Abstract
Terpenoids form the most diversified class of natural products, which have gained application in the pharmaceutical, food, transportation, and fine and bulk chemical industries. Extraction from naturally occurring sources does not meet industrial demands, whereas chemical synthesis is often associated with poor enantio-selectivity, harsh working conditions, and environmental pollutions. Microbial cell factories come as a suitable replacement. However, designing efficient microbial platforms for isoprenoid synthesis is often a challenging task. This has to do with the cytotoxic effects of pathway intermediates and some end products, instability of expressed pathways, as well as high enzyme promiscuity. Also, the low enzymatic activity of some terpene synthases and prenyltransferases, and the lack of an efficient throughput system to screen improved high-performing strains are bottlenecks in strain development. Metabolic engineering and synthetic biology seek to overcome these issues through the provision of effective synthetic tools. This review sought to provide an in-depth description of novel strategies for improving cell factory performance. We focused on improving transcriptional and translational efficiencies through static and dynamic regulatory elements, enzyme engineering and high-throughput screening strategies, cellular function enhancement through chromosomal integration, metabolite tolerance, and modularization of pathways.
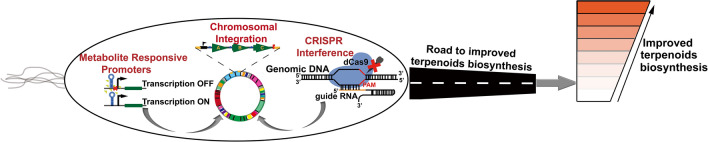
Graphical Abstract
Keywords: Terpenoids, Protein engineering, Dynamic regulation, Promoter engineering, RBS engineering, Cellular tolerance, Chromosomal integration, Modular engineering
Introduction
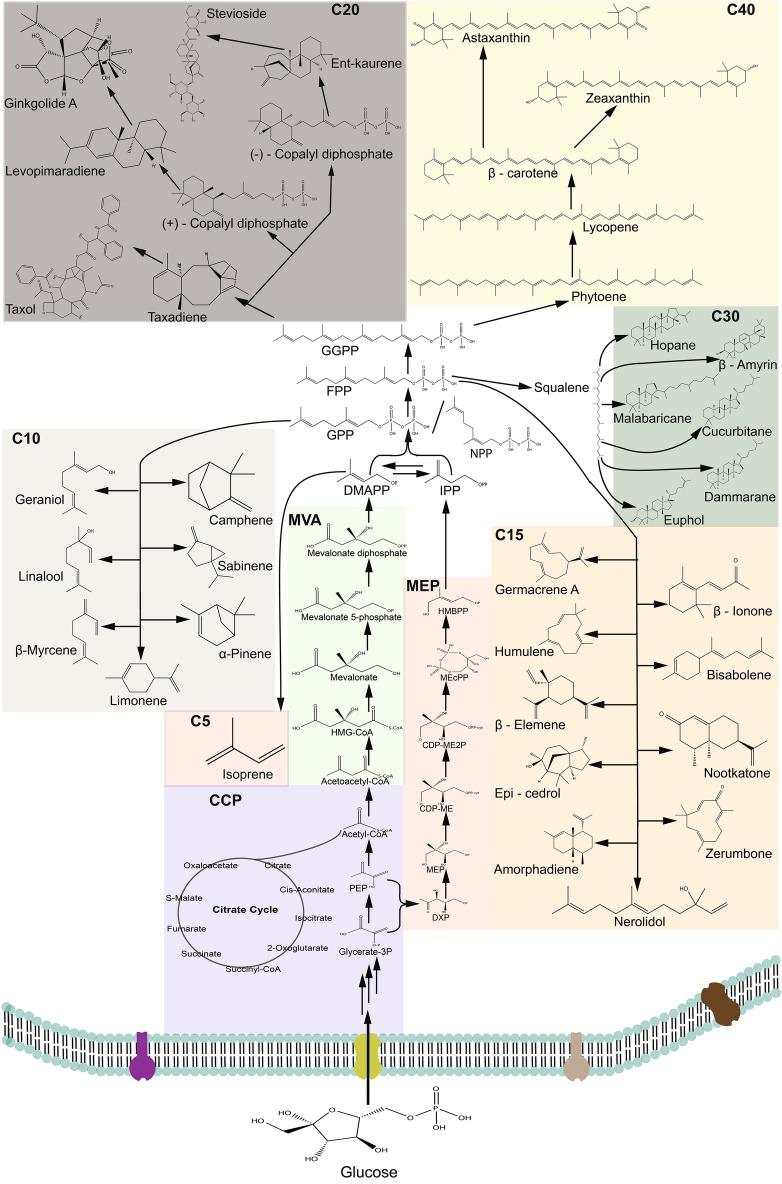
Terpenoids form the most complex class of natural products in chemical and structural terms (Gershenzon and Dudareva 2007; McGarvey and Croteau 1995). Terpenoids (isoprenoids) constitute over 80,000 identified natural compounds, making them the largest known natural compounds (Tetali 2019; Tholl 2015). Terpenoids have a wide distribution in plants, microorganisms, insects, and marine invertebrates (Bian et al. 2018; Chen et al. 2011; Huber et al. 2015; Yamada et al. 2015). Plants exhibit a vast array of isoprenoids; hemiterpenoids (C5), monoterpenoids (C10), sesquiterpenoids (C15), diterpenoids (C20), sesterterpenoids (C25), triterpenoids (C30), and tetraterpenoids (C40) (Fig. 1). These naturally occurring products can be harnessed into useful compounds in the pharmaceutical, food, agricultural, and chemical industries due to their many different properties. Terpenoids are derived from the five-carbon (C5) intermediary units isopentenyl diphosphate (IPP) and its double-bond isomer dimethylallyl diphosphate (DMAPP) from two major universal pathways: the mevalonate (MVA) pathway and the 2-C-methyl-d-erythritol 4-phosphate (MEP) pathway. These are subsequently converted into the plethora of isoprenoids by their respective prenyltransferases and terpene synthases (Tholl 2015) (Fig. 1). This has led to the synthesis of numerous drugs, health care, cosmetic products, flavor and fragrant agents, and biofuels (Tetali 2019).
Fig. 1.
The MVA pathway and MEP pathway for isoprenoids biosynthesis. The isoprenoid biosynthetic pathway can be grouped into the central carbon pathway, upstream isoprenoid pathway, and downstream isoprenoid pathway. PEP, phosphoenolpyruvate; HMG-CoA, S-3-hydroxy-3-methylglutaryl-CoA; DXP, 1-deoxy-d-xylulose 5-phosphate; MEP, 2-C-methyl-d-erythritol 4-phosphate; CDP-ME, 4-(cytidine 5ʹ-diphospho)-2-C-methyl-d-erythritol; CDP-ME2P, 2-phospho-4-(cytidine 5ʹ-diphospho)-2-C-methyl-d-erythritol; MEcPP, 2-C-methyl-d-erythritol 2,4-cyclodiphosphate; HMBPP, 1-hydroxy-2-methyl-2-butenyl 4-diphosphate; IPP, isopentenyl diphosphate; DMAPP, dimethylallyl diphosphate; GPP, geranyl pyrophosphate; NPP, neryl pyrophosphate; FPP, farnesyl pyrophosphate; GGPP, geranylgeranyl pyrophosphate; CCP, central carbon pathway; C5, hemiterpenoids; C10, monoterpenoids; C15, sesquiterpenoids; C20, diterpenoids; C30, triterpenoids; C40, tetraterpenoids
Given their potential benefits in various sectors, the demand for terpenoids is rapidly increasing. Low yields and high costs restrict the direct extraction from plants and other naturally occurring sources. Synthetic production, an alternative source for terpenoid synthesis, produces several isomers. These synthetic chemicals come with several health issues that restrict their direct application due to the possible transfer of by-products and intermediates. However, “wild-type” microbes are unsuitable for commercial purposes since they produce low yields. Designing ideal industrial cell factories is an alternative to producing terpenoids and their derivatives that meet safety and economic concerns. Eliminating the negative feedbacks in the precursor pathways is one of the strategies to increase the pool of precursors. Since wild-type strains are inefficient for industrial applications, it is therefore expedient to metabolically engineer them for high-producing strains.
To ensure a smooth transition to a bio-based economy devoid of complications associated with natural and artificial biosynthetic pathways, developing new, versatile, industrial microbial platforms is required (Jakočiūnas et al. 2020). Since cells have extensive metabolic networks with hard-wired, strictly regulated molecular pathways that resist resource redirection, turning them into successful factories is difficult. (Nielsen and Keasling 2016). The emergence of synthetic biology has facilitated the production of chemicals that, in the past, could not be produced in desired hosts by simply expressing and fine-tuning exogenous pathway genes. To boost productivity, metabolic pathways in these production hosts are frequently regulated by several genetic regulatory tools (Fordjour et al. 2019; Ward et al. 2018). Nevertheless, microbial metabolism is extremely complicated that achieving sufficient yield of a target product from engineered microbe demands careful studies and understanding, as well as the availability of essential tools to manipulate the host strain. The expression of heterologous genes normally results in a metabolic burden on the host strain, affecting the yield of target products. The goal of synthetic biology is to create predictable biological systems. However, bacteria, for example, are very complicated organisms, making it tough to comprehend all of the cell’s functions at the same time and to predict the perturbation outcomes (Jervis et al. 2019).
To overcome these metabolic hurdles and increase the production of metabolites, several strategies have been adopted including: (i) DNA-based engineering, for example, promoter engineering; (ii) RNA engineering including synthetic RNA switches; (iii) protein and cofactor engineering; (iv) metabolic pathway engineering, including modular pathway engineering and compartmentalization engineering; (v) genome-wide engineering; (vi) cell engineering, including transporter engineering (Chen et al. 2018b). This review analyzes recent progress in microbial biosynthesis of terpenoids and examines the critical issues and challenges confronting engineering of cell factories for commercial purposes. In this review, we summarize recent synthetic biology and metabolic engineering strategies to address these challenges and carefully construct a suitable chassis for industrial purposes. We focused on how transcriptional and translational efficiencies through static and dynamic regulatory elements have been harnesses for cell factory development. Also, enzyme engineering and high-throughput screening strategies, cellular function enhancement through chromosomal integration, cell tolerance, and modularization of pathways have been discussed here.
Strategies for developing cell factories for terpenoid biosynthesis
Protein engineering
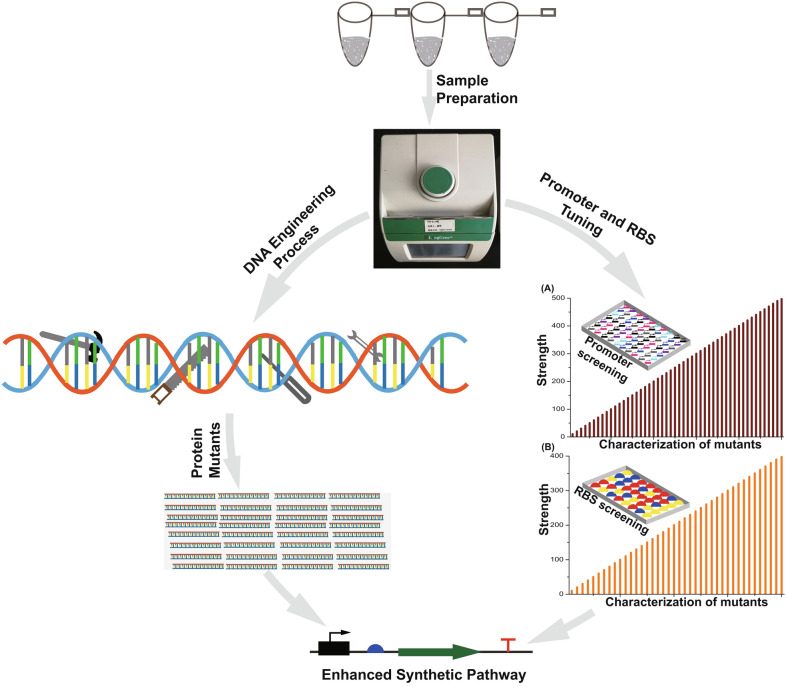
The lack of a centralized database with adequate functionally annotated sequence data has impeded a thorough study of terpene synthases and prenyltransferases in terms of product specificity. In the metabolic process of terpenoid synthesis, terpenoid synthases and prenyltransferases catalyze the synthesis of biologically important terpenoid compounds (Keeling and Bohlmann 2006) (Fig. 1). As is especially true in the case of the terpenoid pathway, various features of a metabolic pathway are not associated exclusively with increasing the concentration of enzymes. Enzymes in their natural states do not show the stability, specificity, or catalytic efficiency required for particular processes (Kokkonen et al. 2019). Protein engineering has been successfully applied to optimize the catalytic efficiencies of rate-limiting enzymes. This has happened through directed evolution and structure-guided engineering using structural information as a tool for enzyme engineering. The lack of a thorough understanding of the structure/function relationship of these terpene synthases and prenyltransferases becomes a deficiency in how to engineer them for high isoprenoids production. To improve properties, protein engineering techniques such as de novo design, directed evolution, rational design, and analytical techniques can be used (Fig. 4). To ensure efficient screening of variants, several high-throughput screening approaches have been developed (Zeng et al. 2020). A lycopene-dependent color high-throughput screening method has already been developed. Adopting this screening strategy, an improved variant form of IDI bearing triple-mutation (L141H/Y195F/W256C) with a catalytic activity of 2.53-fold higher than the wild-type was selected after a directed evolution and site-saturation mutagenesis process. The final strain expressing the mutated enzyme produced more than 1.2 g/L of lycopene, a 2.8-fold increment as compared to the wild-type (Chen et al. 2018a). Wang et al. also developed a novel high-throughput screening method based on DMAPP toxicity to screen for enhanced isoprene synthases (ISPS). Error-prone PCR was used to generate ISPS variants that were cloned into an already constructed DMAPP high-producing strain. A combinatorial mutant with a double mutation (A570T/F340L) was developed to produce isoprene threefold higher than the wild-type strain (Wang et al. 2017).
Fig. 4.
Schematic diagram for producing enzyme, promoter, and RBS library construction and screening. Directed evolution, site-directed mutagenesis, DNA shuffling can be employed to enhance enzyme and regulatory elements efficiency. Engineered promoters and RBS can be used to fine-tune biosynthetic pathways
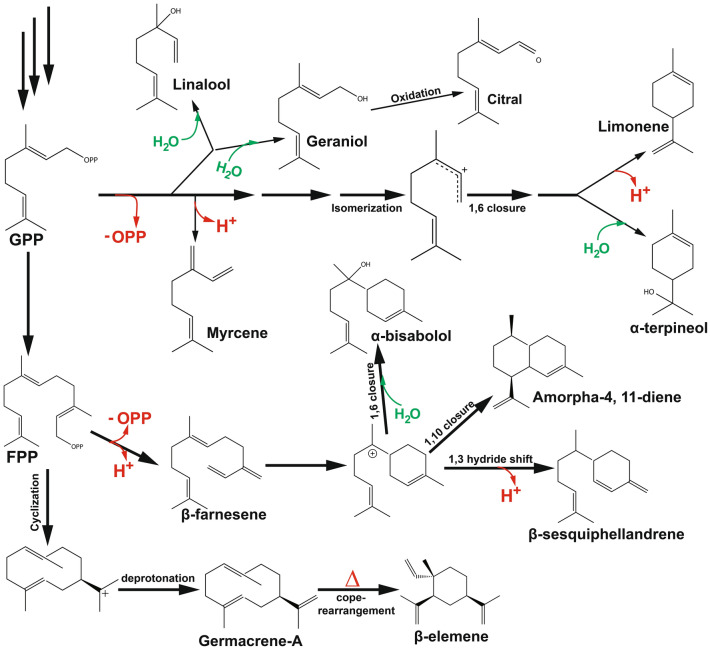
Terpenoid synthases are known to generate intermediates in enzyme-bound carbocation, to achieve structural and functional diversity. This happens after they go through a series of reconfigurations and carbocation quenching (Fig. 2). The process of enzyme carbocation is very important when dealing with protein engineering and has been extensively highlighted by Hong et al. (2020), Huang et al. (2021), Ker et al. (2020), Leferink et al. (2019), McClelland (2008), Raz et al. (2020), Salmon et al. (2015), Tantillo (2010, 2017). Readers are therefore urged to consult the suggestions for further reading. Such functional promiscuity is associated with poor catalytic properties and undesirable product creation (Nobeli et al. 2009). Though enzyme promiscuity is known to provide organisms with genome plasticity to thrive in extreme environmental conditions by altering and reprogramming their metabolic pathways or suppression of undesirable activity (Guzmán et al. 2019), these cyclization reactions often produce “impure” compounds with undesirable products. This increases the cost of production as undesirable by-products have to be removed to ensure a clean commercial product. But the unavailability of enough information about cyclization type and active site sequence (Chen et al. 2011; Christianson 2006) and a holistic and predictive understanding of structural and stability hampers the use of rational engineering to build models of proteins with desirable properties for this purpose. Levopimaradiene (LP), a diterpenoid, is a metabolic product of Levopimaradiene synthase (LPS) via its complex reaction cascade of cyclization, rearrangement, and proton transfers using geranylgeranyl pyrophosphate (GGPP) as its substrate. LPS is known for its promiscuity as it produces isomeric side products such as abietadiene, sandaracopimaradiene, and neoabietadiene (Peters et al. 2000; Ravn et al. 2000). A combinatorial mutation engineering was used to screen for LPS variants with enhanced diterpenoid productivity and selectivity towards LP. The final strain produced an approximately 2600-fold increase in LP. An approximate 700 mg/L of LP was produced in a bench-scale bioreactor under a controlled condition (Leonard et al. 2010).
Fig. 2.
Proposed mechanism of terpenoid carbonation. Terpenoids undergo a wide range of cyclization and rearrangements to ensure final product synthesis
Taxol (paclitaxel) has been a potent chemotherapeutic drug in recent years. However, microbial production is normally hampered by the decomposition of its pathway intermediates to produce undesired products as less than 10% of taxadiene has been speculated to be converted to taxadien-5α-ol by the taxadiene-5-α-hydroxylase (CYP725A4). To ensure product specificity and reduce by-product formation, an alternative pathway was created by mutating taxadiene synthase through site-saturated mutagenesis to yield a 2.4-fold improvement in taxa-4(20)-11(12)-diene which was subsequently hydroxylated to taxadien-α5-ol by CYP725A4 (Edgar et al. 2017). Another strategy for improving protein activity is via enzyme fusion. The fusion of protein has gained increasing recognition in creating novel protein therapeutics and enhancing the performance of engineered strains for biomolecules synthesis. Enzyme fusion is a technique of joining enzymes to produce a recombinant protein with combined characteristics of parental proteins (Uhlen et al. 1992). Enzyme fusion has found application in various areas of biotechnology such as protein purification (Terpe 2003), imaging (Yuste 2005), biopharmaceuticals (Berger et al. 2015), and facilitating downstream fermentation processes (Uhlen et al. 1992). Several reasons for constructing these artificially fused enzymes include improved catalytic activity, activated substrate channeling due to proximity to biocatalysts, higher stability, and cheaper production processes (Elleuche 2015).
In metabolic engineering, one of the main objectives of fusing enzymes is to ensure active sites are close to ensuring intermediates are channeled from active site A to active site B while also preventing competition for these intermediates (Fig. 3B). The direct fusion of enzymes without spacers or linkers, which are indispensable components in building stable, bioactive fused proteins, could result in poor protein expression and reduced catalytic activity (Chen et al. 2013b). Hence, to maintain the functionality of fused enzymes, linkers or spacers are needed. Biosynthesis of isoprene is gaining enough ground because of its associated commercial application (Liu et al. 2019). To increase isoprene production in Cyanobacteria, an isoprene synthase was fused with a highly expressed native protein, cpcB, with or without a linker. A strain with a seven amino acid linker generated 28.9 µg/L/h of isoprene, a 27-fold higher than the strain bearing the unfused enzymes. The study concluded that the relative folding of two enzymes with respect to one another facilitates their catalytic activity (Chaves et al. 2017). Therefore, linkers are an important component in recombinant fusion protein technology. The close active sites of fused proteins guarantee substrate utilization, prevent intermediate diffusion, and alleviate feedback inhibition (Dale et al. 2003). Enzyme fusion has also been proven to remove competition for substrates (Camagna et al. 2019). In S. cerevisiae, geranyl pyrophosphate (GPP), the primary precursor for monoterpenes production, is also a precursor for FPP production. To improve the availability and efficient utilization of GPP for geraniol production, Erg20WW was fused with truncated geraniol synthase in both forward and reverse form. There was 15% increment in geraniol production after 120 h of fermentation compared to the unfused strain (Jiang et al. 2017).
Fig. 3.
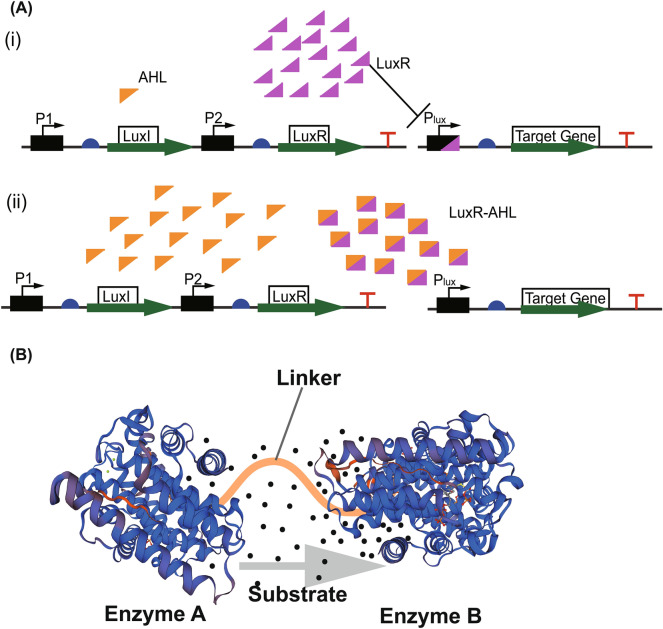
A Illustration of dynamic control of LuxI/LuxR Quorum Sensing (QS) system. (i) At low cell density, the transcriptional regulator, LuxR, binds the Plux promoter to repress the transcription of the target gene. (ii) At high cell density, LuxI protein synthesizes acyl-homoserine lactone (AHL) which binds the transcriptional regulator resulting in dissociation from the promoter. The target gene is subsequently expressed. B Graphical representation of putative enzymes joined with a linker. Sequential pathway enzymes can be modified through enzyme fusion to improve enzymatic reaction. The fusion of enzymes ensures substrates are channelled from one active site to the other
Notable properties of these linkers like secondary structure, length, hydrophobicity, amino acid composition, protease sensitivity, and potential interactions with fused proteins are essential determinants in enzyme fusion technology (Yu et al. 2015). The synthesis of carotenoids begins with the production of phytoene from GGPP catalyzed by phytoene synthase (crtB). Phytoene desaturase (crtI) then catalyzes the synthesis of lycopene, which is finally cyclized to form carotenoids by the lycopene cyclase (crtY). To facilitate an improved production of carotenoid, crtI and crtB domains were joined using two possible configurations with natural and synthetic linkers. A tridomain enzyme comprising crtB, crtI, and crtY was also created. The results from this study demonstrated that the domain order and linker properties affect the expression and stability of the fused proteins, which affects their catalytic effects (Rabeharindranto et al. 2019). In practice, directed evolution, site-directed evolution, enzyme truncation, the fusion of enzymes with other enzymes and or tags are suitable strategies to improve the activities of enzymes. Remarkable advances have been made in enzyme restructure and modification via protein engineering. These could be found in Table 1. This prevents the accumulation of precursors that have resulted in host toxicity. However, the lack of structural–functional relationships that accompanies the fused enzymes becomes a significant challenge as fused enzymes may encounter misfolding, impaired catalytic activity, and low expression level, affecting their desired intentions. Investigations are required to decipher domain–linker interactions and domain–domain interactions to advance the designing of suitable cell factories.
Table 1.
Typical protein engineering strategies for terpenoid biosynthesis
| Strain | Product | Description | Outcome | Refs. |
|---|---|---|---|---|
| Protein engineering | ||||
| E. coli | Viridiflorol | Random mutation by error-prone PCR and enzyme truncation on viridiflorol synthase | 25.7 g/L | Shukal et al. (2019) |
| E. coli | Levopimaradiene | Combinatorial evolution in geranylgeranyl diphosphate synthase and levopimaradiene synthase | 700 mg/L | Leonard et al. (2010) |
| E. coli | Taxa-4(20)-11(12)-diene | Site-saturated mutagenesis on taxadiene synthase | 2.4-fold | Edgar et al. (2017) |
| E. coli | Amorpha-4,11-diene | Mutability landscape engineering of amorpha-4,11-diene synthase | Fourfold | Abdallah et al. (2018) |
| E. coli | Acetyl-CoA | Site-directed mutagenesis of citrate synthase (gltA) | 0.24 g/g glucose | Tovilla-Coutiño et al. (2020) |
| E. coli | 8-epi-cedrol | Fusion of farnesyl pyrophosphate and Epi-cedrol synthase | 1.084 mg/L | Navale et al. (2019) |
| E. coli | Astaxanthin | Fusion of crtZ and crtW | 1.4-fold increment | Nogueira et al. (2019) |
| E. coli | Steviol glycosides | Fusion of CYP714A2 and P450 reductase 2 (AtCPR2) from A. thaliana | 38.4 ± 1.7 mg/L | Moon et al. (2020) |
| E. coli | Lycopene | Enzyme engineering of mevalonate 3-kinase via single amino acid mutations into 5-phosphomevalonate 3-kinase | N. A | Motoyama et al. (2019) |
| E. coli | Albaflavenol and (1R)-6β-hydroxycineole | Fusion of terpene synthases and cytochrome P450 | 90- and 5.4-fold improvement, respectively | Wang et al. (2021d) |
| E. coli | Geraniol | Fusion tagging of geraniol synthase with a laboratory evolved N-terminal of a chloramphenicol acetyltransferase leader sequence | 2124.1 mg/L | Wang et al. (2021c) |
| E. coli | (−)-Carvone | Optimization of pathway enzymes with quantification concatemer (QconCat) method | 15-fold | Yoshida et al. (2021) |
| E. coli | Pinene | Computer-aided directed evolution of geranyl diphosphate and pinene synthase | N. A | Chen et al. (2021) |
| E. coli | Nerol and borneol | Site-directed mutagenesis of truncated bornyl diphosphate synthase from Lippia dulcis | 966.55 mg/L and 87.20 mg/L, respectively | Lei et al. (2021) |
| E. coli | (1R)-6β-Hydroxycineole and albaflavenone | Fusion of 1,8-cineole synthase and cytochrome P450 enzyme CYP176A1 for as well as fusion of epi-isozizaene synthase with CYP170A1 | 5.4-, 2.3-fold, respectively | Wang et al. (2021d) |
| E. coli | Isopentenol | Improved activity of phosphomevalonate decarboxylase (PMD) in an isopentenyl diphosphate (IPP)-bypass | 2.4-fold | Kang et al. (2017) |
| S. cerevisiae | Geranylgeraniol | Overexpression of fused geranylgeranyl pyrophosphate synthase from Pantoea agglomerans and farnesyl diphosphate synthase (PaGGPPS-Erg20) and fused geranylgeranyl pyrophosphate synthase from Pantoea agglomerans and diacylglycerol diphosphate phosphate (PaGGPPS-DPP1) | 5.07 g/L | Wang et al. (2021b) |
| S. cerevisiae | Miltiradiene | Engineering a chimeric protein through the fusion of terpene synthase 1 from Coleus forskohlii and kaurene synthases-like from Salvia miltiorrhia | 3.5 g/L | Hu et al. (2020a) |
| S. cerevisiae | Isoprene | Directed evolution of isoprene synthase | 3.7 g/L | Wang et al. (2017) |
| S. cerevisiae | Lycopene | Inactivation of lycopene cyclase activity of crtYB via directed evolution and improved activity of crtE | 1.61 g/L | Xie et al. (2015) |
| S. cerevisiae | Lycopene | Directed evolution on isopentenyl diphosphate isomerase (idi) | 1.2 g/L | Chen et al. (2018a) |
| S. cerevisiae | Lycopene and β-carotene | Screening of efficient crt variants from Sporidiobolus pararoseus TBRC-BCC 63403 | Four and sevenfold, respectively | Watcharawipas et al. (2021) |
| S. cerevisiae | Carotenoid | A bidomain fusion of crtI and crtB and a tridomain fusion of crtI, crtB, and crtY | Twofold | Rabeharindranto et al. (2019) |
| S. cerevisiae | α-farnesene | Screening of an α-farnesene synthase with enhanced catalytic activity | 10.4 g/L | Wang et al. (2021a) |
| S. cerevisiae | d-Limonene | Fusion of truncated limonene synthase from Citrus limon and truncated neryl diphosphate synthase | 23.7 mg/L | Hu et al. (2020b) |
| S. cerevisiae | Diterpenoids | Fusion of Erg20WW, Erg20 and BTS1 | 3.2- and 2.3-fold | Dong et al. (2020a) |
| S. cerevisiae | Geraniol | Fusion of Erg20WW and geraniol synthase | 15% improvement | Jiang et al. (2017) |
| S. cerevisiae | Germacrene A | Fusion of farnesyl pyrophosphate synthase and germacrene A synthase | 190.7 mg/L | Hu et al. (2017) |
| S. cerevisiae | ( +)-Nootkatone | Fusion of (+)-valencene synthase and farnesyl diphosphate synthase | 59.78 mg/L | Meng et al. (2020a) |
| S. cerevisiae | Zerumbone | Fusion of cytochrome P450 reductase from A. thaliana and α-humulene 8-hydroxylase (CYP71BA1) | 40 mg/L | Zhang et al. (2018a) |
| S. cerevisiae | Sabinene | N-terminal truncation of sabinene synthase | 96% improvement | Jia et al. (2020) |
| S. cerevisiae | Linalool | N-terminal SKIK tagging of engineered linalool synthase and RIAD/RIDD linking of SKIK linalool synthase and ERG20WW | 42% improvement | Zhou et al. (2021) |
| S. cerevisiae | Lutein | Localization of lycopene ε-cyclase to the cell membrane through fusion technology | N. A | Bian et al. (2021) |
| S. cerevisiae | Violaxanthin | N-terminal truncation of zeaxanthin epoxidases | 7.3 mg/g DCW | Cataldo et al. (2020) |
| S. cerevisiae | α-terpineol | Fusion of truncated α-terpineol synthase with Erg20WW | 21.88 mg/L | Zhang et al. (2019) |
| Y. lipolytica | (+)-nootkatoone | Enzyme fusion of truncated NADPH-cytochrome P450 reductase and (+)-nootkatoone synthase coupled with another optimization | 20.5-fold improvement | Guo et al. (2018) |
| Y. lipolytica | Gibberellin (GA3) | Protein engineering and overexpression of pathway enzymes | 12 mg/L | Kildegaard et al. (2021) |
| Y. lipolytica | α-farnesene | Fusion of Erg20 and farnesyl synthase with lipid as the carbon source | 10.2 g/L | Liu et al. (2021d) |
| Pantoea ananatis | Linalool | Improved soluble expression of (S)-linalool synthase by N-terminal fusion with halophilic β-lactamase hexahistidine | 10.9 g/L | Nitta et al. (2021) |
| C. glutamicum | Astaxanthin | Fusion of membrane-bound crtW and crtZ | 3.1 mg/g DCW | Henke and Wendisch (2019) |
| Bacillus subtilis | Amorphadiene | Site-directed mutagenesis of amorphadiene synthase with a one-plasmid CRISPR–Cas9 editing system | 116 mg/L | Song et al. (2021) |
| Cyanobacteria | Isoprene | Fusion isoprene synthase with the native promoter cpcB | 28.9 µg/L/h | Chaves et al. (2017) |
Dynamic pathway regulation
The use of static regulatory elements results in cellular perturbations which can only be addressed through a comprehensive fine-tuning of the various regulatory parts. Static regulation often results in metabolic imbalances that affect a cells’ productivity. Dynamic control of metabolic pathways is crucial in debugging bottlenecks at various points of enzymatic reactions. Taking cues from the natural regulatory metabolic network that respond to intracellular conditions, dynamic regulators have been engineered to manage the production of metabolites and cell growth (Holtz and Keasling 2010). Various dynamic regulatory mechanisms including global regulators (Farmer and Liao 2000), environmental cues (Harder et al. 2018; Yin et al. 2017; Zhao et al. 2018), and chemical cues (Ge et al. 2020) to regulate pathway expression have been developed to ensure a careful balance between the production of biomass and metabolites. Recently, a configurable responsive genetic circuit that genetically controls the activation and repression of pathway genes was developed to control intracellular pyruvate concentration (Xu et al. 2020). In a related study, Shen et al. (2016) adopting feedback responsive promoter enhanced the zeaxanthin synthesis by dynamically regulating the mevalonate pathway to prevent the accumulation of toxic precursors. To enhance the synthesis of monoterpenes, the Erg20 was degron-tagged to control the downstream flux which competes with GPP accumulation, the main precursor for monoterpenes synthesis (Peng et al. 2018).
Dynamic regulation, as an approach, helps microorganisms to thrive in changing environmental conditions and regulate homeostasis, and metabolic flux. One major known method for dynamically regulating pathways is found in the “two-staged metabolic control system”. This involves decoupling the growth and production stages into two to maximize biomass and the production of metabolites (Hartline et al. 2021). One of such regulatory mechanisms is quorum sensing (QS), a mechanism of cell-to-cell communication dependent on cell density in several species of microorganisms, particularly in bacteria (Papenfort and Bassler 2016). This intercellular communication enables bacteria to make a collective decision based on their population. The QS generates, releases, and detects auto-inducers at a certain threshold of cell density (Ge et al. 2020) (Fig. 3A). Knowing the mechanisms at the molecular level of this naturally occurring cell–cell communication system lays a foundation for the engineering of living cells to perform specified and unique tasks. Adopting the two-component QS system luxI–luxR from Vibrio fischeri, Kim et al., achieved 44% increment in bisabolene production from their previous work. In this work, seven variants of the sensor plasmid, carrying luxI–luxR genes, and four variants of the Response plasmid under the control of PluxI promoter, carrying pathway genes to produce bisabolene, were designed to improve the biosynthesis of bisabolene. To avert the problems associated with the plasmid-associated pathway expression system, the QS-based bisabolene pathway was integrated into the E. coli strain resulting in a 1.1 g/L of bisabolene production (Kim et al. 2017). Examples of QS system-associated regulation of metabolic pathways for terpenoids production can be seen in Table 2.
Table 2.
Dynamic and static regulatory strategies used to enhance the cell factory productivities
| Strain | Product | Description | Outcome | Refs. |
|---|---|---|---|---|
| Dynamic regulation | ||||
| E. coli | Bisabolene | An inducer-free Lux QS system | 1.1 g/L | Kim et al. (2018) |
| E. coli | Lycopene | Engineering the Ntr regulon to control intracellular metabolites | 18-fold | Farmer and Liao (2000) |
| E. coli | Zeaxanthin | IPP/FPP-responsive promoter to regulate tuneable intergenic regions (TIGRs) | 2.1-fold | Shen et al. (2016) |
| S. cerevisiae | Linalool and Limonene | An N-degron-dependent protein degradation strategy to downregulate Erg20p | 18 and 76 mg/L, respectively | Peng et al. (2018) |
| S. cerevisiae | Amorpha-4,11-diene | Ergosterol-responsive promoters to regulate Erg9 transcription | 350 mg/L | Yuan and Ching (2015) |
| S. cerevisiae | Lycopene | Growth-phase-dependent dynamic regulation | 1.48 g/L | Su et al. (2020) |
| S. cerevisiae | α-Santalene | Dynamic regulation of Erg9 expression with HXT1 | 92 mg/L | Scalcinati et al. (2012) |
| S. cerevisiae | Nerolidol | An auxin-inducible protein degradation system to decouple growth and production | 3.5 g/L | Lu et al. (2021) |
| S. cerevisiae | Nerolidol | An endoplasmic reticulum-associated protein degradation of Erg9p to redirect flux towards sesquiterpene production | 86% improvement | Peng et al. (2017) |
| B. subtilis | Menaquinone-7 | A bifunctional and modular Phr-60-Rap-60-Spo0A QS system regulated by two endogenous promoters PabrB and PspoiiA | 400-fold | Cui et al. (2019) |
| CRISPR interference (CRISPRi) | ||||
| E. coli | Isoprene, α-bisabolol and lycopene | Development of CRISPRi system for pathway regulation | 2.6-, 10.6-, 8.0-fold increment, respectively | Kim et al. (2016) |
| E. coli | Isopentenol | Combinatorial knockdown of competing pathways with CRISPRi | 98% improvement | Tian et al. (2019) |
| P. putida | Mevalonate | CRISPRi-mediated regulation of glpR, responsible for glycerol utilization | 237 g/L | Kim et al. (2020) |
| C. glutamicum | Decaprenoxanthin | CRISPRi to identify regulatory genes for carotenoid biosynthesis | 43- and ninefold | Göttl et al. (2021) |
| C. glutamicum | Squalene | CRISPRi-mediated repression of competing target genes | 5.2-fold | Park et al. (2019) |
| Synechocystis sp. PCC 6803 | Valencene | Downregulation of crtE with CRISPRi to decrease carotenoid production combined with fusion of ispA and CnVS | 19 mg/g DCW | Dietsch et al. (2021) |
| Methylorubrum extorquens | Carotenoid | CRISPRi-mediated gene mining of phytoene desaturase as well as squalene-hopene cyclase gene repression | 1.9-fold | Mo et al. (2020) |
| Promoter and RBS design | ||||
| E. coli | Geraniol | Optimization of GPP synthase with RBS | 1119 mg/L | Zhou et al. (2015) |
| E. coli | β-carotene | Regulation of atoB, mvaS, and Hmg1 with artificial regulatory parts, MI-46, M-37, and M1-93 | 51% increment | Ye et al. (2016) |
| E. coli | Viridiflorol and Amorphadiene | Transcription and translational optimization of enzymes | 25.7 g/L and 30 g/L, respectively | Shukal et al. (2019) |
| E. coli | Amorphadiene | Combinatorial screening of RBS for translation of pathway enzymes | Fivefold increase | Nowroozi et al. 2014) |
| E. coli | Violaxanthin | RBS optimization of zeaxanthin epoxidase | 231 µg/g DW | Takemura et al. (2019) |
| E. coli | α-Santalene | Promoter replacement to fine-tune the expression of iridoid synthase | 599 g/L | Wang et al. (2021e) |
| E. coli | Steviol | Engineering of 5-UTR and N-terminal of pathway enzymes | 38.4 ± 1.7 mg/L | Moon et al. (2020) |
| E. coli | Salicylate | A combinatorial screening of RBS sequences | 123% | Qian et al. (2019) |
| S. cerevisiae | Sabinene | Downregulating ERG20 with the glucose dependent weak promoter PHXT | 19.4 mg/L | Jia et al. (2020) |
| S. cerevisiae | Squalene-type triterpenoids | Expression of CYP505D13 from Ganoderma lucidum on a yeast expression vector for squalene-type triterpenoids | 3.28 mg/L, 13.77 mg/L, and 12.23 mg/L | Song et al. (2019) |
| S. cerevisiae | Linalool | Downregulating squalene production by replacing the endogenous ERG20 promoter with the sterol-responsive promoter ERG1 | Threefold increment | Zhou et al. (2021) |
| S. cerevisiae | β-amyrin | Employing short synthetic terminators to regulate pathway | 3.16-fold improvement | Ahmed et al. (2019) |
| S. cerevisiae | Lutein | Regulation of pathway enzymes with constitutive promoters as well as temperature-sensitive variant of transcriptional activator Gal4M9 | N. A | Bian et al. (2021) |
| S. cerevisiae | Lycopene | Gal promoter screening | 3.28 g/L | Shi et al. (2019) |
| Y. lipolytica | α-farnesene | Promoter optimization of Sc-tHMG1, IDI and OptFSLERG20 | 2.57 g/L | Liu et al. (2020d) |
| Aspergillus oryzae | Nepetalactol | Promoter replacement to fine-tune the expression of iridoid synthase | 7.2 mg/L | Duan et al. (2021) |
| Rhodobacter capsulatus | Bisabolene | Promoter screening coupled with other pathway engineering strategies | 9.8 g/L | Zhang et al. (2021b) |
| Rhodobacter sphaeroides | Pinene | RBS optimization coupled with fusion of geranyl diphosphate synthase and pinene synthase | N. A | Wu et al. (2021) |
| C. glutamicum | Astaxanthin | Combinatorial RBS, spacer, and start codon library for crtW and crtZ translation | 0.4 mg/L/h | Henke et al. (2016) |
| P. putida | Mevalonate | Development of an inducible CRISPR activation (CRISPRa) system to regulate promoters | 40-fold | Kiattisewee et al. (2021) |
| Chlamydomonas reinhardtii | Carotenoids | Overexpression of wild-type and mutant form of the plant regulatory protein ORANGE under a strong light inducible promoter | Two and threefold, respectively | Yazdani et al. (2021) |
| Synechococcus elongatus UTEX 2973 | Limonene | Fine-tuning GPP synthase expression with synthetic RBS with varying translation rates coupled with crtE mutagenesis | 16.4 mg/L | Lin et al. (2021) |
QS systems that are completely orthogonal forestall the unexpected interference of the two components involved. Such systems are both signal and promoter orthogonal. Recently, the tra QS system from Agrobacterium tumefaciens and the las QS system from Pseudomonas aeruginosa were constructed into a complete orthogonality. To achieve this the EsaI was chosen to synthesize the inducer, N-(3-oxo-hexanoyl)-l-homoserine lactone (3OC6HSL), for the tra system. This system can be employed to metabolically regulate pathway genes (Jiang et al. 2020b). Other QS systems have recently been developed which could be applied to metabolically enhance cell factories’ productivity. For example, a homologous QS regulatory circuit system (hQSRC), a dual-input genetic controller that operates in three modes: (1) a constitutive model for high expression; (2) a tightly repressed mode, and (3) an inducible mode regulated by arabinose and autoinducer-2 was developed for QS-mediated protein expression in Escherichia coli (E. coli) (Hauk et al. 2020). Since QS controls processes that are touted as “expensive public goods” (Schuster et al. 2013), it will be costly and unproductive for a single cell to undertake such a process. Decoupling microbial biosynthesis into growth and production phases improves cell density that subsequently translates into high product formation. Since dynamic regulation predominantly supports a sufficiently dense population, there is a coordinate expression of the target gene when the population is at the large response. Also, dynamic regulation that ensures close interaction between cell density and gene expression could regulate pathway expression.
CRISPR interference (CRISPRi)
Recent advancement in genome engineering has made it possible for biological researchers to directly delete, insert and modify DNA sequences of cells or organisms to elucidate their functions. A number of genomic editing technologies like zinc-finger nucleases (ZFN) based on eukaryotic transcription factors (Miller et al. 2007; Sander et al. 2011; Wood et al. 2011), transcription activator-like effector nucleases (TALENS) from Xanthomonas bacteria (Reyon et al. 2012; Sanjana et al. 2012; Wood et al. 2011; Zhang et al. 2011) and the most recent RNA-guided CRISPR–Cas nuclease system (Cho et al. 2013; Cong et al. 2013; Horvath and Barrangou 2010) have been employed in investigating genomic editing. Clustered regularly interspaced short palindromic repeats (CRISPR) with the CRISPR-associated (Cas) proteins system is RNA-mediated adaptive immune system in prokaryotes that protects them against bacteriophage and plasmid invasion (Barrangou et al. 2007; Barrangou and Marraffini 2014; McGinn and Marraffini 2016).
The CRISPR–Cas9 system has also been harnessed for genome regulation via the inactivation of the Cas9 protein (dCas9) (Qi et al. 2013; Schultenkämper et al. 2020). CRISPR interference (CRISPRi) or dead Cas9 (dCas9) is made possible by mutating the active region of the two domains of Cas9, RuvC and HNH, D10A and H840A, respectively, to attenuate the Cas9, yet retaining its binding ability (Bikard et al. 2013; Ma et al. 2015; Qi et al. 2013). This catalytically dead endonuclease hinders the transcriptional elongation of target genes. CRISPRi is an efficient promising tool to balance and modulate terpenoid production and cell growth as it precisely and predictably binds to fine-tune and repress target pathway genes instead of the traditional gene knockout strategy. This CRISPRi has been employed in pathway engineering to drive the flux towards the production of numerous terpenoids. More recently, CRISPRi-based system has been used to downregulate competing pathways for isopentenol (Tian et al. 2019), valencene (Dietsch et al. 2021). Table 2 provides a list of CRISPRi-mediated systems for regulating terpenoid pathways.
Promoter and ribosome binding site (RBS) designs
Synthetic biology is a gene-combinatorial approach to combining pathway genes from different sources into a proposed metabolic pathway (Yadav et al. 2012). The conditional regulation of gene expression has for years been the object of scientific research. Gene expression can be tuned from transcription initiation, post-translational protein processing through the interaction between transcriptional and translational factors, repressors, activators, or enhancers. Post-transcriptional regulation always affects the level of translated proteins as the level of mRNA transcribed does not always correspond to translated proteins (Jeong et al. 2016; McManus et al. 2014). Promoters and RBS when with known functional characteristics, form an indispensable component of synthetic biology as they establish a baseline for transcription and translation of pathways to ensure optimized native and heterologous pathways (Lee and Trinh 2019). While the construction of well-characterized biological parts involves robust synthetic circuits, the incomplete characterization of the regulators in the construct result in an unstable output in a distinct genetic context. In a recent work to produce β-carotene in E. coli, Ye et al. regulated the expression of the endogenous acetyl-CoA acetyltransferase (atoB) as well as the exogenous enzymes 3-hydroxy-3-methyl-glutaryl-coenzyme A (HMG-CoA) synthase (mvaS) and HMG-CoA reductase (Hmg1) with artificial regulatory parts (MI-46, M-37, M1-93) possessing a characteristic constitutive strength and an RBS library, respectively. This ensured a 51% increment in β-carotene production (Ye et al. 2016).
Optimization of metabolic networks requires a quantitatively characterized pool of the regulatory elements for controlling the expression of target genes (Liu et al. 2018) (Fig. 4). However, several limitations hinder the generation and measurement of a large set of regulatory elements such as trial-and-error, making the processing time-consuming. Hence, a simple model was developed to determine the strength of a promoter based on RNA levels while RBS strength is determined by the efficiency of translation (Kosuri et al. 2013). To produce the sesquiterpene viridiflorol, the biosynthetic pathway was divided into three modules and regulated by a T7 promoter with varying strength. After a careful permutation to transcribe the modules to prevent the accumulation of intermediates and ensure high productivity, the best strain produced 283 mg/L of viridiflorol. RBS was also designed to control the translation of viridiflorol synthase (VS) which increased productivity to 511 ± 37 mg/L, ~ 50% increment (Shukal et al. 2019). In a similar related study, a combinatorial approach involving RBS with varying strength was used to fine-tune the production of amorphadiene. This led to a fivefold increase in amorphadiene accumulation with a subsequent reduction in toxic intermediate metabolites (Nowroozi et al. 2014). Violaxanthin is a carotenoid with numerous pharmaceutical and industrial applications. In the quest to enhance violaxanthin, Takemura et al. modified and designed RBS sequences to regulate Capsicum annuum zeaxanthin epoxidase (CaZEP), the enzyme responsible for catalyzing violaxanthin production from antheraxanthin. A 231 µg/g DW of violaxanthin was achieved (Takemura et al. 2019). Also, a convolutional neural network on cross-RBSs was used in fine-tuning biosensors’ dynamic range (Ding et al. 2020). This shows the importance of promoter and RBS engineering as powerful tools to enhance biosynthetic gene clusters found in transcriptionally silent natural products. The choice (inducible or constitutive) and strength affect the timing and expression level of target proteins. Hence, biological engineers focus on increasing size and complexity to precisely change the degree of expression of several different genes in a pathway (Han et al. 2019) (Table 2).
Regulating expression levels in multi-gene biosynthetic pathways represents a significant challenge for the building of microbial platforms due to a lack of effective control elements and resources (Wei et al. 2018). The cellular burden associated with multi-gene pathways could be minimized by altering each enzyme’s translation initiation rate (TIR) in the pathway of interest to maximizing titer and yield of the desired chemical (Kent and Dixon 2020). In a recent study to improve the supply of GPP for geraniol biosynthesis, the GPP synthase expression was optimized with RBS of increasing TIR. Optimizing the RBS strength resulted in a sixfold increment in geraniol production (Zhou et al. 2015). Though gene deactivation has been applied to block cellular stress response (Sharma et al. 2020), as well as amino acid supplementation to ensure efficient recombinant protein production (Kumar et al. 2020), various degrees of inducible and constitutive promoters have been employed to overcome the metabolic hurdles associated with transcription (Ye et al. 2016; Zhou et al. 2019). To avert the heterogeneities of cellular response that is associated with the use of chemical inducers and the need to regulate inducer concentrations, constitutive promoters have been used in the biosynthesis of the keto-carotenoids astaxanthin and canthaxanthin (Chou et al. 2019; Menin et al. 2019; Nora et al. 2019).
Machine learning has also been applied to predicting promoter sequences (Meng et al. 2017), identification of RNA/DNA binding proteins (Alipanahi et al. 2015), and predicting RBS sequence for improved chemical production (Jervis et al. 2019). In two related studies, machine learning was applied in the modeling pathway for limonene, bisabolene, and pinene synthesis (Dudley et al. 2020; Jervis et al. 2019). Native promoters could also be used to express biosynthetic pathways (Meng et al. 2016; Sengupta et al. 2019; Yuan and Ching 2015). However, they are frequently restricted by the host regulatory network. Synthetic promoters with enhanced strength and orthogonal to intrinsic control networks have been developed (Mordaka and Heap 2018; Redden and Alper 2015) to avert this problem. Since the titer of metabolites typically correlates with gene expression, synthetic biologists have sought the need to expand the synthetic biology toolbox by providing diversified regulatory parts for the construction of efficient endogenous or exogenous pathways (Table 2). However, the fitness of cell factories cannot be overlooked when designing pathway regulators (Bienick et al. 2014).
Cellular tolerance engineering
Saccharomyces cerevisiae and Escherichia coli have been a “laboratory household name” as they continue to fascinate and bedazzle metabolic engineers and system biologists. They have been employed in building a fortified bio-economy. Nevertheless, as the demand for “go green” surges, metabolic engineers and system biologists have sought to enhance the cellular tolerance of these laboratory “workhorses” and other microorganisms to ensure improved cellular exportation for maximum product yield. Mitigation of toxicity effects of both end products and pathway intermediates is a major concern for designing cell factories for both research and industrial purposes. Isoprenoids and their associated intermediate products are of no exemption as excessive accumulation interferes with growth and metabolism. Hence, the need to explore membrane transporters for this purpose. The efflux system has been well studied and has been shown to enhance the biosynthesis of isoprenoids and other chemicals when effectively utilized. Efflux pumps or transporters are membrane proteins devoted to maintaining homeostasis by extruding toxic compounds from the intracellular environment (Jones et al. 2015; Putman et al. 2000). Transporter engineering ensures efficient exportation of compounds to alleviate product feedback inhibition and cytotoxicity (Eggeling and Sahm 2003). One possible alternative to prevent cellular toxicity at the early stages of fermentation is to express pathway genes under a tightly regulated promoter. The Gal promoter is a tightly inducible promoter that is induced by galactose and strongly repressed by glucose (Adams 1972). To alleviate S. cerevisiae’s toxicity to geraniol, pathway genes were expressed under the Gal promoter system (Jiang et al. 2017). Enhancing microbial tolerance coupled with other metabolic engineering has the propensity to increase the production of terpenoids in both model and non-model microorganisms. Some common transporter and tolerance engineering strategies that have been implicated in the biosynthesis of terpenoids are provided in Table 3.
Table 3.
Cellular engineering strategies used to enhance the cell factory productivities
| Strain | Product | Description | Outcome | Refs. |
|---|---|---|---|---|
| Cellular tolerance | ||||
| E. coli | Amorphadiene and kaurene | Overexpression of native AcrAB-TolC, MdtEF-TolC and the exogenous pump mexAB-OprM | 118% and 104% improvement, respectively | Wang et al. (2013) |
| E. coli | Amorphadiene | Overexpression of lipopolysaccharide transport system | N. A | Zhang et al. (2016) |
| E. coli | α-Pinene | Overexpression of AcrB, AcrAB, and TtgB | 1.9-fold | Niu et al. (2018) |
| E. coli | Sabinene | Overexpression of the genes scpA, ygiZ and ybcK via ALE | 191.76 mg/L | Wu et al. (2020a) |
| E. coli | Isoprenol | Genome-wide knockout was employed to identify enzymes associated with isoprenol transport | NA | Wang et al. (2015) |
| E. coli | β-carotene | Development of an artificial membrane vesicle transport system | 24-fold improvement | Wu et al. (2019) |
| E. coli | β-carotene | Engineering membrane bending proteins and membrane synthesis pathway | 44.2 mg/g DCW | Wu et al. (2017) |
| E. coli | Lycopene | Membrane engineering via exogenous of and endogenous expression of almgs, plsB, plsC and dgka, respectively | 36.4 mg/g DCW | Wu et al. (2018) |
| E. coli | Rainbow colorant | Membrane engineering via inner- and outer-membrane vesicle formation and cell morphology engineering | Varying | Yang et al. (2021) |
| E. coli | Squalene | Membrane engineering via membrane proteins overexpression | 612 mg/L | Meng et al. (2020b) |
| S. cerevisiae | Astaxanthin | Atmospheric and room-temperature plasma as well as UV for strain improvement | 404.78 mg/L | Jiang et al. (2020a) |
| S. cerevisiae | Crocetin | Development of a temperature-responsive strain coupled with chromosomal integration of pathway genes | 139.64 ± 2.24 µg/g DCW | Liu et al. (2020e) |
| S. cerevisiae | β-carotene | Comparative proteomic and transcriptional analysis of ABC transporters | 4.04-fold (secretion), 1.33-fold (intracellular) | Bu et al. (2020) |
| S. cerevisiae | Alkane | Exogenous expression of ABC transporters from Y. lipolytica | 80-fold improvement | Chen et al. (2013a) |
| S. cerevisiae | Triterpenoids | Deletion of phosphatidic acid phosphatase (PAH1) for endoplasmic reticulum expansion | 6-, 8-, 16-folds | Arendt et al. (2017) |
| S. cerevisiae | Geraniol | Immobilization of MVA pathway enzymes on yeast surface for in vitro fermentation | 7.55 mg/L | Luo et al. (2021) |
| Y. lipolytica | α-, β-, γ-bisabolene | Exogenously expressing AcrB of the AcrAB-TolC system from E. coli and ABC-G1 from Grosmania clavigera under the constitutive promoter | 2.7-, 8.5-, 1.2-fold, respectively | Zhao et al. (2021) |
| Y. lipolytica | β-carotene | Morphological engineering by deletion of CLA4 and MHY1 genes to convert mycelium form to the yeast form in addition with chromosomal integration | 139% improvement | Liu et al. (2021c) |
| Phaffia rhodozyma | Astaxanthin | Combined atmospheric and room-temperature and UV mutagenesis | 88.57 mg/L | (Zhuang et al. 2020) |
| Phaffia rhodozyma | Carotenoid | Application of magnetic field for improved cellular concentration | 1146.39 ± 26.18 µg/L | (Silva et al. 2020) |
| Chromosomal integration | ||||
| E. coli | Astaxanthin | A plasmid-free strain | 1.4 mg/g CDW | Lemuth et al. (2011) |
| E. coli | β-carotene | Integration of pathway genes | 2.0 g/L | Li et al. (2015) |
| E. coli | β-carotene | Integration of two modules of MVA into the chromosome | 26% improvement | Ye et al. (2016) |
| E. coli | Salvianic acid A | A plasmid-free strain | 5.6 g/L | Zhou et al. (2017) |
| E. coli | Mevalonate | Integration of atoB, mvaS, and mvaE at adhE and ldhA loci | 30 g/L | Wang et al. (2016) |
| E. coli | Bisabolene | Integration of sucrose utilizing operon and MVA pathway | Fivefold improvement | Alonso-Gutierrez et al. (2018) |
| S. cerevisiae | Geraniol | Integration of truncated geraniol synthase | 236.34 mg/L | Jiang et al. (2017) |
| S. cerevisiae | Abscisic acid | Integration of the abscisic gene cluster coupled with plasmid expression | 4.1-fold | Otto et al. (2019) |
| S. cerevisiae | Zerumbone | A multicopy integration of pathway genes | 40 mg/L | Zhang et al. (2018a) |
| S. cerevisiae | Lycopene | Chromosomal integration of engineered crtEB | 41.8 mg/g DCW | Hong et al. (2019) |
| S. cerevisiae | β-carotene | Chromosomal integration of β-carotene biosynthetic pathway genes from Xanthophylomyces dendrorhous | 46.5 mg/g DCW | Fathi et al. (2021) |
| S. cerevisiae | 8-hydroxygeraniol | Development of a plasmid-free strain | 227 mg/L | Yee et al. (2019) |
| S. cerevisiae | Glycyrrhetinic acid and 11-oxo-β-amyrin | Chromosomal integration of glycyrrhetinic acid biosynthetic pathway in two representative strains, haploid and diploid | 18.9 ± 2.0 mg/L and 108.1 ± 4.6 mg/L, respectively | Zhu et al. (2018) |
| S. cerevisiae | (−)-eremophilene | Genomic integration of an Ocimum sanctum sesquiterpene synthase | 34.6 g/L | Deng et al. (2022) |
| Y. lipolytica | β-carotene | Integration of codon-optimized carRA and carB coupled with pathway optimization | 1.7 g/L | Liu et al. (2021b) |
| Y. lipolytica | β-carotene | Multiple chromosomal integration of pathway enzymes under strong promoters | 4 g/L | Gao et al. (2017) |
| Y. lipolytica | Isoprene | Genomic integration of codon-optimized isoprene synthase from Pueraria montana coupled with overexpression of pathway enzymes | ~ 500 µg/L | Shaikh and Odaneth (2021) |
| Bacillus subtilis | Amorphadiene | Chromosomal integration of a fused amorphadiene synthase and green fluorescent protein | 416 ± 15 mg/L | Pramastya et al. (2021) |
| Modularization | ||||
| E. coli | Isoprene | An inter- and intra-module of pathway | 4.7-fold increment | Lv et al. (2016a) |
| E. coli | Taxadiene-5α-ol and Taxadiene | A multivariate-modular pathway | 2400- and 15,000-fold increment, respectively | Ajikumar et al. (2010) |
| E. coli | Pinene | Modular co-culture of MVA pathway and a TIGR-mediated gene cluster | 166.5 mg/L | Niu et al. (2018) |
| S. cerevisiae | Squalene and protopanaxadiol | Engineering of the endoplasmic reticulum as a special compartment triggered a global rewiring of metabolic pathway | 71- and 8-fold, respectively | Kim et al. (2019) |
| S. cerevisiae | Isoprene | Dual regulation of the mitochondrial and endoplasmic reticulum compartments | 2527 mg/L | Lv et al. (2016b) |
| S. cerevisiae | Ginsenoside compound K | Localization of pathway enzymes and metabolic intermediates to lipid droplets | 5 g/L | Shi et al. (2021b) |
| S. cerevisiae | Squalene | Peroxisomal and cytoplasmic engineering | 11 g/L | Liu et al. (2020a) |
| S. cerevisiae | Squalene | A combinatorial engineering of both cytoplasm and mitochondria to alleviate MVA pathway-related toxicity | 21.1 g/L | Zhu et al. (2021) |
| Y. lipolytica | β-ionone | Enhancing cytosolic acetyl-CoA and MVA flux supply via modular engineering and fed-batch fermentation | 0.98 g/L | Lu et al. (2020) |
| Y. lipolytica | Astaxanthin | Subcellular organelle compartmentalization of fused β-carotene ketolase and hydroxylase | 858 mg/L | Ma et al. (2021) |
| Pichia pastoris X33 | α-farnesene | Peroxisomal and cytoplasmic engineering | 2.56 ± 0.04 g/L | Liu et al. (2021a) |
| Bacillus subtilis | Amorphadiene | Modularization of amorphadiene biosynthesis pathway including terpene synthase module, branch pathway module and central metabolic pathway module | 116 mg/L | Song et al. (2021) |
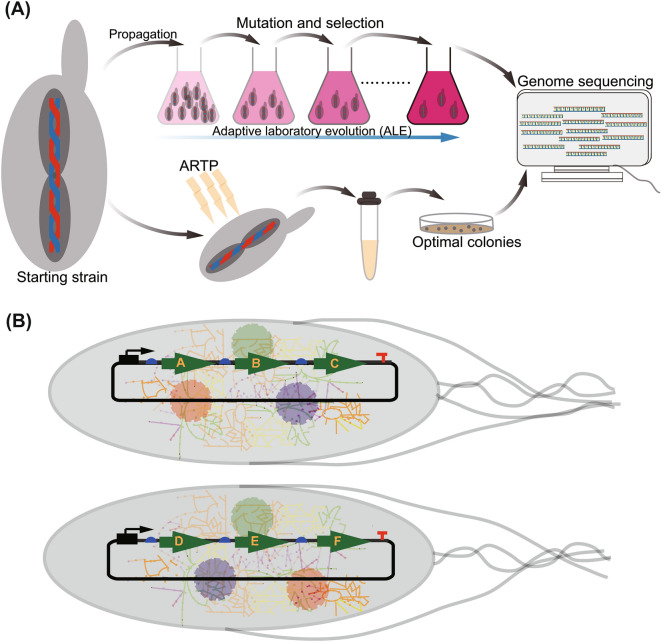
The overexpression of genes associated with efflux pumps has yielded positive results in the production of monoterpenes, sesquiterpenes, and diterpenes (Niu et al. 2018; Wang et al. 2013). Replacing the native promoter of acrAB in E. coli with the P37 strong promoter ensured improved tolerance to pinene (Niu et al. 2018). The cytotoxic effect of terpenoids and their precursors may be due to the damage to organelles, denaturation of proteins, disruption of biological processes, and damage to DNA and the lipid membrane (Nicolaou et al. 2010). Microorganisms have therefore developed a range of defensive strategies to react to these cytotoxic stresses, including overexpression of efflux pumps, activation of stress response genes, and changes in membrane structure (Ramos et al. 2002; Schalck et al. 2021). To build tolerant cell factories, evolutionary adaptation, whole-genome hybridization, genome shuffling, or random mutagenesis may be used, preceded by screening for enhanced variants under the appropriate conditions (David and Siewers 2015; Jullesson et al. 2015) (Fig. 5A). Adopting this strategy to improve the tolerance level of E. coli BL21 for the bicyclic monoterpene, sabinene, the strain was subjected to a gradually increasing concentration of sabinene to drive the evolution process. The mutant strain exhibited an 8.43-fold increase in sabinene production with total production reaching 191.76 mg/L. Transcriptome analysis revealed the overexpression of the genes methyl malonyl-CoA mutase (scpA), the protein that codes for the inner membrane (ygiZ), and the DLP12 prophage family (ybcK) that have been touted to enhance terpene tolerance (Wu et al. 2020a). Bu et al. adopting both comparative proteomics and transcriptional analysis identified five suitable ABC transporters that efficiently transport β-carotene in Saccharomyces cerevisiae. This ensured a 4.04- and 1.33-fold increase in the secretion and intracellular production of β-carotene, respectively (Bu et al. 2020).
Fig. 5.
Schematic illustration of adaptive laboratory evolution (ALE), atmospheric and room-temperature plasma (ARTP), and modular co-culture. A Microorganisms are exposed to a desired selective mechanism and or environment for an iterative period enabling natural selection to optimize variants with enhanced fitness. Genome sequencing and transcriptome can be used to analyze mutant variants. B Modular co-culture engineering. Segregating pathway into modules ensures a holistic assessment of each part for efficient optimization and improvement
In the process to develop solvent tolerance, Gram-negative bacteria develop: (i) an effective change in the composition of membrane fatty acids and phospholipid headgroups; (ii) vesicles containing toxic substances and (iii) toxic organic solvents are exported to the extracellular environment through a resistance-nodulation-cell division (RND) family, an active efflux pump that is energy-dependent (Ramos et al. 2002). The inability of industrial microorganisms to excrete metabolites hinders normal cellular function, which further affects productivity. The biosynthesis of hydrophobic compounds poses a major threat to cell physiological functioning. Carotenoids are large hydrophobic molecules that are not easily excreted by microbial transport systems resulting in intracellular accumulation. An artificial membrane vesicle transport system (AMVTS) was constructed that utilizes membrane lipids to transport hydrophobic compounds. The application of the AMVTS in a β-carotene hyper-producing strain led to a 24-fold increase of secreted β-carotene and a 61% increase in specific production from 27.7 to 44.8 mg/g DCW (Wu et al. 2019). The plasma membrane ABC efflux system is one of the many promising areas for building suitable cell factories for industrial application. To improve the biosynthesis of terpenoids, an improvement in terpenoid-associated transporters must be considered. Moreover, the efflux system is known to be broadly ranged. However, solvent-specific exporters are also known to exist. Hence, it is imperative to carefully study the efflux system to elucidate how they recognize and transport molecules to help engineer product-specific pumps.
Chromosomal integration
Plasmids have been used as an expression system for both endogenous and exogenous pathway genes as it is easy to use or manipulate and incorporate into the host strain, portable, and high copy numbers per cell (Gu et al. 2015; Karim et al. 2013). However, they come with segregational and structural instability, aside from the requirement of antibiotics and sometimes inducer chemicals, resulting in a high cost of production (Friehs 2004). The plasmid-mediated expression system also tends to increase the metabolic burden, especially when using high-copy plasmids. This is due to the channeling of energy towards the keeping and replication of plasmids resulting in poor growth of host cells, leading to low productivity (Wu et al. 2016). Genomic integration provides a way to stably insert pathway genes in the chromosome of desired hosts, which could function with or without antibiotics and inducers. Capitalizing on its stable expression of pathway genes, chromosomal integration was used to enhance geraniol production when a truncated geraniol synthase was integrated into S. cerevisiae’s genome. This resulted in a 23% increment in production, 236.34 mg/L (Jiang et al. 2017).
Plasmid-free cell factories have also been employed in the production of astaxanthin (Lemuth et al. 2011), β-carotene (Li et al. 2015), and many other terpenoids found in Table 3. However, in most cases, the total yield does not meet the industrial requirements. To enhance the expression of pathway genes on the chromosome, several strategies including (Ou et al. 2018): (i) increasing copy numbers of the target gene; (ii) chromosomal loci for integration, and (iii) optimization of the target gene on the chromosome through static and dynamic regulators, have been adopted. The ability to support the expression of pathway genes differs significantly from different chromosomal loci, and these loci ought to be highly expressed, conserved, well-characterized as well as non-essential (Bryant et al. 2014; Yin et al. 2015). To accomplish this, different strategies and methods have been used to stably integrate target genes into chromosomal loci while addressing the issue of copy numbers. To construct an industrial strain for the production of zerumbone, a multicopy integration of the pathway enzymes including cytochrome P450 and a type III membrane protein (ICE2) were integrated into the S. cerevisiae chromosome resulting in a 134-fold 8-hydroxy-α-humulene production. A subsequent multicopy integration of the zerumbone synthase variant (ZSD1S144A) yielded 20.6 mg/L of zerumbone and 40 mg/L in a 5-L bioreactor (Zhang et al. 2018a). Being a precursor for the biosynthesis of isoprenoids (Liu et al. 2019; Marsafari and Xu 2020), mevalonate is also an important industrial component (Xiong et al. 2014). To alleviate the metabolic burden and genetic instability associated with the plasmid expression system while reducing the cost of production to ensure industrial application, two copies of the mevalonate pathway (atoB–mvaS–mvaE) under the expression of a strong constitutive promoter was integrated into E. coli chromosome to replace the adhE and ldhA genes for mevalonate production. Coupled with another pathway engineering (the deletion of sucA and atpFH genes), the final strain produced 30 g/L of mevalonate from 61 g/L glucose in a fed-batch fermentation (Wang et al. 2016). The flippase from the yeast 2-μm plasmid was effectively used to increase the product output and stability using chromosomal integration of gene(s) with multiple copies (CIGMC) (Gu et al. 2015). Another novel strategy involving λ and φ80 bacteriophage site-specific recombination and integration systems have also been used to insert operons of 7.5 and 14 kb into the chromosome with subsequent duplication of target genes (Igonina et al. 2020). Other strategies include chemically inducible chromosomal evolution (CIChE) system (Tyo et al. 2009), clonetegration—a one-step cloning and chromosomal integration of DNA (St-Pierre et al. 2013), a homologous recombination-based method that involves a linear DNA fragment flanked by homologous arms (Storici et al. 2003). A delta integration CRISPR–Cas9 method that ensures a multicopy, highly efficient, one-step and marker-less integration of DNA constructs at the delta sites of the S. cerevisiae chromosome was also developed. This method ensures that fragments spanning from 8 to 24 kb are seamlessly integrated (Shi et al. 2016).
The creation of chromosomally engineered strains is suitable for commercial production, but the relatively low expression on the chromosome has sometimes resulted in inefficient production of target chemicals. However, the expression of exogenous pathway genes using chromosomal integration is highly stable compared to the plasmid-mediated expression system. In line with developing highly efficient platforms for terpenoid production for industrial usage, targeted chromosomal engineering can be coupled with pathway optimization for a precursor supply and an efficient dynamic regulatory system.
Modularization of synthetic pathways
Metabolic pathways involving a large number of genes are notably associated with flux imbalance. Grouping the pathway into suitable modules represents an efficient solution to addressing this issue (Pfleger and Prather 2015; Smanski et al. 2014). Modular co-culture engineering, which involves the partition of pathways into modules and integrating them into separate expression hosts, has also been used to synthesize compounds (Fig. 5B). This system has some advantages of (i) lowering each host metabolic burden; (ii) delivering a variety of cellular conditions in which the various regulatory genes can act; (iii) limiting unintended interaction between various pathways; (iv) adjusting the strain-to-strain ratio to balance the biosynthetic pathway among independent pathway modules; (v) maximizing the performance of complex structures with multiple active substrates; and (vi) facilitating the biosynthesis of a variety of target compounds in a plug-and-play manner (Zhang and Wang 2016). The terpenoid pathway involves several multi-step enzymatic reactions. Hence, constructing cell factories often requires extensive metabolism regulations to ensure an enhanced synthesis of the desired end products. To achieve this goal, several impediments such as (i) tuning the metabolic flux towards target products; (ii) balancing and coordinating corresponding pathways and enzymes to prevent pathway perturbations; (iii) increasing the supply of precursors especially if the strain is growing on glucose to prevent the Crabtree effect; (iv) implementing strategies to optimize the entire pathway (Qin et al. 2015), have to be addressed. To enhance the biosynthesis of isoprene, the isoprene synthetic pathway was divided into two modules; the upstream endogenous MEP pathway and the downstream isoprene pathway made up of the isoprene synthase. An intra-module protein engineering strategy was used to improve the rate-limiting dxs/dxr/idi, while inter-module engineering involving promoter replacement and inducer adjustment was conducted to enhance isoprene synthesis. The final strain achieved a 4.7-fold increment in isoprene as compared to the wild-type strain (Lv et al. 2016a).
In a multi-enzyme synthetic pathway, simple overexpression of rate-limiting enzymes is often associated with an imbalanced pathway that results in the accumulation of toxic intermediate metabolites (Sivy et al. 2011). Recently, a multi-modular engineering approach involving alleviating feedback inhibition, and other pathway engineering strategies were adopted for tyrosol and salidroside overproduction in S. cerevisiae (Liu et al. 2020b). The multidimensional heuristic process (MHP) is a modular pathway optimization approach that assembles and screens multiple repositories of clearly defined transcription factors as well as main enzyme variants in a high-dimensional combinatorial approach to create high-producing strains (Zhang et al. 2018b). This system was used to produce nerolidol, linalool, and astaxanthin from E. coli by partitioning the pathway into three and four modules, respectively (Zhang et al. 2018b). In another related study, the complete β-ionone pathway was divided into three modules: module one responsible for enhancing acetyl-CoA supply comprises the exogenous phosphoketolase from Bifidobacterium bifidum and phosphotransacetylase from Bacillus subtilis; module two contains the endogenous MVA pathway; while the module three consists of the exogenous β-ionone module made up of phytoene dehydrogenase (carB), phytoene synthase/lycopene cyclase (carRP) and carotenoid cleavage dioxygenase (ccD1). These modules were sequentially divided into Yarrowia lipolytica. After pathway optimization augmented with medium and fed-batch optimization, 0.98 g/L of β-ionone was produced after 17 days (Lu et al. 2020). In other instances, in the yeast cells, terpenoid pathways have been segregated into the mitochondria, peroxisomes, endoplasmic reticulum, and cytoplasm compartments to ensure precursor availability for target product synthesis. Table 3 lists several conventional modular approaches that have been used in developing industrial microorganisms.
Modularization of pathways results in a targeted rectification of bottlenecks. Segregating the multi-gene pathway is an efficient way to optimize the expression of a biosynthetic pathway. This provides an effective way to manipulate individual expression levels as it reduces pathway complexity and provides avenues for future uncertainties.
Future perspective for designing efficient microbial platforms
Synthetic biology and metabolic engineering and have demonstrated great capability in ensuring the engineered platforms for the biosynthesis of terpenoids and other essential compounds. The tremendous structural diversity of terpenoids resulting in the vast chemicals for various industries can be linked to terpene synthases’ promiscuous behavior (Fig. 1). Taking into account the biochemical and physiochemical properties of terpenoids, process engineering is a requisite factor when designing cell factories for isoprenoids production. This will allay the inhibitory and toxic effects of isoprenoids and their precursors. Terpenoids inhibition effect is a result of metabolite cytotoxicity in production strains above its threshold. High concentrations of terpenoids and their intermediate products impede cell growth with a subsequent effect on total production. Hence, a suitable method to remove the metabolites at lower concentrations is to enhance the productivity of the biocatalyst. In situ product removal (ISPR) has enhanced higher titers as it prevents product accumulation in culture media and interactions with cells (Alonso-Gutierrez et al. 2013; Brennan et al. 2012; Dong et al. 2020b; Rolf et al. 2020; Schewe et al. 2015). Hence, the need to investigate suitable ISPR mechanisms. The implementation of ISPR is a technical approach to tackling toxicity-associated low productivity and minimizing product loss (Freeman et al. 1993; Salas-Villalobos et al. 2021). Compounds’ hydrophobicity, molecular weight, charge, volatility, and specific binding properties play a major factor in the choice of a compound for the ISPR approach (Freeman et al. 1993). In the two-liquid phase system, one common separation method, two aqueous solutions form a fine emulsion which is correlated to the agitation speed of the reactor. Since the receiving phase is a hydrocarbon, its biocompatibility or toxic effect on the cell factory should be considered. This can be computed with its log Poct (Laane et al. 1987). To expedite the downstream extraction of astaxanthin while taking into consideration the techno economic analysis, an alternative method was established in Haematococcus pluvialis that involves extraction with ethyl acetate from zoospores when growth conditions are restored. This method yielded an 85% extraction rate with 3.5 g of astaxanthin oleoresin produced (Bauer and Minceva 2021). Recently, a pulsed electric field technology, an electroporation method of treating cell biomass before subsequent treatment with traditional solvents or supercritical carbon dioxide was applied to improve the extraction of carotenoids (Martínez et al. 2018, 2020; Saini and Keum 2018). In the downstream processing of industrial-scale bioprocesses, the organic phase becomes a significant practical concern. Microorganisms can also be engineered to enhance their resilience on these hydrophobic chemicals or through the use of solvent-tolerant microbes.
Because all essential information is derived via experimental data rather than meticulously obtained and introduced by domain specialists, a machine learning-based technique allows for the speedier construction of predictive pathway dynamics models. Machine learning (ML), the application of data-driven algorithms could be used to predict the contribution of each particular gene to a specific trait that may be used to analyze, optimize, and develop metabolic or neural networks (Lawson et al. 2021; Mowbray et al. 2021), to improve microbial growth and product synthesis. For several terpene synthases, prenyltransferases that lack structural data, molecular modeling via ML could be used to predict the 3D structures of enzymes, which can then be combined with enzyme–substrate docking studies to enhance several properties such as stability, activity, and specificity (Mazurenko et al. 2020; Singh et al. 2021; Yang et al. 2019). ML can be useful in analyzing the effectiveness of microbial factories via transitional genome-scale modeling, predicting cell phenotypes, and characterizing cell growth (Culley et al. 2020). ML-based modeling has the potential to successfully help design efficient cell factories in the future without knowing comprehensive metabolic regulation pathways. Nevertheless, high-quality quantitative data in multiple situations are required to help address the issues of enzyme engineering, transcription factor binding sites, translation control, ribosomal binding sites, and growth optimization (Helmy et al. 2020).
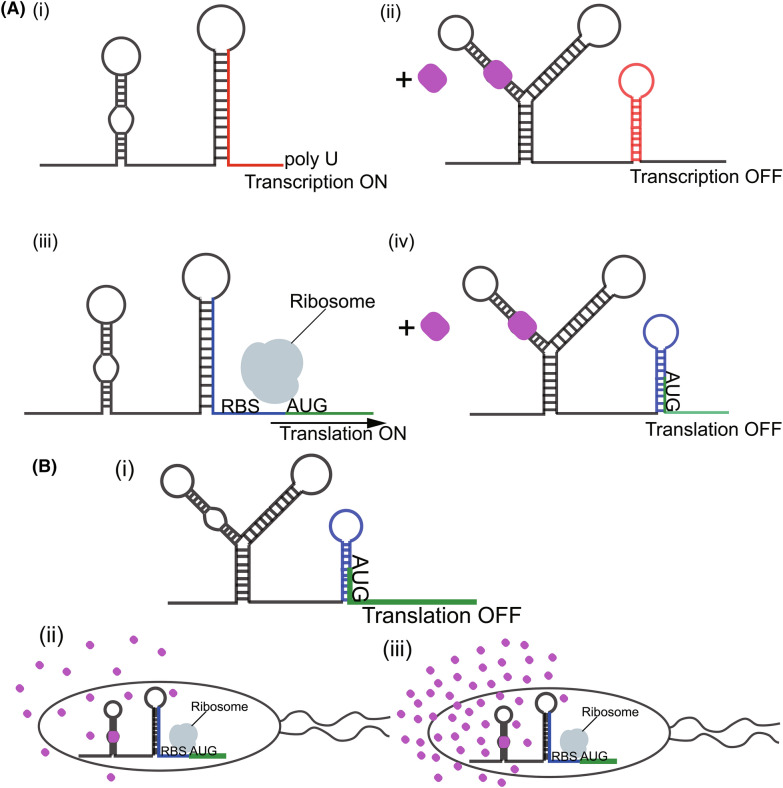
Analysis of metabolic flux is a significant indicator of productive cells. However, cellular metabolites are mainly measured through LC–MS and or GC–MS, a time-consuming throughput for screening in microbial engineering. The inability to precisely quantify and regulate metabolite concentration-related genetic variants especially for most terpenoids, becomes a bottleneck in metabolic engineering (Liu et al. 2017). Metabolite biosensors have gained tremendous recognition in metabolic engineering as these RNA sensors or genetically encoded proteins interact with metabolites to generate detectable phenotypes through the modulation of protein expression (Liu et al. 2015). In addition, due to their quick, precise, and effective mode of action and ease of processing and engineering, biological sensors can respond to different environmental stimuli creating a molecular network. Transcriptional factor-based biosensors are powerful tools that can be used as a high-throughput screening method to develop high-producing strains (Yu et al. 2019) as they present otherwise-obscured intracellular states to a screenable output (Fig. 6B). One such example is a riboswitch which can be turned on and off to regulate expression (Page et al. 2018) (Fig. 6A). In a recent related study, the carotenoid biosynthesis regulator, crtR, in C. glutamicum was engineered to measure the intracellular GGPP concentration during growth (Henke et al. 2020). These sensors could be employed as a high-throughput to screen for isoprenoid-producing strains. Not only do biosensors serve as a high-throughput screening method, but they can also be engineered to function as a dynamic tool. Though it has found its application in developing strains for terpenoid production, adaptive laboratory evolution (ALE), a laboratory “natural selection” process as well as UV and atmospheric and room-temperature plasma (ARTP) holds a brighter prospect of improving cell’s performance, product and intermediates tolerance, and growth rate (Fig. 5A). This random mutation and selection process causes a global disturbance in the genome that will provide additional insights into the regulatory and metabolic circuitry, subsequently providing a platform for developing high-performing strains. Mutants can then be thoroughly screened by a suitable high-throughput method.
Fig. 6.
Mechanism of action of riboswitches. A (i, ii) Binding of ligand to a riboswitch triggers the formation of a hairpin loop that terminates transcription. A (iii, iv) Binding of ligand to a riboswitch generates the formation of a helix that sequesters the RBS to inhibit the translational process. B (i, ii, iii) Application of riboswitches as biosensors. Riboswitches can be linked to colorimetric reporters (for example GFP) to screen for high-producing strains depending on the concentration of the compound of interest. There is a high expression of the reporter gene when the concentration of a ligand is high (B ii) and vice versa
An emerging synthetic pathway that is demonstrating as a promising alternative to the inherently constrained native MVA and MEP pathways termed isopentenyl utilizing pathway (IUP) is gaining grounds. This non-canonical pathway that utilizes isopentenol isomers or prenols as substrate is made up of four genes for monoterpenes synthesis as against the lengthy enzymatic MVA–MEP pathways (Ward et al. 2019). This pathway has been implicated in cell free synthesis of mono-, sesqui-, di-terpenes (Ward et al. 2019). IUP has also been employed in the biosynthesis of various types of terpenoids including linalool (Ferraz et al. 2021), geraniol (Clomburg et al. 2019), nerol, citronellol, lycopene (Chatzivasileiou et al. 2019; Lund et al. 2019; Luo et al. 2020). In addition, a lepidopteran mevalonate (LMVA) pathway has been constructed in E. coli by linking the LMVA pathway with the promiscuous phosphate, NudB. Deletion of the endogenous thiolase genes yielded 390 mg/L of C6-isoprenol (Pang et al. 2021). Likewise, construction of a pyruvate dehydrogenase (PDH) by-pass coupled with gene deletions for enhanced acetyl-CoA flux through the MVA pathway resulted in 2.23 g/L limonene production in S. cerevisiae from a fed-batch shake-flask fermentation (Zhang et al. 2021a). Employing Acinetobacter baylyi ADP1 that catabolizes lignin-derived aromatic substrates couple with gene inactivation and fermentation optimization, Arvay et al. (2021), were able to produce 1014 mg/L of mevalonate through the β-ketoadipate pathway.
Other key strategies include the identification of key regulatory enzymes that could ensure the upregulation of key pathway enzymes for the accumulation of metabolites. This strategy was used in the overproduction of astaxanthin in Xanthophyllomyces dendrorhous when 6-benzylaminopurine was overexpressed (Pan et al. 2020). Also, overexpression of key pathway enzymes have yielded a 1.16-fold increment of sabinene in S. cerevisiae (Jia et al. 2020), 731.18 mg/L of squalene in Y. lipolytica (Tang et al. 2021) and 180 mg/L of bisabolene in Synechocystis sp. PCC 6803 (Rodrigues and Lindberg 2021). Pathway engineering via deletion of competing pathways in the central carbon pathway for acetyl-CoA accumulation while increasing the availability of NADPH has also proven positive as seen in β-carotene production (Wu et al. 2020b). Also, a reduction in lipid biosynthesis through the inactivation of diacylglycerol acyltransferases produced 22.8 g/L of β-farnesene in Y. lipolytica (Shi et al. 2021a), whereas inactivation of phytoene synthase (dr0862) in Deinococcus radiodurans yielded 3.2 ± 0.2 mg/L of pinene (Helalat et al. 2021). Optimization of fermentation parameters, fermentation medium, and terpenoid pathway could improve biomass production while increasing the synthesis of target metabolites (Dai et al. 2021; Liu et al. 2020c, d; Lv et al. 2020; Walls et al. 2020). More so, microorganisms or host organisms with endogenous high-flux isoprenoid pathways, as well as highly tolerant strains like P. putida (Ramos et al. 2015) that have stringent responses to organic solvents could be explored for the biosynthesis of terpenoids (Ankenbauer et al. 2020; Mishra et al. 2020).
Conclusion
Enormous advances have been accomplished in the past decade to engineer microbial platforms for terpenoid biosynthesis; however, these strains still face challenges owing to the intricacy of the terpenoid pathway and tight regulatory networks. Here, we demonstrate the viable approaches for improving the biosynthesis of terpenoids. Overall, efforts towards ensuring suitable microbial platforms have been discussed here for the industrial production of terpenoids. We urge metabolic engineers and synthetic biologists that with the increasing developments in the field, more progress should be made in designing chassis for the biosynthesis of terpenoids.
Acknowledgements
The authors are grateful to the funding support.
Abbreviations
- MVA
Mevalonate
- MEP
2-C-Methyl-d-erythritol 4-phosphate
- IPP
Isopentenyl diphosphate
- DMAPP
Dimethylallyl diphosphate
- QS
Quorum sensing
- AHL
Acyl homoserine lactose
- RBS
Ribosome binding site
- LC–MS
Liquid chromatograph–mass spectrometer
- GC–MS
Gas chromatograph–mass spectrometer
- UTR
Untranslated region
- RND
Resistance nodulation-cell division
- CIGMC
Chromosomal integration of gene(s) with multiple copies
- CIChE
Chemically inducible chromosomal evolution
- AMVTS
Artificial membrane vesicle transport system
- MHP
Multidimensional heuristic process
- CRISPR
Clustered regular interspaced palindromic repeats
- CRISPRi
CRISPR interference
- ALE
Adaptive laboratory evolution
- ARTP
Atmospheric and room-temperature plasma
- ISPR
In situ product removal (ISPR)
- ML
Machine learning
Authors’ contributions
All authors helped in the designing and writing of manuscript. EF, EOM and YH wrote the manuscript. YY, XL and YL contributed in designing of figures. CLL and ZB commented on the manuscript and reviewed it. All authors read and approved the final manuscript.
Funding
This work was supported by the Natural Science Foundation of Jiangsu Province (BK20190610), the China Postdoctoral Science Foundation funded project (2019M651696), Fundamental Research Funds for the Central Universities (JUSRP11963), the 111 Project (111-2-06).
Availability of data and materials
All datasets used and analyzed are available on reasonable request.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
All authors consent to publishing the manuscript in Bioresources and Bioprocessing. There is no conflict of interest for any of the authors regarding the submission of this manuscript.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Chun-Li Liu, Email: liuchunli201774@126.com.
Zhonghu Bai, Email: baizhonghu@jiangnan.edu.cn.
References
- Abdallah II, van Merkerk R, Klumpenaar E, Quax WJ. Catalysis of amorpha-4,11-diene synthase unraveled and improved by mutability landscape guided engineering. Sci Rep. 2018;8(1):9961. doi: 10.1038/s41598-018-28177-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams BG. Induction of galactokinase in Saccharomyces cerevisiae: kinetics of induction and glucose effects. J Bacteriol. 1972;111(2):308. doi: 10.1128/jb.111.2.308-315.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed MS, Ikram S, Rasool A, Li C. Design and construction of short synthetic terminators for β-amyrin production in Saccharomyces cerevisiae. Biochem Eng J. 2019;146:105–116. doi: 10.1016/j.bej.2019.03.011. [DOI] [Google Scholar]
- Ajikumar PK, Xiao W-H, Tyo KEJ, Wang Y, Simeon F, Leonard E, Mucha O, Phon TH, Pfeifer B, Stephanopoulos G. Isoprenoid pathway optimization for Taxol precursor overproduction in Escherichia coli. Science. 2010;330(6000):70–74. doi: 10.1126/science.1191652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alipanahi B, Delong A, Weirauch MT, Frey BJ. Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nat Biotechnol. 2015;33(8):831–838. doi: 10.1038/nbt.3300. [DOI] [PubMed] [Google Scholar]
- Alonso-Gutierrez J, Chan R, Batth TS, Adams PD, Keasling JD, Petzold CJ, Lee TS. Metabolic engineering of Escherichia coli for limonene and perillyl alcohol production. Metab Eng. 2013;19:33–41. doi: 10.1016/j.ymben.2013.05.004. [DOI] [PubMed] [Google Scholar]
- Alonso-Gutierrez J, Koma D, Hu Q, Yang Y, Chan LJG, Petzold CJ, Adams PD, Vickers CE, Nielsen LK, Keasling JD, Lee TS. Toward industrial production of isoprenoids in Escherichia coli: lessons learned from CRISPR–Cas9 based optimization of a chromosomally integrated mevalonate pathway. Biotechnol Bioeng. 2018;115(4):1000–1013. doi: 10.1002/bit.26530. [DOI] [PubMed] [Google Scholar]
- Ankenbauer A, Schäfer RA, Viegas SC, Pobre V, Voß B, Arraiano CM, Takors R. Pseudomonas putida KT2440 is naturally endowed to withstand industrial-scale stress conditions. Microb Biotechnol. 2020;13(4):1145–1161. doi: 10.1111/1751-7915.13571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arendt P, Miettinen K, Pollier J, De Rycke R, Callewaert N, Goossens A. An endoplasmic reticulum-engineered yeast platform for overproduction of triterpenoids. Metab Eng. 2017;40:165–175. doi: 10.1016/j.ymben.2017.02.007. [DOI] [PubMed] [Google Scholar]
- Arvay E, Biggs BW, Guerrero L, Jiang V, Tyo K. Engineering Acinetobacter baylyi ADP1 for mevalonate production from lignin-derived aromatic compounds. Metab Eng Commun. 2021;13:e00173. doi: 10.1016/j.mec.2021.e00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R, Marraffini LA. CRISPR-Cas systems: prokaryotes upgrade to adaptive immunity. Mol Cell. 2014;54(2):234–244. doi: 10.1016/j.molcel.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315(5819):1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- Bauer A, Minceva M. Techno-economic analysis of a new downstream process for the production of astaxanthin from the microalgae Haematococcus pluvialis. Bioresour Bioprocess. 2021;8(1):111. doi: 10.1186/s40643-021-00463-6. [DOI] [Google Scholar]
- Berger S, Lowe P, Tesar M. Fusion protein technologies for biopharmaceuticals: applications and challenges: Editor Stefan R Schmidt. Mabs. 2015;7(3):456–460. doi: 10.1080/19420862.2015.1019788. [DOI] [Google Scholar]
- Bian G, Ma T, Liu T. In vivo platforms for terpenoid overproduction and the generation of chemical diversity. Methods Enzymol. 2018;608:97–129. doi: 10.1016/bs.mie.2018.04.025. [DOI] [PubMed] [Google Scholar]
- Bian Q, Zhou P, Yao Z, Li M, Yu H, Ye L. Heterologous biosynthesis of lutein in S. cerevisiae enabled by temporospatial pathway control. Metab Eng. 2021;67:19–28. doi: 10.1016/j.ymben.2021.05.008. [DOI] [PubMed] [Google Scholar]
- Bienick MS, Young KW, Klesmith JR, Detwiler EE, Tomek KJ, Whitehead TA. The interrelationship between promoter strength, gene expression, and growth rate. PLoS ONE. 2014;9(10):e109105. doi: 10.1371/journal.pone.0109105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bikard D, Jiang W, Samai P, Hochschild A, Zhang F, Marraffini LA. Programmable repression and activation of bacterial gene expression using an engineered CRISPR–Cas system. Nucleic Acids Res. 2013;41(15):7429–7437. doi: 10.1093/nar/gkt520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan TC, Turner CD, Krömer JO, Nielsen LK. Alleviating monoterpene toxicity using a two-phase extractive fermentation for the bioproduction of jet fuel mixtures in Saccharomyces cerevisiae. Biotechnol Bioeng. 2012;109(10):2513–2522. doi: 10.1002/bit.24536. [DOI] [PubMed] [Google Scholar]
- Bryant JA, Sellars LE, Busby SJ, Lee DJ. Chromosome position effects on gene expression in Escherichia coli K-12. Nucleic Acids Res. 2014;42(18):11383–11392. doi: 10.1093/nar/gku828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu X, Lin J-Y, Cheng J, Yang D, Duan C-Q, Koffas M, Yan G-L. Engineering endogenous ABC transporter with improving ATP supply and membrane flexibility enhances the secretion of β-carotene in Saccharomyces cerevisiae. Biotechnol Biofuels. 2020;13(1):168. doi: 10.1186/s13068-020-01809-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camagna M, Grundmann A, Bär C, Koschmieder J, Beyer P, Welsch R. Enzyme fusion removes competition for geranylgeranyl diphosphate in carotenogenesis. Plant Physiol. 2019;179(3):1013. doi: 10.1104/pp.18.01026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cataldo VF, Arenas N, Salgado V, Camilo C, Ibáñez F, Agosin E. Heterologous production of the epoxycarotenoid violaxanthin in Saccharomyces cerevisiae. Metab Eng. 2020;59:53–63. doi: 10.1016/j.ymben.2020.01.006. [DOI] [PubMed] [Google Scholar]
- Chatzivasileiou AO, Ward V, Edgar SM, Stephanopoulos G. Two-step pathway for isoprenoid synthesis. Proc Natl Acad Sci USA. 2019;116(2):506–511. doi: 10.1073/pnas.1812935116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves JE, Rueda-Romero P, Kirst H, Melis A. Engineering isoprene synthase expression and activity in cyanobacteria. ACS Synth Biol. 2017;6(12):2281–2292. doi: 10.1021/acssynbio.7b00214. [DOI] [PubMed] [Google Scholar]
- Chen F, Tholl D, Bohlmann J, Pichersky E. The family of terpene synthases in plants: a mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J. 2011;66(1):212–229. doi: 10.1111/j.1365-313X.2011.04520.x. [DOI] [PubMed] [Google Scholar]
- Chen B, Ling H, Chang MW. Transporter engineering for improved tolerance against alkane biofuels in Saccharomyces cerevisiae. Biotechnol Biofuels. 2013;6(1):21. doi: 10.1186/1754-6834-6-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Zaro JL, Shen WC. Fusion protein linkers: property, design and functionality. Adv Drug Deliv Rev. 2013;65(10):1357–1369. doi: 10.1016/j.addr.2012.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Li M, Liu C, Zhang H, Xian M, Liu H. Enhancement of the catalytic activity of isopentenyl diphosphate isomerase (IDI) from Saccharomyces cerevisiae through random and site-directed mutagenesis. Microb Cell Fact. 2018;17(1):65. doi: 10.1186/s12934-018-0913-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Gao C, Guo L, Hu G, Luo Q, Liu J, Nielsen J, Chen J, Liu L. DCEO Biotechnology: tools to design, construct, evaluate, and optimize the metabolic pathway for biosynthesis of chemicals. Chem Rev. 2018;118(1):4–72. doi: 10.1021/acs.chemrev.6b00804. [DOI] [PubMed] [Google Scholar]
- Chen F, Cheng H, Zhu J, Wang S, Zhang L, Dong H, Liu G, Chen H. Computer-aid directed evolution of GPPS and PS enzymes. BioMed Res Int. 2021;2021:6653500. doi: 10.1155/2021/6653500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31(3):230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- Chou YL, Ko CY, Yen CC, Chen LO, Shaw JF. Multiple promoters driving the expression of astaxanthin biosynthesis genes can enhance free-form astaxanthin production. J Microbiol Methods. 2019;160:20–28. doi: 10.1016/j.mimet.2019.03.012. [DOI] [PubMed] [Google Scholar]
- Christianson DW. Structural biology and chemistry of the terpenoid cyclases. Chem Rev. 2006;106(8):3412–3442. doi: 10.1021/cr050286w. [DOI] [PubMed] [Google Scholar]
- Clomburg JM, Qian S, Tan Z, Cheong S, Gonzalez R. The isoprenoid alcohol pathway, a synthetic route for isoprenoid biosynthesis. Proc Natl Acad Sci USA. 2019;116(26):12810–12815. doi: 10.1073/pnas.1821004116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui S, Lv X, Wu Y, Li J, Du G, Ledesma-Amaro R, Liu L. Engineering a bifunctional Phr60-Rap60-Spo0A quorum-sensing molecular switch for dynamic fine-tuning of menaquinone-7 synthesis in Bacillus subtilis. ACS Synth Biol. 2019;8(8):1826–1837. doi: 10.1021/acssynbio.9b00140. [DOI] [PubMed] [Google Scholar]
- Culley C, Vijayakumar S, Zampieri G, Angione C. A mechanism-aware and multiomic machine-learning pipeline characterizes yeast cell growth. Proc Natl Acad Sci USA. 2020;117(31):18869–18879. doi: 10.1073/pnas.2002959117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Z, Pomraning KR, Panisko EA, Hofstad BA, Campbell KB, Kim J, Robles AL, Deng S, Magnuson JK. Genetically engineered oleaginous yeast Lipomyces starkeyi for sesquiterpene α-Zingiberene production. ACS Synth Biol. 2021;10(5):1000–1008. doi: 10.1021/acssynbio.0c00503. [DOI] [PubMed] [Google Scholar]
- Dale GE, Oefner C, D'Arcy A. The protein as a variable in protein crystallization. J Struct Biol. 2003;142(1):88–97. doi: 10.1016/s1047-8477(03)00041-8. [DOI] [PubMed] [Google Scholar]
- David F, Siewers V. Advances in yeast genome engineering. FEMS Yeast Res. 2015;15(1):1–14. doi: 10.1111/1567-1364.12200. [DOI] [PubMed] [Google Scholar]
- Deng X, Shi B, Ye Z, Huang M, Chen R, Cai Y, Kuang Z, Sun X, Bian G, Deng Z, Liu T. Systematic identification of Ocimum sanctum sesquiterpenoid synthases and (−)-eremophilene overproduction in engineered yeast. Metab Eng. 2022;69:122–133. doi: 10.1016/j.ymben.2021.11.005. [DOI] [PubMed] [Google Scholar]
- Dietsch M, Behle A, Westhoff P, Axmann IM. Metabolic engineering of Synechocystis sp. PCC 6803 for the photoproduction of the sesquiterpene valencene. Metab Eng Commun. 2021;13:e00178. doi: 10.1016/j.mec.2021.e00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding N, Yuan Z, Zhang X, Chen J, Zhou S, Deng Y. Programmable cross-ribosome-binding sites to fine-tune the dynamic range of transcription factor-based biosensor. Nucleic Acids Res. 2020;48(18):10602–10613. doi: 10.1093/nar/gkaa786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Chen S, Zhu J, Gao K, Zha W, Lin P, Zi J. Enhance production of diterpenoids in yeast by overexpression of the fused enzyme of ERG20 and its mutant mERG20. J Biotechnol. 2020;307:29–34. doi: 10.1016/j.jbiotec.2019.10.019. [DOI] [PubMed] [Google Scholar]
- Dong Y, Li X, Duan J, Qin Y, Yang X, Ren J, Li G. Improving the yield of xenocoumacin 1 enabled by in situ product removal. ACS Omega. 2020;5(32):20391–20398. doi: 10.1021/acsomega.0c02357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Y, Liu J, Du Y, Pei X, Li M. Aspergillus oryzae biosynthetic platform for de novo iridoid production. J Agric Food Chem. 2021;69(8):2501–2511. doi: 10.1021/acs.jafc.0c06563. [DOI] [PubMed] [Google Scholar]
- Dudley QM, Karim AS, Nash CJ, Jewett MC. In vitro prototyping of limonene biosynthesis using cell-free protein synthesis. Metab Eng. 2020;61:251–260. doi: 10.1016/j.ymben.2020.05.006. [DOI] [PubMed] [Google Scholar]
- Edgar S, Li FS, Qiao K, Weng JK, Stephanopoulos G. Engineering of taxadiene synthase for improved selectivity and yield of a key Taxol biosynthetic intermediate. ACS Synth Biol. 2017;6(2):201–205. doi: 10.1021/acssynbio.6b00206. [DOI] [PubMed] [Google Scholar]
- Eggeling L, Sahm H. New ubiquitous translocators: amino acid export by Corynebacterium glutamicum and Escherichia coli. Arch Microbiol. 2003;180(3):155–160. doi: 10.1007/s00203-003-0581-0. [DOI] [PubMed] [Google Scholar]
- Elleuche S. Bringing functions together with fusion enzymes—from nature’s inventions to biotechnological applications. Appl Microbiol Biotechnol. 2015;99(4):1545–1556. doi: 10.1007/s00253-014-6315-1. [DOI] [PubMed] [Google Scholar]
- Farmer WR, Liao JC. Improving lycopene production in Escherichia coli by engineering metabolic control. Nat Biotechnol. 2000;18(5):533–537. doi: 10.1038/75398. [DOI] [PubMed] [Google Scholar]
- Fathi Z, Tramontin LRR, Ebrahimipour G, Borodina I, Darvishi F. Metabolic engineering of Saccharomyces cerevisiae for production of β-carotene from hydrophobic substrates. FEMS Yeast Res. 2021;21(1):foaa068. doi: 10.1093/femsyr/foaa068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraz CA, Leferink NGH, Kosov I, Scrutton NS. Isopentenol utilization pathway for the production of linalool in Escherichia coli using an improved bacterial linalool/nerolidol synthase. ChemBioChem. 2021;22(13):2325–2334. doi: 10.1002/cbic.202100110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fordjour E, Adipah FK, Zhou S, Du G, Zhou J. Metabolic engineering of Escherichia coli BL21 (DE3) for de novo production of l-DOPA from d-glucose. Microb Cell Fact. 2019;18(1):74. doi: 10.1186/s12934-019-1122-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman A, Woodley JM, Lilly MD. In situ product removal as a tool for bioprocessing. Bio/technology. 1993;11(9):1007–1012. doi: 10.1038/nbt0993-1007. [DOI] [PubMed] [Google Scholar]
- Friehs K. Plasmid copy number and plasmid stability. Adv Biochem Eng Biotechnol. 2004;86:47–82. doi: 10.1007/b12440. [DOI] [PubMed] [Google Scholar]
- Gao S, Tong Y, Zhu L, Ge M, Zhang Y, Chen D, Jiang Y, Yang S. Iterative integration of multiple-copy pathway genes in Yarrowia lipolytica for heterologous β-carotene production. Metab Eng. 2017;41:192–201. doi: 10.1016/j.ymben.2017.04.004. [DOI] [PubMed] [Google Scholar]
- Ge C, Sheng H, Chen X, Shen X, Sun X, Yan Y, Wang J, Yuan Q. Quorum sensing system used as a tool in metabolic engineering. Biotechnol J. 2020 doi: 10.1002/biot.201900360. [DOI] [PubMed] [Google Scholar]
- Gershenzon J, Dudareva N. The function of terpene natural products in the natural world. Nat Chem Biol. 2007;3(7):408–414. doi: 10.1038/nchembio.2007.5. [DOI] [PubMed] [Google Scholar]
- Göttl VL, Schmitt I, Braun K, Peters-Wendisch P, Wendisch VF, Henke NA. CRISPRi-library-guided target identification for engineering carotenoid production by Corynebacterium glutamicum. Microorganisms. 2021;9(4):670. doi: 10.3390/microorganisms9040670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu P, Yang F, Su T, Wang Q, Liang Q, Qi Q. A rapid and reliable strategy for chromosomal integration of gene(s) with multiple copies. Sci Rep. 2015;5(1):9684. doi: 10.1038/srep09684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Sun J, Li D, Lu W. Heterologous biosynthesis of (+)-nootkatone in unconventional yeast Yarrowia lipolytica. Biochem Eng J. 2018;137:125–131. doi: 10.1016/j.bej.2018.05.023. [DOI] [Google Scholar]
- Guzmán GI, Sandberg TE, LaCroix RA, Nyerges Á, Papp H, de Raad M, King ZA, Hefner Y, Northen TR, Notebaart RA, Pál C, Palsson BO, Papp B, Feist AM. Enzyme promiscuity shapes adaptation to novel growth substrates. Mol Syst Biol. 2019;15(4):e8462. doi: 10.15252/msb.20188462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L, Cui W, Suo F, Miao S, Hao W, Chen Q, Guo J, Liu Z, Zhou L, Zhou Z. Development of a novel strategy for robust synthetic bacterial promoters based on a stepwise evolution targeting the spacer region of the core promoter in Bacillus subtilis. Microb Cell Fact. 2019;18(1):96. doi: 10.1186/s12934-019-1148-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harder BJ, Bettenbrock K, Klamt S. Temperature-dependent dynamic control of the TCA cycle increases volumetric productivity of itaconic acid production by Escherichia coli. Biotechnol Bioeng. 2018;115(1):156–164. doi: 10.1002/bit.26446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartline CJ, Schmitz AC, Han Y, Zhang F. Dynamic control in metabolic engineering: theories, tools, and applications. Metab Eng. 2021;63:126–140. doi: 10.1016/j.ymben.2020.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauk P, Stephens K, Virgile C, VanArsdale E, Pottash AE, Schardt JS, Jay SM, Sintim HO, Bentley WE. Homologous quorum sensing regulatory circuit: a dual-input genetic controller for modulating quorum sensing-mediated protein expression in E. coli. ACS Synth Biol. 2020;9(10):2692–2702. doi: 10.1021/acssynbio.0c00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helalat SH, Jers C, Bebahani M, Mohabatkar H, Mijakovic I. Metabolic engineering of Deinococcus radiodurans for pinene production from glycerol. Microb Cell Fact. 2021;20(1):187. doi: 10.1186/s12934-021-01674-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmy M, Smith D, Selvarajoo K. Systems biology approaches integrated with artificial intelligence for optimized metabolic engineering. Metab Eng Commun. 2020;11:e00149. doi: 10.1016/j.mec.2020.e00149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henke NA, Wendisch VF. Improved astaxanthin production with Corynebacterium glutamicum by application of a membrane fusion protein. Mar Drugs. 2019;17(11):621. doi: 10.3390/md17110621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henke NA, Heider SAE, Peters-Wendisch P, Wendisch VF. Production of the marine carotenoid astaxanthin by metabolically engineered Corynebacterium glutamicum. Mar Drugs. 2016;14(7):124. doi: 10.3390/md14070124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henke NA, Austermeier S, Grothaus IL, Götker S, Persicke M, Peters-Wendisch P, Wendisch VF. Corynebacterium glutamicum CrtR and its orthologs in actinobacteria: conserved function and application as genetically encoded biosensor for detection of geranylgeranyl pyrophosphate. Int J Mol Sci. 2020;21(15):5482. doi: 10.3390/ijms21155482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtz WJ, Keasling JD. Engineering static and dynamic control of synthetic pathways. Cell. 2010;140(1):19–23. doi: 10.1016/j.cell.2009.12.029. [DOI] [PubMed] [Google Scholar]
- Hong J, Park SH, Kim S, Kim SW, Hahn JS. Efficient production of lycopene in Saccharomyces cerevisiae by enzyme engineering and increasing membrane flexibility and NAPDH production. Appl Microbiol Biotechnol. 2019;103(1):211–223. doi: 10.1007/s00253-018-9449-8. [DOI] [PubMed] [Google Scholar]
- Hong B, Luo T, Lei X. Late-stage diversification of natural products. ACS Cent Sci. 2020;6(5):622–635. doi: 10.1021/acscentsci.9b00916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327(5962):167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- Hu Y, Zhou YJ, Bao J, Huang L, Nielsen J, Krivoruchko A. Metabolic engineering of Saccharomyces cerevisiae for production of germacrene A, a precursor of beta-elemene. J Ind Microbiol Biotechnol. 2017;44(7):1065–1072. doi: 10.1007/s10295-017-1934-z. [DOI] [PubMed] [Google Scholar]
- Hu T, Zhou J, Tong Y, Su P, Li X, Liu Y, Liu N, Wu X, Zhang Y, Wang J, Gao L, Tu L, Lu Y, Jiang Z, Zhou YJ, Gao W, Huang L. Engineering chimeric diterpene synthases and isoprenoid biosynthetic pathways enables high-level production of miltiradiene in yeast. Metab Eng. 2020;60:87–96. doi: 10.1016/j.ymben.2020.03.011. [DOI] [PubMed] [Google Scholar]
- Hu Z, Lin L, Li H, Li P, Weng Y, Zhang C, Yu A, Xiao D. Engineering Saccharomyces cerevisiae for production of the valuable monoterpene d-limonene during Chinese Baijiu fermentation. J Ind Microbiol Biot. 2020 doi: 10.1007/s10295-020-02284-6. [DOI] [PubMed] [Google Scholar]
- Huang Z-Y, Ye R-Y, Yu H-L, Li A-T, Xu J-H. Mining methods and typical structural mechanisms of terpene cyclases. Bioresour Bioprocess. 2021;8(1):66. doi: 10.1186/s40643-021-00421-2. [DOI] [Google Scholar]
- Huber T, Weisheit L, Magauer T. Synthesis of xenia diterpenoids and related metabolites isolated from marine organisms. Beilstein J Org Chem. 2015;11:2521–2539. doi: 10.3762/bjoc.11.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igonina O, Samsonov V, Ublinskaya A, Hook C, Malykh E, Kozaeva E, Sycheva E, Stoynova N. A novel one-step method for targeted multiplication of DNA fragments from the Escherichia coli chromosome mediated by coordinated functioning of λ and φ80 bacteriophage recombination systems. J Microbiol Methods. 2020;170:105842. doi: 10.1016/j.mimet.2020.105842. [DOI] [PubMed] [Google Scholar]
- Jakočiūnas T, Klitgaard AK, Kontou EE, Nielsen JB, Thomsen E, Romero-Suarez D, Blin K, Petzold CJ, Gin JW, Tong Y, Gotfredsen CH, Charusanti P, Frandsen RJN, Weber T, Lee SY, Jensen MK, Keasling JD. Programmable polyketide biosynthesis platform for production of aromatic compounds in yeast. Synth Syst Biotechnol. 2020;5(1):11–18. doi: 10.1016/j.synbio.2020.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong Y, Kim J-N, Kim MW, Bucca G, Cho S, Yoon YJ, Kim B-G, Roe J-H, Kim SC, Smith CP, Cho B-K. The dynamic transcriptional and translational landscape of the model antibiotic producer Streptomyces coelicolor A3(2) Nat Commun. 2016;7(1):11605. doi: 10.1038/ncomms11605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jervis AJ, Carbonell P, Vinaixa M, Dunstan MS, Hollywood KA, Robinson CJ, Rattray NJW, Yan C, Swainston N, Currin A, Sung R, Toogood H, Taylor S, Faulon J-L, Breitling R, Takano E, Scrutton NS. Machine learning of designed translational control allows predictive pathway optimization in Escherichia coli. ACS Synth Biol. 2019;8(1):127–136. doi: 10.1021/acssynbio.8b00398. [DOI] [PubMed] [Google Scholar]
- Jia H, Chen T, Qu J, Yao M, Xiao W, Wang Y, Li C, Yuan Y. Collaborative subcellular compartmentalization to improve GPP utilization and boost sabinene accumulation in Saccharomyces cerevisiae. Biochem Eng J. 2020;164:107768. doi: 10.1016/j.bej.2020.107768. [DOI] [Google Scholar]
- Jiang GZ, Yao MD, Wang Y, Zhou L, Song TQ, Liu H, Xiao WH, Yuan YJ. Manipulation of GES and ERG20 for geraniol overproduction in Saccharomyces cerevisiae. Metab Eng. 2017;41:57–66. doi: 10.1016/j.ymben.2017.03.005. [DOI] [PubMed] [Google Scholar]
- Jiang G, Yang Z, Wang Y, Yao M, Chen Y, Xiao W, Yuan Y. Enhanced astaxanthin production in yeast via combined mutagenesis and evolution. Biochem Eng J. 2020;156:107519. doi: 10.1016/j.bej.2020.107519. [DOI] [Google Scholar]
- Jiang W, He X, Luo Y, Mu Y, Gu F, Liang Q, Qi Q. Two completely orthogonal quorum sensing systems with self-produced autoinducers enable automatic delayed cascade control. ACS Synth Biol. 2020 doi: 10.1021/acssynbio.0c00370. [DOI] [PubMed] [Google Scholar]
- Jones CM, Hernández Lozada NJ, Pfleger BF. Efflux systems in bacteria and their metabolic engineering applications. Appl Microbiol Biotechnol. 2015;99(22):9381–9393. doi: 10.1007/s00253-015-6963-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jullesson D, David F, Pfleger B, Nielsen J. Impact of synthetic biology and metabolic engineering on industrial production of fine chemicals. Biotechnol Adv. 2015;33(7):1395–1402. doi: 10.1016/j.biotechadv.2015.02.011. [DOI] [PubMed] [Google Scholar]
- Kang A, Meadows CW, Canu N, Keasling JD, Lee TS. High-throughput enzyme screening platform for the IPP-bypass mevalonate pathway for isopentenol production. Metab Eng. 2017;41:125–134. doi: 10.1016/j.ymben.2017.03.010. [DOI] [PubMed] [Google Scholar]
- Karim AS, Curran KA, Alper HS. Characterization of plasmid burden and copy number in Saccharomyces cerevisiae for optimization of metabolic engineering applications. FEMS Yeast Res. 2013;13(1):107–116. doi: 10.1111/1567-1364.12016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling CI, Bohlmann J. Genes, enzymes and chemicals of terpenoid diversity in the constitutive and induced defence of conifers against insects and pathogens. New Phytol. 2006;170(4):657–675. doi: 10.1111/j.1469-8137.2006.01716.x. [DOI] [PubMed] [Google Scholar]
- Kent R, Dixon N. Contemporary tools for regulating gene expression in bacteria. Trends Biotechnol. 2020;38(3):316–333. doi: 10.1016/j.tibtech.2019.09.007. [DOI] [PubMed] [Google Scholar]
- Ker DS, Chan KG, Othman R, Hassan M, Ng CL. Site-directed mutagenesis of β sesquiphellandrene synthase enhances enzyme promiscuity. Phytochemistry. 2020;173:112286. doi: 10.1016/j.phytochem.2020.112286. [DOI] [PubMed] [Google Scholar]
- Kiattisewee C, Dong C, Fontana J, Sugianto W, Peralta-Yahya P, Carothers JM, Zalatan JG. Portable bacterial CRISPR transcriptional activation enables metabolic engineering in Pseudomonas putida. Metab Eng. 2021;66:283–295. doi: 10.1016/j.ymben.2021.04.002. [DOI] [PubMed] [Google Scholar]
- Kildegaard KR, Arnesen JA, Adiego-Pérez B, Rago D, Kristensen M, Klitgaard AK, Hansen EH, Hansen J, Borodina I. Tailored biosynthesis of gibberellin plant hormones in yeast. Metab Eng. 2021;66:1–11. doi: 10.1016/j.ymben.2021.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SK, Han GH, Seong W, Kim H, Kim S-W, Lee D-H, Lee S-G. CRISPR interference-guided balancing of a biosynthetic mevalonate pathway increases terpenoid production. Metab Eng. 2016;38:228–240. doi: 10.1016/j.ymben.2016.08.006. [DOI] [PubMed] [Google Scholar]
- Kim E-M, Woo HM, Tian T, Yilmaz S, Javidpour P, Keasling JD, Lee TS. Autonomous control of metabolic state by a quorum sensing (QS)-mediated regulator for bisabolene production in engineered E. coli. Metab Eng. 2017;44:325–336. doi: 10.1016/j.ymben.2017.11.004. [DOI] [PubMed] [Google Scholar]
- Kim SK, Kim SH, Subhadra B, Woo SG, Rha E, Kim SW, Kim H, Lee DH, Lee SG. A genetically encoded biosensor for monitoring isoprene production in engineered Escherichia coli. ACS Synth Biol. 2018;7(10):2379–2390. doi: 10.1021/acssynbio.8b00164. [DOI] [PubMed] [Google Scholar]
- Kim JE, Jang IS, Son SH, Ko YJ, Cho BK, Kim SC, Lee JY. Tailoring the Saccharomyces cerevisiae endoplasmic reticulum for functional assembly of terpene synthesis pathway. Metab Eng. 2019;56:50–59. doi: 10.1016/j.ymben.2019.08.013. [DOI] [PubMed] [Google Scholar]
- Kim SK, Yoon PK, Kim SJ, Woo SG, Rha E, Lee H, Yeom SJ, Kim H, Lee DH, Lee SG. CRISPR interference-mediated gene regulation in Pseudomonas putida KT2440. Microb Biotechnol. 2020;13(1):210–221. doi: 10.1111/1751-7915.13382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokkonen P, Bednar D, Pinto G, Prokop Z, Damborsky J. Engineering enzyme access tunnels. Biotechnol Adv. 2019;37(6):107386. doi: 10.1016/j.biotechadv.2019.04.008. [DOI] [PubMed] [Google Scholar]
- Kosuri S, Goodman DB, Cambray G, Mutalik VK, Gao Y, Arkin AP, Endy D, Church GM. Composability of regulatory sequences controlling transcription and translation in Escherichia coli. Proc Natl Acad Sci USA. 2013;110(34):14024–14029. doi: 10.1073/pnas.1301301110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar J, Chauhan AS, Shah RL, Gupta JA, Rathore AS. Amino acid supplementation for enhancing recombinant protein production in E. coli. Biotechnol Bioeng. 2020;117(8):2420–2433. doi: 10.1002/bit.27371. [DOI] [PubMed] [Google Scholar]
- Laane C, Boeren S, Vos K, Veeger C. Rules for optimization of biocatalysis in organic solvents. Biotechnol Bioeng. 1987;30(1):81–87. doi: 10.1002/bit.260300112. [DOI] [PubMed] [Google Scholar]
- Lawson CE, Martí JM, Radivojevic T, Jonnalagadda SVR, Gentz R, Hillson NJ, Peisert S, Kim J, Simmons BA, Petzold CJ, Singer SW, Mukhopadhyay A, Tanjore D, Dunn JG, Garcia Martin H. Machine learning for metabolic engineering: a review. Metab Eng. 2021;63:34–60. doi: 10.1016/j.ymben.2020.10.005. [DOI] [PubMed] [Google Scholar]
- Lee J-W, Trinh CT. Microbial biosynthesis of lactate esters. Biotechnol Biofuels. 2019;12(1):226. doi: 10.1186/s13068-019-1563-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leferink NGH, Ranaghan KE, Karuppiah V, Currin A, van der Kamp MW, Mulholland AJ, Scrutton NS. Experiment and simulation reveal how mutations in functional plasticity regions guide plant monoterpene synthase product outcome. ACS Catal. 2019;8(5):3780–3791. doi: 10.1021/acscatal.8b00692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei D, Qiu Z, Wu J, Qiao B, Qiao J, Zhao G-R. Combining metabolic and monoterpene synthase engineering for de novo production of monoterpene alcohols in Escherichia coli. ACS Synth Biol. 2021;10(6):1531–1544. doi: 10.1021/acssynbio.1c00081. [DOI] [PubMed] [Google Scholar]
- Lemuth K, Steuer K, Albermann C. Engineering of a plasmid-free Escherichia coli strain for improved in vivo biosynthesis of astaxanthin. Microb Cell Fact. 2011;10(1):29. doi: 10.1186/1475-2859-10-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard E, Ajikumar PK, Thayer K, Xiao WH, Mo JD, Tidor B, Stephanopoulos G, Prather KL. Combining metabolic and protein engineering of a terpenoid biosynthetic pathway for overproduction and selectivity control. Proc Natl Acad Sci USA. 2010;107(31):13654–13659. doi: 10.1073/pnas.1006138107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Lin Z, Huang C, Zhang Y, Wang Z, Tang YJ, Chen T, Zhao X. Metabolic engineering of Escherichia coli using CRISPR–Cas9 meditated genome editing. Metab Eng. 2015;31:13–21. doi: 10.1016/j.ymben.2015.06.006. [DOI] [PubMed] [Google Scholar]
- Lin P-C, Zhang F, Pakrasi HB. Enhanced limonene production in a fast-growing cyanobacterium through combinatorial metabolic engineering. Metab Eng Commun. 2021;12:e00164. doi: 10.1016/j.mec.2021.e00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Evans T, Zhang F. Applications and advances of metabolite biosensors for metabolic engineering. Metab Eng. 2015;31:35–43. doi: 10.1016/j.ymben.2015.06.008. [DOI] [PubMed] [Google Scholar]
- Liu Y, Liu Y, Wang M. Design, optimization and application of small molecule biosensor in metabolic engineering. Front Microbiol. 2017;8:2012. doi: 10.3389/fmicb.2017.02012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Mao Z, Guo J, Wei L, Ma H, Tang Y, Chen T, Wang Z, Zhao X. Construction, model-based analysis, and characterization of a promoter library for fine-tuned gene expression in Bacillus subtilis. ACS Synth Biol. 2018;7(7):1785–1797. doi: 10.1021/acssynbio.8b00115. [DOI] [PubMed] [Google Scholar]
- Liu C, Bi H-R, Bai Z, Fan L-H, Tan T-W. Engineering and manipulation of a mevalonate pathway in Escherichia coli for isoprene production. Appl Microbiol Biotechnol. 2019;103:239–250. doi: 10.1007/s00253-018-9472-9. [DOI] [PubMed] [Google Scholar]
- Liu G-S, Li T, Zhou W, Jiang M, Tao X-Y, Liu M, Zhao M, Ren Y-H, Gao B, Wang F-Q, Wei D-Z. The yeast peroxisome: a dynamic storage depot and subcellular factory for squalene overproduction. Metab Eng. 2020;57:151–161. doi: 10.1016/j.ymben.2019.11.001. [DOI] [PubMed] [Google Scholar]
- Liu H, Tian Y, Zhou Y, Kan Y, Wu T, Xiao W, Luo Y. Multi-modular engineering of Saccharomyces cerevisiae for high-titre production of tyrosol and salidroside. Microb Biotechnol. 2020 doi: 10.1111/1751-7915.13667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Wang F, Deng L, Xu P (2020c) Optimizing mevalonate pathway for squalene production in Yarrowia lipolytica. bioRxiv:2020.05.03.075259. 10.1101/2020.05.03.075259
- Liu SC, Liu Z, Wei LJ, Hua Q. Pathway engineering and medium optimization for α-farnesene biosynthesis in oleaginous yeast Yarrowia lipolytica. J Biotechnol. 2020;319:74–81. doi: 10.1016/j.jbiotec.2020.06.005. [DOI] [PubMed] [Google Scholar]
- Liu T, Dong C, Qi M, Zhang B, Huang L, Xu Z, Lian J. Construction of a stable and temperature-responsive yeast cell factory for crocetin biosynthesis using CRISPR–Cas9. Front Bioeng Biotechnol. 2020;8:653–653. doi: 10.3389/fbioe.2020.00653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Chen SL, Xu JZ, Zhang WG. Dual regulation of cytoplasm and peroxisomes for improved α-farnesene production in recombinant Pichia pastoris. ACS Synth Biol. 2021 doi: 10.1021/acssynbio.1c00186. [DOI] [PubMed] [Google Scholar]
- Liu L, Qu YL, Dong GR, Wang J, Hu CY, Meng YH. Elevated β-carotene production using codon-adapted CarRA&B and metabolic balance in engineered Yarrowia lipolytica. Front Microbiol. 2021;12:627150. doi: 10.3389/fmicb.2021.627150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Zhang J, Ye J, Qi Q, Hou J. Morphological and metabolic engineering of Yarrowia lipolytica to increase β-carotene production. ACS Synth Biol. 2021 doi: 10.1021/acssynbio.1c00480. [DOI] [PubMed] [Google Scholar]
- Liu Y, Wang Z, Cui Z, Qi Q, Hou J. α-Farnesene production from lipid by engineered Yarrowia lipolytica. Bioresour Bioprocess. 2021;8(1):78. doi: 10.1186/s40643-021-00431-0. [DOI] [Google Scholar]
- Lu Y, Yang Q, Lin Z, Yang X. A modular pathway engineering strategy for the high-level production of β-ionone in Yarrowia lipolytica. Microb Cell Fact. 2020;19(1):49. doi: 10.1186/s12934-020-01309-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Peng B, Ebert BE, Dumsday G, Vickers CE. Auxin-mediated protein depletion for metabolic engineering in terpene-producing yeast. Nat Commun. 2021;12(1):1051. doi: 10.1038/s41467-021-21313-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund S, Hall R, Williams GJ. An artificial pathway for isoprenoid biosynthesis decoupled from native hemiterpene metabolism. ACS Synth Biol. 2019;8(2):232–238. doi: 10.1021/acssynbio.8b00383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z, Liu N, Lazar Z, Chatzivasileiou A, Ward V, Chen J, Zhou J, Stephanopoulos G. Enhancing isoprenoid synthesis in Yarrowia lipolytica by expressing the isopentenol utilization pathway and modulating intracellular hydrophobicity. Metab Eng. 2020;61:344–351. doi: 10.1016/j.ymben.2020.07.010. [DOI] [PubMed] [Google Scholar]
- Luo B, Jin MM, Li X, Makunga NP, Hu X. Yeast surface display for in vitro biosynthetic pathway reconstruction. ACS Synth Biol. 2021;10(11):2938–2946. doi: 10.1021/acssynbio.1c00175. [DOI] [PubMed] [Google Scholar]
- Lv X, Gu J, Wang F, Xie W, Liu M, Ye L, Yu H. Combinatorial pathway optimization in Escherichia coli by directed co-evolution of rate-limiting enzymes and modular pathway engineering. Biotechnol Bioeng. 2016;113(12):2661–2669. doi: 10.1002/bit.26034. [DOI] [PubMed] [Google Scholar]
- Lv X, Wang F, Zhou P, Ye L, Xie W, Xu H, Yu H. Dual regulation of cytoplasmic and mitochondrial acetyl-CoA utilization for improved isoprene production in Saccharomyces cerevisiae. Nat Commun. 2016;7(1):12851. doi: 10.1038/ncomms12851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv PJ, Qiang S, Liu L, Hu CY, Meng YH. Dissolved-oxygen feedback control fermentation for enhancing β-carotene in engineered Yarrowia lipolytica. Sci Rep. 2020;10(1):17114. doi: 10.1038/s41598-020-74074-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Naseri A, Reyes-Gutierrez P, Wolfe SA, Zhang S, Pederson T. Multicolor CRISPR labeling of chromosomal loci in human cells. Proc Natl Acad Sci USA. 2015;112(10):3002–3007. doi: 10.1073/pnas.1420024112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Li J, Huang S, Stephanopoulos G. Targeting pathway expression to subcellular organelles improves astaxanthin synthesis in Yarrowia lipolytica. Metab Eng. 2021;68:152–161. doi: 10.1016/j.ymben.2021.10.004. [DOI] [PubMed] [Google Scholar]
- Marsafari M, Xu P. Debottlenecking mevalonate pathway for antimalarial drug precursor amorphadiene biosynthesis in Yarrowia lipolytica. Metab Eng Commun. 2020;10:e00121. doi: 10.1016/j.mec.2019.e00121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez JM, Delso C, Angulo J, Álvarez I, Raso J. Pulsed electric field-assisted extraction of carotenoids from fresh biomass of Rhodotorula glutinis. Innov Food Sci Emerg Technol. 2018;47:421–427. doi: 10.1016/j.ifset.2018.04.012. [DOI] [Google Scholar]
- Martínez JM, Schottroff F, Haas K, Fauster T, Sajfrtová M, Álvarez I, Raso J, Jaeger H. Evaluation of pulsed electric fields technology for the improvement of subsequent carotenoid extraction from dried Rhodotorula glutinis yeast. Food Chem. 2020;323:126824. doi: 10.1016/j.foodchem.2020.126824. [DOI] [PubMed] [Google Scholar]
- Mazurenko S, Prokop Z, Damborsky J. Machine learning in enzyme engineering. ACS Catal. 2020;10(2):1210–1223. doi: 10.1021/acscatal.9b04321. [DOI] [Google Scholar]
- McClelland RA (2008) Carbocations. In: Organic reaction mechanisms. 2005. pp 179–211
- McGarvey DJ, Croteau R. Terpenoid metabolism. Plant Cell. 1995;7(7):1015–1026. doi: 10.1105/tpc.7.7.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinn J, Marraffini LA. CRISPR-Cas systems optimize their immune response by specifying the site of spacer integration. Mol Cell. 2016;64(3):616–623. doi: 10.1016/j.molcel.2016.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus CJ, May GE, Spealman P, Shteyman A. Ribosome profiling reveals post-transcriptional buffering of divergent gene expression in yeast. Genome Res. 2014;24(3):422–430. doi: 10.1101/gr.164996.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng J, Wang B, Liu D, Chen T, Wang Z, Zhao X. High-yield anaerobic succinate production by strategically regulating multiple metabolic pathways based on stoichiometric maximum in Escherichia coli. Microb Cell Fact. 2016;15(1):141. doi: 10.1186/s12934-016-0536-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng H, Ma Y, Mai G, Wang Y, Liu C. Construction of precise support vector machine based models for predicting promoter strength. Quant Biol. 2017;5(1):90–98. doi: 10.1007/s40484-017-0096-3. [DOI] [Google Scholar]
- Meng X, Liu H, Xu W, Zhang W, Wang Z, Liu W. Metabolic engineering Saccharomyces cerevisiae for de novo production of the sesquiterpenoid (+)-nootkatone. Microb Cell Fact. 2020;19(1):21. doi: 10.1186/s12934-020-1295-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Y, Shao X, Wang Y, Li Y, Zheng X, Wei G, Kim S-W, Wang C. Extension of cell membrane boosting squalene production in the engineered Escherichia coli. Biotechnol Bioeng. 2020;117(11):3499–3507. doi: 10.1002/bit.27511. [DOI] [PubMed] [Google Scholar]
- Menin B, Lami A, Musazzi S, Petrova AA, Santabarbara S, Casazza AP. A comparison of constitutive and inducible non-endogenous keto-carotenoids biosynthesis in Synechocystis sp. PCC 6803. Microorganisms. 2019;7(11):501. doi: 10.3390/microorganisms7110501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JC, Holmes MC, Wang J, Guschin DY, Lee YL, Rupniewski I, Beausejour CM, Waite AJ, Wang NS, Kim KA, Gregory PD, Pabo CO, Rebar EJ. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol. 2007;25(7):778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- Mishra S, Pandey P, Dubey AP, Zehra A, Chanotiya CS, Tripathi AK, Mishra MN. Engineering a carotenoid-overproducing strain of Azospirillum brasilense for heterologous production of geraniol and amorphadiene. Appl Environ Microbiol. 2020;86(17):e0414-20. doi: 10.1128/aem.00414-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo XH, Zhang H, Wang TM, Zhang C, Zhang C, Xing XH, Yang S. Establishment of CRISPR interference in Methylorubrum extorquens and application of rapidly mining a new phytoene desaturase involved in carotenoid biosynthesis. Appl Microbiol Biotechnol. 2020;104(10):4515–4532. doi: 10.1007/s00253-020-10543-w. [DOI] [PubMed] [Google Scholar]
- Moon JH, Lee K, Lee JH, Lee PC. Redesign and reconstruction of a steviol-biosynthetic pathway for enhanced production of steviol in Escherichia coli. Microb Cell Fact. 2020;19(1):20. doi: 10.1186/s12934-020-1291-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mordaka PM, Heap JT. Stringency of synthetic promoter sequences in Clostridium revealed and circumvented by tuning promoter library mutation rates. ACS Synth Biol. 2018;7(2):672–681. doi: 10.1021/acssynbio.7b00398. [DOI] [PubMed] [Google Scholar]
- Motoyama K, Sobue F, Kawaide H, Yoshimura T, Hemmi H. Conversion of mevalonate 3-kinase into 5-phosphomevalonate 3-kinase by single amino acid mutations. Appl Environ Microbiol. 2019;85(9):e00256-19. doi: 10.1128/AEM.00256-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowbray M, Savage T, Wu C, Song Z, Cho BA, Del Rio-Chanona EA, Zhang D. Machine learning for biochemical engineering: a review. Biochem Eng J. 2021;172:108054. doi: 10.1016/j.bej.2021.108054. [DOI] [Google Scholar]
- Navale G, Sharma P, Said M, Ramkumar S, Dharne M, Thulasiram HV, Shinde S. Enhancing epi -cedrol production in Escherichia coli by fusion expression of farnesyl pyrophosphate synthase and e pi -cedrol synthase. Eng Life Sci. 2019 doi: 10.1002/elsc.201900103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolaou SA, Gaida SM, Papoutsakis ET. A comparative view of metabolite and substrate stress and tolerance in microbial bioprocessing: from biofuels and chemicals, to biocatalysis and bioremediation. Metab Eng. 2010;12(4):307–331. doi: 10.1016/j.ymben.2010.03.004. [DOI] [PubMed] [Google Scholar]
- Nielsen J, Keasling JD. Engineering cellular metabolism. Cell. 2016;164(6):1185–1197. doi: 10.1016/j.cell.2016.02.004. [DOI] [PubMed] [Google Scholar]
- Nitta N, Tajima Y, Yamamoto Y, Moriya M, Matsudaira A, Hoshino Y, Nishio Y, Usuda Y. Fermentative production of enantiopure (S)-linalool using a metabolically engineered Pantoea ananatis. Microb Cell Fact. 2021;20(1):54. doi: 10.1186/s12934-021-01543-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu F-X, He X, Wu Y-Q, Liu J-Z. Enhancing production of pinene in Escherichia coli by using a combination of tolerance, evolution, and modular co-culture engineering. Front Microbiol. 2018;9:1623–1623. doi: 10.3389/fmicb.2018.01623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobeli I, Favia AD, Thornton JM. Protein promiscuity and its implications for biotechnology. Nat Biotechnol. 2009;27(2):157–167. doi: 10.1038/nbt1519. [DOI] [PubMed] [Google Scholar]
- Nogueira M, Enfissi EMA, Welsch R, Beyer P, Zurbriggen MD, Fraser PD. Construction of a fusion enzyme for astaxanthin formation and its characterisation in microbial and plant hosts: a new tool for engineering ketocarotenoids. Metab Eng. 2019;52:243–252. doi: 10.1016/j.ymben.2018.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora LC, Wehrs M, Kim J, Cheng JF, Tarver A, Simmons BA, Magnuson J, Harmon-Smith M, Silva-Rocha R, Gladden JM, Mukhopadhyay A, Skerker JM, Kirby J. A toolset of constitutive promoters for metabolic engineering of Rhodosporidium toruloides. Microb Cell Fact. 2019;18(1):117. doi: 10.1186/s12934-019-1167-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowroozi FF, Baidoo EE, Ermakov S, Redding-Johanson AM, Batth TS, Petzold CJ, Keasling JD. Metabolic pathway optimization using ribosome binding site variants and combinatorial gene assembly. Appl Microbiol Biotechnol. 2014;98(4):1567–1581. doi: 10.1007/s00253-013-5361-4. [DOI] [PubMed] [Google Scholar]
- Otto M, Teixeira PG, Vizcaino MI, David F, Siewers V. Integration of a multi-step heterologous pathway in Saccharomyces cerevisiae for the production of abscisic acid. Microb Cell Fact. 2019;18(1):205. doi: 10.1186/s12934-019-1257-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou B, Garcia C, Wang Y, Zhang W, Zhu G. Techniques for chromosomal integration and expression optimization in Escherichia coli. Biotechnol Bioeng. 2018;115(10):2467–2478. doi: 10.1002/bit.26790. [DOI] [PubMed] [Google Scholar]
- Page K, Shaffer J, Lin S, Zhang M, Liu JM. Engineering riboswitches in vivo using dual genetic selection and fluorescence-activated cell sorting. ACS Synth Biol. 2018;7(9):2000–2006. doi: 10.1021/acssynbio.8b00099. [DOI] [PubMed] [Google Scholar]
- Pan X, Wang B, Duan R, Jia J, Li J, Xiong W, Ling X, Chen C, Huang X, Zhang G, Lu Y. Enhancing astaxanthin accumulation in Xanthophyllomyces dendrorhous by a phytohormone: metabolomic and gene expression profiles. Microb Biotechnol. 2020;13(5):1446–1460. doi: 10.1111/1751-7915.13567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang B, Li J, Eiben CB, Oksen E, Barcelos C, Chen R, Englund E, Sundstrom E, Keasling JD. Lepidopteran mevalonate pathway optimization in Escherichia coli efficiently produces isoprenol analogs for next-generation biofuels. Metab Eng. 2021;68:210–219. doi: 10.1016/j.ymben.2021.10.007. [DOI] [PubMed] [Google Scholar]
- Papenfort K, Bassler BL. Quorum sensing signal–response systems in gram-negative bacteria. Nat Rev Microbiol. 2016;14(9):576–588. doi: 10.1038/nrmicro.2016.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Yu BJ, Choi J-i, Woo HM. Heterologous production of squalene from glucose in engineered Corynebacterium glutamicum using multiplex CRISPR interference and high-throughput fermentation. J Agric Food Chem. 2019;67(1):308–319. doi: 10.1021/acs.jafc.8b05818. [DOI] [PubMed] [Google Scholar]
- Peng B, Plan MR, Chrysanthopoulos P, Hodson MP, Nielsen LK, Vickers CE. A squalene synthase protein degradation method for improved sesquiterpene production in Saccharomyces cerevisiae. Metab Eng. 2017;39:209–219. doi: 10.1016/j.ymben.2016.12.003. [DOI] [PubMed] [Google Scholar]
- Peng B, Nielsen LK, Kampranis SC, Vickers CE. Engineered protein degradation of farnesyl pyrophosphate synthase is an effective regulatory mechanism to increase monoterpene production in Saccharomyces cerevisiae. Metab Eng. 2018;47:83–93. doi: 10.1016/j.ymben.2018.02.005. [DOI] [PubMed] [Google Scholar]
- Peters RJ, Flory JE, Jetter R, Ravn MM, Lee HJ, Coates RM, Croteau RB. Abietadiene synthase from grand fir (Abies grandis): characterization and mechanism of action of the “pseudomature” recombinant enzyme. Biochemistry. 2000;39(50):15592–15602. doi: 10.1021/bi001997l. [DOI] [PubMed] [Google Scholar]
- Pfleger BF, Prather KL. Biological synthesis unbounded? Nat Biotechnol. 2015;33(11):1148–1149. doi: 10.1038/nbt.3399. [DOI] [PubMed] [Google Scholar]
- Pramastya H, Xue D, Abdallah II, Setroikromo R, Quax WJ. High level production of amorphadiene using Bacillus subtilis as an optimized terpenoid cell factory. New Biotechnol. 2021;60:159–167. doi: 10.1016/j.nbt.2020.10.007. [DOI] [PubMed] [Google Scholar]
- Putman M, van Veen HW, Konings WN. Molecular properties of bacterial multidrug transporters. Microbiol Mol Biol Rev. 2000;64(4):672–693. doi: 10.1128/mmbr.64.4.672-693.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152(5):1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian S, Li Y, Cirino PC. Biosensor-guided improvements in salicylate production by recombinant Escherichia coli. Microb Cell Fact. 2019;18(1):18. doi: 10.1186/s12934-019-1069-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Zhou Y, Krivoruchko A, Huang M, Liu L, Khoomrung S, Siewers V, Jiang B, Nielsen J. Modular pathway rewiring of Saccharomyces cerevisiae enables high-level production of L-ornithine. Nat Commun. 2015;8:8224. doi: 10.1038/ncomms9224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabeharindranto H, Castaño-Cerezo S, Lautier T, Garcia-Alles LF, Treitz C, Tholey A, Truan G. Enzyme-fusion strategies for redirecting and improving carotenoid synthesis in S. cerevisiae. Metab Eng Commun. 2019;8:e00086. doi: 10.1016/j.mec.2019.e00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos JL, Duque E, Gallegos MT, Godoy P, Ramos-Gonzalez MI, Rojas A, Teran W, Segura A. Mechanisms of solvent tolerance in gram-negative bacteria. Annu Rev Microbiol. 2002;56:743–768. doi: 10.1146/annurev.micro.56.012302.161038. [DOI] [PubMed] [Google Scholar]
- Ramos JL, Sol Cuenca M, Molina-Santiago C, Segura A, Duque E, Gómez-García MR, Udaondo Z, Roca A. Mechanisms of solvent resistance mediated by interplay of cellular factors in Pseudomonas putida. FEMS Microbiol Rev. 2015;39(4):555–566. doi: 10.1093/femsre/fuv006. [DOI] [PubMed] [Google Scholar]
- Ravn MM, Coates RM, Flory JE, Peters RJ, Croteau R. Stereochemistry of the cyclization-rearrangement of (+)-copalyl diphosphate to (-)-abietadiene catalyzed by recombinant abietadiene synthase from Abies grandis. Org Lett. 2000;2(5):573–576. doi: 10.1021/ol991230p. [DOI] [PubMed] [Google Scholar]
- Raz K, Driller R, Dimos N, Ringel M, Brück T, Loll B, Major DT. The impression of a nonexisting catalytic effect: the role of CotB2 in guiding the complex biosynthesis of cyclooctat-9-en-7-ol. J Am Chem Soc. 2020;142(51):21562–21574. doi: 10.1021/jacs.0c11348. [DOI] [PubMed] [Google Scholar]
- Redden H, Alper HS. The development and characterization of synthetic minimal yeast promoters. Nat Commun. 2015;6(1):7810. doi: 10.1038/ncomms8810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyon D, Tsai SQ, Khayter C, Foden JA, Sander JD, Joung JK. FLASH assembly of TALENs for high-throughput genome editing. Nat Biotechnol. 2012;30(5):460–465. doi: 10.1038/nbt.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues JS, Lindberg P. Metabolic engineering of Synechocystis sp. PCC 6803 for improved bisabolene production. Metab Eng Commun. 2021;12:e00159. doi: 10.1016/j.mec.2020.e00159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolf J, Julsing MK, Rosenthal K, Lütz S. A gram-scale limonene production process with engineered Escherichia coli. Molecules. 2020;25(8):1881. doi: 10.3390/molecules25081881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini RK, Keum Y-S. Carotenoid extraction methods: a review of recent developments. Food Chem. 2018;240:90–103. doi: 10.1016/j.foodchem.2017.07.099. [DOI] [PubMed] [Google Scholar]
- Salas-Villalobos U, Gómez-Acata R, Castillo J, Aguilar O. In-situ product recovery as a strategy for bioprocess integration and depletion of inhibitory products. J Chem Techn Biotechnol. 2021 doi: 10.1002/jctb.6797. [DOI] [Google Scholar]
- Salmon M, Laurendon C, Vardakou M, Cheema J, Defernez M, Green S, Faraldos JA, O'Maille PE. Emergence of terpene cyclization in Artemisia annua. Nat Commun. 2015;6:6143. doi: 10.1038/ncomms7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander JD, Dahlborg EJ, Goodwin MJ, Cade L, Zhang F, Cifuentes D, Curtin SJ, Blackburn JS, Thibodeau-Beganny S, Qi Y, Pierick CJ, Hoffman E, Maeder ML, Khayter C, Reyon D, Dobbs D, Langenau DM, Stupar RM, Giraldez AJ, Voytas DF, Peterson RT, Yeh JR, Joung JK. Selection-free zinc-finger-nuclease engineering by context-dependent assembly (CoDA) Nat Methods. 2011;8(1):67–69. doi: 10.1038/nmeth.1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Cong L, Zhou Y, Cunniff MM, Feng G, Zhang F. A transcription activator-like effector toolbox for genome engineering. Nat Protoc. 2012;7(1):171–192. doi: 10.1038/nprot.2011.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scalcinati G, Knuf C, Partow S, Chen Y, Maury J, Schalk M, Daviet L, Nielsen J, Siewers V. Dynamic control of gene expression in Saccharomyces cerevisiae engineered for the production of plant sesquiterpene α-santalene in a fed-batch mode. Metab Eng. 2012;14(2):91–103. doi: 10.1016/j.ymben.2012.01.007. [DOI] [PubMed] [Google Scholar]
- Schalck T, Bergh BVd, Michiels J. Increasing solvent tolerance to improve microbial production of alcohols, terpenoids and aromatics. Microorganisms. 2021;9(2):249. doi: 10.3390/microorganisms9020249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schewe H, Mirata MA, Schrader J. Bioprocess engineering for microbial synthesis and conversion of isoprenoids. Adv Biochem Eng Biotechnol. 2015;148:251–286. doi: 10.1007/10_2015_321. [DOI] [PubMed] [Google Scholar]
- Schultenkämper K, Brito LF, Wendisch VF. Impact of CRISPR interference on strain development in biotechnology. Biotechnol Appl Biochem. 2020;67(1):7–21. doi: 10.1002/bab.1901. [DOI] [PubMed] [Google Scholar]
- Schuster M, Sexton DJ, Diggle SP, Greenberg EP. Acyl-homoserine lactone quorum sensing: from evolution to application. Annu Rev Microbiol. 2013;67:43–63. doi: 10.1146/annurev-micro-092412-155635. [DOI] [PubMed] [Google Scholar]
- Sengupta A, Sunder AV, Sohoni SV, Wangikar PP. Fine-tuning native promoters of Synechococcus elongatus PCC 7942 to develop a synthetic toolbox for heterologous protein expression. ACS Synth Biol. 2019;8(5):1219–1223. doi: 10.1021/acssynbio.9b00066. [DOI] [PubMed] [Google Scholar]
- Shaikh KM, Odaneth AA. Metabolic engineering of Yarrowia lipolytica for the production of isoprene. Biotechnol Prog. 2021 doi: 10.1002/btpr.3201. [DOI] [PubMed] [Google Scholar]
- Sharma AK, Shukla E, Janoti DS, Mukherjee KJ, Shiloach J. A novel knock out strategy to enhance recombinant protein expression in Escherichia coli. Microb Cell Fact. 2020;19(1):148–148. doi: 10.1186/s12934-020-01407-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H-J, Cheng B-Y, Zhang Y-M, Tang L, Li Z, Bu Y-F, Li X-R, Tian G-Q, Liu J-Z. Dynamic control of the mevalonate pathway expression for improved zeaxanthin production in Escherichia coli and comparative proteome analysis. Metab Eng. 2016;38:180–190. doi: 10.1016/j.ymben.2016.07.012. [DOI] [PubMed] [Google Scholar]
- Shi S, Liang Y, Zhang MM, Ang EL, Zhao H. A highly efficient single-step, markerless strategy for multi-copy chromosomal integration of large biochemical pathways in Saccharomyces cerevisiae. Metab Eng. 2016;33:19–27. doi: 10.1016/j.ymben.2015.10.011. [DOI] [PubMed] [Google Scholar]
- Shi B, Ma T, Ye Z, Li X, Huang Y, Zhou Z, Ding Y, Deng Z, Liu T. Systematic metabolic engineering of Saccharomyces cerevisiae for lycopene overproduction. J Agric Food Chem. 2019;67(40):11148–11157. doi: 10.1021/acs.jafc.9b04519. [DOI] [PubMed] [Google Scholar]
- Shi T, Li Y, Zhu L, Tong Y, Yang J, Fang Y, Wang M, Zhang J, Jiang Y, Yang S. Engineering the oleaginous yeast Yarrowia lipolytica for β-farnesene overproduction. Biotechnol J. 2021;16(7):2100097. doi: 10.1002/biot.202100097. [DOI] [PubMed] [Google Scholar]
- Shi Y, Wang D, Li R, Huang L, Dai Z, Zhang X. Engineering yeast subcellular compartments for increased production of the lipophilic natural products ginsenosides. Metab Eng. 2021;67:104–111. doi: 10.1016/j.ymben.2021.06.002. [DOI] [PubMed] [Google Scholar]
- Shukal S, Chen X, Zhang C. Systematic engineering for high-yield production of viridiflorol and amorphadiene in auxotrophic Escherichia coli. Metab Eng. 2019;55:170–178. doi: 10.1016/j.ymben.2019.07.007. [DOI] [PubMed] [Google Scholar]
- Silva PGPd, Prescendo Júnior D, Sala L, Burkert JFdM, Santos LO. Magnetic field as a trigger of carotenoid production by Phaffia rhodozyma. Process Biochem. 2020;98:131–138. doi: 10.1016/j.procbio.2020.08.001. [DOI] [Google Scholar]
- Singh N, Malik S, Gupta A, Srivastava KR. Revolutionizing enzyme engineering through artificial intelligence and machine learning. Emerg Top Life Sci. 2021;5(1):113–125. doi: 10.1042/etls20200257. [DOI] [PubMed] [Google Scholar]
- Sivy T, Fall R, Rosenstiel T. Evidence of isoprenoid precursor toxicity in Bacillus subtilis. Biosci Biotechnol Biochem. 2011;75:2376–2383. doi: 10.1271/bbb.110572. [DOI] [PubMed] [Google Scholar]
- Smanski MJ, Bhatia S, Zhao D, Park Y, Woodruff LBA, Giannoukos G, Ciulla D, Busby M, Calderon J, Nicol R, Gordon DB, Densmore D, Voigt CA. Functional optimization of gene clusters by combinatorial design and assembly. Nat Biotechnol. 2014;32(12):1241–1249. doi: 10.1038/nbt.3063. [DOI] [PubMed] [Google Scholar]
- Song X, Xiao H, Luo S, Wang X, Wang W, Lin S. Biosynthesis of squalene-type triterpenoids in Saccharomyces cerevisiae by expression of CYP505D13 from Ganoderma lucidum. Bioresour Bioprocess. 2019;6(1):19. doi: 10.1186/s40643-019-0256-6. [DOI] [Google Scholar]
- Song Y, He S, Abdallah II, Jopkiewicz A, Setroikromo R, van Merkerk R, Tepper PG, Quax WJ. Engineering of multiple modules to improve amorphadiene production in Bacillus subtilis using CRISPR–Cas9. J Agric Food Chem. 2021;69(16):4785–4794. doi: 10.1021/acs.jafc.1c00498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storici F, Durham CL, Gordenin DA, Resnick MA. Chromosomal site-specific double-strand breaks are efficiently targeted for repair by oligonucleotides in yeast. Proc Natl Acad Sci USA. 2003;100(25):14994–14999. doi: 10.1073/pnas.2036296100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Pierre F, Cui L, Priest DG, Endy D, Dodd IB, Shearwin KE. One-step cloning and chromosomal integration of DNA. ACS Synth Biol. 2013;2(9):537–541. doi: 10.1021/sb400021j. [DOI] [PubMed] [Google Scholar]
- Su B, Song D, Yang F, Zhu H. Engineering a growth-phase-dependent biosynthetic pathway for carotenoid production in Saccharomyces cerevisiae. J Ind Microbiol Biotechnol. 2020;47(4–5):383–393. doi: 10.1007/s10295-020-02271-x. [DOI] [PubMed] [Google Scholar]
- Takemura M, Kubo A, Higuchi Y, Maoka T, Sahara T, Yaoi K, Ohdan K, Umeno D, Misawa N. Pathway engineering for efficient biosynthesis of violaxanthin in Escherichia coli. Appl Microbiol Biotechnol. 2019;103(23):9393–9399. doi: 10.1007/s00253-019-10182-w. [DOI] [PubMed] [Google Scholar]
- Tang W-Y, Wang D-P, Tian Y, Fan X, Wang C, Lu X-Y, Li P-W, Ji X-J, Liu H-H. Metabolic engineering of Yarrowia lipolytica for improving squalene production. Bioresour Technol. 2021;323:124652. doi: 10.1016/j.biortech.2020.124652. [DOI] [PubMed] [Google Scholar]
- Tantillo DJ. The carbocation continuum in terpene biosynthesis—where are the secondary cations? Chem Soc Rev. 2010;39(8):2847–2854. doi: 10.1039/B917107J. [DOI] [PubMed] [Google Scholar]
- Tantillo DJ. Importance of inherent substrate reactivity in enzyme-promoted carbocation cyclization/rearrangements. Angew Chem Int Ed. 2017;56(34):10040–10045. doi: 10.1002/anie.201702363. [DOI] [PubMed] [Google Scholar]
- Terpe K. Overview of tag protein fusions: from molecular and biochemical fundamentals to commercial systems. Appl Microbiol Biotechnol. 2003;60(5):523–533. doi: 10.1007/s00253-002-1158-6. [DOI] [PubMed] [Google Scholar]
- Tetali SD. Terpenes and isoprenoids: a wealth of compounds for global use. Planta. 2019;249(1):1–8. doi: 10.1007/s00425-018-3056-x. [DOI] [PubMed] [Google Scholar]
- Tholl D. Biosynthesis and biological functions of terpenoids in plants. Adv Biochem Eng Biotechnol. 2015;148:63–106. doi: 10.1007/10_2014_295. [DOI] [PubMed] [Google Scholar]
- Tian T, Kang JW, Kang A, Lee TS. Redirecting metabolic flux via combinatorial multiplex CRISPRi-mediated repression for isopentenol production in Escherichia coli. ACS Synth Biol. 2019;8(2):391–402. doi: 10.1021/acssynbio.8b00429. [DOI] [PubMed] [Google Scholar]
- Tovilla-Coutiño DB, Momany C, Eiteman MA. Engineered citrate synthase alters acetate accumulation in Escherichia coli. Metab Eng. 2020 doi: 10.1016/j.ymben.2020.06.006. [DOI] [PubMed] [Google Scholar]
- Tyo KE, Ajikumar PK, Stephanopoulos G. Stabilized gene duplication enables long-term selection-free heterologous pathway expression. Nat Biotechnol. 2009;27(8):760–765. doi: 10.1038/nbt.1555. [DOI] [PubMed] [Google Scholar]
- Uhlen M, Forsberg G, Moks T, Hartmanis M, Nilsson B. Fusion proteins in biotechnology. Curr Opin Biotechnol. 1992;3(4):363–369. doi: 10.1016/0958-1669(92)90164-e. [DOI] [PubMed] [Google Scholar]
- Walls LE, Malcı K, Nowrouzi B, Li RA, d'Espaux L, Wong J, Dennis JA, Semião AJC, Wallace S, Martinez JL, Keasling JD, Rios-Solis L. Optimizing the biosynthesis of oxygenated and acetylated Taxol precursors in Saccharomyces cerevisiae using advanced bioprocessing strategies. Biotechnol Bioeng. 2020 doi: 10.1002/bit.27569. [DOI] [PubMed] [Google Scholar]
- Wang JF, Xiong ZQ, Li SY, Wang Y. Enhancing isoprenoid production through systematically assembling and modulating efflux pumps in Escherichia coli. Appl Microbiol Biotechnol. 2013;97(18):8057–8067. doi: 10.1007/s00253-013-5062-z. [DOI] [PubMed] [Google Scholar]
- Wang C, Yang L, Shah AA, Choi E-S, Kim S-W. Dynamic interplay of multidrug transporters with TolC for isoprenol tolerance in Escherichia coli. Sci Rep. 2015;5(1):16505. doi: 10.1038/srep16505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Niyompanich S, Tai YS, Wang J, Bai W, Mahida P, Gao T, Zhang K. Engineering of a highly efficient Escherichia coli strain for mevalonate fermentation through chromosomal integration. Appl Environ Microb. 2016;82(24):7176–7184. doi: 10.1128/aem.02178-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Lv X, Xie W, Zhou P, Zhu Y, Yao Z, Yang C, Yang X, Ye L, Yu H. Combining Gal4p-mediated expression enhancement and directed evolution of isoprene synthase to improve isoprene production in Saccharomyces cerevisiae. Metab Eng. 2017;39:257–266. doi: 10.1016/j.ymben.2016.12.011. [DOI] [PubMed] [Google Scholar]
- Wang J, Jiang W, Liang C, Zhu L, Li Y, Mo Q, Xu S, Chu A, Zhang L, Ding Z, Shi G. Overproduction of α-farnesene in Saccharomyces cerevisiae by farnesene synthase screening and metabolic engineering. J Agric Food Chem. 2021;69(10):3103–3113. doi: 10.1021/acs.jafc.1c00008. [DOI] [PubMed] [Google Scholar]
- Wang J, Zhu L, Li Y, Xu S, Jiang W, Liang C, Fang Y, Chu A, Zhang L, Ding Z, Shi G. Enhancing geranylgeraniol production by metabolic engineering and utilization of isoprenol as a substrate in Saccharomyces cerevisiae. J Agric Food Chem. 2021;69(15):4480–4489. doi: 10.1021/acs.jafc.1c00508. [DOI] [PubMed] [Google Scholar]
- Wang X, Chen J, Zhang J, Zhou Y, Zhang Y, Wang F, Li X. Engineering Escherichia coli for production of geraniol by systematic synthetic biology approaches and laboratory-evolved fusion tags. Metab Eng. 2021;66:60–67. doi: 10.1016/j.ymben.2021.04.008. [DOI] [PubMed] [Google Scholar]
- Wang X, Pereira JH, Tsutakawa S, Fang X, Adams PD, Mukhopadhyay A, Lee TS. Efficient production of oxidized terpenoids via engineering fusion proteins of terpene synthase and cytochrome P450. Metab Eng. 2021;64:41–51. doi: 10.1016/j.ymben.2021.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhou S, Liu Q, Jeong S-H, Zhu L, Yu X, Zheng X, Wei G, Kim S-W, Wang C. Metabolic engineering of Escherichia coli for production of α-santalene, a precursor of sandalwood oil. J Agric Food Chem. 2021;69(44):13135–13142. doi: 10.1021/acs.jafc.1c05486. [DOI] [PubMed] [Google Scholar]
- Ward VCA, Chatzivasileiou AO, Stephanopoulos G. Metabolic engineering of Escherichia coli for the production of isoprenoids. FEMS Microbiol Lett. 2018;365(10):fny079. doi: 10.1093/femsle/fny079. [DOI] [PubMed] [Google Scholar]
- Ward VCA, Chatzivasileiou AO, Stephanopoulos G. Cell free biosynthesis of isoprenoids from isopentenol. Biotechnol Bioeng. 2019;116(12):3269–3281. doi: 10.1002/bit.27146. [DOI] [PubMed] [Google Scholar]
- Watcharawipas A, Sansatchanon K, Phithakrotchanakoon C, Tanapongpipat S, Runguphan W, Kocharin K. Novel carotenogenic gene combinations from red yeasts enhanced lycopene and beta-carotene production in Saccharomyces cerevisiae from the low-cost substrate sucrose. FEMS Yeast Res. 2021 doi: 10.1093/femsyr/foab062. [DOI] [PubMed] [Google Scholar]
- Wei L, Xu N, Wang Y, Zhou W, Han G, Ma Y, Liu J. Promoter library-based module combination (PLMC) technology for optimization of threonine biosynthesis in Corynebacterium glutamicum. Appl Microbiol Biotechnol. 2018;102(9):4117–4130. doi: 10.1007/s00253-018-8911-y. [DOI] [PubMed] [Google Scholar]
- Wood AJ, Lo TW, Zeitler B, Pickle CS, Ralston EJ, Lee AH, Amora R, Miller JC, Leung E, Meng X, Zhang L, Rebar EJ, Gregory PD, Urnov FD, Meyer BJ. Targeted genome editing across species using ZFNs and TALENs. Science. 2011;333(6040):307. doi: 10.1126/science.1207773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Yan Q, Jones JA, Tang YJ, Fong SS, Koffas MAG. Metabolic burden: cornerstones in synthetic biology and metabolic engineering applications. Trends Biotechnol. 2016;34(8):652–664. doi: 10.1016/j.tibtech.2016.02.010. [DOI] [PubMed] [Google Scholar]
- Wu T, Ye L, Zhao D, Li S, Li Q, Zhang B, Bi C, Zhang X. Membrane engineering—a novel strategy to enhance the production and accumulation of β-carotene in Escherichia coli. Metab Eng. 2017;43(Pt A):85–91. doi: 10.1016/j.ymben.2017.07.001. [DOI] [PubMed] [Google Scholar]
- Wu T, Ye L, Zhao D, Li S, Li Q, Zhang B, Bi C. Engineering membrane morphology and manipulating synthesis for increased lycopene accumulation in Escherichia coli cell factories. 3 Biotech. 2018;8(6):269–269. doi: 10.1007/s13205-018-1298-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, Li S, Ye L, Zhao D, Fan F, Li Q, Zhang B, Bi C, Zhang X. Engineering an artificial membrane vesicle trafficking system (AMVTS) for the excretion of β-carotene in Escherichia coli. ACS Synth Biol. 2019;8(5):1037–1046. doi: 10.1021/acssynbio.8b00472. [DOI] [PubMed] [Google Scholar]
- Wu T, Liu J, Li M, Zhang G, Liu L, Li X, Men X, Xian M, Zhang H. Improvement of sabinene tolerance of Escherichia coli using adaptive laboratory evolution and omics technologies. Biotechnol Biofuels. 2020;13(1):79. doi: 10.1186/s13068-020-01715-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Yan P, Li Y, Liu X, Wang Z, Chen T, Zhao X. Enhancing β-carotene production in Escherichia coli by perturbing central carbon metabolism and improving the NADPH supply. Front Bioeng Biotechnol. 2020;8:585–585. doi: 10.3389/fbioe.2020.00585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Ma G, Liu C, Qiu XY, Min L, Kuang J, Zhu L. Biosynthesis of pinene in purple non-sulfur photosynthetic bacteria. Microb Cell Fact. 2021;20(1):101. doi: 10.1186/s12934-021-01591-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Lv X, Ye L, Zhou P, Yu H. Construction of lycopene-overproducing Saccharomyces cerevisiae by combining directed evolution and metabolic engineering. Metab Eng. 2015;30:69–78. doi: 10.1016/j.ymben.2015.04.009. [DOI] [PubMed] [Google Scholar]
- Xiong M, Schneiderman DK, Bates FS, Hillmyer MA, Zhang K. Scalable production of mechanically tunable block polymers from sugar. Proc Natl Acad Sci USA. 2014;111(23):8357–8362. doi: 10.1073/pnas.1404596111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Li X, Liu Y, Zhu Y, Li J, Du G, Chen J, Ledesma-Amaro R, Liu L. Pyruvate-responsive genetic circuits for dynamic control of central metabolism. Nat Chem Biol. 2020;16(11):1261–1268. doi: 10.1038/s41589-020-0637-3. [DOI] [PubMed] [Google Scholar]
- Yadav VG, De Mey M, Lim CG, Ajikumar PK, Stephanopoulos G. The future of metabolic engineering and synthetic biology: towards a systematic practice. Metab Eng. 2012;14(3):233–241. doi: 10.1016/j.ymben.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada Y, Kuzuyama T, Komatsu M, Shin-Ya K, Omura S, Cane DE, Ikeda H. Terpene synthases are widely distributed in bacteria. Proc Natl Acad Sci USA. 2015;112(3):857–862. doi: 10.1073/pnas.1422108112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang KK, Wu Z, Arnold FH. Machine-learning-guided directed evolution for protein engineering. Nat Methods. 2019;16(8):687–694. doi: 10.1038/s41592-019-0496-6. [DOI] [PubMed] [Google Scholar]
- Yang D, Park SY, Lee SY. Production of rainbow colorants by metabolically engineered Escherichia coli. Adv Sci. 2021 doi: 10.1002/advs.202100743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazdani M, Croen MG, Fish TL, Thannhauser TW, Ahner BA. Overexpression of native ORANGE (OR) and OR mutant protein in Chlamydomonas reinhardtii enhances carotenoid and ABA accumulation and increases resistance to abiotic stress. Metab Eng. 2021;68:94–105. doi: 10.1016/j.ymben.2021.09.006. [DOI] [PubMed] [Google Scholar]
- Ye L, Zhang C, Bi C, Li Q, Zhang X. Combinatory optimization of chromosomal integrated mevalonate pathway for β-carotene production in Escherichia coli. Microb Cell Fact. 2016;15(1):202. doi: 10.1186/s12934-016-0607-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee DA, DeNicola AB, Billingsley JM, Creso JG, Subrahmanyam V, Tang Y. Engineered mitochondrial production of monoterpenes in Saccharomyces cerevisiae. Metab Eng. 2019;55:76–84. doi: 10.1016/j.ymben.2019.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J, Wang H, Fu XZ, Gao X, Wu Q, Chen GQ. Effects of chromosomal gene copy number and locations on polyhydroxyalkanoate synthesis by Escherichia coli and Halomonas sp. Appl Microbiol Biotechnol. 2015;99(13):5523–5534. doi: 10.1007/s00253-015-6510-8. [DOI] [PubMed] [Google Scholar]
- Yin X, Shin HD, Li J, Du G, Liu L, Chen J. Pgas, a low-pH-induced promoter, as a tool for dynamic control of gene expression for metabolic engineering of Aspergillus niger. Appl Environ Microbiol. 2017;83(6):e03222-16. doi: 10.1128/aem.03222-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida E, Kojima M, Suzuki M, Matsuda F, Shimbo K, Onuki A, Nishio Y, Usuda Y, Kondo A, Ishii J. Increased carvone production in Escherichia coli by balancing limonene conversion enzyme expression via targeted quantification concatemer proteome analysis. Sci Rep. 2021;11(1):22126. doi: 10.1038/s41598-021-01469-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K, Liu C, Kim B-G, Lee D-Y. Synthetic fusion protein design and applications. Biotechnol Adv. 2015;33(1):155–164. doi: 10.1016/j.biotechadv.2014.11.005. [DOI] [PubMed] [Google Scholar]
- Yu H, Wang N, Huo W, Zhang Y, Zhang W, Yang Y, Chen Z, Huo Y-X. Establishment of BmoR-based biosensor to screen isobutanol overproducer. Microb Cell Fact. 2019;18(1):30. doi: 10.1186/s12934-019-1084-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Ching C-B. Dynamic control of ERG9 expression for improved amorpha-4,11-diene production in Saccharomyces cerevisiae. Microb Cell Fact. 2015;14:38–38. doi: 10.1186/s12934-015-0220-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuste R. Fluorescence microscopy today. Nat Methods. 2005;2(12):902–904. doi: 10.1038/nmeth1205-902. [DOI] [PubMed] [Google Scholar]
- Zeng W, Guo L, Xu S, Chen J, Zhou J. High-throughput screening technology in industrial biotechnology. Trends Biotechnol. 2020 doi: 10.1016/j.tibtech.2020.01.001. [DOI] [PubMed] [Google Scholar]
- Zhang H, Wang X. Modular co-culture engineering, a new approach for metabolic engineering. Metab Eng. 2016;37:114–121. doi: 10.1016/j.ymben.2016.05.007. [DOI] [PubMed] [Google Scholar]
- Zhang F, Cong L, Lodato S, Kosuri S, Church G, Arlotta P. LETTErs efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat Biotechnol. 2011;29:149–153. doi: 10.1038/nbt.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Chen X, Stephanopoulos G, Too HP. Efflux transporter engineering markedly improves amorphadiene production in Escherichia coli. Biotechnol Bioeng. 2016;113(8):1755–1763. doi: 10.1002/bit.25943. [DOI] [PubMed] [Google Scholar]
- Zhang C, Liu J, Zhao F, Lu C, Zhao GR, Lu W. Production of sesquiterpenoid zerumbone from metabolic engineered Saccharomyces cerevisiae. Metab Eng. 2018;49:28–35. doi: 10.1016/j.ymben.2018.07.010. [DOI] [PubMed] [Google Scholar]
- Zhang C, Seow VY, Chen X, Too H-P. Multidimensional heuristic process for high-yield production of astaxanthin and fragrance molecules in Escherichia coli. Nat Commun. 2018;9(1):1858. doi: 10.1038/s41467-018-04211-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Li M, Zhao G-R, Lu W. Alpha-Terpineol production from an engineered Saccharomyces cerevisiae cell factory. Microb Cell Fact. 2019;18(1):160. doi: 10.1186/s12934-019-1211-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Liu X, Meng Y, Zhang L, Qiao J, Zhao G-R. Combinatorial engineering of Saccharomyces cerevisiae for improving limonene production. Biochem Eng J. 2021;176:108155. doi: 10.1016/j.bej.2021.108155. [DOI] [Google Scholar]
- Zhang Y, Song X, Lai Y, Mo Q, Yuan J. High-yielding terpene-based biofuel production in Rhodobacter capsulatus. ACS Synth Biol. 2021 doi: 10.1021/acssynbio.1c00146. [DOI] [PubMed] [Google Scholar]
- Zhao EM, Zhang Y, Mehl J, Park H, Lalwani MA, Toettcher JE, Avalos JL. Optogenetic regulation of engineered cellular metabolism for microbial chemical production. Nature. 2018;555(7698):683–687. doi: 10.1038/nature26141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Zhu K, Li J, Zhao Y, Li S, Zhang C, Xiao D, Yu A. High-efficiency production of bisabolene from waste cooking oil by metabolically engineered Yarrowia lipolytica. Microb Biotechnol. 2021 doi: 10.1111/1751-7915.13768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wang C, Yang L, Choi ES, Kim SW. Geranyl diphosphate synthase: an important regulation point in balancing a recombinant monoterpene pathway in Escherichia coli. Enzyme Microb Technol. 2015;68:50–55. doi: 10.1016/j.enzmictec.2014.10.005. [DOI] [PubMed] [Google Scholar]
- Zhou L, Ding Q, Jiang GZ, Liu ZN, Wang HY, Zhao GR. Chromosome engineering of Escherichia coli for constitutive production of salvianic acid A. Microb Cell Fact. 2017;16(1):84. doi: 10.1186/s12934-017-0700-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S, Lyu Y, Li H, Koffas MAG, Zhou J. Fine-tuning the (2S)-naringenin synthetic pathway using an iterative high-throughput balancing strategy. Biotechnol Bioeng. 2019;116(6):1392–1404. doi: 10.1002/bit.26941. [DOI] [PubMed] [Google Scholar]
- Zhou P, Du Y, Fang X, Xu N, Yue C, Ye L. Combinatorial modulation of linalool synthase and farnesyl diphosphate synthase for linalool overproduction in Saccharomyces cerevisiae. J Agric Food Chem. 2021;69(3):1003–1010. doi: 10.1021/acs.jafc.0c06384. [DOI] [PubMed] [Google Scholar]
- Zhu M, Wang C, Sun W, Zhou A, Wang Y, Zhang G, Zhou X, Huo Y, Li C. Boosting 11-oxo-β-amyrin and glycyrrhetinic acid synthesis in Saccharomyces cerevisiae via pairing novel oxidation and reduction system from legume plants. Metab Eng. 2018;45:43–50. doi: 10.1016/j.ymben.2017.11.009. [DOI] [PubMed] [Google Scholar]
- Zhu Z-T, Du M-M, Gao B, Tao X-Y, Zhao M, Ren Y-H, Wang F-Q, Wei D-Z. Metabolic compartmentalization in yeast mitochondria: Burden and solution for squalene overproduction. Metab Eng. 2021;68:232–245. doi: 10.1016/j.ymben.2021.10.011. [DOI] [PubMed] [Google Scholar]
- Zhuang Y, Jiang G-L, Zhu M-J. Atmospheric and room temperature plasma mutagenesis and astaxanthin production from sugarcane bagasse hydrolysate by Phaffia rhodozyma mutant Y1. Process Biochem. 2020;91:330–338. doi: 10.1016/j.procbio.2020.01.003. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All datasets used and analyzed are available on reasonable request.