Graphical abstract
Keywords: Elaidic acid, Linoelaidic acid, Serum lipidomics, Liver inflammation, Gut microbiota
Highlights
-
•
EA and LEA gavage may induce liver inflammation via NF-κB / MAPK signaling pathways.
-
•
LEA intake significantly promoted TAG accumulation in liver and serum.
-
•
Specific SM/TAG molecules can be used as biomarkers following EA/LEA intake.
-
•
LEA gavage significantly changed fecal microbiota by enriching Lactobacillus.
Abstract
This work aims to study the effects of oral gavage (0.2 mg/g body weight) of elaidic acid (C18:1–9 t, EA) and linoelaidic acid (C18:2–9 t,12 t, LEA) on lipid metabolism, inflammation and gut homeostasis of mice. Results showed that both EA and LEA gavage significantly increased LDL-c, TC and oxidative stress levels in the liver and serum and may stimulate liver inflammation via NF-κB and MAPK signaling pathway. Compared with EA, LEA gavage significantly promoted TAG accumulation and inflammatory signaling. Serum lipidomics revealed that LEA intake significantly increased the concentration of ∼50 TAGs, while EA gavage primarily caused significant decreases in several SMs. 16S rRNA demonstrated that LEA ingestion markedly changed fecal microbiota by enriching Lactobacillus (phylum Firmicutes), however, EA treatment did not affect it. Overall, LEA gavage has more severe consequences on TAG accumulation, inflammation and microbial structure than EA, highlighting that the number of trans double bonds affects these processes.
1. Introduction
Trans fatty acids (TFAs) are fatty acids with one or more double bonds in the trans configuration, which are found naturally in meat and dairy products at limited levels (Kuhnt et al., 2011). But they are present at much higher levels in processed food, e.g., margarine, shortenings, bakery products, and deep-fried foods made with hydrogenated vegetable oils. This kind of TFA is known as industrial TFA. In partial hydrogenation, up to 50 % of double bonds were transformed to trans form (Micha & Mozaffarian, 2009), which had a longer shelf life, pleasant mouthfeel, and were less expensive. However, accumulating epidemiological and nutritional evidence suggested that increased intake of industrial TFA was associated with cardiovascular disease, liver disease, diabetes, and others (Guo et al., 2023, Oteng et al., 2019, Li et al., 2023).
Elaidic acid (C18:1–9 t, EA) was the predominant industrial TFA formed in partially hydrogenated vegetable oils, accounting for 20–30 % of the total (Kuhnt et al., 2011). Intake of hydrogenated vegetable oils rich in EA would lead to an elevated low density lipoprotein cholesterol (LDL-c) level, inflammation, oxidative stress, etc., which were confirmed by the in vitro (Iwata et al., 2011, Liu et al., 2022, Shao and Ford, 2014), animal (Cassagno et al., 2005, Jeyapal et al., 2018, Monguchi et al., 2017), and human studies (Mensink and Katan, 1990, Lopez-Garcia et al., 2005). For example, compared with oleic acid, mice fed with EA induced significantly higher TNF-α and IL-1β protein levels in serum as well as reactive oxygen species production in mouse aorta, which were subsequently confirmed by an in-vitro experiment using smooth muscle cells (Monguchi et al., 2017). In support, plasma C-reactive protein and IL-6 levels in healthy men were significantly increased after consumption of a diet containing 8 % energy from TFA (mainly EA) versus oleic acid (Baer et al., 2004). Unlike EA, linoelaidic acid (C18:2–9 t,12 t, LEA) was mainly induced by the heating process, such as frying, baking, and the deodorization step during oil refining (Guo et al., 2023, Xu et al., 2022, Li et al., 2013). Both LEA and EA were negatively associated with human health, clinical studies have shown that LEA possessed a greater risk to cardiovascular health (Lemaitre et al., 2006). However, the consequences of a specific TFA like EA on serum lipoprotein level, inflammation, etc., are, therefore, still scarcely studied. No study has compared the effect of LEA versus EA on lipid metabolism and inflammation, via animal models. Therefore, it is of great significance to investigate the health effects of LEA and EA separately to accurately reflect the impacts of a specific industrial TFA on the regulation of lipid profile and inflammatory status of mice.
The gut microbiota (GM) plays an important role in maintaining the health of the host. As a result, the abnormal microbiota profile can lead to several metabolic syndromes, such as obesity, diabetes, and dyslipidemia (Delzenne et al., 2015, Shen et al., 2013). Although previous studies have shown that a high-TFA (mainly EA) diet containing hydrogenated oil (Carvalho et al., 2018, Ge et al., 2019) or shortening (Okamura et al., 2021) can alter the GM composition, little is known about the influence of LEA on gut homeostasis. In addition, there has been little effort trying to understand the regulation effects of GM on dyslipidemia and inflammation induced by EA or LEA. Within this context, this work aims to investigate (1) the effects of EA and LEA gavage on the regulation of lipid metabolism, inflammation, as well as the GM structure of C57BL/6J mice, (2) the molecular mechanisms underlying the proinflammatory effects of EA and LEA, and (3) the correlations between the GM profile and dyslipidemia or inflammation caused by EA and LEA.
2. Materials and methods
2.1. Animals and treatment
All animal experiments were performed as approved by the Animal Ethics Review Committee at Southwest University. Six-week-old C57BL/6J mice (male, 18 ∼ 22 g) were purchased from SPF Biotechnology Co., Ltd. (Beijing, China) and housed in specific pathogen-free (SPF) conditions with 6 mice per cage (temperature of 25 ± 1 ℃, humidity of 60 ± 3 %, and light of 12 h day/night cycle). After 1 week of acclimation, animals were randomly divided into 3 groups: the normal control (NC) group, elaidic acid (EA) group, and linolelaidic acid (LEA) group (n = 6/group). Mice in the EA and LEA groups received 0.2 mg/g body weight by oral gavage of elaidic acid and linolelaidic acid dissolved in olive oil (30 mg/mL). Another group (NC) of mice was gavaged with olive oil. Mice were given free access to chow diet (Byrness Weil Biotech Ltd., Chongqing, China) and water for 8 weeks. Body weights were recorded every week. Elaidic acid (>99 %) and linolelaidic acid (>99 %) were obtained from Nu Chek Prep, Inc (Elysian, MN, USA).
When the mice reached 14 weeks of age, they were anesthetized by isoflurane and euthanized by cervical dislocation following blood collection via the orbital puncture. After setting on ice for 15 min, plasma was collected by centrifugation (2000 × g, 5 min, 4 ℃). The liver and small intestine were removed, weighted and one portion of them was cut and fixed in 4 % formalin for histological analysis. The day before the mice were euthanized, fresh fecal samples were collected in sterile tubes and immediately placed on dry ice. All samples were stored at −80 °C until analysis.
2.2. Biochemical assays
The contents of triglyceride (TAG), total cholesterol (TC), high-density lipoprotein cholesterol (HDL-c), and low-density lipoprotein cholesterol (LDL-c) in the serum and liver were assayed by their respective commercial kits (Nanjing Jiancheng Bioengineering Institute, Nanjing, China) according to the manufacturer’s instructions. For liver samples, the tissue was homogenized in ice-cold anhydrous ethanol (1:9, w/v) and then centrifugated at 10,000 × g for 10 min at 4 °C. The protein concentration in the resulting supernatant was determined by a BCA protein assay kit (Beijing Dingguo Changsheng Biotechnology Co. Ltd., Beijing, China). TAG, TC, HDL-c, and LDL-c concentrations in the serum and liver were expressed as mmol/L and mmol/g protein, respectively.
2.3. Histological analysis
The liver and small intestine were fixed in 4 % formalin for 48 h, embedded in paraffin, and cut into 5-μm-thick sections. Subsequently, the sections were stained with hematoxylin and eosin (H&E). Finally, images of stained sections were photographed using a light microscope (Nikon Eclipse TE2000-U, Nikon, Tokyo, Japan).
2.4. Inflammatory cytokines measurements
The levels of tumor necrosis factor-α (TNF-α), interleukin 6 (IL-6), and interleukin-1β (IL-1β) were analyzed by their respective commercial kits (Tianjin Anorikang Biotechnology Co., Tianjin, China) using serum or supernatants of liver obtained by homogenizing with saline (1:9, w/v) followed by centrifugation at 10,000 × g for 10 min at 4 °C. The protein concentration in the tissue was determined by a BCA protein assay kit (Beijing Dingguo Changsheng Biotechnology Co. Ltd., Beijing, China), and the result was expressed as pg/mg protein.
2.5. Oxidative stress determination
The contents of nitric oxide (NO), malondialdehyde (MDA), and reduced glutathione (GSH) and the activities of inducible nitric oxide synthase (iNOS), superoxide dismutase (SOD), and myeloperoxidase (MPO) in the liver and small intestine were assessed by corresponding commercial kits (Nanjing Jiancheng Bioengineering Institute, Nanjing, China) using the supernatant of the tissue obtained by homogenizing with saline (1:9, w/v) followed by centrifugation at 10,000 × g for 10 min at 4 °C. The protein concentration in the tissue was determined by a BCA protein assay kit (Beijing Dingguo Changsheng Biotechnology Co. Ltd., Beijing, China), and the result was expressed as μmol/g protein or nmol/g protein or U/mg protein. Plasma levels of iNOS, NO, and GSH were evaluated by the same method.
2.6. Western blotting analysis
The total proteins in the liver were prepared by homogenizing the tissue in an ice-cold lysis buffer containing 0.1 % protease (v/v) and 1 % phosphatase inhibitor (v/v) (Beyotime Biotechnology, Shanghai, China); the cytoplasmic proteins and nuclear proteins in the liver were separated and collected using a commercial kit (Keygene Biotechnology, Nanjing, China). The tissue slurry was centrifugated at 13,000 × g for 15 min at 4 ℃. The BCA protein assay kit (Beijing Dingguo Changsheng Biotechnology Co. Ltd., Beijing, China) was used to measure the protein concentration. Equal amounts of protein were separated by 10 % or 12 % sodium dodecyl sulfate–polyacrylamide gel electrophoresis after denaturation at 95 ℃ for 10 min and then transferred onto polyvinylidene difluoride (PVDF) membranes. The membranes were blocked with 5 % skim milk (w/v) diluted in TBST at room temperature for 2 h and incubated overnight at 4 °C with primary antibodies against β-actin (1:5000), TLR4 (1:1000), IKKα/β (1:1000), p-IKKα/β (1:1000), IkBα (1:5000), p-IkBα (1:5000), p65 (1:5000), p-p65 (1:1000), LaminB (1:5000), JNK (1:2000), p-JNK (1:1000), ERK (1:5000), p-ERK (1:2000), p38(1:1000), and p-p38 (1:1000). All antibodies were purchased from Cell Signaling Technology, Inc (Danvers, MA, USA). After incubation, the membranes were washed with TBST and incubated with secondary antibodies at room temperature for 2 h. Finally, the immunoreactive bands were visualized by the ChemiScope 6000 Pro Imaging system (Clinx Science Instruments Co., Ltd., Shanghai, China).
2.7. Plasma lipidomics
Lipids were extracted from mice serum using a modified Bligh and Dyer's method and dried in the SpeedVac under OH mode. Before analysis, lipid extracts were reconstituted in chloroform: methanol (1:1, v/v) containing appropriate internal standards. Lipidomic analyses were conducted at LipidALL Technologies Company Limited (Changzhou, Jiangsu, China) by liquid chromatography-mass spectrometry (LC-MS). The LC-MS analyses were performed on an ExionLC-AD UPLC system coupled with a Sciex QTRAP 6500 Plus system (SCIEX, Framingham, USA). All samples were analyzed in the ESI mode with the following parameters: curtain gas of 20 mL/min, ion spray voltage of 5500 V, the temperature of 400 ℃, ion source gas 1 of 35 mL/min, and ion source gas 2 of 35 mL/min.
In brief, polar lipids were separated by normal phase (NP)-HPLC using a TUP-HB silica column (150 × 2.1 mm × 3 µm) (Lam et al., 2014, Lam et al., 2018). The mobile phase A and B consisted of chloroform, methanol, and ammonium hydroxide (89.5:10:0.5, v/v/v) and chloroform, methanol, ammonium hydroxide, and water (55:39:0.5:5.5, v/v/v), respectively. Multiple reaction monitoring (MRM) transitions were set up for comparative analysis of various polar lipids. Glycerol lipids including diacylglycerols (DAG) and triacylglycerols (TAG) were analyzed using a modified version (Shui et al., 2010, Song et al., 2020) of reverse phase HPLC/MRM. Separation of neutral lipids was achieved on a Phenomenex Kinetex-C18 column (4.6 × 100 mm × 2.6 µm) using an isocratic mobile phase containing chloroform: methanol: 0.1 M ammonium acetate 100:100:4 (v/v/v). Free cholesterols (Cho) and cholesteryl esters (CE) were analyzed under atmospheric pressure chemical ionization (APCI) mode on a Jasper HPLC coupled to Sciex 4500 MD as described previously (Shui et al., 2011).
Individual lipid species were quantified by referencing to spiked internal standards of the same lipid class. Polar lipids were calculated using internal standards including d9-PC32:0(16:0/16:0), d7-PE33:1(15:0/18:1), d31-PS, d7-PA33:1(15:0/18:1), d7-PG33:1(15:0/18:1), d7-PI33:1(15:0/18:1), d7-PA33:1(15:0/18:1), d7-LPC18:1, d7-Cer d18:1/15:0, d8-SMd18:1/18:1, d9-PC36:1(18:0/18:1), d9-PE36:1(18:0/18:1), d9-SM d18:1/ 18:1, C8-GluCer, etc., which were purchased from Avanti Polar Lipids (Alabaster, Alabama, USA). Short-, medium- and long-chain TAGs were quantified by referencing internal standards of d5-TAG(14:0)3, d5-TAG(16:0)3, and d5-TAG(18:0)3, respectively, which were obtained from CDN isotopes (Pointe-Claire, Quebec, Canada). DAGs were quantified using d5-DAG17:0/17:0 and d5-DAG18:1/18:1 as internal standards (Avanti Polar Lipids). Free Cho and CE were calculated using d6-Cho and d6-C18:0 CE (CDN isotopes) as internal standards. D31-16:0 (Sigma-Aldrich) and d8-20:4 (Cayman Chemicals) were used for free fatty acids (FFA) quantitation, while d3-16:0-acylcarnitine (Cayman Chemicals) was used for acyl-carnitines calculation.
2.8. Fecal microbiota sequencing
Total DNA in mice fecal samples was extracted using the Qiagen QIAmpDNA stool kit (Qiagen, USA) following the manufacturer’s instruction. The quality and concentration of the isolated DNA were verified by gel electrophoresis and by using a NanoDrop spectrophotometer (Thermo Scientific, USA). PCR amplification of the bacterial 16S rRNA genes V3-V4 region was performed using the forward primer 338F (5′-ACTCCTACGGGAGGCAGCA-3′) and the reverse primer 806R (5′-GGACTACHVGGGTWTCTAAT-3′). PCR amplicons were purified with Agencourt AMPure Beads (Beckman Coulter, Indianapolis, IN) and quantified using the PicoGreen dsDNA Assay Kit (Invitrogen, Carlsbad, CA, USA).
Sequencing was performed using the Illumina MiSeq platform with MiSeq Reagent Kit v3 at Shanghai Personal Biotechnology Co., Ltd (Shanghai, China). The Quantitative Insights into Microbial Ecology (QIIME, v1.8.0) pipeline was employed to process the sequencing data as previously reported (Caporaso et al., 2010). Briefly, after low-quality sequences were filtered, the remaining high-quality sequences were clustered into operational taxonomic units (OTUs) at 97 % sequence identity by UCLUST (Edgar, 2010). The representative sequences for each OTU were selected through the Greengenes Database (DeSantis et al., 2006). OTU-level alpha diversity indices, such as Chao1, ACE, Shannon diversity index, and Simpson index, were calculated using QIIME. PLS-DA was generated using an R package based on weighted UniFrac distances.
2.9. Data analysis
The data were expressed as mean ± standard deviation and analyzed using Prism 8 (GraphPad, San Diego, CA, USA). The differences among the three experimental groups were analyzed with SPSS 22.0 (IBM Corporation, Armonk, NY, USA) by one-way analysis of variance (ANOVA) with Tukey as post hoc test. Differences were considered statistically significant at the p < 0.05 level.
3. Results
3.1. EA and LEA gavage induced dyslipidemia without weight gain
After gavage with 0.2 mg/g body weight EA, LEA (both in olive oil), or olive oil for 8 weeks, no statistical differences in mice body weight (Fig. 1a) or weight gain (Fig. 1b) were found among the three groups. The same result was confirmed by Ge et al.(2019), who showed that intake of low-fat diet with partially hydrogenated soybean oil (containing 57.4 % EA and 1.9 % LEA) did not induce weight changes in male C57BL/6J mice. Nevertheless, the evidence in the literature was still controversial as to the correlation between iTFA consumption and obesity (Pipoyan et al., 2021). The liver (Fig. 1c) and small intestine (Fig. 1d) index of the LEA-gavage group were significantly higher than those of the control (NC) group. A similar trend was observed between the EA and NC group, yet only the small intestine index reached a statistically significant difference (p < 0.05). H&E staining (Fig. 1e) indicated that the liver of the mice in the NC group had normal hepatic architecture without any evidence of inflammation and steatosis. While EA and LEA gavage resulted in hepatocytes with loose arrangements and inflammatory infiltration (marked with black arrows). In addition, there were swollen cells in the liver tissue of the LEA group. Upon histological analysis of the small intestine (Fig. 1f), the EA and LEA groups exhibited the destruction of villi compared to the NC group, showing the proliferation of lymphocytes (marked with black arrows) and a decreased number of goblet cells. The fasting glucose level (Fig. 1o) did not differ among the groups.
Fig. 1.
EA and LEA gavage increased TAG and LDL-c levels in serum and liver of C57BL/6J mice. (a) Body weight and (b) weight gain; liver (c) and small intestine (d) index; histological analysis of liver (e) and small intestine (f) tissue; serum TC (g), TAG (h), LDL-c (i), and HDL-c (j) concentrations; hepatic TC (k), TAG (l), LDL-c (m), and HDL-c (n) levels; (o) fasting glucose concentrations at the end of the experiment.
Serum TC (Fig. 1g), TAG (Fig. 1h), and LDL-c (Fig. 1i) concentrations were significantly greater in the LEA group than in the NC group. Similar trends were observed between the EA and NC group except for the TAG content (p > 0.05). Meanwhile, compared with the NC group, the serum HDL-c (Fig. 1j) concentration of the LEA group was significantly lower (p < 0.05). Nevertheless, this was not the case for the EA group (p > 0.05). The TC (Fig. 1k), TAG (Fig. 1l), LDL-c (Fig. 1m), and HDL-c (Fig. 1n) levels in the liver followed a similar changing trend as those in the serum, except that EA treatment notably decreased the hepatic HDL-c level compared with the NC group (0.0054 vs. 0.0099 mmol/g prot, p < 0.05). Moreover, EA treatment resulted in a significantly lower hepatic LDL-c level than LEA treatment (0.024 vs. 0.037 mmol/g prot, p < 0.05).
3.2. EA and LEA gavage induced oxidative stress
Compared to the NC group, both the EA and LEA groups induced serum oxidative stress as evidenced by a significant increase and decrease in NO (Fig. 2a) and GSH (Fig. 2b) concentration, respectively, but LEA treatment led to a more dramatic increase in the former (1.9-fold compared with EA, p < 0.05). In line with this, serum TNOS activity (Fig. 2c) was significantly enhanced in the EA and LEA groups as compared to the NC group. LEA gavage also promoted liver oxidative stress by notably raising the level of NO (Fig. 2d) and MDA (Fig. 2f) (5.2- and 2.1-fold compared to other groups, respectively, p < 0.05) as well as the activity of TNOS (Fig. 2g), decreasing that of GSH (Fig. 2e), and down-regulating the activities of TSOD (Fig. 2h) and MPO (Fig. 2i). Whereas EA gavage only led to significant decreases in GSH content and MPO activity in the liver as compared to the NC group. Intestinal levels of NO (Fig. 2j) and MDA (Fig. 2l) were significantly higher in the EA- and LEA-gavage groups than those in the control group, but LEA treatment resulted in a more dramatic increase in the MDA level (1.6-fold compared with EA, p < 0.05). Though a higher intestinal TNOS activity (Fig. 2m) was observed in both the EA and LEA group (vs. the NC group), a statistical difference was only reached for the latter. Compared with the NC group, the intestinal GSH level (Fig. 2k) and the activities of TSOD (Fig. 2n) and MPO (Fig. 2o) were significantly reduced in the EA and LEA groups. Consequently, both EA and LEA administration induced oxidative stress in the small intestine of C57BL/6J mice.
Fig. 2.
EA and LEA gavage increased oxidative stress in serum, liver, and small intestine of C57BL/6J mice. Serum NO (a), GSH (b), and TNOS (c) concentrations; NO (d), GSH (e), MDA (f), TNOS (g), TSOD (h) and MPO (i) levels in the liver; NO (j), GSH (k), MDA (l), TNOS (m), TSOD (n) and MPO (o) levels in the small intestine. All data are expressed as mean ± SD (n = 4 mice per group).
3.3. EA and LEA gavage may stimulate liver inflammation via NF-κB and MAPK signaling pathways
Serum (Fig. 3a-c) and liver (Fig. 3d-f) levels of inflammatory cytokines (TNF-α, IL-6, and IL-1β) increased in the following order: NC < EA < LEA except for the plasma concentration of TNF-α (NC < EA ≈ LEA). Therefore, EA and LEA gavage evoked inflammation in both serum and the liver. This agreed well with the inflammatory state of the liver tissue in the trans group (Fig. 1e). More importantly, the above results indicate that LEA gavage leads to significantly more severe inflammation.
Fig. 3.
EA and LEA gavage may induce inflammatory responses in serum and liver of C57BL/6J mice via MAPK and NF-κB signaling pathway. Serum TNF-α (a), IL-6 (b), and IL-1β (c) concentrations; liver TNF-α (d), IL-6 (e), and IL-1β (f) levels; (g) western blot analysis of p-ERK, p-JNK, p-p38, and β-actin in the liver; (h) western blot analysis of TLR-4, p-IKK α/β, p-IkBα, p-p65, and β-actin in the liver; (i) western blot analysis of cytosolic and nuclear p-p65 in the liver. All data are expressed as mean ± SD (n = 4 mice per group).
The nuclear factor-κB (NF-κB) and mitogen-activated protein kinase (MAPK) pathways are the two major pathways involved in the regulation of transcript and protein expression for cytokines such as TNF-α, IL-6, and IL-1β. Therefore, we determined the expression levels of proteins related to these two pathways to further explore the molecular mechanism of liver inflammation caused by TFAs. As shown in Fig. 3g, the western blot experiment reveals that the expression levels of phosphorylated protein toll-like receptor 4 (TLR4), IKKα/β, IκBα, and p65 in the liver increased in the following order: NC < EA < LEA, suggesting that EA and LEA gavage activate the NF-κB pathway. Furthermore, the activation of the NF-κB pathway was significantly more augmented in the LEA vs. EA group. Fig. 3h indicated that EA and LEA administration resulted in the translocation of NF-κB p65 into the nucleus of hepatocytes. Likewise, this effect was more pronounced in the LEA group (vs. EA group). LEA treatment also activated the MAPK pathway (Fig. 3i), as evidenced by a significantly enhanced phosphorylation of ERK and JNK, and p38 in relation to the NC group. A similar trend was observed between the EA and NC group, however, only the expression levels of ERK and JNK reached statistically significant differences. Similar to the NF-κB pathway, the activation of the MAPK pathway was also significantly more intense in the LEA vs. EA group.
3.4. EA and LEA gavage modified serum lipid profiles
A total of 600 lipid molecules from 5 lipid categories (glycerolipids, glycerophospholipids, sphingolipids, fatty acyls, and glycosphingolipids, Fig. 4h-l) and 22 subclasses (Fig. 4a) were detected by the untargeted lipidomics approach. As shown in Fig. 4a, the number of identified TAG molecules (1 4 8) was the highest, accounting for 24.7 % of the total detected lipid species, followed by 73 kinds of phosphatidylcholine (PC, 12.2 %) and 60 kinds of phosphatidylethanolamine (PE, 10.0 %). PC1 and PC2 collectively explained above 50 % of the total variation present in the data, but no clear separation between the control and trans groups was observed (Fig. 4b). Nevertheless, relative to the EA group (Fig. 4c), the LEA group (Fig. 4d) was better discriminated with the control. Volcano plots highlighted 30 identified features that were significantly different in the EA group (Fig. 4e) and 88 in the LEA group (Fig. 4f) when compared to the control, which was consistent with the increased divergence of PCA ellipses between the LEA and NC groups. The highest number of significantly different identified features was visualized between the EA and LEA groups (Fig. 4g). Compared to the NC group, 5/6 features were significantly down-regulated in the EA group whereas an opposite trend was found in the LEA group. Notably, almost all identified lipid features were significantly up-regulated after 8 weeks of LEA gavage, which was indicative of a dramatic serum lipid accumulation.
Fig. 4.
Serum lipidomic analysis of C57BL/6J mice after gavage with EA and LEA for 8 weeks. (a) The quantity and relative percentage of serum lipids identified in positive and negative ion modes; (b) principal component analysis (PCA) plot of serum lipid profiling among all groups; PCA plot comparing the NC and EA groups (c) and the NC and LEA groups (d); volcano plots highlighting significantly upregulated (marked with red dots) and downregulated (marked with green dots) lipid species between the EA and NC group (e), the LEA and NC group (f), and the LEA and NC group (g); (h) quantification analysis of significantly differential lipid species using internal standards; heatmap displaying the significantly differential FFA (i), LysoPC (j), SM (k), PC (l), plasamalogenPC (m), PE(n), and TAG (o) based on ANOVA. In figures E-G, significantly differential lipid species were selected out using the criteria of FC > 1.5 or < 1/1.5 and VIP > 1.
Quantitative analysis based on internal standards showed that the top 3 abundant serum lipid subcategories were PC (Fig. 4i), cholesterol ester (CE, Fig. 4h), and free fatty acids (FFA, Fig. 4k), with total concentrations ranging from 1711.9 to 2258.9 nmol/L, 1673.3 to 2269.8 nmol/L, and 1718.8 to 1879.3 nmol/L, respectively. There were 6 lipid subclasses that vary significantly among the three groups, e.g., TAG, plasmalogenPC, PC, PE, lysophosphatidylcholine (LPC), and sphingomyelin (SM). The total TAG concentration of the LEA group (626.38 ± 243.31 nmol/L) was remarkably higher than that of the EA (237.61 ± 61.38 nmol/L) and NC (243.75 ± 93.29 nmol/L) groups, which agreed well with the serum TAG level determined by commercial kits. PE revealed a similar changing pattern to TAG.
The specific changes in the abundance of specific molecular species of these subclasses were comprehensively analyzed and the results were presented in Fig. 4m-t. Accordingly, we saw the highest number (71) of significantly different features representing TAG, followed by SM (18) and PE (14). As shown in Fig. 4m, compared with the control, EA gavage significantly reduced serum concentrations of long-chain saturated fatty acids (C16:0 and C18:0). Compared to the NC and EA groups, there were significant increases of 4 DAG species (Fig. 4r) following LEA gavage, including the highly abundant DAG 36:2 (18:1/18:1). In TAG species (Fig. 4t), LEA treatment remarkably increased the long-chain species with 52–58 carbons, driven mainly by the increase of TAG 54:3(C18:1), TAG 54:4(18:1), and TAG 54:4(18:2) when compared to the NC and EA groups. Notably, almost all of the significantly increased individual species were polyunsaturated. All listed SM species (Fig. 4o) were significantly decreased following EA gavage (vs. control), including the highly abundant SM d18:1/24:1 and SM d18:1/22:0. While LEA gavage did not affect SM at all (except for SM d18:1/23:1). Interestingly, we observed that 8 kinds of SM were significantly more abundant in the LEA vs. EA group. The changing pattern of plasmalogenPC species (Fig. 4q) was similar to that of SM. In response to EA exposure, five PC subclasses with 34–36 carbons and 1–2 double bonds were significantly decreased (Fig. 4p). This decrease was driven by a notable decrease of the highly abundant PC 34:2 and PC 34:1. In contrast, LEA exposure led to a marked increase in 5 PC species with longer (36–40) and more unsaturated (3–7 double bonds) chain lengths. These data suggest the opposing roles of EA and LEA in the selective alteration of serum PC species. It was interesting to note that all listed PC species (Fig. 4p) were significantly more abundant in the LEA versus the EA group except for PC36:1 (18:0/18:1). All identified significantly changed LPC species (Fig. 4n) were decreased with EA gavage (vs. control, except for LPC 14:0 e), driven mainly by the 16:0- and 18:0-containing species. While LEA barely affected the individual LPCs except for LPC 20:4. The response of individual PE species (Fig. 4s) to EA and LEA treatment revealed disparities.
Finally, combing the fold change analysis (FC > 3 or < 0.5) and VIP scores (VIP > 1), five serum lipids with a p-value < 0.05 (Dunn) and a FC value < 0.5, e.g., SM d18:1/26:1, GM3 d18:1/25:1, SM d18:0/24:0, SM d18:1/22:0, and SM d18:1/21:0, were selected as the potential biomarkers following EA gavage. While 19 TAGs, including TAG 56:5(20:1), TAG 56:5(18:2), TAG 56:4(20:1), TAG 54:3(16:0), TAG 56:4(18:2), TAG 54:4(16:1), TAG 56:4(18:1), TAG 56:5(20:2), TAG 54:3(16:1), TAG 54:2(18:2), TAG 56:2(20:1), TAG 54:4(18:0), TAG 46:3(18:2), TAG 54:7(18:2), TAG 56:3(20:1), TAG 54:5(18:2), TAG 52:5(16:2), TAG 54:5(16:1), and TAG 50:4(18:2), together with PE 36:4(18:2/18:2) and PI 36:4 (18:2/18:2), were chosen as potential biomarkers (all with a p-value < 0.05 and a FC value > 3) following LEA gavage.
3.5. LEA gavage changed the gut microbiota structure
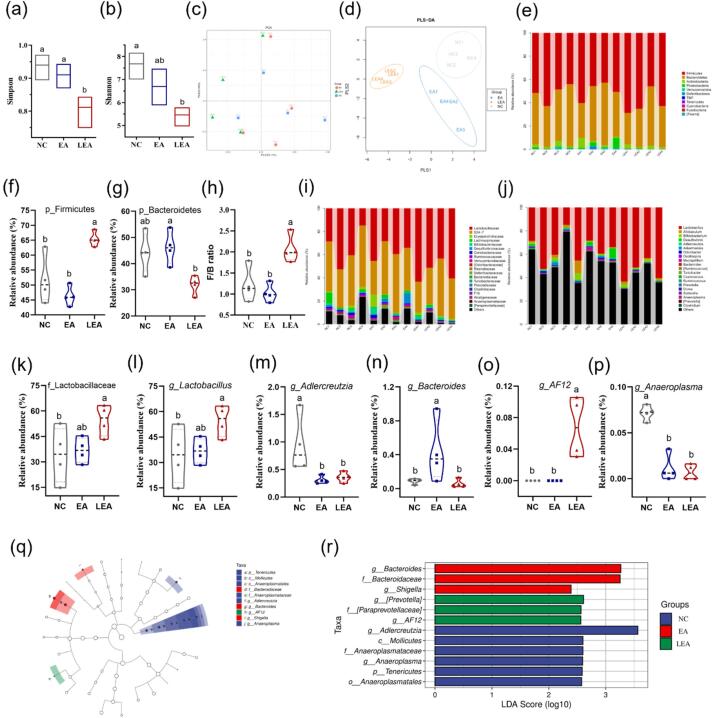
A total of 1,095,329 valid sequences, representing a total of 10,783 operational taxonomic units (OTUs), were detected in 12 stool samples. The Venn diagram reveals that the three groups shared 1939 OTUs, and the number of unique OTUs was 463, 272, and 167 for the NC, EA, and LEA groups, respectively. Accordingly, the Simpson (Fig. 5a) and Shannon (Fig. 5b) indices were the lowest in the LEA group, suggesting that LEA gavage significantly decreases fecal bacterial diversity and richness. Nevertheless, EA gavage did not affect it. Besides, we observed a significantly higher Simpson index in the EA versus the LEA group. Beta diversity analysis based on the PCA score plot (Fig. 5c) revealed that the fecal microbiota of the LEA group can be largely separated from the NC and LEA groups, suggesting that LEA gavage has a significant effect on the microbial structure. The supervised PLS-DA score plot (Fig. 5d) further shows that the difference in the fecal microbiota induced by LEA gavage was apparently greater than that by EA gavage when compared to the control.
Fig. 5.
Changes in fecal microbial composition of C57BL/6J mice after gavage EA and LEA for 8 weeks. α-Diversity analysis indicated by the Simpson (a) and Shannon index (b); PCA (c) and PLS-DA (d) plot of fecal microbial profiling among all groups; microbiota taxonomic profiles at the phylum (e), family (i), and genus (j) level; relative abundances of Firmicutes (f) and Bacteroidetes (g); (h) The ratio of Firmicutes to Bacteroidetes; (k) relative abundance of Lactobacillaceae at the family level; (l-p) Significantly changed bacterial communities at the genus level. LEfSe analysis (q) and LDA score (> 2, r) of bacterial taxa with significant differences from phylum to genus among the three experimental groups.
At the phylum level (Fig. 5e), the relative abundance of Firmicutes in the LEA group (65.4 ± 2.5 %, Fig. 5f) was significantly higher and that in the control group (51.8 ± 8.0 %). Following LEA gavage, mice showed a lower relative proportion of Bacteroidetes (31.9 ± 3.5 %) than the control (44.2 ± 7.5 %), nevertheless, it did not establish a statistically significant difference (p > 0.05, Fig. 5g). Whereas the EA and NC groups had a comparable abundance of Bacteroidetes (46.1 % vs. 44.2 %). Consequently, compared with the NC and EA groups, LEA gavage resulted in a significantly higher F/B ratio (Fig. 5h). At the family level (Fig. 5i), the intestinal microbiota was dominated by Lactobacillaceae and Bacteroidales_S24-7 group belonging to Firmicutes and Bacteroidetes, respectively, which collectively accounted for 77–90 % of the total flora. As shown in Fig. 5k, Lactobacillaceae was significantly more abundant in the LEA group (54.6 ± 9.1 %) than in the NC (34.1 ± 16.2 %) and EA (36.8 ± 7.3 %) groups. At the genus level, Lactobacillus (Fig. 5l) in the family of Lactobacillaceae and AF 12 (Fig. 5o) in the family of Rikenellaceae (phylum Bacteroidetes) were significantly enriched in the LEA group when compared to the NC or EA group. While Adlercreutzia (Fig. 5m) in the family of Coriobacteriaceae (phylum Actinobacteria) and Anaeroplasma (Fig. 5p) in the family of Anaeroplasmataceae (phylum Tenericutes) were significantly less abundant in the trans group than in the control group. Notably, we observed that EA gavage significantly expanded the relative proportion of Bacteroides (Fig. 5n) belonging to family Bacteroidaceae. Lefse analysis (Fig. 5q and r) further reveals the dominant bacterial communities in each group. Accordingly, Adlercreutzia and Anaeroplasma (class Mollicutes; order Anaeroplasmatales; phylum Tenericutes) were specific to the NC group; Bacteroides and Shigella were enriched in the EA group; while AF 12 together with [Prevotella] in the family of [Paraprevotellaceae] dominated the LEA group. Based on the study conducted by Mohammadi et al. (2023), it could be concluded that changes in gut microbiota structure were highly correlated with the position of double bonds in TFAs.
3.6. Correlation of genus abundance with metabolic parameters
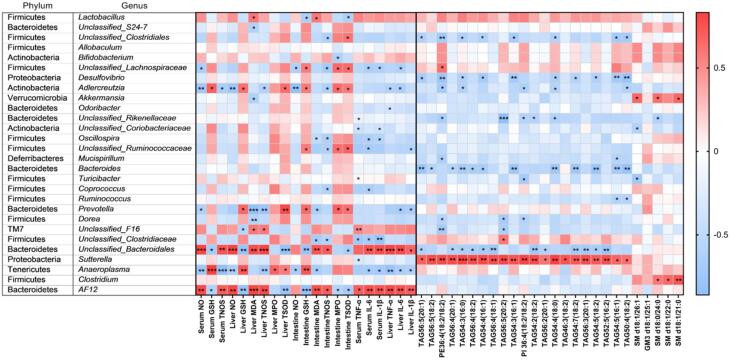
Spearman’s correlation analyses were performed to examine the connections between the abundance of specific genera and metabolic parameters, including serum NO, GSH, and TNOS, hepatic/intestinal NO, GSH, MDA, TNOS, MPO, and TSOD, serum/hepatic TNF-α, IL-6, and IL-1β, as well as 26 kinds of potential serum lipid biomarkers. The correlation heat map (Fig. 6) showed that AF12 and Unclassified_Bacteroidales had significantly positive correlations with parameters related to serum/hepatic/intestinal oxidative stress and serum/hepatic inflammation, while Anaeroplasma and Adlercreutzia (except for serum inflammation) inversely correlated with these parameters (p < 0.05). Likewise, Prevotella displayed a significantly negative relationship with hepatic/intestinal oxidative stress and hepatic inflammation. In addition, we observed a significant negative correlation between the relative abundance of Unclassified_Lachnospiraceae and Unclassified_Clostridiaceae with serum inflammation. Upon correlation analysis between intestinal bacteria genera and potential serum lipid biomarkers, the heat map (Fig. 6) reveals that Unclassified_Clostridiales, Desulfovibrio, Adlercreutzia, Bacteroides, and Unclassified_Bacteroidales were negatively correlated with some lipid biomarkers related to LEA gavage (p < 0.05), while only Sutterella showed a strong positive relationship with all of them (p < 0.05). Additionally, we observed that Akkermansia and Clostridium presented a significantly positive correlation with 3 serum lipid biomarkers related to EA gavage.
Fig. 6.
The potential mechanism of EA and LEA gavage affecting the intestinal microbiota, inflammation and oxidative stress of C57BL/6J mice.
4. Discussion
The health consequences of industrial TFA intake have been extensively researched since the 1950 s. The object of these studies was mainly hydrogenated fats derived from diets, consisting of multiple TFA. While the damaging effects of a particular TFA, e.g., EA or LEA, were rarely investigated using animal models. Our work attempted to achieve this by gavaging mice with pure EA or LEA. With a gavage dose of 0.2 mg/g body weight and an average food intake of 2 g/d per mice (energy content of the feed was 3.6 kcal/g), the energy ratio supplied by EA or LEA was estimated to be ∼ 0.5 %, which falls in the recommended intake (<1 %) proposed by The World Health Organization (WHO). Therefore, the TFA intake was considered normal in this experiment.
In this work, we found the consistent effects of EA and LEA gavage on the physiological processes of C57BL/6J mice, which includes: (1) long-term normal intake of EA or LEA induced increased level of LDL-c, TC and oxidative stress in the liver and serum of mice. This was consistent with former studies, and similar results have been reported by Cassagno et al. (2005), who fed C57BL/6J mice with a low amount of EA-enriched diet. (2) Both EA and LEA gavage induced inflammation in liver and serum, more importantly, we found that liver inflammation may stimulate via NF-κB and MAPK signaling pathway. Though at the cellular level, it has been demonstrated that both EA and LEA were associated with NF-κB activation in human endothelial cells (Iwata et al., 2011), and EA activated the MAPK signaling pathway in Kuper cells resulting in the release of the inflammatory factors IL-1β and IL-18 (Liu et al., 2022). To the best of our knowledge, this was the first study reveals that EA and LEA may stimulate hepatic inflammation via NF-κB and MAPK signaling pathway in an animal model.
More importantly, our results revealed that EA and LEA behave differently in changing lipogenesis, serum lipid species and gut microbial structure, which includes: (1) LEA gavage significantly promoted TAG biosynthesis in liver and serum, resulting in marked heavier livers of mice in the LEA group. This may be due to the fact that LEA gavage increases the expression levels of genes related to lipogenesis and/or changes enzyme/substrate affinities (Du et al., 2011). In contrast, EA gavage did not affect the TAG content either in liver or serum. (2) more specifically, serum lipidomics revealed that the concentration of ∼ 50 kinds of long-chain polyunsaturated TAG were significantly increased by LEA gavage, while EA gavage primarily reduced the level of several SM. (3) compared with EA, LEA intake significantly increased MAPK and NF-κB signaling in liver. This may correlate with increased NO production following LEA gavage, besides, differences in the way EA and LEA were incorporated into cell membrane and lipid rafts may account for this discrepancy (Iwata et al., 2011). (4) LEA gavage significantly enriched genera Lactobacillus from phylum Firmicutes, however, overall, EA treatment did not alter the gut microbial profile. Although Lactobacillus is generally considered to be beneficial to human health, in our study, it’s overgrowth (accounts for 54.6 % of the population) may cause a decrease in the proportion of other beneficial communities, resulting in a dysbiosis of the intestinal homeostasis. This was supported by the fact that LEA gavage resulted in a significant decrease and increase in α diversity and F/B ratio, respectively. The fact that EA gavage barely changes the GM profile agrees with Hua et al. (2020), who reported that intake of high fat diet with 1 % EA (vs. diet-induced obesity group) did not modify the microbial structure. LEA gavage significantly altered the fecal microbial structure while EA gavage did not, this may relate to the augmented TAG accumulation in the LEA group. TAG accumulation may cause oxidative stress and intestinal inflammation, leading to changed intestinal environment and gut microbiota. Actually, LEA treatment stimulated significantly higher inflammatory responses and intestinal MDA and TNOS levels than EA. Besides, metabolites of TAG like free fatty acids may affect the growth and metabolic processes of intestinal flora. Additionally, EA and LEA gavage may affect the absorption extent of fatty acids in the meal. This directly determines the fatty acids entering the large intestine and may account for the discrepancy in the effects of EA and LEA on fecal microbiota.
Furthermore, combining all the results, we conclude that LEA gavage has more severe consequences on lipid metabolism, inflammation, and microbial structure than EA, suggesting that the number of trans double bonds probably plays a significant role in determining the extent to which iTFA were harmful to the metabolism of mice. In support, a population-based study showed that LEA intake was associated with a 3-fold higher risk of primary cardiac arrest than EA (Lemaitre et al., 2002). Nevertheless, the underlying mechanism is unclear for studies concerned with the effect of increased trans double bonds on inflammation and intestinal homeostasis are scarce, therefore further research is urgently required. Another interesting finding demonstrated by serum lipidomics was that LEA gavage remodeled the metabolism of TAG, whereas EA mainly impacted the metabolism of SM by decreasing their concentrations. And specific TAG and SM species were selected as potential biomarkers following LEA and EA intake, respectively. LEA and EA have been reported to incorporate primarily into TAG and phosphohipids of plasma, respectively (Du et al., 2011, Moore et al., 1980). As a result, LEA gavage may have the greatest effect on TAG. While the reasons for the decrease in serum SM concentration induced by EA gavage remain to be investigated. Last but not least, the sample size of this work was relatively small given the cost of EA and LEA by gavage. Further studies with enlarged sample size and clinical trials are advised. Despite of this, our study could still serve as a reference for researching the effects of a particular TFA on inflammation and gut microbiota in mice.
5. Conclusions
In summary, the present study demonstrates that long-term normal intake of two common industrial TFA (EA and LEA) may induce liver inflammation in mice via NF-κB and MAPK signaling pathway. Besides, our work highlights a differential effect of EA and LEA and that the number of trans double bonds affects TAG accumulation, inflammation, and gut microbiota. Compared to EA, LEA gavage significantly promoted TAG accumulation in liver and serum, increased MAPK and NF-κB signaling in liver, and enhanced the proliferation of genera Lactobacillus. Consequently, LEA intake has a much greater disturbing effect on lipid metabolism and gut microbial structure than EA. Moreover, serum lipidomics reveal that specific TAG and SM molecules could be used as potential biomarkers following LEA and EA consumption, respectively. We believe that these differences caused by EA and LEA could affect their health effects in humans and this definitely requires further investigation.
Ethical statement
All animal experiments were performed as approved by the Animal Ethics Review Committee at Southwest University, and animals used in our experiment were followed by the Guidelines in the Care and Use of Animal.
CRediT authorship contribution statement
Liting Wan: Writing – original draft, Formal analysis, Data curation, Conceptualization. Tian Li: Writing – review & editing, Methodology, Investigation, Conceptualization. Mengying Yao: Software, Formal analysis, Data curation, Conceptualization. Baoshun Zhang: Resources, Project administration, Methodology, Funding acquisition. Weimin Zhang: Writing – review & editing, Supervision, Funding acquisition, Conceptualization. Jiachao Zhang: Writing – review & editing, Supervision, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was funded by the National Natural Science Foundation of China (32060579, 31660495).
Contributor Information
Weimin Zhang, Email: zhwm1979@163.com.
Jiachao Zhang, Email: zhjch321123@163.com.
Data availability
The data that has been used is confidential.
References
- Baer D.J., Judd J.T., Clevidence B.A., Tracy R.P. Dietary fatty acids affect plasma markers of inflammation in healthy men fed controlled diets: A randomized crossover study. American Journal of Clinical Nutrition. 2004;79(6):969–973. doi: 10.1093/ajcn/79.6.969. [DOI] [PubMed] [Google Scholar]
- Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Pẽa A.G., Goodrich J.K., Gordon J.I., Huttley G.A., Kelley S.T., Knights D., Koenig J.E., Ley R.E., Lozupone C.A., McDonald D., Muegge B.D., Pirrung M., Knight R. QIIME allows analysis of high-throughput community sequencing data. Nature Methods. 2010;5(7):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho G.C.B.C., Moura C.S., Roquetto A.R., Barrera-Arellano D., Yamada A.T., dos Santos A., Saad M.J.A., Amaya-Farfan J. Impact of trans-fats on heat-shock protein expression and the gut microbiota profile of mice. Journal of Food Science. 2018;83(2):489–498. doi: 10.1111/1750-3841.13997. [DOI] [PubMed] [Google Scholar]
- Cassagno N., Palos-Pinto A., Costet P., Breilh D., Darmon M., Bérard A.M. Low amounts of trans 18: 1 fatty acids elevate plasma triacylglycerols but not cholesterol and alter the cellular defence to oxidative stress in mice. British Journal of Nutrition. 2005;94(3):346–352. doi: 10.1079/bjn20051512. [DOI] [PubMed] [Google Scholar]
- Delzenne N.M., Cani P.D., Everard A., Neyrinck A.M., Bindels L.B. Gut microorganisms as promising targets for the management of type 2 diabetes. Diabetologia. 2015;10(58):2206–2217. doi: 10.1007/s00125-015-3712-7. [DOI] [PubMed] [Google Scholar]
- DeSantis T.Z., Hugenholtz P., Larsen N., Rojas M., Brodie E.L., Keller K., Huber T., Dalevi D., Hu P., Andersen G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Applied and Environmental Microbiology. 2006;72(7):5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z.Y., Degrace P., Gresti J., Loreau O., Clouet P. Vaccenic and elaidic acid equally esterify into triacylglycerols, but differently into phospholipids of fed rat liver cells. Lipids. 2011;46(7):647–657. doi: 10.1007/s11745-011-3569-6. [DOI] [PubMed] [Google Scholar]
- Edgar R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Ge Y., Liu W., Tao H., Zhang Y., Liu L., Liu Z., Qiu B., Xu T. Effect of industrial trans-fatty acids-enriched diet on gut microbiota of C57BL/6 mice. European Journal of Nutrition. 2019;58(7):2625–2638. doi: 10.1007/s00394-018-1810-2. [DOI] [PubMed] [Google Scholar]
- Guo Q., Li T., Qu Y., Liang M., Ha Y., Zhang Y., Wang Q. New research development on trans fatty acids in food: Biological effects, analytical methods, formation mechanism, and mitigating measures. Progress in Lipid Research. 2023;89 doi: 10.1016/j.plipres.2022.101199. [DOI] [PubMed] [Google Scholar]
- Hua Y., Fan R., Zhao L., Tong C., Qian X., Zhang M., Xiao R., Ma W. Trans-fatty acids alter the gut microbiota in high-fat-diet-induced obese rats. British Journal of Nutrition. 2020;124(12):1251–1263. doi: 10.1017/S0007114520001841. [DOI] [PubMed] [Google Scholar]
- Iwata N.G., Pham M., Rizzo N.O., Cheng A.M., Maloney E., Kim F. Trans fatty acids induce vascular inflammation and reduce vascular nitric oxide production in endothelial cells. PLoS ONE. 2011;6(12):e29600. doi: 10.1371/journal.pone.0029600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeyapal S., Putcha U.K., Mullapudi V.S., Ghosh S., Sakamuri A., Kona S.R., Vadakattu S.S., Madakasira C., Ibrahim A. Chronic consumption of fructose in combination with trans fatty acids but not with saturated fatty acids induces nonalcoholic steatohepatitis with fibrosis in rats. European Journal of Nutrition. 2018;57(6):2171–2187. doi: 10.1007/s00394-017-1492-1. [DOI] [PubMed] [Google Scholar]
- Kuhnt K., Baehr M., Rohrer C., Jahreis G. Trans fatty acid isomers and the trans-9/trans-11 index in fat containing foods. European Journal of Lipid Science and Technology. 2011;113(10):1281–1292. doi: 10.1002/ejlt.201100037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam S.M., Tong L., Duan X., Petznick A., Wenk M.R., Shui G. Extensive characterization of human tear fl uid collected using different techniques unravels the presence of novel lipid amphiphiles. Journal of Lipid Research. 2014;55(2):289–298. doi: 10.1194/jlr.M044826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam S.M., Wang R., Miao H., Li B., Shui G. An integrated method for direct interrogation of sphingolipid homeostasis in the heart and brain tissues of mice through postnatal development up to reproductive senescence. Analytica Chimica Acta. 2018;1037:152–158. doi: 10.1016/j.aca.2018.01.015. [DOI] [PubMed] [Google Scholar]
- Lemaitre R.N., King I.B., Mozaffarian D., Sotoodehnia N., Rea T.D., Kuller L.H., Tracy R.P., Siscovick D.S. Plasma phospholipid trans fatty acids, fatal ischemic heart disease, and sudden cardiac death in older adults: The cardiovascular health study. Circulation. 2006;114(3):209–215. doi: 10.1161/CIRCULATIONAHA.106.620336. [DOI] [PubMed] [Google Scholar]
- Lemaitre R.N., King I.B., Raghunathan T.E., Pearce R.M., Weinmann S., Knopp R.H., Copass M.K., Cobb L.A., Siscovick D.S. Cell membrane trans-fatty acids and the risk of primary cardiac arrest. Circulation. 2002;105(6):697–701. doi: 10.1161/hc0602.103583. [DOI] [PubMed] [Google Scholar]
- Li A., Yuan B., Li W., Wang F., Ha Y. Thermally induced isomerization of linoleic acid in soybean oil. Chemistry and Physics of Lipids. 2013;166(1):55–60. doi: 10.1016/j.chemphyslip.2012.12.003. [DOI] [PubMed] [Google Scholar]
- Li X., Wu F., Guenther S., Looso M., Kuenne C., Zhang T., Wiesnet M., Klatt S., Zukunft S., Leming I., Poschet G., Wietelmann A., Atzberger A., Potente M., Yuan X.J., Braun T. Inhibition of fatty acid oxidation enables heart regeneration in adult mice. Nature. 2023;622:619–626. doi: 10.1038/s41586-023-06585-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Nan B., Yang C., Li X., Yan H., Yuan Y. Elaidic acid induced NLRP3 inflammasome activation via ERS-MAPK signaling pathways in kupffer cells. Biochimica et Biophysica Acta - Molecular and Cell Biology of Lipids. 2022;1867(1) doi: 10.1016/j.bbalip.2021.159061. [DOI] [PubMed] [Google Scholar]
- Lopez-Garcia E., Schulze M.B., Meigs J.B., Manson J.E., Rifai N., Stampfer M.J., Willett W.C., Hu F.B. Consumption of trans fatty acids is related to plasma biomarkers of inflammation and endothelial dysfunction. Journal of Nutrition. 2005;135(3):562–566. doi: 10.1093/jn/135.3.562. [DOI] [PubMed] [Google Scholar]
- Micha R., Mozaffarian D. Trans fatty acids: Effects on metabolic syndrome, heart disease and diabetes. nature reviews. Endocrinology. 2009;6(5) doi: 10.1038/nrendo.2009.79. [DOI] [PubMed] [Google Scholar]
- Mensink R.P., Katan M.B. Effect of dietary trans fatty acids on high-density and low-density lipoprotein cholesterol levels in healthy subjects. New England Journal of Medicine. 1990;323(7):439–445. doi: 10.1056/NEJM199008163230703. [DOI] [PubMed] [Google Scholar]
- Mohammadi F., Green M., Tolsdorf E., Greffard K., Leclercq M., Bilodeau J.-F., Droit A., Foster J., Bertrand N., Rudkowska I. Industrial and ruminant trans-fatty acids-enriched diets differentially modulate the microbiome and fecal metabolites in C57BL/6 mice. Nutrients. 2023;15:1433. doi: 10.3390/nu15061433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monguchi T., Hara T., Hasokawa M., Nakajima H., Mori K., Toh R., Irino Y., Ishida T., Hirata K., Ichi, Shinohara M. Excessive intake of trans fatty acid accelerates atherosclerosis through promoting inflammation and oxidative stress in a mouse model of hyperlipidemia. Journal of Cardiology. 2017;70(2):121–127. doi: 10.1016/j.jjcc.2016.12.012. [DOI] [PubMed] [Google Scholar]
- Moore C., Alfin-Slater R., Aftergood L. Incorporation and disappearance of trans fatty acids in rat tissues. The American journal of clinical nutrition. 1980;33(11):2318–2323. doi: 10.1093/ajcn/33.11.2318. [DOI] [PubMed] [Google Scholar]
- Okamura T., Hashimoto Y., Majima S., Senmaru T., Ushigome E., Nakanishi N., Asano M., Yamazaki M., Takakuwa H., Hamaguchi M., Fukui M. Trans fatty acid intake induces intestinal inflammation and impaired glucose tolerance. Frontiers in Immunology. 2021;12 doi: 10.3389/fimmu.2021.669672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oteng A.B., Loregger A., van Weeghel M., Zelcer N., Kersten S. Industrial trans fatty acids stimulate SREBP2-mediated cholesterogenesis and promote non-alcoholic fatty liver disease. Molecular Nutrition and Food Research. 2019;63(19):1900385. doi: 10.1002/mnfr.201900385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pipoyan D., Stepanyan S., Stepanyan S., Beglaryan M., Costantini L., Molinari R., Merendino N. The effect of trans fatty acids on human health: Regulation and consumption patterns. Foods. 2021;10(10):2452. doi: 10.3390/foods10102452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao F., Ford D.A. Elaidic acid increases hepatic lipogenesis by mediating sterol regulatory element binding protein-1c activity in HuH-7 cells. Lipids. 2014;49(5):403–413. doi: 10.1007/s11745-014-3883-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J., Obin M.S., Zhao L. The gut microbiota, obesity and insulin resistance. Molecular Aspects of Medicine. 2013;34(1):39–58. doi: 10.1016/j.mam.2012.11.001. [DOI] [PubMed] [Google Scholar]
- Shui G., Cheong W.F., Jappar I.A., Hoi A., Xue Y., Fernandis A.Z., Tan B.K.H., Wenk M.R. Derivatization-independent cholesterol analysis in crude lipid extracts by liquid chromatography/mass spectrometry: Applications to a rabbit model for atherosclerosis. Journal of Chromatography A. 2011;1218(28):4357–4365. doi: 10.1016/j.chroma.2011.05.011. [DOI] [PubMed] [Google Scholar]
- Shui G., Guan X.L., Low C.P., Chua G.H., Goh J.S.Y., Yang H., Wenk M.R. Toward one step analysis of cellular lipidomes using liquid chromatography coupled with mass spectrometry: Application to Saccharomyces cerevisiae and Schizosaccharomyces pombe lipidomics. Molecular BioSystems. 2010;6(6):1008–1017. doi: 10.1039/b913353d. [DOI] [PubMed] [Google Scholar]
- Song J.W., Lam S.M., Fan X., Cao W.J., Wang S.Y., Tian H., Chua G.H., Zhang C., Meng F.P., Xu Z., Fu J.L., Huang L., Xia P., Yang T., Zhang S., Li B., Jiang T.J., Wang R., Wang Z., Shui G. Omics-driven systems interrogation of metabolic dysregulation in COVID-19 pathogenesis. Cell Metabolism. 2020;32(2):188–202. doi: 10.1016/j.cmet.2020.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Liu S., Shen M., Xie J., Yang J. Evaluation of trans fatty acids, carbonyl compounds and bioactive minor components in commercial linseed oils. Food Chemistry. 2022;369 doi: 10.1016/j.foodchem.2021.130930. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that has been used is confidential.