Abstract
Immune checkpoint inhibitors (ICIs) that target programmed cell death protein 1 (PD-1) and programmed death-ligand 1 (PD-L1) have shown modest activity as monotherapies for the treatment of ovarian cancer (OC). The rationale for using these therapies in combination with poly (ADP-ribose) polymerase inhibitors (PARP-Is) has been described, and their in vivo application will benefit from ex vivo platforms that aid in the prediction of patient response or resistance to therapy. This study examined the effectiveness of detecting patient-specific immune-related activity in OC using three-dimensional (3D) spheroids. Immune-related cell composition and PD-1/PD-L1 expression status were evaluated using cells dissociated from fresh OC tissue from two patients prior to and following 3D culture. The patient sample with the greatest increase in the proportion of PD-L1 + cells also possessed more activated cytotoxic T cells and mature DCs compared to the other patient sample. Upon cytokine stimulation, patient samples demonstrated increases in cytotoxic T cell activation and DC major histocompatibility complex (MHC) class-II expression. Pembrolizumab increased cytokine secretion, enhanced olaparib cytotoxicity, and reduced spheroid viability in a T cell-dependent manner. Furthermore, durvalumab and olaparib combination treatment increased cell death in a synergistic manner. This work demonstrates that immune cell activity and functional modulation can be accurately detected using our ex vivo 3D spheroid platform, and it presents evidence for their utility to demonstrate sensitivity to ICIs alone or in combination with PARP-Is in a preclinical setting.
Supplementary information
The online version contains supplementary material available at (10.1007/s00262-021-02849-z)
Keywords: Immune checkpoint inhibitors, PARP inhibitors, Ovarian cancer, Spheroid, 3D cultures
Introduction
Ovarian cancer (OC) is the leading cause of death for women with gynecologic cancer in the USA [1]. Surgical debulking followed by chemotherapy is the current standard of care, yet most patients become resistant resulting in a five-year survival rate below 50%. To elicit long-term disease remission, the incorporation of new therapies into the current treatment paradigm and personalized testing methods to define patient therapy usage are under considerable investigation.
Immunotherapies have revolutionized the treatment of many solid tumors and there exists a rationale for their use in OC. OC patients with tumor-infiltrating lymphocytes (TILs) display a significant improvement in five-year survival compared to patients without TILs. This positive correlation between survival and immune cell recruitment to the tumor provides compelling evidence that antitumor immune surveillance is an important determinant for OC clinical outcomes [2–4] and suggests the immunogenic nature of OC could be exploited as a treatment option by using immune checkpoint inhibitors (ICIs), such as those that target programmed cell death protein 1 (PD-1) and programmed death-ligand 1 (PD-L1). Unfortunately, the reports of OC patient response to ICI therapy have generally been underwhelming with response rates less than 10% and no current FDA approval [5, 6]. It is unclear if this drug class is simply ineffective against OC or if the preclinical research to date is hindering the translation of ICI efficacy to the clinic. This may be remedied by the development of more complex in vitro models that facilitate better understanding of the microenvironment and improved drug testing [7].
Despite the evidence against their clinical utility in OC, both preclinical and clinical studies of ICI combinations with drugs such as poly(ADP-ribose) polymerase inhibitors (PARP-Is) continue [8–10]. PARP-Is have shown impressive clinical activities for OC patients [11]. However, intrinsic and acquired resistance often limit their effectiveness as monotherapies [12]. The role PARP-Is play as immune modulators to enhance checkpoint blockade efficacy has recently emerged [11, 13, 14]. A phase I/II clinical trial demonstrated that the PARP-I, niraparib, in combination with pembrolizumab produced complete or partial responses in 18% of patients with recurrent platinum-resistant OC compared to less than a 5% response rate with niraparib alone [9]. Further understanding of the immune modulatory capacity of anti-PD-1/PD-L1 inhibitors alone and in combination with PARP-Is will enhance our knowledge of what drives sensitivity for solid tumor indications, including OC.
To extend immunotherapy research to a broader range of solid tumors, we have modified an existing ex vivo OC 3D spheroid assay, EV3D™, to detect the potential synergy between anti-PD-1/PD-L1 inhibitors, pembrolizumab or durvalumab, in combination with the PARP-I, olaparib [15]. This work builds upon previous studies through the inclusion and characterization of autologous immune cells. Immune composition and function were evaluated prior to monitoring therapy-related changes in spheroid phenotypes and viability. Overall patient-specific differences in immune composition and drug response were examined.
Methods
Generation of 3D spheroids
Written informed consent was obtained from patients in accordance with the Institutional Review Board (IRB) approved biology protocols by Prisma Health Cancer Institute (IRB-Committee C). Live tissue was received within 24 h of surgery and dissociated to single cells via mechanical and enzymatic dissociation performed according to EV3D™ assay protocols (KIYATEC, Inc., South Carolina, USA). Briefly, mechanical and enzymatic dissociation preceded agitation over 1–2 h. The process was completed by filtration, and, if necessary, red blood cell lysis. Cells were cryopreserved until ready for use. Spheroids were generated as previously described [15]. Briefly, cells were seeded in KIYA PREDICT™ media (KIYATEC, Inc., South Carolina, USA) in 384-well round-bottom, ultra-low attachment plates (Corning Inc., New York, USA) and centrifuged at 500 × g for five minutes then placed in a 37 °C incubator at 5% CO2. KIYA PREDICT™ media includes DMEM, fetal bovine serum, and penicillin/streptomycin, without specific growth factor supplementation.
Spheroid drug response assay
Given the prevalence of drug resistance and altered drug penetration for 3D cultures [16, 17], tested drug concentrations for experiments were in the micromolar or microgram per milliliter range. For pembrolizumab and olaparib combination studies, 100 μg/mL pembrolizumab (SelleckChem, Texas, USA), 50 μM or 100 μM olaparib (MedChemExpress, New Jersey, USA) for OVC33 or OVC45, respectively, were added alone or together with KIYA PREDICT™ media as no treatment control or 0.2% DMSO as vehicle control. Viability was determined after 48 h. For durvalumab (Selleckchem, Texas, USA) and olaparib combination studies, spheroids were treated with olaparib for 48 h, followed by durvalumab for an additional 72 h. For direct pembrolizumab treatment of T cells, CD3 + cells were separated from dissociated bulk tumor cells using the EasySep CD3 + Selection kit II (StemCell Technologies, Vancouver, Canada) and incubated in the presence or absence of 300 μg/mL pembrolizumab to saturate all PD-1 sites. T cells were then added to the bulk cells and seeded for 3D spheroid culture. Viability was determined after 48 h. Viability readouts were conducted using CellTiter-Glo® 3D Cell Viability Assay (Promega, Wisconsin, USA), and relative luminescence units (RLUs) were recorded using a TECAN infinite M1000pro (TECAN, Mannedorf, Switzerland).
Flow cytometry
Spheroids were resuspended and incubated in ACCUTASE™ (StemCell Technologies, Vancouver, Canada) to facilitate dissociation. Dissociated cells were washed in PBS and resuspended in FACs buffer (2% FBS, 2 mM EDTA, in PBS). Antibodies and dilutions used are listed in Table 1. Antibodies were added and incubated for 10 min at 4 °C. Samples were washed, centrifuged then resuspended in FACs buffer. DRAQ 7 (BD Pharmingen™, New Jersey, USA) dead cell dye was added for dead cell detection and exclusion. Samples were analyzed using the CytoFLEX LX flow cytometer and software (Beckman Coulter, California, USA). Percent of parent was graphed and evaluated for statistics using GraphPad Prism (GraphPad Software, California, USA).
Table 1.
Antibodies used in present study
| Antibody | Supplier | Catalog Number | Application | Dilution |
|---|---|---|---|---|
| CD45-FITC | Miltenyi | 130–110-769 | FCM | 1:100 |
| PD-L1-PE | BD Biosciences | 557,924 | FCM | 1:20 |
| EpCAM-PerCP-Vio700 | Miltenyi | 130–111-120 | FCM | 1:100 |
| CD3-APC | Miltenyi | 130–113-135 | FCM | 1:100 |
| CD4-FITC | Miltenyi | 130–114-531 | FCM | 1:100 |
| CD8-PerCP-Vio700 | Miltenyi | 130–110-682 | FCM | 1:100 |
| CD25-PE | Miltenyi | 130–115-534 | FCM | 1:100 |
| CD69-APC-Vio770 | Miltenyi | 130–112-616 | FCM | 1:100 |
| PD-1-PE-Vio770 | Miltenyi | 130–117-698 | FCM | 1:100 |
| CD11 c-APC-Vio770 | Miltenyi | 130–114-111 | FCM | 1:100 |
| HLA-DR-APC | Miltenyi | 130–111-943 | FCM | 1:100 |
| CD103-PE | Miltenyi | 130–111-985 | FCM | 1:100 |
| REA control FITC | Miltenyi | 130–113-437 | FCM | 1:100 |
| mouse IgG1k PE | BD Biosciences | 555,749 | FCM | 1:20 |
| REA control (S) PerCP- | Miltenyi | 130–113-441 | FCM | 1:100 |
| Vio700 | ||||
| REA control (S) APC | Miltenyi | 130–113-434 | FCM | 1:100 |
| REA control (S) FITC | Miltenyi | 130–113-437 | FCM | 1:100 |
| REA control (S) PerCP- | Miltenyi | 130–113-441 | FCM | 1:100 |
| Vio700 | ||||
| REA control (S) PE | Miltenyi | 130–113-438 | FCM | 1:100 |
| REA control (S) APC- | Miltenyi | 130–113-435 | FCM | 1:100 |
| Vio770 | ||||
| mouse IgG2b PE-Vio 770 | Miltenyi | 130–096-825 | FCM | 1:20 |
| REA control (S) APC- | Miltenyi | 130–113-435 | FCM | 1:100 |
| Vio770 | ||||
| REA control (S) APC | Miltenyi | 130–113-434 | FCM | 1:100 |
| REA control (S) PE | Miltenyi | 130–113,438 | FCM | 1:100 |
| pan Cytokeratin | Abcam | ab86134 | IHC | 1:500 |
| CD11c | Abcam | ab52632 | IHC | 1:50 |
| PD-1 | Abcam | ab170190 | IHC | 1:100 |
| CD8 | Abcam | ab17147 | IHC | 1:50 |
| PD-L1 | Abcam | ab210931 | IHC | 1:1000 |
| mouse IgG1 kappa | Abcam | ab91353 | IHC | 1:50 |
| recombinant rabbit IgG | Abcam | ab172730 | IHC | 1:50 |
| mouse IgG1 kappa | Abcam | ab170190 | IHC | 1:100 |
| mouse IgG2a kappa | Abcam | ab18415 | IHC | 1:1000 |
| CD8 | Abcam | ab4055 | IF | 1:100 |
| CD11c | Abcam | ab254183 | IF | 1:50 |
| PD-L1 | Abcam | ab205921 | IF | 1:200 |
| Alexa fluor 594 | Life Technologies | A11012 | IF | 1:500 |
| Alexa fluor 488 | Life Technologies | A11029 | IF | 1:500 |
Immunohistochemistry
Upon receipt of fresh tissue, a portion was removed during mechanical dissociation and immediately fixed in formalin for 48 h and processed as previously described [15]. The fixed tissue was embedded in paraffin and 10-µm sections were mounted onto glass slides. Following hematoxylin and eosin staining, slides were cover-slipped using Permount medium. According to antibody specifications, rehydration and antigen retrieval were performed using citrate buffer pH 6.0 (Abcam, Cambridge, UK) or Tris–EDTA buffer pH 9.0 (Abcam, Cambridge, UK). Antibodies and dilutions used are listed in Table 1. Antibody staining was visualized using Mouse and Rabbit Specific HRP/DAB IHC Detection Kit-Micro-polymer (Abcam, Cambridge, UK). Bright-field images were acquired at 40X using Invitrogen™ EVOS™ M7000 Imaging System (Thermo Fisher Scientific, Massachusetts, USA).
Immunofluorescence
Spheroids were fixed in 3.7% formaldehyde, washed in FACs buffer, and cytospun to adhere cells to glass slides. Cells were permeabilized using 0.3% Triton X-100 in PBS, incubated in blocking buffer (0.1% bovine serum albumin, 0.2% Triton X-100, 10% goat serum and 0.05% Tween 20) for one hour followed by primary antibody in a humidifier at 4 °C overnight. Antibodies and dilutions used are listed in Table 1. Following primary antibody incubation, cells were washed with blocking buffer and then incubated with secondary antibodies in the dark for one hour. Cells were washed with blocking buffer, nuclei were stained, and slides were mounted with a cover slip using Fluoroshield mounting medium with DAPI (Abcam, Cambrdige, UK).
Cytokine stimulation
T cell conditioned media (T cell CM) was used as a source of cytokines to stimulate immune-related functions. Separated CD3 + cells were expanded in ImmunoCult-XF T cell Expansion Medium (StemCell Technologies, Vancouver, Canada) according to manufacturer’s recommendations. Briefly, for the initiation of T cell expansion, ImmunoCult Human CD3/CD28 T cell Activator (StemCell Technologies, Vancouver, Canada) was added to growth medium with 10 ng/mL interleukin-2 (IL-2) (Sigma, Missouri, USA). Expanded T cells were pelleted, and the T cell CM was aliquoted and stored at -20 °C. For 3D spheroid stimulation, spheroids were formed overnight, and the T cell CM was added at a 1:1 ratio the following day and incubated for 48 or 72 h.
Cytokine detection
Human Discovery Immunotherapy Fixed Panel Magnetic Luminex performance assay was purchased from R&D Systems (Minnesota, USA). Supernatant was collected at day three of spheroid culture and stored at −80 °C. Samples were processed according to manufacturer’s recommendations, and the assay plate was run on Bio-Rad Luminex® BioPLEX200™ System (Bio-Rad Laboratories, California, USA). Analyte concentrations were determined by interpolation of assay standards using GraphPad Prism (GraphPad Software, California, USA). Fold change analyte secretions were determined for pembrolizumab, olaparib, and combination by comparing them to vehicle control samples.
Statistical analysis
Statistical analysis was performed using GraphPad Prism software version 8.2.1 (GraphPad Software, California, USA). Results are expressed as the mean ± standard deviation (SD). Unpaired t tests were used to determine significance between two groups. Unpaired one-way ANOVA with multiple comparisons was used to determine significance across three or more groups. Combenefit software generated concentration responses and Loewe synergy indices from calculated percent viability normalized to vehicle control [18].
Results
Patient tumor tissues display a non-desert phenotype
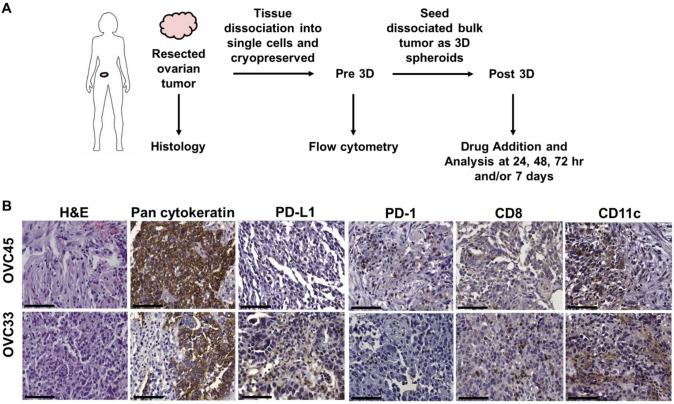
Two newly diagnosed, treatment-naïve serous OC patient samples matched in stage (IIIC) and grade (high) were chosen for testing. Patient samples were characterized from tissue resection through 3D spheroid culture. Cells were characterized following spheroid culture (Post 3D) and compared back to the original cell composition found Pre 3D (Fig. 1a). Histological analysis of the tissues verified the immune composition for both patient samples tested, OVC45 and OVC33 (Fig. 1a and supplementary Fig. 1). Both samples were composed primarily of tumor cells as identified by pan cytokeratin staining. PD-1 + and CD8 + cells were distributed throughout both tissues along with CD11c + dendritic cells (DCs). While OVC33 stained more positive for PD-L1 expression compared to OVC45, the staining in general was very diffuse and faint. Given detection of both cytotoxic T cells and DCs, both tissues were classified as non-desert.
Fig. 1.
Tissue processing schematic and characterization. (a) Schematic of tissue processing for characterization. Flow cytometry was conducted to evaluate cell composition following tissue dissociation and prior to 3D culture. Immunofluorescence and flow cytometry were conducted at different timepoints to evaluate 3D spheroid culture and drug treatment effects on cell populations. (b) Representative images of formalin-fixed paraffin embedded tissues. Pan-cytokeratin was used to determine the presence of epithelial cells within the tumor tissues. Additional immune-related markers selected to assess infiltration were PD-L1, PD-1, CD8, and CD11c. Representative images are from two independent experiments. Scale bar = 75 μm
An increased proportion of PD-L1 + tumor cells were detected following ex vivo 3D spheroid culture
To determine patient-specific similarities and the effects of 3D cell culture, the Pre 3D cellular composition was compared to the Post 3D cellular composition. Live tumor cells were assessed via flow cytometry for EpCAM expression after dead cell exclusion within a gating region defined as the “tumor gate” (supplementary Fig. 2c/d). The percentage of EpCAM + cells was normalized to the “tumor gate.” Thus, the data reveal an abundance of EpCAM- cells present within the “tumor gate” which could include EpCAM- tumor cells or other cell types. Importantly, there was no significant change in the proportion of total EpCAM+ cells Post 3D, indicating no potentially negative impact on tumor cell presence within the 3D cultures (supplementary Fig. 3). When EpCAM + cells were further analyzed for PD-L1 expression, OVC45 had more Pre 3D EpCAM+/PD-L1- cells compared to OVC33 (Fig. 2a). Interestingly, both patient samples had significant increases in EpCAM+/PD-L1+tumor cells following spheroid culture, with OVC33 demonstrating the greatest increase in this population.
Fig. 2.
Characterization of PD-L1 expression Pre 3D and Post 3D. (a) Representative data of tumor cells assessed using EpCAM as a marker. Quantification of PD-L1 negative tumor cells and PD-L1+tumor cells identified Pre 3D and Post 3D for OVC45 and OVC33 is shown in the right panel. (b) Representative data of immune cells assessed using CD45 as a marker. CD45+ cells were detected within the large gate defined for tumor cells. Quantification of PD-L1 negative immune cells and PD-L1+ immune cells identified Pre 3D and Post 3D for both tumor tissues is shown in the right panel. (c) Representative data of immune cells assessed using CD45 as a marker within the lymphocyte gate. Quantification of PD-L1 negative immune cells and PD-L1+ immune cells identified Pre 3D and Post 3D for both patient samples is shown in the right panel. All percentages are expressed as a portion of the “tumor gate” or “lymphocyte gate,” respectively. Two independent experiments and three independent experiments were conducted for Pre 3D and Post 3D, respectively. Post 3D for all data shown = 48 h in 3D culture. Isotype controls (black events) were subtracted from marker values (OVC45 = blue events; OVC33 = green events). Unpaired t tests were used for sample comparison using GraphPad. Error bars reflect SD. *p < 0.05, **p < 0.01
To examine this phenotype further, the impact of T cells on EpCAM+/PD-L1+ cells was tested by culturing OVC45 and OVC33 following T cell depletion using CD3+ selection (supplementary Fig. 4). Dual EpCAM+/PD-L1+ cells were reduced by approximately 50% for OVC33 when cultured without T cells (supplementary Fig. 4b). A decrease in IFNγ levels was also detected Post 3D for OVC33 when T cells were depleted. This was not observed for OVC45 (supplementary Fig. 4c). These data suggest a T cell-dependent impact on the microenvironment within the 3D culture platform.
When evaluating the immune population, after dead cell exclusion, live CD45+ cells were detected in both the large gate defined for tumor cells and within the smaller lymphocyte gate (supplementary Fig. 2). OVC33 had significantly more Pre 3D immune cells compared to OVC45 (Fig. 2b). Approximately 20% of the CD45+ cells within the tumor gate for OVC33 were also found to be PD-L1+. The majority (greater than 95%) of CD45+ cells within the lymphocyte gate were found to be PD-L1- for both patient samples (Fig. 2c).
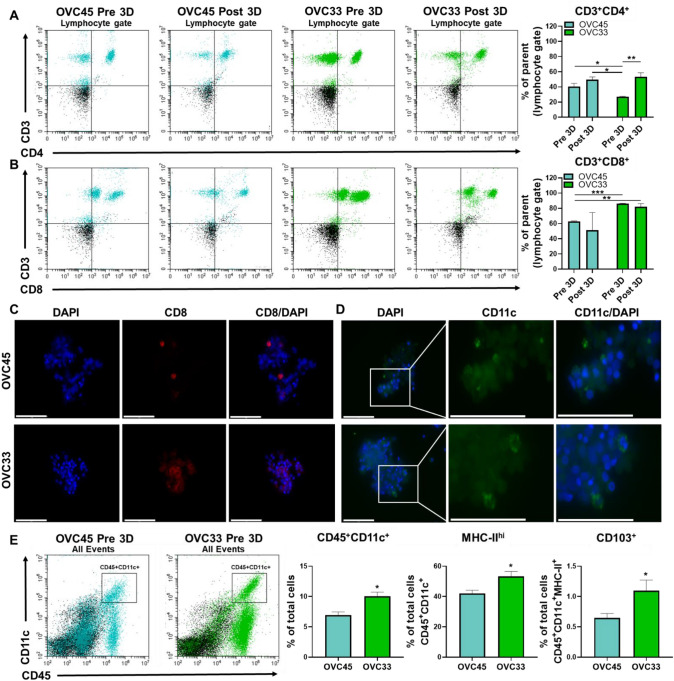
The inter-patient proportion of T cell subpopulations was determined next. An evaluation of total CD3+ cells revealed small shifts in relative amounts with spheroid culture (supplementary Fig. 3). However, OVC45 had significantly more helper T cells (CD3+/CD4+) compared to OVC33 Pre 3D (Fig. 3a) with no significant change following 3D culture for OVC45, but a significant increase for OVC33. While no significant change in cytotoxic T cells (CD3+/CD8+) following 3D culture was observed for either patient sample, OVC33 did have significantly more CD3+/CD8+T cells than OVC45 (Fig. 3b). The presence of CD8+T cells in both tissues following 3D culture was confirmed using immunofluorescence (Fig. 3c). OVC33 had clusters of CD8+cells, a morphological phenotype associated with activated T cells [19, 20].
Fig. 3.
T cell and DC populations within the 3D spheroid culture system. (a) Representative data and quantification of CD4+T cells (CD3+/CD4+). (b) Representative data and quantification of CD8+T cells (CD3+/CD8+). (c) Representative images verify the presence of CD8+cells as determined by immunofluorescence following 48 h in 3D spheroid culture. Scale bars = 75 μm. Two independent experiments and three independent experiments were conducted for Pre 3D and Post 3D, respectively. (d) The presence of DCs following 48 h in 3D spheroid culture was determined via immunofluorescence. Scale bars = 75 μm. Image inset shows a zoomed version of the same magnification. (e) Representative data for DCs defined by dual CD45+/CD11c+. The percent of dual CD45/CD11c events were determined for OVC45 and OVC33. Isotype controls (black events) were subtracted from marker values (OVC45 = blue events, OVC33 = green events). Expression of MHC-II and CD103 expression was determined for DC populations across the patient samples. Two independent experiments were conducted for OVC45, and three independent experiments were conducted for OVC33. Isotype controls were subtracted from marker values. Error bars reflect SD. Not significant = n.s., *p < 0.05, **p < 0.01, ***p < 0.005
Since T cell activation has been shown to be regulated by DCs in the tumor immune microenvironment (TIME), and DCs have been shown to play a critical role in ICI efficacy [21, 22], the presence of tumor-associated CD45+/CD11c+DCs was examined [23–25]. DCs were detected in both OVC45 and OVC33 following 3D culture confirming the ability of the spheroid system to maintain them in culture (Fig. 3d). OVC33 contained more DCs (Fig. 3e) that were found to express both higher levels of MHC class-II (MHC-II), indicating higher antigen-presenting machinery and CD103 found on DCs with a potent stimulatory impact on effector T cell priming. These results demonstrate that immune-related patient-specific differences can be detected within our spheroid system and may shift through the course of 3D cell culture in a patient-specific manner.
Differential immune cell populations are detected within ex vivo 3D spheroids
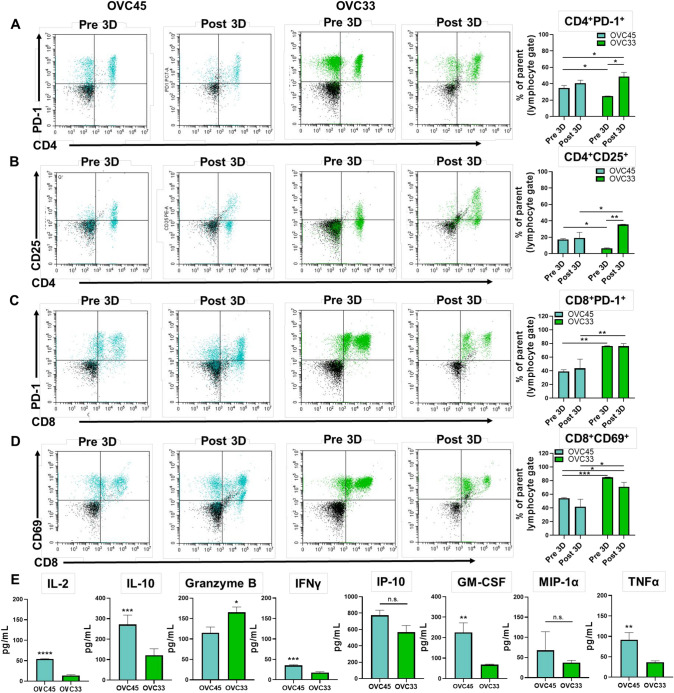
The presence of different T cell populations and markers of activation were characterized. OVC45 had greater CD4+/PD-1+cells and Tregs (CD4+/CD25+) Pre 3D (Fig. 4a) compared to OVC33 (Fig. 4b). Conversely, OVC33 had more CD8+/PD-1+ cells and activated cytotoxic T cells (CD8+/CD69+) (Fig. 4c) compared to OVC45 (Fig. 4d). The patient-specific T cell populations were proportionally stable in 3D culture for OVC45, while OVC33 had a significant increase in CD4+/PD-1+ cells and Tregs Post 3D.
Fig. 4.
Characterization of T cell populations Pre 3D and Post 3D. (a) Representative data of CD4+T cells evaluated for activation and exhaustion via PD-1 expression. (b) Tregs were identified by dual CD4+/CD25+. (c) CD8+ cytotoxic T cells were examined by analyzing expression levels of PD-1. (d) The activation status of CD8+ cytotoxic T cells was evaluated via CD69 expression. Two independent experiments were conducted for OVC45 and Pre 3D OVC33, and three independent experiments were conducted for OVC33 Post 3D. Post 3D for all data shown = 72 h in culture. Isotype controls (black events) were subtracted from marker values (OVC45 = blue events, OVC33 = green events). Unpaired t tests were used for sample comparison using GraphPad. (e) Cytokines were evaluated from supernatants collected following 72 h in 3D culture from three independent experiments for OVC45 and OVC33. Unpaired t tests were used for sample comparison using GraphPad. Error bars reflect SD. Not significant = n.s., *p < 0.05, **p < 0.01, ***p < 0.005, ****p < 0.001
The presence of cytokines known to be secreted by immune-related cells was also examined (Fig. 4e). OVC45 secreted significantly greater amounts of IL-2, IL-10, and IFNγ compared to OVC33; however, there was no significant difference in the amount of IFNγ-induced protein 10 (IP-10). Interleukin-10 (IL-10) is an immune-suppressive cytokine known to be produced by Tregs [26]. Despite the observed increase in Tregs in OVC33 Post 3D, the proportion of Tregs in OVC45 Pre 3D was greater than that of OVC33 (Fig. 4b). These results suggest the Pre 3D immune composition may be a better reflection of the detected cytokine secretion. OVC33 had significantly greater granzyme B compared to OVC45 potentially reflecting the higher proportion of activated cytotoxic T cells found in OVC33 throughout 3D culture (Fig. 4d). The presence of granulocyte–macrophage colony-stimulating factor (GM-CSF) indicates T cell activation for both patient samples [27]. Finally, the detection of macrophage inflammatory protein-1 alpha (MIP-1α) and tumor necrosis factor alpha (TNFα) in both samples may provide evidence of the presence of macrophages in the spheroids [28, 29].
Immune cell function can be enhanced through cytokine stimulation in ex vivo 3D spheroids
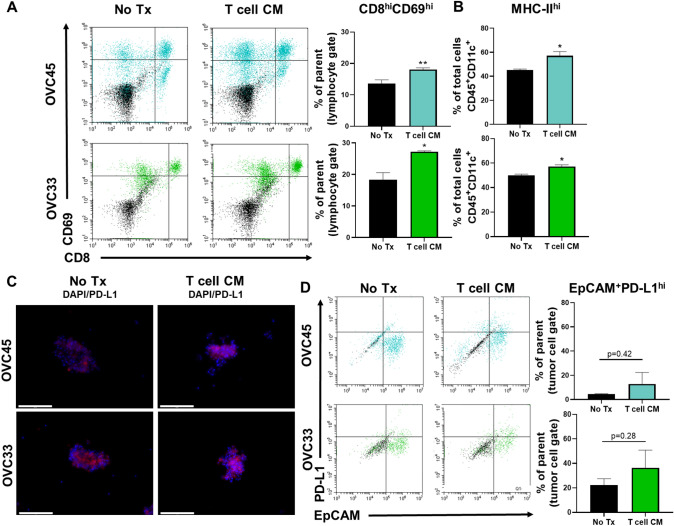
To demonstrate the ability of the immune cells within the 3D spheroids to modulate their activation status, spheroids were cultured after formation in conditioned T cell expansion medium (T cell CM). Activated T cells rapidly divide and secrete key cytokines to promote immune responses [30]. Thus, conditioned medium from the expansion of primary OC TILs was used as a source of cytokines for stimulation of the T cells resident in the 3D spheroids. By using this cytokine cocktail, different cell types and different activation mechanisms following a single treatment were evaluated. Treatment with T cell CM induced significant increases in activated cytotoxic T cells (CD8hiCD69hi) for both patient samples (Fig. 5a). T cell CM also resulted in an increase in MHC-II expression on CD45+/CD11c+ DCs from both samples (Fig. 5b). Given the detected increases in T cell activation and DC maturation, PD-L1 expression was examined. Increased PD-L1+ expression was detected by immunofluorescence for both tissues following T cell CM treatment (Fig. 5c), and this increase was associated with tumor cells specifically as an upward trend in dual EpCAM+/PD-L1+ cells was detected for both tissues (Fig. 5d). These results demonstrate that the immune cells are active in the ex vivo 3D spheroid cultures, and their function can be enhanced through treatment modulation.
Fig. 5.
Cytokine stimulation enhances immune-related function in 3D spheroid cultures. (a) Representative data and quantification of the activation status of cytotoxic T cells was defined by dual CD8 high and CD69 high positivity following no treatment or T cell CM treatment for 48 h. Quantification of activated cytotoxic T cells from three independent experiments for OVC45 and two independent experiments for OVC33. (b) Quantification of MHC-II high expression from two independent experiments. OVC45 and OVC33 is shown in blue or green, respectively. (c) Representative images of PD-L1 expression for OVC45 and OVC33 following 48 h of no treatment or T cell CM treatment. Images were taken at the same exposure. Scale bars = 125 μm. (d) Representative data showing PD-L1 expression on tumor cells defined by dual PD-L1 and EpCAM positivity. Quantification of PD-L1+ tumor cells from two independent experiments. Isotype controls (black events) were subtracted from marker values (OVC45 = blue events, OVC33 = green events). Unpaired t tests were used for sample comparison using GraphPad. Error bars reflect SD. *p < 0.05, **p < 0.01
Pembrolizumab alters T cell function, enhances olaparib efficacy, and induces T cell-dependent reduction in spheroid viability
To determine the effects of checkpoint inhibitors upon the 3D spheroids, pembrolizumab-related changes in cytokine secretion and cell viability were examined. No changes in secreted cytokines were detected in OVC45, while OVC33 had increases in granzyme B, MIP-1α, and TNFα (Fig. 6a). Spheroid viability was tested following treatment with pembrolizumab and the PARP-I, olaparib, alone or in combination. Pembrolizumab treatment alone did not result in a change in spheroid viability for either sample (Fig. 6b). OVC33 was more sensitive to olaparib monotherapy compared to OVC45. Reduced spheroid viability for OVC45 occurred only when treated with combination pembrolizumab and olaparib, and this was further evident upon spheroid visualization (Fig. 6c). OVC33 spheroids appeared less dense with less cell contact following olaparib or combination treatment (Fig. 6c). To enhance pembrolizumab efficacy, direct incubation of the T cells with pembrolizumab prior to spheroid incorporation was tested via T cell separation from the Pre 3D bulk cell suspension. For these experiments, all PD-1 sites were saturated with drug. The maximum testing concentration of pembrolizumab was selected based upon pembrolizumab’s relatively high half-life (approximately 27 days) ultimately resulting in a gradual approach to steady state in vivo. An intravenous dosing frequency of 10 mg/kg once every two weeks has a predicted pembrolizumab maximum serum concentration of approximately 200 μg/mL for advanced solid tumor cancer patients [31]. OVC33 had a significant reduction in spheroid viability only when spheroids were treated with T cells incubated with pembrolizumab (Fig. 6d). This result demonstrates that pembrolizumab treatment can reduce spheroid viability and that its efficacy is T cell dependent.
Fig. 6.
Pembrolizumab alters T cell function, enhances olaparib efficacy, and induces T cell-dependent spheroid death. (a) Cytokine secretion was determined from supernatants collected for both patient samples following VC or pembrolizumab treatment for 48 h. Supernatant was collected from three independent experiments. Depicted is the fold change in cytokine concentrations as determined by normalizing pembrolizumab treatment to VC. (b) 3D spheroids were treated with media (No Tx), VC, olaparib, pembrolizumab, or both olaparib and pembrolizumab (Combo) for 48 h. (c) Representative images of 3D spheroids following 48 h of treatment. Scale bars = 650 μm. (d) Pre 3D bulk T cells were separated and incubated with pembrolizumab. Pre 3D bulk cells were then either cultured with or without treated T cells, and after 48 h, spheroid viability was determined. Spheroid viability for all experiments was determined using CellTiter-Glo® Glo. Unpaired one-way ANOVA with multiple comparison was conducted on the mean of three independent experiments. Error bar = SD. Not significant = n.s., *p < 0.05, **p < 0.01
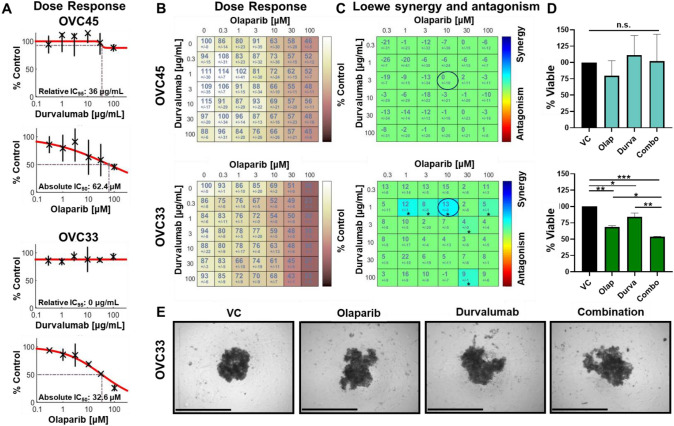
Durvalumab and olaparib synergistically reduce OVC33 spheroid viability
Since enrichment of PD-L1+ tumor cells was detected for both patient samples, the sensitivity of these samples to the anti-PD-L1 antibody durvalumab was evaluated. Clinical study data have suggested some improvement in disease control rates in OC when treated with combination durvalumab and olaparib [32]. Durvalumab in combination with olaparib was tested by sequential dosing to better mimic clinical dosing strategies and determine if drug order has an impact on ICI/PARP-I combination studies. OVC33 remained more sensitive to single agent olaparib than OVC45 (Fig. 7a). Durvalumab treatment alone did not result in a dose-dependent reduction in spheroid viability. The cross dose–response of both drugs was compared, and whether the percent viability following treatments was synergistic was examined (Fig. 7b) [18]. Six combination treatments were deemed significantly synergistic for OVC33, including 10 µM olaparib and 1 µg/mL durvalumab (Fig. 7c). Significant changes in spheroid viability were not detected for OVC45 following this same drug treatment (Fig. 7d). The half-life of durvalumab is relatively high, resulting in predicted achievable serum concentration levels greater than 10 μg/mL with a twice weekly intravenous dosing regimen [33]. These data suggest our findings may be clinically achievable. Representative images of OVC33 show decreased spheroid density and a loss of compactness and cell contact following combination therapy (Fig. 7e). Ultimately, synergistic efficacy was detected between durvalumab and olaparib treatment using our 3D spheroid culture and the response is patient specific.
Fig. 7.
Durvalumab and olaparib synergistically reduce OVC33 spheroid viability. (a) OVC45 and (b) OVC33 were treated with a range of olaparib or durvalumab, and viability was determined. Percent viability was determined by normalizing to VC. Dose–response curves and relative IC95 or absolute IC50 was determined across two independent experiments using Combenefit software. (c) Depicted are the mean and error of percent spheroid viability following olaparib and durvalumab cross dose–response treatment for OVC45 and OVC33 normalized to VC. (d) Loewe synergy and antagonism was determined from the change in spheroid viability following olaparib and durvalumab cross dose–response across two independent experiments. Synergistic combinations (blue) or antagonistic combinations (red) are only color-coded if there is statistical significance. Synergy and antagonism heat maps and significance were calculated and generated by Combenefit software. * p < 0.05. (e) A single combination is highlighted by the circled data set on the synergy heatmap for OVC45 and OVC33. (f) Representative spheroid images of OVC33 are shown post-treatment. Scale bar = 650 μm
Discussion
In this study, we have demonstrated that patient-specific, non-expanded, autologous tumor cells and immune cells can be incorporated ex vivo into 3D spheroids and monitored over the course of a week for changes in immune cell composition, activation, cytokine secretion, and drug response. Significantly, an increase in the EpCAM+/PD-L1+ population and shifts in Tregs were observed. When comparing two patient samples, significant differences in immune cell composition were also reflected in their cytokine secretion profiles and responses to olaparib, pembrolizumab, and durvalumab. These data show the ability of our immune adapted ex vivo 3D spheroid platform, EV3D™, to model patient-specific response to PARP-I/ICI combination therapy relative to each patient’s TIME.
Effective immunotherapy requires understanding the TIME as it often drives therapy response [34]. Taking advantage of the relationship between the quality and character of the TIME and response to immunotherapy has been proposed as a personalized approach for the treatment of cancer [34–36]. Generally, OC demonstrates low to modest somatic mutational burden which may explain the overall limited antitumor activity detected with ICI monotherapy in the clinic [37–40]. Yet if the OC TIME is immunologically “hot” or T cell inflammed, there is a moderate to high probability of response to anti-PD-1/PD-L1 treatment [40]. Recent reports show that OC positive for PD-L1 expression correlated with higher response to pembrolizumab [6, 41]. In our study, the patient sample with the highest PD-L1 expression, OVC33, significantly responded to anti-PD-1/PD-L1 treatment and in a T cell-dependent manner. OVC33′s TIME composition may reflect T cell exhaustion and dysfunction as it had lower levels of detected cytokines compared to OVC45, and although OVC33 had more activated cytotoxic T cells, they were predominately PD-1hi expressors. The low levels of cytokine secretion by OVC33 were found to be reversible by pembrolizumab treatment. Given its T cell profile, CD103+ DC population, and PD-L1+ cells, OVC33 may have the “right” immune composition to be reinvigorated by a PD-1/PD-L1 inhibitor. More patient samples will have to be evaluated within our 3D model to make any potential correlation between Pre 3D immune composition and response to an ICI.
PARP-Is can induce synthetic lethality in BRCA1/2-deficient OC. Interestingly, non-BRCA1/2 mutant OCs that are classified as possessing “BRCAness” qualities respond to PARP-Is [42, 43]. We did not address the role of BRCA in these 3D spheroid studies. However, olaparib has been reported to increase immune cell infiltration [2, 4]. Due to limited control of self-assembly during spheroid formation and the alterations in the original distribution of the tumor/immune landscape, we believe other 3D culture models, such as microtumor models, are more ideal to monitor therapy-induced immune cell infiltration [44]. Yet, we propose it is the TIME character and functional capabilities, not solely the spatial arrangement that may be reflective of therapeutic response making a relatively fast and high-throughput 3D spheroid system ideal for both preclinical research and future, potential clinical applications.
Despite the limitations of 3D spheroids, they provide compelling advantages for preclinical research. Previous 3D immune cultures have often relied on the incorporation of allogenic immune cells from healthy donors. The phenotype of tumor-associated immune cells has been shown to be functionally different from those found in the periphery [45], and the use of allogeneic immune cells incorporates potential issues related to non-HLA matching. Our model incorporates all cells found within a patient’s TIME making the model system ideal for preclinical research including basic biology questions and immuno-oncology drug development.
This study provides evidence for the utility of our 3D spheroid models for solid tumor immune cell research. It is our ultimate goal to translate this knowledge of two patients into a larger study comparing immune-modified EV3D™ response with clinical response in patients treated with ICIs to identify patients who will truly benefit from these often high-cost drugs. Placing a patient’s tumor cells in direct contact with a selected therapy provides a more direct response prediction than the use of more detached biomarkers such as protein expression and mutational analysis. This study provides proof of concept data for the ability of our immune-modified EV3D™ platform to measure response to single agent and combination PARP-I and ICI through the direct interaction between a patient’s cells and drug.
Conclusion
This work furthers efforts to expand in vitro testing of immuno-oncology agents and ex vivo detection methods of ICI sensitivity in solid tumors. The need for combination therapies to overcome monotherapy resistance often limits ICI utility for many tumor types. We hope to harness the power of patient-specific TIME to identify signatures relating cell composition and function to therapy response to find biomarkers that predict drug sensitivity. Ultimately, the methods developed here will be translated for the personalized, clinical prediction of ICI response to improve patient response rates and aid in the enrollment and stratification of patients in future clinical trials.
Supplementary information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors would like to thank the patients for their participation in this study. We would also like to extend our appreciation to the ITOR Biorepository at Prisma Health for their support.
Abbreviations
- 3D
Three-dimensional
- BRCA1/2
Breast-related cancer antigens 1 and 2
- DC
Dendritic cells
- GM-CSF
Granulocyte–macrophage colony-stimulating factor
- ICI
Immune checkpoint inhibitor
- IRB
Institutional Review Board
- IFNγ
Interferon gamma
- IL-2
Interleukin-2; IL-10: Interleukin-10
- IL-10
IFNγ-induced protein 10
- MIP-1α
Macrophage inflammatory protein
- MHC
Major histocompatibility complex
- MHC-II
Major histocompatibility complex class-II
- OC
Ovarian cancer
- PBMCs
Peripheral blood mononuclueated cells
- PARP-I
Poly (ADP-ribose) polymerase inhibitors
- PD-1
Programmed cell death protein 1
- PD-L1
Programmed death-ligand 1
- RLUs
Relative luminescence units
- SD
Standard deviation
- T cell CM
T cell expansion media
- TILs
Tumor-infiltrating lymphocytes
- TIME
Tumor immune microenvironment
- TNFα
Tumor necrosis factor alpha
- Tregs
Regulatory T cells
- VC
Vehicle control
Author contributions
KMA designed the study, performed the experiments and data analysis, and wrote the manuscript; AKE performed the experiments, and wrote the manuscript; KL performed flow cytometry and immunofluorescence for cytokine stimulation experiments, and provided experimental design assistance; SS provided experimental design assistance and data review; LMH provided facilities/logistics for experimental performance; TMD directed the project, provided data review, and wrote the manuscript. All authors read and approved the final manuscript.
Funding
All work for this study was funded by KIYATEC, Inc.
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Compliance with ethical standards
Conflict of interest
Dr. Appleton, Ms. Elrod, Ms. Lassahn, Mr. Shuford, Ms. Holmes, and Dr. DesRochers are current employees of KIYATEC, Inc.
Ethical approval
Written informed consent was obtained from patients in accordance with the Institutional Review Board (IRB)-approved biology protocols by Prisma Health, formally known as Greenville Health System, Cancer Institute (IRB-Committee C).
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 2.Pantelidou C, Sonzogni O, De Oliveria TM, Mehta AK, Kothari A, Wang D, et al. PARP inhibitor efficacy depends on CD8(+) T-cell recruitment via intratumoral STING pathway activation in BRCA-deficient models of triple-negative breast cancer. Cancer Discov. 2019;9(6):722–737. doi: 10.1158/2159-8290.CD-18-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang L, Conejo-Garcia JR, Katsaros D, Gimotty PA, Massobrio M, Regnani G, et al. Intratumoral T cells, recurrence, and survival in epithelial ovarian cancer. N Engl J Med. 2003;348(3):203–213. doi: 10.1056/NEJMoa020177. [DOI] [PubMed] [Google Scholar]
- 4.Zhang G, Liu C, Bai H, Cao G, Cui R, Zhang Z. Combinatorial therapy of immune checkpoint and cancer pathways provides a novel perspective on ovarian cancer treatment. Oncol Lett. 2019;17(3):2583–2591. doi: 10.3892/ol.2019.9902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ghisoni E, Imbimbo M, Zimmermann S, Valabrega G. Ovarian cancer immunotherapy: turning up the heat. Int J Mol Sci. 2019;20(12):2927. doi: 10.3390/ijms20122927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matulonis UA, Shapira-Frommer R, Santin AD, Lisyanskaya AS, Pignata S, Vergote I, et al. Antitumor activity and safety of pembrolizumab in patients with advanced recurrent ovarian cancer: results from the phase II KEYNOTE-100 study. Ann Oncol. 2019;30(7):1080–1087. doi: 10.1093/annonc/mdz135. [DOI] [PubMed] [Google Scholar]
- 7.Shuford S, Wilhelm C, Rayner M, Elrod A, Millard M, Mattingly C, et al. Prospective validation of an Ex Vivo, patient-derived 3d spheroid model for response predictions in newly diagnosed ovarian cancer. Scientific Reports. 2019;9(1):11153. doi: 10.1038/s41598-019-47578-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Z, Sun K, Xiao Y, Feng B, Mikule K, Ma X, et al. Niraparib activates interferon signaling and potentiates anti-PD-1 antibody efficacy in tumor models. Sci Rep. 2019;9(1):1853. doi: 10.1038/s41598-019-38534-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Konstantinopoulos PA, Waggoner S, Vidal GA, Mita M, Moroney JW, Holloway R, et al. Single-arm phases 1 and 2 trial of niraparib in combination with pembrolizumab in patients with recurrent platinum-resistant ovarian carcinoma. JAMA Oncol. 2019;5(8):1141–1149. doi: 10.1001/jamaoncol.2019.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farkkila A, Gulhan DC, Casado J, Jacobson CA, Nguyen H, Kochupurakkal B, et al. Immunogenomic profiling determines responses to combined PARP and PD-1 inhibition in ovarian cancer. Nat Commun. 2020;11(1):1459. doi: 10.1038/s41467-020-15315-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boussios S, Karihtala P, Moschetta M, Karathanasi A, Sadauskaite A, Rassy E, et al. Combined Strategies with Poly (ADP-Ribose) Polymerase (PARP) Inhibitors for the treatment of ovarian cancer: a literature review. Diagnostics (Basel) 2019;9(3):87. doi: 10.3390/diagnostics9030087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang X, Li X, Li W, Bai H, Zhang Z. PARP inhibitors in ovarian cancer: Sensitivity prediction and resistance mechanisms. J Cell Mol Med. 2019;23(4):2303–2313. doi: 10.1111/jcmm.14133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ding L, Kim HJ, Wang Q, Kearns M, Jiang T, Ohlson CE, et al. PARP inhibition elicits sting-dependent antitumor immunity in brca1-deficient ovarian cancer. Cell Rep. 2018;25(11):2972–2980. doi: 10.1016/j.celrep.2018.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zimmer AS, Nichols E, Cimino-Mathews A, Peer C, Cao L, Lee MJ, et al. A phase I study of the PD-L1 inhibitor, durvalumab, in combination with a PARP inhibitor, olaparib, and a VEGFR1-3 inhibitor, cediranib, in recurrent women's cancers with biomarker analyses. J Immunother Cancer. 2019;7(1):197. doi: 10.1186/s40425-019-0680-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shuford S, Wilhelm C, Rayner M, Elrod A, Millard M, Mattingly C, et al. Prospective validation of an ex vivo, patient-derived 3d spheroid model for response predictions in newly diagnosed ovarian cancer. Sci Rep. 2019;9(1):11153. doi: 10.1038/s41598-019-47578-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Riedl A, Schlederer M, Pudelko K, Stadler M, Walter S, Unterleuthner D, et al. Comparison of cancer cells in 2D vs 3D culture reveals differences in AKT-mTOR-S6K signaling and drug responses. J Cell Sci. 2017;130(1):203–218. doi: 10.1242/jcs.188102. [DOI] [PubMed] [Google Scholar]
- 17.Kapalczynska M, Kolenda T, Przybyla W, Zajaczkowska M, Teresiak A, Filas V, et al. 2D and 3D cell cultures-a comparison of different types of cancer cell cultures. Arch Med Sci. 2018;14(4):910–919. doi: 10.5114/aoms.2016.63743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Di Veroli GY, Fornari C, Wang D, Mollard S, Bramhall JL, Richards FM, et al. Combenefit: an interactive platform for the analysis and visualization of drug combinations. Bioinformatics. 2016;32(18):2866–2868. doi: 10.1093/bioinformatics/btw230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ingulli E, Mondino A, Khoruts A, Jenkins MK. In vivo detection of dendritic cell antigen presentation to CD4(+) T cells. J Exp Med. 1997;185(12):2133–2141. doi: 10.1084/jem.185.12.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Norbury CC, Malide D, Gibbs JS, Bennink JR, Yewdell JW. Visualizing priming of virus-specific CD8+ T cells by infected dendritic cells in vivo. Nat Immunol. 2002;3(3):265–271. doi: 10.1038/ni762. [DOI] [PubMed] [Google Scholar]
- 21.Mayoux M, Roller A, Pulko V, Sammicheli S, Chen S, Sum E, et al. Dendritic cells dictate responses to PD-L1 blockade cancer immunotherapy. Sci Transl Med. 2020;12(534):eaav7431. doi: 10.1126/scitranslmed.aav7431. [DOI] [PubMed] [Google Scholar]
- 22.Versteven M, Van den Bergh JMJ, Marcq E, Smits ELJ, Van Tendeloo VFI, Hobo W, et al. Dendritic cells and programmed death-1 blockade: a joint venture to combat cancer. Front Immunol. 2018;9:394. doi: 10.3389/fimmu.2018.00394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flies DB, Higuchi T, Harris JC, Jha V, Gimotty PA, Adams SF. Immune checkpoint blockade reveals the stimulatory capacity of tumor-associated CD103(+) dendritic cells in late-stage ovarian cancer. Oncoimmunology. 2016;5(8):e1185583. doi: 10.1080/2162402X.2016.1185583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Broz ML, Binnewies M, Boldajipour B, Nelson AE, Pollack JL, Erle DJ, et al. Dissecting the tumor myeloid compartment reveals rare activating antigen-presenting cells critical for T cell immunity. Cancer Cell. 2014;26(6):938. doi: 10.1016/j.ccell.2014.11.010. [DOI] [PubMed] [Google Scholar]
- 25.Salmon H, Idoyaga J, Rahman A, Leboeuf M, Remark R, Jordan S, et al. Expansion and activation of CD103(+) dendritic cell progenitors at the tumor site enhances tumor responses to therapeutic PD-L1 and BRAF inhibition. Immunity. 2016;44(4):924–938. doi: 10.1016/j.immuni.2016.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dennis KL, Blatner NR, Gounari F, Khazaie K. Current status of interleukin-10 and regulatory T-cells in cancer. Curr Opin Oncol. 2013;25(6):637–645. doi: 10.1097/CCO.0000000000000006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi Y, Liu CH, Roberts AI, Das J, Xu G, Ren G, et al. Granulocyte-macrophage colony-stimulating factor (GM-CSF) and T-cell responses: what we do and don't know. Cell Res. 2006;16(2):126–133. doi: 10.1038/sj.cr.7310017. [DOI] [PubMed] [Google Scholar]
- 28.Parameswaran N, Patial S. Tumor necrosis factor-alpha signaling in macrophages. Crit Rev Eukaryot Gene Expr. 2010;20(2):87–103. doi: 10.1615/CritRevEukarGeneExpr.v20.i2.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maurer M, von Stebut E. Macrophage inflammatory protein-1. Int J Biochem Cell Biol. 2004;36(10):1882–1886. doi: 10.1016/j.biocel.2003.10.019. [DOI] [PubMed] [Google Scholar]
- 30.Han Q, Bagheri N, Bradshaw EM, Hafler DA, Lauffenburger DA, Love JC. Polyfunctional responses by human T cells result from sequential release of cytokines. Proc Natl Acad Sci U S A. 2012;109(5):1607–1612. doi: 10.1073/pnas.1117194109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ahamadi M, Freshwater T, Prohn M, Li CH, de Alwis DP, de Greef R, et al. Model-based characterization of the pharmacokinetics of pembrolizumab: a humanized anti-pd-1 monoclonal antibody in advanced solid tumors. CPT Pharmacometrics Syst Pharmacol. 2017;6(1):49–57. doi: 10.1002/psp4.12139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee JM, Cimino-Mathews A, Peer CJ, Zimmer A, Lipkowitz S, Annunziata CM, et al. Safety and clinical activity of the programmed death-ligand 1 inhibitor durvalumab in combination with poly (ADP-Ribose) polymerase inhibitor olaparib or vascular endothelial growth factor receptor 1–3 inhibitor cediranib in women's cancers: a dose-escalation. Phase I Study J Clin Oncol. 2017;35(19):2193–2202. doi: 10.1200/JCO.2016.72.1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baverel PG, Dubois VFS, Jin CY, Zheng Y, Song X, Jin X, et al. Population pharmacokinetics of durvalumab in cancer patients and association with longitudinal biomarkers of disease status. Clin Pharmacol Ther. 2018;103(4):631–642. doi: 10.1002/cpt.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Merlano MC, Abbona A, Denaro N, Garrone O. Knowing the tumour microenvironment to optimise immunotherapy. Acta Otorhinolaryngol Ital. 2019;39(1):2–8. doi: 10.14639/0392-100X-2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Binnewies M, Roberts EW, Kersten K, Chan V, Fearon DF, Merad M, et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med. 2018;24(5):541–550. doi: 10.1038/s41591-018-0014-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Filipovic A, Miller G, Bolen J. Progress toward identifying exact proxies for predicting response to immunotherapies. Front Cell Dev Biol. 2020;8:155. doi: 10.3389/fcell.2020.00155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cai DL, Jin LP. Immune cell population in ovarian tumor microenvironment. J Cancer. 2017;8(15):2915–2923. doi: 10.7150/jca.20314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heong V, Ngoi N, Tan DS. Update on immune checkpoint inhibitors in gynecological cancers. J Gynecol Oncol. 2017;28(2):e20. doi: 10.3802/jgo.2017.28.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature. 2013;500(7463):415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maleki VS. High and low mutational burden tumors versus immunologically hot and cold tumors and response to immune checkpoint inhibitors. J Immunother Cancer. 2018;6(1):157. doi: 10.1186/s40425-018-0479-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Varga A, Piha-Paul S, Ott PA, Mehnert JM, Berton-Rigaud D, Morosky A, et al. Pembrolizumab in patients with programmed death ligand 1-positive advanced ovarian cancer: analysis of KEYNOTE-028. Gynecol Oncol. 2019;152(2):243–250. doi: 10.1016/j.ygyno.2018.11.017. [DOI] [PubMed] [Google Scholar]
- 42.Turner N, Tutt A, Ashworth A. Hallmarks of 'BRCAness' in sporadic cancers. Nat Rev Cancer. 2004;4(10):814–819. doi: 10.1038/nrc1457. [DOI] [PubMed] [Google Scholar]
- 43.Jiang X, Li W, Li X, Bai H, Zhang Z. Current status and future prospects of PARP inhibitor clinical trials in ovarian cancer. Cancer Manag Res. 2019;11:4371–4390. doi: 10.2147/CMAR.S200524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Di Modugno F, Colosi C, Trono P, Antonacci G, Ruocco G, Nistico P. 3D models in the new era of immune oncology: focus on T cells, CAF and ECM. J Exp Clin Cancer Res. 2019;38(1):117. doi: 10.1186/s13046-019-1086-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schnell A, Schmidl C, Herr W, Siska PJ. The peripheral and intratumoral immune cell landscape in cancer patients: a proxy for tumor biology and a tool for outcome prediction. Biomedicines. 2018;6(1):25. doi: 10.3390/biomedicines6010025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.