Summary
Antibiotics inhibiting the fatty acid synthesis pathway (FASII) of the major pathogen Staphylococcus aureus reach their enzyme targets, but bacteria continue growth by using environmental fatty acids (eFAs) to produce phospholipids. We assessed the consequences and effectors of FASII-antibiotic (anti-FASII) adaptation. Anti-FASII induced lasting expression changes without genomic rearrangements. Several identified regulators affected the timing of adaptation outgrowth. Adaptation resulted in decreased expression of major virulence factors. Conversely, stress responses were globally increased and adapted bacteria were more resistant to peroxide killing. Importantly, pre-exposure to peroxide led to faster anti-FASII-adaptation by stimulating eFA incorporation. This adaptation differs from reports of peroxide-stimulated antibiotic efflux, which leads to tolerance. In vivo, anti-FASII-adapted S. aureus killed the insect host more slowly but continued multiplying. We conclude that staphylococcal adaptation to FASII antibiotics involves reprogramming, which decreases virulence and increases stress resistance. Peroxide, produced by the host to combat infection, favors anti-FASII adaptation.
Subject areas: Microbiology, Microbial metabolism
Graphical abstract
Highlights
-
•
Anti-FASII bypass via fatty acid incorporation reprograms S. aureus proteins
-
•
Anti-FASII-adaptation leads to increased stress tolerance and reduced virulence
-
•
Peroxide accelerates fatty acid incorporation and anti-FASII adaptation
-
•
Membrane phospholipids are proposed as primary triggers for fitness adjustments
Microbiology; Microbial metabolism
Introduction
Staphylococcus aureus is a gram-positive opportunistic bacterium that remains a major cause of disease and mortality in humans and animals. The unsolved crisis of non-treatable infections, notably due to methicillin-resistant S. aureus (MRSA), underlines the need for alternative treatments, especially in compromised patients.1,2,3,4
Enzymes of the fatty acid (FA) synthesis pathway (FASII) are longtime candidate targets for drug development against S. aureus infections.5,6 FabI, enoyl-ACP reductase, was a preferred target as a narrow spectrum inhibitor that would not disrupt the gut microbiome during treatment.7 However, while FASII inhibitors (anti-FASII) targeting FabI effectively reached their targets in S. aureus, bacterial growth continued in in vitro or mouse bacteremia models. In a process termed FASII bypass, bacteria incorporate environmental FAs (eFAs), which are abundant in host biotopes, and use them directly to produce Firmicute pathogen membrane phospholipids8,9,10,11 (Figure S1 schematizes FASII and FASII bypass pathways). Exposure to host lipids during septicemic infection thus favors S. aureus adaptation to anti-FASII treatment.10 The considerable investment in developing these drugs overlooked the risks of accumulating anti-FASII-adapted bacterial populations in lipid-rich host organs, which apply to streptococcal, enterococcal, and staphylococcal infections.8,9,10,11 New anti-FASII drugs are in continued development against these Firmicutes without systematic vetting of their limitations.12,13,14,15 Characterizing the anti-FASII-adapted S. aureus populations that might persist in the host after treatment is thus crucial for future use.
We report here that anti-FASII treatment results in the emergence of a unique S. aureus fitness state. Anti-FASII-adapted bacteria produce less virulence factors. In contrast, stress responses are activated, and adapted bacteria show greater peroxide tolerance. Pre-exposure to peroxides accelerates anti-FASII adaptation by enhancing FA incorporation, thus identifying a new strategy by which peroxide priming facilitates antibiotic survival. These findings elucidate the state of anti-FASII-adapted bacteria that persist in the host after anti-FASII treatment. The present study provides the groundwork for reframing the uses of anti-FASII drugs for any future uses.
Results
Anti-FASII treatment leads to long-term S. aureus adaptation without detectable chromosomal rearrangements
Our previous studies ruled out a role for point mutations or indels in S. aureus adaptation to anti-FASII in serum-containing medium; studies were performed using either of two potent FabI inhibitors, triclosan16 or AFN-1252, a pipeline antimicrobial with high FabI specificity,7 giving equivalent results.10 As large chromosomal rearrangements are not detected by mi-Seq,10 we resorted to nanopore sequencing of four anti-FASII (AFN-1252)-adapted (AD) and two non-treated (NT) cultures. No modifications specific to anti-FASII adaptation were detected, showing that anti-FASII-adaptation occurs in the absence of chromosomal rearrangements (including inversions, deletions or amplifications), or point mutations as verified previously. Once adapted to anti-FASII, growth was robust in SerFA liquid medium (BHI supplemented with serum and fatty acids); however, bacteria grew poorly in FA-free medium (Figure S2). We conclude that alterations leading to anti-FASII adaptation are phenotypic, and not genotypic. For example, epigenetic modifications, and/or activation of a positive feedback loop might maintain anti-FASII-adapted bacteria in the adapted state; these possibilities remain to be investigated.
Anti-FASII adaptation involves massive protein reprogramming
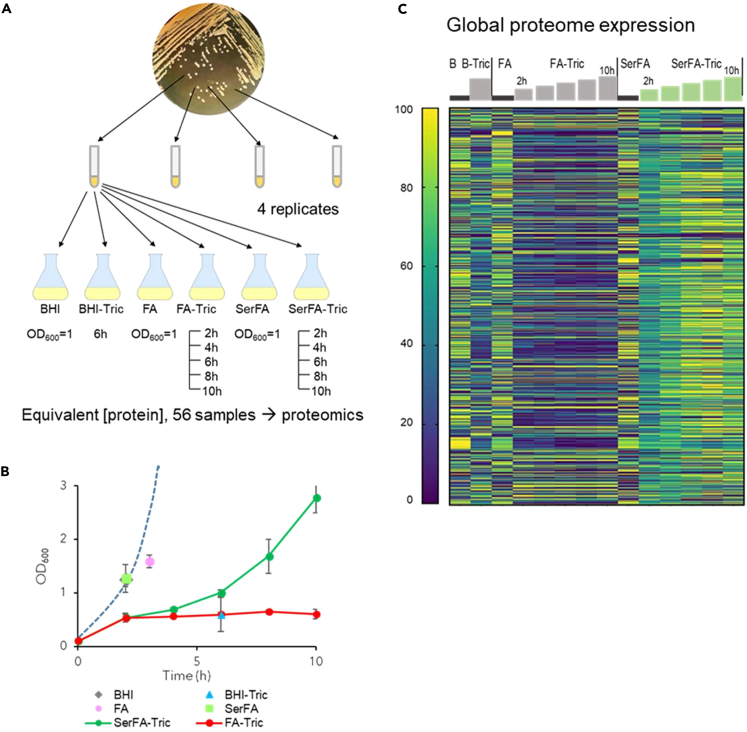
A proteomics approach was used to elucidate changes occurring during anti-FASII adaptation. FASII inhibition is overcome by eFAs provided in the presence of serum. Serum protects bacteria from toxic free FAs, and promotes FASII bypass.10 A complete kinetics study on S. aureus USA300 used the anti-FASII triclosan, both without and with added serum, at 2, 4, 6, 8, and 10 h post-anti-FASII addition (Figures 1A–1C respectively, show strategy, growth kinetics, and proteome results; Tables S1 subtabs 1 and 2 give complete results according to cluster analyses). As expected, protein abundance was mainly lower in triclosan medium (FA-Tric) without serum, where adaptation occurs by mutation emergence.11 In contrast, massive protein level changes in adaptation (SerFA-Tric) medium suggest metabolic activity in latency phase prior to full outgrowth (compare SerFA and SerFA-Tric, Figure 1C).
Figure 1.
Proteomic kinetics and global expression changes related to S. aureus USA300 strain adaptation to anti-FASII
(A) Schematics of sample preparation. Samples (4 biological replicates) were grown and prepared in conditions and harvesting times (or OD600 for controls) as indicated below flasks.
(B) Sample growth. Means and standard deviations are indicated. Points correspond to sampling times. See Table S1 for details. Dashed blue line represents typical growth in SerFA medium determined separately. S. aureus adapts to anti-FASII within 10h on SerFA-Tric, but not on FA-Tric, after a latency period (double arrow).
(C) Heatmap of proteins showing altered levels in at least 1 condition relative to other samples. All sample conditions are shown. Sampling times (h) previous steps correspond to 2, 4, 6, 8, and 10 h for FA-Tric and SerFA-Tric. The heatmap shows global protein changes, and is determined relative to weighted value for each protein, as on scale at left (navy, down-represented; yellow, up-represented; see STAR Methods for analyses).
We first exploited the data to screen for proteins whose levels selectively increased in the presence of specific signals related to FAs and/or anti-FASII (Figure S3). Highest expression of several Type 7 secretion system proteins (comprised between SAUSA300_0278 and SAUSA300_0289) occurred in FA conditions, as reported.19 The FarE FA efflux pump was also upregulated, presumably to avoid accumulation and toxicity of free FAs;20 as we reported, induction was alleviated in serum, which lowers toxicity.10 Overall, serum markedly affects S. aureus responses to anti-FASII, thus highlighting its important role in adaptation.
Proteins related to FASII and phospholipid metabolism mainly increased in SerFA-Tric as compared to all other conditions, notably in FA-Tric without serum (Figure S4). This suggests that the major regulator FapR is in a depressed state during anti-FASII adaptation.21 PlsC (1-acyl-sn-glycerol-3-phosphate acyltransferase), is required for phospholipid synthesis and anti-FASII adaptation. After an initial decrease, PlsC levels were restored during adaptation outgrowth, in accordance with results using a reporter fusion.21 This result further underlines the importance of serum in enabling FASII bypass.
We focus in the following text on the changes occurring in serum-supplemented medium containing anti-FASII (SerFA), in which FASII bypass occurs without genetic modification. Three categories of proteins, regulators, stress response proteins, and virulence factors, are analyzed.
Anti-FASII adaptation triggers shifts in regulatory protein expression and phosphorylation
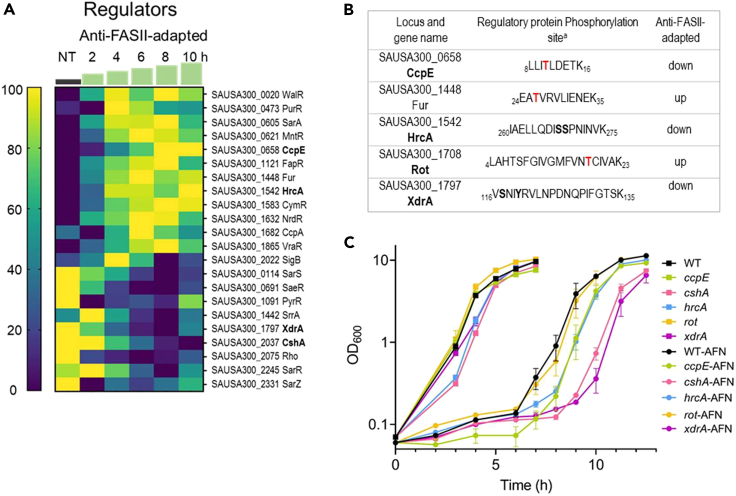
Regulatory proteins are likely implicated in the massive changes during anti-FASII adaptation. We detected 22 hypothetical or confirmed regulators whose expression decreased (9) or increased (13) during adaptation, affecting stress response, virulence, nutritional immunity and metabolism, or combinations of these factors (Figure 2A). Phosphorylation is a frequent post-translational modification that alters functions of numerous regulatory proteins.22,23,24 We performed a phosphoproteomics analysis with a focus on this protein class. Five regulators showed altered phosphorylation patterns during transition or upon adaptation to anti-FASII (Figure 2B; Figure S5; Table S2 for full results). Based on both omics results, we selected CcpE, CshA, HrcA, Rot, and XdrA as showing high expression and/or phosphorylation changes at or just prior to adaptation outgrowth (8–10 h) (Figures 2A and 2B) for further study.
Figure 2.
Anti-FASII affects pools and phosphorylation status of regulatory proteins
(A) Expression of known or putative regulators that differ in at least one time point during anti-FASII adaptation (SerFA-Tric) compared to non-treated (NT) SerFA sample. Changes in putative regulatory protein levels are shown in anti-FASII adaptation SerFA medium (see Table S1 for all test conditions). Gene loci and names are at right. Times (h) of sampling are indicated above green steps. Heatmap (scale at left) is determined relative to weighted value for each protein (navy, down-represented; yellow, up-represented; see STAR Methods for analyses).
(B) Phosphorylated regulatory proteins with altered expression during anti-FASII adaptation. Peptides showing differential phosphorylation at 10 h post-anti-FASII adaptation are indicated (see Table S2 for complete information). a Peptide positions in respective protein sequences are indicated. Phosphorylated amino acids are in bold red or in bold black when there is an ambiguity.
(C) Effects of regulator gene inactivation on anti-FASII adaptation. Kinetics of anti-FASII-adaptation of USA300 (WT) and insertional mutants, based on regulators with altered protein and/or phosphorylation levels (A and/or B; selected loci in bold), were compared to that of the parental strain. Cultures were grown without (squares) and with anti-FASII AFN-1252 (circles). Growth curves and standard deviations are based on biological triplicates.
To determine how the selected regulators affected S. aureus anti-FASII adaptation, we compared growth kinetics of USA300 and the corresponding mutant strains17 (Figure 2C). Adaptation was delayed by ∼2 h in the cshA mutant. CshA controls the supply of acetyl-CoA; its interruption results in phospholipids enriched in saturated FAs (Khemici et al., 2020). We propose that the membrane alteration induced by cshA inactivation might interfere with eFA incorporation.
The xdrA mutant also showed a 2 h adaptation delay. Both protein and phosphorylation levels of XdrA decreased during anti-FASII treatment. The putative FA degradation (Fad) gene fadX is reportedly upregulated in an xdrA mutant.25 Fad might transiently compete for eFAs and limit their availability for FASII bypass. Additionally, XdrA DNA binding activity overlaps that of the CodY regulator at the capsule synthesis locus.26 We recently showed that a codY mutant is delayed in adaptation, which we proposed relates to a positive regulatory effect of CodY on Acc expression.21 The delay in anti-FASII adaptation of the xdrA mutant might relate to a coordinate role between XdrA and CodY. Finally, hrcA and ccpE mutants showed minor changes in anti-FASII adaptation kinetics, while rot had no detectable change (Figure 2C).
Thus, while regulatory mutants individually affected anti-FASII-adaptation efficacy, no single tested regulator acted as a master controller. In Escherichia coli, stress response regulators respond to discrete extracytoplasmic signals, each of which independently contributes to stress adaptation.27 Similarly, differentially expressed regulators may respond to different stimuli consequent to membrane perturbation by anti-FASII, which cooperatively promote adaptation.
Reduced levels and activities of virulence factors in anti-FASII-adapted S. aureus
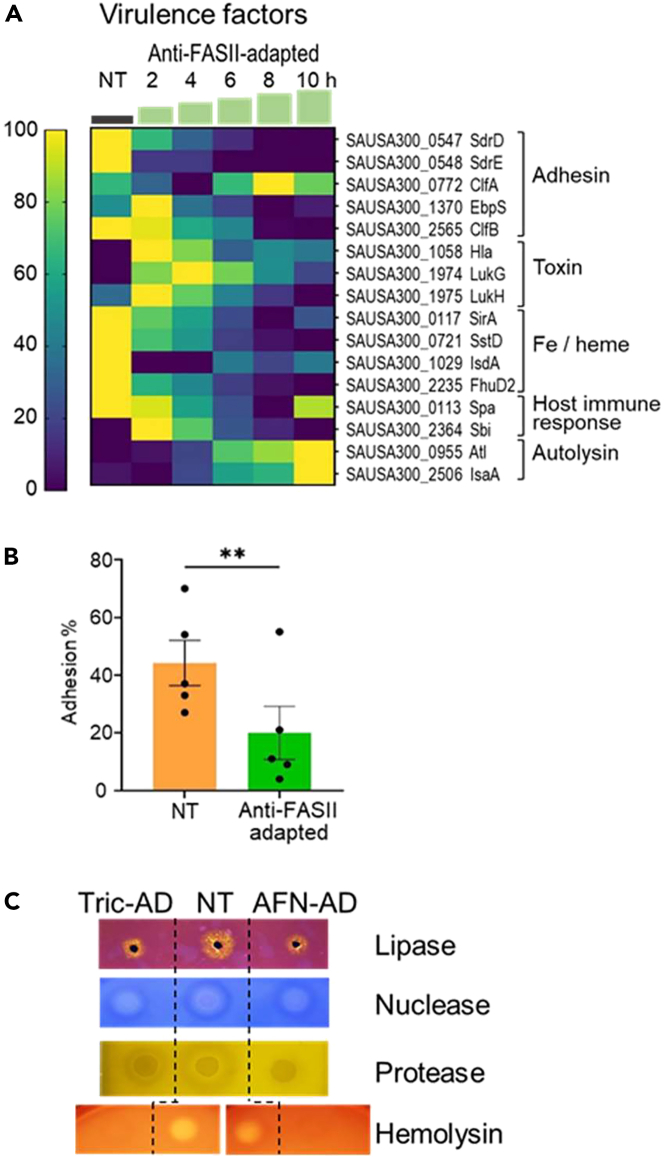
Levels of 10 out of 16 detected virulence-related proteins decreased transiently or durably during anti-FASII adaptation compared to non-treated S. aureus cultures (Figure 3A). Several adhesins showed marked decreases, including ClfB, which is required for virulence;28 iron receptor protein and toxin levels were also overall reduced. In contrast, two peptidoglycan hydrolases, Atl and IsaA, showed increased levels. Hydrolases are implicated in housekeeping, and also in virulence;29 their increased expression upon adaptation outgrowth might resolve cell wall thickening in the latency phase (PW, AG, JAM, in preparation). We asked whether the overall decreased levels of adhesion factors in adapted S. aureus (i.e., SdrD, SdrE, EbpS, and ClfB) might affect adhesion to host cells. Anti-FASII-adapted S. aureus (using AFN-1252) adhered poorly to human THP-1 macrophages compared to non-adapted bacteria (20% versus 44%, p < 0.01; Figure 3B). Secreted virulence factors were not detected by proteomics. We thus evaluated lipase, nuclease, protease, and hemolysin in non-treated versus anti-FASII-adapted S. aureus cultures (Figures 3C and S6). Activities of all exoproteins except nuclease were visibly lower in triclosan- or AFN-1252- adapted cultures. Altogether, these results suggested that anti-FASII-adapted S. aureus populations may be less fit for virulence.
Figure 3.
Expression changes during anti-FASII treatment
(A) Heat maps of known or putative virulence factors. Samples shown are in anti-FASII adaptation medium (SerFA; see Table S1 for results in all test conditions). Kinetics (in h) of sampling is indicated above green steps. Gene loci, protein names, and functional categories are at right. Correspondence between color and expression in heatmap (scale at left) is determined relative to weighted value for each protein (navy, down-represented; yellow, up-represented see STAR Methods for analyses).
(B) Differential NT and anti-FASII-adapted S. aureus adhesion to macrophage. Non-treated (NT) or AFN-1252-treated S. aureus USA300 (3x105) were added to of THP-1 macrophage monolayers (3x105 cells per well), and incubated for 1 h at 4°C. Colony forming units (CFU) of bacteria that adhered to macrophage were determined on five independent bacterial samples; means and standard errors are shown (p = 0.003; see STAR Methods).
(C) Secreted virulence factor activities. Non-treated (NT) and anti-FASII-adapted cultures treated with triclosan (Tric-ad) or AFN-1252 (AFN-ad) were grown overnight, and reached similar OD600 values (=13, 9 and 9 respectively for NT, Tric-ad, and AFN-ad). Cultures (for protease detection) and culture supernatants (lipase, nuclease, and hemolysin detection) were prepared (see STAR Methods) and spotted on appropriate detection medium. Representative results of 3 biologically independent replicates are shown (Figure S6 for replicate results).
Anti-FASII-adapted S. aureus produce increased levels of stress response proteins, with concomitant resistance to peroxide stress
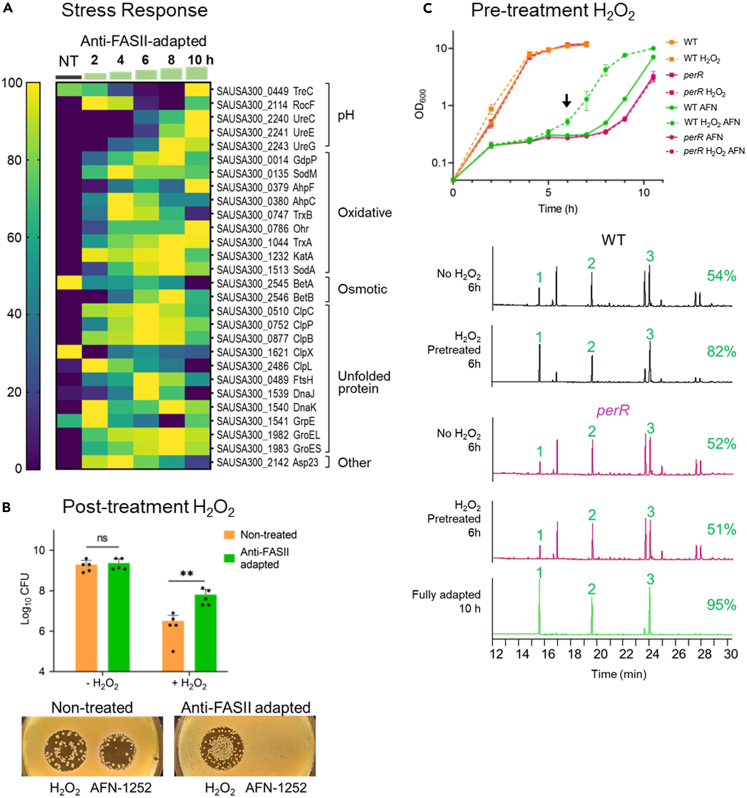
Levels of 26 of 28 assessed stress-related proteins, involved in pH, oxidative, osmotic, and unfolded protein responses, showed transient or lasting increases in anti-FASII-adapted S. aureus compared to the non-treated control. Among them, 18 proteins showed pronounced expression changes during adaptation outgrowth, at 8 to 10 h (Figure 4A).
Figure 4.
Anti-FASII adaptation confers oxidative stress resistance, and is accelerated by prior exposure to peroxide stress
(A) Stress response heatmap. Results are shown for anti-FASII adaptation medium (SerFA; see Table S1 for all test conditions). Sampling times (h) are indicated above green steps. Gene names and functional categories are at right. Heatmap (scale at left) is determined relative to weighted value for each protein (navy, down-represented; yellow, up-represented; see STAR Methods for analyses).
(B) S. aureus anti-FASII adaptation confers increased H2O2 resistance. Upper: USA300 non-treated (NT, orange bar) and AFN-1252-adapted overnight cultures (AD, green bar) were challenged with 0.5 mM H2O2 for 5 h, and CFUs were determined; means and standard errors are shown. ∗∗, p < 0.01. Lower: Lawns of NT and AD cultures (100 μL of dilutions adjusted to OD600 = 0.1) were prepared on SerFA solid medium, and plates were spotted with 1.5 mm H2O2 and 4 nm (1.5 μg) AFN-1252, and photographed after 48 h incubation at 37°C. Representative of 3 independent assays.
(C) Priming S. aureus with H2O2 accelerates anti-FASII adaptation and requires PerR. Upper: USA300 (WT) and perR (SAUSA300_1842) mutant SerFA cultures were grown overnight without or with 0.5 mM H2O2. Cultures were diluted (OD600 = 0.1) in SerFA without or with 0.5 μg/mL AFN-1252, and growth was monitored. Results are shown for biological triplicates. H2O2 –primed samples, without or with AFN addition at T0 are as indicated. Lower: FA profiles of indicated strains harvested at 6 h post-anti-FASII treatment (arrow in “C”). In green, FA profile of fully adapted H2O2-pretreated cultures at 10 h, shown here for perR and non-distinguishable from WT. 1, 2, and 3, eFAs present in SerFA medium (respectively C14, C16, and C18:1). At left of each profile, proportions of incorporated eFAs (%) are the average of two independent measurements (<3% difference between replicates).
Host production of reactive oxidative species (ROS) is a documented defense against pathogen infection.30 We asked whether increased levels of stress-related factors in anti-FASII-adapted S. aureus affected tolerance to ROS. To test this, non-treated and anti-FASII-adapted cultures were subjected or not to 0.5 mM H2O2 for 5 h, followed by plating for colony forming units (CFUs) on solid medium. Anti-FASII-adapted S. aureus CFUs were ∼20-fold higher than for non-treated bacteria (p < 0.01; Figure 4B upper). We also prepared lawns of non-treated and anti-FASII-adapted bacteria on solid SerFA agar, and spotted them with H2O2, or with AFN-1252 as reference (Figure 4B lower). Anti-FASII-adapted S. aureus displayed enhanced growth within the H2O2 inhibitory zone was stimulated compared to non-treated cultures. As expected, only anti-FASII-adapted S. aureus are entirely refractory to AFN-1252. Both approaches confirm that anti-FASII-adapted bacteria grow better when suddenly exposed to peroxide, which might confer an advantage during infection.
Priming with peroxides prior to anti-FASII challenge stimulates adaptation by enhancing eFA incorporation
Stress response induction by anti-FASII led us to ask the contrary, whether stress priming prior to antibiotic challenge would stimulate anti-FASII bypass. A ROS priming effect reportedly induces tolerance to various antibiotics in E. coli.31,32,33 The effect of ROS priming on anti-FASII adaptation in S. aureus was tested by subjecting cultures to overnight growth without or with H2O2 (0.5 mM). Cultures were then diluted in H2O2-free medium to compare kinetics of anti-FASII (AFN-1252) adaptation (Figure 4C upper). H2O2 priming shortened the adaptation time by 1.5–1.7 h compared to non-primed WT cultures; similar effects were obtained by priming with phenazine-methosulfate (PMS, a redox cycling compound; 20–50 μM;31 Table 1). Conversely, treating the WT strain with reducing agents (Na citrate 10 mM, or vitamin C 5.7 mM) retarded adaptation by 3.5 and 1.7 h, respectively; doubling the vitamin C concentration led to a > 16 h delay (Table 1). The S. aureus oxidation state is thus implicated in anti-FASII adaptation.
Table 1.
ROS pretreatment accelerates S. aureus adaptation to anti-FASII; treatment with reducing agents slows adaptation
| Straina | Additiveb | Time (h) to exit (SD) | Δ anti-FASII exit (h) compared to WTc | N | p valued |
|---|---|---|---|---|---|
| WT | - | 8.2 ± 0.26 | – | 12 | – |
| WT | H2O2 | 6.4 ± 0.51 | +1.8 | 12 | 0.0001 |
| WT | PMS | 6.6 ± 0.41 | +1.6 | 5 | 0.0003 |
| katA | – | 7.0 ± 0 | +1.2 | 3 | 0.0022 |
| katA | H2O2 | 5.7 ± 0.29 | +2.5 | 3 | 0.0022 |
| perR | - | 9.5 ± 0 | −1.3 | 3 | 0.0022 |
| perR | H2O2 | 9.5 ± 0 | −1.3 | 3 | 0.0022 |
| sufD∗ | – | 8.1 ± 0.25 | −0.1 | 4 | Ns |
| sufD∗ | H2O2 | 7.3 ± 0.24 | +0.8 | 4 | 0.0005 |
| WT | Na citrate | 11.5 ± 0.71 | −3.3 | 2 | ND |
| WT | VitC 1 | 9.8 ± 0.35 | −1.6 | 2 | ND |
| WT | VitC 2 | >24 h | > −16 | 2 | ND |
Strains USA300 (WT) or mutant derivatives katA SAUSA300_1232, perR SAUSA300_1842, or sufD∗ intergenic between sufC (SAUSA300_0818) and sufD (SAUSA300_0819), at position 898566,17,18 are used.
Oxidizing agents were added only to SerFA precultures: H2O2, 0.5 mM; phenazine-methosulfate (PMS), 20 (n = 2), 30 (n = 1), 40 (n = 1), 50 (n = 1) μg/mL, gave equivalent results. Reducing agents were added at the same time as anti-FASII, here AFN-1252 (SerFA-AFN): Na citrate, 10 mM; vitamin C (VitC 1, 5.7 mM or 1 mg/mL, or VitC2, 2 mg/mL).
Bacterial outgrowth for USA300 in SerFA-AFN (top line) without additives is used as reference. Differences (Δ) are: +, early exit from latency (h); -, delayed exit (h). SD, standard deviation; N, number of independent experiments.
p values indicate the statistical difference between growth of SerFA-AFN WT (top row) and the indicated mutants and/or conditions, determined by the Mann-Whitney test, using GraphPad Prism 9.5.1 software. p values <0.05 are indicated. Nd, not determined; Ns, not significant.
We asked how H2O2 priming affects eFA incorporation during adaptation. Cultures prepared as aforementioned were harvested for FA profile analysis 6 h after anti-FASII treatment (using AFN-1252), i.e., during the latency phase. H2O2 priming increased eFA incorporation efficiency compared to non-primed cultures (respectively, 82% versus 54% eFAs; Figure 4C lower). We conclude that H2O2 priming enhances eFA incorporation, which accelerates adaptation to the FASII antibiotic.
PerR is required for H2O2 priming of anti-FASII adaptation
PerR (SAUSA300_1842) is a conserved metal-dependent H2O2 sensor and regulator protein.34 As PerR senses H2O2, we examined its implication in H2O2-primed anti-FASII adaptation. Unlike the USA300 parent, perR mutant adaptation was not stimulated by H2O2 priming (Figure 4C upper). This indicates that one or more PerR-controlled functions are involved in H2O2 priming.
The PerR regulon is annotated as comprising suf operon genes. Suf, an H2O2-induced iron-sulfur cluster repair system, leads to antibiotic efflux via the AcrAB efflux pump and antibiotic tolerance in E. coli.31,33 However, anti-FASII efflux would lead to anti-FASII-sensitivity; in contrast, the aforementioned results show that eFA incorporation was even more efficient in peroxide-primed compared to non-primed cultures (Figure 4C lower). To exclude involvement of suf in the H2O2 priming response and anti-FASII adaptation, we compared WT responses to those of a transposon insertion inactivating the suf homolog in S. aureus.18 Anti-FASII adaptation was stimulated in priming of both WT and suf strains, ruling out a critical role for Suf in adaptation (Table 1).
PerR also represses katA, a catalase that degrades H2O2; a katA mutant lacks catalase and would accumulate H2O2. Anti-FASII adaptation in katA was ∼1 h shorter than in the WT parental strain. H2O2 priming of the katA mutant further shortened the adaptation time by ∼2 h relative to WT without peroxide (Table 1). katA is presumably induced in the perR mutant, which would annihilate the peroxide priming effect, as consistent with results (Figure 4C).
These results link peroxide priming and KatA repression by PerR, to faster eFA incorporation and anti-FASII adaptation. They clearly distinguish a tolerance state involving antibiotic efflux and persistence as shown previously,31,32,33 from anti-FASII adaptation, where the antibiotic reaches its target without preventing bacterial growth.
Anti-FASII-adapted S. aureus are less virulent, yet persist in a Galleria mellonella infection model
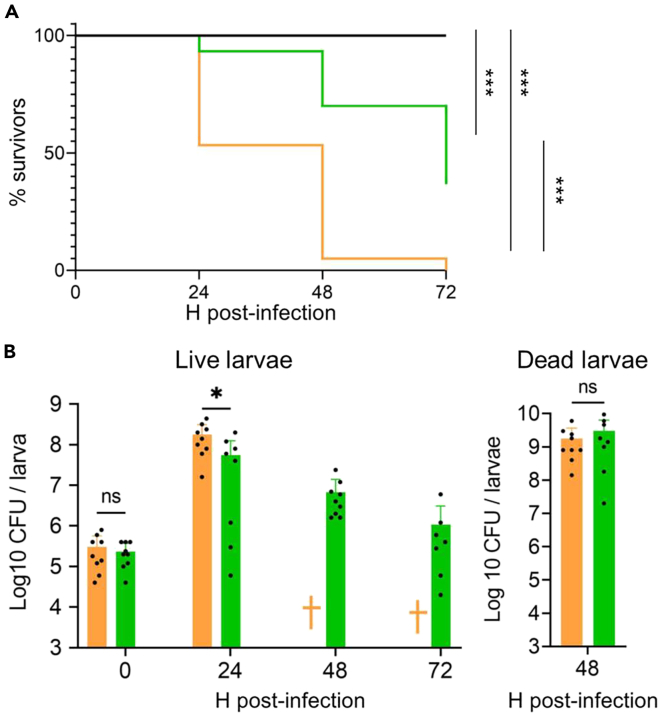
Lower virulence factor production but greater stress resistance due to anti-FASII adaptation raises questions on the outcome of anti-FASII-adapted S. aureus infection. We compared anti-FASII-adapted versus non-treated S. aureus in an insect G. mellonella model, which allows the use of a large cohort. Moreover, larval hemocoel, like blood serum, is lipid-rich.35 AFN-1252 was used for this study, as it is non-toxic in animals,36,37 and we showed previously that AFN-1252 treatment did not stop infection in a mouse model.10 Non-treated and AFN-1252-adapted S. aureus USA300 showed equivalent growth kinetics in SerFA in vitro (Figure S2). Insects infected by 106 anti-FASII-adapted bacteria were killed more slowly than those infected by equivalent CFUs of untreated bacteria, as monitored over 72 h post-infection (Figure 5A). At T48, 95% of insects were killed by untreated S. aureus, compared to 30% killing with anti-FASII-adapted S. aureus. At T72, when all larvae infected by untreated S. aureus were dead, anti-FASII-adapted S. aureus killed over 60% of the insects. CFUs were undetected in only two larvae infected by anti-FASII-adapted bacteria (one at 24 h and one at 72 h), which may have cleared infection. Overall, killing by anti-FASII-adapted S. aureus was delayed but not stopped.
Figure 5.
Comparison of untreated and anti-FASII-treated S. aureus in a G. mellonella infection model
Insects were injected with 106 CFU S aureus USA300 that were either non-treated (NT) or preadapted to anti-FASII AFN-1252 (AD).
(A) Insect mortality in NT and AD S. aureus. Survival was plotted using Kaplan-Meier with pooled values of biologically independent triplicates (60 insects per condition). Survival curves between treatment groups were analyzed by log rank (Mantel-Cox) tests, which showed that both NT and AD survival kinetics were significantly different from the PBS control group and from each other p values <0.001 (∗∗∗). The Cox proportional hazard model, conducted between NT and AD infected larvae, confirms that NT-infected larvae had 8.77-fold higher hazard than AD-infected larvae infected (p value = 0.001). (B) legend: “Left, bacterial CFUs in surviving larvae. CFUs were determined on insects infected as described in “A” and at the same time intervals. For NT, standard deviation (std dev) values were based on 9 insects at both 0 and 24 h points. For AD, the means and standard deviation were based on 9, 9, 9, and 8 for respective consecutive time points. †, no surviving insects. As all NT-infected larvae died by 48h, significance between NT and AD was only determined at 0 h and 24 h. This was performed by non-parametric Mann-Whitney test. Further experimentation is needed to analyze the significance of declining CFUs in the AD-treated insects. Right, bacterial CFUs in dead infected insects (9 NT- and 8 AD-infected larvae) at 48 h. Analyses were done using the non-parametric Mann-Whitney test (GraphPad Prism 9.5.1 software). ∗, p = 0.02; ns, non-significant.
CFUs from insects infected by anti-FASII-adapted bacteria were about 3-fold lower at 24 h compared to those infected by non-treated bacteria. Greater larval killing may be due to the higher number of CFUs and/or to the greater virulence of non-treated compared to anti-FASII-adapted bacteria. The underlying reasons for insect death, i.e., greater CFUs, and/or differential expression in the NT and AD states, will require further investigation. CFUs continued to decrease in surviving insects infected by AD (Figure 5B left). In contrast, CFUs in dead larvae at T48 were comparable for both groups, indicating that anti-FASII-adapted bacteria multiplied as well as non-treated bacteria in the insects they killed (Figure 5B right).
To determine whether anti-FASII-adapted bacteria remained adapted during infection, CFU platings as aforementioned were done in parallel on SerFA and SerFA-AFN solid media. Only anti-FASII-adapted bacteria formed colonies on SerFA-AFN solid medium (Table S3). Among surviving insects infected with adapted bacteria, only one out of nine contained bacteria that returned to the non-adapted state at 72 h; this indicates that bacteria remained mainly adapted during insect infection. Altogether, these results show that anti-FASII-adapted S. aureus are less virulent, but can multiply in the insect host. Greater stress resistance capacity may help withstand host conditions and can explain bacterial persistence in the insect host.
Discussion
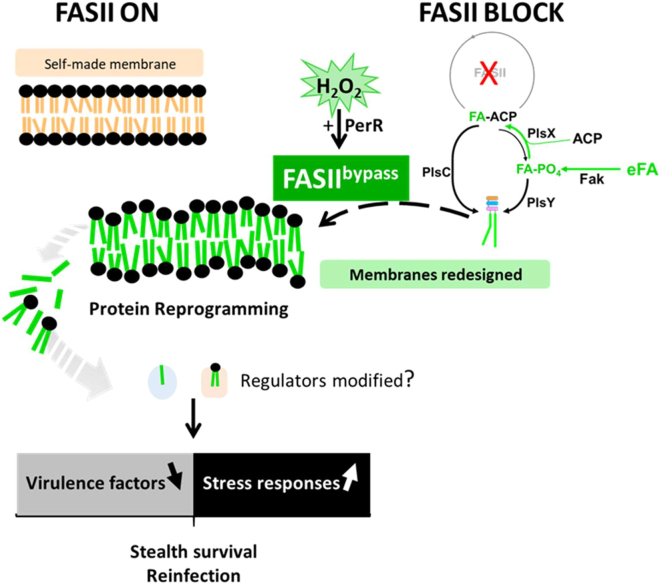
eFA incorporation during FASII-antibiotic-induced bypass redesigns S. aureus membrane phospholipids, leading to massive shifts in protein expression, and a prolonged adaptation state without detectable genomic rearrangements or mutations (see Figure 6 model). These changes, which begin before bacterial outgrowth, alter the S. aureus fitness state toward reduced virulence and greater stress tolerance. Importantly, peroxide priming accelerates adaptation; rather than inducing efflux and antibiotic tolerance,31,32,33 priming stimulates eFA incorporation and a bona fide adaptation. The in vitro phenotypes of anti-FASII-adapted bacteria are consistent with their insect infection capacity, where they kill insects more slowly but continue multiplying in the host. We propose that membrane lipid alterations, as provoked by anti-FASII, comprise the primary signal leading to the massive changes in protein profiles.
Figure 6.
Summary model of anti-FASII adaptation and S. aureus fitness
S. aureus synthesizes fatty acids (FAs) to produce membrane phospholipids (upper left, FAs in orange). Anti-FASII treatment in SerFA medium promotes FASII bypass, during which exogenous FAs (eFAs, in green) are incorporated and constitute the membrane phospholipid FAs.10 FASII bypass is accelerated by H2O2 priming, which requires PerR, but is lower if KatA is present. Anti-FASII adaptation is accompanied by massive changes in protein expression. Membrane perturbation in the new phospholipid environment is proposed as the primary signal for protein reprogramming; as reported, membrane FAs or phospholipids may also shed internally and bind to regulatory proteins to modulate their function.38,39,40,41 Decreased virulence factor production may help bacteria escape host immune surveillance.42 Up-regulation of stress response protein levels confers greater ROS tolerance, and could facilitate survival during infection.43 Peroxide priming accelerates FA incorporation and anti-FASII adaptation by a novel PerR-dependent process. Anti-FASII treatment would favor emergence of S. aureus populations that are transiently less infectious, but that may persist in the host.
Various regulators are implicated in anti-FASII adaptation, e.g., XdrA or CshA, and to lesser extents HrcA or CcpE. However, no single regulator could be attributed a “master” role. We hypothesize that membrane perturbations, as caused by anti-FASII, are primary signals leading to reprogramming. Signaling lipids may also be liberated, which could modulate regulator activities. For example, FAs and cardiolipin reportedly affect activities of the S. aureus two-component system SaeRS.38,39 FAs may regulate virulence gene expression by binding to regulator proteins, as shown in Vibrio cholerae.40 Interactions between regulators and lipid species released during anti-FASII treatment may be a rapid means to adjust regulator functions for FASII bypass (Figure 6).
This study establishes a connection between oxidative stress and FASII antibiotic bypass, by showing that ROS priming stimulates eFA incorporation to boost adaptation (Figure 4). This priming mechanism is distinct from the antibiotic tolerance mechanism described in E. coli, where ROS priming favors antibiotic exclusion such that bacteria remain antibiotic sensitive.31,32,33 H2O2-priming stimulates anti-FASII adaptation by increasing eFA incorporation, which rules out a role for antibiotic efflux.
Peroxides may have a direct role in stimulating FASII bypass: H2O2-mediated oxidation of FASII initiation enzymes could diminish competition with the FASII bypass system (Figure 6). A perR mutant, which activates KatA to degrade H2O2, lengthens adaptation time, and abolishes the H2O2 priming effect. Conversely, a katA mutant, which expectedly accumulates H2O2, shortens adaptation time. Direct or indirect H2O2 targets could be involved in disabling FASII to favor FASII bypass; this question is currently under study. Host macrophages and neutrophils generate peroxides presumably to control infection.44,45 S. aureus adaptation to anti-FASII would be potentiated by peroxides, which needs to be considered for future anti-FASII drug development.
Lower virulence, greater stress response, and exclusive eFA utilization during in anti-FASII-adapted staphylococci may provide benefits for survival. Interestingly, streptococci and enterococci preferentially incorporate eFAs, which repress FASII genes even without antibiotics.8,46,47 Although staphylococci and streptococci use distinct FASII regulation strategies (feedback versus feedforward in S. aureus47,48), they have common infection biotopes. FASII bypass may provide an energy gain and be advantageous during infection. Parallels also exist between anti-FASII-adapted S. aureus, and small colony variants (SCVs): both emerge more efficiently in oxidative stress,49,50 and both produce less virulence factors,42,51 which might facilitate bacterial escape from host immune surveillance. In both cases, bacterial reservoirs might lead to re-infection (Figure 6).
The use of anti-FASII in Firmicutes raises the risks of accumulating anti-FASII-adapted bacterial reservoirs in lipid-rich host biotopes.8,9,10,11,52 Remarkably, this has not stopped development of anti-FASII drugs to treat Firmicute infections, mainly against S. aureus.12,13,14 Triclosan, an anti-FASII with non-specific side effects, was widely used as an antiseptic for decades before being partially banned. The emergence of triclosan-resistant point mutants and adaptation to triclosan are documented; effects on mammalian cells were also reported.9,53,54,55,56 Residual S. aureus populations that persist after anti-FASII treatment would provide a reservoir for chronic infections.
While anti-FASII does not eliminate S. aureus when compensatory eFAs are available, we showed that it does reduce virulence factor production. Combining anti-FASII with synergistic treatments that prevent adaptation could potentiate its efficacy.21 As anti-oxidants delay anti-FASII adaptation (Table 1), they may offer perspectives for a bi-therapy approach that exploits reduced virulence in anti-FASII-adapted bacteria by slowing or stopping outgrowth.
Limitations of the study
This study gives insights into S. aureus responses during adaptation to FASII-directed antibiotics in selected conditions relevant to the host environment, namely by adding serum and FA to medium. It will be interesting to determine whether other FASII antibiotics and modified environments, e.g., different sera, lipids, and organ extracts, would induce other changes in expression during ant-FASII adaptation. This would be of particular importance when evaluating oxidative stress and virulence factors produced during S. aureus adaptation to FASII antibiotics. CFU comparisons in surviving larvae were limited to 0 and 24 h time points, as all insects in the NT group were dead at 48 h. Greater larval killing may be due to higher CFUs and/or to the greater virulence of NT compared to AD bacteria. Further experimentation and deeper statistical analyses, which are beyond the scope of this work, will be needed to analyze the correlations between insect death and CFUs remaining in insects. More evolved animal models should be used to confirm results in the insect infection model, and pursue the possibility of persistent infection by anti-FASII-adapted S. aureus.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial strains | ||
| USA300 FPR3757 JE2 (referred to as USA300) | BEI Resources Nebraska Library |
https://doi.org/10.1128/mBio.00537-12 |
| Transposon insertions in USA300 FPR3757 | ||
| SAUSA300_0658 (ccpE) | ||
| position 898566 between SAUSA300_0818 (sufC) and SAUSA300_0819 (sufD) | ||
| SAUSA300_1232 (katA) | ||
| SAUSA300_1542 (hrcA) | ||
| SAUSA300_1708 (rot) | ||
| SAUSA300_1797 (xdrA) | ||
| SAUSA300_1842 (perR) | ||
| SAUSA300_2037 (cshA) | ||
| Biological samples | ||
| Newborn calf serum | Sigma | 12023C-500ML |
| Sheep blood (5%) agar plates | Bio-Mérieux | 43041 |
| Powdered skim milk | Regilait | Supermarket |
| Chemicals, peptides, and recombinant proteins | ||
| Fatty acids C14, C16 | Sigma | CAS # and product #: 544-63-8 & M3128; 57-10-3 & P0500 |
| Fatty acid C18:1cis | Larodan, Sweden | CAS # & product #: 112-80-1 & 10-1801 |
| Triclosan (Irgasan) | Sigma | CAS # & product #: 3380-34-5 & 72779 |
| AFN-1252 | MedChem Express | CAS # & product #: 620175-39-5 & HY-16911 |
| Hydrogen peroxide (H2O2) | Sigma | CAS # & product #: 7722-84-1 & H1009 |
| Phenazine-methosulfate (PMS) | Sigma | CAS # & product # 299-11-6 & P9625 |
| Rhodamine B (for lipase detection) | Sigma | CAS # 81-88-9 |
| Deposited data | ||
| Nanopore DNA genome sequence | Mendeley | https://doi.org/10.17632/grt4htck9k.1 |
| Proteomics (14 conditions in quadruplicate) | Pride partner repository |
http://proteomecentral.proteome.xchange.org Identifier PPXD034256 |
| Phosphoproteomics data | Center for Computational Mass Spectrometry 395 repository University of California, San Diego |
http://massive.ucsd.edu/ProteoSAFe/status.jsp?task=db3f5002e32e4a47a71996358bc6ae8c Identifier MassIVE MSV000089781 |
| Experimental models: Cell lines | ||
| THP-1 differentiated macrophages | Institut Cochin collection | https://doi.org/10.1101/2022.12.02.518834 |
| Experimental models: Organisms/strains | ||
| Galleria mellonella | Micalis institute Facilities | |
| Oligonucleotides | ||
| Primer 5'-3' | Oligo name | Locus |
| GCAGGCACATATTGGTGAAGT | ccpE_F | SA300_0658 |
| GATCCACTGTAATAGTTGCATGG | ccpE_R | |
| GAAGAAGGATATGAATGGGT | sufD∗_F | position 898566 between SAUSA300_0818 and SAUSA300_0819 |
| GGCTTTAGAATAATCAACAAG | sufD∗_R | |
| GAACGCAACAAGGTATTGAA | katA_F | SAUSA300_1232 |
| CCTCAGTCATTACTTGAATAT | katA_R | |
| GAGCGACATAACTTGAATGTTAG | hrcA_F | SAUSA300_1542 |
| GCATAGCTGTAGGTCCAATCA | hrcA_R | |
| GCATTGCTGTTGCTCTACTTGC | rot_F | SAUSA300_1708 |
| CGACACTGTATTTGGAATTTTGCA | rot_R | |
| GGTTCGTATAGAGGCTGGTTA | xdrA_F | SAUSA300_1797 |
| GTACCGAAAATTGGTTGGTTATCT | xdrA_R | |
| CCAATATCTTTAAACACTCT | perR_F | SAUSA300_1842 |
| CATCATTGCGACAAGCAGGC | perR_R | |
| GGTCTGCTTCCACCGCTGC | cshA_F | SAUSA300_2037 |
| ACTTTTACAAGAGTTAGTAGAAGC |
cshA_R |
|
| Software and algorithms | ||
| D-Genies |
Dot plot large Genomes |
https://dgenies.toulouse.inra.fr/ |
| GraphPad Prism 9.5.1 | GraphPad Software Kaplan-Meier Log rank Mantel-Cox Cox proportional hazard Non-parametric Mann-Whitney |
https://www.graphpad.com/ |
| Rstudio | K-means clustering analysis | https://www.rstudio.com/products/rstudio/download/#download |
| X!TandemPipeline C++, MassChroQ, MCQR | Free resource proteomics analyses | https://forgemia.inrae.fr/pappso |
| Other | ||
| DNeasy® Blood & Tissue Kit | Qiagen | Cat No./ID: 69504 |
| HaltTM Protease & Phosphatase Inhibitor Cocktail (100X) |
Thermo Scientific, Fr | Référence: 78430 |
| Nuclease detection medium | DNase agar (Oxoid, Thermo Scientific, Fr) |
CM0321 |
Resource availability
Lead contact
Added information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Alexandra Gruss (alexandra.gruss@inrae.fr).
Materials availability
This study did not generate new unique reagents.
Data and code availability
All relevant data are within the manuscript, supporting information files, and depositories. Nanopore genome sequencing, Proteomic data, and phosphoproteomic data were respectively deposited in Mendeley, Pride partner repository, and Center for Computational Mass Spectrometry repository (U. California, San Diego). Accession numbers are listed in the key resources table. All data are publicly available as of publication. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Method details
Strains, media, and growth conditions
Experiments were performed using S. aureus SAUSA300_FPR3757 JE2 strain, referred to as USA300, and transposon insertion derivative strains from the Nebraska mutant library (17; generously supplied by BEI Resources NIAID, NIH, USA). All strains were confirmed for transposon insertion by PCR (see Table S4 for strains and primers used). Cultures were grown aerobically at 37°C. Solid and liquid growth media were based on BHI as follows: no additives (BHI), containing 0.5 mM FAs (BHI-FA, with an equimolar mixture of 0.17 mM each C14, C16, C18:1 [Larodan, Sweden]), and BHI-FA containing 10% newborn calf serum (Ser-FA; Eurobio Scientific, France [Fr]). Where specified, the anti-FASII triclosan16 was added at 0.25 μg/ml in media without serum, and at 0.5 μg/ml in media containing serum, to respectively give BHI-Tric, FA-Tric, or SerFA-Tric, as described.10,11,52 H2O2 (0.5 mM final concentration) was added to SerFA precultures when indicated. The anti-FASII AFN-12527 was used in SerFA at 0.5 μg/ml (SerFA-AFN) as described.10 For most experiments, S. aureus USA300 was streaked on solid BHI medium, and independent colonies were used to inoculate overnight SerFA pre-cultures. For proteomic and phosphoproteomic studies, cultures were inoculated at a starting OD600 = 0.1.
Nanopore sequencing
Six single colonies of USA00 were resuspended in SerFA and grown overnight. Cultures were then diluted in SerFA, SerFA-AFN-1252, 10% mouse serum, or 10% mouse serum containing AFN-1252, and grown to OD600 = 1. Whole chromosomal DNA was prepared from the independent cultures as described, and outsourced for nanopore sequencing (Eurofins, Germany). Whole genome DNA sequences were presented as circularized genomes, and entered in the Mendeley database https://doi.org/10.17632/grt4htck9k.1. Sequences were compared by performing full genome alignments https://dgenies.toulouse.inra.fr/ 09/02/2023.
Proteomics preparation
Adaptation to anti-FASII varies with growth media: BHI-Tric-grown and FA-Tric-grown S. aureus do not adapt in the time periods tested, (high frequency adaptive mutations arise with a delay in FA-Tric;11), whereas SerFA-Tric-grown S. aureus adapt without mutation after an initial latency period (6-8 hours, depending on growth conditions).10 Kinetics experiments were performed on USA300 to determine the protein changes associated with FA-Tric and SerFA-Tric anti-FASII adaptation. Cultures for each condition were prepared as independent quadruplicates. For each sample, BHI precultures were diluted and shifted to the specified medium starting cultures at OD600 = 0.1. Control cultures in BHI, BHI-FA and BHI-SerFA were grown to OD600 = ∼1. BHI-Tric cultures (no added FAs) were collected at 6 h. USA300 samples grown in FA-Tric and SerFA-Tric were collected at 2, 4, 6, 8 and 10 h post-antibiotic addition (see Figure 1A; Table S1 for growth conditions and complete data). For each sample, 20 OD600 units culture equivalent was collected and centrifuged for 10 min at 4°C at 8000 rpm. Pellets were washed twice in Tris 10mM pH7.0 containing 0.02% TritonX-100, and Halt™ Protease & Phosphatase Inhibitor Cocktail (100X) (Thermo Scientific, Fr). Pellets were then resuspended in 650 μl washing buffer, mixed with 0.1 mm silica beads and subjected to 3 cycles of vigorous shaking (Fast-Prep-24, MP-Bio, Fr). After 10 min centrifugation at 10000 rpm, supernatants were recovered and stored at -80°C prior to analyses.
Protein extractions, LC-MS/MS analyses, and bioinformatics, and statistical data analyses were done as described in detail.57 The reference genome GenBank Nucleotide accession code NC_007793.1 was used for protein annotation. The bioinformatic tools used for proteomics analysis (X!TandemPipeline C++, MassChroQ, MCQR) are open and free resources available in the following repository: https://forgemia.inrae.fr/pappso. The mass spectrometry proteomics data was deposited in the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the dataset identifier PXD034256.
Proteome data analyses
Protein abundance differences were detected by ANOVA tests for all methods used (spectral counting, SC; extracted ion chromatograms, XIC; and peak counting, PC). The abundance of a protein was considered significantly variable when the adjusted p-value was <0.05. To generate heat maps, data analyses were done as detailed previously.57,58 Temporal proteomic profiles were constructed using the relative protein abundances obtained from SC, XIC, and PC (Spectral counting, extract ion chromatogram, peak counting), using proteins showing significant abundance variation. These values were scaled using self-organizing tree algorithm (SOTA) clustering. Protein quantification, statistical methods and data analysis were conducted in R 3.3.2 using the following packages: ade4, clValid, ggplot2, lattice, lme4, made4, nlme and reshape2.58 The data was then normalized with GraphPad Prism 9.5.1, so that for each protein, values are between 0% and 100%, on a linear scale; protein levels appear as relative values. The numerical data is presented in Table S1, and primary data is entered in the PRIDE database (identifier: PXD034256). False discovery rates (FDR) were determined using X!tandemPipeline, which is based on the calculation of peptide and protein e-values, and filters on this (peptide e-value is 0.01 and protein e-value is 0.0001). The FDR is calculated with a reverse database in X!TandemPipeline. FDR obtained by this method was compared to the standards used in proteomics (< 1% FDR).57 Proteins whose abundancies were significantly different in one or more conditions were manually curated and classified according to functional groups.
Phosphoproteome preparation
USA300 cultures were prepared in BHI and SerFA and harvested at OD600 = ∼1. SerFA-Tric cultures were harvested at 6h and 10 h post-treatment. Samples were prepared independently from those in the proteomics study, as independent biological triplicates, and treated as in proteomics studies, except that we collected the equivalent of 50 OD600 units. Bacteria were processed as for proteome extraction except that Tris was replaced by triethylammonium bicarbonate (50 mM; n.b. Tris interferes with dimethyl tag labelling) containing antiprotease and antiphosphatase at the recommended concentrations. Lysed bacteria after Fast-Prep were centrifuged 15 min at 12000 rpm supernatants containing soluble proteins were kept at -80°C before use. Protein concentrations were determined by the Bradford method.
One mg protein samples were evaporated and resuspended in 1 ml 5 % formic acid and dimethyl-tag labeled as described.59 Briefly, differential on-column labeling of peptide amine groups (NH2) created dimethyl labels leading to mass shifts of +28.0313 Da for the peptides from SerFA 3 h samples, +32.0564 Da for the peptides from SerFA-Tric 6 h samples and +36.0757 for the peptides from SerFA-Tric 10 h samples respectively. The three samples were mixed and then submitted to six rounds of phosphopeptide enrichment with 5mg TiO2 beads/mg of protein (Titansphere Phos-TiO, GL Sciences Inc., Netherlands) as described.60
LC-MS/MS analyses of samples were done using an Ultimate 3000 nano-RSLC coupled on line with a Q Exactive HF mass spectrometer (Thermo Scientific, San Jose California). 1 μL of each sample was loaded on a C18 Acclaim PepMap100 trap-column 300 μm inner diameter (ID) x 5 mm, 5 μm, 100Å, (Thermo Scientific) for 3.0 minutes at 20 μL/min with 2% acetonitrile (ACN), 0.05% TFA in H2O and then separated on a C18 Acclaim Pepmap100 nano-column, 50 cm x 75 μm ID, 2 μm, 100 Å (Thermo Scientific) with a 100 minute linear gradient from 3.2% to 20% buffer B (A: 0.1% FA in H2O, B: 0.1% FA in ACN), from 20 to 32% of B in 20 min and then from 32 to 90% of B in 2 min, hold for 10 min and returned to the initial conditions. The flow rate was 300 nL/min.
Labeled peptides were analyzed with top15 higher energy collisional dissociation (HCD) method: MS data were acquired in a data-dependent strategy selecting fragmentation events based on the 15 most abundant precursor ions in the survey scan (m/z range from 350 to 1650). Resolution of the survey scan was 120,000 at m/z 200 Th and for MS/MS scans the resolution was set to 15,000 at m/z 200 Th. For HCD acquisition, the collision energy = 27 and isolation width = 1.4 m/z. Precursors with unknown charge state, charge state of 1 and 5 or greater than 5 were excluded. Peptides selected for MS/MS acquisition were then placed on an exclusion list for 20 s using the dynamic exclusion mode to limit duplicate spectra. The mass spectrometry proteomics data are deposited to the Center for Computational Mass Spectrometry repository (Univ. California, San Diego) via the MassIVE tool, dataset identifier MassIVE MSV000089781: http://massive.ucsd.edu/ProteoSAFe/status.jsp?task=db3f5002e32e4a47a71996358bc6ae8c.
Phosphoproteome data analyses
Proteins were identified by database searching using SequestHT with Proteome Discoverer 2.5 software (Thermo Scientific, Fr) against the Uniprot S. aureus USA300 database (2020-01 release, 2607 sequences). Precursor mass tolerance was set at 10 ppm and fragment mass tolerance was set at 0.02 Da, and up to 2 missed cleavages were allowed. Oxidation (M), acetylation (Protein N-terminus), and phosphorylation (S, T, Y) were set as variable modifications. The differentially dimethyl-labeled peptides in primary amino groups K and N-ter (see above), and carbamidomethylation (C) were set as fixed modifications. Peptides and proteins were filtered with a false discovery rate (FDR) at 1% using the Percolator tool.61 Protein quantitation was performed with precursor ions quantifier node in Proteome Discoverer 2.5 software, peptide and protein quantitation based on pairwise ratios and t-test statistical validation.
Macrophage adhesion
Confluent cell lawns of THP-1 macrophages were prepared as described to obtain confluent cell lawns of 3x105 cells per well.62 S. aureus was pre-cultured in SerFA medium and then subcultured overnight in SerFA medium without or with AFN-1252. The next day, bacteria were subcultured in the same corresponding fresh media to OD600 =1-2. Bacteria were then washed twice with PBS buffer, and diluted in RPMI GluMax (Gibco, France) to obtain an MOI = 1. One ml of the dilution was added in each well in 24-well plates. Plates were centrifuged at 1000 rpm for 5 minutes to sediment bacteria, and then incubated at 4°C for 1h. After incubation, wells were washed 3 times with PBS, then 1ml of sterile cold water was added to each well and left for 5min at room temperature. Finally, the cells and bacteria were scraped from the surface and CFU were determined on SerFA agar plates. Controls without macrophage were done in parallel. Results are derived from 5 biological replicates, and are presented as the mean ± standard error of the mean (SEM) using GraphPad Prism 9.5.1 (San Diego, Ca). CFU counts were compared by paired t-test (P<0.05).
Exoprotein activity assays
SerFA day precultures were diluted into SerFA without or with either triclosan (SerFA-Tric) and AFN-1252 (SerFA-AFN). Solid medium as prepared for exoprotein detection were then spotted with resulting overnight saturated cultures or culture supernatants as indicated. For nuclease detection, DNase agar (Oxoid, Thermo Scientific, Fr) containing toluidine blue O 0.05 g/L (Sigma, Fr) was prepared as described.63 Culture supernatants from test samples were heated to 80°C for 10 m, and then spotted (10 μl) on plates. Photographs were taken after overnight incubation at 37°C. Contrast was uniformly enhanced by Photoshop to visualize pink halos indicating nuclease activity. To measure protease activity, powdered skim milk 50 g/L was added to autoclaved 1% non-nutrient agar (Invitrogen) as described.64 Cultures were spotted (10 μl) on plates and allowed to dry, then incubated at 37°C for 72 h and photographed. Lipase activity was assayed in medium comprising 1% non-nutrient agar (Invitrogen) to which was added 2.5% olive oil and 0.001% Rhodamine B (starting from a Rhodamine B stock solution of 1 mg per ml in water; Sigma-Aldrich, Fr) as described.65 To improve olive oil emulsification, NaCl was added to medium (1M final concentration), followed by vigorous shaking just prior to plate preparation. Supernatants from overnight cultures were sterile-filtered through a 0.2 μm membrane syringe filter (Pall Corporation, Michigan). Supernatants (10 μl) were deposited in holes pierced in solid medium. After 24 h incubation at 37°C plates were visualized under UV light at 312 nm and photographed. Hemolytic activity was assayed on S. aureus sterile-filtered supernatants that were prepared from overnight cultures. Aliquots (10 μl) were spotted on 5% sheep blood agar plates (BioMérieux SA, Fr). After drying, plates were incubated at 37°C overnight, and photographed.
H2O2 resistance assays
S. aureus non-treated or AFN-adapted cells (see above) were diluted to OD600= 0.005 in SerFA and grown for 1 h at 37°C, after which H2O2 (0.5mM final concentration) was added or not to cultures, and allowed to grow for 5 h. Growth kinetics was followed by OD600 determinations, and CFUs were determined at the final time point. Plates were photographed after overnight incubation at 37°C. Lawns of non-treated or AFN-adapted cells prepared as above were prepared on SerFA solid medium (100 μl of dilutions adjusted to OD600 = 0.1). Plates were spotted with 1.5 mm H2O2 and 4 nm (1.5 μg) AFN-1252 and photographed after 48 h incubation at 37°C.
H2O2 and PMS pre-treatment assays, and treatments by reducing agents
For pre-adaptation in hydrogen peroxide or in PMS, S. aureus were precultured in SerFA and then subcultured in SerFA containing or not 0.5 mM H2O2 (final concentration) or PMS (20-50 μM) for 16 h at 37°C. Cultures were diluted to OD600 = 0.1 in SerFA or SerFA medium containing AFN-1252 (SerFA-AFN), and growth was monitored.
For effects of reducing agents on anti-FASII adaptation, S. aureus were precultured in SerFA, and then diluted to OD600 = 0.1 in SerFA or SerFA medium containing AFN-1252 (SerFA-AFN), which were supplemented or not with NaCitrate (10 mM) or Vitamin C (5.7 mM), and growth was monitored. SerFA-AFN outgrowth time was assessed according to oxidizing and reducing conditions by the Mann-Whitney test, using GraphPad Prism 9.5.1 software. P-values <0.05 are indicated.
G. mellonella infection setups
Killing capacities and CFUs of anti-FASII-adapted and untreated S. aureus USA300 were compared in the G. mellonella insect model.66 G. mellonella larvae were reared on beeswax and pollen in sealed containers with a wire mesh lid permitting aeration. The rearing container was stored at 27°C temperature in a humidified incubator in our laboratory facilities. Fifth instar larvae weighing ∼250 mg were subjected to starvation for 24 h at 27°C and 1-2 h at 37°C prior to infection. S. aureus SerFA cultures were subcultured in SerFA, and SerFA plus AFN-1252 (0.5 μg/ml; SerFA-AFN) for overnight growth. Resulting cultures were then diluted to 0.1 and grown in respective media to OD600 = 1. Based on previous CFU determinations, each culture was pelleted, washed once in PBS, and then resuspended in PBS to obtain 108 CFU/ml. Ten μl (106 CFUs) was administered by injection in the fourth proleg. To follow larvae mortality following S. aureus infection, 20 larvae were inoculated using 3 independent cultures per culture condition (totaling 60 insects injected with SerFA, and 60 with SerFA-AFN), and 20 larvae were inoculated with PBS buffer for the control. Surviving larvae were counted every 24 h for 72 hours. To monitor S. aureus bacterial counts post-infection, the experiment above was repeated, and surviving larvae (9 total per condition, treated in 3 groups from independent biological replicates) were sacrificed until there were no surviving larvae left. CFUs were also determined from newly-dead larvae. To determine S. aureus CFUs, surviving larvae were chilled in ice for 30 min, and then crushed in 1 ml sterile deionized water. The resulting content was vortexed for ∼30 sec. Five μl of 10-fold dilutions were prepared in sterile water and spotted on SerFA and SerFA-AFN solid media. CFUs were determined after overnight incubation at 37°C. The detection threshold was 103 CFU per insect. Results of the 3 biologically independent experiments were pooled for presentation of insect mortality, and similarly for CFUs as done in the second independent experiment. Note that G. mellonella naturally harbor enterococci.67 In contrast to S. aureus, enterococci are catalase-negative on routine plate tests (n.b., some enterococci produce catalase upon heme addition). S. aureus were thus differentiated from enterococci on plates used for CFU enumeration by their catalase-positive response (i.e., bubble formation) upon application of a 3% hydrogen peroxide solution. Enterococci do not produce bubbles in these conditions.
Results of G. mellonella infection were analyzed for insect mortality and CFUs using GraphPad Prism 9.5.1, as specified in Figure 5 legend. Insect mortality was represented on a Kaplan-Meier plot, and survival between treatment groups including the PBS-injected reference was analyzed by log rank (Mantel-Cox) tests. The Cox proportional hazard model was also used to compare insect survival when infected by non-treated or anti-FASII-adapted bacteria. CFU comparisons in surviving and in dead larvae were done by non-parametric Mann-Whitney only at time points where there were sufficient individuals in both infected cohorts (i.e., at 0 and 24 h in live insects, and at 48 h in dead insects).
Acknowledgments
The Nebraska Transposon Mutant Library strains were generously provided by BEI Resources, NIAID, NIH, USA, and by Dr. P. Fey (Nebraska Medical Center, Omaha, USA). We acknowledge Micalis colleagues V. Sanchis, C. Nielson-LeRoux, and C. Buisson for valuable advice on the G. mellonella infection model. We gratefully acknowledge our colleagues for their skillful technical assistance: C. Buisson (Micalis) for insectarium and insect model, L. Dupont (Micalis) for proteomic sample preparations, J. Guignot (Institut Cochin, Paris) for adhesion experiments, and F. Delolme (Université de Lyon) for phosphoproteome studies. We thank P. Gaudu and V. Leguillier (Micalis), and P. Bouloc (I2BC, Gif-sur-Yvette) for valuable discussions.
We gratefully acknowledge funding support from the French National Research Agency StaphEscape project ANR project 16CE150013 (A.G., C.G., A.F.), the JPIAMR antimicrobial resistance project (JPIAMR2022-070; France, Germany, Spain) (A.G.), the Fondation pour la Recherche Médicale (DBF20161136769, France) (A.G.), and DIM One Health (RPH17043DJA, France) (A.F.). P.W. received a Franco-Thai scholarship from Campus France and Khon Kaen University, Thailand. J.P.L. and C.G. acknowledge financial support from the CNRS, France and the ITMO Cancer AVIESAN (Alliance Nationale pour les Sciences de la Vie et de la Santé, National Alliance for Life Sciences and Health, France) within the framework of the cancer plan for Orbitrap mass spectrometer funding. Phosphoproteomes were performed at the Protein Science Facility, SFR BioSciences CNRS UAR3444, INSERM US8, UCBL, ENS de Lyon, 50 Avenue Tony Garnier, 69007 Lyon, Fr.
Author contributions
Project conceptualization, P.W., J.A.M., A.G.; methodology, D.H., P.W., J.U., J.A.M., K.G., A.P., G.K., A.O.M., J.P.L., and C.L.; data analysis, P.W., J.A.M., A.G., A.F., J.U., D.H., C.H., A.O.M., A.P., J.P.L., and C.G.; data interpretation, all authors; manuscript writing – original draft, and figures, P.W., A.G., C.H., A.F., A.O.M., D.H., J.P.L., and C.G.; manuscript review and editing, all authors. All authors read and approved the final draft of the manuscript.
Declaration of interests
The authors declare that no competing interests exist.
Published: March 16, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.109505.
Supplemental information
References
- 1.Cusumano J.A., Klinker K.P., Huttner A., Luther M.K., Roberts J.A., LaPlante K.L. Towards precision medicine: Therapeutic drug monitoring-guided dosing of vancomycin and beta-lactam antibiotics to maximize effectiveness and minimize toxicity. Am. J. Health Syst. Pharm. 2020;77:1104–1112. doi: 10.1093/ajhp/zxaa128. [DOI] [PubMed] [Google Scholar]
- 2.Lakhundi S., Zhang K. Methicillin-Resistant Staphylococcus aureus: Molecular Characterization, Evolution, and Epidemiology. Clin. Microbiol. Rev. 2018;31 doi: 10.1128/CMR.00020-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lowy F.D. Staphylococcus aureus infections. N. Engl. J. Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 4.Rasigade J.P., Vandenesch F. Staphylococcus aureus: a pathogen with still unresolved issues. Infect. Genet. Evol. 2014;21:510–514. doi: 10.1016/j.meegid.2013.08.018. [DOI] [PubMed] [Google Scholar]
- 5.Wang J., Soisson S.M., Young K., Shoop W., Kodali S., Galgoci A., Painter R., Parthasarathy G., Tang Y.S., Cummings R., et al. Platensimycin is a selective FabF inhibitor with potent antibiotic properties. Nature. 2006;441:358–361. doi: 10.1038/nature04784. [DOI] [PubMed] [Google Scholar]
- 6.Wright H.T., Reynolds K.A. Antibacterial targets in fatty acid biosynthesis. Curr. Opin. Microbiol. 2007;10:447–453. doi: 10.1016/j.mib.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karlowsky J.A., Kaplan N., Hafkin B., Hoban D.J., Zhanel G.G. AFN-1252, a FabI inhibitor, demonstrates a Staphylococcus-specific spectrum of activity. Antimicrob. Agents Chemother. 2009;53:3544–3548. doi: 10.1128/AAC.00400-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brinster S., Lamberet G., Staels B., Trieu-Cuot P., Gruss A., Poyart C. Type II fatty acid synthesis is not a suitable antibiotic target for Gram-positive pathogens. Nature. 2009;458:83–86. doi: 10.1038/nature07772. [DOI] [PubMed] [Google Scholar]
- 9.Gloux K., Guillemet M., Soler C., Morvan C., Halpern D., Pourcel C., Vu Thien H., Lamberet G., Gruss A. Clinical Relevance of Type II Fatty Acid Synthesis Bypass in Staphylococcus aureus. Antimicrob. Agents Chemother. 2017;61 doi: 10.1128/AAC.02515-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kénanian G., Morvan C., Weckel A., Pathania A., Anba-Mondoloni J., Halpern D., Gaillard M., Solgadi A., Dupont L., Henry C., et al. Permissive Fatty Acid Incorporation Promotes Staphylococcal Adaptation to FASII Antibiotics in Host Environments. Cell Rep. 2019;29:3974–3982.e4. doi: 10.1016/j.celrep.2019.11.071. [DOI] [PubMed] [Google Scholar]
- 11.Morvan C., Halpern D., Kénanian G., Hays C., Anba-Mondoloni J., Brinster S., Kennedy S., Trieu-Cuot P., Poyart C., Lamberet G., et al. Environmental fatty acids enable emergence of infectious Staphylococcus aureus resistant to FASII-targeted antimicrobials. Nat. Commun. 2016;7 doi: 10.1038/ncomms12944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fage C.D., Lathouwers T., Vanmeert M., Gao L.J., Vrancken K., Lammens E.M., Weir A.N.M., Degroote R., Cuppens H., Kosol S., et al. The Kalimantacin Polyketide Antibiotics Inhibit Fatty Acid Biosynthesis in Staphylococcus aureus by Targeting the Enoyl-Acyl Carrier Protein Binding Site of FabI. Angew. Chem., Int. Ed. Engl. 2020;59:10549–10556. doi: 10.1002/anie.201915407. [DOI] [PubMed] [Google Scholar]
- 13.Parker E.N., Cain B.N., Hajian B., Ulrich R.J., Geddes E.J., Barkho S., Lee H.Y., Williams J.D., Raynor M., Caridha D., et al. An Iterative Approach Guides Discovery of the FabI Inhibitor Fabimycin, a Late-Stage Antibiotic Candidate with In Vivo Efficacy against Drug-Resistant Gram-Negative Infections. ACS Cent. Sci. 2022;8:1145–1158. doi: 10.1021/acscentsci.2c00598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pishchany G., Mevers E., Ndousse-Fetter S., Horvath D.J., Jr., Paludo C.R., Silva-Junior E.A., Koren S., Skaar E.P., Clardy J., Kolter R. Amycomicin is a potent and specific antibiotic discovered with a targeted interaction screen. Proc. Natl. Acad. Sci. USA. 2018;115:10124–10129. doi: 10.1073/pnas.1807613115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yao J., Rock C.O. Resistance Mechanisms and the Future of Bacterial Enoyl-Acyl Carrier Protein Reductase (FabI) Antibiotics. Cold Spring Harbor Perspect. Med. 2016;6:a027045. doi: 10.1101/cshperspect.a027045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McMurry L.M., Oethinger M., Levy S.B. Triclosan targets lipid synthesis. Nature. 1998;394:531–532. doi: 10.1038/28970. [DOI] [PubMed] [Google Scholar]
- 17.Fey P.D., Endres J.L., Yajjala V.K., Widhelm T.J., Boissy R.J., Bose J.L., Bayles K.W. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. mBio. 2013;4:e00537-12. doi: 10.1128/mBio.00537-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roberts C.A., Al-Tameemi H.M., Mashruwala A.A., Rosario-Cruz Z., Chauhan U., Sause W.E., Torres V.J., Belden W.J., Boyd J.M. The Suf Iron-Sulfur Cluster Biosynthetic System Is Essential in Staphylococcus aureus, and Decreased Suf Function Results in Global Metabolic Defects and Reduced Survival in Human Neutrophils. Infect. Immun. 2017;85 doi: 10.1128/IAI.00100-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lopez M.S., Tan I.S., Yan D., Kang J., McCreary M., Modrusan Z., Austin C.D., Xu M., Brown E.J. Host-derived fatty acids activate type VII secretion in Staphylococcus aureus. Proc. Natl. Acad. Sci. USA. 2017;114:11223–11228. doi: 10.1073/pnas.1700627114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alnaseri H., Arsic B., Schneider J.E.T., Kaiser J.C., Scinocca Z.C., Heinrichs D.E., McGavin M.J. Inducible Expression of a Resistance-Nodulation-Division-Type Efflux Pump in Staphylococcus aureus Provides Resistance to Linoleic and Arachidonic Acids. J. Bacteriol. 2015;197:1893–1905. doi: 10.1128/JB.02607-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pathania A., Anba-Mondoloni J., Gominet M., Halpern D., Dairou J., Dupont L., Lamberet G., Trieu-Cuot P., Gloux K., Gruss A. (p)ppGpp/GTP and Malonyl-CoA Modulate Staphylococcus aureus Adaptation to FASII Antibiotics and Provide a Basis for Synergistic Bi-Therapy. mBio. 2021;12:e03193-20. doi: 10.1128/mBio.03193-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Derouiche A., Bidnenko V., Grenha R., Pigonneau N., Ventroux M., Franz-Wachtel M., Nessler S., Noirot-Gros M.F., Mijakovic I. Interaction of bacterial fatty-acid-displaced regulators with DNA is interrupted by tyrosine phosphorylation in the helix-turn-helix domain. Nucleic Acids Res. 2013;41:9371–9381. doi: 10.1093/nar/gkt709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garcia-Garcia T., Poncet S., Derouiche A., Shi L., Mijakovic I., Noirot-Gros M.F. Role of Protein Phosphorylation in the Regulation of Cell Cycle and DNA-Related Processes in Bacteria. Front. Microbiol. 2016;7:184. doi: 10.3389/fmicb.2016.00184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mijakovic I., Grangeasse C., Turgay K. Exploring the diversity of protein modifications: special bacterial phosphorylation systems. FEMS Microbiol. Rev. 2016;40:398–417. doi: 10.1093/femsre/fuw003. [DOI] [PubMed] [Google Scholar]
- 25.McCallum N., Hinds J., Ender M., Berger-Bächi B., Stutzmann Meier P. Transcriptional profiling of XdrA, a new regulator of spa transcription in Staphylococcus aureus. J. Bacteriol. 2010;192:5151–5164. doi: 10.1128/JB.00491-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lei M.G., Lee C.Y. Repression of Capsule Production by XdrA and CodY in Staphylococcus aureus. J. Bacteriol. 2018;200 doi: 10.1128/JB.00203-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bury-Moné S., Nomane Y., Reymond N., Barbet R., Jacquet E., Imbeaud S., Jacq A., Bouloc P. Global analysis of extracytoplasmic stress signaling in Escherichia coli. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dyzenhaus S., Sullivan M.J., Alburquerque B., Boff D., van de Guchte A., Chung M., Fulmer Y., Copin R., Ilmain J.K., O'Keefe A., et al. MRSA lineage USA300 isolated from bloodstream infections exhibit altered virulence regulation. Cell Host Microbe. 2023;31:228–242.e8. doi: 10.1016/j.chom.2022.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stapleton M.R., Horsburgh M.J., Hayhurst E.J., Wright L., Jonsson I.M., Tarkowski A., Kokai-Kun J.F., Mond J.J., Foster S.J. Characterization of IsaA and SceD, two putative lytic transglycosylases of Staphylococcus aureus. J. Bacteriol. 2007;189:7316–7325. doi: 10.1128/JB.00734-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu G.Y., Essex A., Buchanan J.T., Datta V., Hoffman H.M., Bastian J.F., Fierer J., Nizet V. Staphylococcus aureus golden pigment impairs neutrophil killing and promotes virulence through its antioxidant activity. J. Exp. Med. 2005;202:209–215. doi: 10.1084/jem.20050846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gerstel A., Zamarreño Beas J., Duverger Y., Bouveret E., Barras F., Py B. Oxidative stress antagonizes fluoroquinolone drug sensitivity via the SoxR-SUF Fe-S cluster homeostatic axis. PLoS Genet. 2020;16 doi: 10.1371/journal.pgen.1009198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mosel M., Li L., Drlica K., Zhao X. Superoxide-mediated protection of Escherichia coli from antimicrobials. Antimicrob. Agents Chemother. 2013;57:5755–5759. doi: 10.1128/AAC.00754-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu Y., Vulić M., Keren I., Lewis K. Role of oxidative stress in persister tolerance. Antimicrob. Agents Chemother. 2012;56:4922–4926. doi: 10.1128/AAC.00921-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ji C.J., Kim J.H., Won Y.B., Lee Y.E., Choi T.W., Ju S.Y., Youn H., Helmann J.D., Lee J.W. Staphylococcus aureus PerR Is a Hypersensitive Hydrogen Peroxide Sensor using Iron-mediated Histidine Oxidation. J. Biol. Chem. 2015;290:20374–20386. doi: 10.1074/jbc.M115.664961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kazek M., Kaczmarek A., Wrońska A.K., Boguś M.I. Dodecanol, metabolite of entomopathogenic fungus Conidiobolus coronatus, affects fatty acid composition and cellular immunity of Galleria mellonella and Calliphora vicina. Sci. Rep. 2021;11 doi: 10.1038/s41598-021-95440-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hunt T., Kaplan N., Hafkin B. Safety, tolerability and pharmacokinetics of multiple oral doses of AFN-1252 administered as immediate release (IR) tablets in healthy subjects. J. Chemother. 2016;28:164–171. doi: 10.1179/1973947815Y.0000000075. [DOI] [PubMed] [Google Scholar]
- 37.Parsons J.B., Frank M.W., Subramanian C., Saenkham P., Rock C.O. Metabolic basis for the differential susceptibility of Gram-positive pathogens to fatty acid synthesis inhibitors. Proc. Natl. Acad. Sci. USA. 2011;108:15378–15383. doi: 10.1073/pnas.1109208108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schurig-Briccio L.A., Parraga Solorzano P.K., Lencina A.M., Radin J.N., Chen G.Y., Sauer J.D., Kehl-Fie T.E., Gennis R.B. Role of respiratory NADH oxidation in the regulation of Staphylococcus aureus virulence. EMBO Rep. 2020;21 doi: 10.15252/embr.201845832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yeo W.S., Dyzenhaus S., Torres V.J., Brinsmade S.R., Bae T. Regulation of Bacterial Two-Component Systems by Cardiolipin. Infect. Immun. 2023;91 doi: 10.1128/iai.00046-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lowden M.J., Skorupski K., Pellegrini M., Chiorazzo M.G., Taylor R.K., Kull F.J. Structure of Vibrio cholerae ToxT reveals a mechanism for fatty acid regulation of virulence genes. Proc. Natl. Acad. Sci. USA. 2010;107:2860–2865. doi: 10.1073/pnas.0915021107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang L., Matsuo M., Calderon C., Fan S.H., Ammanath A.V., Fu X., Li N., Luqman A., Ullrich M., Herrmann F., et al. Molecular Basis of Rhodomyrtone Resistance in Staphylococcus aureus. mBio. 2022;13 doi: 10.1128/mbio.03833-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tuchscherr L., Löffler B., Proctor R.A. Persistence of Staphylococcus aureus: Multiple Metabolic Pathways Impact the Expression of Virulence Factors in Small-Colony Variants (SCVs) Front. Microbiol. 2020;11:1028. doi: 10.3389/fmicb.2020.01028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.DeCoursey T.E. During the respiratory burst, do phagocytes need proton channels or potassium channels, or both? Sci. STKE. 2004;2004:pe21. doi: 10.1126/stke.2332004pe21. [DOI] [PubMed] [Google Scholar]
- 44.Cole J., Aberdein J., Jubrail J., Dockrell D.H. The role of macrophages in the innate immune response to Streptococcus pneumoniae and Staphylococcus aureus: mechanisms and contrasts. Adv. Microb. Physiol. 2014;65:125–202. doi: 10.1016/bs.ampbs.2014.08.004. [DOI] [PubMed] [Google Scholar]
- 45.Mayer-Scholl A., Averhoff P., Zychlinsky A. How do neutrophils and pathogens interact? Curr. Opin. Microbiol. 2004;7:62–66. doi: 10.1016/j.mib.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 46.Lambert C., Bachmann C., Gaillard M., Hautcoeur A., Gloux K., Guilbert T., Méhats C., Prost B., Solgadi A., Abreu S., et al. The double-edged role of FASII regulator FabT in Streptococcus pyogenes infection. bioRxiv. 2023 doi: 10.1101/2023.06.07.543851. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lambert C., Poyart C., Gruss A., Fouet A. FabT, a Bacterial Transcriptional Repressor That Limits Futile Fatty Acid Biosynthesis. Microbiol. Mol. Biol. Rev. 2022;86 doi: 10.1128/mmbr.00029-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Albanesi D., Reh G., Guerin M.E., Schaeffer F., Debarbouille M., Buschiazzo A., Schujman G.E., de Mendoza D., Alzari P.M. Structural basis for feed-forward transcriptional regulation of membrane lipid homeostasis in Staphylococcus aureus. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Painter K.L., Strange E., Parkhill J., Bamford K.B., Armstrong-James D., Edwards A.M. Staphylococcus aureus adapts to oxidative stress by producing H2O2-resistant small-colony variants via the SOS response. Infect. Immun. 2015;83:1830–1844. doi: 10.1128/IAI.03016-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peyrusson F., Nguyen T.K., Najdovski T., Van Bambeke F. Host Cell Oxidative Stress Induces Dormant Staphylococcus aureus Persisters. Microbiol. Spectr. 2022;10 doi: 10.1128/spectrum.02313-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sendi P., Proctor R.A. Staphylococcus aureus as an intracellular pathogen: the role of small colony variants. Trends Microbiol. 2009;17:54–58. doi: 10.1016/j.tim.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 52.Morvan C., Halpern D., Kénanian G., Pathania A., Anba-Mondoloni J., Lamberet G., Gruss A., Gloux K. The Staphylococcus aureus FASII bypass escape route from FASII inhibitors. Biochimie. 2017;141:40–46. doi: 10.1016/j.biochi.2017.07.004. [DOI] [PubMed] [Google Scholar]
- 53.Cherednichenko G., Zhang R., Bannister R.A., Timofeyev V., Li N., Fritsch E.B., Feng W., Barrientos G.C., Schebb N.H., Hammock B.D., et al. Triclosan impairs excitation-contraction coupling and Ca2+ dynamics in striated muscle. Proc. Natl. Acad. Sci. USA. 2012;109:14158–14163. doi: 10.1073/pnas.1211314109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yueh M.F., Taniguchi K., Chen S., Evans R.M., Hammock B.D., Karin M., Tukey R.H. The commonly used antimicrobial additive triclosan is a liver tumor promoter. Proc. Natl. Acad. Sci. USA. 2014;111:17200–17205. doi: 10.1073/pnas.1419119111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ciusa M.L., Furi L., Knight D., Decorosi F., Fondi M., Raggi C., Coelho J.R., Aragones L., Moce L., Visa P., et al. A novel resistance mechanism to triclosan that suggests horizontal gene transfer and demonstrates a potential selective pressure for reduced biocide susceptibility in clinical strains of Staphylococcus aureus. Int. J. Antimicrob. Agents. 2012;40:210–220. doi: 10.1016/j.ijantimicag.2012.04.021. [DOI] [PubMed] [Google Scholar]
- 56.Westfall C., Flores-Mireles A.L., Robinson J.I., Lynch A.J.L., Hultgren S., Henderson J.P., Levin P.A. The widely used antimicrobial triclosan induces high levels of antibiotic tolerance in vitro and reduces antibiotic efficacy up to 100-fold in vivo. Antimicrob. Agents Chemother. 2019;63 doi: 10.1128/AAC.02312-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bednarz B., Millan-Oropeza A., Kotowska M., Świat M., Quispe Haro J.J., Henry C., Pawlik K. Coelimycin Synthesis Activatory Proteins Are Key Regulators of Specialized Metabolism and Precursor Flux in Streptomyces coelicolor A3(2) Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.616050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Millan-Oropeza A., Henry C., Lejeune C., David M., Virolle M.J. Expression of genes of the Pho regulon is altered in Streptomyces coelicolor. Sci. Rep. 2020;10:8492. doi: 10.1038/s41598-020-65087-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boersema P.J., Raijmakers R., Lemeer S., Mohammed S., Heck A.J.R. Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat. Protoc. 2009;4:484–494. doi: 10.1038/nprot.2009.21. [DOI] [PubMed] [Google Scholar]
- 60.Soufi B., Täumer C., Semanjski M., Macek B. Phosphopeptide Enrichment from Bacterial Samples Utilizing Titanium Oxide Affinity Chromatography. Methods Mol. Biol. 2018;1841:231–247. doi: 10.1007/978-1-4939-8695-8_16. [DOI] [PubMed] [Google Scholar]
- 61.The M., MacCoss M.J., Noble W.S., Käll L. Fast and Accurate Protein False Discovery Rates on Large-Scale Proteomics Data Sets with Percolator 3.0. J. Am. Soc. Mass Spectrom. 2016;27:1719–1727. doi: 10.1007/s13361-016-1460-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bourrel A.S., Picart A., Fernandez J.C., Hays C., Saubamea B., Mignon V., Poyart C., Tazi A., Guignot J. Specific interaction between Group B Streptococcus CC17 hypervirulent clone and phagocytes. bioRxiv. 2022 doi: 10.1101/2022.12.02.518834. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zierdt C.H., Golde D.W. Deoxyribonuclease-positive Staphylococcus epidermidis strains. Appl. Microbiol. 1970;20:54–57. doi: 10.1128/am.20.1.54-57.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Park J.H., Lee J.H., Cho M.H., Herzberg M., Lee J. Acceleration of protease effect on Staphylococcus aureus biofilm dispersal. FEMS Microbiol. Lett. 2012;335:31–38. doi: 10.1111/j.1574-6968.2012.02635.x. [DOI] [PubMed] [Google Scholar]
- 65.Araiza-Villanueva M., Avila-Calderón E.D., Flores-Romo L., Calderón-Amador J., Sriranganathan N., Qublan H.A., Witonsky S., Aguilera-Arreola M.G., Ruiz-Palma M.D.S., Ruiz E.A., et al. Proteomic Analysis of Membrane Blebs of Brucella abortus 2308 and RB51 and Their Evaluation as an Acellular Vaccine. Front. Microbiol. 2019;10:2714. doi: 10.3389/fmicb.2019.02714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sheehan G., Dixon A., Kavanagh K. Utilization of Galleria mellonella larvae to characterize the development of Staphylococcus aureus infection. Microbiology (Read.) 2019;165:863–875. doi: 10.1099/mic.0.000813. [DOI] [PubMed] [Google Scholar]
- 67.Allonsius C.N., Van Beeck W., De Boeck I., Wittouck S., Lebeer S. The microbiome of the invertebrate model host Galleria mellonella is dominated by Enterococcus. Anim. Microbiome. 2019;1:7. doi: 10.1186/s42523-019-0010-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data are within the manuscript, supporting information files, and depositories. Nanopore genome sequencing, Proteomic data, and phosphoproteomic data were respectively deposited in Mendeley, Pride partner repository, and Center for Computational Mass Spectrometry repository (U. California, San Diego). Accession numbers are listed in the key resources table. All data are publicly available as of publication. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.