Abstract
Several cell membrane proteins have been identified as herpes simplex virus (HSV) entry mediators (Hve). HveA (formerly HVEM) is a member of the tumor necrosis factor receptor family, whereas the poliovirus receptor-related proteins 1 and 2 (PRR1 and PRR2, renamed HveC and HveB) belong to the immunoglobulin superfamily. Here we show that a truncated form of HveC directly binds to HSV glycoprotein D (gD) in solution and at the surface of virions. This interaction is dependent on the native conformation of gD but independent of its N-linked glycosylation. Complex formation between soluble gD and HveC appears to involve one or two gD molecules for one HveC protein. Since HveA also mediates HSV entry by interacting with gD, we compared both structurally unrelated receptors for their binding to gD. Analyses of several gD variants indicated that structure and accessibility of the N-terminal domain of gD, essential for HveA binding, was not necessary for HveC interaction. Mutations in functional regions II, III, and IV of gD had similar effects on binding to either HveC or HveA. Competition assays with neutralizing anti-gD monoclonal antibodies (MAbs) showed that MAbs from group Ib prevented HveC and HveA binding to virions. However, group Ia MAbs blocked HveC but not HveA binding, and conversely, group VII MAbs blocked HveA but not HveC binding. Thus, we propose that HSV entry can be mediated by two structurally unrelated gD receptors through related but not identical binding with gD.
The entry of herpes simplex virus (HSV) into mammalian cells consists of a chain of events involving several of the 10 viral envelope glycoproteins (49, 58). The initial attachment of virus through glycoprotein C (gC) and/or gB to cell surface heparan sulfate proteoglycans (21, 22, 28) is not sufficient for virus penetration (4, 16, 26, 43). Fusion of viral envelope with cell plasma membrane requires gD, gB, and the gH-gL complex (49). These glycoproteins presumably act in concert (19, 20) and induce fusion only upon interaction with one or more specific cellular receptors (5, 25, 26, 28).
A number of cellular proteins have been postulated as HSV-specific surface receptors based on potential interactions with gD (2, 27). More recently, expression cloning led to the isolation of HVEM/HveA (herpesvirus entry mediator A), a lymphotoxin receptor (31) and member of the tumor necrosis factor receptor family, which allows entry of many strains of HSV type 1 (HSV-1) and HSV-2 into the normally nonpermissive Chinese hamster ovary (CHO) cells (32). A truncated form of HveA expressed by a recombinant baculovirus interacts with gD in vitro and on purified virions (36, 53). The HveA-gD interaction requires native structure but not N-glycosylation of gD and leads to the formation of a complex with a 2:1 molar ratio (53).
Although HveA meets all of the criteria to be a cellular receptor for gD mediating HSV entry, it could not be used by three infectious HSV strains, rid1, rid2, and ANG (32). gDs from these strains have one (rid1 and -2) or three (ANG) amino acid substitutions in the ectodomain compared to gD from the HSV-1 KOS strain (11, 24) and were unable to bind HveA in vitro (53).
Recently expression cloning led to the identification and isolation of two other cell surface proteins allowing HSV entry into CHO cells independently of HveA (18, 52). The genes coding for these proteins were cloned several years ago and named poliovirus receptor-related protein 1 (PRR1) (30) and polio virus receptor-related protein 2 (PRR2) (13). Both are members of the immunoglobulin (Ig) superfamily closely related to the poliovirus receptor (Pvr). Based on their ability to promote entry of herpesviruses into cells, PRR1 and PRR2 were renamed HveC and HveB, respectively (18, 52). PRR2/HveB was shown to enhance entry of a restricted number of mutant strains of HSV-1 (those carrying mutations in gD, such as rid1, rid2, and ANG), some HSV-2 strains, and pseudorabies virus (PRV) into CHO cells (52). PRR1/HveC was active as an entry mediator for all alphaherpesviruses tested so far (HSV-1, HSV-2, PRV, and bovine herpesvirus 1 [BHV-1]) (18). The third member of this Ig subfamily, Pvr-HveD, allowed entry of PRV and BHV-1 into nonpermissive cells but did not function for HSV (18). The cellular function of PRR1/HveC, like that of Pvr-HveD or PRR2/HveB, remains unknown; however, a recent report suggests that a murine poliovirus receptor-related protein (mPRR2) (33) could act as a homophilic intercellular adhesion molecule (1). HveC is a 518-amino-acid type I membrane glycoprotein (30). The clone used in this study encodes a variant protein of 517 amino acids in size with a substitution of residues 194 to 205 and carrying an additional N-linked glycosylation site at position 202 (18). This sequence, isolated from a placenta cDNA, is similar to sequences found in HeLa cells or brain tissue cDNA (18). The HveC extracellular region, like that of Pvr-HveD and HveB, consists of three Ig-like domains classified as V-C2-C2 from the most distal to the membrane-proximal domain (13, 55). HveC mRNA appears to be expressed ubiquitously in human tissues and cell lines (18, 30).
In our initial study, transfection of nonpermissive CHO cells with HveC enhanced entry of all HSV-1 and HSV-2 strains tested. Moreover, it could be used for entry by other alphaherpesviruses such as BHV and PRV (18). A soluble truncated form of HveC consisting of the ectodomain and bearing a C-terminal histidine tag, called HveCt, proved to be an efficient inhibitor of HSV infection of CHO cells stably expressing full-length HveC. More importantly, it could block viral entry in several neuron-like cell lines (IMR5, SY5Y, and NT-2) more efficiently than soluble truncated HveA (HveAt) (18).
In this study, we identified HSV gD as the viral ligand for HveC. We found that HveCt and truncated forms of gD interacted in direct binding assays and that HveCt bound to gD on purified virions. When the gD-HveC interaction was compared with the gD-HveA interaction, we observed a number of similarities but also several significant differences.
MATERIALS AND METHODS
Cells and virus.
Spodoptera frugiperda Sf9 cells (GIBCO BRL) were maintained in suspension in Sf900II medium (GIBCO BRL) or as a monolayer in supplemented Grace’s medium (GIBCO BRL).
Baculovirus construction.
Plasmid pBG38, containing the complete human HveC open reading frame, was used as the template in the PCR. A fragment of HveC corresponding to amino acids Gln31 to His346 was amplified. The upstream primer, 5′-GCGTGATCAGGTGGTCCAGGTGAACGACTCCATGTAT-3′, added a BclI restriction site overlapping the codon for Gln31. The downstream primer, 5′-CGGTGATCAATGATGATGATGATGATGTTCGGGAGGAGACGGGGTGTA-3′, added five histidine codons following His346, a stop codon, and a BclI site. The 979-bp fragment was digested with BclI, gel purified, and ligated to the vector pVT-Bac (51) which had been previously digested with BamHI and dephosphorylated. In that construct the HveC signal peptide was replaced by a mellitin signal sequence. Due to the cloning strategy, an extra aspartic acid residue was added to the N terminus of HveC. The generation of recombinant baculovirus has been described previously (46, 56). Briefly, the resulting plasmid pCK285 was cotransfected with Baculogold DNA (Pharmingen) into Sf9 cells. Recombinant baculoviruses were purified through two rounds of plaque selection on Sf9 cell monolayers. Plaques were tested for HveCt expression by Western blotting using the anti-HveC peptide antibody R145 (see below) and amplified. The recombinant baculovirus was named bac-HveC(346t), and the recombinant protein was designated HveCt or HveC(346t).
Purification of HveCt.
Sf9 cells in 3-liter suspension cultures (New Brunswick Celligen Plus Bioreactor) were infected with bac-HveC(346t) at a multiplicity of infection of 4 PFU per cell. After 48 h, cells were removed by centrifugation at 2,000 × g for 30 min at 4°C. The supernatant fluid was filtered through a 0.22-μm-pore-size membrane and concentrated to 1 liter, and the medium was exchanged against phosphate-buffered saline (PBS), using tangential flow filtration with a 10-kDa-cutoff membrane (Millipore). Five milliliters of Ni-nitrilotriacetic acid resin (Qiagen) preequilibrated with PBS was added per 3-liter culture and incubated overnight at 4°C on a rotary shaker. The resin was pelleted at 500 rpm, for 10 min at 4°C, transferred to a column, and washed with PBS. The bound protein was eluted with increasing concentrations of imidazole (10, 25, 50, 250, and 500 mM) in 0.02 M phosphate buffer (pH 7.5)–0.5 M NaCl. The 250 mM imidazole fraction was dialyzed against PBS and concentrated (10-kDa-cutoff centrifugation membrane; Millipore). Typically 6 to 7 mg was purified from each liter of culture.
Antibodies.
A synthetic peptide (AVLRAKKGQDDKVLVATC, corresponding to amino acids 155 to 172 of HveC) was coupled to keyhole limpet hemocyanin as previously described (7) and used to immunize two rabbits. The anti-HveC peptide antiserum used here is referred to as R145. Polyclonal antiserum R154, was generated by immunizing a rabbit with HveCt purified from culture supernatant of recombinant baculovirus-infected cells as described above. Polyclonal antibody R7 was raised against HSV-2 gD isolated from infected mammalian cells (23). Generation of rabbit polyclonal sera R46 and R47 directed against gC, R69 directed against gB, and R137 directed against gH-gL was described previously (14, 38). In the cosedimentation assay, the following antibodies were used: against gB, monoclonal antibodies (MAbs) SS10, DL16, and DL21 (41) and polyclonal serum R69 (14); against gC, MAbs MP1, MP5 (42), and 1C8 (17) and rabbit polyclonal serum R46 (14); against gD, MAbs 1D3 (17), DL2 (8), and DL11 (8, 34) and polyclonal serum R7 (23); against gH-gL, MAbs LP11 (3), 53S (45), and H6 (12) and polyclonal serum R137 (38). Anti-capsid protein VP5 MAb NC1 (9) was used in Western blotting.
Glycoproteins.
The production and purification of gD-1(306t)KOS, gD-2(306t)333, gD-1(QAAt), gD-1(▿34t), gD-1(▿126t), gD-1(▿243t), gD-1(▵290-299t), gD-1(306t)rid1, gD-1(306t)ANG, gC-1(457t), and HVEM(200t)/HveA(200t) have been described elsewhere (35, 37, 46, 50, 53). Construction and purification of gD-1(234t), gD-1(275t), and gD-1(285t) are described elsewhere (40). The gH(792t)-gL complex was isolated from a mouse L-cell line (HL7) stably transfected with plasmids pCMV3gH(792) and pCMVgL-1 as described previously (38). gB(724t) is a truncated form of gB-1 lacking the transmembrane domain and cytoplasmic tail produced in the baculovirus expression system (54).
SDS-PAGE and enzymatic digestion.
Precast Tris-glycine gels (Novex) were used to separate purified glycoproteins under denaturing and reducing conditions as described previously (53). Enzymatic digestion of purified protein with peptide N-glycosidase F (New England Biolabs) or endoglycosidase H (Boehringer Mannheim) prior to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed according to the manufacturer’s instructions.
Mass spectrometry.
Matrix-assisted laser desorption ionization mass spectrometry was performed as previously described (39) on a sample of HveCt dissolved in 50% acetonitrile containing 1% trifluoroacetic acid and diluted with 2-(4-hydroxyphenylazo)benzoic acid (Aldrich).
Enzyme-linked immunosorbent assay (ELISA).
Soluble receptor protein HveA(200t) or HveC(346t) in PBS was bound to microtiter plates overnight at 4°C. Plates were washed with 0.1% Tween 20 in PBS (PBS-Tween) and incubated in PBS with 5% milk and 0.2% Tween 20 (blocking solution) for 30 min at room temperature (RT). Plates were washed with PBS-Tween and incubated with various concentrations of the soluble HSV glycoproteins to be tested in blocking solution for 2 hours at RT. Plates were washed with PBS-Tween and incubated in blocking solution containing the appropriate antiserum for 30 min at RT. After being washed with PBS-Tween, the plates were incubated with horseradish peroxidase-conjugated secondary antibody diluted 1,000-fold in blocking solution for 30 min at RT. Plates were then washed with PBS-Tween and with 20 mM citrate buffer (pH 4.5). The horseradish peroxidase substrate [2,2′-azinobis(3-ethylbenzthiazolinesulfonic acid); Moss, Inc.] in citrate buffer (pH 4.5) was added, and the A405 was read with a microtiter plate reader (Bio-Tek). Results are presented after subtracting background signal obtained from parallel mock-coated well.
Gel filtration.
Purified soluble protein HveCt or gD-1(Δ290-299t), alone or in combination, was diluted in PBS and incubated overnight at 4°C. A volume of 200 μl was applied to a calibrated Superdex 200 column (Pharmacia HR 10/30). Fractions of 500 μl were collected and analyzed by Western blotting using rabbit polyclonal serum R7 to detect gD-1(Δ290-299t) and R145 to visualize HveCt.
Binding of HveCt to virions.
Sucrose gradient-purified KOS virions (107 PFU corresponding to an estimate of 5 × 108 particles) were incubated with 150 μg of HveCt at 4°C for 2 h. Samples were loaded on top of a 10-30-60% sucrose discontinuous gradient and centrifuged for 4.5 h at 16,000 × g, using an SW41 swinging-bucket rotor (Beckman). The virus band at the 30%-60% interface was collected and concentrated by centrifugation for 1 h at 35,000 × g in an SW50.1 rotor (Beckman). Viral pellets were dissolved in SDS sample buffer, boiled, and subjected to SDS-PAGE and Western blotting. Membranes were probed with antibody NC1 to detect capsid protein VP5 together with serum R154 to detect HveCt. In competition assays adapted from Nicola et al. (36), viruses were first incubated with a cocktail of antiglycoprotein antibodies or with anti-gD MAbs for 1 h at 37°C prior to incubation with soluble HveCt.
Nucleotide sequence accession numbers.
The HveC sequence GenBank accession no. is AF060231. The HVEM/HveA sequence GenBank accession no. is U70321.
RESULTS
Production and characterization of baculovirus-expressed HveCt.
A large quantity of HveCt was produced in the baculovirus expression system (Fig. 1). The same system was successfully used to produce soluble forms of the HVEM/HveA ectodomain and HSV glycoproteins, all of which displayed properties similar to those of proteins synthesized in mammalian cells (37, 46, 53, 56). We consistently obtained 6 to 7 mg of purified HveCt per liter of cell supernatant.
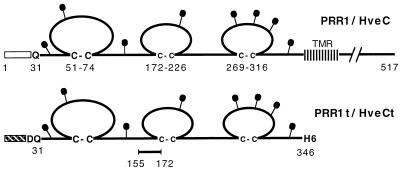
FIG. 1.
Schematic representation of PRR1/HveC proteins. The 517-amino-acid human PRR1/HveC is represented with residues numbered from methionine 1. The open box indicates the HveC signal peptide, and the transmembrane region is abbreviated TMR. The putative N-linked carbohydrates are shown as black lollipops. In the baculovirus construct, the mellitin signal peptide (hatched box) replaced the natural signal peptide (amino acids 1 to 30) from HveC. An additional N-terminal aspartic acid residue was inserted due to the cloning strategy. HveCt was truncated after His346, and five histidine residues were added to generate a six-His tag at the C terminus of HveCt. The synthetic peptide (amino acids 155 to 172) was used to generate rabbit antiserum R145.
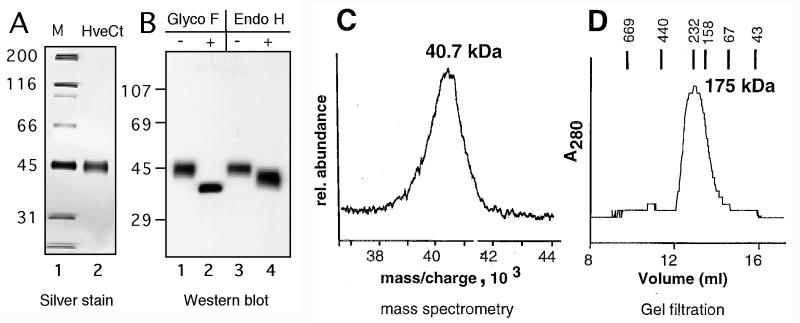
After purification by nickel chromatography, HveCt was analyzed by SDS-PAGE under denaturing and reducing conditions (Fig. 2A and B). The purified protein as revealed by silver staining migrated as a thick band with a size of 45 kDa (Fig. 2A). On Western blotting the same band reacted with antipeptide rabbit serum R145 (Fig. 2B, lanes 1 and 3). The primary amino acid sequence of HveC contains eight consensus sites for N-linked glycosylation in the extracellular domain (Fig. 1) (18, 30). Purified HveCt was treated with glycopeptidase F or endoglycosidase H to determine the presence and structure of the N-linked carbohydrates. Treatment with glycopeptidase F yielded a sharper and faster-migrating band on a Western blot (Fig. 2B, lane 2). The apparent size of the protein core (36 kDa) was consistent with the calculated molecular weight of HveCt. Endoglycosidase H digestion resulted in the appearance of a broad band between 45 and 38 kDa (Fig. 2B, lane 4). An increased amount of enzyme in the reaction did not alter this pattern, suggesting that digestion was complete under these conditions (data not shown). This finding indicated that several glycosylation sites of HveCt generated in insect cells were used; however, the number of complex or high-mannose-type carbohydrates on each protein was variable. The isoelectric point of HveCt as determined by isoelectric focusing gel analysis was 6.6, which correlates with the theoretical value of 6.39 (data not shown).
FIG. 2.
Biochemical characterization of HveCt. (A) Silver stain of HveCt after nickel chromatography purification and SDS-PAGE under denaturing and reducing conditions. Sizes of the molecular weight markers (M) are indicated in kilodaltons. (B) After SDS-PAGE in denaturing and reducing conditions, proteins were transferred to nitrocellulose and detected with antipeptide serum R145. Lanes 1 and 3, purified HveCt used as mock-digested controls. In lane 2, N-linked carbohydrates of the purified protein were digested by glycopeptidase (Glyco) F; in lane 4, purified HveCt was treated with endoglycosidase (Endo) H. (C) Mass spectrometric analysis of purified HveCt. The calculated mass of singly charged species is indicated in kilodaltons. (D) Purified HveCt (24 μM in PBS) was loaded on a Superdex 200 size exclusion column and eluted with PBS. Elution profiles monitored by A280 is shown. Calculated size is based on positions of molecular size standards, indicated in kilodaltons.
By mass spectrometric analysis, the molecular mass of the HveCt glycoprotein was 40.7 kDa, but the broadness of the peak suggested considerable heterogeneity (Fig. 2C), probably reflecting the variability of HveCt glycosylation. The same variability was observed when HveCt was separated on a size exclusion column (Fig. 2D). In such experiments, the size of the eluted protein was 176 kDa, suggesting that it can oligomerize in solution. The observed molecular size was consistent with the presence of an oligomer made up of four HveCt molecules.
Interaction between purified gD and HveC in vitro.
We previously showed that HveC expression by CHO cells allowed HSV entry into these otherwise nonpermissive cells (18). This observation is reminiscent of the role played by HveA in the same system (32). Since HveC allowed entry into CHO cells in the absence of coexpression of HveA, it was hypothesized to play a similar role as a gD-binding cellular receptor.
To more precisely analyze the molecular interaction underlying the biological activity of HveCt, we performed ELISA to study the direct binding between HveCt and virion glycoproteins, and in particular gD. In a preliminary experiment 96-well plates were coated with increasing amounts of HveCt and then incubated with various concentrations of soluble gD(306t) [in this study, gD(306t) refers to the truncated protein derived from the HSV-1 KOS strain unless stated otherwise]. Saturation of the plate, based on maximal gD binding, was achieved at an HveCt concentration of 200 nM (data not shown). This concentration was used in subsequent experiments described below.
Specificity of gD for different viral receptors.
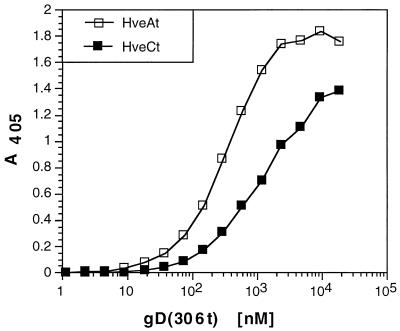
We previously showed that HveA bound to gD in vitro (ELISA) and to viral particles (36, 53). This interaction was specific since soluble forms of neither the human immunodeficiency virus receptor CD4, the Rous sarcoma virus receptor Tva, nor the mannose-6-phosphate receptor bound to gD (53). Here we found that gD directly interacted with HveC in vitro (Fig. 3). In this experiment, ELISA plates were saturated with HveCt or HveAt and incubated with gD at concentrations ranging from 1 nM to 20 μM. Binding of HveCt with gD was saturable at a concentration of 10 to 20 μM gD, whereas in the case of HveAt, saturation was achieved at a lower gD concentration (2 to 3 μM) as reported previously (53). The apparent affinity, based on half-maximal binding, seemed to be slightly higher for gD(306t) binding to HveAt (KD = 0.3 μM) than to HveCt (KD = 1 μM). In addition, the slopes of these two curves were different, which might reflect differences in complex formation. These observations led us to perform experiments to further delineate the differences as well as the similar aspects of binding of HveC and HveA to gD.
FIG. 3.
Comparison of gD binding to HveCt and HveAt. ELISA plates coated with HveCt or HveAt at 200 nM in PBS were incubated with increasing concentrations of gD(306t). Bound gD was detected with antiserum R7 followed by peroxidase-conjugated secondary antibody and substrate. Absorbance was read at 405 nm.
Characterization of the interaction between gD and HveC in vitro. (i) Specificity for gD.
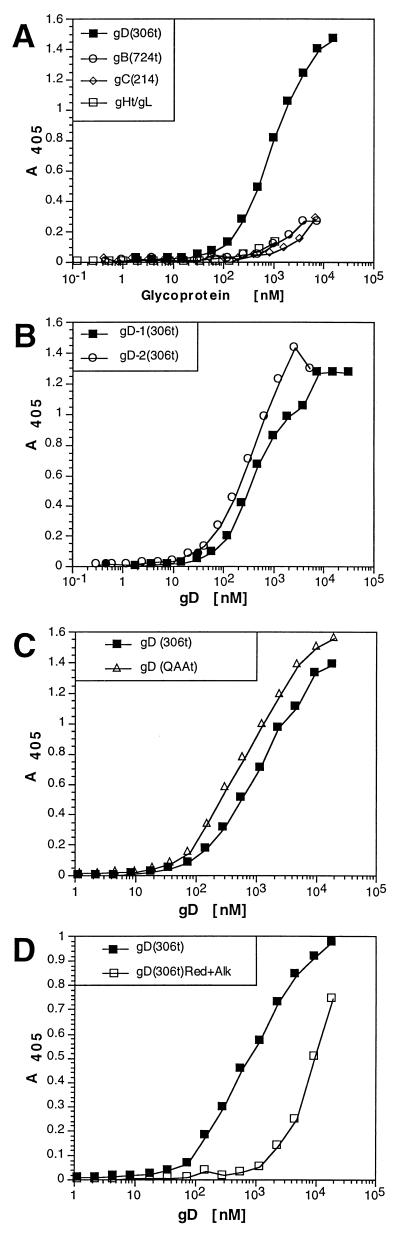
Several HSV glycoproteins are involved in virion entry into cells. To explore the possibility that HveCt could bind to other HSV-1 glycoproteins, we performed ELISA using soluble forms of several other envelope glycoproteins (i.e., gB, gC, and gH-gL). These immunoaffinity-purified proteins display native immunoreactivity when tested with several MAbs which recognize conformational epitopes (12, 37, 38). HveCt-coated plates were incubated with increasing concentrations of gD-1(306t), gC-1(457t), gB-1(724t), or gH(792t)-gL, and bound glycoproteins were detected with specific antibodies. Only gD displayed significant binding to HveCt (Fig. 4A).
FIG. 4.
Analysis of binding of gD to HveCt by ELISA. Ninety-six-well plates were saturated with 50 μl of 200 nM HveCt in PBS and incubated with variable concentrations of purified HSV glycoproteins. (A) Glycoproteins bound to immobilized HveC were detected with specific antibodies (R7 for gD, R47 for gC, R69 for gB, and R137 for gH-gL) followed by peroxidase-conjugated secondary antibody and substrate. (B) gD-1(306t)KOS and gD-2(306t)333 at various concentrations were incubated on HveCt-coated plates. (C) Mutant gD (QAAt) lacking N-CHO was compared to the glycosylated control gD(306t) for binding to immobilized HveCt. (D) Purified gD-1(306t) was reduced and alkylated prior to incubation on the HveCt-coated plate. Rabbit polyclonal serum R7 was used to detect any type of gD.
(ii) gD-1 versus gD-2.
The external domain of gDs from HSV-1 strain KOS and HSV-2 strain 333 share 88% identity, with 35 differences in amino acid sequence scattered throughout the gD ectodomain. Binding of baculovirus-produced gD-2(306t) to HveCt was compared to binding of gD-1(306t) by ELISA (Fig. 4B). Despite considerable sequence difference, the two glycoproteins bound to HveCt equally well, consistent with the ability of both HSV-1 and HSV-2 to utilize HveCt to enter cells (18).
(iii) Glycosylation.
Sodora et al. (47) constructed a triple mutant of gD-1 KOS, named gD(QAA), in which the three signals for addition of N-CHO were eliminated. Virus carrying this mutated gD displayed normal infectivity in vitro and in vivo (48, 50). In addition, the truncated form gD(QAAt) expressed in baculovirus interacted with HveA as well as the glycosylated gD(306t) (53). Here we used gD(QAAt) to assess the implication of N-linked carbohydrates in gD binding to HveCt (Fig. 4C). The interaction of N-CHO-free protein gD(QAAt) to HveCt was not significantly altered compared with the glycosylated counterpart gD(306t).
(iv) Native structure.
When gD(306t) was denatured by heating in the presence of a reducing agent and alkylated to prevent refolding, its binding to HveCt was significantly reduced (Fig. 4D). The denatured gD binding curve suggested a different kind of interaction, presumably less specific or possibly restricted to a linear portion of HveC resulting in a lower-affinity binding. This result correlates with the observations that maintenance of the gD structure is necessary for biological activity (29, 37). These data highlighted several similarities in the binding of HveCt and HveAt (53) to gD. Both receptors interacted with gD-1 and gD-2; in both cases, binding was not affected by N-glycosylation of gD and the native structure of gD was required for efficient binding.
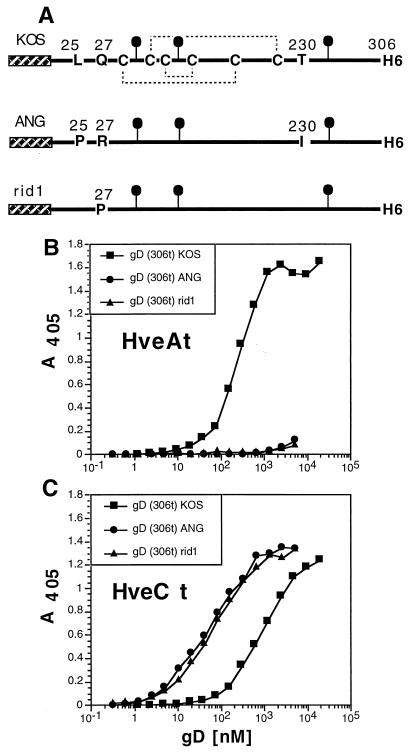
Comparison of variant gDs binding to HveC and HveA. (i) ANG and rid1 mutants.
HSV-1 entry into CHO cells was enhanced by expression of HveC regardless of the HSV strain tested (18). In contrast, HveA expression did not render CHO cells permissive to infection with an HSV strain such as rid1, rid2, or ANG (32). The first two strains carry a single mutation at amino acid 27 of gD (Q27P or Q27R, respectively), allowing these viruses to escape from gD KOS-mediated interference (11). HSV-1(ANG) gD has three mutations in its ectodomain (L25P, Q27R, and T230I) as well as several in the cytoplasmic tail (24). Truncated forms of gD(rid1t) and gD(ANGt) were produced in the baculovirus expression system and antigenically characterized (37). These forms of gD, renamed gD(306t)rid1 and gD(306t)ANG for coherence, were tested for their binding to HveA and HveC in comparison with gD(306t)KOS (Fig. 5). Consistent with the infection data, neither form of gD interacted with HveA, as shown previously (reference 53 and Fig. 5B). In striking contrast, these two proteins showed an enhanced ability to bind HveCt compared with binding of the control gD(306t)KOS (Fig. 5C). The shapes of the two curves were similar, suggesting that all three forms of gD interacted similarly with HveC.
FIG. 5.
Binding of gD(306t) from HSV strains KOS, ANG, and rid1 to HveCt. (A) gDs from both HSV (ANG) and HSV(rid1) were expressed in baculovirus as truncated forms and affinity purified. Cysteine residues on gD(306t) from KOS wild-type strain as well as mutated residues are indicated. The hatched box represents the mellitin signal peptide, and the black lollipops represent N-linked carbohydrates. gDs from these strains were compared to gD KOS for binding to immobilized HveAt (B) or HveCt (C). Antiserum R7 was used to detect bound gD in these ELISAs.
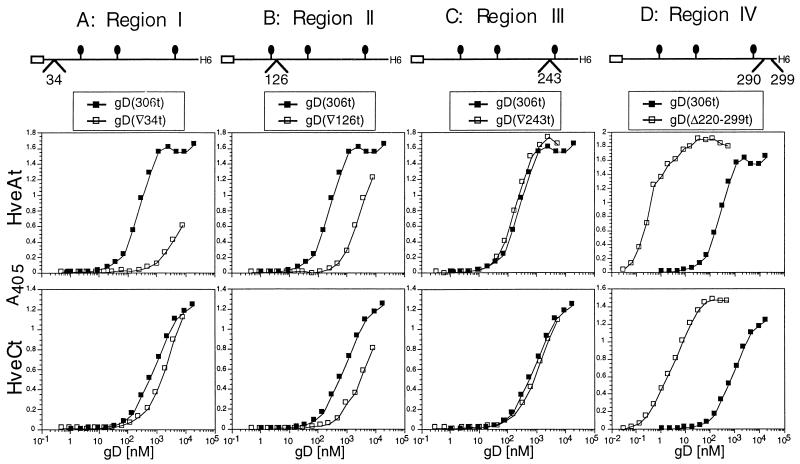
(ii) gD mutated in functional regions.
Four functional regions on gD have been identified by linker insertion mutagenesis based on the inability of the mutated full-length glycoprotein to restore infectivity of a gD-null virus (6). Linker insertions after amino acids 34, 126, and 243 disrupted functional regions I, II, and III, respectively. Functional region IV was altered by the substitution of amino acids 290 to 299 with the linker. These mutated gDs were expressed by recombinant baculoviruses (37) and tested for binding to HveAt (57) and HveCt. Disruption of functional region I in gD(▿34t) very significantly reduced gD binding to HveAt, whereas this mutation marginally affected binding to HveCt (Fig. 6A). The mutation in functional region II harbored by gD(▿126t) induced a 10-fold decrease in binding to either HveCt or HveAt (Fig. 6B). The gD(▿243t) mutant (functional region III) displayed normal binding to both receptors (Fig. 6C). Thus, the inability of gD(▿243) to perform its role during infection does not correlate with a defect in interaction with either receptor. gD(▵290-299t), altered in functional region IV, was previously shown to exhibit enhanced binding to HveAt (40, 53, 57). Here we found that gD(▵290-299t) was similarly enhanced in its binding to HveC (Fig. 6D). For both receptors, binding was increased approximately 100-fold compared to wild-type gD(306t).
FIG. 6.
Effects of linker insertions in functional regions of gD on binding to HveCt and HveAt. ELISA plates were saturated with HveAt (top panels) or HveCt (bottom panels) and incubated with various concentrations of purified mutant gDs. Binding of each mutant is compared to binding of wild-type gD(306t) (black squares). gD(▿34t) is mutated in functional region I (A), gD(▿126t) is mutated in region II (B), gD(▿243t) is mutated in region III (C), and gD(Δ290-299) is mutated in functional region IV (D). The position of the linker insertion is schematically represented for each mutant. Antiserum R7 was used to detect bound gD.
Taken together, the results show that three of the four changes in gD had similar effects on binding to both cellular proteins. In contrast, the mutation in functional region I had very different effects on binding to HveA versus HveC, confirming that integrity of this region is crucial for HveA binding (32, 36, 53) but is not necessary for HveC binding.
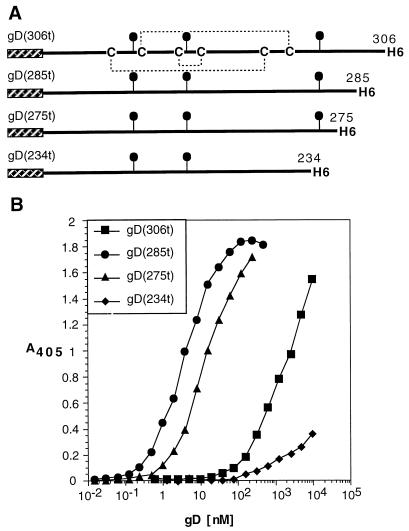
(iii) C-terminal gD truncations.
To further map the sites on gD involved in binding to HveCt, larger C-terminal truncations of HSV-1 gD (Fig. 7A) were generated (40) and tested for HveCt binding by ELISA. Mutant gD(285t) lacking part of functional region IV showed an enhanced binding capacity compared to gD(306t) (Fig. 7B). Similarly, the shorter version, gD(275t), bound HveCt better than gD(306t) (Fig. 7B). In contrast, truncation after amino acid 234 significantly decreased the ability of the shorter gD(234t) to bind HveCt (Fig. 7B). These data indicated that the region between amino acids 234 and 275 was crucial for binding to HveC, whereas the region downstream of amino acid 285 altered binding with HveC and affected the affinity of the interaction. Similar observations are made concerning the ability of HveA to bind with these truncated forms of gD (40).
FIG. 7.
Effect of C-terminal truncation on binding to HveC. (A) Shorter versions of gD-1 KOS were produced in the baculovirus expression system, purified, and tested for binding to HveC. (B) ELISA was performed with HveCt bound to the plate and incubated with variable amounts of gD. Bound gD was detected with antiserum R7.
Interaction of soluble HveCt with gD at the surface of viral particles.
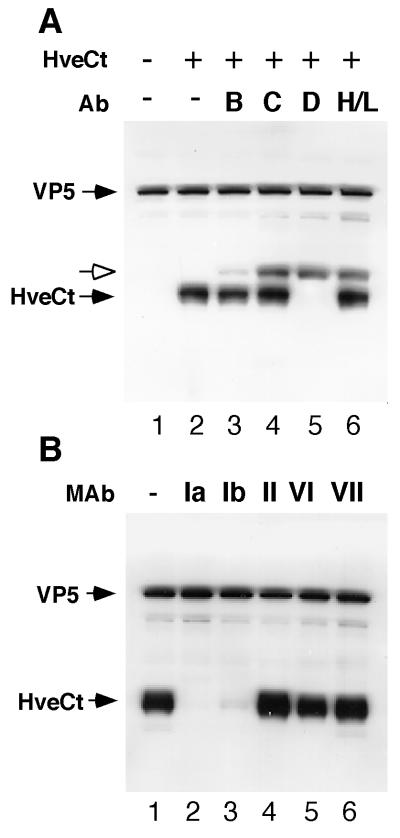
After demonstrating binding of HveCt with truncated gD by ELISA, we analyzed the interaction of HveCt with gD on the surface of viral particles. We previously showed that incubation of purified HSV-1 KOS virions with soluble HveCt could block virus entry (18). To assess direct binding of soluble HveCt to viral particles, we used the cosedimentation assay developed by Nicola et al. (36) for studying the HveAt-HSV interaction. Here HveCt was cosedimented with purified virions through a sucrose step gradient. The virus band at the 30%-60% sucrose boundary was collected and analyzed by Western blotting (Fig. 8). Presence of the virus in this fraction was demonstrated by probing the blot for the major capsid protein VP5. HveCt was also detected in this fraction when incubated with the virus prior to centrifugation (Fig. 8A, lane 2), indicating that HveCt bound directly to virions. To confirm that gD is the target for HveC binding to virions, we attempted to block the HveCt-virion interaction with antibodies directed against several HSV-1 glycoproteins. Identical aliquots of purified virions were pretreated separately with cocktails of monoclonal and polyclonal antibodies directed against gB, gC, gD, or gH-gL (lanes 3 to 6). Only anti-gD antibodies prevented cosedimentation of HveC with virus, as revealed by the absence of HveC in the virus fraction (lane 5). Antibodies directed against gB, gC, or gH-gL did not compete with HveCt. Thus, gD is the target for HveCt binding on the viral envelope.
FIG. 8.
Binding of HveCt to HSV particles is blocked by anti-gD antibodies. (A) Purified HSV-1 KOS virions (107 PFU) were incubated at 4°C for 2 h with (lane 2) or without (lane 1) HveCt (150 μg) and loaded onto a sucrose gradient. The viral band was collected and analyzed by SDS-PAGE and Western blotting. Membranes were probed for presence of VP5 and HveCt (R154 serum). In blocking experiments, virions were preincubated with cocktails of antibodies (Ab) specific for HSV glycoproteins: for gB, SS10 (0.5 μl of ascites), DL16 (5 μg of IgG), DL21 (5 μg of IgG), and R69 (0.5 μl of serum) (lane 3); for gC, MP1 (0.5 μl of ascites), MP5 (5 μg of IgG), 1C8 (5 μg of IgG), and R46 (0.5 μl of serum) (lane 4); for gD, 1D3 (0.5 μl of ascites), DL2 (5 μg of IgG), DL11 (5 μg of IgG), and R7 (0.5 μl of serum) (lane 5); for gH-gL, LP11 (0.5 μl of ascites), 53S (5 μg of IgG), H6 (5 μg of IgG), and R137 (0.5 μl of serum) (lane 6). Rabbit Ig heavy chain is detected by goat anti-rabbit secondary antibody and is indicated with a white arrow. (B) Prior to cosedimentation with HveCt, purified HSV-1 KOS virions (107 PFU) were preincubated with 50 μg of a monoclonal IgG (HD1 [group Ia], DL11 [group Ib], DL6 [group II], DL2 [group VI], or 1D3 [group VII]) during 1 h at 37°C. Untreated control is shown in lane 1.
The same cosedimentation assay was used to define which regions of gD were important for HveC binding. Monoclonal IgGs that recognize distinct antigenic sites of the gD molecule were used to pretreat virions before incubation with HveCt (Fig. 8B). MAbs HD1 and DL11, from antigenic groups Ia and Ib, respectively, prevented attachment of HveCt to viral particles (lanes 2 and 3). In contrast, MAbs DL6, DL2, and 1D3, mapping to antigenic sites II, VI, and VII, respectively, did not affect HveCt binding to purified virions (lanes 4 to 6). This finding suggests that antigenic sites Ia and Ib on gD overlap regions important for HveC binding. The pattern of blocking by this panel of MAbs is different from that observed previously for HveAt in that HveAt binding to virion was blocked by MAbs in groups Ib and VII but not Ia (36).
gD-HveCt complex formation in solution.
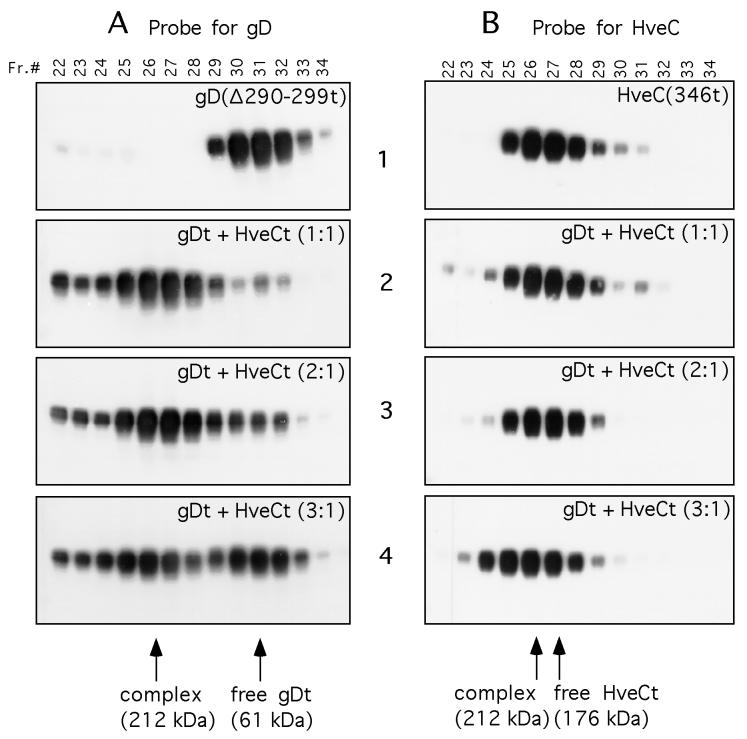
We next investigated the capacity of purified gDt and HveCt to form a complex in solution. In this assay, the high-affinity gD(Δ290-299t) and HveCt were incubated together in PBS overnight at 4°C prior to loading on a gel filtration column (Superdex 200). Eluted fractions were collected and subjected to Western blot analysis. Membranes were probed for gD (Fig. 9A) or HveC (Fig. 9B). Since anti-gD polyclonal serum R7 is much more sensitive than anti-HveC peptide serum R145, direct quantitative comparison of band intensities between Fig. 9A and B is not possible. When gD(Δ290-299t) was run alone on the Superdex column, it eluted with an apparent size of 61 kDa (Fig. 9A1) confirming the formation of a dimer in solution (15). When equimolar amounts of HveC and gD were mixed, gD eluted at a higher molecular mass (176 kDa) and coeluted with HveC (Fig. 9A2 and B2). This shift in gD elution probably reflected the formation of a gD-HveC complex in solution as reported for other protein-protein interactions (10).
FIG. 9.
Gel filtration chromatography of the HveC-gD complex. Purified HveCt and gD(Δ290-299t) were loaded independently or mixed at the indicated ratio on a Superdex 200 column. Elution was performed with PBS and monitored by measuring UV absorption at 280 nm. Fractions (Fr.) of 0.5 ml were collected and analyzed by SDS-PAGE in denaturing and reducing conditions. After protein transfer, blots were probed with serum R7 to detect gD (A) or R145 to detect HveC (B). Sizes of complexes were calculated according to elution of standards used to calibrate the column. Purified HveCt (A1) or gD(Δ290-299t) (B1) was diluted to 20 μM in PBS and loaded on the column. Panels A2 and B2 show protein elution from a column loaded with gD(Δ290-299t) (20 μM) and HveCt (20 μM) premixed overnight at 4°C in PBS. The initial molar ratio of gDt monomer to HveCt monomer is 1:1. Panels A3 and B3 show protein elution from a column loaded with gD(Δ290-299t) (20 μM) and HveCt (10 μM) mixed overnight at 4°C in PBS. The initial molar ratio of gD to HveC is 2:1. Panels A4 and B4 show protein elution from a column loaded with gD(Δ290-299t) (30 μM) and HveCt (10 μM) mixed overnight at 4°C in PBS. The initial molar ratio of gD to HveC is 3:1.
To begin to address the question of stoichiometry of both components in this complex, we followed the same approach used to analyze the gD-HveA soluble complex (53). Here HveCt and gD(Δ290-299t) were mixed at various molar ratios prior to gel filtration, and column fractions were analyzed by Western blotting. When gD(Δ290-299t) was mixed with HveCt at a 2:1 molar ratio (Fig. 9A3 and B3), the majority of the gD again coeluted with HveC at a higher molecular mass. A limited amount of free gDt (dimer) was also detected by Western blotting, but no gD peak (A280) was visible on the elution profile (data not shown). In contrast, when the molar ratio of gDt to HveCt was 3:1 (Fig. 9A4 and B4), a significant excess of free gD dimers was detected in fractions 30 to 33 (Fig. 9A4) corresponding to a now visible A280 peak of free gDt (data not shown). A simple interpretation of the data is that at an initial ratio of two gDt molecules to one HveCt molecule the maximal amount of gD that could be incorporated in the complex was present, leading to a minimal amount of free gDt (10). This would suggest a stoichiometry close to 2 gDt:1 HveCt in the complex formed in solution. Quantification of silver-stained proteins in the complex (fractions 26 in Fig. 9A3, A4, B3, and B4) after SDS-PAGE indicated a gD/HveCt molar ratio of 1.6:1 (data not shown). This finding indicated that as many as two gD molecules might bind on each HveC molecule, although saturation of the receptor in solution was difficult to achieve under these conditions. In any case, this ratio was very different from the 1 gD:2 HveA ratio determined previously for the interaction of gD with HveA under similar conditions (53). One puzzling observation was that the apparent size of the gD-HveCt complex was not significantly larger than that of HveCt alone unless gD was in excess. One possibility is that HveCt conformation and/or oligomerization was altered by gD binding such that its elution properties changed. For instance, a two-dimer complex of HveCt (176 kDa) (Fig. 9B1) could be disrupted upon binding to gD to form a complex of one HveCt dimer and two gD dimers with a calculated size of 210 kDa (212 kDa observed) (Fig. 9A4 and B4). Since HveC is highly glycosylated, it could also be possible that these posttranslational modifications have different effects on HveC elution (44) in the presence or absence of gD.
DISCUSSION
Recently, three proteins that could mediate entry of HSV into normally nonpermissive CHO cells have been identified. The first isolated protein, HVEM/HveA, is a member of the tumor necrosis factor receptor family (32). PRR1/HveC and PRR2/HveB belong to the Ig superfamily (13, 18, 30, 52). HveC allowed entry of HSV into CHO cells in absence of HveA expression, suggesting functional similarity (18). We showed that HveC, although structurally unrelated to HveA, also bound to HSV gD. We used 40-kDa HveCt purified from the culture supernatant of recombinant baculovirus-infected cells to analyze the interaction between HveC and gD by ELISA, on the virion, and in solution. Our results were compared with the HveA-gD binding data (36, 40, 53, 57) and revealed both similarities and differences in HveC and HveA interaction with HSV gD.
HveCt forms a complex with gD.
We demonstrated direct, specific, and saturable binding of gD(306t) to HveCt by ELISA. Among other glycoproteins involved in viral entry, gD was shown to be the target for HveCt binding to HSV particles since only anti-gD antibodies were able to block cosedimentation of HveCt with KOS virus. We also showed in vitro that N-CHO on gD were irrelevant for binding to HveCt whereas the native structure of gD was critical for this interaction. Despite a number of amino acid differences in their extracellular domains, gDs from HSV-1 and HSV-2 were able to bind similarly to HveCt, reflecting the ability of both HSV-1 and HSV-2 to use HveC during viral entry (18).
Although these general characteristics of the gD-HveCt interaction are shared by the gD-HveAt interaction (36, 53), other aspects of these interactions were not identical. First, ELISA binding curves were different when gD(306t) was tested with each receptor, and the affinity for gD(306t) appeared to be three to four times higher in the case of HveA. Second, there were differences in the slopes of the binding curves which might also reflect differences in complex formation. Third, saturation of HveA occurred at a lower concentration of gD, suggesting that HveA might bind less gD in the complex than HveC.
Binding of HveC to gD from HSV strains ANG and rid1.
Some HSV-1 strains, such as rid1 and ANG, were able to enter CHO cells expressing HveC, but they could not use HveA for entry into these cells (18, 32). Consistent with this finding, gD(306t)ANG and gD(306t)rid1 from these strains could not bind to HveAt in vitro (53) but did bind to HveCt. Interestingly, Nicola et al. (35) found that gD(306t) from the ANG or rid1 strain displayed an enhanced blocking of HSV KOS infection of Vero cells compared to the homologous gD(306t)KOS. This enhanced inhibitory effect was explained by a possible stronger binding of the variant forms of gD to a common receptor used by all three strains on Vero cells. Our present ELISA results suggest that HveC could be such a receptor; however, its expression on primate cells has not yet been analyzed.
Domains of gD involved in HveC or HveA interaction.
The N-terminal region of gD turned out to be important for HveA but not HveC binding. This was demonstrated by the study of both natural (ANG and rid1) and artificial [gD(▿34t)] mutants.
Competition experiments with anti-gD MAbs also pointed out that the N-terminal region of gD was not directly involved in HveC binding as it was shown to be in HveA binding. A group VII MAb, 1D3, which recognizes amino acid residues 11 to 19, blocked the interaction between HveAt and HSV KOS virions (36). The same MAb did not block the cosedimentation of HveCt with KOS viral particles. A group Ia MAb did not interfere with HveA binding to HSV particles, suggesting that epitope Ia overlaps with the HveC binding site but not with the HveA binding site.
Binding of a MAb (DL11) of group Ib, on the contrary, was able to block both HveA and HveC interaction with virions (reference 36 and this study), indicating that there might be a common domain involved in the interaction of both receptors with gD. Together with previous studies on HveA (36, 40), these data indicate that such a domain might be part of the DL11 epitope located between amino acids 234 and 275. Direct binding competition studies using both soluble receptors are in progress to determine if their binding sites within the group Ib epitope are overlapping.
gD-HveC complex formation.
Previous results showed that the proportion of gD to HveAt in the complex in solution was 1:2 (53). We performed similar gel filtration experiments to begin to determine the stoichiometry of the gD-HveC complex in solution. By Western blotting, we detected residual free gD when the initial ratio was 2:1, whereas excess of free gD was present when the initial ratio was 3:1. This finding suggested a maximum stoichiometry of two gD molecules per HveC molecule in the complex. Quantification of each protein in the column fraction containing the complex was carried out by SDS-PAGE and silver staining using purified standards of each protein. The ratio of gDt to HveCt was 1.6 (data not shown). The fact that the ratio was less than 2 indicated that saturation of HveC by gD in solution was difficult to achieve under our conditions. Whether HSV penetration requires complete receptor saturation remains to be examined.
Estimation of the binding of soluble HveCt and gDt in solution was complicated due to HveC oligomerization. HveCt appeared to form a four-molecule complex in solution, based on the size of 176 kDa (4 × 44 kDa) observed in our gel filtration experiments. Surprisingly, addition of gD to the HveC double dimer did not drastically increase the size of the complex as expected. A possible explanation is that binding of gDt in solution disrupted the HveCt double dimer. A complex containing one HveCt dimer and two gDt dimers, the size of which would be approximately 210 kDa (176/2 + 2 × 61), is consistent with the 212-kDa complex observed in the presence of excess gD. The closely related molecule mPRR2 (33), the murine homolog of PRR2/HveB, was shown to play a role in cell adhesion by self attachment (1). Aoki et al. (1) propose that mPRR2 from different cells associate in inverted positions. By analogy, truncated HveCt might consist of two dimers bound in inverted position. On the cell surface HveC might be dimeric, where it would be targeted by HSV gD without a need for gD to break a tetrameric complex. Experiments to refine our data on complex formation (affinity, stoichiometry, and kinetics) are now in progress.
In this study we showed that a new HSV receptor, HveC, allows viral entry by directly interacting with gD, as shown previously for HveA. These structurally unrelated gD receptors differ in formation and stoichiometry of the complex with gD, as well as in regions on gD involved in the interaction with each receptor. However, these two receptors possibly share a common binding domain and display similar alterations in their binding to several mutated gDs. Further experiments are required to determine if the mechanism of action of HveA and HveC is interchangeable in a unique process leading to viral entry or if the observed differences account for two distinct entry pathways.
ACKNOWLEDGMENTS
This investigation was supported by Public Health Service grants NS-30606 from the National Institute of Neurological Diseases and Stroke (R.J.E. and G.H.C.) and AI-18289 (G.H.C. and R.J.E.), AI-07325 (A.V.N.), AI-30040 (J.D.L.), and AI-36293 (P.G.S) from the National Institute of Allergy and Infectious Diseases.
We thank William Moore and Lynn Spruce from the protein chemistry laboratory of the School of Medicine of the University of Pennsylvania, supported by core grants of the Diabetes and Cancer Centers (DK-19525 and CA-16520), for their help for mass spectrometric analysis and peptide synthesis. We are grateful to Tao Peng for the purified gH-gL proteins and to Manuel Ponce de Leon and Charline Peng for excellent technical assistance. We also thank Sharon Willis and Ann Rux for critical reading of the manuscript.
REFERENCES
- 1.Aoki J, Koike S, Asou H, Ise I, Suwa H, Tanaka T, Miyasaka M, Nomoto A. Mouse homolog of poliovirus receptor-related gene 2 product, mPRR2, mediates homophilic cell aggregation. Exp Cell Res. 1997;235:374–384. doi: 10.1006/excr.1997.3685. [DOI] [PubMed] [Google Scholar]
- 2.Brunetti C R, Burke R L, Hoflack B, Ludwig T, Dingwell K S, Johnson D C. Role of mannose-6-phosphate receptors in herpes simplex virus entry into cells and cell-to-cell transmission. J Virol. 1995;69:3517–3528. doi: 10.1128/jvi.69.6.3517-3528.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buckmaster E A, Gompels U, Minson A. Characterisation and physical mapping of an HSV-1 glycoprotein of approximately 115 × 10(3) molecular weight. Virology. 1984;139:408–413. doi: 10.1016/0042-6822(84)90387-8. [DOI] [PubMed] [Google Scholar]
- 4.Cai W, Gu B, Person S. Role of glycoprotein B of herpes simplex virus type 1 in viral entry and cell fusion. J Virol. 1988;62:2596–2604. doi: 10.1128/jvi.62.8.2596-2604.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campadelli-Fiume G, Arsenakis M, Farabegoli F, Roizman B. Entry of herpes simplex virus 1 in BJ cells that constitutively express viral glycoprotein D is by endocytosis and results in degradation of the virus. J Virol. 1988;62:159–167. doi: 10.1128/jvi.62.1.159-167.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiang H-Y, Cohen G H, Eisenberg R J. Identification of functional regions of herpes simplex virus glycoprotein gD by using linker-insertion mutagenesis. J Virol. 1994;68:2529–2543. doi: 10.1128/jvi.68.4.2529-2543.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen G H, Dietzschold B, Ponce de Leon M, Long D, Golub E, Varrichio A, Pereira L, Eisenberg R J. Localization and synthesis of an antigenic determinant of herpes simplex virus glycoprotein D that stimulates production of neutralizing antibody. J Virol. 1984;49:102–108. doi: 10.1128/jvi.49.1.102-108.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohen G H, Isola V J, Kuhns J, Berman P W, Eisenberg R J. Localization of discontinuous epitopes of herpes simplex virus glycoprotein D: use of a nondenaturing (“native” gel) system of polyacrylamide gel electrophoresis coupled with Western blotting. J Virol. 1986;60:157–166. doi: 10.1128/jvi.60.1.157-166.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohen G H, Ponce de Leon M, Diggelmann H, Lawrence W C, Vernon S K, Eisenberg R J. Structural analysis of the capsid polypeptides of herpes simplex virus types 1 and 2. J Virol. 1980;34:521–531. doi: 10.1128/jvi.34.2.521-531.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cunningham B C, Ultsch M, de Vos A M, Mulkerrin M G, Clauser K R, Wells J A. Dimerization of the extracellular domain of the human growth hormone receptor by a single hormone molecule. Science. 1991;254:821–825. doi: 10.1126/science.1948064. [DOI] [PubMed] [Google Scholar]
- 11.Dean H J, Terhune S S, Shieh M, Susmarski N, Spear P G. Single amino acid substitutions in gD of herpes simplex virus 1 confer resistance to gD-mediated interference and cause cell-type-dependent alterations in infectivity. Virology. 1994;199:67–80. doi: 10.1006/viro.1994.1098. [DOI] [PubMed] [Google Scholar]
- 12.Dubin G, Jiang H. Expression of herpes simplex virus type 1 glycoprotein L (gL) in transfected mammalian cells: evidence that gL is not independently anchored to cell membranes. J Virol. 1995;69:4564–4568. doi: 10.1128/jvi.69.7.4564-4568.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eberlé F, Dubreuil P, Mattei M-G, Devilard E, Lopez M. The human PRR2 gene, related to the poliovirus receptor gene (PVR), is the true homolog of the murine MPH gene. Gene. 1995;159:267–272. doi: 10.1016/0378-1119(95)00180-e. [DOI] [PubMed] [Google Scholar]
- 14.Eisenberg R J, Ponce de Leon M, Friedman H M, Fries L F, Frank M M, Hastings J C, Cohen G H. Complement component C3b binds directly to purified glycoprotein C of herpes simplex virus types 1 and 2. Microb Pathog. 1987;3:423–435. doi: 10.1016/0882-4010(87)90012-x. [DOI] [PubMed] [Google Scholar]
- 15.Eisenberg R J, Ponce de Leon M, Pereira L, Long D, Cohen G H. Purification of glycoprotein gD of herpes simplex virus types 1 and 2 by use of monoclonal antibody. J Virol. 1982;41:1099–1104. doi: 10.1128/jvi.41.3.1099-1104.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forrester A, Farrell H, Wilkinson G, Kaye J, Davis-Poynter N, Minson T. Construction and properties of a mutant of herpes simplex virus type 1 with glycoprotein H coding sequences deleted. J Virol. 1992;66:341–348. doi: 10.1128/jvi.66.1.341-348.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedman H M, Cohen G H, Eisenberg R J, Seidel C A, Cines D B. Glycoprotein C of herpes simplex virus 1 acts as a receptor for the C3b complement component on infected cells. Nature (London) 1984;309:633–635. doi: 10.1038/309633a0. [DOI] [PubMed] [Google Scholar]
- 18.Geraghty R J, Krummenacher C, Eisenberg R J, Cohen G H, Spear P G. Entry of alphaherpesviruses mediated by poliovirus receptor related protein 1 and poliovirus receptor. Science. 1998;280:1618–1620. doi: 10.1126/science.280.5369.1618. [DOI] [PubMed] [Google Scholar]
- 19.Handler C G, Cohen G H, Eisenberg R J. Cross-linking of glycoprotein oligomers during herpes simplex virus type 1 entry. J Virol. 1996;70:6076–6082. doi: 10.1128/jvi.70.9.6076-6082.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Handler C G, Eisenberg R J, Cohen G H. Oligomeric structure of glycoproteins in herpes simplex virus type 1. J Virol. 1996;70:6067–6075. doi: 10.1128/jvi.70.9.6067-6070.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herold B C, Visalli R J, Sumarski N, Brandt C, Spear P G. Glycoprotein C-independent binding of herpes simplex virus to cells requires cell surface heparan sulfate and glycoprotein B. J Gen Virol. 1994;75:1211–1222. doi: 10.1099/0022-1317-75-6-1211. [DOI] [PubMed] [Google Scholar]
- 22.Herold B C, WuDunn D, Soltys N, Spear P G. Glycoprotein C of herpes simplex virus type 1 plays a principal role in the adsorption of virus to cells and in infectivity. J Virol. 1991;65:1090–1098. doi: 10.1128/jvi.65.3.1090-1098.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Isola V J, Eisenberg R J, Siebert G R, Heilman C J, Wilcox W C, Cohen G H. Fine mapping of antigenic site II of herpes simplex virus glycoprotein D. J Virol. 1989;63:2325–2334. doi: 10.1128/jvi.63.5.2325-2334.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Izumi K M, Stevens J G. Molecular and biological characterization of a herpes simplex virus type 1 (HSV-1) neuroinvasiveness gene. J Exp Med. 1990;172:487–496. doi: 10.1084/jem.172.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson D C, Burke R L, Gregory T. Soluble forms of herpes simplex virus glycoprotein D bind to a limited number of cell surface receptors and inhibit virus entry into cells. J Virol. 1990;64:2569–2576. doi: 10.1128/jvi.64.6.2569-2576.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson D C, Ligas M W. Herpes simplex viruses lacking glycoprotein D are unable to inhibit virus penetration: quantitative evidence for virus-specific cell surface receptors. J Virol. 1988;62:4605–4612. doi: 10.1128/jvi.62.12.4605-4612.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaner R J, Baird A, Mansukhani A, Basilico C, Summers B D, Florkiewicz R Z, Hajjar D P. Fibroblast growth factor receptor is a portal of cellular entry for herpes simplex virus type 1. Science. 1990;248:1410–1413. doi: 10.1126/science.2162560. [DOI] [PubMed] [Google Scholar]
- 28.Lee W C, Fuller A O. Herpes simplex virus type 1 and pseudorabies virus bind to a common saturable receptor on Vero cells that is not heparan sulfate. J Virol. 1993;67:5088–5097. doi: 10.1128/jvi.67.9.5088-5097.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Long D, Wilcox W C, Abrams W R, Cohen G H, Eisenberg R J. Disulfide bond structure of glycoprotein D of herpes simplex virus types 1 and 2. J Virol. 1992;66:6668–6685. doi: 10.1128/jvi.66.11.6668-6685.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lopez M, Eberlé F, Mattei M-G, Gabert J, Birg F, Bardin F, Maroc C, Dubreuil P. Complementary DNA characterization and chromosomal localization of a human gene related to the poliovirus receptor-encoding gene. Gene. 1995;155:261–265. doi: 10.1016/0378-1119(94)00842-g. [DOI] [PubMed] [Google Scholar]
- 31.Mauri D N, Ebner R, Kochel K D, Montgomery R I, Cheung T C, Yu G-L, Murphy M, Eisenberg R J, Cohen G H, Spear P G, Ware C F. LIGHT, a new member of the TNF superfamily, and lymphotoxin (LT) α are ligands for herpesvirus entry mediator (HVEM) Immunity. 1998;8:21–30. doi: 10.1016/s1074-7613(00)80455-0. [DOI] [PubMed] [Google Scholar]
- 32.Montgomery R I, Warner M S, Lum B J, Spear P G. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell. 1996;87:427–436. doi: 10.1016/s0092-8674(00)81363-x. [DOI] [PubMed] [Google Scholar]
- 33.Morrison M E, Racaniello V R. Molecular cloning and expression of a murine homolog of the human poliovirus receptor gene. J Virol. 1992;66:2807–2813. doi: 10.1128/jvi.66.5.2807-2813.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muggeridge M I, Isola V J, Byrn R A, Tucker T J, Minson A C, Glorioso J C, Cohen G H, Eisenberg R J. Antigenic analysis of a major neutralization site of herpes simplex virus glycoprotein D, using deletion mutants and monoclonal antibody-resistant mutants. J Virol. 1988;62:3274–3280. doi: 10.1128/jvi.62.9.3274-3280.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nicola A V, Peng C, Lou H, Cohen G H, Eisenberg R J. Antigenic structure of soluble herpes simplex virus (HSV) glycoprotein D correlates with inhibition of HSV infection. J Virol. 1997;71:2940–2946. doi: 10.1128/jvi.71.4.2940-2946.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nicola A V, Ponce de Leon M, Xu R, Hou W, Whitbeck J C, Krummenacher C, Montgomery R I, Spear P G, Eisenberg R J, Cohen G H. Monoclonal antibodies to distinct sites on the herpes simplex virus (HSV) glycoprotein D block HSV binding to HVEM. J Virol. 1998;72:3595–3601. doi: 10.1128/jvi.72.5.3595-3601.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nicola A V, Willis S H, Naidoo N N, Eisenberg R J, Cohen G H. Structure-function analysis of soluble forms of herpes simplex virus glycoprotein D. J Virol. 1996;70:3815–3822. doi: 10.1128/jvi.70.6.3815-3822.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peng T, Ponce de Leon M, Jiang H, Dubin G, Lubinski J, Eisenberg R J, Cohen G H. The gH-gL complex of herpes simplex virus (HSV) stimulates neutralizing antibody and protects mice against HSV type 1 challenge. J Virol. 1998;72:65–72. doi: 10.1128/jvi.72.1.65-72.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rux A H, Moore W T, Lambris J D, Abrams W R, Peng C, Friedman H M, Cohen G H, Eisenberg R J. Disulfide bond structure determination and biochemical analysis of glycoprotein C from herpes simplex virus. J Virol. 1996;70:5455–5465. doi: 10.1128/jvi.70.8.5455-5465.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rux A H, Willis S H, Nicola A V, Hou W, Peng C, Lou H, Cohen G H, Eisenberg R J. Functional region IV of glycoprotein D from herpes simplex virus modulates glycoprotein binding to the herpesvirus entry mediator. J Virol. 1998;72:7091–7098. doi: 10.1128/jvi.72.9.7091-7098.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Samanta S, Eisenberg R J, Cohen G H. Abstract of the 19th International Herpesvirus Workshop, Vancouver, British Columbia. 1994. Studies of monomeric and oligomeric forms of HSV gB, abstr. 29. [Google Scholar]
- 42.Seidel-Dugan C, Ponce de Leon M, Friedman H M, Fries L F, Frank M M, Cohen G H, Eisenberg R J. C3b receptor activity on transfected cells expressing glycoprotein C of herpes simplex virus types 1 and 2. J Virol. 1988;62:4027–4036. doi: 10.1128/jvi.62.11.4027-4036.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shieh M-T, WuDunn D, Montgomery R I, Esko J D, Spear P G. Cell surface receptors for herpes simplex virus are heparan sulfate proteoglycans. J Cell Biol. 1992;116:1273–1281. doi: 10.1083/jcb.116.5.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shire S J. Analytical ultracentrifugation and its use in biotechnology. In: Schuster T M, Laue T M, editors. Modern analytical ultracentrifugation. Boston, Mass: Birkhäuser; 1994. pp. 261–297. [Google Scholar]
- 45.Showalter S D, Zweig M, Hampar B. Monoclonal antibodies to herpes simplex virus type 1 proteins, including the immediate-early protein ICP 4. Infect Immun. 1981;34:684–692. doi: 10.1128/iai.34.3.684-692.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sisk W P, Bradley J D, Leipold R J, Stoltzfus A M, Ponce de Leon M, Hilf M, Peng C, Cohen G H, Eisenberg R J. High-level expression and purification of secreted forms of herpes simplex virus type 1 glycoprotein gD synthesized by baculovirus-infected insect cells. J Virol. 1994;68:766–775. doi: 10.1128/jvi.68.2.766-775.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sodora D L, Cohen G H, Muggeridge M I, Eisenberg R J. Absence of asparagine-linked oligosaccharides from glycoprotein D of herpes simplex virus type 1 results in a structurally altered but biologically active protein. J Virol. 1991;65:4424–4431. doi: 10.1128/jvi.65.8.4424-4431.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sodora D L, Eisenberg R J, Cohen G H. Characterization of a recombinant herpes simplex virus which expresses a glycoprotein D lacking asparagine-linked oligosaccharides. J Virol. 1991;65:4432–4441. doi: 10.1128/jvi.65.8.4432-4441.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spear P G. Membrane fusion induced by herpes simplex virus. In: Bentz J, editor. Viral fusion mechanisms. Boca Raton, Fla: CRC Press, Inc.; 1993. pp. 201–232. [Google Scholar]
- 50.Tal-Singer R, Eisenberg R J, Valyi-Nagy T, Fraser N W, Cohen G H. N-linked oligosaccharides on herpes simplex virus glycoprotein gD are not essential for establishment of viral latency or reactivation in the mouse eye model. Virology. 1994;202:1050–1053. doi: 10.1006/viro.1994.1437. [DOI] [PubMed] [Google Scholar]
- 51.Tessier D C, Thomas D Y, Khouri H E, Laliberte F, Vernet T. Enhanced secretion from insect cells of a foreign protein fused to the honeybee melittin signal peptide. Gene. 1991;98:177–183. doi: 10.1016/0378-1119(91)90171-7. [DOI] [PubMed] [Google Scholar]
- 52.Warner, M. S., W. Martinez, R. J. Geraghty, R. I. Montgomery, J. C. Whitbeck, R. Xu, R. J. Eisenberg, G. H. Cohen, and P. G. Spear. A cell surface protein with herpesvirus entry activity (HveB) confers susceptibility to infection by herpes simplex virus type 2, mutants of herpes simplex virus type 1 and pseudorabies virus. Virology, in press. [DOI] [PubMed]
- 53.Whitbeck J C, Peng C, Lou H, Xu R, Willis S H, Ponce de Leon M, Peng T, Nicola A V, Montgomery R I, Warner M S, Soulika A M, Spruce L A, Moore W T, Lambris J D, Spear P G, Cohen G H, Eisenberg R J. Glycoprotein D of herpes simplex virus (HSV) binds directly to HVEM, a member of the tumor necrosis factor receptor superfamily and a mediator of HSV entry. J Virol. 1997;71:6083–6093. doi: 10.1128/jvi.71.8.6083-6093.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Whitbeck, J. C., G. H. Cohen, and R. J. Eisenberg. 1998. Unpublished data.
- 55.Williams A. A year in the life of the immunoglobulin superfamily. Immunol Today. 1987;8:298–303. doi: 10.1016/0167-5699(87)90016-8. [DOI] [PubMed] [Google Scholar]
- 56.Willis S H, Peng C, Ponce de Leon M, Nicola A V, Rux A H, Cohen G H, Eisenberg R J. Expression and purification of secreted forms of herpes simplex virus glycoproteins from baculovirus-infected insect cells. In: Brown M S, MacLean A R, editors. Methods in molecular medicine: herpes simplex virus protocols. Vol. 10. Totowa, N.J: Humana Press; 1997. pp. 131–156. [DOI] [PubMed] [Google Scholar]
- 57.Willis S H, Rux A H, Peng C, Whitbeck J C, Nicola A V, Lou H, Hou W, Salvador L, Eisenberg R J, Cohen G H. Examination of the kinetics of herpes simplex virus glycoprotein D binding to the herpesvirus entry mediator, using surface plasmon resonance. J Virol. 1998;72:5937–5947. doi: 10.1128/jvi.72.7.5937-5947.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.WuDunn D, Spear P G. Initial interaction of herpes simplex virus with cells is binding to heparan sulfate. J Virol. 1989;63:52–58. doi: 10.1128/jvi.63.1.52-58.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]