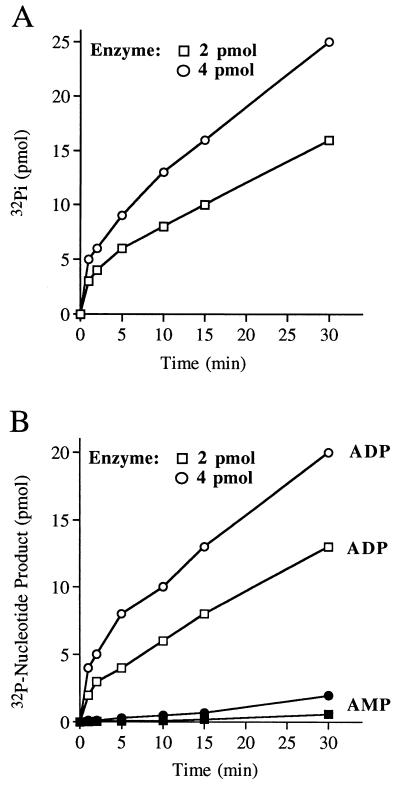
A kinetic analysis of the hydrolysis of 10 μM [γ-32P]ATP by 2 and 4 pmol of the 9.5S enzyme (glycerol gradient fraction 6) revealed an initial burst of 32Pi formation after 1 min of reaction; the amplitudes of the burst were proportional to input enzyme and nearly stoichiometric to the molar amount of 23-kDa protein included in the reaction. A slower linear phase of Pi formation followed the burst (Fig. 6A). The observed rate of Pi release during the slow phase was proportional to input enzyme. The steady-state rate of Pi formation from 10 μM [γ-32P]ATP was 0.2 per min per 23-kDa protomer. Analysis of the hydrolysis of 10 μM [α-32P]ATP by 2 and 4 pmol of fraction 6 revealed an initial burst of [α-32P]ADP formation after 1 min; the amplitudes of the burst were nearly stoichiometric with the molar amount of input 23-kDa protein. A slower linear phase of ADP formation was seen after the burst (Fig. 6B). The steady-state rate of ADP formation was ∼0.17 molecule/min/23-kDa protomer. These results suggest that the initial chemical step of ATP hydrolysis to ADP and Pi by the 9.5S oligomeric enzyme is fast compared to a subsequent step that is limiting in the steady state. We noted that low levels of [α-32P]AMP were formed at late times during the reaction; we surmise that the AcNPV phosphatase catalyzes the hydrolysis of the β-phosphate of the [α-32P]ADP product formed during the reaction.
FIG. 6.
Kinetics of ATP hydrolysis by the 9.5S phosphatase. (A) Reaction mixtures containing (per 10 μl) 10 μM [γ-32P]ATP and either 2 or 4 pmol of 23-kDa protein from glycerol gradient fraction 6 were incubated at 30°C. Aliquots (10 μl) were withdrawn at 1, 2, 5, 10, 15, and 30 min and quenched immediately by mixing with 1 μl of 1 M formic acid. The reaction products were analyzed by PEI-cellulose TLC. The extent of 32Pi formation is plotted as function of reaction time. (B) Reaction mixtures containing (per 10 μl) 10 μM [α-32P]ATP and either 2 or 4 pmol of 23-kDa protein from fraction 6 were incubated at 30°C. The extents of ADP and AMP formation are plotted as function of reaction time.
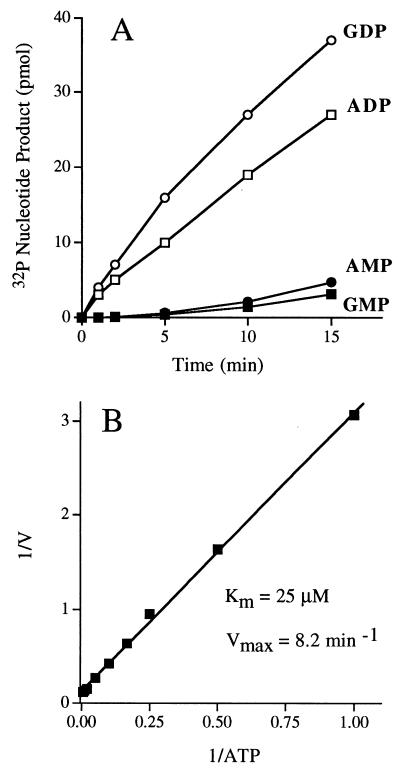
The kinetics of hydrolysis of 10 μM [α-32P]ATP and 10 μM [α-32P]GTP by 1 pmol of glycerol gradient fraction 16 are shown in Fig. 7A. The 2.5S form of the enzyme did not display the slow secondary phase of product formation seen with the 9.5S enzyme; rather, ADP and GDP accumulated smoothly at rates of 2 and 3 pmol/min/23-kDa protomer, respectively. Low levels of [α-32P]AMP and [α-32P]GMP were detected at late times during the reaction, after substantial conversion to ADP had already occurred. We did not detect any release of 32Pi from [α-32P]ATP or [α-32P]GTP. Kinetic parameters were determined for the 2.5S enzyme by measuring the extent of [α-32P]ADP formation from [α-32P]ATP during a 15-min reaction as a function of input [α-32P]ATP concentration in the range of 1 to 200 μM. From a double-reciprocal plot of the data (Fig. 7B), we calculated a Km of 25 μM ATP and a Vmax of 8.2 min−1.
FIG. 7.
Characterization of the NTPase activity of the 2.5S phosphatase. (A) ATP and GTP hydrolysis. Reaction mixtures containing (per 10 μl) 10 μM [α-32P]ATP or [α-32P]GTP and 1 pmol of 23-kDa protein from glycerol gradient fraction 16 were incubated at 30°C. Aliquots (10 μl) were withdrawn at the times specified and quenched immediately by mixing with 1 μl of 1 M formic acid. The reaction products were analyzed by PEI-cellulose TLC. The extents of ADP or GDP formation and AMP or GMP formation are plotted as function of reaction time. (B) Steady-state kinetic parameters of ATP hydrolysis. Reaction mixtures (10 μl) containing 1 pmol of 23-kDa protein from glycerol gradient fraction 16 and either 1, 2, 4, 6, 10, 20, 50, 100, or 200 μM [α32P]ATP were incubated at 30°C for 15 min. The extent of ADP formation (in picomoles) was determined by TLC analysis of the reaction products. A double-reciprocal plot of the rate of ADP formation (min−1 = pmol of ADP formed/15) versus [ATP] is shown.
Hydrolysis of ATP by the 9.5S and 2.5S enzymes was optimal in the absence of a divalent cation; addition of 1 mM MgCl2 inhibited the ATPase activities by ≥95% (data not shown). The C119A mutant protein displayed no detectable ATPase activity (data not shown). We conclude that the RNA triphosphatase and ATPase functions are mediated by the same active site on the AcNPV enzyme.
DISCUSSION
Is triphosphate-terminated RNA a true substrate for baculovirus phosphatase?
The presumptive role of the baculovirus RNA triphosphatase activity would be to hydrolyze the phosphoanhydride bond between the β and γ phosphates of baculovirus late and very late mRNAs, thus preparing them for capping by an RNA guanylyltransferase. For this model to be plausible, the catalytic activity of AcNPV protein must be robust enough to execute this function. Our data argue that this is the case. The heavy and light forms of the AcNPV protein release 0.3 and 0.9 molecules of Pi from 1 μM triphosphate-terminated poly(A) per 23-kDa monomer per s, respectively. This is comparable to the steady-state turnover number of 1 to 2 molecules of Pi released per s per enzyme determined for the mouse RNA triphosphatase domain on the same poly(A) substrate (10). The mouse and baculovirus phosphatases are both members of the superfamily of phosphatases that act via a cysteinyl phosphate intermediate. The turnover number of the baculovirus RNA triphosphatase is also quite close to the value of 0.5 to 0.8 s−1 reported for the RNA triphosphatase component of the vaccinia virus capping enzyme (17). The vaccinia virus enzyme is structurally unrelated to the AcNPV and mouse enzymes and acts via a different mechanism which requires a divalent action and appears not to involve a covalent phosphoenzyme intermediate. The baculovirus RNA triphosphatase activity, like that of the mouse and nematode RNA triphosphatases (10, 27), is lower in the presence of magnesium, which is an essential cofactor for the guanylyltransferase step of the cap synthetic pathway. However, because γ-phosphate cleavage is generally much faster than RNA guanylylation, an order of magnitude rate decrement in the triphosphatase step in the presence of magnesium is unlikely to limit the overall rate of cap formation (23). We surmise, based on comparisons to other RNA triphosphatases for which the connection to RNA capping is well established, that the baculovirus enzyme is sufficiently active to engage in cap formation.
Unlike the C. elegans RNA triphosphatase, which was reported not to hydrolyze ATP or GTP at low micromolar substrate concentrations (27), the baculovirus phosphatase did hydrolyze ATP to ADP and GTP to GDP. The maximal rate of ATP hydrolysis at saturating ATP concentration (≥100 μM) by the more active 2.5S component was 8.2 min−1. This value is substantially less than the turnover number of 52 min−1 observed for the hydrolysis of 1 μM triphosphate-terminated poly(A). Note that the rate of hydrolysis of 1 μM ATP was ∼0.3 min−1. These data argue that the AcNPV enzyme prefers RNA 5′ triphosphates as a substrate over free NTPs. The phosphohydrolase activity is apparently not restricted to NTPs, insofar as we could detect low levels of conversion of GDP to GMP and ADP to AMP during the course of reactions in which a substantial percent of the input NTP had been converted to NDP. We did not attempt to directly assay the kinetic parameters for NMP formation in reactions containing exclusively 32P-NDP substrates.
Three groups of investigators had previously documented that the AcNPV protein possesses phosphomonoesterase activity with para-nitrophenyl phosphate (PNPP) as a substrate (9, 12, 20). Hakes et al. (9) noted that the specific activity of the AcNPV enzyme on PNPP was 2.5% of that of the vaccinia virus H1 phosphatase; the vaccinia virus enzyme is the prototype of the dual-specificity protein phosphatase subfamily. We estimate from kinetic data presented by Sheng and Charbonneau (20), who measured phosphatase activity in the presence of 50 mM PNPP, that AcNPV protein released ∼0.6 molecule of phosphate per enzyme per s. The turnover number for a nonspecific substrate is similar to what we find for Pi release from a triphosphate-terminated poly(A) substrate included at a concentration of 1 μM. Although previous investigators were able to detect release of phosphate from peptide substrates containing either phosphoserine or phosphotyrosine, the protein phosphatase activity was extremely feeble. Hakes et al. (9) found that the specific activities of the AcNPV enzyme against phosphotyrosine- and phosphoserine-containing protein substrates were 1.8 and 0.04% of the specific activities of the vaccinia virus H1 protein. Sheng and Charbonneau (20) measured the hydrolysis of defined phosphotyrosine-containing peptides at substrate concentrations of 2 to 10 μM. From their data, we calculate catalytic rates of 0.006 to 0.6 molecule of phosphate release per enzyme per h. Their data regarding dephosphorylation of a serine/threonine-containing substrate translates into a rate of 0.003 phosphates released per enzyme per h. These catalytic rates in hydrolysis of phosphopeptides are 3 to 5 orders of magnitude slower than what we observed for cleavage of triphosphate-terminated poly(A). Previously, Sheng and Charbonneau (20) suggested that the baculovirus phosphatase may have a unique substrate specificity. Our findings, together with the earlier data, imply that the baculovirus enzyme is an RNA triphosphatase rather than a protein phosphatase.
Two physical forms of the AcNPV phosphatase.
Sedimentation analysis revealed the existence of two physical states of the baculovirus phosphatase: a discrete 9.5S homo-oligomeric form and a 2.5S form (a putative monomer). The sedimentation coefficients and distributions of the two components were not affected by increasing the ionic strength of the protein solution and the sedimentation buffer to 1 M NaCl. Thus, we conclude that the 9.5S oligomer is fairly stable. The two physical forms of the phosphatase were functionally distinct. The specific activities of the 2.5S form on RNA 5′ triphosphate and NTP substrates was severalfold higher than the specific activity of the heavy form. The 2.5S enzyme displayed typical steady-state kinetics in ATP hydrolysis, with a Km of 25 μM ATP. In contrast, the 9.5S enzyme displayed a transient burst of product formation roughly stoichiometric to input enzyme, followed by a slow linear phase. We surmise that the oligomeric enzyme is subject to a rate-limiting step in the steady state (other than reaction chemistry) and that this step is either distinct from or slower than the rate-limiting step for the monomeric enzyme. Conceivably, the oligomeric phosphatase is subject to negative allosteric regulation by occupancy of some of the substrate or product binding sites. For example, the slower steady-state rate (normalized for concentration of the 23-kDa protomer) may arise if only a fraction of the phosphatase active sites within the oligomeric protein are functioning at any given time. To analyze this interesting phenomenon further, it will be helpful to define the structural moieties on the enzyme responsible for subunit-subunit interaction and then assess the functional consequences of mutations that prevent oligomerization.
Biological implications.
Baculovirus early genes are transcribed by cellular RNA polymerase II (pol II). The 5′ ends of early mRNAs are presumably formed by the host cell capping apparatus, which is targeted to the pol II elongation complex by direct physical interaction with the phosphorylated carboxyl-terminal domain of the largest pol II subunit (10, 15, 32). Baculovirus late and very late mRNAs are transcribed by a novel amanitin-resistant RNA polymerase that is uniquely induced in virus-infected cells (2). Thus, in order for late and very late mRNAs to be capped, either the novel polymerase must interact with the cellular capping enzymes or the virus must encode its own capping machinery.
The notion that baculoviruses would encode an RNA triphosphatase involved in mRNA cap formation gains currency in light of the purification and characterization by Guarino et al. (8) of the AcNPV RNA polymerase responsible for the transcription of late and very late genes. The polymerase consists of four equimolar subunits encoded by the virus: Lef-8, Lef-4, Lef-9, and p47 (8). The 464-amino-acid Lef-4 polypeptide contains seven sequence motifs (referred to as motifs I, III, IIIa, IV, V, Vc, and VI) that are conserved in the same order and with similar spacing in the RNA guanylyltransferases of Saccharomyces cerevisiae, Schizosaccharomyces pombe, Candida albicans, Chlorella virus, C. elegans, and mouse (7, 25, 31). All of these proteins catalyze a two-step capping reaction in which the enzyme reacts first with GTP to form a covalent enzyme-GMP intermediate in which GMP is linked via a phosphoramidate (P—N) bond to the invariant lysine of motif I (KxDG). The GMP is then transferred to diphosphate-terminated RNA to form the GpppN cap structure. Guarino et al. (8) have shown that the AcNPV RNA polymerase reacts with GTP to form a covalent protein-GMP adduct with GMP bound to the Lef-4 subunit. We have found that recombinant Lef-4 purified from bacteria reacts with [α-32P]GTP to form a covalent protein-guanylate complex (7).
Lef-4 is essential for baculovirus replication and for the expression of baculovirus late and very late genes in vivo (4, 18). In contrast, the RNA triphosphatase is dispensable; i.e., virus replication still occurs in cultured insect cells when the phosphatase gene is deleted (13). However, the deletion mutant displays a cell line-dependent defect in the production of occluded virus, which may reflect reduced rates of protein synthesis at very late times in the semipermissive cells (13). The host cell variability of the phosphatase deletion phenotype may arise if the host RNA triphosphatase substitutes to a greater or lesser degree for the baculovirus-encoded enzyme or if there are redundant virus-encoded enzymes capable of removing the γ phosphate of viral mRNAs. The last model may well apply, insofar as Lef-4 itself appears to possess intrinsic RNA triphosphatase and ATPase activities (7, 8a).
REFERENCES
- 1.Ahola T, Kaarianen L. Reaction in alphavirus mRNA capping: formation of a covalent complex of nonstructural protein nsP1 with 7-methyl-GMP. Proc Natl Acad Sci USA. 1995;92:507–511. doi: 10.1073/pnas.92.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beniya H, Funk C J, Rohrmann G F, Weaver R F. Purification of a virus-induced RNA polymerase from Autographa californica nuclear polyhedrosis virus-infected Spodoptera frugiperda cells that accurately initiates late and very late transcription in vitro. Virology. 1996;216:12–19. doi: 10.1006/viro.1996.0029. [DOI] [PubMed] [Google Scholar]
- 3.Bisaillon M, Lemay G. Viral and cellular enzymes involved in synthesis of mRNA cap structures. Virology. 1997;236:1–7. doi: 10.1006/viro.1997.8698. [DOI] [PubMed] [Google Scholar]
- 4.Carstens E B, Chan H, Yu H, Williams G V, Casselman R. Genetic analyses of temperature-sensitive mutations in baculovirus late expression genes. Virology. 1994;204:323–337. doi: 10.1006/viro.1994.1537. [DOI] [PubMed] [Google Scholar]
- 5.Denu J M, Stuckey J A, Saper M A, Dixon J E. Form and function in protein dephosphorylation. Cell. 1996;87:361–364. doi: 10.1016/s0092-8674(00)81356-2. [DOI] [PubMed] [Google Scholar]
- 6.Fuchs L Y, Woods M S, Weaver R F. Viral transcription during Autographa californica nuclear polyhedrosis virus infection: a novel RNA polymerase induced in infected Spodoptera frugiperda cells. J Virol. 1983;67:3773–3776. doi: 10.1128/jvi.48.3.641-646.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gross, C. H., and S. Shuman. Unpublished data.
- 8.Guarino, L. A., B. Xu, J. Jin, and W. Dong. A virus-encoded RNA polymerase purified from baculovirus-infected cells. J. Virol., in press. [DOI] [PMC free article] [PubMed]
- 8a.Guarino, L. A. Unpublished data.
- 9.Hakes D J, Martell K J, Zhao W G, Massung R F, Esposito J J, Dixon J E. A protein phosphatase related to the vaccinia virus VH1 is encoded in the genomes of several orthopoxviruses and a baculovirus. Proc Natl Acad Sci USA. 1993;90:4017–4021. doi: 10.1073/pnas.90.9.4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ho C K, Sriskanda V, McCracken S, Bentley D, Schwer B, Shuman S. The guanylyltransferase domain of mammalian mRNA capping enzyme binds to the phosphorylated carboxyl-terminal domain of RNA polymerase II. J Biol Chem. 1998;273:9577–9585. doi: 10.1074/jbc.273.16.9577. [DOI] [PubMed] [Google Scholar]
- 11.Ho C K, Van Etten J L, Shuman S. Expression and characterization of an RNA capping enzyme encoded by Chlorella virus PBCV-1. J Virol. 1996;70:6658–6664. doi: 10.1128/jvi.70.10.6658-6664.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim D, Weaver R F. Transcription mapping and functional analysis of the protein tyrosine/serine phosphatase (PTPase) gene of the Autographa californica nuclear polyhedrosis virus. Virology. 1993;195:587–595. doi: 10.1006/viro.1993.1410. [DOI] [PubMed] [Google Scholar]
- 13.Li Y, Miller L K. Properties of a baculovirus mutant defective in the protein phosphatase gene. J Virol. 1995;69:4533–4537. doi: 10.1128/jvi.69.7.4533-4537.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mao X, Schwer B, Shuman S. Yeast mRNA cap methyltransferase is a 50-kDa protein encoded by an essential gene. Mol Cell Biol. 1995;15:4167–4174. doi: 10.1128/mcb.15.8.4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCracken S, Fong N, Rosonina E, Yankulov K, Brothers G, Siderovski D, Hessel A, Foster S, Shuman S, Bentley D L. 5′ capping enzymes are targeted to pre-mRNA by binding to the phosphorylated C-terminal domain of RNA polymerase II. Genes Dev. 1997;11:3306–3318. doi: 10.1101/gad.11.24.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moss B. Poxviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, et al., editors. Fields virology. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2637–2671. [Google Scholar]
- 17.Myette J R, Niles E G. Domain structure of the vaccinia virus mRNA capping enzyme: expression in Escherichia coli of a subdomain possessing the RNA 5′ triphosphatase and guanylyltransferase activities and a kinetic comparison to the full-size enzyme. J Biol Chem. 1996;271:11936–11944. doi: 10.1074/jbc.271.20.11936. [DOI] [PubMed] [Google Scholar]
- 18.Passarelli A L, Miller L K. Identification of genes encoding late expression factors located between 56.0 and 65.4 map units of the Autographa californica nuclear polyhedrosis virus genome. Virology. 1993;197:704–714. doi: 10.1006/viro.1993.1646. [DOI] [PubMed] [Google Scholar]
- 19.Pena L, Yanez J, Revilla Y, Vinuela E, Salas M L. African swine fever virus guanylyltransferase. Virology. 1992;193:319–328. doi: 10.1006/viro.1993.1128. [DOI] [PubMed] [Google Scholar]
- 20.Sheng Z, Charbonneau H. The baculovirus Autographa californica encodes a protein tyrosine phosphatase. J Biol Chem. 1993;268:4728–4733. [PubMed] [Google Scholar]
- 21.Shi S, Krug R M. Surprising function of the three influenza viral polymerase proteins: selective protection of viral mRNAs against the cap-snatching reaction catalyzed by the same polymerase proteins. Virology. 1996;226:430–435. doi: 10.1006/viro.1996.0673. [DOI] [PubMed] [Google Scholar]
- 22.Shibagaki Y, Itoh N, Yamada H, Nagata S, Mizumoto K. mRNA capping enzyme: isolation and characterization of the gene encoding mRNA guanylyltransferase subunit from Saccharomyces cerevisiae. J Biol Chem. 1992;267:9521–9528. [PubMed] [Google Scholar]
- 23.Shuman S. Capping enzyme in eukaryotic mRNA synthesis. Prog Nucleic Acid Res Mol Biol. 1995;50:101–129. doi: 10.1016/s0079-6603(08)60812-0. [DOI] [PubMed] [Google Scholar]
- 24.Shuman S. A proposed mechanism of mRNA synthesis and capping by vesicular stomatitis virus. Virology. 1997;271:1–6. doi: 10.1006/viro.1996.8305. [DOI] [PubMed] [Google Scholar]
- 25.Shuman S, Schwer B. RNA capping enzyme and DNA ligase—a superfamily of covalent nucleotidyl transferases. Mol Microbiol. 1995;17:405–410. doi: 10.1111/j.1365-2958.1995.mmi_17030405.x. [DOI] [PubMed] [Google Scholar]
- 26.Shuman S, Surks M, Furneaux H, Hurwitz J. Purification and characterization of a GTP:pyrophosphate exchange activity from vaccinia virions. J Biol Chem. 1980;255:11588–11598. [PubMed] [Google Scholar]
- 27.Takagi T, Moore C R, Diehn F, Buratowski S. An RNA 5′-triphosphatase related to the protein tyrosine phosphatases. Cell. 1997;89:867–873. doi: 10.1016/s0092-8674(00)80272-x. [DOI] [PubMed] [Google Scholar]
- 28.Tsukamoto T, Shibagaki Y, Imajoh-Ohmi S, Murakoshi T, Suzuki M, Nakamura A, Gotoh H, Mizumoto K. Isolation and characterization of the yeast mRNA capping enzyme β subunit gene encoding RNA 5′-triphosphatase, which is essential for cell viability. Biochem Biophys Res Commun. 1997;239:116–122. doi: 10.1006/bbrc.1997.7439. [DOI] [PubMed] [Google Scholar]
- 29.Venkatesan S, Gershowitz A, Moss B. Modification of the 5′ end of mRNA: association of RNA triphosphatase with the RNA guanylyltransferase-RNA (guanine-7-) methyltransferase complex from vaccinia virus. J Biol Chem. 1980;255:903–908. [PubMed] [Google Scholar]
- 30.Wang S P, Shuman S. Structure-function analysis of the mRNA cap methyltransferase of Saccharomyces cerevisiae. J Biol Chem. 1997;272:14683–14689. doi: 10.1074/jbc.272.23.14683. [DOI] [PubMed] [Google Scholar]
- 31.Wang S P, Deng L, Ho C K, Shuman S. Phylogeny of mRNA capping enzymes. Proc Natl Acad Sci USA. 1997;94:9573–9578. doi: 10.1073/pnas.94.18.9573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yue Z, Maldonado E, Pillutla R, Cho H, Reinberg D, Shatkin A J. Mammalian capping enzyme complements mutant S. cerevisiae lacking mRNA guanylyltransferase and selectively binds the elongating form of RNA polymerase II. Proc Natl Acad Sci USA. 1997;94:12898–12903. doi: 10.1073/pnas.94.24.12898. [DOI] [PMC free article] [PubMed] [Google Scholar]