Abstract
Glycoprotein D (gD) of herpes simplex virus (HSV) is essential for virus entry and has four functional regions (I to IV) important for this process. We previously showed that a truncated form of a functional region IV variant, gD1(Δ290-299t), had an enhanced ability to block virus entry and to bind to the herpesvirus entry mediator (HveAt; formerly HVEMt), a cellular receptor for HSV. To explore this phenotype further, we examined other forms of gD, especially ones with mutations in region IV. Variant proteins with deletions of amino acids between 277 and 300 (region IV), as well as truncated forms lacking C-terminal residues up to amino acid 275 of gD, were able to block HSV entry into Vero cells 1 to 2 logs better than wild-type gD1(306t). In contrast, gD truncated at residue 234 did not block virus entry into Vero cells. Using optical biosensor technology, we recently showed that gD1(Δ290-299t) had a 100-fold-higher affinity for HveAt than gD1(306t) (3.3 × 10−8 M versus 3.2 × 10−6 M). Here we found that the affinities of other region IV variants for HveAt were similar to that of gD1(Δ290-299t). Thus, the affinity data follow the same hierarchy as the blocking data. In each case, the higher affinity was due primarily to a faster kon rather than to a slower koff. Therefore, once the gDt-HveAt complex formed, its stability was unaffected by mutations in or near region IV. gD truncated at residue 234 bound to HveAt with a lower affinity (2.0 × 10−5 M) than did gD1(306t) due to a more rapid koff. These data suggest that residues between 234 and 275 are important for maintaining stability of the gDt-HveAt complex and that functional region IV is important for modulating the binding of gD to HveA. The binding properties of any gD1(234t)-receptor complex could account for the inability of this form of gDt to block HSV infection.
The envelope of herpes simplex virus (HSV) contains at least 11 viral glycoproteins (39). The attachment of HSV to cells is mediated by interaction of virion envelope glycoprotein C (gC) and/or gB with cell surface glycosaminoglycans (12, 13, 46). This is followed by the interaction of gD with cellular receptors (2, 17, 18, 22, 26, 42). Then, pH-independent fusion occurs between virus envelope and the host cell plasma membrane (45); gB, gD, and the gH-gL complex have all been implicated in this step (39).
Using a panel of gD mutants and complementation analysis, we previously identified regions of gD which are important for virus entry (3). Mutations which did not globally alter gD antigenic structure yet resulted in a protein that failed to complement the infectivity of a gD-null virus were grouped into four noncontiguous functional regions, I through IV. To address why these proteins do not function in infection, we used baculovirus vectors to overexpress one truncated, soluble variant from each of the four functional regions (33) as well as wild-type gD from the KOS strain, gD1(306t) (38). Three of the mutants showed diminished or no ability to block infection compared to wild-type gD. Unexpectedly, a variant with a deletion in functional region IV, gD1(Δ290-299t), had a markedly enhanced inhibitory effect on HSV type 1 (HSV-1) infectivity and cell-to-cell spread compared to wild-type gD (33).
Recently, expression cloning was used to identify a HeLa cell gene product which, upon expression in normally nonpermissive Chinese hamster ovary (CHO) cells, allows for entry of many HSV strains (26). The gene product, the herpesvirus entry mediator (HveA; formerly HVEM), is a type I integral membrane protein and is a member of the tumor necrosis factor receptor superfamily (15, 20, 23, 24, 26). Subsequently, we showed that truncated, soluble gD (gDt) from the KOS strain bound directly to a soluble, truncated form of HveA (HveAt) in vitro (42). This binding is dependent on the native conformation of gD (42, 44). We demonstrated that purified HSV-1 KOS virions bound directly to HveAt in the absence of any other cell-associated components and that antibodies specific for gD, but not the other entry glycoproteins (gB, gC, or the gH-gL complex), completely block HSV binding to HveA (32). From these data, we believe that HveA mediates HSV entry by serving as a receptor for the virus and that virion gD is the principal ligand of HSV binding to HveA.
Recently, we used optical biosensor technology which employs surface plasmon resonance (SPR) detection to show that gD1(Δ290-299t) has a 100-fold-higher affinity (KD) for HveAt than does wild-type gDt (44). This is due primarily to a 40-fold-higher rate of complex formation (kon) for gD1(Δ290-299t) binding to HveAt compared to gD1(306t) binding to HveAt. Thus, this functional region IV variant protein exhibits an enhanced ability both to block HSV infection and to bind to receptor compared to that of the wild-type protein. In this study, our goal was to further explore the properties of region IV of gD. Our approach was to study the properties of purified, soluble forms of several region IV variants as well as proteins truncated at or prior to this region. We used assays which measured blocking of HSV entry into Vero cells as well as direct binding to HveAt.
MATERIALS AND METHODS
Cells and viruses.
African green monkey kidney (Vero) cells were grown in Dulbecco’s minimal essential medium supplemented with 5% fetal calf serum and penicillin-streptomycin solution (100 U/ml; GIBCO/BRL) at 37°C under 5% CO2. Spodoptera frugiperda Sf9 insect cells (GIBCO/BRL) used for producing recombinant baculoviruses and recombinant glycoproteins were propagated in Sf900II medium (GIBCO/BRL) at 27°C. HSV-1 strain KOS was propagated and titers were determined on Vero cells. HSV-1(hrR3) was propagated on D14 cells (11), and titers were determined on Vero cells. The hrR3 strain of HSV-1 KOS and D14 cells were kindly provided by S. Weller.
Construction of recombinant baculoviruses. (i) Baculoviruses expressing gD1(285t), gD1(275t), and gD1(234t).
Plasmid pRE4 (6), which contains the full-length gD open reading frames of HSV-1 (Patton strain), was used to produce baculovirus recombinants expressing gD truncated at amino acids 285, 275, and 234. DNA fragments were generated by PCR using plasmid pRE4 and the same 5′ primer as that used for the construction of the recombinant virus bac-gD-1(306t) (38). The 3′ primer for gD1(285t) was CGGGAATTCAGTGGTGGTGGTGGTGGTGCGTGCCTACGGGGTCCTCCAAGA. The 3′ primer for gD1(275t) was AAAACTGCAGTTAATGATGATGATGATGATGATCCTCGGGGTCTTC. The 3′ primer for gD1(234t) was AAAACTGCAGTTAATGATGATGATGATGATGGTATACGGCGACGGT. The 3′ primers added six histidine codons and a stop codon to the truncated gD open reading frame, as well as a PstI site. The PCR-amplified products were digested with BamHI and PstI and ligated with DNA from the transfer plasmid pVTBac (41), which had also been digested with BamHI and PstI. The ligated plasmids pWF291, pCP280, and pCP281 were used to transform Escherichia coli XL-2 Blue competent cells (Stratagene). Each of these was recombined into baculovirus (Autographa californica nuclear polyhedrosis virus) by cotransfection with Baculogold DNA (Pharmingen). Plaques were picked and amplified. Culture supernatants were screened for gD expression by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and Western immunoblotting. The resultant recombinant viruses are designated bac-gD1(285t), bac-gD1(275t), and bac-gD1(234t). The protein products are designated gD1(285t), gD1(275t), and gD1(234t). The strategy used to produce gD1(306t) and gD1(Δ290-299t) has been previously described (33, 38).
(ii) Baculoviruses expressing internal deletion mutants gD1(Δ277-290t) and gD1(Δ277-299t).
DNA fragments containing the gD1(Δ277-290t) and gD1(Δ277-299t) genes were generated by PCR using plasmids pHC238 and pHC239 (3) as templates and the primers described previously for construction of the recombinant baculovirus expressing gD1(306t) (38). The PCR products were each ligated into the transfer vector pVTBac to produce plasmids pAR273 and pAR274, respectively. Fragments cloned into pVTBac were then sequenced by the Sanger dideoxynucleotide chain termination method as modified for Taq polymerase cycle sequencing, using an ABI 373A automated DNA sequencer. Both strands of the portion of the gD coding region containing the mutation were sequenced. Sequence data were analyzed by using the GeneWorks software package (IntelliGenetics, Inc.). pAR273 and pAR274 were each recombined into baculovirus as described above and resulted in viruses designated bac-gD1(Δ277-290t) and bac-gD1(Δ277-299t). The protein products are designated gD1(Δ277-290t) and gD1(Δ277-299t). gD1(Δ277-290t) has amino acids 277 to 290 deleted, G replacing A at 277, and amino acids KIFL inserted after G. gD1(Δ277-299t) has amino acids 277 to 299 deleted, G replacing A at 277, and amino acids KIF inserted after G. gD1(Δ290-299t) has amino acids 290 to 299 deleted, R replacing I at residue 290, and amino acids KIFL inserted after R.
Production and purification of gDt.
The production and purification of gDt have been previously described (38, 43). In short, Sf9 cells were grown in suspension cultures and infected with recombinant baculovirus at a multiplicity of infection of 4 PFU/cell. At 48 h postinfection, cells were pelleted by centrifugation and the supernatant was passed over an affinity column. gD1(306t) and gD1(Δ290-299t) were purified on a monoclonal antibody (MAb) DL6 column as previously described (33, 38), and gD1(275t), gD1(Δ277-290t) and gD1(Δ277-299t) were purified on a MAb 1D3 column, using the same methodology. gD1(285t) and gD1(234t) were purified on a nickel-nitriloacetic acid resin column, using a stepwise imidazole gradient as described previously for HveAt (42). The yields of purified proteins were approximately 5 mg/liter of infected cell supernatant for gD1(Δ277-299t) and gD1(Δ277-290t), 1 to 3 mg/liter for gD1(275t) and gD1(234t), and 6 mg/liter for gD1(285t).
Production and purification of HveAt.
Mature HveA is 245 amino acids long (26). A soluble form of HveA truncated at amino acid 200, just prior to the transmembrane region (HveAt), was produced from recombinant baculovirus-infected insect cells and purified by nickel affinity chromatography as previously described (42).
Polyclonal and monoclonal antibodies.
Rabbit anti-gD serum R7 (16) was used for Western immunoblotting. Rabbit anti-gB (R69) and anti-gC (R46) sera (9) were used in immunoperoxidase assays. Anti-gD MAb DL6 (antigenic group IIb), which recognizes a continuous epitope from residues 272 to 279 (8, 16), and anti-gD MAb ID3 (group VII) (10, 21), which recognizes a continuous epitope from residues 11 to 19 (4, 7), were used for immunoaffinity purification and for analysis of antigenic activity. Anti-gD MAbs HD1 (group Ia) (27, 35), DL11 (group Ib) (5, 27), VID (group IIIa) (28, 36), ABD (group IIIb) (36), 45S (group IV) (37), DL2 (group VI) (5), D4 (group IX) (14), and AP7 (group XII) (3, 25) recognize discontinuous epitopes and were used for analysis of antigenic structure. HD1, DL11, and DL2 were also used for analysis of thermal stability.
SDS-PAGE analysis.
SDS-PAGE under reducing conditions and Western blot (immunoblot) analysis were performed as previously described (31).
ELISA.
Antigenic analysis of gDt mutants by enzyme-linked immunosorbent assay (ELISA) was performed by coating 96-well microtiter plates with gDt and probing with twofold serial dilutions of ascites fluids of anti-gD MAb DL11, 1D3, D4, HD1, DL2, ABD, DL6, AP7, or VID as previously described (33).
Thermal denaturation analysis of gDt mutants was carried out as previously described (33). Briefly, 80 μg of each protein per ml in phosphate-buffered saline (PBS) was brought to 37°C and then to various temperatures from 37 to 100°C for 5 min in a DNA thermal cycler (Perkin-Elmer Cetus). The samples were cooled on ice, diluted to 8 μg/ml in PBS, and coated onto 96-well plates. ELISA was then performed with MAb ascites fluid dilutions ranging from 1:400 to 1:10,000.
Binding of gDt to HveAt was accomplished by ELISA as previously described (42). Inhibition of HSV-1 entry into Vero cells by soluble gDt was carried out as previously described (34).
HSV plaque inhibition assay.
The effect of purified forms of gDt on HSV-1 plaque formation on Vero cells was assayed as previously described (17), with modifications as specified by Nicola et al. (33).
Measurement of binding of gDt to HveAt with an optical biosensor.
All SPR experiments were carried out on a Biacore X or Biacore 2000 (Biacore AB) with active temperature control at 25°C as previously described (44). The running buffer was PBS (0.1 M sodium phosphate, 0.15 M NaCl [pH 7.0]; Pierce) containing 0.005% Tween 20 (PBS/T), to act as a surfactant to decrease nonspecific sticking to flow cells. Approximately 2,000 resonance units (RU) of purified HveAt was coupled to flow cell 1 (Fc1) of a CM5 sensor chip via primary amines according to the manufacturer’s specifications. To compensate for nonspecific binding to the dextran matrix and changes in refractive index upon addition of sample, Fc2 was activated and then blocked without the addition of protein. To characterize the binding of gDt variants to HveAt, the flow path was set to include both flow cells, the flow rate was set to 50 μl/min, and the data collection rate was set to high. Protein samples were serially diluted in PBS/T. Each gDt sample was injected for 2 min to monitor association. Then the sample was replaced with buffer flow, and the dissociation phase was monitored for 2 min. To regenerate the HveAt surface, any remaining gDt was removed by injecting brief pulses of 0.2 M sodium carbonate (pH 9.5) until the response signal returned to baseline.
SPR data were analyzed with BIAevaluation software, version 3.0, which employs global fitting. The global fitting analysis allows simultaneous fitting of both the association and dissociation phases of the sensorgram to all curves of the working set. This improves the robustness and stability of the fitting procedure (1). All sensorgrams were corrected for nonspecific binding and refractive index changes by subtracting the control sensorgram (Fc2) from the HveAt surface sensorgram (Fc1). Model curve fitting was done by using a 1:1 Langmuir interaction with drifting baseline. For gD1(234t), a maximum koff was estimated by using the equation ln(R0/Rn) = koff(tn − t0), where R0 is the response at time zero (t0) of dissociation and Rn is the response at time n (tn) (19).
RESULTS
Production of gDt proteins.
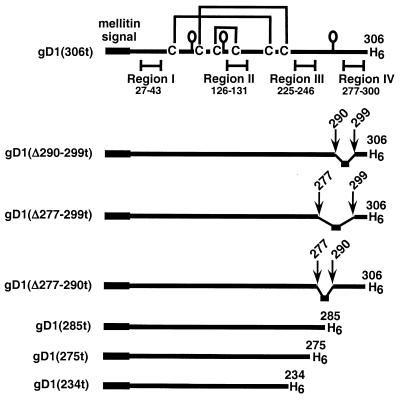
The gDt variants used in this study are depicted in Fig. 1. Each protein is truncated prior to the transmembrane region at the indicated amino acid (e.g., 306, 285, 275, or 234) and has a six-histidine tail at the C terminus. In addition to gD1(Δ290-299t), two variants having insertion-deletions in functional region IV were cloned into baculovirus in order to more closely map the activities of functional region IV. The first protein, gD1(Δ277-299t), lacks amino acids 290 to 299 like gD1(Δ290-299t) does, but in addition lacks residues 277 to 289. The second protein, gD1(Δ277-290t), lacks amino acids 277 to 290, just upstream of the Δ290-299 mutation. All three proteins are truncated at residue 306. DNA sequencing of pAR273 and pAR274 confirmed the expected sequence for gD1(Δ277-299t) and gD1(Δ277-290t). Three other C-terminally truncated forms of gD were cloned into baculovirus. gD1(285t) is truncated within region IV, while gD1(275t) is truncated just prior to region IV and gD1(234t) is truncated within functional region III. Each purified protein ran as a single silver-stained band on SDS-PAGE and reacted with anti-gD polyclonal antiserum (R7) on a Western blot (data not shown).
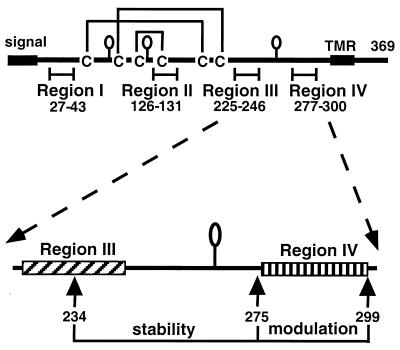
FIG. 1.
Schematic representations of truncated HSV gD. Each baculovirus-derived form of gD is truncated prior to the C-terminal transmembrane region at the indicated residue. The honeybee mellitin signal peptide replaces the wild-type signal and is cleaved in the fully processed protein. Each mature protein has amino acids DP (not shown) at the N terminus and a six-histidine tag at the C terminus. Each mutant contains all six cysteines and all three N-CHO consensus sites (shown as balloons) except for gD1(234t), which lacks the N-CHO site at residue 262.
Antigenic analysis of gDt.
We used an ELISA to examine the antigenic reactivity of each variant protein with a panel of gD-specific MAbs which span the length of the protein. ELISA reactivity was quantitated relative to that of gD1(306t) (Table 1) as previously described (31). Most of the MAbs bound well to the truncated gD proteins. However, each variant exhibited some differences in reactivity with at least one MAb, consistent with the fact that the proteins have been mutated. As expected, each protein reacted well with 1D3, which binds to a linear epitope (residues 11 to 19) (4, 7) in the N-terminal region of gD. As expected, gD1(275t) and gD1(234t) did not react with DL6 since this MAb binds to a continuous epitope within residues 272 to 279 (8, 16). gD1(Δ277-290t) and gD1(Δ277-299t) reacted weakly with DL6. As expected from previous studies with full-length mammalian forms of gD (3), MAb AP7 bound poorly to gD1(Δ277-290t) and not at all to the other variants. DL11 recognized gD1(234t), although reactivity was markedly less than that of gD1(306t). Although it is shorter than the other variants in this study, gD1(234t) had wild-type reactivity with conformation-dependent MAbs HD1, 45S, and DL2 and decreased reactivity with MAbs DL11, ABD, and D4. Thus, while antigenic differences were evident among the variants studied, when viewing the panel of MAbs as a whole rather than individually, we conclude that the variants are not globally altered in structure relative to gD1(306t). In addition, the antigenic structures of the baculovirus-expressed proteins are similar to those of the analogous mammalian-produced forms of gD (3, 5).
TABLE 1.
Antigenic analysis of variant gDt by ELISA
| Protein | MAb bindinga at antigenic site:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Ia (HD1) | Ib (DL11) | IIb (DL6)c | IIIa (VID) | IIIb (ABD) | IV (45S) | VI (DL2) | VII (1D3)c | IX (D4) | XII (AP7) | |
| gD1(306t) | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| gD1(Δ290-299t)b | 1.1 | 1.1 | 1.0 | 0.8 | 1.0 | 0.8 | 0.7 | 1.0 | 0.6 | 0.0 |
| gD1(Δ277-299t) | 0.7 | 0.7 | 0.4 | 0.8 | 1.0 | 0.5 | 1.0 | 1.0 | 0.3 | 0.0 |
| gD1(Δ277-290t) | 1.0 | 1.0 | 0.2 | 0.9 | 1.0 | 1.0 | 1.0 | 1.0 | 0.9 | 0.3 |
| gD1(285t) | 2.0 | 1.0 | 1.0 | 1.0 | 0.7 | 1.0 | 1.0 | 1.5 | 0.7 | 0.0 |
| gD1(275t) | 0.9 | 2.2 | 0.0 | 1.0 | 1.0 | 0.6 | 1.0 | 1.0 | 0.2 | 0.0 |
| gD1(234t) | 1.2 | 0.5 | 0.0 | NDd | 0.9 | 1.0 | 1.1 | 0.7 | 0.4 | 0.0 |
Values shown represent the 50% saturation concentration for MAb binding relative to gD1(306t) reactivity, which is normalized to 1.0. Each value represents the mean of at least two independent experiments done in duplicate.
ELISA reactivity of gD1(Δ290-299t) with these MAbs was reported previously (31) and is shown here for comparison.
Recognizes continuous epitopes. All other MAbs recognize discontinuous (conformation-dependent) epitopes.
ND, not done.
Thermal denaturation of gDt.
Previously, we measured the effect of mutation on structure by determining the thermal denaturation profile of gDt (33). In this study, individual samples of each protein were incubated for 5 min at temperatures ranging from 37 to 100°C and then chilled to 4°C. The reactivity of each sample with several MAbs was then determined by ELISA. Tm, the temperature at which each protein is 50% reactive with the indicated MAb, is an indicator of protein stability (Table 2). gD1(234t), which showed the largest antigenic changes from wild type, had Tm values of approximately 52°C, whereas gD1(306t) had Tm values of approximately 58°C. The remaining mutants, which were antigenically more similar to gD1(306t), had Tm values of 56°C. Thus, the antigenic structure analysis and thermal denaturation profiles of the mutant proteins are in agreement.
TABLE 2.
Thermal denaturation of gDt variants
| Protein |
Tm (°C)
|
|||
|---|---|---|---|---|
| Determined by ELISA with MAb (group):
|
Avg | |||
| HD1 (Ia) | DL11 (Ib) | DL2 (VI) | ||
| gD1(306t) | 59 | 58 | 57 | 58 |
| gD1(Δ290-299t) | 57 | 55 | 56 | 56 |
| gD1(Δ277-299t) | 56 | 54 | 57 | 56 |
| gD1(Δ277-290t) | 58 | 54 | 57 | 56 |
| gD1(275t) | 56 | 56 | 55 | 56 |
| gD1(234t) | 53 | NDa | 51 | 52 |
ND, not determined.
Effect of gDt on virus entry.
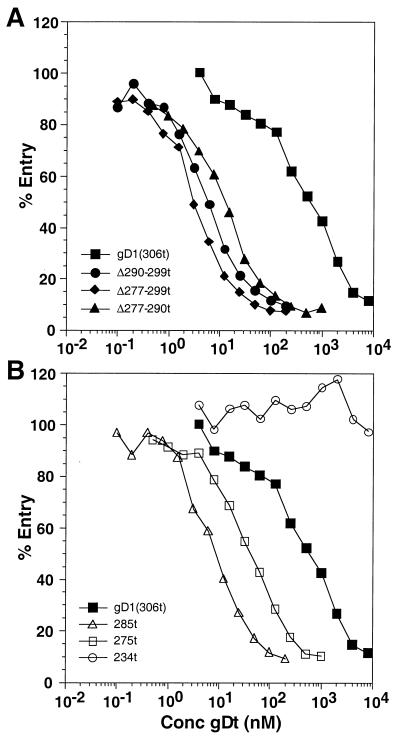
Soluble gD inhibits HSV infection (17, 34, 40), and gD1(Δ290-299t) has an enhanced blocking effect on HSV-1 entry and plaque formation compared to the effect of the wild type, gD1(306t) (31, 33). Is this enhanced-inhibition phenotype limited to gD variants lacking residues 290 to 299, or is it a general characteristic of mutants with deletions in region IV? To determine the inhibitory effects of our gD variants on HSV infection, we used an entry assay employing HSV-1(hrR3) which contains the lacZ gene under control of the ICP6 promoter (11, 34). The level of β-galactosidase activity induced by 5 h postinfection is determined and used as a measure of virus entry. Figure 2A shows that gD1(Δ277-299t) and gD1(Δ277-290t) were each more effective at blocking HSV-1 entry into Vero cells than was gD1(306t) and were similar in blocking ability to gD1(Δ290-299t). Thus, all three region IV mutants exhibited an enhanced ability to inhibit HSV entry.
FIG. 2.
Effect of gDt on HSV-1 entry into Vero cells. Confluent cells on 96-well plates were incubated with various concentrations of purified gDt at 4°C for 90 min. HSV-1(hrR3) was added at an multiplicity of infection of 0.5 PFU/cell, and the plate was incubated for another 90 min at 4°C. Plates were then shifted to 37°C for 5 h. Cells were lysed, and β-galactosidase activity was measured on aliquots of the cytoplasmic extract, using the substrate chlorophenol red-β-d-galactopyranoside and measuring the increase in absorbance over 50 min at 570 nm; 100% entry corresponds to no added inhibitor. (A) Blocking of entry with gD1(306t) compared to that of the region IV mutants; (B) blocking of entry with gD1(306t) (same curve as in panel A) compared to that of the other truncation mutants.
We next tested for the blocking phenotype of gD variants that were truncated further in from the C terminus than gD1(306t). gD1(285t) is truncated within functional region IV, gD1(275t) is truncated just prior to region IV, and gD1(234t) is truncated within functional region III. Figure 2B shows that gD1(285t) and gD1(275t) both inhibited HSV entry into Vero cells better than gD1(306t). In contrast, gD1(234t) did not inhibit HSV entry at all. Thus, deletion of portions or all of functional region IV enhanced the ability of gD to block infection, while a truncation that also removed part of region III had a deleterious effect on blocking. The gDt variants demonstrated the same trends for blocking of HSV plaque formation on Vero cells as for blocking of HSV entry (plaque formation data not shown).
Region IV modulates binding of gDt to HveAt. (i) ELISA.
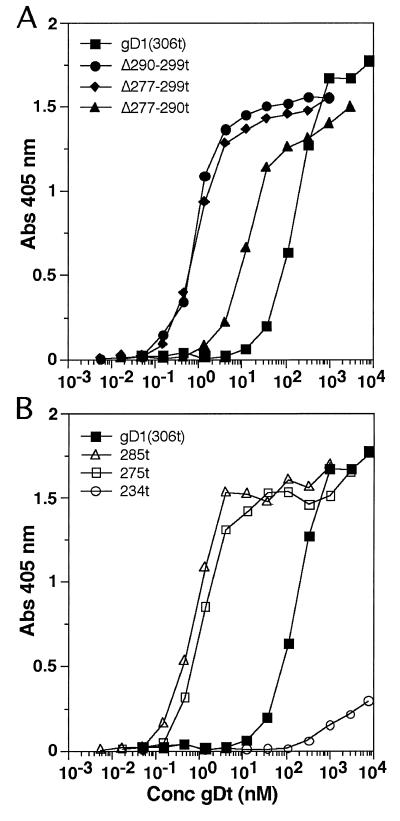
We next examined the ability of each of the variants to bind to HveAt by ELISA. As previously shown (42), gD1(Δ290-299t) exhibited enhanced binding to HveAt over gD1(306t) (Fig. 3A). gD1(Δ277-299t) bound to HveAt as well as gD1(Δ290-299t). gD1(Δ277-290t) bound to HveAt about 10-fold better than gD1(306t) but 10-fold less well than the other two region IV mutants. Figure 3B shows that forms of gD truncated at 285 and 275 also bound 100-fold better to HveAt than gD1(306t). In contrast, gD1(234t) bound very poorly to HveAt by ELISA, much less than gD1(306t). In summary, soluble gDt binds to HveAt in the following order: gD1(Δ277-299t) = gD1(Δ290-299t) = gD1(285t) = gD1(275t) > gD1(Δ277-290t) > gD1(306t) > gD1(234t).
FIG. 3.
Binding of gDt to HveAt. ELISA plates were coated with 50 μl of 200 nM HveAt in PBS, blocked, and incubated with various concentrations of gDt. Bound gDt was detected with rabbit antiserum R7, followed by peroxidase-conjugated secondary antibody and substrate. The data are the averages of results for duplicate wells, and each experiment was repeated twice with similar results. (A) Binding of gD1(306t) compared with that of the three region IV mutants. (B) Binding of gD1(306t) (same curve as in panel A) compared with that of the other truncation mutants. Abs, absorbance.
(ii) Optical biosensor technology.
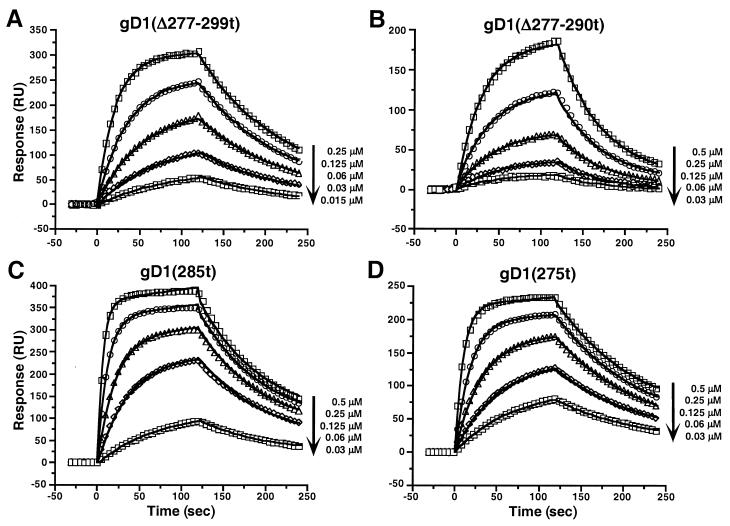
To better characterize the binding of gDt to HveAt, we used optical biosensor technology (19, 29). The Biacore is an optical biosensor which detects changes in SPR to quantitatively measure the direct interactions between biomolecules in real time without the need for labels. While ELISA gives affinity rankings, Biacore gives quantitative affinities (KD [equilibrium dissociation constant]) as well as on (kon) and off (koff) rates for the formation of each gDt-HveAt complex. To calculate kinetic and thermodynamic binding parameters for the gDt-HveAt interactions, twofold serial dilutions of each gDt mutant were injected onto a chip with HveAt covalently bound to it. The data are presented as a sensorgram, which shows response plotted as a function of time. Figure 4 shows a group of sensorgram overlays for the binding of a series of concentrations of gD1(Δ277-299t), gD1(Δ277-290t), gD1(285t), and gD1(275t) to immobilized HveAt. The solid lines represent the best global fits to a simple 1:1 Langmuir binding model with drifting baseline, assuming that gDt is a dimer in solution (42, 44). Using this model, the χ2 values (a standard statistical measure of the closeness of fit) were all below 5 (Table 3), and the residuals, which correspond to the difference between the actual and fitted data, are within ±4 RU, indicating a good fit (1) (not shown). Assessment of fit is important since the on and off rates are calculated from the fitted data.
FIG. 4.
Overlay of sensorgrams showing the interaction of decreasing concentrations of the indicated gDt variants with immobilized HveAt. Data points were collected every 0.2 or 0.4 s, but for simplicity selected points at every 5 s are shown as open symbols. The solid lines are the best global fits to the simple bimolecular model.
TABLE 3.
Kinetic and affinity values for gDt-HveAt complex formation
| Protein | kon (103 s−1 M−1) | koff (10−2 s−1) | KD (10−6 M) (koff/kon)e |
|---|---|---|---|
| gD1(306t)a | 6.1 ± 0.6b | 2.0 ± 0.2b | 3.2 ± 0.6 |
| gD1(Δ290-299t)a | 240 ± 70b | 0.78 ± 0.1b | 0.033 ± 0.01 |
| gD1(Δ277-299t) | 160 | 0.97 | 0.061 |
| gD1(Δ277-290t) | 31 | 1.5 | 0.48 |
| gD1(285t) | 300 | 1.1 | 0.037 |
| gD1(275t) | 180 | 0.74 | 0.041 |
| gD1(234t) | NDc | 20d | 20f |
Values for gD1(306t) and gD1(Δ290-299t) were reported previously (44) and are shown here for comparison.
Average ± standard deviation from at least three separate experiments.
ND, not determined.
Estimated maximum koff (see Fig. 5B).
χ2 values for the global fits ranged from 1.1 to 4.5.
From Scatchard analysis (see Fig. 5C).
gD1(Δ290-299t), gD1(Δ277-299t), gD1(285t), and gD1(275t) each had 10−8 M affinities (KD) for HveAt, gD1(Δ277-290t) had 10−7 M affinity, and gD1(306t) had 10−6 M affinity (Table 3). The same trend was seen in the ELISA data (Fig. 2). Also, gD1(285t) without the histidine tag had the same affinity for HveAt as did gD1(285t) with the histidine tag (data not shown). Hence, the tag does not account for the differences in affinities among variants. Examination of the kinetic parameters (Table 3) shows that the differences in KD among the mutants arose primarily from variations (almost 100-fold) in the rates of complex formation (kon), while less variation (threefold) was detected in the off rates. These results support the idea that region IV down-modulates the rate of the gDt-HveAt complex formation, with residues 290 to 299 having the greatest effect.
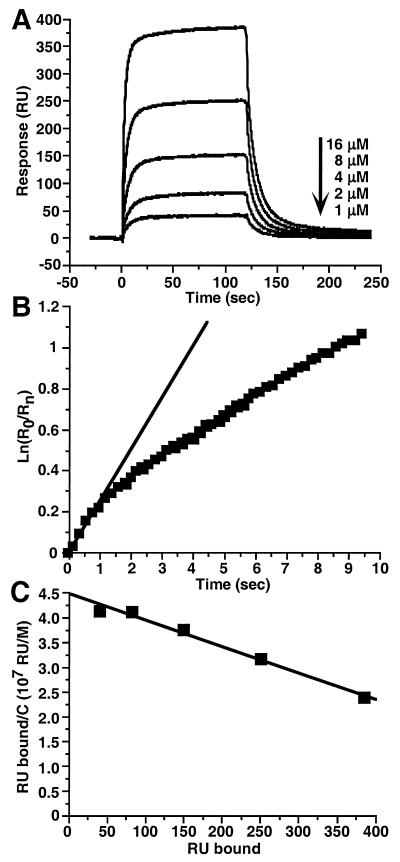
The sensorgrams in Fig. 5A show that gD1(234t) bound to HveAt since there was an increase in response with increasing concentrations of gDt. Since these data failed to fit a 1:1 Langmuir binding model, we estimated a maximum koff for the gD1(234t)-HveAt complex by plotting ln(R0/Rn) versus time, using only the initial dissociation data for gD1(234t) (Fig. 5B). The slope of the line gave a maximum koff of 0.2 s−1, which is 10-fold faster than for the other gDt-HveAt complexes. These results indicate that the gD1(234t)-HveAt complex was unstable and dissociated more rapidly than the other complexes. The increased rate at which the gD1(234t) sensorgrams (Fig. 5A) reached a plateau compared with sensorgrams of the other forms of gDt (Fig. 4) indicates that the gD1(234t)-HveAt complex came to equilibrium faster than the other complexes. Scatchard analysis (Fig. 5C) gave an equilibrium dissociation constant (KD) of 2 × 10−5 M for the gD1(234t)-HveAt complex. This is a sixfold-lower affinity than that observed for the gD1(306t)-HveAt complex (Table 3).
FIG. 5.
Binding of gD1(234t) to immobilized HveAt. (A) Sensorgram overlays. (B) Maximum koff determination using the dissociation phase of the sensorgram at 16 μM gD1(234t). Squares show the data for the first 10 s of dissociation, and the straight line is the fit to the initial second of data. The slope of the line gives a maximum koff of 2 × 10−1 s−1. r2 for the linear fit is 0.989. (C) Scatchard analysis. Equilibrium binding levels (RU bound) were obtained from sensorgrams in panel A. C is the molar concentration of gD1(234t). The negative inverse of the slope of the line gives a KD of 2 × 10−5 M. r2 for the linear fit is 0.978.
DISCUSSION
Linker-insertion mutagenesis followed by complementation and antigenic analysis identified four functional regions in gD (3). Proteins with mutations in any of these four regions failed to rescue the infectivity of a gD-null virus. Subsequently, we showed that truncated forms of gD carrying mutations in one of these four regions exhibited widely varying abilities to block HSV infection. Surprisingly, an insertion-deletion variant lacking amino acids 290 to 299 of gD region IV was able to block infection up to 400 times better than gD1(306t), depending on the strain of HSV used (31, 33). The ability of the functional region variants to block infection was paralleled by their ability to bind to HveAt. Specifically, we found that the region IV variant bound to HveAt 100-fold better than gD1(306t) (42, 44).
To explore the reasons for the unusual properties of gD1(Δ290-299t), additional variants with deletions of amino acids in region IV were examined. The phenotypes of the region IV variants examined in this study are summarized in Table 4. Each of the region IV variants failed to complement the infectivity of a gD-null virus (3). However, the present study shows that soluble forms of these variants were better able than wild-type gDt to block virus entry and to bind to HveAt.
TABLE 4.
Phenotypes of gD variants
| Proteina | Phenotype
|
|||
|---|---|---|---|---|
| Complemen- tation of virusb | Blockage of HSV-1 entry | Blockage of plaque formation | Binding to HveAt | |
| gD1(wild type) | + | + | + | + |
| gD1(Δ290-299) | − | +++ | +++ | +++ |
| gD1(Δ277-299) | − | +++ | +++ | +++ |
| gD1(Δ277-290) | − | +++ | +++ | ++ |
| gD1(285t) | NAc | +++ | NDd | +++ |
| gD1(275t) | NA | +++ | +++ | +++ |
| gD1(234t) | NA | − | − | ± |
For the complementation assays, protein refers to gD in the virion envelope. For all other experiments, protein refers to truncated forms of gD produced in a baculovirus expression system.
From reference 3.
NA, not applicable.
ND, not done.
Effect of gDt on HSV entry into Vero cells.
In addition to the region IV variants, gD with C-terminal truncations up to amino acid 275 demonstrated an enhanced ability to block HSV-1 entry compared to gD1(306t). Thus, it appears that the absence of amino acids between 275 and 300 actually enhances the blocking activity of the protein. In contrast, gD truncated at amino acid 234 did not block HSV infection. Although gD1(234t) lacks the N-linked oligosaccharide (N-CHO) at amino acid 262, this feature is not responsible for its failure to block entry since gDt lacking signals for all three N-CHOs inhibited HSV infection to the same extent as gD1(306t) (30). Thus, some or all of the residues between 234 and 275 are important for the blocking of HSV infection.
Binding of gDt to HveAt.
As previously demonstrated for gD1(Δ290-299t) (42, 44), gDt variants with either insertion-deletions or truncations in or near region IV had higher affinities for HveAt than gD1(306t). gD1(Δ277-290t) was intermediate in binding to HveAt in that its affinity was about 10-fold higher than that of gD1(306t) but 10-fold lower than those of the other region IV variants. Only gD1(306t) and gD1(Δ277-290t) contain residues 291 to 299; thus, the absence of residues 291 to 299 may affect the conformation of gDt in such a way as to increase the affinity of gDt for HveAt. The enhanced affinity of the region IV variants for HveAt was due primarily to higher (as much as 40-fold) on rates rather than to lower off rates. Thus, mutations in region IV of gD affect gDt-HveAt complex formation more than complex stability (44).
gD1(234t) is the shortest of the gD truncation variants examined which retains some antigenic conformation (5). gD1(234t) has a lower Tm than gD1(306t), suggesting that it is less stable. The SPR data showed that gD1(234t) bound to HveAt but that the complex dissociated rapidly, indicating that gD residues prior to 234 are sufficient for gDt-HveAt complex formation, but that additional residues are needed to stabilize the complex once formed. Interestingly, of all the gDt-HveAt complexes studied to date by SPR, the gD1(234t)-HveAt complex is the only one that exhibits a markedly higher off rate (44). The low affinity of gD1(234t) for HveAt is likely due to the lower stability of the complex compared to the other gDt-HveAt complexes. Thus, it appears that residues between 234 and 275 (overlapping functional region III) are involved in stabilizing the gDt-HveAt interaction (Fig. 6).
FIG. 6.
Top, schematic drawing of gD from HSV-1 with the four functional regions. The balloons represent the N-CHOs. TMR is the transmembrane region. Bottom, functional regions III and IV of gD. Residues between 234 and 275 help to stabilize the gD-HveA complex, while residues between 275 and 299 down-modulate the binding of gD to HveA.
Comparison of blocking of HSV entry with gDt binding directly to HveAt.
The HSV entry blocking data parallel the direct binding data except for gD1(234t), which bound to HveAt but did not block HSV entry into Vero cells. Assuming that soluble gDt blocks HSV entry by competing with virion-associated gD for a cellular receptor, it is possible that the affinity of gD1(234t) for a receptor(s) on Vero cells is not high enough to block HSV infection. Alternatively, the virus may use a receptor on Vero cells that differs from HveA and that does not bind gD1(234t) at all. Further experiments to identify specific HSV receptors on Vero and other cells are needed.
Functional region IV of gD does not contain the HveA binding domain.
Several observations suggest that functional region IV of gD does not contain the HveAt binding domain. First, gD1(275t) binds to HveAt, and this variant of gDt lacks region IV and all downstream residues. Second, the stability (koff) of the gD1(275t)-HveAt complex is very similar to that of the gD1(306t)-HveAt complex, suggesting that most of the gD residues involved in maintaining the complex are prior to 275. Consistent with these observations, the group II MAb DL6, which binds to a linear epitope (amino acids 272 to 279) overlapping functional region IV, coimmunoprecipitates gDt with HveAt and does not block binding of virus to HveAt (32). Thus, as gDt is able to bind both DL6 and HveAt simultaneously, residues 272 to 279 are not necessary for maintaining the interaction between gDt and HveAt.
Role of gD domains in binding to HveA.
In this study, we found that residues downstream of 234 are not critical for the initial binding of gD to HveA but appear to be important for maintaining the stability of the complex. Residues downstream of amino acid 275 (including region IV) are important for modulation of binding (Fig. 6) but appear not to be contact residues for HveA. Thus, the data are most consistent with the idea that the HveA binding residues are upstream of amino acid 234 on gD. Earlier studies implicated the N terminus of gD (region I) as being directly involved in binding to HveA. First, Whitbeck et al. (42) demonstrated that residue 27 of gDt (within functional region I) is involved in HveAt binding since a variant of gDt (from the rid1 strain of HSV-1) with a point mutation at residue 27 did not bind to HveAt. The rid1 strain of HSV-1 was also unable to utilize HveA to enter CHO cells (26). Second, binding to HveAt was greatly diminished by a linker-insertion mutation in functional region I (44). Third, binding of gDt to HveAt could be blocked by MAbs in antigenic group VII which recognize a linear epitope within residues 11 to 19 (7, 32).
MAbs in group Ib, which map to residues both upstream and downstream of amino acid 234, also blocked HveAt binding to gDt (32). One possibility is that the group Ib MAbs block the ability of residues downstream of 234 to stabilize the gDt-HveAt interaction, thereby disrupting the complex. Another possibility is that group Ib MAbs block upstream residues that are contact residues for HveAt binding to gDt. Clearly, more experiments need to be done to map the HveA binding residues on gD.
Our current concept is that functional region IV acts to modulate the binding of gD to HveA. This still leaves open the question of why variants with portions of region IV deleted are defective in complementation assays. We previously suggested that a tighter association between gD and receptor might alter the ability of virion gD to trigger subsequent steps in entry (42). Based on the results of this study, this hypothesis can be modified to state that the more rapid association of gD and receptor exhibited by region IV variants might prevent virion gD from triggering or participating in a subsequent step. If this hypothesis is correct, then the complementation-negative phenotype with enhanced affinity might be dominant. One way to test this would be to cotransfect cells with plasmids expressing mutant and wild-type forms of gD and then superinfect them with a gD-null virus. One would then test the infectivity of the progeny virus.
Some interesting questions remain to be answered: Does the interaction between gD and HveA involve a conformational change in gD? Does a conformational change occur when region IV is deleted? Further studies of the gD-HveA interaction are needed to address these questions.
ACKNOWLEDGMENTS
This investigation was supported by Public Health Service grants AI-18289 from the National Institute of Allergy and Infectious Diseases and NS-36731 and NS-30606 from the National Institute of Neurological Diseases and Stroke. S.H.W. and A.H.R. received support from Public Health Service grant AI-07324, and A.V.N. received support from Public Health Service grant AI-07325. We thank the Schools of Dental and Veterinary Medicine at the University of Pennsylvania for supplying funds for the purchase of the Biacore X.
We thank Gabriela Canziani, Manager of the Biosensor/Interaction Analysis Core Facility, and Irwin Chaiken and Sheng-jiun Wu, Medical School of the University of Pennsylvania, for help with SPR training and data analysis. We thank Claude Krummenacher, J. Charles Whitbeck, and John J. Rux for critical reading of the manuscript. Automated DNA sequencing was provided by the Biopolymer Analysis Laboratory of the School of Dental Medicine at the University of Pennsylvania.
REFERENCES
- 1.Biacore AB. BIAevaluation software handbook, version 3.0. Uppsala, Sweden: Biacore AB; 1997. [Google Scholar]
- 2.Campadelli-Fiume G, Arsenakis M, Farabegoli F, Roizman B. Entry of herpes simplex virus 1 in BJ cells that constitutively express viral glycoprotein D is by endocytosis and results in degradation of the virus. J Virol. 1988;62:159–167. doi: 10.1128/jvi.62.1.159-167.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chiang H-Y, Cohen G H, Eisenberg R J. Identification of functional regions of herpes simplex virus glycoprotein gD by using linker-insertion mutagenesis. J Virol. 1994;68:2529–2543. doi: 10.1128/jvi.68.4.2529-2543.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen G H, Dietzschold B, Ponce de Leon M, Long D, Golub E, Varrichio A, Pereira L, Eisenberg R J. Localization and synthesis of an antigenic determinant of herpes simplex virus glycoprotein D that stimulates production of neutralizing antibody. J Virol. 1984;49:102–108. doi: 10.1128/jvi.49.1.102-108.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen G H, Isola V J, Kuhns J, Berman P W, Eisenberg R J. Localization of discontinuous epitopes of herpes simplex virus glycoprotein D: use of a nondenaturing (“native” gel) system of polyacrylamide gel electrophoresis coupled with Western blotting. J Virol. 1986;60:157–166. doi: 10.1128/jvi.60.1.157-166.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen G H, Wilcox W C, Sodora D L, Long D, Levin J Z, Eisenberg R J. Expression of herpes simplex virus type 1 glycoprotein D deletion mutants in mammalian cells. J Virol. 1988;62:1932–1940. doi: 10.1128/jvi.62.6.1932-1940.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dietzschold B, Eisenberg R J, Ponce de Leon M, Golub E, Hudecz F, Varrichio A, Cohen G H. Fine structure analysis of type-specific and type-common antigenic sites of herpes simplex virus glycoprotein D. J Virol. 1984;52:431–435. doi: 10.1128/jvi.52.2.431-435.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eisenberg R J, Long D, Ponce de Leon M, Matthews J T, Spear P G, Gibson M G, Lasky L A, Berman P, Golub E, Cohen G H. Localization of epitopes of herpes simplex virus type 1 glycoprotein D. J Virol. 1985;53:634–644. doi: 10.1128/jvi.53.2.634-644.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eisenberg R J, Ponce de Leon M, Friedman H M, Fries L F, Frank M M, Hastings J C, Cohen G H. Complement component C3b binds directly to purified glycoprotein C of herpes simplex virus types 1 and 2. Microb Pathog. 1987;3:423–435. doi: 10.1016/0882-4010(87)90012-x. [DOI] [PubMed] [Google Scholar]
- 10.Friedman H M, Cohen G H, Eisenberg R J, Seidel C A, Cines D B. Glycoprotein C of herpes simplex virus 1 acts as a receptor for the C3b complement component on infected cells. Nature (London) 1984;309:633–635. doi: 10.1038/309633a0. [DOI] [PubMed] [Google Scholar]
- 11.Goldstein D J, Weller S K. An ICP6::LacZ insertional mutagen is used to demonstrate that the UL52 gene of herpes simplex virus type 1 is required for virus growth and DNA synthesis. J Virol. 1988;62:2970–2977. doi: 10.1128/jvi.62.8.2970-2977.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herold B C, Visalli R J, Sumarski N, Brandt C, Spear P G. Glycoprotein C-independent binding of herpes simplex virus to cells requires cell surface heparan sulfate and glycoprotein B. J Gen Virol. 1994;75:1211–1222. doi: 10.1099/0022-1317-75-6-1211. [DOI] [PubMed] [Google Scholar]
- 13.Herold B C, WuDunn D, Soltys N, Spear P G. Glycoprotein C of herpes simplex virus type 1 plays a principal role in the adsorption of virus to cells and in infectivity. J Virol. 1991;65:1090–1098. doi: 10.1128/jvi.65.3.1090-1098.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Highlander S L, Sutherland S L, Gage P J, Johnson D C, Levine M, Glorioso J C. Neutralizing monoclonal antibodies specific for herpes simplex virus glycoprotein D inhibit virus penetration. J Virol. 1987;61:3356–3364. doi: 10.1128/jvi.61.11.3356-3364.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsu S, Solovyev I, Colombero A, Elliott R, Kelley M, Boyle W J. ATAR, a novel tumor necrosis factor receptor family member, signals through TRAF2 and TRAF5. J Biol Chem. 1997;272:13471–13474. doi: 10.1074/jbc.272.21.13471. [DOI] [PubMed] [Google Scholar]
- 16.Isola V J, Eisenberg R J, Siebert G R, Heilman C J, Wilcox W C, Cohen G H. Fine mapping of antigenic site II of herpes simplex virus glycoprotein D. J Virol. 1989;63:2325–2334. doi: 10.1128/jvi.63.5.2325-2334.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson D C, Burke R L, Gregory T. Soluble forms of herpes simplex virus glycoprotein D bind to a limited number of cell surface receptors and inhibit virus entry into cells. J Virol. 1990;64:2569–2576. doi: 10.1128/jvi.64.6.2569-2576.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson D C, Ligas M W. Herpes simplex viruses lacking glycoprotein D are unable to inhibit virus penetration: quantitative evidence for virus-specific cell surface receptors. J Virol. 1988;62:4605–4612. doi: 10.1128/jvi.62.12.4605-4612.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karlsson R, Michaelsson A, Mattson A. Kinetic analysis of monoclonal antibody-antigen interactions with a new biosensor based analytical system. J Immunol Methods. 1991;145:229–240. doi: 10.1016/0022-1759(91)90331-9. [DOI] [PubMed] [Google Scholar]
- 20.Kwon B S, Tan K B, Ni J, Kwi-Ok-Oh, Lee Z H, Kim K K, Kim Y-J, Wang S, Gentz R, Yu G-L, Harrop J, Lyn S D, Silverman C, Porter T G, Truneh A, Young P R. A newly identified member of the tumor necrosis factor receptor superfamily with a wide tissue distribution and involvement in lymphocyte activation. J Biol Chem. 1997;272:14272–14276. doi: 10.1074/jbc.272.22.14272. [DOI] [PubMed] [Google Scholar]
- 21.Lasonder E, Schellenkens G A, Koedijk D G, Damhof R A, Welling-Wester S, Feijlbrief M, Scheffer A J, Welling G W. Kinetic analysis of synthetic analogues of linear-epitope peptides of glycoprotein D of herpes simplex virus type 1 by surface plasmon resonance. Eur J Biochem. 1996;240:209–214. doi: 10.1111/j.1432-1033.1996.0209h.x. [DOI] [PubMed] [Google Scholar]
- 22.Lee W C, Fuller A O. Herpes simplex virus type 1 and pseudorabies virus bind to a common saturable receptor on Vero cells that is not heparan sulfate. J Virol. 1993;67:5088–5097. doi: 10.1128/jvi.67.9.5088-5097.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marsters S A, Ayres T M, Skubatch M, Gray C L, Rothe M, Ashkenazi A. Herpes virus entry mediator, a member of the tumor necrosis factor receptor (TNFR) family, interacts with members of the TNFR-associated factor family and activates the transcription factors NF-κB and AP-1. J Biol Chem. 1997;272:14029–14032. doi: 10.1074/jbc.272.22.14029. [DOI] [PubMed] [Google Scholar]
- 24.Mauri D N, Ebner R, Kochel K D, Montgomery R I, Cheung T C, Yu G-L, Murphy M, Eisenberg R J, Cohen G H, Spear P G, Ware C F. LIGHT, a new member of the TNF superfamily, and lymphotoxin (LT) α are ligands for herpesvirus entry mediator (HVEM) Immunity. 1998;8:21–30. doi: 10.1016/s1074-7613(00)80455-0. [DOI] [PubMed] [Google Scholar]
- 25.Minson A C, Hodgman T C, Digard P, Hancock D C, Bell S E, Buckmaster E A. An analysis of the biological properties of monoclonal antibodies against glycoprotein D of herpes simplex virus and identification of amino acid substitutions that confer resistance to neutralization. J Gen Virol. 1986;67:1001–1013. doi: 10.1099/0022-1317-67-6-1001. [DOI] [PubMed] [Google Scholar]
- 26.Montgomery R I, Warner M S, Lum B J, Spear P G. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell. 1996;87:427–436. doi: 10.1016/s0092-8674(00)81363-x. [DOI] [PubMed] [Google Scholar]
- 27.Muggeridge M I, Isola V J, Byrn R A, Tucker T J, Minson A C, Glorioso J C, Cohen G H, Eisenberg R J. Antigenic analysis of a major neutralization site of herpes simplex virus glycoprotein D, using deletion mutants and monoclonal antibody-resistant mutants. J Virol. 1988;62:3274–3280. doi: 10.1128/jvi.62.9.3274-3280.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muggeridge M I, Roberts S R, Isola V J, Cohen G H, Eisenberg R J. Herpes simplex virus. In: Van Regenmortel M H V, Neurath A R, editors. Immunochemistry of viruses. II. The basis for serodiagnosis and vaccines. Amsterdam, The Netherlands: Elsevier Biochemical Press; 1990. pp. 459–481. [Google Scholar]
- 29.Myszka D G. Kinetic analysis of macromolecular interactions using surface plasmon resonance biosensors. Curr Opin Biotechnol. 1997;8:50–57. doi: 10.1016/s0958-1669(97)80157-7. [DOI] [PubMed] [Google Scholar]
- 30.Nicola, A. V., R. J. Eisenberg, and G. H. Cohen. 1997. Unpublished data.
- 31.Nicola A V, Peng C, Lou H, Cohen G H, Eisenberg R J. Antigenic structure of soluble herpes simplex virus glycoprotein D correlates with inhibition of HSV infection. J Virol. 1997;71:2940–2946. doi: 10.1128/jvi.71.4.2940-2946.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nicola A V, Ponce de Leon M, Xu R, Hou W, Whitbeck J C, Krummenacher C, Montgomery R I, Spear P G, Eisenberg R J, Cohen G H. Monoclonal antibodies to distinct sites on the herpes simplex virus (HSV) glycoprotein D block HSV binding to HVEM. J Virol. 1998;72:3595–3601. doi: 10.1128/jvi.72.5.3595-3601.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nicola A V, Willis S H, Naidoo N N, Eisenberg R J, Cohen G H. Structure-function analysis of soluble forms of herpes simplex virus glycoprotein D. J Virol. 1996;70:3815–3822. doi: 10.1128/jvi.70.6.3815-3822.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peng T, Ponce de Leon M, Jiang H, Dubin G, Lubinski J, Eisenberg R J, Cohen G H. The gH-gL complex of herpes simplex virus (HSV) stimulates neutralizing antibody and protects mice against HSV type 1 challenge. J Virol. 1998;72:65–72. doi: 10.1128/jvi.72.1.65-72.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pereira L, Klassen T, Baringer J R. Type-common and type-specific monoclonal antibody to herpes simplex virus type 1. Infect Immun. 1980;29:724–732. doi: 10.1128/iai.29.2.724-732.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seigneurin J M, Desgranges C, Seigneurin D, Paire J, Renversez J C, Jacquemont B, Micouin C. Herpes simplex virus glycoprotein D: human monoclonal antibody produced by bone marrow cell line. Science. 1983;221:173–175. doi: 10.1126/science.6304881. [DOI] [PubMed] [Google Scholar]
- 37.Showalter S D, Zweig M, Hampar B. Monoclonal antibodies to herpes simplex virus type 1 proteins, including the immediate-early protein ICP4. Infect Immun. 1981;34:684–692. doi: 10.1128/iai.34.3.684-692.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sisk W P, Bradley J D, Leipold R J, Stoltzfus A M, Ponce de Leon M, Hilf M, Peng C, Cohen G H, Eisenberg R J. High-level expression and purification of secreted forms of herpes simplex virus type 1 glycoprotein gD synthesized by baculovirus-infected insect cells. J Virol. 1994;68:766–775. doi: 10.1128/jvi.68.2.766-775.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spear P G. Entry of alphaherpesviruses into cells. Semin Virol. 1993;4:167–180. [Google Scholar]
- 40.Tal-Singer R, Peng C, Ponce de Leon M, Abrams W R, Banfield B W, Tufaro F, Cohen G H, Eisenberg R J. Interaction of herpes simplex virus glycoprotein gC with mammalian cell surface molecules. J Virol. 1995;69:4471–4483. doi: 10.1128/jvi.69.7.4471-4483.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tessier D C, Thomas D Y, Khouri H E, Laliberte F, Vernet T. Enhanced secretion from insect cells of a foreign protein fused to the honeybee melittin signal peptide. Gene. 1991;98:177–183. doi: 10.1016/0378-1119(91)90171-7. [DOI] [PubMed] [Google Scholar]
- 42.Whitbeck J C, Peng C, Lou H, Xu R, Willis S H, Ponce de Leon M, Peng T, Nicola A V, Montgomery R I, Warner M S, Soulika A M, Spruce L A, Moore W T, Lambris J D, Spear P G, Cohen G H, Eisenberg R J. Glycoprotein D of herpes simplex virus (HSV) binds directly to HVEM, a member of the TNFR superfamily and a mediator of HSV entry. J Virol. 1997;71:6083–6093. doi: 10.1128/jvi.71.8.6083-6093.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Willis S H, Peng C, Ponce de Leon M, Nicola A V, Rux A H, Cohen G H, Eisenberg R J. Expression and purification of secreted forms of herpes simplex virus glycoproteins from baculovirus-infected insect cells. In: Brown M S, MacLean A R, editors. Methods in molecular medicine: herpes simplex virus protocols. Vol. 10. Clifton, N.J: Humana Press; 1997. pp. 131–156. [DOI] [PubMed] [Google Scholar]
- 44.Willis S H, Rux A H, Peng C, Whitbeck J C, Nicola A V, Lou H, Hou W, Salvador L, Eisenberg R J, Cohen G H. Examination of the kinetics of herpes simplex virus glycoprotein D binding to the herpesvirus entry mediator, using surface plasmon resonance. J Virol. 1998;72:5937–5947. doi: 10.1128/jvi.72.7.5937-5947.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wittels M, Spear P G. Penetration of cells by herpes simplex virus does not require a low pH-dependent endocytic pathway. Virus Res. 1990;18:271–290. doi: 10.1016/0168-1702(91)90024-p. [DOI] [PubMed] [Google Scholar]
- 46.WuDunn D, Spear P G. Initial interaction of herpes simplex virus with cells is binding to heparan sulfate. J Virol. 1989;63:52–58. doi: 10.1128/jvi.63.1.52-58.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]