Abstract
Background
Glioblastomas (GBMs) in patients harboring somatic or germinal mutations of mismatch-repair (MMR) genes exhibit a hypermutable phenotype. Here, we describe a GBM patient with increased tumor mutational burden and germline MMR mutations, treated using anti-PD1 therapy.
Methods
A woman with newly diagnosed GBM (nGBM) was treated by surgery, radiotherapy, and temozolomide. The tumor recurred after 13 months leading to a second surgery and treatment with nivolumab. Whole-exome sequencing was performed on the nGBM, recurrent GBM (rGBM), and blood. Immune infiltration was investigated by immunohistochemistry and the immune response in the blood during treatment was analyzed by flow cytometry.
Results
High density of infiltrating CD163 + cells was found in both GBM specimens. Large numbers of CD3 + and CD8 + T cells were homogeneously distributed in the nGBM. The infiltration of CD4 + T cells and a different CD8 + T cell density were observed in the rGBM. Both GBM shared 12,431 somatic mutations, with 113 substitutions specific to the nGBM and 1,683 specific to the rGBM. Germline variants included pathogenic mutation in the MSH2 (R359S) gene, suggesting the diagnosis of Lynch syndrome. Systemic immunophenotyping revealed the generation of CD8 + T memory cells and persistent activation of CD4 + T cells. The patient is still receiving nivolumab 68 months after the second surgery.
Conclusions
Our observations indicate that the hypermutator phenotype associated with germinal mutations of MMR genes and abundant T-cell infiltration contributes to a durable clinical benefit sustained by a persistent and robust immune response during anti-PD1 therapy.
Electronic supplementary material
The online version of this article (10.1007/s00262-020-02769-4) contains supplementary material, which is available to authorized users.
Keywords: Glioblastoma (GBM), Mismatch repair (MMR), Tumor mutational burden (TMB), Lynch syndrome (LS), Nivolumab/anti-PD1 therapy, Tumor infiltration lymphocytes (TILs)
Introduction
DNA mutations are at the basis of tumor formation. The majority of DNA mutations can arise stochastically in dividing cells, such as stem cells. A strong correlation has been found between the number of stem cells in an organ and the lifelong chance of developing cancer in that organ [1]. In a fraction of cases, cancer might depend on environmental factors or several intrinsic sources, including defective DNA proofreading due to mutations in the DNA replicative polymerases Polδ1 and Polε or DNA replication repair mutations [as in cancer predisposition syndromes, such as constitutional or biallelic mismatch repair (MMR) deficiency (CMMRD), Lynch syndrome (LS), and polymerase proofreading-associated polyposis (PPAP)]. Carcinomas arise in patients with LS, secondary to the acquisition of a somatic hit in the alternative wild-type allele of the same MMR gene that carries the germline alteration, thus inducing deficiency in the DNA repair machinery, resulting in the accumulation of frameshift mutations (i.e., hypermutated tumors) that can generate neoantigens. Although data gathered by the International Biallelic Mismatch Repair Deficiency consortium revealed that all malignant cancers are hypermutant in CMMRD [2], early evidence should be confirmed in the case of premalignant LS lesions [3]. This may help to explain why in some cancers with constitutive deficiency of mismatch repair genes, such as colon cancers, the number of mutations is exponentially higher than that in their counterparts with no such deficiency (hypermutations) [4].
There are nine major MMR genes: MutS homologs (MSH2, MSH3, MSH4, MSH5, and MSH6), MutL homologs (MLH1 and MLH3), and human postmeiotic segregation genes (PMS1 and PMS2). Four of these genes (i.e., MLH1, PMS2, MSH2, and MSH6) play a key role during DNA replication. MMR deficiency in tumors results in the accumulation of DNA defects that can bring a hypermutable phenotype with a high tumor mutational burden (TMB). Moreover, polymorphisms in DNA repair genes such as MLH1 might constitute glioblastoma (GBM) susceptibility factors [5]. Germinal mutations in MMR genes are typically described as causative factors of LS and CMMRD [6, 7]. LS exhibits autosomal dominant inheritance and is comprised of multiple cancers, particularly those involving the colorectum, stomach, and endometrium. Furthermore, an increased risk for other cancers, including GBM, has been described [8]. The cumulative risk of brain tumors has been estimated to be approximately 1–6% in LS patients, with the risk higher in subjects carrying pathogenic MSH2 mutations (compared to MLH1 and MSH6 mutations), and brain tumors in individuals with LS are rarely associated with microsatellite instability [9].
In approximately 20% of recurrent GBM (rGBM) cases, hypermutations are detected after alkylating chemotherapy with temozolomide (TMZ); these hypermutations are associated with somatic mutations or the decreased expression of MMR genes [10]. Recently, a high TMB was also described in newly diagnosed GBM (nGBM) without other known (e.g., POLε, MMR) germline mutations [11].
In addition to standard treatments [surgery, radiotherapy (RT), alkylating chemotherapy], different immunotherapy (IT) approaches for the treatment of GBM, such as dendritic cell (DC) vaccination, which has already been approved for other solid tumors, are under investigation [12]. Inhibition of immune “checkpoints”, such as PD-1, CTLA-4, and PD-L1, by immune checkpoint inhibitors (ICIs) has provided important advances in the treatment of melanoma and other cancers. Moreover, some positive predictive factors for the ICI response, such as a high TMB, T cell infiltration of the tumor microenvironment (TME), and the expression and engagement of immune checkpoints within the TME and HLA setting, were recently detected [13]. In July 2017, the US FDA granted accelerated approval of the anti-PD1 antibody nivolumab for the treatment of metastatic colorectal cancer with mismatch repair or microsatellite instability [14].
Here, we report a case of rGBM carrying germline MSH2 mutation with a high TMB, and with a massive infiltration of CD8 + T cells treated with anti-PD1 therapy.
Materials and methods
Patient and monitoring
At first GBM recurrence, the patient was enrolled in the CheckMate 143 study (NCT02017717) [15]. After written informed consent was obtained, bimonthly clinical assessment and brain magnetic resonance imaging (MRI) with gadolinium were performed. IT treatment was scheduled with nivolumab, as planned by the study. Disease progression was defined according to Response-Assessment in Neuro-Oncology (RANO) criteria [16], and confirmed by our pathologist (BP) according to the World Health Organization (WHO) classification.
Sequencing and mapping
Total DNA was isolated from peripheral blood and Formalin-Fixed Paraffin-Embedded (FFPE) samples and exome sequencing was performed in collaboration with BGI Genomics (China). High-quality reads for these samples were mapped by BWA [17] to the hg19 human genome assembly with default parameters. All mapped reads were then marked for duplicates by Picard to eliminate potential duplications. Sanger validation of the germline MMR mutations was performed.
Somatic mutations
Somatic mutations from whole-exome sequencing (WES) data from samples from patients with GBM were identified by applying the variant-calling software SAVI2 (statistical algorithm for variant frequency identification) [18], which is based on an empirical Bayesian method. Specifically, a list of candidate variants was generated by successively eliminating positions without variant reads, positions with a low sequencing depth, positions that were biased for one strand, and positions that contained only low-quality reads. Then, the numbers of high-quality reads for forward-strand reference alleles, reverse-strand reference alleles, forward-strand nonreference alleles, and reverse-strand nonreference alleles were calculated at the remaining candidate positions to build the prior and posterior distribution of mutation allele fraction. Finally, somatic mutations were identified on the basis of the posterior distribution of differences in mutation allele fraction between normal and tumor samples. SAVI2 was used to assess mutations by simultaneously considering multiple tumor samples, as well as their corresponding RNA samples, if available. VarScan2 was used to call indels in comparison with TCGA samples.
HLA Class I genotyping data and T cell receptor (TCR) β-chain sequencing and analysis
High-resolution HLA class I genotyping and TCR analysis of germline and GBM specimens were performed as extensively reported by Chowell [13].
Immunohistochemistry (IHC) and quantification of immune infiltration
IHC was performed on both tumor specimens to investigate PD-L1 expression, immune infiltrates, and MMR status. Paraffin-embedded 3-μm-thick sections were processed using AutostainerPlus (Dako, Agilent, Denmark), and antigen’s retrieval was performed in a PT Link pretreatment module (Dako, Agilent, Denmark) when requested. Briefly, slides were first blocked in 3% H2O2 (Sigma-Aldrich, USA), and then they were incubated with normal goat serum (Dako, Agilent, Denmark) and with the primary antibodies against the following: PD-L1 (1:100), MSH6 (dilution 1:100), MSH2 (dilution 1: 100), PMS2 (dilution 1:100), MLH1 (dilution 1:100), CD3 (dilution 1:50), CD8 (dilution 1:50), and CD4 (dilution 1:50), (Dako, Agilent, Denmark), CD163/MRQ-26 for microglia/macrophages (pre-diluted 1:2), Cell Marque, Sigma Aldrich USA), and FOXP3 (dilution 1:50, Santa Cruz Biotechnology, USA). Sections were subsequently incubated with anti-mouse or anti-rabbit Envision conjugated with peroxidase conjugated (Dako, Denmark) as a secondary antibody. Finally, slides were reacted with diaminobenzidine (DAB Substrate Chromogen System, Dako, Denmark) and counterstained with hematoxylin.
Aperio Cs2 scanScope and ImageScope software were used to quantitative evaluation of CD163 staining levels. Digital slides were acquired, and automated image analysis was used to quantify the percentage of infiltrating cells positive for CD163 (% positivity), based on a binary distribution of the 3,3′ diaminobenzidine (DAB) signal (positive/negative). Manual quantification was used to define the total number of CD3 + , CD8 + , and CD4 + tumor infiltrating lymphocytes (TILs) in nGBM specimen and rGBM specimen. The quantitative evaluation was performed by examining five different high-power fields (40 × objective) for each section stained for CD3, CD8 and CD4. CD3 + , CD8 + , and CD4 + TILs in each image were manually and independently counted three times by two investigators (NDI and SP) and confirmed by an experienced neuropathologist (BP).
Immune monitoring
Immune cell subsets were monitored by flow cytometry using anti-CD45-PerCP, anti-CD3-VioBlue, anti-CD4-PE-Cy7, anti-CD8-PE, and monoclonal antibodies (Miltenyi Biotec). Acquisition analyses were performed using a MACSQuant analyzer and MACSQuantify Software (Miltenyi Biotec). The T-cell activation status was investigated by using anti-CD3-FITC, CD38-APC, and HLA-DR-VioBlue. The memory status of T cells was evaluated using anti- CD3-Vioblue, anti-CD45RA-APC-Vio770, and anti-CCR7-PE-Vio770. Peripheral blood lymphocytes (PBLs) (frozen after each treatment), were thawed and grown in RPMI 1640 medium containing 10% heat-inactivated FBS, 100 U/mL penicillin, 100 U/mL streptomycin, 100 μg/mL glutamine, 0.1 mM nonessential amino acids, 1 mM sodium pyruvate, 50 μM β-mercaptoethanol, and 10 U/mL IL-2 (Roche). PBLs were stimulated for 3 h with 1.0 μM ionomycin and 50 ng/mL phorbol myristate acetate (PMA) and for 1 h with brefeldin in the presence of 30 U/mL IL-2 (Roche). The IFN-γ expression was analyzed by intracellular staining. Cell surface antigens were stained prior to fixation. Cells were then fixed and permeabilized using the Cytofix/Cytoperm solution (BD Biosciences) and intracellularly stained with an anti-IFN-γ antibody (Miltenyi Biotec) according to the manufacturer’s instructions.
Molecular analysis
O6-methylguanine-DNA-methyltransferase (MGMT) promoter methylation status was evaluated by methylation-specific PCR (MSP) with a standard protocol [19].
Results
Clinical story and treatment
The patient is a 33-year-old woman with no history of any diseases until a GBM diagnosis in 2013.
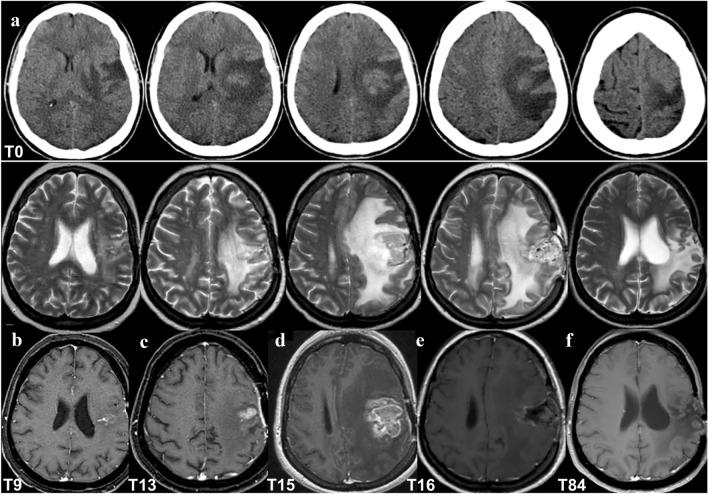
On July 2013 (Time 0), she underwent her first neurosurgery for a frontal parenchymal mass (Fig. 1a), with the diagnosis of GBM with unmethylated MGMT promoter and wild-type Isocitrate Dehydrogenase 1 (IDH1) and 2 (IDH2). Then she was treated with RT and TMZ as the standard of care [20] with transitory disease stability (Fig. 1b). On August 2014 (Time 13; Fig. 1c) radiological recurrence was suggested, and recurrence was confirmed by MRI two months later (Time 15; Fig. 1d). Gross total resection (GTR) was performed (November 2014, Time 16; Fig. 1e), confirming the GBM diagnosis.
Fig. 1.
Patient brain neuroimaging. a July 2013 (Time 0): pre-surgery brain computed tomography (CT) showing an intra-axial frontal expansive lesion with edema and mass effect. b–f Brain MRI, top T2 and bottom T1 with contrast agent weighted imaging, respectively; b April 2014 (Time 9): MRI after gross total resection (GTR) of the lesion, slight gliosis without mass effect; c, d August–October 2014 (Time 13–Time 15): recurrence with T1 enhancing lesion with increasing size, edema and mass effect; e November 2014 (Time 16): early brain MRI post-second surgery and GTR of the rGBM, showing persisting edema and mass effect; f June 2020 (Time 84): brain MRI shows gliosis and ex-vacuum right lateral ventricle enlargement, with no signs of recurrence
The patient started IT with nivolumab, which is still ongoing. There were no signs of clinical or radiological progression (stable disease of nontarget left frontal) from January 2015 (Time 17) until June 2020 (Time 84) (Fig. 1f), and the patient is alive at 84 months after nGBM diagnosis. During treatment, she developed anemia, a known adverse event of therapy. Moreover, based on the concomitant hematochezia (May 2017, Time 46), fecal occult blood testing was performed, which gave positive results. Then, the patients underwent colonoscopy, and one polyp was detected in the transverse colon. On June 2017 (Time 47) the intestinal lesion was removed and diagnosed as an adenoma with high-grade dysplasia.
Histological and molecular characterization
Because of the dramatic response to the treatment, we studied the case by histological review, analysis of MMR-related alterations on tumor and blood DNA and characterization of the immune response.
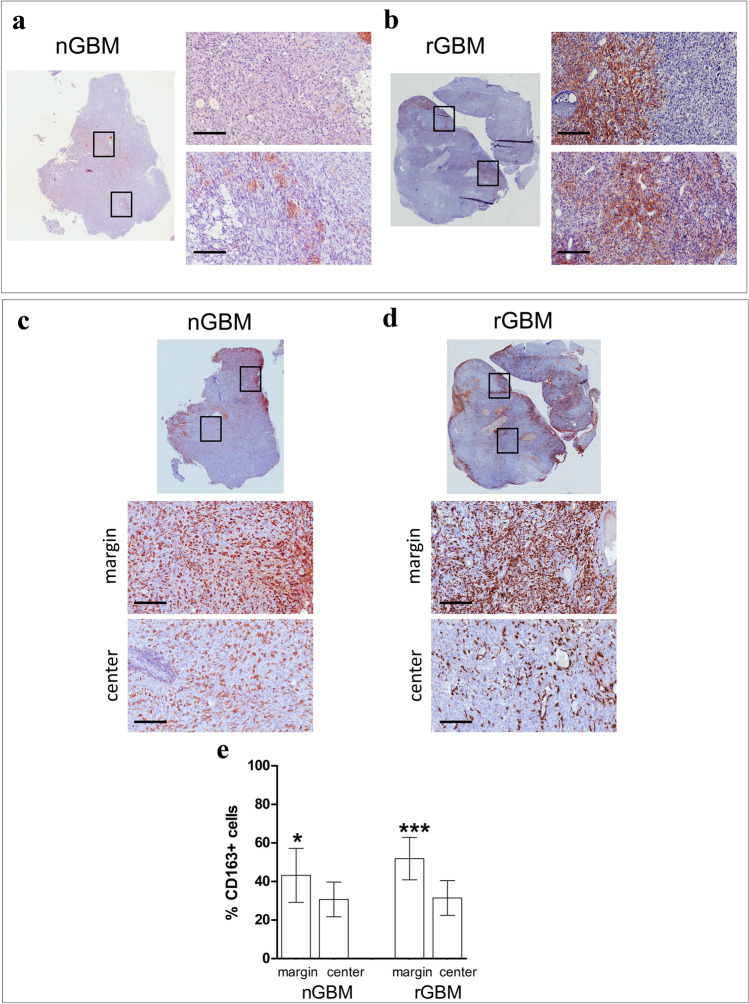
PD-L1 expression by IHC was, respectively, absent in the nGBM specimen and present in 10–15% of the cells in the rGBM specimen (Fig. 2a, b). Histological analysis revealed the presence of immune infiltrates in both nGBM and rGBM specimens. CD163 + cells were abundant in both specimens (Fig. 2c, d), but significantly more frequent in the tumor margins compared to the tumor center (Fig. 2e).
Fig. 2.
Expression and distribution of PD-L1 and CD163 in the nGBM and rGBM specimens. a, b Entire sections (magnification 0 ×), and representative areas (black rectangles) of nGBM (a) and rGBM (b) show that PD-L1 is only expressed after recurrence, and positive cells are mainly distributed at the margin of the specimen (scale bar 200 μm). c, d Entire section and representative marginal and central areas display that CD163 + infiltrating cells are found around the vessels and scattered in the tumor center of the specimens (scale bar 200 μm). e An automated quantification analysis revealed that the percentage of CD163 + cells, calculated with respect to the total nucleated cells, was higher in the margin compared to the tumor center of nGBM (43.2 ± 4.0% vs. 30.7 ± 9.7%, respectively; P = 0.04), as well as rGBM specimen (51.9 ± 9.0% vs. 31.4 ± 11.5%, respectively; P = 0.0008)
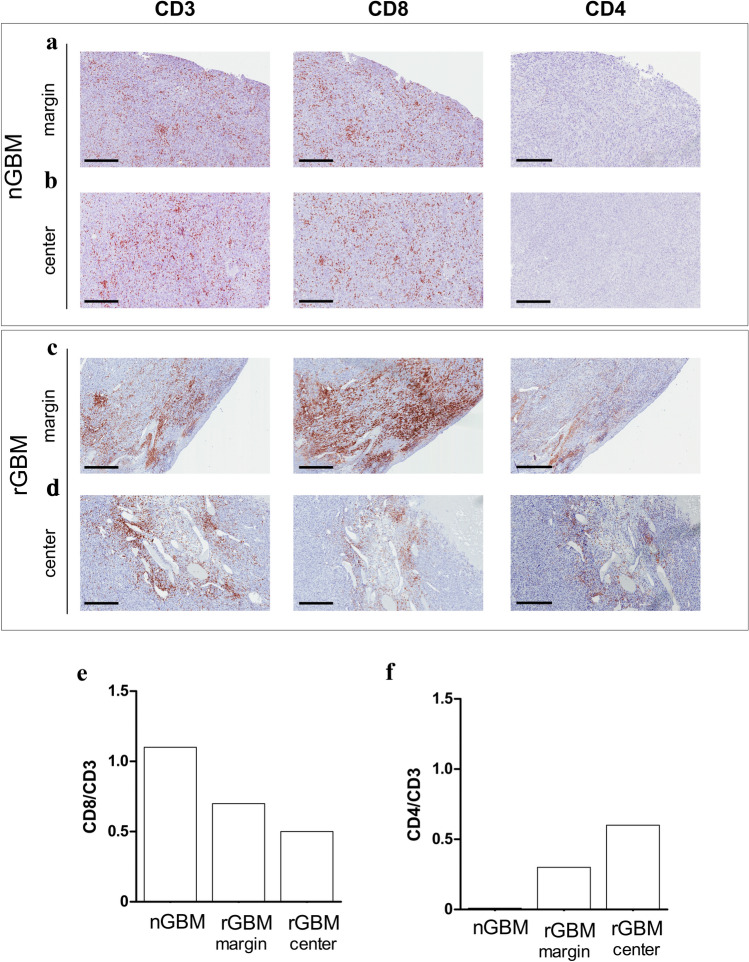
A high density of infiltrating CD8 + T cells was homogeneously distributed in the whole nGBM specimen, including the tumor center and margins. CD8 + T cells were shown to colocalize with CD3 + T cells when the same regions of adjacent tumor sections were investigated, whereas CD4 + T cells were not totally absent (Fig. 3a, b). Some regions of the rGBM specimen were enriched in CD4 + T cells (Fig. 3c, d). Few of these cells, which were preferentially distributed at the periphery of the specimen, expressed FOXP3 (Supplementary Fig. 1). Due to their heterogeneous distribution, we separately counted tumor infiltrating lymphocytes (TILs) in both the tumor center and tumor margins and found a significant difference in the distribution of CD3 + and CD8 + TILs between the tumor margins and tumor center (P < 0.01, Fig. 3e, Supplementary Table 1). The absolute numbers are reported in Supplementary Table S1.
Fig. 3.
CD8 + cell infiltration are abundant in both nGBM and rGBM specimens. a, b Representative adjacent sections of the nGBM specimen showing a high distribution of CD3 + and CD8 + and total absence of CD4 + TILs. High density of CD3 + and CD8 + TILs was observed in both tumor center (a) and tumor margins (b) of the nGBM specimen. c, d Changes in CD3 + and CD8 + T cell distribution and increased of CD4 + TILs were found in the rGBM. A manual count performed in the center area and tumor margins separately confirmed a significant higher density of CD3 + CD8 + TILs at the tumor margins (Scale Bar 300 μm)
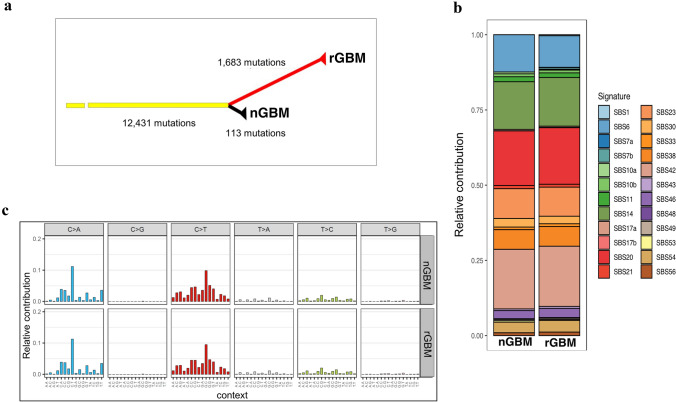
Molecular analysis identified 34,167 germinal single nucleotide polymorphisms (SNPs). One of them is a missense mutation in the MSH2 gene (rs63751617) causing the amino acid change p.Arg359Ser, confirmed by Sanger sequencing. rs63751617 was reported to be a pathogenic variant associated with Lynch syndrome by the International Society for Gastrointestinal Hereditary Tumours (InSiGHT) [21]. The MSH2 A359S mutation was shown to be pathogenic by SIFT prediction. Tumors had lost the MSH2 and MSH6 protein expression and were positive for other MMR proteins (MLH1 and PMS2), as determined by IHC (Supplementary Fig. 2). The nGBM and rGBM specimens had 12,431 SNPs in common, while 113 SNPs were specific to the first tumor, and 1,683 were specific to the second tumor (Fig. 4a). One of the common SNPs is the splice site mutation c.2458 + 1G > A, which is in the most conserved splice donor site in exon 14 of MSH2. The distribution of variant allele fraction (VAF) provided indication that the majority of the variants were clonal (Supplementary Fig. 3).
Fig. 4.
a Phylogenetic tree of the patient. Branch lengths are proportional to the number of mutations detected. b Mutation signature analysis. c Stacked bar plot indicating the contribution of each mutational signature in the nGBM and rGBM specimens
Mutation signature analysis highlighted that both specimens were highly enriched with the single base substitution (SBS) signatures SBS6, SBS14, SBS20, and SBS21, which are believed to be associated with defective DNA mismatch repair [22] (Fig. 4b, c). In total, these four signatures contributed to ~ 50% of the single base substitutions in the nGBM and rGBM specimens. Consistent with a previous study of hypermutant gliomas [10], we found the loss of function mutations in the RB pathway (CDKN2A p.W110* and CDKN2A p.A36Rfs*17) and Akt-mTOR pathway (AKT2 p.R371C and MTOR p.L1952F, p.R1301C, p.Q291H, p.C216Y, p.T102I, p.E73K). As shown in a recent report [23], we also found that the BRCA1, CREBBP, NOTCH2, ERBB2, GNAS, EP300, and APC genes were mutated in our samples. The extent of the response to anti-PD1 IT has been shown to be particularly associated with the accumulation of insertion–deletion (indel) mutations in tumors [24]. Using TCGA (The Cancer Genome Atlas) GBM cohort data as the background, we found that the number of indels in this patient was significantly higher than that in most hypermutant GBM patient data in TCGA (Supplementary Fig. 4).
Haplotype analysis revealed that all three HLA-1 loci were heterozygous in both the blood and GBM specimen, while the loss of heterozygosity was detected in HLA-DRB1. Based on the genotyping data, this patient belongs to the B44 supertype [13]. We performed TCR analysis of the tumors (Supplementary Table 2). In the nGBM specimen, we found only 4 different TCR clones with an entropy of 1.33, while in the rGBM specimen, we found 27 different clones (entropy = 3.06) (Supplementary Table 3).
Immunological characterization
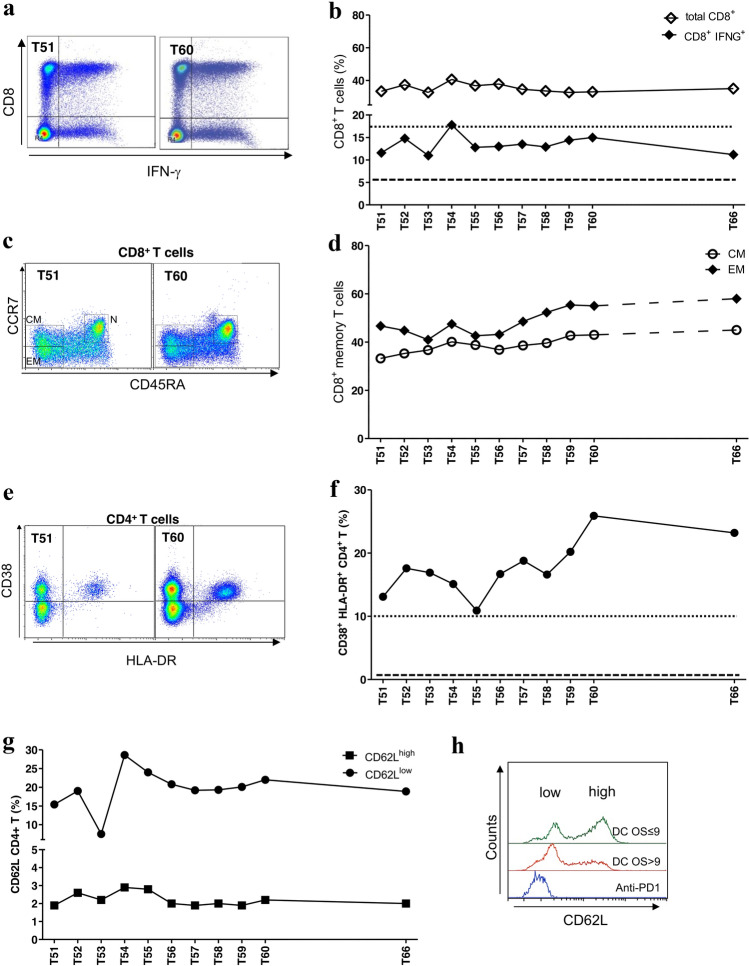
During anti-PD1 therapy, we characterized peripheral CD8 + and CD4 + T cells by flow cytometry at 11 time points (from 51st to 60th month and at 66th month). As control we considered eight rGBM patients treated with DC-IT, that we previously characterized in the study NCT04002804, and named Variant (V)-DENDR2 (V-DENDR2) [25]. In five of these eight patients, the CD8 + and CD4 + T-cell activation and CD8 + T-cell memory formation were significantly related to an improved prognosis [overall survival (OS) > 9 months]. We calculated the mean cell frequency at each vaccination (2nd to 5th) for the two groups of controls (n = 5 V-DENDR2 OS > 9 and n = 3 V-DENDR2 OS ≤ 9), and the median of the observations was used as a reference to determine the presence or the absence of an active immune response in the anti-PD1-treated patient. The frequency of CD8 + T cells expressing IFN-γ, indicative of cytotoxic ability, in the anti-PD1-treated patient was comparable to that measured in the V-DENDR2 OS > 9 group [median = 17.6 (dotted line) vs. anti-PD1, median = 13; ns] (Fig. 5a, b). The memory subsets were defined by assessing the expression of CD45RA and CCR7. In the anti-PD1-treated patient, both CD8 + T central (CM CD45RA- CCR7 +) and effector memory (EM) cells coexisted with naïve T cells, which persisted over time (Fig. 5c, d) and expressed high levels of IFN-γ (Supplementary Fig. 5).
Fig. 5.
Immune monitoring of the peripheral immune response activated by the anti-PD1 therapy. a Representative dot plots showing the CD8 + T-cell positivity for IFN-γ at two different time points (Time 51 and Time 60) in anti-PD1-treated patient. b Kinetics of the frequency of CD3 + CD8 + T cells (white square) and CD8 + T cells expressing IFN-γ (black square) assessed by flow cytometry. The dotted and dashed lines represent the median of the CD8 + T-cell activation evaluated by IFN-γ expression in V-DENDR2 OS > 9 and OS ≤ 9, respectively. c Representative dot plots showing the memory CD8 + T-cell subsets at two different time points. CM central memory, EM effector memory, N naïve. d Time course of the frequency of CD8 + central (CM) and effector memory (EM). e Representative dot plots showing the double positive CD38 and HLA-DR cells at two different time points. f Kinetics of the frequency of CD38+/HLA-DR+-activated cells evaluated in CD45/CD3/CD4+ T cells. The dotted and dashed lines represent the median of the CD8 + T-cell activation evaluated by IFN-γ expression in V-DENDR2 OS > 9 and OS ≤ 9, respectively. g Kinetics of the frequency of CD4 + T cells expressing CD62L as low and high in anti-PD1-treated patient. h Flow cytometry histogram showing the distribution of the CD62L levels in anti-PD1-treated patient compared to control patients (DC OS ≤ 9 and DC OS > 9)
As correlation between peripheral CD4 + T cells and clinical response to anti-PD1 therapy has been described [26], we tested the activation status of CD4 + T cells by evaluating the coexpression of CD38 and HLA-DR. The double-positive subset frequency in the anti-PD1-treated patient was similar to that observed in V-DENDR2 patients OS > 9 (median = 9.6 (dotted line) vs. 16.9 in the anti-PD1-treated patient, P = 0.008) (Fig. 5e, f). We also confirmed the presence of high proportions of CD62Llow CD4 + T cells, which was recently shown to be related to responsiveness and long-term survival in non-small cell lung cancer (NSCLC) patients treated with anti-PD1 therapy [27]. The basal frequency of the CD62Llow CD4 + T-cell subset was significantly higher in patients with an OS > 9 than in patients with an OS ≤ 9 (Supplementary Fig. 6). The frequency of CD62Lhigh CD4 + T cells was significantly lower in patients with an OS > 9 than in patients with an OS ≤ 9 (Fig. 5g). In the anti-PD1-treated patient, the CD62Llow CD4 + T-cell subset was highly prevalent (Fig. 5g), as revealed by a single peak in the flow cytometry histogram (Fig. 5h), and the frequency levels were comparable to those in the V-DENDR2 OS > 9 group. These different CD62L expression profiles are compatible with two distinct memory T-cell subsets, an effector subset in the anti-PD1-treated patient and the V-DENDR2 OS > 9 group, compatible with the clinical outcome, and indicative of a rested effector subset in the V-DENDR2 OS ≤ 9 group.
Notably, we found a history of multiple cancers in first-degree relatives of the patient. The patient’s mother, who was affected by uterine well-differentiated adenocarcinoma at the age of 59 (treated with surgery and local RT and was then stable the following 9 years); the patient’s only brother presented a non-intradermal left occipital Schwannoma at 35 years of age.
Based on family history, genetic data, and the diagnosis of bowel adenoma with high-grade dysplasia, we made the diagnosis of LS based on Amsterdam criteria II for LS. For adenoma no further treatment was necessary. Annual colonoscopies were performed as a follow-up and resulted negative. Additionally, the MSH2 germline mutation was detected also in patient’s mother and brother.
Discussion
The prognosis of GBM patients is dismal and the median survival is 14.6 months [20]. In the clinical trial CheckMate 143, the results of which were recently published [15], nivolumab as monotherapy failed to improve the OS of patients. Here, we have described a patient whose survival is clearly beyond the expectations, considering the median OS of GBM, the median survival (8.3 months) of the subgroup C of patients enrolled in CheckMate 143 [15], which showed an unmethylated MGMT promoter and the absence of steroid treatment use at randomization, and can be considered a “good responder” to ICI treatment. In addition, the present case lacks a positive prognostic factor, such as mutant IDH1/2, as also confirmed by WES in both newly diagnosed and recurrent GBM.
PD-L1 expression was found in only the rGBM specimen from our patient. In general, PD-L1 has a wide range of expression in GBM [28]. The positive correlation between PD-L1 expression and responsiveness to anti-PD1 therapy is still debated [29–31].
We detected a high GBM TMB by exome sequencing [4], while GBM tumors usually have approximately 50 somatic mutations [32]. The patient’s first and second tumors shared 12,431 sequence changes.
Indeed, in the constitutive/germinal DNA, we identified a missense mutation in MSH2 (rs63751617) causing the amino acid change Arg359Ser, shown to be pathogenic by SIFT prediction.
The gene products of MSH2 and MSH6 are involved in mismatch repair and are mutated in LS and CMMRD, two syndromes associated with increased cancer risk [6, 7, 9, 33].
In LS brain tumors mostly develop in MSH2 gene mutation carriers, and these tumors are usually not the first tumor diagnosed [9].Others reported the case of germline MLH1 mutation and PMS2 mutation in long-term GBM survivors but no other clinical signs of LS [34].
MMR deficiency predicts response of solid tumors to PD-1 blockade [35], but very few cases of GBM patients with MMR germline mutations treated with ICI have been reported [2]. Increasing evidence supports that MMR deficiency and hypermutations are not predictive of the response to anti-PD1 therapy in high grade glioma (HGG) [23, 36]. However, the interval between disease onset and recurrence (13 months), and the much longer duration of stability during ICI treatment, suggest that long-term survival can be linked to IT [37]. Here, we highlight several key points to potentially explain the impressive positive response to anti-PD1 therapy. Our case shows missense pathogenic germinal mutation on MSH2 (R359S) and a high TMB, unlike HGG cases with somatic mutations in only MSH6 reported by Ahmed and colleagues [36]. Constitutional MMR mutations can induce greater burden of clonal variants and can also generate an optimal antitumor T-cell response, unlike subclonal variant s which are associated with an immunosuppressive microenvironment and a poor antitumor response [23]. Comparison with 105 patients with hypermutated GBM showed that the number of indel in our patient was significantly higher than that in most of them. Both newly diagnosed and recurrent GBM show the massive infiltration of CD8 + T cells, in agreement with previous data supporting the predictive role of the frequency of CD8 + T cells which is significantly increased in rGBM with a high TBM in comparison with their primary tumors [38].
The rGBM specimen exhibited the infiltration of CD4 + T cell, which was not found in the nGBM specimen, and most of these cells did not express FoxP3. We speculate that TMB can lead to the formation of neoepitopes that can be recognized by CD4 + T cells [6, 7]. This increase in neo-epitopes could contribute to the efficacy of the anti-PD1 therapy.
Our patient did not receive steroids at the randomization (group C based on CheckMate 143-NCT02017717 [15]), leading to a generation of a specific peripheral T-cell response that included both CD8 + and CD4 + T cells. Corticosteroid administration was revealed to have a very negative impact on the survival of the patients, even those with MGMT promoter methylation, which is likely involved in the abrogation of immune response priming and activation.
We analyzed the percentages of immune cells in the peripheral blood at 11 timepoints (ten were consecutive) and considered a group of rGBM patients enrolled in the clinical study DENDR2 as a control. An OS greater than 9 months (OS9) after the second surgery was the survival endpoint [25]. The immune activation measured in the anti-PD1-treated patient was comparable to that observed in V-DENDR2 patients, in whom the immune activation was related to an improved outcome.
A relevant increase in CD4 + T cells coexpressing HLA-DR and CD38 supports the presence of a subset of T helper cells involved in sustaining CD8 + T-effector cells.
Interestingly, we observed a progressive increase in a specific population of activated CD38 + /HLA-DR + CD4 + T cells and a higher frequency of CD8 + CM T cells. Others reported that these CD8 + T cells had an effector-like phenotype (HLA-DR+, CD38 + , Bcl-2lo), expressing high levels of PD-1, with an increased number of phenotypically active NK and PD-1 + CD8 + cells at baseline acting as a positive predictive factor during anti-PD1 treatment in NSCLC [29–31].
A recent study applied mass cytometry analysis to peripheral blood mononuclear cells (PBMCs) derived from patients with melanoma and showed that IT perturbed the T-cell compartment, favoring peripheral T cells with a functionally activated status in responders [39]. Preliminary data in an animal model showed memory T-cell phenotypes and the upregulation of genes related to immunological memory after ICI treatment [40]. A low basal level of CD4 + T cells was correlated with a worse outcome in rGBM patients treated with anti-PD-L1 in a phase I trial [41], and lower baseline peripheral T cell receptor clonality has been described as a positive prognostic marker in the neoadjuvant ICI setting in GBM [42].
Most relevant, the identification of high CD62Llow CD4 + T cells confirmed the key role of this subset in predicting the responsiveness to the therapy, and the possibility of performing immune monitoring in the peripheral blood [27]. Finally, the case presented the B44 supertype which has been described as a positive predictive factor in melanoma [13].
In conclusion, our study identified multiple variables related to the response to anti-PD1 therapy and depicted the distinct evolution of the peripheral immune profile in GBM under IT. While nivolumab treatment in the randomized CheckMate 143 clinical trial recently resulted in a failure [15], our study suggests that some patients might benefit from this therapy.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Silvia Musio for collaborating in flow cytometry acquisitions, Massimo Costanza for critical reading and suggestions, the staff of the SOL Group Spa-Italy for the cryo-management service and the technical assistance, and Sara Murrone for graphic support. The manuscript was edited by American Journal Experts (AJE).
Author contributions
Patient recruitment, treatment, and follow-up were performed by EA with the support of ME. Histological analysis and interpretations were performed by MP and BP. Analysis and quantification of the immune infiltrates and preparation of Figs. 2 and 3 were performed by NDI. Genetic data were generated by RP and TL. JZ and RR contributed to analysis of WES data and manuscript writing. Radiological studies were performed by VC. AI contributed to data interpretation. Supervision of the study and was contributed by GF. Immune monitoring data and interpretations were performed by SP. EA, GF and SP co-wrote the manuscript. All authors have critically reviewed and approved the final manuscript.
Funding
The study was supported by funds provided by no-profit associations, such as Brancatelli ONLUS, il Fondo di Gio ONLUS, and donations of patient families for the development of immunotherapy strategies based on the project entitled: “Immunotherapy in preclinical models of glioma”.
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Compliance with ethical standards
Conflict of interest
There are no competing interests in the report.
Ethics approval and consent to participate
An informed consent was signed for the use of biological material for research purposes.
Consent for publication
Written consent was obtained from the patient for publication of this case report and any accompanying images.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Tomasetti C, Vogelstein B. Cancer etiology. Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science. 2015;347:78–81. doi: 10.1126/science.1260825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bouffet E, Larouche V, Campbell BB, et al. Immune checkpoint inhibition for hypermutant glioblastoma multiforme resulting from germline biallelic mismatch repair deficiency. J Clin Oncol. 2016;34:2206–2211. doi: 10.1200/JCO.2016.66.6552. [DOI] [PubMed] [Google Scholar]
- 3.Chang K, Bai HX, Zhou H, et al. Residual convolutional neural network for the determination of IDH status in low- and high-grade gliomas from MR imaging. Clin Cancer Res. 2018;24:1073–1081. doi: 10.1158/1078-0432.CCR-17-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campbell BB, Light N, Fabrizio D, et al. Comprehensive analysis of hypermutation in human cancer. Cell. 2017;171:1042–1056.e10. doi: 10.1016/j.cell.2017.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rodriguez-Hernandez I, Perdomo S, Santos-Briz A, et al. Analysis of DNA repair gene polymorphisms in glioblastoma. Gene. 2014;536:79–83. doi: 10.1016/j.gene.2013.11.077. [DOI] [PubMed] [Google Scholar]
- 6.Lynch HT, Lanspa S, Shaw T, et al. Phenotypic and genotypic heterogeneity of Lynch syndrome: a complex diagnostic challenge. Fam Cancer. 2018;17:403–414. doi: 10.1007/s10689-017-0053-3. [DOI] [PubMed] [Google Scholar]
- 7.Vasen HFA, Ghorbanoghli Z, Bourdeaut F, et al. Guidelines for surveillance of individuals with constitutional mismatch repair-deficiency proposed by the European Consortium “Care for CMMR-D” (C4CMMR-D) J Med Genet. 2014;51:283–293. doi: 10.1136/jmedgenet-2013-102238. [DOI] [PubMed] [Google Scholar]
- 8.Watson P, Vasen HFA, Mecklin J-P, et al. The risk of extra-colonic, extra-endometrial cancer in the Lynch syndrome. Int J Cancer. 2008;123:444–449. doi: 10.1002/ijc.23508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Therkildsen C, Ladelund S, Rambech E, et al. Glioblastomas, astrocytomas and oligodendrogliomas linked to Lynch syndrome. Eur J Neurol. 2015;22:717–724. doi: 10.1111/ene.12647. [DOI] [PubMed] [Google Scholar]
- 10.Johnson BE, Mazor T, Hong C, et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science. 2014;343:189–193. doi: 10.1126/science.1239947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mahlokozera T, Vellimana AK, Li T, et al. Biological and therapeutic implications of multisector sequencing in newly diagnosed glioblastoma. Neuro Oncol. 2018;20:472–483. doi: 10.1093/neuonc/nox232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finocchiaro G, Pellegatta S. Immunotherapy with dendritic cells loaded with glioblastoma stem cells: from preclinical to clinical studies. Cancer Immunol Immunother. 2016;65:101–109. doi: 10.1007/s00262-015-1754-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chowell D, Morris LGT, Grigg CM, et al. Patient HLA class I genotype influences cancer response to checkpoint blockade immunotherapy. Science. 2018;359:582–587. doi: 10.1126/science.aao4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Overman MJ, McDermott R, Leach JL, et al. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): an open-label, multicentre, phase 2 study. Lancet Oncol. 2017;18:1182–1191. doi: 10.1016/S1470-2045(17)30422-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reardon DA, Brandes AA, Omuro A, et al. Effect of nivolumab vs bevacizumab in patients with recurrent glioblastoma: the checkmate 143 phase 3 randomized clinical trial. JAMA Oncol. 2020 doi: 10.1001/jamaoncol.2020.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wen PY, Chang SM, Van den Bent MJ, et al. Response assessment in neuro-oncology clinical trials. J Clin Oncol. 2017;35:2439–2449. doi: 10.1200/JCO.2017.72.7511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trifonov V, Pasqualucci L, Tiacci E, et al. SAVI: a statistical algorithm for variant frequency identification. BMC Syst Biol. 2013;7(Suppl 2):S2. doi: 10.1186/1752-0509-7-S2-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eoli M, Menghi F, Bruzzone MG, et al. Methylation of O6-methylguanine DNA methyltransferase and loss of heterozygosity on 19q and/or 17p are overlapping features of secondary glioblastomas with prolonged survival. Clin Cancer Res. 2007;13:2606–2613. doi: 10.1158/1078-0432.CCR-06-2184. [DOI] [PubMed] [Google Scholar]
- 20.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 21.Heald B, Hampel H, Church J, et al. Collaborative group of the Americas on Inherited Gastrointestinal Cancer Position statement on multigene panel testing for patients with colorectal cancer and/or polyposis. Fam Cancer. 2020;19:223–239. doi: 10.1007/s10689-020-00170-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alexandrov LB, Kim J, Haradhvala NJ, et al. The repertoire of mutational signatures in human cancer. Nature. 2020;578:94–101. doi: 10.1038/s41586-020-1943-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Touat M, Li YY, Boynton AN, et al. Mechanisms and therapeutic implications of hypermutation in gliomas. Nature. 2020;580:517–523. doi: 10.1038/s41586-020-2209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mandal R, Samstein RM, Lee K-W, et al. Genetic diversity of tumors with mismatch repair deficiency influences anti-PD-1 immunotherapy response. Science. 2019;364:485–491. doi: 10.1126/science.aau0447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eoli M, Corbetta C, Anghileri E, et al. Expansion of effector and memory T cells is associated with increased survival in recurrent glioblastomas treated with dendritic cell immunotherapy. Neurooncol Adv. 2019;1:vds022. doi: 10.1093/noajnl/vdz022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spitzer MH, Carmi Y, Reticker-Flynn NE, et al. Systemic immunity is required for effective cancer immunotherapy. Cell. 2017;168:487–502.e15. doi: 10.1016/j.cell.2016.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kagamu H, Kitano S, Yamaguchi O, et al. CD4+ T-cell immunity in the peripheral blood correlates with response to anti-PD-1 therapy. Cancer Immunol Res. 2020;8:334–344. doi: 10.1158/2326-6066.CIR-19-0574. [DOI] [PubMed] [Google Scholar]
- 28.Nduom EK, Wei J, Yaghi NK, et al. PD-L1 expression and prognostic impact in glioblastoma. Neuro Oncol. 2016;18:195–205. doi: 10.1093/neuonc/nov172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.AlHarbi M, Ali Mobark N, AlMubarak L, et al. Durable response to nivolumab in a pediatric patient with refractory glioblastoma and constitutional biallelic mismatch repair deficiency. Oncologist. 2018;23:1401–1406. doi: 10.1634/theoncologist.2018-0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robert C, Long GV, Brady B, et al. Nivolumab in previously untreated melanoma without BRAF mutation. N Engl J Med. 2015;372:320–330. doi: 10.1056/NEJMoa1412082. [DOI] [PubMed] [Google Scholar]
- 31.Mazzaschi G, Facchinetti F, Missale G, et al. The circulating pool of functionally competent NK and CD8+ cells predicts the outcome of anti-PD1 treatment in advanced NSCLC. Lung Cancer. 2019;127:153–163. doi: 10.1016/j.lungcan.2018.11.038. [DOI] [PubMed] [Google Scholar]
- 32.Vogelstein B, Papadopoulos N, Velculescu VE, et al. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Terui H, Tachikawa T, Kakuta M, et al. Molecular and clinical characteristics of MSH6 germline variants detected in colorectal cancer patients. Oncol Rep. 2013;30:2909–2916. doi: 10.3892/or.2013.2781. [DOI] [PubMed] [Google Scholar]
- 34.Jue TR, Olafson LR, Siddell AH, et al. A case study of a long-term glioblastoma survivor with unmethylated MGMT and hypermutated genotype. Cold Spring Harb Mol Case Stud. 2019 doi: 10.1101/mcs.a003251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Le DT, Durham JN, Smith KN, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357:409–413. doi: 10.1126/science.aan6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ahmad H, Fadul CE, Schiff D, Purow B. Checkpoint inhibitor failure in hypermutated and mismatch repair-mutated recurrent high-grade gliomas. Neurooncol Pract. 2019;6:424–427. doi: 10.1093/nop/npz016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Palmieri G, Colombino M, Cossu A, et al. Genetic instability and increased mutational load: which diagnostic tool best direct patients with cancer to immunotherapy? J Transl Med. 2017;15:17. doi: 10.1186/s12967-017-1119-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Q, Hu B, Hu X, et al. Tumor evolution of glioma-intrinsic gene expression subtypes associates with immunological changes in the microenvironment. Cancer Cell. 2017;32:42–56.e6. doi: 10.1016/j.ccell.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krieg C, Nowicka M, Guglietta S, et al. High-dimensional single-cell analysis predicts response to anti-PD-1 immunotherapy. Nat Med. 2018;24:144–153. doi: 10.1038/nm.4466. [DOI] [PubMed] [Google Scholar]
- 40.Park J, Kim CG, Shim J-K, et al. Effect of combined anti-PD-1 and temozolomide therapy in glioblastoma. Oncoimmunology. 2019;8:e1525243. doi: 10.1080/2162402X.2018.1525243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lukas RV, Rodon J, Becker K, et al. Clinical activity and safety of atezolizumab in patients with recurrent glioblastoma. J Neurooncol. 2018;140:317–328. doi: 10.1007/s11060-018-2955-9. [DOI] [PubMed] [Google Scholar]
- 42.Cloughesy TF, Mochizuki AY, Orpilla JR, et al. Neoadjuvant anti-PD-1 immunotherapy promotes a survival benefit with intratumoral and systemic immune responses in recurrent glioblastoma. Nat Med. 2019;25:477–486. doi: 10.1038/s41591-018-0337-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files.